Abstract
DNA replication occurs in a defined temporal order known as the replication timing (RT) program and is regulated during development, coordinated with 3D genome organization and transcriptional activity. However, transcription and RT are not sufficiently coordinated to predict each other, suggesting an indirect relationship. Here, we exploit genome-wide RT profiles from 15 human cell types and intermediate differentiation stages derived from human embryonic stem cells to construct different types of RT regulatory networks. First, we constructed networks based on the coordinated RT changes during cell fate commitment to create highly complex RT networks composed of thousands of interactions that form specific functional subnetwork communities. We also constructed directional regulatory networks based on the order of RT changes within cell lineages, and identified master regulators of differentiation pathways. Finally, we explored relationships between RT networks and transcriptional regulatory networks (TRNs) by combining them into more complex circuitries of composite and bipartite networks. Results identified novel trans interactions linking transcription factors that are core to the regulatory circuitry of each cell type to RT changes occurring in those cell types. These core transcription factors were found to bind cooperatively to sites in the affected replication domains, providing provocative evidence that they constitute biologically significant directional interactions. Our findings suggest a regulatory link between the establishment of cell-type–specific TRNs and RT control during lineage specification.
During development, specific transcriptional programs and epigenetic landscapes are established that maintain the identities and functionality of the specialized cell types that emerge. Despite characterization of changes in transcriptome and epigenome during development (Gifford et al. 2013; Xie et al. 2013; Roadmap Epigenomics Consortium et al. 2015; Tsankov et al. 2015), little is known about the role of spatiotemporal genome organization in cell fate specification. Changes in gene activity and chromatin 3D organization are coordinated with dynamic changes in the temporal order of genome duplication, known as the replication timing (RT) program (Hiratani et al. 2008, 2010; Rivera-Mulia et al. 2015, 2018a). Spatiotemporal control of RT is conserved in all eukaryotes (Rivera-Mulia and Gilbert 2016a; Solovei et al. 2016; Zhao et al. 2017), and alterations in the RT program are associated with different diseases (Ryba et al. 2012; Gerhardt et al. 2014; Rivera-Mulia et al. 2017, 2019; Sasaki et al. 2017). RT is regulated during development in discrete chromosome units, referred to as replication domains (RDs), that segregate into distinct nuclear compartments (Jackson and Pombo 1998; Ryba et al. 2010; Rivera-Mulia and Gilbert 2016b; Rivera-Mulia et al. 2018a). Hence, we reasoned that RT can be exploited to characterize the gene regulatory relationships established during development.
Previously, we generated a comprehensive database of RT programs during human lineage specification and found that approximately half of the genome undergoes dynamic changes that are closely coordinated with the establishment of transcriptional programs (Rivera-Mulia et al. 2015). Additionally, our previous findings showed that developmentally RT regulated genes are higher in the hierarchy of transcriptional regulatory networks (TRNs), suggesting that this type of gene regulates all other genes during cell fate commitment (Rivera-Mulia et al. 2015). However, strong gene expression was not restricted to early replicating genomic regions, and transcriptional activation during cell fate commitment often preceded RT changes (Rivera-Mulia et al. 2015; Rivera-Mulia and Gilbert 2016b). In fact, although a long-standing correlation between early replication and gene expression has been observed in all eukaryotes, the link between RT and transcriptional activity is complex, and causal relationships have not been established (Solovei et al. 2016; Rivera-Mulia and Gilbert 2016b). Here, we explored the possibility that RT can be regulated by the establishment of complex regulatory circuits of transcription factors rather than by the transcription levels of genes within each RD.
Results
Construction of correlation-based RT networks
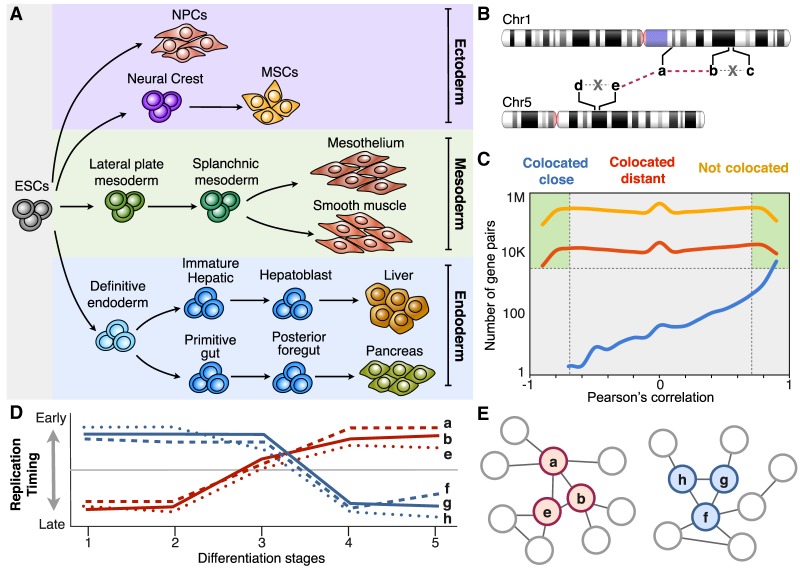
To determine whether the RT program can be used for novel gene regulatory interaction identification, we defined a model that describes the relationship between all possible combinations of gene pairs (nodes), establishing gene interactions (edges) according to their correlated RT patterns during differentiation toward cell types representing the three main germ layers (Fig. 1A). Distinct filters were applied in our model: (1) We included only genes that change RT during cell differentiation (removing RT constitutive genes), and (2) we included edges only between genes separated by at least 500 kb and/or in different chromosomes (Fig. 1B,C; Supplemental Fig. S1). Separation by >500 kb was chosen to remove gene pairs within the same RD, which we have shown vary in size from 0.4 Mb to 0.8 Mb (Hiratani et al. 2008; Pope et al. 2014; Rivera-Mulia et al. 2015), and included only distal gene interactions; (3) we established gene interactions (edges) between significantly correlated gene pairs (statistical significance of gene pair interactions was calculated as Bonferroni's adjusted P-values; see Methods). After applying these filters, gene pairs and interaction edges were obtained (green boxes in Fig. 1C) and RT networks constructed. As expected, most of the genes colocated within 500 kb had correlation values ≥0.9, and only a small fraction showed lack of correlation or negative correlation (blue line in Fig. 1C). In contrast, gene pairs separated by >500 kb, or from distinct chromosomes, showed correlation values ranging from −0.9 to +0.9 (red and orange lines in Fig. 1C). Here, we used gene pairs in trans (from distinct chromosomes) as well as gene pairs in cis but separated by >500 kb (classified as colocated distant) to construct distinct models of RT networks (Fig. 1C). Figure 1, D and E, illustrates hypothetical examples of two distinct RT patterns along a particular cell differentiation lineage, the correlations for which constitute connections between gene pairs exploited to construct the corresponding RT networks. These findings revealed correlated RT changes for thousands of gene pairs during differentiation that are likely to be mediated by common mechanisms related to differentiation. These gene pairs were then used to construct the following networks.
Figure 1.
Coordinated changes in RT can be exploited to construct gene regulatory networks. (A) RT programs of distinct cell types representing intermediate stages of human embryonic stem cell differentiation toward endoderm, mesoderm, and ectoderm were analyzed for the construction of RT regulatory networks. (B) Depiction of different highly RT-correlated genes from distinct chromosomes and the establishment of network interaction edges between them. From all possible combinations of gene pairs, those colocated within 500 kb were removed from the analysis to include exclusively distal gene interactions. Regulatory interactions (edges) between gene pairs are considered only for genes located >500 kb apart (colocated distant) or in distinct chromosomes (not colocated); that is, edges between genes b-c and d-e were not included in the analysis. (C) Number of gene pairs as function of RT correlation for distinct categories of gene pairs: colocated close (within 500 kb), colocated distant (separated by >500 kb), and not colocated (from different chromosomes). Only gene pairs with significant RT correlations and located at least 500 kb apart were considered. (D) RT patterns of distinct hypothetical genes during five distinct differentiation intermediate stages (distinct number of differentiation intermediates were used from each pathway). (E) Construction of RT regulatory networks based on the significant Pearson's correlation (distance between nodes are proportional to the correlation strength).
