Abstract
In Drosophila ovarian somatic cells, PIWI-interacting small RNAs (piRNAs) against transposable elements are mainly produced from the ∼180-kb flamenco (flam) locus. flam transcripts are gathered into foci, located close to the nuclear envelope, and processed into piRNAs in the cytoplasmic Yb bodies. The mechanism of Yb body formation remains unknown. Using RNA fluorescence in situ hybridization, we found that in the follicle cells of ovaries the 5′-ends of flam transcripts are usually located in close proximity to the nuclear envelope and outside of Yb bodies, whereas their extended downstream regions mostly overlap with Yb bodies. In flamKG mutant ovaries, flam transcripts containing the first and, partially, second exons but lacking downstream regions are gathered into foci at the nuclear envelope, but Yb bodies are not assembled. Strikingly, piRNAs from the protein-coding gene transcripts accumulate at higher levels in flamKG ovaries indicating that piRNA biogenesis may occur without Yb bodies. We propose that normally in follicle cells, flam downstream transcript regions function not only as a substrate for generation of piRNAs but also as a scaffold for Yb body assembly, which competitively decreases piRNA production from the protein-coding gene transcripts. By contrast, in ovarian somatic cap and escort cells Yb body assembly does not require flam transcription.
INTRODUCTION
The PIWI-interacting RNA (piRNA) pathway controls expression of transposable elements (TEs) in the somatic and germ cells of Drosophila ovaries. PIWI clade proteins guided by small (24–30 nt) piRNAs recognize complementary TE transcripts resulting in their degradation or transcriptional repression (for a review, see Czech and Hannon, 2016). In the somatic cells of ovaries, piRNA machinery operates via the single nuclear Piwi protein, whereas the germ cells possess also two cytoplasmic proteins of this family—Aubergine (Aub) and Argonaute3 (Ago3), which are involved in the process of posttranscriptional silencing coupled with the piRNA amplification, the ping-pong cycle. Most piRNAs against TEs are transcribed as the long RNA precursors from the piRNA clusters, genome regions containing multiple damaged TE copies (Brennecke et al., 2007). In the germ cells, piRNAs are mainly generated from the convergently transcribed dual-strand clusters, whereas in the somatic cells, piRNA clusters, exemplified by the flam locus, are unidirectionally transcribed (Klattenhoff et al., 2009; Malone et al., 2009; Pane et al., 2011; Zhang et al., 2012). Although both dual- and unistrand piRNA clusters are located predominantly in the heterochromatin, different mechanisms direct their transcripts for processing into piRNAs (for a review, see Huang et al., 2017).
The flam piRNA cluster generates long, capped and alternatively spliced transcripts, with the short first exon represented in all spliced variants (Goriaux et al., 2014; Mohn et al., 2014). flam transcripts are assembled into one or two foci, designated DotCOM/Flam bodies (herein flam foci), located in close proximity to the nuclear envelope and adjacent to the cytoplasmic Yb bodies (Dennis et al., 2013, 2016; Murota et al., 2014), where their processing into piRNAs occurs (Olivieri et al., 2010; Saito et al., 2010; Qi et al., 2011). The components of the exon junction complex (EJC) as well as the Nxf1/Nxt1 exportin complex were shown to participate in the transfer of flam transcripts away from their site of transcription to the nuclear flam foci and for their further export to the Yb body (Dennis et al., 2016). Importantly, Yb body assembly appears to be dependent on the nucleocytoplasmic export of flam transcripts (Dennis et al., 2016). Yb bodies are composed of Yb, Armitage (Armi), Vreteno (Vret), Shutdown (Shu), Sister of Yb (SoYb), and some other proteins required for piRNA biogenesis (Haase et al., 2010; Olivieri et al., 2010, 2012; Saito et al., 2010; Handler et al., 2011; Zamparini et al., 2011). One or two Yb bodies per cell are usually located nearby to mitochondria (Szakmary et al., 2009; Saito et al., 2010), which carries on its surface the Zucchini (Zuc) nuclease, responsible for cleavage of transcripts into piRNAs (Pane et al., 2007; Ipsaro et al., 2012; Nishimasu et al., 2012). Mature Piwi–piRNA complexes then move into the nucleus where they induce transcriptional silencing of TEs.
