Abstract
Low copy repeats (LCRs) are recognized as a significant source of genomic instability, driving genome variability and evolution. The Chromosome 22 LCRs (LCR22s) mediate nonallelic homologous recombination (NAHR) leading to the 22q11 deletion syndrome (22q11DS). However, LCR22s are among the most complex regions in the genome, and their structure remains unresolved. The difficulty in generating accurate maps of LCR22s has also hindered localization of the deletion end points in 22q11DS patients. Using fiber FISH and Bionano optical mapping, we assembled LCR22 alleles in 187 cell lines. Our analysis uncovered an unprecedented level of variation in LCR22s, including LCR22A alleles ranging in size from 250 to 2000 kb. Further, the incidence of various LCR22 alleles varied within different populations. Additionally, the analysis of LCR22s in 22q11DS patients and their parents enabled further refinement of the rearrangement site within LCR22A and -D, which flank the 22q11 deletion. The NAHR site was localized to a 160-kb paralog shared between the LCR22A and -D in seven 22q11DS patients. Thus, we present the most comprehensive map of LCR22 variation to date. This will greatly facilitate the investigation of the role of LCR variation as a driver of 22q11 rearrangements and the phenotypic variability among 22q11DS patients.
Low copy repeats (LCRs), also referred to as segmental duplications, are a driving force in genome evolution, adaptation, and instability. In the diploid human genome, >5% of the reference assembly consists of LCRs (Bailey et al. 2001, 2002; International Human Genome Sequencing Consortium 2004). Duplications have long been recognized as a potential source for the rapid evolution of new genes with novel functions (Bailey et al. 2001; Jiang et al. 2007; Dennis et al. 2017). Recent studies have suggested potential functional roles for genes within LCRs in synaptogenesis, neuronal migration, and neocortical expansion within the human lineage (Charrier et al. 2012; Dennis et al. 2012, 2017; Boyd et al. 2015; Florio et al. 2015). However, these regions are highly enriched for gaps and assembly errors even within the most recent versions of the human reference genome (Bovee et al. 2008; Genovese et al. 2013; Chaisson et al. 2015). This is because LCRs are both highly sequence identical and copy number polymorphic. These features strongly hamper the study of the precise role of LCRs as drivers of human disease or evolution.
High sequence homology between LCR copies is a driver of recurrent genomic rearrangements. Misalignment of homologous chromosomes or sister chromatids can lead to nonallelic homologous recombination (NAHR) (Inoue and Lupski 2002). NAHR between LCRs results in reciprocal deletions, duplications, or inversions, often referred to as genomic disorders (Inoue and Lupski 2002). The 22q11 deletion syndrome (22q11DS) is the most common genomic disorder, with a prevalence of 1 in 3000–6000 live births (McDonald-McGinn et al. 2015). The syndrome has a heterogeneous presentation, often including congenital heart disease, immunodeficiency, palatal anomalies, hypocalcemia, multiple additional congenital anomalies, and psychiatric illness, including 25% with schizophrenia (McDonald-McGinn et al. 2015). Chromosome 22q11.21 contains four LCR22s, often termed, consecutively, LCR22A–D. NAHR occurs most often between LCR22A and LCR22D (89%) or between LCR22A and LCR22B (6%), generating a deletion of, respectively, ∼3 and ∼1.5 Mb (Edelmann et al. 1999). Furthermore, different recurrent translocations between Chromosome 22 and Chromosomes 8, 11, or 17 have been described. The breakpoints of these translocations are located within palindromic AT-rich repeats (PATRRs), with the one on Chromosome 22 localizing to LCR22B (Leach 1994; Lewis et al. 1999; Cunningham et al. 2003; Gotter et al. 2007; Kato et al. 2012).
All LCR22s are composed of different repeat subunits which are present in variable composition, copy number, and orientation (Shaikh et al. 2007). LCR22A and LCR22D are the largest and, in the current genome build, estimated to span 1 Mb and 400 kb, respectively. The size and structure of the LCR22s continues to be variable and inconsistent between various genome assemblies, with the current reference genome still containing sequence gaps within LCR22A. Copy number variations (CNVs) exist within the LCR22s (Guo et al. 2011). In addition, a CNV encompassing PRODH, DGCR6, and DGCR5 (referred to as LCR22A+) was recently mapped within LCR22A (Guo et al. 2018). Nonetheless, the overall architecture of several LCR22s remains unresolved. Because the 22q11DS breakpoints are embedded within these unresolved LCR22s, their exact locations have, despite extensive efforts (Guo et al. 2016), remained elusive. We set out to map these repeats to elucidate the LCR22 structures and their variability and to further refine the 22q11DS rearrangement breakpoint regions.
Results
Subunit-resolution LCR assemblies using fiber FISH
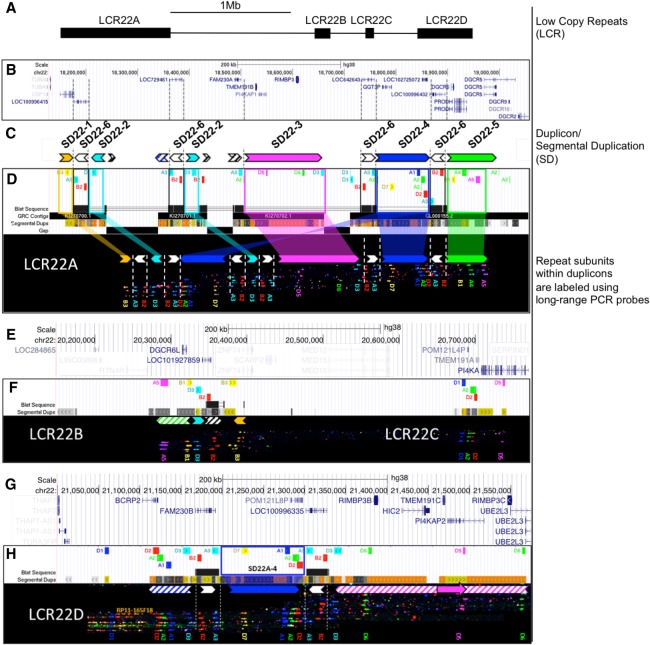
To resolve the LCR22 subunit organization, we first redefined repeat subunits that are present in the LCR22s of human reference genome build 38 (hg38) (Fig. 1A). We aligned the LCR22 sequences to each other, revealing all segments with a sequence similarity >99%. Based on this LCR decomposition, we identified distinct repeat subunits that have a copy number of at least two on Chromosome 22q11.21 (Fig. 1). This resulted in the identification of 20 subunit families, each of which includes a repeat subunit and all of its paralogs on 22q11 (Supplemental Table S1).
Figure 1.
In silico hg38 fiber FISH probe positions compared to duplicon composition of LCR22A. Terminology used to describe individual elements is depicted on the right. (A) Schematic overview of the LCR22s in Chromosome 22q11.21. (B) RefSeq-curated gene set overlapping with the LCR22s. (C) Duplicon decomposition of the hg38 structure of LCR22A. Duplicons were deduced from mapped haplotypes. Filled, colored arrows represent copies of duplicons and hatched arrows represent partial copies of duplicons of the same color. (D) UCSC Genome Browser hg38 reference assembly tracks of Segmental Dups2,24, GRC contigs, gap positions, and fiber FISH probe BLAT positions (white panel). Positions of the latter are aligned with recordings of fiber FISH patterns in LCR22A (black bar). Decomposition of one LCR22A haplotype to duplicons is illustrated using colored (nonwhite) arrows. For the showcased allele, duplicon order centromeric to telomeric is SD22-1, -2, -4, -2, -3, -4, and -5, and the arrow direction represents inverted or direct orientation. Larger duplicons are flanked by copies of SD22-6 (white arrows). Probe identifiers are indicated below the fiber pattern. (E) RefSeq annotated genes overlapping with LCR22B and -C. (F) LCR22B and -C, fiber FISH patterns have the same order and distances as those predicted in hg38 and contain partial duplication of LCR22A duplicons (hatched arrows). (G) RefSeq annotated genes overlapping with LCR22D. (H) All LCR22D molecules present at the same centromeric start, overlapping with predicted hg38 probe positions. The first duplicon displays a partial SD22-4 and SD22-2 (hatched blue arrow), followed by a complete SD22-4 flanked by SD22-6 copies (white arrow). The distal end of LCR22D consists of partial duplications of SD22-3 (hatched magenta arrow). Nested, solid magenta arrow represents probe D5 position variant.
We next used fiber FISH to further resolve the structure of LCR22s and to obtain a more accurate map of these regions than what is available in the reference genome. We designed fluorescent probes for 14 repeat subunits to visualize their order and the distance between them. (Fig. 1; Supplemental Table S2). We used long-range PCR to generate the probes, which were labeled with different colors to obtain distinct signals from adjacent subunits (Fig. 1D, UCSC track). We used BACs flanking each of the LCR22 repeat clusters as probes to anchor them within unique sequence (Supplemental Fig. S1).
