Abstract
Eukaryotic cell survival requires maintenance of plasma membrane (PM) homeostasis in response to environmental insults and changes in lipid metabolism. In yeast, a key regulator of PM homeostasis is target of rapamycin (TOR) complex 2 (TORC2), a multiprotein complex containing the evolutionarily conserved TOR protein kinase isoform Tor2. PM localization is essential for TORC2 function. One core TORC2 subunit (Avo1) and two TORC2-associated regulators (Slm1 and Slm2) contain pleckstrin homology (PH) domains that exhibit specificity for binding phosphatidylinositol-4,5-bisphosphate (PtdIns4,5P2). To investigate the roles of PtdIns4,5P2 and constituent subunits of TORC2, we used auxin-inducible degradation to systematically eliminate these factors and then examined localization, association, and function of the remaining TORC2 components. We found that PtdIns4,5P2 depletion significantly reduced TORC2 activity, yet did not prevent PM localization or disassembly of TORC2. Moreover, truncated Avo1 (lacking its C-terminal PH domain) was still recruited to the PM and supported growth. Even when all three PH-containing proteins were absent, the remaining TORC2 subunits were PM-bound. Revealingly, Avo3 localized to the PM independent of both Avo1 and Tor2, whereas both Tor2 and Avo1 required Avo3 for their PM anchoring. Our findings provide new mechanistic information about TORC2 and pinpoint Avo3 as pivotal for TORC2 PM localization and assembly in vivo.
INTRODUCTION
The plasma membrane (PM) of a cell separates its internal contents from the environment, contains proteins that act as channels, transporters, adhesion molecules, and sensors, and provides a platform for scaffolding many different signaling modules (Lodish et al., 2016). Hence, the ability to maintain proper PM lipid and protein content and to adjust PM composition appropriately in response to changing extracellular and intracellular conditions is critical for cell survival. In budding yeast (Saccharomyces cerevisiae), it has been demonstrated that a multisubunit protein kinase, target of rapamycin (TOR) complex 2 (TORC2), is responsible for controlling PM homeostasis (Eltschinger and Loewith, 2016; Roelants et al., 2017). Cues from the external and internal milieu affect the efficiency with which TORC2 phosphorylates its primary downstream effector, the AGC-family protein kinase Ypk1 (and its paralogue Ypk2) (Kamada et al., 2005; Roelants et al., 2011; Berchtold et al., 2012; Niles et al., 2012; Sun et al., 2012). Ypk1 (and Ypk2), in turn, phosphorylate targets that control various pathways responsible for PM homeostasis (Gaubitz et al., 2016; Roelants et al., 2017).
TORC2 contains a very large, evolutionarily conserved TOR polypeptide. Metazoans possess a single TOR-encoding gene (human mTOR, 2549 residues; González; Saxton and Sabatini, 2017; Tatebe and Shiozaki, 2017), whereas budding yeast (Heitman et al., 1991), fission yeast (Ikai et al., 2011), and other fungal genomes (Eltschinger and Loewith, 2016) encode two TOR proteins, Tor1 and Tor2 (2470 and 2474 residues, respectively, in S. cerevisiae). In TORC2, Tor2 alone suffices as the catalytic subunit (Helliwell et al., 1998a; Loewith et al., 2002, Wedaman et al., 2002; Hill et al., 2018). Tor2 assembles with Lst8, Avo1, Avo2, Avo3/Tsc11, and Bit61 (and/or its paralogue Bit2) to form a large (∼1.4-MDa) heterodimeric complex, in which each protomer contains two copies of each subunit (Wullschleger et al., 2005; Karuppasamy et al., 2017). TORC2 localizes to the PM (Kunz et al., 2000; Sturgill et al., 2008) at multiple, punctate foci that are dynamic (Berchtold and Walther, 2009; Leskoske et al., 2018).
As shown first by two-hybrid interaction (Uetz et al., 2000, Ito et al., 2001) and confirmed by coimmunoprecipitation (Audhya et al., 2004) and in vitro binding assays (Fadri et al., 2005), the Avo2 subunit of TORC2 also associates with two paralogous proteins, Slm1 and Slm2, that are components of very static, furrow-like PM invaginations, called eisosomes (Douglas and Konopka, 2014). Slm1 and Slm2 are required for TORC2-mediated phosphorylation of Ypk1, purportedly because stimuli that elevate TORC2-Ypk1 signaling cause Slm1 and Slm2 to dissociate from eisosomes and associate, instead, with TORC2 (Berchtold et al., 2012; Niles et al., 2012). It was proposed that the role of Slm1/2 is to bind Ypk1 and deliver this substrate to TORC2 (Berchtold et al., 2012; Niles et al., 2012). However, other evidence shows that a conserved region in the middle of the Avo1 subunit of TORC2 is what binds and presents Ypk1 to the active site in Tor2 (Liao and Chen, 2012; Tatebe et al., 2017; Tatebe and Shiozaki, 2017), leaving open the question of how, at the mechanistic level, Slm1 and Slm2 promote TORC2-mediated phosphorylation of Ypk1.
As one approach to investigating how TORC2 serves as a sensor of PM status, it is important to understand how TORC2 physically associates with the PM. Avo1 (1176 residues), one of the essential subunits of TORC2, contains a PH domain (residues 1065–1169) at its C-terminal end, and this isolated domain binds phosphatidylinositol-4,5-bisphosphate (PtdIns4,5P2) in vitro and, when tagged with GFP, localizes at the PM in vivo (Berchtold and Walther, 2009). Similarly, Slm1 (686 residues) and Slm2 (656 residues) each contain a demonstrated ∼110-residue PtdIns4,5P2-binding PH domain toward the C-terminal end (Audhya et al., 2004; Tabuchi et al., 2006; Daquinag et al., 2007; Gallego et al., 2010).
The PM pool of PtdIns4,5P2 is generated by the essential enzyme Mss4, the sole type I PtdIns4P 5-kinase encoded in the S. cerevisiae genome (Boronenkov and Anderson, 1995; Desrivières et al., 1998; Homma et al., 1998; Audhya and Emr, 2003). In response to a variety of stimuli, local and transient changes in PtdIns4,5P2 level affect the efficiency with which critical PH domain–containing signaling proteins and regulators of the actin cytoskeleton associate with the PM (Di Paolo and De Camilli, 2006; Strahl and Thorner, 2007; Janmey et al., 2018). In this regard, it was reported that deletion of the C-terminal PH domain in Avo1 rendered cells inviable, but this inviability could be rescued by expression of an Avo1 construct in which another PM-targeted element (CaaX box) was substituted for its PH domain (Berchtold and Walther, 2009). On this basis, it was inferred, but never directly tested, that Avo1 was critical for mediating PM assembly of TORC2 via the interaction of its PH domain with PM PtdIns4,5P2.
Analysis of the physiological functions of TORC2 have relied on the use of temperature-sensitive (ts) mutations in essential subunits of TORC2 (Avo1, Avo3, Lst8 or Tor2; Helliwell et al., 1998b; Ho et al., 2005; Kamada et al., 2005; Aronova et al., 2008; Liao and Chen, 2012) because, normally, TORC2 is insensitive to inhibition by rapamycin (Gaubitz et al., 2015). However, in the case of TORC2, shifting a heat-sensitive mutant to the restrictive temperature makes the molecular mechanism underlying the ensuing phenotype hard to deconvolve because heat stress itself has been shown to alter PtdIns4,5P2 levels (Desrivières et al., 1998), perturb Slm1 and Slm2 function (Daquinag et al., 2007), activate the yeast MAPKs Hog1 (Winkler et al., 2002) and Slt2/Mpk1 (Kamada et al., 1995; Truman et al., 2007), and even stimulate TORC2-Ypk1 action (Sun et al., 2012). Hence, as an alternative means of assessing the role of PtdIns4,5P2 and the PH domains in Avo1, Slm1, and Slm2, as well as the contributions of these and other TORC2 subunits, in the assembly and PM localization of TORC2, we exploited the recently developed method for specific protein tagging that allows rapid and selective protein degradation induced upon addition of the plant hormone auxin (or a more cell permeable synthetic auxin mimic) (Nishimura et al., 2009; Morawska and Ulrich, 2013). In this way, we were able to obtain novel insights into the contributions that PtdIns4,5P2 and individual subunits make to TORC2 assembly and its anchoring and function at the PM.
RESULTS
Ablating PM PtdIns4,5P2 impairs TORC2 activity but not its PM localization
It has been reported that a C-terminal truncation of Avo1 that removed only its PH domain prevented growth; that a GFP-labeled fragment containing only the PH domain of Avo1, when overexpressed from a GAL promoter, localized uniformly to the entire cell periphery; and that replacement of the PH domain in Avo1 with a different PM-targeting motif (CaaX box) restored viability (Berchtold and Walther, 2009). On these grounds, it was concluded that TORC2 is recruited to the PM via the binding of the Avo1 PH domain to PtdIns4,5P2 in the PM and that TORC2 localization at the PM is sufficient for its function. We had concerns about the first conclusion because, using a plasmid shuffle approach (Boeke et al., 1984) to assess the functionality of various Avo1 derivatives, we found that cells expressing C-terminally truncated Avo1 as the sole source of this TORC2 subunit were still viable, like cells expressing full-length Avo1 in the same manner (Supplemental Figure S1). In further support of our findings, more recent genomewide analysis of yeast gene function by saturation transposon mutagenesis (Michel et al., 2017) demonstrated that insertions within the PH domain sequence of Avo1 were tolerated in all strain backgrounds tested (Supplemental Figure S2).
Therefore, as an alternative approach to assessing the importance of PM PtdIns4,5P2 in TORC2 localization and function, we examined the effect of eliminating Mss4, the sole enzyme responsible for converting PtdIns4P to PtdIns4,5P2 at the PM (Strahl and Thorner 2007). Prior work had shown that a shift of cells carrying an mss4ts allele to the nonpermissive temperature caused a ≤90% reduction in PM PtdIns4,5P2 (Desrivières et al., 1998, Stefan et al., 2002). However, to avoid any other perturbations to the cells that might be elicited by a dramatic temperature shift, and because MSS4 is an essential gene, we fused an auxin-inducible degron (AID*) tag and a 6xHA epitope to the C-terminus of the MSS4 ORF at its endogenous locus on chromosome IV. AID* is the minimal sequence motif required for auxin-dependent recognition by the plant F-box protein TIR1 (Gray et al., 1999), which, when expressed in S. cerevisiae, is able to integrate readily into the yeast Skp1- and Cdc53/Cul1-containing ubiquitin ligase/E3 (SCF complex; Nishimura et al., 2009; Morawska and Ulrich, 2013). Addition of the cell-permeable synthetic auxin 1-naphthaleneacetic acid (1-NAA) to the yeast growth medium results in TIR1 binding to an AID*-containing target, rapid polyubiquitinylation of that protein, and its ensuing efficient degradation by the proteasome. Indeed, as confirmed by immunoblot analysis, addition of 1-NAA to cells expressing Mss4-AID*-6HA and TIR1 led to dramatic loss of this enzyme within 30 min and to its nearly complete removal in less than one cell doubling (∼110 min in synthetic minimal medium; Figure 1A). The 1-NAA-induced degradation of AID*-tagged Mss4 required expression of TIR1 in the same cells (Supplemental Figure S3A). Moreover, as expected for an essential gene, TIR1-containing cells expressing WT MSS4 were viable on plates containing 1-NAA, whereas TIR1-containing cells expressing Mss4-AID*-6HA were unable to grow (Figure 1B).
FIGURE 1:
PtdIns4,5P2 is required for TORC2 activity, but not for PM localization of TORC2 subunits. (A) A culture growing in exponential phase of a strain (yNM706) expressing TIR1 from the TDH3 promoter integrated at the LEU2 locus and expressing MSS4-AID*-6HA from its native promoter at its endogenous locus was treated with 1-NAA (1 mM). At the indicated times, samples were withdrawn and analyzed by SDS–PAGE and immunoblotting with an anti-HA mAb to assess the level of Mss4-AID*-6HA (top panel) and with rabbit polyclonal anti-Pgk1 as a control for loading of equivalent amounts of total sample protein (bottom panel), as described in Materials and Methods. MSS4 TIR1 cells (yIZ082) (denoted “WT”) served as the negative control for antibody specificity. (B) Serial dilutions of cultures of an MSS4 TIR1 (yIZ082) strain and an otherwise isogenic MSS4-AID*-6HA TIR1 strain (yNM706) were spotted onto agar plates of SCD-T medium buffered with 50 mM K2HPO4/KH2PO4 (pH 6) and containing either DMSO alone (-) or 1-NAA (1 mM final concentration) dissolved in an equal volume of the same solvent (+ 1-NAA), incubated for 2 d at 30°C, and photographed. (C) MSS4-AID*-6HA TIR1 cells (yNM706) carrying a CEN plasmid (pGFP-PH-7) expressing GFP-PHPLCδ1 under control of the CUP1 promoter were grown in SCD-T-U treated with either vehicle (DMSO) or 1 mM 1-NAA in the same solvent. After 30 min, GFP-PHPLCδ1 expression was induced by addition of CuSO4 (final concentration 100 μM) and, after further incubation for 90 min, the cells were examined using a conventional epifluorescence microscope, as described in Materials and Methods. Representative cells are shown. Scale bar, 2 μm. (D) An MSS4 TIR1 strain (yIZ082) and an MSS4-AID*-6HA TIR1 strain (yNM706), each carrying a plasmid (pAEA419) expressing Ypk15A-myc from the YPK1 promoter in the vector pRS316, were grown to midexponential phase in SCD-T-U and at time 0 exposed to 1-NAA (final concentration 1 mM) in DMSO. Aliquots of these cultures were withdrawn at the indicated times and lysed, and samples of these extracts containing equivalent amounts of protein were resolved by phosphate-affinity SDS–PAGE and analyzed by immunoblotting (top panel), as described in Materials and Methods. In parallel, the same samples also were resolved by standard SDS–PAGE and analyzed by immunoblotting with appropriate antibodies to confirm depletion of Mss4-AID*-6HA (anti-HA mAb), equivalent Ypk15A expression (anti-myc mAb), and equal sample loading (polyclonal anti-Pgk1 antibody) (bottom panels). MSS4 TIR1 cells (yIZ082) carrying empty vector pRS316 (denoted as -) served as the negative control for antibody specificity. Values below each of the lanes on the right are the relative level of Ypk1 phospho-isoforms (boxed in red), normalized to the Pgk1 loading control, where the value at time 0 before 1-NAA addition was set to 1.00 (one of two independent experiments is shown). (E) Derivatives of an MSS4-AID*-6HA TIR1 strain (yNM706) expressing from their native promoters at their endogenous loci either Tor2-mNG-3HA (yNM986), Avo1-GFP (yNM1073), Avo3-GFP (yNM1065), or Avo2-GFP (yNM1066), as indicated, were grown, treated, and lysed and samples of the resulting extracts were analyzed by immunoblotting, using the same control (“WT”) as in A, except that, where appropriate, anti-GFP antibodies were used to detect GFP-tagged proteins. (F) Three of the same MSS4-AID*-6HA TIR1 strains described in E, namely expressing either Avo1-GFP (yNM1073), Avo3-GFP (yNM1065), or Avo2-GFP (yNM1066), were examined immediately before (0 min) and then 60 and 120 min after their exposure to 1 mM 1-NAA using HiLo fluorescence microscopy, as described in Materials and Methods. Representative cells are shown. Scale bar, 2 μm. (G) An MSS4-AID*-6HA TIR1 strain (yNM1090) simultaneously expressing from their native promoters at their endogenous loci Tor2-mNG-3HA, Slm1-mKate2, and Pil1-BFP were treated and examined as in F. Representative cells are shown. Scale bar, 2 μm.
