Abstract
Blood vessel formation requires endothelial cell (EC) migration that depends on dynamic remodeling of the cytoskeleton. Rudhira/Breast Carcinoma Amplified Sequence 3 (BCAS3) is a cytoskeletal protein essential for EC migration and sprouting angiogenesis during mouse development and is implicated in metastatic disease. Here, we report that Rudhira mediates cytoskeleton organization and dynamics during EC migration. Rudhira binds to both microtubules (MTs) and vimentin intermediate filaments (IFs) and stabilizes MTs. Rudhira depletion impairs cytoskeletal cross-talk, MT stability, and hence focal adhesion disassembly. The BCAS3 domain of Rudhira is necessary and sufficient for MT-IF cross-linking and cell migration. Pharmacologically restoring MT stability rescues gross cytoskeleton organization and angiogenic sprouting in Rudhira-depleted cells. Our study identifies the novel and essential role of Rudhira in cytoskeletal cross-talk and assigns function to the conserved BCAS3 domain. Targeting Rudhira could allow tissue-restricted cytoskeleton modulation to control cell migration and angiogenesis in development and disease.
INTRODUCTION
Cell migration in physiological or pathological contexts depends on coordinated changes in the cytoskeleton and cell-matrix adhesions. Directed endothelial cell (EC) migration is an important prerequisite for developmental as well as pathological angiogenesis. ECs respond to molecular or mechanical cues in the dynamically changing microenvironment as they move to target tissues for sprouting and angiogenic remodeling. Whereas the fundamental cytoskeletal machinery operates in ECs, few EC-specific cytoskeletal modulators are known. Perturbing the cytoskeleton results in a dramatic loss of EC function. For example, noncentrosomal microtubules (MTs) and vimentin intermediate filaments (IFs) have recently been shown to have an indispensable role in sprouting angiogenesis (Dave and Bayless, 2014; Martin et al., 2018). Further, disruption of either plus or minus ends of MTs can inhibit MT-actin cross-talk, adhesion dynamics, and thereby EC sprouting (Bayless and Johnson, 2011). Regulation of cytoskeletal interactions is likely to be important in developmental as well as tumor angiogenesis.
Whereas MTs and IFs can interact directly, several molecules are known to bridge cytoskeletal components. Cytolinkers of the plakin family are well characterized and connect MTs, IFs, actin filaments, and plasma membrane components. The importance of cytoskeletal cross-talk is evident from the early postnatal lethality of mice lacking the prototype cytolinker plectin (Andrä et al., 1997). Many ubiquitously expressed molecules, such as the MT motor kinesin and tumor suppressor APC, are critical for MT-IF cross-linking in fibroblasts and migrating astrocytes, respectively (Gyoeva and Gelfand, 1991; Sakamoto et al., 2013). Recent elegant studies show that whereas MTs are essential for vimentin IF assembly, vimentin IFs provide memory for MT cytoskeleton regrowth, highlighting the significance of the cross-talk and the positive feedback interaction between these two cytoskeletal components (Gan, Ding, Burckhardt, et al., 2016; Leduc and Etienne-Manneville, 2017).
Cytoskeletal components play a critical role in establishing cell polarity, force generation, and regulation of adhesion complex components during cell migration. Close and intricate interactions among actin filaments, MTs, IFs, and cytoskeleton-associated proteins bring about dynamic reorganization of cell shape and focal adhesions (FAs), essential for directed cell migration. Association with vimentin IFs stabilizes MTs against depolymerizing stresses and shrinkage, likely owing to the 10-fold slower turnover rate of vimentin IFs (Gan, Ding, Burckhardt, et al., 2016). In addition, MTs have been proposed to grow along vimentin IFs, although the bridging components in this process remain elusive (Gan, Ding, Burckhardt, et al., 2016). The physiological significance of this interaction is also unclear, in part owing to the full-term survival of the vimentin knockout mouse and the likely redundancy in IF functions (Colucci-Guyon et al., 1994). Several cytoskeleton-associated molecules, such as the MT plus-end tracking proteins (+TIPs) end binding protein (EB1) and CLIP170 and spectraplackin family member ACF7, cross-bridge MTs and actin and guide MT growth to FAs for FA turnover and persistent migration (Stehbens and Wittmann, 2012).
Depletion of individual cytoskeleton components, associated proteins, or cytolinkers often disrupts overall cytoskeleton architecture and dynamics, perturbing cell migration and adhesion. Multiple and/or redundant roles of the molecules involved as well as context-dependent responses make it challenging to decipher global and tissue-specific mechanisms that regulate this process. Identifying additional molecular components could help unravel mechanisms to control or promote cell migration in desired contexts.
Rudhira/BCAS3 (Breast Carcinoma Amplified Sequence 3) is a cytoskeletal protein essential for mouse developmental angiogenesis and implicated in tumor metastasis (Siva et al., 2007; Jain et al., 2012; Shetty, Joshi, et al., 2018). Rudhira binds to MTs and IFs and promotes directional cell migration (Jain et al., 2012). In this study, we examine the mechanism by which Rudhira controls cytoskeletal remodeling during cell migration. We show that Rudhira directly associates with MTs and IFs for MT-IF cross-talk, MT stability and dynamics, and thereby FA turnover and cell migration, through its conserved BCAS3 domain. Our study provides new insights into the mechanism of cytoskeletal cross-linking and reorganization during cell migration, which will help us understand physiological and pathological angiogenesis.
RESULTS
Rudhira is required for gross cytoskeletal organization
We reported earlier that Rudhira/BCAS3 interacts with MTs and IFs and is required for actin reorganization for directional EC migration. Rudhira depletion deregulates several cellular and molecular processes critical for sprouting angiogenesis in vitro and in vivo, including cell adhesion and invasion (Shetty, Joshi, et al., 2018). To explore the possible mechanisms by which Rudhira functions in cell migration, we examined the effect of Rudhira depletion (knockdown, KD) on cytoskeletal organization as compared with the nonsilencing control (NS) in the mouse saphenous vein EC line (SVEC). Immunolocalization showed that unlike in the control, in KD cells MTs were not aligned toward and appeared bent at the cell periphery, whereas vimentin IFs were fewer, occupied less area within the cells, and were not extended but present only in the perinuclear region (Figure 1A). KD cells also had thick actin bundles at the cell cortex (Figure 1B). NS and KD cells expressing vimentin-green fluorescent protein (GFP) also showed similar results (Supplemental Figure S1, A and B). Vimentin IF organization depends on actin organization and MT-dependent transport (Gan, Ding, Burckhardt, et al., 2016; Leduc and Etienne-Manneville, 2017). Depolymerization of actin stress fibers using 10 µM cytochalasin D for 20 min in KD cells resulted in reorganized and extended vimentin IFs, which appeared comparable to those in the control (NS) cells (Supplemental Figure S1, C and D). These data suggest that thick cortical bundles of actin in KD cells may lead to the defective vimentin IF organization. Protein levels of the cytoskeletal components were, however, unaltered (Figure 1C). Overall, these data show that Rudhira depletion results in a grossly disorganized cytoskeleton.
FIGURE 1:
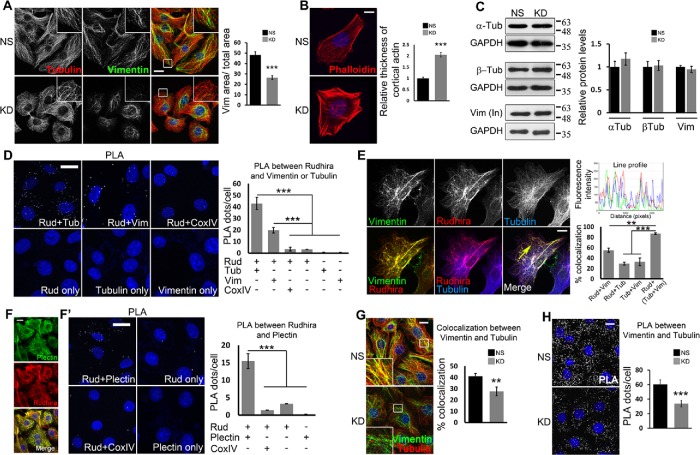
Rudhira interacts with and controls cross-talk between MTs and IFs. (A, B) NS and KD cells were costained for cytoskeleton markers, tubulin and vimentin (A) or phalloidin (B) to detect gross cytoskeleton organization. Boxed regions are magnified in the insets. The graph in A shows the ratio of vimentin IF area to total cell area (11 cells), and the graph in B shows the relative thickness of cortical actin (25 cells). (C) Immunoblots detect the levels of cytoskeletal proteins. Graphs show the quantitation of relative protein levels. (D) Direct interaction between Rudhira and tubulin or vimentin in wild-type SVECs analyzed by PLA. Cells stained with a single antibody or the Rudhira+CoxIV combination were taken as negative controls. The graph shows the quantitation of PLA dots per cell indicating the extent of interaction. (E) Relative localization of vimentin IFs, Rudhira, and MTs was performed by triple immunostaining in wild-type SVECs. The line profile shows the fluorescence intensity peaks for the three colors along the yellow arrow. The graph shows the percent overlap of Rudhira with vimentin or tubulin or vimentin+tubulin pattern, as indicated (15 cells). (F, F’) Relative association of Rudhira with the cytolinker plectin by immunofluorescence (F) or PLA (F’). Cells stained with a single antibody or Rudhira+CoxIV combination were taken as negative controls. The graph shows the quantitation of PLA dots per cell indicating the extent of interaction. (G) NS or KD cells were analyzed for coalignment of vimentin and MTs by coimmunofluorescence. Boxed regions are magnified in the insets. The graph shows the percent overlap between vimentin and tubulin patterns (30 cells). (H) Vimentin and MT association by PLA. The graph shows the quantitation of PLA dots per cell indicating extent of interaction. Error bars indicate SEM. Results shown are a representative of at least three independent experiments with at least three biological replicates. Statistical analysis was carried out using one-way ANOVA. Scale bar: (A, D, F, F’, G, H) 10 μm, (B, E) 20 μm. **p < 0.01, ***p < 0.001.
Rudhira directly interacts with and bridges IFs and MTs
The intricate association of cytoskeletal components is dynamically regulated during cell migration. MTs and vimentin IFs are coaligned in mesenchymal cells for efficient migration. While initially vimentin IFs form along MTs, later these filaments provide a template for MT growth (Gan, Ding, Burckhardt, et al., 2016). Further, IFs organize primarily by MT-dependent transport and actin-dependent flow, suggestive of extensive cytoskeletal cross-talk and cross-regulation during cell migration (Leduc and Etienne-Manneville, 2017). Rudhira interacts with tubulin and vimentin and localizes to MTs and IFs. To test the likely direct interaction of Rudhira with MTs and IFs in vivo, we used proximity ligation assay (PLA), which detects interaction at single-molecule resolution. Rudhira associated with both tubulin and vimentin, suggesting direct interactions of Rudhira with these cytoskeleton components within the cells (Figure 1D). PLA between Rudhira and the mitochondrial protein CoxIV was used as a negative control in addition to single antibody controls (Figure 1D). In addition, triple immunofluorescence analysis showed that Rudhira associates with MTs at sites often overlapping with vimentin Ifs, suggesting that these interactions may be regulated by local factors or tubulin or vimentin properties (Figure 1E, line profile and graph). These data indicate that Rudhira may simultaneously associate with MTs and IFs, and IF association of Rudhira may favor its binding to MTs.
