Abstract
TP53 is the most frequently mutated gene in human cancers, and despite intensive research efforts, genome-scale studies of p53 function in whole animal models are rare. The need for such in vivo studies is underscored by recent challenges to established paradigms, indicating that unappreciated p53 functions contribute to cancer prevention. Here we leveraged the Drosophila system to interrogate p53 function in a postmitotic context. In the developing embryo, p53 robustly activates important apoptotic genes in response to radiation-induced DNA damage. We recently showed that a p53 enhancer (p53RErpr) near the cell death gene reaper forms chromatin contacts and enables p53 target activation across long genomic distances. Interestingly, we found that this canonical p53 apoptotic program fails to activate in adult heads. Moreover, this failure to exhibit apoptotic responses was not associated with altered chromatin contacts. Instead, we determined that p53 does not occupy the p53RErpr enhancer in this postmitotic tissue as it does in embryos. Through comparative RNA-seq and chromatin immunoprecipitation–seq studies of developing and postmitotic tissues, we further determined that p53 regulates distinct transcriptional programs in adult heads, including DNA repair, metabolism, and proteolysis genes. Strikingly, in the postmitotic context, p53-binding landscapes were poorly correlated with nearby transcriptional effects, raising the possibility that p53 enhancers could be generally acting through long distances.
INTRODUCTION
p53 is a transcription factor that coordinates cellular responses to stress, such as cell cycle arrest, DNA repair, senescence, and apoptosis (Vousden and Prives, 2009). TP53 is the most frequently mutated gene in human cancers. Most cancer-associated p53 mutations occur in its DNA-binding domain, suggesting that p53 DNA binding and associated gene regulation are crucial in tumor suppression (Hainaut and Hollstein, 2000). Numerous genome-scale studies have characterized p53 transcriptional programs and, in a comprehensive literature survey, we found a total of 30 peer-reviewed studies that combine binding and expression data sets (Jen and Cheung, 2005; Ceribelli et al., 2006; Wei et al., 2006; Shaked et al., 2008; Smeenk et al., 2008, 2011; Lee et al., 2010; Bandele et al., 2011; Botcheva et al., 2011; Botcheva and McCorkle, 2014; Li et al., 2012a, 2013; Nikulenkov et al., 2012; Kenzelmann Broz et al., 2013; Menendez et al., 2013; Schlereth et al., 2013; Zeron-Medina et al., 2013; Akdemir et al., 2014; Chang et al., 2014; Idogawa et al., 2014; Janky et al., 2014; McDade et al., 2014; Rashi-Elkeles et al., 2014; Sanchez et al., 2014; Kirschner et al., 2015; Sammons et al., 2015; Su et al., 2015; Tonelli et al., 2015; Younger et al., 2015; Tanikawa et al., 2017). Although these have provided powerful insights into general p53 functions, the majority were performed in immortalized/cancer cell lines, which limits insights to proliferative contexts often associated with distortions in the p53 regulatory network itself. In addition, p53 can be activated by diverse stressors, including extended cell culture (Shaked et al., 2008; Botcheva et al., 2011). Among all these comprehensive genome-scale studies, only two compared tissue-specific stress programs in vivo (Tonelli et al., 2015; Tanikawa et al., 2017). Given that p53 mutations are associated with a variety of cancers, in vivo studies are ideal to dissect the mechanisms that modulate p53 function to specific cellular environments, hence tissue-specific actions of p53 are likely important for its potent tumor suppressive capability.
Early studies demonstrated that p53 regulatory axes can be uncoupled from one another. For example, the p53175P point mutation abolishes apoptotic responses but has no effect on p53-mediated cell cycle arrest (Rowan et al., 1996). Recently three key lysine residues have been identified as crucial for p53-mediated apoptosis, cell cycle arrest, and senescence (Li et al., 2012b). Strikingly, the combined abrogation of these three pathways, through p53 lysine mutations or combined knockout of p53 downstream targets, is not sufficient to recapitulate early onset cancer phenotypes observed in p53 null mice (Li et al., 2012b; Valente et al., 2013). Together these observations establish that noncanonical p53 programs are crucial for tumor suppression.
Drosophila has proven to be a powerful in vivo model for investigating p53 biology (Lu et al., 2010; Lunardi et al., 2010; Link et al., 2013; Merlo et al., 2014; Wylie et al., 2016; Ingaramo et al., 2018). In addition to the array of genetic tools that allow for rigorous in vivo studies, p53 function is highly conserved in Drosophila, and it is the only member of this gene family present in the fly genome (Sutcliffe and Brehm, 2004; Belyi and Levine, 2009; Lu et al., 2009). Previously, we and others described p53 stress-programs of gene regulation in embryos (Brodsky et al., 2000, 2004; Sogame et al., 2003; Akdemir et al., 2007). Here we examine the adult Drosophila head as a model for p53 function in tissues that are almost entirely postmitotic and where p53 was previously shown to be active (Merlo et al., 2014). Interestingly, we found that canonical stress-induced apoptotic networks are unresponsive in postmitotic tissue and instead distinct programs are activated. To understand how p53 promotes alternate programs, we established that constitutive occupancy at a pivotal p53 enhancer correlates with apoptogenic outcomes. Furthermore, we mapped genome-wide p53 DNA occupancy in both the developing and the postmitotic tissues and integrated these with RNA-seq profiles. To our knowledge, these analyses constitute the first genome-scale profile integrating p53 binding and transcriptional regulation in Drosophila melanogaster.
RESULTS
Canonical p53 apoptotic programs are unresponsive in postmitotic tissue
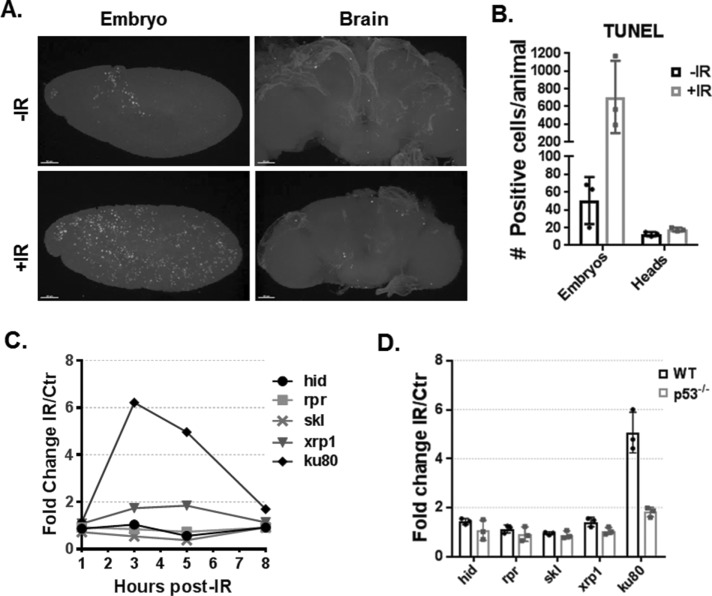
Like its human counterpart, Drosophila p53 can direct transcriptional programs to promote adaptive responses to various cellular stresses, including apoptosis (Brodsky et al., 2000; Ollmann et al., 2000). In Drosophila embryos, ionizing radiation (IR) stress has been characterized as a potent inducer of p53-mediated apoptosis (Sogame et al., 2003). Interestingly, induction of p53-mediated apoptosis appears to be highly sensitive to cell type and cell-cycle modifications, for example, endocycling cells of Drosophila larval salivary glands fail to up-regulate cell death upon DNA damage, underscoring the context specificity of p53-regulated programs (Moon et al., 2005, 2008; Fan et al., 2010; Zhang et al., 2014; Arya and White, 2015; Qi and Calvi, 2016). Furthermore, recent reports have shown that expression of human tau in Drosophila neurons leads to DNA damage and apoptosis (Khurana et al., 2012). Surprisingly, p53 loss enhances tau-mediated apoptosis in Drosophila postmitotic neurons (Merlo et al., 2014). Therefore, we hypothesized that in postmitotic tissue, p53 does not activate cell death in response to IR. To test this hypothesis, we exposed flies to IR and quantified cell death by TUNEL (terminal deoxynucleotidyl transferase dUTP nick end labeling) in both embryos and adult brains (Figure 1, A and B). As expected, in embryos, a robust apoptotic wave can be detected following IR. In brains, by contrast, no significant increase in TUNEL-positive cells was observed. Samples treated with DNase were used as positive control, and samples in which no terminal deoxynucleotidyl transferase (TdT) enzyme was added served as the negative control (Supplemental Figure S1, A and B). These results suggest canonical apoptotic programs triggered by p53 are nonresponsive in postmitotic tissue.The proapoptotic genes Head Involution Defective (hid), Reaper (rpr), and Sickle (skl) are among the most robust stress-induced p53 targets in embryos (Brodsky et al., 2004; Akdemir et al., 2007), and furthermore, when these genes are collectively removed, virtually all stress-dependent and programmed cell death is abolished (White et al., 1994). Therefore, to determine whether the proapoptotic p53 program can be transcriptionally activated in postmitotic tissue, we tested for up-regulation of these apoptogenic genes in Drosophila heads 1, 3, 5, and 8 h post-IR exposure using previously described reverse transcription-droplet digital PCR (RT-ddPCR) assays (Link et al., 2013). Interestingly, all three cell death genes were unresponsive up to 8 h post-IR (Figure 1C) and, as technical controls, we reproduced induction of hid, rpr, and skl on embryonic RNAs, as previously reported (Supplemental Figure S1C) (Brodsky et al., 2004; Akdemir et al., 2007; Link et al., 2013). To exclude the possibility that apoptotic genes are nonresponsive in heads due to a lack of p53 (or low p53), we examined nuclear lysates by performing Western blots. As seen in Supplemental Figure S2B, we found that p53 protein is readily detected in heads even before IR treatment. Therefore, p53 absence does not account for a lack of damage-induced apoptosis in this tissue. We also tested IR activation of other known embryonic p53 targets, xrp1 and ku80. In contrast to embryo tissue, where xrp1 is rapidly induced over 10-fold following IR, no up-regulation above twofold was observed in postmitotic tissue. However, within 3 h of IR challenge, robust induction of ku80 was detected in head tissue. To test whether p53 mediates activation of ku80 in heads, we repeated the RT-ddPCR experiment at the 3-h time point to include p53−/− heads (Figure 1D). Notably, ku80 induction was lost in p53−/− animals and, confirming our time course results, all cell death genes remained unresponsive in wild type (WT) and p53−/−. Therefore, p53 is required for ku80 induction in postmitotic tissue. Together these results establish that the apoptotic program triggered by p53 in irradiated embryos is unresponsive in postmitotic tissue.
