Abstract
Objective:
The incorporation of high atomic number materials such as gold nanoparticles (GNPs) into tumor cells is being tested to enhance the local radiotherapy (RT) dose. It is also known that the radiosensitivity of tumor cells depends on the phase of their cell cycle. Triple combination of GNPs, phase of tumor cell population, and RT for improved outcomes in cancer treatment.
Methods:
We used a double-thymidine block method for synchronization of the tumor cell population. GNPs of diameters 17 and 46 nm were used to capture the size dependent effects. A radiation dose of 2 Gy with 6 MV linear accelerator was used to assess the efficacy of this proposed combined treatment. A triple negative breast cancer cell line, MDA-MB-231 was chosen as the model cell line. Monte Carlo (MC) calculations were done to predict the GNP-mediated cell death using the experimental GNP uptake data.
Results:
There was a 1.5- and 2- fold increase in uptake of 17 and 46 nm GNPs in the synchronized cell population, respectively. A radiation dose of 2 Gy with clinically relevant 6 MV photons resulted in a 62 and 38 % enhancement in cell death in the synchronized cell population with the incorporation of 17 and 46 nm GNPs, respectively. MC data supported the experimental data, but to a lesser extent.
Conclusion:
A triple combination of GNPs, cell cycle synchronization, and RT could pave the way to enhance the local radiation dose while minimizing side effects to the surrounding healthy tissue.
Advances in knowledge:
This is the first study to show that the combined use of GNPs, phase of tumor cell population, and RT could enhance tumor cell death.
Introduction
Nanotechnology has been at the forefront of many biomedical applications. Among other applications, cancer nanotechnology is expected to generate innovations and play a critical role in future cancer therapeutics. Nanoparticle (NP)-based targeted therapeutics has emerged as a promising alternative to conventional treatment approaches and could overcome major issues such as nonspecific distribution and tumor resistance. Among other NP systems, gold nanoparticles (GNPs) have been tested successfully as radiosensitizers in both in vivo and in vitro studies.1–7 The use of GNPs seems more promising in comparison to earlier attempts using iodine since gold has a higher atomic number than iodine and has favorable biocompatibility. During radiation therapy, the presence of GNPs would enhance the cross-sections of low energy electrons and other cell damaging species (such as free radicals which could damage DNA).8–10
Hainfeld -et al. used small GNPs with a diameter of 1.9 nm and demonstrated that EMT-6 mammary tumors implanted in mice, which had received an intravenous injection of 1.35 g GNPs /kg mouse, could be completely eradicated in 30 days following irradiation with 250 kVp X rays.11 However, the GNP concentration used in this study was very high and it is important to reduce the NP concentration necessary for future clinical applications. As a step forward in this direction, it has been demonstrated that the size and the surface properties of GNPs can be tailored to achieve similar radiation dose enhancement effects with low concentrations of GNPs in vitro.12 For example, it was shown that the NPs of diameter 50 nm has a higher uptake and radiation dose enhancement among the size range from 10 to 100 nm.12,13 A recent study showed that both uptake and radiation dose enhancement due to very small GNPs (diameter 5 nm) could be improved by incorporating them in a lipid NP system.14 NPs are mostly internalized via receptor mediated endocytosis process and get trapped in either endosomes or lysosomes before they get excreted from the cell as illustrated in Figure 1a.15,16 A cross-sectional transmission electron microscopy (TEM) image in Figure 1b shows that these NPs are localized within the cytoplasm mostly closer to the perinuclear region (Supplementary Material 1).
Figure 1.
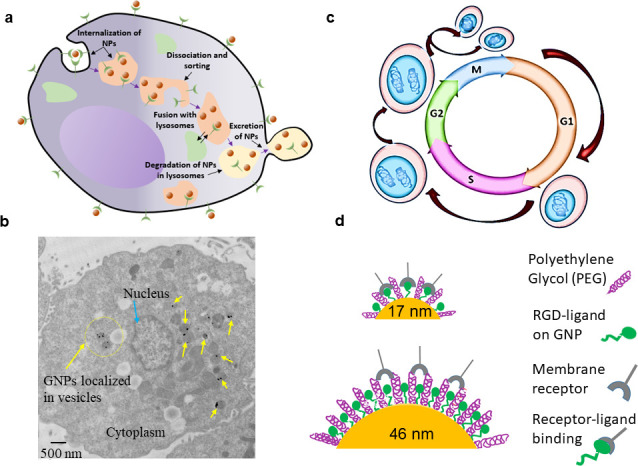
Incorporation of both the size of GNPs and phase of the tumor cell population into current GNP-mediated radiotherapy protocols for further improvement in therapeutic results. (a) Path of GNPs within cells where NPs get trapped in small vesicles within the cytoplasm before their excretion from the cell. (b) Cross-sectional image of a cell showing the localized NPs within small vesicles closer to the perinuclear region. (c) Different phases of a regular cell cycle which include G1, S, G2, and M. (d) Variation of receptor-ligand interaction based on the surface curvature or the size of NPs.
Most of the GNP uptake and resulting radiation dose enhancement studies have been conducted so far using an unsynchronized cell population where cells were in different phases of their cell cycle. However, a recent study has shown that synchronization of tumor cell population could lead to differences in uptake and radiation dose enhancement due to GNPs.14 A eukaryotic cell cycle can be divided into four major phases; G1, S, G2, and M (Figure 1c). The genetic information is duplicated during synthesis phase (S) and the cell divides into two daughter cells during mitosis phase (M). S and M phases are separated by gap phases, G1 and G2. The cell cycle starts with the G1 phase and the size of the cell increases its size during this phase. At the end the cell divides into two daughter cells during the M phase. Cells that have temporarily stopped dividing enter the G0 phase. The progression through the cell cycle is well regulated.17 It has been shown that the cell cycle phase plays a major role in a cell's relative radiosensitivity, with cells being the most radiosensitive in the G2-M phase, less sensitive in the G1 phase, and the least sensitive during the late S phase.18 Hence, one of the major goals of this study is to find the difference in GNP uptake and radiation dose enhancement in a regular (unsynchronized) cell population vs a synchronized cell population. We used thymidine to synchronize the tumor cell population since it is a clinically available pharmacological agent.14,19 In addition, we also considered the size of GNPs in our study as discussed next.