Cytoskeletal components are often connected by cytolinker proteins. Plectin is the most studied cytolinker that connects several cytoskeletal components to each other and to the cell membrane. Recent reports show that plectin links MTs and IFs during directed cell migration (De Pascalis et al., 2018). To characterize the cytoskeletal cross-linking property of Rudhira, we tested its association with plectin. In agreement with its proposed role of bridging cytoskeletal components, we observed high overlap of Rudhira with plectin (Figure 1F). PLA between Rudhira and plectin confirmed their interaction and suggested that they may have similar function in vivo (Figure 1F′). PLA between Rudhira and the mitochondrial protein CoxIV was used as a negative control. Expectedly, MT-IF association in vivo was significantly reduced in Rudhira-depleted cells, as detected by double immunofluorescence (Figure 1G) and confirmed by PLA (Figure 1H).
The loss of plectin phenocopies IF depletion, which, interestingly, is similar to the loss of Rudhira (De Pascalis et al., 2018). Hence, we further tested whether Rudhira and plectin are dependent on each other for their localization. Whereas rudhira KD resulted in a loss of filamentous pattern of plectin (Supplemental Figure S2A), short hairpin RNA (shRNA)-mediated KD of plectin did not grossly affect Rudhira localization and pattern (Supplemental Figure S2, B and C). This is in concordance with our earlier data, which show that Rudhira organization is maintained even when one of the cytoskeletal components, MTs or vimentin IFs, is intact (Jain et al., 2012). Plectin is absent in the cell peripheral regions of KD cells (Supplemental Figure S2A), which may contribute to defects in IF and MT organization and cross-linking at the cell cortex. Rudhira overexpression did not affect tubulin-vimentin colocalization (Supplemental Figure S2D). Consistent with its cytoskeletal cross-linking function, plectin depletion resulted in reduced MT-vimentin IF colocalization (Supplemental Figure S2E). Rudhira overexpression on plectin depletion partially restored MT-vimentin IF association, suggesting that the two function similarly (Supplemental Figure S2E). Combined, these data show that Rudhira is required for gross cytoskeletal organization and is critical for MT-IF bridging in ECs.
Rudhira governs the association and dynamics of MTs and vimentin IFs in ECs
The cross-talk between IFs and MTs is essential for efficient EC migration (Nieuwenhuizen, Nahidiazar, et al., 2015; Gan, Ding, Burckhardt, et al., 2016). Hence, we tested for the association of IFs and MTs in live cells in low density cultures, where cells are in a migratory state. Live imaging of MTs using silico-rhodamine–conjugated docetaxel (SiR-tubulin) showed that MTs grew radially toward the cell periphery and were stabilized there in control cells (NS), whereas KD had fewer MTs at the cell periphery and they often started to bend before reaching the periphery (red asterisk in Supplemental Figure S3A and Supplemental Video S1). To test MT-IF cross-talk in live cells, we transiently expressed vimentin-GFP and incubated cells with SiR-tubulin. vimentin-GFP filaments were less extended in KD cells, resulting in reduced alignment with MTs and perturbed dynamics (Figure 2A and Supplemental Video S2). These data suggest that Rudhira is required for cytoskeletal cross-talk and organization for cell migration. Rudhira may also have a role in promoting the assembly of or stabilizing IFs.
FIGURE 2:
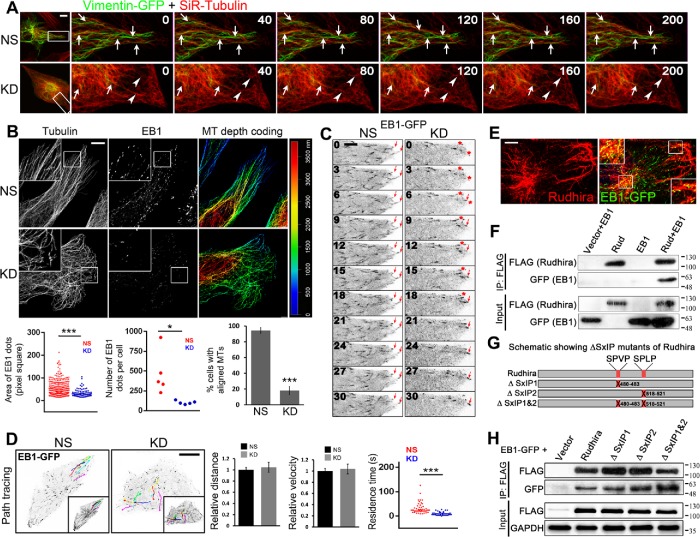
Rudhira is required for MT-vimentin IF association and dynamics in migrating ECs. (A) Time-lapse images of SVEC NS and KD cells transiently transfected with vimentin-GFP and stained with SiR-tubulin, imaged at 10-s intervals. Arrows indicate a persistence of coaligned vimentin IFs and MTs toward the cell periphery, whereas arrowheads indicate the absence of vimentin IFs and bent MTs before reaching the cell periphery in live migrating cells. (B) Superresolution imaging after immunostaining NS and KD cells for β-tubulin and EB1 to detect cell peripheral MT architecture and growing MTs. The panel to the right shows depth coding to detect MTs at cell periphery, the sites near cell-matrix contacts. The graphs show the area of EB1 dots (2329 EB1 dots for NS and 514 EB1 dots for KD over five images each), the number of EB1 dots per cell (five images each), and the percentage of cells with aligned MTs (40 cells for NS and 46 cells for KD). Boxed regions are magnified in the insets. (C) Time-lapse images of SVEC NS and KD cells transiently transfected with EB1-GFP and imaged at 3-s intervals. The arrows indicate persistence of aligned EB1 positive MT growing end in NS and not in KD, whereas the red asterisk indicates a MT end not stabilized at the cell periphery. Twenty live cells each of NS and KD were imaged. (D) Overlay of EB1-GFP tracks and their time-projection (insets) in NS and KD. Time-lapse images of EB1-GFP comets were analyzed manually for calculating relative distance and average velocity (10 EB1-GFP comets from three cells) and residence-time at the cell periphery (50 EB1-GFP comets from five cells) shown in the graphs. (E) SVEC cells transiently transfected with EB1-GFP were counterstained with Rudhira to detect the overlap between the two. (F) HEK293T cells cotransfected with FLAG-tagged Rudhira and EB1-GFP were analyzed for interaction by coimmunoprecipitation with FLAG antibody. (G) Schematic showing the positions and deletions of SxIP-like motifs (marked in red) in Rudhira protein. (H) HEK293T cells cotransfected with FLAG-tagged Rudhira SxIP deletion mutants and EB1-GFP were analyzed for an interaction by coimmunoprecipitation with FLAG antibody. Error bars indicate SEM. Results shown are a representative of at least three independent experiments with at least three biological replicates taken into account. Statistical analysis was carried out using one-way ANOVA. Scale bar: (A, D) 10 μm, (B) 5 μm, (C) 1 μm, (E) 10 μm. *p < 0.05, ***p < 0.001.
Movie S1.
Loss of Rudhira results in unaligned MTs and MT bending before reaching cell periphery (related to Figure 2). Time-lapse images of SVEC NS and KD incubated with SiR-Tubulin monitored for 2 minutes, imaged at 4 second intervals (6 fps). Arrow indicates persistence of radial MT at cell periphery, while red asterisk indicates a MT bending away from the cell periphery. Green asterisk shows a MT travelling parallel to the cell periphery.
Movie S2.
Rudhira bridges microtubules and intermediate filaments for cell migration (related to Figure 2). Time-lapse images of SVEC NS and KD transiently expressing Vimentin-GFP incubated with SiRTubulin monitored for 4 minutes, imaged at 10 second intervals (6 fps). Arrow indicates persistent MT-IF coalignment and their similar dynamics, while arrowhead indicates absence of peripheral IFs and perturbed MT-IF association and dynamics in live migrating cells.
The MT cytoskeleton is a highly dynamic macromolecular assembly, with a turnover rate of 5–15 min. MTs govern cell polarity and along with actin also control IF organization during migration. Controlled MT dynamics and stability contribute to cell migration by the release of cell-ECM contacts and polarized asymmetric distribution of vesicles. Consistent with the earlier observation (Figure 1A), superresolution microscopy of KD cells showed defective MT arrays, with MTs often failing to reach the cell periphery as observed by depth-coding of MTs (Figure 2B). Further, MTs in KD cells seemed to cross over each other, indicating undirected growth, unlike in control cells which displayed aligned MTs near the periphery. In addition, KD cells showed reduced costaining for the +TIP, EB1, indicative of fewer growing MTs (Figure 2B).
To test whether impaired migration of KD cells is due to defects in MT growth, we assessed EB1-GFP transfected control and KD cells by live imaging. Rudhira-depleted cells had fewer EB1–GFP-positive MTs (Figure 2C and Supplemental Video S3). The +TIPs bind to and stabilize MTs at FAs for a time period of more than 15 s. The EB1 comets in KD cells appeared to be smaller in size and shorter-lived than those in control cells (Figure 2, B–D, and Supplemental Video S3). Time-projected (for 60 s) images (see Materials and Methods) also showed that MT growth in KD cells followed a crisscross pattern toward the cell periphery, as compared with the straight linear growth of MTs in control, as judged by EB1-GFP movement in live cells (Figure 2, C and D, and Supplemental Video S4). Unlike MTs in controls, which grew radially toward and were stabilized at the periphery, MTs in KD cells were rarely stabilized and often started to bend before reaching the cell periphery (Figure 2D and Supplemental Videos S3 and S4). This suggests that MTs in KD cells encounter a physical constraint, likely thick actin stress fibers (Figure 1B), which may prevent their growth to the periphery, the site of cell-matrix adhesions. However, the distance traveled or the average velocity of EB1-GFP comets were not significantly affected (Figure 2D).
Movie S3.
Rudhira regulates microtubule growth and alignment towards cell periphery (related to Figure 2). Time-lapse images of the cell peripheral region of SVEC NS and KD transiently transfected with EB1-GFP monitored for 2-3 minutes imaged at 3-4 second intervals (6 fps). Fps: frames per second. Arrows indicate persistence of aligned EB1 positive MT growing end in NS and not in KD while asterisks indicate a MT end not stabilized at the cell periphery. Also note the shorter EB1-GFP comets in KD as compared to NS.