In the somatic cells of ovaries, generation of piRNAs is thought to be defined by the presence of specific nucleotide motifs in piRNA precursors that facilitate binding of the piRNA biogenesis machinery and further recruitment of nucleases for primary piRNA processing (Homolka et al., 2015; Ishizu et al., 2015). Artificial recruitment of Yb or Armi proteins to transcripts was shown to be sufficient for their processing into piRNAs (Pandey et al., 2017), and the cis-regulatory elements targeting the downstream regions of RNAs for piRNA production were recently identified in flam first and second exons as well as in the 3′-untranslated region (3′-UTR) of traffic jam (tj) gene (Homolka et al., 2015; Ishizu et al., 2015). These sequences appear to be bound by Yb protein according to the CLIP assay (Ishizu et al., 2015). However, similar cis-regulatory elements were revealed in the luciferase (Homolka et al., 2015) as well as in the CG9257 (Ishizu et al., 2015) genes indicating their widespread presence in the genome. In agreement with this, a number of gene transcripts in ovarian somatic cells (OSCs) including tj were shown to generate piRNAs (Robine et al., 2009; Saito et al., 2009). It is currently unclear whether these cis-elements are sufficient for the delivery of transcripts into the Yb body and/or for Yb body assembly. Thus, the mechanisms that define selective piRNA production from the piRNA clusters in somatic cells remain underexplored.
Here we demonstrate that in the follicle cells of ovaries, the 5′-ends of flam transcripts are accumulated in close proximity to the nuclear envelope, whereas extended downstream transcript regions overlap with the Yb bodies. The 5′-ends of flam transcripts appear to be sufficient for their aggregation in the flam foci, but not for Yb body assembly. Intriguingly, in flam mutant ovaries lacking Yb bodies, piRNA production from 3′-UTRs of the genic mRNAs was increased. These results indicate that Yb body assembly is not necessary for piRNA biogenesis itself, but is essential for the preferential production of piRNAs against TEs compared with genic transcripts.
RESULTS
The first flam exon and the downstream flam transcript regions are differently positioned relative to Yb bodies and nuclear envelope
Previously, it was shown that in both the follicle cells of ovaries and OSC culture flam transcripts were assembled into one or two DotCOM/Flam bodies juxtaposed with the Yb bodies (Dennis et al., 2013, 2016; Murota et al., 2014). However, there was controversy concerning the localization of these bodies in OSCs and follicle cells of ovaries. In OSCs, Flam bodies were found in the cytoplasm (Murota et al., 2014), whereas in follicle cells of ovaries both nuclear and cytoplasmic flam foci were observed (Dennis et al., 2016). We performed RNA FISH in OSCs with the flam fourth exon probe (#508; Figure 1A; Dennis et al., 2013) coupled with immunostaining of the nuclear envelope (with anti-lamin Dm0 antibody) and Yb bodies (with anti-Yb antibody) and observed FISH signals in both nucleus and cytoplasm (Supplemental Figure S1), similarly to the results previously obtained by Dennis et al. (2016) in the follicle cells of ovaries. Moreover, in a fraction of OSCs we detected the simultaneous presence of flam signals at both sides of the nuclear envelope, with the cytoplasmic portion overlapping with the Yb bodies (Supplemental Figure S1). The doubling of signal likely reflects an intermediate stage of translocation through the nuclear pore of multiple flam transcripts from the point of their assembly within the nucleus to the newly formed Yb body. Clearly, this indicates that flam transcripts in both the follicle cells and OSCs are first gathered in the foci within the nucleus and after that are transported to the cytoplasmic Yb body, the center of their processing into piRNAs.