RT subnetwork communities are composed of genes with specific functional annotations
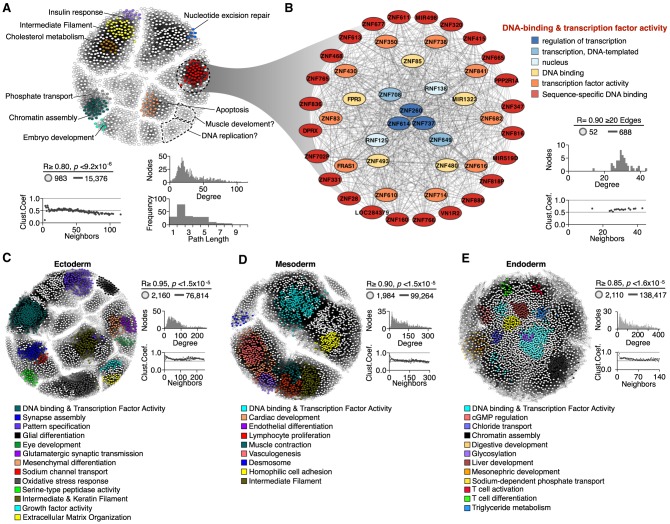
To examine whether correlated RT changes are linked to functional gene regulatory interactions, we constructed RT networks among significantly correlated gene pairs and characterized their connectivity and association to specific functional annotations. First, we constructed a RT network including all significantly RT-correlated gene pairs across all differentiation pathways, positioning each node in a 2D space using the force-directed layout algorithm in Cytoscape (Shannon et al. 2003), with the edge length being proportional to the Pearson's correlation strength (Fig. 2A; Supplemental File 1). The resulting RT network was composed of highly interconnected groups of nodes (with >90% of the nodes having a degree distribution greater than 25), with very short distances (path length less than 3) and high clustering coefficients within each group (Fig. 2A). Next, we identified local neighborhoods (subnetwork communities) within the global networks according to the connectivity and distances between nodes (Blondel et al. 2008). Finally, to validate the biological significance of the novel gene interactions detected within RT networks, we examined their functional organization by performing ontology analysis of each subnetwork community using the spatial analysis of functional enrichment (SAFE) algorithm (Baryshnikova 2016; Costanzo et al. 2016). We found subnetwork communities with highly interconnected nodes of genes involved in specific functions, which were color-coded based on the enrichment of functional ontology annotations (Fig. 2A). Closer inspection of subnetwork communities annotated with specific functions grouped the genes according to their ontology terms (Fig. 2B; Supplemental File 1). These findings confirm that gene regulatory interactions can be identified exploiting the cell-type–specific RT program.
Figure 2.
Functional annotation of RT networks. (A) RT-correlated gene pairs across all differentiation pathways from human ES cells were identified and exploited to construct a RT network. (B) Detailed subnetwork connectivity from the RT network shown in A and its respective node organization per ontology term. (C–E) RT networks and functional subnetwork communities constructed for each differentiation pathway: ectoderm (C), mesoderm (D), and endoderm (E). Interaction edges between gene pairs were established only for significantly correlated nodes (Bonferroni's adjusted P-values with alpha = 0.05/n), and the subset of most connected nodes (>20 edges) was used to visualize RT networks displayed as 2D maps in Cytoscape (Shannon et al. 2003). Pearson's correlation and Bonferroni-corrected P-value thresholds, as well as the connectivity analysis for each network, are shown (degree distribution, path lengths, and clustering coefficients). Highly interconnected subnetwork communities were annotated with functional ontology terms using the SAFE algorithm (Baryshnikova 2016) and are displayed in distinct colors.
Distinct coordinated changes in RT are restricted to specific differentiation pathways (Rivera-Mulia et al. 2015). Thus, to identify lineage-specific gene interactions, we generated RT networks of differentiation toward each germ layer separately by combining the cell types of each lineage (ectoderm, mesoderm, and endoderm). Then we visualized these RT networks as 2D maps and identified subnetwork communities as described above. Consistently, we found RT networks composed by highly interconnected subnetwork communities associated with specific functions relevant to each germ layer (Fig. 2C–E; Supplemental File 1). For ectodermal lineage, we found subnetwork communities for genes associated to synapsis assembly, glial differentiation, eye development, glutamatergic synaptic transmission, and mesenchymal differentiation (Fig. 2C). For mesodermal lineages, we identified communities annotated for cardiac development, endothelial and lymphocyte differentiation, muscle contraction, and vasculogenesis (Fig. 2D). Finally, for endodermal lineages, we obtained subnetwork communities linked to liver, digestive, and mesonephric development, as well as T cell differentiation and activation (Fig. 2E). Complete lists of genes within each subnetwork community and their interactions are shown in the Supplemental File 1. Subnetwork communities without ontology enrichment might identify regulatory relationships that have not been previously annotated. To test this hypothesis, we explored the genes within these communities and identified subnetworks communities that link genes involved in specific processes. For example, we identified a subnetwork community associated with DNA replication that includes GINS1, MCM4, MND1, and NCAPG2, genes that are required for replication, cell cycle regulation, and chromosome condensation (Fig. 2A). We also identified a community linked to muscle development that includes RSPO2, ADAMTS5, ADAMTS1, and MIR490, genes that have been linked to myogenesis control (Han et al. 2011; Stupka et al. 2013; Sun et al. 2013; Du et al. 2017). These findings validate the functional relationships between the nodes of these novel interactions.
To confirm that the coordinated changes in RT define the functional subnetwork communities, we constructed randomized networks and performed the ontology analysis. To do so, we preserved exactly the same gene nodes and number of interactions in each network but randomly shuffled all edges. Ontology analysis of these randomized networks resulted in generic ontology terms (“cellular regulation,” “biological process,” “regulation of development,” and “cellular metabolism”) and did not identified subnetwork communities annotated with more specific terms (Supplemental File 1).
Correlated RT networks vary greatly in size depending on the parameters selected; here we considered only the most significant correlated nodes that switch RT between the very early (>0.3) and very late (<−0.3) replication and with >20 degree count (number of edges). Relaxing the parameters (for early and late replication, significance threshold, and degree count) would produce larger networks with higher number of local neighborhoods (at the expense of computational time). To evaluate the effect of the distinct thresholds on the network size and connectivity, we obtained correlated RT networks using distinct thresholds and quantified the number of nodes and edges obtained. We found that increasing correlation threshold decreases the size of the networks (Supplemental Table S1). Moreover, because the distinct lineages vary in the number of differentiation stages (2–5), we evaluated the effect of the number of time points in the construction of correlated RT networks. To do so, we generated correlated RT networks for pancreas differentiation using two, three, four, and five differentiation stages. We found that higher number of time points increases the network size but reduces its connectivity (Supplemental Table S2). These results suggest that increasing the number of differentiation time points improves the network analysis as correlation values can be estimated more precisely. Additionally, most of the genes that change RT during development switch from early to late replication (EtoL) or from late to early replication (LtoE); however, a small fraction of genes changes RT back and forth during development (Rivera-Mulia et al. 2015). To characterize these types of changes, we constructed correlated LtoEtoL and EtoLtoE RT networks. We identified subnetwork communities annotated with lineage-specific functions (Supplemental Fig. S2). For endodermal cell types, we identified a subnetwork community with LtoEtoL changes associated with regulation of protein dephosphorylation, as well as a subnetwork community with EtoLtoE changes associated with protein phosphorylation (Supplemental Fig. S2). Because RT is coordinated with transcriptional potential, these results suggest the transient induction of phosphatases and down-regulation of kinases correlated with RT changes during endodermal differentiation. For ectodermal cell types, we found that LtoEtoL networks contain subnetwork communities associated with the epithelial–mesenchymal transition and mesenchymal differentiation, whereas EtoLtoE networks included communities linked to neurogenesis and axon development (Supplemental Fig. S2). Overall, our results revealed that dynamic changes in RT can be exploited to characterize the complex regulatory interactions established during cell fate commitment.