Movie S4.
Rudhira regulates microtubule growth towards cell periphery (related to Figure 2). Timelapse images of SVEC NS and KD transiently transfected with EB1-GFP monitored for 1-2 minutes imaged at 3 second intervals and manually tracked EB1-GFP positive comets (6 fps).
EB1 is a regulator of MT dynamics (Akhmanova and Steinmetz, 2010). Interestingly, a recent study had identified Rudhira as an EB1-binding protein in a high-throughput screen of SxIP/SxIP-like motif containing proteins; however, this putative interaction was not validated biochemically (Jiang et al., 2012). We transiently expressed EB1-GFP in SVEC cells and by immunofluorescence observed partial overlap with Rudhira. However, Rudhira appeared filamentous and was not restricted to EB1 comets, which mark MT+ ends (Figure 2E). This is in agreement with our earlier study where we found that Rudhira localizes along the length of MTs (Jain et al., 2012). Coimmunoprecipitation in HEK293T coexpressing Rudhira-FLAG and EB1-GFP confirmed Rudhira and EB1 interaction (Figure 2F). However, it is possible that Rudhira is able to coimmunoprecipitate EB1 through an indirect interaction. Rudhira harbors two SxIP-like motifs, implicated in mediating EB1 interaction, in the C-terminal region (amino acid residues 480–483 and 518–521) (Figure 2G) (Jiang et al., 2012). However, deleting either (∆SxIP1 or ∆SxIP2) or both of these motifs (∆SxIP1&2) did not abolish its interaction with EB1 (Figure 2, G and H), suggesting that SxIP-like motifs of Rudhira are dispensable for this function. Rudhira may, however, interact with EB1 through other motifs/domains which remain unknown. These data show that Rudhira interacts with EB1 and may thereby also control MT dynamics and organization, essential for cell migration.
Rudhira associates with and stabilizes MTs
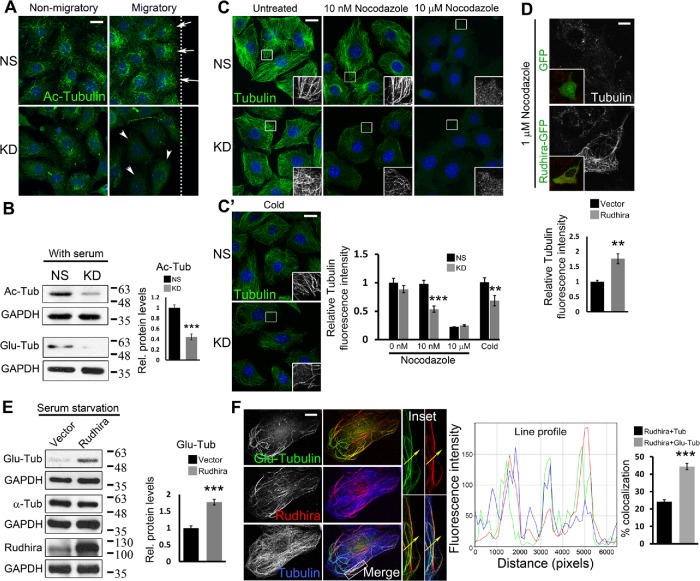
The differential association and dissociation of MT-associated proteins (MAPs) and cytoskeletal components modulate MT stability, essential for cell migration. α-tubulin acetylation or detyrosination (Glu) classically marks stable MTs (Bulinski and Gundersen, 1991). Acetylation also provides mechanical resistance to breakage (Xu et al., 2017). The cytoskeleton in KD cells was grossly disorganized, and our data indicated a role of Rudhira in MT and vimentin IF cross-talk. Because vimentin preferentially associates with stable MTs (Gurland and Gundersen, 1995) and functions to stabilize MTs, we tested whether binding of Rudhira contributes to MT stability. Stable MTs are oriented toward the leading edge of a migrating cell and because of their higher affinity for MT motors in vitro are considered to maintain directional migration by polarized delivery of vesicles (Witte and Bradke, 2008). Immunolocalization for acetylated (Ac) tubulin in a scratched EC monolayer 2 h after wounding showed that compared with control, KD cells had fewer stable MTs that did not localize to the leading edge (Figure 3A). Immunoblot also showed significant decrease in Ac- and Glu- α-tubulin levels, indicating that Rudhira depletion destabilized MTs (Figure 3B). This was confirmed with MT-depolymerizing drug and cold treatment. As compared with controls, MTs in Rudhira-depleted cells were more sensitive to both MT depolymerization stresses (Figure 3, C and C′). Nocodazole (10 µM) caused complete MT depolymerization in both control and KD cells. However, low concentrations of nocodazole (4–400 nM) depolymerize dynamic but not stable MTs in a dose-dependent manner. KD cells were more sensitive to 10 nM nocodazole and showed a dramatic reduction in MT number as compared with controls, which showed a well-organized MT-array with little apparent reduction in MT numbers (Figure 3C).
FIGURE 3:
Rudhira is essential for MT stability. (A) Levels and localization of Ac MTs were analyzed in NS and KD cells by immunostaining a scratch-wound healing assay. Arrows point to Ac MTs toward the leading edge and arrowheads to Ac MTs distributed all over the cell. The dotted line represents the wound margin. Migratory, adjacent to the wound; Nonmigratory, away from the wound. (B) Ac-tubulin, Glu-tubulin levels were analyzed by immunoblot. The graph shows the quantitation of relative Ac-tubulin levels. (C, C’) NS and KD cells were treated with the indicated dosages of nocodazole for 30 min (C) or cold PBS at 4°C (C’), and MTs were analyzed by immunostaining for β-tubulin. Boxed regions (C, C’) are magnified in the insets. The graph shows the quantitation of relative tubulin fluorescence intensity. (D) SVEC cells were transiently transfected with GFP or Rudhira-GFP (inset) and treated with nocodazole, and MTs were analyzed by immunostaining for tubulin. The graph shows the quantitation of relative tubulin fluorescence intensity (five transfected cells). (E) Immunoblot analysis for Glu-tubulin levels post-48 h serum starvation in HEK293 cells overexpressing Rudhira. The graph shows the quantitation of relative Glu-tubulin levels. (F) Relative localization of detyrosinated MTs (Glu-tubulin), Rudhira, and total MTs (tubulin) was performed by triple immunostaining in wild-type SVECs. The line profile shows the fluorescence intensity peaks for the three colors along the yellow arrow in the inset (magnified boxed region). The graph shows the percent overlap of Rudhira with tubulin or Glu-tubulin. Error bars indicate SEM. Results shown are a representative of three independent experiments. Statistical analysis was carried out using one-way ANOVA. Scale bar: (A, C, C’) 20 μm, (D) 5 μm, (F) 10 μm. **p < 0.01, ***p < 0.001.
Rudhira overexpression in HeLa cells did not affect MT growth dynamics significantly (Supplemental Figure S3B). Distance traveled and the average velocity of EB1-GFP comets were also unaltered (Supplemental Figure S3B). However, treatment of cells that overexpress Rudhira with MT-depolymerizing doses of nocodazole (1 µM) showed that their MTs are nocodazole-resistant, as compared with control, where most MTs were depolymerized (Figure 3D; Supplemental Figure S3C). Further, Glu-tubulin levels were increased (Figure 3E) and the stable MTs were often associated with Rudhira as seen by immunolocalization (Supplemental Figure S3C). Triple immunofluorescence analysis showed that Rudhira had a preferential association with detyrosinated MTs (Figure 3F, line profile and graph). Thus, like vimentin IFs, Rudhira binds to and stabilizes MTs and promotes MT-IF association, likely leading to MT stability.
Rudhira-depleted cells have large FAs
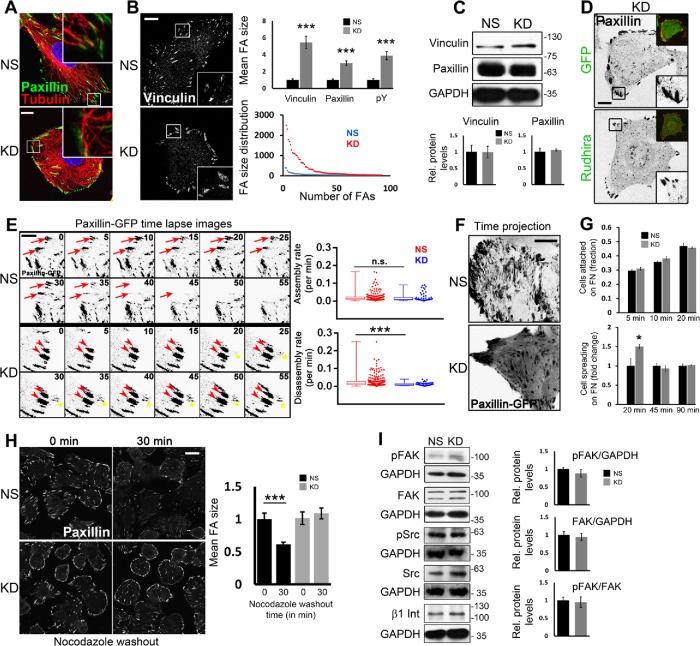
MT dynamics and stability have been well studied in the context of cell migration. Cells adhere to the extracellular matrix (ECM) ligands via FAs assembled on the cell-peripheral ends of actin stress fibers. MT and F-actin recruitment is essential for FA organization and dynamics (Kodama, Karakesisoglou, et al., 2003). Whereas FA assembly is actin-driven, disassembly requires their interaction with MTs and subsequent internalization, resulting in contact dissociation from the ECM. Vimentin IFs have also been shown to directly associate with FA molecule vinculin and control its localization. Additionally, recent reports show that the cytolinker plectin regulates FA dynamics by coupling IFs to FAs (De Pascalis et al., 2018). However, MT targeting of FAs is essential for FA disassembly and thereby cell migration. Expectedly, the bent and unaligned MTs in KD cells were unable to reach FAs as compared with the controls, which efficiently targeted FAs radially (Figure 4A). Immunolocalization of the FA molecules vinculin and paxillin revealed a dramatic increase in the size and reduction in number of FAs on Rudhira depletion as compared with control, suggesting impaired FA dynamics (Figure 4B; Supplemental Figure S4A). This was confirmed by staining the cells with a phospho-tyrosine (pY) antibody because FA proteins are highly tyrosine-phosphorylated (Supplemental Figure S4B). Further, immunoblotting showed that the levels of vinculin and paxillin were not significantly altered (Figure 4C). Conversely, Rudhira overexpression increases migration rate (Jain et al., 2012), and as expected, immunolocalization analysis showed a significant decrease in FA size in Rudhira-overexpressing cells (Rudh2AGFP) as compared with the untransfected or vector controls (Supplemental Figure S4C). Further, transient overexpression of Rudhira in KD cells rescued the FA size phenotype (Figure 4D). Double immunolocalization showed that Rudhira does not colocalize with Paxillin or pY (Supplemental Figure S4, D and D′), suggesting that Rudhira controls cytoskeletal organization and dynamics resulting in modulated downstream FA dynamics and cell migration. Therefore, we hypothesized that Rudhira depletion may regulate the cytoskeleton to increase FA assembly or decrease disassembly or both. Additionally, the loss of filamentous localization of plectin on Rudhira depletion (Supplemental Figure S2A) could also result in the FA phenotype. Expectedly, plectin depletion resulted in increased stress fibers, a phenotype which was already reported in addition to the enlarged FAs (Supplemental Figure S4E). Interestingly, Rudhira overexpression in plectin-depleted conditions reversed the FA size phenotype showing that Rudhira can substitute for plectin, likely by mediating MT-IF cross-talk (Supplemental Figure S4F).