The size of the GNP can play a significant role during the receptor-ligand interaction when they are functionalized with different molecules as illustrated in Figure 1d. In order to include the size variation, we used GNPs of 17 and 46 nm diameter. NPs of this size domain have a higher potential to enter cells and penetrate through the tumor matrix as compared to their smaller or larger counterparts.20 Both of these NP systems were functionalized with polyethylene glycol (PEG) considering their use in future clinical studies.21 In addition, a peptide containing integrin binding domain, RGD, was added as a driving force for internalization of PEGylated GNPs within tumor cells (Figure 1d).22
In summary, this is the first time that the size dependent radiation dose enhancement due to GNPs is compared in a synchronized vs a regular tumor cell population with smaller and larger GNPs along with a clinically approved pharmacologial agent, thymidine. To better understand the underlying mechanism, we further compared our experimental data with theoretical predictions based on Monte Carlo simulations (MC) using TOPAS-nBio.23–25 Hence, our study will lay the foundation to consider not only the size of the GNPs but also the phase of the tumor cell population when designing further investigations to acclerate the use of GNPs in future clinical trials.
methods and Materials
Synthesis and surface modification of gold nanoparticles
The citrate reduction method was used to synthesize GNPs of diameters 17 and 46 nm. 300 µL of 1% HAuCl4•3H2O was added to 30 mL of distilled water and was brought to a boil while continuously stirring. At boiling point, 600 and 300 µL of 1% anhydrous citric acid was added to synthesize GNPs of diameter 17 and 46 nm, respectively. A 1% PEG solution was prepared with thiol-terminated PEG methyl ether of molecular weight 2000 Da. The solution was added to GNP solutions to achieve a grafting density of 1 PEG molecule per nm2. Following PEGylation of GNPs, the peptide sequence containing integrin binding domain, RGD (CKKKKKKGGRGDMFG (RGD-peptide)), was added to solutions to ensure that the number of RGD peptides matched that of the PEG molecules. The molecular weight of the RGD-peptide is 1760 Da.
Synchronization of cells
Breast cancer cells (MDA-MB-231) were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) with 10% Fetal Bovine Serum (FBS) in 6-well tissue culture dishes at 40% confluency (1.3 × 105 cells). The doubling time of the cells was about 37 h.26 We used the double thymidine blocking method for synchronization of the tumor cell population.14 Thymidine inhibits DNA synthesis and arrests cells in S phase. In summary, cells were first washed with Phosphate Buffered Saline (PBS) followed by adding 3 mL of 2 mM thymidine in DMEM/10% FBS/1% penicillin/streptomycin (PS). Cells were washed twice with PBS and regular DMEM after 26 h of incubation with thymidine. The second thymidine block was done using 2 mM thymidine DMEM/10% FBS/1% PS for 24 h.
Preparation of cells for cell cycle analysis
We used propidium iodine (PI) staining to verify the cell cycle.14 The cells were collected in buffer (PBS + 2% FBS) at required time points and washed with PBS twice followed by fixation through re-suspension in 1% paraformeldahyde (PFA)/PBS on ice for 15 mins. After fixation, cells were washed with PBS and re-suspended in a solution containing 0.6 mL PBS and 1.4 mL freezer cold 100% ethanol. The cell were kept in the dark at 4°C for an hour for further dehydration. Cell were then centrifuged at 20°C and the cell pellet was re-suspended in 1 ml PBS/BSA (0.5% BSA-Bovine Serum Albumin). To permeabilize the cell membrane and degrade RNA, the cells were centrifuged at 20°C and the pellet was re-suspended in PBTB (PBS, 0.5% BSA, 0.1% Triton-X 100) followed by addition of RNaseA (100 µg mL−1). The tubes containing cell samples were placed on a shaker at 37°C for 25 min. For quantifying DNA, PI was added (1:100 from 1 mg mL−1) to the tubes and placed on a shaker at 4°C for at least 1 h. As the final step, the cells were centrifuged at 20°C and re-suspended in 1 mL of PBS/BSA. Before running the samples on a BD FACScalibur flow cytometer, the stained cell samples were filtered through a 50 µm cell strainer. Propidium iodide is highly fluorescent at 488 nm with broad emission centered around 600 nm.
Cellular uptake study
Synchronized cell populations were obtained first using the double thymidine blocking method.14 For the NP uptake study, synchronized and unsynchronized MDA-MB-231 cell cultures were incubated with GNPs at a concentration of 0.2 nM for 20 hours. Following NP incubation, cells were washed three times with PBS to remove GNPs not internalized within cells. The cells were then harvested into a single cell suspension using Trypsin 0.25% (HyClone), and the concentration of cells was determined using a Beckman Coulter Z2 Particle Counter and Size Analyzer. For quantification of NP uptake, a known number of cells was processed using Aqua Regia (3:1 ratio of HCl:HNO3) in an oil bath at 200°C until the solution became colourless. The samples were diluted and concentrations of gold (Au) atoms were measured using an Inductively Coupled Plasmon – Mass Spectrometer (ICP-MS) instrument (Thermo X-Series II (X7) quadrupole).
For imaging studies, the cells grown on coverslips were rinsed three times with PBS after NP incubation and fixed with 4% PFA in PBS for 10 min at room temperature followed by washing with PBS. Coverslips were mounted onto glass slides and were dried overnight for microscopy.
Radiation treatment
Synchronized breast cancer cell populations were obtained using the double thymidine blocking method as discussed previously. The synchronized and unsynchronized cells treated without (control) and with GNPs (0.2 nM concentration for 20 h) were given a 2 Gy radiation dose with 6 MeV photons using a linear accelerator (Elekta Oncology Systems, Stockholm, Sweden). The 2 Gy dose was delivered at a dose rate of 600 MU/min (or 6 Gy/min). A radiation dose of 2 Gy was chosen since it is one of most commonly used fractionated dose in the clinic. After the radiation treatment, clonogenic assay and DNA double strand breaks (DSBs) assay were performed as discussed next to assess the effectiveness of the radiation treatment.
