Abstract
In fat and skeletal muscle cells, insulin-responsive amino peptidase (IRAP) along with glucose transporter 4 (Glut4) and sortilin, represents a major component protein of the insulin-responsive vesicles (IRVs). Here, we show that IRAP, similar to Glut4 and sortilin, is retrieved from endosomes to the trans-Golgi network by retromer. Unlike Glut4, retrograde transport of IRAP does not require sortilin, as retromer can directly bind to the cytoplasmic tail of IRAP. Ablation of IRAP in 3T3-L1 adipocytes shifts the endosomal pool of Glut4 to more acidic endosomes, but does not affect IRV targeting, stability, and insulin responsiveness of Glut4.
INTRODUCTION
Insulin-responsive amino peptidase (IRAP (also known as placental leucine amino peptidase [Rogi et al., 1996]) is a type 2 membrane protein with a zinc-dependent amino peptidase domain localized in the extracellular/luminal C-terminal part of the protein (Keller, 2003). In fat and skeletal muscle cells, IRAP represents a major (or the major) protein component of the insulin-responsive membrane vesicles (IRVs) that undergo fusion with the plasma membrane in response to insulin stimulation (Kandror et al., 1994; Keller et al., 1995; Keller, 2003; Kupriyanova et al., 2002). In addition to IRAP, these vesicles compartmentalize insulin-sensitive glucose transporter 4 (Glut4), low-density lipoprotein receptor–related protein 1 (LRP1) sorting receptor sortilin, small amounts of metabolic receptors, such as the transferrin receptor and the IGFII/mannose 6-phosphate receptor, as well as secretory carrier-associated membrane proteins with unknown biological functions and vesicle-associated membrane proteins required for vesicle fusion with the plasma membrane (Bogan and Kandror, 2010; Kandror and Pilch, 2011). The physiological significance of insulin-dependent translocation of IRAP to the plasma membrane is not completely clear but may include proteolytic cleavage and inactivation of vasopressin (Herbst et al., 1997; Habtemichael et al., 2015), oxytocin (Rogi et al., 1996), and other circulating biologically active peptides.
It has also been suggested that IRAP is involved in the regulation of IRV translocation. Thus, the cytoplasmic N-terminus of IRAP can interact with proteins implicated in this process, such as TBC1D1 (Mafakheri et al., 2018), TBC1D4/AS160 (Larance et al., 2005; Peck et al., 2006; Jedrychowski et al., 2010), p115 (Hosaka et al., 2005), tankyrase (Chi and Lodish, 2000; Yeh et al., 2007), and TUG (Habtemichael et al., 2015). Also, it has been shown that microinjection of the peptide corresponding to the cytoplasmic IRAP tail into 3T3-L1 adipocytes causes translocation of Glut4 (Waters et al., 1997), whereas knockdown of IRAP in these cells has a strong inhibitory effect on translocation of Glut4 (Yeh et al., 2007). Still, the role of IRAP in the regulation of IRV translocation remains controversial as another group has reported that knockdown of IRAP in 3T3-L1 adipocytes does not affect insulin-stimulated translocation of Glut4, but may increase its plasma membrane content under basal conditions (Jordens et al., 2010). Also, knockout of IRAP in vivo decreases total protein levels of Glut4 but does not affect its translocation in primary adipocytes (Keller et al., 2002). Finally, our results suggest that insulin responsiveness of the IRVs depend on sortilin, rather than IRAP (Huang et al., 2013). Given these contradictory data, we decided to further investigate trafficking and potential regulatory functions of IRAP in adipose cells in relation to Glut4 stability and translocation.
After insulin-stimulated translocation of the IRVs to the plasma membrane, Glut4 is internalized into endosomes from where it is retrieved to the trans-Golgi network (TGN) in order to escape degradation in lysosomes (Slot et al., 1991; Foster et al., 2001; Shewan et al., 2003). Sortilin plays the key role in Glut4 retrieval by interacting with the first luminal loop of Glut4 and recruiting retromer via its cytoplasmic tail (Pan et al., 2017). IRAP is also localized in the TGN (Shi et al., 2008), but the mechanism of IRAP targeting to this compartment is unknown. Neither is it known whether sortilin plays any role in IRAP trafficking and vice versa. Here, we show that IRAP is retrieved from endosomes to the TGN by retromer. Retrograde traffic of IRAP and sortilin are independent of each other. Neither does IRAP play a major role in endosomal retrieval or insulin-dependent translocation of Glut4.
RESULTS AND DISCUSSION
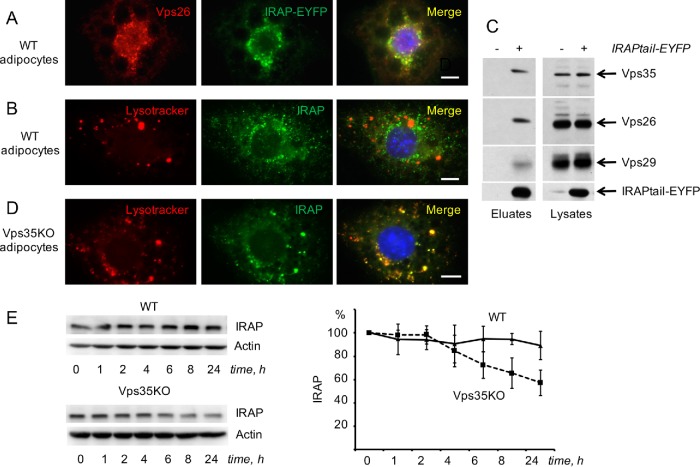
Undifferentiated 3T3-L1 preadipocytes endogenously express IRAP, but not sortilin or Glut4. The latter proteins are induced on days 3 and 4 of differentiation correspondingly (ElJack et al., 1999; Shi and Kandror, 2005; Shi et al., 2008). In undifferentiated preadipocytes, IRAP is localized predominantly in the syntaxin 6–positive perinuclear compartment, likely TGN (Shi et al., 2008). The TGN targeting information is contained in the cytoplasmic tail of IRAP as IRAPtail fused with EYFP has a similar intracellular localization (Shi et al., 2008). Here, we confirm our previous results and demonstrate that both full-length IRAP and IRAPtail (Figure 1A) show a strong colocalization with the retromer subunits Vps35 and Vps26 (Figure 1, B and C; Table 1). At the same time, there is virtually no colocalization between IRAP and lysotracker (Figure 1D; Table 1). Correspondingly, IRAP in preadipocytes is very stable with half-life exceeding 24 h (Figure 1E). Thus, IRAP is capable of interacting with retromer and maintaining TGN localization in the complete absence of either Glut4 or sortilin.
