Abstract
Background
This study aimed to investigate the role of micro-RNA 205-5p (miR-205-5p) in the progression of gastric cancer, and the target of miR-205-5p in human gastric cancer cells in vitro.
Material/Methods
Expression of miR-205-5p and PTEN in gastric cancer tissue samples and adjacent normal gastric tissue from 35 patients was studied using immunohistochemistry and in situ hybridization. SGC-7901 human gastric cancer cells included a normal control (NC) group, a group transfected with empty vector (Vector), a group treated with miR-205-5p inhibitor (miR-inhibitor), and a group treated with miR-205-5p inhibitor and small interfering PTEN mRNA (miR-inhibitor+si-PTEN). Quantitative reverse transcription polymerase chain reaction (qRT-PCR) measured miR-205-5p expression, cell proliferation was measured by MTT assay, cell apoptosis by flow cytometry, transwell and wound healing assays measured cell migration, and transmission electron microscopy (TEM) showed ultrastructural changes in SGC-7901 cells. PTEN, AKT and p-AKT protein expression were measured using Western blot. The correlation between miR-205-5p and PTEN was analyzed using a dual-luciferase reporter assay.
Results
Increased expression of miR-205-5p and PTEN in gastric cancer tissues were correlated with tumor stage. In SGC-7901 cells, miR-205-5p mRNA expression in the miR-inhibitor and miR-inhibitor+si-PTEN groups was significantly lower than that in the NC group (P<0.001). In the miR-inhibitor group, cell proliferation was significantly decreased, and apoptosis was significantly increased (P<0.001).
Conclusions
In gastric cancer, increased expression of miR-205-5p was associated with tumor stage, and in SGC-7901 cells PTEN was a target gene for miR-205-5p.
MeSH Keywords: MicroRNAs, Nitrohydroxyiodophenylacetate, Proto-Oncogene Proteins c-akt, Stomach Neoplasms
Background
Worldwide, more than one million people are diagnosed with gastric cancer each year, and gastric cancer remains the second most common cause of cancer-related death with a mortality rate of approximately 10% [1,2]. Gastric cancer has a complex molecular and genetic pathogenesis, and studies have shown that several oncogenes and oncogene inhibitors are involved in its development and progression [3]. However, the most common molecular mechanisms of gastric cancer remain unclear and continue to be of research interest.
MicroRNAs (miRNAs) are small non-coding nucleotides of 18–25 residues [4], which regulate target gene expression by binding to the 3′-untranslated regions (UTRs) of their mRNAs. Several miRNAs have been reported to be abnormally expressed in different malignant tumor tissues, which influence signaling pathways to produce tumor promoting or inhibiting effects [5]. Recently, accumulating experimental data has suggested critical regulatory effects of miRNAs in the biological behavior of tumor cells, including proliferation, differentiation, migration, and invasion [6,7]. Studies have shown that miRNAs are involved in the ontogeny, development, progression, and metastasis of gastric cancer [8,9], which may have significance for diagnosis and treatment. Identifying the mechanism of action of miRNA in gastric cancer might improve the survival rate and quality of life for patients with gastric cancer.
Previous studies have identified significant differences in miRNA-205-5p expression in ovarian cancer [10], bladder cancer [11], breast cancer [12], prostate cancer [13], and pancreatic cancer [14], but its expression in gastric cancer and its role in the progression of gastric cancer remain unclear. Bioinformatic analysis has shown that the phosphatase and tensin homolog (PTEN) is the target gene of miR-205-5p in gastric cancer.
Therefore, this study aimed to investigate the role of miR-205-5p in the progression of gastric cancer by examining gastric cancer tissue from patients diagnosed with different stages of gastric cancer and adjacent normal gastric tissue, and the target of miR-205-5p in the human gastric cancer cell line, SGC-7901, in vitro.
Material and Methods
Clinical information
This study included 35 patients with primary gastric cancer (20 men and 15 women) who were diagnosed and treated with surgical excision at the First People’s Hospital of Changzhou between June 2016 and May 2017. The patients were aged between 41–74 years, with a median age of 63 years. Among the subjects, 20 and 15 cases were with and without lymph node metastasis, respectively. Based on TNM staging (7th edition), drafted jointly by the Union for International Cancer Control (UICC) and the American Joint Committee on Cancer (AJCC) [15,16], nine and 26 cases were classified as stage I–II and stage III–IV, respectively. All patients received gastric cancer D2 radical surgery, and postoperative histopathological examination of gastrectomy tumor tissue confirmed the diagnosis of primary gastric cancer. The exclusion criteria were concurrent local or systemic infection, concurrent abortive tuberculosis, other tumors, recent use of medication to treat Helicobacter pylori, and preoperative radiotherapy, chemotherapy, or immunotherapy. Also, normal adjacent gastric mucosa tissues >5 cm from the tumor borders were collected as controls from each patient. The tissues were placed in 10% paraformaldehyde, fixed for 48 h, embedded in paraffin wax and sectioned at 4 μm. The study was approved by the Ethics Committee of Wuxi 3rd People’s Hospital (Approval No. 2016042002), and written informed consent was obtained from each patient.