FIGURE 1:
The p53 apoptotic program is unresponsive in postmitotic tissue. (A) TUNEL labeling in Drosophila WT embryos and brains, with and without IR stress (scale bar = 50 µm). (B) Quantification of TUNEL positive cells (mean values represented with SD, n = 3). (C) Expression time course for indicated genes following IR stress measured by RT-ddPCR from WT head tissue (mean values represented with SD, n = 3). Note that all five genes are known p53 targets in IR-treated embryos. (D) RT-ddPCR for indicated genes 3 h post-IR in head tissue of WT and p53−/− animals.
Occupancy at the p53 rpr enhancer predicts activation of apoptotic program
To understand why apoptogenic genes in the reaper region are unresponsive in postmitotic tissue, we focused on a well-characterized p53 response element ∼5 kb upstream of rpr (referred to as p53RErpr) (Brodsky et al., 2000). Genetic ablation of the p53RErpr eliminates stress-induced activation of rpr as well as hid and skl (a genomic region spanning more than 300 kb) (Link et al., 2013). Strikingly, this enhancer is also required for activation of genes in trans such as xrp1 and ku80 (Link, 2011; Link et al., 2013). Long-range regulation of these p53 targets is accomplished through chromatin contacts that link the p53RErpr and the cell death genes (Link et al., 2013). Therefore, we hypothesized that altered or diminished looping of this enhancer might preclude p53-mediated activation of cell death genes at this locus. To test this hypothesis, we examined chromatin folding through the reaper region by applying our previously published digital-3C assay on chromatin prepared from heads (Link et al., 2013). We found that the chromosome conformation pattern through the entire locus was well preserved in heads (Figure 2A) when compared with the pattern seen in embryos (Link et al., 2013). Therefore, failure to activate cell death genes in heads is not explained by substantial alterations of p53 enhancer contacts.
FIGURE 2:
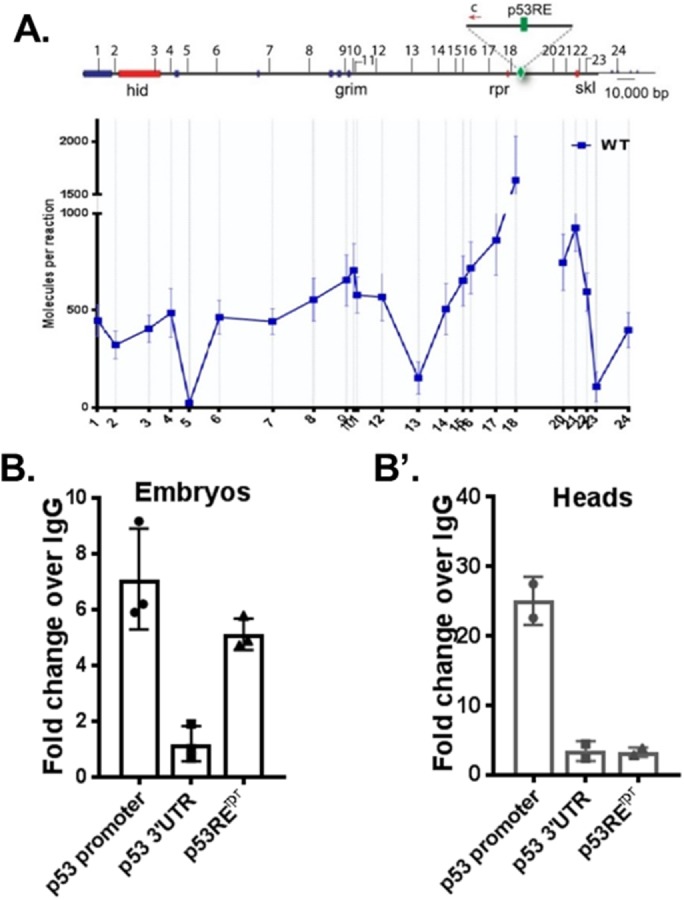
p53 binding at the rpr enhancer is lost in postmitotic tissue. (A) 3C-ddPCR experiments in Drosophila head tissue. Interactions between the p53RErpr (green) and IR induced cell death p53 targets (red) were measured. Note that the p53RErpr looping pattern is very similar to published patterns from embryos (Link et al., 2013). B and B’ show ChIP-ddPCR experiments measuring p53 occupancy at the p53RErpr in embryo and head tissue. Note that patterns were reproduced using ChIP-seq (Figure 4). Positive and negative control regions corresponding to the p53 promoter and p53 3′UTR were included (Merlo et al., 2014). Values represent fold change over immunoprecipitation using IgG. Mean values represented with SD, (A) n = 3, (B) n = 2 (see ChIP methods).
An alternative hypothesis that could explain how the response of reaper region genes is distinct in the two tissues invokes p53 occupancy at the p53RErpr. To test this hypothesis, we measured p53 binding at the p53RErpr through chromatin immunoprecipitation (ChIP)-ddPCR assays on embryo and head samples using established positive and negative regions (p53 promoter and p53 3′UTR, respectively) as our benchmarks (Merlo et al., 2014). Since p53 is often prebound to response elements prior to stress-induced target activation (Lee et al., 2010; Merlo et al., 2014; Tonelli et al., 2015), these ChIP experiments were conducted without perturbation. As seen in Figure 2B, we detected p53 binding to the p53RErpr in embryonic samples consistent with earlier studies (Merlo et al., 2014). However, in stark contrast, the p53RErpr was not bound by p53 in adult heads (Figure 2B’). Taken together, these results indicate that apoptogenic genes may be unresponsive because p53 does not occupy the p53RErpr in postmitotic tissue.
Genome-wide p53 DNA occupancy
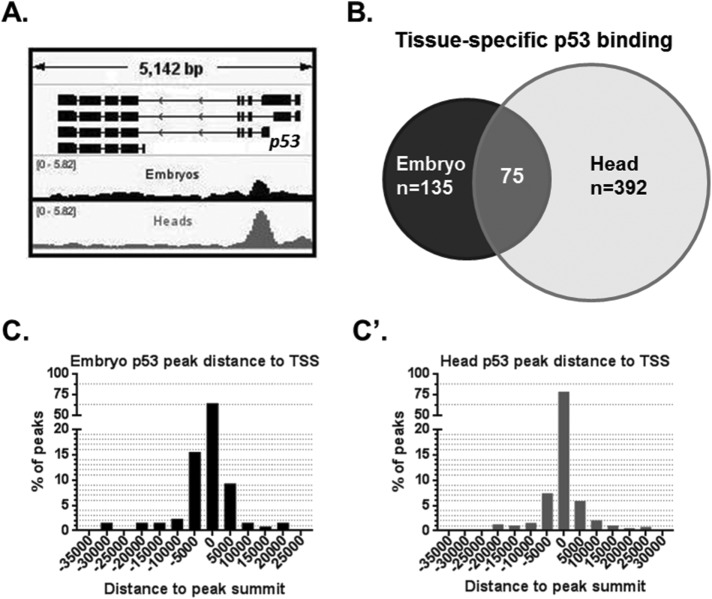
Given that p53RErpr occupancy is distinct in proliferative versus postmitotic tissues, we hypothesized that unique enhancer networks enable p53 to direct tissue-specific responses. To characterize genome-wide p53 DNA binding, we performed ChIP in embryos and heads, followed by high throughput sequencing (ChIP-seq). To ensure the biological validity of p53-enriched regions, we also performed ChIP-seq in corresponding p53−/− samples processed in parallel. From these data sets, authentic p53 peaks were called by comparing ChIP signals in WT and p53−/− tissue. In addition, we confirmed the ChIP-seq quality by assessing the previously described p53-binding site at the p53 gene promoter (Figure 3A) (Merlo et al., 2014). A total of 135 p53-enriched regions in embryos (e-peaks) and 392 in heads (h-peaks) were identified at FDR of 0.05. Interestingly, 75 regions had p53 enrichment in both tissues (Figure 3B). Additionally, a significant portion of called peaks contained the highly conserved p53-binding motif (in heads, 25.8%; embryos, 24.4%) and, as seen in Figure 3, C and C’, a majority of p53-binding sites in both heads (61.5%) and embryos (78.3%) were found within 5 kb of a transcription start site (TSS). We also confirmed these results by directly measuring p53 peaks of varying enrichment using ChIP-ddPCR on newly prepared chromatin from heads and, as seen in Supplemental Figure S3, all peaks tested were validated (N = 6).
FIGURE 3:
p53 genome-wide binding profiles in Drosophila embryos and heads. (A) ChIP-seq detects a published p53-enriched region at the promoter of p53 gene (Merlo et al., 2014). Graph displays fold enrichment over p53−/− samples (for normalized tracks including the p53−/− see Supplemental Figure S3). (B) Venn diagram of ChIP-seq p53 called peaks shared and distinct among the indicated tissues. (C, C’) Distribution of p53 peaks by distance between the peak and the closest TSS (C in embryos and C’ in heads).
Consistent with results from our ChIP-ddPCR assays (Figure 2, B and B’), our ChIP-seq data sets detected p53 binding at the p53RErpr in embryos but not in heads (Figure 4A). In addition, p53 enrichment was detected at the canonical targets hid and xrp1 not only in embryos but, surprisingly, also in heads where these genes fail to activate on IR (Figure 4, B and C). Therefore, p53 binding at each of these two genes is not sufficient to predict stimulus-induced gene activation; consistent with indications that other elements contribute to activation of hid and xrp1 in irradiated embryos (Link et al., 2013).
