Abstract
Marine protists are a polyphyletic group of organisms playing major roles in the ecology and biogeochemistry of the oceans, including performing much of Earth’s photosynthesis and driving the carbon, nitrogen, and silicon cycles. In addition, marine protists occupy key positions in the tree of life, including as the closest relatives of metazoans. Despite all the reasons to better understand them, knowledge of the cell biology of most marine protist lineages is sparse. This is beginning to change thanks to vibrant growth in the development of new model organisms. Here, we survey some recent advances in studying the cell biology of marine protists toward understanding the functional basis of their unique features, gaining new perspectives on universal eukaryotic biology, and for understanding homologous biology within metazoans and the evolution of metazoan traits.
INTRODUCTION
Because most of eukaryotic diversity is represented by unicellular organisms, their inclusion among model organisms is essential to understanding the evolutionary history, structure, and (dys)function of all eukaryotes. Unicellular eukaryotes are often lumped as “protists,” a term that is useful despite its taxonomic irrelevance and origin as a definition by exclusion—a protist being any eukaryote that’s not a plant, animal, or fungus. A few freshwater and terrestrial protists are considered to be model organisms, for example, by the National Science Foundation in their Proposal Classification Form: the chlorophytes Acetabularia and Chlamydomonas, the ciliates Paramecium and Tetrahymena, and the amoebozoan Dictyostelium. Another fairly well-developed model is the apicomplexan Plasmodium. There are no marine protists among the current model organisms, limiting our ability to understand how they achieve their essential functions in powering ocean ecosystems and global biogeochemical cycles (Caron et al., 2011), and to exploit them for new cell biological insights. To address this gap, marine protists have been the focus of recent large and coordinated efforts, funded by the Gordon and Betty Moore Foundation, to build the genomic resources and to develop the methods for genetic manipulation needed to advance key species to the status of tractable model organism (for more information on these programs, see Keeling et al., 2014; Waller et al., 2018).
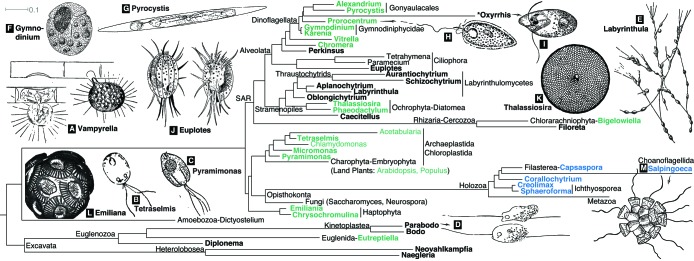
Marine protists include representatives of the diverse morphologies, physiologies, and life histories found among the major eukaryotic lineages. The rhizarian vampyrellids offer an example of the weird and wonderful biology to be found: vampyrellid trophozoites explore their surroundings using a form of rolling locomotion, piercing the cell walls of their chlorophyte prey and extracting the protoplast by phagocytosis, leaving behind an empty shell with one or more small holes; well-fed trophozoites then form digestive cysts, from which the next generation of young trophozoites emerge to resume the rolling hunt (Figure 1A; More et al., 2019). Clarifying the phylogenetic relationships among the deeply divergent protist lineages is an active area of research (e.g., Adl et al., 2019), and we display a view of eukaryotic diversity consistent with current understanding in Figure 1, highlighting marine protists that are developing models along with existing terrestrial and freshwater model protists. Some of the developing model marine protists are in the same class as established freshwater or terrestrial model protists, for example, Micromonas and other chlorophytes (Figure 1, B and C). But most represent major lineages of eukaryotes that otherwise lack tractable model organisms, including the Euglenozoa (Figure 1D), several groups of the stramenopiles-alveolates-rhizarians (often called SAR) lineage (Figure 1, E–K), and haptophytes (Figure 1L).