To ensure that the observed degradation resulted in loss of Mss4 function, we monitored the level of PM PtdIns4,5P2 using a fluorescent probe, a GFP-tagged derivative of the PH domain of human phospholipase Cδ1 (PLCδ1), which we and others have demonstrated has a very high affinity and selectivity for PtdIns4,5P2 in both animal cells (Stauffer et al., 1998; Lemmon, 2008) and yeast (Stefan et al., 2002; Garrenton et al., 2010). Cells expressing Mss4-AID*-6HA and TIR1 were treated with solvent (DMSO) or with 1-NAA in the same solvent for 30 min and then synthesis of GFP-PHPLCδ1 from the CUP1 promoter was induced by the addition of CuSO4. After 90 min (120 min of total 1-NAA treatment), the cells were examined by fluorescence microscopy. In the control cells where Mss4-AID*-6HA was not subjected to auxin-induced degradation, the GFP signal prominently decorated the PM, as expected, whereas in the 1-NAA-treated cells, all of the GFP fluorescence was found in the cytosol (Figure 1C). Thus, degradation of Mss4-AID*-6HA led to very efficient depletion of PtdIns4,5P2 from the PM.
To determine whether depletion of PM PtdIns4,5P2 affected the function of TORC2, we analyzed the state of phosphorylation of its major downstream effector Ypk1 using a derivative, Ypk15A-myc, which we have previously demonstrated can be phosphorylated only at four C-terminal TORC2-specific sites (Leskoske et al., 2017). To assess its phosphorylation status, we used phosphate-affinity (Phos-tag™) SDS–PAGE, a technique in which the degree of retardation of a protein reflects the extent of its phosphorylation (Kinoshita et al., 2009, 2015). As expected, in the absence of Mss4-AID*-6HA degradation, prominent lower-mobility isoforms of Ypk5A-myc were observed, indicative of its TORC2-dependent phosphorylation, whereas after initiation of Mss4-AID*-6HA degradation, these phosphorylated species were substantially reduced (Figure 1D, right). To rule out the possibility, albeit unlikely, that 1-NAA itself is a nonspecific inhibitor of TORC2 kinase activity, we treated cells expressing WT MSS4 with 1-NAA in the same manner and observed no diminution of TORC2-mediated Ypk1 phosphorylation (Figure 1D, left). Thus, depletion of PM PtdIns4,5P2 upon ablation of Mss4 caused a marked decrease in the TORC2-mediated phosphorylation of Ypk1, and a similar result was recently reported after shift of the temperature-sensitive mss4-103 mutant to the restrictive temperature (Riggi et al., 2018). The observed kinetics of loss of the TORC2-dependent Ypk1 phosphoisoforms presumably reflects the rate of turnover of PtdIns4,5P2, once synthesis of this phosphoinositide has been halted by destruction of Mss4, as well as the rate at which the TORC2 sites in Ypk1 are dephosphorylated, once its TORC2-dependent phosphorylation has been compromised. In any event, our results show that the PM pool of PtdIns4,5P2 is needed to maintain optimal TORC2 activity.
One trivial explanation for the loss of TORC2 function upon degradation of Mss4 and depletion of PM PtdIns4,5P2 is that this phosphoinositide is needed to stabilize Avo1 or one of the other TORC2 subunits. However, immunoblot analysis demonstrated that in cells each expressing from its endogenous locus a functional fluorescent protein-tagged version of one of the core (essential) subunits (Avo1, Avo3, and Tor2) and one peripheral (nonessential) subunit that we tested (Avo2), degradation of Mss4-AID*-6HA did not significantly alter the steady-state level of any of these TORC2 constituents (Figure 1E). Avo1, Avo2, and Avo3 were each tagged at the C terminus with GFP(F64L S65T) (eGFP; hereafter GFP; Cormack et al., 1996), as described in Materials and Methods. Tor2 was tagged within its N-terminal HEAT repeat region with mNeonGreen (mNG) and a 3xHA epitope (Shaner et al., 2013), as described in detail elsewhere (Leskoske et al., 2018).
Another possible explanation for the loss of TORC2 function upon Mss4 degradation is that the absence of PtdIns4,5P2 at the PM causes disassembly of TORC2. As described in Materials and Methods and documented elsewhere (Leskoske et al., 2017; Locke and Thorner, 2019), we have developed protocols for complete PM solubilization and immuno-isolation of TORC2. Therefore, we performed coimmunoprecipitation experiments to examine the state of TORC2 assembly in the PtdIns4,5P2-depleted cells. Using a yeast strain expressing MSS4-AID*-6HA, Avo3-3C-3xFLAG, Tor2-mNG-3xHA, and Avo1-6xHA from their endogenous chromosomal loci, we found that immunoprecipitation of Avo3 with anti-FLAG epitope antibodies coimmunoprecitated equivalent amounts of both Tor2 and Avo1, even after Mss4 was degraded to a barely detectable level (Supplemental Figure S3B), a condition under which no detectable PtdIns4,5P2 remains at the PM (Figure 1C). Thus, disassembly of TORC2 was not responsible for the drastic reduction in TORC2-mediated Ypk1 phosphorylation observed in the absence of PtdIns4,5P2.
Given the evidence that association of TORC2 with the PM is required for its function, and given our demonstration that all of the subunits remain intact, we thought that the most likely explanation for the observed loss of TORC2-mediated Ypk1 phosphorylation upon degradation of Mss4 and depletion of PM PtdIns4,5P2 would be dissociation of TORC2 from the PM. Unexpectedly, however, when we degraded Mss4-AID*-6HA in cells endogenously expressing either Avo1-GFP, Avo3-GFP, or Avo2-GFP, we observed no significant diminution of the PM-associated foci that are characteristic of TORC2, even 120 min after 1-NAA treatment (Figure 1F and Supplemental Figure S3C). Similarly, when we degraded Mss4-AID*-6HA in cells endogenously expressing Tor2-mNG-3HA, we did not observe any significant reduction in the overall number of PM-associated foci (Figure 1G and Supplemental Figure S3D). Likewise, in cells in which we degraded Mss4-AID*-6HA that endogenously expressed Bit61-GFP, there was no loss of the GFP-marked PM puncta (M. N. Martinez Marshall, unpublished observations). Most tellingly, we were able to use a fluorescence microscope equipped with a very fast scanner and optics that allowed simultaneous recording of two different fluorescent signals to examine cells coexpressing Mss4-AID*-6HA, Tor2-mNG, and Avo3-mKate2. Despite the fact that others (Berchtold et al., 2012) and we (Leskoske et al., 2018) have observed that the cortical TORC2 puncta are highly dynamic, and the fact that both Avo3 (∼700 molecules per cell) and Tor2 (∼1800 molecules per cell) are proteins of very low abundance (Ho et al, 2018), we were able to analyze the degree of colocalization for these two TORC2 subunits directly. In the confocal images obtained, we found that the majority of the cortical puncta detected were positive for Tor2 and Avo3, both before and after degradation of Mss4 and depletion of PtdIns4,5P2 (Supplemental Figure S3E). Thus, PM localization of the three essential subunits (Tor2, Avo1, and Avo3) and two ancillary subunits (Avo2 and Bit61) of TORC2 were all independent of the PM content of PtdIns4,5P2.
To monitor known PM-associated PtdIns4,5P2-binding proteins in the same cells, we examined Mss4-AID*-6HA TIR1 cells expressing Tor2-mNG-3HA from its endogenous locus and also expressing from their endogenous loci in the same cells both the diagnostic eisosome constituent Pil1, tagged at its C terminus with blue fluorescent protein [BFP; GFP (Y66H Y145F)] (Wachter et al., 1997), and the eisosome-associated protein Slm1, tagged at its C terminus with the red fluorescent protein mKate2 (Shcherbo et al., 2009). In marked contrast to the behavior of Tor2-mNG-3HA, when PM PtdIns4,5P2 was depleted by degradation of Mss4-AID*-6HA, the cortical puncta containing Pil1 were clearly reduced in number and displayed a marked tendency to clump into distinctly larger aggregates (Figure 1G and Supplemental Figure S3D), in agreement with mislocalization of Pil1 observed in a mss4ts mutant at the restrictive temperature (Karotki et al., 2011). Pil1 contains an F-BAR (FCH/Bin-Amphiphysin-Rvs) domain that has been shown to be specific for binding to PtdIns4,5P2 (Karotki et al., 2011). Similarly, the eisosome-associated protein Slm1 exhibited the same behavior as Pil1 (Figure 1G), in agreement with mislocalization of Slm1 upon shift of mss4ts mutants to the nonpermissive temperature reported by others (Audhya et al., 2004; Yu et al., 2004; Fadri et al., 2005). Slm1 contains a PH domain that also has been shown to be specific for binding to PtdIns4,5P2 (Audhya et al., 2004; Tabuchi et al., 2006; Daquinag et al., 2007; Gallego et al., 2010). Given that Slm1 and Slm2 are normally necessary for efficient TORC2-mediated phosphorylation of Ypk1 (Berchtold et al., 2012; Niles et al., 2012), although the mechanism by which they do so is unclear (Leskoske et al., 2018), the Slm1 aggregation observed when PM PtdIns4,5P2 is depleted might be sufficient to explain the reduced activity of TORC2 toward Ypk1 under these conditions.
The PH domain of Avo1 is dispensable for its PM localization
We constructed a TIR1 strain expressing Avo1-AID*-6HA and demonstrated, first, that it was rapidly and efficiently degraded in a 1-NAA-induced manner (Figure 2A) and fully able to support growth when present as the sole source of Avo1 (Figure 2B), unless degraded in a TIR1- and 1-NAA-dependent manner (Figure 2B), as expected for an essential gene. To ensure that the observed degradation had the physiological consequence of disrupting TORC2 function, we monitored one of the known diagnostic phenotypes of normal TORC2 activity, proper polarization of cortical actin patches to the bud (Schmidt et al., 1996; Loewith et al., 2002; Kamada et al., 2005). Gratifyingly, we found that within 60 min after Avo1-AID*-6HA degradation was initiated, the actin patches had become almost completely depolarized (Figure 2C). Likewise, ablation of Avo1-AID*-6HA also compromised TORC2-mediated phosphorylation of Ypk15A (Figure 2D).
FIGURE 2:
The PH domain of Avo1 is dispensable for its PM association and its function. (A) A culture growing in exponential phase of a strain (yNM718) expressing TIR1 from the TDH3 promoter integrated at the LEU2 locus and expressing Avo1-AID*-6HA from its native promoter at its endogenous locus was treated with 1-NAA (1 mM). At the indicated times, samples were withdrawn and analyzed by SDS–PAGE and immunoblotting with an anti-HA mAb to assess the level of Avo1-AID*-6HA (top panel) and with rabbit polyclonal anti-Pgk1 as a control for loading of equivalent amounts of total sample protein (bottom panel), as described in Materials and Methods. AVO1 TIR1 cells (yIZ082) (denoted “WT”) served as the negative control for antibody specificity. (B) Serial dilutions of cultures of an AVO1 TIR1 strain (yIZ082), an AVO1-6HA strain (yNM928), an AVO1-6HA TIR1 strain (yNM927), an AVO1-AID*-6HA strain (yNM784), and an AVO1-AID*-6HA TIR1 strain (yNM718), as indicated, were spotted onto agar plates of SCD-T medium buffered with 50 mM K2HPO4/KH2PO4 (pH 6) and containing either DMSO alone (-) or 1-NAA (1 mM final concentration) dissolved in an equal volume of the same solvent (+ 1-NAA), incubated for 2 d at 30˚C, and photographed. (C) Cells of an AVO1-AID*-6HA TIR1 strain (yNM718) were grown to midexponential phase in SCD-T and treated with 1 mM 1-NAA, and at the indicated times samples were withdrawn, fixed, stained with an F-actin-binding probe, rhodamine-labeled phalloidin, and examined using a conventional epifluorescence microscope, as described in Materials and Methods. Representative cells are shown. Scale bar, 2 μm. (D) An AVO1-AID*-6HA TIR1 strain (yNM718) carrying a plasmid (pAEA419) expressing Ypk15A-myc from the YPK1 promoter in the vector pRS316 was grown to mid-exponential phase in SCD-T-U and at time 0 exposed to 1-NAA (1 mM final concentration) in DMSO. Aliquots of these cultures were withdrawn at the indicated times, lysed, and samples of these extracts containing equivalent amounts of protein were resolved by phosphate-affinity SDS–PAGE and analyzed by immunoblotting (top panel), as described in Materials and Methods. In parallel, the same samples also were resolved by standard SDS–PAGE and analyzed by immunoblotting with appropriate antibodies to confirm depletion of Avo1-AID*-6HA (anti-HA mAb) and equal sample loading (polyclonal anti-Pgk1 antibody) (bottom panels). AVO1 TIR1 cells (yIZ082) carrying empty vector pRS316 (denoted WT) served as the negative control for antibody specificity. Values below each of the lanes on the right are the relative level of Ypk1 phospho-isoforms (boxed in red), normalized to the Pgk1 loading control, where the value at time 0 before 1-NAA addition was set to 1.00 (one of three independent experiments is shown). (E) Cultures of AVO1-AID*-6HA TIR1 cells (yNM718) expressing from the GALS promoter in the vector pRS416, as indicated, either full-length Avo1-mKate2-FLAG (pAEA399) or a truncated version lacking the C-terminal residues 1059–1176 of Avo1, which removes its PH domain (residues 1065–1169), Avo1ΔPH-mKate2-FLAG (pNM160), growing exponentially in SC-T-U medium containing 2% raffinose-0.2% sucrose as the carbon source and buffered with 50 mM K2HPO4/KH2PO4 (pH 6), were treated with 1 mM 1-NAA. After 1 h, galactose was added (2% final concentration) to induce expression of the plasmid-borne Avo1 variants. After 3 h, the cells were harvested and lysed, and samples of the resulting extracts were resolved by SDS–PAGE and analyzed by immunoblotting with anti-HA to confirm removal of Avo1-AID*-6HA, with anti-FLAG to confirm production of the mKate2-FLAG-tagged Avo1 variants, and with anti-Pgk1 to confirm equal sample loading. AVO1 TIR1 cells (yIZ082) carrying empty vector pRS416 (denoted WT) was the negative control for antibody specificity. (F) Samples of the same AVO1-AID*-6HA TIR1 cells expressing the indicated mKate2-FLAG-tagged Avo1 variants, as in E, from the final (4 h) time point were examined by HiLo fluorescence microscopy. Representative cells are shown. Scale bar, 2 μm. (G) Serial dilutions of cultures of an AVO1 TIR1 strain (in this case, TIR1 was inserted at the HIS3 locus; yNM793) carrying an empty LEU2-marked vector (pRS315; denoted -) and an AVO1-AID*-6HA TIR1 strain (yNM786) (also with TIR1 inserted at the HIS3 locus) carrying, as indicated, either the same empty vector (-) or the same plasmid expressing from the native AVO1 promoter either full-length Avo1-9myc (pLZ3) or Avo1(ΔPH)-9myc (pLZ11), were spotted onto agar plates of SCD-T-L medium buffered with 50 mM K2HPO4/KH2PO4 (pH 6) and containing either DMSO alone (-) or 1-NAA (1 mM final concentration) dissolved in an equal volume of the same solvent (+ 1-NAA), incubated for 2 d at 30°C, and photographed.
