Abstract
Cytochrome P450 3A4 isoform (CYP3A4) transcription is controlled by hepatic transcription factors (TFs), but how TFs dynamically interact remains uncertain. We hypothesize that several TFs form a regulatory network with nonlinear, dynamic, and hierarchical interactions. To resolve complex interactions, we have applied a computational approach for estimating Sobol’s sensitivity indices (SSI) under generalized linear models to existing liver RNA expression microarray data (GSE9588) and RNA-seq data from genotype-tissue expression (GTEx), generating robust importance ranking of TF effects and interactions. The SSI-based analysis identified TFs and interacting TF pairs, triplets, and quadruplets involved in CYP3A4 expression. In addition to known CYP3A4 TFs, estrogen receptor α (ESR1) emerges as key TF with the strongest main effect and as the most frequently included TF interacting partner. Model predictions were validated using small interfering RNA (siRNA)/short hairpin RNA (shRNA) gene knockdown and clustered regularly interspaced short palindromic repeats (CRISPR)–mediated transcriptional activation of ESR1 in biliary epithelial Huh7 cells and human hepatocytes in the absence of estrogen. Moreover, ESR1 and known CYP3A4 TFs mutually regulate each other. Detectable in both male and female hepatocytes without added estrogen, the results demonstrate a role for unliganded ESR1 in CYP3A4 expression consistent with unliganded ESR1 signaling reported in other cell types. Added estrogen further enhances ESR1 effects. We propose a hierarchical regulatory network for CYP3A4 expression directed by ESR1 through self-regulation, cross regulation, and TF-TF interactions. We also demonstrate that ESR1 regulates the expression of other P450 enzymes, suggesting broad influence of ESR1 on xenobiotics metabolism in human liver. Further studies are required to understand the mechanisms underlying role of ESR1 in P450 regulation.
SIGNIFICANCE STATEMENT
This study focuses on identifying key transcription factors and regulatory networks for CYP3A4, the main drug metabolizing enzymes in liver. We applied a new computational approach (Sobol’s sensitivity analysis) to existing hepatic gene expression data to determine the role of transcription factors in regulating CYP3A4 expression, and used molecular genetics methods (siRNA/shRNA gene knockdown and CRISPR-mediated transcriptional activation) to test these interactions in life cells. This approach reveals a robust network of TFs, including their putative interactions and the relative impact of each interaction. We find that ESR1 serves as a key transcription factor function in regulating CYP3A4, and it appears to be acting at least in part in a ligand-free fashion.
Introduction
Most abundant among all drug-metabolizing enzymes in the liver, the cytochrome P450 3A4 isoform (CYP3A4) metabolizes 30%–45% of commonly used drugs (Danielson, 2002). Large interperson variability in both basal and induced CYP3A4 expression/activity (Rahmioglu et al., 2011; Achour et al., 2014) strongly influences optimal drug dosage, efficacy, and toxicity (Werk and Cascorbi, 2014), but the underlying causes remain largely unknown. Early studies had estimated that genetic factors constitute 80%–90% of basal and 66% of induced CYP3A4 variability (Penno et al., 1981; Ozdemir et al., 2000; Rahmioglu et al., 2011), but genetic factors contributing to CYP3A4 variability remain poorly defined. The frequency of CYP3A4 coding region variants is low (Wang and Sadee, 2012). We have identified a cis-acting regulatory intronic variant named CYP3A4*22 (minor allele frequency 5% in Europeans), which reduces CYP3A4 expression via aberrant splicing (Wang and Sadee, 2016) and has been included in clinical biomarker panels predicting CYP3A4 activity (Wang et al., 2011). However, CYP3A4*22 cannot account for a large portion of CYP3A4 variability that likely involves trans-acting transcription factors (TFs) and epigenetic processes.
Transcription of CYP3A4 genes, located in a cluster of CYP3A4, CYP3A5, CYP3A7, and CYP3A43, is regulated by dynamic interactions of numerous TFs (Ourlin et al., 1997; Jover et al., 2001; Rodríguez-Antona et al., 2003; Tirona et al., 2003; Martínez-Jiménez et al., 2007; Istrate et al., 2010; Thomas et al., 2013). While genetic TF variants can affect CYP3A expression in trans, the effect size of each single TF variant is limited by the complex interplay between several TFs and environmental conditions that impact TF expression. Thus, TF-TF interactions and epistasis between TF variants need to be considered to account for CYP3A4 variability.
A previous study proposed that the combined effect of sex and genetic variants in ABCB1 and several TFs accounts for 24.6% of the variation in hepatic CYP3A4 expression, using multiple linear regression analysis (Lamba et al., 2010), but the P value increased with each predictor added to the model, and the final model was not statistically significant (P = 0.1). Another study (Yang et al., 2010), incorporating drug-metabolizing activity, whole genome expression, and single-nucleotide polymorphism data from 466 human livers, has provided a comprehensive view of the functionality, genetic control, and interactions of TFs and P450 enzymes; yet the specific roles of each TF and dynamic interactions with other TFs remain unclear. Specifically, the expression levels of ESR1 (estrogen receptor α ESR1) and other known P450 regulators (such as pregnane X receptor [NR1I2], constitutive androstane receptor [NR1I3], hepatocyte nuclear factor 4α [HNF4A]) were correlated with CYP3A4 and other measured enzyme activity levels, but these TFs failed to be included in a higher order regulatory network (Yang et al., 2010). The possible reason might be that conventional methods failed to control for the correlation between TFs (the input variables) that are known to be dependent on each other because of cross-expression regulation (Odom et al., 2004).
This raises the question of the validity of traditional coexpression based methods to predict functional interactions. To overcome this issue, we have developed a novel computational approach for estimating Sobol’s sensitivity indices (SSI) under generalized linear models (Lu et al., 2018), the potential utility of which was illustrated with a computed network of TFs regulating CYP3A4 expression. SSI provides a unified way of quantifying output sensitivity with respect to any subset of the input variables, based on the variance decomposition of the response variable with applications in analyzing various complex systems (Xu and Gertner, 2008; Li et al., 2010; Chastaing et al., 2012, 2015; Mara and Tarantola, 2012).
Unlike a weighted coexpression network algorithm (Zhang and Horvath, 2005), which uses the absolute value of pairwise expression correlation to define gene expression similarity, SSI analysis allows us to build the coexpression network using a relatively smaller data set (fewer parameters need to be estimated) and to potentially avoid accumulating analytic biases introduced from piling up multiple different modeling techniques in a pipeline.
Compared with other multivariate regression and P value based analyses, our SSI based analysis has these features: 1) It can generate robust importance ranking of the input variables without first requiring the development of a model of the complete dynamic system. 2) It can tolerate irrelevant inputs without affecting the importance ranking of other relevant inputs. 3) It allows for dependence among input variables. Thus, SSI is well suited for analyzing TF–CYP3A4 interactions with an observational data set of modest size. Moreover, our simulation studies have shown SSI-based analysis outperforms ANOVA P values and multivariate regression P values in feature selection (Lu et al., 2018).
In this study, we applied SSI analysis to existing liver gene expression microarray data (GSE9588) (Yang et al., 2010), as used in our preliminary SSI analysis (Lu et al., 2018), and RNA-seq data from genotype-tissue expression (GTEx) (Battle et al., 2017), to test key TFs and their interactions critical for CYP3A4 expression. Then we used gene knockdown (KD) or clustered regularly interspaced short palindromic repeats (CRISPR)–cas9 mediated TF gene transcriptional activation (TA) to evaluate the predicted effects of TFs on CYP3A4 expression. The results define TF interactions and support the hypothesis that unliganded ESR1 is a master regulator among several other TFs of CYP3A4. Because these TFs also affect other P450 enzymes in human liver, we extended the analysis to determine other P450 enzymes that are similarly affected by ESR1 and their TF network.
Materials and Methods
SSI Analysis of TF Interactions with CYP3A4.
The mRNA data sets used are published microarray data (GSE9588) from 427 livers (Yang et al., 2010) and RNA-seq data from GTEx (153 livers). We selected 45 liver enriched transcription factors (TF) (Yang et al., 2010). Seventy-eight probes were used in microarray data because some of the TFs were measured by more than one probe. In the first-order analysis, we estimated the SSI of each single TF by fitting a univariate polynomial model of degree 3.
The main effect Sobol’s indices were estimated by empirical variances of the best fitting polynomial expressions. Ranking of these Sobol’s indices can be interpreted as ranking the proportion of CYP3A4 (or other P450 enzymes) variability that can be explained by individual candidate TFs.
In the second-order analysis (to test the interactions between two TFs), we estimated the main effect Sobol’s indices of TF pairs by fitting full cubic polynomial model that contains all pairwise product terms in addition to the terms used in the first-order analysis. A similar method was used for the third-order analysis (to test the interactions between three TFs). For fourth-order analysis (to test the interactions between four TFs), because the total number of possible TF quadruplets was too large (>1.4 million), we selected only TFs that appeared in the top 200 triplets (the total number of possible quadruplets was 194,580).