FIGURE 4:
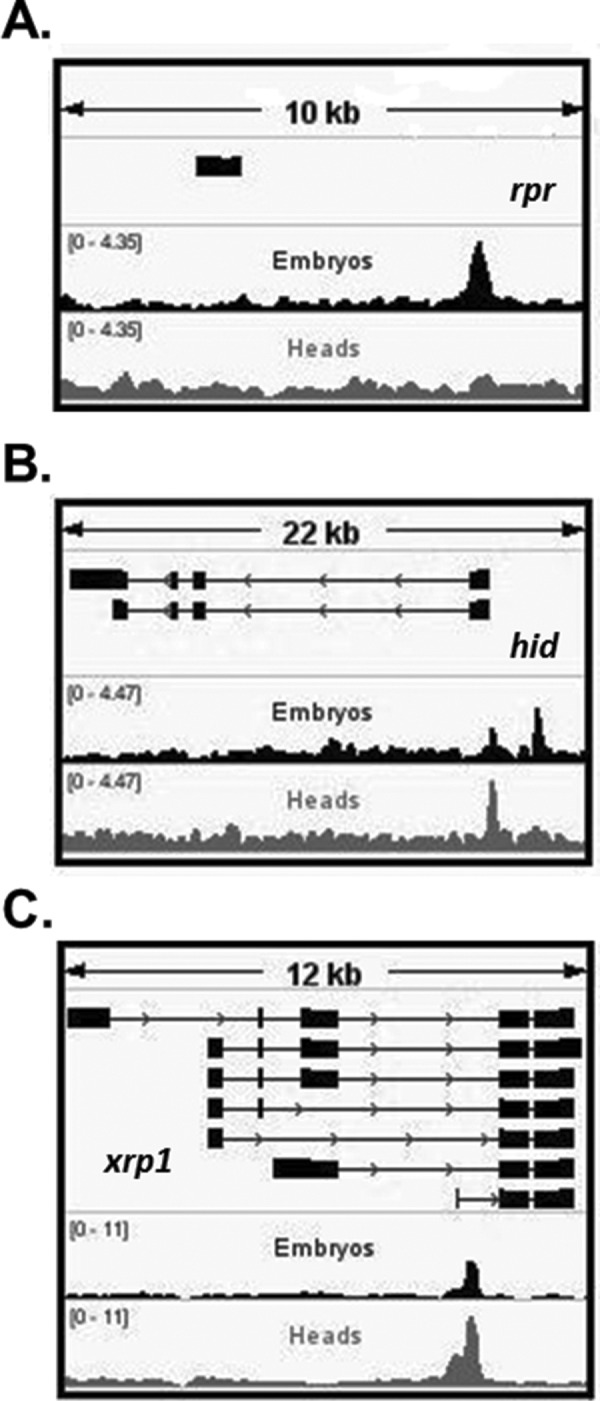
Binding at rpr is lost in postmitotic tissue among canonical stress-induced embryonic targets. (A) ChIP-seq confirms that p53 occupies the p53RErpr in embryos but not in heads. (B, C) p53-enriched regions in hid and xrp1 are detected in both tissues, although these genes are responsive only in embryos. All graphs display fold enrichment over p53−/− samples (for normalized tracks including the p53−/−, see Supplemental Figure S3).
To infer programs potentially engaged by p53, we identified genes within 5 kb of a p53-enriched region and performed pathway enrichment analyses using the gene ontology enRIchment analysis (GOrilla) web tool (Supplemental Tables S1 and S2) (Eden et al., 2009). The 5-kb interval was chosen based on distance between reaper and the p53RErpr enhancer. Apoptotic pathway components were enriched near embryonic peaks (Supplemental Table S1), but this was not seen for p53-binding profiles in heads (Supplemental Table S2).
p53 promotes alternate IR-induced programs in postmitotic tissue
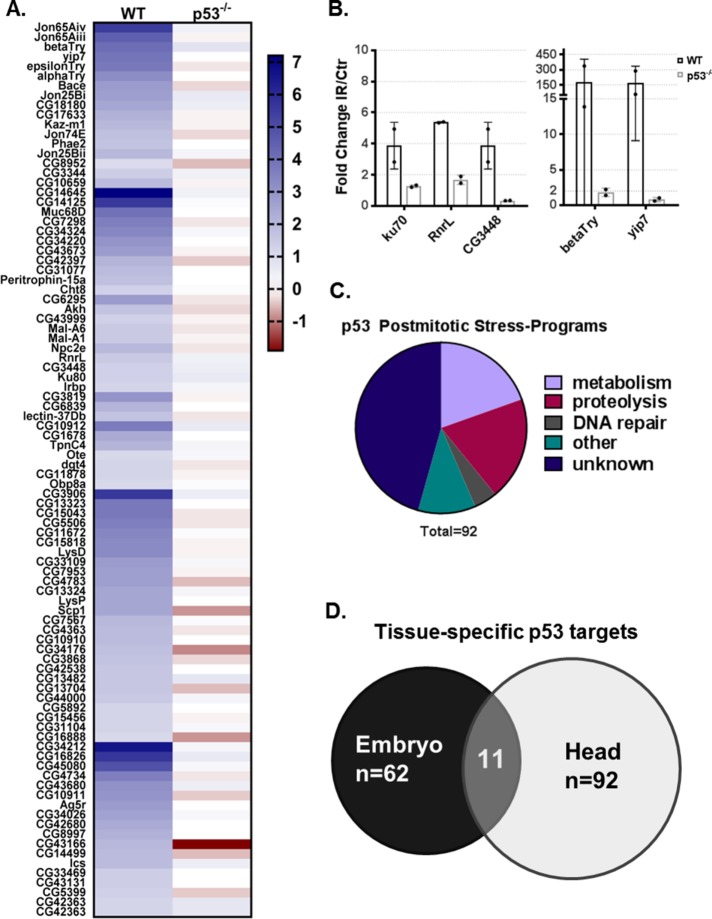
Although p53 does not induce stress-responsive apoptosis in postmitotic tissue, we observed p53-dependent induction of the DNA repair gene ku80 (Figure 1, C and D) and p53 binding in 392 regions (Figure 3B). Therefore, we hypothesized that p53 directs DNA damage response programs in postmitotic tissues. To determine the full scope of p53 transcriptional effects, we measured genome-wide expression changes provoked by IR in WT and p53−/− Drosophila heads through paired-end RNA-sequencing experiments. We observed a robust genome-scale p53 transcriptional effect in response to radiation and as expected, p53 IR-dependent gene activation was significantly more prevalent than IR-dependent repression (Supplemental Figure S5, A and B). Strikingly, in postmitotic tissue, nearly all stress-induced gene activation was p53-dependent. After performing differential gene expression analyses, we uncovered 92 head radiation-induced p53-dependent (hRIPD) protein-coding genes (Figure 5A). To validate the RNA-seq findings, we selected a set of five genes that have predicted human orthologues (Supplemental File 1) implicated in disease and confirmed p53-mediated IR induction for each using RT-ddPCR (Figure 5B). These assays clearly verified patterns observed in the RNA-seq data sets although the induction amplitudes varied, consistent with the stochastic nature of this perturbation. Next, we annotated the hRIPD genes for the biological process in which they function. Interestingly, the top three biological processes represented by hRIPD genes are metabolism, proteolysis, and DNA repair, including Ku80 and Ku70, which are central components of the nonhomologous end joining (NHEJ) pathway. However, most hRIPD genes are uncharacterized (Figure 5C); 47% have no functional annotation, 37% have predicted functions, and only 16% have a tested function. Interestingly, we identified a possible novel Drosophila DNA repair gene, CG3448, which contains a XRCC4-like domain. In humans, XRCC4 binds the NHEJ ligase, Lig4, and this complex is responsible for the ligation step of NHEJ. p53-regulated metabolic genes include Adipokinetic hormone (Akh), a fly-functional homolog of mammalian glucagon; Akh regulates metabolism of carbohydrates, lipids, and glycogen (Galikova et al., 2015). Intriguingly, studies in mice demonstrated that on starvation, p53 is required for gluconeogenesis and amino acid catabolism (Prokesch et al., 2017). Furthermore, p53 has been implicated in lipid metabolism in the mammalian system (Goldstein and Rotter, 2012). These observations establish the Drosophila head as an important in vivo model to study p53 stress-responsive programs.
FIGURE 5:
Stimulus-dependent p53 transcriptional programs in postmitotic tissue. (A) Heat map of novel p53 postmitotic targets (genes are grouped by biological process), IR fold change as log2 is colored according to legend. (B) RT-ddPCR validation of selected targets from RNA-seq (mean values represented with SD, n = 2). (C) Pie chart showing hRIPD genes by functional annotation. (D) The Venn diagram shows distinct and overlapping p53 targets identified in embryos and heads.
Originally, embryonic stress-induced profiles were described through microarray methods (Brodsky et al., 2004; Akdemir et al., 2007). To achieve a more comprehensive analysis and to enable direct comparisons with data sets reported here, we performed similar paired-end RNA-sequencing experiments on control and irradiated embryos using previously standardized protocols (Brodsky et al., 2004; Akdemir et al., 2007). After applying the same cutoffs, we identified embryonic radiation-induced p53-dependent (eRIPD) genes (Supplemental Figure S6). As expected, previously reported p53 embryonic targets were detected in our assay, including the proapoptotic program (rpr, skl, and hid). Interestingly, of the 92 hRIPD genes and the 62 eRIPD genes, only a small subset of 11 genes was shared by both tissues (Figure 5D). Notably, within this shared gene set, the DNA repair pathway is prominent. Relevant in this regard are the ku80 and ku70 (Irbp) genes as well as CG3448, predicted to engage in the ligation step of double-strand break repair. Together, these observations indicate that DNA repair is a major stress-induced p53 program commonly shared by irradiated embryos and heads.
Basal p53 genome-wide DNA occupancy does not predict IR-responsive targets in postmitotic tissue
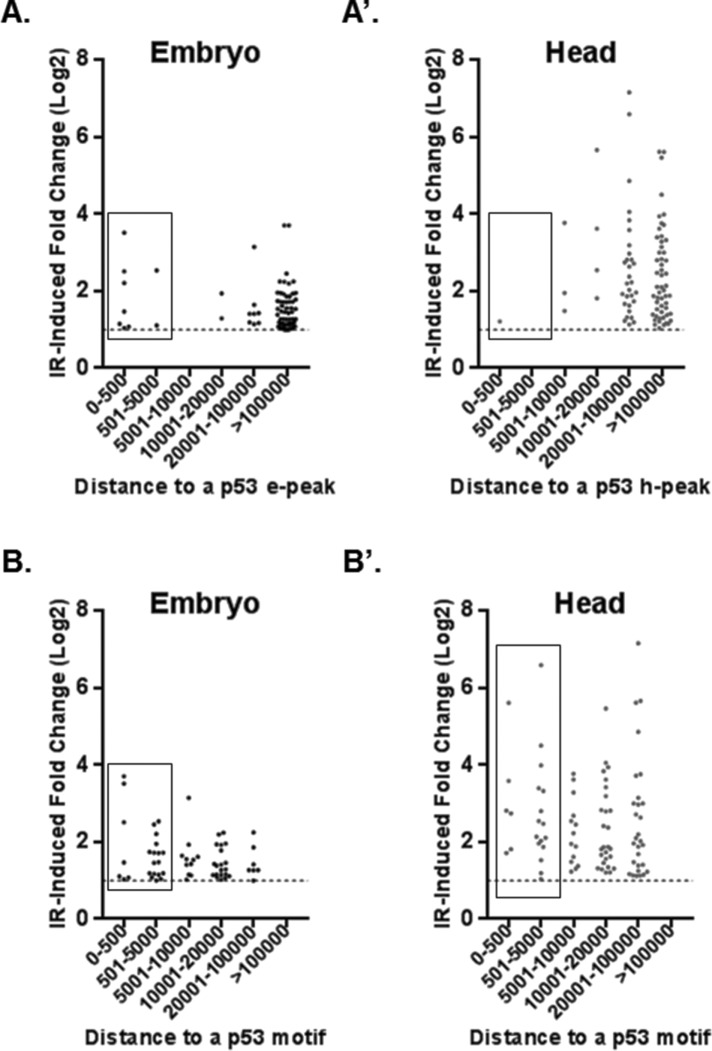
p53 can either be prebound or be recruited to stress-response target genes (Shaked et al., 2008; Tonelli et al., 2015).Therefore, to systematically examine constitutive p53 DNA occupancy and p53 IR-induced gene activation, we integrated our ChIP-seq and RNA-seq data sets (Supplemental File 2). We found that in embryos, only 14.5% of eRIPD genes contained TSSs within 5 kb of an e-peak (Figure 6A) and, as expected, some of the top IR targets including the apoptotic program were among genes near a p53-binding site. More surprisingly, in heads, only one hRIPD gene (CG15456) was associated with a h-peak within 5kb of the TSS (Figure 6A’). In addition, we examined whether p53 peaks correlated with constitutive gene expression differences between WT and p53−/− (Supplemental Figure S7). In embryos only ∼2% of 572 genes basally affected by p53 loss were within 5kb of a p53 e-peak (these include genes with expression twofold higher or lower in p53−/− compared with WT). Likewise, applying the same analyses in heads, we found only ∼3.4% of 583 genes affected by loss of p53−/− within 5 kb of a h-peak. Notably, however, in both embryos and heads, most p53-binding sites were indeed found within 5 kb of a TSS (61.5 and 78.3%, respectively) (Figure 3, C and C’). Therefore, despite a tendency for binding near genes, only a small portion of p53 peaks in both embryos and heads could predict p53-dependent gene expression.