FIGURE 4:
MT-mediated FA disassembly is impaired on Rudhira depletion. (A) Immunostaining for tubulin and paxillin to detect the association of cell peripheral MTs with FAs. (B, C) NS or rudhira KD SVECs were analyzed by immunostaining (B) or by immunoblot of cell lysates (C) to detect FAs marked by vinculin, paxillin, and pY, as indicated (also see Supplemental Figure S4, A and B). Graphs in B show the quantitation of FA size, and the size and number distribution for FAs (10, 8, and 6 images for analysis of vinculin, paxillin and pY, respectively), and graphs in C show the quantitation of relative vinculin and paxillin levels. (D) KD cells were transiently transfected with Rudhira-IRES-GFP or IRES-GFP only vector control and analyzed for FA size by immunostaining for paxillin. (E) Time-lapse images of NS and KD cells transiently transfected with paxillin-GFP monitored for 2–3 h and shown at 5-min intervals. Red arrows indicate a FA getting disassembled. Red arrowheads indicate a FA persisting over time and not getting disassembled. Yellow asterisks indicate a FA getting assembled over time. Graphs show the quantitation of FA assembly and disassembly rates computed using FAAS (see Materials and Methods), represented as whisker plots combined with scatter plots to show the distribution of individual FA. Five to seven optical slices were taken at 5-min intervals for 2–3 h. FAAS identified 225 and 240 FA in NS cells for assembly and disassembly, respectively, and 57 and 58 FA in KD cells for assembly and disassembly, respectively. Eight to ten live cells each for NS and KD were imaged and analyzed. (F) Time-projected images of NS and KD cells imaged live after transient transfection with paxillin-GFP and projected over 2 h to show the dynamics of FAs. (G) Quantitation of attachment and spreading profiles of cells on fibronectin with time, as indicated. (H) Recovery after nocodazole treatment and immunostaining of fixed cells to detect FAs marked by paxillin. The graph shows the quantitation of FA size. Boxed regions in panels A, B, and D are magnified in the insets. Inset (D, top) also shows transfected cell expressing GFP. (I) Immunoblot of NS or KD cell lysates to detect the levels of FA signaling proteins Src, pSrc, FAK, pFAK, and β1 Integrin. Graphs show the quantitation of relative FAK and pFAK levels. Error bars indicate SEM. Results shown are a representative of at least three independent experiments. Statistical analysis was carried out using one-way ANOVA. Scale bar: (A, B, F, H) 10 μm, (D) 20 μm, (E) 1 μm. *p < 0.05, ***p < 0.001.
Rudhira depletion impairs MT-dependent FA disassembly
Directional cell migration requires continuous coordinated removal and formation (turnover) of FAs at the leading edge and release of attachment at the rear. Defects in the process of FA assembly or disassembly are both detrimental to cell migration. We examined the steady state dynamics of FAs in control and KD cells transiently transfected with Paxillin-GFP using time-lapse live imaging (Figure 4, E and F, and Supplemental Video S5). Our observations and analysis of the time-lapse images by the FA Analysis Server (FAAS; see Materials and Methods) (Berginski and Gomez, 2013) showed that FA assembly was not affected by Rudhira depletion (0.0219 min-1 in NS and 0.0213 min-1 in KD), whereas disassembly was reduced to half of that in the control cells (0.0269 min-1 in NS and 0.0129 min-1 in KD) (Figure 4E and Supplemental Video S5). Time-projection of the live images also showed highly dynamic FAs in control cells, whereas KD FAs appeared to be immobile (Figure 4F and Supplemental Video S5).
Movie S5.
Rudhira regulates focal adhesion disassembly (related to Figure 4). Time-lapse images of SVEC NS and KD transiently transfected with Paxillin-GFP monitored for 2-3 hours imaged at 5-minute intervals (4 fps). Arrow indicates a FA getting disassembled, while arrowhead indicates a FA persisting over time and not getting disassembled.
To confirm these results, we used specialized molecular and cellular functional assays. Rudhira-depleted cells did not show a significant difference in attachment but spread earlier than controls on fibronectin matrix, indicating that FA assembly is not impaired (Figure 4G). The early initial spreading of rudhira KD cells could be due to the persistence of FAs even after the 20 min in suspension, within which time FAs disassemble in control cells. Treatment with the MT depolymerizing agent, nocodazole, inhibits FA disassembly because MTs are not recruited to FA (Ezratty et al., 2005). Upon nocodazole treatment, whereas control cells showed FA disassembly after 30 min of nocodazole washout, Rudhira KD ECs continued to show large FAs that failed to turn over (Figure 4H and Supplemental Figure S5A). Taken together, these data suggest that Rudhira functions in MT-mediated FA disassembly. It is unlikely, however, that Rudhira is a FA-relaxing molecule, since MTs are dispensable for gross localization of Rudhira (Jain et al., 2012).
FA disassembly requires FA kinase (FAK) phosphorylation, subsequent MT-targeting of FAs and Dynamin2-mediated FA internalization (Ezratty et al., 2005). Immunolocalization and immunoblotting showed that Rudhira depletion does not alter levels of the components involved in FA-mediated signaling, namely FAK, pFAK, and β1 Integrin; Src and pSrc; and the early events of FA disassembly (Figure 4I; Supplemental Figure S5, B and C). These data validate that Rudhira has a primary function at the cytoskeleton, downstream to which it promotes FA turnover and cell migration.
Rudhira-dependent MT stability is essential for cytoskeletal organization
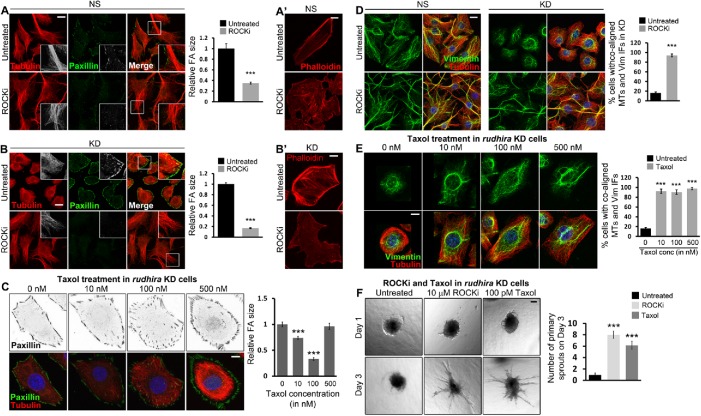
We next asked whether cytoskeletal cross-talk regulating MT stability was the primary mode of action of Rudhira. The Rho GTPase RhoA acts through Rho-associated kinase (ROCK) to disassemble MTs and IFs. Treatment with ROCK inhibitor can restore MT assembly and stability, IF extension, as well as FA dynamics. Expectedly, we observed the loss of FAs and stress fibers and extended MTs in ROCKi-treated control (NS) cells (Figure 5, A inset and A′). Interestingly, rudhira KD cells treated with ROCKi also showed almost complete rescue of phenotypes as observed by the recovery of MT organization and cell-peripheral alignment and reduced FA size (Figure 5B). Further, cortical actin bundles and stress fibers were dramatically reduced (Figure 5B′). MTs did not bend and could reach the cell periphery, possibly because they were not impeded by the thick cortical actin (Figure 5, B inset and B′). These data suggest that the primary function of Rudhira is to provide physiological stability to MTs, and the loss of Rudhira can be compensated for by stabilizing MTs or inhibiting MT disassembly pharmacologically. However, MTs may also reorganize in response to ROCKi-induced cell shape changes, which cannot be ruled out. It is also possible that Rudhira depletion deregulates Rho GTPase effectors, such as mDia and Tau, to affect MT stability indirectly.
FIGURE 5:
Rudhira stabilizes MTs for cytoskeletal organization and angiogenesis. (A, A’, B, B’) Control NS (A, A’) or rudhira KD (B, B’) cells were kept untreated or treated with 10 µM ROCKi and analyzed for FA size and MT organization by coimmunostaining for paxillin and tubulin (A, B) or actin using phalloidin (A’, B’). Graphs in panels A and B show the quantification of relative FA size in ROCKi treated or untreated cells. Boxed regions in panels A and B are magnified in the insets. (C) Rudhira KD cells were treated with different concentrations of Taxol (paclitaxel), as indicated, and analyzed for FA size and MT stabilization by coimmunostaining for paxillin and tubulin. Note the increasing fluorescence intensity of tubulin and bundling (stability) of MTs with the increase in Taxol concentration. The graph shows the quantification of relative FA size in KD cells treated with different Taxol concentrations compared with the untreated. (D) NS or KD cells were kept untreated or treated with ROCKi and analyzed for MT-IF association by coimmunostaining for vimentin and tubulin. The graph shows the percentage of cells with coaligned MTs and IFs. (E) KD cells were treated with a range of Taxol concentrations, as indicated, and analyzed for MT-IF association by coimmunostaining for vimentin and tubulin. The graph shows the percentage of cells with coaligned MTs and IFs in each condition. (F) Spheroids formed from Rudhira KD cells were taken for collagen-based spheroid sprouting assay in the presence or absence of ROCKi or Taxol, as indicated. The graph shows the quantification of the number of primary sprouts formed on day 3 on each treatment. Error bars indicate SEM. Results shown are a representative of at least three independent experiments. Statistical analysis was carried out using one-way ANOVA. Scale bar: (A, B, D) 20 μm, (A’, B’, C, E) 10 μm, (F) 100 μm. ***p < 0.001.
