Abstract
Condensins are highly conserved proteins that are important for chromosome maintenance in nearly all forms of life. Although many organisms employ two forms of the condensin complex, the condensin genes in Tetrahymena have expanded even further. Here we report a form of condensin that is specifically active during sexual reproduction. This complex, condensin D, is composed of the core condensin proteins, Smc2 and Smc4, and two unique subunits, the kleisin Cph5 and Cpd2. Cpd2 is also found in somatic nuclei in vegetative cells, but is dispensable for growth and nuclear division. Immunoprecipitation experiments show that condensin D interacts with a putative member of a chromatin-remodeling complex during development. Condensin D is required for sexual reproduction and for endoreplication and genome reduction of the progeny’s somatic nuclei. Altogether, Tetrahymena possesses at least four forms of condensin to fulfill the needs of maintaining chromosomes in two different nuclei containing the somatic and germline genomes.
INTRODUCTION
SMC (structural maintenance of chromosomes) complexes contain evolutionarily conserved proteins that play a central role in chromosome biology (Uhlmann, 2016). Diversification of these proteins in eukaryotes has produced condensin, cohesin, and SMC5/6, three distinct types of SMC complexes with partly overlapping roles in chromosome condensation, sister chromatid cohesion, and DNA repair. Both condensin and cohesin bind DNA topologically within the ring formed by the two SMC proteins linked by a kleisin subunit (Ivanov and Nasmyth, 2005; Cuylen et al., 2011). Both proteins are also proposed to function by creating chromatin loops (Ganji et al., 2018; van Ruiten and Rowland, 2018). However, cohesin is primarily responsible for holding sister chromatids together and for maintaining interphase nuclear architecture, whereas condensin mainly functions to condense and separate chromatids before chromosome divisions (Nasmyth and Haering, 2009; Yuen and Gerton, 2018). Understanding how SMC complexes shape chromosomes and the nuclear landscape is critical to many biological disciplines, including development, cancer, and reproduction research.
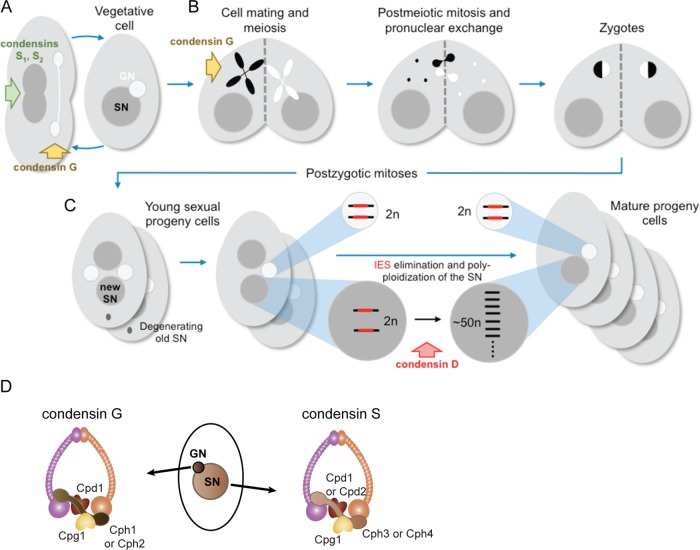
Tetrahymena thermophila is a free-living, unicellular freshwater ciliate that has long been used in the study of chromosome dynamics (Ruehle et al., 2016). Nuclear dualism, in which two distinct genomes are maintained within one cell, is a central feature of ciliate biology and makes Tetrahymena an interesting model for such research (Figure 1A; Karrer, 2000). The germline nucleus (GN) contains a diploid genome of five pairs of chromosomes and divides by mitosis and meiosis, but is transcriptionally silent during vegetative growth (Cole and Sugai, 2012). In contrast, the polyploid, transcriptionally active somatic nucleus (SN) divides by splitting and is regenerated during each cycle of sexual reproduction, which is initiated by starvation conditions (Figure 1B; Cole and Sugai, 2012). After meiosis and fertilization, the zygotic nucleus divides twice and two of the resulting products differentiate into new SN (Figure 1C). Supplemental Figure S1 shows cytological images of meiosis and various stages of postmeiotic development. The genome is drastically remodeled during SN development: germline chromosomes are fragmented into approximately 225 pieces, telomeres are added, and thousands of germline-limited sequences are removed in an RNA-directed process of DNA elimination (Noto and Mochizuki, 2017). These internal eliminated sequences (IESs) are remnants of ancient transposons and make up nearly a third of the germline genome (Chalker and Yao, 2011). The final step of nuclear development is endoreplication of the fragmented chromosomes, which continues after the cells return to vegetative growth, until the total copy number reaches ∼50 N (Allis et al., 1987).
FIGURE 1:
Growth, sexual reproduction, and nuclear differentiation in Tetrahymena. (A–C) Schematic diagram of the Tetrahymena vegetative and sexual life cycles and nuclear dualism. (A) In vegetative reproduction, the GN divides mitotically with the help of condensin G, and the polyploid SN employs condensin S1 to evenly distribute chromosomes to the daughter cells. (B) In sexual reproduction, starved cells of different mating types pair and the GN of both cells undergo meiosis (which requires condensin G) to generate four meiotic products, one of which is chosen as the gamete. The gamete in each cell is duplicated and the partners exchange gametic nuclei, which fuse to form the zygotic nucleus. (C) After two rounds of postzygotic mitoses, the progeny cells contain four nuclei, two of which develop into new SN, while the old parental SN is degraded. Condensin D is required for SN development, a process that includes chromosome breakage, IES elimination, and chromosomal endoreplication. (D) GN and SN condensin complexes in Tetrahymena.
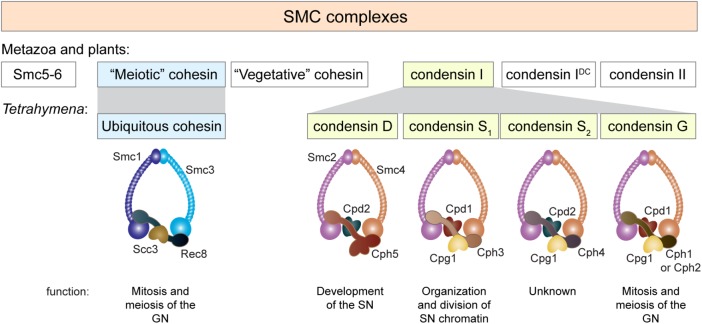
Both the GN and SN employ a set of condensins to compact and disentangle the chromosomes; this process is essential for chromosome segregation (Howard-Till and Loidl, 2018). The two nuclei have very different modes of division, and some Tetrahymena condensin genes appear to have multiplied and diversified to encode components of specialized complexes with specific functions in each nucleus (Figure 1, A and D; Howard-Till and Loidl, 2018). In short, the GN employs a condensin composed of the common Smc2 and Smc4 SMC proteins, partnered with one of two germline-limited kleisin proteins, Cph1 or Cph2, along with two additional proteins that are also found in the SN, Cpd1 (formerly Cpdt1) and Cpg1. The condensin responsible for SN division differs in the kleisin component: it contains Cph3 instead of Cph1/2.
In this study, we identify and investigate the function of an additional condensin complex during sexual reproduction and nuclear differentiation in Tetrahymena.
RESULTS
Cpd2 and Cph5 are components of a novel condensin complex
We previously showed that HA-tagged Cpd2 expressed from the endogenous locus localizes specifically to the SN of Tetrahymena (Howard-Till and Loidl, 2018). Because CPD2 expression levels peak in the late stages of conjugation (http://tfgd.ihb.ac.cn; Xiong et al., 2013), we hypothesized that the encoded protein may be most active in the newly generated SN of mating cells.
To better understand how Cpd2 condensin contributes to SN development, we immunoprecipitated Cpd2-HA from vegetative and late-stage mating cell extracts to identify interacting proteins. Mass spectrometry (MS) analysis of Cpd2-HA–binding proteins identified other proteins in the condensin complex, including Smc2, Smc4, Cph4, and Cpg1, indicating that Cpd2 is a functional condensin protein (Table 1; Figure 1D). Cpd2-HA immunoprecipitation (IP) from the mating cell extract identified yet another putative condensin complex member, TTHERM_00384720p, a previously unidentified Cph/kleisin homologue, which we called Cph5. The predicted protein consists of 2151 aa, which is more than twice the size of other Cph proteins. The N-terminal half of Cph5 aligns with the other four Tetrahymena Cph proteins, showing the highest homology in the first few hundred amino acids (Supplemental Figure S2). To determine whether the Cph4 and Cph5 kleisin proteins participate in distinct condensin complexes, ectopically expressed HA-Cph4 was immunoprecipitated from late-stage mating cell extracts. HA-Cph4 was found to interact with Smc2, Smc4, Cpg1, and Cpd2, but not with Cph5 (Table 1). This suggests that Cph4 and Cph5 form separate complexes with Cpd2: one in the vegetative SN and the other in the developing SN.