We first assayed the LCR22-specific fiber FISH probe pattern on DNA fibers generated from haploid cell line CHM1 and HapMap cell line GM12878. These genomes have been well characterized and were included in the Platinum Genome Project (Eberle et al. 2017). The haploid state of CHM1 significantly reduced mapping complexity of the repeat clusters. We hybridized our custom probe set on fibers of CHM1 and GM12878 and detected more than 100 informative fibers, each >200 kb in size. We then tiled the clustered fibers, enabling the de novo assembly of the subunit order over more than 1 Mb.
We compared the assembled subunit patterns to the in silico determined subunit positions in hg38. Probe patterns of LCR22B and LCR22C were in agreement with those in hg38 for both cell lines (Fig. 1E,F). In contrast, the observed LCR22A and LCR22D patterns diverged from hg38 to different extents (Fig. 1G,H; Supplemental Figs. S2–S4). LCR22D structure was identical in CHM1 and GM12878. This structure mostly matched hg38, except for the position of a single probe, D5, as shown (Supplemental Fig. S2B). The de novo assembled LCR22A allele in CHM1 was larger than the one predicted by hg38 (Supplemental Fig. S3). Similarly, GM12878 also had two distinct LCR22A alleles, both of which were different from the hg38 predicted allele (Supplemental Fig. S4). Based on the distance between probe signals, LCR22A alleles in GM12878 were estimated to be ∼1.20 and ∼0.65 Mb, respectively. The CHM1 allele was also estimated to be ∼1.20 Mb, however, it differed in its composition from the GM12878 allele of the same size.
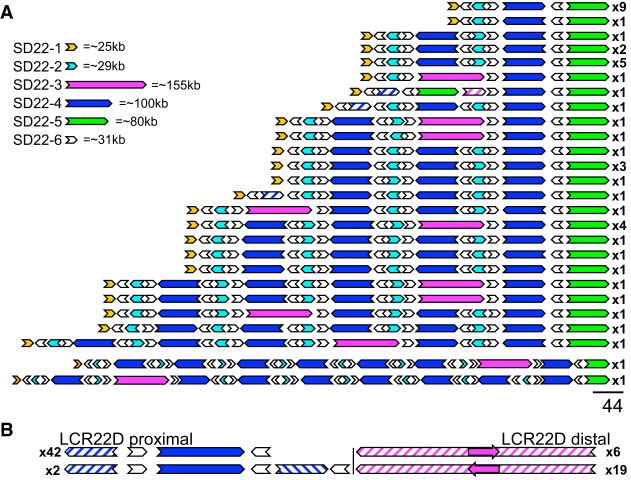
Because the first three observed LCR22A assemblies differed substantially from the reference and from one another, we wondered whether those alleles were exceptional. We assembled fiber patterns in 31 additional cell lines (Supplemental Fig. S5). As observed previously, the LCR22B and LCR22C patterns were identical and in agreement with hg38 in all individuals tested. However, we assembled 41 additional LCR22A alleles that showed a surprising level of variation (Fig. 2A). Based on fiber FISH assembly of a total of 44 LCR22A alleles in 33 samples (Supplemental Fig. S5), we observed 25 distinct haplotypes varying in length from ∼300 kb to >2 Mb (Fig. 2; Supplemental Fig. S6). No individual in this subset was homozygous for the structure of LCR22A. In contrast, LCR22D displayed less variability. We observed three haplotypes for LCR22D (Fig. 2B; Supplemental Fig. S2), with allelic variation including different positions of probe D5 (Supplemental Fig. S2B) and a duplication of probes A1-A2-D2-A3-B2-D3 (Supplemental Fig. S2C).
Figure 2.
Fiber FISH-mapped haplotypes of LCR22A and LCR22D observed in a cohort of 33 cell lines. (A) Twenty-six haplotypes observed for LCR22A. Haplotypes are aligned at the distal unique anchor of LCR22A. (B) Proximal and distal haplotypes observed for LCR22D. Filled, colored arrows represent copies of duplicons, and hatched arrows represent partial copies of duplicons of the same color. Size estimates of individual SD22s are shown (upper left). Frequencies of haplotypes are depicted on the right (i.e., x9, x1, etc.).
LCR22A fiber patterns identify core duplicons
Despite the observed scale of variation within LCR22A, we observed a nonrandom pattern of probe clusters within the mapped haplotypes. We predicted that these probe clusters represent segmental duplications or duplicons within the LCR22s, and the observed differences in LCR22 architecture is driven by the copy number variation of a small set of such duplicons. We visually deduced a minimal set of different probe clusters, which were designated as SD22s and are henceforth referred to as duplicons (Fig. 1).
We identified six probe clusters to define six LCR22 duplicons, designated SD22-1 to SD22-6 (Fig. 1C). All 44 fiber FISH mapped alleles of LCR22A presented a conserved proximal and distal end, represented by SD22-1 and SD22-5, respectively (Fig. 2A). In contrast to the proximal and distal anchors, SD22-2, -3, -4, and -6 were copy number variable among alleles, with SD22-3 being absent in some (Fig. 2A). A majority of the duplicons maintain their structural integrity when comparing various LCR22A alleles. We rarely observed partial copies of any given duplicon, except in three LCR22A haplotypes, which had partial copies of SD22-4 and -2.
This analysis revealed that every LCR22A allele mapped by fiber FISH was composed of SD22-1 to SD22-6 in different orders, copy numbers, and orientations. We never observed a tandem array of any single duplicon as any two subsequent copies of SD22-2, -3, -4, or -5 were always flanked by a paralog of SD22-6. Moreover, the orientation of SD22-6 relative to its surrounding duplicons was conserved. Although, most alleles have a single copy of SD22-1 and -5 at the centromeric and telomeric end of assembled LCR22A haplotypes, respectively, we did occasionally observe copy number variants of SD22-5 (Fig. 2). In a previous study on a cohort of 15,579 normal individuals, Guo et al. (2018) identified a deletion (0.3%) and reciprocal duplication (1.3%) embedded in LCR22A. Of the 33 cell lines we analyzed by fiber FISH, one was from an individual carrying the duplication embedded within LCR22A (Supplemental Fig. S7A, Family 5; Guo et al. 2018). Probe patterns of this individual's LCR22A confirmed a SD22-5 duplication within LCR22A.
Sequence and gene content of LCR22A duplicons
To determine the sequence content of each of the duplicons within LCR22A, we compared the observed probe patterns of the duplicons to the expected positions in the reference genome (Fig. 1B–D). In silico fiber FISH probe patterns of reference contigs KI270701.1, KI270702.1, and the proximal 150 kb of reference contig GL000155.2 individually matched duplicons SD22-2, -3, and -4, respectively (Fig. 1C,D). SD22-2 did not contain any genes, whereas SD22-3 contained TMEM191B, RIMBP3, and PI4KAP1, and SD22-4 contained GGT3PA. Further, SD22-1 contained USP18 and SD22-5 contained PRODH, DGCR5, and DGCR6. SD22-6 corresponded to a ∼31 kb repeat in hg38, which was present five times in the reference LCR22A with sequence similarities of 97% and higher (Fig. 1C,D, BLAT track and white arrows). The SD22-6 hg38 sequence contains paralogs of a lincRNA with sequence similarity to FAM230C. Each of these paralogs contains copies of the translocation breakpoint type A (TBTA, AB261997.1), which consists of an unstable palindromic AT-rich repeat (PATRR). Thus, the different alleles of LCR22A contained a different copy number of the genes and other sequences based on the respective copy number of the duplicons (Supplemental Fig. S8).
Bionano optical mapping confirms fiber FISH assemblies
To evaluate the fiber FISH assemblies with an orthogonal technology, we performed Bionano assays on a total of eight cell lines: the haploid cell line CHM1, GM12878, and two trios, containing 22q11DS patients and their parents. Because a certain degree of paralogous variation between segmental duplications is missed by fiber FISH, we expected some mismatch when comparing individual label sites (Bailey et al. 2002). However, other than this expected paralogous variation, de novo assembled LCR22 duplicon order and orientation should be consistent between both data sets. We compared the fiber FISH and Bionano results by first converting the fiber FISH duplicon order and orientation information into sequences, stitching together the duplicon sequences from the reference genome. We then in silico labeled these sequences to convert them to Bionano optical map format (Supplemental Fig. S9B,E,H,K,N,Q,T,W,BB,EE,HH,KK, green bar) and then compared them to the observed Bionano assemblies in the same individual (Supplemental Fig. S9B,E,H,K,N,Q,T,W,BB,EE,HH,KK, blue bar). For CHM1, GM12878, and the two 22q11DS families, we generated Bionano data using the Direct Label and Stain (DLS) enzyme. After examination of single molecules from these samples at both LCR22A and LCR22D, we generated a list of partial haplotypes with strong single molecule support for each sample (Supplemental Fig. S9). We observed that the cluster of SD22-4 and its flanking SD22-6 duplicons contained five DLS labels that were polymorphic between paralogs (Supplemental Fig. S10). These polymorphisms allowed us to stitch the partial haplotypes into end-to-end haplotypes of LCR22A and LCR22D.