Materials
Normal human gastric epithelial cells (GES-1) and the human gastric cancer cell lines SGC-7901, MGC-803, MKN45, AGS, BGC-823, MKN-4, and MKN-28 were purchased from the Shanghai Cell Bank, Chinese Academy of Sciences. Materials included in situ hybridization (ISH) reagent, miR-205 probe (5′-CAGACTCCGGTGGAATGAAGGA-3′) and miR-205 blocking oligo (5′-TCCTTCATTCCACCGGAGTCTG-3′), which were obtained from Wuhan Boster Biological Technology (Wuhan, China). Lipofectamine 2000 was obtained from Life Technologies (Gaithersburg, MD, USA), a real-time quantitative polymerase chain reaction (qRT-PCR) kit and SYBR fluorochrome kit were obtained from Takara (Minato-ku, Tokyo, Japan), and the miR-205-5p inhibitor (5′-CAGACUCCGGUGGAAUGAAGA-3′) and miR-205-5p negative control (5′-CAGUACUUUUGUGUAGUACAA-3′) were obtained from Shanghai GenePharma Co., Ltd (Shanghai, China). PTEN short interfering RNA (siRNA) was obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA), and thiazolyl blue 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), the Annexin-V-fluorescein isothiocyanate (Annexin V-FITC)/propidium iodide (PI) apoptosis detection kit and the polyvinylidene fluoride (PVDF) membrane were obtained from Sigma-Aldrich (St. Louis MO, USA). The bicinchoninic acid (BCA) disodium protein test kit was obtained from Nanjing KeyGen Biotech Co., Ltd. (Nanjing, China). Antibodies to PTEN (ab170941), p-AKT (ab131443), AKT (ab179463) and GAPDH (ab181602) were obtained from Abcam (Cambridge, UK). The luciferase reporter gene testing kit and the miR-205-5p luciferase reporter gene plasmid were obtained from Promega (Madison, WI, USA).
Histology using hematoxylin and eosin (H&E)
Paraffin sections were dried and dewaxed with xylene and dehydrated in graded ethanol, stained with hematoxylin for 10 min, processed with 1% hydrochloric acid for 30 s, neutralized with 1% ammonium hydroxide, stained with 0.5% eosin for 5 min, dehydrated in graded ethanol, treated with xylene, sealed with neutral gum and finally examined microscopically at ×100 and ×400.
Immunohistochemistry
After dewaxing, tissue sections were incubated with 3% H2O2 at ambient temperature to inhibit endogenous peroxidase, followed by antigen recovery and blocking with goat serum. Sections were then treated with a primary antibody to PTEN (1: 1000) at 4°C overnight, followed by washing with phosphate-buffered saline (PBS) and incubation with the secondary antibody for 2 h. Diaminobenzidine (DAB) was used for visualization. Sections were counterstained with hematoxylin, sealed, and photographed using a light microscope at ×100 and ×400. Image-Pro Plus Software version X (Media Cybernetics, Silver Springs, MD, USA) was used to analyze PTEN protein expression.
In situ hybridization
Paraffin slices were dewaxed and digested using pepsin treatment solution (ambient temperature for 30 min, with 20 μl of pre-hybridization solution in droplets for hybridization, followed by adding hybridization solution containing miR-205-5p oligonucleotide probe (1: 1000) on a tissue chip for overnight hybridization in a 37°C water bath. Sections were then washed three times (10 min each) with 2×saline-sodium citrate scrubbing solution for and processed with confining liquid for 30 min. Further, biotinylated anti-rat digoxin was added and incubated at 37°C for 60 min, and then washed four times with PBS for 5 min each. DAB was used to visualize the location of the probe, and the reaction was terminated by washing with water. Sections were then dehydrated with ethanol, treated with xylene and sealed with neutral gum. Samples were examined by light microscopy at ×100 and ×400 and photographed. Image-Pro Plus software, version X (Media Cybernetics, Silver Springs, MD, USA) was used to analyze miR-205-5p expression.
Cell culture
All cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% fetal bovine serum (FBS) (Gibco, Thermofisher Scientific, Waltham, MA, USA) at 5% CO2 and 37°C. Cells were trypsinized at 80–90% confluence for routine sub-culture. Cells in the logarithmic growth phase were used for subsequent experiments.
Cell transfection
SGC-7901 gastric cancer cells were divided into normal control (NC), vector, miR-205-5p inhibitor transfection (miR-inhibitor) and miR-205-5p inhibitor and small interfering PTEN mRNA joint inhibition (miR-inhibitor+si-PTEN) groups. NC cells were cultured normally, and cells in the vector group were transfected with control Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), cells in miR-inhibitor group were transfected with 10-nM miR-205-5p inhibitor in Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), and those in miR-inhibitor+si-PTEN group were transfected with 10-nM miR-205 inhibitor and 10-nM si-PTEN using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). Cells were treated for 48 h before they were used in subsequent experiments.
Extraction of total cellular mRNA and quantitative reverse transcription PCR
Cells were lysed in 500 μl of lysis buffer. TRIzol lysate was used to extract total mRNA for reverse transcription. Further, miR-205-5p was amplified by PCR using U6 as an internal reference. Reverse transcription of 100 ng of total RNA was performed at 37°C for 15 min followed by heating at 95°C for 5 min. PCR was performed according to the manufacturer’s instructions. The mRNA expression level was calculated using 2−ΔΔCT, and experiments were performed in triplicate.
The primers for miR-205-5p were:
Forward: 5′-CCTTCATTCCACCGGAGT-3′, and
Reverse: 5′-GTCCAGTTTTTTTTTTTTTTTCAGACT-3′.
The primers for U6 were:
Forward: 5′-TCCTTCGGCAGCACATATAG-3′, and
Reverse: 5′-AGGGGCATGGTAATCTTCT-3′.