For the detailed SSI method description, please refer to Lu et al. (2018). The relevant functions for SSI estimation under general linear models with identity, log, and logit links are implemented and available in the R package “Sobol sensitivity” (CRAN repository; also see Supplemental Methods for the link to the R script).
Cell Culture Conditions.
Experiments were performed on two cell types expressing CYP3A4: Huh7 from biliary epithelial cells and primary human hepatocytes. Primary human hepatocytes (Lonza, Basel, Switzerland) (see Supplemental Table 1 for demographics) were incubated in serum-free William’s E medium supplemented with penicillin/streptomycin/Fungizone (100 U/100 µg/0.25 µg/ml), 100 nM dexamethasone, 2 mM l-glutamine, 15 mM HEPES, and ITS (0.55 mg/ml human transferrin, 1 mg/ml bovine insulin, and 0.5 µg/ml sodium selenite, from Sigma-Aldrich). Cells were cultured on Matrigel Basement Membrane Matrix to create a 2D culture condition.
Hepatocytes from all six donors were used for estrogen-stimulation experiments. Hepatocytes only from donor Hum-1, Hum-3, and Hum-6 were used for small interfering RNA (siRNA) experiments. Huh7 cells were cultured at 37°C in a humidified incubator at 5% CO2 in Dulbecco’s modified Eagle’s medium supplemented with 10% charcoal-stripped serum (Sigma-Aldrich), 100 U/ml penicillin, and 100 µg/ml streptomycin.
Gene KD of ESR1 and Select TFs with shRNA and/or siRNA.
Short hairpin RNA (shRNA) targeting ESR1, NR1I3, and progesterone receptor membrane component 1 (PGRMC1) were designed as reported (Huang and Sinicrope, 2010) (Supplemental Table 2). DNA fragments were inserted into Lentiviral-based shRNA vector PSIH1 (ID#26597; Addgene) using BamH1 and EcoRI sites. Lentiviral particles were prepared as described by Wang et al. (2015) and transduced into Huh7 cells. The cells were harvested 72 hours later. Accell siRNAs targeting ESR1 (Accell siESR1 SMART pool) was purchased from GE Healthcare Dharmacon (Lafayette, CO). Silencer siRNAs targeting ESR1 (#4098), forkhead box protein A2 (FOXA2, #5941), NR1I2 (#6638), HNF4A (#290203), PGRMC1 (#108079), NR1I3 (#5535), peroxisome proliferator activated receptor α (PPARA, #5348), and nontargeting control were purchased from Thermo Fisher Scientific (Waltham, MA).
Accell siRNAs were introduced into Huh7 cells by incubating with 1 µM siRNA in the delivery medium. For Silencer siRNAs, lipofectamine RNAiMAX reagent was used. After incubation for 72 hours (Huh7 cells) or 6 days (hepatocytes), the cells were harvested for total RNA preparation, reverse transcription, and real-time polymerase chain reaction (PCR) quantification. The shESR1 stable cell lines were established using puromycin selection.
CRISPR-Mediated Gene TA.
By fusing VP64 (the universal transcription activator) with dcas9 (an inactive mutant cas9 protein), the dcas9-VP64 fusion protein specifically activates gene transcription when tethered by guide RNA (gRNA) to a target gene promotor (Konermann et al., 2015). We used lentiviral based vectors—Lentiviral-VP64-dcas9 (#61425; addgene) and LentisgRNA vector (#61427; Addgene)—for the VP64-dcas9 fusion protein and gRNA delivery. The gRNA sequences were from Konermann et al. (2015) (Supplemental Table 2); three gRNA for each promoter of the TFs are listed in Supplemental Table 2.
Three promoter domains were targeted for ESR1: isoforms 001 (NM_000125), 201(NM_001122742), and 202 (NM_001122740). These isoforms are ubiquitously expressed, share the same coding sequence, but differ at their 5′ untranslated region. For transient expression, lentiviral particles containing expression vectors for VP64-dcas9 and a mixture of three gRNA targeting a specific gene promoter were incubated with Huh7 in the presence of 8 µg/ml SureEntry transduction reagent (Qiagen, Valencia, CA) for 24 hours. Cells were harvested 72 hours after transduction. Huh7 cell lines stably expressing VP64-dcas and gRNA targeting ESR1 promoters were established by blasticidin and Zeocin double selection.
RNA Preparation and RNA Expression Analysis.
Total RNA was prepared using RNA mini prep kits from Zymo Research (Irvine, CA). RNA was reverse transcribed into cDNA using RTIII reverse transcriptase (Life Technologies, Carlsbad, CA) and measured using real-time PCR with specific primers (Supplemental Table 2) and SYBR Green PCR master mix (Life Technologies), using GAPDH as the internal control (Wang et al., 2011). Human liver samples were obtained from the Cooperative Human Tissue Network (CHTN, Bethesda, MD) (see Supplemental Table 3 for the demographics of liver donors).
CYP3A4 Protein Analyses Using Capillary Western Blot Technology.
Human hepatocytes in six-well plates were lysed with 100 µl of radioimmunoprecipitation assay lysis buffer supplemented with protease inhibitor cocktail (Roche, South San Francisco, CA). Total protein concentrations were measured using Bradford method (Thermo Fisher Scientific).
Capillary Western analyses were performed using the Protein Simple Jess system (Bio-Techne, San Jose, CA) according to manufacturer’s protocol. Briefly, cell lysates were diluted with 0.1 × sample buffer to concentration of 1 mg/ml. Then four parts of diluted samples were combined with one part 5 × fluorescent master mix (containing 5 × sample buffer, 5 × fluorescent standard, and 200 mM dithiothreitol) and heated at 95°C for 5 minutes. Then the denatured samples, blocking reagent, mouse anti-CYP3A4 antibody (clone 946002, at 1:50 dilution; R&D Systems, Minneapolis, MN), rabbit anti-β-actin antibody (ab8227, 1:50; Abcam, Cambridge, MA), horseradish peroxidase–conjugated anti-mouse secondary antibody (1:20), fluorescent-conjugated near infrared anti-rabbit secondary antibody (1:20), and chemiluminescent substrate were dispensed into designated wells in an assay plate. A biotinylated ladder provided the molecular weight standard for each assay. After plate loading, the separation, electrophoresis, and immunodetection steps take place in the fully automated capillary system.
Data Analysis.
Data are expressed as mean ± S.D. Statistical analysis was performed using Prism (GraphPad Software, San Diego, CA). The University of Florida Biosafety Committee and the institutional review board committee approved the lentivirus study and human tissue study, respectively.
Results
SSI Analysis Identifies ESR1 as a Main Regulator of CYP3A4 Expression.
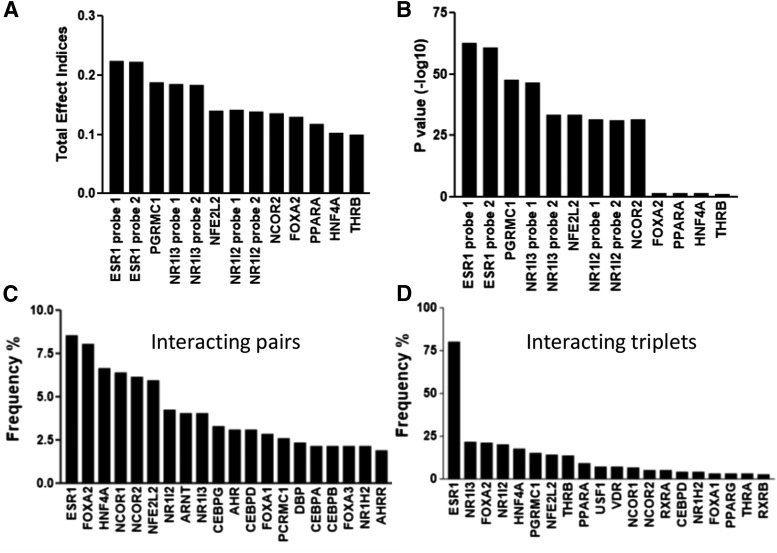
We used published microarray data (GSE9588) from 427 livers and selected 45 liver-enriched candidate TFs (Yang et al., 2010) for the SSI analysis. Some TFs were measured by more than one probe. First-order analysis identified 10 TFs with the highest SSI values (Fig. 1, A and B). As expected, these TFs were similar to the previous results with regression analysis (Yang et al., 2010) using the same data set. Two probes (both target coding regions) for ESR1 scored with the highest first-order total effect index and the lowest P value (Fig. 1, A and B).
Fig. 1.