Survival fractions of irradiated cells (clonogenic assay)
Clonogenic assay was performed after the radiation treatment as we have done previously.12 The cells were washed with PBS three times followed by addition of trypsin for removal of the cells from the petri dish bottom. After incubation with trypsin for five minutes, the cells were diluted to form a single-cell suspension followed by counting for seeding in 60 mm tissue culture dishes. From the non-irradiated cell suspension, 200 cells were plated while from the cell suspension irradiated with 2 Gy, 400 cells were plated. The number of cells seeded for the irradiated sample was higher since their survival was less. It is also important to have at least 50 colonies at the end of the experiment. The number of cells seeded was smaller for the control (non-irradiated) to avoid overlapping of colonies since their survival was higher. The cells were kept in the 37°C humidified incubator with 5% CO2 for 14 days for colony formation. Colonies were stained and fixed with 0.1% of methylene blue (BioShop) in 70% ethyl alcohol.12 The control dishes (with no treatment) were counted first to determine the plating efficiency (PE).14 The plating efficiency (PE) was obtained although the following formula:
The survival farction for the treated cells were obtained using the following formula14 :
Immunofluorescence assay
Coverslips were placed in 6-well dishes and cells were plated to about 70% confluency.12 Once the cell were adhered to coverslips, NPs were introduced at 0.2 nM concentration and incubated for 20 h. A radiation dose of 2 Gy was given using 6 MV linear accelerator after the NP incubation period. The DNA DSBs were probed 24 h after the radiation treatment since we wanted to account for the permanent damage to DNA. Most of the damage to DNA is repaired in less than 24 h.12 Hence, after 24 h of the radiation treatment, cells were fixed with 2% PFA/0.2% Triton-X 100 for 20 min at room temperature (Sigma-Aldrich) followed by washing with PBS three times. Cells were then treated with 0.1% Triton-X for 20 min at room temperature followed by washing with PBS three times.12 Before incubation with primary antibodies, cells were treated with 3% BSA (Bovine Serum Albumin) for one hour. The coverslips were placed upside down on a 50 µL drop of γH2AX primary antibody (dilution factor: 1:800; Ser 139. Millipore 05–363 Lot 2276332) overnight at 4°C followed by washing with 0.5% BSA/0.175% Tween 20/PBS three times. Similar to primary antibody incubation, the fluorescently labelled secondary antibody (dilution factor: 1:500; anti mouse IgG Alexa 647. Life Technologies Ref A31571, Lot423849) was done for 45 min. After the incubation with secondary antibody, coverslips were washed with 0.5% BSA/0.175% Tween 20/PBS three times followed by washing with PBS three times. For imaging studies, coverslips were mounted on microscope glass.
Theoretical estimation of dose enhancement using Monte Carlo simulations
Monte Carlo (MC) simulations was used to determine the GNP-mediated local dose enhancement and the expected decrease in cell survival.27,28 In summary, radial dose distributions of secondary electrons resulting from single GNPs when they were irradiated by a 6 MV photons was calculated. We used a 6 MV spectrum from a Varian TrueBeam as particle source for simulations in the TOPAS-nBio MC system version 3.0.1.14,23,29 The modified local effect model (LEM) was used to determine the extent of GNP-mediated lethal damage within a cell nucleus based on the size, number of GNPs present, and their distribution within the cell.30 The geometry of the MDA-MB-231 cells was chosen as a 2 µm thick ellipsoid with minor/major axis diameters of 8.5/18.5 µm.31 In order to calculate survival factions, we used α = 0.019 and β = 0.052 as our linear quadratic model parameters.6
Results and discussion
Characterization of GNP complexes
We used GNPs of diameter 17 and 46 nm for this study. Both types of NPs were functionalized with PEG and a peptide containing integrin binding domain, RGD (RGD-peptide). The core diameter and surface functionalization of NPs were verified using multiple techniques (Figure 2 and Supplementary Material 1). The core diameter of GNPs was characterized using Scanning Electron Microscopy (SEM) (Figure 2a,b). The surface plasmon absorption of GNPs was measured using Ultraviolet-Visible (UV-Vis) spectroscopy to further justify the core size, homogeneity, and functionalization. As shown in Figure 2c, Supplementary Material 1, the UV-Vis peak wavelength of as-made 17 nm and 46 GNPs was at 520 and 532 nm32,33 and it was red shifted to 523 and 535 nm after functionalization with PEG and RGD peptide, respectively. The ζ potential of 17 nm GNPs was changed from −50.50 to 4.60 mV after functionalization with both PEG and RGD while it was shifted from −36.30 to 8.0 mV for 46 nm GNPs (Figure 2c, Supplementary Material 1). Since our NPs were not optically labeled, we used Cytoviva’s enhanced dark-field and Hyper-Spectral Imaging (HSI) to optically observe them. This technique allows spectral mapping of NPs within the image plane. For example, Figure 2d,e show the dark field images of 17 and 46 nm GNPs while Figure 2f,g show the reflectance spectra collected from HSI images corresponding to NPs of 17 and 46 nm, respectively. The two spectra collected from the background of Figure 2d is almost flat, as shown in Figure 2f, while reflectance spectra from GNPs shows a broader peak within a range of 500 to 700 nm. We used the same technique- to visualize GNPs localized within cells as discussed later in this paper.
Figure 2.