TABLE 1:
Proteins associated with Cpd1, Cpd2, and Cph4.
| Number of peptides | |||||
|---|---|---|---|---|---|
| Gene ID | Name/function | Cpd2 Veg. | Cpd1 Veg. | Cpd2 14 h | Cph4 8 h |
| TTHERM_00392760 | Cpd2 | 426 | 2 | 263 | 486 |
| TTHERM_00502600 | Parp6, WGR domain protein | 152 | 0 | 3 | 5 |
| TTHERM_00812950 | Smc2 | 141 | 159 | 110 | 558 |
| TTHERM_00446400 | Smc4 | 133 | 180 | 109 | 585 |
| TTHERM_01299730 | Cph4, condensin complex subunit 2 | 90 | 0 | 30 | 104 |
| TTHERM_00919690 | Cpg1, AT-hook motif protein | 79 | 33 | 7 | 157 |
| TTHERM_00784600 | Kinesin motor catalytic domain protein | 53 | 0 | 0 | 0 |
| TTHERM_00024320 | Chromosome condensation regulator repeat protein | 52 | 0 | 0 | 0 |
| TTHERM_00268010 | Hypothetical protein | 46 | 0 | 0 | 0 |
| TTHERM_00355820 | Hypothetical protein | 43 | 0 | 0 | 0 |
| TTHERM_000354599 | IQ calmodulin-binding motif protein | 37 | 0 | 0 | 0 |
| TTHERM_00703920 | EF-hand pair protein | 36 | 1 | 0 | 0 |
| TTHERM_00069230 | Kinesin motor catalytic domain protein | 32 | 0 | 0 | 0 |
| TTHERM_00941400 | IQ calmodulin-binding motif protein | 31 | 0 | 0 | 0 |
| TTHERM_01151610 | Normocyte-binding protein 2a | 29 | 0 | 0 | 0 |
| TTHERM_00001120 | Hypothetical protein | 24 | 0 | 0 | 0 |
| TTHERM_00384720 | Cph5, hypothetical protein* | 0 | 0 | 101 | 0 |
| TTHERM_00670660 | Serine/threonine kinase domain* | 0 | 0 | 40 | 0 |
| TTHERM_00222270 | Nmp1, PHD zinc finger protein* | 0 | 0 | 31 | 0 |
| TTHERM_00554600 | Cph3 | 0 | 25 | 0 | 0 |
| TTHERM_00540340 | Cph2 | 0 | 23 | 0 | 0 |
| TTHERM_01013300 | NADPH cytochrome P450 family reductase | 0 | 19 | 0 | 0 |
| TTHERM_00728870 | Cph1 | 0 | 17 | 0 | 0 |
Proteins were included in the table if more than 15 peptides were present in any given sample and fewer than two peptides were present in the WT control IP. Bold text denotes previously identified condensin subunits. Proteins specific to the 14-h mating stage IPs are marked with *.
Veg., vegetative.
Two additional proteins were identified as specifically associating with Cpd2 in late-stage mating cells: TTHERM_00222270p, called Nmp1 for New Mac PHD finger protein; and TTHERM_00670660p, a predicted serine/threonine kinase. Nmp1-HA localized to the SN in vegetative cells and in mating cells at all stages (Supplemental Figure S3). MS analysis of Nmp1-HA IPs from 14-h mating cell extracts confirmed the interaction with Cpd2 and Cph5, and identified several additional proteins containing zinc finger domains, SET domains, or domains with histone acetyltransferase homology (Supplemental Table S1). Cpd2-HA IPs from late-stage mating cell extracts also contained substantially fewer Cpg1 peptides compared with vegetative cell samples, which may indicate that Cpg1 is not part of the developmental condensin complex, condensin D (Table 1). Several proteins were identified only in vegetative cell samples, the most abundant of which was PARP6 (encoded by TTHERM_00502600; Table 1). This result suggests that Cpd2-containing condensin complexes might have an additional, albeit nonessential, role in the SN during vegetative growth.
To determine whether Cpd2-associated proteins in vegetative cells specifically associate with Cpd2-condensin, Cpd1-HA IPs were also performed on vegetative cell extracts. In addition to other condensin proteins, this experiment identified a completely different set of interacting proteins, suggesting that Cpd1 and Cpd2 condensin complexes are unlikely to have redundant functions in the vegetative SN (Table 1; see Supplemental files for complete data sets).
Condensin D localizes to SN during sexual reproduction and is excluded from elimination bodies during SN development
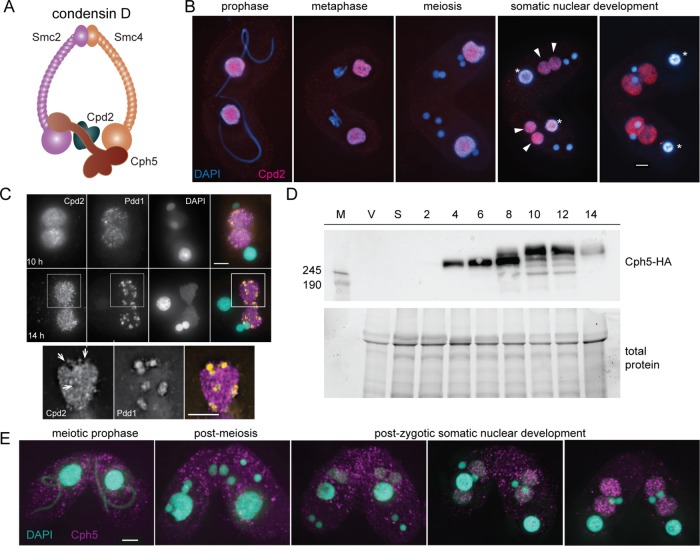
Based on the IP results, it seems likely that Cpd2 and Cph5 participate in a specific condensin complex (which we will call condensin D) that is active in late development (Figure 2A). The localization patterns of Cpd2 and Cph5 support this hypothesis. We find Cpd2 in the parental SN in mating cells during meiosis and fertilization stages and in the developing SN of progeny cells shortly after postzygotic nuclear divisions (Figure 2B). In later stages of SN development (14–16 h after initiation of mating), Cpd2 forms granular structures and is not uniformly distributed throughout the nuclei (Figure 2C). At this stage, DNA sequences destined for removal are aggregated into elimination bodies (Madireddi et al., 1996; Kataoka and Mochizuki, 2015). Heterochromatin marks consisting of trimethylated histone H3 lysine 9 (H3K9me3) and lysine 27 (H3K27me3) can be found in these bodies, as well as the HP1-like heterochromatin-binding protein, Pdd1 (Liu et al., 2007). Costaining of Cpd2-HA expressing cells with an anti-Pdd1 antibody showed that Cpd2 appears to be excluded from the elimination bodies (Figure 2C).
FIGURE 2:
Cpd2 and Cph5 localize to SN. (A) A developmental form of condensin (condensin D) is probably composed of Smc2, Smc4, Cpd2, and Cph5. (B) Cpd2-HA immunostaining (red) shows that Cpd2 localizes to paternal SNs during the early stages of mating. After fertilization and postzygotic divisions, Cpd2 appears in the differentiating SNs as they begin to swell (arrowheads), and remains in the developing nuclei until the late stages of mating, whereas it is lost from parental SNs (asterisks). (C) Costaining of Cpd2-HA and Pdd1 (orange) in developing SNs shows that Cpd2 is excluded from elimination bodies. Projected images of 3D stacks at 10 and 14 h. Enlarged images of a selected slice from the cell shows regions that lack Cpd2 staining (arrows) contain a Pdd1 signal in the merged image. (D) Western blots of whole cell protein extracts show that Cph5-HA is expressed as early as 4 h after initiation of mating, which corresponds to meiotic prophase. However, at later time points corresponding to SN development (8–14 h), additional Cph5-HA bands suggest that posttranslational modifications (probably phosphorylation) have occurred. M, molecular weight marker; V, vegetative cells; S, starved cells; 2–14, cells at the given number of hours after initiation of mating. (E) Immunostaining of Cph5-HA expressing cells fixed at different stages of mating shows that Cph5 specifically localizes to SN at late stages of development. Scale bars, 5 μm.