Reexpression of any Avo1 mutant of interest after removal of the endogenously expressed Avo1-AID*-6HA by 1-NAA treatment allowed us to investigate the function and PM association of that Avo1 derivative. Given that we found that Avo1-GFP still localized to the PM in cells depleted of PtdIns4,5P2 (Figure 1F), we used the reexpression approach to ascertain directly the role of its C-terminal PH domain in Avo1 PM localization, which had not heretofore been examined. To this end, we constructed CEN plasmids that expressed from the galactose-inducible GALS promoter (Mumberg et al., 1994) either full-length Avo1 tagged at its C terminus with mKate2 and a FLAG epitope, or an otherwise identical construct from which the last 117 residues (1059–1176) of the AVO1 ORF had been deleted, thereby removing its 105-residue PH domain (1065–1169). We elected to use the GALS promoter (a partially crippled derivative of the GAL1 promoter) because, although nicely galactose-inducible, it drives a significantly lower level of gene expression than any native GAL promoter (Mumberg et al., 1994). Indeed, using this strategy, we were able to reexpress these Avo1 derivatives in cells after complete depletion of Avo1-AID*-6HA (Figure 2E). Revealingly, when these same cells were examined by fluorescence microscopy, Avo1(∆PH)-mKate2-FLAG mutant decorated the PM in the characteristic puncta to an extent equivalent to that observed for full-length Avo1-mKate2-FLAG (Figure 2F). As noted by others (Higuchi-Sanabria et al., 2016), at the extended exposure times necessary to image mKate2-tagged proteins of low abundance, background autofluorescence from the vacuole becomes detectable.
Moreover, in agreement with our plasmid shuffle results (Supplemental Figure S1) and the data obtained by saturation transposon mutagenesis (Supplemental Figure S2), we found that both full-length Avo1-9myc and its PH domain–less derivative Avo1(∆PH)-9myc, each expressed from the native AVO1 promoter, were able to support cell growth when present as the sole source of Avo1 after removal of Avo1-AID*-6HA (Figure 2G). Hence, the conclusions of a prior report (Berchtold and Walther, 2009) that the PH domain is required for the essential function of Avo1 and that the PH domain is necessary for PM recruitment of Avo1 (and, by inference, for PM recruitment and function of TORC2) are incorrect.
Avo1 is dispensable for PM localization of other TORC2 subunits
To assess the role of Avo1 in PM recruitment of other TORC2 constituents, we induced the TIR1- and 1-NAA-dependent degradation of Avo1-AID*-6HA in cells in which functional fluorescent protein-tagged derivatives of other TORC2 subunits were each expressed from their respective endogenous chromosomal locus. We found that both of the other two essential TORC2 components (Tor2-mNG-3HA and Avo3-GFP), as well as two more peripheral (nonessential) TORC2 components (Avo2-GFP and Bit61-GFP), all remained associated with the PM at a normal level (Figure 3A) and remained stable in the cells even when the amount of Avo1-AID*-6HA was reduced to a barely detectable level (Figure 3B). Furthermore, coimmunoprecipitation analysis demonstrated that the other two core TORC2 components (Tor2 and Avo3) remained associated, even after Avo1 was degraded to a barely detectable level (Figure 3C). Thus, Avo1 is not required for the other subunits of TORC2 to assemble and associate with the PM.
FIGURE 3:
PH domain–containing proteins Avo1, Slm1, and Slm2 are dispensable for PM localization of TORC2 subunits. (A) Exponentially growing cultures of AVO1-AID*-6HA TIR1 cells also expressing from their native promoter at their endogenous locus either Tor2-mNG-3HA (yNM977), Avo3-GFP (yNM888), Avo2-GFP (yNM847), or Bit61-GFP (yNM886), as indicated, were examined by HiLo fluorescence microscopy immediately before (0 min) or at the indicated times after treatment of the cultures with 1 mM 1-NAA. Representative cells are shown. Scale bar, 2 μm. (B) Extracts from the same cells as in A were resolved by SDS–PAGE and analyzed by immunoblotting with anti-HA antibodies to confirm removal of Avo1-AID*-6HA and level of expression of Tor2-mNG-3HA, with anti-GFP antibodies to assess the level of expression of Avo3-GFP, Avo2-GFP, and Bit61-GFP, and with anti-Pgk1 to confirm equal sample loading. AVO1 TIR1 cells (yIZ082) (denoted WT) were used as the negative control for specificity of the immunoblots. (C) An Avo1-AID*-6HA TIR1 strain expressing Tor2-mNG-3HA from its native promoter at its endogenous locus (yNM992), an otherwise isogenic strain also expressing Avo3-3C-3FLAG from its endogenous locus (yNM994), and an AVO1 AVO3 TOR2 TIR1 control strain (yIZ082), were grown on phosphate-buffered SCD-T to mid-exponential phase and then treated with either solvent (DMSO) alone (0) or 1 mM 1-NAA in the same solvent for 60 min, as indicated. Cells were then harvested and lysed, and FLAG-tagged proteins were immuno-isolated from the extracts using agarose beads coated with anti-FLAG antibodies as described in Materials and Methods. Samples of the bound proteins were solubilized, resolved by SDS–PAGE on an 8% gel, and analyzed by immunoblotting with anti-HA and anti-FLAG antibodies (as well as with anti-Pgk1 antibodies to confirm equal protein loading). (D) Exponentially growing cultures of Avo1-AID*-6HA Slm1-AID*-9myc slm2Δ TIR1 cells expressing from their native promoter at their endogenous locus either Avo3-GFP (yNM1026) or Avo2-mKate2 (yNM862) were examined by HiLo fluorescence microscopy immediately before (0 min) or at the indicated times after treatment of the cultures with 1 mM 1-NAA. Representative cells are shown. Scale bar, 2 μm. (E) Extracts from the same cells as in C, and the same control for antibody specificity as in B, were resolved by SDS–PAGE and analyzed by immunoblotting with anti-HA antibodies to confirm removal of Avo1-AID*-6HA, with anti-myc antibodies to confirm removal of Slm1-AID*-9myc, and with anti-Pgk1 to confirm equal sample loading.
However, in addition to Avo1, two proteins (Slm1 and Slm2) that reportedly associate transiently with TORC2 (Berchtold et al., 2012; Niles et al., 2012), mainly through their interaction with the Avo2 and Bit61 subunits of TORC2 (Fadri et al., 2005; Karuppasamy et al., 2017), also possess PH domains that have been shown to localize these molecules to the PM via binding to PtdIns4,5P2 (Audhya et al., 2004; Tabuchi et al., 2006; Daquinag et al., 2007; Gallego et al., 2010). Thus, we considered the possibility that Slm1 and/or Slm2 might contribute to tethering the other TORC2 components to the PM even in the absence of Avo1. To examine this issue, we generated TIR1 cells expressing both Avo1-AID*-6HA and Slm1-AID*-9myc endogenously and carrying a slm2∆ mutation, as well as expressing tagged versions of other TORC2 components of interest. Here too, we observed that a core TORC2 subunit (Avo3-GFP) and an ancillary subunit (Avo2-mKate2), each of which we examined in this genetic background, still remained associated with the PM (Figure 3D), even though both AID*-tagged PH domain–containing proteins were reduced to barely detectable levels, as judged by immunoblotting with the appropriate antibodies (Figure 3E). Hence, neither Avo1 nor the Slm proteins are necessary for localization of other TORC2 subunits to the PM.
Avo3 localizes to the PM independent of Tor2
The N-terminal half of Tor2 comprises dozens of HEAT repeats in two blocks (Perry and Kleckner, 2003) that form two α-helical solenoid superstructures (Karuppasamy et al., 2017), which have been implicated in TORC2 association with the PM (Kunz et al., 2000). Hence, it seemed reasonable that, as the largest subunit in TORC2, Tor2 is the protein that serves as the PM anchor for the other subunits. To test this hypothesis, we expressed from the endogenous TOR2 locus in TIR1-containing cells a functional Tor2 derivative, in which an AID* tag and a mNG-3HA cassette were inserted between residues N321 and T322 of the TOR2 ORF, constructed by the same approach we used to generate Tor2-mNG-3HA (lacking the AID* tag), which is described in detail elsewhere (Leskoske et al., 2018). As observed for all of the other AID*-containing proteins we constructed for this study, Tor2-AID*-mNG-3HA was rapidly degraded after addition of 1-NAA (Figure 4A) and, concomitantly, TORC2-mediated Ypk1 phosphorylation was markedly reduced (Figure 4A).
FIGURE 4:
PM localization of Avo3 does not require Tor2, but PM localization of Tor2 and other TORC2 subunits requires Avo3. (A) A culture of a strain (yAEA346) expressing TIR1 from the TDH3 promoter integrated at the LEU2 locus, TOR2-AID*-mNG-3HA from its native promoter at its endogenous locus, and Ypk15A-myc from plasmid pAEA419, was grown in SCD-T-U at 30°C to midexponential phase and then treated with 1-NAA (1 mM). At the indicated times, aliquots of these cultures were withdrawn and lysed, and samples of these extracts containing equivalent amounts of protein were resolved by phosphate-affinity SDS–PAGE and analyzed by immunoblotting (top panel), as described in Materials and Methods. In parallel, the same samples were also resolved by standard SDS–PAGE and analyzed by immunoblotting with appropriate antibodies to confirm depletion of Tor2-AID*-3HA (anti-HA mAb) and equal sample loading (polyclonal anti-Pgk1 antibody; bottom panels). TOR2 TIR1 cells (yIZ082) carrying empty vector pRS316 (denoted WT) were used as the negative control for specificity of the immunoblots. (B) Exponentially growing cultures of TOR2-AID*-mNG-3HA TIR1 cells also expressing from their native promoter at their endogenous locus either Avo3-mKate2 (yNM1035) (top panels) or Avo2-mKate2 (yNM1034) (bottom panels) were examined by HiLo fluorescence microscopy immediately before (0 min) or at the indicated times after treatment of the cultures with 1 mM 1-NAA. Representative cells are shown. Scale bar, 2 μm. (C) A culture of a strain (yNM858) expressing TIR1 from the TDH3 promoter integrated at the LEU2 locus, AVO3-AID*-6HA from its native promoter at its endogenous locus, and Ypk15A-myc from plasmid pAEA419 was grown in SCD-T-U at 30°C to midexponential phase and then treated with 1-NAA (1 mM). At the indicated times, aliquots of these cultures were withdrawn and lysed, and samples of these extracts containing equivalent amounts of protein were resolved by phosphate-affinity SDS–PAGE and analyzed by immunoblotting (top panel), as described in Materials and Methods. In parallel, the same samples also were resolved by standard SDS–PAGE and analyzed by immunoblotting with appropriate antibodies to confirm depletion of Avo3-AID*-3HA (anti-HA mAb) and equal sample loading (polyclonal anti-Pgk1 antibody; bottom panels). TOR2 TIR1 cells (yIZ082) carrying empty vector pRS316 (denoted WT) were used as the negative control for antibody specificity. (D) Exponentially growing cultures of AVO3-AID*-6HA TIR1 cells also expressing from their native promoter at their endogenous locus either Tor2-mNG-3HA (yNM975), Avo1-GFP (yNM998), Avo2-GFP (yNM1031), or Bit61-GFP (yNM884), as indicated, were examined by HiLo fluorescence microscopy immediately before (0 min) or at the indicated times after treatment of the cultures with 1 mM 1-NAA. Representative cells are shown. Scale bar, 2 μm. (E) Extracts from the same cells as in D were resolved by SDS–PAGE and analyzed by immunoblotting with anti-HA antibodies to confirm removal of Avo3-AID*-6HA and level of expression of Tor2-mNG-3HA, with anti-GFP antibodies to assess the level of expression of Avo1-GFP, Avo2-GFP and Bit61-GFP, and with anti-Pgk1 to confirm equal sample loading. AVO1 TIR1 cells (yIZ082) (denoted WT) were used as the negative control for antibody specificity.
If Tor2 is the essential PM anchor for all other TORC2 components, we expected that, after degradation of Tor2-AID*-mNG-3HA, each subunit would no longer exhibit the cortical foci characteristic of TORC2. Indeed, this was the case for Avo2-mKate2 expressed in the same cells (Figure 4B). Strikingly, however, when Avo3-mKate2 was expressed in the same cells, it remained as PM-associated puncta, even when the amount of Tor2-AID*-mNG-3HA in the cells was undetectable by either immunoblotting (Figure 4A) or imaging of the mNG signal (Figure 4B). This result was surprising because Avo3 is an essential subunit, binds tightly to Tor2, and is required for the overall integrity of TORC2 (Wullschleger et al., 2005; Ho et al., 2008). Nevertheless, Tor2 is not required for the PM localization of Avo3.
Avo3 is required for PM localization of all other TORC2 subunits
To examine the converse relationship, we expressed from the endogenous AVO3 locus in TIR1-containing cells a functional Avo3 derivative carrying in-frame at its C-terminus an AID* tag and a 6HA epitope. As expected, Avo3-AID*-6HA was rapidly degraded after addition of 1-NAA (Figure 4C) and, concomitantly, TORC2-mediated Ypk1 phosphorylation was decreased (Figure 4C).
Most importantly, upon degradation of Avo3-AID*-6HA, Tor2-mNG-3HA expressed in the same cells was completely displaced from the PM within 60 min after treatment of the cells with 1-NAA, as judged by fluorescence microscopy (Figure 4D), yet the Tor2-mNG-3HA remained stable in the cytosol, as judged by immunoblotting (Figure 4E). The same was observed for Avo1, the other essential core subunit of TORC2; when Avo1-GFP was expressed in the same cells, it was completely displaced from the PM after Avo3-AID*-6HA degradation (Figure 4D), yet it remained stable in the cytosol (Figure 4E). Likewise, both Avo2-GFP and Bit61-GFP expressed in the same cells also were displaced from the PM (Figure 4D) and remained readily detectably in the cytosol (Figure 4E). Thus, Avo3 is the TORC2 subunit responsible for PM retention of all the other TORC2 components.
