Abstract
Filopodia are actin-filled membrane protrusions that play essential roles in cell motility and cell–cell communication and act as precursors of dendritic spines. IRSp53 is an essential regulator of filopodia formation, which couples Rho-GTPase signaling to actin cytoskeleton and membrane remodeling. IRSp53 has three major domains: an N-terminal inverse-BAR (I-BAR) domain, a Cdc42- and SH3-binding CRIB-PR domain, and an SH3 domain that binds downstream cytoskeletal effectors. Phosphorylation sites in the region between the CRIB-PR and SH3 domains mediate the binding of 14-3-3. Yet the mechanism by which 14-3-3 regulates filopodia formation and dynamics and its role in cell migration are poorly understood. Here, we show that phosphorylation-dependent inhibition of IRSp53 by 14-3-3 counters activation by Cdc42 and cytoskeletal effectors, resulting in down-regulation of filopodia dynamics and cancer cell migration. In serum-starved cells, increased IRSp53 phosphorylation triggers 14-3-3 binding, which inhibits filopodia formation and dynamics, irrespective of whether IRSp53 is activated by Cdc42 or downstream effectors (Eps8, Ena/VASP). Pharmacological activation or inhibition of AMPK, respectively, increases or decreases the phosphorylation of two of three sites in IRSp53 implicated in 14-3-3 binding. Mutating these phosphorylation sites reverses 14-3-3-dependent inhibition of filopodia dynamics and cancer cell chemotaxis.
INTRODUCTION
Cell migration is an essential process in tissue and organ development, and it is also the source of pathological conditions such as inflammation and cancer metastasis (Friedl and Gilmour, 2009). Migrating cells project filopodia, which are thin, rod-like membrane protrusions that dynamically extend and retract as a result of the polymerization/depolymerization of parallel bundles of actin filaments in their interior (Mattila and Lappalainen, 2008; Yang and Svitkina, 2011; Fischer et al., 2018). Filopodia act as sensory organs of the cell, which in response to chemical gradients or tension changes mediate the crosstalk of cells with their neighbors and the environment (Chan and Odde, 2008; Jacquemet et al., 2015). As a result, filopodia play a central role in cell migration and the establishment of cell polarity (Meyen et al., 2015). However, the mechanisms by which cells regulate filopodia formation and dynamics in response to signaling and mechanical cues are not fully understood.
IRSp53 (also known as BAIAP2) is a key regulator of filopodia formation and cell motility that couples Rho GTPase and kinase signaling pathways with actin cytoskeleton remodeling and membrane dynamics (Miki et al., 2000; Krugmann et al., 2001; Disanza et al., 2006, 2013; Disanza and Scita, 2008; Suetsugu et al., 2006; Goh et al., 2012; Kast et al., 2014; Carman and Dominguez, 2018). IRSp53 plays essential roles in a number of developmental processes, such as eye lens formation (Chauhan et al., 2009), myoblast fusion (Segal et al., 2016), and dendritic spine formation (Soltau et al., 2002, 2004). Dysregulation of IRSp53 function is a contributing factor in tumorigenesis (Funato et al., 2004; Liu et al., 2010; Yan et al., 2017) and several neurological disorders (Kang et al., 2016), including learning defects (Bobsin and Kreienkamp, 2016), attention deficit disorder (Ribases et al., 2009), autism spectrum disorder (Toma et al., 2011), schizophrenia (Fromer et al., 2014), and Alzheimer’s disease (Wan et al., 2013). The N-terminal ∼230 amino acids of IRSp53 consist of an inverse BAR (I-BAR) domain, which mediates membrane binding/deformation (Mattila et al., 2007) and antiparallel dimerization (Millard et al., 2005; Lee et al., 2007b). Following the I-BAR domain, IRSp53 has a Cdc42-binding motif that overlaps with a proline-rich sequence and is thus referred to as the CRIB-PR domain. C-terminal to the CRIB-PR domain, IRSp53 contains an SH3 domain, which mediates the interactions with a large number of cytoskeletal effectors, including Eps8 (Funato et al., 2004; Disanza et al., 2006), Ena/VASP (Krugmann et al., 2001; Disanza et al., 2013; Oikawa et al., 2013), N-WASP (Lim et al., 2008), WAVE (Miki et al., 2000; Nakagawa et al., 2003; Abou-Kheir et al., 2008), mDia (Fujiwara et al., 2000; Goh et al., 2012), PSD-95 (Bockmann et al., 2002; Soltau et al., 2004; Choi et al., 2005; Barilari and Dente, 2010) and Shank3 (Bockmann et al., 2002; Soltau et al., 2002). In the inactive state, the SH3 domain of IRSp53 binds intramolecularly to the CRIB-PR domain, partially inhibiting IRSp53’s ability to induce plasma membrane remodeling in cells (Kast et al., 2014). IRSp53 is activated by the binding of either Cdc42 to the CRIB-PR domain and/or cytoskeletal effectors to the SH3 domain (Kast et al., 2014), leading to increased formation of membrane ruffles and filopodia-like protrusions (Krugmann et al., 2001; Soltau et al., 2002; Disanza et al., 2006, 2013; Lim et al., 2008; Kast et al., 2014).
The ∼80–amino acid region between the CRIB-PR and SH3 domains (residues 292–374) hosts multiple phosphorylation sites that have been implicated in IRSp53 regulation (Heung et al., 2008; Robens et al., 2010; Cohen et al., 2011; Kast and Dominguez, 2019). In particular, three of these sites (T340, T360, and S366) have been implicated in the binding of 14-3-3 (Robens et al., 2010; Cohen et al., 2011; Kast and Dominguez, 2019). 14-3-3 proteins constitute a conserved family of seven isoforms, denoted β, γ, ε, ζ, η, σ, and θ (also called τ), that function as phospho-S/T adaptors to regulate the activities of a large number of proteins in a phosphorylation-dependent manner (Yaffe, 2002). Four 14-3-3 isoforms (γ, θ, ζ, and σ) have been shown to bind to IRSp53 (Jin et al., 2004; Mackie and Aitken, 2005; Robens et al., 2010; Cohen et al., 2011). Numerous ligands of 14-3-3 are proteins implicated in the regulation of cell migration (Freeman and Morrison, 2011), whereas little is known about the functional role of 14-3-3 binding to phosphorylated IRSp53 (pIRSp53), or the kinases implicated in this regulatory mechanism. The one exception is the phosphorylation of IRSp53 residue S366, known from chemical genetics screening to be mediated by AMPK and AMPK-related kinases (Banko et al., 2011; Cohen et al., 2011; Schaffer et al., 2015). Binding of 14-3-3 to this site leads to loss of cell spreading and cell polarity (Cohen et al., 2011). While the kinases that directly phosphorylate T340 and T360 are unknown, it has been shown that 14-3-3 binding to these sites inhibits interactions with Cdc42 and cytoskeletal effectors (Robens et al., 2010; Kast and Dominguez, 2019). Yet the functional consequences of the interaction of 14-3-3 with pIRSp53 on the formation and dynamics of filopodia and its effect on directed cell motility remain largely unexplored.
Here, we show that 14-3-3 counters activation of phosphorylated IRSp53 by Cdc42, Eps8 and VASP, resulting in impaired filopodia formation and dynamics and cancer cell migration. We further show that phosphorylation of two of the three sites that mediate the binding of 14-3-3 to IRSp53 (T360 and S366) increases with serum starvation or phenformin treatment and decreased with dorsomorphin treatment, thus implicating AMPK in their phosphorylation. Collectively, the results demonstrate that IRSp53 is a chemotactic signal transducer that in response to AMPK-dependent phosphorylation and 14-3-3 binding down-regulates cytoskeleton and plasma membrane remodeling to inhibit cell motility.
RESULTS
14-3-3 inhibits IRSp53- and Cdc42-dependent filopodia dynamics
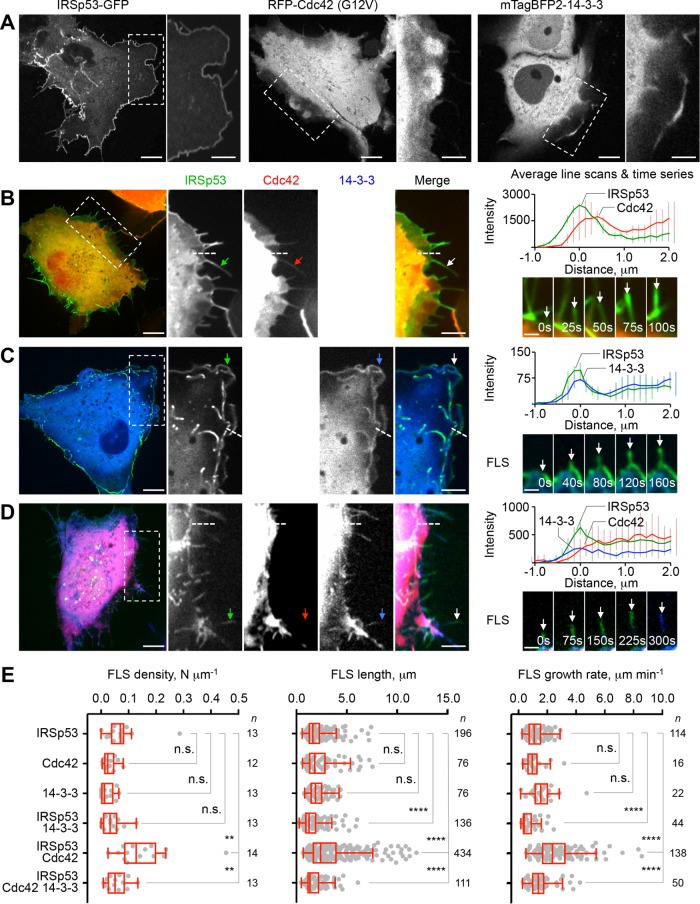
Phosphorylation of IRSp53 has been shown to trigger its interaction with 14-3-3 (Jin et al., 2004; Mackie and Aitken, 2005; Robens et al., 2010; Cohen et al., 2011; Kast and Dominguez, 2019). However, it remains to be demonstrated whether this interaction regulates the activity of IRSp53, and specifically its role in filopodia formation and cell migration activated by interactions with Cdc42 through its CRIB-PR domain or downstream cytoskeletal effectors (such as Eps8 and VASP) through its SH3 domain. We used spinning-disk confocal microscopy to determine the density, length and growth rate of filopodia-like structures (FLS) in live COS-7 cells expressing IRSp53-GFP, RFP-Cdc42 (constitutively active mutant, G12V) and mTagBFP2-14-3-3θ (hereafter referred to as 14-3-3) individually or together (Figure 1 and Supplemental Figure S1). In these experiments, cells were serum-starved, which we have found enhances IRSp53 phosphorylation at 14-3-3 binding sites (Kast and Dominguez, 2019). Because the range of cellular protrusions observed was morphologically diverse, we define FLS as nontapered dynamic protrusions, 0.2–0.5 μm in width and extending at least 0.5 μm from the cell periphery.