The CPH5 gene is developmentally transcribed, with a peak of expression at ∼8 h after initiation of mating (data from the Tetrahymena functional genomics database; http://tfgd.ihb.ac.cn; Xiong et al., 2013). To monitor protein expression and localization, Cph5 was C-terminally tagged with a single HA epitope. Western blotting of protein extracts from Cph5-HA expressing cells showed the protein first appearing at 4 h after initiation of mating, which corresponds to meiotic prophase (Figure 2D). However, anti-HA staining of fixed Cph5-HA cells showed that the protein only localizes to the new SN at ∼8 h after initiation of mating (Figure 2E). At this time point, we also saw an electrophoretic mobility shift by Western blotting, suggesting posttranscriptional modification of Cph5 (Figure 2D). By 10 h, the shifted form had become the predominant band. The HA-tagged protein is only expressed from the maternal nucleus, which begins to be degraded as the new SN are developing. Therefore, any Cph5 expression occurring at later time points will come from the untagged gene in the new SN and is therefore undetectable in this system.
Depletion of condensin D components prevents development of new SN in mating cells
RNA interference (RNAi) against CPD2 (cpd2i for short) did not result in any obvious phenotype in vegetative or meiotic division. However, mating cpd2i cells arrested before degradation of one of the two new GN, resulting in cells with two GN and two SN (Table 2). This phenotype is often associated with a failure to complete SN development (Coyne et al., 1999; Nikiforov et al., 1999; Mochizuki et al., 2002; Malone et al., 2005; Liu et al., 2007). RNAi efficiency is often variable from cell to cell and may not be completely penetrant against genes involved in late development. Therefore, we generated germline knockouts of the CPD2 gene using CRISPR–Cas9-mediated genome editing (Suhren et al., 2017). This approach resulted in the deletion of 11 bases (604–614) near the beginning of the CPD2 open reading frame, causing a frame shift and premature translation stop codon after 173 aa. Subsequent crosses of this strain produced homozygous lines carrying the mutant allele in both the GN and SN.
TABLE 2:
Viability and completion of mating for cpd2 mutant or RNAi cells.
| Genotype | Viability (%)a | Mating completion (%)b |
|---|---|---|
| CPD2hpc | nd | 97 |
| cpd2i | nd | 12 |
| B2086d× CU428d | 81 | 94 |
| CU428 × cpd2cr-4 | 96 | 89 |
| B2086 × cpd2cr-6 | 85 | 91 |
| cpd2cr-4 × 6 | 2 | 4 |
| cpd2cr + CPD2-HA (4–1) × (6–4) | 97 | 85 |
aViability was assayed by isolating single pairs and scoring for growth after 2 d (n = 88 pairs). nd, not determined.
bTo check for the completion of mating, cells were fixed at 24–26 h after initiation of mating. Slides were DAPI stained and nuclear morphology was evaluated (n = 100 cells). Cells containing two SN and one GN were scored as having completed mating.
cNoninduced RNAi strain.
dB2086 and CU428 are the WT strains.
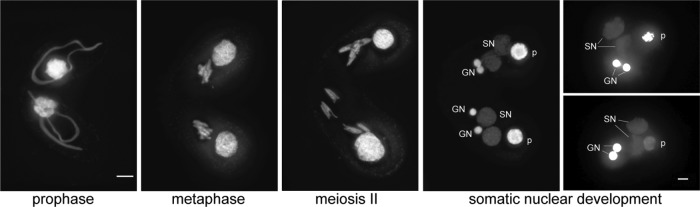
Matings of the cpd2 CRISPR cell lines (cpd2cr) phenocopied the cpd2i matings, that is, the strains failed to produce progeny and arrested at the two GN/two SN stage with low DNA content in the new SN (Table 2; Figures 3 and 4A). To ensure that no off-target mutations were responsible for the observed phenotype, the mutant lines were rescued by transformation with a functional HA-tagged CPD2 gene. Matings of the rescued lines showed that they had normal viability, and 4′,6-diamidino-2-phenylindole (DAPI) staining of fixed cells showed that most cells completed conjugation by 24 h, similar to the wild type (WT; Table 2).
FIGURE 3:
cpd2cr mutants complete meiotic divisions but arrest at a late stage of SN development. Chromosome condensation, meiotic divisions, and fertilization occur normally, and SN differentiation begins after the postzygotic divisions. However, after the mating pairs break apart, the cells arrest with two SN and two GN, and often do not completely degrade the parental SN (p). Scale bars, 5 μm.
FIGURE 4:
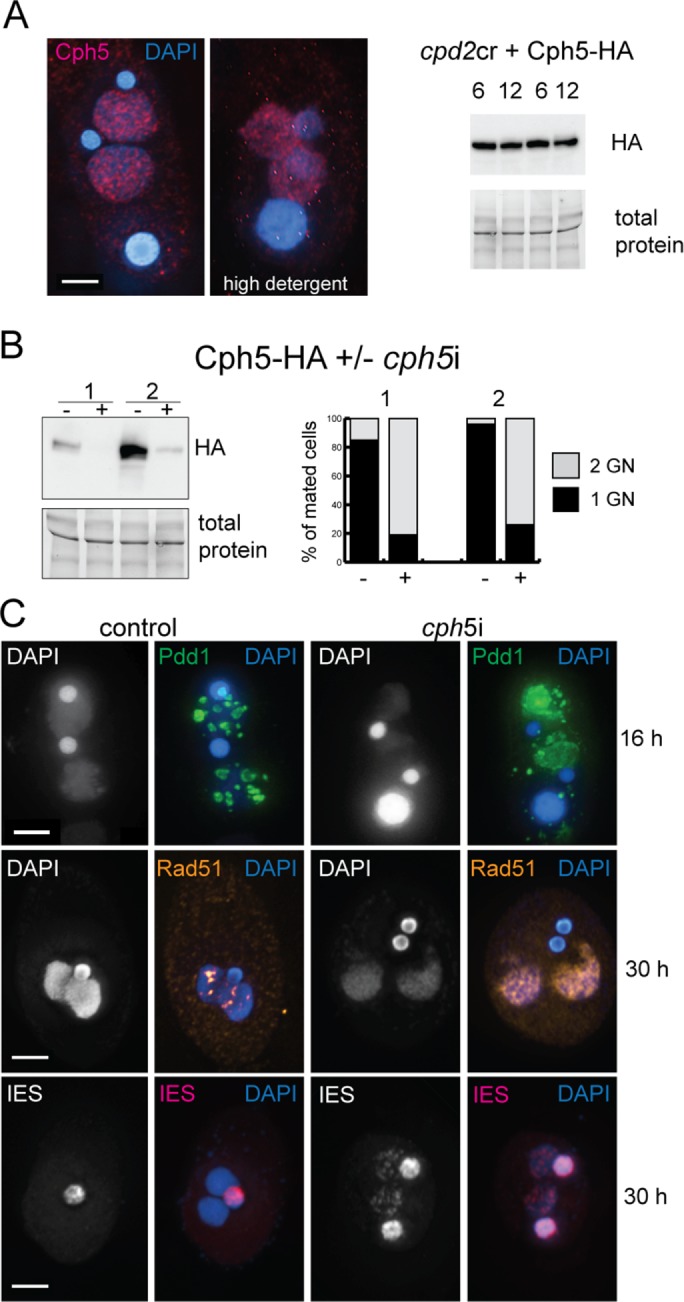
Cph5 is required to complete SN development. (A) In cpd2cr cells, Cph5 localizes to developing SN but is no longer modified at later developmental stages. Western blotting shows protein extracts taken from two independent experiments at 6 and 12 h after initiation of mating. (B) RNAi depletion of Cph5 in Cph5-HA expressing cells. The Cph5-HA level is strongly reduced after induction of RNAi, and mating cells do not complete sexual reproduction. The graph shows the number of cell pairs that complete mating in two independent experiments, assayed by counting the cells showing two SN/two GN vs. one SN/two GN at 30 h after initiation of mating. (C) Immunostaining of Rad51 and Pdd1, and FISH analysis of IESs in cph5i cells show that elimination structures do not form normally in these cells, DNA damage repair is delayed, and IESs are not completely eliminated.