To dissect the effect of Rudhira depletion on MT stability from other properties leading to defective FA turnover, such as cell shape changes, we transiently stabilized MTs in KD cells with paclitaxel and scored for FA size. We observed a dose-dependent decrease in FA size with an increasing concentration of paclitaxel (Taxol, 10–100 nM) (Figure 5C). This suggests that drug-mediated MT stabilization can partially rescue the FA phenotype resulting from loss of Rudhira. However, at higher concentrations of paclitaxel, FA size increased again, possibly due to drastic loss of MT dynamics, which could impede FA turnover. More interestingly, transient treatment with ROCKi led to the reorganization of vimentin IFs and their coassociation with MTs in KD cells, which was comparable to that in NS cells (Figure 5D). Importantly, treatment of KD cells with different concentrations of Taxol (10–500 nM) resulted in the extension of vimentin IFs and their association with MTs (Figure 5E). Taxol binding stabilizes MTs resulting in an accumulation of detyrosinated/Ac MTs, which have a higher affinity for vimentin IFs (Gurland and Gundersen, 1995). Thus, Taxol treatment in KD cells may enhance MT stability or MT detyrosination leading to MT-vimentin IF association. These data indicate that pharmacologically stabilizing MTs while still maintaining their dynamics in Rudhira-depleted cells is sufficient to restore normal cytoskeletal organization. These data suggest that the primary role of Rudhira is to stabilize MTs in vivo, likely by cross-linking MTs and IF components in ECs. Actin fibers are known to push vimentin IFs toward the cell center (Jiu et al., 2015; Leduc and Etienne-Manneville, 2017). The extension of vimentin IFs to the cell periphery post-ROCKi treatment (Supplemental Figure S6, A and B) may also be due to the loss of thick cortical actin fibers, similar to what was observed with Cytochalasin D treatment (Supplemental Figure S1, C and D).
To further identify whether Rudhira affects FA size through its MT-vimentin IF cross-linking property, we tested the effect of Rudhira modulation on FAs and vimentin IFs when MTs are depolymerized and on FAs and MTs when vimentin IFs are aggregated. Compared with the NS controls, the KD cells had larger FAs, reduced vimentin IFs which were not extended to the cell periphery, and disorganized MTs (Supplemental Figure S7A), as seen earlier (Figures 1A and 4B). On MT depolymerization, FA size was increased in NS, as expected, but not changed significantly in KD (Supplemental Figure S7B). Vimentin IF organization in both NS and KD cells was comparable to their untreated counterparts (Supplemental Figure S7, A and B). Similarly, vimentin IF aggregation using either cycloheximide or acrylamide did not alter MT organization or FA size in KD (Supplemental Figure S7, A, C, and D). On the other hand, Rudhira overexpression resulted in smaller FAs and unaltered MT and vimentin IF organization as compared with the untransfected internal controls (Supplemental Figure S7E). Interestingly, Rudhira-overexpressing cells had smaller FAs as compared with the untransfected internal controls, even with MT depolymerization (Supplemental Figure S7F) or vimentin IF aggregation (Supplemental Figure S7G). In either case, the other cytoskeletal component was unaltered and comparable to the untreated controls (Supplemental Figure S7, E–G). These data show that Rudhira regulates MT and vimentin IF organization and may compensate for MTs/IFs for maintaining FA size. Further, Rudhira overexpression results in reduced FA size on depletion of MTs or vimentin Ifs, suggesting that its MT-vimentin IF cross-linking role is not necessary for this function.
Rudhira regulates MT stability for angiogenic sprouting
Rudhira functions in EC migration during angiogenesis. In mice, loss of Rudhira causes midgestation lethality due to severe cardiovascular patterning defects and the loss of angiogenic sprouting (Shetty, Joshi, et al., 2018). As both MTs and vimentin IFs have critical roles in endothelial sprouting (Bayless and Johnson, 2011), we hypothesized that the loss of sprouting in KD cells was due to a loss in MT-IF association and reduced physiological stability of MTs. ROCKi and Taxol treatment are widely used to modulate sprouting angiogenesis (Kroll, Epting, et al., 2009; Martin et al., 2018). Whereas ROCK inhibition promotes sprouting, low-dose Taxol (100 pM) is either inhibitory (in normoxia) or ineffective (in hypoxia) in normal cells. Taxol treatment could stabilize MTs and promote MT-IF association at both high and low concentrations, such as ROCKi treatment. However, for functional rescue, we used a low Taxol concentration because both the dynamics and the stability of MTs are essential for sprouting. KD cells, which otherwise fail to sprout as reported earlier (Shetty, Joshi, et al., 2018), when treated with either ROCKi or low-dose (100 pM) Taxol rescued sprouting angiogenesis (Figure 5F). This suggests that Rudhira is essential for MT stability, cytoskeletal cross-linking, organization, and dynamics during developmental vascular remodeling.
The C-terminal BCAS3 domain is necessary and sufficient for cytoskeletal organization and cell migration
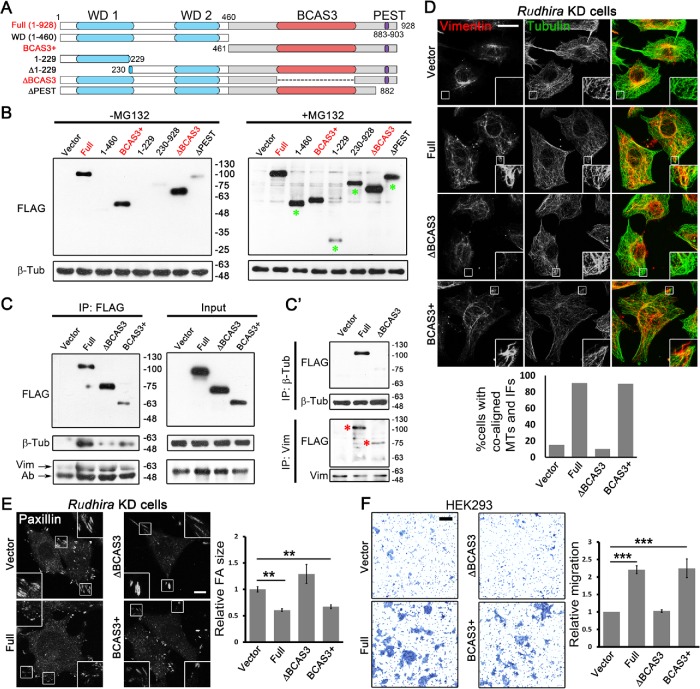
To elucidate how the organization of Rudhira protein mediates its function, we undertook a deletion analysis. Rudhira is reported to have predicted WD40-like structural domains, involved in protein interactions, at the N-terminal region and an uncharacterized BCAS3 domain in the C-terminal region (Siva and Inamdar, 2006). Using multiple bioinformatics domain analysis servers and on the basis of high confidence score, we mapped the limits of these domains (Supplemental Figure S8A; also see Materials and Methods). Rudhira also encodes multiple isoforms, and a shorter isoform of unknown function that lacks the initial 229 residues is reported. The protein structure prediction tool Phyre2 predicted one β-propeller (maximum 99.8% confidence and 17% identity) near the N-terminus (residues 92–434) (Supplemental Figure S8B) and RaptorX predicted the presence of two β-propellers (residues 57–350, 351–582) (Supplemental Figure S8B′). Interestingly, the C-terminal region did not align to any structure and is considered to be highly disordered (Supplemental Figure S8, B and B′). A PEST motif (signal for protein degradation, residues 883–903) was also identified in the C-terminal region (Figure 6A; Supplemental Figure S8A).
FIGURE 6:
BCAS3 domain of Rudhira is necessary and sufficient for cytoskeletal cross-talk, organization, and cell migration. (A) Schematic showing the deletion mutants of different regions of Rudhira protein based on putative motifs/domains identified using bioinformatics analyses (see Materials and Methods). WD1, WD2: WD40 domains (blue); BCAS3: BCAS3 domain (red); PEST: PEST motif (purple). The dotted line indicates the region deleted in ∆BCAS3. (B) Validation of the expression of FLAG-tagged Rudhira full-length or deletion mutants overexpressed in HEK293T cells by immunoblot, with or without MG132, as indicated. (C, C’) FLAG-tagged Rudhira full-length BCAS3 or BCAS3+ or ∆BCAS3 fragments were analyzed for interaction with β-tubulin or vimentin by coimmunoprecipitation with FLAG antibody (C), confirmed by reverse coimmunoprecipitation with β-tubulin or vimentin antibody (C’). (D, E) Rudhira full-length or fragments (cloned in pIRES2-EGFP vector) were transiently transfected in KD cell line to test for the rescue of MT, vimentin IF organization, MT-vimentin IF association, and FA organization by double immunostaining for tubulin and vimentin (D), tubulin and actin (see Supplemental Figure S8F), or FA marker Paxillin (E). Boxed regions in panels D and E are magnified in the insets and show cell peripheral regions. Graphs show the quantitation of the percentage of cells showing coaligned MTs and vimentin IFs from 10 cells and Paxillin FA size from at least 13 cells. (F) Rudhira full-length or fragments were overexpressed in HEK293 cells and tested for function using a transwell-migration assay, quantified in the graph. Error bars indicate SEM. Results shown are a representative of at least three independent experiments. Statistical analysis was carried out using one-way ANOVA. Scale bar: (D) 20 μm, (E) 10 μm, (F) 100 μm. **p < 0.01, ***p < 0.001.
Whereas the WD40 domains function in protein–protein interactions, the BCAS3 domain is reported in proteins expressed in breast cancer and implicated in the progression of breast cancer (Bärlund, Monni, et al., 2002). Interestingly, a majority of the Rudhira posttranslational modifications (PTMs) identified in high-throughput mass spectrometric screens was present in the C-terminal half of the protein (BCAS3+ fragment), which houses the BCAS3 domain, typically 229–245 amino acids long (Supplemental Figure S8C). Analysis with MAPanalyzer (Microtubule-associated Protein Analyzer; http://systbio.cau.edu.cn/mappred/) (Zhou et al., 2015) showed putative MT-interacting motifs in Rudhira distributed along the entire length of the protein (Supplemental Figure S8D). Structural prediction using RaptorX suggested that the N-terminal 1–460 fragment containing WD40 domains would form a six-bladed β-propeller and ∆BCAS3 (∆527–805) would form two β-propellers as in the full-length protein, whereas the BCAS3+ fragment (461-928) would be mostly disordered (Supplemental Figure S8E). On the basis of this information, we generated deletion mutants harboring/lacking the putative domains or isoforms (Figure 6A; see Materials and Methods) and expressed them in HEK293T cells.
Some of the deletion mutants expressed poorly, suggesting that the fragments may be unstable. Treatment with the proteasomal inhibitor MG132 stabilized these fragments (Figure 6B). However, the WD40 domain containing fragment devoid of the BCAS3 domain (∆BCAS3) and the BCAS3 domain containing fragment (BCAS3+) both expressed at levels similar to those of the full-length protein (Full). Also, the WD40 and the BCAS3 domains were predicted at high confidence by multiple bioinformatics tools, as compared with other domains/motifs (Supplemental Figure S8A). Hence, further molecular and functional analysis was limited to these to avoid the possible differences due to varied expression levels.