FIGURE 6:
Most IR induced p53-dependent genes are distant from p53-binding sites. (A, A’) Distances spanning from the TSS of indicated RIPD gene set and the nearest p53 ChIP peak are plotted. (B, B’) Distances spanning from the TSS of the indicated RIPD gene set and a canonical p53 unsplit binding motif (associated with IR activation [Tonelli et al., 2015]). Note that genes having a peak/motif within 5 kb are boxed.
We also sought to assess de novo recruitment of p53 binding in response to IR but, unfortunately, the capacity of our irradiator precluded treatment of sufficient numbers of adults flies needed to prepare high quality chromatin suitable for ChIP-seq studies. However, Tonelli et al. (2015) found that IR-activated genes bound by p53 are more likely to contain the canonical motif with no spacer between the two decameric half-sites (Tonelli et al., 2015). Therefore, as an alternative approach, we probed the promoters of eRIPD and hRIPD genes for the presence of the unsplit p53 consensus (within 5kb of TSSs) (Figure 6B). We found that 30.5% of eRIPD genes contain the p53 motif compared with 14.5% RIPD genes that are prebound by p53. In heads, 23.9% of hRIPD genes have the motif versus 1.09% that are prebound. These analyses raise the possibility that de novo p53 occupancy may be an important driver of stress-induced gene activation, especially in postmitotic tissue.
DISCUSSION
We interrogated p53 function using RNA-seq and ChIP-seq studies to comparatively examine the action of p53 in postmitotic and embryonic tissues. In contrast with developing embryos—and despite significant DNA damage marked by γ-H2Av (Supplemental Figure S8A)—p53 did not trigger apoptotic programs in the adult fly head after radiation exposure (Figures 1 and 5). Mechanisms that specify distinct stimulus-dependent p53 programs in these tissues are not understood, but different levels and/or types of IR-induced DNA damage could be a contributing factor. We had previously shown in embryos that genetic loss of a single p53 enhancer (p53RErpr) upstream of rpr abrogates p53 IR activation of not only rpr but also genes far away, such as hid, skl, xrp1, and egr (Link, 2011; Link et al., 2013). We had also shown that the p53RErpr enhancer forms long-range chromatin contacts to its targets (Link et al., 2013). Interestingly, our ChIP-seq data in embryos revealed p53 binding not only at rpr but also at hid, xrp1, and egr (Figure 4 and Supplemental Figure S4). Strikingly, in heads, where all of these genes are unresponsive (Figures 1 and 5), p53 binding is maintained at each of these sites with the exception of rpr, where p53 occupancy at the p53RErpr was clearly lost (Figure 4). Therefore, p53 binding at sites near hid, egr and xrp1 is not sufficient to induce p53 target activation at these genes. Furthermore, since long range contacts between the p53RErpr and sites at hid and skl were unchanged in heads relative to embryos (Figure 2A), chromatin folding does not appear to determine target activation either. Furthermore, the combined observation that p53RErpr loops are maintained in heads, while p53 binding to p53RErpr is lost, corroborates our previous observations that p53 DNA binding is not required for chromatin looping of the p53RErpr (Link et al., 2013). Instead, our results suggest that p53 occupancy at the p53RErpr may act as the tissue-specific feature that licenses responsive loci within the reaper region. Consistent with this model, Zhang et al. (2008a) reported that this genomic region is compacted by H3K9me3 as embryonic development proceeds. Furthermore, this epigenetic silencing of p53-regulated apoptotic genes is also detected in endoreplicative tissues of third-instar larvae (Zhang et al., 2014).
Our RNA-seq studies revealed that in response to IR stress, Drosophila p53 can activate a diverse set of genes in adult heads (Figure 5 and Supplemental Figure S5). A significant portion of these genes, 88%, are unique to postmitotic tissue (Figure 5D). Pathway-enrichment analyses determined that p53 activates DNA repair, metabolism and proteolysis genes as well as many uncharacterized genes (Figure 5C). Importantly, many of the novel p53 target genes uncovered here have predicted human orthologues and some are associated with disease, including cancer (see Supplemental File 1). Interestingly, DNA repair appears to be a p53-driven program shared by proliferating and postmitotic tissues. The precise advantages of p53-mediated induction of DNA repair genes remain to be rigorously investigated (Jassim et al., 2003) but may be critical for tumor suppression (Janic et al., 2018). We observed that bulk γ-H2Av phosphorylation showed similar kinetics in WT and p53−/− heads up to 3 h postradiation (Supplemental Figure S8B), but defective DNA repair phenotypes beyond the detection limits of our assay or independent of the marker used are clearly possible (Yuan et al., 2010). Intriguingly, human orthologues of several p53 stress-responsive targets (Figure 5, A and B) are members of the Kallikrein (KLK) family of serine proteases (fly genes are alphaTry, betaTry, yip7, and Jon25Bi). KLK dysregulation and/or mislocalization have been associated with carcinogenesis and other pathologies including neurological disease (Kryza et al., 2016), but they have not been defined as p53 targets. KLK3 (yip7 and jon25Bi), also known as prostate-specific antigen, is a prominent biomarker for prostate cancer and may promote cancer evasion through interference in the p53 pathway (Niu et al., 2008; Filippou et al., 2016). Additionally, KLKs have been implicated with other hallmarks of cancer, such as energy metabolism, angiogenesis, cancer invasion, and metastasis (Filippou et al., 2016). It will be interesting to determine whether KLKs are p53 targets in mammals and whether KLKs are impacted by mutant p53 alleles. Another interesting target uncovered by these analyses is CG3448, which contains a XRCC4-like domain. In mice, XRCC4 loss leads to embryonic lethality associated with massive apoptosis of early postmitotic neurons (Yan et al., 2006). Given the types of targets we uncovered through RNA-seq, it appears that p53 postmitotic programs are adaptive for cell survival and, therefore, studying p53 in this context could reveal mechanisms employed by p53 alleles that are thought to have oncogenic properties (Lavigueur et al., 1989; Harvey et al., 1995; Lang et al., 2004; Olive et al., 2004; Hanel et al., 2013). Consistent with this, recent studies have uncovered a neuroprotective role of p53 in a Drosophila model of tauopathy (Merlo et al., 2014). Intriguingly, the IR response found here is distinct from the pathways reported in those studies, underscoring the context specificity of the p53 regulatory network in vivo.
We also examined genome-wide p53 DNA occupancy and integrated these data sets with corresponding RNA-seq data sets to determine how preconfigured enhancer networks may dictate tissue-specific responses. A powerful advantage in these studies is that all p53 peaks were called in relation to p53−/− ChIP-seq data sets produced in parallel. As expected, p53 was heavily enriched close to TSSs with 135 e-peaks (in embryos) and 393 h-peaks (in heads) (Figure 3). In embryos, p53 was poised at apoptotic genes as well as several top IR-induced targets (e.g., rpr, hid, and eiger). However, unlike the p53RErpr enhancer where tissue-specific binding predicted transcriptional regulation of the proximal gene rpr, p53-mediated gene expression could not be generally inferred from the DNA-binding location alone and, in heads, only one IR-responsive gene was prebound by p53. This gene, CG15456, has not been functionally studied but, notably, a human orthologue designated MIEN1 (migration and invasion enhancer 1) is an oncogene associated with breast, colorectal, and oral cancers (Katz et al., 2010; Dong et al., 2015; Rajendiran et al., 2015).
As seen in other models, our data exposed relatively low correlations between p53 binding and nearby p53-dependent gene expression (Tonelli et al., 2015). Several explanations could account for the discordance between p53 binding and IR-activated genes. First, transcriptional regulation could be imposed by looping of long distance enhancers. In mammalian cells and in Drosophila, for example, regulatory contacts spanning 430 kb and even across chromosomes have been documented (Link et al., 2013; Melo et al., 2013). Second, stimulus-dependent binding could govern regulatory targets and, while technical limitations prevented us from profiling p53 occupancy after radiation (discussed above), our computational evidence is certainly consistent with this (Figure 6B). Third, some targets could represent indirect responses. Taken together, these analyses underscore the danger of solely relying on p53 binding to predict gene regulation.
Interestingly, when we performed pathway-enrichment analyses to determine programs bound by p53 in postmitotic tissue (genes within 5 kb of a ChIP peak), we detected many developmental genes. Given that p53 function in development has been reported (Tedeschi and Di Giovanni, 2009; Contreras et al., 2018), it will be interesting to study whether occupancy at these sites represents residual binding from p53 functions performed during development. Together, these genome-wide analyses establish the Drosophila head as a useful in vivo model to dissect p53 stress responses in postmitotic tissues.
MATERIALS AND METHODS
Fly stocks
Flies were kept at 18–25°C and fed standard medium. The p53ns allele (Sogame et al., 2003) and the parental wild-type strain w1118 were used for experiments in Figures 1, C and D, and 2A. All other experiments were performed using the p535A-1-4 allele (Xie and Golic, 2004) paired with the parental wild-type yw strain. Flies were 1–2 wk old; genotypes were age-matched in each experiment.
TUNEL
Staged yw embryos (3–4 h) and adult yw flies (5–10 d) were treated with 40 Gy of IR and recovered for 1.5 h. Brains were dissected in cold HL-3 buffer (Stewart et al., 1994) and fixed for 1 h at room temperature in 4% paraformaldehyde PBST (1× PBS and 0.3% Triton X-100). Three washes were performed (10 min each) in PBST. Fixed brains were kept overnight in cold 1× PBS.