To test whether Cph5 localization is dependent on Cpd2, the cpd2cr strains were transformed with the Cph5-HA construct. In the absence of Cpd2, Cph5 remains localized to the developing SN, even under high detergent fixation conditions, indicating that it is probably still chromatin bound (Figure 4A). Western blots of protein extracts from mating cells harvested at 6 and 12 h showed that Cph5-HA is expressed but, unlike in WT cells, the protein has the same electrophoretic mobility at both time points (Figure 4A). This result suggests that posttranslational modifications of Cph5 that normally occur during SN development do not occur in the absence of Cpd2.
RNAi against CPH5 (cph5i) produced a phenotype similar to that of cpd2i or cpd2cr cells. Meiotic events, gametic exchange, and fertilization occurred normally, but cells did not complete mating and arrested at the two GN/two SN stage (Figure 4B). In addition, the new SN appeared to have low DNA content with retained IES sequences (as assayed by FISH), a persistent Rad51 (DNA damage marker) signal, and abnormal Pdd1 localization during SN development (Figure 4C).
cpd2cr mutants fail to complete DNA elimination in new SN
cpd2cr cells appear to complete meiosis and initiate SN differentiation normally (Figure 3). Therefore, the failure to produce sexual progeny may be due to a defect in completing SN development. An important part of the developmental program is elimination of the transposon-like IESs (Noto and Mochizuki, 2017). To determine whether IES elimination is affected in cpd2cr cells, we performed FISH against a multicopy IES sequence. In WT cells IES FISH signal was found only in the GN, whereas in cpd2cr cells the FISH signal was also found in the new SN (Figure 5A). PCR analysis showed higher levels of IES retention in cpd2cr cells than in WT cells at all IES loci (Figure 5B). Although these results could reflect the lack of amplification of SN DNA (see below), when combined with the FISH data they suggest that IES elimination is defective in cpd2cr cells.
FIGURE 5:
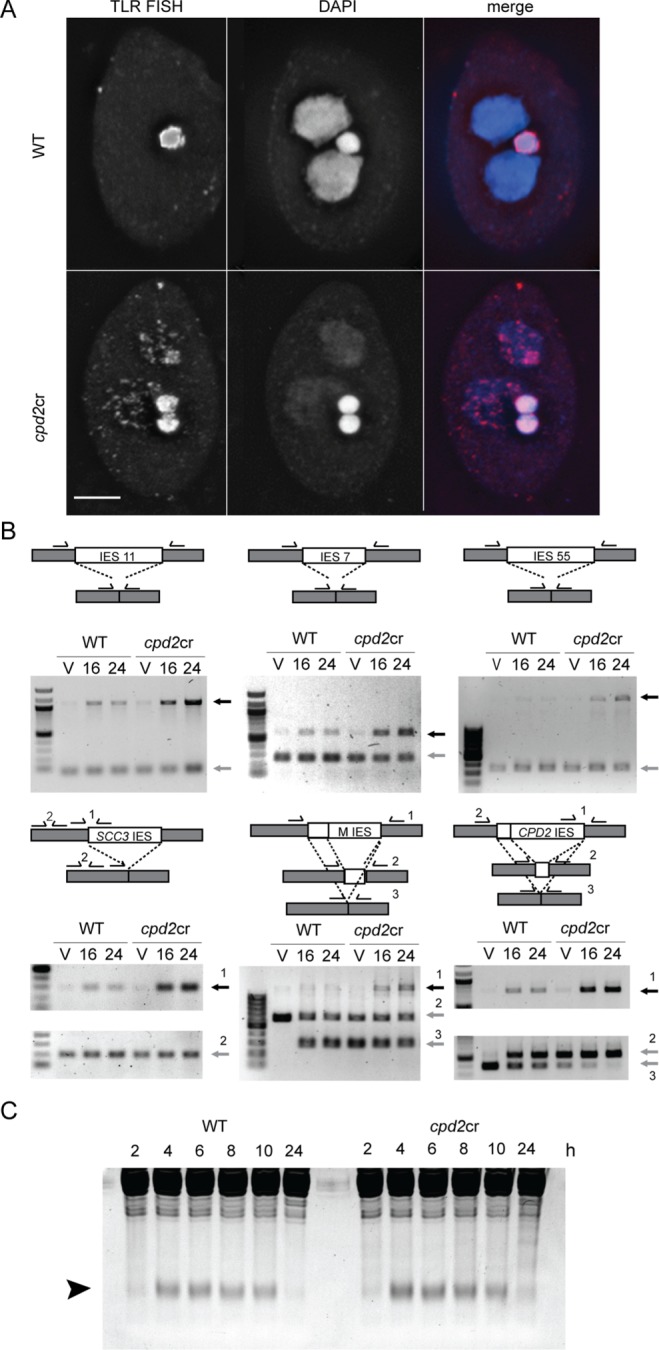
cpd2cr cells retain sequences that are normally eliminated during development. (A) FISH against a repetitive sequence normally found only in the GN (TLR, red) shows its abnormal localization in the developing SN of cpd2cr cells. Mutant cells also retain both GN, one of which is normally degraded at the end of the sexual cycle, and show weak SN DAPI staining, indicating developmental arrest. (B) PCR analysis of IES elimination shows increased levels of multiple IESs in cpd2cr cells. Diagrams show the amplification strategy used for each reaction. Gray boxes represent retained sequences, and white boxes the eliminated sequences. Primers are indicated by half arrows above the boxes. Black arrows indicate the PCR product from the germline form; gray arrows indicate the somatic or common form. The larger sizes of the IESs adjacent to the SCC3 and CPD2 genes necessitated two PCRs, one to amplify a region spanning one border of the IES, and a control reaction for the IES flank. (C) Small RNA production is normal in cpd2cr cells. Total RNA from mating cells was collected at the indicated time points after initiation of mating and analyzed by SDS–PAGE, followed by ethidium bromide staining. The arrowhead indicates small RNAs.
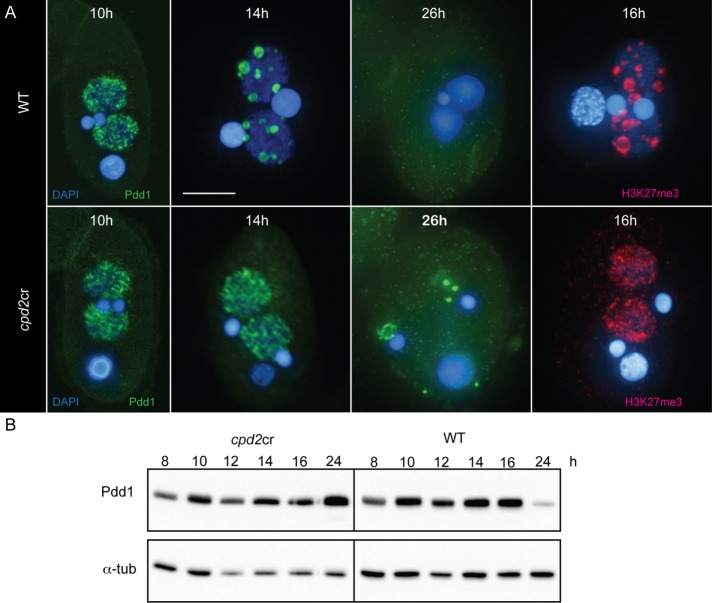
Next, we analyzed several key steps in IES elimination to investigate which step was blocked. An early step in the elimination process is the production of small RNAs from germline-limited sequences that are destined to be eliminated from the SN (Mochizuki et al., 2002). Scan RNA production appeared similar in both cpd2cr and WT cells (Figure 5C). Subsequent steps in the pathway require chromatin marks (including H3K9me3 and H3K27me3) to be placed on IES sequences in the developing SN; these marks are recognized and bound by the chromodomain protein, Pdd1 (Taverna et al., 2002; Liu et al., 2007). H3K27me3 and Pdd1 immunostaining patterns were similar (Liu et al., 2007). In WT cells, H3K27me3 and Pdd1 localize to the developing SN as they begin to swell, and form foci that become progressively larger and fewer in number (Figure 6A; Smothers et al., 1997; Liu et al., 2007). These structures have been previously shown to be centers of DNA elimination (Chalker, 2008). In the cpd2cr cells, H3K27me3 and Pdd1 localize to developing SN and form numerous small, dispersed foci, but these foci fail to coalesce into large elimination bodies (Figure 6A). In addition, whereas WT cells show almost no Pdd1 staining at 26 h after initiation of mating, the mutants show aberrant cytoplasmic Pdd1 foci, and a large amount of Pdd1 is occasionally associated with the new SN (Figure 5A). Western blot analysis supports the cytological data, by showing that Pdd1 expression persists in cpd2cr cells at 24 h, whereas Pdd1 levels are greatly reduced in WT cells at this time point (Figure 6B).