FIGURE 1:
14-3-3 inhibits IRSp53- and Cdc42-induced FLS in live cells. (A) COS-7 cells individually expressing IRSp53-GFP, RFP-Cdc42 or mTagBFP2-14-3-3 (as indicated). Scale bars throughout this figure represent 10 and 5 μm in whole-cell and inset images, respectively. (B–D) COS-7 cells coexpressing IRSp53-GFP with RFP-Cdc42 or mTagBFP2-14-3-3. Shown on the right are the average fluorescence intensities from 10 line scans across the cell edge (as exemplified by dashed lines in the insets) and representative time series of growing FLS (scale bar = 5 μm). Arrows point to examples of IRSp53, Cdc42 and 14-3-3 localization to cell protrusions. (E) Quantification of the density, length and growth rate of FLS. The statistical significance of the measurements was determined using the Mann–Whitney rank sum test, based on the indicated number of observations (n) recorded from 12–14 cells and two independent transfections (n.s., nonsignificant; **p < 0.01; ****p < 0.0001).
The density, length and growth rate of FLS was similar for cells expressing IRSp53, Cdc42 or 14-3-3 individually (Figure 1, A and E, and Supplemental Movie S1A), albeit the number of FLS formed in cells expressing these proteins was greater than in untransfected cells (Supplemental Figure S1, B and C). As previously described (Kast et al., 2014), the majority to FLS in IRSp53-expressing cells were filled with actin (Supplemental Figure S1B). When expressed alone, IRSp53 was mainly found at the plasma membrane, which is likely due to its affinity for PI(3,4)P2 and PI(4,5)P2 (Mattila et al., 2007), which are naturally abundant in the plasma membrane. In contrast, Cdc42 and 14-3-3 displayed mostly a diffused localization, with some accumulation at dynamic plasma membrane ruffles. Coexpression of IRSp53 and Cdc42 produced many, long, and highly dynamic FLS (Figure 1, B and E, and Supplemental Movie S1B), consistent with previous observations (Krugmann et al., 2001; Disanza et al., 2006, 2013; Lim et al., 2008; Kast et al., 2014). IRSp53 and Cdc42 colocalized throughout the cell, and were specifically enriched in FLS. In contrast, coexpression of IRSp53 and 14-3-3 did not significantly change the density of FLS compared with cells expressing these two proteins individually, but did produce a modest reduction in both the length and growth rate of FLS (Figure 1, C and E, and Supplemental Movie S1C). Interestingly, 14-3-3 was less diffused in these cells than when expressed alone and often colocalized with IRSp53 to dynamic membrane ruffles at the cell periphery (Supplemental Movie S1C). Compared to cells coexpressing IRSp53 and Cdc42, the additional expression of 14-3-3 markedly reduced the density, mean length and growth rate of FLS (Figure 1, D and E, and Supplemental Movie S1D) and this inhibition was significantly more pronounced in serum-starved than in fed cells (Supplemental Figure S1D). Membrane ruffles were rarely observed in cells coexpressing the three proteins, and while IRSp53 and 14-3-3 colocalized along FLS, Cdc42 was mostly excluded from cell protrusions (Figure 1D, insets). In growing protrusions, 14-3-3 appeared to trail IRSp53 (Figure 1D, FLS insets, and Supplemental Movie S1D). Together, these results suggest that 14-3-3 negatively regulates IRSp53- and Cdc42-dependent FLS formation and dynamics, and that colocalization of IRSp53 and 14-3-3 at cell protrusions displaces Cdc42, consistent with these two proteins binding competitively to IRSp53.
Movie S1.
14-3-3 inhibits IRSp53- and Cdc42-induced FLS in live cells. This movie corresponds to cell images shown in Figure 1, and illustrates serum starved COS-7 cells expressing the following proteins: (A) IRSp53-GFP (left), RFP-Cdc42 (center), and mTagBFP2-14-3-3 (right), (B) IRSp53-GFP and RFP-Cdc42, (C) IRSp53-GFP and mTagBFP2-14-3-3, (D) IRSp53-GFP, RFP-Cdc42 and mTagBFP2-14-3-3. Scale bars throughout this movie represent 10 m and 5 m in whole-cell and inset images, respectively.
14-3-3 inhibits IRSp53- and effector-dependent filopodia dynamics
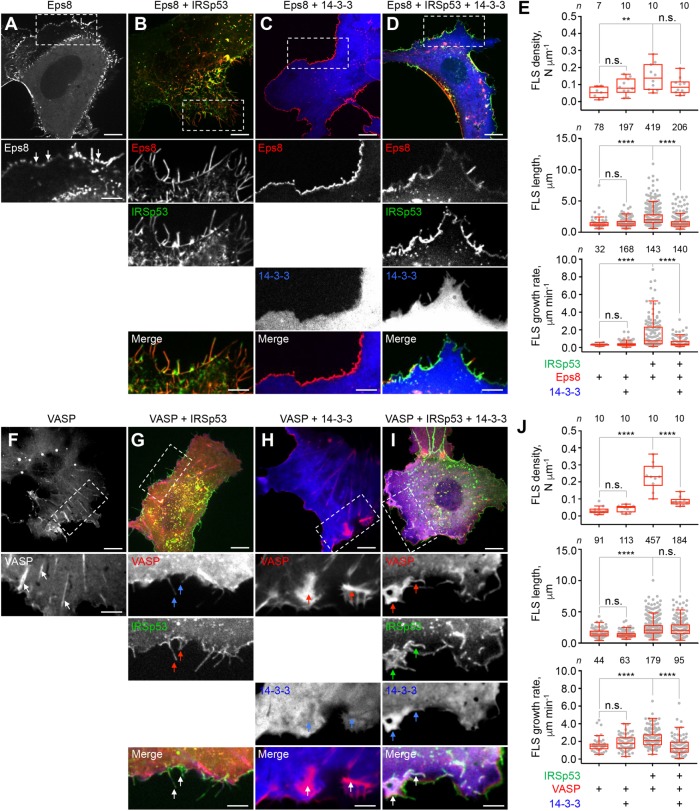
Downstream cytoskeletal effectors that bind to the SH3 domain can also activate filopodia formation by IRSp53 (Disanza et al., 2006, 2013; Kast et al., 2014). Thus, we asked whether 14-3-3 also inhibits effector-induced FLS formation and dynamics. To address this question, we studied two well-characterized downstream effectors of IRSp53: the oncoprotein Eps8 (Funato et al., 2004; Disanza et al., 2006; Liu et al., 2010; Robens et al., 2010; Kast et al., 2014) and the actin filament elongation factor VASP (Vaggi et al., 2011; Disanza et al., 2013).
Similar to Cdc42, expression of GFP-Eps8 alone had little effect on FLS number, size, or dynamics (Figure 2, A and E, and Supplemental Movie S2A). However, distinct from the diffused localization of Cdc42 (Figure 1A), Eps8 was enriched near the cell periphery, and despite relatively low ectopic expression levels (Supplemental Figure S2), it promoted the formation of microspikes extending <0.5 μm from the edge of the cell (arrows in Figure 2A, inset). Similar to Cdc42 (Figure 1B), coexpression of Eps8 and IRSp53 dramatically increased the density, length and growth rate of FLS (Figure 2, B and E, and Supplemental Movie S2B), consistent with previous observations (Disanza et al., 2006; Kast et al., 2014). In these cells, the cytosolic distribution of IRSp53 and Eps8 was virtually eliminated; both proteins colocalized along the length of FLS and on patches at the plasma membrane. Coexpression of Eps8 and 14-3-3 did not have any significant effect on FLS behavior (Figure 2, C and E, and Supplemental Movie S2C). These two proteins appeared mostly segregated, with Eps8 restricted to the cell periphery and 14-3-3 diffused throughout the cytosol. Compared to cells coexpressing IRSp53 and Eps8, the additional expression of 14-3-3 had a dramatic inhibitory effect on the length and growth rate of FLS (Figure 2, D and E, and Supplemental Movie S2D), analogous to the effect of 14-3-3 on FLS formed by IRSp53 and Cdc42 (Figure 1D).
FIGURE 2:
14-3-3 Inhibits IRSp53, EPS8, and VASP-induced FLS in live cells. (A–D) Serum-starved COS-7 cells coexpressing GFP-Eps8 with IRSp53-mTagBFP2 or mCherry-14-3-3 (individually or together). Arrows point to examples of IRSp53, Eps8, and 14-3-3 localization to cell protrusions. Scale bars throughout this figure represent 10 and 5 μm in whole-cell and inset images, respectively. (E) Quantification of the density, length, and growth rate of FLS. The statistical significance of the measurements was determined using the Mann–Whitney rank sum test, based on the indicated number of observations (n) reordered from 7–10 cells and two independent transfections (n.s., nonsignificant; **p < 0.01; ****p < 0.0001). (F–I) Serum-starved COS-7 cells coexpressing GFP-VASP with IRSp53-mTagBFP2 or mCherry-14-3-3 (individually or together). Arrows point to examples of VASP, IRSp53 and 14-3-3 localization. Notice that the density of 14-3-3 drops in VASP-enriched areas (H), whereas 14-3-3 is enriched in areas positive for both IRSp53 and VASP (I). (J) Quantification of the density, length, and growth rate of FLS. The statistical significance of the measurements was determined using the Mann–Whitney rank sum test, based on the indicated number of observations (n) reordered from 7–10 cells and two independent transfections (n.s., nonsignificant; ****p < 0.0001).
Movie S2.
14-3-3 inhibits IRSp53- and Eps8- or VASP-induced FLS in live cells. This movie corresponds to cell images shown in Figure 2, and illustrates serum starved COS-7 cells expressing the following proteins: (A) GFP-Eps8, (B) GFP-Eps8 and IRSp53-mTagBFP2, (C) GFP-Eps8 and mCherry-14-3-3, (D) GFP-Eps8, IRSp53-mTagBFP2 and mCherry-14-3-3, (E) GFP-VASP, (F) GFP-VASPand IRSp53-mTagBFP2, (G) GFP-VASPand mCherry-14-3-3, (H) GFP-VASP, IRSp53-mTagBFP2 and mCherry-14-3-3. Scale bars throughout this movie represent 10 μm and 5 μm in whole-cell and inset images, respectively.