Directional RT networks identify regulatory interactions of cell fate commitment
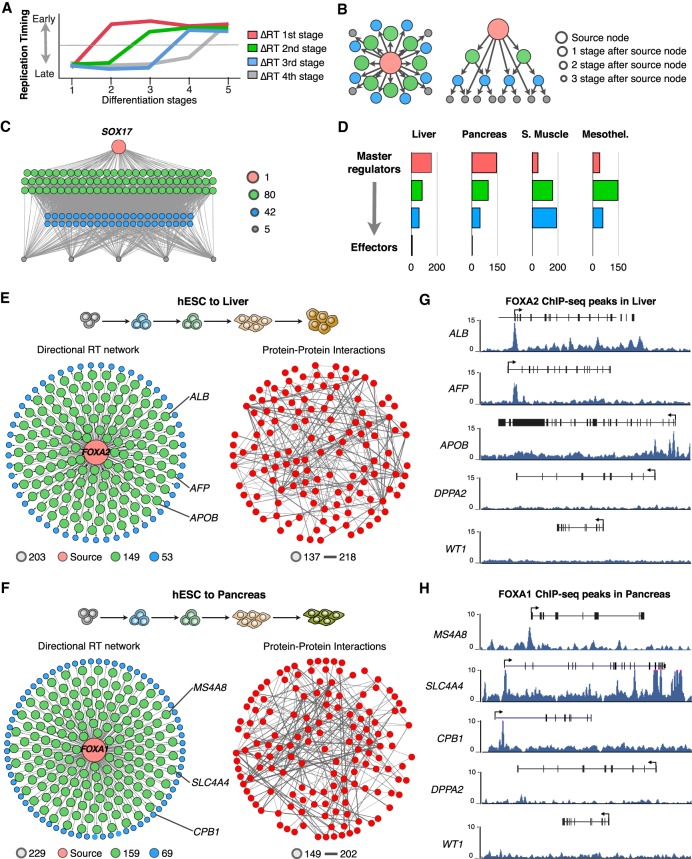
The RT networks described above (Fig. 2) identified subnetwork communities enriched for specific functional annotations. However, these correlated RT networks are not directional and, as such, cannot characterize the hierarchical relationships or identify potential targets for regulators. Hence, here we analyzed whether RT can be also exploited to characterize the hierarchical gene regulatory interactions during lineage commitment. To do so, we took advantage of the data collected at multiple intermediate differentiation stages (more than three differentiation intermediates) toward pancreas, liver, smooth muscle, and mesothelium and constructed directional RT networks for each of the specific differentiation pathways. First, we filtered all genes that change RT significantly (and classified them according to the order of RT changes during each differentiation pathway (Fig. 3A), identified those that change during the earliest cell fate transition, assigned directional edges to genes that changed RT in the subsequent differentiation stage, and repeated this for each stage (see Methods). Then directional RT networks were displayed either in 2D maps or in a hierarchical arrangement, and nodes were color/size-coded according to the order of the changes in RT during distinct differentiation pathways (Fig. 3B). Construction of directional RT networks constitutes a novel approach to identify the genes with earliest RT changes during cell fate commitment (red nodes in Fig. 3C) as well as their downstream relationships in terms of temporal ordering of RT change (green, blue, and gray nodes in Fig. 3C). To test this approach and its value for identifying novel gene regulatory interactions, we constructed directed RT networks for known transcription factors that are key regulators for either pluripotency or cell differentiation. Early replication correlates with transcriptional induction, and changes from EtoL are associated with gene down-regulation (Rivera-Mulia et al. 2016b). Consistently, the earliest EtoL RT changes during differentiation are enriched in genes associated with pluripotency regulation (Rivera-Mulia et al. 2015). To characterize this type of gene relationship, we constructed EtoL directional networks for each differentiation pathway using as source nodes genes involved in pluripotency and maintenance of early progenitors (Supplemental Fig. S3). Next, to characterize the establishment of gene interactions associated with cell fate commitment, we focused on changes from LtoE and constructed LtoE directional RT networks using as source nodes key regulators of cell fate commitment for each differentiation pathway. Consistently, known downstream targets were identified among the downstream targets predicted by the directed RT networks (Supplemental Fig. S4). Hence, we classified the nodes in hierarchy levels according to the order of RT changes: “Master regulators” were defined as the genes that change RT in the earliest differentiation transition with the largest degree of connectivity (red node in Fig. 3C), whereas downstream nodes were classified as “managers” and “effectors” according to the time during differentiation when they change RT. Manager genes were defined as those that change RT in intermediate differentiation stages and effector genes as those that change in the latest differentiation stage (Fig. 3C). Because endodermal lineages have more differentiation stages than mesodermal lineages, genes from the two intermediate stages were classified as managers. Establishment of RT networks occurs differently for each germ layer. For endoderm cell types (liver and pancreas), most of the changes occur very early during differentiation, with 51% of genes behaving as master regulators (switching RT during the earliest transitions), 47% as managers, and only 2% as effectors (Fig. 3D). In contrast, for mesodermal cell types (smooth muscle and mesothelium), fewer master regulators were connected with an increased number of downstream nodes. Only 11%–17% of the genes were classified as master regulators, 40%–60% as managers, and 24%–48% as effectors.
Figure 3.
Directional RT networks. (A) Distinct genes change RT at different time points during cell fate commitment, and the order of RT changes can be used to construct directional RT networks. Red genes change during the first transition between differentiation stages, whereas gray genes change at the last differentiation transition. (B) 2D maps and hierarchical displays of a directional RT networks were constructed based on a source gene (central node) and downstream connected nodes. (C) An exemplary directional RT network for liver differentiation is shown. The central node is SOX17, and all downstream nodes were connected based on temporal times during differentiation at which they change RT. (D) Classification of gene nodes according to their hierarchy in directed RT networks. Master regulators were operationally defined as those genes that change RT in the earliest differentiation transition and have the largest degree of connectivity (red nodes). Green and blue nodes represent “manager” nodes that are connected to the final “effector” nodes at the lowest level of the network (gray nodes). Node distribution in each of the hierarchical levels for each differentiation pathway is shown. (E,F) Directed RT networks of FOXA2 and FOXA1 during liver and pancreatic differentiation, respectively. FOXA2 and FOXA1 (red node) change from late to early replication during the earliest stages of differentiation; potential downstream genes were identified as those genes that change in subsequent differentiation stages (green and blue nodes). Sixty-five percent or more of the genes within the directed RT networks interact with each other at the protein level (protein–protein interactions were obtained from the STRING database; nodes were mapped using the same layout as the directional RT network; and edge thickness indicates the strength of the data supporting each interaction). (G,H) ChIP-seq signals show the binding of FOXA2 and FOXA1 at the promoters of predicted hepatic-specific and pancreas-specific downstream genes (ALB, AFP, and APOB for liver and CPB1, MS4A8, and SLC4A4 for pancreas). A pluripotency gene (DPPA2) and a mesodermal-specific gene (WT1) are shown as a control.
The presence of known targets for key differentiation regulators between the predicted downstream targets in the directed RT networks suggests that the interactions identified by this approach reflect functional gene relationships. To validate these gene interactions, we constructed directional networks for transcription factors key for the differentiation control toward liver and pancreas. Then, we obtained the protein–protein interactions from the STRING database (Szklarczyk et al. 2019) and identified the known interactions between the nodes in our networks (Fig. 3E,F). We found that 65%–75% of the genes within the directional RT networks interact at the protein level with at least other gene, with ≥40% genes interacting with other five to 20 in the network (Fig. 3F; Supplemental Fig. S4). Moreover, during the preparation of this paper, ChIP-seq data sets for transcription factors binding became available for liver and pancreas (Diaferia et al. 2016; Davis et al. 2018; Wang et al. 2018), permitting us to further validate these downstream targets predicted by the novel RT networks. We constructed directed RT networks for liver and pancreatic differentiation using FOXA2 and FOXA1 as source nodes, respectively (Fig. 3E,F), and calculated the enrichment of ChIP-seq peaks of these transcription factors at the predicted downstream targets. We found moderate enrichment of FOXA2 peaks and a highly significant enrichment of FOXA1 peaks at the promoters of predicted downstream targets for liver and pancreas development, respectively (Supplemental Table S3). Exemplary target genes and ChIP-seq signals for downstream nodes of the FOXA2 and FOXA1–directed RT networks are shown in Figure 3G,H. These findings validate the ability of the directed RT networks to identify novel directional regulatory mechanisms and place them into their temporal order during cell fate commitment.
RT network edges overlap with known transcriptional regulatory interactions
To determine the extent to which RT networks capture the regulatory interactions characterized by other methods, we analyzed their overlap with TRNs using a previously described set of cell-type–specific networks of TFs (Neph et al. 2012). First, we identified the cell types that most closely match the TRNs to our RT networks, as follows: hESC-derived hepatocytes were compared to TRNs from HepG2, a liver cancer cell line that retains morphological and functional hepatocyte properties (Knowles et al. 1980; Berger et al. 2015); hESC-derived mesothelial cells were compared to TRNs from HCF cells, cardiac fibroblasts that during development and in vitro differentiation can be derived from mesothelial cells (Mutsaers 2004); and hESC-derived neural precursors were compared to TRNs from the SK-N-SH cell line after treatment with retinoic acid. SK-N-SH cells were derived from a neuroblastoma and retinoic acid causes differentiation to neural phenotype (Preis et al. 1988). Next, we constructed RT networks using only the subset of genes present in the TRNs (475 TFs) that change RT and are significantly correlated in their RT patterns in each differentiation pathway. Finally, we identified the number of common and unique edges between RT networks and TRNs. We found that in all three cases there was a highly significant overlap compared with the expected overlap by randomly selecting the same number of edges (Fig. 4A). In fact, significant overlap was also observed when all cell types from both RT networks and TRNs were classified per germ layer (Supplemental Table S4), and common edges were identified for ectoderm and mesoderm, even when distinct cell types were used for each germ layer. These results confirm a high overlap between RT and transcriptional networks and further validate the gene regulatory interactions identified using the RT program.
Figure 4.
RT and transcriptional networks overlap and can be combined into composite networks. (A) Overlap analysis of RT and TRNs interaction edges. RT networks were constructed for matching cell types in the TRNs (Neph et al. 2012), and common and unique interaction edges were identified. Only genes within the TRNs were used (475 transcription factors), maintaining the filters for significant correlation and gene pairs in trans (separated by >500 kb or in distinct chromosomes). A hypergeometric test was performed to test the overlap significance (P-values are shown). (B,C) RT networks are enriched in motifs with multiple nodes. (B) The most enriched motif for each differentiation pathway is shown. (C) Motif frequency distribution at distinct thresholds in Pearson's correlation values (each bar represents a distinct correlation threshold –R). (D) Construction of composite networks by combining RT and transcriptional networks. RT networks were used to define the “base network,” which includes all nodes of RT-correlated genes; interaction edges between RT nodes were extracted from TRNs. Composite networks included all RT edges (black undirected lines), transcriptional edges within RT network motifs (directed solid red arrows), and transcriptional edges outside RT network motifs (directed dashed gray arrows). Exemplary composite networks for liver (E) and mesothelium differentiation (F) are shown. Pearson's correlation and Bonferroni's corrected P-values thresholds, as well as the number of nodes and edges, are shown.