FIGURE 6:
cpd2cr cells do not form elimination bodies. (A) In WT cells, Pdd1-bound, heterochromatin-marked IESs coalesce into large foci at 14–16 h after induction of mating, as indicated by immunofluorescence staining with anti-Pdd1 or anti-H3K27me3 antibodies. In cpd2cr cells, these foci do not form and Pdd1 persists as abnormal cellular aggregates. Scale bar, 5 μm. (B) Western blotting also shows abnormal persistence of Pdd1 expression. Protein extracts were made from aliquots of mating cpd2cr or WT cells processed at the indicated time points after induction of mating.
cpd2cr mutants fail to undergo DNA amplification in new SN
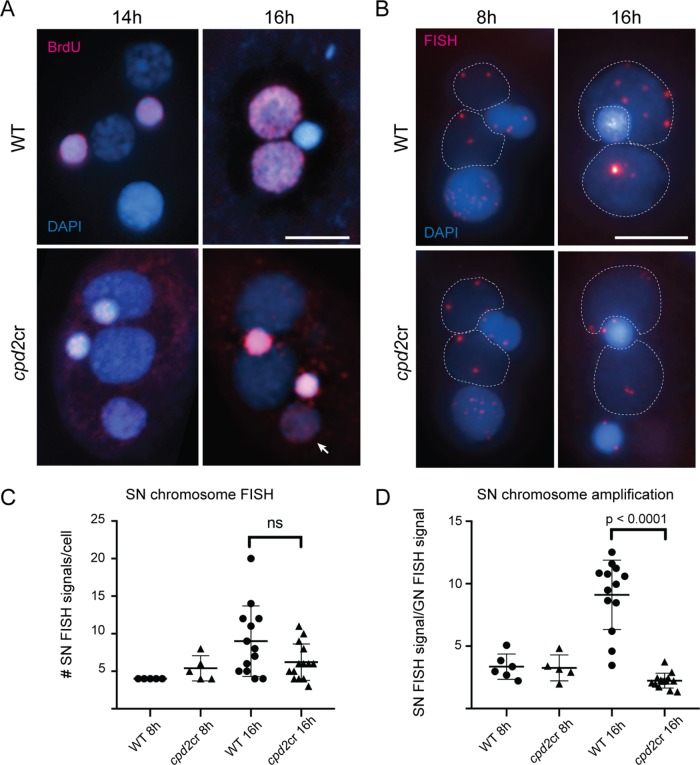
Failure to eliminate IESs was previously reported to occur concomitantly with a failure to endoreplicate chromosomes in the new SN (Nikiforov et al., 1999; Malone et al., 2005), and we found the same for cpd2cr cells. 5-Bromo-2′-deoxyuridine (BrdU) incorporation in WT cells showed that replication in the new SN occurs between 14 and 16 h after induction of mating (Figure 7A), whereas in cpd2cr cells, BrdU incorporation was never observed in the developing SN. Analysis of the number and intensity of FISH signals suggested that the chromosomal copy number increases approximately twofold in WT cells at 16 h after induction of mating, but remains unchanged in the mutant (Figure 7, B–D). In WT cells, the number of FISH signals in late development was often similar to the number in early development, but the increased size and intensity indicated that replication had occurred (Figure 7B). In contrast, in cpd2cr cells, the foci were of similar intensity at both time points. These results show that cpd2cr cells do not amplify DNA in new SN. However, whether this is a consequence of the overall developmental block is unknown. IES elimination is known to occur independently of replication (Nikiforov et al., 1999); therefore, failure to replicate is unlikely to cause a failure in elimination. BrdU signals in the GN of cpd2cr cells appear brighter than in WT cells, and a BrdU signal can also be seen in the degenerating parental SN (Figure 7A). This result could indicate abnormal replication in these nuclei, which might be a consequence of the disrupted developmental cycle.
FIGURE 7:
Chromosomes are not replicated in the new SN of cpd2cr mutants. (A) In WT cells, BrdU incorporation (red) occurs throughout the SN in cells midway through development. In cpd2cr cells, BrdU staining was found only in the GN, which served as an internal positive control, and in the old SN (arrow). (B) FISH analysis of a 79-kb chromosome shows that foci increase in size and intensity between 8 and 16 h after initiation of mating in the developing SN (dotted lines) in WT cells but remain unchanged in cpd2cr cells. (A, B) Scale bar, 5 μm. (C) FISH signals in SN were counted in cells fixed at 8 or 16 h after initiation of mating. At 16 h, more signals, on average, are seen in WT cells than in cpd2cr cells, but the difference was not statistically significant (unpaired t test, p = 0.06). (D) Quantitation of FISH signal intensity shows that the somatic copy number increases to about two times the germline copy number in WT cells, but remains unchanged in cpd2cr cells (unpaired t test, p < 0.0001).
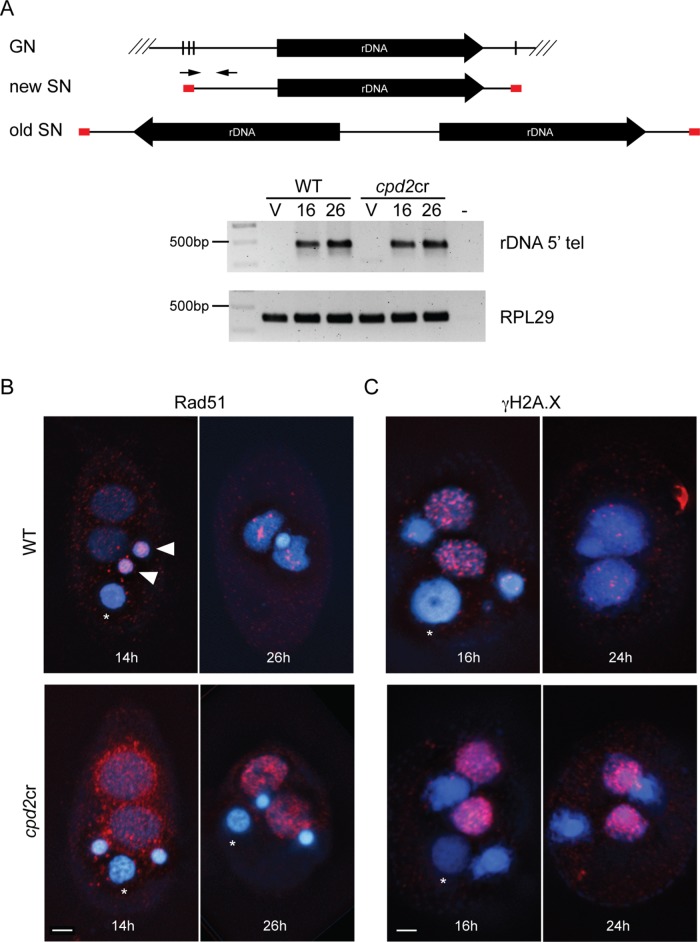
Other major events during SN development are the breakage of chromosomes at specified chromosome breakage sites (CBSs) and the de novo addition of telomeres (Fan and Yao, 1996; Karrer, 2012). To assay whether chromosome breakage and telomere addition occur in cpd2cr cells, PCR was performed using a telomere-specific primer and a primer specific to a 5′ upstream sequence of the rRNA genes (rDNA) (Lin et al., 2012). These primers only generate a product from the developing SN because in mature SN the rDNA is converted to a palindromic form (Figure 8A). Analysis of both WT and cpd2cr cells showed a PCR product from processed rDNA at 16 and 24 h after initiation of mating (Figure 8A). This result suggests that breakage and telomere addition is occurring, at least at some CBSs, in the cpd2cr mutant.
FIGURE 8:
New SN in cpd2cr cells show signs of DNA lesions. (A) PCR analysis of chromosome breakage at the rDNA chromosome shows normal breakage and telomere addition in cpd2cr cells. Diagrams show the germline and somatic rDNA forms in old and new SNs. Primers (arrows) corresponding to rDNA telomere sequence (red boxes) and the 5′ upstream region only amplify rDNA in the new SN, which is formed directly after cleavage and telomere addition. DNA samples were made from WT or cpd2cr vegetative (V) or mating cells at 16 and 24 h after initiation of mating. The new rDNA form was amplified from DNA samples from both WT and mutant mating cells, but not from vegetative cells. (B) Abnormal Rad51 (red) accumulated in developing SN of cpd2cr cells at 14 and 26 h after initiation of mating. Asterisks indicate the degrading parental nucleus; closed arrowheads indicate germline Rad51 staining at the 14 h time point. (C) The γH2A.X signal (red) was stronger at 16 h in cpd2cr than in WT cells and persisted to 24 h (when it was mostly lost in WT cells). Scale bars, 5 μm.