FIGURE 1:
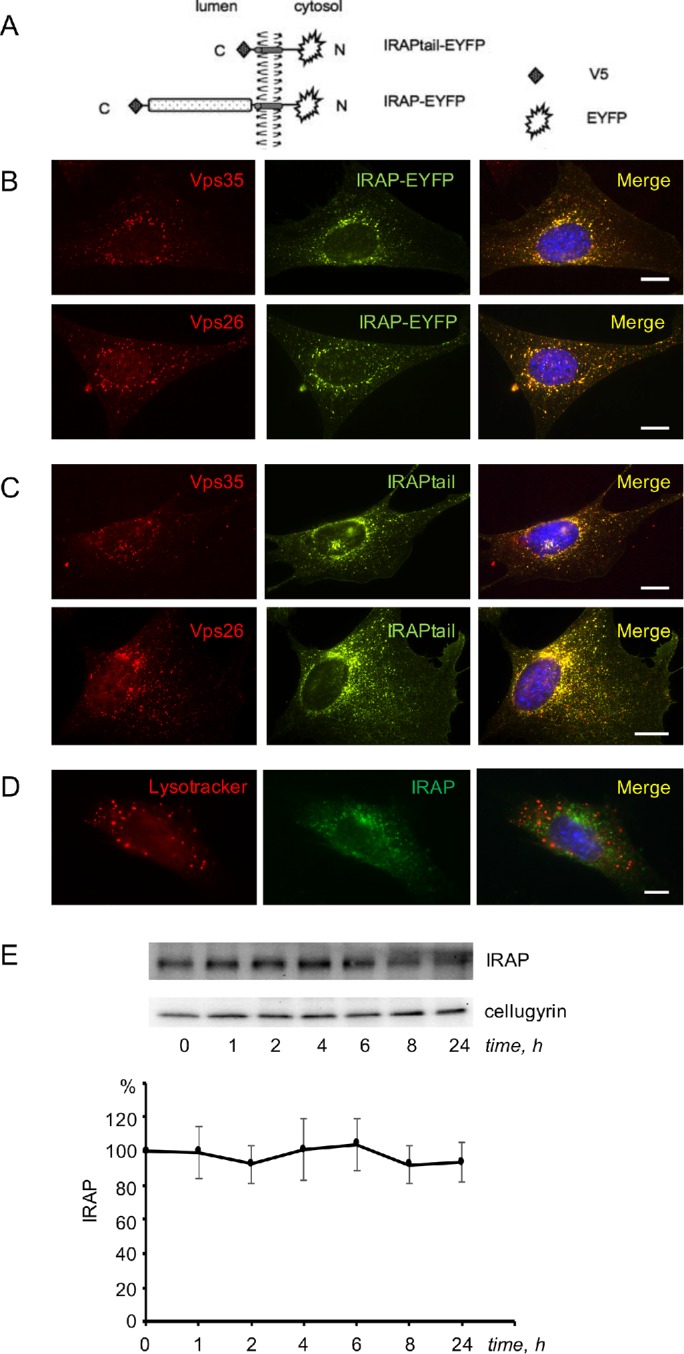
In undifferentiated 3T3-L1 cells, IRAP interacts with retromer and has a long half-life. (A) Constructs used in the study. (B, C) The 3T3-L1 preadipocytes stably expressing full-length IRAP-EYFP (B) and IRAPtail-EYFP (C) were fixed, stained with the polyclonal anti-Vps35 antibody or anti-Vps26 antibody, as indicated on the panels, and analyzed under a fluorescence microscope. (D) Wild-type 3T3-L1 preadipocytes were incubated with lysotracker, fixed, stained with the monoclonal anti-IRAP antibody, and analyzed under a fluorescence microscope. (E) Wild-type 3T3-L1 preadipocytes were incubated with emetine for the indicated periods of time, and total lysates (80 µg) were analyzed by Western blotting. The graph shows quantification of the data. Error bars represent SE.
TABLE 1:
Quantitative analysis of protein colocalization.
| # | Cell type | Expressed construct(s) | Staining 1 | Staining 2 | Corresponding | Pearson’s coefficienta |
|---|---|---|---|---|---|---|
| 1 | Preadipocyte | IRAP-EYFP | Vps35 | EYFP | Figure 1B (top) | 0.94 ± 0.02 |
| 2 | Preadipocyte | IRAP-EYFP | Vps26 | EYFP | Figure 1B (bottom) | 0.94 ± 0.01 |
| 3 | Preadipocyte | IRAPtail-EYFP | Vps35 | EYFP | Figure 1C (top) | 0.94 ± 0.02 |
| 4 | Preadipocyte | IRAPtail-EYFP | Vps26 | EYFP | Figure 1C (bottom) | 0.92 ± 0.03 |
| 5 | Preadipocyte | None | LysoTracker | IRAP | Figure 1E | 0.38 ± 0.12 |
| 6 | Adipocyte | IRAP-EYFP | Vps26 | EYFP | Figure 2A | 0.80 ± 0.06 |
| 7 | Adipocyte | None | LysoTracker | IRAP | Figure 2B | 0.52 ± 0.11 |
| 8 | Adipocyte | Vps35 CRISPR | LysoTracker | IRAP | Figure 2D | 0.93 ± 0.03 |
| 9 | Adipocyte | Sortilin shRNA | LysoTracker | IRAP | Figure 4A | 0.60 ± 0.13 |
| 10 | Adipocyte | None | LysoTracker | Glut4 | Figure 7A (top) | 0.52 ± 0.11 |
| 11 | Adipocyte | IRAP CRISPR | LysoTracker | Glut4 | Figure 7A (bottom) | 0.71 ± 0.13 |
| 12 | Adipocyte | None | Vps35 | Glut4 | Figure 7B (top) | 0.82 ± 0.09 |
| 13 | Adipocyte | IRAP CRISPR | Vps35 | Glut4 | Figure 7B (bottom) | 0.64 ± 0.07 |
aPearson’s coefficient was determined by ImageJ analysis of 12–14 randomly selected cells.