Embryos were dechorionated using 50% bleach, followed by a thorough wash with distilled water and fixed for 20 min at interface of n-heptane and 4% formaldehyde in 0.1 M PO4 (pH 7.2) (72% Na2HPO4, 28% NaH2PO4) on a shaker platform in glass tubes. The vitelline membrane was removed using the methanol cracking method. The next day, embryos were transferred to PBST through gradual methanol series (75, 50, and 25% MeOH PBST, 5 min each wash). Then three washes were performed (10 min each) in PBST.
TUNEL labeling was performed using Millipore’s kit FragEL DNA Fragmentation Detection Kit, Fluorescent. Briefly, samples were treated with proteinase k (1:100 in PBST), followed by four quick washes in PBST. A postfixation step was performed using 4% formaldehyde in PBST for 20 min at room temperature, followed by three washes (5 min each) in PBST. Tissue was equilibrated in TdT labeling solution for 30 min at 37°C without enzyme. Next, 30 µl of TdT labeling solution containing 3 µl of TdT enzyme was added to samples, and an incubation was performed at 37°C for 2 h. Finally, four washes were performed using PBST (15 min each) at room temperature (diamidino-2-phenylindole [DAPI] stain performed in the third wash). Brains were mounted with tape spacers in RapiClear (SunJin lab) and imaged using a Leica Sp8 (25× water lens) with 2-µm sections through the whole brain. Resulting stacks were analyzed using Imaris (Bitplane). After Gaussian smoothing, the Imaris Spots function was used to quantify TUNEL-positive cells in each brain.
RT-PCR
For IR stimulus, flies were treated with 40 Gy of γ-radiation. Total RNA was extracted from the tissue of interest using Invitrogen’s TRIzol reagent according to the manufacturer’s protocol. Next, samples were treated with Ambion’s TURBO DNA-free kit. cDNA synthesis was performed with Bio-Rad’s iScript Reverse Transcription Supermix for RT-qPCR. Droplet digital PCR (ddPCR) was performed using Bio-Rad’s EvaGreen system. Gene expression of each RIPD gene was normalized to rp49 expression, and fold induction was calculated comparing IR and mock samples in each genotype (primer sequences are listed in Table 1). All graphs containing error bars constitute two to three biological replicates; each biological replicate represents 15–50 animals homogenized together. Statistical significance was determined through multiple t test calculated using GraphPad Prism Software. Alternatively, conventional PCR using Promega goTaq DNA polymerase was used to assay gene expression by gel electrophoresis.
TABLE 1:
RT-PCR primer sequences.
| Target | Primer sequence |
|---|---|
| rp49 FWDa | ATG ACC ATC CGC CCA GCA TAC A |
| rp49 REVa | CGT AAC CGA TGT TGG GCA TCA GAT ACT |
| hid FWDa | GAT GGG GAT TCG AGT TCG GAT TCG GAT |
| hid REVa | CAC TGC CCA CCG ACC AAG TGC TAT A |
| rpr FWDa | GTG TGC GCC AGC AAC AAA GAA CTA |
| rpr REVa | TTG CGA TGG CTT GCG ATA TTT GCC |
| skl FWDa | GAG AGA ATG AGC GAG ACA GTG ACA GAG A |
| skl REVa | TCG ATT TGA AAA CTA GCG ACT GCT TAC A |
| xrp1 FWDa | CAT TAC CAA CAT CAA GCG TTC TGC TCC G |
| xrp1 REVa | TGT TGC TGG TGC TGG TAC TGG TAC TT |
| ku80 FWDa | TGT GTG GCG GAG ATT CTT AAG GA |
| ku80 REVa | ATC CTC GCA GGC TGT CTT ATT CAC A |
| ku70 FWD | AGG GCA AGG AGT TCG AGT TT |
| ku70 REV | GGA AGG CGT CCA GTT CGA TA |
| RnrL FWD | TAA GAG AGA TGG CAG GCA GG |
| RnrL REV | CCA TTG ATG ACT TGC AGG GTG |
| CG3448 FWD | ACT TCA ACG CTC TCA GCT CTC |
| CG3448 REV | CGT CGT CCA TCC ATT TGC TTC |
| BetaTry FWD | CCT CCT ATG GCT ACG GAA ACC |
| Beta Try REV | CAG CAC ATC CGT ATC CCC AG |
| Yip7 FWD | CCA TCA TCG GAA ACG AGT GGG |
| Yip7 REV | CTT GGG TGA ACT CGG GGC TA |
aPublished in Link et al. (2013).
RNA-seq
Approximately 100 fly heads were homogenized together per condition (yw and p535A-1-4, mock/IR treated). Embryos of the same genotypes were collected in standard grape juice agar plates and staged 3–4 h when IR/mock treatment was applied; animals were then allowed to recover for 1.5 h and were dechorionated using 50% bleach. RNA was extracted following the same Trizol protocol described above. After treatment with Ambion’s TURBO DNase kit, an isopropanol precipitation was performed. Next, 1 µg of total RNA was used for library preparation following standard Illumina protocols for poly (a)-stranded paired-end RNA-seq (reading length of 150 base pairs). One collection was processed and sequenced for each condition/genotype.
Sequenced read pairs were preprocessed to remove adapters using Cutadapt (Martin, 2011) and low-quality reads or bases with Prinseq (Schmieder and Edwards, 2011). Sequence alignment was to the Drosophila genome dm6 (Illumina) using Tophat2 and the parameters -p 10—mate-inner-dist = 200—mate-std-dev = 40—library-type fr-firststrand—no-coverage-search (Kim et al., 2013). The open-source Picard toolkit was used to mark PCR duplicates, which were removed using SAMtools along with low-quality alignments (quality score below 25) prior to downstream analyses (Li et al., 2009; Picard, 2017).
Differential gene expression analyses were performed using the Cuffdiff program (Trapnell et al., 2010) through the UT Southwestern Medical Center’s BioHPC Galaxy Service (galaxy.biohpc.swmed.edu) (Afgan et al., 2016). The library normalization method was geometric with blind dispersion estimation, and bias correction was performed. For analyses of p53 target activation, genes with expression values below 2 in all data sets were excluded as well as noncoding RNAs. A pseudocount of 1 was added to all gene expression values. The fold change was calculated between IR and mock IR samples, and a cutoff of 2′ fold change was used. For basal gene expression analyses, fold change was calculated between p53−/− and WT, and the cutoff used equaled 2′. We annotated biological functions of genes of interest using the batch download tool at Flybase (FB2017_02) (Gramates et al., 2017).
GOrilla (Eden et al., 2009)
We performed GO analyses using the two unranked lists of genes mode of GOrilla. Below is the description from results provided by the GOrilla web tool:
p Value is the enrichment p value computed according to the mHG or HG model (not corrected for multiple testing of 6948 GO terms).
FDR q value is the correction of the above p value for multiple testing using the Benjamini and Hochberg (1995) method. Namely, for the ith term (ranked according to p value), the FDR q value is (p value * number of GO terms)/i.
ChIP
We adapted previously published ChIP protocols (Chanas et al., 2004; Negre et al., 2006). Starting with ∼30 ml of adult flies, Drosophila heads were separated by flash-freezing on liquid nitrogen, followed by vigorous vortexing and sieve-sorting (Hogentogler, number 30 on the top and number 40 on the bottom). Next, heads were homogenized while being fixed in 10 ml of 1% formaldehyde in 60 mM KCl, 15 mM NaCl, 4 mM MgCl2, 15 mM HEPES (pH 7.6), 0.5% Triton X-100, and freshly added 0.5 mM dithiothreitol (DTT) and EDTA-free protease inhibitors (Roche). Tissue was mechanically disrupted in a ground glass homogenizer (five strokes) and then in a Douncer with type A, loose, pestles (10 strokes). Fixation step together with homogenization totaled 15 min. Fixation was stopped with the addition of glycine to 225 mM and 5 min incubation on ice. Nuclei were recovered by centrifugation at 4000 × g for 5 min at 4°C. Nuclei were washed three times with 3 ml of the same buffer used during fixation without formaldehyde. Next, nuclei were washed once with 3 ml of lysis buffer (140 mM NaCl, 15 mM HEPES, pH 7.6, 1 mM EDTA, 0.5 mM EGTA [ethylene glycol tetraacetic acid], 0.1% sodium deoxycholate, 1% Triton X-100 and freshly added 0.5 mM DTT, and protease inhibitors). Then, nuclei were resuspended in 900 µl of sonication buffer (lysis buffer + 0.1% SDS and 0.5% N-lauroylsarcosine). Samples were incubated for 10 min while rotating at 4°C. Sonication was performed in three 1.5-ml Eppendorf tubes (300 µl of sample each) with the Diagenode Bioruptor for 45 min at high, 0.5 min on/off. After sonication, samples were again incubated while rotating for 10 min at 4°C. Next, debris was spun down for 5 min at 4000 × g at 4°C and the supernatant was transferred to a clean tube. The pellet was resuspended in 900 μl of sonication buffer and incubated while rotating for 10 min at 4°C. Samples were pelleted again and the supernatant was transferred and combined with supernatant from the previous step. The combined supernatant was centrifuged two more times at max speed for 10 min each time. One percent of the sample was kept for the input control, and the rest was split evenly for immunoprecipitation (using 2 µg of Drosophila anti-p53 d200 from Santa Cruz and normal rabbit IgG also from Santa Cruz). Immunoprecipitation was performed overnight on a nutator at 4°C. The next day, 60 µl of Santa Cruz Protein A/G beads slurry (rinsed with lysis buffer) was incubated with samples for 4 h on a nutator at 4°C. Beads were washed three times with lysis buffer 5–10 min each and once with TE buffer on a nutator at 4°C. Beads were eluted with 100 µl of elution buffer (20 mM Tris–HCl, pH 7.5, 5 mM EDTA, and 50 mM NaCl and freshly added 1% SDS, 50 µg/ml Proteinase K, and 20 µg/ml RNase A) for 10 min at 65°C in a thermoshaker at 900 rpm twice. Eluate was kept at 65°C in a thermoshaker at 900 rpm overnight for de-cross-linking. The next day, a standard phenol/chloroform extraction was performed, followed by isopropanol precipitation of the ChIP DNA with added glycogen.
Embryos were collected and staged to 4–6 h, dechorionated with 50% bleach, followed by a thorough wash with distilled water. Embryos were then prepared according to the ChIP protocol described above.
ChIP enrichment was quantified by Bio-Rad’s ddPCR with EvaGreen system, following manufacture’s guidelines. Primers used are listed in Table 2. Statistical significance was determined through paired t test, calculated using GraphPad Prism Software.