Influence of the top-scoring liver-enriched transcription factors (from a total of 45 selected TFs) obtained from SSI analysis, using microarray gene expression data from 427 livers. (A and B) Results from the first-order analysis, total effect indices (A) and P value (B). (C and D) Results from the higher order analysis, frequencies of TFs appearing in the top 1000 interacting pairs (C) and triplets (D). TFs measured by more than one microarray probe yielded similar results.
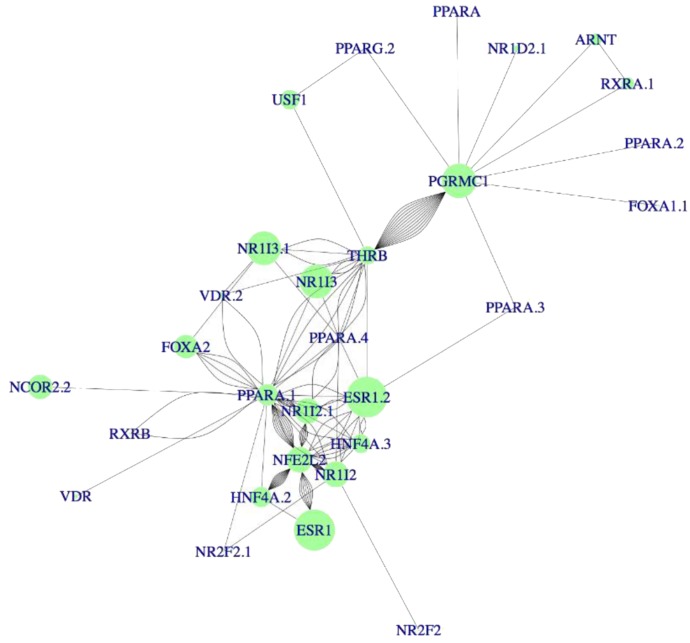
Because the RNA data sets included liver tissues from both male and female donors, we tested whether the role of ESR1 was driven by female livers, and ESR1 was the strongest predictor for CYP3A4 expression regardless of the sex of the donors. Subsequent higher order analysis to test the interactions between two, three, or four TFs identified interacting pairs, triplets, and quadruplets critical to CYP3A4 expression (Fig. 1, C and D; Fig. 2). Again, ESR1 appeared most frequently in the top 1000 interacting pairs and triplets (Fig. 1, C and D) with the strongest main effects (Fig. 2), suggesting ESR1 coordinates CYP3A4 expression by interacting with other TFs.
Fig. 2.
Key transcription factors and their interactions affecting CYP3A4 expression identified with SSI analysis. Dot sizes represent the main effect of each TF on CYP3A4 expression: the larger the dot, the greater the regulatory effect of that TF had on CYP3A4 expression. The lines represent the interactions between the connected TFs, and the distance of the line indicates the effect size of the interactions: the shorter the distance, the higher the SSI value of the interaction. The main quadruples are formed by different combinations of critical TFs: ESR1, HNF4A, PPARA, NR1I2, NR1I3, NFE2L2, NCOR, and THRB. TFs measured by more than one microarray probe yielded similar results.
Most of the TFs identified in first-order analysis were consistently identified also in the higher order network analysis. This was in contrast to previous study (Yang et al., 2010) where the TFs identified in regression analysis failed to be the top regulator or hub genes in the network analysis. To replicate this result, we downloaded RNA-seq data from GTEx. Because of the smaller sample size (153 livers), the full higher order SSI analysis was not feasible, so we applied first-order analysis to select the best predictors. Again, ESR1 emerged as a predictor for CYP3A4 expression (Table 1), consistent with the results from the microarray data. Moreover, NR1I3, NR1I2, and FOXA2 were also recognized as regulators of CYP3A4 in the GTEx data.
TABLE 1.
Gene expression regulators for drug-metabolizing P450 enzymes predicted by Sobol’s sensitivity indices analysis using microarray and GTEx data
| Gene | Microarray Data | GTEx Data |
|---|---|---|
| CYP3A4 | ESR1, NR1I3, PGRMC1, FOXA2, NR1I2 | ESR1, NR1I3, FOXA2, NR1I2 |
| CYP3A5 | ESR1, NR1I3, PGRMC1 | ESR1, NR1I2, NR1I3, NR1H3 |
| CYP3A7 | ESR1, NR1I3, PGRMC1 | ESR1, NR1I2, NR1I3, NR3C1 |
| CYP3A43 | ESR1, NR1I3, PGRMC1 | No clear predictors |
| CYP2B6 | ESR1, NR1I3, PGRMC1 | NR1I2, NR1I3 |
| CYP2C9 | NR1I3, PGRMC1, ESR1 | NR1I3, ESR1, PGRMC1, NR1H3, PPARD |
| CYP2C19 | NR1I3, PGRMC1, ESR1, NCOR2 | ESR1, NR3C1, NR1I2 |
| CYP2A6 | No clear predictors | No clear predictors |
| CYP2D6 | No clear predictors | No clear predictors |
| CYP1A1 | No clear predictors | No clear predictors |
| CYP1A2 | No clear predictors | No clear predictors |
| CYP1B1 | No clear predictors | No clear predictors |
Test SSI Results Using Gene KD and CRISPR-Mediated Gene TA.
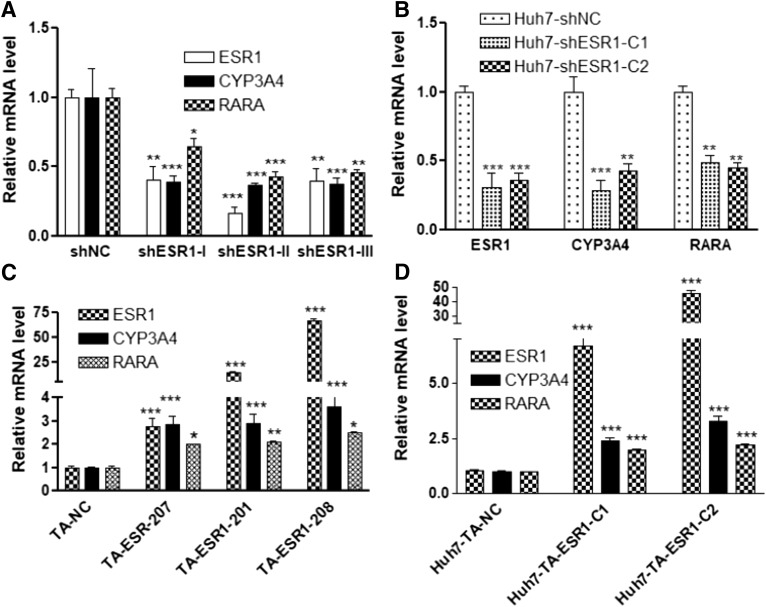
For ESR1 KD, we used three different shRNAs to knock down ESR1 in Huh7 cells (see Supplemental Table 2 for the shRNA sequence). All three shRNAs reduced ESR1 expression to 17%–40% of the negative control levels, resulting in >60% reduction of CYP3A4 expression (Fig. 3A).
Fig. 3.
Effects of ESR1 KD (A and B) or ESR1 TA (C and D) on the expression of CYP3A4 in Huh7 cells. (A) Transient ESR1 KD using shRNA. (B) Two stable ESR1 KD Huh7 cell lines (Huh7-shESR1-C1 and Huh7-shESR1-C2) established using shRNA. (C) Transient expression of ESR1 TA gRNA. (D) ESR1 TA gRNA expression in stable Huh7 cell lines: Huh7-TA-ESR1-C1 (201) and Huh7-TA-ESR1-C2 (208). RARA serves as the positive control for ESR1 KD or TA. Data are mean ± S.D., compared with negative control (NC): *P < 0.05; **P < 0.01; ***P < 0.001, ANOVA with Bonferroni comparisons of selected pairs post test.
To replicate this result, we used siRNA. Similar to shRNA, siRNA from two different sources (Silencer and Accell siRNA) both decreased ESR1 mRNA levels ≥50%, leading to ∼50% reduction in CYP3A4 expression (Supplemental Fig. 1). To test the long-term effect of ESR1 KD on CYP3A4 expression, we established stably transfected cell lines using shRNA. Compared with the negative controls (Huh7-shNC), the two shESR1 Huh7 cell lines (Huh7-shESR1-C1 and Huh7-shESR-C2) expressing lower ESR1 levels also had substantially lower CYP3A4 expression (Fig. 3B). As a positive control, we also tested the expression level of RARA, a nuclear receptor previously shown to be responsive to ESR1 depletion in MCF7 cells (Caizzi et al., 2014). As expected, RARA expression decreased upon ESR1 KD.