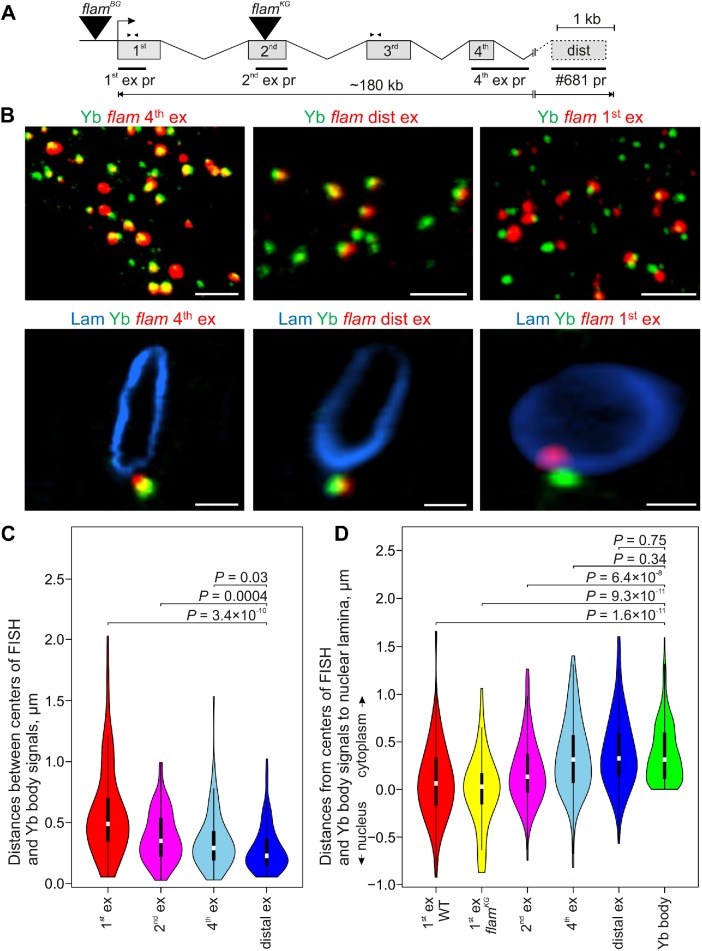
FIGURE 1:
The fourth and distal exons of flam transcripts are mostly localized in Yb bodies, whereas the flam first exon is not. (A) Scheme of the flam locus with the designation of exon/intron structure (according to Goriaux et al. [2014] and FlyBase r6), sites of TE insertions in flamBG and flamKG mutations (black triangles), RNA FISH probes (black rectangles), and primer pairs used for RT-qPCR (arrowheads). (B) RNA FISH (red) with flam fourth exon probe (left panels), distal exon probe (middle panels), or first exon probe (right panels) coupled with immunostaining of Yb bodies with anti-Yb antibody (green; top panels) and nuclear envelope with anti-lamin Dm0 antibody (blue; enlarged view of representative nuclei; bottom panels) in the follicle cells of stage 12 egg chambers. Full Z-series projections of confocal image stacks are shown in the top panels, and single optical sections are shown in the bottom panels. Scale bars are 5 μm in the top panels and 1.5 μm in the bottom panels. (C) Violin plots showing the distribution of distances between centers of Yb bodies and flam RNA FISH signals for the first (red), second (magenta), fourth (blue), or distal (dark blue) exon probes in the follicle cells of early egg chambers. Distances were measured with IMARIS software for 40–50 cells in two biological replicates for each probe. (D) Violin plots showing the distribution of distances from the centers of flam RNA FISH signals and Yb bodies to the nuclear lamina. Distances for 40–50 cells in two biological replicates (for first, second, fourth, and distal exons in WT ovaries), in one replicate (for first exon in flamKG/Df ovaries), or in eight replicates (for Yb bodies) were measured with IMARIS software. In C and D M-W U-test was used for the pairwise comparison of distributions between the distances of the first exon-Yb, the second exon-Yb, and the fourth exon-Yb (C), or between the distances of the first, second, fourth, and distal exons and Yb bodies to the nuclear lamina (D).