Table 2:
ChIP-PCR primer sequences.
| Target | Primer sequence |
|---|---|
| p53 promoter FWDa | CGCTTGTACTTGCATCATTCG |
| p53 promoter REVa | GCGCCTTGGCTGGATAAAC |
| 3′ UTR FWDa | GTGGCAGCCGGTCGAA |
| 3′ UTR REVa | CAGCCAAAGCGGATGCA |
| p53RErpr FWDa | CGGAAAACTGATATGGCGATAAG |
| p53RErpr REVa | CGGTCCCTCAGTCTCCAAGTC |
| CG3967 FWD | GGC ATT GAA ATA CTT TTT GCG GTC |
| CG3967 REV | TCG TTT GCG ATC GTT CCG TT |
| corp FWD | TTG TTG CTC TAC GCC AAG CG |
| corp REV | ATT AAA CTC GTG CCA CCC CA |
| CG13204 FWD | GTG TGC ATG CAG CTC TCG |
| CG13204 REV | ATC GGA ATC TGC CAA CCG TC |
| Mhc FWD | GTT GTG TCG GAA CTC ATC CCT |
| Mhc REV | AGA TGA GCT GCG GTT GAT TGA |
| lok FWD | TTG AAA AGT GCG TTC CTA GCG |
| lok REV | AGT TCT TGA TGG CTC AGG CG |
| RpL10Aa FWD | AGT GCA GGA GTC TGC CCA TA |
| RpL10Aa REV | TTC TCT GTT GTG GGT GTC GC |
aPrimer sequences were first published in Merlo et al. (2014).
ChIP-seq
ChIP DNA was quantified using the Promega’s QuantiFluor ONE dsDNA System according to the manufacturer’s protocol. Then, 10 ng of ChIP DNA was used to prepare next-generation sequencing (NGS) libraries following previously published protocols (Liu and Kraus, 2017; Quail et al., 2008). Libraries were amplified with eight PCR cycles. ChIP libraries were sent for sequencing on Illumina NextSeq 500 at the McDermott Center NGS Core at UT Southwestern Medical Center. One collection (30 ml adults) was processed sequenced for each condition/genotype.
Sequencing read pairs were preprocessed to remove adapters using Cutadapt (Martin, 2011) and low-quality reads or bases with Prinseq (Schmieder and Edwards, 2011). Sequence alignment was to the Drosophila genome dm6 using Bowtie2 (Langmead and Salzberg, 2012). The open-source Picard toolkit was used to mark PCR duplicates, and downstream analyses were performed on uniquely mapped reads (Picard, 2017). MACS2 was used to call peaks with the –nomodel and –ratio flags (Zhang et al., 2008b). The NCIS scaling ratio was calculated for each negative control and ChIP comparison using the NCIS R package (Liang and Keles˛, 2012; R Core Team, 2017). Fold enrichment bedGraphs were created using the MACS2 bdgcmp subroutine using the procedure outlined in the MACS2 documentation and converted to bigwig format with the UCSC bedGraphToBigWig program (Kent et al., 2010). Individual signal tracks were created in deeptools using bamCoverage (–binSize 10 –smoothLength 30 –normalizeUsing CPM –extendReads 200) (Ramírez et al., 2016). Distance to nearest MACS2 peak was determined for all unique TSS of protein-coding genes in the RefSeq annotation of the Drosophila genome using BEDtools (Quinlan and Hall, 2010). Motif search was performed using Homer with the custom p53 motif matrix (Heinz et al., 2010):
| >GGACATGCCCAGACATGCCC dp53(p53) 8 | |||
| 0.33 | 0.1736 | 0.33 | 0.1664 |
| 0.275 | 0.108 | 0.421 | 0.198 |
| 0.481 | 0.052 | 0.334 | 0.135 |
| 0.019 | 0.826 | 0.07 | 0.086 |
| 0.657 | 0.118 | 0.056 | 0.17 |
| 0.221 | 0.028 | 0.099 | 0.653 |
| 0.002 | 0.003 | 0.99 | 0.006 |
| 0.077 | 0.406 | 0.021 | 0.498 |
| 0.15 | 0.535 | 0.078 | 0.238 |
| 0.168 | 0.36 | 0.132 | 0.341 |
| 0.348 | 0.137 | 0.341 | 0.175 |
| 0.241 | 0.078 | 0.526 | 0.156 |
| 0.499 | 0.022 | 0.405 | 0.076 |
| 0.007 | 0.987 | 0.004 | 0.003 |
| 0.652 | 0.1 | 0.029 | 0.22 |
| 0.165 | 0.054 | 0.108 | 0.674 |
| 0.083 | 0.046 | 0.854 | 0.018 |
| 0.126 | 0.343 | 0.057 | 0.476 |
| 0.211 | 0.416 | 0.109 | 0.266 |
| 0.19 | 0.349 | 0.138 | 0.325 |
Chromosome Conformation Capture (3C)
Tissue preparation.
Starting with 5 ml of whole flies, Drosophila heads were separated from bodies by flash-freezing on liquid nitrogen, followed by vigorous vortexing and sieve-sorting. The intact heads were cross-linked in 1 ml of the following buffer: 2% formaldehyde, 50 mM HEPES, pH 7.6, 100 mM NaCl, 0.1 mM EDTA, and 0.5 mM EGTA during 15 min at room temperature in the vortex (gentle mixing). Formaldehyde was quenched by rinsing tissue in 1 ml of 1× PBS, 0.01% Triton X-100, and 0.125 M glycine two times. Next, tissue was incubated on ice for 15 min in lysis buffer (10 mM Tris, pH 8.0, 10 mM NaCl, 0.2% NP40, fresh protease inhibitors). Then, mechanical lysis was performed with a glass homogenizer with type A, loose, pestles (10 strokes). Lysate was spun down for 5 min at 4000 × g in 4°C to recover nuclei. The supernatant was discarded.
Digestion.
Nuclear pellets were resuspended in 1 ml of 1.2× HindIII digestion buffer. Then, each sample was split into four tubes and the final volume of each tube was brought up to 350 μl of 1.2× HindIII digestion buffer along with 0.3% SDS. Samples were incubated at 65°C for 10 min while shaking at 1100 rpm. Then, 2% Triton was added and samples were incubated at 37°C for 15 min shaking at 1100 rpm. Digestion was performed with 700 U of HindIII enzyme at 37°C overnight shaking at 1100 rpm. The following day, HindIII was inactivated using 1.6% SDS with incubation for 30 min at 65°C shaking at 1100 rpm.
Ligation.
Ligation was performed in final volume of 16 ml of the following buffer: 1× T4 DNA ligase buffer, 1% Triton X-100, and water. Samples were allowed to stand at bench for 30 min before adding ligase to allow Triton to sequester SDS. Next, 8000 U of T4 DNA ligase was added to samples, and incubation was performed at 16°C overnight.
Reverse cross-linking.
The following was added to reverse cross-link DNA: 0.2 M NaCl, 20 µg/ml RNase, and 120 µg/ml Proteinase K. Samples were incubated at 65°C for at least 4 h.
DNA purification.
To purify the 3C DNA, phenol/chloroform extraction was followed by ethanol precipitation. Last, samples were put through Invitrogen PCR clean-up columns to increase DNA purity. DNA quality was checked with gel electrophoresis and ddPCR dilution curves.
ddPCR.
Interaction frequency between the p53RErpr and our previously characterized regions was assayed by ddPCR according to our published protocol (probes and primers) (Link et al., 2013). Controls for normalization and background assessment spanned a gene desert genomic locus, also from our previously published assay (Link et al., 2013).
Western blot
Whole cell or nuclear lysates were prepared from tissue of interest. The protein was quantified using the Bio-Rad Protein Assay according to the manufacturer’s instructions with a bovine serum albumin standard curve. The protein lysate was separated by SDS–PAGE and transferred to Immobilon-P membranes (Millipore) using a standard wet transfer protocol. The membrane was blocked for 1 h at room temperature or overnight at 4°C while rocking (5% milk, 0.5% Tween-20, and 1× TBS). The membrane was rinsed twice and washed once for 15 min on a rocker (0.2% milk, 0.2% Tween-20, and 1× TBS). The primary antibody was diluted in 1% milk, 0.2% Tween-20, and 1× TBS and was incubated for 1 h at room temperature or overnight at 4°C on a rocker (1:500 anti-drosophilap53 [C-11] from Santa Cruz, 1:1000 anti-γ-H2Av [UNC93-5.2.1] from Development Studies Hybridoma Bank [DSHB], and 1:5000, anti-tubulin [E7] from DSHB). The membrane was rinsed in wash buffer twice and washed once for 15 min on a rocker at room temperature. Horseradish peroxidase (HRP)-conjugated secondary antibody diluted in 1% milk, 0.2% Tween-20, and 1× TBS was incubated for 30–60 min on a rocker at room temperature. Next, the membrane was rinsed twice with wash buffer and washed three times for 10 min each on a rocker at room temperature. HRP was detected using chemiluminescence according to the manufacturer’s protocol. Membrane signal was capture and developed in GeneMate Blue Autoradiography Film. Last, films were scanned and analyzed in ImageJ software.
Supplementary Material
Acknowledgments
We thank Lee Kraus, Ray McDonald, Yasmin Vasquez, and Shino Murakami for guidance and resources for ChIP protocols; Michael Buszczak, Beatriz Fontoura, and Kim McKim for intellectual input; Hugo Bellen for resources for TUNEL experiments; Jessica Alatorre for technical support; Vivek Tiwari for guidance in unshown experiments; BioHPC at UT Southwestern and Galaxy, for bioinformatics services; microscopy support from the Neurovisualization core at Baylor College of Medicine (IDDRC, 1U54 HD083092 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development); FlyBase (NIH-5U41HG000739-24 and British Medical Research Council MR/N030117/1) for integrating Drosophila specific bioinformatics resources, including genome browsing; the Integrative Genomics Viewer (IGV) from the Broad Institute and UC San Diego for Software to facilitate visualization of NGS data; and the Developmental Studies Hybridoma Bank for antibodies. Bloomington Drosophila Stock Center (NIH-P40OD018537) provided stocks used in this study. This work was supported by the National Institutes of Health (1F31GM108472-04 and 4T32CA124334-10 to P.K., F32NS092270 to N.L., 1F31CA189691-01 to A.W., R01GM120196 and R01EY010199 to H.K., and R01GM072124, R01GM115682 to J.M.A.), the American Cancer Society (128847-PF-15-160-01-DDC to A.E.J.), the Cancer Prevention and Research Institute of Texas (RP170086 to J.M.A.), and the Welch Foundation (I-1865 to J.M.A.).
Abbreviations used:
- Akh
adipokinetic hormone
- ChIP
chromatin immunoprecipitation
- DAPI
diamidino-2-phenylindole
- ddPCR
droplet digital PCR
- DSHB
Development Studies Hybridoma Bank
- DTT
dithiothreitol
- EGTA
ethylene glycol tetraacetic acid
- eRIPD
embryonic radiation-induced p53 dependent
- GOrilla
gene ontology enRIchment analysis
- hid
Head Involution Defective
- hRIPD
head radiation-induced p53 dependent
- HRP
horseradish peroxidase
- IR
ionizing radiation
- KLK
Kallikrein
- NGS
next-generation sequencing
- NHEJ
nonhomologous end joining
- rpr
Reaper
- RT
reverse transcription
- skl
Sickle
- TdT
terminal deoxynucleotidyl transferase
- TSS
transcription start site
- TUNEL
terminal deoxynucleotidyl transferase dUTP nick end labeling.