We aligned the observed optical maps from CHM1, GM12878, and all LCR22A alleles in the two families to those converted from the fiber FISH results (Supplemental Figs. S3, S4, S9). All alignments showed strong agreement between in silico and observed optical maps after accounting for the expected paralogous variation. The copy number, order, and orientation of detected duplicons were identical between the two techniques. Overall, the Bionano optical maps confirmed the fiber FISH assemblies with minimal discrepancies.
Bionano optical mapping reveals population-specific LCR22 variation
To determine the prevalence of different variants in LCR22A and LCR22D, both within and between populations, we mapped the variability, by Bionano optical mapping using the Nt.BspQI enzyme, in a cohort of 154 phenotypically normal individuals from 26 populations spanning five superpopulations: African (AFR), American (AMR), East Asian (EAS), European (EUR), and South Asian (SAS) (Levy-Sakin et al. 2019).
Structural variation at LCR22A
We generated distinct LCR22A configurations by collapsing optical map assembled contigs that overlapped LCR22A and had single molecule support (Fig. 3A–D). Not all assembled contigs were able to span the entire LCR22A region from end to end, due to factors such as insufficient molecular coverage, short molecule lengths, and/or longer LCR22A haplotype lengths. Additionally, the presence of two Nt.BspQI nicking sites, on opposite strands, in close proximity to one another near the beginning of SD22-3 created a fragile site on the DNA that consistently interrupted molecules in that location (Supplemental Fig. S11A). Although this catalog of LCR22A haplotypes is likely not comprehensive, it captures a substantial amount of large-scale structural variation at this locus.
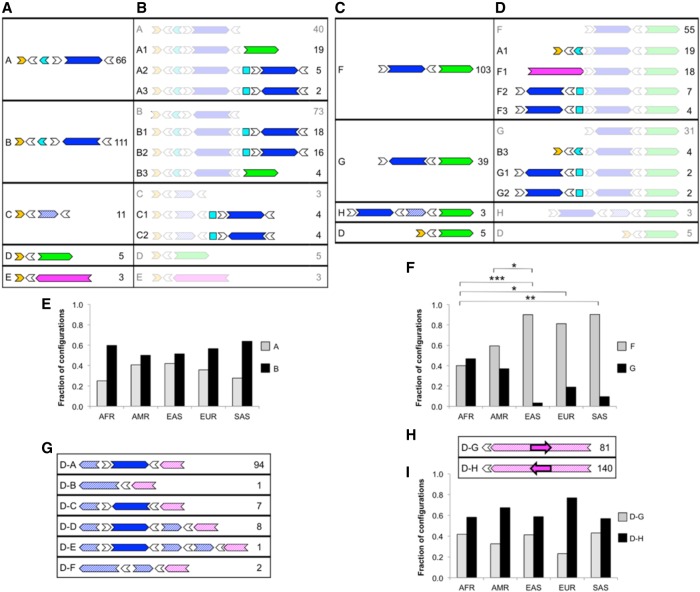
Figure 3.
LCR22A and LCR22D configurations across a diverse control data set observed using Bionano optical mapping. Diagrams depict order and orientation of observed duplicons as defined in Figure 1. Minimal (A) and extended (B) configurations anchored in unique sequences upstream of LCR22A. Minimal (C) and extended (D) configurations anchored in unique sequence downstream from LCR22A. (E) Observed occurrences for upstream-anchored LCR22A configurations A and B in different populations. (F) Observed occurrences of downstream-anchored LCR22A configurations F and G in different populations. (G) Configurations anchored in unique sequence upstream of LCR22D. (H) Configurations anchored in unique sequence downstream from LCR22D. (I) Observed occurrences of downstream-anchored LCR22D configurations D–G and D–H in different populations. For each configuration in A–D and G–H, the ID is on the left, and the number of times that configuration was observed in the data set is on the right. Duplicons for which an orientation could not be determined are represented as squares. (AFR) African; (AMR) American; (EAS) East Asian; (EUR) European; (SAS) South Asian; (*) P < 0.05; (**) P < 0.01; (***) P < 0.001, Fisher's exact test, adjusted. Pairs of populations without asterisks in E, F, and I were not significantly different at P < 0.05.
We identified a total of 16 nonredundant partial and complete LCR22A configurations in the set of 154 individuals (Fig. 3). As seen in the fiber FISH results, the majority of the variation involved variable copy number and orientation of SD22-4 (Fig. 3A–D). One notable configuration, not observed in the fiber FISH data set, harbored a deletion of almost the entire locus, with a minimal composition of SD22-1 to inverted SD22-6 to SD22-5 (Fig. 3, configuration D).
We next evaluated the prevalence of each configuration within the data set (for details, see Methods) and observed clear differences in prevalence between different configurations (Fig. 3). For minimal configurations anchored upstream of LCR22A (Fig. 3A), the most common duplicon to follow the initial cluster of SD22-1, SD22-2, and flanking SD22-6 copies was an inverted copy of SD22-4 (Fig. 3A, configuration B), which accounted for 111/196 (57%) and 25/44 (57%) of the observed configurations in this group, in optical map and fiber FISH data sets, respectively. The next most common configuration was a copy of SD22-4 in the direct orientation (Fig. 3A, configuration A), which accounted for 66/196 (34%) of the observed configurations in the optical map data and 14/44 (32%) in the fiber FISH data. Configuration A is also the structure corresponding to the beginning of both the hg19 and hg38 reference haplotypes. These results indicated that neither of the two most recent reference genomes represented the major allele at this locus.
Among the minimal configurations anchored downstream from LCR22A (Fig. 3C), a direct copy of SD22-4 preceded SD22-5 in 103/150 (69%) observed configurations (Fig. 3C, configuration F), which is consistent with the reference genomes. The next most common configuration (Fig. 3C, configuration G) accounted for 39/150 (26%) of observed configurations, had SD22-5 preceded by an inverted SD22-4. In the fiber FISH data set, 39/44 (89%) of chromosomes displayed configuration F, but only 5/44 (11%) showed SD22-5 preceded by an inverted SD22-4. The fiber FISH samples were taken exclusively from individuals of European descent, and the distal portion of LCR22A differed significantly among various ethnicities (Fig. 3F). Among European samples in the Bionano data set, configuration F accounted for 26/33 (79%) of observed configurations, whereas configuration G accounted for the remaining 7/33 (21%), values which were more concordant with the fiber FISH results in individuals of European descent. For both groups of minimal configurations, anchored upstream of or downstream from LCR22A, the shortest end-to-end haplotype containing SD22-1 to inverted SD22-6 to SD22-5 (configuration D) comprised a small minority of cases, accounting for ∼3% of the observed configurations.
The extended configuration (Fig. 3B,D) was observed in fewer samples because not all single molecules that matched the minimal configurations were long enough to extend into an additional duplicon. Nevertheless, this smaller data set illustrated several distinctive patterns. The three most common upstream configurations observed, each representing ∼20%–24% of the extended upstream alleles, followed the anchoring SD22-1 and SD22-2 with (1) direct SD22-4 and SD22-5, that is, the hg19 haplotype (Fig. 3B, configuration A1); (2) tandem copies of indirect SD22-4 (configuration B1); and (3) an indirect and then a direct copy of SD22-4 (configuration B2). The remaining 34% of cases comprised seven configurations, each accounting for 3%–6% of observed configurations.
Among the downstream extended configurations (Fig. 3D), the hg19 haplotype (configuration A1) and configuration F1, which matched the distal end of the hg38 haplotype, that is, SD22-3, SD22-4, and SD22-5, were the two predominant configurations observed—each representing 28%–30% of the observed configurations. Although the latter configuration was also common in the fiber FISH data (20% of alleles) (Fig. 2A), the nine end-to-end haplotypes containing configuration F1 showed only one match to the exact hg38 structure, instead representing a total of six haplotypes of varying lengths and copy number of SD22-4, suggesting that this configuration class in the optical map data is likely to also represent a wide range of end-to-end haplotypes.
We next wanted to determine whether the observed configurations differed by population. Configurations A, B, F, and G (Fig. 3A,C) were the only observed configurations that provided an adequate sample size for this population-based analysis. We observed substantial differences between the superpopulations for configurations F and G (P < 0.05, Fisher's exact test), with the largest difference occurring between the African and EAS populations (Fig. 3F). Thus, at the distal end of LCR22A, SD22-4 in the direct orientation (configuration F) was more common overall, but it accounted for only 16/41 (39%) of the observed configurations in Africans, compared to 24/27 (89%) of the configurations observed in EASs (Fig. 3F). At the proximal end of LCR22A, SD22-4 in an inverted orientation (configuration B) was observed more frequently than SD22-4 in the direct orientation (configuration A) in every population (Fig. 3E).