Building blocks of RT networks are motifs with multiple nodes
Previous studies explored the architecture of gene regulatory relationships by analyzing either transcriptional or protein interactions and found that complex cellular networks are constituted by sets of small network motifs, such as interactions between transcription factors and their targets (Zhang et al. 2005; Alon 2007). Here, we performed a topology characterization of RT networks constructed with the subset of genes (475 TFs) present in the TRNs (Neph et al. 2012) to explore the most overrepresented patterns of connectivity. We computed all possible network motifs composed by two to four nodes and identified the motifs with high enrichment in each RT network constructed per differentiation pathway (Supplemental Fig. S5–S7). Statistical significance (P-value <0.01) of each motif occurrence in the RT networks was calculated by comparing to the frequency of the same motif in randomized networks (Milo et al. 2002; Baiser et al. 2016; Elhesha and Kahveci 2016). The most frequent motifs with three and four nodes in each differentiation pathway are shown in Figure 4B. Previous observations in transcriptional and protein regulatory networks that suggest that gene regulatory networks are composed mainly by small motifs with two to three nodes (Yeger-Lotem et al. 2004; Alon 2007). In contrast, we found that the most enriched motifs are the motifs with higher number of nodes (Fig. 4C). In fact, motif enrichment increases with higher number of nodes, and this distribution is maintained at distinct Pearson's correlation thresholds and in all differentiation pathways analyzed (Fig. 4C). These findings suggest a higher connectivity for RT-correlated genes than that previously observed in transcriptional and protein regulatory networks. Additionally, because significant overlap between RT and transcriptional networks was observed in all differentiation pathways (Fig. 4A), we examined the presence of transcriptional edges within the RT networks motifs. We found that transcriptional edges were present in most of the motifs, suggesting an interconnectivity between TRNs and RT networks (Supplemental Figs. S5–S7).
Composite networks: combining RT and TRNs
To better understand how the regulatory circuitries are established during cell fate commitment, we constructed a model of composite regulatory networks by merging RT and TRNs. Previous studies showed that distinct types of interactions (such as protein–protein and transcription regulation) can be combined to explore more complex cellular circuitries (Yeger-Lotem et al. 2004; Vidal et al. 2011). Here, we combined the regulatory interactions observed in RT networks with those identified in the TRNs between TFs detected for the closest cell types. First, we obtained all gene nodes present in both correlated RT and TFs networks (Fig. 4D). Then we combined all the interactions between RT nodes with the transcriptional interactions in the TRNs from matching cell types and constructed composite networks (Fig. 4D). RT networks for each differentiation pathway are constituted by multiple unconnected motifs of four or fewer nodes; however, the addition of transcriptional edges revealed more complex and highly interconnected networks with all nodes interacting with at least three other nodes (Fig. 4B–D). Exemplary composite networks for liver and mesothelium show the connectivity of known key regulators for each differentiation pathway (Fig. 4E,F).
Bipartite networks reveal transcription factors as regulators of RT
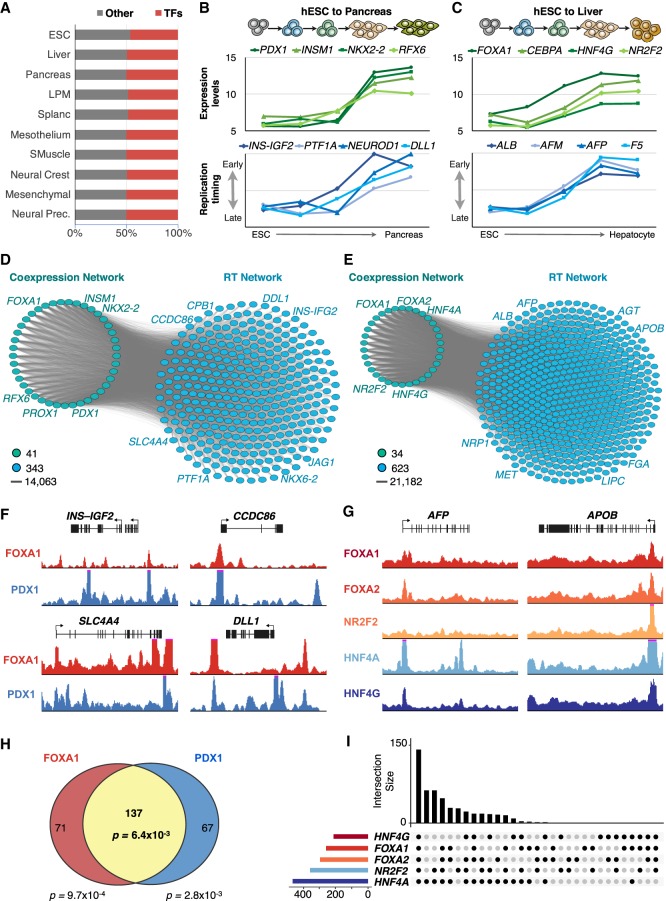
To further characterize the gene regulatory interactions established during cell fate commitment, we analyzed the cell-type–specific transcriptomes and their relationships with RT. First, we exploited the genome-wide transcriptome data that we obtained previously from the same cell types as for RT (Rivera-Mulia et al. 2015). Our highly comprehensive characterization of gene expression, including multiple replicates for each differentiation stage, allowed us to identify with confidence the genes that are differentially expressed during cell fate commitment toward each cell type and the genes that better distinguish each intermediate stage. Coexpressed genes were identified by weighted correlation network analysis (Langfelder and Horvath 2008). Strong correlations between gene expression levels are widely used to identify regulatory interactions (D'haeseleer et al. 2000; Li 2002; Allocco et al. 2004; Novak and Jain 2006; Horvath and Dong 2008; Laurenti et al. 2013; Gabr et al. 2015); thus, we constructed coexpressed networks for each differentiation pathway. To decrease the complexity of the data to a computationally manageable size, we focused on the top 100 genes that are significantly coexpressed in specific cell types/intermediate differentiation stages. In all differentiation pathways and for each differentiation stage, we found that transcription factors were among the most significant genes distinguishing each cell type (Fig. 5A). Moreover, ontology analysis (Ashburner et al. 2000; The Gene Ontology Consortium 2015) using the different subsets of genes revealed strong enrichment of genes regulating the specification of each cell type (Supplemental Table S5).
Figure 5.
Bipartite networks. (A) Transcription factor expression patterns distinguish each cell type/differentiation intermediate. Cell-type–specific expression patterns were analyzed to identify the most significant differentially coexpressed genes, and then the resulting genes were classified as TFs or other type. (B) Expression patterns of exemplary key TFs of pancreas development correlate with the RT of downstream regulators of pancreatic differentiation. (C) Expression patterns of exemplary key TFs of liver development correlate with the RT of downstream regulators of hepatic differentiation. (D,E) Bipartite networks of pancreas (D) and liver (E) differentiation. Bipartite networks were constructed based on the correlation between transcriptional levels of the genes in the TRN side and RT changes of genes in the RT side. TRNs were constructed from the top genes coexpressed in pancreas and liver, respectively. RT network sides were constructed by identification of genes with RT patterns highly correlated with the expression levels of genes at the TRN side. Only edges between TRN and RT networks are shown (with each gene in the RT side connected with at least 50% of the nodes in the TRN side), as all nodes within each network are interconnected with all others. (F) ChIP-seq signals show the co-occupancy of FOXA1 and PDX1 at exemplary pancreatic-specific downstream targets predicted by the bipartite network shown in D. (G) ChIP-seq signals show the co-occupancy of FOXA1, FOXA2, NR2F2, HNF4A, and HNF4G at hepatic-specific downstream targets predicted by the bipartite networks shown in E. (H,I) Co-occupancy analysis for the pancreatic-specific and hepatic-specific TFs at the promoters of downstream targets.
Because we found that (1) subnetwork communities associated with transcription factor activity were identified in all RT-correlated networks (Fig. 2), (2) interactions between TFs in TRNs significantly overlap with RT networks (Fig. 4), and (3) TF expression patterns distinguish each cell type (Fig. 5A), we hypothesize that gene regulation by TFs might be critical not only for cell-type–specific transcriptional program establishment but also for RT program control during cell fate specification. To analyze the potential role of TF in RT regulation, we constructed bipartite networks in which we identified the correlated patterns of RT that correlated with the expression levels of the TRNs. First, we identified the genes whose RT patterns are correlated with the expression levels of the top 100 genes that distinguish each cell type. Exemplary gene expression levels for a subset of TFs critical for pancreas and liver differentiation are shown in Figure 5, B and C, as well as the corresponding genes with correlated patterns for RT regulation. Then, we constructed bipartite networks that consist of two independent but interconnected networks: “coexpression networks” that contain genes coexpressed in specific developmental stages/cell types and the RT network contains genes whose RT changes were highly correlated with the expression patterns from the coexpression network. These gene interactions are in trans and cannot be explained by the “simple” correlation between RT and transcription but correlated transcriptional changes with remote, unlinked RT changes. Hence, the regulatory interactions described here cannot be explained by the previously recognized correlation. Interaction edges between each side of the bipartite networks were established for nodes in the RT side that correlate (R ≥ 0.75) with the transcriptional levels of at least 50% of the genes in the coexpression network. Bipartite networks for pancreatic and liver differentiation are shown in Figure 5, D and E, respectively. TFs that regulate these specific differentiation pathways are highlighted in each bipartite network at the transcriptional side (Fig. 5D,E), and known pancreatic-specific and liver-specific downstream genes were found in for each bipartite network at the RT side (Fig. 5D,E). These results suggest that establishment of TRNs during cell differentiation might be required for RT control.