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-12-0791) on March 20, 2019.
REFERENCES
- Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Eberhard C, et al (2016). The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res , W3–W10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akdemir F, Christich A, Sogame N, Chapo J, Abrams JM. (2007). p53 directs focused genomic responses in Drosophila. Oncogene , 5184–5193. [DOI] [PubMed] [Google Scholar]
- Akdemir KC, Jain AK, Allton K, Aronow B, Xu X, Cooney AJ, Li W, Barton MC. (2014). Genome-wide profiling reveals stimulus-specific functions of p53 during differentiation and DNA damage of human embryonic stem cells. Nucleic Acids Res , 205–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arya R, White K. (2015). Cell death in development: signaling pathways and core mechanisms. Semin Cell Dev Biol , 12–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandele OJ, Wang X, Campbell MR, Pittman GS, Bell DA. (2011). Human single-nucleotide polymorphisms alter p53 sequence-specific binding at gene regulatory elements. Nucleic Acids Res , 178–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyi VA, Levine AJ. (2009). One billion years of p53/p63/p73 evolution. Proc Natl Acad Sci USA , 17609–17610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botcheva K, McCorkle SR. (2014). Cell context dependent p53 genome-wide binding patterns and enrichment at repeats. PLoS One , e113492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botcheva K, McCorkle SR, McCombie WR, Dunn JJ, Anderson CW. (2011). Distinct p53 genomic binding patterns in normal and cancer-derived human cells. Cell Cycle , 4237–4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky MH, Nordstrom W, Tsang G, Kwan E, Rubin GM, Abrams JM. (2000). Drosophila p53 binds a damage response element at the reaper locus. Cell , 103–113. [DOI] [PubMed] [Google Scholar]
- Brodsky MH, Weinert BT, Tsang G, Rong YS, McGinnis NM, Golic KG, Rio DC, Rubin GM. (2004). Drosophila melanogaster MNK/Chk2 and p53 regulate multiple DNA repair and apoptotic pathways following DNA damage. Mol Cell Biol , 1219–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceribelli M, Alcalay M, Vigano MA, Mantovani R. (2006). Repression of new p53 targets revealed by ChIP on chip experiments. Cell Cycle , 1102–1110. [DOI] [PubMed] [Google Scholar]
- Chanas G, Lavrov S, Iral F, Cavalli G, Maschat F. (2004). Engrailed and polyhomeotic maintain posterior cell identity through cubitus-interruptus regulation. Dev Biol , 522–535. [DOI] [PubMed] [Google Scholar]
- Chang GS, Chen XA, Park B, Rhee HS, Li P, Han KH, Mishra T, Chan-Salis KY, Li Y, Hardison RC, et al (2014). A comprehensive and high-resolution genome-wide response of p53 to stress. Cell Rep , 514–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras EG, Sierralta J, Glavic A. (2018). p53 is required for brain growth but is dispensable for resistance to nutrient restriction during Drosophila larval development. PLoS One , e0194344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Huang Y, Kong L, Li J, Kou J, Yin L, Yang J. (2015). C35 is overexpressed in colorectal cancer and is associated tumor invasion and metastasis. Biosci Trends , 117–121. [DOI] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. (2009). GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics , 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Lee TV, Xu D, Chen Z, Lamblin AF, Steller H, Bergmann A. (2010). Dual roles of Drosophila p53 in cell death and cell differentiation. Cell Death Differ , 912–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippou PS, Karagiannis GS, Musrap N, Diamandis EP. (2016). Kallikrein-related peptidases (KLKs) and the hallmarks of cancer. Crit Rev Clin Lab Sci , 277–291. [DOI] [PubMed] [Google Scholar]
- Galikova M, Diesner M, Klepsatel P, Hehlert P, Xu Y, Bickmeyer I, Predel R, Kuhnlein RP. (2015). Energy homeostasis control in drosophila adipokinetic hormone mutants. Genetics , 665–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein I, Rotter V. (2012). Regulation of lipid metabolism by p53 - fighting two villains with one sword. Trends Endocrinol Metab , 567–575. [DOI] [PubMed] [Google Scholar]
- Gramates LS, Marygold SJ, Santos GD, Urbano JM, Antonazzo G, Matthews BB, Rey AJ, Tabone CJ, Crosby MA, Emmert DB, et al (2017). FlyBase at 25: looking to the future. Nucleic Acids Res , D663–D671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainaut P, Hollstein M. (2000). p53 and human cancer: the first ten thousand mutations. Adv Cancer Res , 81–137. [DOI] [PubMed] [Google Scholar]
- Hanel W, Marchenko N, Xu S, Yu SX, Weng W, Moll U. (2013). Two hot spot mutant p53 mouse models display differential gain of function in tumorigenesis. Cell Death Differ , 898–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey M, Vogel H, Morris D, Bradley A, Bernstein A, Donehower LA. (1995). A mutant p53 transgene accelerates tumour development in heterozygous but not nullizygous p53-deficient mice. Nat Genet , 305–311. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell , 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idogawa M, Ohashi T, Sasaki Y, Maruyama R, Kashima L, Suzuki H, Tokino T. (2014). Identification and analysis of large intergenic non-coding RNAs regulated by p53 family members through a genome-wide analysis of p53-binding sites. Hum Mol Genet , 2847–2857. [DOI] [PubMed] [Google Scholar]
- Ingaramo MC, Sanchez JA, Dekanty A. (2018). Regulation and function of p53: A perspective from Drosophila studies. Mech Dev , 82–90. [DOI] [PubMed] [Google Scholar]
- Janic A, Valente LJ, Wakefield MJ, Di Stefano L, Milla L, Wilcox S, Yang H, Tai L, Vandenberg CJ, Kueh AJ, et al (2018). DNA repair processes are critical mediators of p53-dependent tumor suppression. Nat Med , 947–953. [DOI] [PubMed] [Google Scholar]
- Janky R, Verfaillie A, Imrichova H, Van de Sande B, Standaert L, Christiaens V, Hulselmans G, Herten K, Naval Sanchez M, Potier D, et al (2014). iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS Comput Biol , e1003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jassim OW, Fink JL, Cagan RL. (2003). Dmp53 protects the Drosophila retina during a developmentally regulated DNA damage response. EMBO J , 5622–5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jen KY, Cheung VG. (2005). Identification of novel p53 target genes in ionizing radiation response. Cancer Res , 7666–7673. [DOI] [PubMed] [Google Scholar]
- Katz E, Dubois-Marshall S, Sims AH, Faratian D, Li J, Smith ES, Quinn JA, Edward M, Meehan RR, Evans EE, et al (2010). A gene on the HER2 amplicon, C35, is an oncogene in breast cancer whose actions are prevented by inhibition of Syk. Br J Cancer , 401–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Zweig AS, Barber G, Hinrichs AS, Karolchik D. (2010). BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics , 2204–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenzelmann Broz D, Spano Mello S, Bieging KT, Jiang D, Dusek RL, Brady CA, Sidow A, Attardi LD. (2013). Global genomic profiling reveals an extensive p53-regulated autophagy program contributing to key p53 responses. Genes Dev , 1016–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana V, Merlo P, DuBoff B, Fulga TA, Sharp KA, Campbell SD, Gotz J, Feany MB. (2012). A neuroprotective role for the DNA damage checkpoint in tauopathy. Aging Cell , 360–362. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol , R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner K, Samarajiwa SA, Cairns JM, Menon S, Perez-Mancera PA, Tomimatsu K, Bermejo-Rodriguez C, Ito Y, Chandra T, Narita M, et al (2015). Phenotype specific analyses reveal distinct regulatory mechanism for chronically activated p53. PLoS Genet , e1005053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryza T, Silva ML, Loessner D, Heuzé-Vourc´h N, Clements JA. (2016). The kallikrein-related peptidase family: Dysregulation and functions during cancer progression. Biochimie , 283–299. [DOI] [PubMed] [Google Scholar]
- Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, Valentin-Vega YA, Terzian T, Caldwell LC, Strong LC, et al (2004). Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell , 861–872. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods , 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavigueur A, Maltby V, Mock D, Rossant J, Pawson T, Bernstein A. (1989). High incidence of lung, bone, and lymphoid tumors in transgenic mice overexpressing mutant alleles of the p53 oncogene. Mol Cell Biol , 3982–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, Li M, Michalowski AM, Zhang X, Liao H, Chen L, Xu Y, Wu X, Huang J. (2010). A genomewide study identifies the Wnt signaling pathway as a major target of p53 in murine embryonic stem cells. Proc Natl Acad Sci USA , 69–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. (2009). The sequence alignment/map format and SAMtools. Bioinformatics , 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, He Y, Dubois W, Wu X, Shi J, Huang J. (2012a). Distinct regulatory mechanisms and functions for p53-activated and p53-repressed DNA damage response genes in embryonic stem cells. Mol Cell , 30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Kon N, Jiang L, Tan M, Ludwig T, Zhao Y, Baer R, Gu W. (2012b). Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell , 1269–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Liu J, McLaughlin N, Bachvarov D, Saifudeen Z, El-Dahr SS. (2013). Genome-wide analysis of the p53 gene regulatory network in the developing mouse kidney. Physiol Genomics , 948–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang K, Keles˛ S. (2012). Normalization of ChIP-seq data with control. BMC Bioinformatics , 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link N. (2011). Communal Cell Death and p53 Mediated Transcriptional Control in Drosophila Melanogaster. PhD Thesis Dallas: University of Texas Southwestern Medical Center. [Google Scholar]
- Link N, Kurtz P, O’Neal M, Garcia-Hughes G, Abrams JM. (2013). A p53 enhancer region regulates target genes through chromatin conformations in cis and in trans. Genes Dev , 2433–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Kraus WL. (2017). Catalytic-independent functions of PARP-1 determine Sox2 pioneer activity at intractable genomic loci. Mol Cell , 589–603 e589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu WJ, Amatruda JF, Abrams JM. (2009). p53 ancestry: gazing through an evolutionary lens. Nat Rev Cancer , 758–762. [DOI] [PubMed] [Google Scholar]
- Lu WJ, Chapo J, Roig I, Abrams JM. (2010). Meiotic recombination provokes functional activation of the p53 regulatory network. Science , 1278–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunardi A, Di Minin G, Provero P, Dal Ferro M, Carotti M, Del Sal G, Collavin L. (2010). A genome-scale protein interaction profile of Drosophila p53 uncovers additional nodes of the human p53 network. Proc Natl Acad Sci USA , 6322–6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal , 10–12. [Google Scholar]
- McDade SS, Patel D, Moran M, Campbell J, Fenwick K, Kozarewa I, Orr NJ, Lord CJ, Ashworth AA, McCance DJ. (2014). Genome-wide characterization reveals complex interplay between TP53 and TP63 in response to genotoxic stress. Nucleic Acids Res , 6270–6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo CA, Drost J, Wijchers PJ, van de Werken H, de Wit E, Oude Vrielink JA, Elkon R, Melo SA, Leveille N, Kalluri R, et al (2013). eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol Cell , 524–535. [DOI] [PubMed] [Google Scholar]
- Menendez D, Nguyen TA, Freudenberg JM, Mathew VJ, Anderson CW, Jothi R, Resnick MA. (2013). Diverse stresses dramatically alter genome-wide p53 binding and transactivation landscape in human cancer cells. Nucleic Acids Res , 7286–7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlo P, Frost B, Peng S, Yang YJ, Park PJ, Feany M. (2014). p53 prevents neurodegeneration by regulating synaptic genes. Proc Natl Acad Sci USA , 18055–18060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon NS, Di Stefano L, Morris EJ, Patel R, White K, Dyson NJ. (2008). E2F and p53 induce apoptosis independently during Drosophila development but intersect in the context of DNA damage. PLoS Genet , e1000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon NS, Frolov MV, Kwon EJ, Di Stefano L, Dimova DK, Morris EJ, Taylor-Harding B, White K, Dyson NJ. (2005). Drosophila E2F1 has context-specific pro- and antiapoptotic properties during development. Dev Cell , 463–475. [DOI] [PubMed] [Google Scholar]
- Negre N, Hennetin J, Sun LV, Lavrov S, Bellis M, White KP, Cavalli G. (2006). Chromosomal distribution of PcG proteins during Drosophila development. PLoS Biol , e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikulenkov F, Spinnler C, Li H, Tonelli C, Shi Y, Turunen M, Kivioja T, Ignatiev I, Kel A, Taipale J, et al (2012). Insights into p53 transcriptional function via genome-wide chromatin occupancy and gene expression analysis. Cell Death Differ , 1992–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y, Yeh S, Miyamoto H, Li G, Altuwaijri S, Yuan J, Han R, Ma T, Kuo HC, Chang C. (2008). Tissue prostate specific antigen (PSA) facilitates refractory prostate tumor progression via enhancing ARA70-regulated androgen receptor transactivation. Cancer Res , 7110–7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, Crowley D, Jacks T. (2004). Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell , 847–860. [DOI] [PubMed] [Google Scholar]
- Ollmann M, Young LM, Di Como CJ, Karim F, Belvin M, Robertson S, Whittaker K, Demsky M, Fisher WW, Buchman A, et al (2000). Drosophila p53 is a structural and functional homolog of the tumor suppressor p53. Cell , 91–101. [DOI] [PubMed] [Google Scholar]
- Picard. (2017). Picard ToolKit. http://broadinstitute.github.io/picard.