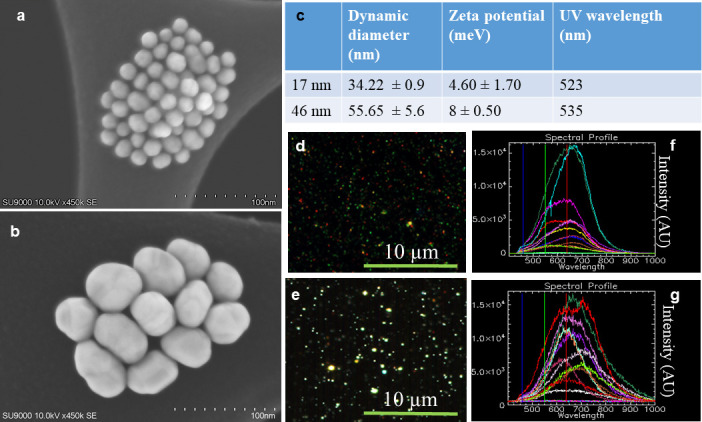
Characterization of gold nanoparticles (GNPs). (a-b) SEM images of GNPs of diameter 17 and 46 nm, respectively. (c) Hydrodynamic diameter, ζ potential, and UV peak wavelength of polyethylene glycol and RGD peptide modified GNPs. (d-e) Dark-field and hyperspectral imaging spectra of 17 and 46 nm GNPs, respectively. (f-g) Reflectance spectra collected bright spots the HSI images in d & e, respectively.
Cellular uptake of GNPs in synchronized cell populations
A cell spends almost half of its life time in the G1 phase as illustrated in the schematic Figure 1d, which is one of the reasons a higher fraction of cells in G1 phase is seen. The distribution of the cell cycle phase in an unsynchronized cell population is given in Figure 3a. Most of the GNP-mediated uptake and radiation dose enhancement studies are done so far using an unsynchronized tumour cell population. In order to first understand how uptake can vary in a synchronized tumor cell population, we tested both serum starvation and double thymidine block method for synchronization of the tumor cell population.34,35 As illustrated in Figure 3b, the thymidine block method was more effective in synchronizing the majority of the tumor cell population and we proceeded with that method for our uptake and radiation dose enhancement studies. In addition, the thymidine block method is also clinically feasible since thymidine is an approved pharmacological agent.19 Thymidine is a DNA synthesis inhibitor that can arrest cells at the G1/S boundary, prior to DNA replication (Figure 3b).
Figure 3.
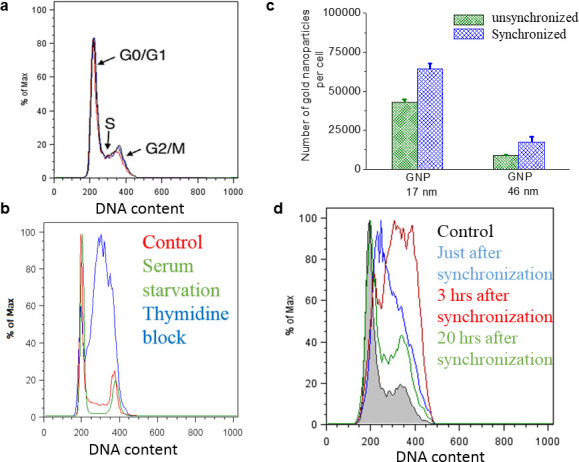
Comparison of cellular uptake of GNPs in a synchronized vs an unsynchronized cell population. (A) Distribution of the cell cycle Phase in a control (unsynchronized) population. (B) Comparison of two cell synchronization methods: thymidine blocking and serum starvation. (C) Cellular uptake of 17 and 46 nm GNPs in a synchronized vs an unsynchronized cell population. (D) Dynamics of the cell cycle phase after synchronization using thymidine blocking method.
Our ultimate goal was to develop a protocol that can be transferable for future in vivo work. There was a 1.5- and 2-fold increase in uptake of 17 and 46 nm GNPs in synchronized vs unsynchrinized cell populations for MDA-MB-231 (Figure 3c). GNPs used for this study were functionalized with PEG and RGD peptide as explained in the previous section. The higher uptake of 17 nm GNPs vs 46 nm in both synchronized and unsynchronized cell populations could be explained by the variation in receptor ligand interaction based on their surface curvatures as illustrated in Figure 1d. For example, the higher surface curvature of smaller NP exposed the RGD peptide on the GNP surface to the surface receptors on the cell membrane better while the lower surface curvature of larger NP allowed PEG to screen the RGD peptide, considering that the PEG molecule is slightly larger than the RGD peptide.22 We imaged the distribution of GNPs within cells using darkfield and hyperspectral imaging technology. Figure 4a,b shows the GNP distribution and their spectral information in an unsynchronized tumor cell population while Figure 4c,d show similar information for a synchronized tumour cell population. There was an apparent increase in GNP clusters in synchronized cells further supporting our quantitative data in Figure 3a (also see Supplementary Material 1). In order to understand the increase in uptake in the synchronized cell population for both NP sizes, we looked at the variation in cell cycle as a fuction of time (Figure 3d). Originally cells were mostly populated in S phase just after synchronization and the population was shifted towards G2/M within 3 h. The cells in G2/M phase have the longest exposure time to NPs since their initial division as compared to cells in either S or G1 phase. At the 20 h time point, when cells were collected to determine the variation of uptake in synchronized vs unsynchronized, there was a higher population of cells still in G2/M phase as compared to the unsynchronized (control) population. Hence, the increase in GNP uptake could be due the the presence of more cells in G2/M which had more time to take up NPs before their divison. We also tested this result in a head and neck cancer cell line, CAL-33, with a similar cell doubling time (see Supplementary Material 1). A similar increase in uptake was seen in the synchronized tumor cell population.
Figure 4.

Qualitative comparison of cellular uptake of 17 nm diameter GNPs in a synchronized vs an unsynchronized cell population. a-b & c-d) Dark-field image and corresponding reflectance spectra collected from few GNPs clusters within a synchronized vs an unsynchronized MDA-MD-231 cells, respectively.