During the preparation of this manuscript, ChIP-seq data sets for transcription factors binding became available for liver and pancreas (Diaferia et al. 2016; Davis et al. 2018; Wang et al. 2018). Hence, to further analyze the relationship of TFs whose expression strongly correlates with the RT of downstream genes in trans, we analyzed ChIP-seq data for known TFs required for pancreatic and liver cell fate commitment (Fig. 5F,G). For pancreatic differentiation, we analyzed whether downstream targets predicted at the RT side of the bipartite network (Fig. 5D), contain binding sites for FOXA1 (endoderm-specific TF) and PDX1 (pancreatic-specific TF). We found ChIP-seq peaks for both TFs in close proximity to the promoters (<20 kb) of the downstream targets predicted in the bipartite network (Fig. 5F). In fact, 80% of the predicted downstream genes were bound by at least one of the two TFs, and 40% presented highly significant co-occupancy (P-value = 6.4 × 10−3) for both TFs at the promoters (Fig. 5H). For liver differentiation, we obtained ChIP-seq data for five different TFs required for the regulation of hepatic differentiation (FOXA1, FOXA2, NR2F2, HNF4A, and HNF4G), which allowed us to test for co-occupancy of these key regulators at the predicted targets. We found that 81% of the predicted downstream genes were bound by at least one of the five TFs, and 23% presented co-occupancy of all five TFs at their promoters (Fig. 5G,I). Moreover, binding of these TFs at the promoters of the predicted targets (Fig. 5I) was highly significant for all combinations of TFs co-occupancy compared with expected occurrence (Supplemental Table S6). These findings suggest a regulatory link between the establishment of cell-type–specific transcriptional networks and RT control during lineage specification.
Discussion
In this study, we introduced a new approach to construct gene regulatory networks, exploiting the dynamic changes in DNA RT during lineage specification. RT is cell-type–specific (Hiratani et al. 2010; Ryba et al. 2011; Rivera-Mulia et al. 2015); regulation of RT is critical to maintain genome stability (Donley et al. 2013; Neelsen et al. 2013; Alver et al. 2014); and abnormal RT is observed in disease (Ryba et al. 2012; Gerhardt et al. 2014; Rivera-Mulia et al. 2017, 2018b; Sasaki et al. 2017). RT is closely related to the spatiotemporal organization of the genome with early and late replicating domains segregating to distinct nuclear compartments (Pope et al. 2014; Rivera-Mulia and Gilbert 2016b). Cell fate commitment is accompanied by dynamic changes in RT that are globally coordinated with transcriptional activity (Rivera-Mulia et al. 2015; Rivera-Mulia and Gilbert 2016b). Hence, RT constitutes a functional readout of genome organization that is linked to gene regulation during cell fate commitment. However, despite a significant correlation of early RT with transcriptional activity, RT and transcription cannot predict each other, suggesting an indirect relationship. Here, we constructed RT networks based on RT changes across 15 cell types and differentiation intermediates derived from human embryonic stem cells. We identified thousands of genes from different chromosomes that are correlated in RT during cell differentiation (Fig. 1C), and constructed distinct RT network models based on their dynamic changes. Our results suggest an intimate regulatory link between RT and cell-type–specific TRNs that is not revealed by association with transcription in cis but is uncovered by RT networks.
Directional RT networks were able to identify sets of unlinked genes whose RT changes coordinately during differentiation, the master regulators of RT changes, and their temporally downstream targets. To validate our model of directional RT networks, we showed that the gene interactions identified by our novel networks could predict the downstream targets of known regulators of specific differentiation pathways for which ChIP-seq data were available (Fig. 3). The algorithms to construct these RT networks that we present here can be applied to explore the interactions of any gene of interest (for detailed information on the computational pipeline, see Methods).
Combining transcriptional and RT networks into composite and bipartite networks revealed new insights into gene regulation during cell fate commitment. First, we found that there is a significant overlap between TRNs and RT networks of TFs (Fig. 4A) and that transcriptional edges are present in most of the motifs identified in correlated RT networks (Supplemental Figs. S5–S7). These findings suggest an interconnectivity between transcriptional and RT networks, which we confirmed by constructing composite networks that combine transcriptional and RT interactions. Composite networks revealed more complex circuitries in which transcriptional edges connected otherwise separated RT motifs (Fig. 4D,E). Finally, an important question in the DNA replication field is what regulates the RT program. Correlations with chromatin features have been described for decades (Rivera-Mulia and Gilbert 2016b), but we do not understand yet how RT is regulated and how it is remodeled during cell fate specification. Recent studies suggest a causal link between transcriptional activity and RT control, as targeting strong artificial transcriptional activators or histone acetyltransferases is sufficient to advance RT in some middle-late replication loci (Goren et al. 2008; Hassan-Zadeh et al. 2012; Koryakov et al. 2012; Therizols et al. 2014; Blin et al. 2019). However, it is not clear whether these changes are dependent on transcriptional activity, histone acetylation, or other chromatin changes associated with transcription. Moreover, individual knockout/knockdown or overexpression of many transcription and chromatin structure regulators (including TFs such as MYC [also known as C-MYC], MYCN [also known as N-MYC], MYOD1, and PAX5) has no effect on RT (Dileep et al. 2015), and combinatorial coregulation of multiple TFs might be required to control TRNs (Novershtern et al. 2011; Gerstein et al. 2012). Indeed, in budding yeast, binding of Fkh1 and Fkh2 TFs near a class of replication origins is necessary for early replication, but this function is separable from its transcription activity (Ostrow et al. 2017). Moreover, recently discovered cis elements that are necessary for control of RT in pluripotent cells have been shown to contain sites of POU5F1, SOX2, and NANOG co-occupancy, TFs that are central to the pluripotency TRN (Sima et al. 2019). Thus, it is possible that core TRN TFs may independently regulate mammalian RT, perhaps to coordinate cell-type–specific changes in RT with changes in transcription.
Here, we identified an intriguing relationship of TFs whose expression strongly correlates with the RT of downstream genes in trans (Fig. 5B,C) and constructed a novel class of bipartite networks (Fig. 5D,E). Bipartite networks allowed us to identify hundreds of RT-correlated genes correlated with expression levels of coexpressed TFs within the same differentiation pathways. This is of particular significance to our understanding of RT control because our findings suggest that establishment of complex circuitries/complete regulatory TFs networks, rather than transcriptional induction of specific downstream targets, might be required to shape the RT program during development. Because the chromatin is assembled at the replication fork and different types of chromatin are assembled at different times during S phase (Lande-Diner et al. 2009), a change in a transcriptional network that stimulates an RT change would alter the chromatin structure of an entire RD and all the genes within that domain contributing to the regulation of nuclear function and organization during cell fate commitment. Overall, the RT networks suggest a novel hypothesis that can be tested to unveil the mechanisms for RT regulation during lineage specification. Experimental manipulation of TFs followed by differentiation protocols would provide novel insights of whether TFs regulate RT independently of their transcriptional roles, whether all TFs are able to regulate RT or only a specific class of TFs have this property, and whether a complex combinatorial co-occupancy of several TFs and/or binding to superenhancers is required to remodel the RT program.
Methods
Extraction of RT values at the TSS of NCBI RefSeq genes
RT data from multiple cell types and intermediate differentiation stages derived from human embryonic stem cells (Rivera-Mulia et al. 2015) were used to extract the RT values at the transcription start sites (TSSs) of all RefSeq genes. All RT data sets are publicly available in the Gilbert laboratory database at http://www.replicationdomain.org, as well as in the ENCODE portal (Davis et al. 2018). Briefly, average RT profiles were obtained from multiple replicates, and RT values were predicted at the TSS from the loess smoothed RT profiles (Ryba et al. 2011). These data consist of RT values at the TSSs of all RefSeq genes from 15 cell types derived from hESCs representing three main germ layers: ectoderm, mesoderm, and endoderm (Fig. 1A).