MTT proliferation assay
Log-phase SGC-7901 cells were trypsinized to produce single-cell suspensions and seeded into a 96-well plate at a density of 1×103 cells/well. After 7 d of culture, 20 μl of MTT solution was added, and the plate incubated for 4–6 h at 37°C. Further, a sterile straw was used to remove the supernatant, and 150 μl dimethyl sulfoxide (DMSO) was added to each well. The mixture was shaken for 10 min at ambient temperature to dissolve crystals, during which the mixture was patted and mixed 2–3 times. Absorbance (A) was measured at 490 nm and used to calculate cell proliferation rates in each group. The experiment was performed in triplicate.
Cell apoptosis detection by flow cytometry
Cells were digested with ethylenediaminetetraacetic acid (EDTA)-free pancreatin and then washed with supernatant culture to stop the reaction. Cells were collected after 5 min and centrifuged at 4°C and washed three times with PBS. In total, 1×105 cells were collected and resuspended in 500 μl of 1×buffer. Then, 5 μl of Annexin-V–fluorescein isothiocyanate (FITC) and 10 μl of propidium iodide (PI) were added to the suspensions and incubated in the dark for 15 min at ambient temperature before flow cytometry was performed to detect apoptosis.
Transwell invasion assay
Cells were digested with pancreatin and re-suspended in serum-free culture solution before use. A culture insert was placed into a culture plate, which contained culture medium supplemented with fetal calf serum (FCS). Cells (2×104/ml) were then added and cultured for 48 h. The insert was then removed and fixed with methanol, stained with crystal violet for 15 min and rubbed with a swab to carefully remove the cells on the upper side of the microporous film. The cells on the lower side of the microporous film were photographed using a fluorescence microscope, with four random fields photographed at ×100 to count the cell numbers. The experiment was performed in triplicate.
Wound healing assay
Cells in the experimental groups were harvested and then seeded into 6-well plates at a density of approximately 1–3×105 cells/well. At 80% cell confluence, a 20 μl pipette tip was used to score the middle of the plate. The dislodged cells were removed by washing with PBS, and the remaining cells cultured for 24 h for cell fusion. Photographs were obtained at ×100 and at 0 and 24 h and used to determine wound healing rates.
Transmission electron microscopy examination
Cells in the experimental groups were digested and collected by centrifugation, washed twice with PBS and fixed with 2.5% glutaraldehyde in PBS for 24 h. Cells were washed three times with PBS (10 min each), and fixed with 1% osmic acid for 1 h. After washing three times with PBS (10 min each) cells were dehydrated using graded acetone at 50%, 70%, 90% and 100% for 10 min in each, immersed in epoxy resin (Epon 812) for 24 h and embedded for 24 h. Blocks were sectioned using a LKB-III ultra-microtome into 500–700 Å ultrathin sections and stained with lead nitrate and uranyl nitrate before examination at ×20000 and photographed using a Hitachi H-600 transmission electron microscope (Hitachi, Tokyo, Japan).
Western blot
Cells were collected and washed twice with ice-cold PBS. Cells (1×107) were lysed in 100 μl of lysis buffer containing 10 mM Tris HCl, pH 7. 6, 100 mM NaCl, 1 mM EDTA, 1 μg/ml aprotinin and 100 μg/ml phenylmethylsulfonyl fluoride (PMSF), for 1 h on ice. After centrifuging for 10 min at 12000 rpm, the supernatant was assayed for protein content using the Bradford method with the solvent as the blank and bovine serum albumin (BSA) as the standard, and absorbance was measured at 570 nm using a spectrophotometer. Then, 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed and proteins were transferred to a PVDF membrane and blocked with 20 mM Tris HCl, 137 mM NaCl, and 0.1% Tween 20 (TBST) containing 5% BSA for 1 h. Membranes were incubated overnight incubation at 4°C with primary antibody and washed three times with TBST for 5 min each before incubation with secondary antibody for 1 h. Blots were then washed three times with TBST for 5 min each and visualized using a chemiluminescence reagent and a gel scanner.
Dual luciferase reporter assay
The target binding site of miR-205-5p at the PTEN-3′-UTR was predicted using online databases (TargetScan, miRbase and miRanda). Wild-type (WT) pGL3-PTEN-3′-UTR and mutant (MUT) pGL3-CYLD-3′-UTR luciferase vectors of PTEN-3′-UTR were constructed. Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) was used to transfect pGL3-PTEN-3′-UTR and pGL3-CYLD-3′-UTR luciferase vectors in miR-250-5p mimic or miR-250-5p mimic control SGC-7901 cells. The luciferase activity of cells was assessed using a luciferase kit 48 h after transfection.
Statistical analysis
Data analysis was performed using SPSS version 21.0 statistical software. Data were presented as the mean ± standard deviation (SD). A Fisher least significant difference (LSD) test was used for comparisons between the groups. P<0.05 was considered to be statistically significant.