Order of assembly of TORC2 components at the PM
As documented in this study, Avo1 is dispensable for localization of all other TORC2 components at the PM (Figure 3, A and B), Tor2 is not required for localization of Avo3 at the PM (Figure 4A), whereas Avo3 is required for retention of Tor2 at the PM (Figure 4D). These interrelationships suggest an order of dependency in which Avo3 associates with the PM first and is critical for recruitment of Tor2, which is then, in turn, necessary for the association of Avo1 and the other TORC2 components.
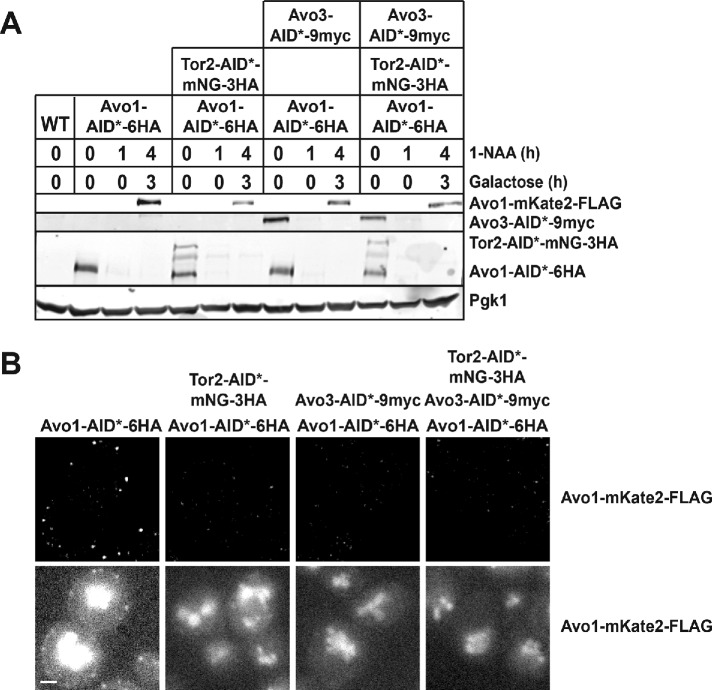
To test this scenario directly, we first degraded Avo1-AID*-6HA in TIR1-expressing cells in which we could simultaneously degrade Tor2-AID*-mNG-3HA, Avo3-AID*-9myc, or both, and then reexpressed Avo1-mKate2-FLAG to determine whether it was capable of associating with the PM under each of these conditions. In addition, in these same cells, we coexpressed Ypk1D242A, a variant that bypasses the need for its TORC2 phosphorylation (Roelants et al., 2011), to avoid any detrimental effects of depleting essential TORC2 subunits (i.e., growth arrest, aberrant morphology) during the longer incubation time required for galactose-induced gene expression. We found, as before (Figure 2, E and F), that Avo1-mKate2-FLAG expressed after degradation of Avo1-AID*-6HA in otherwise WT cells (Figure 5A) was fully capable of forming the PM-associated puncta diagnostic of TORC2, which was revealed best using the image analysis software Fiji (Schindelin et al., 2012) to subtract the nonspecific cellular background signal (Figure 5B). In marked contrast, and consistent with the order of assembly that our other data suggested, in the absence of either Tor2 or Avo3 (or both), no Avo1-mKate2-FLAG was capable of associating with the PM (Figure 5B), even though it was stably expressed at a readily detectable level (Figure 5A).
FIGURE 5:
Absence of either Avo3 or Tor2 prevents recruitment of Avo1 to the PM. (A) Cultures of AVO1-AID*-6HA TIR1 cells (yNM718) expressing from the GALS promoter Avo1-mKate2-FLAG (pAEA399) in the vector pRS416, AVO1-AID*-6HA TOR2-AID*-mNG-3HA TIR1 cells (yNM1040) carrying pAEA399, AVO1-AID*-6HA AVO3-AID*-9MYC TIR1 cells (yNM1064) carrying pAEA399, and AVO1-AID*-6HA TOR2-AID*-mNG-3HA AVO3-AID*-9MYC TIR1 cells (yNM1056) carrying pAEA399, as indicated, grown to midexponential phase in SC-T-U medium containing 2% raffinose–0.2% sucrose as the carbon source and buffered with 50 mM K2HPO4/KH2PO4 (pH 6), were treated with 1 mM 1-NAA. After 1 h, galactose was added (2% final concentration) to induce expression of the plasmid-borne Avo1-mKate2-FLAG. After 3 h, the cells were harvested and lysed, and samples of the resulting extracts were resolved by SDS–PAGE and analyzed by immunoblotting with anti-HA to confirm removal of Avo1-AID*-6HA and Tor2-AID*-mNG-3HA, with anti-myc to confirm removal of Avo3-AID*-9myc, with anti-FLAG to confirm production of the mKate2-FLAG-tagged Avo1, and with anti-Pgk1 to confirm equal sample loading. AVO1 TIR1 cells (yIZ082) carrying empty vector pRS416 (denoted WT) served as the negative control for antibody specificity. (B) Samples of the same cells as in A from the final (4 h) time point were examined by HiLo fluorescence microscopy with (top panels) and without (bottom panels) subtraction of the nonspecific cellular background signal. Representative cells are shown. Scale bar, 2 μm.
To further test our model, we degraded Avo3-AID*-9myc in TIR1- and Ypk1D242A-expressing cells in which we could simultaneously degrade either Avo1-AID*-6HA or Tor2-AID*-mNG-3HA, or both, and then reexpressed Avo3-mKate2-FLAG to determine whether it was capable of associating with the PM under each of these conditions. Revealingly, we found that Avo3-mKate2-FLAG expressed after degradation of Avo3-AID*-9myc (Figure 6A) was fully capable of reforming the PM-associated puncta diagnostic of TORC2 in otherwise WT cells, in cells lacking Avo1, in cells lacking Tor2, or in cells lacking both Avo1 and Tor2 (Figure 6B). These observations demonstrate unequivocally that Avo3 is the subunit responsible for the PM anchoring of TORC2.
FIGURE 6:
PM recruitment of Avo3 is independent of both Avo1 and Tor2. (A) Cultures of AVO3-AID*-9MYC TIR1 cells (yNM1057) expressing from the GALS promoter Avo3-mKate2-FLAG (pAEA400) in the vector pRS416, AVO3-AID*-9MYC AVO1-AID*-6HA TIR1 cells (yNM1064) carrying pAEA400, AVO3-AID*-9MYC TOR2-AID*-mNG-3HA TIR1 cells (yNM1062) carrying pAEA400, and AVO3-AID*-9MYC AVO1-AID*-6HA TOR2-AID*-mNG-3HA TIR1 cells (yNM1056) carrying pAEA400, as indicated, grown to midexponential phase in SC-T-U medium containing 2% raffinose–0.2% sucrose as the carbon source and buffered with 50 mM K2HPO4/KH2PO4 (pH 6), were treated with 1 mM 1-NAA. After 1 h, galactose was added (2% final concentration) to induce expression of the plasmid-borne Avo1-mKate2-FLAG. After 2 h, the cells were harvested and lysed, and samples of the resulting extracts resolved by SDS–PAGE and analyzed by immunoblotting with anti-myc to confirm removal of Avo3-AID*-9myc, with anti-HA to confirm removal of Avo1-AID*-6HA and Tor2-AID*-mNG-3HA, with anti-FLAG to confirm production of the mKate2-FLAG-tagged Avo3, and with anti-Pgk1 to confirm equal sample loading. AVO3 TIR1 cells (yIZ082) carrying empty vector pRS416 (denoted WT) served as the negative control for antibody specificity. (B) Samples of the same cells as in A from the final (3 h) time point were examined by HiLo fluorescence microscopy with (top panels) and without (bottom panels) subtraction of the nonspecific cellular background signal. Representative cells are shown. Scale bar, 2 μm.
N-terminal Armadillo repeats of Avo3 are responsible for its membrane anchoring
To discern whether any specific sequence elements in Avo3 mediate its PM association, we constructed plasmids that expressed from the GALS promoter a series of N-terminal truncations of Avo3-mKate2-FLAG in which successive regions of Avo3 were systematically removed, as well as an internal deletion and a corresponding fragment (Figure 7A). We then introduced this set of plasmids into AVO3-AID*-9myc TIR1 cells. To examine which of these constructs was able to form the PM-associated puncta diagnostic of TORC2, we first degraded Avo3-AID*-9myc in the TIR1-expressing cells by addition of 1-NAA and then, after 1 h, we induced expression of each Avo3-mKate2-FLAG construct and, after an additional 2 h, examined the cells by fluorescence microscopy to determine whether it was capable of associating with the PM. Revealingly, we found that full-length Avo3-mKate2-FLAG, Avo3(∆2-300)-mKate2-FLAG and Avo3(∆2-500)-mKate2-FLAG were capable of reforming the PM-associated puncta diagnostic of TORC2 in these cells (Figure 7B), whereas the next largest truncation, Avo3(∆2-700)-mKate2-FLAG, and all the others were not (Figure 7B), even though all of the constructs were expressed, as judged by immunoblotting (Supplemental Figure S4). In this regard, we found consistently that Avo3(∆2-500)-mKate2-FLAG was expressed at a significantly lower level than the other constructs, likely explaining its apparently weaker PM localization. The internal deletion Avo3(∆300-500)-mKate2-FLAG was also unable to associate with the PM (Figure 7B), indicating that this region is necessary either for PM binding and/or the proper folding of the remainder of Avo3. Indeed, the corresponding fragment Avo3(300-500)-Kate2-FLAG did not localize to the PM, suggesting that, by itself, this segment of Avo3 is not sufficient for PM binding (Figure 3B). Taken together, these results suggest that sequence elements lying within the N-terminal portion (especially residues 500–700) of the Armadillo repeat region of Avo3 (Supplemental Figure S5) mediate its PM tethering. However, neither an Avo3(300–500) fragment nor a larger Avo3(300–700) fragment tagged with a different fluorescent marker (mNG) was able to localize to the PM (Supplemental Figure S6, B and C), evidence again that this region of Avo3 is not sufficient for PM binding and/or is unable to fold properly in the absence of the rest of Avo3.
FIGURE 7:
N-terminal ARM repeats in Avo3 are required for its PM association. (A) Schematic depiction of full-length Avo3 and the set of N-terminal truncations, one internal deletion, and one internal fragment, all tagged at their C-terminus with mKate2-FLAG (not shown), that were examined in this study. Dark gray, ARM repeat domain. (B) An AVO3-AID*-9MYC TIR1 strain (yNM1057) was transformed with empty vector pRS416, or plasmids expressing from the GALS promoter either full-length Avo3-mKate2-FLAG (pAEA400), Avo3(Δ2-300)-mKate2-FLAG (pNM166), Avo(3Δ2-500)-mKate2-FLAG (pNM165), Avo3(Δ2-700)-mKate2-FLAG (pNM164), Avo3(Δ2-810)-mKate2-FLAG (pNM167), Avo3(Δ2-910)-mKate2-FLAG (pNM163), Avo3(Δ2-1100)-mKate2-FLAG (pNM162), Avo3(Δ300-500)-mKate2-FLAG (pNM170), or Avo3(300-500)-mKate2-FLAG (pNM168), as indicated. The cultures were grown in phosphate-buffered SCRS-T-U to mid-exponential phase and treated with 1 mM 1-NAA to induce Avo3-AID*-9myc degradation. After 1 h, induction of each plasmid-borne mKate2-FLAG construct was induced by addition of galactose (2% final concentration). After incubation for 2 h, the cells were examined by HiLo fluorescence microscopy, as described in Materials and Methods. Scale bars, 2 μm. (C) Samples of the same cells as in B expressing either empty vector, full-length Avo3, or the set of N-terminal truncations depicted in A were spotted on phosphate-buffered SCRS-T-U plates containing DMSO alone (-) (left) or 1 mM 1-NAA in same volume of DMSO (+ 1-NAA) (right). After 2 d, the plates were photographed. (D) As in C, for the same cells as in B expressing either empty vector, full-length Avo3, the internal deletion, or the fragment depicted in A.
Consistent with all of the other data presented in this study to show that Avo3 is the subunit responsible for PM anchoring of TORC2 (and that TORC2 must be associated with the PM to be functional), only full-length Avo3-mKate2-FLAG and Avo3(∆2-300)-mKate2-FLAG were capable of maintaining cell viability when present as the sole source of Avo3 (Figure 7, C and D; Supplemental Figure S6D) and able to colocalize Tor2 to the PM (Supplemental Figure S6, E and F).
DISCUSSION
In this study, we used chemogenetically induced protein degradation under otherwise unperturbed conditions and fluorescence microscopy, immunoblotting, and immunoprecipitation as approaches for understanding the interrelationships that govern the membrane targeting, assembly, and function of the multiprotein complex TORC2 in vivo. Imaging was challenging because the current best estimates for the abundance of the core (essential) TORC2 subunits are remarkably low (in molecules per cell) (Tor2 [∼1800]; Avo1 [∼1000]; Avo3 [∼700]; and Lst8 [∼3000]), in comparison to, for example, core eisosome components, which have an abundance that is at least an order of magnitude higher (Pil1 [∼40,000] and Lsp1 [∼30,000]) (Ho et al., 2018). Lst8, a toroidal β-propeller protein, is about twice as abundant as Tor2 because it has the role of binding tightly to and stabilizing the kinase domain fold of any TOR (Yang et al., 2013; Baretic´ et al., 2016), and therefore it associates with both Tor1 (∼1,500 molecules per cell; Ho et al., 2018) and Tor2 and is found in both TORC1 and TORC2 (Loewith et al., 2002; Wedaman et al., 2002). Despite their low abundance, we were able to label each of these subunits and other TORC2 components with fluorescent protein and epitope tags (without or with an AID* degron) in a manner that preserved their biological function when each was expressed from its native promoter at its endogenous chromosomal locus and to visualize their subcellular location at this level of expression using, in most experiments, a microscope outfitted to perform highly inclined and laminated optical sheet (HiLo) fluorescence microscopy (Tokunaga et al., 2008). In this regard, we noted that the bulk of Tor2-mNG-3HA was found in discrete foci at the cell periphery, although a readily detectable fluorescent signal was also associated with the vacuolar membrane. Thus, even in cells expressing Tor1, which can only populate TORC1 (Loewith et al., 2002; Wedaman et al., 2002; Hill et al., 2018), the complex that localizes to the vacuole membrane (Péli-Gulli et al., 2015; Noda, 2017), some fraction of TORC1 contains Tor2, in agreement with prior evidence that Tor2, and not Tor1, is essential for yeast cell viability (Heitman et al., 1991; Kunz et al., 1993; Helliwell et al., 1994). Most importantly, cells expressing each core TORC2 subunit and essential gene product tagged with the AID* degron were unable to grow on plates containing 1-NAA and, reassuringly, this inviability on 1-NAA–containing medium was rescued in all of these strains when they expressed the constitutively active Ypk1D242A allele (Supplemental Figure S7), which is able to bypass the need for functional TORC2 (Roelants et al., 2011; Leskoske et al., 2017).