In differentiated adipocytes, IRAP continues to demonstrate a significant colocalization with retromer (Figure 2A; Table 1) but not with lysotracker (Figure 2B; Table 1). Using GFP-Trap immunoprecipitation with (Figure 2C) and without (Supplemental Figure S1) cross-linking, we show that IRAPtail not just colocalizes with retromer in 3T3-L1 adipocytes but also may interact with it directly. Apparently, retromer not just binds to the cytoplasmic tail of IRAP but also plays an important role of IRAP retrieval from endosomes to TGN. To this end, we have shown that knockout of one of the retromer subunits, Vps35, using CRISPR/Cas9 approach (described in detail in Pan et al., 2017) strongly increases colocalization of IRAP with lysotracker (Figure 2D; Table 1) and decreases stability of IRAP (Figure 2E) in a Bafilomycin A–sensitive manner (unpublished data).
FIGURE 2:
In differentiated 3T3-L1 adipocytes, IRAP is retrieved to the perinuclear compartment by retromer. (A) The 3T3-L1 adipocytes stably expressing full-length IRAP-EYFP were fixed, stained with anti-Vps26 antibody, and analyzed under a fluorescence microscope. (B) Wild-type 3T3-L1 adipocytes were incubated with lysotracker, fixed and stained with the monoclonal anti-IRAP antibody, and analyzed under a fluorescence microscope. (C) The 3T3-L1 adipocytes stably expressing IRAPtail-EYFP were cross-linked with DSP and the lysates were subjected to GFP-Trap immunoprecipitation (see Materials and Methods); the eluates and the input (lysates) were immunoblotted with the antibodies indicated on the right; IRAPtail-EYFP was stained with anti-GFP antibody. All samples had been analyzed in the same gel, and irrelevant lanes were spliced out. A representative result of three independent experiments is shown. (D) Vps35-depleted 3T3-L1 adipocytes were incubated with lysotracker, fixed and stained with the monoclonal anti-IRAP antibody, and analyzed under a fluorescence microscope. (E) Wild-type and Vps35-depleted 3T3-L1 adipocytes were incubated with emetine for the indicated periods of time, and total lysates (80 µg) were analyzed by Western blotting. The graph shows quantification of the data. Error bars represent SE.
To further investigate the effect of Vps35 knockout on IRAP traffic at the biochemical level, we have separated adipocyte extracts obtained from wild-type and CRISPR/Cas9-edited 3T3-L1 adipocytes by differential centrifugation into the heavy membrane fraction (P1), which includes endoplasmic reticulum, plasma membrane, as well as large endosomes and TGN membranes, and the vesicular fraction (P2), which includes the IRVs and small transport vesicles (Figure 3A). In wild-type 3T3-L1 adipocytes, ca. 15–20% of total IRAP is localized in the vesicular fraction, whereas most syntaxin 6 (a TGN resident protein) is recovered in heavy membranes (Figure 3B). Knockout of Vps35 decreases total amounts of IRAP, which is consistent with its increased presence in lysosomes and decreased half-life as seen in Figure 2. In addition, knockout of Vps35 virtually eliminates IRAP presence in small vesicles, whereas the relative amount of IRAP in the heavy membrane fraction is increased (Figure 3B). Note that neither total expression levels nor the biochemical distribution of syntaxin 6 and actin is altered in Vps35 knockout cells (Figure 3A). These results suggest that knockout of Vps35 suppresses retrograde traffic of IRAP by blocking IRAP exit from endosomes into small endosome-to-TGN transport carriers (Mari et al., 2008).
FIGURE 3:
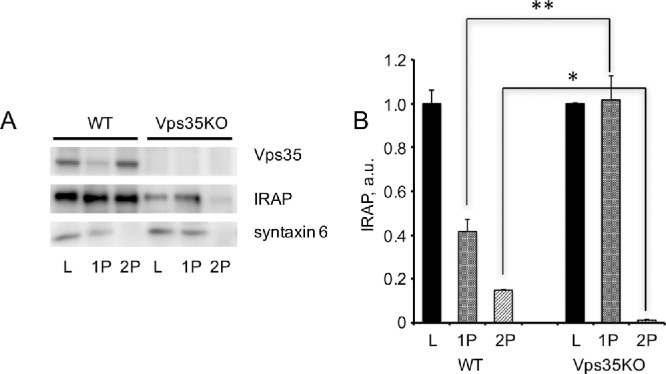
Knockout of Vps35 in 3T3-L1 adipocytes inhibits formation of small IRAP-containing transport vesicles. (A) Total lysates (L) prepared from wild type and Vps35-depleted (KO) 3T3-L1 adipocytes were fractionated into heavy membrane pellet (P1) and vesicular fraction (P2) that were analyzed by Western blotting (20 µg per lane). (B) A relative distribution of IRAP between fractions. The presence of each protein in total lysates (L) of wild type and Vps35-depleted 3T3-L1 adipocytes is 100%. **p < 0.01; *p < 0.05. Results of three independent experiments were analyzed by Student’s t test.
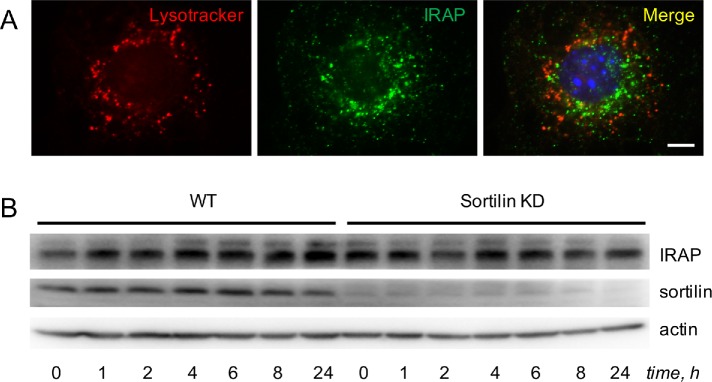
We have recently shown that retrograde traffic of Glut4 depends not only on retromer but also on sortilin that acts as a transmembrane scaffold coupling Glut4 to retromer (Pan et al., 2017). According to our model, diminished levels of the Glut4 protein in fat tissue in obesity, insulin resistance, and diabetes (Shepherd and Kahn, 1999; Graham and Kahn, 2007; Huang and Czech, 2007; Bogan, 2012) may be caused by decreased expression of sortilin (Keller et al., 2008; Kaddai et al., 2009). However, knockdown of sortilin with short hairpin RNA (shRNA) does not significantly increase colocalization between IRAP and lysotracker (Figure 4A; Table 1) or affect stability of IRAP (Figure 4B). On the basis of these results, we conclude that retrieval of IRAP to the TGN takes place in a retromer-dependent (Figures 1–3) and sortilin-independent (Figure 4) manner. In agreement with our results, overall protein levels of IRAP in type 2 diabetes do not change (Maianu et al., 2001) in spite of reduced expression of sortilin.