Similar results were obtained in serum-starved COS-7 cells expressing GFP-VASP (Figure 2, F–J, and Supplemental Movie S2, E–H), another effector of IRSp53 that drives actin filament elongation when clustered by IRSp53 at the cell periphery (Disanza et al., 2013). When expressed alone, VASP localized mainly to focal adhesions and near the cell periphery (arrows in Figure 2F, inset). Although GFP-VASP was expressed at approximately ninefold higher levels than endogenous VASP (Supplemental Figure S2), it had no significant effect on the behavior of cell protrusions (Figure 2, F and J, and Supplemental Movie S2E). The coexpression of VASP with IRSp53 dramatically increased the number and dynamics of FLS (Figure 2, G and J, and Supplemental Movie S2F), consistent with previous observations (Disanza et al., 2013). However, VASP’s coexpression with 14-3-3 had no apparent effect on the behavior or appearance of FLS compared with cells expressing VASP alone (Figure 2, H and J, and Supplemental Movie S2G). In these cells, 14-3-3 was diffusely localized and mostly excluded from VASP clusters (arrows in Figure 2H, insets). The coexpression of 14-3-3 with VASP and IRSp53 dramatically reduced the number and growth rate of FLS, although it did not significantly change their average length (Figure 2, I and J, and Supplemental Movie S2H). In this case, 14-3-3 accumulated in patches where VASP and IRSp53 also colocalized (Figure 2I, arrows in insets). Together, these observations suggest that 14-3-3-dependent inhibition of filopodia formation and growth dynamics involves a direct interaction with IRSp53 and occurs independent of whether IRSp53 is activated by the binding of Cdc42 to the CRIB-PR domain or cytoskeletal effectors to the SH3 domain.
AMPK-dependent IRSp53 phosphorylation inhibits filopodia dynamics
Based on phosphoproteomic studies compiled in PhosphoSitePlus (Hornbeck et al., 2015), IRSp53 is most frequently phosphorylated in the S/T-rich region that links the CRIB-PR and SH3 domains. 14-3-3 binding to this region inhibits the binding of Cdc42 to the CRIB-PR and cytoskeletal effectors to the SH3 domain (Robens et al., 2010; Cohen et al., 2011; Kast and Dominguez, 2019). Consistently, phosphorylation of IRSp53 at T340 and T360, possibly by a kinase downstream of GSK3β although not GSK3β itself (Robens et al., 2010), and S366 by AMPK (Banko et al., 2011; Schaffer et al., 2015) or the AMPK-related kinases MARK1 (Goodwin et al., 2014) and MARK2 (Cohen et al., 2011), have been linked to 14-3-3 binding. While these studies point to different kinases and phosphorylation sites, in a recent study we show that 14-3-3 binds to two pairs of phosphorylation sites that share one site in common (pT340/pT360 and pT340/pS366), thus reconciling previous findings (Kast and Dominguez, 2019). However, disagreement persists about the specific kinase(s) responsible for IRSp53 phosphorylation in connection with 14-3-3 binding, albeit AMPK-related kinases emerge as the most likely candidate (Banko et al., 2011; Cohen et al., 2011; Goodwin et al., 2014; Schaffer et al., 2015). Consistent with these studies, we have found that serum starvation, a condition that enhances AMPK-dependent phosphorylation (Pirkmajer and Chibalin, 2011), increases the amount of 14-3-3 that coimmunoprecipitates with IRSp53-FLAG from cells (Kast and Dominguez, 2019).
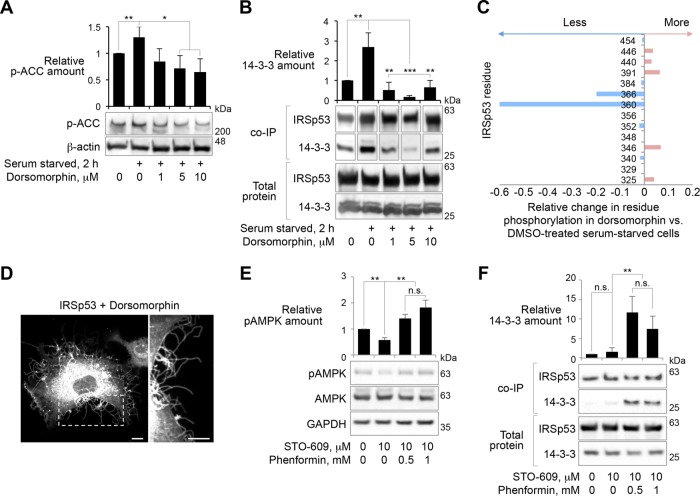
Here, we tested the role of AMPK-related kinases on 14-3-3-dependent regulation of IRSp53. In control experiments, 2-h serum starvation increased the phosphorylation of acetyl-CoA carboxylase (ACC), a known AMPK substrate (Munday et al., 1988), and ACC phosphorylation was reduced by treatment with dorsomorphin (also known as compound C), a specific inhibitor of AMPK-related kinases (Zhou et al., 2001) (Figure 3A). Neither serum starvation nor dorsomorphin treatment affected the expression levels of IRSp53 or 14-3-3 (Figure 3B). Yet, analogous to ACC, serum starvation led to an approximately threefold increase in the total amount of 14-3-3 that coimmunoprecipitates with IRSp53-FLAG, and this effect was strongly attenuated by dorsomorphin treatment (Figure 3B). Furthermore, dorsomorphin treatment dramatically reduced the phosphorylation of IRSp53 at residues T360 and S366, as revealed by phosphopeptide fingerprinting using mass spectrometry (Figure 3C and Supplemental Tables S1 and S2). Consistent with these results, IRSp53-expressing cells treated with dorsomorphin displayed increased FLS formation (Figure 3D and Supplemental Movie S3), presumably due to reduced binding and inhibition by endogenous 14-3-3 that is expressed at high levels (Sluchanko and Gusev, 2017). Furthermore, insulin treatment postserum starvation, a condition known to increase filopodia formation (Heung et al., 2008), significantly reduced the amount of 14-3-3 that coimmunoprecipitates with IRSp53 (Supplemental Figure S3). Conversely, treatment of cells with phenformin, an AMPK-specific activator (Hawley et al., 2003), led to a significant increase in the association of 14-3-3 with IRSp53 (Figure 3, E and F). Phenformin specifically activates AMPK, and not other AMPK-related kinases (Lizcano et al., 2004; Sakamoto et al., 2004). Phenformin-dependent activation depends on the phosphorylation of the AMPK catalytic (α) subunit at residue T172, which is basally phosphorylated by calmodulin-dependent protein kinase kinase (CAMKK) (Vincent et al., 2015). Therefore, to monitor the activation of AMPK in fed HEK293T cells, we examined the extent of T172 phosphorylation in response to phenformin treatment after reducing the basal phosphorylation of AMPK by treatment with STO-609, a CAMKK inhibitor (Tokumitsu et al., 2002). On STO-609 treatment, the addition of 0.5 or 1.0 mM phenformin greatly enhanced the phosphorylation of T172 (Figure 3E), whereas the expression levels of IRSp53 and 14-3-3 appeared unaffected (Figure 3F). Importantly, phenformin treatment led to a 7- to 11-fold increase in the total amount of 14-3-3 coimmunoprecipitating with IRSp53-FLAG (Figure 3F). Combined, these results support the notion that phosphorylation of at least two of the three 14-3-3 binding sites in IRSp53 depends either directly or indirectly on the activity of AMPK.
FIGURE 3:
AMPK-mediated phosphorylation controls the binding of 14-3-3 to IRSp53. (A) Effect of serum starvation and dorsomorphin-dependent inhibition of AMPK-related kinases on the phosphorylation of the canonical AMPK substrate acetyl-CoA carboxylase (ACC) in HEK293T cells. (B) Amount of 14-3-3 that coimmunoprecipitates with IRSp53-FLAG from HEK293T cells. Note that neither serum starvation nor dorsomorphin treatment affects the expression of endogenous 14-3-3, whereas starvation increases and dorsomorphin decreases the amount of 14-3-3 that coimmunoprecipitates with IRSp53-FLAG. Abundance is reported relative to fed cells expressing IRSp53-FLAG. (C) Relative change in the fraction of phosphorylated IRSp53 peptides detected by tandem MS/MS in DMSO-treated vs. 5 μM dorsomorphin-treated starved cells. (D) COS-7 cell expressing IRSp53-GFP treated with 5 μM dorsomorphin. Scale bars represent 10 and 5 μm in whole-cell and inset images, respectively. (E) Effect of phenformin treatment on the phosphorylation of residue T172 of the catalytic subunit of AMPK (kinase activation) in HEK293T cells. Cells were pretreated with the Ca2+-CAMKK inhibitor STO-609 to suppress basal phosphorylation of AMPK before treatment with the indicated concentrations of phenformin. (F) Amount of 14-3-3 that coimmunoprecipitates with IRSp53-FLAG from HEK293T cells treated with phenformin. Note that neither STO-609 nor phenformin treatment affect the expression of IRSp53-FLAG or endogenous 14-3-3, whereas phenformin dramatically increases the amount of 14-3-3 that coimmunoprecipitates with IRSp53-FLAG. Abundance is reported relative to untreated fed cells expressing IRSp53-FLAG. Throughout this figure, error bars are ± SD from three independent experiments. The statistical significance of all the measurements was determined using an unpaired t test based on three independent experiments (n.s., nonsignificant; *p < 0.05, **p < 0.01, ***p < 0.001).
Movie S3.
14-3-3 binding to IRSp53 depends on AMPK phosphorylation and abrogation of this interaction results in aberrant FLS formation. This movie corresponds to cell images shown in Figures 3, 4 and S4, and illustrates serum starved COS-7 cells expressing the following proteins: (A) M234-GFP (IRSp53 mutant T340A/T360A/S366A), (B) IRSp53-GFP treated with dorsomorphin, (C) M234-GFP and RFP-Cdc42, (D) M234-GFP and mTagBFP2-14-3-3, (E) M234-GFP, RFP-Cdc42 andmTagBFP2-14-3-3, (F) M234-GFP, mCherry-Eps8 and mTagBFP2-14-3-3. Scale bars throughout this movie represent 10 μm.