Size dependent radiation dose enhancement due to GNPs in a synchronized vs an unsynchronized cell population
One of the major problems in radiation therapy is the toxicity to the surrounding healthy tissue. In order to enhance the local radiation dose without causing much damage to surrounding healthy tissue, high-Z material systems such as GNPs are being incorporated into current radiation therapy protocols. This involves designing GNP complexes suitable for optimum delivery at both in vivo and in vitro levels.20,26,36 The interactions of radiation with GNPs causing dose enhancement are Compton scattering, photoelectric effect, and Auger cascades (Supplementary Material 1). The electrons produced in these processes can interact with water molecules and create reactive chemical species, such as free radicals,37 that can damage DNA within the cell nucleus.38,39
We measured the effect of GNP-mediated radiation dose enhancement in a synchronized vs an unsynchronized cell population using clonogenic assay and DNA double strand breaks (DSBs) assay. As illustrated in Figures 3c, 5a, the presence of higher number of 17 nm GNPs within cells did not lead to higher sensitization in the unsynchronized tumor cell population. Moreover, 46 nm GNPs had a higher sensitization even though there was a smaller number of them within the cell. This clearly shows radiation dose enhancement is dependent on both the size and the number of GNPs present. For example, the synchronization of tumor cell populations led to a more significant increase in uptake of 17 nm GNPs as compared to 46 nm GNPs which resulted in a significantly higher radiation dose enhancement for 17 nm GNPs after delivering 2 Gy of radiation dose with a 6 MV linear accelerator (Figure 6a). There was a 61.5 and 31% increase in cell death in the synchronized cell population for 17 and 46 nm GNPs, respectively.
Figure 5. .
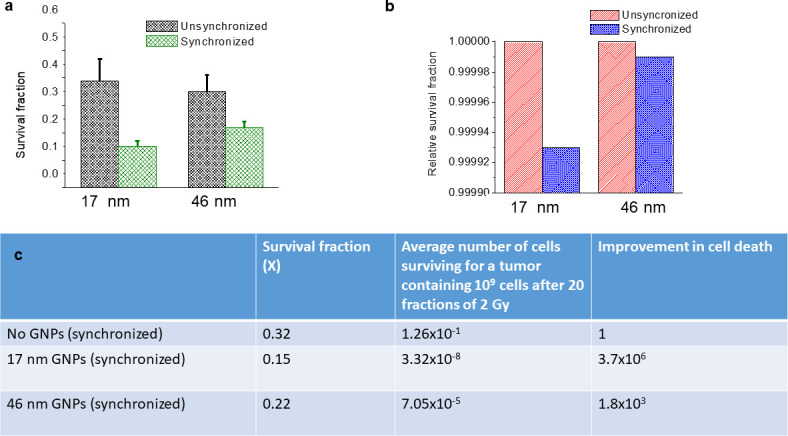
GNP-mediated RT was size and cell cycle dependent. (a-b) Comparison of survival fractions derived from experiment and theory for 17 and 46 nm GNPs in synchronized vs unsynchronized cells. (c) A prediction for a clinically used protocol based on the experimental data in (a). The adopted protocol was 20 fractions of 2 Gy with 6 MV photons.
Figure 6. .

Triple combination of RT, GNPs, and cell cycle synchronization. (a) Quantitative data showing the enhancement in cell death in synchronized tumor cell population. Inset figure shows the linear accelerator set up used for the study. (b) Quantitative data showing that there is no significant increase in DNA DSBs in synchronized tumor cell population after the radiation treatment. (c) Optical images showing that there was no apparent increase in DNA DSBs in cells treated with 17 (top panel) and 46 nm (bottom panel) GNPs. Nuclei and DNA DSBs are marked in blue and red, respectively.
It is shown previously that maximum radiosensitivity is usually observed in the G2-M phase of the cell cycle.40,41 Our cell cycle analysis data in Figure 3d demonstrates that our synchronized population had a higher percent of cells still in the G2-M phase at the time of radiation treatment. According to the Figure 6b, the synchronized cell population with no NPs had a sensitization effect as a result of the fact that a higher population of cells were at the G2-M at the time of irradiation. Our study demonstrates that both the size of the NPs and the phase of the tumor cell population can play a big role in future radiation therapy. However, there was no significant increase in DNA DSBs in synchronized vs unsynchronized tumor cells after the radiation treatment (Figure 6b,c). Therefore, we believe that further enhancement in cell death in synchronized tumor cell population could be due to other mechanisms of cell death which includes damage to mitochondria.42 We will pursue these studies in the near future.
Monte Carlo simulation of the experimental data
Monte Carlo (MC) simulations were also performed to estimate the reduction in cell survival in a synchronized vs an unsynchronized cell population using different cellular distributions of GNPs (Figure 7a). MC simulation studies have shown that microscopic dose enhancement for keV photons can be increased by a factor of up to 3000 as compared to doses originating from a hypothetical water NP at distances of approximately 10 µm.43,44 The dominant interaction with GNPs for MeV photon beams is through Compton scattering with a cross-section three orders of magnitude lower than the photoelectric cross-section for keV photons. However, MeV photons still contain an increasing percent of keV photons and electrons with increasing treatment depth. We used the experimental uptake data in Figure 3b for MC simulations, i.e. 42,811 and 8,700 GNPs of 17 and 46 nm in unsynchronized vs 64,215 and 17, 400 GNPs of 17 and 46 nm in the synchronized cell population, respectively. A schematic representation of the cell and GNP geometries used for our MC calculations is shown in Figure 7a. For example, individual GNPs or vesicle (an average of 8 GNPs per vesicle) distributions were randomly selected either uniformly within the cytoplasm or using an exponentially decreasing likelihood with distance from the nucleus. Our optical and TEM images of cells suggest that most of the vesicles containing GNPs are closer to the nucleus perinuclear region of the cells (Figure 7b).
Figure 7. .
Monte-Carlo modelling of radiation dose enhancement. (a) Schematic representation of the four geometric scenarios simulated, a 8 µm diameter nucleus at the center of an oval representation of a MDA-MB-231 cell (axes 11.5 µm and 15.5 µm) with different distributions of GNPs: (i) GNPs freely distributed uniformly within the cytoplasm, (ii) a likelihood distribution exponentially decreasing with distance from the nucleus, (iii) GNPs constrained to vesicles and distributed uniformly within the cytoplasm, and (iv) GNPs constrained to vesicles and the vesicles are closer to the nucleus. (b) TEM image showing that GNPs are localized in vesicles closer to the nucleus. (c-d) Survival fractions after a radiation dose of 2 Gy fraction with 6 MV photons from a Varian Linac for 17 and 46 nm GNPs, respectively.