FIGURE 4:
In differentiated 3T3-L1 adipocytes, IRAP is retrieved to the perinuclear compartment in a sortilin-independent manner. (A) 3T3-L1 adipocytes stably transfected with shRNA against sortilin were incubated with lysotracker, fixed, stained with the monoclonal anti-IRAP antibody and analyzed under a fluorescence microscope. (B) Wild-type 3T3-L1 adipocytes and 3T3-L1 adipocytes stably transfected with shRNA against sortilin (KD) were incubated with emetine for the indicated periods of time, and total lysates (80 µg) were analyzed by Western blotting. A representative result of three independent experiments is shown.
Once IRAP is retrieved to the TGN, it is recruited into the IRVs via luminal interactions with sortilin and Glut4 (Shi et al., 2008) and becomes a prominent protein in this compartment (Kandror et al., 1994; Keller et al., 1995; Keller, 2003; Kupriyanova et al., 2002). Several lines of evidence (see Introduction) suggest that IRAP may play a role in IRVs translocation. To investigate this question, we knocked down IRAP in differentiated adipocytes with the help of small interfering RNA (siRNA) used in a previous publication (Yeh et al., 2007). As is seen in Figure 5A, we were able to achieve ca. 70% reduction in the IRAP protein compared to cells transfected with scrambled siRNA. However, neither the total protein levels of Glut4 and sortilin (Figure 5A) nor the efficiency of their recruitment into the IRVs (Figure 5B) changed with IRAP knockdown. Depletion of IRAP did not affect insulin-induced phosphorylation of Akt (unpublished data) or insulin-stimulated glucose uptake (Figure 5C).
FIGURE 5:
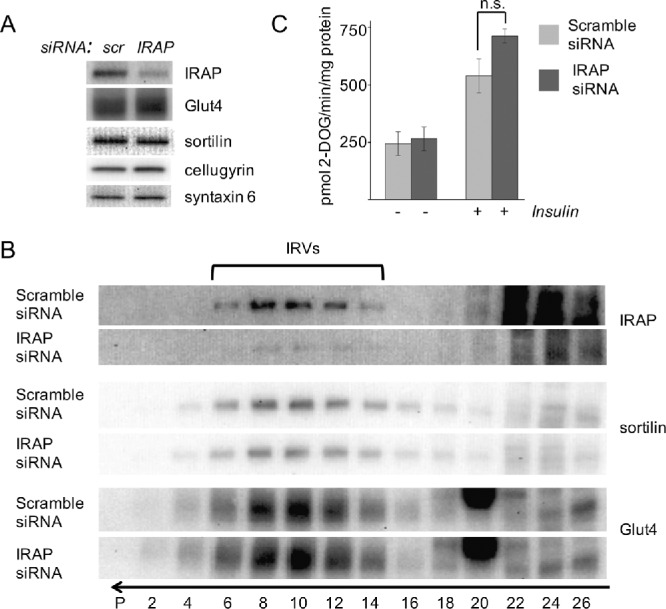
Knockdown of IRAP using siRNA does not affect formation of the IRVs and insulin-stimulated glucose uptake in 3T3-L1 adipocytes. (A, B) The 3T3-L1 adipocytes were treated with siRNA against IRAP, and total lysates were analyzed by Western blotting (A) and continuous sucrose gradient centrifugation (B). The arrow shows the direction of sedimentation. The position of the IRVs is indicated. (C) Insulin-stimulated glucose uptake was measured in wild type and IRAP-depleted 3T3-L1 adipocytes. Error bars represent SD; n.s., not significant.
To confirm these results, we knocked out IRAP from 3T3-L1 adipocytes using the CRISPR/Cas9 technology and performed further experiments in three single-cell clones and in a pooled clone of IRAP-depleted cells. Figure 6A shows a representative result of this work. Basically, we conclude that depletion of IRAP in cultured 3T3-L1 adipocytes does not affect expression levels of Glut4 and sortilin. Additional experiments did not reveal any changes in half-lives of Glut4 or sortilin caused by depletion of IRAP (unpublished data). By the same token, CRISPR/Cas9-mediated depletion of IRAP did not change insulin-induced phosphorylation of Akt and AS160 (unpublished data) and insulin-stimulated glucose uptake (Figure 6B) in cultured adipocytes. Thus, down-regulation of IRAP does not significantly change stability of the Glut4 protein or its insulin-dependent translocation to the plasma membrane regardless of the experimental technique used.
FIGURE 6:
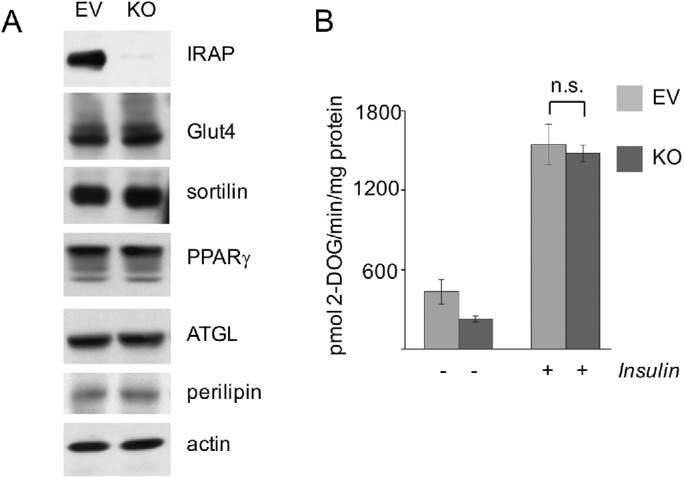
Knockout of IRAP using CRISPR/Cas9 does not affect insulin-stimulated glucose uptake in 3T3-L1 adipocytes. (A) Total lysates (80 µg) of CRISPR/Cas9-edited and empty lentiCRISPR plasmid-infected 3T3-L1 adipocytes were analyzed by Western blotting. Pooled clone of IRAP-depleted and control cells are shown, three individual single-cell clones of IRAP-depleted cells produced similar results. (B) Insulin-stimulated glucose uptake was measured in wild type and IRAP-depleted 3T3-L1 adipocytes. Error bars represent SD. For the effect of insulin on the control 3T3-L1 adipocytes, p = 0.011; for IRAP-depleted, p < 0.0005; n.s., not significant.