FIGURE 1:
Overview of approximate phylogenetic relationships among developing marine protist models and established freshwater model systems, displayed at the genus level, as well as illustrations of select marine protists. On the phylogeny, marine protists are in bold, with marine models used to explore multicellularity and photosynthesis in blue and green, respectively. We include a subset of genera from Waller et al. (2018) as well as additional cases to highlight lineages discussed herein; see Supplemental Table 1 for a complete list. Small subunit rRNA alignment r132 from SILVA (Quast et al., 2012) was used in IQ-TREE (Schmidt et al., 2014) to produce the phylogeny, including an archaeal outgroup. Scale bar indicates average number of substitutions per site. Some lineages were rearranged in TreeGraph (Stöver and Müller, 2010) to reflect literature consensus. Drawings A–M (not to scale) are adapted from the following sources: Tetraselmis (Stokes, 1888), Pyramimonas and Pyrocystis (West, 1916), and Prorocentrum (Calkins, 1926). Emiliana huxleyi is adapted from a scanning electron micrograph image (CC BY 2.5; Taylor, 2011). Thalassiosira is adapted from microphotos of a fossil prepared by Anne Gleich (CC BY 2.0; Picturepest). The Salpingoeca rosetta colony is redrawn from a microscope image (CC BY-SA 3.0; Mark Dayel; redrawn by Tiago Pratas). Labyrinthula cells are illustrated (E) moving along the shared ectoplasmic network (Moore, 1911). Not shown on the phylogeny, Vampyrella is illustrated (A) boring into and sucking out (left) and completely emptying (right) a Spirogyra cell (Verworn, 1899, p. 148). All other drawings were adapted from Calkins (1901).
Cell biologists push their field forward via two general strategies: increasing depth and increasing breadth. Going deeper into long-studied biological systems such as Saccharomyces cerevisiae is often achieved by technical innovations that allow testing of previously inaccessible hypotheses. Gaining a broader perspective on long-studied processes is achieved by exploring biological diversity to identify different twists on known mechanisms as well as entirely new biology. Here, we highlight three aspects of marine protist cell biology that illustrate how the “breadth” strategy is yielding fresh insights. First, the cell biology of early-diverging holozoan lineages is being used to explore the evolution of multicellularity in animals. Second, some marine protists have evolved atypical mechanisms for performing essential and otherwise highly conserved tasks. Third, many marine protist lineages have unique cell biology that offers windows into the evolutionary heritage of all eukaryotes.
INSIGHTS INTO ORIGIN OF MULTICELLULARITY
The availability of more, and more diverse, protist genomes and transcriptomes has revealed that machinery previously thought to be unique to metazoans is not. For example, genes encoding lamin-like proteins have now been detected in diverse protists including choanoflagellates and the marine rhizarian Corallomyxa (now Filoreta) tenera (Kollmar, 2015). Choanoflagellates are of particular interest to developmental biologists because they are one of the closest living relatives of animals in many molecular phylogenetic analyses (see Figure 1M) and share with other holozoans a variety of genes required for embryogenesis in animals (e.g., cadherins, tyrosine kinases, and Myc; Booth et al., 2018). Taking a genome-wide approach, Richter et al. (2018) identified hundreds of additional gene families formerly considered to be animal-specific that instead are present in the common ancestor of metazoans and choanoflagellates. This includes examples that will be familiar to many cell biologists, including Notch signaling pathway receptors and ligands, components of the innate immune system including Toll-like receptors, and enzymes responsible for the hydrolytic cleavage of glycosaminoglycans in the extracellular matrix. The opportunity to gain insight into the ancestral functions of such genes is now becoming a reality with the development of transformation methods for the ichthyosporean Creolimax fragrantissima (Suga and Ruiz-Trillo, 2013) and the choanoflagellate Salpingoeca rosetta (Figure 1M). Booth et al. (2018) introduced recombinant septin and tubulin genes into S. rosetta, and localization of the fluorescently tagged proteins to the basal poles of cells in rosette-shaped colonies was consistent with the behavior of homologous genes in fungi and metazoan epithelial cells, suggesting conserved functions in establishing and maintaining cell polarity.
The critical role of the microbiome in determining the morphology and function of animals and plants is now widely recognized (McFall-Ngai et al., 2013; Vandenkoornhuyse et al., 2015). Free-living bacteria may also have important impacts on diverse eukaryotes, with examples from marine systems including animals, seaweeds, and protists. For example, rosette formation in S. rosetta occurs in response to several bacterial compounds released in outer membrane vesicles, and sexual reproduction is triggered by bacterial chondroitin lyase (Woznica and King, 2018). With continued development of additional tools, marine protists offer new models for investigating bacteria-mediated development.
NEW WAYS TO SOLVE OLD PROBLEMS
Some marine protists have unusual cell biological features that reveal alternative, derived solutions for conserved tasks. Perhaps the classic example is the dinokaryon fibrillar chromosomes of dinoflagellates (Figure 1, F–H) that are always condensed, replacing the nucleosomal histones found in other eukaryotes (Iwamoto et al., 2016). Dinoflagellates utilize a nucleoprotein derived from a viral homologue as a bulk protein for packing DNA, even while retaining divergent (and not highly expressed) but still recognizable versions of the ancestral histone genes (Gornik et al., 2012). The acquisition of this virus-derived gene corresponds with a tremendous increase of genome size in dinoflagellates relative to their alveolate relatives, and may enable them to manage so much DNA. Perhaps relatedly, dinoflagellates have so far proved refractory to genetic manipulation, and the development of such tools would enable deeper study of this intriguing phenomenon.