- Prokesch A, Graef FA, Madl T, Kahlhofer J, Heidenreich S, Schumann A, Moyschewitz E, Pristoynik P, Blaschitz A, Knauer M, et al (2017). Liver p53 is stabilized upon starvation and required for amino acid catabolism and gluconeogenesis. FASEB J , 732–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi S, Calvi BR. (2016). Different cell cycle modifications repress apoptosis at different steps independent of developmental signaling in Drosophila. Mol Biol Cell , 1885–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail MA, Kozarewa I, Smith F, Scally A, Stephens PJ, Durbin R, Swerdlow H, Turner DJ. (2008). A large genome center’s improvements to the Illumina sequencing system. Nat Methods , 1005–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics , 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendiran S, Kpetemey M, Maji S, Gibbs LD, Dasgupta S, Mantsch R, Hare RJ, Vishwanatha JK. (2015). MIEN1 promotes oral cancer progression and implicates poor overall survival. Cancer Biol Ther , 876–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T. (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res , W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashi-Elkeles S, Warnatz HJ, Elkon R, Kupershtein A, Chobod Y, Paz A, Amstislavskiy V, Sultan M, Safer H, Nietfeld W, et al (2014). Parallel profiling of the transcriptome, cistrome, and epigenome in the cellular response to ionizing radiation. Sci Signal , rs3. [DOI] [PubMed] [Google Scholar]
- R Core Team. (2017). R: A Language and Environment for Statistical Computing, Vienna, Austria: R Foundation for Statistical Computing. Retrieved from www.R-project.org.
- Rowan S, Ludwig RL, Haupt Y, Bates S, Lu X, Oren M, Vousden KH. (1996). Specific loss of apoptotic but not cell-cycle arrest function in a human tumor derived p53 mutant. EMBO J , 827–838. [PMC free article] [PubMed] [Google Scholar]
- Sammons MA, Zhu J, Drake AM, Berger SL. (2015). TP53 engagement with the genome occurs in distinct local chromatin environments via pioneer factor activity. Genome Res , 179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Y, Segura V, Marin-Bejar O, Athie A, Marchese FP, Gonzalez J, Bujanda L, Guo S, Matheu A, Huarte M. (2014). Genome-wide analysis of the human p53 transcriptional network unveils a lncRNA tumour suppressor signature. Nat Commun , 5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlereth K, Heyl C, Krampitz AM, Mernberger M, Finkernagel F, Scharfe M, Jarek M, Leich E, Rosenwald A, Stiewe T. (2013). Characterization of the p53 cistrome–DNA binding cooperativity dissects p53’s tumor suppressor functions. PLoS Genet , e1003726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R, Edwards R. (2011). Quality control and preprocessing of metagenomic datasets. Bioinformatics , 863–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaked H, Shiff I, Kott-Gutkowski M, Siegfried Z, Haupt Y, Simon I. (2008). Chromatin immunoprecipitation-on-chip reveals stress-dependent p53 occupancy in primary normal cells but not in established cell lines. Cancer Res , 9671–9677. [DOI] [PubMed] [Google Scholar]
- Smeenk L, van Heeringen SJ, Koeppel M, Gilbert B, Janssen-Megens E, Stunnenberg HG, Lohrum M. (2011). Role of p53 serine 46 in p53 target gene regulation. PLoS One , e17574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeenk L, van Heeringen SJ, Koeppel M, van Driel MA, Bartels SJ, Akkers RC, Denissov S, Stunnenberg HG, Lohrum M. (2008). Characterization of genome-wide p53-binding sites upon stress response. Nucleic Acids Res , 3639–3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogame N, Kim M, Abrams JM. (2003). Drosophila p53 preserves genomic stability by regulating cell death. Proc Natl Acad Sci USA , 4696–4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart BA, Atwood HL, Renger JJ, Wang J, Wu CF. (1994). Improved stability of Drosophila larval neuromuscular preparations in haemolymph-like physiological solutions. J Comp Physiol A , 179–191. [DOI] [PubMed] [Google Scholar]
- Su D, Wang X, Campbell MR, Song L, Safi A, Crawford GE, Bell DA. (2015). Interactions of chromatin context, binding site sequence content, and sequence evolution in stress-induced p53 occupancy and transactivation. PLoS Genet , e1004885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe JE, Brehm A. (2004). Of flies and men; p53, a tumour suppressor. FEBS Lett , 86–91. [DOI] [PubMed] [Google Scholar]
- Tanikawa C, Zhang YZ, Yamamoto R, Tsuda Y, Tanaka M, Funauchi Y, Mori J, Imoto S, Yamaguchi R, Nakamura Y, et al (2017). The transcriptional landscape of p53 signalling pathway. EBioMedicine , 109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedeschi A, Di Giovanni S. (2009). The non-apoptotic role of p53 in neuronal biology: enlightening the dark side of the moon. EMBO Rep , 576–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonelli C, Morelli MJ, Bianchi S, Rotta L, Capra T, Sabo A, Campaner S, Amati B. (2015). Genome-wide analysis of p53 transcriptional programs in B cells upon exposure to genotoxic stress in vivo. Oncotarget , 24611–24626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol , 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente LJ, Gray DH, Michalak EM, Pinon-Hofbauer J, Egle A, Scott CL, Janic A, Strasser A. (2013). p53 efficiently suppresses tumor development in the complete absence of its cell-cycle inhibitory and proapoptotic effectors p21, Puma, and Noxa. Cell Rep , 1339–1345. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C. (2009). Blinded by the light: the growing complexity of p53. Cell , 413–431. [DOI] [PubMed] [Google Scholar]
- Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, et al (2006). A global map of p53 transcription-factor binding sites in the human genome. Cell , 207–219. [DOI] [PubMed] [Google Scholar]
- White K, Grether ME, Abrams JM, Young L, Farrell K, Steller H. (1994). Genetic control of programmed cell death in Drosophila. Science , 677–683. [DOI] [PubMed] [Google Scholar]
- Wylie A, Jones AE, D’Brot A, Lu WJ, Kurtz P, Moran JV, Rakheja D, Chen KS, Hammer RE, Comerford SA, et al (2016). p53 genes function to restrain mobile elements. Genes Dev , 64–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie HB, Golic KG. (2004). Gene deletions by ends-in targeting in Drosophila melanogaster. Genetics , 1477–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan CT, Kaushal D, Murphy M, Zhang Y, Datta A, Chen C, Monroe B, Mostoslavsky G, Coakley K, Gao Y, et al (2006). XRCC4 suppresses medulloblastomas with recurrent translocations in p53-deficient mice. Proc Natl Acad Sci USA , 7378–7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younger ST, Kenzelmann-Broz D, Jung H, Attardi LD, Rinn JL. (2015). Integrative genomic analysis reveals widespread enhancer regulation by p53 in response to DNA damage. Nucleic Acids Res , 4447–4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Adamski R, Chen J. (2010). Focus on histone variant H2AX: to be or not to be. FEBS Lett , 3717–3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeron-Medina J, Wang X, Repapi E, Campbell MR, Su D, Castro-Giner F, Davies B, Peterse EF, Sacilotto N, Walker GJ, et al (2013). A polymorphic p53 response element in KIT ligand influences cancer risk and has undergone natural selection. Cell , 410–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Mehrotra S, Ng WL, Calvi BR. (2014). Low levels of p53 protein and chromatin silencing of p53 target genes repress apoptosis in Drosophila endocycling cells. PLoS Genet , e1004581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lin N, Carroll PM, Chan G, Guan B, Xiao H, Yao B, Wu SS, Zhou L. (2008. a). Epigenetic blocking of an enhancer region controls irradiation-induced proapoptotic gene expression in Drosophila embryos. Dev Cell , 481–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al (2008. b). Model-based analysis of ChIP-Seq (MACS). Genome Biol , R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.