Further, we analyzed the distribution of flam transcripts in the ovaries using several RNA FISH probes to different flam regions. In the follicle cells of germaria and early egg chambers, FISH with the flam fourth exon probe revealed the frequent overlap of FISH signals with Yb bodies (Figure 1B, left panels). The signals of #681 probe (Dennis et al., 2013), corresponding to the distal flam exon (Figure 1A), also usually overlapped with Yb bodies (Figure 1B, middle panels). However, in some follicle cells distal flam exon signals were not revealed, which may be either because they are not included in all flam splice variants or because they are more rapidly cleaved into piRNAs. Strikingly, when we employed the probe for the first flam exon, the overlap of RNA FISH signals with the Yb bodies was drastically reduced (Figure 1B, right panels). The probe for the extended part of the second exon was intermediately overlapped with the Yb bodies. Accordingly, median distances between the centers of Yb bodies and RNA FISH signals were ∼0.2 µm for the distal exon, ∼0.3 µm for the fourth exon, ∼0.4 µm for the second exon, and ∼0.5 µm for the first exon (Figure 1C). These findings indicate that the 5′-end of flam RNA (the first exon) is mainly located out of the Yb body, whereas the long downstream region of flam transcript is associated with the Yb body. Moreover, the median three-dimensional (3D) distances to the nuclear lamina were notably different between the first and the downstream flam exons (Figure 1D). Whereas the first flam exon was mostly positioned at the nuclear lamina, the downstream transcript regions protruded further into the cytoplasm (at 0.3 µm median distances), and the second exon occupied an intermediate position (at 0.1 µm median distance). Thus, the downstream flam transcripts are more localized in the Yb body than the 5′-end of flam RNA.
Interestingly, we failed to detect flam transcripts simultaneously containing both first and third exons by the conventional RT-PCR, whereas these transcripts were revealed by more sensitive RT-PCR with an exon-specific primer (Supplemental Figure S2). Therefore, the continuous flam transcripts with both first and third exons seem to be very short-lived, whereas first and third exons mostly exist as the separate RNAs.
The presence of the first and a part of the second flam exon is sufficient for the formation of flam foci but not for the Yb body assembly
It was shown that flamBG and flamKG mutations are caused by artificial transposon insertions in the promoter region or in the second exon of the flam locus, respectively (Figure 1A), resulting in a more than 10-fold decrease of flam transcript level (Brennecke et al., 2007; Mével-Ninio et al., 2007). In the flamBG line, the TE insertion disrupts the binding site for the Cubitus Interruptus transcription factor located upstream of TSS, thus impairing flam transcription (Goriaux et al., 2014). However, in the case of flamKG mutation the mechanism of flam transcription disturbance was less clear. To address this and further questions we analyzed mutant ovaries bearing flamBG or flamKG chromosomes combined with the X chromosome deletion covering the whole flam locus (Df), which allowed us to exclude the influence of genetic background. In addition, flamBG/Df and flamKG/Df ovaries exhibited more normal morphology than the homozygous mutants. We hypothesized that in flamKG mutant the termination of flam transcription may occur at the TE sequence, resulting in the retention of the first exon and part of the second exon but not of the downstream transcript regions. Indeed, by RT-qPCR analysis we revealed the preservation of the flam first exon transcripts and the lack of the third exon transcripts in flamKG/Df ovaries, whereas both transcript regions were absent in flamBG/Df ovaries (Figure 2A).