Structural variation at LCR22D
LCR22D was substantially less polymorphic and complex than LCR22A, but it nonetheless harbored some large-scale structural variation (Supplemental Fig. S2). Following the same procedure as above, we compiled configurations for LCR22D from the optical map data from 154 individuals. We observed six upstream-anchored configurations (D-A to -F), five of which involved the paralog of SD22-4 that is present in the proximal half of LCR22D (Fig. 3G). In the downstream-anchored half of LCR22D, we only observed a 64-kb inversion (Fig. 3H). Because these two regions were distant from one another, we analyzed them separately to minimize the length of the molecules required to identify each configuration. Among the upstream-anchored configurations, the most predominant was configuration D-A, which represents the configuration observed in the reference genome, accounting for 94/113 (83%) of all observed configurations (compared to 95% [42/44] in the fiber FISH data) (Fig. 2B). The next most common configurations were a full inversion or partial duplication of SD22-4 (configurations D-C and D-D), accounting for 7/113 (6%) and 8/113 (7%) of the observed configurations, respectively (Fig. 3G). In the fiber FISH data, we observed a partial duplication (D-D) in 2/44 (4.5%) alleles, although we did not see any inversions of SD22-4. We detected no population-based differences among these upstream-anchored LCR22D configurations.
Within the observed downstream-anchored configurations of LCR22D, configuration D-H accounted for 140/221 (63%) of the observed configurations. Thus, we observed configuration D-H, with an inversion, more frequently than configuration D-G, which represents the configuration observed in the reference genome. Configuration D-H accounted for 30/39 (77%) of the observed configurations in Europeans, which was consistent with the fiber FISH data from European samples in which 19/25 (76%) individuals carried the inverted D-H configuration (Fig. 2B). In the optical map cohort, we observed the D-H variant more frequently in all five superpopulations, with no statistically significant differences between populations (Fig. 3I).
Thus, Bionano optical mapping not only confirmed the fiber FISH assemblies, but also extended these findings demonstrating large-scale structural variation in LCR22A and LCR22D in a cohort of 154 individuals from five superpopulations.
The 22q11DS rearrangement breakpoints are localized within SD22-4 and flanking SD22-6
The mapping of 22q11DS rearrangement breakpoints within the LCR22s has so far remained elusive. To refine the rearrangement breakpoints and to identify potential variability of the rearrangements, we generated assemblies using either a combination of fiber FISH and optical mapping or fiber FISH only in eight 22q11DS patients and their parents (Supplemental Fig. S7). To reduce complexity and assure correct assembly of the rearranged LCR22s, we generated fiber FISH maps for the probands in Families 1 and 2 from lymphocyte-derived somatic cell hybrids containing only the del(22)(q11.21). For both of these probands, patterns of the rearranged LCR22s were reflected identically in the somatic hybrids and EBV cell lines (Supplemental Fig. S12). In all families, we confirmed the parent of origin of the deletion by STR marker analysis (STR_DATA_FAMILIES.zip in Supplemental Material).
Seven of the probands tested had the typical LCR22A-D deletion, and one proband carried the smaller LCR22A-B deletion (Fig. 4; Supplemental Fig. S7). In Family 1, the individual with the 22q11DS presented with five LCR22 patterns. Four were indicative of complete alleles of LCR22A, -B, -C, and -D. Moreover, these were all identical to one of the mother's LCR22 structures, and thus represented the non-rearranged allele (Fig. 4B,D; Supplemental Fig. S7B,D). The fifth LCR22 pattern initially presented with a duplicon order and orientation identical to one of the father's LCR22A alleles (Fig. 4A). Distally from the third copy of SD22-4, the allele transitioned to a pattern identical to one of the LCR22D alleles in the same parent (Fig. 4C). The probe pattern suggested that LCR22A and LCR22D had been merged into one LCR, with a breakpoint either in SD22-4 or in one of its flanking SD22-6 paralogs.
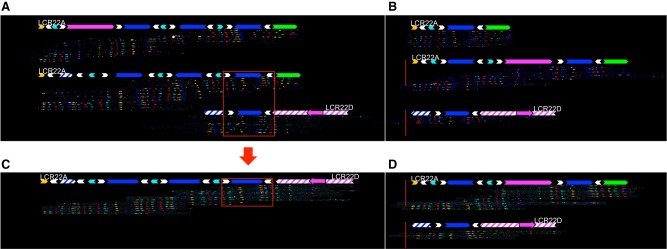
Figure 4.
Analysis of a 22q11.21 deletion in a proband and parents using fiber FISH. The family shown here is Family 1 in Supplemental Figure S8. Fiber FISH assemblies are aligned with a duplicon representation as defined in Figure 1. Both alleles of LCR22A and LCR22D are shown for each individual in the family trio. All individuals had the same configuration at both alleles of LCR22D, which is thus shown only once for each. (A) Parent of origin. The shared region between LCR22A and LCR22D is marked by a red box. (B) Other parent. Non-rearranged alleles of LCR22A and LCR22D transmitted to the proband are shown inside the red box. Rearranged (C) and non-rearranged (D) alleles observed in the proband. Red arrow marks duplicon(s) that were involved in recombination and define the breakpoint region.
Within the probands, seven out of eight of the rearranged alleles overlapped with the parental alleles at the longest shared region between LCR22A and LCR22D (Figs. 1, 4; Supplemental Fig. S7), suggesting this as the likely location for the rearrangement breakpoints. This region comprises SD22-4 and two flanking SD22-6 duplicons, together forming a 160-kb stretch of homologous sequence. These constitute a large direct repeat in LCR22A and LCR22D, which has been previously proposed to contain rearrangement breakpoints (Guo et al. 2016). In all individuals, one copy of SD22-4 was present in LCR22D in a direct orientation (Supplemental Fig. S7), whereas it is found in variable copy numbers and orientations in LCR22A. In all seven rearranged alleles, SD22-5 was deleted, thereby generating hemizygosity for PRODH, DGCR5, and DGCR6.
We also mapped the LCR22A-B rearrangement in one family (Family 7) (Supplemental Fig. S7). One of the father's LCR22A alleles was identical to his offspring's rearranged chromosome, up to the last distal direct paralog of SD22-6 (white arrow). At this position, the patient's rearranged LCR22 transitioned to the last two probes of LCR22B. This pattern suggested that a NAHR occurred at SD22-6. Assembly of a second individual with an LCR22A-B deletion supports this breakpoint location distally in the fragment of duplicon SD22-6 (Supplemental Fig. S13). Thus, we have further refined the LCR22A-D deletion breakpoints in multiple 22q11DS patients within a 160-kb duplicon containing SD22-4 and SD22-6, and LCR22A-B deletion breakpoints within SD22-6, which contains the highly recombinogenic palindromic AT-rich repeats (PATRRs).
LCR22s in nonhuman primate genomes
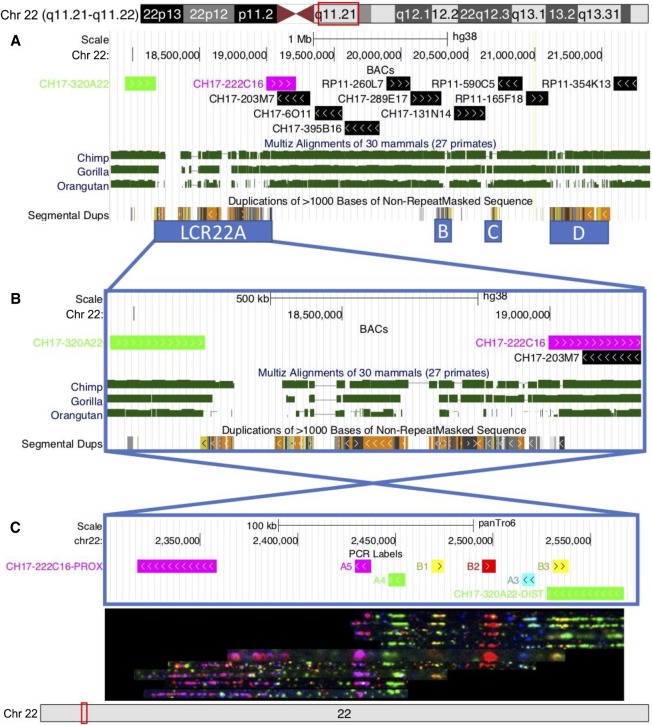
To determine the evolutionary origin of the LCR22s, we first looked at the conservation of the region in the chimpanzee, gorilla, and orangutan genomes (Supplemental Fig. S14). Considering that the human genome contains gaps and even differs in LCR22 structure between hg19 and hg38, it was not surprising that the conservation between species is low within the LCR22s (Fig. 5A). In addition, the syntenic regions show incomplete sequence contigs with gaps within the LCRs (Supplemental Fig. S14). As an initial analysis of LCR22s in nonhuman primates using our methods, we used fiber FISH to analyze LCR22A in the chimpanzee. We first identified syntenic regions flanking the human LCR22A in the most recent chimpanzee reference genome (January 2018; Clint_PTRv2/panTro6). BAC sequences adjacent to the human LCR22A (CH17-320A22 proximal and CH17-203M7 distal from LCR22A) and fiber FISH probe sequences within the LCR22s were aligned to this reference genome to delineate the LCR22A region. In this reference prediction, BAC probe CH17-222C16 is located proximal and BAC probe CH17-320A22 is located distal from LCR22A, an inversion compared with human (Fig. 5C). The probes located in the region are A5 (magenta), A4 (green), B1 (yellow), B2 (red), A3 (cyan), and B3 (yellow) (Fig. 5A). We set out to confirm and/or compare this reference genome with another chimpanzee genome (AG 06939A). Hybridization of combed chimpanzee DNA with the human LCR22 probes confirmed the LCR22A structure predicted by the chimpanzee reference genome (Fig. 5B). This duplicon composition is identical to the smallest duplicon pattern observed in humans (configuration D in Fig. 3A,C). This suggests that the expansion and variability of LCR22A may be human specific.