For the TA experiments, we used CRISPR-mediated gene TA to increase the ESR1 expression in Huh7 cells. We designed TA gRNA targeting three ESR1 promoters (Supplemental Table 2) that drive the ubiquitous expression of three ESR1 isoforms: ESR1-201 (ENST00000206249), ESR-207 (ENST00000440973), and ESR1-208 (ENST00000443427), respectively. These three ESR1 isoforms encode the same ESR1 protein but differ in their 5′ untranslated region. Compared with the negative control (TA-NC), TA gRNA targeting different ESR1 promoters increased ESR1 expression from 3- to 67-fold (Fig. 3C). All these conditions consistently increased CYP3A4 expression up to 3-fold regardless of the wide range of increases in ESR1 expression (Fig. 3C).
Similar results were obtained with two TA gRNA expressing stable cell lines—Huh7-TA-ESR1-C1 (ESR1-201) and Huh7-TA-ESR1-C2 (ESR1-208)—in which ESR1 mRNA levels increased 7- and 46-fold while CYP3A4 expression increased 2- and 3-fold, respectively (Fig. 3D). These results demonstrate a key role of ESR1 in CYP3A4 expression.
Given the low expression of CYP3A4 mRNA in Huh7 compared with hepatocytes, it is possible that a 3-fold increase represents a ceiling level in Huh7 cells. Alternatively, ESR1 has many splice isoforms (Flouriot et al., 2000; Poola et al., 2000; Perlman et al., 2005), some of which display constitutive or dominant negative activities (Flouriot et al., 2000; Hattori et al., 2016). Different ESR1 expression profiles in Huh7 cells compared with human liver may limit its trans-activating effects on CYP3A4 expression. Indeed, Huh7 with TA-ESR1 expresses many ESR1 splice variants missing different coding exons, such as exon 2, exon 4, exon5, and exon 6. These exon-missing variants encode either nonfunctional protein or C-terminal truncated ESR1 proteins (Hattori et al., 2016). Therefore, ESR1 TA may not generate sufficient levels of functional ESR1 protein.
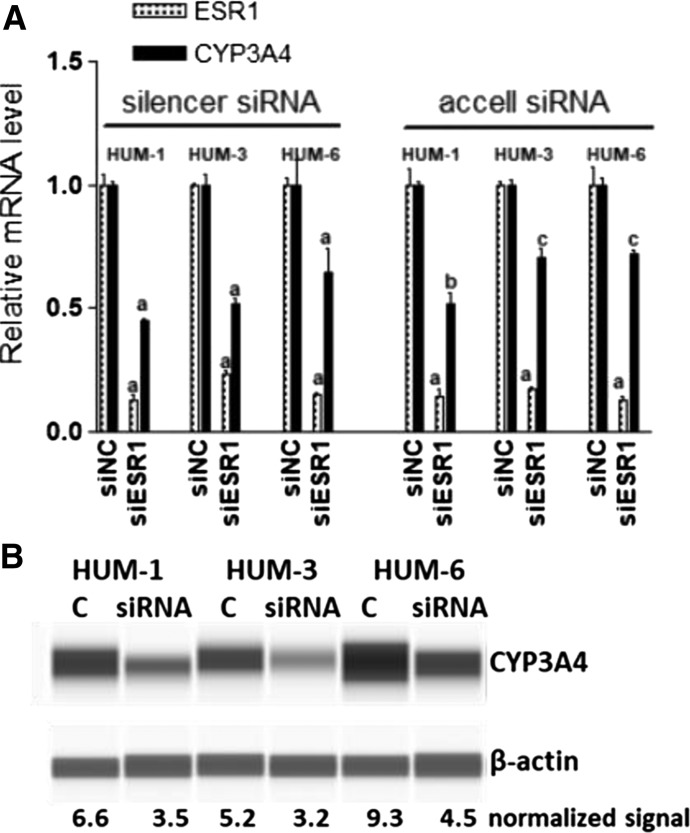
Moreover, it is unclear whether those C-terminal truncated isoforms have different trans-activity or dominant negative effects on full-length ESR1. The finding that expression of the positive control RARA also increased upon all TA experiments to a similar extent as CYP3A4 supports this notion. To test whether ESR1 regulates CYP3A4 expression in human primary culture hepatocytes, we treated hepatocytes cultured on Matrigel overlay (2D culture) with ESR1 siRNA for 6 days. This reduced the ESR1 expression over 80%, and decreased the CYP3A4 expression ∼50% (Fig. 4A), consistent with the results from Huh7 cells. The expression of RARA mRNA also decreased to 65% with ESR1 siRNA (mean ± S.D., 65.9% ± 9.3%, P < 0.05). Consistently, the average CYP3A4 protein expression also reduced to 55% of control (mean ± S.D., 54.8% ± 6.9%) after ESR1 KD (Fig. 4B).
Fig. 4.
Effects of ESR1 KD with silencer siRNA or Accell siRNA on the expression of CYP3A4 in primary culture hepatocytes from three donors. (A) ESR1 and CYP3A4 mRNA expression. Compared with negative control (siNC): aP < 0.001; bP < 0.01; cP < 0.05, ANOVA with Bonferroni comparison of selected pairs post test. (B) Image of CYP3A4 protein expression from capillary Western blotting.
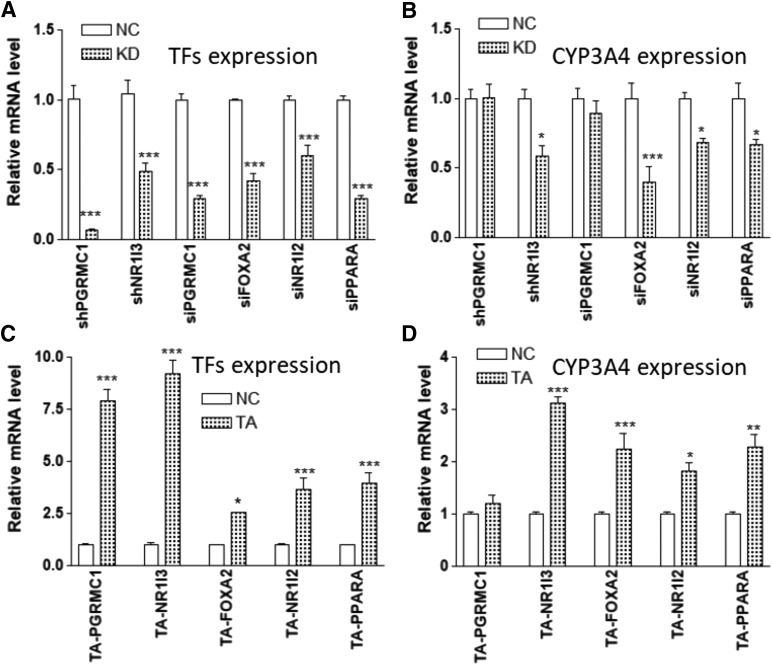
SSI analysis identified PGRMC1 as a potential TF in CYP3A4 expression, directing a small network independent of ESR1 in the microarray data set (Fig. 2). KD of PGRMC1 using shRNA or siRNA (over 70% reduction) or TA of PGRMC1 expression (up to 8-fold) with TA gRNA did not change CYP3A4 expression (Fig. 5), indicating that PGRMC1 does not regulate CYP3A4 expression.
Fig. 5.
Effects of gene KD (using shRNA or siRNA) (A and B) or CRISPR-mediated gene TA (C and D) of indicated transcription factors on CYP3A4 expression in Huh7 cells. (A and C) The mRNA levels of indicated transcription factors. (B and D) The mRNA levels of CYP3A4. Compared with negative control (NC): *P < 0.05; **P < 0.01; ***P < 0.001, ANOVA with Bonferroni comparison of selected pairs post test.
Other TFs that repeatedly appear as CYP3A4 regulators in SSI analysis, such as NR1I3, NR1I2, and FOXA2, are known CYP3A4 transcriptional regulators. Manipulation by KD and TA of the expression of these TFs all significantly changed the expression of CYP3A4, as expected (Fig. 5).
PPARA is a known CYP3A4 regulator, and KD PPARA in hepatocytes reduced the CYP3A4 expression (Klein et al., 2012). Although not detected in the first-order analysis, PPARA appears in network analyses (Fig. 2) and frequently interacts with ESR1 triplets (Fig. 1D), indicating PPARA is part of the ESR1 network. Consistent with the previous results, KD PPARA reduced CYP3A4 expression (Fig. 5).
Cross-Regulation between ESR1 and Other Key TFs.
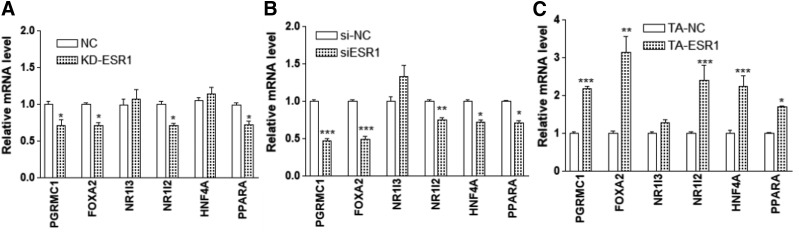
We then tested whether changes in ESR1 expression would affect the expression of the TFs tested in this study or other known CYP3A4 regulators. KD of ESR1 in Huh7 (76% reduction in ESR1) decreased PGRMC1, FOXA2, NR1I2, and PPARA expression (Fig. 6A). Similarly, KD of ESR1 in the primary culture hepatocytes (82% reduction in ESR1) decreased the expression of PGRMC1, FOXA2, NR1I2, PPARA, and HNF4A (Fig. 6B), but had no effect on NR1I3 in either cell type.