Disrupting 14-3-3 binding to IRSp53 leads to aberrant FLS formation
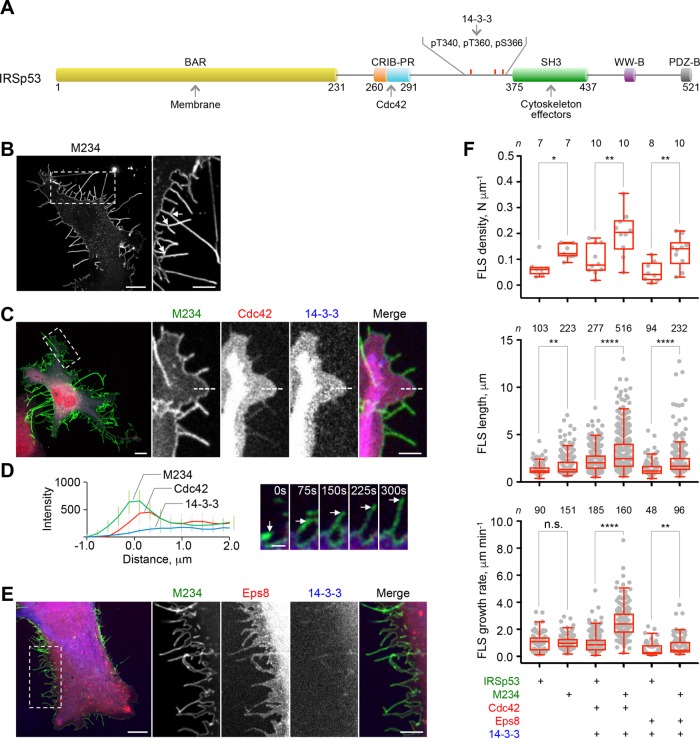
To specifically test the role of IRSp53 phosphorylation and 14-3-3 binding on the inhibition of FLS formation and dynamics, we generated a mutant in which the three phosphorylation sites implicated in 14-3-3 binding, located between the CRIB-PR and SH3 domains (Robens et al., 2010; Cohen et al., 2011; Kast and Dominguez, 2019), were simultaneously mutated to alanine (Figure 4A). For consistency with our recent study (Kast and Dominguez, 2019), we call this mutant M234 (IRSp53 mutant T340A/T360A/S366A). When expressed alone and approximately to the same extent as WT IRSp53 (compare Supplemental Figures S1A and S4A), M234 significantly increased the number of FLS (Figure 4, B and F, and Supplemental Movie S3A), potentially reflecting the lack of IRSp53 inhibition by endogenous 14-3-3. Importantly, FLS formed with mutant M234 were mostly filled with actin (Supplemental Figure S4B) but were frequently branched, although the cause of this phenotype is unclear (arrows in Figure 4B, inset). Coexpression of M234 with Cdc42 further increased the number, length, and growth rate of FLS (Supplemental Figure S4C and Supplemental Movie S3C), whereas coexpression of M234 with 14-3-3 did not appreciably inhibit FLS formation (Supplemental Figure S4D and Supplemental Movie S3D). The protrusions formed under these conditions, however, could not be reliably quantified, as they were highly branched and curved.
FIGURE 4:
Phosphoinhibitory mutations in IRSp53 abrogate 14-3-3-dependent inhibition of FLS dynamics. (A) Domain diagram of IRSp53 showing the three residues that on phosphorylation mediate the binding of 14-3-3 and which were mutated to alanine in mutant M234 (B) COS-7 cell expressing M234-GFP (IRSp53 mutant T340A/T360A/S366A). Scale bars throughout this figure represent 10 and 5 μm in whole-cell and inset images, respectively. (C) COS-7 cell expressing M234-GFP, RFP-Cdc42, and mTagBFP2-14-3-3. (D) Average fluorescence intensities from 10 line scans across the cell edge (as exemplified by dashed lines in the insets in C) and a representative time series of a growing FLS (indicated by arrows). Scale bar represents 5 μm. (E) COS-7 cell expressing M234-GFP, mCherry-Eps8 and mTagBFP2-14-3-3. (F) Quantification of the density, length, and growth rate of FLS. The statistical significance of the measurements was determined using the Mann–Whitney rank sum test, based on the indicated number of observations (n) recorded from 7–10 cells and two independent transfections (n.s., nonsignificant; *p < 0.05; **p < 0.01; ****p < 0.0001).
Compared to WT IRSp53, cells coexpressing M234, Cdc42, and 14-3-3 displayed a markedly increased number of FLS, which were also longer and more dynamic (Figure 4, C and F, and Supplemental Movie S3E). 14-3-3 was largely excluded from these protrusions (Figure 4, C and D), which is consistent with the lack of binding to IRSp53. In support of this interpretation, FLS from cells expressing M234 were not as sensitive to serum starvation as those from cells expressing IRSp53 (compare Supplemental Figures S1D and S4E). When coexpressed with Eps8 and 14-3-3, M234 also produced more FLS than WT IRSp53, and these protrusions were significantly longer and somewhat more dynamic (Figure 4, E and F, and Supplemental Movie S3F). 14-3-3 was also largely absent from FLS in these cells. Interestingly, however, both WT IRSp53 and M234 produced more dynamic FLS when activated by Cdc42 than by Eps8 (Figure 4F), suggesting that Cdc42 plays roles in both FLS initiation and dynamics, whereas Eps8 only impacts initiation. These results allow us to conclude that the sole mutation of three S/T residues implicated in 14-3-3 binding in IRSp53 leads to dramatic effects in FLS formation and dynamics, likely due to the lack of inhibition by 14-3-3.
14-3-3 binding to IRSp53 inhibits cancer cell chemotaxis
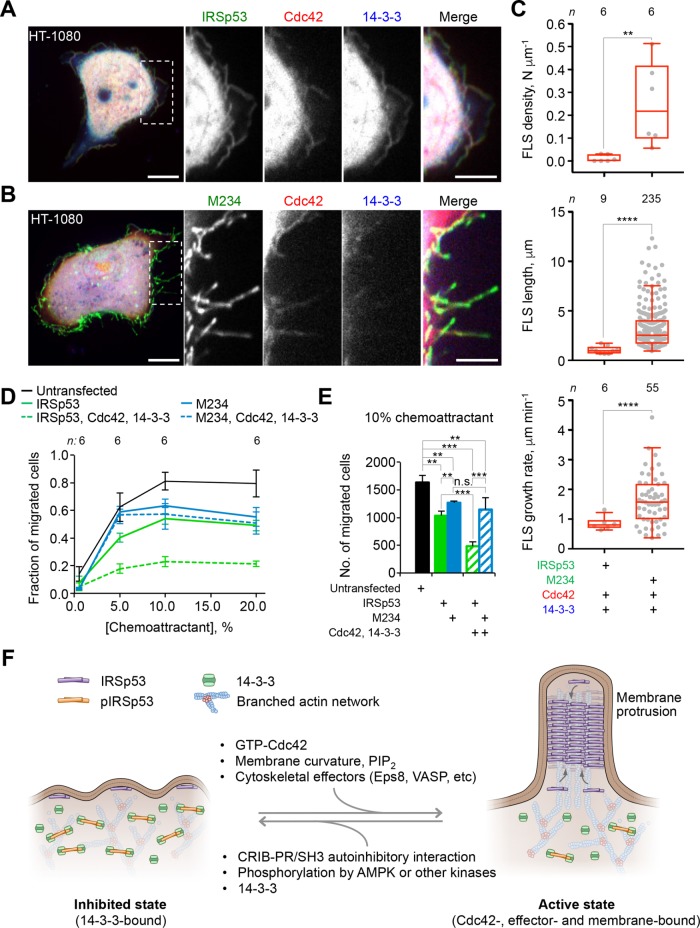
Dynamic filopodia are required to establish cell polarity and drive-directed cell migration in vivo (Meyen et al., 2015). Since 14-3-3 inhibits IRSp53-dependent FLS formation and dynamics, we asked whether it also regulates cell motility and polarity through phosphorylation-dependent interaction with IRSp53. We analyzed the chemotactic migration of human fibrosarcoma HT-1080 cells as a function of IRSp53 or M234 coexpression with Cdc42 and 14-3-3. Unlike COS-7 cells (Figure 1, D and E), HT-1080 cells coexpressing IRSp53, Cdc42, and 14-3-3 produced a small number of FLS, and these FLS were typically short and had a low growth rate (Figure 5, A and C, and Supplemental Movie S4A). In contrast, a large number of dynamic FLS were observed in cells coexpressing M234, Cdc42, and 14-3-3 (Figure 5, B and C, and Supplemental Movie S4B). Similar to COS-7 cells (Figure 4, B–D), M234 localized mainly to the plasma membrane and FLS, and Cdc42 tended to colocalize with M234, whereas 14-3-3 was mostly absent from FLS (Figure 5B and Supplemental Movie S4B).
FIGURE 5:
14-3-3 binding to IRSp53 inhibits cancer cell chemotaxis. (A, B) Serum-starved fibrosarcoma HT-1080 cells expressing RFP-Cdc42, mTagBFP2-14-3-3, and IRSp53-GFP (or M234-GFP). Scale bars represent 10 and 5 μm in whole-cell and inset images, respectively. (C) Quantification of the density, length, and growth rate of FLS. The statistical significance of the measurements was determined using the Mann–Whitney rank sum test, based on the indicated number of observations (n) reordered from six cells and two independent transfections (**p < 0.01; ****p < 0.0001). (D) Quantification of the fraction of HT-1080 cells expressing RFP-Cdc42, mTagBFP2-14-3-3, and either IRSp53-GFP (green traces) or M234-GFP (blue traces) that migrate from the top plate (0.5% FBS) to the bottom plate (0.5–20% FBS) of a 96-well IncuCyte chemotaxis system as a function chemoattractant concentration. Chemotaxis plates were calibrated by quantifying the time at which the highest fraction of control cells (untransfected cells, black lines) had migrated to the bottom plate, corresponding to 33 h for IRSp53 and 42 h for M234. (E) Statistical analyses of cell chemotaxis were performed at the optimal chemoattractant concentration of 10% FBS (determined from D). Error bars are ± SD from two independent experiments per expression condition, each consisting of three replicas. The statistical significance of the measurements was determined using an unpaired t test based on three independent experiments (n.s., nonsignificant; **p < 0.01; ***p < 0.001). (F) The role of IRSp53 in cell protrusion dynamics. IRSp53 exists in two states: active (Cdc42-, membrane-, and cytoskeletal effector-bound) and inactive (autoinhibited through intramolecular interaction of the CRIB-PR and SH3 domains and additionally locked on phosphorylation-dependent binding of 14-3-3). AMPK is directly and/or indirectly responsible for the phosphorylation of two of the three 14-3-3 binding sites (T360 and S366). In the active state, IRSp53 forms coats at the membrane, which dictates the curvature of the membrane (Carman and Dominguez, 2018), and recruits cytoskeletal proteins that promote actin assembly to induce the formation of dynamic cell protrusions. Cell protrusion formation is inhibited in the inactive state.
Movie S4.
Abrogation of 14-3-3 binding to IRSp53 results in FLS formation in fibrosarcoma HT-1080 cells. This movie corresponds to cell images shown in Figure 5, and illustrates serum starved HT-1080 cells expressing the following proteins: (A) IRSp53-GFP, RFP-Cdc42 and mTagBFP2-14-3-3, (B) M234-GFP (IRSp53 mutant T340A/T360A/S366A), RFP-Cdc42 and mTagBFP2-14-3-3. Scale bars throughout this movie represent 10 μm and 5 μm in whole-cell and inset images, respectively.