The first and unexpected conclusion we reached using these methods was that, contrary to a prior report (Berchtold and Walther, 2009), the PtdIns4,5P2-binding PH domain of Avo1 is not required for the in vivo function of Avo1, or for Avo1 recruitment to the PM, or for tethering the other components of TORC2 at the PM. That the PH domain of Avo1 is dispensable for cell viability, and thus for TORC2 function, was revealed by three independent methods (plasmid shuffle, saturation transposon mutagenesis, and rescue of the growth of cells in which Avo1-AID*-6HA was continuously degraded). In the prior study, the criterion used to assess the essentiality of the Avo1 PH domain was inviability of the spores deduced to carry a mutant avo1(∆PH) locus that these investigators generated. Although difficult, it is possible to reconcile our results with the prior work, if the PH domain of Avo1 has some unique function in the germination process that it does not have in mitotically growing cells. Alternatively, the PH domain in Avo1 may play a strictly regulatory role. For example, it has been reported recently that under conditions of hyperosmotic shock, TORC2 becomes sequestered at PM invaginations enriched in PtdIns4,5P2 (Riggi et al., 2018); however, the role of the PH domain of Avo1 in that process per se was not directly examined by these researchers.
We also explored the role of PtdIns4,5P2 in TORC2 function and localization by degrading Mss4, the sole PtdIns4P 5-kinase in S. cerevisiae, which very effectively depleted the PM pool of PtdIns4,5P2 within one cell-doubling time, indicating that the PtdIns4,5P2 5-phosphatases responsible for turnover of this phosphoinositide are very active (Srinivasan et al., 1997; Stolz et al., 1998). Consistent with our results with Avo1 and its PH domain, the precipitous loss of Ptdns4,5P2 did not result in displacement of any TORC2 component from the PM, and, likewise, removal of Avo1 and both of the other two PH domain–containing proteins (Slm1 and Slm2) that associate more transiently with TORC2 did not prevent localization of the other TORC2 components to the PM. Nonetheless, depletion of PM PtdIns4,5P2 significantly decreased TORC2 activity, monitored by the state of phosphorylation of TORC2 sites in its substrate Ypk1. Unlike TORC2, we found that Pil1, a core component of eisosomes (Karotki et al., 2011), and Slm1, which is primarily an eisosome-associated protein (Kamble et al., 2011; Olivera-Couto et al., 2011), formed large cortical aggregates when the PM PtdIns4,5P2 was depleted by Mss4 degradation. Because Slm1 and Slm2 are normally necessary for efficient TORC2-mediated phosphorylation of Ypk1 (Berchtold et al., 2012, Niles et al., 2012), this aggregation might be sufficient to explain the reduction in TORC2-mediated phosphorylation of Ypk1 under these conditions. However, our data suggest an alternative explanation, based on analogy to an observation made about mTORC2. It has been reported that binding of PtdIns3,4,5P3 to the PH domain of mSin1 (the Avo1 orthologue) in mTORC2 alleviates an inhibitory constraint that the mSin1 PH domain imposes on mTORC2 activity (Liu et al., 2015). Thus, another possible explanation for why the loss of PtdIns4,5P2 markedly reduced yeast TORC2 activity, yet had no effect on the localization or state of assembly of TORC2, is that the role of PtdIns4,5P2 binding to the PH domain of Avo1 is to alleviate a negative (perhaps steric) constraint that the PH domain exerts (perhaps intramolecular occlusion of the CRIM domain in Avo1, which would prevent efficient binding of Ypk1). In other words, the role of PtdIns4,5P2 is as a positive allosteric effector. This scenario readily explains why deletion of the Avo1 PH domain is well tolerated.
In the same way, Avo2 (∼1000 molecules per cell), Slm1 (∼4000 molecules per cell), and Slm2 (∼1000 molecules per cell) (Ho et al., 2018) reportedly associate (Uetz et al., 2000, Ito et al., 2001, Audhya et al., 2004, Fadri et al., 2005), and we (Leskoske et al., 2018) and others (Bartlett et al., 2015) have observed that a fraction of the cellular Avo2 pool colocalizes with eisosomes. However, in the studies described here, when either Tor2 or Avo3 was degraded, PM-localized Avo2-containing puncta were reduced to an undetectable level, even though Avo2 was readily detectable in extracts of the same cells. Hence, it appears that cortical localization of Avo2 is primarily through its association with TORC2 and not eisosomes.
The most striking new conclusions drawn from our studies are that Avo3 is able to associate with the PM in the absence of either Tor2 or Avo1, that the N-terminal portion of the long conserved ARM repeat region in Avo3 is necessary for its PM recruitment, and that during de novo assembly of TORC2, Avo3 is most likely the first subunit to arrive at the PM and is responsible for anchoring Tor2, which, in turn, then recruits Avo1. Indeed, when Avo3-AID*-9myc was degraded to undetectable levels, the amounts of both Tor2 and Avo1 remained stable in the cytosol, whereas the peripherally associated TORC2 subunits Avo2 and Bit61 gradually decreased, indicating that in the absence of core TORC2 assembly these ancillary subunits are less stable. Thus, our findings provide new mechanistic insight about TORC2 and pinpoint Avo3 as pivotal for TORC2 PM localization and assembly in vivo.
Avo3 contains about 20 ARM repeats (Supplemental Figure S5; ∼each ∼40 residues long and each comprising a small three-helix bundle); in other proteins with multiple tandem ARM motifs, the repeats pack against each other in a superhelical manner to form an alpha-solenoid (Peifer et al., 1994; Groves and Barford, 1999; Reichen et al., 2014). Our observations about the essential role of Avo3 in PM anchoring of Tor2 and all other TORC2 components are compatible with a recently obtained cryo-EM structure of yeast TORC2 (Karuppasamy et al., 2017). In this model, the deduced location of the N-terminal portion of the Avo3 alpha-solenoid structure, corresponding to the N-terminal part of its entire ARM repeat array (residues 285-1250), is prominently solvent-exposed. By contrast, available evidence indicates that the C-terminal end of Avo3 is in close association with the FRB domain of Tor2 (Gaubitz et al., 2015, Karuppasamy et al., 2017), an element in Tor2 that overhangs its active site (Yang et al., 2013, Baretic´ et al., 2016). Among the first proteins in which the remarkable alpha-solenoid fold was observed at atomic resolution was mammalian β-catenin/Drosophila Armadillo (Huber et al., 1997). As judged using the structure prediction program Phyre2 (Kelley et al., 2015), overall, and as in β-catenin, the cylindrical alpha-solenoid structure in Avo3 contains a long groove lined with many positively charged residues. Whether this feature permits Avo3 to associate with the negatively charged head groups of PM phospholipids directly, or whether Avo3 associates with the acidic surface of an as yet unidentified integral PM membrane protein that acts as a dock for Avo3, is not yet known, but provides a fruitful avenue for further study.
MATERIALS AND METHODS
Yeast strains and growth conditions
All S. cerevisiae strains used in this study are listed in Table 1. All plasmids used in this study are listed in Table 2. Unless noted otherwise, yeast cells were grown at 30°C in synthetic complete (SC) medium (Sherman, 2002) lacking tryptophan and containing 2% glucose/dextrose as the carbon source (SCD-T). When necessary, SC medium also lacked the nutrients necessary to maintain selection for plasmids and/or was supplemented with appropriate nutrients to satisfy the growth requirements of auxotrophic mutations. Auxin-induced degradation of AID*-tagged proteins was initiated in TIR1-expressing cells (Nishimura et al., 2009, Morawska and Ulrich, 2013) growing in SCD-T buffered with 50 mM K2HPO4/KH2PO4 (pH 6) by addition of 1-NAA (1 mM final concentration), a cell-permeable synthetic auxin (Robert et al., 2010), dissolved in DMSO. Standard PCR-based methods (Janke et al., 2004) and conventional yeast transformation and recombinational insertion (Sherman, 2002) were used to generate AID*- and epitope-tagged fusion proteins, chromosomal integrations, and gene deletions. The template used for the AID* sequence was a plasmid described by Morawska and Ulrich (2013). TIR1-expressing strains were created by integration of TIR1 from plasmid pTIR4 (encoding OsTIR1 under control of the TDH3 promoter) into the LEU2 locus, as described by Eng et al. (2014). The C-termini of the chromosomal AVO1, AVO2, AVO3, BIT61, and SLM1 ORFs were tagged by in-frame integration of the coding sequences for eGFP (Janke et al., 2004) or mKate2 (Lee et al., 2013); the C-terminus of the chromosomal PIL1 ORF was tagged in the same way with mTagBFP (Malcova et al., 2016). The endogenous TOR2 locus was tagged internally by in-frame insertion between N321 and T322 (Sturgill et al., 2008) with either a cassette encoding mNG (Shaner et al., 2013) followed by a 3HA epitope or with an AID*-mNG-3HA cassette, as described in detail elsewhere (Leskoske et al., 2018). To create an eGFP-tagged version of Avo1 that retained function, the coding sequence for eGFP was fused in-frame to a modified AVO1 ORF that had been extended by appending in-frame the sequence encoding a 111-residue segment (residues P321 to L431) of the cytosolic C-terminal tail of the Ste2 in which 7 Lys was mutated to Arg (Ballon et al., 2006) and an Sla1 recognition site (GPFSD) was mutated to GPAAD. Proper construction of all tagged TORC2 subunits was verified by direct DNA sequence analysis, expression of the corresponding protein confirmed by immunoblotting, and retention of biological functional validated by the ability of each integrated construct (expressed from its endogenous promoter at its normal chromosomal locus) to support viability and Tor2-dependent phosphorylation of Ypk1.
TABLE 1:
Yeast strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| BY4742 | MATα his3∆1 leu2∆0 lys2∆0 ura3∆0 | Research Genetics |
| yKL2 | BY4742 avo1∆::kanMX | This study |
| yIZ082 | MATa his3∆200 leu2-3,112 ura3-52 TIR1::LEU2 | Ibarlucea-Benitez et al., 2018 |
| yNM706 | yIZ082 TIR1::LEU2 MSS4-AID*-6HA::hphNT1 | This study |
| yNM986 | MATa his3∆200 leu2-3,112 ura3-52 TIR1::LEU2 MSS4-AID*-6HA::hphNT1 TOR2-N321-mNG-3HA AVO3-mKate2::SpHis5 | This study |
| yNM1065 | MATa his3∆200 leu2-3,112 ura3-52 TIR1::LEU2 MSS4-AID*-6HA::hphNT1 AVO3-GFP::kanMX4 AVO2-mKate2::SpHis5 | This study |
| yNM1066 | MATa his3∆200 leu2-3,112 ura3-52 TIR1::LEU2 MSS4-AID*-6HA::hphNT1 AVO2-GFP::kanMX4 AVO3-mKate2::SpHis5 | This study |
| yNM1073 | MATa his3∆200 leu2-3,112 ura3-52 lys2-801 TIR1::LEU2 MSS4-AID*-6HA::hphNT1 AVO1-GFP | This study |
| yNM1090 | MATa his3∆200 leu2-3,112 ura3-52 lys2-801 TIR1::LEU2 MSS4-AID*-6HA::hphNT TOR2-N321-mNG-3HA PIL1-BFP::kanMX4 SLM1-mKate2::SpHis5 | This study |
| yNM788 | MATα his3∆200 leu2-3,112 ura3-52 MSS4-AID*-6HA::hphNT1 | This study |
| yNM1119 | MATa his3∆200 leu2-3,112 ura3-52 TIR1::LEU2 MSS4-AID*-6HA::hphNT1 Avo1-6HA::hphNT1 | This study |
| yNM1122 | MATa his3∆200 leu2-3,112 ura3-52 TIR1::LEU2 MSS4-AID*-6HA::hphNT1 Avo3-3C-3FLAG:: kanMX4 | This study |
| yNM718 | MATa his3∆200 leu2-3,112 ura3-52 TIR1::LEU2 AVO1-AID*-6HA::hphNT1 | This study |
| yNM927 |
MATa
his3∆200 leu2-3,112 ura3-52 lys2-801 TIR1::LEU2
AVO1-6HA::hphNT1 |
This study |
| yNM928 | MATα his3∆200 leu2-3,112 ura3-52 lys2-801 AVO1-6HA::hphNT1 | This study |
| yNM784 | MATα his3∆200 leu2-3,112 ura3-52 lys2-801 AVO1-AID*-6HA::hphNT1 | This study |
| yNM847 | MATa his3∆200 leu2-3,112 ura3-52 TIR1::LEU2 AVO1-AID*-6HA::hphNT1 AVO2-GFP::kanMX4 | This study |
| yNM886 | MATa his3∆200 leu2-3,112 ura3-52 TIR1::LEU2 AVO1-AID*-6HA::hphNT1 BIT61-GFP::kanMX4 AVO2-mKate2::SpHis5 | This study |
| yNM888 | MATa his3∆200 leu2-3,112 ura3-52 TIR1::LEU2 AVO1-AID*-6HA::hphNT1 AVO3-GFP::kanMX4 AVO2-mKate2::SpHis5 | This study |
| yNM977 | MATa his3∆200 leu2-3,112 ura3-52 TIR1::LEU2 AVO1-AID*-6HA::hphNT1 TOR2-N321-mNG-3HA AVO2-mKate2::SpHis5 | This study |
| yNM992 | MATa his3∆200 leu2-3,112 ura3-52 TIR1::LEU2 Avo1-AID*-6HA::hphNT1 Tor2-N321-mNeonGreen-3HA | This study |
| yNM994 | MATa his3∆200 leu2-3,112 ura3-52 TIR1::LEU2 Avo1-AID*-6HA:: hphNT1 Tor2-N321-mNeonGreen-3HA Avo3-3C-3FLAG:: kanMX4 | This study |
| yNM1026 |
MATa
his3∆200 leu2-3,112 ura3-52 TIR1::LEU2 AVO1-AID*-6HA::hphNT1 SLM1-AID*-9MYC::kanMX4 slm2∆::CaURA3 AVO3-GFP::kanMX4 |
This study |
| yNM862 |
MATa
his3∆200 leu2-3,112 ura3-52 TIR1::LEU2 AVO1-AID*-6HA::hphNT1 SLM1-AID*-9MYC::kanMX4 slm2∆::CaURA3 AVO2-mKate2::SpHis5 |
This study |
| yNM786 | MATα TIR1::HIS3 AVO1-AID*-6HA::hphNT1 | This study |
| yNM793 | MATα TIR1::HIS3 | This study |
| yNM884 | MATα his3∆200 leu2-3,112 ura3-52 TIR1::LEU2 AVO3-AID*-6HA::hphNT1 BIT61-GFP::kanMX4 AVO2-mKate2::SpHis5 | This study |
| yNM975 | MATa his3∆200 leu2-3,112 ura3-52 TIR1::LEU2 AVO3-AID*-6HA::hphNT1 TOR2-N321-mNG-3HA AVO2-mKate2::SpHis5 | This study |
| yNM998 | MATα his3∆200 leu2-3,112 ura3-52, TIR1::LEU2 AVO3-AID*-6HA::hphNT1 AVO1-GFP | This study |
| yNM1031 | MATα his3∆200 leu2-3,112 ura3-52 TIR1::LEU2 AVO3-AID*-6HA::hphNT1 AVO2-GFP::kanMX4 | This study |
| yNM858 | MATα his3∆200 leu2-3,112 ura3-52 TIR1::LEU2 AVO3-AID*-6HA::hphNT1 | This study |
| yAEA346 | MATa TIR1::LEU2 his3∆200 leu2-3,112 ura3-52 TOR2-N321-AID*-mNG-3HA | This study |
| yNM1035 | MATa his3∆200 leu2-3,112 ura3-52 TIR1::LEU2 TOR2-N321-AID*-mNG-3HA AVO3-mKate2::SpHis5 | This study |
| yNM1034 | MATa his3∆200 leu2-3,112 ura3-52 TIR1::LEU2 TOR2-N321-AID*-mNG-3HA AVO2-mKate2::SpHis5 | This study |
| yNM1056 | MATa his3∆200 leu2-3,112 ura3-52 TIR1::LEU2 AVO1-AID*-6HA::hphNT1 AVO3-AID*-9myc::kanMX4 TOR2-N321-AID*-mNG-3HA | This study |
| yNM1057 | MATa his3∆200 leu2-3,112 ura3-52 TIR1::LEU2 Avo3-AID*-9myc::kanMX4 | This study |
| yNM1062 | MATα his3∆200 leu2-3,112 ura3-52 TIR1::LEU2 AVO3-AID*-9myc::kanMX4 TOR2-N321-AID*-mNG-3HA | This study |
| yNM1064 | MATα his3∆200 leu2-3,112 ura3-52 TIR1::LEU2 AVO1-AID*-6HA::hphNT1 AVO3-AID*-9myc::kanMX4 | This study |
| yNM1040 | MATa his3∆200 leu2-3,112 ura3-52 TIR1::LEU2 AVO1-AID*-6HA::hphNT1 TOR2-N321-AID*-mNG-3HA | This study |
| yNM721 | MATa his3∆200 leu2-3,112 ura3-52 TIR1::LEU2 AVO1-AID*-6HA::hphNT1 SLM1-AID*-GFP:: hphNT1 slm2∆::kanMX | This study |
| yNM739 | MATα his3∆200 leu2-3,112 ura3-52 lys2-801 TIR1::LEU2 LST8-AID*-6HA::hphNT1 AVO2-mRuby2::SpHis5 | This study |
| yNM983 | MATa his3∆200 leu2-3,112 ura3-52 TIR1::LEU2 Avo3-AID*-6HA::hphNT1 Tor2-N321-mNeonGreen-3HA | This study |
TABLE 2:
Plasmids used in this study.