Figure 5.
Conservation of the 22q11 region. (A) UCSC Genome Browser hg38 view of BACs that were evaluated covering the region, with LCR22A flanking BACs highlighted in green (CH17-320A22) and magenta (CH17-222C16). MULTIZ alignments (Blanchette et al. 2004) of chimpanzee (Pan_tro 3.0/panTro5), gorilla (GSMRT3/gorGor5), and orangutan (WUGSC 2.0.2/ponAbe2) genomes to the human genome identify high conservation in the unique 22q11 region, but lower conservation in the LCR22s. (B) A zoomed in view of LCR22A. (C) Fiber FISH de novo assembly performed on DNA from a chimpanzee cell line. The observed probe pattern is complementary with the predicted probe composition based on the chimpanzee reference genome. The LCR22A region is present in an opposite orientation compared to the human reference.
Discussion
The LCR22 reference sequences have contained gaps since the first human genome assembly was released (Cole et al. 2008; Schneider et al. 2017). Although whole-genome short-read sequencing is now routine, alignment of short sequencing reads to the human reference sequence generally fails to detect and assemble large structural variants and repetitive regions like the LCR22s. Because of the length of the duplications, even assemblies using longer-range technologies like Pacific Biosciences (PacBio) and 10x Genomics linked reads have been unable to assemble these regions (Berlin et al. 2015; Weisenfeld et al. 2017). To resolve these gaps, we combined fiber FISH and Bionano optical mapping, and show that an astounding level of inter-individual variability of LCR22A, and to a lesser extent LCR22D, has likely impeded the assembly of a complete reference sequence for these LCRs. These maps revealed at least 25 different alleles of LCR22A and six variants of LCR22D. LCR22A alleles ranged in size from ∼250 to ∼2000 kb. Most of these alleles could be decomposed into six core duplicons (SD22-1 to SD22-6), with duplicons presenting in different orientations and at variable positions within the LCR. The most frequent LCR22A haplotype had the following structure; SD22-1, SD22-6 (inverted orientation), SD22-2 (inverted orientation), SD22-6 (direct orientation), SD22-4 (direct orientation), SD22-6 (inverted orientation), SD22-5, which made up ∼25% of all mapped alleles. Its structure is very similar to one of the first LCR22A sequences proposed (Shaikh et al. 2000), a haplotype which was presented in hg19. Thus far, only one smaller haplotype was detected, in which SD22-1 was directly followed by SD22-6 (indirect orientation) and SD22-5. This might indicate the requirement of a minimal haplotype to maintain a viable gene dosage.
None of the 19 normal, diploid parents in the cohort were homozygous for LCR22A, which suggests the existence of a high number of different haplotypes in humans. Consequently, any homologous recombination between two (different or identical) alleles of LCR22A will likely generate a novel allele with a duplicon composition different from the parent of origin (Supplemental Fig. S15). LCR22s are known to be sites with an increased recombination rate when compared to their surrounding loci (Frazer et al. 2007; Torres-Juan et al. 2007). Thus, LCR22s are likely to be “hotspots” for the introduction of novel structural variants in the population. Furthermore, configurations of LCR22A and LCR22D varied in frequency among populations. Some of these configurations might be more vulnerable to NAHR than others. Consequently, variation in the frequency of the 22q11DS among populations (Botto et al. 2003; McDonald-McGinn et al. 2005) may result from frequency differences of LCR22A and LCR22D configurations and their respective vulnerability to NAHR. Because our sample size is relatively small, we expect that the alleles we observed are likely to be a small subset of all haplotypes that may exist in the population. Although the reference genomes of other hominoids including the chimpanzee, gorilla, and orangutan contain gaps within LCR22s, a fiber FISH based analysis of LCR22A showed that the chimpanzee LCR22A corresponds to the smallest observed human allele. This suggests that the LCR22A expansion and inter-individual variability is human-specific. However, to assess LCR22 evolution and variability, a more detailed analysis of multiple individuals from hominoid and other primate species is warranted.
Studies on genome-wide LCR diversity have identified numerous LCR clusters, mainly in pericentromeric and subtelomeric regions (Goidts et al. 2006b). However, none of those come close to the level of complexity and the number of haplotypes found in the LCR22s. A few studies using either WGS read depth–based predictions, digital droplet PCR, or custom BAC arrays have revealed copy number variability between individuals within regions containing LCRs (Sudmant et al. 2010, 2015; Handsaker et al. 2015; Dennis and Eichler 2016). Eight distinct haplotypes have been described for the LCR clusters on Chromosome 17q21.31, ranging in size from 1.08 to 1.49 Mb (Steinberg et al. 2012). Similarly, the 1000 Genomes Project observed copy number variation ranging from 2 to 11 copies of a ∼900-kb region (Chr 15: 20,353,991–27,802,370) in 15q11-q12 (Siva 2008; Sudmant et al. 2010). Such repeat expansions have mainly been found to be human-specific when compared to their orthologs in great ape genomes (Goidts et al. 2006a). Moreover, significant variation between different human populations suggests that these genomic rearrangements happened recently or are still ongoing (Dennis and Eichler 2016). However, a majority of these studies are based on short-read whole-genome sequencing data, which are not as reliable for determining true copy number and complex architecture of regions containing LCRs.
Our approach has also allowed us to further refine the localization of recurrent rearrangements to specific modules within LCR22. In seven out of eight families, the rearrangement occurred within a 160-kb core module, containing SD22-4 and SD22-6, which is present within both LCR22A and LCR22D in the vast majority of haplotypes. A previous study had predicted that NAHR occurred within a duplicon referred to as BCRP2 (Guo et al. 2016). Our analysis of the same two trios (Family 7 and 8) (Supplemental Fig. S7) confirmed the presence of paralogous repeats of BCRP2 at fiber FISH probe D2 in the LCR22s and further showed that NAHR regions overlap at one BCRP2 locus embedded in the SD22-4 duplicon. In two probands with LCR22A-B deletions, the breakpoint region was further narrowed to a 31-kb subunit of SD22-6, which contained palindromic AT-repeats (PATRRs). Paralogs of SD22-6 flank copies of every other duplicon in the assembled alleles, and each of these paralogs in hg38 contained PATRRs. Thus, we hypothesize that the PATRRs could be driving the rearrangements at this locus. PATRRs are known to form cruciform structures, which are prone to double-strand breaks (Leach 1994; Lobachev et al. 2002). If these breaks occur simultaneously at multiple loci in the genome, these are often resolved by nonhomologous end joining (NHEJ), thereby rearranging the genome in some cases (Kurahashi and Emanuel 2001; Kato et al. 2008).
Although 22q11DS is the most frequent microdeletion syndrome, the underlying cause for the wide spectrum and variability of phenotypes observed has not been fully elucidated. Variation of genes embedded in copy number variable regions like LCR22s has been so far ignored. We suggest that copy number variable genes embedded in the LCR22s could explain some of the phenotypic variability observed in individuals with the 22q11DS and human in general. SD22-3 contains at least two known active genes (TMEM191B and RIMBP3) (Fig. 1B). TMEM191B is expressed in brain tissue (The GTEx Consortium 2013; Fagerberg et al. 2014). Not every allele of LCR22A features this duplicon, but neither is it observed to be present in more than one copy. Both TMEM191B and RIMBP3 have paralogs in LCR22D (TMEM191C, RIMBP3B, and -C). The genes PRODH, DGCR5, and DGCR6 reside in SD22-5 (Guo et al. 2018), which was retained in even the smallest mapped allele of LCR22A. In all mapped individuals with the typical 22q11 deletion, this duplicon is deleted, confirming previous observations of its hemizygosity in most patients (Liu et al. 2002; Jacquet et al. 2003; Michaelovsky et al. 2012; Guo et al. 2016, 2018). Additionally, the presence of pseudogenes (PI4KAP1 and P2, GGT3P, DGCR6L, BCRP2, GGT2) and lncRNAs (FAM230A, FAM230B, and at least seven noncharacterized paralogs) in different copy numbers could influence gene expression. However, in the absence of an unambiguous reference sequence, it remains challenging to investigate the gene activity of each duplicon. Moreover, the observed size differences of LCR22A might exert a spatial effect on chromatin looping in the cell, thereby altering topologically associated domains (Kleinjan and van Heyningen 2005; De Laat and Duboule 2013; Weischenfeldt et al. 2013). The phenotypic effect of variable repeat architecture could be minor for intact alleles but could alter gene expression completely when the LCR22s are rearranged. Hence, with these LCR22 assemblies, we envision future work to further elucidate the effect of multicopy genes at this locus.