Interestingly, the full-length protein as well as the BCAS3+ fragment could efficiently coimmunoprecipitate tubulin. However, tubulin interaction was dramatically reduced with ∆BCAS3 (Figure 6C), suggesting that the BCAS3 domain of Rudhira, and not the WD40 domain, is necessary and sufficient for tubulin/MT interaction. In addition, all three fragments could coimmunoprecipitate vimentin. This was confirmed by reverse coimmunoprecipitation with tubulin and vimentin, wherein tubulin could efficiently pull down the full-length protein but not ∆BCAS3 mutant, whereas vimentin could pull down the full length as well as ∆BCAS3, although the interaction with ∆BCAS3 was slightly reduced as compared with the full length (Figure 6C′). This suggested that whereas Rudhira contained multiple vimentin-binding regions, tubulin-binding regions were present mainly in the BCAS3+ fragment. It is important to note that, in ∆BCAS3, the region from 527 to 805 was deleted (instead of 521–792) because of the high sequence conservation in the BCAS3 domain until the 805 residue and to avoid the deletion of the overlapping SxIP motif (518–521). Also, the BCAS3+ fragment (461–928) had WD40 domains deleted but retained the BCAS3 domain and phosphorylation sites reported at high frequency.
To test whether the interactions with tubulin and vimentin correlated with function, we overexpressed Rudhira full-length or deletion mutants in KD cells and assayed for rescue of the KD phenotypes, namely reduced MT-vimentin IF association, enlarged FAs, and increased actin stress fibers. The full-length protein or BCAS3+ restored MT and vimentin organization, MT-vimentin IF association (Figure 6D), FA size, and actin organization (Figure 6E; Supplemental Figure S8, F and G; Supplemental Video S6). However, ∆BCAS3-expressing KD cells continued to show disorganized actin and MTs (Supplemental Figure S8F), reduced and less extended vimentin IFs, loss of IF-MT alignment (Figure 6D), and large FAs (Figure 6E; Supplemental Figure S8G; Supplemental Video S6). To test the functional relevance of the BCAS3 domain of Rudhira, we checked the effect of its presence on cell migration in a transwell assay. Overexpression of the full-length protein or the BCAS3+ in HEK293 cells resulted in an increase in migration, whereas ∆BCAS3 did not (Figure 6F). Together, these data show that Rudhira–cytoskeleton interactions leading to MT-IF cross-talk mediated by the BCAS3 domain are essential for regulating cytoskeleton architecture. Further, the BCAS3 domain is necessary and sufficient for Rudhira function.
Movie S6.
BCAS3 domain of Rudhira is essential for MT dynamic organization (related to Figure 6). Time-lapse images of SVEC KD cell line transiently overexpressing Rudhira or RudhiraΔBCAS3, incubated with SiR-Tubulin monitored for 2 minutes, imaged at 4 second intervals (6 fps). Arrow indicates persistence of radial MT at cell periphery, while asterisk indicates a MT bending before reaching cell periphery. Note the MTs criss-crossing and not aligned towards cell periphery (red box) in vector/ΔBCAS3 transfected KD cells, as compared to aligned MTs (green box) in Rudhira transfected KD cells.
DISCUSSION
In a dynamically regulated system such as the vasculature, controlled EC migration and sprouting angiogenesis are key to ensuring blood supply during development and tissue repair. Defects in this process can lead to developmental anomalies or even embryonic death. The cycle of EC proliferation, migration, and sprouting angiogenesis is influenced by several physiological and pathological cues. Cytoskeletal remodeling underlies all of these processes and mediates molecular cross-talk to ensure a calibrated response over a range of signals. Cell type-specific components ensure an appropriate response to the dynamic cues from circulation as well as the tissue microenvironment. Rudhira is a dynamically regulated molecule with tissue-specific roles in regulating the cytoskeleton in endothelial migration and sprouting angiogenesis (Jain et al., 2012; Shetty, Joshi, et al., 2018). Here we investigated the molecular mechanism by which Rudhira regulates the cytoskeleton and found that Rudhira cross-links IF and MT cytoskeleton, stabilizes MTs, and directs MTs for FA disassembly, mediated by its BCAS3 domain.
Vimentin IFs act as a template for MT growth and stabilize MTs. Our data suggest a major role of Rudhira in MT-IF cross-talk. The cytolinker function of Rudhira sufficiently explains the molecular and cellular phenotypes observed on its depletion. However, it is also likely and probable that Rudhira functions with other cytolinkers and cytoskeleton-associated molecules for coupling MTs and IFs.
The primary function of Rudhira appears to be binding to MTs and vimentin IFs, providing physiological stability to MTs and promoting vimentin IF extension to aid cytoskeletal organization and downstream processes. It is, however, possible that the restoration of cytoskeletal architecture and sprouting observed on treatment with ROCKi or Taxol was not through their effect on MTs but rather due to probable effects on other cytoskeletal components including Rudhira, vimentin, or actin. We reported earlier that rudhira KD cells have dramatically reduced soluble vimentin (Shetty, Joshi, et al., 2018). ROCKi and Taxol are also known to solubilize vimentin, which may also rescue the loss of Rudhira. It is also possible that ROCKi or Taxol treatment may stabilize MTs or prevent their disassembly and simultaneously lead to the formation of secondary sites for MT nucleation, which may lead to the rescue of Rudhira depletion phenotypes and sprouting. Nocodazole-mediated MT depolymerization leads to increased RhoA activity, which acts through ROCK to enhance stress fiber and FA assembly. Sensitivity to nocodazole-mediated MT depolymerization in KD cells could be a result of high RhoA activity, which is restored with ROCKi treatment. This is in agreement with our earlier study, where we find that active RhoA is able to override the molecular functions of Rudhira (Jain et al., 2012). Loss of Rudhira results in embryonic lethality in mouse with gross cardiovascular patterning defects. It is unlikely that pharmacologically restoring MT stability would completely override the effect of loss of Rudhira. However, controlled restoration of MT stability and dynamics or the expression of another cytolinker in rudhira knockout may be useful in delineating and restoring the primary molecular function of Rudhira in vivo.
Aberrant cell-matrix adhesion is the underlying cause of defective migration in a variety of contexts (Palecek et al., 1997). FA turnover is a complex process. Molecules that regulate FA components are known; however, local interactions that direct MTs to FAs remain unclear. The +TIP proteins, such as CLASPs, localize to FAs as well as to MTs, thereby bridging the two for FA disassembly (Stehbens et al., 2014). Rudhira, on the other hand, binds MTs and IFs but not FAs, highlighting its role as a targeted regulator of the cytoskeleton in this process. The effect of Rudhira depletion on FA disassembly and thereby cell migration appears to be downstream of its more direct role in aligning growing MTs toward the cell periphery. Hence the role of Rudhira is to organize the MT cytoskeleton downstream of FAK phosphorylation to bring about FA disassembly. In addition, the unaligned growth and fewer growing MTs in KD cells suggest a role for Rudhira before MTs encounter actin stress fibers at the cell periphery. Rudhira-mediated control of FA and MT dynamics is unlikely to be independent, owing to the strong correlation between the organization of the two, as suggested in the literature as well as in our study (Stehbens and Wittmann, 2012).
The MT-organizing center does not realign in rudhira KD cells (Jain et al., 2012), which may also lead to disorganized MT growth and architecture (Jain et al., 2012). Additionally, the involvement of independent molecular pathways controlling FA and MT organization cannot be ignored. Rudhira interacts with EB1 and may thereby govern MT growth toward FAs. This also raises the interesting possibility that Rudhira may lay down tracks for MT growth. Identification of further molecular interactors of Rudhira will delineate its position in the molecular pathway governing MT growth and recruitment to FAs.
Rudhira has conserved WD40 and BCAS3 domains; however, the relevance of its organization to normal in vivo function is not known. WD40 domain-containing proteins assume a beta propeller structure that is thought to act as a scaffold for multiple protein interactions. However, our experiments suggest that the C terminal fragment bearing the BCAS3 domain, rather than the N-terminal WD40 domain-containing fragment, could bind tubulin and, importantly, restore function. In addition to the BCAS3 domain, the BCAS3+ fragment includes a PEST domain, SxIP motifs, some of the C-terminal region, and frequently reported phosphorylation sites, which may also contribute to its function. However, these additional features require the BCAS3 domain, as removing the BCAS3 alone caused loss of BCAS3+ function. Some of the fragments, including the alternative isoform, the PEST motif deletion, and others, showed increased susceptibility to ubiquitin–proteasome-mediated degradation, suggesting complex regulation. Transient and dynamic expression of some of these isoforms could provide temporal or context-dependent control of Rudhira function, a possibility that merits further investigation. Our analysis also assigns molecular function to the conserved BCAS3 domain whose functions can now be tested in several autophagy-related and cancer-expressed proteins that harbor it.
In addition to its primary role in ECs and angiogenesis, Rudhira/BCAS3 is misexpressed in several cancers and associated with coronary artery disease, making it a principal target in these diseases. Our finding, that the BCAS3 domain is required for promoting cytoskeleton cross-talk, and maintaining MT architecture and FA dynamics, will help devise strategies for controlled alteration of cytoskeletal architecture to correct aberrant cell migration, tissue malignancy, or degeneration.
MATERIALS AND METHODS
Cell culture
Mouse SVEC was obtained from K. Rau (National Centre for Biological Sciences, Bangalore, India), and HEK293, HEK293T, and HeLa cells were from the American Type Culture Collection. Cells were cultured in DMEM (ThermoFisher Scientific) supplemented with 10% fetal bovine serum (FBS) (Life Technologies, ThermoFisher Scientific). Serum starvation was performed for 48 h in DMEM. Generation of KD lines is reported elsewhere (Shetty, Joshi, et al., 2018). Briefly, rudhira/BCAS3 shRNA vectors (715, 716) and scrambled (NS) control vector (TR30015) (Origene) were microporated into SVEC according to the manufacturer's instructions (Neon, Life Technologies). The transfected cells were selected in puromycin-containing media (1 μg/ml) for 7 d and >99% RFP positive population was obtained by fluorescence-associated cell sorting using a BD Aria II (BD Pharmingen). Rudhira/BCAS3 KD was validated by qRT-PCR and Western blot analysis. The control cell line is referred to as NS and rudhira as KD (Jain et al., 2012).