Unlike Glut4, expression of IRAP is not limited to adipocytes and skeletal myocytes. In several cell types, IRAP has been found in distinct vesicles with biochemical parameters (size, sedimentation coefficient, and buoyant density) similar to those in the IRVs but lacking insulin responsiveness (Thoidis and Kandror, 2001; Liao et al., 2006; Fernando et al., 2007). These observations also suggest that the presence of IRAP per se does not confer insulin responsiveness to membrane vesicles. We cannot exclude the possibility, however, that IRAP may play a role in the formation and/or retention of the IRVs in certain lines of 3T3-L1 cells with different expression levels of other IRV proteins with redundant functions in the vesicular traffic.
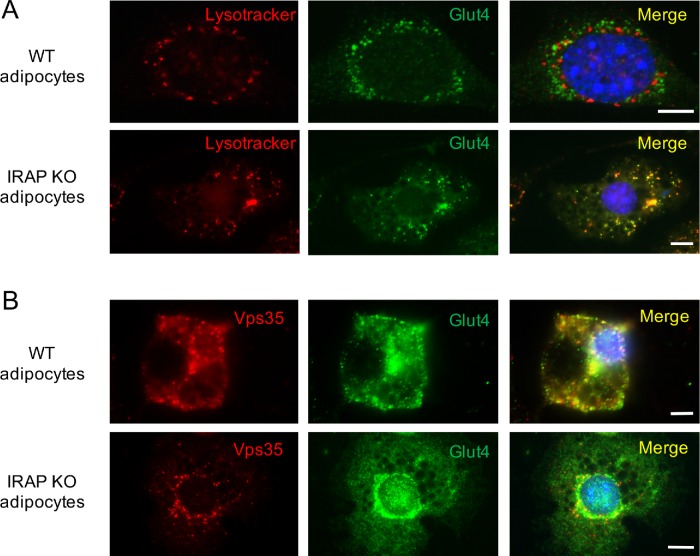
Interestingly, depletion of IRAP from 3T3-L1 adipocytes increases colocalization between Glut4 and lysotracker (Figure 7A; Table 1), suggesting that, in IRAP-null cells, Glut4 may populate late endosomes with low luminal pH values. This is consistent with the recent report by Babdor et al. (2017), who have shown that knockout of IRAP accelerates maturation of early endosomes into lysosomes. At the same time, Glut4 continues to colocalize with retromer even in IRAP-null adipocytes, although the extent of colocalization is decreased somewhat (Figure 7B; Table 1). Given that stability of Glut4 in these cells does not change, we suggest that in cultured adipocytes, Glut4 can be efficiently retrieved from more acidic late endosomes. This, however, may not be the case in IRAP-null primary adipocytes as knockout of IRAP in vivo decreases Glut4 protein levels without affecting its mRNA (Keller et al., 2002). Thus, IRAP may help to maintain steady levels of the Glut4 protein in primary adipocytes by stabilizing Glut4-containing endosomes and slowing their transition to lysosomes. Alternatively, genetic ablation of IRAP in vivo may have a noncell autonomous effect on Glut4 expression.
FIGURE 7:
Knockout of IRAP using CRISPR/Cas9 causes relocalization of Glut4 to more acidic endosomes where it is still colocalizes with retromer. (A) Wild-type and IRAP-depleted adipocytes were incubated with lysotracker, fixed, and stained with the rabbit polyclonal antibody against Glut4. (B) Wild-type and IRAP-depleted adipocytes were fixed and stained with the rabbit polyclonal antibody against Glut4 and mouse monoclonal antibodies against Vps35.
MATERIALS AND METHODS
Reagents and antibodies
Insulin was purchased from Sigma-Aldrich (St, Louis, MO); bovine serum (BS) and fetal bovine serum (FBS) were purchased from Atlanta Biologicals (Lawrenceville, GA). 3H2-deoxyglucose was purchased from PerkinElmer (Waltham, MA). Mouse monoclonal antibodies against actin and IRAP (3E1), rabbit polyclonal antibodies against Akt, phospho-Akt (Thr308), AS160, phospho-AS160 (Thr642), ATGL, PPARγ (81B8), and perilipin-1 (D1D8) were purchased from Cell Signaling Technology (Danvers, MA). Mouse monoclonal antibody against Glut4 (1F8) and rabbit polyclonal antibody against IRAP were a gift of P. Pilch (Boston University, Boston, MA). Rabbit polyclonal antibodies against Glut4, sortilin, and Vps26 were purchased from Abcam (Cambridge, MA); against GFP was purchased from GeneTex (Irvine, CA); mouse monoclonal antibodies against sortilin, syntaxin 6 was purchased from BD Bioscience Pharmingen (San Diego, CA); and against Vps29 and Vps35 was purchased from Santa Cruz Biotechnology (Dallas, TX). Goat polyclonal antibody against Vps35 was from Imgenex (San Diego, CA). Horseradish peroxidase (HRP)-conjugated anti-mouse and anti-rabbit immunoglobulin G (IgG) were from Cell Signaling Technology (Danvers, MA). HRP-conjugated anti-goat IgG, fluorescein isothiocyanate–conjugated donkey anti-rabbit antibody, Texas Red–conjugated donkey anti-mouse antibody, Alexa Fluor647–conjugated donkey anti-mouse antibody, Alexa Fluor546–conjugated donkey anti-goat antibody, LysoTracker Red DND-99, ProLong Gold antifade reagent with DAPI, Halt Protease and Phosphatase Inhibitor Cocktail, DMEM, Opti-MEM, and Dulbecco’s phosphate-buffered saline (PBS) were from Thermo Fisher Scientific (Waltham, MA).
Stable cell lines
Preparation and culturing of 3T3-L1 cells stably expressing IRAP-EYFP and IRAPtail-EYFP (Figure 1A) were described in Shi et al. (2008), and 3T3-L1 cells depleted of sortilin with the help of pBabe-shRNA were described previously in Shi and Kandror (2005). For adipose differentiation, cells were grown in DMEM containing 10% calf bovine serum. Two days after confluence, cells were transferred to the differentiation medium (DMEM with 10% FBS, 0.174 μM insulin, 1 μM dexamethasone, and 0.5 mM 3-isobutyl-1-methylxanthine). After 48 h, differentiation medium was replaced with DMEM containing 10% FBS.