| Plasmid | Description | Source |
|---|---|---|
| pRS313 | ARS-CEN, HIS3 vector | Sikorski and Hieter, 1989 |
| pRS315 | ARS-CEN, LEU2 vector | Sikorski and Hieter, 1989 |
| pRS316 | ARS-CEN, URA3 vector | Sikorski and Hieter, 1989 |
| pRS416 | ARS-CEN, URA3 vector | Sikorski and Hieter, 1989 |
| pGFP-PH-7 | pRS316 CUP1prom::GFP-PH PLCδ1 | Stauffer et al., 1998; Stolz, 2001 |
| pAEA399 | pRS416 GALSprom::AVO1-mKate2-FLAG | This study |
| pAEA400 | pRS416 GALSprom::AVO3-mKate2-FLAG | This study |
| pAEA403 | pRS313 GALSprom::YPK1D242A | This study |
| pAEA419 | pRS316 GALSprom::YPK1S51A S71A T504A S644A T662A-myc [Ypk15A] | This study |
| pNM160 | pRS416 GALSprom::AVO1Δ1059-1176-mKate2-FLAG [AVO1(ΔPH)] | This study |
| pAM66 | pRS316 YPK1prom::YPK1 | Roelants et al., 2004 |
| pFR273 | pRS316 YPK1prom::YPK1D242A | Roelants et al., 2011 |
| pLZ1 | pRS316 AVO1prom::AVO1 | This study |
| pLZ2 | pRS316 AVO1prom::9myc | This study |
| pLZ3 | pRS315 AVO1prom::AVO1-9myc | This study |
| pLZ4 | pRS315 AVO1prom::AVO1(Δ2-200)-9myc | This study |
| pLZ5 | pRS315 AVO1prom::AVO1(Δ2-400)-9myc | This study |
| pLZ6 | pRS315 AVO1prom::AVO1(Δ2-600)-9myc | This study |
| pLZ7 | pRS315 AVO1prom::AVO1(Δ200-400)-9myc | This study |
| pLZ8 | pRS315 AVO1prom::AVO1(Δ400-600)-9myc | This study |
| pLZ9 | pRS315 AVO1prom::AVO1(Δ600-830)-9myc [AVO1(ΔCRIM)] | This study |
| pLZ10 | pRS315 AVO1prom::AVO1(Δ831-920)-9myc [AVO1(ΔRBD)] | This study |
| pLZ11 | pRS315 AVO1prom::AVO1(Δ1059-1176)-9myc [AVO1(ΔPH)] | This study |
| pNM162 | pRS416 GALSprom::AVO3(Δ2-1100)-mKate2-FLAG | This study |
| pNM163 | pRS416 GALSprom::AVO3(Δ2-910)-mKate2-FLAG | This study |
| pNM164 | pRS416 GALSprom::AVO3(Δ2-700)-mKate2-FLAG | This study |
| pNM165 | pRS416 GALSprom::AVO3(Δ2-500)-mKate2-FLAG | This study |
| pNM166 | pRS416 GALSprom::AVO3(Δ2-300)-mKate2-FLAG | This study |
| pNM167 | pRS416 GALSprom::AVO3(Δ2-810)-mKate2-FLAG | This study |
| pNM168 | pRS416 GALSprom::AVO3(300-500)-mKate2-FLAG | This study |
| pNM169 | pRS416 GALSprom::AVO3(300-500)-mNeonGreen-FLAG | This study |
| pNM170 | pRS416 GALSprom::AVO3(∆300-500)-mKate2-FLAG | This study |
| pNM173 | pRS416 GALSprom::AVO3(300-700)-mNeonGreen-FLAG | This study |
| pNM174 | pRS416 GALSprom::AVO3-mNeonGreen-FLAG | This study |
To express genes under control of the galactose-inducible promoter GALS, derived by truncation of the GAL1 promoter (Mumberg et al., 1994), cells were pregrown in SC-T medium containing 2% raffinose–0.2% sucrose (SCRS-T), and then galactose was added (final concentration 2%) and the cells were incubated for an additional 2–3 h, depending on the experiment. For spot assays to assess cell growth and viability, cell cultures were pregrown overnight to saturation in SCD-T buffered with 50 mM K2HPO4/KH2PO4 (pH 6) and diluted to A600 nm = 0.125 in a 96-well microtiter plate, and then those wells were subjected to serial fivefold dilutions. Samples of each dilution then were spotted, using a Steers-type multipronged inoculator, onto agar plates containing phosphate-buffered SCD-T medium (supplemented with appropriate nutrients to select for plasmids, when necessary) and containing either 1 mM 1-NAA in DMSO or an equivalent volume of DMSO alone (control plates). Plates were incubated at 30°C and photographed after 2 d. In the case of cells carrying plasmids expressing GALS-driven genes, cultures were pregrown overnight in SCRS-T and plated on agar plates of phosphate-buffered SCRS-T also containing 2% galactose.
Preparation of cell extracts and immunoblotting
Samples (2 ml) of exponentially growing cultures (generally A600 nm = 0.8) were harvested by brief centrifugation in a microfuge (Eppendorf Model 5415D) at maximum rpm and the resulting cell pellets stored at –80°C. For analysis, the pellets were thawed on ice and lysed in 150 μl 1.85 M NaOH containing 7.4% β-mercaptoethanol, and proteins in the resulting lysate were precipitated by addition of 150 μl 50% trichloroacetic acid on ice for 10 min. Precipitated proteins were collected by centrifugation for 2 min in the microfuge at maximum rpm and washed twice with ice-cold acetone to remove excess trichloroacetic acid. The precipitated protein was solubilized in a volume (typically, 83 μl) of 5% SDS–0.1 M Tris base to yield a final concentration representing an amount of the initial cell culture of 0.02 A600 nm units per μl, and then 6× SDS–PAGE sample buffer (typically 17 μl) was added.
The resulting solubilized protein was heated at 65°C for 15 min and samples (typically 10 μl) were resolved by SDS–PAGE at 160 V on 8% gels with a 29:1::acrylamide:bis-acrylamide ratio. For phosphate-affinity SDS–PAGE, samples of the same solubilized protein also were resolved in 8% gels containing 35 μM Phos-tag reagent (Kinoshita et al., 2015; Wako Chemicals USA, Richmond, VA). Resolved proteins were transferred electrophoretically to a nitrocellulose membrane and the resulting protein blots incubated for 1 h in Odyssey blocking buffer (LI-COR, Lincoln, NE) that was diluted 1:1 with PBS containing 0.1% Tween-20 (PBS+Tw). After blocking, the membranes were incubated overnight in the same blocking buffer with an appropriate primary antibody (at the indicated dilution): mouse anti-c-myc mAb 9E10 (Evan et al., 1985) (1:100; Monoclonal Antibody Facility, Cancer Research Laboratory, University of California, Berkeley); rabbit anti-DYKDDDDK (FLAG) tag antibody (1:1000; Cell Signalling, binds to same epitope as Sigma’s Anti-FLAG M2 antibody); mouse anti-HA.11 (1:1000; BioLegend); or rabbit polyclonal anti-Pgk1 antiserum (1:20,000; Baum et al., 1978). After washing with PBS+Tx (three times with ≥10 ml), filter-bound immune-complexes were detected by incubation with an appropriate infrared dye–labeled secondary antibody—CF770-conjugated goat anti-mouse IgG (Biotium), IRDye800CW-conjugated goat anti-rabbit IgG (Li-Cor), or IRDye680RD-conjugated goat anti-mouse IgG (Li-Cor)—that was diluted 1:10,000 in 1:1::Odyssey blocking buffer:PBS+Tw. After final washing, the resulting immunoblots were analyzed using an infrared imaging system (Odyssey, LI-COR).
Analysis of TORC2 complexes by coimmunoprecipitation
To analyze the state of assembly of TORC2 complexes before and after 1-NAA-induced degradation of a component of interest, proteins coimmunoprecipitating with Avo3-3C-3xFLAG were examined in two different strains. In one case, the cells coexpressed from their endogenous loci Mss4-AID*-6HA, Avo3-3C-3xFLAG, Tor2-mNG-3HA, and Avo1-6HA (the HA-tagged proteins were readily resolved from each other by their distinct differences in molecular mass: Tor2, ∼290,000; Avo1, ∼132,000; Mss4, ∼90,000). In the other case, the cells coexpressed from their endogenous loci Avo1-AID*-6HA, Avo3-3C-3xFLAG, and Tor2-mNG-3HA. In each case, cultures (50 ml) were grown in phosphate-buffered SCD-T medium to midexponential phase, incubated with solvent (DMSO) alone or 1 mM 1-NAA in the same solvent for 60–120 min, harvested by centrifugation, and frozen in liquid N2. Frozen cell pellets (equivalent to an initial cell culture density of 50 A600 nm units) were resuspended in 300 μl ice-cold 2× TNEGT buffer (300 mM NaCl, 20% glycerol, 0.24% Tergitol [Type NP-40, Sigma NP40S], 2 mM EDTA, 1 mM phenylmethanesulfonyl fluoride [PMSF], 100 mM Tris, pH 7.6, adjusted to contain 1× Sigma fungal and yeast protease inhibitor cocktail [Sigma-Aldrich, P8215] and 2× Roche cOmplete protease inhibitor tablet [Roche, Basel, Switzerland]). An equivalent volume of dry chilled glass beads was added and cells were lysed with six 30-s pulses of rapid agitation using a high-speed Fast Prep FP120 cell-disrupting homogenizer (BIO101/Savant) with 1-min cooling periods on ice in between pulses. After lysis, 300 μl of 1× TNEGT buffer (150 mM NaCl, 10% glycerol, 0.12% Tergitol, 1 mM EDTA, 1 mM phenylmethanesulfonyl fluoride [PMSF], 50 mM Tris pH 7.6 containing 1× Sigma fungal and yeast protease inhibitor cocktail) was added. The resulting diluted lysate was separated from the glass beads through the conical tip of a plastic microfuge tube punctured with a 25-gauge hypodermic needle by centrifugation in a microfuge (Model 5424; Eppendorf) at 500 × g for 1 min into a fresh tube. To ensure complete membrane solubilization, the recovered liquid was incubated for 30 min at 4°C on a roller drum. The resulting solubilized extract was clarified by centrifugation at 10,000 × g in a microfuge and the resulting supernatant fraction was withdrawn and mixed with 20 μl of a slurry of agarose beads covalently coated with anti-DYKDDDDK (FLAG epitope) antibody (Biotool, B23101) previously equilibrated in 1× TNEGT. After incubation for 2 h at 4°C on a roller drum, the beads were rinsed six times with 1 ml of 1× TNEGT buffer and the remaining bound proteins were eluted with 30 μl of 1× SDS–PAGE sample buffer, resolved by SDS–PAGE on 8% gels, analyzed by immunoblotting with the appropriate antibodies, and visualized using our infrared imaging system (Odyssey CLx; Li-Cor).