Construction of RT networks based on coordinated changes in RT
To construct correlated RT networks, we denoted the set of genes as V = {g1, g2, …, gn} and the set of cell types as {c1, c2, …, cs}. Then three filters were applied to include exclusively genes that change RT and are correlated in during differentiation. First, we removed all genes that do not change RT during cell differentiation because all genes that have the same (or very similar) RT values across all cell types will yield high correlation values regardless of their RT values. Thus, we removed all RT constitutive genes and included only the genes that change RT during cell differentiation between the very early (>0.3) and very late (<−0.3) replication. It is worth noting that this is an aggressive filter as these parameters would consider some genes as constitutive even if they may have high variation in RT, although it is always early or late in replication. Second, we removed gene pairs with correlated RT patterns that are located within the same RD. If two genes are located close to each other on the same chromosome, their RT values are expected to be highly correlated owing to a natural outcome of the DNA replication process. Such correlations are extensively characterized and not as relevant for this work compared with those among physically distant genes, for the correlations between distant genes provide hints about the existence of complex interactions that regulate the order in which genes are replicated. Because we have shown that RDs vary in size from 0.4 Mb to 0.8 Mb (Hiratani et al. 2008; Pope et al. 2014; Rivera-Mulia et al. 2015), we removed all gene pairs separated by <500 kb (distance threshold denoted as µ). Thus, for all possible gene pairs in V, we obtained the locations of gi and gj on the DNA. If they are on the same chromosome, we classified them as colocated; if they are in different chromosomes, we classified them as not colocated. If the two genes gi and gj are colocated, the distance between their TSS positions d(gi, gj) was calculated, and all genes with d < µ were removed from the analysis. Finally, we established interaction edges exclusively for gene pairs that are significantly correlated. To do so, for each gene gi ∈ V, we constructed a vector wi with an entry of the RT of gi in cell type cx. Then, the model of correlated RT network was defined as G = (V, E), where V and E denote the set of nodes and edges, respectively. For all pairs of genes gi, gj ∈ G, we computed the Pearson's correlation coefficient between their vectors wi and wj. Statistical significance for the Pearson's correlation of each gene pair was calculated as follows:
where the P-value being 2 × P( T > t), r is the correlation coefficient and n is the number of observations. Next, we applied Bonferroni correction to the P-values and drawn an edge between gi and gj when
Pij is the Bonferroni-corrected P-value, a is the statistical significance threshold, and m is the number of tests conducted. For each such significant correlation, an edge (gi, gj) is added to the set E.
Correlated RT networks were constructed across all samples, as well as for distinct subsets of cell types focusing on the three major germ layers: ectoderm, mesoderm, and endoderm. In our correlated RT network models, each node represents a gene, and each edge represents a relationship between correlated RT of the corresponding two genes in the network.
RT network visualization
To visualize our RT network, we focused on the subset of highly connected nodes (>20 degree count). First, we constructed a 2D map of the RT-correlated networks using the force-directed layout algorithm in Cytoscape (Shannon et al. 2003), with the edge length being proportional to the Pearson's correlation strength (Fig. 2). Next, we detected subnetwork communities by running a Louvain community detection algorithm (Blondel et al. 2008). This method is a heuristic method that is based on modularity optimization. Finally, we used SAFE algorithm to annotate functional attributes for communities. SAFE (Baryshnikova 2016) is an automated network annotation algorithm. Given a biological network and a set of functional groups or quantitative features of interest, SAFE performs local enrichment analysis to determine which regions of the network are significantly overrepresented for each group of features. Thus, local neighborhoods were identified, and functional attributes were annotated based on the Gene Ontology (GO) terms (Baryshnikova 2016).
Construction of directional RT networks
Directional RT networks were generated for each differentiation pathway for changes in LtoE and EtoL. In these directed RT network models, we do not consider the correlated RT patterns but the temporal order of RT changes between two corresponding genes. For differentiation pathway ESCs (the earliest stage) → lateral plate mesoderm → splanchnic mesoderm → smooth muscle (the latest stage) of a LtoE directed RT network, we draw a directed edge from a gene that switches in earlier stage to a gene that switches in later stage only if the difference of switching stage is one or two. For example, if LtoE pattern of gene g_1 is L → E → E → E in the differentiation pathway ESCs → lateral plate mesoderm → splanchnic mesoderm → smooth muscle and LtoE pattern of gene g_2 is L → L → E → E in the same pathway, we draw an directed edge from g_1 to g_2 as g_1 switches in lateral plate mesoderm stage (earlier) and g_2 switches in splanchnic mesoderm stage (later), assuming the change from LtoE of gene g_1 could be causally linked to the change of gene g_2 in the next stage. Similarly, we constructed EtoL directed networks considering g_1 changes E → L → L → L and identified g_2 genes with patterns E → E → L → L.
RT network edges overlap with known transcriptional regulatory interactions
We compared the topologies of the RT networks with those of TRNs. Particularly, we used the TRNs constructed using TFs (Neph et al. 2012). To do that, for each cell lineage, we counted the number of edges common to its RT network and TRN by focusing on the nodes/TFs that are common to both networks (i.e., at least have one edge in each network). An undirected edge in the RT network overlaps with a directed edge in the TRN if the gene pairs corresponding to an edge are same in the RT network and the TRN. By using the number of common edges in the two networks, we calculate the P-value of the overlap from their hypergeometric distributions. To do so, we denoted the number of nodes (i.e., genes) in the given TRN with n. The total number of possible edges (excluding self-edges) in a directed complete graph with n nodes is M = C(n, 2) × 2 = n × (n−1), where C(x, y) denotes x choose y (i.e., the number of y-element combinations of a set with x elements). Let us also denote the number of edges present in the actual TRN with m, the number of edges in the undirected RT network with K, and the number of common edges between TRN and RT networks with k. Then we computed the probability that the number of common edges between the two networks is equal to a specific value (say i) as
The numerator in this probability mass function (PMF) describes the number of ways to pick exactly i edges from the RT network in m draws from a complete graph, without replacement. The denominator shows the number of alternative network topologies with the same nodes as the RT network, which has m edges. By using this PMF, we calculate the P-value of having more than or equal to k common edges between the RT network and the TRN as . Thus, significant P-values signify unexpectedly large number of common edges between the two networks.
RT network motif identification
Network motifs are defined as recurrent and statistically significant subgraphs or patterns. We identified the most frequent motifs in the RT networks by creating all possible shapes of connected nodes (in terms of undirected edges) for two, three, and four node subgraphs. The analysis was limited to motifs of four or fewer nodes: first, because it has been shown that the fundamental regulatory subnetwork patterns consist of a small number of nodes networks (Milo et al. 2002; Yeger-Lotem et al. 2004; Baiser et al. 2016; Elhesha and Kahveci 2016) and, second, because the number of possible motif topologies grow exponentially with number of nodes, making it impossible to test larger motif sizes. To identify the most frequent motifs in the RT networks, we counted the number of matching motifs between the RT network and randomized networks. We also created multiple shuffled networks that have the same number of nodes and edges with the RT network and set a z-score as 2.54 for a subgraph to be considered as a motif present in the network. Is important to note that the motif frequency (i.e., the number of times a given motif appears in a given network) does not monotonically decrease or increase with the motif size, and the statistical significance of the motif abundance is independent of the motif size and topology (Elhesha and Kahveci 2016). This is because motif count does not have downward closure property, and a motif is considered as abundant in a given network only if its frequency is significantly higher than the number of times the same motif appears in randomized networks (P-value <0.01).
Construction of composite RT and gene expression networks
The composite network model merges the interactions observed in RT networks (using only genes present in the networks of Neph et al. 2012) with the interactions observed in TRNs. To construct the composite networks, we used as a base network the motifs (all subgraphs consisting in connected two, three, and four nodes) detected in the RT networks constructed from the subset of genes present in the TRNs. We then combined the RT and TRNs by taking the union of their edge sets. To do so, we draw the transcriptional edges between the RT nodes according to the interactions identified in the TRNs. Composite networks were visualized in Cytoscape (Shannon et al. 2003). Transcriptional edges are directional, and they were differentiated according to the connectivity within/outside RT motifs; that is, if a TRN interaction was present within an RT network motif, it was visualized as a directed solid edge, and if the interaction occurs between nodes from different RT network motifs, it was visualized as a directed dashed edge.
Construction of bipartite RT and transcriptional networks
A bipartite network is a graph with two components. Each component is a set of nodes. In our model, first component is based on the expression patterns of genes, and the second component is based in the RT of genes. For our analysis, we started with the list of genes coexpressed in specific cell types that constitute the first component. We append an edge between the first component and the second component if the RT of a gene in the second component is correlated with expression of a gene in the first component with more than a certain correlation threshold. Next we removed a gene in the second component if the number of edges of this gene (correlation with coexpressed genes) is less than a specified ratio of total number of genes in the first component.
ChIP-seq data analysis
ChIP-seq peaks and aligned reads (hg19 genome assembly) for the FOXA1 (ENCSR735KEY), FOXA2 (ENCSR310NYI), NR2F2 (ENCSR338MMB), HNF4A (ENCSR601OGE), and HNF4G (ENCSR297GII) transcription factors in liver tissue were downloaded from the ENCODE data portal (Davis et al. 2018). FOXA1 transcription factor-specific peaks and raw reads in pancreas tissue were collected from Diaferia et al. (2016) (GSE64557), and PDX1-specific ChIP-seq data in pancreas tissue was downloaded from Wang et al. (2018) (GSE106949). Aligned reads for FOXA1 and PDX1 in the pancreas tissue on hg19 genome assembly were generated using the Bowtie 2 alignment program (Langmead and Salzberg 2012). PDX1-specific peak calling against the respective input was performed using the MACS2 program (Zhang et al. 2008) with the following parameters: “-g hs –q 0.05”. All the significant transcription factor peaks (FDR < 0.05) were retained for downstream analysis, and the raw ChIP-seq signal tracks were scaled to the signal track with minimum coverage for visualization purpose. Individual transcription factor peaks and their respective combinations in liver and pancreas were mapped to the annotated (Harrow et al. 2012) hg19 TSSs (±20 kb) using the “bedtools map” function (Quinlan and Hall 2010) with default parameters. Overlap significance and enrichment of transcription factor binding in RT network–specific genes were measured using a Fisher's exact test by comparing against a similar number of random sets of non-RT network genes. Realigning our data to hg38 (GRCh38) would not significantly affect our conclusions as the major improvements of hg38 assembly are in the annotation of centromeric and other repetitive genomic regions (Guo et al. 2017), which are not included in our analysis.