Results
Clinical pathology, PTEN protein expression, and miR-205-5p expression
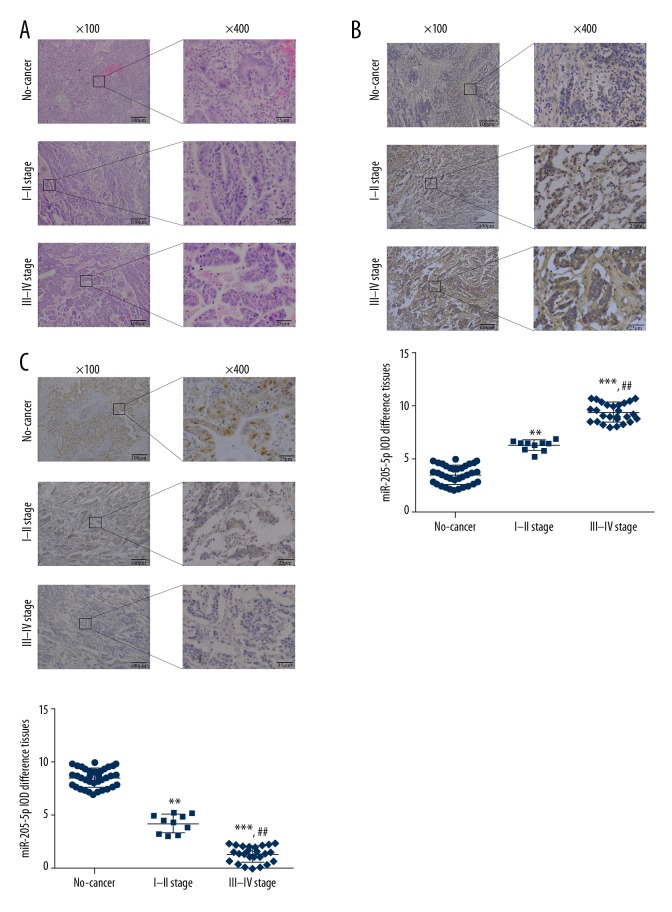
Histology was used to confirm that the normal adjacent gastric tissue samples were free from tumor, but some samples contained foci of mild atypical hyperplasia or intestinal metaplasia. Gastric cancer tissue tissues showed gastric cancer with tumor invasion (Figure 1A). The results of in situ hybridization (ISH) results showed that miR-205-5p was mainly expressed in the cytoplasm and cell nucleus. The gastric cancer tissues from patients with stage I–II and III–IV gastric cancer showed a significantly increased level of miR-205-5p expression when compared with normal gastric tissues (P<0.01 and P<0.001, respectively) (Figure 1B). Also, a significant difference in miR-205-5p expression levels was observed in gastric cancer tissue from patients with stage I–II and stage III–IV gastric cancer (P<0.01) (Figure 1B). Immunohistochemistry showed that gastric cancer tissue from patients with stage I–II and stage III–IV gastric cancer showed significantly reduced expression levels of PTEN compared with the normal adjacent gastric tissues (P<0.01 and P<0.001, respectively) (Figure 1C). PTEN protein expression differed significantly between gastric cancer tissue from patients with stage I–II and stage III–IV gastric cancer (P<0.01) (Figure 1C).
Figure 1.
Histology of gastric cancer tissue and adjacent normal gastric tissue, PTEN protein expression, and expression of micro-RNA 205-5p (miR-205-5p). (A) Adjacent normal gastric tissue, gastric cancer tissue from patients with stage I–II, and stage III–IV gastric cancer. Hematoxylin and eosin (H&E). Magnification, ×100 or ×400. (B) miR-205-5p expression by in situ hybridization. Magnification, ×100 or ×400. ** P<0.01 and *** P<0.001 compared with normal; ## P<0.01 compared with stage I–II. (C) PTEN protein expression by immunohistochemistry. Magnification, ×100 or ×400. ** P<0.01 and *** P<0.001 compared with Normal; ## P<0.01, compared with stage I–II.
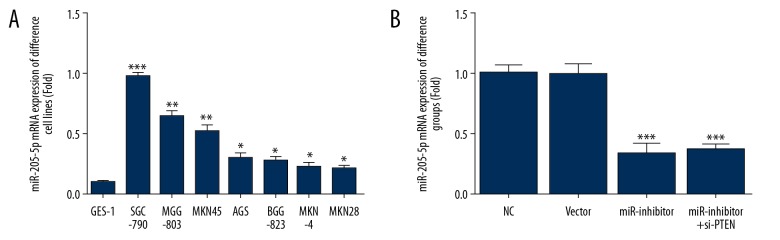
miR-205-5p mRNA expression
Quantitative reverse transcription PCR (qRT-PCR) showed a significant increase in miR-205-5p RNA expression level in different gastric cancer cell lines (P<0.05, P<0.01, or P<0.001) (Figure 2A), with the highest expression level in SGC-7901 cells. Also, miR-205-5p inhibitor transfection significantly inhibited miR-205-5p expression levels in the miR-inhibitor and miR-inhibitor+siPTEN groups compared with the NC group (P<0.001, respectively) (Figure 2B).
Figure 2.
micro-RNA 205-5p (miR-205-5p) expression in human gastric cancer cell lines. (A) miR-205-5p expression in different gastric cancer cell lines. * P<0.05, ** P<0.01 and *** P<0.001 compared with GES-1, which is a human gastric mucosal cell line. (B) miR-205-5p gene expression in different groups of AGS-7901 cells. *** P<0.001 compared with the normal control (NC) group.