We tracked the movement of HT-1080 cells expressing WT IRSp53 and M234 using a 96-well chemotaxis plate setup, where cells in the upper chamber migrate toward a lower reservoir containing the chemoattractant. Over a range of chemoattractant concentrations (0.5–20% fetal bovine serum [FBS]), cells expressing either IRSp53 or M234 performed chemotaxis less efficiently than untransfected cells (Figure 5D). For both transfected and untransfected cells, the fraction of migrated cells peaked at 10% chemoattractant, which was selected as the optimal concentration for statistical analysis (Figure 5E). Cells expressing WT IRSp53 displayed lower overall chemotaxis efficiency than cells expressing M234, particularly at lower chemoattractant concentrations (≤10%). However, the difference between IRSp53 and M234 became far more pronounced when coexpressed with Cdc42 and 14-3-3, which had no effect on the migration of M234-expressing cells but severely reduced the migration of IRSp53-expressing cells (Figure 5, D and E). These results suggest that phosphorylation-dependent inhibition of IRSp53 by 14-3-3 regulates polarized cell migration.
DISCUSSION
This study establishes IRSp53 as a chemotactic signal transducer that in response to AMPK-dependent phosphorylation (and likely phosphorylation by other kinases) binds 14-3-3, which in turn down-regulates cytoskeleton and plasma membrane remodeling, filopodia formation, and cell motility (Figure 5F). Owing to its unique multidomain architecture, IRSp53 acts as a signaling hub, capable of integrating activatory (GTP-Cdc42, membrane curvature and phospholipids, downstream cytoskeleton effectors) and inhibitory (intramolecular CRIB-PR/SH3 interaction, phosphorylation, and 14-3-3 binding) inputs. Cytoskeleton and membrane remodeling effects are further enhanced through the formation of two-dimensional coats at the membrane via lateral oligomerization, as observed for IRSp53 as well as other members of the I-BAR protein family (Mattila et al., 2007; Saarikangas et al., 2009; Pykalainen et al., 2011; Prevost et al., 2015; Carman and Dominguez, 2018).
The live-cell studies described here show that 14-3-3 inhibits IRSp53-dependent FLS formation and dynamics irrespective of whether it is activated by interaction with Cdc42 via its CRIB-PR domain or with cytoskeletal effectors such as Eps8 and VASP via its SH3 domain (Figures 1 and 2). Phosphoproteomics and coimmunoprecipitation of 14-3-3 from serum-starved, dorsomorphin-, or phenformin-treated cells clearly implicate AMPK (and possibly AMPK-related kinases) in the phosphorylation of at least two of the sites responsible for 14-3-3 binding (T360 and S366) (Figure 3). Of these, S366 is known to be directly phosphorylated by AMPK and AMPK-related kinases from chemical genetics screening studies (Banko et al., 2011; Cohen et al., 2011; Schaffer et al., 2015), whereas the effect of AMPK on the phosphorylation of T360 could be indirect. We further show that dorsomorphin treatment leads to a dramatic increase in the number of FLS (Figure 3D and Supplemental Movie S3B), further implicating AMPK in the regulation of cell protrusion formation and consistent with its role in the control of actin dynamics (Moser et al., 2010), cell polarity (Lee et al., 2007a), and cell motility (Hurtado de Llera et al., 2012; Yan et al., 2015; Aparicio et al., 2016; Kumar et al., 2018). Whereas the kinase(s) implicated in the phosphorylation of T340 is still unknown, one thing seems clear – different kinases converge on IRSp53 to control 14-3-3 binding, offering alternative routes of regulation.
Perhaps more striking is the explosive increase in FLS formation observed with mutant M234 when expressed alone or in conjunction with Cdc42 (or Eps8) and 14-3-3 (Figure 4 and Supplemental Figure S4). Of note, only three S/T residues of the 521 residues of IRSp53 were substituted by alanine in this mutant (T340A, T360A, and S366A). These three mutations are unlikely to affect folding since they fall within the 83-aa region between the CRIB-PR and SH3 domains, which is predicted unstructured with three types of amino acids (S/T/P) accounting for 36% of the sequence. These observations highlight at least three points: 1) the intramolecular autoinhibitory interaction between the CRIB-PR and SH3 domains (Krugmann et al., 2001; Lim et al., 2008; Kast et al., 2014) is insufficient to maintain IRSp53 in a fully inactive state. This likely explains why the autoinhibited conformation can be competitively reversed in vitro and in cells by interactions with the plasma membrane, Cdc42, and cytoskeletal effectors (Kast et al., 2014). 2) Complete IRSp53 inhibition depends on phosphorylation-dependent interaction with 14-3-3. 3) It conclusively demonstrates that other than through membrane binding, IRSp53 contributes to membrane remodeling through its ability to recruit cytoskeletal effectors. Indeed, when expressed alone, the I-BAR domain of IRSp53 increases the number of FLS to approximately the same extent as full-length IRSp53 activated by Cdc42, but this effect is through membrane deformation alone (decoupled from cytoskeleton assembly) and, as a result, some of these filopodia are not actin-filled (Kast et al., 2014). In contrast, M234 produces many more filopodia than full-length IRSp53 when coexpressed with Cdc42 (or Eps8) and 14-3-3, and these protrusions are filled with actin to a similar extent as for WT IRSp53 (Supplemental Figure S4B). Such a dramatic increase in protrusion formation can be explained only by the combined contribution of IRSp53’s other domains, including recruitment to Cdc42 patches at the membrane via its CRIB-PR domain and recruitment of downstream actin assembly factors via its SH3 domain. Curiously, the FLS formed by M234 are often branched and much longer than with WT IRSp53 (Figure 4B and Supplemental Figure S4C). The reason for this phenotype is unclear, but it could be due to increased accumulation of unregulated M234 along the plasma membrane (Kast and Dominguez, 2019).
The results also provide additional insights into previous observations that the lifetime of FLS increases when two of the three 14-3-3 binding sites are removed in IRSp53 (Robens et al., 2010). Our observed increase in the length of FLS on expression of M234 is in principle consistent with longer FLS lifetimes. However, we also observed that the growth rate of FLS was similar for IRSp53- and M234-expressing cells, which suggests that the longer lifetimes of FLS may be due to a deficiency with the removal of M234 from the plasma membrane by 14-3-3. Consistent with this interpretation, IRSp53 becomes more abundant in the cytosol with serum starvation, whereas M234 remains enriched at plasma membrane (Kast and Dominguez, 2019). These results suggest that 14-3-3 enhances filopodia dynamics by controlling the abundance of IRSp53 at the plasma membrane.
We also show that 14-3-3 inhibits chemotaxis of IRSp53-expressing cancer cells, but has no effect on those cells expressing the mutant M234 (Figure 5). 14-3-3 proteins are a conserved family of phospho-S/T adaptors, known primarily for their roles in the control of cell-cycle progression and apoptosis (Gardino and Yaffe, 2011) and the regulation of cell metabolism (Kleppe et al., 2011). Yet 14-3-3 also interacts with several cytoskeletal proteins, and like AMPK (see above) has been implicated in the regulation of actin cytoskeleton and membrane dynamics (Gohla and Bokoch, 2002; Jin et al., 2004; Rosenberg et al., 2008; Robens et al., 2010; Cohen et al., 2011; Freeman and Morrison, 2011; Boudreau et al., 2013) and in countless examples of cell migration regulation (Toyo-oka et al., 2003; Preisinger et al., 2004; Tak et al., 2007; Yu et al., 2008; O’Toole et al., 2011; Goc et al., 2012; Tsigkari et al., 2012; Wu et al., 2018). Whether and how 14-3-3 and AMPK work together in these processes is unknown, since in most cases the kinases that phosphorylate 14-3-3 binding sites are unknown. This study provides strong support for a cellular mechanism whereby 14-3-3 and AMPK (or AMPK-related kinases) work hand-in-hand to control cell morphology and motility, using IRSp53 as an intermediate.
MATERIALS AND METHODS
Antibodies
Mouse monoclonal anti-IRSp53 (catalogue number: sc-136470; 1:500 dilution), mouse monoclonal anti–pan-14-3-3 (catalogue number: sc-133233; 1:500 dilution), mouse monoclonal anti–β-actin (catalogue number: sc-47778; 1:1000 dilution), and mouse monoclonal anti-GAPDH (catalogue number: sc-32223; 1:1000 dilution) were purchased from Santa Cruz Biotechnology (Dallas, TX). Rabbit monoclonal anti-Cdc42 (catalogue number: 2462S; 1:1000 dilution), rabbit monoclonal anti-Eps8 (catalogue number: 43114S; 1:1000 dilution), rabbit monoclonal anti-VASP (catalogue number: 2795S; 1:1000 dilution), rabbit monoclonal anti-AMPKα1 (catalogue number: 2795S; 1:1000 dilution), rabbit monoclonal anti–phospho-AMPKα1 (T172) (catalogue number: 2535S; 1:1000 dilution), rabbit monoclonal anti-ACC (phospho-acetyl-CoA carboxylase; catalogue number: 11818S; 1:1000 dilution), and horseradish peroxidase (HRP)-linked anti-mouse and anti-rabbit immunoglobulin G secondary antibodies (catalogue numbers: 7076S and 7074S; 1:5000 dilution) were purchased from Cell Signaling Technology (Danvers, MA).
Protein cloning, expression, and purification
Mammalian cell expression constructs.
Full-length human IRSp53 (UniProt: Q9UQB8, isoform 4) was cloned into vectors pEGFP-N1 and mTagBFP2-N1. For Co-IPs and protein purification, IRSp53 was PCR-extended to include a C-terminal FLAG-tag and cloned into vector pEGFP-C1 using restriction sites NheI and SalI. IRSp53 mutations T340A, T360A, and S366A were introduced using the QuickChange II XL mutagenesis kit (Agilent Technologies, Santa Clara, CA; catalogue number: 200521). Full-length human Eps8 (UniProt: Q12929) and human 14-3-3θ (UniProt: P27348) were cloned into vectors mTagBFP2-C1, pEGFP-C1, and mCherry-C1 for live-cell imaging. Both human RFP-Cdc42 (constitutively active mutant G12V) and human GFP-VASP were gifts from Tatyana Svitkina (University of Pennsylvania). Constructs were generated using the primers and/or oligos reported in Supplemental Table S3, and inserted genes in all expression constructs were fully sequenced (Supplemental Table S4).
Mammalian cell protein expression and purification.