Data access
Normalized data of RT and gene expression values from all the hESC-derived cell types analyzed are available in the Supplemental Material, including a Cytoscape session with all gene networks shown in this paper's Supplemental File 1. Additionally, the source code and documentation to reproduce the gene networks shown in this paper are available in the Supplemental Code, as well as on GitHub (https://github.com/sebkim/RTNet).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grant GM083337 (D.M.G.).
Author contributions: J.C.R.-M., T.K., and D.M.G. conceived and designed the study; J.C.R.-M., S.K., H.G., A.C., and F.A. performed data analysis and interpretation; J.C.R.-M. and D.M.G. wrote the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.247049.118.
References
- Allocco DJ, Kohane IS, Butte AJ. 2004. Quantifying the relationship between co-expression, co-regulation and gene function. BMC Bioinformatics 5: 18 10.1186/1471-2105-5-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon U. 2007. Network motifs: theory and experimental approaches. Nat Rev Genet 8: 450–461. 10.1038/nrg2102 [DOI] [PubMed] [Google Scholar]
- Alver RC, Chadha GS, Blow JJ. 2014. The contribution of dormant origins to genome stability: from cell biology to human genetics. DNA Repair (Amst) 19: 182–189. 10.1016/j.dnarep.2014.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene Ontology: tool for the unification of biology. Nat Genet 25: 25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baiser B, Elhesha R, Kahveci T. 2016. Motifs in the assembly of food web networks. Oikos 125: 480–491. 10.1111/oik.02532 [DOI] [Google Scholar]
- Baryshnikova A. 2016. Systematic functional annotation and visualization of biological networks. Cell Syst 2: 412–421. 10.1016/j.cels.2016.04.014 [DOI] [PubMed] [Google Scholar]
- Berger E, Vega N, Weiss-Gayet M, Géloën A. 2015. Gene network analysis of glucose linked signaling pathways and their role in human hepatocellular carcinoma cell growth and survival in HuH7 and HepG2 cell lines. Biomed Res Int 2015: 821761 10.1155/2015/821761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin M, Le Tallec B, Nähse V, Schmidt M, Brossas C, Millot GA, Prioleau M-N, Debatisse M. 2019. Transcription-dependent regulation of replication dynamics modulates genome stability. Nat Struct Mol Biol 26: 58–66. 10.1038/s41594-018-0170-1 [DOI] [PubMed] [Google Scholar]
- Blondel VD, Guillaume J-L, Lambiotte R, Lefebvre E. 2008. Fast unfolding of communities in large networks. J Stat Mech 2008: P10008 10.1088/1742-5468/2008/10/P10008 [DOI] [Google Scholar]
- Costanzo M, VanderSluis B, Koch EN, Baryshnikova A, Pons C, Tan G, Wang W, Usaj M, Hanchard J, Lee SD, et al. 2016. A global genetic interaction network maps a wiring diagram of cellular function. Science 353: aaf1420 10.1126/science.aaf1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CA, Hitz BC, Sloan CA, Chan ET, Davidson JM, Gabdank I, Hilton JA, Jain K, Baymuradov UK, Narayanan AK, et al. 2018. The encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res 46: D794–D801. 10.1093/nar/gkx1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'haeseleer P, Liang S, Somogyi R. 2000. Genetic network inference: from co-expression clustering to reverse engineering. Bioinformatics 16: 707–726. 10.1093/bioinformatics/16.8.707 [DOI] [PubMed] [Google Scholar]
- Diaferia GR, Balestrieri C, Prosperini E, Nicoli P, Spaggiari P, Zerbi A, Natoli G. 2016. Dissection of transcriptional and cis-regulatory control of differentiation in human pancreatic cancer. EMBO J 35: 595–617. 10.15252/embj.201592404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dileep V, Rivera-Mulia JC, Sima J, Gilbert DM. 2015. Large-scale chromatin structure-function relationships during the cell cycle and development: insights from replication timing. Cold Spring Harb Symp Quant Biol 80: 53–63. 10.1101/sqb.2015.80.027284 [DOI] [PubMed] [Google Scholar]
- Donley N, Stoffregen EP, Smith L, Montagna C, Thayer MJ. 2013. Asynchronous replication, mono-allelic expression, and long range cis-effects of ASAR6. PLoS Genet 9: e1003423 10.1371/journal.pgen.1003423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Shih C-H, Wosczyna MN, Mueller AA, Cho J, Aggarwal A, Rando TA, Feldman BJ. 2017. Macrophage-released ADAMTS1 promotes muscle stem cell activation. Nat Commun 8: 669 10.1038/s41467-017-00522-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhesha R, Kahveci T. 2016. Identification of large disjoint motifs in biological networks. BMC Bioinformatics 17: 408 10.1186/s12859-016-1271-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabr H, Rivera-Mulia JC, Gilbert DM, Kahveci T. 2015. Computing interaction probabilities in signaling networks. EURASIP J Bioinform Syst Biol 2015: 10 10.1186/s13637-015-0031-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium. 2015. Gene Ontology Consortium: going forward. Nucleic Acids Res 43: D1049–D1056. 10.1093/nar/gku1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt J, Tomishima MJ, Zaninovic N, Colak D, Yan Z, Zhan Q, Rosenwaks Z, Jaffrey SR, Schildkraut CL. 2014. The DNA replication program is altered at the FMR1 locus in fragile X embryonic stem cells. Mol Cell 53: 19–31. 10.1016/j.molcel.2013.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan K-K, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, et al. 2012. Architecture of the human regulatory network derived from ENCODE data. Nature 489: 91–100. 10.1038/nature11245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford CA, Ziller MJ, Gu H, Trapnell C, Donaghey J, Tsankov A, Shalek AK, Kelley DR, Shishkin AA, Issner R, et al. 2013. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell 153: 1149–1163. 10.1016/j.cell.2013.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goren A, Tabib A, Hecht M, Cedar H. 2008. DNA replication timing of the human β-globin domain is controlled by histone modification at the origin. Genes Dev 22: 1319–1324. 10.1101/gad.468308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Dai Y, Yu H, Zhao S, Samuels DC, Shyr Y. 2017. Improvements and impacts of GRCh38 human reference on high throughput sequencing data analysis. Genomics 109: 83–90. 10.1016/j.ygeno.2017.01.005 [DOI] [PubMed] [Google Scholar]
- Han XH, Jin Y-R, Seto M, Yoon JK. 2011. A WNT/β-catenin signaling activator, R-spondin, plays positive regulatory roles during skeletal myogenesis. J Biol Chem 286: 10649–10659. 10.1074/jbc.M110.169391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. 2012. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 22: 1760–1774. 10.1101/gr.135350.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan-Zadeh V, Chilaka S, Cadoret J-C, Ma MK-W, Boggetto N, West AG, Prioleau M-N. 2012. USF binding sequences from the HS4 insulator element impose early replication timing on a vertebrate replicator. PLoS Biol 10: e1001277 10.1371/journal.pbio.1001277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Yokochi T, Schwaiger M, Chang C-W, Lyou Y, Townes TM, Schübeler D, Gilbert DM. 2008. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol 6: e245 10.1371/journal.pbio.0060245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Rathjen J, Kulik M, Papp B, Fussner E, Bazett-Jones DP, Plath K, Dalton S, et al. 2010. Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Res 20: 155–169. 10.1101/gr.099796.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Dong J. 2008. Geometric interpretation of gene coexpression network analysis. PLoS Comput Biol 4: e1000117 10.1371/journal.pcbi.1000117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Pombo A. 1998. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol 140: 1285–1295. 10.1083/jcb.140.6.1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles BB, Howe CC, Aden DP. 1980. Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science 209: 497–499. 10.1126/science.6248960 [DOI] [PubMed] [Google Scholar]
- Koryakov DE, Pokholkova GV, Maksimov DA, Belyakin SN, Belyaeva ES, Zhimulev IF. 2012. Induced transcription results in local changes in chromatin structure, replication timing, and DNA polytenization in a site of intercalary heterochromatin. Chromosoma 121: 573–583. 10.1007/s00412-012-0382-9 [DOI] [PubMed] [Google Scholar]
- Lande-Diner L, Zhang J, Cedar H. 2009. Shifts in replication timing actively affect histone acetylation during nucleosome reassembly. Mol Cell 34: 767–774. 10.1016/j.molcel.2009.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurenti E, Doulatov S, Zandi S, Plumb I, Chen J, April C, Fan J-B, Dick JE. 2013. The transcriptional architecture of early human hematopoiesis identifies multilevel control of lymphoid commitment. Nat Immunol 14: 756–763. 10.1038/ni.2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K-C. 2002. Genome-wide coexpression dynamics: theory and application. Proc Natl Acad Sci 99: 16875–16880. 10.1073/pnas.252466999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U. 2002. Network motifs: simple building blocks of complex networks. Science 298: 824–827. 10.1126/science.298.5594.824 [DOI] [PubMed] [Google Scholar]