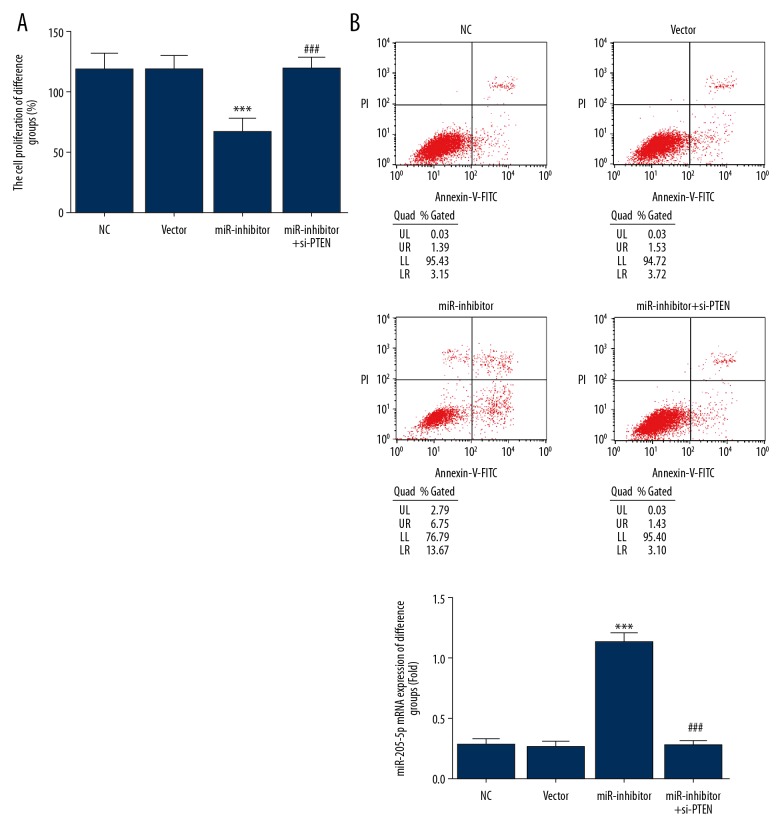
Analysis of cell proliferation and apoptosis
The miR-inhibitor group exhibited a significantly lower cell proliferation rate than the NC group (P<0.001) (Figure 3A). However, cotransfection with si-PTEN significantly increased cell proliferation in the miR-inhibitor+si-PTEN group compared with the miR-inhibitor group (P<0.001) (Figure 3A). The miR-inhibitor group exhibited a significantly higher cell apoptosis rate than the NC control (P<0.001) (Figure 3B), but was significantly decreased following si-PTEN co-transfection in the miR-inhibitor+si-PTEN group (P<0.001) (Figure 3B). The NC and vector groups did not differ significantly in terms of cell proliferation or apoptosis, indicating that no damage had occurred during transfection.
Figure 3.
Cell proliferation and apoptosis in human gastric cancer cell lines. (A)Cell proliferation rates in different cell groups assessed by the MTT assay. *** P<0.001 compared with the normal control (NC) group; ### P<0.001 compared with miR-inhibitor group. (B) Cell apoptosis rates of different groups assessed by flow cytometry. *** P<0.001 compared with the NC group; ### P<0.001 compared with the miR-inhibitor group.
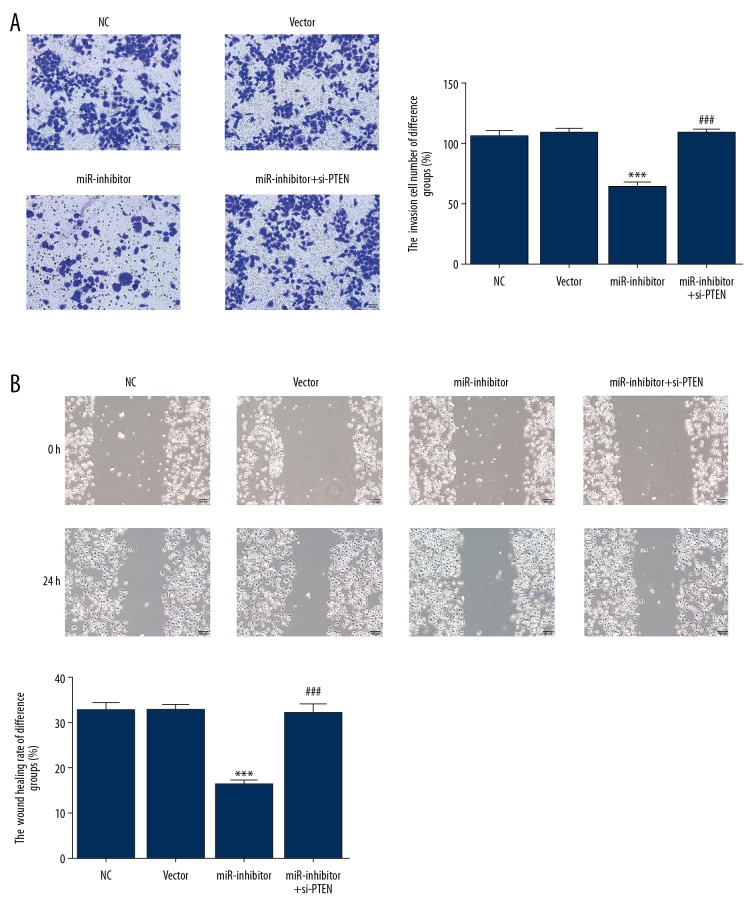
Cell migration
The transwell assay showed significantly lower numbers of invasive cells in the miR-inhibitor group when compared with the NC group (P<0.001) (Figure 4A). However, significantly higher numbers of migrating cells were observed in the miR-inhibitor+si-PTEN group than in the miR-inhibitor group (P<0.001). Similarly, the wound healing rate in the miR-inhibitor group was lower than that in the NC group (P<0.001) (Figure 4B), but the miR-inhibitor+si-PTEN group showed a significantly higher wound healing rate than the miR-inhibitor group (P<0.001).
Figure 4.
Cell migration of human gastric cancer cells. (A) Cell migration of GSC-7901 cells in the different groups assessed by the transwell assay. *** P<0.001 compared with the normal control (NC) group; ### P<0.001 compared with the miR-inhibitor group. (B) Wound healing rates of different groups assessed with a wound healing assay. *** P<0.001 compared with the NC group; ### P < 0.001 compared with the miR-inhibitor group.