Fig. 6.
Effects of ESR1 KD (A and B) or CRISPR-mediated gene TA (C) on the expression of transcription factors as indicated in Huh7 cells (A and C) or hepatocytes (B). Compared with negative control (NC): *P < 0.05; **P < 0.01; ***P < 0.001, ANOVA with Bonferroni comparison of selected pairs post test.
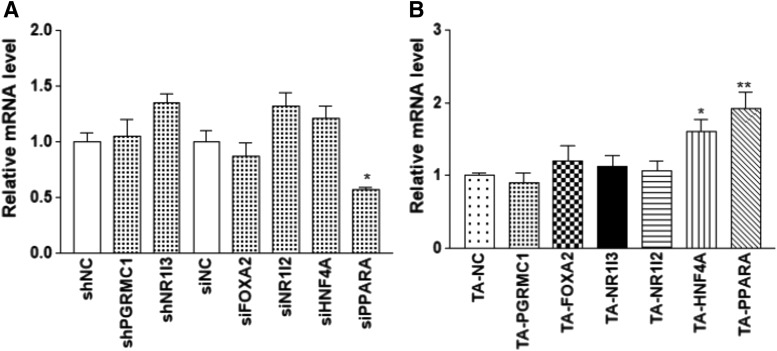
Consistent with the KD experiments, TA of ESR1 in Huh7 (15-fold increase in ESR1 [ESR1-201 isoform]) increased the expression of PGRMC1, FOXA2, NR1I2, PPARA, and HNF4A (Fig. 6C) but not NR1I3. On the other hand, KD or TA of PGRMC1, FOXA2, NR1I3, and NR1I2 (see Fig. 5, A and C for changes in the expression of these TFs) had no effect on the ESR1 expression (Fig. 7). KD PPARA (68% reduction) reduced the ESR1 expression, but TA PPARA or HNF4A (5- and 4-fold increase in PPARA and HNF4A, respectively) increased the ESR1 expression, indicating cross-regulation between ESR1 and other TFs.
Fig. 7.
Effects of gene KD (A) or CRISPR-mediated gene TA (B) of indicated transcription factors on the expression of ESR1 in Huh7 cells. Compared with negative control (NC): *P < 0.05; **P < 0.01, ANOVA with Dunnett comparison with control post test.
Effect of Estrogen on the Expression of ESR1 and CYP3A4.
One of the important features of a master regulator (or core regulator) is self-regulation for sustainable expression. To test whether hepatocyte ESR1 can be regulated by its own ligand estrogen, we treated hepatocytes with 1 µM 17β-estradiol (E2) every 6 hours as reported elsewhere (Choi et al., 2013) to maintain an effective level of E2 inside the hepatocyte, since E2 is actively metabolized by CYP3A4. We measured a hepatic-specific ESR1 isoform that is driven by a promoter ∼150 kb upstream of the canonical promoter for ESR1 (Flouriot et al., 1998).
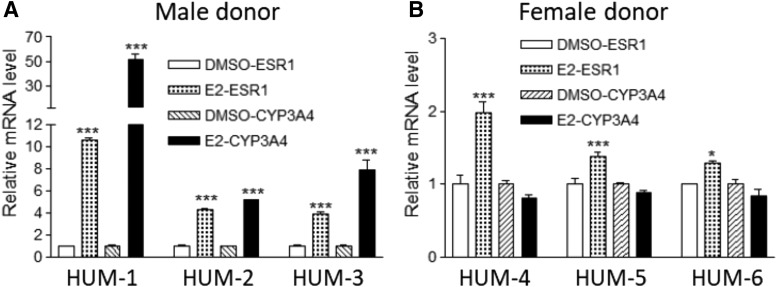
In all hepatocytes tested, the hepatic-specific ESR1 isoform mRNA increased after E2 treatment (Fig. 8). The changes in ESR1 expression were more profound in hepatocytes from male than female donors (two-way ANOVA, P < 0.001). CYP3A4 mRNA increased substantially after ESR1 treatment only in the cells from male donors (Fig. 8). The different effects of estrogen treatment between the male and female donors could have resulted from increased basal levels of ESR1 in females (Flouriot et al., 1998).
Fig. 8.
Effects of estrogen treatment on the expression of ESR1 and CYP3A4 in hepatocytes from male (A) and female donors (B). Hepatocytes were treated with 1 µM 17β -estradiol (E2) for 48 hours, and the mRNA levels of liver-specific ESR1 isoforms and CYP3A4 were measured. Data represent results of hepatocytes from six donors (HUM1–6, three females and three males), each in triplicate. Compared with DMSO: *P < 0.05; ***P < 0.001, ANOVA with Bonferroni comparison of selected pairs post test.
Regulation of Other P450 Enzymes by ESR1.
We then asked whether ESR1 regulates the expression of other P450 enzymes. We used first-order SSI analysis to scan the predictors for additional 11 P450 enzymes using microarray and GTEx data sets. Shown in Table 1, ESR1 appears as a regulator for CYP3A5, CYP3A7, CYP2C9, and CYP2C19, but not for CYP2D6, CYP1A1, CYP1A2, or CYP1B1 in both the microarray and GTEx data sets. Consistently, KD of ESR1 (∼80% reduction) in hepatocytes decreases expression of CYP3A5, CYP3A7, CYP2C9, and CYP2C19 but did not change the expression of CYP2D6, CYP1A1, CYP1A2, or CYP1B1 (Fig. 9). However, the KD of ESR1 also reduced the expression of CYP2A6, even though the SSI analysis failed to identify clear predictors for CYP2A6 in either data set (Fig. 9; Table 1).
Fig. 9.
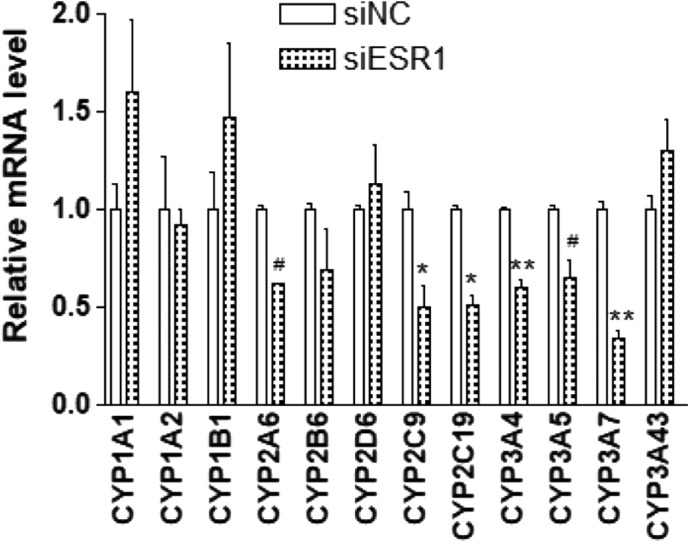
Effects of ESR1 KD on the expression of P450 enzymes as indicated in primary culture hepatocytes. Hepatocytes cultured with Matrigel overlay were treated with siRNA for 6 days; the mRNA levels of indicated P450 enzymes then were measured using real-time PCR with CYBR green. Compared with negative control (NC): *P < 0.05; **P < 0.01, ANOVA with Bonferroni comparison of selected pairs post test. # P<0.05 without post test.
Consistent with the KD results, in a cohort of 84 livers the expression levels of CYP3A4, 3A7, 2C9, and 2C19 were strongly correlated with that of liver-specific ESR1, while the expression levels of CYP2D6 and CYP3A43 were not (Table 2). The expression level of CYP3A5 was not correlated with ESR1 in the whole cohort, probably due to the frequent genetic variant CYP3A5*3 which causes aberrant splicing and non-sense-mediated RNA decay. After the CYP3A5 nonexpressors (CYP3A5*3 homozygous carriers) were excluded, the level of CYP3A5 was strongly correlated with ESR1 as expected (Table 2).
TABLE 2.
Correlation between mRNA expression levels of several P450 enzymes and liver-specific ESR1 isoform in 83 human livers
| Gene | Correlation Coefficient | P Value |
|---|---|---|
| CYP3A4 | 0.446 | <0.001 |
| CYP3A5 | 0.108 | 0.409 |
| CYP3A7 | 0.347 | 0.007 |
| CYP3A43 | 0.012 | 0.931 |
| CYP2C9 | 0.327 | 0.005 |
| CYP2C19 | 0.485 | <0.001 |
| CYP2D6 | −0.012 | 0.929 |
| CYP3A5 (n = 13)a | 0.693 | 0.009 |
Excluding CYP3A5*3 homozygous carriers.