New SN in cpd2cr cells show hallmarks of persistent DNA damage
In addition to the other developmental abnormalities in cpd2cr cells, the developing SN of cpd2cr cells show evidence of DNA lesions. Immunostaining against the DNA repair protein Rad51 showed aberrant Rad51 accumulation in and around the developing SN at 14 h after initiation of mating, which persisted until at least the 26-h time point (Figure 8B). Immunostaining was also performed to detect γH2A.X, another marker of DNA breaks or DNA damage. Both WT and cpd2cr cells showed γH2A.X signals in the developing SN at 16 h (Figure 8C). This is likely due to DNA breakage and repair that occurs as a normal part of IES and CBS processing. However, the signals were brighter in the mutant cells and persisted for longer, which may be due to a defect in IES excision and repair. The persistent DNA damage may lead to the eventual degradation of the new SN DNA in the cpd2cr cells.
A possible explanation of both the persistent DNA breaks and failure to remove IESs is that although cleavage of one or both ends of some IESs occurs, the SN-destined sequences are not properly rejoined. Alternatively, elevated Rad51 and γH2A.X signaling could be caused by replication stress or replication fork stalling as a result of blocked DNA replication (Gonzalez-Prieto et al., 2013).
DISCUSSION
Composition and regulation of Cpd2-containing condensin complexes
We previously showed that the GN and SN employ distinct condensin complexes to mediate chromosome condensation and segregation (Howard-Till and Loidl, 2018). Here, we show that at least two additional condensin complexes are present in SN during growth and development, and that these contain a specialized homologue of the CapD2 subunit, Cpd2 (Figure 9). Cpd2 is one of two CapD2 proteins expressed in Tetrahymena but, unlike the other (Cpd1), it is dispensable for vegetative growth. MS analysis of Cpd1 or Cpd2 complexes in vegetative cells showed that both contain the core condensin components Smc2, Smc4, and Cpg1 (Table 1). However, the two complexes differ in the kleisin subunit: Cpd1 associates with Cph1, 2, or 3, whereas Cpd2 associates with Cph4 (Figure 9). Cpd2 also strongly associates with PARP6, suggesting a function in transcriptional regulation or DNA repair in vegetative cells (Schreiber et al., 2006). However, because Cpd2 was not found to be required for vegetative growth, we focused on its essential role in SN development.
FIGURE 9:
Reduction and radiation of SMC complexes in Tetrahymena. Three types of SMC complexes have evolved in fungi, metazoa, and plants: the DNA repair complex Smc5/6, cohesin, and condensin. Cohesins are further classified into complexes specialized for either meiosis or vegetative growth. Condensin has also duplicated and diverged to form condensins I and II, and in the case of C. elegans, condensin IDC. Tetrahymena may have started with a reduced set of SMC complexes, containing only cohesin and condensin I. Its cohesin more closely resembles the meiotic cohesin of the other eukaryote groups and did not further diverge. The non-SMC subunits of the condensin complex subsequently multiplied into diverse forms with different and specific nuclear functions. The distinct complexes seem to be largely defined by the kleisin/Cph subunit. The GN employs condensin G, with Cph1 and Cph2 being partially redundant. Although the localization of Cph3, Cph4, and Cph5 is overlapping in the SN, Cph3 (condensin S1) is required for chromosome division, Cph4 (condensin S2) has an as-yet unknown function, and Cph5 (condensin D) participates in development of the SN.
In late-mating cells, at the stage when DNA elimination occurs and chromosomes are fragmented and amplified, Cpd2 associates with yet another kleisin subunit, which we have designated Cph5. At the same stage, Cpd2 association with Cph4 and Cpg1 is reduced, suggesting that Cpd2 primarily forms a development-specific complex (condensin D) consisting of Smc2, Smc4, and Cph5 (Figure 9). The Cph5 protein is approximately twice the size of the other Cph homologues, with the C-terminal half showing little homology to other known proteins. In most condensin complexes, the two HEAT repeat subunits, CapG and CapD, associate with the kleisin subunit to form a bridge between the ATPase domains of the SMC subunits (Kschonsak et al., 2017). The additional bulk of the C-terminal domain of Cph5 may preclude its binding by Cpg1; alternatively, that space may be occupied by other proteins associating with the complex. Cph5 appears to be posttranslationally modified, possibly phosphorylated, later in development, and the modification is dependent on Cpd2. It is tempting to speculate that the putative phosphorylation may be mediated by the kinase encoded by TTHERM_00670660, which was found to associate with Cpd2 during development. Further investigation of this modification is critical because it may be important for regulating DNA binding or interacting with its protein partners. Phosphorylation of the Rec8 kleisin subunit of Tetrahymena cohesin is linked with its chromosomal association and activation (Ali et al., 2018); therefore, a similar mechanism might regulate Cph5 in condensin.
A specialized condensin complex is active in developing SN
Cpd2 and its partner Cph5 define a distinct developmental condensin complex (condensin D). Unlike canonical condensin complexes, this specialized condensin does not appear to function in chromosome segregation. Instead, it is essential for completion of sexual reproduction and for the developmental program of the SN. Several aspects of SN development are affected in the absence of condensin D, including DNA elimination and polyploidization. Mating cpd2 or cph5 mutants arrest with very low SN DNA content and with signs of DNA damage or stalled replication in the new SN. However, the exact nature of the developmental block remains unclear. Condensin D appears to be excluded from DNA elimination bodies, making it unlikely that the complex participates directly in the deletion process. However, elimination bodies do not form properly in the absence of Cpd2 or Cph5, suggesting that condensin D may help to organize the genome to promote IES elimination. Such a role is not without precedent: similarly, the double-strand break repair protein Ku80 (Tku80 in Tetrahymena) does not localize to elimination bodies but is required for the aggregation and elimination of IESs (Lin et al., 2012; Noto and Mochizuki, 2017). Condensin D could drive the compaction of retained sequences, and factors that help determine IES borders could act as insulators to partition IES DNA from SN-destined sequences. This would resemble the action of cohesin in forming topologically associating domains (TADs) or of the specialized Caenorhabditis elegans condensin (condensin IDCC) in restructuring the inactive X chromosome into structural units similar to TADs (Crane et al., 2015; Fudenberg et al., 2016; Schwarzer et al., 2017). Loss of condensin D could disrupt genome reorganization in the SN, thus preventing IES compartmentalization into elimination bodies, where factors needed for elimination reside. Alternatively, lack of condensin D might simply cause developmental arrest before IES elimination and DNA amplification. Further research into the molecular mechanism of condensin D is needed to rule out this possibility.
Association of condensin D with chromatin remodelers in the developing SN
The PHD finger protein Nmp1 associates with Cpd2 and Cph5 during development, and Nmp1 IP identified several other predicted DNA-binding or chromatin-remodeling factors containing PHD fingers, zinc fingers, SET domains, or AT-hook motifs (Supplemental Table S1). In particular, three predicted members of the SAGA transcriptional coactivator complex were pulled down—Aap6, Ada2, and Hat2 (Gcn5); these have been previously identified as transcriptional regulators in Tetrahymena (Helmlinger and Tora, 2017; Saettone et al., 2018). By analogy with other model systems, these factors might help in condensin loading (Toselli-Mollereau et al., 2016; Robellet et al., 2017).
Functional radiation of SMC complexes in Tetrahymena
SMC complexes have evolved in prokaryotes for organizing DNA in nucleoids and facilitating the segregation of sister DNA molecules (Nolivos and Sherratt, 2014; Uhlmann, 2016). In eukaryotes, they have undergone strong diversification to form the Smc5/6 complex, which functions in DNA repair, and condensins and cohesins, which have distinct and overlapping functions in gene regulation, chromosome organization and condensation, sister chromatid cohesion, and DNA repair (Uhlmann, 2016; Verver et al., 2016). Cohesins have further specialized into vegetative and meiotic versions, whereas condensins have diverged to form condensin I, which functions in metaphase chromosome compaction, and condensin II, which has a broader function in chromatin and chromosome structural maintenance. In C. elegans, a modified form of condensin I (condensin IDC) forms part of the dosage compensation complex, which has a role in female X chromosomal gene repression (Meyer, 2010). In contrast, ciliates may originally have possessed a limited set of SMC complexes. In Tetrahymena, the Smc5/6 complex has probably been lost, and only a single cohesin complex retained (Howard-Till et al., 2013; Ali et al., 2018). Condensin II was also lost (Hirano, 2012), but condensin I underwent secondary diversification to form at least four versions, which are distinguished by their kleisin/Cph subunit and HEAT repeat-binding proteins (Figure 9; Howard-Till and Loidl, 2018). Such diversification in Tetrahymena may have been necessitated by nuclear dualism, as the functional differences between the SN and GN required functionally distinct condensins to generate and maintain the two genomes.