Immunostaining, antibodies, and small molecule treatment
Cells were fixed in 4% paraformaldehyde at room temperature for 15 min or 100% methanol at –20°C for 10 min and processed for immunostaining using standard procedure. Primary antibodies used were against Rudhira (Jain et al., 2012), Vinculin, α-tubulin, Ac-tubulin (Sigma Chemical Co.), β-tubulin (Developmental Studies Hybridoma Bank [DSHB]; ThermoFisher Scientific; Abcam), Paxillin, FAK, β1 Integrin (Merck), vimentin, Glu-tubulin, EB1 (Abcam), plectin (Santa Cruz Biotechnology), GFP (ThermoFisher Scientific), pFAK, and pY (Cell Signaling Technologies). Secondary antibodies were coupled to Alexa-Fluor 488 or Alexa-Fluor 568 or Alexa-Fluor 633 (Molecular Probes). Phalloidin was conjugated to Alexa-Fluor 633 (Molecular Probes). Nocodazole, ROCK inhibitor (ROCKi, Y27632), and Taxol (paclitaxel) were from Sigma Chemical. Cells were treated with ROCKi or Taxol for 1 h and processed for immunostaining with Paxillin, tubulin or vimentin antibodies, or Phalloidin, as indicated (Figure 5, A–E).
Fluorescence microscopy, live-cell imaging, and analysis
Confocal microscopes (LSM 510 Meta, LSM 880 with Airy Scan from Zeiss, FV3000 from Olympus), Spinning Disc Microscope (Perkin Elmer with Yokogawa camera attachment), or a motorized inverted microscope with fluorescence attachment (IX81, Olympus) were used for fluorescence microscopy and time-lapse imaging. Line profile of fluorescence intensities (Figures 1E and 3F) and three-dimensional shadow and surface reconstruction of cells (Supplemental Figure S1B) were generated in ZEN Blue software from Zeiss. Superresolution microscopy was performed by imaging in the Airy Scan image acquisition and processing mode of the LSM 880 (Zeiss). For live-cell imaging, a sample heater (37°C) and CO2 incubation chamber (Tokai Hit) were used to control temperature and CO2 levels during live-cell imaging. All images in a set were adjusted equally for brightness and contrast using Adobe Photoshop CS2, where required. Rudhira NS or KD cells were transiently transfected with EB1-GFP and seeded on fibronectin-coated glass-bottom dishes. Live imaging for EB1-GFP was carried out 24 h postseeding for 3 min at 4-s intervals to determine MT growth and alignment. EB1-GFP live images were time-projected in ImageJ (National Institutes of Health [NIH]) to represent MT growth. EB1-GFP tracks were generated manually and residence time was calculated manually using ImageJ (NIH) with the manual tracking plug-in. Rudhira NS or KD cells were transiently transfected with Paxillin-GFP and seeded on fibronectin-coated glass-bottom dishes. Live imaging for Paxillin-GFP was carried out 24 h postseeding for 2–3 h at 5-min intervals to determine FA assembly and disassembly rates under steady state. The images were processed for estimation of various parameters using FAAS (Berginski and Gomez, 2013). Paxillin-GFP, EB1-GFP live images were time-projected in ImageJ (NIH) to represent FA and MT growth dynamics, respectively. Rudhira NS or KD cells were transiently transfected with vimentin-GFP and seeded on fibronectin-coated glass-bottom dishes. Cells were incubated with 250 nM SiR-tubulin for 2 h before live imaging was carried out for 4 min at 10-s intervals to test coalignment of MTs and IFs.
In situ PLA (Duolink assay)
In situ PLA reaction was performed on SVEC cell lines. The cells were cultured, fixed, permeabilized, and stained with primary antibodies as indicated. Thereafter, the protocol for PLA as recommended by the manufacturer (Duolink) was followed. Post-PLA, nuclei were counterstained with 4’, 6-diamidino-2-phenylindole (DAPI).
Cell attachment and spreading assays
The assays and quantitation were carried out as mentioned in Uchil et al. (2014), who cited Humphries (2001), with a few modifications. Briefly, 96-well plates were coated with fibronectin (10 µg/ml) for 60 min at room temperature and blocked with heat-denatured filter-sterilized bovine serum albumin for 30 min at room temperature. Cells were put in suspension in warm medium at 37°C, 5% CO2 to disassemble already formed FA. Thereafter, 20,000 (for attachment) or 10,000 (for spreading) cells were seeded per well and allowed to attach or spread for the indicated times. Floating or loosely attached cells were removed by washing twice with phosphate-buffered saline (PBS) and then fixed with 4% paraformaldehyde. For spreading assay, the extent of spreading was quantified in ImageJ (NIH) from RFP (expressed from the NS or KD vector) images. For attachment assay, cells were stained with 0.1% crystal violet for 60 min at room temperature and washed three times with water, the dye was solubilized in 100 µl 10% acetic acid, and the absorbance was measured at 570 nm using a plate reader. The total number of cells attached at 3 h was set to 100% for both NS and KD lines.
Nocodazole and cold treatment
For MT stability experiments (Figure 3, C, C′, and D), cells were treated with indicated dosages of nocodazole or cold PBS (4°C) for 30 min, washed twice with cold PBS, and fixed in ice-cold methanol at -20°C. For MT recovery experiments (Figure 4H; Supplemental Figure S5, A and B), cells were treated with nocodazole (10 µM) for 30 min in complete medium at 37°C, 5% CO2, washed twice with PBS, and incubated with fresh culture medium for desired time intervals as indicated. Thereafter, cells were fixed and taken for immunostaining of Vinculin, F-actin (Phalloidin), and α-tubulin, as indicated.
SiR-tubulin labeling of MTs
SiR-tubulin was as used in Lukinavicius, Reymond, et al. (2014) and was a kind gift from S. Agasti (Jawaharlal Nehru Centre for Advanced Scientific Research [JNCASR]). MT labeling was performed as indicated in Lukinavicius, Reymond, et al. (2014). Briefly, cells were incubated with 250 nM of SiR-tubulin for 2 h in complete medium at 37°C, 5% CO2, washed twice with fresh culture medium, and taken for live-cell imaging.
Coimmunoprecipitation and Western blot analysis
Lysate (50 µg) was used for Western blot analysis by standard protocols. Primary antibodies used were as indicated earlier. Horseradish peroxidase (HRP)-conjugated secondary antibodies against appropriate species were used and signal developed using Clarity Western ECL substrate (Bio-Rad). Western blot intensities were normalized to GAPDH and quantification was carried out using ImageJ (NIH). For coimmunoprecipitation assays, 500 µg lysate of HEK293T cells overexpressing Rudhira fragments were incubated overnight with 10 µl of FLAG M2 beads (Sigma Chemical), 10 µl of β-tubulin antibody (DSHB), or vimentin antibody (Sigma Chemical), captured on Protein G-sepharose beads (Sigma Chemical), washed three times in lysis buffer, and analyzed by immunoblotting with anti-β-tubulin (Abcam), vimentin (Sigma Chemical; Abcam), or FLAG (Sigma Chemical) antibody.
Spheroid sprouting and transwell migration assay
The assays and quantitation were carried out as described previously (Jain et al., 2012; Shetty, Joshi, et al., 2018). Briefly, for spheroid sprouting, 750 cells each of the KD line were taken for spheroid formation in a round-bottom nonadherent 96-well dish (Costar) in 1% carboxy methyl cellulose in 10% FBS in DMEM. The spheroids formed were transferred to collagen gels (Rat tail, Type I; ThermoFisher Scientific) with a final concentration of 2.5 mg/ml, with or without ROCKi or Taxol. Gels were overlaid with 200 µl of 10% FBS in DMEM and the sprouting was monitored for 3 d. For transwell migration, 24 h after transfection with desired plasmid vectors, cells were serum-starved for 12 h and 20,000 cells were plated onto the upper chamber of the transwell filter inserts with 8 μm pore size, 24-well format (Costar). Serum medium (10%) was added to the lower chamber to serve as a chemoattractant. After 24 h, cells were fixed in 4% paraformaldehyde for 10 min at room temperature. Cells on the top of the filter were removed using a cotton swab. Cells that had migrated to the bottom were fixed and stained with 0.5% crystal violet for 10 min at room temperature. The dye was extracted in methanol and absorbance was measured spectrophotometrically at 570 nm.
Rudhira in silico analysis, deletion mutant cloning, plasmid constructs, and transfection
Domain/motif prediction analysis of mouse Rudhira protein sequence (Uniprot Id Q8CCN5.2) was performed using various bioinformatics tools, namely Superfamily (https://supfam.org/SUPERFAMILY/), Motif Scan (https://myhits.isb-sib.ch/cgi-bin/motif_scan), Pfam (https://pfam.xfam.org/), NCBI-CDD (www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi?INPUT_TYPE=precalc&SEQUENCE=33300978), Motif Finder (www.genome.jp/tools/motif/), Interpro (www.ebi.ac.uk/interpro/), and epestfind in the EMBOSS package of ExPASy (www.expasy.org/tools/). PTMs in Rudhira were identified using PhosphoSitePlus (www.phosphosite.org/homeAction.action). MT-binding regions were predicted using MAPanalyzer (http://systbio.cau.edu.cn/mappred/). Rudhira full-length protein or deletion mutant structure prediction was performed using Phyre2 (www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index) and RaptorX (http://raptorx.uchicago.edu/).
pCMV-Rudh-IRES2-EGFP (Figures 4D and 6, D–F; Supplemental Figures S7, E–G, and S8, F and G), pCAG-Rudh-2A-GFP (Supplemental Figure S4C), and pCMV-RudhFL-FLAG (Figures 2, F and H, and 6, B, C, and C′) were described earlier (Jain et al., 2012). Rudhira ORF was subcloned from pCAG-Rudh-2A-GFP vector into pEGFP-N3 vector (Clontech) using NheI-SacII sites to obtain pCAG-Rudhira-GFP (Figure 3D; Supplemental Figure S3C). Rudhira ORF was subcloned from pCMV-RudhFL-FLAG vector into pSNAPf vector (New England Biolabs) using NheI-EcoRI sites to obtain Rudhira-SNAP (Supplemental Figure S3B). For deletion mutant cloning, the regions to be cloned were PCR amplified from pCMV-RudhFL-FLAG vector and cloned in pCMV-Tag2B vector (Stratagene) (Figures 2H and 6, B, C, and C′). The desired fragments from pCMV-Tag2B vectors were digested using EcoRI-XhoI and subcloned into the compatible EcoRI-SalI sites of pIRES2-EGFP vector (Clontech), to obtain GFP fluorescent reporter plasmids (Figures 4D and 6, D–F; Supplemental Figures S7, E–G, and S8, F and G). Vimentin-GFP plasmid was a kind gift from S. Etienne-Manneville (Institut Pasteur, Paris), EB1-GFP plasmid was a kind gift from Y. Mimori-Kiyosue (Riken Kobe, Japan), and paxillin-GFP was a kind gift from R. Horwitz (University of Virginia). Control and plectin shRNAs were purchased from the ShRNA Resource Centre (Department of Microbiology and Cell Biology, Indian Institute of Science, Bangalore, India) and were from the MISSION Human shRNA Library (Sigma Chemical Co.). The shRNAs were transfected in SVEC, selected in 1 µg/ml puromycin. The target sequence of plectin shRNA was CGATGAGGAGATGAACGAGAT. HEK293 and HEK293T cells were transfected using the calcium phosphate method, whereas SVEC and HeLa cells were transfected using Lipofectamine 2000 (ThermoFisher Scientific).