Discussion
Using SSI analysis, we have identified ESR1, as well as several TFs (FOXA2, NR1I3, and NR1I2) as regulators for CYP3A4 expression in the human adult liver, an organ with sparse studies of estrogen receptor functions. Although previous multivariate regression analyses had similarly identified several overlapping TFs, including ESR1 (Yang et al., 2010), the SSI approach provides robust quantitative assessments of ranking order, placing ESR1 as a main regulator.
Although FOXA2, NR1I3, and NR1I2 were known CYP3A4 regulators, a role for ESR1 in CYP3A4 expression had yet to be studied in detail. Moreover, SSI has enabled the evaluation of dynamic interactions between TFs, also highlighting ESR1 as an interactive partner listed most frequently in TF doublets, triplets, and quadruplets, thus placing it on top of a hierarchical TF network. These relationships appear to apply to both males and females, consistent with a role for unliganded ESR1, which has been shown to bind to numerous domains throughout the genome (Caizzi et al., 2014).
The results were tested experimentally using gene KD (with shRNA and/or siRNA) and CRISPR-mediated TA. KD or TA ESR1 not only changed the CYP3A4 expression but also changed the expression of other CYP3A4 TF regulators. In turn, ESR1 expression was regulated by its own ligand, estrogen, and by other CYP3A4 TFs, namely PPARA and HNF4A. These results support a hierarchical regulatory network for CYP3A4 expression, directed by ESR1, involving self-regulation, cross-regulation, and TF-TF interactions. Moreover, SSI analysis and gene KD in hepatocytes also support a regulatory role of ESR1 in the expression of additional drug-metabolizing P450 enzymes, suggesting broad regulatory functions in hepatic P450 expression.
Unliganded ESR1 as a Master Regulator for CYP3A4 Expression?
Although ESR1 was known to mediate estrogen-induced expression of several P450 enzymes (Choi et al., 2013; Go et al., 2015), a role for unliganded ESR1 had yet to be reported. Our study, with no estrogen added in KD or TA experiments, further supports a role of unliganded ESR1 (without estrogen) in CYP3A4 expression in hepatic cells and also in the expression of several other P450 enzymes (2A6, 2C9, 2C19, 3A5, and 3A7) (Fig. 9). These results are consistent with recent studies demonstrating that unliganded ESR1 regulates gene expression (Stellato et al., 2016) in other cell types.
This conclusion is strengthened by chromatin immunoprecipitation followed by next generation sequencing (ChIP-seq) data showing binding of unliganded ESR1 to a subset of ESR1 binding sites associated with developmental functions in MCF7 cells (Caizzi et al., 2014). Moreover, unliganded ESR1 binding is facilitated by other TFs (Caizzi et al., 2014), some of which are known to regulate hepatic CYP3A4 expression in the liver (such as FOXA, HNF4α, and PPAR). Our study also used SSI analysis to identify these TFs as interactive ESR1 partners. We therefore propose a hierarchical regulatory network for CYP3A4 expression centered on ESR1.
ESR1 is situated at a high hierarchical level, interacting directly with FOXA2, HNF4A, NR1I2, and NR1I3. By contrast, TFs at a lower hierarchical order in this network (e.g., PPARA) interact with upstream high-order TFs but do not interact directly with downstream partners. This is consistent with PPARA appearing more frequently in interacting triplets than in pairs (Fig. 1, C and D). These multilayered interactions could account for the limited observed effects of genetic variants of single TF on the expression of CYP3A4 (Wang and Sadee, 2012).
Unliganded ESR1 as Regulator of Multiple P450 Enzymes.
In addition to CYP3A4, our results have shown that ESR1 also regulates other cytochrome P450 drug-metabolizing enzymes—CYP2C9, CYP2C19, CYP3A4, CYP3A5, and CYP3A7 (Fig. 9)—which share common regulators with CYP3A4 such as NR1I2, NR1I3, FOXA2, which themselves interact with ESR1 (Achour et al., 2014). These results support the notion that ESR1 is situated at a higher hierarchical order of regulatory networks. The involvement of ESR1 in P450 enzyme expression is further supported by the reduced hepatic expression of many P450 enzymes in the cyp3a, cyp2a, cyp2b, cyp2c, and cyp2d families in ESR1 knockout mice (Bryzgalova et al., 2006), supporting broad functions for ESR1 in liver maturation, differentiation, and tissue-specific gene expression including P450 enzymes.
Regulation of ESR1 by Estrogen in Human Liver.
Although estrogen down-regulates ESR1 in breast cancer MCF cells and reproductive tissues (Shupnik et al., 1989; Flouriot et al., 1998), our results showed estrogen to up-regulate a liver-specific ESR1 isoform (Flouriot et al., 1998), driven by a promoter ∼150 kb upstream of the canonical ESR1 promoter, in hepatocytes. This result is consistent with the presence of ESR1 binding sites at this distal promoter that was shown to be activated by estrogen (Flouriot et al., 1998).
Estrogen-mediated ESR1 up-regulation was highly variable, with stronger effects in the hepatocytes from male donors compared with female donors, which was possibly caused by the different levels of preexposure estrogen between the males and females and among different individuals. This result suggests ESR1 is self-regulated by its own ligand in the liver, which is a common feature for core regulators (Saint-André et al., 2016) and an important mechanism for their sustainable expression. The result also suggests that ESR1 is up-regulated by estrogen in the female liver, consistent with the higher levels of both ESR1 (Flouriot et al., 1998) and CYP3A4 in female livers (Wolbold et al., 2003; Wang et al., 2011).
Application of SSI Analysis.
SSI analysis was applied to select regulators for CYP3A4 and an additional 11 drug-metabolizing enzymes using both microarray and GTEx data. Of 12 P450 enzymes analyzed (Table 1), ESR1 repeatedly appeared as a regulator for five P450 enzymes (CYP2C9, CYP2C19, CYP3A4, CYP3A5, CYP3A7) in both the microarray and GTEx data sets. ESR1 KD in hepatocytes validated the regulatory roles of ESR1 in the expression of these five P450 enzymes. Similarly, the SSI analysis showed no clear regulators for CYP2D6, CYP1A1, CYP1A2, or CYP1B1 expression in either data set, consistent with a lack of effect on the expression of these P450 enzymes with ESR1 KD.
These results support a highly predictive value of SSI analysis when results are consistent in the two data sets, with one exception. Whereas SSI analysis failed to show ESR1 effects on CYP2A6, the KD of ESR1 reduced CYP2A6 expression in hepatocytes. This experimental result is consistent with a recent study showing the regulation of CYP2A6 by ESR1 (Kao et al., 2017) and in ESR1 knockout mice (Bryzgalova et al., 2006). Possibly, the discrepancy may have been caused by confounding factors, such as CYP2A6 copy number variation (gene duplication and deletion) or strong up-regulation by CYP2A6 inducers (Fukami et al., 2007; Mwenifumbo et al., 2008).
Although the expression of CYP1 family members is up-regulated by estrogen treatment (Go et al., 2015), the expression of these P450 enzymes was not affected by ESR1 KD, suggesting different regulatory roles of unbound ESR1 and estrogen-bound ESR1. These results also indicate that any possible residual estrogen in the cell culture serum cannot account for the observed ESR1-mediated effect under our current study conditions. This is consistent with no differences in CYP3A4 and ESR1 expression in Huh7 cells cultured with normal fetal bovine serum (10%) or charcoal-stripped serum (data not shown). These results further support the unliganded ESR1 effects observed in our study.
SSI Analysis of PGRMC1.
SSI analysis of the CYP3A4 microarray data revealed a small TF network independent of ESR1, centered around PGRMC1 (Fig. 2). PGRMC1 is known to bind directly to P450 proteins including CYP3A4 and to modulate P450 activity (Oda et al., 2011), with multiple functions in substrate binding, steroid signaling, and more (Rohe et al., 2009; Li et al., 2016); however, its role in P450 expression had not been reported. Our KD or TA results suggested PGRMC1 does not play a role in transcriptional CYP3A4 regulation. Since PGRMC1 expression level correlates with that of ESR1, we have considered that the positive result of SSI analysis may have been caused by this coexpression. Alternatively, because the SSI analysis using GTEx data did not reveal PGRMC1 as a predictor for CYP3A4 and other P450 enzymes, it is also possible that a positive prediction for PGRMC1 is confounded by some artifacts in the microarray data set.