Although much more work is needed to fully understand the molecular mechanism of condensin D, this novel condensin clearly plays an interesting role in the highly complex process of nuclear differentiation. This new function for condensin, in addition to its previously described roles in the two distinct nuclei of Tetrahymena, emphasizes the value of this model system for studying condensin biology specifically and chromatin dynamics in general.
MATERIALS AND METHODS
Strains and growth conditions
WT strains of Tetrahymena thermophila, including B2086, CU427, and CU428, were obtained from the Tetrahymena stock center (Cornell University). Cells were grown at 30°C in Neff's medium (Orias et al., 2000). For mating, cells of different mating types were starved overnight in 10 mM Tris-Cl (pH, 7.4) and then mixed at equal concentrations ([2–5] × 105 cells/ml).
Protein tagging
C-terminal HA tagging of proteins was performed using a gene knock-in strategy to preserve expression from the endogenous promoter (Howard-Till et al., 2013). Homologous regions corresponding to the 3′ coding sequence and 3′ untranslated region of the genes were amplified using primers listed in Supplemental Table S2. These regions were combined with the HA-tagging sequence and Neo4 selection cassette of pHA-Neo using the NEBuilder HiFi DNA assembly master mix (New England Biolabs, Frankfurt, Germany). Strains were transformed by biolistic particle bombardment as previously described (Cassidy-Hanley et al., 1997; Bruns and Cassidy-Hanley, 2000).
Immunofluorescence
Immunofluorescence was performed as previously described (Ali et al., 2018). For most experiments, cells were suspended in 10 mM Tris, pH 7.4, and fixed with formaldehyde (4% final concentration) and Triton X-100 (0.5% final concentration) for 30 min. Cells were then resuspended in 4% paraformaldehyde and 3.4% sucrose solution and spread onto slides. Slides were stained with appropriate antibodies and mounted with Vectashield anti-fading agent (Vector Laboratories, Burlingame, CA) supplemented with 0.5 μg/ml DAPI. For visualization of HA-tagged proteins, either monoclonal mouse anti-HA (1:1000) or polyclonal rabbit anti-HA (1:200; Sigma, St. Louis, MO) was used. Other antibodies used for immunofluorescence were rabbit anti-Pdd1 (1:1000; Abcam, Cambridge, UK) and rabbit anti-H3K27me3 (1:1000; Millipore/Merck, Burlington, MA), monoclonal mouse anti-Dmc1/Rad51, clone 51RAD01 (1:50; Neomarkers, Fremont, CA), and monoclonal mouse anti-phosphorylated H2A.X (1:200; BioLegend, San Diego, CA).
To detect replication during development, cells were incubated in BrdU for 1 h and then fixed for immunostaining (as described above). Before incubation with anti-BrdU antibody (1:40; Abcam, Cambridge, UK), samples were denatured as previously described (Shodhan et al., 2017).
Protein analysis
Preparation of protein extracts and IPs were performed as previously described (Ali et al., 2018). HA-Cph4 expressing cells (Howard-Till and Loidl, 2018) were harvested at 8 h after initiation of mating. Extracts from Cpd2-HA expressing cells were prepared from growing cells or mating cells at 14 h after initiation of mating. MS analyses were performed by the MFPL Mass Spectrometry Facility using the VBCF instrument pool. The following antibodies were used for Western blotting: monoclonal mouse anti-HA (1:1000; Sigma, St. Louis, MO), rabbit anti-Pdd1 (1:10,000; Abcam, Cambridge, UK), and monoclonal mouse anti-tubulin alpha antibody-2 (DM1A; 1:10,000; Neomarkers, Fremont, CA).
RNAi
RNAi constructs were created and introduced into cells as previously described for pREC8hpCYH (Howard-Till et al., 2013). Primers used to amplify gene fragments are listed in Supplemental Table S1. RNAi in mating cells was performed by adding 0.05 μg/ml CdCl2 to cells during overnight starvation. Induced, starved cells were then mixed to initiate mating.
CRISPR–Cas9-mediated mutation of CPD2
The cpd2cr strain was created using a previously described CRISPR–Cas9 editing system adapted for use in Tetrahymena (Suhren et al., 2017). To create the CPD2 editing construct, a 20–base pair target sequence matching the consensus (GN19NGG; 5′-gcatttgagaagagtaaaag-3′) near the 5′ end of the gene was chosen. Corresponding sense and antisense oligos were annealed and ligated into the BbsI restriction site of the guide RNA template of the pC9T vector (gift of Kazufumi Mochizuki, Le Centre National de la Recherche Scientifique, Montpellier, France). Starved cells of strain B2086 were transformed with the gene-editing construct by biolistic particle bombardment (Cassidy-Hanley et al., 1997; Bruns and Cassidy-Hanley, 2000). Transformants were recovered in Neff's medium and selected with 100 μg/ml paromomycin, with 1 μg/ml CdCl2 added during selection to induce Cas9 expression. Transformant lines were PCR screened using a GN-specific primer to amplify the target region, which was subsequently sequenced to identify a single line with an 11–base pair deletion. The deletion strain was then crossed further to produce progeny of different mating types in which both the GN and SN carried the deletion. The complete absence of the WT gene was confirmed by PCR, reverse transcriptase-PCR, and cDNA sequencing.
Fluorescence in situ hybridization
FISH to visualize IESs in cells fixed at 24 h after initiation of mating was performed using probes hybridizing to the TLR family of repetitive IESs (Loidl and Scherthan, 2004; Noto et al., 2010; Kataoka and Mochizuki, 2015). FISH signal intensity was quantified using Icy image analysis software (http://icy.bioimageanalysis.org). For each cell, the total signal intensity for the SN was divided by the signal intensity of the GN to estimate the copy number in the SN relative to the GN.
Supplementary Material
Acknowledgments
This study was supported by the Austrian Science Fund (FWF), Grant no. P 28336-B28. We thank Markus Hartl (Max F. Perutz Laboratories [MFPL], Vienna, Austria) and the MFPL Mass Spectrometry Facility for help with protein analysis. We also thank Kazufumi Mochizuki (Le Centre National de la Recherche Scientifique, Montpellier, France) and Kensuke Kataoka (National Institute for Basic Biology, Okazaki, Japan) for tagging and CRISPR constructs, as well as for helpful discussions.
Abbreviations used:
- CBS
chromosome breakage site
- FISH
fluorescence in situ hybridization
- GN
germline nucleus/nuclei
- IES
internal eliminated sequence
- IP
immunoprecipitation
- MS
mass spectrometry
- RNAi
RNA interference
- SN
somatic nucleus/nuclei
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-08-0487) on March 20, 2019.