IRSp53-FLAG for pull-down studies was expressed in HEK293 cells grown to ∼40% confluence in 15-cm plates, transiently transfected with 10 μg plasmid DNA in 30 μl of a 1 mg ml–1 polyethylenimine solution (Polysciences, Warrington, PA; catalogue number: 23966). After 48 h expression, IRSp53 phosphorylation was increased by serum-depriving cells for 2 h (see below). Cells were then washed and incubated in phosphate-buffered saline (PBS) with the addition of 0.1 μM Calyculin-A (Cell Signaling Technology, Danvers, MA; catalogue number: 9902) and 1× Halt Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific, Waltham, MA; catalogue number: 87796) for 15 min at 37°C. Cells were harvested, pelleted, and resuspended in lysis buffer composed of PBS supplemented with 1 mM EDTA, 5% glycerol, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride (PMSF), 0.1 μM Calyculin A, 1× cOmplete protease inhibitor cocktail (MilliporeSigma, St. Louis, MO; catalogue number: 11697498001), and 1× Halt Phosphatase Inhibitor Cocktail. Cells were then incubated in lysis buffer on ice for 30 min and freeze–thawed three times. Insoluble cellular components and nuclei were removed by centrifugation at 12,000 × g for 10 min. Clarified lysates were incubated with 25 μl anti-FLAG antibody conjugated to magnetic beads (MilliporeSigma; catalogue number: M8823-1ML) for 2 h at 4°C and washed three times with 250 μl of lysis buffer (without Triton X-100). IRSp53 was eluted with 50 μl wash buffer supplemented with 300 mM NaCl and 0.5 mg ml–1 3xFLAG peptide (Bimake, Jupiter, FL; catalogue number: B23112). Proteins were visualized by Western blot using Immobilon-P polyvinylidene difluoride (PVDF) membranes (MilliporeSigma; catalogue number: IPVH00010). The relative abundance of proteins was determined by densitometry analysis using the program Fiji (https://imagej.net/Fiji). All blots presented in the figures were prepared from the unmodified blots shown in Supplemental Figure S5.
Cell lines, media, transfections, activators, and inhibitors
HEK293T cells (HEK293T/17; catalogue number: CRL-11268) and COS-7 cell (catalogue number: CRL-1651) were purchased from the American Type Culture Collection (Manassas, VA). HT-1080 cells were from Essen BioScience (Ann Arbor, MI). Cells were authenticated and verified to be free of mycoplasma contamination by the manufacturer, and additional testing for mycoplasma contamination was not performed. All cell lines were maintained in DMEM GlutaMAX supplemented with 10% FBS and antibiotic-antimycotic (Thermo Fisher Scientific; catalogue number: 15240062). Plasmids were transfected immediately after cell seeding using FuGENE 6 (Promega, Madison, WI; catalogue number: E2691). In some cases, actin was labeled using SiR-actin (Cytoskeleton, Denver, CO; catalogue number: CY-SC001) by incubating COS-7 cells 12 h after seeding with 10 μM verapamil (Cytoskeleton catalogue number: CY-SC001) and 1 μM SiR-actin. When indicated, cells were serum-starved by incubation for 2 h in growth medium lacking FBS. For pharmacological activation of AMPK, HEK293T cells expressing IRSp53-FLAG were pretreated with the CaMKK2 inhibitor STO-609 (Santa Cruz Biotechnology; catalogue number: sc-202820) for 1 h before replacing the culture media with fresh media that include the indicated concentrations of the AMPK activator phenformin hydrochloride (Santa Cruz Biotechnology; catalogue number: sc-219590) for 1 h before lysis. AMPK was inhibited by incubating IRSp53-FLAG-expressing cells in cell culture media (with or without 10% FBS) supplemented with the indicated concentrations of dorsomorphin dihydrochloride (Santa Cruz Biotechnology; catalogue number: sc-361173) for 2 h. In some of the experiments (as indicated), serum-starved cells were additionally treated with 0.1 μM insulin (Santa Cruz Biotechnology; catalogue number: sc-360248) for 10 min before imaging or lysis.
Live cell imaging
All cells were imaged 18–24 h posttransfection using either an Olympus IX81 inverted microscope equipped with a 100× (1.4 NA) Plan-Apo oil immersion objective and a Yokogawa spinning disk confocal attached to an Andor iXon3 EMCCD camera (Figures 1–5 and Supplemental Figure S4, C and D) or a Nikon Ti2 inverted microscope equipped with a 100× (1.4 NA) Plan-Apo oil immersion objective and a Yokogawa CSU-W1 spinning disk confocal attached to a Hamamatsu ORCA-FLASH4.0 CMOS camera (Supplemental Figures S1B and S4B). Cells in DMEM FluoroBrite (Thermo Fisher Scientific; catalogue number: A1896701) medium supplemented with 0.5% FBS were imaged at 37°C and 5% CO2. Images were captured at 2- or 5-s intervals for 5 min at 14-bit resolution using the Metamorph software (Molecular Devices, San Jose, CA). The analysis of videos and images, including measurements of cell protrusion density, length, and growth rate, was performed with the software Fiji (National Institutes of Health [NIH]).
Phosphoproteomics
Coomassie Blue-stained gels were cut into 1 × 1-mm pieces, destained with 50% methanol and 1.25% acetic acid, reduced with 5 mM dithiothreitol (DTT), and alkylated with 20 mM iodoacetamide. The pieces were then washed with 20 mM ammonium bicarbonate and dehydrated with acetonitrile. Proteins were proteolyzed overnight at 37°C with the addition of 5 ng ml–1 trypsin in 20 mM ammonium bicarbonate. Peptides were extracted with 0.3% trifluoroacetic acid, followed by a 50% acetonitrile/water solution. Tryptic peptides were separated by reverse-phase high-performance liquid chromatography using a 75 μm i.d. × 25 cm Acclaim PepMap nano LC column and eluted peptides were analyzed by LC-MS/MS on a Q Exactive HF mass spectrometer (Thermo Fisher Scientific). The mass spectrometer was set to repetitively scan the mass-to-charge ratio in the range 300–1400 (resolution of 240,000), followed by data-dependent MS/MS scans (resolution of 15,000) on the 20 most abundant ions, with a minimum automatic gain control (AGC) target of 1e4. Dynamic exclusion was enabled with a repeat count of 1 and a repeat duration of 30 s. The Fourier transform mass spectrometry full scan AGC target value was set to 3e6, while MSn AGC was 1e5. The MSn injection time was set to 160 ms, and microscans were not averaged. FT preview mode charge state screening and monoisotopic precursor selection were all enabled, with rejection of unassigned and 1+ charge states.
Database searching was performed using the Proteome Discoverer software v1.4 (Thermo Fisher Scientific) and against the UniProt human database appended with common contaminants using the SEQUEST algorithm. SEQUEST was searched with a fragment ion mass tolerance of 0.020 Da and a parent ion tolerance of 10.0 PPM. Carbamidomethyl of cysteine was specified as a fixed modification. Methionine oxidation, N-terminal acetylation, and serine phosphorylation were specified as variable modifications. The program Scaffold v4.6.1 (Proteome Software, Portland, OR) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability by the Scaffold Local FDR algorithm, whereas protein identifications were accepted if they could be established at greater than 95.0% probability and contained at least one identified peptide. Protein probabilities were assigned by the Protein Prophet algorithm (Nesvizhskii et al., 2003). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Proteins sharing significant peptide evidence were grouped into clusters. The localization of the phosphorylation sites was validated using the Ascore algorithm (Beausoleil et al., 2006) in Scaffold PTM v3.2.0. All identified peptides, posttranslational modifications, and peptides are reported in Supplemental Tables 1 and 2.
Chemotaxis
Cell migration analysis was performed using an IncuCyte ClearView 96-Well Chemotaxis Plate System (Essen BioScience) by tracking the migration of HT-1080 cells from a 96-well upper plate to a lower plate containing the chemoattractant (FBS). The upper plate was coated with 10 μg ml–1 fibronectin and seeded with ∼2000 cells. Cells were either untransfected or transiently transfected with constructs IRSp53-GFP, M234-GFP, RFP-Cdc42, and/or mCherry-14-3-3, as indicated (Figure 5). Cells in the upper plate were incubated for 3 h in DMEM (Thermo Fisher Scientific) supplemented with 0.5% FBS to allow for their attachment before loading the lower plate containing DMEM supplemented with 0.5–20% FBS. Chemotaxis plates were calibrated for parameters that could affect migration time, including fibronectin coating, chemoattractant concentration, and imaging delay, by determining the time where the maximum number of untransfected cells had migrated from the top plate to the bottom plate. A total of six bright field green and red fluorescence images were collected per well (top and bottom) every 2 h and for a period of 63 h. Images were analyzed using the IncuCyte Chemotaxis Software (Essen BioScience) using either bright field images for untransfected cells or green and red fluorescence images for transfected cells. Each experimental condition was repeated at least four times.
Quantification and statistical analysis
Filopodia statistics.
Live cell images were sampled randomly for FLS analysis. Randomly motorized stage coordinates (x and y), within the viewable area of the 35-mm FluoroDish (World Precision Instruments, Sarasota, FL; catalogue number: FD35), were generated and used in the automated image collection macro of the program MetaMorph (Molecular Devices) designed to move the stage, focus, and perform image recordings. Nonoverlapping FLS, defined as dynamic, nontapered protrusions 0.2–0.5 μm in width and extending at least 0.5 μm from the cell periphery, were measured manually using the program Fiji (NIH) in images containing attached, nonblebbing and nondividing cells. FLS density was determined as the ratio between the number of FLS and the total perimeter of the cell. FLS length was determined by measuring the maximum length of dynamic protrusions observed over at least 5 min of an image sequence. FLS length was measured in the green (or blue) channel for cells expressing IRSp53-GFP (or IRSp53-mTagBFP2) or M234-GFP. For cells expressing only RFP-Cdc42, mCherry-VASP, mCherry-Eps8, mTAGBFP2-14-3-3θ, or cells treated with SiR-actin, FLS were quantified using the respective emission channels. These proteins are typically found along the length of FLS (Jacquemet et al., 2019), and measuring FLS lengths in these channels has a negligible effect (<10%) on the length distribution of FLS. FLS growth rate was determined as the maximum extension speed of a FLS growing uninterruptedly for 30 s or more. FLS growth rate was not measured for static FLS or for FLS that moved out of the field of view within 30 s. Statistical analysis was performed on the aggregated number of observations.
Statistical methods.
The data are presented as the mean ± SD. The statistical significance of the measurements was determined, using the program Prism 7c (GraphPad Software, La Jolla, CA), using either the unpaired two-sided Student’s t test or Mann–Whitney’s rank sum test (as indicated). A p value ≤ 0.05 was considered significant.
Supplementary Material
Acknowledgments
This research was funded by National Institutes of Health grant R01-MH087950 to R.D. D.J.K. was in part supported by American Cancer Society grant PF-13-033-01-DMC.
Abbreviations used:
- ACC
acetyl-CoA carboxylase
- AGC
automatic gain control
- CAMKK
calmodulin-dependent protein kinase kinase
- FBS
fetal bovine serum
- FLS
filopodia-like structures.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-09-0600) on March 20, 2019.