In summary, high-resolution optical mapping has allowed us to reveal an extraordinary level of variability within LCR22s. Our map of this genomic region is, to date, the most comprehensive for the LCR22s in the human genome reference sequence. Further, this map provides a framework for the alignment of both short and long read sequences which will ultimately close the remaining reference gaps and enable sequence-based analysis of the LCRs. Understanding the LCR variation will shed light on the mechanisms leading to 22q11 rearrangements and the different frequencies of the variation among populations. This knowledge will likely guide future prenatal counseling and testing for 22q11-related disorders. The LCR22s have rapidly expanded during hominoid evolution (Guo et al. 2011) and, considering the region encompasses nine known active genes and comprises 54 different RNAs, it seems plausible that the region influences important human traits. Thus, it is likely that the LCR variability has phenotypic consequences, which may play a role in phenotypic variability in 22q11DS and affect other traits in the normal population. The ability to visualize and reconstruct complete and intact LCR haplotypes will greatly enhance our ability to start unraveling these important correlations.
Methods
Patients and EBV cell lines
Patients with the 22q11DS were diagnosed using either a FISH assay with TUPLE1/ARSA probes (Abbot Molecular), the MLPA SALSA P250 DiGeorge diagnostic probe kit (MRC-Holland), or with the CytoSure Constitutional v3 (4 × 180k) (OGT). All individuals in the study were informed of the project's outlines and gave written consent for their EBV cell lines and DNA to be used for sequencing and genotyping purposes. The study was approved by the Medical Ethics committee of the University hospital/KU Leuven (S52418), the Institutional Review Board approved research protocol (COMIRB 07-0386) at the University of Colorado Denver, School of Medicine, and the Children's Hospital of Philadelphia under Institutional Review Board (IRB) protocol 07-005352. Fiber FISH mapping was performed on Epstein Barr virus transformed lymphoblastoid cell lines from peripheral blood from probands and their parents. EBV cell lines were established as described (Hui-Yuen et al. 2011). Eleven patients were recruited during routine consultations in the hospital of Leuven, one at the Children's Hospital of Philadelphia, and two at the Albert Einstein College of Medicine (IRB: 1999-201-047). HapMap control cell lines and chimpanzee fibroblast cell line AG 06939A were obtained from the Coriell Cell Repository and cultured according to standard protocols.
In silico characterization of repeat subunits in LCR22s
All segmental duplication track positions were downloaded in BED format from UCSC in the region Chr 22: 18,000,000–25,500,000 (hg38), including paralogous LCRs located elsewhere in the genome. These were merged with BEDTools v2.17.0 (Quinlan and Hall 2010), and sequences were retrieved with the UCSC Table Browser (Kent et al. 2002; Karolchik et al. 2004, 2014). These were then self-aligned using BLASTN v2.2.28+ (Altschul et al. 1990) and filtered for reciprocal BLAST hits, alignments <100bp, and alignments <99% identity. If multiple queries aligned to the same subject segmental duplication at different positions, the segmental duplication was split into multiple units. Unit positions were converted to BED format, sequences retrieved through the UCSC Table Browser, self-aligned again, and similarly filtered. Clusters of units aligning to each other were each considered a subunit family (Supplemental Table S1).
BAC DNA, long-range PCR probe design, and labeling
Using the subunit sequences library, 14 fluorescent probes were designed (Supplemental Table S2).
For each of these 14 subunits, long-range PCR primer pairs were designed, producing amplicons between 2946 and 9794 bp (Supplemental Table S2). PCR reactions were performed with the TAKARA LA v2 kit (Takara Bio) using the standard gDNA protocol. Template gDNA was extracted from the same cell line for all reactions, to reduce amplicon variation between batches.
BAC clones were obtained from BacPac Resources (CHORI) as E. coli stab cultures, which were grown according to recommendations. BAC DNA was extracted using the Nucleobond Xtra BAC kit (Macherey-Nagel).
Subunit PCR Amplicons and BAC DNA were purified and antibody-labeled by random prime amplification (BioPrime DNA Labeling System; Invitrogen). An indirect detection system with primary labels Biotin-dUTP, Digoxigenin-dUTP, and Fluorescein-dUTP was used. The use of three labels allowed production of six detectable probe colors: three of each label separately and three of each pairwise combination.
DNA combing, FISH, and fiber pattern assembly
DNA fibers were stretched using the Genomic Vision extraction kit and combing system for a total of 33 human and one chimpanzee cell line (Supplemental Table S3) using standard methodology (for details, see Supplemental Methods).
Coverslips with combed DNA were hybridized with the designed probe pattern and washed using the manufacturer's standard protocol. Probes were detected by indirect labeling with BV480 Streptavidin (pseudocolored red; BD Biosciences; 564876), Cy3 IgG Fraction Monoclonal Mouse Anti-Fluorescein (pseudocolored green; Jackson Immunoresearch; 200-162-037), and Alexa Fluor 647 IgG Fraction Monoclonal Mouse Anti-Digoxigenin (pseudocolored blue; Jackson Immunoresearch; 200-602-156). Probe mixes produced pseudocolors cyan, magenta, and yellow. Slides with labeled DNA were mounted in the provided scanner adapters and scanned at three excitation channels on a customized automated fluorescence microscope (Genomic Vision).
Images were compiled to one complete slide recording and visualized in FiberStudio (Genomic Vision). Slides were manually screened, and fiber signals were cropped to single image files. Individual images were visually aligned based on matching colors and distances between different probes. Fibers were tiled to complete alleles for LCR22A, B, C, and D, and compared to hg38 probe positions in the UCSC Genome Browser. Chimeric fiber patterns and false positive signals caused by noise were eliminated by filtering for overlapping patterns identical in color sequence and spacing
Assembly of artificial LCR22 reference sequences
To confirm the fiber FISH assemblies, Bionano assays were performed on an overlapping cohort of seven individuals. To compare results from the two methods, fiber FISH results were converted in silico into the optical map format. Using the hg38 reference genome sequences of SD22-1 to SD22-6 and LCR22D, the sequence of each allele was predicted based on the orientation and copy number of subunits detected in the fiber FISH assemblies. Those sequences were then in silico labeled at recognition sites of the enzyme used for Bionano optical mapping, generating CMAP data files for all LCR22 repeats.
Bionano genome mapping and assembly
High-molecular-weight DNA was extracted and processed for Bionano genome mapping using standard methods and protocols provided by the vendor (Bionano Genomics). The DNA was labeled using the Bionano Prep Early Access Direct Labeling and Staining (DLS) Kit (Bionano Genomics). The DNA was loaded onto the Bionano Genomics Saphyr Chip and linearized and visualized using the Saphyr system (for details, see Supplemental Methods).
Detection of structural variation within LCR22s
Structural variation in the LCR22s was evaluated in Bionano genome map data labeled using the Nt.BspQI nickase enzyme from 154 individuals representing 26 diverse populations from five superpopulations (Levy-Sakin et al. 2019). Assembled contigs mapping to LCR22s were realigned to Chromosome 22 using RefAligner from Bionano Solve 3.1 (for details, see Supplemental Methods).
LCR22 haplotype identification from Bionano data
A catalog of configurations for each locus was generated by compiling the configurations observed in the assembled Bionano contigs from the full data set derived from normal individuals and verifying that each entry was supported by single molecules in at least one sample. Configurations were first grouped into categories in which the members were mutually exclusive, so that longer configurations would be analyzed separately from those that were subsets of them. Using this approach, a “minimal” set of configurations that were anchored upstream of or downstream from the repetitive region were constructed for LCR22A (Fig. 3A,C). An “extended” set was also created that expanded on the minimal configurations, where available (Fig. 3B,D). LCR22D contained a variable proximal region, as well as a distal region that contained a single structural variant. These two regions were analyzed separately (Fig. 3G–I).
A package called OMGenSV (Supplemental Code) was used to genotype each set of configurations in all the samples. In silico labeled representations of each configuration were created in Bionano CMAP format using OMGenSV's get_cmap_subsets.py and add_cmap_files.py scripts to combine 1 Mb of flanking unique region from the reference chromosome with representative assembled contigs observed in the normal population-based samples. For all configurations in a given group, their CMAP representations were kept as consistent between one another as possible, that is, containing the same flanking areas. For each grouped set of configurations described above, single molecules from each sample were used to determine which configuration(s) the sample contained (for details, see Supplemental Methods). Each observed configuration in a given sample was counted once, and the overall prevalence of any configuration was calculated by dividing the number of times that particular configuration was observed by the total number of all configurations observed for that locus in the relevant group.
LCR22 haplotype reconstruction in trios
Full haplotypes at LCR22A and LCR22D were reconstructed for Families 1 and 2 from Bionano genome map data labeled with the DLS enzyme as follows. For each proband, the molecule-to-contig alignments for all local contigs were examined to break apart the contigs into local configurations that had strong molecule support. To identify any additional configurations that had not been assembled, the local molecules (identified as described above) were aligned to the reference Chromosome 22 using OMBlastMapper with the following parameters: --alignmentjoinmode 3 --indelp 10 --invp 10 --transp 10 --closeref 4000000 --closefrag 4000000 --filtermode 1 --minmatch 4 --maxclusteritem 1 --trimmode 1 --minscore 30. The resulting alignment was visualized using OMView and manually inspected for additional configurations (Leung et al. 2017).