Another case of innovation in marine protists is in the regulation of nonreceptor tyrosine kinase signaling in the ichthyosporean C. fragrantissima. Universally, Src kinase is negatively regulated by Csk, but in C. fragrantissima the Csk gene has been has lost (Suga and Miller, 2018). Transient overexpression of Src in transfected C. fragrantissima prevented growth, whereas coexpression of Src with one of the C. fragrantissima protein–tyrosine phosphatases rescued the phenotype, showing that another protein–tyrosine phosphatase has been coopted in place of Csk. An intriguing additional detail is that the Src kinases of C. fragrantissima and other symbiotic (with marine invertebrates) ichthyosporeans have a bulky gatekeeper residue in the active site cleft, which mimics that of drug-resistant mammalian Src kinases and may have ecological relevance.
UNIQUE CELL BIOLOGY
Other marine protists possess cell biology that has no obvious counterpart in plants or animals, such as the vampyrellids described earlier. Perhaps the most widely known example is the siliceous cell walls, or frustules, of diatoms (Figure 1K), which come in a bewildering array of species-specific, finely detailed, hierarchically porous variants. There is much interest in understanding the ontogeny of diatom cell walls not only for biological insight, but for biotechnological applications. There has been a great deal of progress in identifying components of diatom frustule biosynthesis, and this knowledge has recently been applied to support in vitro synthesis of hierarchically porous silica (Pawolski et al., 2018). The essentiality of the frustule in diatoms has presented a challenge to in vivo manipulation. The closest relatives of diatoms, the parmales, do not require their silica shells and the biosynthesis of the silica plates in Triparma laevis can be controlled by simply manipulating silicate availability. Yamada et al. (2019) demonstrated that the early steps of silica wall formation in T. laevis are homologous to the process of frustule formation in diatoms, confirming that parmales provide an alternative system in which to study frustule formation.
Diatoms and other photosynthetic stramenopiles and alveolates offer independent examples from the chlorophytes of the integration of a photosynthetic organelle into the heterotrophic physiology of the host (see Figure 1), and thus insight into other ways that complex evolutionary event might be accomplished. Despite the convergence of photoautotrophy, diatoms use pathways for circadian regulation of gene expression more like those of mammals than plants (Annunziata et al., 2018), and have distinct mechanisms to coordinate mitochondrial and plastid metabolism (Murik et al., 2019).
Another unique aspect of cell biology exists in a group of nonphotosynthetic stramenopiles, the Labyrinthulomycetes (Figure 1E). These organisms have a unique organelle, the bothrosome (sagenogenetosome), that is responsible for the production of an ectoplasmic network involved in cell motility and the search for and attachment to food sources (Fossier Marchan et al., 2018; Iwata and Honda, 2018; Hamamoto and Honda, 2019). Iwata et al. (2017) suggest that the bothrosome may be related to the endoplasmic reticulum–plasma membrane junctions with diverse functions in eukaryotic cell biology (Stefan, 2018). Genetic tools are being developed for some Labyrinthulomycetes (e.g., Okino et al., 2018), and promise to help uncover the molecular mechanisms of such novel biology.
THE FUTURE OF MARINE PROTIST CELL BIOLOGY
The few examples mentioned here of recent progress in the cell biology of marine protists only touch the surface of the questions that further work on these organisms will open to investigation. For example, it appears that many interspecific interactions among marine protists (e.g., host–parasite and predator–prey) are highly specific, but very little is known about either the cell biological mechanisms underlying such specificity or their ecological and evolutionary ramifications. Two promising model species among the euglenozoan phagotrophic marine flagellates, the kinetoplastid Parabodo caudatus (Figure 1D; Gomaa et al., 2017) and the diplonemid Diplonema papillatum (Kaur et al., 2018), have recently been transformed, opening the door to investigating how these micropredators hunt and choose their prey. Continued progress along the “breadth” pathway to advancing cell biology will require developing these and other protists as model systems.