Quantification and statistical analyses
Statistical significance analyses were performed using one-way analysis of variance (ANOVA) in the data analysis package in Microsoft Excel or Student’s t test; p < 0.05 was considered significant. All relevant data are within the article and the Supplemental Information files.
Supplementary Material
Acknowledgments
We thank Y. Mimori-Kiyosue, Riken Kobe, Japan, for EB1-GFP plasmid; R. Horwitz for Paxillin-GFP plasmid; S. Etienne-Manneville, Institut Pasteur, Paris, for vimentin-GFP plasmid; S. Agasti and Ranjan Sasmal, JNCASR, for SiR-tubulin; P. Jindal, A. Sinha, and A. Sam for Bioinformatics analyses and generating deletion mutants; the JNCASR Imaging facility, NCBS Central Imaging and Flow Facility, and laboratory members for fruitful discussions. This work was funded by grants from the Department of Biotechnology, Government of India (Sanction no. BT/PR11246/BRB/10/644/2008 dated 29.09.2009), the Wellcome Trust, UK (094879/B/10/Z); and intramural funds from the Life Science Research Education and Training at JNCASR, India (LSRET/JNC).
Abbreviations used:
- Ac
acetylated
- BCAS3
Breast Carcinoma Amplified Sequence 3
- EB1
end binding protein
- EC
endothelial cell
- ECM
extracellular matrix
- FA
focal adhesion
- FAAS
focal adhesion analysis server
- FAK
FA kinase
- FBS
fetal bovine serum
- GFP
green fluorescent protein
- IF
intermediate filament
- KD
knockdown
- MAPS
microtubule-associated proteins
- MT
microtubule
- NS
nonsilencing control
- PBS
phosphate-buffered saline
- PLA
proximity ligation assay
- PTMs
posttranslational modifications
- pY
phospho-tyrosine
- ROCK
Rho-associated kinase
- Rudh
Rudhira
- SiR-tubulin
silico-rhodamine–conjugated docetaxal
- SVEC
saphenous vein endothelial cell line
- Tub
tubulin.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-08-0484) on April 17, 2019.
REFERENCES
Boldface names denote co–first authors.
- Akhmanova A, Steinmetz MO. (2010). Microtubule +TIPs at a glance. J Cell Sci (Pt 20), 3415–3419. [DOI] [PubMed] [Google Scholar]
- Andrä K, Lassmann H, Bittner R, Shorny S, Fässler R, Propst F, Wiche G. (1997). Targeted inactivation of plectin reveals essential function in maintaining the integrity of skin, muscle, and heart cytoarchitecture. Genes Dev , 3143–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bärlund M, Monni O, Weaver JD, Kauraniemi P, Sauter G, Heiskanen M, Kallioniemi OP, Kallioniemi A. (2002). Cloning of BCAS3 (17q23) and BCAS4 (20q13) genes that undergo amplification, overexpression, and fusion in breast cancer. Genes Chromosomes Cancer , 311–317. [DOI] [PubMed] [Google Scholar]
- Bayless KJ, Johnson GA. (2011). Role of the cytoskeleton in formation and maintenance of angiogenic sprouts. J Vasc Res , 369–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berginski ME, Gomez SM. (2013). The Focal Adhesion Analysis Server: a web tool for analyzing focal adhesion dynamics. F1000Res . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulinski JC, Gundersen GG. (1991). Stabilization and post-translational modification of microtubules during cellular morphogenesis. Bioessays , 285–293. [DOI] [PubMed] [Google Scholar]
- Colucci-Guyon E, Portier MM, Dunia I, Paulin D, Pournin S, Babinet C. (1994). Mice lacking vimentin develop and reproduce without an obvious phenotype. Cell , 679–694. [DOI] [PubMed] [Google Scholar]
- Dave JM, Bayless KJ. (2014). Vimentin as an integral regulator of cell adhesion and endothelial sprouting. Microcirculation , 333–344. [DOI] [PubMed] [Google Scholar]
- De Pascalis C, Perez-Gonzalez C, Seetharaman S, Boeda B, Vianay B, Burute M, Leduc C, Borghi N, Trepat X, Etienne-Manneville S. (2018). Intermediate filaments control collective migration by restricting traction forces and sustaining cell-cell contacts. J Cell Biol , 3031–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezratty EJ, Partridge MA, Gundersen GG. (2005). Microtubule-induced focal adhesion disassembly is mediated by dynamin and focal adhesion kinase. Nat Cell Biol , 581–590. [DOI] [PubMed] [Google Scholar]
- Gan Z, Ding L, Burckhardt CJ, Lowery J, Zaritsky A, Sitterley K, Mota A, Costigliola N, Starker CG, Voytas DF, et al. (2016). Vimentin intermediate filaments template microtubule networks to enhance persistence in cell polarity and directed migration. Cell Syst , 252–263. e258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurland G, Gundersen GG. (1995). Stable, detyrosinated microtubules function to localize vimentin intermediate filaments in fibroblasts. J Cell Biol , 1275–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyoeva FK, Gelfand VI. (1991). Coalignment of vimentin intermediate filaments with microtubules depends on kinesin. Nature , 445–448. [DOI] [PubMed] [Google Scholar]
- Humphries MJ. (2001). Cell-substrate adhesion assays. Curr Protoc Cell Biol 9.1. 1–9.1. 11. [DOI] [PubMed] [Google Scholar]
- Jain M, Bhat GP, Vijayraghavan K, Inamdar MS. (2012). Rudhira/BCAS3 is a cytoskeletal protein that controls Cdc42 activation and directional cell migration during angiogenesis. Exp Cell Res , 753–767. [DOI] [PubMed] [Google Scholar]
- Jiang K, Toedt G, Montenegro Gouveia S, Davey NE, Hua S, van der Vaart B, Grigoriev I, Larsen J, Pedersen LB, Bezstarosti K, et al. (2012). A Proteome-wide screen for mammalian SxIP motif-containing microtubule plus-end tracking proteins. Curr Biol , 1800–1807. [DOI] [PubMed] [Google Scholar]
- Jiu Y, Lehtimaki J, Tojkander S, Cheng F, Jaalinoja H, Liu X, Varjosalo M, Eriksson JE, Lappalainen P. (2015). Bidirectional interplay between vimentin intermediate filaments and contractile actin stress fibers. Cell Rep , 1511–1518. [DOI] [PubMed] [Google Scholar]
- Kodama A, Karakesisoglou I, Wong E, Vaezi A, Fuchs E. (2003). ACF7: an essential integrator of microtubule dynamics. Cell , 343–354. [DOI] [PubMed] [Google Scholar]
- Kroll J, Epting D, Kern K, Dietz CT, Feng Y, Hammes H-P, Wieland T, Augustin HG. (2009). Inhibition of Rho-dependent kinases ROCK I/II activates VEGF-driven retinal neovascularization and sprouting angiogenesis. Am J Physiol Heart Circ Physiol , H893–H899. [DOI] [PubMed] [Google Scholar]
- Leduc C, Etienne-Manneville S. (2017). Regulation of microtubule-associated motors drives intermediate filament network polarization. J Cell Biol , 1689–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukinavicius G, Reymond L, D’Este E, Masharina A, Gottfert F, Ta H, Guther A, Fournier M, Rizzo S, Waldmann H, et al. (2014). Fluorogenic probes for live-cell imaging of the cytoskeleton. Nat Methods , 731. [DOI] [PubMed] [Google Scholar]
- Martin M, Veloso A, Wu J, Katrukha EA, Akhmanova A. (2018). Control of endothelial cell polarity and sprouting angiogenesis by non-centrosomal microtubules. eLife , e33864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwenhuizen RP, Nahidiazar L, Manders EM, Jalink K, Stallinga S, Rieger B. (2015). Co-orientation: quantifying simultaneous co-localization and orientational alignment of filaments in light microscopy. PLoS One , e0131756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palecek SP, Loftus JC, Ginsberg MH, Lauffenburger DA, Horwitz AF. (1997). Integrin-ligand binding properties govern cell migration speed through cell-substratum adhesiveness. Nature , 537–540. [DOI] [PubMed] [Google Scholar]
- Sakamoto Y, Boeda B, Etienne-Manneville S. (2013). APC binds intermediate filaments and is required for their reorganization during cell migration. J Cell Biol , 249–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty R, Joshi D, Jain M, Vasudevan M, Paul JC, Bhat G, Banerjee P, Abe T, Kiyonari H, VijayRaghavan K, et al. (2018). Rudhira/BCAS3 is essential for mouse development and cardiovascular patterning. Sci Rep , 5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siva K, Inamdar MS. (2006). Rudhira is a cytoplasmic WD40 protein expressed in mouse embryonic stem cells and during embryonic erythropoiesis. Gene Expr Patterns , 225–234. [DOI] [PubMed] [Google Scholar]
- Siva K, Venu P, Mahadevan A, Shankar S, Inamdar MS. (2007). Human BCAS3 expression in embryonic stem cells and vascular precursors suggests a role in human embryogenesis and tumor angiogenesis. PLoS One , e1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehbens SJ, Paszek M, Pemble H, Ettinger A, Gierke S, Wittmann T. (2014). CLASPs link focal adhesion-associated microtubule capture to localized exocytosis and adhesion site turnover. Nat Cell Biol , 561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehbens S, Wittmann T. (2012). Targeting and transport: how microtubules control focal adhesion dynamics. J Cell Biol , 481–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchil PD, Pawliczek T, Reynolds TD, Ding S, Hinz A, Munro JB, Huang F, Floyd RW, Yang H, Hamilton WL, et al. (2014). TRIM15 is a focal adhesion protein that regulates focal adhesion disassembly. J Cell Sci , 3928–3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte H, Bradke F. (2008). The role of the cytoskeleton during neuronal polarization. Curr Opin Neurobiol , 479–487. [DOI] [PubMed] [Google Scholar]
- Xu Z, Schaedel L, Portran D, Aguilar A, Gaillard J, Marinkovich MP, Théry M, Nachury MV. (2017). Microtubules acquire resistance from mechanical breakage through intralumenal acetylation. Science , 328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Yang S, Mao T, Zhang Z. (2015). MAPanalyzer: a novel online tool for analyzing microtubule-associated proteins. Database . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.