Fluorescence microscopy and image analysis
For live-cell imaging, cells were grown to midexponential phase (A600 nm ∼ 0.8) at 30°C in SCD-T, unless otherwise noted, and samples of the cultures deposited onto the surface of concanavalin A-coated (0.1 μg/ml) coverslips. In most instances, fluorescence microscopy was performed at room temperature using a Carl Zeiss Elyra PS.1 structured illumination fluorescence microscope (Carl Zeiss, Jena, Germany) operating in HiLo mode (Tokunaga et al., 2008), equipped with a Zeiss 100× PlanApo 1.46NA TIRF objective, a main focus drive of the AxioObserver Z1 Stand, a WSB PiezoDrive 08 for superresolution, and an Andor 512 × 512 EM-CCD camera (100 × 100 nm pixel size; Andor Technology, South Windsor, CT). In HiLo mode, the cells are examined using the laser source set at near the TIRF angle to illuminate only a section of the sample (so the excitation is as bright as possible), but with an illumination plane not limited to the surface close to the coverslip, which, together, give a wide-field image in which out-of-focus fluorescence is decreased (thereby reducing the background), resulting in better signal-to-noise contrast. To visualize TORC2 subunits tagged with either mNG (excitation λmax 506 nm; emission λmax 517) or eGFP (excitation λmax 488 nm; emission λmax 507), cell samples were excited with an argon laser at 488 nm at 2.3% power (100 mW) and emission was captured in a 495- to 550-nm window using a bandpass filter. For TORC2 subunits tagged with mKate2 (excitation λmax 588 nm; emission λmax 633), excitation was at 561 nm at 2.3% power (100 mW) and emission was monitored in a 570- to 620-nm window using a different bandpass filter; and for proteins tagged with mTagBFP (excitation λmax 399 nm; emission λmax 456), excitation was at 405 nm at 2.3% power (100 mW) and emission was monitored in a 420- to 480-nm window using another bandpass filter. Images (average of eight scans; 400 ms/scan for mNG, eGFP, or BFP, and 600 ms/scan for mKate2) were processed using ZE software (Zeiss) and analyzed using Fiji (National Institutes of Health, Bethesda, MD; Schindelin et al., 2012). To avoid changes in image quality due to occasional fluctuations in laser intensity, all panels shown in any given figure represent experiments performed on the same day, and scaled and adjusted identically for brightness using Fiji (Schindelin et al., 2012).
Alternatively, where indicated, cells were viewed under a conventional epifluorescence microscope (Olympus, Model BH-2; Olympus America, Center Valley, PA) equipped with a 100× objective, Solalight source (Lumencor, Beaverton, OR), and appropriate bandpass filters (Chroma Technology, Rockingham, VT). Images were collected using an EM-CCD camera (Photometrics, Tucson, AZ) and processed with μManager (Edelstein et al., 2010) and Photoshop (Adobe Systems, San Jose, CA).
Cells of a strain (yNM986) coexpressing Tor2-mNeonGreen-3HA, Avo3-mKate2, and Mss4-AID*-6xHA, before and after 1-NAA-induced degradation of Mss4, were examined under an inverted Carl Zeiss LSM 880 confocal laser-scanning microscope using an Airyscan detector equipped with a Gallium Arsenide Phosphide (GaAsP) photomultiplier tube, a Zeiss Plan-Aprochromat 63×/1.4 oil NA objective, and an Axio Observer 7 stand. For visualization, a small volume (0.5 μl) of the cell samples, which were grown in phosphate-buffered synthetic medium, was spotted directly on the surface of a fresh Swiss Glass slide (Fisherfinest Premium), covered with a 22 × 22 mm coverslip (VMR), and viewed immediately. Images were processed using ZEN software (Zeiss) and analyzed using Fiji (NIH, Bethesda, MD; Schindelin et al., 2012).
Labeling of F-actin with rhodamine-phalloidin
Decoration of filamentous actin with rhodamine–phalloidin was performed by minor modifications of a previously described protocol (Pringle et al., 1989). Briefly, the cells to be examined were fixed in 3.7% formaldehyde for 30 min at room temperature. The resulting fixed cells were then washed twice with PBS containing 0.1% Triton X-100 and stained with gentle agitation in the dark with a final concentration of 1.5 µM rhodamine–phalloidin (Molecular Probes/Thermo Fisher Scientific, Eugene, OR) from a stock dissolved in methanol. After 30 min, the cells were washed three times in PBS, resuspended in PBS containing 60% glycerol, and visualized using a conventional epifluorescence microscope, as above, using an FITC bandpass filter (Chroma).
Reproducibility
All results reported reflect findings made repeatedly in at least three independent trials of each experiment shown.
Supplementary Material
Acknowledgments
This work was supported by Erwin Schroedinger Fellowship J3787-B21 from the Austrian Science Fund (to A. E.-A.), by NIH Predoctoral Traineeship GM07232 and by a UC Berkeley MacArthur & Lakhan-Pal Graduate Fellowship (to K.L.L.), and by NIH R01 Research Grant GM21841 (to J.T.). This work was also aided, in part, by National Institutes of Health (USA) S10 Equipment Grant OD018136 (to Steven E. Ruzin, Director, UC Berkeley Biological Imaging Facility) for a Zeiss Elyra PS.1 structured illumination microscope. We thank Holly Aaron, Steve Ruzin, and Denise Schichnes (UC Berkeley) and Samantha Fore (Carl Zeiss Microscopy, Pleasanton, CA) for their invaluable help and advice about fluorescence microscopy. We are grateful to Françoise Roelants for advice and critical reading of the manuscript. We thank all the other members of the Thorner lab for helpful discussions and support. We thank the Drubin–Barnes and Koshland laboratories (UC Berkeley) for provision of essential research materials and Leslie Stolz and John D. York (then at Duke University) for the gift of plasmid pGFP-PH-7.
Abbreviations used:
- AID*
minimal sequence motif required for auxin-dependent recognition by the plant F-box protein TIR1
- ARM
a structural motif (3-helix bundle) first described in Armadillo (Drosophila β-catenin)
- BFP
blue fluorescent protein ( Aequorea victoria green fluorescent protein variant carrying Y66H Y145F substitutions)
- CaaX box
consensus motif for protein prenylation
- eGFP
A. victoria green fluorescent protein variant carrying F64L S65T substitutions
- EM-CCD
electron-multiplying charge-coupled device
- FITC
fluorescein isothiocyanate
- 5-FOA
5-fluoro-orotic acid
- GFP
A. victoria green fluorescent protein
- HEAT repeat
a structural motif comprising two alpha helices connected by a short loop that is tandemly repeated in many copies to form an alpha-solenoid, named after the four proteins (Huntingtin, translation elongation factor 3, phosphoprotein phosphatase 2A, and yeast Tor1) in which this extended superhelical structure was first identified
- HiLo
highly inclined and laminated optical sheet fluorescence microscopy
- mAb
monoclonal antibody
- MAPK
mitogen-activated protein kinase
- mKate2
Entacmaea quadricolor red fluorescent protein variant
- mNG
Branchiostoma lanceolatum monomeric fluorescent protein Neon Green
- mTORC2
mammalian TORC2
- 1-NAA
1-naphthaleneacetic acid (a cell-permeable synthetic auxin)
- ORF
open reading frame
- PBS+Tw
phosphate-buffered saline containing 0.1% Tween-20 detergent
- PH
pleckstrin homology
- PLCδ1
human phospholipase Cδ1
- PM
plasma membrane
- PtdIns4P
phosphatidylinositol-4-phosphate
- PtdIns4,5P2
phosphatidylinositol-4,5- bisphosphate
- SCF
Skp1-, Cdc53/Cul1- and F-box protein containing ubiquitin ligase (E3)
- TIRF
total internal reflection fluorescence
- TORC2
target of rapamycin complex 2
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-10-0682) on April 10, 2019.
REFERENCES
- Aronova S, Wedaman K, Aronov PA, Fontes K, Ramos K, Hammock BD, Powers T. (2008). Regulation of ceramide biosynthesis by TOR complex 2. Cell Metab , 148–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audhya A, Emr SD. (2003). Regulation of PI4,5P2 synthesis by nuclear-cytoplasmic shuttling of the Mss4 lipid kinase. EMBO J , 4223–4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audhya A, Loewith R, Parsons AB, Gao L, Tabuchi M, Zhou H, Boone C, Hall MN, Emr SD. (2004). Genome-wide lethality screen identifies new PI4,5P2 effectors that regulate the actin cytoskeleton. EMBO J , 3747–3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballon DR, Flanary PL, Gladue DP, Konopka JB, Dohlman HG, Thorner J. (2006). DEP-domain-mediated regulation of GPCR signaling responses. Cell , 1079–1093. [DOI] [PubMed] [Google Scholar]
- Baretić D, Berndt A, Ohashi Y, Johnson CM, Williams RL. (2016). Tor forms a dimer through an N-terminal helical solenoid with a complex topology. Nat Commun , 11016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett K, Gadila SK, Tenay B, McDermott H, Alcox B, Kim K. (2015). TORC2 and eisosomes are spatially interdependent, requiring optimal level of phosphatidylinositol 4,5-bisphosphate for their integrity. J Biosci , 299–311. [DOI] [PubMed] [Google Scholar]
- Baum P, Thorner J, Honig L. (1978). Identification of tubulin from the yeast Saccharomyces cerevisiae . Proc Natl Acad Sci USA , 4962–4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchtold D, Piccolis M, Chiaruttini N, Riezman I, Riezman H, Roux A, Walther TC, Loewith R. (2012). Plasma membrane stress induces relocalization of Slm proteins and activation of TORC2 to promote sphingolipid synthesis. Nat Cell Biol , 542–547. [DOI] [PubMed] [Google Scholar]
- Berchtold D, Walther TC. (2009). TORC2 plasma membrane localization is essential for cell viability and restricted to a distinct domain. Mol Biol Cell , 1565–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD, LaCroute F, Fink GR. (1984). A positive selection for mutants lacking orotidine-5’-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet , 345–346. [DOI] [PubMed] [Google Scholar]
- Boronenkov IV, Anderson RA. (1995). The sequence of phosphatidylinositol-4-phosphate 5-kinase defines a novel family of lipid kinases. J Biol Chem , 2881–2884. [DOI] [PubMed] [Google Scholar]
- Cormack BP, Valdivia RH, Falkow S. (1996). FACS-optimized mutants of the green fluorescent protein (GFP). Gene , 33–38. [DOI] [PubMed] [Google Scholar]
- Daquinag A, Fadri M, Jung SY, Qin J, Kunz J. (2007). The yeast PH domain proteins Slm1 and Slm2 are targets of sphingolipid signaling during the response to heat stress. Mol Cell Biol , 633–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrivières S, Cooke FT, Parker PJ, Hall MN. (1998). MSS4, a phosphatidylinositol-4-phosphate 5-kinase required for organization of the actin cytoskeleton in Saccharomyces cerevisiae . J Biol Chem , 15787–15793. [DOI] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P. (2006). Phosphoinositides in cell regulation and membrane dynamics. Nature , 651–657. [DOI] [PubMed] [Google Scholar]
- Douglas LM, Konopka JB. (2014). Fungal membrane organization: the eisosome concept. Annu Rev Microbiol , 377–393. [DOI] [PubMed] [Google Scholar]
- Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. (2010). Computer control of microscopes using µManager. Curr Protoc Mol Biol Chapter 14: Unit 14.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltschinger S, Loewith R. (2016). TOR complexes and the maintenance of cellular homeostasis. Trends Cell Biol , 148–159. [DOI] [PubMed] [Google Scholar]
- Eng T, Guacci V, Koshland D. (2014). ROCC, a conserved region in cohesin’s Mcd1 subunit, is essential for the proper regulation of the maintenance of cohesion and establishment of condensation. Mol Biol Cell , 2351–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan GI, Lewis GK, Ramsay G, Bishop JM. (1985). Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol , 3610–3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadri M, Daquinag A, Wang S, Xue T, Kunz J. (2005). The pleckstrin homology domain proteins Slm1 and Slm2 are required for actin cytoskeleton organization in yeast and bind phosphatidylinositol-4,5-bisphosphate and TORC2. Mol Biol Cell , 1883–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego O, Betts MJ, Gvozdenovic-Jeremic J, Maeda K, Matetzki C, Aguilar-Gurrieri C, Beltran-Alvarez P, Bonn S, Fernández-Tornero C, Jensen LJ, et al (2010). A systematic screen for protein-lipid interactions in Saccharomyces cerevisiae . Mol Syst Biol , 430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrenton LS, Stefan CJ, McMurray MA, Emr SD, Thorner J. (2010). Pheromone-induced anisotropy in yeast plasma membrane phosphatidylinositol-4,5-bisphosphate distribution is required for MAPK signaling. Proc Natl Acad Sci USA , 1805–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaubitz C, Oliveira TM, Prouteau M, Leitner A, Karuppasamy M, Konstantinidou G, Rispal D, Eltschinger S, Robinson GC, Thore S, et al (2015). Molecular basis of the rapamycin insensitivity of Target of Rapamycin complex 2. Mol Cell , 977–988. [DOI] [PubMed] [Google Scholar]
- Gaubitz C, Prouteau M, Kusmider B, Loewith R. (2016). TORC2 structure and function. Trends Biochem Sci , 532–545. [DOI] [PubMed] [Google Scholar]
- González A, Hall MN. (2017). Nutrient sensing and TOR signaling in yeast and mammals. EMBO J , 397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, del Pozo JC, Walker L, Hobbie L, Risseeuw E, Banks T, Crosby WL, Yang M, Ma H, Estelle M. (1999). Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana . Genes Dev , 1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves MR, Barford D. (1999). Topological characteristics of helical repeat proteins. Curr Opin Struct Biol , 383–389. [DOI] [PubMed] [Google Scholar]
- Heitman J, Movva NR, Hall MN. (1991). Targets for cell cycle arrest by the immuno-suppressant rapamycin in yeast. Science , 905–909. [DOI] [PubMed] [Google Scholar]
- Helliwell SB, Howald I, Barbet N, Hall MN. (1998a). TOR2 is part of two related signaling pathways coordinating cell growth in Saccharomyces cerevisiae . Genetics , 99–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell SB, Schmidt A, Ohya Y, Hall MN. (1998b). The Rho1 effector Pkc1, but not Bni1, mediates signalling from Tor2 to the actin cytoskeleton. Curr Biol , 1211–1214. [DOI] [PubMed] [Google Scholar]
- Helliwell SB, Wagner P, Kunz J, Deuter-Reinhard M, Henriquez R, Hall MN. (1994). TOR1 and TOR2 are structurally and functionally similar but not identical phosphatidylinositol kinase homologues in yeast. Mol Biol Cell , 105–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi-Sanabria R, Garcia EJ, Tomoiaga D, Munteanu EL, Feinstein P, Pon LA. (2016). Characterization of fluorescent proteins for three- and four-color live-cell imaging in S. cerevisiae . PLoS One , e0146120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill A, Niles B, Cuyegkeng A, Powers T. (2018). Redesigning TOR kinase to explore the structural basis for TORC1 and TORC2 assembly. Biomolecules , E36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho B, Baryshnikova A, Brown GW. (2018). Unification of protein abundance datasets yields a quantitative Saccharomyces cerevisiae proteome. Cell Syst , 192–205. [DOI] [PubMed] [Google Scholar]