There are many challenges along that road. Moving even fairly standard and universal methods into new species can be challenging for many reasons, for marine protists including typically small cell size, the need for seawater, and diverse cell wall composition. Progress can be slow with too few people focusing on each species to provide critical mass or shared resources. We encourage cell biologists experienced in working on well-established model organisms to team up with investigators developing new models for processes of interest to them; the synergy of such collaborations promises to expand an already golden age of cell biology.
Supplementary Material
Acknowledgments
We thank the Gordon and Betty Moore Foundation for their financial support of both our work and our travel to broaden our understanding of the growing field of marine protist cell biology.
Abbreviation used:
- SAR
stramenopiles-alveolates-rhizarians
Footnotes
REFERENCES
- Adl SM, Bass D, Lane CE, Lukeš J, Schoch CL, Smirnov A, Agatha S, Berney C, Brown MW, Burki F, et al. (2019). Revisions to the classification, nomenclature, and diversity of eukaryotes. J Eukaryot Microbiol , 4–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annunziata R, Ritter A, Fortunato AE, Cheminant-Navarro S, Agier N, Huysman MJJ, Winge P, Bones A, Bouget F-Y, Cosentino Lagomarsino M, et al. (2018). A bHLH-PAS protein regulates light-dependent diurnal rhythmic processes in the marine diatom Phaeodactylum tricornutum . BioRxiv 271445. [Google Scholar]
- Booth DS, Szmidt-Middleton H, King N, Kellogg D. (2018). Transfection of choanoflagellates illuminates their cell biology and the ancestry of animal septins. Mol Biol Cell , 3026–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calkins GN. (1901). Marine protozoa from Woods Hole. Bull US Fish Comm , 415–468. Accessed via The Freshwater and Marine Image Bank. Available at https://content.lib.washington.edu/fishweb/ (accessed 31 January 2019). [Google Scholar]
- Calkins GN. (1926). The Biology of the Protozoa. Philadelphia and New York: Lea and Febiger, p. 269. [Google Scholar]
- Caron DA, Countway PD, Jones AC, Kim DY, Schnetzer A. (2011). Marine protistan diversity. Annu Rev Mar Sci , 467–493. [DOI] [PubMed] [Google Scholar]
- Fossier Marchan L, Lee Chang KJ, Nichols PD, Mitchell WJ, Polglase JL, Gutierrez T. (2018). Taxonomy, ecology and biotechnological applications of thraustochytrids: a review. Biotechnol Adv , 26–46. [DOI] [PubMed] [Google Scholar]
- Gomaa F, Garcia PA, Delaney J, Girguis PR, Buie CR, Edgcomb VP. (2017). Toward establishing model organisms for marine protists: Successful transfection protocols for Parabodo caudatus (Kinetoplastida: Excavata). Environ Microbiol , 3487–3499. [DOI] [PubMed] [Google Scholar]
- Gornik SG, Ford KL, Mulhern TD, Bacic A, McFadden GI, Waller RF. (2012). Loss of nucleosomal DNA condensation coincides with appearance of a novel nuclear protein in dinoflagellates. Curr Biol , 2303–2312. [DOI] [PubMed] [Google Scholar]
- Hamamoto Y, Honda D. (2019). Nutritional intake of Aplanochytrium (Labyrinthulea, Stramenopiles) from living diatoms revealed by culture experiments suggesting the new prey–predator interactions in the grazing food web of the marine ecosystem. PLoS One , e0208941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto M, Hiraoka Y, Haraguchi T. (2016). Uniquely designed nuclear structures of lower eukaryotes. Curr Opin Cell Biol , 66–73. [DOI] [PubMed] [Google Scholar]
- Iwata I, Honda D. (2018). Nutritional intake by ectoplasmic nets of Schizochytrium aggregatum (Labyrinthulomycetes, Stramenopiles). Protist , 727–743. [DOI] [PubMed] [Google Scholar]
- Iwata I, Kimura K, Tomaru Y, Motomura T, Koike K, Koike K, Honda D. (2017). Bothrosome formation in Schizochytrium aggregatum (Labyrinthulomycetes, Stramenopiles) during zoospore settlement. Protist , 206–219. [DOI] [PubMed] [Google Scholar]
- Kaur B, Valach M, Peña-Diaz P, Moreira S, Keeling PJ, Burger G, Lukeš J, Faktorová D. (2018). Transformation of Diplonema papillatum, the type species of the highly diverse and abundant marine microeukaryotes Diplonemida (Euglenozoa). Environ Microbiol , 1030–1040. [DOI] [PubMed] [Google Scholar]