Data access
Fiber FISH and STR data are available in the Supplemental Material, and cell lines used to map repeats are available upon request. Bionano optical mapping data from this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA418343. Scripts used in this study are available at https://github.com/yuliamostovoy/OMGenSV and as Supplemental Code.
Supplementary Material
Acknowledgments
Excellent technical assistance was provided by O. Tran and A. Jin. The molar cell line CHM1 was kindly provided by E. Eichler. W.D. is a fellow at Katholieke Universiteit Leuven supported by Agentschap voor Innovatie door Wetenschap en Technologie (131625). This work was made possible by grants from the Katholieke Universiteit Leuven Programmafinanciering Vlaanderen (PFV/10/016) SymBioSys and project 1665 Fondation Jérôme Lejeune to J.R.V. and Geconcerteerde Onderzoeksacties (GOA/12/015) to J.R.V. and Koen Devriendt by the National Institute of Mental Health (5U01MH101723-02), by the National Institute of General Medical Sciences (GM120772 to T.H.S. and P.-Y.K.; GM125757 to B.S.E. and M.X.) and the Belgian Science Policy Office Interuniversity Attraction Poles (BELSPO-IAP) program through the project IAP P7/43-BeMGI.
Author contributions: W.D. led conception of the work and experimental design using fiber FISH. T.H.S. and P.-Y.K. conceived the study and analysis using Bionano optical mapping. A.S. and E.V. informed parents and patients during hospital follow-up and collected blood samples. C.R.C. coordinated the study and subject enrollment. E.A.G. and S.K.C. carried out sample processing and experimentation. Data were collected by W.D., Y.M., L.V., F.Y., and M.S.H.; W.D., Y.M., L.V., and F.Y. performed data analysis and interpretation. W.D., Y.M., J.R.V., L.V., T.H.S., B.S.E., D.M.-M., and B.M. drafted the article, which was critically revised by J.R.V., B.M., B.S.E., T.H.S., A.S., M.X., M.S.H., and D.M.-M. Final approval of the version to be published was given by W.D., Y.M., F.Y., L.V., S.P., M.S.H, A.S., E.V., E.A.G., C.R.C., S.K.C., D.M.-M., B.M., P.-Y.K., M.X., B.S.E., T.H.S, and J.R.V.
Footnotes
[Supplemental material is available for this article.]
Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.248682.119.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Bailey JA, Yavor AM, Massa HF, Trask BJ, Eichler EE. 2001. Segmental duplications: organization and impact within the current human genome project assembly. Genome Res 11: 1005–1017. 10.1101/gr.GR-1871R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JA, Gu Z, Clark RA, Reinert K, Samonte R V, Schwartz S, Adams MD, Myers EW, Li PW, Eichler EE. 2002. Recent segmental duplications in the human genome. Science 297: 1003–1007. 10.1126/science.1072047 [DOI] [PubMed] [Google Scholar]
- Berlin K, Koren S, Chin CS, Drake JP, Landolin JM, Phillippy AM. 2015. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat Biotechnol 33: 623–630. 10.1038/nbt.3238 [DOI] [PubMed] [Google Scholar]
- Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AF, Roskin KM, Baertsch R, Rosenbloom K, Clawson H, Green ED, et al. 2004. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res 14: 708–715. 10.1101/gr.1933104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botto LD, May K, Fernhoff PM, Correa A, Coleman K, Rasmussen SA, Merritt RK, O'Leary LA, Wong LY, Elixson EM, et al. 2003. A population-based study of the 22q11.2 deletion: phenotype, incidence, and contribution to major birth defects in the population. Pediatrics 112: 101–107. 10.1542/peds.112.1.101 [DOI] [PubMed] [Google Scholar]
- Bovee D, Zhou Y, Haugen E, Wu Z, Hayden HS, Gillett W, Tuzun E, Cooper GM, Sampas N, Phelps K, et al. 2008. Closing gaps in the human genome with fosmid resources generated from multiple individuals. Nat Genet 40: 96–101. 10.1038/ng.2007.34 [DOI] [PubMed] [Google Scholar]
- Boyd JL, Skove SL, Rouanet JP, Pilaz LJ, Bepler T, Gordân R, Wray GA, Silver DL. 2015. Human-chimpanzee differences in a FZD8 enhancer alter cell-cycle dynamics in the developing neocortex. Curr Biol 25: 772–779. 10.1016/j.cub.2015.01.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson MJP, Huddleston J, Dennis MY, Sudmant PH, Malig M, Hormozdiari F, Antonacci F, Surti U, Sandstrom R, Boitano M, et al. 2015. Resolving the complexity of the human genome using single-molecule sequencing. Nature 517: 608–611. 10.1038/nature13907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrier C, Joshi K, Coutinho-Budd J, Kim JE, Lambert N, De Marchena J, Jin WL, Vanderhaeghen P, Ghosh A, Sassa T, et al. 2012. Inhibition of SRGAP2 function by its human-specific paralogs induces neoteny during spine maturation. Cell 149: 923–935. 10.1016/j.cell.2012.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole CG, McCann OT, Collins JE, Oliver K, Willey D, Gribble SM, Yang F, McLaren K, Rogers J, Ning Z, et al. 2008. Finishing the finished human chromosome 22 sequence. Genome Biol 9: R78 10.1186/gb-2008-9-5-r78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham LA, Coté AG, Cam-Ozdemir C, Lewis SM. 2003. Rapid, stabilizing palindrome rearrangements in somatic cells by the center-break mechanism. Mol Cell Biol 23: 8740–8750. 10.1128/MCB.23.23.8740-8750.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Laat W, Duboule D. 2013. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature 502: 499–506. 10.1038/nature12753 [DOI] [PubMed] [Google Scholar]
- Dennis MY, Eichler EE. 2016. Human adaptation and evolution by segmental duplication. Curr Opin Genet Dev 41: 44–52. 10.1016/j.gde.2016.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis MY, Nuttle X, Sudmant PH, Antonacci F, Graves TA, Nefedov M, Rosenfeld JA, Sajjadian S, Malig M, Kotkiewicz H, et al. 2012. Evolution of human-specific neural SRGAP2 genes by incomplete segmental duplication. Cell 149: 912–922. 10.1016/j.cell.2012.03.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis MY, Harshman L, Nelson BJ, Penn O, Cantsilieris S, Huddleston J, Antonacci F, Penewit K, Denman L, Raja A, et al. 2017. The evolution and population diversity of human-specific segmental duplications. Nat Ecol Evol 1: 0069 10.1038/s41559-016-0069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle MA, Fritzilas E, Krusche P, Källberg M, Moore BL, Bekritsky MA, Iqbal Z, Chuang HY, Humphray SJ, Halpern AL, et al. 2017. A reference data set of 5.4 million phased human variants validated by genetic inheritance from sequencing a three-generation 17-member pedigree. Genome Res 27: 157–164. 10.1101/gr.210500.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann L, Pandita RK, Spiteri E, Funke B, Goldberg R, Palanisamy N, Chaganti RS, Magenis E, Shprintzen RJ, Morrow BE. 1999. A common molecular basis for rearrangement disorders on chromosome 22q11. Hum Mol Genet 8: 1157–1167. 10.1093/hmg/8.7.1157 [DOI] [PubMed] [Google Scholar]
- Fagerberg L, Hallström BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, et al. 2014. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics 13: 397–406. 10.1074/mcp.M113.035600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florio M, Albert M, Taverna E, Namba T, Brandl H, Lewitus E, Haffner C, Sykes A, Wong FK, Peters J, et al. 2015. Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science 347: 1465–1470. 10.1126/science.aaa1975 [DOI] [PubMed] [Google Scholar]
- Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, et al. 2007. A second generation human haplotype map of over 3.1 million SNPs. Nature 449: 851–861. 10.1038/nature06258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese G, Handsaker RE, Li H, Altemose N, Lindgren AM, Chambert K, Pasaniuc B, Price AL, Reich D, Morton CC, et al. 2013. Using population admixture to help complete maps of the human genome. Nat Genet 45: 406–414. 10.1038/ng.2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goidts V, Armengol L, Schempp W, Conroy J, Nowak N, Müller S, Cooper DN, Estivill X, Enard W, Szamalek JM, et al. 2006a. Identification of large-scale human-specific copy number differences by inter-species array comparative genomic hybridization. Hum Genet 119: 185–198. 10.1007/s00439-005-0130-9 [DOI] [PubMed] [Google Scholar]
- Goidts V, Cooper DN, Armengol L, Schempp W, Conroy J, Estivill X, Nowak N, Hameister H, Kehrer-Sawatzki H. 2006b. Complex patterns of copy number variation at sites of segmental duplications: an important category of structural variation in the human genome. Hum Genet 120: 270–284. 10.1007/s00439-006-0217-y [DOI] [PubMed] [Google Scholar]