Radial dose distributions were superimposed to calculate radiation dose to the nucleus and predict cell survival fractions. Our calculations showed a lower survival fraction when GNPs were closer to the nucleus (see configurations (ii) and (iv) in Figure 7a) as expected. In agreement with experimental data shown in Figures 5a, 17 nm GNPs were more effective in causing more radiation damage as compared to 46 nm ones. However, the experimental survival fractions are much lower than the ones predicted by MC calculations. This could be due to the fact that some biological mechanisms were not yet incorporated into the MC simulations which includes sensitization effects due to synchronization of cell cycle. In addition, the simulations were purely based on localized physical dose enhancement ignoring the potentially dominant effects GNPs have in the generation of reactive oxygen species and other cell stressors. Overall, there is still a large gap between the simulated (physics) events and the biological outcome. However, we find that the trends of our simulations are consistent with the experiments suggesting that the biological effects observed are at least in part caused by the physical dose enhancement as simulated.
Conclusion
The size of GNPs and phase of the tumor cell population play a very important role in their uptake and resulting radiation dose enhancement properties. There was a 1.5 and 2.0 fold increase in uptake of 17 and 46 nm GNPs in synchronized tumor cell population, respectively. Similarly, there was a 61.5 and 31% increase in cell death in the synchronized cell population for 17 and 46 nm GNPs, respectively (Figure 5a). Our MC calculations showed a similar trend based on the increase in number of GNPs present in synchronized cells (Figure 5b). Most importantly, we have demonstrated this GNP-mediated radiation dose enhancement using a clinically relevant fractionated radiation dose of 2 Gy and 6 MV photon beam.
It is generally recognized that in vitro data cannot be extrapolated directly to in vivo or clinical settings since in vitro assays do not account for tumour microenvironmental factors and the fact that tumours may contain clonogenic subpopulations of cells with different sensitivity to the radiation of interest.45 However, we used a model calculation introduced by Hill and Bristow45 to calculate the predicted dose enhancement for a treatment with n multiple fractions based on the survival fraction data for a single fraction (X). The model calculation assumes that each treatment generates a survival fraction value X which is equally effective throughout the fractionated treatment. According to the model calculation, the survival following n treatments is given by . Since the limit of clinical detection of most solid tumours is approximately 1 gram of tissue (corresponding to approximately 109 cells), we used that number along with the model calculation to determine the final number of cells that survived after 20 fractions as illustrated in Figure 5c. For example, over the course of radiation treatment, GNPs of size 17 nm are 1000-fold better as a radiosensitizer than 46 nm GNPs in a synchronized tumor cell population. The principle significance comes from the fact that a small difference in survival can translate into large differences over multiple treatment fractions, which can lead to a significant difference in tumour control probability.
Finally, we believe that introduction of GNPs into cell-cycle-controlled tumors would generate significant improvement in radiation therapy. Considering that thymidine is a clinically approved agent, the biocompatibility of GNPs could accelerate the translation of such applications to the clinical context in the near future. For example, a few clinical trials are being actively carried out for the approval of gold nanoparticles for cancer diagnostics and therapy while one phase one clinical trial has been successfully completed.3,21,46 Hence, GNP-mediated cancer nanotechnology will have a significant impact in enhancing local radiation dose while minimizing damage to the surrounding tissue.
Footnotes
Acknowledgment: Arthur Blackburn, PhD for his assistance in imaging. The authors would like acknowledge Canada Foundation for Innovation (CFI), Natural Sciences and Engineering Research Council of Canada (NSERC) and University of Victoria for their financial support. KR was supported by NSERC graduate scholarship. We also acknowledge support from the NIH/NCI [grant no. R01CA187003 (‘‘TOPAS-nBio: a Monte Carlo tool for radiation biology research’’) to JS.
Contributor Information
Kristy Rieck, Email: krieck@uvic.ca.
Kyle Bromma, Email: kbromma@uvic.ca.
Wonmo Sung, Email: WSUNG1@mgh.harvard.edu.
Aaron Bannister, Email: gargoyle@uvic.ca.
Jan Schuemann, Email: JSCHUEMANN@mgh.harvard.edu.
Devika Basnagge Chithrani, Email: devikac@uvic.ca.
REFERENCES
- 1. Sztandera K , Gorzkiewicz M , Klajnert-Maculewicz B . Gold nanoparticles in cancer treatment . Mol Pharm 2019. ; 16 : 1 – 23 . doi: 10.1021/acs.molpharmaceut.8b00810 [DOI] [PubMed] [Google Scholar]
- 2. Jelveh S , Chithrani DB . Gold nanostructures as a platform for combinational therapy in future cancer therapeutics . Cancers 2011. ; 3 : 1081 – 110 . doi: 10.3390/cancers3011081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schuemann J , Berbeco R , Chithrani DB , Cho SH , Kumar R , McMahon SJ , et al. . Roadmap to clinical use of gold nanoparticles for radiation sensitization . Int J Radiat Oncol Biol Phys 2016. ; 94 : 189 – 205 . doi: 10.1016/j.ijrobp.2015.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Le Goas M , Paquirissamy A , Gargouri D , Fadda G , Testard F , Aymes-Chodur C , et al. . Irradiation effects on polymer-grafted gold nanoparticles for cancer therapy . ACS Applied Bio Materials 2019. ; 2 : 144 – 54 . doi: 10.1021/acsabm.8b00484 [DOI] [PubMed] [Google Scholar]