- Mutsaers SE. 2004. The mesothelial cell. Int J Biochem Cell Biol 36: 9–16. 10.1016/S1357-2725(03)00242-5 [DOI] [PubMed] [Google Scholar]
- Neelsen KJ, Zanini IMY, Mijic S, Herrador R, Zellweger R, Ray Chaudhuri A, Creavin KD, Blow JJ, Lopes M. 2013. Deregulated origin licensing leads to chromosomal breaks by rereplication of a gapped DNA template. Genes Dev 27: 2537–2542. 10.1101/gad.226373.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neph SS, Stergachis ABA, Reynolds AA, Sandstrom RR, Borenstein EE, Stamatoyannopoulos JAJ. 2012. Circuitry and dynamics of human transcription factor regulatory networks. Cell 150: 1274–1286. 10.1016/j.cell.2012.04.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak BA, Jain AN. 2006. Pathway recognition and augmentation by computational analysis of microarray expression data. Bioinformatics 22: 233–241. 10.1093/bioinformatics/bti764 [DOI] [PubMed] [Google Scholar]
- Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, Habib N, Yosef N, Chang CY, Shay T, et al. 2011. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell 144: 296–309. 10.1016/j.cell.2011.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrow AZ, Kalhor R, Gan Y, Villwock SK, Linke C, Barberis M, Chen L, Aparicio OM. 2017. Conserved forkhead dimerization motif controls DNA replication timing and spatial organization of chromosomes in S. cerevisiae. Proc Natl Acad Sci 114: E2411–E2419. 10.1073/pnas.1612422114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope BD, Ryba T, Dileep V, Yue F, Wu W, Denas O, Vera DL, Wang Y, Hansen RS, Canfield TK, et al. 2014. Topologically associating domains are stable units of replication-timing regulation. Nature 515: 402–405. 10.1038/nature13986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preis PN, Saya H, Nádasdi L, Hochhaus G, Levin V, Sadée W. 1988. Neuronal cell differentiation of human neuroblastoma cells by retinoic acid plus herbimycin A. Cancer Res 48: 6530–6534. [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Mulia JC, Gilbert DM. 2016a. Replicating large genomes: divide and conquer. Mol Cell 62: 756–765. 10.1016/j.molcel.2016.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Mulia JC, Gilbert DM. 2016b. Replication timing and transcriptional control: beyond cause and effect—part III. Curr Opin Cell Biol 40: 168–178. 10.1016/j.ceb.2016.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Mulia JC, Buckley Q, Sasaki T, Zimmerman J, Didier RA, Nazor K, Loring JF, Lian Z, Weissman S, Robins AJ, et al. 2015. Dynamic changes in replication timing and gene expression during lineage specification of human pluripotent stem cells. Genome Res 25: 1091–1103. 10.1101/gr.187989.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Mulia JC, Desprat R, Trevilla-García C, Cornacchia D, Schwerer H, Sasaki T, Sima J, Fells T, Studer L, Lemaitre JM, et al. 2017. DNA replication timing alterations identify common markers between distinct progeroid diseases. Proc Natl Acad Sci 114: E10972–E10980. 10.1073/pnas.1711613114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Mulia JC, Dimond A, Vera D, Trevilla-García C, Sasaki T, Zimmerman J, Dupont C, Gribnau J, Fraser P, Gilbert DM. 2018a. Allele-specific control of replication timing and genome organization during development. Genome Res 28: 800–811. 10.1101/gr.232561.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Mulia JC, Schwerer H, Besnard E, Desprat R, Trevilla-García C, Sima J, Bensadoun P, Zouaoui A, Gilbert DM, Lemaitre JM. 2018b. Cellular senescence induces replication stress with almost no affect on DNA replication timing. Cell Cycle 17: 1667–1681. 10.1080/15384101.2018.1491235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Mulia JC, Sasaki T, Trevilla-García C, Nakamichi N, Knapp D, Hammond C, Chang B, Tyner JW, Devidas M, Zimmerman J, et al. 2019. Replication timing alterations in leukemia reflect stable clinically-relevant changes in genome architecture. bioRxiv 10.1101/549196 [DOI] [PMC free article] [PubMed]
- Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al. 2015. Integrative analysis of 111 reference human epigenomes. Nature 518: 317–330. 10.1038/nature14248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, Schulz TC, Robins AJ, Dalton S, Gilbert DM. 2010. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res 20: 761–770. 10.1101/gr.099655.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T, Hiratani I, Sasaki T, Battaglia D, Kulik M, Zhang J, Dalton S, Gilbert DM. 2011. Replication timing: a fingerprint for cell identity and pluripotency. PLoS Comput Biol 7: e1002225 10.1371/journal.pcbi.1002225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T, Battaglia D, Chang BH, Shirley JW, Buckley Q, Pope BD, Devidas M, Druker BJ, Gilbert DM. 2012. Abnormal developmental control of replication-timing domains in pediatric acute lymphoblastic leukemia. Genome Res 22: 1833–1844. 10.1101/gr.138511.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Rivera-Mulia JC, Vera D, Zimmerman J, Das S, Padget M, Nakamichi N, Chang BH, Tyner J, Druker BJ, et al. 2017. Stability of patient-specific features of altered DNA replication timing in xenografts of primary human acute lymphoblastic leukemia. Exp Hematol 51: 71–82.e3. 10.1016/j.exphem.2017.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sima J, Chakraborty A, Dileep V, Michalski M, Klein KN, Holcomb NP, Turner JL, Paulsen MT, Rivera-Mulia JC, Trevilla-Garcia C, et al. 2019. Identifying cis elements for spatiotemporal control of mammalian DNA replication. Cell 176: 816–830.e18. 10.1016/j.cell.2018.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovei I, Thanisch K, Feodorova Y. 2016. How to rule the nucleus: divide et impera. Curr Opin Cell Biol 40: 47–59. 10.1016/j.ceb.2016.02.014 [DOI] [PubMed] [Google Scholar]
- Stupka N, Kintakas C, White JD, Fraser FW, Hanciu M, Aramaki-Hattori N, Martin S, Coles C, Collier F, Ward AC, et al. 2013. Versican processing by a disintegrin-like and metalloproteinase domain with thrombospondin-1 repeats proteinases-5 and -15 facilitates myoblast fusion. J Biol Chem 288: 1907–1917. 10.1074/jbc.M112.429647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Chen D, Cao L, Zhang R, Zhou J, Chen H, Li Y, Li M, Cao J, Wang Z. 2013. MiR-490-3p modulates the proliferation of vascular smooth muscle cells induced by ox-LDL through targeting PAPP-A. Cardiovasc Res 100: 272–279. 10.1093/cvr/cvt172 [DOI] [PubMed] [Google Scholar]
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, et al. 2019. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47: D607–D613. 10.1093/nar/gky1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therizols P, Illingworth RS, Courilleau C, Boyle S, Wood AJ, Bickmore WA. 2014. Chromatin decondensation is sufficient to alter nuclear organization in embryonic stem cells. Science 346: 1238–1242. 10.1126/science.1259587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankov AM, Gu H, Akopian V, Ziller MJ, Donaghey J, Amit I, Gnirke A, Meissner A. 2015. Transcription factor binding dynamics during human ES cell differentiation. Nature 518: 344–349. 10.1038/nature14233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal M, Cusick ME, Barabási A-L. 2011. Interactome networks and human disease. Cell 144: 986–998. 10.1016/j.cell.2011.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Sterr M, Burtscher I, Chen S, Hieronimus A, Machicao F, Staiger H, Häring H-U, Lederer G, Meitinger T, et al. 2018. Genome-wide analysis of PDX1 target genes in human pancreatic progenitors. Mol Metab 9: 57–68. 10.1016/j.molmet.2018.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Schultz MD, Lister R, Hou Z, Rajagopal N, Ray P, Whitaker JW, Tian S, Hawkins RD, Leung D, et al. 2013. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell 153: 1134–1148. 10.1016/j.cell.2013.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeger-Lotem E, Sattath S, Kashtan N, Itzkovitz S, Milo R, Pinter RY, Alon U, Margalit H. 2004. Network motifs in integrated cellular networks of transcription-regulation and protein–protein interaction. Proc Natl Acad Sci 101: 5934–5939. 10.1073/pnas.0306752101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LV, King OD, Wong SL, Goldberg DS, Tong AHY, Lesage G, Andrews B, Bussey H, Boone C, Roth FP. 2005. Motifs, themes and thematic maps of an integrated Saccharomyces cerevisiae interaction network. J Biol 4: 6 10.1186/jbiol23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based Analysis of ChIP-Seq (MACS). Genome Biol 9: R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao PA, Rivera-Mulia JC, Gilbert DM. 2017. Replication domains: genome compartmentalization into functional replication units. Adv Exp Med Biol 1042: 229–257. 10.1007/978-981-10-6955-0_11 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.