REFERENCES
- Ali EI, Loidl J, Howard-Till RA. (2018). A streamlined cohesin apparatus is sufficient for mitosis and meiosis in the protist Tetrahymena. Chromosoma , 421–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allis CD, Colavito-Shepanski M, Gorovsky MA. (1987). Scheduled and unscheduled DNA synthesis during development in conjugating Tetrahymena. Dev Biol , 469–480. [DOI] [PubMed] [Google Scholar]
- Bruns PJ, Cassidy-Hanley D. (2000). Biolistic transformation of macro- and micronuclei. Methods Cell Biol , 501–512. [DOI] [PubMed] [Google Scholar]
- Cassidy-Hanley D, Bowen J, Lee JH, Cole E, VerPlank LA, Gaertig J, Gorovsky MA, Bruns PJ. (1997). Germline and somatic transformation of mating Tetrahymena thermophila by particle bombardment. Genetics , 135–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker DL. (2008). Dynamic nuclear reorganization during genome remodeling of Tetrahymena. Biochim Biophys Acta , 2130–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker DL, Yao MC. (2011). DNA elimination in ciliates: transposon domestication and genome surveillance. Annu Rev Genet , 227–246. [DOI] [PubMed] [Google Scholar]
- Cole E, Sugai T. (2012). Developmental progression of Tetrahymena through the cell cycle and conjugation. Methods Cell Biol , 177–236. [DOI] [PubMed] [Google Scholar]
- Coyne RS, Nikiforov MA, Smothers JF, Allis CD, Yao MC. (1999). Parental expression of the chromodomain protein Pdd1p is required for completion of programmed DNA elimination and nuclear differentiation. Mol Cell , 865–872. [DOI] [PubMed] [Google Scholar]
- Crane E, Bian Q, McCord RP, Lajoie BR, Wheeler BS, Ralston EJ, Uzawa S, Dekker J, Meyer BJ. (2015). Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature , 240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuylen S, Metz J, Haering CH. (2011). Condensin structures chromosomal DNA through topological links. Nat Struct Mol Biol , 894–901. [DOI] [PubMed] [Google Scholar]
- Fan Q, Yao M. (1996). New telomere formation coupled with site-specific chromosome breakage in Tetrahymena thermophila. Mol Cell Biol , 1267–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. (2016). Formation of chromosomal domains by loop extrusion. Cell Rep , 2038–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganji M, Shaltiel IA, Bisht S, Kim E, Kalichava A, Haering CH, Dekker C. (2018). Real-time imaging of DNA loop extrusion by condensin. Science , 102–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Prieto R, Munoz-Cabello AM, Cabello-Lobato MJ, Prado F. (2013). Rad51 replication fork recruitment is required for DNA damage tolerance. EMBO J , 1307–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmlinger D, Tora L. (2017). Sharing the SAGA. Trends Biochem Sci , 850–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. (2012). Condensins: universal organizers of chromosomes with diverse functions. Genes Dev , 1659–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard-Till R, Loidl J. (2018). Condensins promote chromosome individualization and segregation during mitosis, meiosis, and amitosis in Tetrahymena thermophila. Mol Biol Cell , 466–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard-Till RA, Lukaszewicz A, Novatchkova M, Loidl J. (2013). A single cohesin complex performs mitotic and meiotic functions in the protist tetrahymena. PLoS Genet , e1003418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov D, Nasmyth K. (2005). A topological interaction between cohesin rings and a circular minichromosome. Cell , 849–860. [DOI] [PubMed] [Google Scholar]
- Karrer KM. (2000). Tetrahymena genetics: two nuclei are better than one. Methods Cell Biol , 127–186. [DOI] [PubMed] [Google Scholar]
- Karrer KM. (2012). Nuclear dualism. Methods Cell Biol , 29–52. [DOI] [PubMed] [Google Scholar]
- Kataoka K, Mochizuki K. (2015). Phosphorylation of an HP1-like protein regulates heterochromatin body assembly for DNA elimination. Dev Cell , 775–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kschonsak M, Merkel F, Bisht S, Metz J, Rybin V, Hassler M, Haering CH. (2017). Structural basis for a safety-belt mechanism that anchors condensin to chromosomes. Cell , 588–600.e524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin IT, Chao JL, Yao MC. (2012). An essential role for the DNA breakage-repair protein Ku80 in programmed DNA rearrangements in Tetrahymena thermophila. Mol Biol Cell , 2213–2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Taverna SD, Muratore TL, Shabanowitz J, Hunt DF, Allis CD. (2007). RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena. Genes Dev , 1530–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl J, Scherthan H. (2004). Organization and pairing of meiotic chromosomes in the ciliate Tetrahymena thermophila. J Cell Sci , 5791–5801. [DOI] [PubMed] [Google Scholar]
- Madireddi MT, Coyne RS, Smothers JF, Mickey KM, Yao MC, Allis CD. (1996). Pdd1p, a novel chromodomain-containing protein, links heterochromatin assembly and DNA elimination in Tetrahymena. Cell , 75–84. [DOI] [PubMed] [Google Scholar]
- Malone CD, Anderson AM, Motl JA, Rexer CH, Chalker DL. (2005). Germ line transcripts are processed by a Dicer-like protein that is essential for developmentally programmed genome rearrangements of Tetrahymena thermophila. Mol Cell Biol , 9151–9164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer BJ. (2010). Targeting X chromosomes for repression. Curr Opin Genet Dev , 179–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA. (2002). Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell , 689–699. [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH. (2009). Cohesin: its roles and mechanisms. Annu Rev Genet , 525–558. [DOI] [PubMed] [Google Scholar]
- Nikiforov MA, Smothers JF, Gorovsky MA, Allis CD. (1999). Excision of micronuclear-specific DNA requires parental expression of pdd2p and occurs independently from DNA replication in Tetrahymena thermophila. Genes Dev , 2852–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolivos S, Sherratt D. (2014). The bacterial chromosome: architecture and action of bacterial SMC and SMC-like complexes. FEMS Microbiol Rev , 380–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noto T, Kurth HM, Kataoka K, Aronica L, DeSouza LV, Siu KW, Pearlman RE, Gorovsky MA, Mochizuki K. (2010). The Tetrahymena argonaute-binding protein Giw1p directs a mature argonaute-siRNA complex to the nucleus. Cell , 692–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noto T, Mochizuki K. (2017). Whats, hows and whys of programmed DNA elimination in Tetrahymena. Open Biol , 170172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orias E, Hamilton EP, Orias JD. (2000). Tetrahymena as a laboratory organism: useful strains, cell culture, and cell line maintenance. Methods Cell Biol , 189–211. [DOI] [PubMed] [Google Scholar]
- Robellet X, Vanoosthuyse V, Bernard P. (2017). The loading of condensin in the context of chromatin. Curr Genet , 577–589. [DOI] [PubMed] [Google Scholar]
- Ruehle MD, Orias E, Pearson CG. (2016). Tetrahymena as a unicellular model eukaryote: genetic and genomic tools. Genetics , 649–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saettone A, Garg J, Lambert JP, Nabeel-Shah S, Ponce M, Burtch A, Thuppu Mudalige C, Gingras AC, Pearlman RE, Fillingham J. (2018). The bromodomain-containing protein Ibd1 links multiple chromatin-related protein complexes to highly expressed genes in Tetrahymena thermophila. Epigenet Chromatin , 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber V, Dantzer F, Ame JC, de Murcia G. (2006). Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol , 517–528. [DOI] [PubMed] [Google Scholar]
- Schwarzer W, Abdennur N, Goloborodko A, Pekowska A, Fudenberg G, Loe-Mie Y, Fonseca NA, Huber W, C HH, Mirny L, Spitz F. (2017). Two independent modes of chromatin organization revealed by cohesin removal. Nature , 51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shodhan A, Kataoka K, Mochizuki K, Novatchkova M, Loidl J. (2017). A Zip3-like protein plays a role in crossover formation in the SC-less meiosis of the protist Tetrahymena. Mol Biol Cell , 825–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smothers JF, Mizzen CA, Tubbert MM, Cook RG, Allis CD. (1997). Pdd1p associates with germline-restricted chromatin and a second novel anlagen-enriched protein in developmentally programmed DNA elimination structures. Development , 4537–4545. [DOI] [PubMed] [Google Scholar]
- Suhren JH, Noto T, Kataoka K, Gao S, Liu Y, Mochizuki K. (2017). Negative regulators of an RNAi-heterochromatin positive feedback loop safeguard somatic genome integrity in Tetrahymena. Cell Rep , 2494–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna SD, Coyne RS, Allis CD. (2002). Methylation of histone h3 at lysine 9 targets programmed DNA elimination in Tetrahymena. Cell , 701–711. [DOI] [PubMed] [Google Scholar]
- Toselli-Mollereau E, Robellet X, Fauque L, Lemaire S, Schiklenk C, Klein C, Hocquet C, Legros P, N'Guyen L, Mouillard L, et al. (2016). Nucleosome eviction in mitosis assists condensin loading and chromosome condensation. EMBO J , 1565–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F. (2016). SMC complexes: from DNA to chromosomes. Nat Rev Mol Cell Biol , 399–412. [DOI] [PubMed] [Google Scholar]
- van Ruiten MS, Rowland BD. (2018). SMC complexes: universal DNA looping machines with distinct regulators. Trends Genet , 477–487. [DOI] [PubMed] [Google Scholar]
- Verver DE, Hwang GH, Jordan PW, Hamer G. (2016). Resolving complex chromosome structures during meiosis: versatile deployment of Smc5/6. Chromosoma , 15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J, Lu Y, Feng J, Yuan D, Tian M, Chang Y, Fu C, Wang G, Zeng H, Miao W. (2013). Tetrahymena functional genomics database (TetraFGD): an integrated resource for Tetrahymena functional genomics. Database (Oxford) , bat008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen KC, Gerton JL. (2018). Taking cohesin and condensin in context. PLoS Genet , e1007118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.