- Gotter AL, Nimmakayalu MA, Jalali GR, Hacker AM, Vorstman J, Duffy DC, Medne L, Emanuel BS. 2007. A palindrome-driven complex rearrangement of 22q11.2 and 8q24.1 elucidated using novel technologies. Genome Res 17: 470–481. 10.1101/gr.6130907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The GTEx Consortium. 2013. The Genotype-Tissue Expression (GTEx) project. Nat Genet 45: 580–585. 10.1038/ng.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Freyer L, Morrow B, Zheng D, Index F. 2011. Characterization of the past and current duplication activities in the human 22q11.2 region. BMC Genomics 12: 71 10.1186/1471-2164-12-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Delio M, Haque N, Castellanos R, Hestand MS, Vermeesch JR, Morrow BE, Zheng D. 2016. Variant discovery and breakpoint region prediction for studying the human 22q11.2 deletion using BAC clone and whole genome sequencing analysis. Hum Mol Genet 25: 3754–3767. 10.1093/hmg/ddw221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T, Diacou A, Nomaru H, McDonald-McGinn DM, Hestand M, Demaerel W, Zhang L, Zhao Y, Ujueta F, Shan J, et al. 2018. Deletion size analysis of 1680 22q11.2DS subjects identifies a new recombination hotspot on chromosome 22q11.2. Hum Mol Genet 27: 1150–1163. 10.1093/hmg/ddy028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handsaker RE, Van Doren V, Berman JR, Genovese G, Kashin S, Boettger LM, Mccarroll SA. 2015. Large multiallelic copy number variations in humans. Nat Genet 47: 296–303. 10.1038/ng.3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui-Yuen J, McAllister S, Koganti S, Hill E, Bhaduri-McIntosh S. 2011. Establishment of Epstein-Barr virus growth-transformed lymphoblastoid cell lines. J Vis Exp e3321 10.3791/3321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Lupski JR. 2002. Molecular mechanisms for genomic disorders. Annu Rev Genomics Hum Genet 3: 199–242. 10.1146/annurev.genom.3.032802.120023 [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. 2004. Finishing the euchromatic sequence of the human genome. Nature 431: 931–945. 10.1038/nature03001 [DOI] [PubMed] [Google Scholar]
- Jacquet H, Berthelot J, Bonnemains C, Simard G, Saugier-Veber P, Raux G, Campion D, Bonneau D, Frebourg T. 2003. The severe form of type I hyperprolinaemia results from homozygous inactivation of the PRODH gene. J Med Genet 40: e7 10.1136/jmg.40.1.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z, Tang H, Ventura M, Cardone MF, Marques-Bonet T, She X, Pevzner PA, Eichler EE. 2007. Ancestral reconstruction of segmental duplications reveals punctuated cores of human genome evolution. Nat Genet 39: 1361–1368. 10.1038/ng.2007.9 [DOI] [PubMed] [Google Scholar]
- Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ. 2004. The UCSC Table Browser data retrieval tool. Nucleic Acids Res 32: D493–D496. 10.1093/nar/gkh103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Barber GP, Casper J, Clawson H, Cline MS, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L, Haeussler M, et al. 2014. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res 42: D764–D770. 10.1093/nar/gkt1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Inagaki H, Kogo H, Ohye T, Yamada K, Emanuel BS, Kurahashi H. 2008. Two different forms of palindrome resolution in the human genome: deletion or translocation. Hum Mol Genet 17: 1184–1191. 10.1093/hmg/ddn008 [DOI] [PubMed] [Google Scholar]
- Kato T, Kurahashi H, Emanuel BS. 2012. Chromosomal translocations and palindromic AT-rich repeats. Curr Opin Genet Dev 22: 221–228. 10.1016/j.gde.2012.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. 2002. The Human Genome Browser at UCSC. Genome Res 12: 996–1006. 10.1101/gr.229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinjan DA, van Heyningen V. 2005. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet 76: 8–32. 10.1086/426833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurahashi H, Emanuel BS. 2001. Long AT-rich palindromes and the constitutional t(11;22) breakpoint. Hum Mol Genet 10: 2605–2617. 10.1093/hmg/10.23.2605 [DOI] [PubMed] [Google Scholar]
- Leach DRF. 1994. Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. Bioessays 16: 893–900. 10.1002/bies.950161207 [DOI] [PubMed] [Google Scholar]
- Leung AK, Jin N, Yip KY, Chan TF. 2017. OMTools: a software package for visualizing and processing optical mapping data. Bioinformatics 33: 2933–2935. 10.1093/bioinformatics/btx317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy-Sakin M, Pastor S, Mostovoy Y, Li L, Leung AK, McCaffrey J, Young E, Lam ET, Hastie AR, Wong KH, et al. 2019. Genome maps across 26 human populations reveal population-specific patterns of structural variation. Nat Commun 10: 1025 10.1038/s41467-019-08992-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis S, Akgun E, Jasin M. 1999. Palindromic DNA and genome stability: further studies. Ann N Y Acad Sci 870: 45–57. 10.1111/j.1749-6632.1999.tb08864.x [DOI] [PubMed] [Google Scholar]
- Liu H, Heath SC, Sobin C, Roos JL, Galke BL, Blundell ML, Lenane M, Robertson B, Wijsman EM, Rapoport JL, et al. 2002. Genetic variation at the 22q11 PRODH2/DGCR6 locus presents an unusual pattern and increases susceptibility to schizophrenia. Proc Natl Acad Sci 99: 3717–3722. 10.1073/pnas.042700699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev KS, Gordenin DA, Resnick MA. 2002. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108: 183–193. 10.1016/S0092-8674(02)00614-1 [DOI] [PubMed] [Google Scholar]
- McDonald-McGinn DM, Minugh-Purvis N, Kirschner RE, Jawad A, Tonnesen MK, Catanzaro JR, Goldmuntz E, Driscoll D, LaRossa D, Emanuel BS, et al. 2005. The 22q11.2 deletion in African-American patients: an underdiagnosed population? Am J Med Genet 134A: 242–246. 10.1002/ajmg.a.30069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald-McGinn DM, Sullivan KE, Marino B, Philip N, Swillen A, Vorstman JA, Zackai EH, Emanuel BS, Vermeesch JR, Morrow BE, et al. 2015. 22q11.2 deletion syndrome. Nat Rev Dis Prim 1: 15071 10.1038/nrdp.2015.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelovsky E, Frisch A, Carmel M, Patya M, Zarchi O, Green T, Basel-Vanagaite L, Weizman A, Gothelf D. 2012. Genotype-phenotype correlation in 22q11.2 deletion syndrome. BMC Med Genet 13: 122 10.1186/1471-2350-13-122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider VA, Graves-Lindsay T, Howe K, Bouk N, Chen H-C, Kitts PA, Murphy TD, Pruitt KD, Thibaud-Nissen F, Albracht D, et al. 2017. Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res 27: 849–864. 10.1101/gr.213611.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh TH, Kurahashi H, Saitta SC, O'Hare AM, Hu P, Roe BA, Driscoll DA, McDonald-McGinn DM, Zackai EH, Budarf ML, et al. 2000. Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: genomic organization and deletion endpoint analysis. Hum Mol Genet 9: 489–501. 10.1093/hmg/9.4.489 [DOI] [PubMed] [Google Scholar]
- Shaikh TH, O'Connor RJ, Pierpont ME, McGrath J, Hacker AM, Nimmakayalu M, Geiger E, Emanuel BS, Saitta SC. 2007. Low copy repeats mediate distal chromosome 22q11.2 deletions: sequence analysis predicts breakpoint mechanisms. Genome Res 17: 482–491. 10.1101/gr.5986507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siva N. 2008. 1000 Genomes project Nat Biotechnol 26: 256 10.1038/nbt0308-256b [DOI] [PubMed] [Google Scholar]
- Steinberg KM, Antonacci F, Sudmant PH, Kidd JM, Campbell CD, Vives L, Malig M, Scheinfeldt L, Beggs W, Ibrahim M, et al. 2012. Structural diversity and African origin of the 17q21.31 inversion polymorphism. Nat Genet 44: 872–880. 10.1038/ng.2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant PH, Kitzman JO, Antonacci F, Alkan C, Malig M, Tsalenko A, Sampas N, Bruhn L, Shendure J, Eichler EE. 2010. Diversity of human copy number variation and multicopy genes. Science 330: 641–646. 10.1126/science.1197005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant PH, Mallick S, Nelson BJ, Hormozdiari F, Krumm N, Huddleston J, Coe BP, Baker C, Nordenfelt S, Bamshad M, et al. 2015. Global diversity, population stratification, and selection of human copy-number variation. Science 349: aab3761 10.1126/science.aab3761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Juan L, Rosell J, Sánchez-de-la-Torre M, Fibla J, Heine-Suñer D. 2007. Analysis of meiotic recombination in 22q11.2, a region that frequently undergoes deletions and duplications. BMC Med Genet 8: 14 10.1186/1471-2350-8-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischenfeldt J, Symmons O, Spitz F, Korbel JO. 2013. Phenotypic impact of genomic structural variation: insights from and for human disease. Nat Rev Genet 14: 125–138. 10.1038/nrg3373 [DOI] [PubMed] [Google Scholar]
- Weisenfeld NI, Kumar V, Shah P, Church DM, Jaffe DB. 2017. Direct determination of diploid genome sequences. Genome Res 27: 757–767. 10.1101/gr.214874.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.