Immunofluorescence
Undifferentiated and differentiated 3T3-L1 cells were grown on coverslips coated with polylysine (Neuvitro, Vancouver, WA). Serum-starved cells were fixed with 4% paraformaldehyde in PBS (pH 7.4) for 10 min and permeabilized with 0.2% Triton X-100 for 3 min at room temperature. After blocking with 5% donkey serum for 1 h, cells were stained overnight at 4°C with primary antibodies, followed by incubation with secondary antibody for 1 h at room temperature. In several experiments, cells prior to fixing were incubated with lysotracker (0.1–0.4 μM) for 40–60 min at 37°C. Antifade solution was used for mounting cells on slides. Slides were examined with the help of the Axio Observer Z1 fluorescence microscope equipped with the Hamamatsu digital camera C10600/ORCA-R2 and Axiovision 4.8.1 program (Carl Zeiss, Thornwood, NY). Each scale bar is 5 μm. Each image shows a representative result of at least three independent experiments. Quantification of images using the ImageJ program is shown in Table 1.
Cross-linking and immunoprecipitation
Wild-type 3T3-L1 adipocytes and 3T3-L1 adipocytes expressing IRAPtail-EYFP (Figure 1A) in 15-cm dishes were washed twice with PBS and once with Krebs-Ringer phosphate buffer (12.5 mM HEPES, 120 mM NaCl, 6 mM KCl, 1.2 mM MgSO4, 1.0 mM CaCl2, 0.6 mM Na2HPO4, 0.4 mM NaH2PO4, 2.5 mM d-glucose, pH 7.4), and dithiobis[succinimidyl propionate] (DSP) was added to the final concentration 2 mM for 30 min. Cross-linking reaction was stopped with 50 mM Tris (pH 7.4), and cells were incubated for 10 min in quenching buffer (50 mM Tris, 150 mM NaCl, pH 7.4) and washed once with the same buffer supplemented with the protease and phosphatase inhibitor cocktail. Cells were harvested, pelleted by microcentrifugation for 2 min at 3000 × g, and frozen. All the following procedures were carried out on ice or in a cold room. The cells were resuspended in cold PBS supplemented with 5% glycerol, the protease and phosphatase inhibitor cocktail, and 0.5% Triton X-100. The suspension was incubated for 10 min and passed three times through a 21-G needle, and the lysates were clarified for 10 min at 16,000 × g in a microcentrifuge. Supernatants were adjusted to equal protein concentration and volume (400–450 µl) and added to GFP-Trap_MA magnetic beads (ChromoTek GmbH, Germany) washed with the same buffer (25 µl of the original suspension per sample). The beads were incubated on a rotator for 70–80 min and washed four times with PBS supplemented with 5% glycerol and 0.2% Triton X-100. The immunoprecipitated proteins were eluted from the beads by 10-min incubation in 2 × Laemmli loading buffer at 97°C.
Sucrose gradient centrifugation
For velocity gradient centrifugation, samples (∼0.2 ml, ∼1 mg) were loaded onto a 4.6-ml linear 10–30% (wt/vol) sucrose gradient in 20 mM HEPES with 1 mM EDTA and the protease and phosphatase inhibitor cocktail, pH 7.4, and centrifuged for 55 min in a SW55 rotor (Beckman Coulter, Fullerton, CA) at 48,000 rpm. Each gradient was separated into 22–26 fractions starting from the bottom of the tube. Fractions were further analyzed by SDS–PAGE and Western blotting.
siRNA-mediated knockdown of IRAP
The anti-IRAP siRNA 5′-GCCCUGUUCCAGACAAACCTT-3′ (Yeh et al., 2007) was obtained from Integrated DNA Technologies (Coralville, IA), and scrambled siRNA was from Ambion (Austin, TX). siRNAs (30 nM of each) were transfected into 3T3-L1 adipocytes using the DeliverX Plus delivery kit from Panomics (Fremont, CA). Cells were analyzed 48 h after transfection.
CRISPR-Cas9 genome editing
Depletion of Vps35 with the help of the CRISPR/Cas9 approach was described previously (Pan et al., 2017). Depletion of IRAP in 3T3-L1 preadipocytes was performed using the CRISPR/Cas9 system according to Sanjana et al. (2014). Briefly, two single-guide RNAs (sgRNAs) targeting exon 2 of the IRAP gene (5′-AATATGAGCCCCGAGGTTCG-3′, 5′-ACAAATCGCCGTTGTTGCCC-3′ correspondingly) were designed using online software (http://crispr.mit.edu and http://chopchop.cbu.uib.no). The lentiCRISPR v2 plasmid (Addgene, #52961) was used to express sgRNAs and the Cas9 protein along with the envelope plasmid pCMV-VSV-G and the packaging plasmid psPAX2. Cells infected with lentiviruses were treated with puromycin (3 μg/ml) for the selection of stable transfectants; single-cell colonies were isolated and grown in the medium with 3 μg/ml puromycin. The genomic region surrounding the sgRNA target site was amplified by PCR and sequenced. The primers used for the PCR were 1F, AGGTCCTGGCAGAGGGTAAT; 1R, ATCGGCTTCAGCTTCCAAGG; 2F, GCAGCAGAGCACAGAACAAG; 2R, GCTCTTCAAGACACACGGGA.
Measurements of protein stability
Aqueous solutions of emetine (10 μM) with or without Bafilomycin A (100 nM) were added to plates at time 0. At the indicated time intervals, the cells were rinsed three times with PBS and harvested in ice-cold RIPA buffer (Millipore, Billerica, MA) with the protease inhibitor cocktail. Cell lysates were vortexed, rotated at 4°C for 30 min, and spun for 20 min at 16,000 × g in a microcentrifuge at 4°C. The presence of the individual proteins in supernatants was analyzed by Western blotting.
[3H] 2-deoxyglucose uptake
This assay was performed in six-well plates. Cells were washed three times with serum-free DMEM, starved for 4 h, washed twice with warm Krebs-Ringer-HEPES (KRH) buffer without glucose (121 mM NaCl, 4.9 mM KCl, 1.2 mM MgSO4, 0.33 mM CaCl2, 12 mM HEPES, pH 7.4), and treated with either 100 nM insulin or carrier (5 µM HCl) at 37°C for 15 min. Radioactive 2-deoxyglucose (0.1 mM, 0.625 μCi/ml) was added to cells for 5 min. The assay was terminated by aspirating the radioactive media, and the cells were washed three times with 2 ml of ice-cold KRH containing 25 mM D-glucose. Each well was then extracted with 400 μl of 0.1% SDS in KRH buffer without glucose, and 300 μl of aliquots were removed for determination of radioactivity by liquid scintillation counting. Measurements were made in triplicates and corrected for specific activity and nonspecific diffusion (as determined in the presence of 5 μM cytochalasin B), which was <5% of the total uptake. The protein concentration was determined using the BCA protein assay kit (Thermo Fisher Scientific, Waltham, MA) and was used to normalize counts.