- Ho H-L, Lee H-Y, Liao H-C, Chen M-Y. (2008). Involvement of Saccharomyces cerevisiae Avo3p/Tsc11p in maintaining TOR complex 2 integrity and coupling to downstream signaling. Eukaryot Cell , 1328–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho H-L, Shiau Y-S, Chen M-Y. (2005). Saccharomyces cerevisiae TSC11/AVO3 participates in regulating cell integrity and functionally interacts with components of the Tor2 complex. Curr Genet , 273–288. [DOI] [PubMed] [Google Scholar]
- Homma K, Terui S, Minemura M, Qadota H, Anraku Y, Kanaho Y, Ohya Y. (1998). Phosphatidylinositol-4-phosphate 5-kinase localized on the plasma membrane is essential for yeast cell morphogenesis. J Biol Chem , 15779–15786. [DOI] [PubMed] [Google Scholar]
- Huber AH, Nelson WJ, Weis WI. (1997). Three-dimensional structure of the Armadillo repeat region of β-catenin. Cell , 871–882. [DOI] [PubMed] [Google Scholar]
- Ibarlucea-Benitez I, Ferro LS, Drubin DG, Barnes G. (2018). Kinesins relocalize the chromosomal passenger complex to the midzone for spindle disassembly. J Cell Biol , 1687–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikai N, Nakazawa N, Hayashi T, Yanagida M. (2011). The reverse, but coordinated, roles of Tor2 (TORC1) and Tor1 (TORC2) kinases for growth, cell cycle and separase-mediated mitosis in Schizosaccharomyces pombe . Open Biol , 110007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. (2001). A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA , 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, Knop M. (2004). A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast , 947–962. [DOI] [PubMed] [Google Scholar]
- Janmey PA, Bucki R, Radhakrishnan R. (2018). Regulation of actin assembly by PI(4,5)P2 and other inositol phospholipids: An update on possible mechanisms. Biochem Biophys Res Commun , 307–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y, Fujioka Y, Suzuki NN, Inagaki F, Wullschleger S, Loewith R, Hall MN, Ohsumi Y. (2005). Tor2 directly phosphorylates the AGC kinase Ypk2 to regulate actin polarization. Mol Cell Biol , 7239–7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y, Jung US, Piotrowski J, Levin DE. (1995). The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genes Dev , 1559–1571. [DOI] [PubMed] [Google Scholar]
- Kamble C, Jain S, Murphy E, Kim K. (2011). Requirements of Slm proteins for proper eisosome organization, endocytic trafficking and recycling in the yeast Saccharomyces cerevisiae . J Biosci , 79–96. [DOI] [PubMed] [Google Scholar]
- Karotki L, Huiskonen JT, Stefan CJ, Ziółkowska NE, Roth R, Surma MA, Krogan NJ, Emr SD, Heuser J, Grünewald K, Walther TC. (2011). Eisosome proteins assemble into a membrane scaffold. J Cell Biol , 889–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karuppasamy M, Kusmider B, Oliveira TM, Gaubitz C, Prouteau M, Loewith R, Schaffitzel C. (2017). Cryo-EM structure of Saccharomyces cerevisiae Target of Rapamycin complex 2. Nat Commun , 1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg JE. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc , 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita E, Kinoshita-Kikuta E, Koike T. (2009). Separation and detection of large phosphoproteins using Phos-tag SDS-PAGE. Nat Protoc , 1513–1521. [DOI] [PubMed] [Google Scholar]
- Kinoshita E, Kinoshita-Kikuta E, Koike T. (2015). Advances in Phos-tag-based methodologies for separation and detection of the phosphoproteome. Biochim Biophys Acta , 601–608. [DOI] [PubMed] [Google Scholar]
- Kunz J, Henriquez R, Schneider U, Deuter-Reinhard M, Movva NR, Hall MN. (1993). Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell , 585–596. [DOI] [PubMed] [Google Scholar]
- Kunz J, Schneider U, Howald I, Schmidt A, Hall MN. (2000). HEAT repeats mediate plasma membrane localization of Tor2p in yeast. J Biol Chem , 37011–37020. [DOI] [PubMed] [Google Scholar]
- Lee S, Lim WA, Thorn KS. (2013). Improved blue, green, and red fluorescent protein tagging vectors for S. cerevisiae . PLoS One , e67902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA. (2008). Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol , 99–111. [DOI] [PubMed] [Google Scholar]
- Leskoske KL, Roelants FM, Emmerstorfer-Augustin A, Augustin C, Si EP, Hill JM, Thorner J. (2018). Phosphorylation by the stress-activated MAPK Slt2 down-regulates the yeast TOR complex 2. Genes Dev , 1576–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leskoske KL, Roelants FM, Marshall MNM, Hill JM, Thorner J. (2017). The stress-sensing TORC2 complex activates yeast AGC-family protein kinase Ypk1 at multiple novel sites. Genetics , 179–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H-C, Chen M-Y. (2012). Target of rapamycin complex 2 signals to downstream effector yeast protein kinase 2 (Ypk2) through adheres-voraciously-to-Target-of-Rapamycin-2 protein 1 (Avo1) in Saccharomyces cerevisiae . J Biol Chem , 6089–6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Gan W, Chin YR, Ogura K, Guo J, Zhang J, Wang B, Blenis J, Cantley LC, Toker A, et al (2015). PtdIns(3,4,5)P3-dependent activation of the mTORC2 kinase complex. Cancer Discov , 1194–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke MN, Thorner J. (2019). Rab5 GTPases are required for optimal TORC2 function. J Cell Biol , 961–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodish H, Berk A, Kaiser CA, Krieger M, Bretscher A, Ploegh H, Amon A, Martin KC. (2016). Molecular Cell Biology, 8th Ed, New York: W.H. Freeman/Macmillan Learning. [Google Scholar]
- Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, Hall MN. (2002). Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell , 457–468. [DOI] [PubMed] [Google Scholar]
- Malcova I, Farkasovsky M, Senohrabkova L, Vasicova P, Hasek J. (2016). New integrative modules for multicolor-protein labeling and live-cell imaging in Saccharomyces cerevisiae . FEMS Yeast Res , fow027. [DOI] [PubMed] [Google Scholar]
- Michel AH, Hatakeyama R, Kimmig P, Arter M, Peter M, Matos J, De Virgilio C, Kornmann B. (2017). Functional mapping of yeast genomes by saturated transposition. Elife , e23570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morawska M, Ulrich HD. (2013). An expanded tool kit for the auxin-inducible degron system in budding yeast. Yeast , 341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Müller R, Funk M. (1994). Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res , 5767–5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niles BJ, Mogri H, Hill A, Vlahakis A, Powers T. (2012). Plasma membrane recruitment and activation of the AGC kinase Ypk1 is mediated by Target of Rapamycin complex 2 (TORC2) and its effector proteins Slm1 and Slm2. Proc Natl Acad Sci USA , 1536–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. (2009). An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods , 917–922. [DOI] [PubMed] [Google Scholar]
- Noda T. (2017). Regulation of autophagy through TORC1 and mTORC1. Biomolecules , E52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivera-Couto A, Graña M, Harispe L, Aguilar PS. (2011). The eisosome core is composed of BAR domain proteins. Mol Biol Cell , 2360–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M, Berg S, Reynolds AB. (1994). A repeating amino acid motif shared by proteins with diverse cellular roles. Cell , 769–791. [DOI] [PubMed] [Google Scholar]
- Péli-Gulli MP, Sardu A, Panchaud N, Raucci S, De Virgilio C. (2015). Amino acids stimulate TORC1 through Lst4-Lst7, a GTPase-activating protein complex for the Rag family GTPase Gtr2. Cell Rep , 1–7. [DOI] [PubMed] [Google Scholar]
- Perry J, Kleckner N. (2003). The ATRs, ATMs, and TORs are giant HEAT repeat proteins. Cell , 151–155. [DOI] [PubMed] [Google Scholar]
- Pringle JR, Preston RA, Adams AE, Stearns T, Drubin DG, Haarer BK, Jones EW. (1989). Fluorescence microscopy methods for yeast. Methods Cell Biol , 357–435. [DOI] [PubMed] [Google Scholar]
- Reichen C, Hansen S, Plückthun A. (2014). Modular peptide binding: from a comparison of natural binders to designed Armadillo repeat proteins. J Struct Biol , 147–162. [DOI] [PubMed] [Google Scholar]
- Riggi M, Niewola-Staszkowska K, Chiaruttini N, Colom A, Kusmider B, Mercier V, Soleimanpour S, Stahl M, Matile S, Roux A, Loewith R. (2018). Decrease in plasma membrane tension triggers PtdIns(4,5)P2 phase separation to inactivate TORC2. Nat Cell Biol , 1043–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert S, Kleine-Vehn J, Barbez E, Sauer M, Paciorek T, Baster P, Vanneste S, Zhang J, Simon S, Čovanová M, et al (2010). ABP1 mediates auxin inhibition of clathrin-dependent endocytosis in Arabidopsis. Cell , 111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelants FM, Breslow DK, Muir A, Weissman JS, Thorner J. (2011). Protein kinase Ypk1 phosphorylates regulatory proteins Orm1 and Orm2 to control sphingolipid homeostasis in Saccharomyces cerevisiae . Proc Natl Acad Sci USA , 19222–19227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelants FM, Leskoske KL, Martinez Marshall MN, Locke MN, Thorner J. (2017). The TORC2-dependent signaling network in the yeast Saccharomyces cerevisiae . Biomolecules , E66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelants FM, Torrance PD, Thorner J. (2004). Differential roles of PDK1- and PDK2-phosphorylation sites in the yeast AGC kinases Ypk1, Pkc1 and Sch9. Microbiology , 3289–3304. [DOI] [PubMed] [Google Scholar]
- Saxton RA, Sabatini DM. (2017). mTOR signaling in growth, metabolism, and disease. Cell , 960–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al (2012). Fiji—an open source platform for biological image analysis. Nat Methods , 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Kunz J, Hall MN. (1996). TOR2 is required for organization of the actin cytoskeleton in yeast. Proc Natl Acad Sci USA , 13780–13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Lambert GG, Chammas A, Ni Y, Cranfill PJ, Baird MA, Sell BR, Allen JR, Day RN, Israelsson M, et al (2013). A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum . Nat Methods , 407–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbo D, Murphy CS, Ermakova GV, Solovieva EA, Chepurnykh TV, Shcheglov AS, Verkhusha VV, Pletnev VZ, Hazelwood KL, Roche PM, et al (2009). Far-red fluorescent tags for protein imaging in living tissues. Biochem J , 567–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. (2002). Getting started with yeast. Methods Enzymol , 3–41. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae . Genetics , 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S, Seaman M, Nemoto Y, Daniell L, Suchy SF, Emr SD, De Camilli P, Nussbaum R. (1997). Disruption of three phosphatidylinositol-polyphosphate 5-phosphatase genes from Saccharomyces cerevisiae results in pleiotropic abnormalities of vacuole morphology, cell shape, and osmohomeostasis. Eur J Cell Biol , 350–360. [PubMed] [Google Scholar]
- Stauffer TP, Ahn S, Meyer T. (1998). Receptor-induced transient reduction in plasma membrane PtdIns(4,5)P2 concentration monitored in living cells. Curr Biol , 343–346. [DOI] [PubMed] [Google Scholar]
- Stefan CJ, Audhya A, Emr SD. (2002). The yeast synaptojanin-like proteins control the cellular distribution of phosphatidylinositol (4,5)-bisphosphate. Mol Biol Cell , 542–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz LE. (2001). Genetic and Biologic Characterization of a Family of Inositol Polyphosphate 5-Phosphatases in Saccharomyces cerevisiae. PhD Thesis Durham, NC: Duke University. [Google Scholar]
- Stolz LE, Huynh CV, Thorner J, York JD. (1998). Identification and characterization of an essential family inositol polyphosphate 5-phosphatases (INP51, INP52 and INP53 gene products) in the yeast Saccaromyces cerevisiae . Genetics , 1715–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl T, Thorner J. (2007). Synthesis and function of membrane phosphoinositides in budding yeast Saccharomyces cerevisiae. Biochim Biophys. Acta , 353–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill TW, Cohen A, Diefenbacher M, Trautwein M, Martin DE, Hall MN. (2008). TOR1 and TOR2 have distinct locations in live cells. Eukaryot Cell , 1819–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Miao Y, Yamane Y, Zhang C, Shokat KM, Takematsu H, Kozutsumi Y, Drubin DG. (2012). Orm protein phosphoregulation mediates transient sphingolipid biosynthesis response to heat stress via the Pkh-Ypk and Cdc55-PP2A pathways. Mol Biol Cell , 2388–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi M, Audhya A, Parsons AB, Boone C, Emr SD. (2006). The phosphatidylinositol 4,5-bisphosphate and TORC2 binding proteins Slm1 and Slm2 function in sphingolipid regulation. Mol Cell Biol , 5861–5875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatebe H, Murayama S, Yonekura T, Hatano T, Richter D, Furuya T, Kataoka S, Furuita K, Kojima C, Shiozaki K. (2017). Substrate specificity of TOR complex 2 is determined by a ubiquitin-fold domain of the Sin1 subunit. Elife , e19594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatebe H, Shiozaki K. (2017). Evolutionary conservation of the components in the TOR signaling pathways. Biomolecules , E77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga M, Imamoto N, Sakata-Sogawa K. (2008). Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat Methods , 159–161. [DOI] [PubMed] [Google Scholar]
- Truman AW, Millson SH, Nuttall JM, Mollapour M, Prodromou C, Piper PW. (2007). In the yeast heat shock response, Hsf1-directed induction of Hsp90 facilitates the activation of the Slt2 (Mpk1) mitogen-activated protein kinase required for cell integrity. Eukaryot Cell , 744–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al (2000). A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae . Nature , 623–627. [DOI] [PubMed] [Google Scholar]
- Wachter RM, King BA, Heim R, Kallio K, Tsien RY, Boxer SG, Remington SJ. (1997). Crystal structure and photodynamic behavior of the blue emission variant Y66H/Y145F of green fluorescent protein. Biochemistry , 9759–9765. [DOI] [PubMed] [Google Scholar]
- Wedaman KP, Reinke A, Anderson S, Yates J, McCaffery JM, Powers T, Gilmore R. (2002). Tor Kinases are in distinct membrane-associated protein complexes in Saccharomyces cerevisiae . Mol Biol Cell , 1204–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler A, Arkind C, Mattison CP, Burkholder A, Knoche K, Ota I. (2002). Heat stress activates the yeast high-osmolarity glycerol mitogen-activated protein kinase pathway, and protein tyrosine phosphatases are essential under heat stress. Eukaryot. Cell , 163–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Oppliger W, Hall MN. (2005). Molecular organization of Target of Rapamycin complex 2. J Biol Chem , 30697–30704. [DOI] [PubMed] [Google Scholar]
- Yang H, Rudge DG, Koos JD, Vaidialingam B, Yang HJ, Pavletich NP. (2013). mTOR kinase structure, mechanism and regulation. Nature , 217–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JW, Mendrola JM, Audhya A, Singh S, Keleti D, DeWald DB, Murray D, Emr SD, Lemmon MA. (2004). Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol Cell , 677–688. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.