- Keeling PJ, Burki F, Wilcox HM, Allam B, Allen EE, Amaral-Zettler LA, Armbrust EV, Archibald JM, Bharti AK, Bell CJ, et al. (2014). The marine microbial eukaryote transcriptome sequencing project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol , e1001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollmar M. (2015). Polyphyly of nuclear lamin genes indicates an early eukaryotic origin of the metazoan-type intermediate filament proteins. Sci Rep , 10652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, Domazet-Lošo T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, et al. (2013). Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA , 3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore T. (1911). “Labyrinthulidea.” In: Encyclopedia Britannica (11th ed.), Vol. , 34. [Google Scholar]
- More K, Simpson AGB, Hess S. (2019). Two new marine species of Placopus (Vampyrellida, Rhizaria) that perforate the theca of Tetraselmis (Chlorodendrales, Viridiplantae). J Eukaryot Microbiol, 10.1111/jeu.12698. [DOI] [PubMed] [Google Scholar]
- Murik O, Tirichine L, Prihoda J, Thomas Y, Araújo WL, Allen AE, Fernie AR, Bowler C. (2019). Downregulation of mitochondrial alternative oxidase affects chloroplast function, redox status and stress response in a marine diatom. New Phytol , 1303–1316. [DOI] [PubMed] [Google Scholar]
- Okino N, Wakisaka H, Ishibashi Y, Ito M. (2018). Visualization of endoplasmic reticulum and mitochondria in Aurantiochytrium limacinum by the expression of EGFP with cell organelle-specific targeting/retaining signals. Mar Biotechnol , 182–192. [DOI] [PubMed] [Google Scholar]
- Pawolski D, Heintze C, Mey I, Steinem C, Kröger N. (2018). Reconstituting the formation of hierarchically porous silica patterns using diatom biomolecules. J Struct Biol , 64–74. [DOI] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Gerken J, Peplies J, Yarza P, Yilmaz P, Schweer T, Glöckner FO. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res , D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter DJ, Fozouni P, Eisen MB, King N. (2018). Gene family innovation, conservation and loss on the animal stem lineage. eLife , e34226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HA, Minh BQ, von Haeseler A, Nguyen L-T. (2014). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol , 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan CJ. (2018). Building ER-PM contacts: keeping calm and ready on alarm. Curr Opin Cell Biol , 1–8. [DOI] [PubMed] [Google Scholar]
- Stokes AC. (1888). A preliminary contribution toward a history of the fresh-water infusoria of the United States. J Trenton Nat Hist Archive , PI. III, fig. 4, 118–119. [Google Scholar]
- Stöver BC, Müller KF. (2010). TreeGraph 2: Combining and visualizing evidence from different phylogenetic analyses. BMC Bioinf , 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga H, Miller WT. (2018). Src signaling in a low-complexity unicellular kinome. Sci Rep , 5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga H, Ruiz-Trillo I. (2013). Development of ichthyosporeans sheds light on the origin of metazoan multicellularity. Dev Biol , 284–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AR. (2011). Issue Image. PLoS Biol Available at https://journals.plos.org/plosbiology/article?id=10.1371/image.pbio.v09.i06 (accessed 31 January 2019). [Google Scholar]
- Vandenkoornhuyse P, Quaiser A, Duhamel M, Le Van A, Dufresne A. (2015). The importance of the microbiome of the plant holobiont. New Phytol , 1196–1206. [DOI] [PubMed] [Google Scholar]
- Verworn M. (1899). General Physiology: An Outline of the Science of Life, London and New York: Macmillan and Company. [Google Scholar]
- Waller RF, Cleves PA, Rubio-Brotons M, Woods A, Bender SJ, Edgcomb V, Gann ER, Jones AC, Teytelman L, von Dassow P, et al. (2018). Strength in numbers: collaborative science for new experimental model systems. PLoS Biol , e2006333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West GS. (1916). Algae. Vol. 1. Myxophyceae, Peridinieae, Bacillarieae, Chlorophyceae Together with a Brief Summary of the Occurrence and Distribution of Freshwater Algae, London and Edinburgh: Cambridge University Press. [Google Scholar]
- Woznica A, King N. (2018). Lessons from simple marine models on the bacterial regulation of eukaryotic development. Curr Opin Microbiol , 108–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Katsura H, Noël M-H, Ichinomiya M, Kuwata A, Sato S, Yoshikawa S. (2019). Ontogenetic analysis of siliceous cell wall formation in Triparma laevis f. inornata (Parmales, Stramenopiles). J Phycol , 196–203. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.