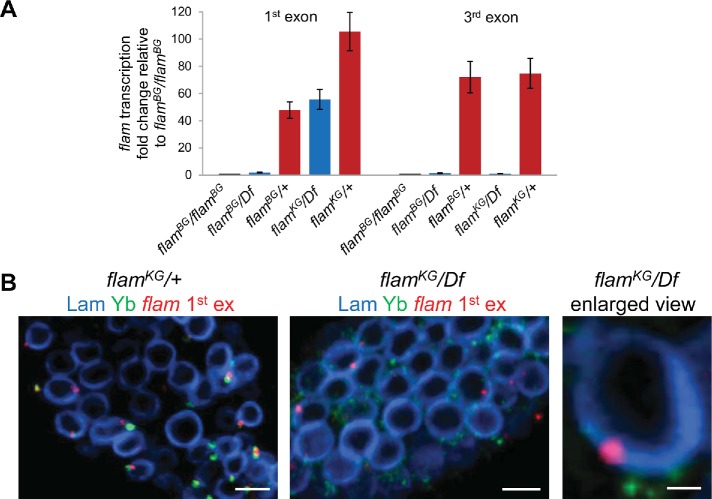
FIGURE 2:
flam first exon transcripts are retained and form perinuclear foci in flamKG/Df ovaries. (A) RT-qPCR analysis of flam transcription in flamBG and flamKG mutant ovaries using primer pairs for the first or third exons. Fold change of flam transcription in the ovaries with indicated genotypes relative to flamBG/flamBG ovaries, normalized on the Adh transcript levels, is indicated along the Y-axis (mean ± SD for two replicates). The flam third exon transcripts were barely detected in flamBG and flamKG mutants, but the flam first exon transcripts are retained in flamKG mutant. (B) RNA FISH (red) with flam first exon probe coupled with immunostaining of Yb bodies with anti-Yb antibody (green) and nuclear envelope with anti-lamin Dm0 antibody (blue) in the follicle cells of the stage 2 egg chamber of flamKG/+ (left panel), flamKG/Df (central panel), and enlarged view of flamKG/Df (right panel). Scale bars are 5 μm in the left and central panels and 0.7 μm in the right panel.
Next, we assayed by RNA FISH whether the transcripts bearing only the first and part of the second flam exon in the flamKG/Df ovaries are assembled into foci. Although less efficient than in wild type (WT), the assembly of flam foci was detected using the first exon probe in flamKG/Df but not in flamBG/Df ovaries (Figure 2B and Supplemental Figure S3). At the same time, flam fourth exon FISH signals were not revealed in either flamBG/Df, or in flamKG/Df ovaries (Supplemental Figure S3). Furthermore, median 3D distances from flam first exon signals to the nuclear lamina in flamKG/Df ovaries were similar to that seen in WT ovaries (Figure 1D), suggesting the sufficiency of this region for the aggregation of transcripts into foci near the nuclear envelope.
Recently, Dennis et al. (2016) have found that the complete absence of flam transcripts in flamBG ovaries or the blocking of their nuclear export in the nxf1 or nxt1 mutants resulted in Yb body disintegration. Additionally, it was shown that the first and second exons of flam transcripts contain the cis-regulatory signals required for Yb protein binding and for the production of piRNAs from downstream sequences (Homolka et al., 2015; Ishizu et al., 2015). We therefore investigated whether the presence of only the first and part of the second flam exon transcripts in flamKG ovaries is sufficient for the assembly and/or maintenance of the Yb bodies. Immunostaining with anti-Yb antibody revealed the disintegration of Yb bodies in the follicle cells of flamBG and flamKG mutants (Figure 3, A and B). We note that in many cases Yb bodies were almost completely dispersed in the cytoplasm of flamBG as well as flamKG ovaries, resulting in the underestimation of quantitative effects of mutations in Figure 3B. Moreover, even when incompletely disintegrated, Yb body remnants were not juxtaposed with RNA FISH signals of the flam first exon (Figure 2B). Another component of Yb body, the Armi protein, became dispersed as well in flam mutants (Supplemental Figure S4A). Therefore, whereas the transcripts corresponding to the first flam exon are assembled into flam foci, the presence of flam downstream sequence(s) and/or the longer transcripts are necessary for the formation and maintenance of the nearby Yb bodies. Of note, the intact Yb bodies were observed in piwi mutant follicle cells (Supplemental Figure S4A; Olivieri et al., 2010), ruling out the possibility that the disintegration of Yb bodies in flam mutants is an indirect effect of the loss of piRNAs or of the activation of TEs.