Cell ultrastructure
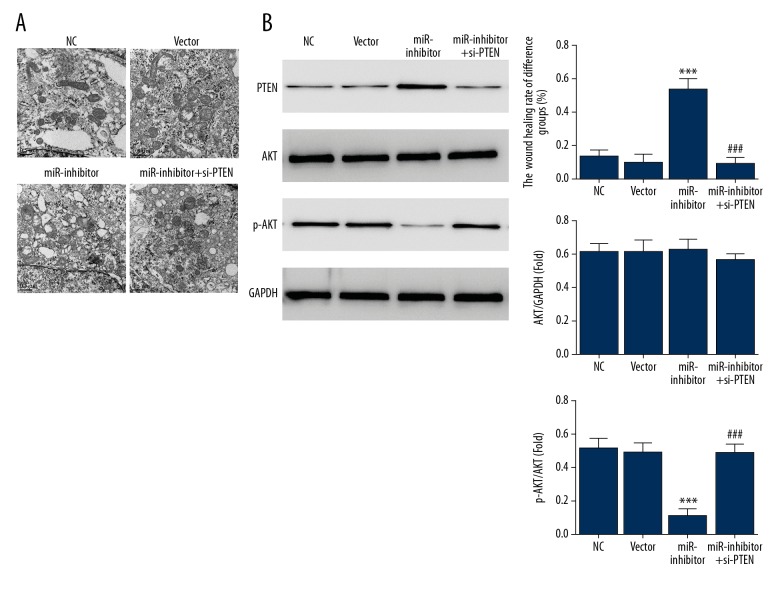
Compared with SGC-7901 cells in the miR-inhibitor group, cells in the NC and vector groups were irregular in shape, had abundant microvilli, small cytoplasm, large and irregular nuclei with notches and deformities, with a high nuclear to cytoplasm ratio, and large and dense nucleoli (Figure 5A). Some cells had more than one nucleolus. Within the cytoplasm, there were vacuoles and fewer mitochondria, which had an irregular shape and fewer cristae. The rough endoplasmic reticulum was underdeveloped and less abundant. Polyribosomes were more frequently observed than free ribosomes.
Figure 5.
Cell ultrastructure and protein-related changes in SGC-7901 human gastric cancer cells. (A) SGC-7901 cell ultrastructure of different groups assessed by transmission electron microscopy (TEM). (B) The relative proteins expression of different groups assessed by Western blot. *** P<0.001 compared with the normal control group; ### P<0.001 compared with the miR-inhibitor group.
In contrast, the miR-inhibitor group showed SGC-7901 cells with fewer microvilli, more cytoplasm, a lower nuclear to cytoplasmic ratio, round and regular nuclei with lesser deformities and denser nucleoli or more intra-nucleolus vacuoles than the NC and vector groups. Some cell nuclei showed degeneration, and cells formed glandular cavities and intercellular connections. Several organelles and more swollen mitochondria with cristae were observed and rough endoplasmic reticulum, smooth endoplasmic reticulum were present. The endoplasmic reticulum showed expansion, edema and vacuolar degeneration. Mature secretory granules and lipid droplets mixed with single ribosomes and polyribosomes were observed within the cytoplasm. The results showed that miR-205-5p inhibition induced SGC-7901 cell differentiation. Following miR-205-5p inhibitor and si-PTEN co-transfection, cellular ultrastructure was restored to normal.
Protein expression in SGC-7901 cells
Western blot showed a significant difference between the miR-inhibitor and NC groups in terms of PTEN and p-AKT protein expression levels (P<0.001, respectively) (Figure 5B). Compared with the miR-inhibitor group, PTEN protein expression decreased significantly and p-AKT protein expression increased significantly in the miR-inhibitor+si-PTEN group (P<0.001, respectively) (Figure 5B). No significant difference was detected in AKT protein expression among the four groups.
Prediction and verification of miR-205-5p
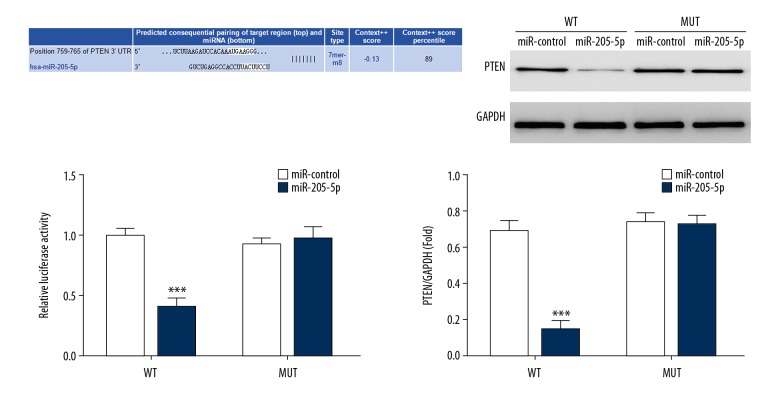
Luciferase activity in the miR-205-5p mimic group transfected with pGL3-PTEN-3′-UTR was significantly lower than that in the miR-205-5p mimic control group transfected with the pGL3-PTEN-3′-UTR and pGL3-CYLD-3′-UTR. Therefore, following PTEN-3′-UTR transfection with the luciferase vectors, miR-205-5p addition resulted in the inhibition of luciferase activity. However, when there was a mutation in the PTEN-3′UTR and miR-205-5p binding site, the luciferase activity was not inhibited, and miR-205-5p could significantly inhibit luciferase reporter gene activity in the WT PTEN-3′-UTR region. Western blot showed that the miR-205-5p mimic group had lower PTEN protein expression levels than those in the blank and miR-205-5p mimic control groups. These results suggest that PTEN was the target gene for miR-205-5p in gastric cancer cells (Figure 6).