REFERENCES
- Abou-Kheir W, Isaac B, Yamaguchi H, Cox D. (2008). Membrane targeting of WAVE2 is not sufficient for WAVE2-dependent actin polymerization: a role for IRSp53 in mediating the interaction between Rac and WAVE2. J Cell Sci , 379–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio IM, Espino J, Bejarano I, Gallardo-Soler A, Campo ML, Salido GM, Pariente JA, Pena FJ, Tapia JA. (2016). Autophagy-related proteins are functionally active in human spermatozoa and may be involved in the regulation of cell survival and motility. Sci Rep , 33647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banko MR, Allen JJ, Schaffer BE, Wilker EW, Tsou P, White JL, Villen J, Wang B, Kim SR, Sakamoto K, et al. (2011). Chemical genetic screen for AMPKalpha2 substrates uncovers a network of proteins involved in mitosis. Mol Cell , 878–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barilari M, Dente L. (2010). The neuronal proteins CIPP, Cypin and IRSp53 form a tripartite complex mediated by PDZ and SH3 domains. Biol Chem , 1169–1174. [DOI] [PubMed] [Google Scholar]
- Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. (2006). A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol , 1285–1292. [DOI] [PubMed] [Google Scholar]
- Bobsin K, Kreienkamp HJ. (2016). Severe learning deficits of IRSp53 mutant mice are caused by altered NMDA receptor-dependent signal transduction. J Neurochem , 752–763. [DOI] [PubMed] [Google Scholar]
- Bockmann J, Kreutz MR, Gundelfinger ED, Bockers TM. (2002). ProSAP/Shank postsynaptic density proteins interact with insulin receptor tyrosine kinase substrate IRSp53. J Neurochem , 1013–1017. [DOI] [PubMed] [Google Scholar]
- Boudreau A, Tanner K, Wang D, Geyer FC, Reis-Filho JS, Bissell MJ. (2013). 14-3-3sigma stabilizes a complex of soluble actin and intermediate filament to enable breast tumor invasion. Proc Natl Acad Sci USA , E3937–E3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman PJ, Dominguez R. (2018). BAR domain proteins-a linkage between cellular membranes, signaling pathways, and the actin cytoskeleton. Biophys Rev , 1587–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CE, Odde DJ. (2008). Traction dynamics of filopodia on compliant substrates. Science , 1687–1691. [DOI] [PubMed] [Google Scholar]
- Chauhan BK, Disanza A, Choi SY, Faber SC, Lou M, Beggs HE, Scita G, Zheng Y, Lang RA. (2009). Cdc42- and IRSp53-dependent contractile filopodia tether presumptive lens and retina to coordinate epithelial invagination. Development , 3657–3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Ko J, Racz B, Burette A, Lee JR, Kim S, Na M, Lee HW, Kim K, Weinberg RJ, Kim E. (2005). Regulation of dendritic spine morphogenesis by insulin receptor substrate 53, a downstream effector of Rac1 and Cdc42 small GTPases. J Neurosci , 869–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen D, Fernandez D, Lazaro-Dieguez F, Musch A. (2011). The serine/threonine kinase Par1b regulates epithelial lumen polarity via IRSp53-mediated cell-ECM signaling. J Cell Biol , 525–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disanza A, Bisi S, Winterhoff M, Milanesi F, Ushakov DS, Kast D, Marighetti P, Romet-Lemonne G, Muller HM, Nickel W, et al. (2013). CDC42 switches IRSp53 from inhibition of actin growth to elongation by clustering of VASP. EMBO J , 2735–2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disanza A, Mantoani S, Hertzog M, Gerboth S, Frittoli E, Steffen A, Berhoerster K, Kreienkamp HJ, Milanesi F, Di Fiore PP, et al. (2006). Regulation of cell shape by Cdc42 is mediated by the synergic actin-bundling activity of the Eps8-IRSp53 complex. Nat Cell Biol , 1337–1347. [DOI] [PubMed] [Google Scholar]
- Disanza A, Scita G. (2008). Cytoskeletal regulation: coordinating actin and microtubule dynamics in membrane trafficking. Curr Biol , R873–R875. [DOI] [PubMed] [Google Scholar]
- Fischer RS, Lam PY, Huttenlocher A, Waterman CM. (2018). Filopodia and focal adhesions: an integrated system driving branching morphogenesis in neuronal pathfinding and angiogenesis. Dev Biol, DOI: 10.1016/j.ydbio.2018.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman AK, Morrison DK. (2011). 14-3-3 Proteins: diverse functions in cell proliferation and cancer progression. Semin Cell Dev Biol , 681–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P, Gilmour D. (2009). Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol , 445–457. [DOI] [PubMed] [Google Scholar]
- Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, Georgieva L, Rees E, Palta P, Ruderfer DM, et al. (2014). De novo mutations in schizophrenia implicate synaptic networks. Nature , 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Mammoto A, Kim Y, Takai Y. (2000). Rho small G-protein-dependent binding of mDia to an Src homology 3 domain-containing IRSp53/BAIAP2. Biochem Biophys Res Commun , 626–629. [DOI] [PubMed] [Google Scholar]
- Funato Y, Terabayashi T, Suenaga N, Seiki M, Takenawa T, Miki H. (2004). IRSp53/Eps8 complex is important for positive regulation of Rac and cancer cell motility/invasiveness. Cancer Res , 5237–5244. [DOI] [PubMed] [Google Scholar]
- Gardino AK, Yaffe MB. (2011). 14-3-3 proteins as signaling integration points for cell cycle control and apoptosis. Semin Cell Dev Biol , 688–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goc A, Abdalla M, Al-Azayzih A, Somanath PR. (2012). Rac1 activation driven by 14-3-3zeta dimerization promotes prostate cancer cell-matrix interactions, motility and transendothelial migration. PLoS One , e40594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh WI, Lim KB, Sudhaharan T, Sem KP, Bu W, Chou AM, Ahmed S. (2012). mDia1 and WAVE2 proteins interact directly with IRSp53 in filopodia and are involved in filopodium formation. J Biol Chem , 4702–4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohla A, Bokoch GM. (2002). 14-3-3 regulates actin dynamics by stabilizing phosphorylated cofilin. Curr Biol , 1704–1710. [DOI] [PubMed] [Google Scholar]
- Goodwin JM, Svensson RU, Lou HJ, Winslow MM, Turk BE, Shaw RJ. (2014). An AMPK-independent signaling pathway downstream of the LKB1 tumor suppressor controls Snail1 and metastatic potential. Mol Cell , 436–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley SA, Boudeau J, Reid JL, Mustard KJ, Udd L, Makela TP, Alessi DR, Hardie DG. (2003). Complexes between the LKB1 tumor suppressor, STRAD alpha/beta and MO25 alpha/beta are upstream kinases in the AMP-activated protein kinase cascade. J Biol , 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heung MY, Visegrady B, Futterer K, Machesky LM. (2008). Identification of the insulin-responsive tyrosine phosphorylation sites on IRSp53. Eur J Cell Biol , 699–708. [DOI] [PubMed] [Google Scholar]
- Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E. (2015). PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res , D512–D520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado de Llera A, Martin-Hidalgo D, Gil MC, Garcia-Marin LJ, Bragado MJ. (2012). AMP-activated kinase AMPK is expressed in boar spermatozoa and regulates motility. PLoS One , e38840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemet G, Hamidi H, Ivaska J. (2015). Filopodia in cell adhesion, 3D migration and cancer cell invasion. Curr Opin Cell Biol , 23–31. [DOI] [PubMed] [Google Scholar]
- Jacquemet G, Stubb A, Saup R, Miihkinen M, Kremneva E, Hamidi H, Ivaska J. (2019). Filopodome mapping identifies p130Cas as a mechanosensitive regulator of filopodia stability. Curr Biol , 202–216.e207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Smith FD, Stark C, Wells CD, Fawcett JP, Kulkarni S, Metalnikov P, O’Donnell P, Taylor P, Taylor L, et al. (2004). Proteomic, functional, and domain-based analysis of in vivo 14-3-3 binding proteins involved in cytoskeletal regulation and cellular organization. Curr Biol , 1436–1450. [DOI] [PubMed] [Google Scholar]
- Kang J, Park H, Kim E. (2016). IRSp53/BAIAP2 in dendritic spine development, NMDA receptor regulation, and psychiatric disorders. Neuropharmacology , 27–39. [DOI] [PubMed] [Google Scholar]
- Kast DJ, Dominguez R. (2019). Mechanism of IRSp53 inhibition by 14-3-3. Nat Commun , 483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kast DJ, Yang C, Disanza A, Boczkowska M, Madasu Y, Scita G, Svitkina T, Dominguez R. (2014). Mechanism of IRSp53 inhibition and combinatorial activation by Cdc42 and downstream effectors. Nat Struct Mol Biol , 413–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleppe R, Martinez A, Doskeland SO, Haavik J. (2011). The 14-3-3 proteins in regulation of cellular metabolism. Semin Cell Dev Biol , 713–719. [DOI] [PubMed] [Google Scholar]
- Krugmann S, Jordens I, Gevaert K, Driessens M, Vandekerckhove J, Hall A. (2001). Cdc42 induces filopodia by promoting the formation of an IRSp53:Mena complex. Curr Biol , 1645–1655. [DOI] [PubMed] [Google Scholar]
- Kumar D, Rahman H, Tyagi E, Liu T, Li C, Lu R, Lum D, Holmen SL, Maschek JA, Cox JE, et al. (2018). Aspirin suppresses PGE2 and activates AMP kinase to inhibit melanoma cell motility, pigmentation and selective tumor growth in vivo. Cancer Prev Res (Phila) , 629–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Kerff F, Chereau D, Ferron F, Klug A, Dominguez R. (2007b). Structural basis for the actin-binding function of missing-in-metastasis. Structure , 145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Koh H, Kim M, Kim Y, Lee SY, Karess RE, Lee SH, Shong M, Kim JM, Kim J, Chung J. (2007a). Energy-dependent regulation of cell structure by AMP-activated protein kinase. Nature , 1017–1020. [DOI] [PubMed] [Google Scholar]
- Lim KB, Bu W, Goh WI, Koh E, Ong SH, Pawson T, Sudhaharan T, Ahmed S. (2008). The Cdc42 effector IRSp53 generates filopodia by coupling membrane protrusion with actin dynamics. J Biol Chem , 20454–20472. [DOI] [PubMed] [Google Scholar]
- Liu PS, Jong TH, Maa MC, Leu TH. (2010). The interplay between Eps8 and IRSp53 contributes to Src-mediated transformation. Oncogene , 3977–3989. [DOI] [PubMed] [Google Scholar]