Taken together, we have identified unliganded ESR1 as a master regulator for transcriptional regulation of CYP3A4. ESR1 appears to regulate the transcription of several other P450 enzymes. Moreover, we have demonstrated the utility of SSI analysis for predictor selection and for higher order regulatory network construction. Because ESR1 expression is regulated by multiple factors such as diet (Gao et al., 2012; Cordero et al., 2013), drugs (Killer et al., 2009), malignancy (Hishida et al., 2013), oxidative stress (Mahalingaiah et al., 2015), genetics, and DNA methylation (Maekawa et al., 2016), the network of interacting ESR1 and other TFs identified in this study may serve as a guide for integrating the combined effects of genetic, epigenetic, and nongenetic factors in TF coding genes on expression of P450 enzymes, and in particular CYP3A4.
This approach has the potential to account for a substantial portion of genetic variability in the expression of drug-metabolizing P450 enzymes. Future studies will focus on testing the association between ESR1 genetic variants and the expression of P450 enzymes, and on understanding the mechanisms underlying ESR1 regulation in the liver, including ESR1 expression profiles, ESR1 genomic occupancy, physical interactions between ESR1 and other TFs, and the involvement of long noncoding RNA and microRNA in ESR1 regulatory networks.
Acknowledgments
We acknowledge Dr. Feng Zhang for the gift of Addgene plasmids #61427 and #61427, and Dr. Frank Sinicrope for the gift of Addgene plasmid #26597. Human liver samples were provided by the Cooperative Human Tissue Network, which is funded by the National Cancer Institute.
Abbreviations
- ESR1
estrogen receptor α
- FOXA2
forkhead box protein A2
- gRNA
guide RNA
- GTEx
genotype-tissue expression
- HNF4A
hepatocyte nuclear factor 4α
- KD
knockdown
- NC
negative control
- NR1I2
pregnane X receptor
- NR1I3
constitutive androstane receptor
- P450
cytochrome P450 enzyme
- PCR
polymerase chain reaction
- PGRMC1
progesterone receptor membrane component 1
- PPARA
peroxisome proliferator activated receptor α
- shRNA
short hairpin RNA
- siRNA
small interfering RNA
- SSI
Sobol’s sensitivity indices
- TA
transcriptional activation
- TF
transcription factor
Authorship Contributions
Participated in research design: Wang, Sadee.
Conducted experiments: Wang.
Contributed new reagents or analytic tools: Lu, Rempala.
Performed data analysis: Wang, Lu.
Wrote or contributed to the writing of the manuscript: Wang, Lu, Sadee.
Footnotes
This study was supported by National Institutes of Health National Institute of General Medical Science [Grant R01GM120396], and Institute of General Medical Science Pharmacogenetics Research Network [Grant U01 GM092655]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the National Center for Research Resources.
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Achour B, Barber J, Rostami-Hodjegan A. (2014) Expression of hepatic drug-metabolizing cytochrome p450 enzymes and their intercorrelations: a meta-analysis. Drug Metab Dispos 42:1349–1356. [DOI] [PubMed] [Google Scholar]
- Battle A, Brown CD, Engelhardt BE, Montgomery SB, GTEx Consortium; Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group; Statistical Methods groups—Analysis Working Group; Enhancing GTEx (eGTEx) groups; NIH Common Fund; NIH/NCI; NIH/NHGRI; NIH/NIMH; NIH/NIDA; Biospecimen Collection Source Site—NDRI; Biospecimen Collection Source Site—RPCI; Biospecimen Core Resource—VARI; Brain Bank Repository—University of Miami Brain Endowment Bank; Leidos Biomedical—Project Management; ELSI Study; Genome Browser Data Integration &Visualization—EBI; Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz; Lead analysts; Laboratory, Data Analysis &Coordinating Center (LDACC); NIH program management; Biospecimen collection; Pathology; eQTL manuscript working group (2017) Genetic effects on gene expression across human tissues [published correction appears in Nature (2018) 553:530]. Nature 550:204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryzgalova G, Gao H, Ahren B, Zierath JR, Galuska D, Steiler TL, Dahlman-Wright K, Nilsson S, Gustafsson JA, Efendic S, et al. (2006) Evidence that oestrogen receptor-alpha plays an important role in the regulation of glucose homeostasis in mice: insulin sensitivity in the liver. Diabetologia 49:588–597. [DOI] [PubMed] [Google Scholar]
- Caizzi L, Ferrero G, Cutrupi S, Cordero F, Ballaré C, Miano V, Reineri S, Ricci L, Friard O, Testori A, et al. (2014) Genome-wide activity of unliganded estrogen receptor-α in breast cancer cells. Proc Natl Acad Sci USA 111:4892–4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastaing G, Gamboa F, Prieur C. (2012) Generalized Hoeffding-School decomposition for dependent variables-application to sensitivity analysis. Electron J Stat 6:2420–2448. [Google Scholar]
- Chastaing G, Gamboa F, Prieur C. (2015) Generalized Sobol sensitivity indices for dependent variables: numerical methods. J Stat Comput Simul 85:1306–1333. [Google Scholar]
- Choi SY, Koh KH, Jeong H. (2013) Isoform-specific regulation of cytochromes P450 expression by estradiol and progesterone. Drug Metab Dispos 41:263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero P, Campion J, Milagro FI, Martinez JA. (2013) Transcriptomic and epigenetic changes in early liver steatosis associated to obesity: effect of dietary methyl donor supplementation. Mol Genet Metab 110:388–395. [DOI] [PubMed] [Google Scholar]
- Danielson PB. (2002) The cytochrome P450 superfamily: biochemistry, evolution and drug metabolism in humans. Curr Drug Metab 3:561–597. [DOI] [PubMed] [Google Scholar]
- Flouriot G, Brand H, Denger S, Metivier R, Kos M, Reid G, Sonntag-Buck V, Gannon F. (2000) Identification of a new isoform of the human estrogen receptor-alpha (hER-alpha) that is encoded by distinct transcripts and that is able to repress hER-alpha activation function 1. EMBO J 19:4688–4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flouriot G, Griffin C, Kenealy M, Sonntag-Buck V, Gannon F. (1998) Differentially expressed messenger RNA isoforms of the human estrogen receptor-alpha gene are generated by alternative splicing and promoter usage. Mol Endocrinol 12:1939–1954. [DOI] [PubMed] [Google Scholar]
- Fukami T, Nakajima M, Yamanaka H, Fukushima Y, McLeod HL, Yokoi T. (2007) A novel duplication type of CYP2A6 gene in African-American population. Drug Metab Dispos 35:515–520. [DOI] [PubMed] [Google Scholar]
- Gao R, Ding Y, Liu X, Chen X, Wang Y, Long C, Li S, Guo L, He J. (2012) Effect of folate deficiency on promoter methylation and gene expression of Esr1, Cdh1 and Pgr, and its influence on endometrial receptivity and embryo implantation. Hum Reprod 27:2756–2765. [DOI] [PubMed] [Google Scholar]
- Go RE, Hwang KA, Choi KC. (2015) Cytochrome P450 1 family and cancers. J Steroid Biochem Mol Biol 147:24–30. [DOI] [PubMed] [Google Scholar]
- Hattori Y, Ishii H, Munetomo A, Watanabe H, Morita A, Sakuma Y, Ozawa H. (2016) Human C-terminally truncated ERα variants resulting from the use of alternative exons in the ligand-binding domain. Mol Cell Endocrinol 425:111–122. [DOI] [PubMed] [Google Scholar]
- Hishida M, Nomoto S, Inokawa Y, Hayashi M, Kanda M, Okamura Y, Nishikawa Y, Tanaka C, Kobayashi D, Yamada S, et al. (2013) Estrogen receptor 1 gene as a tumor suppressor gene in hepatocellular carcinoma detected by triple-combination array analysis. Int J Oncol 43:88–94. [DOI] [PubMed] [Google Scholar]
- Huang S, Sinicrope FA. (2010) Sorafenib inhibits STAT3 activation to enhance TRAIL-mediated apoptosis in human pancreatic cancer cells. Mol Cancer Ther 9:742–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Istrate MA, Nussler AK, Eichelbaum M, Burk O. (2010) Regulation of CYP3A4 by pregnane X receptor: the role of nuclear receptors competing for response element binding. Biochem Biophys Res Commun 393:688–693. [DOI] [PubMed] [Google Scholar]
- Jover R, Bort R, Gómez-Lechón MJ, Castell JV. (2001) Cytochrome P450 regulation by hepatocyte nuclear factor 4 in human hepatocytes: a study using adenovirus-mediated antisense targeting. Hepatology 33:668–675. [DOI] [PubMed] [Google Scholar]