Subcellular fractionation of 3T3-L1 cells
The 3T3-L1 adipocytes or preadipocytes were washed three times with serum-free DMEM, warmed to 37°C, and starved in the same media for 2 h. Cells were treated with 100 nM insulin or carrier (5 mM HCl at 1000× dilution) in DMEM for 15 min at 37°C. Cells were then washed three times with cold HES buffer (250 mM sucrose, 20 mM HEPES, 1 mM EDTA, pH 7.4) with protease inhibitor cocktail and harvested in the same buffer (0.3–1 ml/10-cm dish). Homogenization was performed in a ball-bearing homogenizer (Isobiotec, Heidelberg, Germany) with a 12 µM clearance by 10 strokes for adipocytes and 15 strokes for preadipocytes. Homogenates were centrifuged at 14,000 × g for 20 min (P1). Membrane vesicles in supernatants (P2) were concentrated by pelleting at 200,000 × g for 90 min. Pellets were resuspended in 0.1–0.25 ml of HES buffer.
Gel electrophoresis and Western blotting
Proteins were separated in SDS–polyacrylamide gels according to Laemmli (Laemmli, 1970) and transferred to Immobilon-P membranes (Millipore, Bedford, MA). Following transfer, the membrane was blocked with 5% BSA in PBS with 0.5% Tween 20 for 1 h. Blots were probed overnight with specific primary antibodies at 4°C followed by 1 h incubation at room temperature with HRP-conjugated secondary antibodies. Protein bands were detected with the enhanced chemiluminescence substrate kit (PerkinElmer Life Sciences, Boston, MA) using a Bio-Rad ChemiDoc XRS+ System (Hercules, CA).
Supplementary Material
Acknowledgments
This work was supported by research grants DK52057 and DK107498 from the National Institutes of Health (NIH) to K.V.K. X.P. was supported by institutional training grant 2T32DK007201 from the NIH.
Abbreviations used:
- BS
bovine serum
- DSP
dithiobis[succinimidyl propionate]
- FBS
fetal bovine serum
- Glut4
glucose transporter 4
- IRAP
insulin-responsive amino peptidase
- IRV
insulin-responsive vesicles
- KRH
Krebs-Ringer-HEPES
- LRP1
low-density lipoprotein receptor–related protein 1
- PBS
phosphate-buffered saline
- sgRNA
single-guide RNA
- TGN
trans-Golgi network.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-12-0792) on April 3, 2019.
REFERENCES
- Babdor J, Descamps D, Adiko AC, Tohme M, Maschalidi S, Evnouchidou I, Vasconcellos LR, De Luca M, Mauvais FX, Garfa-Traore M, et al. (2017). IRAP(+) endosomes restrict TLR9 activation and signaling. Nat Immunol , 509–518. [DOI] [PubMed] [Google Scholar]
- Bogan JS. (2012). Regulation of glucose transporter translocation in health and diabetes. Annu Rev Biochem , 507–532. [DOI] [PubMed] [Google Scholar]
- Bogan JS, Kandror KV. (2010). Biogenesis and regulation of insulin-responsive vesicles containing GLUT4. Curr Opin Cell Biol , 506–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi NW, Lodish HF. (2000). Tankyrase is a golgi-associated mitogen-activated protein kinase substrate that interacts with IRAP in GLUT4 vesicles. J Biol Chem , 38437–38444. [DOI] [PubMed] [Google Scholar]
- ElJack A, Kandror KV, Pilch PF. (1999). Formation of an insulin-responsive vesicular compartment is an early event in 3T3-L1 adipocyte differentiation. Mol Biol Cell , 1581–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando RN, Luff SE, Albiston AL, Chai SY. (2007). Sub-cellular localization of insulin-regulated membrane aminopeptidase, IRAP to vesicles in neurons. J Neurochem , 967–976. [DOI] [PubMed] [Google Scholar]
- Foster LJ, Li D, Randhawa VK, Klip A. (2001). Insulin Accelerates Inter-endosomal GLUT4 traffic via phosphatidylinositol 3-kinase and protein kinase B. J Biol Chem , 44212–44221. [DOI] [PubMed] [Google Scholar]
- Graham TE, Kahn BB. (2007). Tissue-specific alterations of glucose transport and molecular mechanisms of intertissue communication in obesity and type 2 diabetes. Horm Metab Res , 717–721. [DOI] [PubMed] [Google Scholar]
- Habtemichael EN, Alcazar-Roman A, Rubin BR, Grossi LR, Belman JP, Julca O, Loffler MG, Li H, Chi NW, Samuel VT, Bogan JS. (2015). Coordinated regulation of vasopressin inactivation and glucose uptake by action of TUG protein in muscle. J Biol Chem , 14454–14461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst JJ, Ross SA, Scott HM, Bobin SA, Morris NJ, Lienhard GE, Keller SR. (1997). Insulin stimulates cell surface aminopeptidase activity toward vasopressin in adipocytes. Am J Physiol , E600–E606. [DOI] [PubMed] [Google Scholar]
- Hosaka T, Brooks CC, Presman E, Kim SK, Zhang Z, Breen M, Sztul E, Pilch PF. (2005). p115 Interacts with the GLUT4 vesicle protein, IRAP, and plays a critical role in insulin-stimulated GLUT4 translocation. Mol Biol Cell , 2882–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Buckler-Pena D, Nauta T, Singh M, Asmar A, Shi J, Kim JY, Kandror KV. (2013). Insulin responsiveness of glucose transporter 4 in 3T3-L1 cells depends on the presence of sortilin. Mol Biol Cell , 3115–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Czech MP. (2007). The GLUT4 glucose transporter. Cell Metab , 237–252. [DOI] [PubMed] [Google Scholar]
- Jedrychowski MP, Gartner CA, Gygi SP, Zhou L, Herz J, Kandror KV, Pilch PF. (2010). Proteomic analysis of GLUT4 storage vesicles reveals LRP1 to be an important vesicle component and target of insulin signaling. J Biol Chem , 104–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordens I, Molle D, Xiong W, Keller SR, McGraw TE. (2010). IRAP is a key regulator of GLUT4 Trafficking by controlling the sorting of GLUT4 from endosomes to specialized insulin-regulated vesicles. Mol Biol Cell , 2034–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaddai V, Jager J, Gonzalez T, Najem-Lendom R, Bonnafous S, Tran A, Le Marchand-Brustel Y, Gual P, Tanti JF, Cormont M. (2009). Involvement of TNF-alpha in abnormal adipocyte and muscle sortilin expression in obese mice and humans. Diabetologia , 932–940. [DOI] [PubMed] [Google Scholar]