- 5. Cabello G , Nwoko KC , Mingarelli M , McLaughlin AC , Trembleau L , Feldmann J , et al. . Physicochemical tools: toward a detailed understanding of the architecture of targeted radiotherapy nanoparticles . ACS Applied Bio Materials 2018. ; 1 : 1639 – 46 . doi: 10.1021/acsabm.8b00476 [DOI] [PubMed] [Google Scholar]
- 6. Jain S , Coulter JA , Hounsell AR , Butterworth KT , McMahon SJ , Hyland WB , et al. . Cell-specific radiosensitization by gold nanoparticles at megavoltage radiation energies . Int J Radiat Oncol Biol Phys 2011. ; 79 : 531 – 9 . doi: 10.1016/j.ijrobp.2010.08.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Khoo AM , Cho SH , Reynoso FJ , Aliru M , Aziz K , Bodd M , et al. . Radiosensitization of prostate cancers in vitro and in vivo to erbium-filtered orthovoltage x-rays using actively targeted gold nanoparticles . Sci Rep 2017. ; 7 : 18044 . doi: 10.1038/s41598-017-18304-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carter JD , Cheng NN , Qu Y , Suarez GD , Guo T . Nanoscale energy deposition by X-ray absorbing nanostructures . J Phys Chem B 2007. ; 111 : 11622 – 5 . doi: 10.1021/jp075253u [DOI] [PubMed] [Google Scholar]
- 9. Zheng Y , Hunting DJ , Ayotte P , Sanche L . Radiosensitization of DNA by gold nanoparticles irradiated with high-energy electrons . Radiat Res 2008. ; 169 : 19 – 27 . doi: 10.1667/RR1080.1 [DOI] [PubMed] [Google Scholar]
- 10. Butterworth KT , McMahon SJ , Currell FJ , Prise KM . Physical basis and biological mechanisms of gold nanoparticle radiosensitization . Nanoscale 2012. ; 4 : 4830 – 8 . doi: 10.1039/c2nr31227a [DOI] [PubMed] [Google Scholar]
- 11. Hainfeld JF , Slatkin DN , Smilowitz HM . The use of gold nanoparticles to enhance radiotherapy in mice . Phys Med Biol 2004. ; 49 : N309 – N315 . doi: 10.1088/0031-9155/49/18/N03 [DOI] [PubMed] [Google Scholar]
- 12. Chithrani DB , Jelveh S , Jalali F , van Prooijen M , Allen C , Bristow RG , et al. . Gold nanoparticles as radiation sensitizers in cancer therapy . Radiat Res 2010. ; 173 : 719 – 28 . doi: 10.1667/RR1984.1 [DOI] [PubMed] [Google Scholar]
- 13. Chithrani BD , Ghazani AA , Chan WCW . Determining the size and shape dependence of gold nanoparticle uptake into mammalian cells . Nano Lett 2006. ; 6 : 662 – 8 . doi: 10.1021/nl052396o [DOI] [PubMed] [Google Scholar]
- 14. Bromma K , Rieck K , Kulkarni J , O’Sullivan C , Sung W , Cullis P , et al. . Use of a lipid nanoparticle system as a Trojan horse in delivery of gold nanoparticles to human breast cancer cells for improved outcomes in radiation therapy . Cancer Nanotechnology 2019. ; 10 : 1 . doi: 10.1186/s12645-019-0046-z [DOI] [Google Scholar]
- 15. Ng CT , Tang FM , Li JJ , Ong C , Yung LL , Bay BH . Clathrin-mediated endocytosis of gold nanoparticles in vitro. anatomical record . Anat Rec 2015. ; 298 : 418 – 27 . [DOI] [PubMed] [Google Scholar]
- 16. Chithrani DB . Intracellular uptake, transport, and processing of gold nanostructures . Mol Membr Biol 2010. ; 27 : 299 – 311 . doi: 10.3109/09687688.2010.507787 [DOI] [PubMed] [Google Scholar]
- 17. Meyers RA . Encyclopedia of molecular cell biology and molecular medicine . 2nd . Weinheim, Germany: : The British Institute of Radiology. ; 2004. . [Google Scholar]
- 18. Pawlik TM , Keyomarsi K . Role of cell cycle in mediating sensitivity to radiotherapy . Int J Radiat Oncol Biol Phys 2004. ; 59 : 928 – 42 . doi: 10.1016/j.ijrobp.2004.03.005 [DOI] [PubMed] [Google Scholar]
- 19. Heinemann L , Simpson GR , Annels NE , Vile R , Melcher A , Prestwich R , et al. . The effect of cell cycle synchronization on tumor sensitivity to reovirus oncolysis . Mol Ther 2010. ; 18 : 2085 – 93 . doi: 10.1038/mt.2010.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yohan D , Cruje C , Lu X , Chithrani DB . Size-dependent gold nanoparticle interaction at Nano–Micro interface using both monolayer and multilayer (tissue-like) cell models . Nanomicro Lett 2016. ; 8 : 44 – 53 . doi: 10.1007/s40820-015-0060-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Libutti SK , Paciotti GF , Byrnes AA , Alexander HR , Gannon WE , Walker M , et al. . Phase I and pharmacokinetic studies of CYT-6091, a novel PEGylated colloidal gold-rhTNF nanomedicine . Clin Cancer Res 2010. ; 16 : 6139 – 49 . doi: 10.1158/1078-0432.CCR-10-0978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cruje C , Yang C , Uertz J , van Prooijen M , Chithrani BD . Optimization of PEG coated nanoscale gold particles for enhanced radiation therapy . RSC Adv. 2015. ; 5 : 101525 – 32 . doi: 10.1039/C5RA19104A [DOI] [Google Scholar]