- Lizcano JM, Goransson O, Toth R, Deak M, Morrice NA, Boudeau J, Hawley SA, Udd L, Makela TP, Hardie DG, Alessi DR. (2004). LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J , 833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie S, Aitken A. (2005). Novel brain 14-3-3 interacting proteins involved in neurodegenerative disease. FEBS J , 4202–4210. [DOI] [PubMed] [Google Scholar]
- Mattila PK, Lappalainen P. (2008). Filopodia: molecular architecture and cellular functions. Nat Rev Mol Cell Biol , 446–454. [DOI] [PubMed] [Google Scholar]
- Mattila PK, Pykalainen A, Saarikangas J, Paavilainen VO, Vihinen H, Jokitalo E, Lappalainen P. (2007). Missing-in-metastasis and IRSp53 deform PI(4,5)P2-rich membranes by an inverse BAR domain-like mechanism. J Cell Biol , 953–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyen D, Tarbashevich K, Banisch TU, Wittwer C, Reichman-Fried M, Maugis B, Grimaldi C, Messerschmidt EM, Raz E. (2015). Dynamic filopodia are required for chemokine-dependent intracellular polarization during guided cell migration in vivo. Elife , e05279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H, Yamaguchi H, Suetsugu S, Takenawa T. (2000). IRSp53 is an essential intermediate between Rac and WAVE in the regulation of membrane ruffling. Nature , 732–735. [DOI] [PubMed] [Google Scholar]
- Millard TH, Bompard G, Heung MY, Dafforn TR, Scott DJ, Machesky LM, Futterer K. (2005). Structural basis of filopodia formation induced by the IRSp53/MIM homology domain of human IRSp53. EMBO J , 240–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser TS, Jones RG, Thompson CB, Coyne CB, Cherry S. (2010). A kinome RNAi screen identified AMPK as promoting poxvirus entry through the control of actin dynamics. PLoS Pathog , e1000954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munday MR, Campbell DG, Carling D, Hardie DG. (1988). Identification by amino acid sequencing of three major regulatory phosphorylation sites on rat acetyl-CoA carboxylase. Eur J Biochem , 331–338. [DOI] [PubMed] [Google Scholar]
- Nakagawa H, Miki H, Nozumi M, Takenawa T, Miyamoto S, Wehland J, Small JV. (2003). IRSp53 is colocalised with WAVE2 at the tips of protruding lamellipodia and filopodia independently of Mena. J Cell Sci , 2577–2583. [DOI] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. (2003). A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem , 4646–4658. [DOI] [PubMed] [Google Scholar]
- Oikawa T, Okamura H, Dietrich F, Senju Y, Takenawa T, Suetsugu S. (2013). IRSp53 mediates podosome formation via VASP in NIH-Src cells. PLoS One , e60528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole TE, Bialkowska K, Li X, Fox JE. (2011). Tiam1 is recruited to beta1-integrin complexes by 14-3-3zeta where it mediates integrin-induced Rac1 activation and motility. J Cell Physiol , 2965–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirkmajer S, Chibalin AV. (2011). Serum starvation: caveat emptor. Am J Physiol Cell Physiol , C272–C279. [DOI] [PubMed] [Google Scholar]
- Preisinger C, Short B, De Corte V, Bruyneel E, Haas A, Kopajtich R, Gettemans J, Barr FA. (2004). YSK1 is activated by the Golgi matrix protein GM130 and plays a role in cell migration through its substrate 14-3-3zeta. J Cell Biol , 1009–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevost C, Zhao H, Manzi J, Lemichez E, Lappalainen P, Callan-Jones A, Bassereau P. (2015). IRSp53 senses negative membrane curvature and phase separates along membrane tubules. Nat Commun , 8529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pykalainen A, Boczkowska M, Zhao H, Saarikangas J, Rebowski G, Jansen M, Hakanen J, Koskela EV, Peranen J, Vihinen H, et al. (2011). Pinkbar is an epithelial-specific BAR domain protein that generates planar membrane structures. Nat Struct Mol Biol , 902–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribases M, Bosch R, Hervas A, Ramos-Quiroga JA, Sanchez-Mora C, Bielsa A, Gastaminza X, Guijarro-Domingo S, Nogueira M, Gomez-Barros N, et al. (2009). Case-control study of six genes asymmetrically expressed in the two cerebral hemispheres: association of BAIAP2 with attention-deficit/hyperactivity disorder. Biol Psychiatry , 926–934. [DOI] [PubMed] [Google Scholar]
- Robens JM, Yeow-Fong L, Ng E, Hall C, Manser E. (2010). Regulation of IRSp53-dependent filopodial dynamics by antagonism between 14-3-3 binding and SH3-mediated localization. Mol Cell Biol , 829–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg MM, Yang F, Giovanni M, Mohn JL, Temburni MK, Jacob MH. (2008). Adenomatous polyposis coli plays a key role, in vivo, in coordinating assembly of the neuronal nicotinic postsynaptic complex. Mol Cell Neurosci , 138–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarikangas J, Zhao H, Pykalainen A, Laurinmaki P, Mattila PK, Kinnunen PK, Butcher SJ, Lappalainen P. (2009). Molecular mechanisms of membrane deformation by I-BAR domain proteins. Curr Biol , 95–107. [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Goransson O, Hardie DG, Alessi DR. (2004). Activity of LKB1 and AMPK-related kinases in skeletal muscle: effects of contraction, phenformin, and AICAR. Am J Physiol Endocrinol Metab , E310–E317. [DOI] [PubMed] [Google Scholar]
- Schaffer BE, Levin RS, Hertz NT, Maures TJ, Schoof ML, Hollstein PE, Benayoun BA, Banko MR, Shaw RJ, Shokat KM, Brunet A. (2015). Identification of AMPK phosphorylation sites reveals a network of proteins involved in cell invasion and facilitates large-scale substrate prediction. Cell Metab , 907–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal D, Dhanyasi N, Schejter ED, Shilo BZ. (2016). Adhesion and fusion of muscle cells are promoted by filopodia. Dev Cell , 291–304. [DOI] [PubMed] [Google Scholar]
- Sluchanko NN, Gusev NB. (2017). Moonlighting chaperone-like activity of the universal regulatory 14-3-3 proteins. FEBS J , 1279–1295. [DOI] [PubMed] [Google Scholar]
- Soltau M, Berhorster K, Kindler S, Buck F, Richter D, Kreienkamp HJ. (2004). Insulin receptor substrate of 53 kDa links postsynaptic shank to PSD-95. J Neurochem , 659–665. [DOI] [PubMed] [Google Scholar]
- Soltau M, Richter D, Kreienkamp HJ. (2002). The insulin receptor substrate IRSp53 links postsynaptic shank1 to the small G-protein cdc42. Mol Cell Neurosci , 575–583. [DOI] [PubMed] [Google Scholar]
- Suetsugu S, Kurisu S, Oikawa T, Yamazaki D, Oda A, Takenawa T. (2006). Optimization of WAVE2 complex-induced actin polymerization by membrane-bound IRSp53, PIP(3), and Rac. J Cell Biol , 571–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tak H, Jang E, Kim SB, Park J, Suk J, Yoon YS, Ahn JK, Lee JH, Joe CO. (2007). 14-3-3epsilon inhibits MK5-mediated cell migration by disrupting F-actin polymerization. Cell Signal , 2379–2387. [DOI] [PubMed] [Google Scholar]
- Tokumitsu H, Inuzuka H, Ishikawa Y, Ikeda M, Saji I, Kobayashi R. (2002). STO-609, a specific inhibitor of the Ca(2+)/calmodulin-dependent protein kinase kinase. J Biol Chem , 15813–15818. [DOI] [PubMed] [Google Scholar]
- Toma C, Hervas A, Balmana N, Vilella E, Aguilera F, Cusco I, del Campo M, Caballero R, De Diego-Otero Y, Ribases M, et al. (2011). Association study of six candidate genes asymmetrically expressed in the two cerebral hemispheres suggests the involvement of BAIAP2 in autism. J Psychiatr Res , 280–282. [DOI] [PubMed] [Google Scholar]
- Toyo-oka K., Shionoya A, Gambello MJ, Cardoso C, Leventer R, Ward HL, Ayala R, Tsai LH, Dobyns W, Ledbetter D, et al. (2003). 14-3-3epsilon is important for neuronal migration by binding to NUDEL: a molecular explanation for Miller-Dieker syndrome. Nat Genet , 274–285. [DOI] [PubMed] [Google Scholar]
- Tsigkari KK, Acevedo SF, Skoulakis EM. (2012). 14-3-3epsilon is required for germ cell migration in Drosophila. PLoS One , e36702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaggi F, Disanza A, Milanesi F, Di Fiore PP, Menna E, Matteoli M, Gov NS, Scita G, Ciliberto A. (2011). The Eps8/IRSp53/VASP network differentially controls actin capping and bundling in filopodia formation. PLoS Comput Biol , e1002088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent EE, Coelho PP, Blagih J, Griss T, Viollet B, Jones RG. (2015). Differential effects of AMPK agonists on cell growth and metabolism. Oncogene , 3627–3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Zuo X, Zhuo Y, Zhu M, Danziger SA, Zhou Z. (2013). The functional role of SUMO E3 ligase Mms21p in the maintenance of subtelomeric silencing in budding yeast. Biochem Biophys Res Commun , 746–752. [DOI] [PubMed] [Google Scholar]
- Wu Q, Zhu J, Liu F, Liu J, Li M. (2018). Downregulation of 14-3-3beta inhibits proliferation and migration in osteosarcoma cells. Mol Med Rep , 2493–2500. [DOI] [PubMed] [Google Scholar]
- Yaffe MB. (2002). How do 14-3-3 proteins work?—Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett , 53–57. [DOI] [PubMed] [Google Scholar]
- Yan G, Chen V, Lu X, Lu S. (2017). A signal-based method for finding driver modules of breast cancer metastasis to the lung. Sci Rep , 10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Tsukamoto O, Nakano A, Kato H, Kioka H, Ito N, Higo S, Yamazaki S, Shintani Y, Matsuoka K, et al. (2015). Augmented AMPK activity inhibits cell migration by phosphorylating the novel substrate Pdlim5. Nat Commun , 6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Svitkina T. (2011). Filopodia initiation: focus on the Arp2/3 complex and formins. Cell Adh Migr , 402–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Han W, Shi T, Lv B, He Q, Zhang Y, Li T, Zhang Y, Song Q, Wang L, Ma D. (2008). PTPIP51, a novel 14-3-3 binding protein, regulates cell morphology and motility via Raf-ERK pathway. Cell Signal , 2208–2220. [DOI] [PubMed] [Google Scholar]
- Zhou G, Myers R, Li Y, Chen Y, Shen X, Fenyk-Melody J, Wu M, Ventre J, Doebber T, Fujii N, et al. (2001). Role of AMP-activated protein kinase in mechanism of metformin action. J Clin Invest , 1167–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.