- Kandror KV, Pilch PF. (2011). The sugar is sIRVed: sorting Glut4 and its fellow travelers. Traffic , 665–671. [DOI] [PubMed] [Google Scholar]
- Kandror KV, Yu L., Pilch PF. (1994). The major protein of GLUT4-containing vesicles, gp160, has aminopeptidase activity. J Biol Chem , 30777–30780. [PubMed] [Google Scholar]
- Keller MP, Choi Y, Wang P, Davis DB, Rabaglia ME, Oler AT, Stapleton DS, Argmann C, Schueler KL, Edwards S, et al. (2008). A gene expression network model of type 2 diabetes links cell cycle regulation in islets with diabetes susceptibility. Genome Res , 706–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller SR. (2003). The insulin-regulated aminopeptidase: a companion and regulator of GLUT4. Front Biosci , s410–s420. [DOI] [PubMed] [Google Scholar]
- Keller SR, Davis AC, Clairmont KB. (2002). Mice deficient in the insulin-regulated membrane aminopeptidase show substantial decreases in glucose transporter GLUT4 Levels but maintain normal glucose homeostasis. J Biol Chem , 17677–17686. [DOI] [PubMed] [Google Scholar]
- Keller SR, Scott HM, Mastick CC, Aebersold R, Lienhard G. (1995). Cloning and characterization of a novel insulin-regulated membrane aminopeptidase from GLUT4 vesicles. J Biol Chem , 23612–23618. [DOI] [PubMed] [Google Scholar]
- Kupriyanova TA, Kandror V, Kandror KV. (2002). Isolation and characterization of the two major intracellular Glut4 storage compartments. J Biol Chem , 9133–9138. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature , 680–685. [DOI] [PubMed] [Google Scholar]
- Larance M, Ramm G, Stockli J, van Dam EM, Winata S, Wasinger V, Simpson F, Graham M, Junutula JR, Guilhaus M, James DE. (2005). Characterization of the role of the Rab GTPase-activating protein AS160 in insulin-regulated GLUT4 trafficking. J Biol Chem , 37803–37813. [DOI] [PubMed] [Google Scholar]
- Liao H, Keller SR, Castle JD. (2006). Insulin-regulated aminopeptidase marks an antigen-stimulated recycling compartment in mast cells. Traffic , 155–167. [DOI] [PubMed] [Google Scholar]
- Mafakheri S, Florke RR, Kanngiesser S, Hartwig S, Espelage L, De Wendt C, Schonberger T, Hamker N, Lehr S, Chadt A, Al-Hasani H. (2018). AKT and AMP-activated protein kinase regulate TBC1D1 through phosphorylation and its interaction with the cytosolic tail of insulin-regulated aminopeptidase IRAP. J Biol Chem , 17853–17862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maianu L, Keller SR, Garvey WT. (2001). Adipocytes exhibit abnormal subcellular distribution and translocation of vesicles containing glucose transporter 4 and insulin-regulated aminopeptidase in type 2 diabetes mellitus: implications regarding defects in vesicle trafficking. J Clin Endocrinol Metab , 5450–5456. [DOI] [PubMed] [Google Scholar]
- Mari M, Bujny MV, Zeuschner D, Geerts WJ, Griffith J, Petersen CM, Cullen PJ, Klumperman J, Geuze HJ. (2008). SNX1 defines an early endosomal recycling exit for sortilin and mannose 6-phosphate receptors. Traffic , 380–393. [DOI] [PubMed] [Google Scholar]
- Pan X, Zaarur N, Singh M, Morin P, Kandror KV. (2017). Sortilin and retromer mediate retrograde transport of Glut4 in 3T3-L1 adipocytes. Mol Biol Cell , 1667–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck GR, Ye S, Pham V, Fernando RN, Macaulay SL, Chai SY, Albiston AL. (2006). Interaction of the Akt substrate, AS160, with the glucose transporter 4 vesicle marker protein, insulin-regulated aminopeptidase. Mol Endocrinol , 2576–2583. [DOI] [PubMed] [Google Scholar]
- Rogi T, Tsujimoto M, Nakazato H, Mizutani S, Tomoda Y. (1996). Human placental leucine aminopeptidase/oxytocinase. A new member of type II membrane-spanning zinc metalloprotease family. J Biol Chem , 56–61. [DOI] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F. (2014). Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods , 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd PR, Kahn BB. (1999). Glucose transporters and insulin action. Implications for insulin resistance and diabetes mellitus. N Engl J Med , 248–257. [DOI] [PubMed] [Google Scholar]
- Shewan AM, van Dam EM, Martin S, Luen TB, Hong W, Bryant NJ, James DE. (2003). GLUT4 recycles via a trans-Golgi network (TGN) subdomain enriched in Syntaxins 6 and 16 but not TGN38: involvement of an acidic targeting motif. Mol Biol Cell , 973–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Huang G, Kandror KV. (2008). Self-assembly of Glut4 storage vesicles during differentiation of 3T3-L1 adipocytes. J Biol Chem , 30311–30321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Kandror KV. (2005). Sortilin is essential and sufficient for the formation of Glut4-storage vesicles in 3T3-L1 adipocytes. Dev Cell , 99–108. [DOI] [PubMed] [Google Scholar]
- Slot SW, Geuze HJ, Gigengack S, Lienhard GE, James DE. (1991). Immuno-localization of the insulin-regulatable glucose transporter in brown adipose tissue of the rat. J Cell Biol , 123–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoidis G, Kandror KV. (2001). A Glut4-vesicle marker protein, insulin-responsive aminopeptidase, is localized in a novel vesicular compartment in PC12 cells. Traffic , 577–587. [DOI] [PubMed] [Google Scholar]
- Waters SB, D’Auria M, Martin SS, Nguyen C, Kozma LM, Luskey KL. (1997). The amino terminus of insulin-responsive aminopeptidase causes Glut4 translocation in 3T3-L1 adipocytes. J Biol Chem , 23323–23327. [DOI] [PubMed] [Google Scholar]
- Yeh TY, Sbodio JI, Tsun ZY, Luo B, Chi NW. (2007). Insulin-stimulated exocytosis of GLUT4 is enhanced by IRAP and its partner tankyrase. Biochem J , 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.