Figure 6.
Prediction and verification of micro-RNA 205-5p (miR-205-5p) using luciferase vectors. *** P<0.001 compared with the miR mimic control transfected with the wild-type luciferase vector.
Discussion
Increasingly sophisticated studies have identified the roles of microRNA (miRNA) and have shown that miRNA is abnormally expressed in several tumors and is involved in the ontogeny, development, invasion, and progression of tumors. The target genes of miRNAs determine whether they function as oncogenes or antioncogenes and a single miRNA may regulate thousands of genes, but its expression and functions are influenced by multiple factors, such as gene sequence, phosphorylation levels, and enzymatic modification.
Several miRNAs are involved in the ontogeny and development of gastric cancer through different mechanisms. For instance, miR-25, miR-106, and miR-93b in the miR-106-25 family show significantly increased expression in gastric cancer tissues and cell lines, and they may participate in the ontogeny of gastric cancer [17]. Overexpression of miR-650 promotes gastric cancer by affecting tumor growth inhibiting factor 4 [18], whereas down-regulation of miR-27 can suppress gastric cancer cell proliferation [19]. increased expression of miR-9 has been shown in gastric cancer when compared with normal adjacent gastric tissues [20]. Therefore, these miRNAs function as oncogenes in gastric cancer, but some miRNAs can inhibit oncogenes. For example, miR-34a suppresses gastric cancer cell proliferation and migration by inhibiting its target, the platelet-derived growth factor receptor (PDGFR) gene [21].
Furthermore, miR-21 expression is increased in gastric cancer tissues associated with lymph node metastasis when compared with normal lymph nodes [22], indicating that its expression may be associated with gastric cancer metastasis. In the present study, we used in situ hybridization (ISH) to detect miR-205-5p expression in gastric cancer tissue and normal adjacent gastric tissues from patients with different stages of gastric cancer. The findings showed that gastric cancer tissues showed significantly increased expression of miR-205-5p when compared with normal adjacent gastric tissues, and this expression increased further in tumors from patients with an increased tumor stage. These findings suggest that miR-205-5p is an oncogene involved in gastric cancer. A dual-luciferase reporter assay confirmed that the PTEN-3′-UTR site sequence could hybridize with miR-205-5p and that miR-205-5p might directly regulate PTEN expression and activity.
PTEN is a homolog of protein-tyrosine-phosphatase and is located on human chromosome 10q2, and it functions as an antitumor factor in several cancers [23]. PTEN expression can affect cell apoptosis, proliferation, and migration in vitro and can affect intracellular mitochondria and suppress tumor growth [24–26]. The results of the present study showed that stimulation of PTEN expression by miR-205-5p inhibition significantly inhibited the activity of SGC-7901 cells and inhibited AKT phosphorylation. Ultrastructural analysis also showed that inhibition of miR-205-5p prevented damage to mitochondria, reduced the nuclear to cytoplasmic ratio, promoted the formation of connecting structures between cells, and increased the number of organelles and free ribosomes.
Previous in vitro studies have shown that AKT signaling activated gastric cancer cell proliferation and migration [27]. However, the effect of the PTEN/AKT signaling pathway in the progression of gastric cancer remains unclear. In addition to participating in cell apoptosis, the PTEN/AKT signaling pathway may be involved in cell cycle regulation in tumor resistance and angiogenesis [28]. The results of the present study indicate that PTEN may have an anticancer effect by inhibiting AKT phosphorylation.
Conclusions
Because overexpression of micro-RNA 205-5p (miR-205-5p) may be a factor in the ontogeny and development of gastric cancer, the detection of the expression levels in gastric tumor tissue may be a promising diagnostic and prognostic biomarker. This study showed that inhibition of miR-205-5p expression activated PTEN expression and inhibited AKT activation, and was associated with lower tumor stage or tumor progression.
Footnotes
Source of support: Departmental sources
Conflict of interest
None.