- 23. Schuemann J , McNamara AL , Ramos-Mendez J , Perl J , KH;Paganetti H , Incerti, S H , et al. . Topas-nbio: an extension to the TOPAS simulation toolkit for cellular and sub-cellular radiobiology . Radiation research 2019. ; In press . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McNamara AL , Ramos-Méndez J , Perl J , Held K , Dominguez N , Moreno E , et al. . Geometrical structures for radiation biology research as implemented in the TOPAS-nBio toolkit . Phys Med Biol 2018. ; 63 : 175018 . doi: 10.1088/1361-6560/aad8eb [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ramos-Méndez J , Perl J , Schuemann J , McNamara A , Paganetti H , Faddegon B . Monte Carlo simulation of chemistry following radiolysis with TOPAS-nBio . Phys Med Biol 2018. ; 63 : 105014 . doi: 10.1088/1361-6560/aac04c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yohan D , Cruje C , Lu X , Chithrani D . Elucidating the uptake and distribution of nanoparticles in solid tumors via a multilayered cell culture model . Nanomicro Lett 2015. ; 7 : 127 – 37 . doi: 10.1007/s40820-014-0025-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lin Y , McMahon SJ , Paganetti H , Schuemann J , Yuting L , Stephen JM , Harald P , Jan S . Biological modeling of gold nanoparticle enhanced radiotherapy for proton therapy . Phys Med Biol 2015. ; 60 : 4149 . doi: 10.1088/0031-9155/60/10/4149 [DOI] [PubMed] [Google Scholar]
- 28. Sung W , Ye S-J , McNamara AL , McMahon SJ , Hainfeld J , Shin J , et al. . Dependence of gold nanoparticle radiosensitization on cell geometry . Nanoscale 2017. ; 9 : 5843 – 53 . doi: 10.1039/C7NR01024A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McNamara AL , Ramos-Méndez J , Perl J , Held K , Dominguez N , Moreno E , et al. . Geometrical structures for radiation biology research as implemented in the TOPAS-nBio toolkit . Phys Med Biol 2018. ; 63 : 175018 . doi: 10.1088/1361-6560/aad8eb [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lin Y , McMahon SJ , Paganetti H , Schuemann J . Biological modeling of gold nanoparticle enhanced radiotherapy for proton therapy . Phys Med Biol 2015. ; 60 : 4149 – 68 . doi: 10.1088/0031-9155/60/10/4149 [DOI] [PubMed] [Google Scholar]
- 31. Liu Z , Lee Y , Jang Jhee , Li Y , Han X , Yokoi K , et al. . Microfluidic cytometric analysis of cancer cell transportability and invasiveness . Sci Rep 2015. ; 5 : 14272 . doi: 10.1038/srep14272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jain PK , Lee KS , El-Sayed IH , El-Sayed MA . Calculated absorption and scattering properties of gold nanoparticles of different size, shape, and composition: applications in biological imaging and biomedicine . J Phys Chem B 2006. ; 110 : 7238 – 48 . doi: 10.1021/jp057170o [DOI] [PubMed] [Google Scholar]
- 33. Link S , El-Sayed MA . Spectral properties and relaxation dynamics of surface plasmon electronic oscillations in gold and silver nanodots and nanorods . J Phys Chem B 1999. ; 103 : 8410 – 26 . doi: 10.1021/jp9917648 [DOI] [Google Scholar]
- 34. Chen G , Deng X . Cell synchronization by double thymidine block . Bio Protoc 2018. ; 8 : e2994 . doi: 10.21769/BioProtoc.2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim JA , Åberg C , Salvati A , Dawson KA . Role of cell cycle on the cellular uptake and dilution of nanoparticles in a cell population . Nature Nanotechnology 2012. ; 7 : 62 – 8 . doi: 10.1038/nnano.2011.191 [DOI] [PubMed] [Google Scholar]
- 36. Yang C , Bromma K , Chithrani D . Peptide mediated in vivo tumor targeting of nanoparticles through optimization in single and multilayer in vitro cell models . Cancers 2018. ; 10 : 84 . doi: 10.3390/cancers10030084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Carter JD , Cheng NN , Qu Y , Suarez GD , Guo T . Nanoscale energy deposition by X-ray absorbing nanostructures . J Phys Chem B 2007. ; 111 : 11622 – 5 . doi: 10.1021/jp075253u [DOI] [PubMed] [Google Scholar]
- 38. Zheng Y , Sanche L . Low energy electrons in nanoscale radiation physics: relationship to radiosensitization and chemoradiation therapy . Reviews in Nanoscience and Nanotechnology 2013. ; 2 : 1 – 28 . doi: 10.1166/rnn.2013.1022 [DOI] [Google Scholar]
- 39. Butterworth KT , McMahon SJ , Taggart LE , Prise KM . Radiosensitization by gold nanoparticles: effective at megavoltage energies and potential role of oxidative stress . Translational Cancer Research 2013. ; 2 : 269 – 79 . [Google Scholar]
- 40. Otani K , Naito Y , Sakaguchi Y , Seo Y , Takahashi Y , Kikuta J , et al. . Cell-cycle-controlled radiation therapy was effective for treating a murine malignant melanoma cell line in vitro and in vivo . Sci Rep 2016. ; 6 : 30689 . doi: 10.1038/srep30689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hennequin C , Favaudon V . Biological basis for chemo-radiotherapy interactions . Eur J Cancer 2002. ; 38 : 223 – 30 . doi: 10.1016/S0959-8049(01)00360-4 [DOI] [PubMed] [Google Scholar]
- 42. Taggart LE , McMahon SJ , Currell FJ , Prise KM , Butterworth KT . The role of mitochondrial function in gold nanoparticle mediated radiosensitisation . Cancer Nanotechnol 2014. ; 5 : 5 . doi: 10.1186/s12645-014-0005-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lin Y , McMahon SJ , Scarpelli M , Paganetti H , Schuemann J . Comparing gold nano-particle enhanced radiotherapy with protons, megavoltage photons and kilovoltage photons: a Monte Carlo simulation . Phys Med Biol 2014. ; 59 : 7675 – 89 . doi: 10.1088/0031-9155/59/24/7675 [DOI] [PubMed] [Google Scholar]
- 44. Jones BL , Krishnan S , Cho SH . Estimation of microscopic dose enhancement factor around gold nanoparticles by Monte Carlo calculations . Med Phys 2010. ; 37 ( 7Part1 ): 3809 – 16 . doi: 10.1118/1.3455703 [DOI] [PubMed] [Google Scholar]
- 45. Hill RPB , Robert B . The scientific basis of radiotherapy . : Tannock R. P , Bristrow R. G , The basic science of oncology, I. F. H . Toronto: : The British Institute of Radiology. ; 2008. . . 289 – 321 . [Google Scholar]
- 46. El-Sayed IH . Nanotechnology in head and neck cancer: the race is on . Curr Oncol Rep 2010. ; 12 : 121 – 8 . doi: 10.1007/s11912-010-0087-2 [DOI] [PMC free article] [PubMed] [Google Scholar]