- Kao TL, Chen YL, Kuan YP, Chang WC, Ho YC, Yeh S, Jeng LB, Ma WL. (2017) Estrogen-estrogen receptor α signaling facilitates bilirubin metabolism in regenerating liver through regulating cytochrome P450 2A6 expression. Cell Transplant 26:1822–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killer N, Hock M, Gehlhaus M, Capetian P, Knoth R, Pantazis G, Volk B, Meyer RP. (2009) Modulation of androgen and estrogen receptor expression by antiepileptic drugs and steroids in hippocampus of patients with temporal lobe epilepsy. Epilepsia 50:1875–1890. [DOI] [PubMed] [Google Scholar]
- Klein K, Thomas M, Winter S, Nussler AK, Niemi M, Schwab M, Zanger UM. (2012) PPARA: a novel genetic determinant of CYP3A4 in vitro and in vivo. Clin Pharmacol Ther 91:1044–1052. [DOI] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, et al. (2015) Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517:583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamba V, Panetta JC, Strom S, Schuetz EG. (2010) Genetic predictors of interindividual variability in hepatic CYP3A4 expression. J Pharmacol Exp Ther 332:1088–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Rabitz H, Yelvington PE, Oluwole OO, Bacon F, Kolb CE, Schoendorf J. (2010) Global sensitivity analysis for systems with independent and/or correlated inputs. J Phys Chem A 114:6022–6032. [DOI] [PubMed] [Google Scholar]
- Li X, Rhee DK, Malhotra R, Mayeur C, Hurst LA, Ager E, Shelton G, Kramer Y, McCulloh D, Keefe D, et al. (2016) Progesterone receptor membrane component-1 regulates hepcidin biosynthesis. J Clin Invest 126:389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Wang D, Wang M, Rempala GA. (2018) Estimation of Sobol’s sensitivity indices under generalized linear models. Commun Stat Theory Methods 47:5163–5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa R, Sato S, Okada M, Lee L, Tamura I, Jozaki K, Kajimura T, Asada H, Yamagata Y, Tamura H, et al. (2016) Tissue-specific expression of estrogen receptor 1 is regulated by DNA methylation in a T-DMR. Mol Endocrinol 30:335–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahalingaiah PK, Ponnusamy L, Singh KP. (2015) Chronic oxidative stress causes estrogen-independent aggressive phenotype, and epigenetic inactivation of estrogen receptor alpha in MCF-7 breast cancer cells. Breast Cancer Res Treat 153:41–56. [DOI] [PubMed] [Google Scholar]
- Mara T, Tarantola S. (2012) Variance-based sensitivity indices for models with dependent inputs. Reliab Eng Syst Saf 107:115–121. [Google Scholar]
- Martínez-Jiménez CP, Jover R, Donato MT, Castell JV, Gómez-Lechón MJ. (2007) Transcriptional regulation and expression of CYP3A4 in hepatocytes. Curr Drug Metab 8:185–194. [DOI] [PubMed] [Google Scholar]
- Mwenifumbo JC, Al Koudsi N, Ho MK, Zhou Q, Hoffmann EB, Sellers EM, Tyndale RF. (2008) Novel and established CYP2A6 alleles impair in vivo nicotine metabolism in a population of Black African descent. Hum Mutat 29:679–688. [DOI] [PubMed] [Google Scholar]
- Oda S, Nakajima M, Toyoda Y, Fukami T, Yokoi T. (2011) Progesterone receptor membrane component 1 modulates human cytochrome p450 activities in an isoform-dependent manner. Drug Metab Dispos 39:2057–2065. [DOI] [PubMed] [Google Scholar]
- Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, Schreiber J, Rolfe PA, Gifford DK, et al. (2004) Control of pancreas and liver gene expression by HNF transcription factors. Science 303:1378–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ourlin JC, Jounaïdi Y, Maurel P, Vilarem MJ. (1997) Role of the liver-enriched transcription factors C/EBP alpha and DBP in the expression of human CYP3A4 and CYP3A7. J Hepatol 26 (Suppl 2):54–62. [DOI] [PubMed] [Google Scholar]
- Ozdemir V, Kalow W, Tang BK, Paterson AD, Walker SE, Endrenyi L, Kashuba AD. (2000) Evaluation of the genetic component of variability in CYP3A4 activity: a repeated drug administration method. Pharmacogenetics 10:373–388. [DOI] [PubMed] [Google Scholar]
- Penno MB, Dvorchik BH, Vesell ES. (1981) Genetic variation in rates of antipyrine metabolite formation: a study in uninduced twins. Proc Natl Acad Sci USA 78:5193–5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman WR, Matsumoto M, Beltaifa S, Hyde TM, Saunders RC, Webster MJ, Rubinow DR, Kleinman JE, Weickert CS. (2005) Expression of estrogen receptor alpha exon-deleted mRNA variants in the human and non-human primate frontal cortex. Neuroscience 134:81–95. [DOI] [PubMed] [Google Scholar]
- Poola I, Koduri S, Chatra S, Clarke R. (2000) Identification of twenty alternatively spliced estrogen receptor alpha mRNAs in breast cancer cell lines and tumors using splice targeted primer approach. J Steroid Biochem Mol Biol 72:249–258. [DOI] [PubMed] [Google Scholar]
- Rahmioglu N, Heaton J, Clement G, Gill R, Surdulescu G, Zlobecka K, Hodgkiss D, Ma Y, Hider RC, Smith NW, et al. (2011) Genetic epidemiology of induced CYP3A4 activity. Pharmacogenet Genomics 21:642–651. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Antona C, Bort R, Jover R, Tindberg N, Ingelman-Sundberg M, Gómez-Lechón MJ, Castell JV. (2003) Transcriptional regulation of human CYP3A4 basal expression by CCAAT enhancer-binding protein alpha and hepatocyte nuclear factor-3 gamma. Mol Pharmacol 63:1180–1189. [DOI] [PubMed] [Google Scholar]
- Rohe HJ, Ahmed IS, Twist KE, Craven RJ. (2009) PGRMC1 (progesterone receptor membrane component 1): a targetable protein with multiple functions in steroid signaling, P450 activation and drug binding. Pharmacol Ther 121:14–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-André V, Federation AJ, Lin CY, Abraham BJ, Reddy J, Lee TI, Bradner JE, Young RA. (2016) Models of human core transcriptional regulatory circuitries. Genome Res 26:385–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shupnik MA, Gordon MS, Chin WW. (1989) Tissue-specific regulation of rat estrogen receptor mRNAs. Mol Endocrinol 3:660–665. [DOI] [PubMed] [Google Scholar]
- Stellato C, Porreca I, Cuomo D, Tarallo R, Nassa G, Ambrosino C. (2016) The “busy life” of unliganded estrogen receptors. Proteomics 16:288–300. [DOI] [PubMed] [Google Scholar]
- Thomas M, Burk O, Klumpp B, Kandel BA, Damm G, Weiss TS, Klein K, Schwab M, Zanger UM. (2013) Direct transcriptional regulation of human hepatic cytochrome P450 3A4 (CYP3A4) by peroxisome proliferator-activated receptor alpha (PPARα). Mol Pharmacol 83:709–718. [DOI] [PubMed] [Google Scholar]
- Tirona RG, Lee W, Leake BF, Lan LB, Cline CB, Lamba V, Parviz F, Duncan SA, Inoue Y, Gonzalez FJ, et al. (2003) The orphan nuclear receptor HNF4α determines PXR- and CAR-mediated xenobiotic induction of CYP3A4. Nat Med 9:220–224. [DOI] [PubMed] [Google Scholar]
- Wang D, Guo Y, Wrighton SA, Cooke GE, Sadee W. (2011) Intronic polymorphism in CYP3A4 affects hepatic expression and response to statin drugs. Pharmacogenomics J 11:274–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Papp AC, Sun X. (2015) Functional characterization of CYP2D6 enhancer polymorphisms. Hum Mol Genet 24:1556–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Sadee W. (2012) The making of a CYP3A biomarker panel for guiding drug therapy. J Pers Med 2:175–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Sadee W. (2016) CYP3A4 intronic SNP rs35599367 (CYP3A4*22) alters RNA splicing. Pharmacogenet Genomics 26:40–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werk AN, Cascorbi I. (2014) Functional gene variants of CYP3A4. Clin Pharmacol Ther 96:340–348. [DOI] [PubMed] [Google Scholar]
- Wolbold R, Klein K, Burk O, Nüssler AK, Neuhaus P, Eichelbaum M, Schwab M, Zanger UM. (2003) Sex is a major determinant of CYP3A4 expression in human liver. Hepatology 38:978–988. [DOI] [PubMed] [Google Scholar]
- Xu C, Gertner G. (2008) Uncertainty and sensitivity analysis for models with correlated parameters. Reliab Eng Syst Saf 93:1563–1573. [Google Scholar]
- Yang X, Zhang B, Molony C, Chudin E, Hao K, Zhu J, Gaedigk A, Suver C, Zhong H, Leeder JS, et al. (2010) Systematic genetic and genomic analysis of cytochrome P450 enzyme activities in human liver. Genome Res 20:1020–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Horvath S. (2005) A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 4 (1), doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]