References
- 1.Ren J, Niu G, Wang X, et al. Overexpression of FNDC1 in gastric cancer and its prognostic significance. J Cancer. 2018;9:4586–95. doi: 10.7150/jca.27672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Japanese Gastric Cancer Association. Japanese gastric cancer treatment guidelines 2014 (ver.4) Gastric Cancer. 2017;20:1–19. doi: 10.1007/s10120-016-0622-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cavatorta O, Scida S, Miraglia C, et al. Epidemiology of gastric cancer and risk factors. Acta Biomed. 2018;89:82–87. doi: 10.23750/abm.v89i8-S.7966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shin VY, Ng EK, Chan VW, et al. A three-miRNA signature as promising non-invasive diagnostic marker for gastric cancer. Mol Cancer. 2015;14:202. doi: 10.1186/s12943-015-0473-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cortez MA, Anfossi S, Ramapriyan R, et al. Role of miRNAs in immune responses and immunotherapy in cancer. Genes Chromosomes Cancer. 2019;58(4):244–53. doi: 10.1002/gcc.22725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang W, Zang J, Jing X, et al. Identification of candidate miRNA biomarkers from miRNA regulatory network with application to prostate cancer. J Transl Med. 2014;12:66. doi: 10.1186/1479-5876-12-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nalluri JJ, Barh D, Azevedo V, Ghosh P. miRsig: A consensus-based network inference methodology to identify pan-cancer miRNA-miRNA interaction signatures. Sci Rep. 2017;7:39684. doi: 10.1038/srep39684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nicoloso MS, Spizzo R, Shimizu M, et al. MicroRNAs – the micro steering wheel of tumour metastases. Nat Rev Cancer. 2009;9:293–302. doi: 10.1038/nrc2619. [DOI] [PubMed] [Google Scholar]
- 9.Ueda T, Volinia S, Okumura H, et al. Relation between microRNA expression and progression and prognosis of gastric cancer: A microRNA expression analysis. Lancet Oncol. 2010;11:136–46. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi X, Xiao L, Mao X, et al. miR-205-5p mediated downregulation of PTEN contributes to cisplatin resistance in C13K human ovarian cancer cells. Front Genet. 2018;9:555. doi: 10.3389/fgene.2018.00555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghorbanmehr N, Gharbi S, Korsching E, et al. miR-21-5p, miR-141-3p, and miR-205-5p levels in urine-promising biomarkers for the identification of prostate and bladder cancer. Prostate. 2019;79:88–95. doi: 10.1002/pros.23714. [DOI] [PubMed] [Google Scholar]
- 12.De Cola A, Lamolinara A, Lanuti P, et al. MiR-205-5p inhibition by locked nucleic acids impairs metastatic potential of breast cancer cells. Cell Death Dis. 2018;9:821. doi: 10.1038/s41419-018-0854-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li L, Li S. miR-205-5p inhibits cell migration and invasion in prostatic carcinoma by targeting ZEB1. Oncol Lett. 2018;16:1715–21. doi: 10.3892/ol.2018.8862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaudhary AK, Mondal G, Kumar V, et al. Chemosensitization and inhibition of pancreatic cancer stem cell proliferation by overexpression of microRNA-205. Cancer Lett. 2017;402:1–8. doi: 10.1016/j.canlet.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amin MB, Greene FL, Edge SB, et al. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. Cancer J Clin. 2017;67:93–99. doi: 10.3322/caac.21388. [DOI] [PubMed] [Google Scholar]
- 16.Sobin LH, Gospodarowicz MK, Wittekind C. TNM classification of malignant tumors. 7th ed. Hoboken: John Wiley & Sons; 2009. pp. 239–48. [Google Scholar]
- 17.Petrocca F, Visone R, Onelli MR, et al. E2F1-regulated microRNAs impair TGF beta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–86. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 18.Zhang X, Zhu W, Zhang J, et al. MicroRNA-650 targets ING4 to promote gastric cancer tumorigenicity. Biochem Biophys Res Commun. 2010;395:275–80. doi: 10.1016/j.bbrc.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 19.Liu T, Tang H, Lang Y, et al. MicroRNA-27a functions as an oncogene in gastric adenocarcinoma by targeting prohibitin. Cancer Lett. 2009;273:233–42. doi: 10.1016/j.canlet.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 20.Tsai KW, Liao YL, Wu CW, et al. Aberrant hypermethylation of miR-9 genes in gastric cancer. Epigenetics. 2011;6:1189–97. doi: 10.4161/epi.6.10.16535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng Y, Gao JJ, Liu YM, Wu XL. MicroRNA-34A inhibits the growth, invasion and metastasis of gastric cancer by targeting PDGFR and MET expression. Biosci Rep. 2014;34 doi: 10.1042/BSR20140020. pii: e00112. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Xu Y, Sun J, Xu J, et al. miR-21 is a promising novel biomarker for lymph node metastasis in patients with gastric cancer. Gastroenterol Res Pract. 2012;2012 doi: 10.1155/2012/640168. 640168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Juric D, Castel P, Griffith M, et al. Convergent loss of PTEN leads to clinical resistance to a PI(3) Kalpha inhibitor. Nature. 2015;518:240–44. doi: 10.1038/nature13948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu LF, Wu ZP, Chen Y, et al. MicroRNA-21 (miR-21) regulates cellular proliferation, invasion, migration, and apoptosis by targeting PTEN, RECK, and Bcl-2 in lung squamous carcinoma, Gejiu City, China. PLoS One. 2014;9:e103698. doi: 10.1371/journal.pone.0103698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maxwell PJ, Neisen J, Messenger J, Waugh DJ. Tumor-derived CXCL8 signaling augments stroma- derived CCL2- promoted proliferation and CXCL12-mediated invasion of PTEN-deficient prostate cancer cells. Oncotarget. 2014;5:4895–908. doi: 10.18632/oncotarget.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen X, Wang W, Zhang J, Li S, et al. Involvement of caspase-3/PTEN signaling pathway in isoflurane-induced decrease of self-renewal capacity of hippocampal neural precursor cells. Brain Res. 2015;1625:275–86. doi: 10.1016/j.brainres.2015.08.047. [DOI] [PubMed] [Google Scholar]
- 27.Choi JH, Lee GH, Kim HG, et al. Suppression of PMA-induced human fibrosarcoma HT-1080 invasion and metastasis by kahweol via inhibiting Akt/JNK1/2/p38 MAPK signal pathway and NF-κB dependent transcriptional activities. Food Chem Toxicol. 2018;125:1–9. doi: 10.1016/j.fct.2018.12.038. [DOI] [PubMed] [Google Scholar]
- 28.Li J, Gong X, Jiang R, et al. Fisetin inhibited growth and metastasis of triple-negative breast cancer by reversing epithelial-to-mesenchymal transition via PTEN/Akt/GSK3β signal pathway. Front Pharmacol. 2018;9:772. doi: 10.3389/fphar.2018.00772. [DOI] [PMC free article] [PubMed] [Google Scholar]