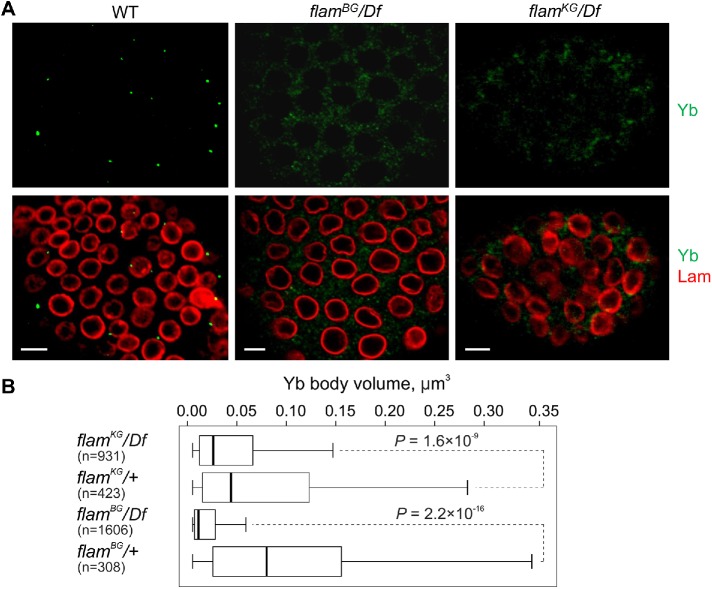
FIGURE 3:
Disintegration of Yb bodies in flam mutants. (A) Immunostaining of follicle cells from fourth stage egg chambers in flamBG/+ (WT), flamBG/flamBG, and flamKG/Df ovaries with anti-Yb (green) and anti-lamin Dm0 (red) antibodies. Scale bars are 7 μm. (B) Box plots showing a distribution of volumes of Yb bodies from egg chambers of stages 3–5 measured with IMARIS software in flam mutants in comparison with the corresponding controls. The number of counted signals is indicated for each genotype. M-W U-test was used for the pairwise comparison of distributions between mutant and control ovaries.
piRNA production from the gene transcripts is enhanced in flamKG ovaries
Interestingly, in spite of the lack of Yb bodies, the total amount of Yb protein remained nearly unchanged in flam mutants (Supplemental Figure S4B). Moreover, we observed some smaller aggregates of colocalized Yb and Armi proteins in the cytoplasm of follicle cells of flam mutants (Supplemental Figure S4C) that potentially may be involved in piRNA biogenesis. It is known that loading of piRNAs into Piwi is required for its nuclear import (Olivieri et al., 2010; Saito et al., 2010). In follicle cells of flam mutants, we revealed the unaltered nuclear localization of Piwi (Supplemental Figure S5) indicating the robust piRNA generation in these cells.
Next, to examine piRNA production in the follicle cells of flam mutant ovaries, we performed deep sequencing of small RNA libraries obtained from flamKG/Df and flamKG/+ ovaries. In flamKG mutant, piRNA production from the flam locus was greatly reduced along its entire length (Figure 4A), including the first exon and part of the second exon that are presented in the flamKG line (Supplemental Figure S6). This observation suggests that the 5′-end of flam transcripts is likely not exported to the cytoplasm for piRNA processing in flamKG mutant. Then we evaluated the effect of flam mutation on the piRNAs generated from the known genic piRNA clusters operating in somatic cells (Robine et al., 2009; Wen et al., 2014). The abundance of piRNAs derived from the 3′-UTR of tj mRNA and from other genic piRNA clusters from somatic cells was significantly increased in flamKG mutant compared with control ovaries (Figure 4, A–C, and Supplemental Data S1). Of note, the levels of piRNAs generated from other piRNA clusters which are predominantly germline-specific (Malone et al., 2009; Mohn et al., 2014), as well as the levels of microRNAs abundant in OSCs, were not noticeably altered (Figure 4A). Our analysis of the publicly available small RNA libraries (Malone et al., 2009; which, however, were sequenced drastically less deeply than our libraries) confirmed the enhanced generation of genic piRNAs in flamKG mutant (Supplemental Data S1). Thus, in follicle cells of ovaries, piRNA production from the protein-coding gene transcripts becomes even more effective when Yb body assembly is impaired, indicating that this process can be mediated by piRNA machinery components dispersed in the cytoplasm. Collectively, these data imply that the assembly of Yb bodies in follicle cells makes piRNA processing preferable toward the flam transcripts relative to the protein-coding gene transcripts.
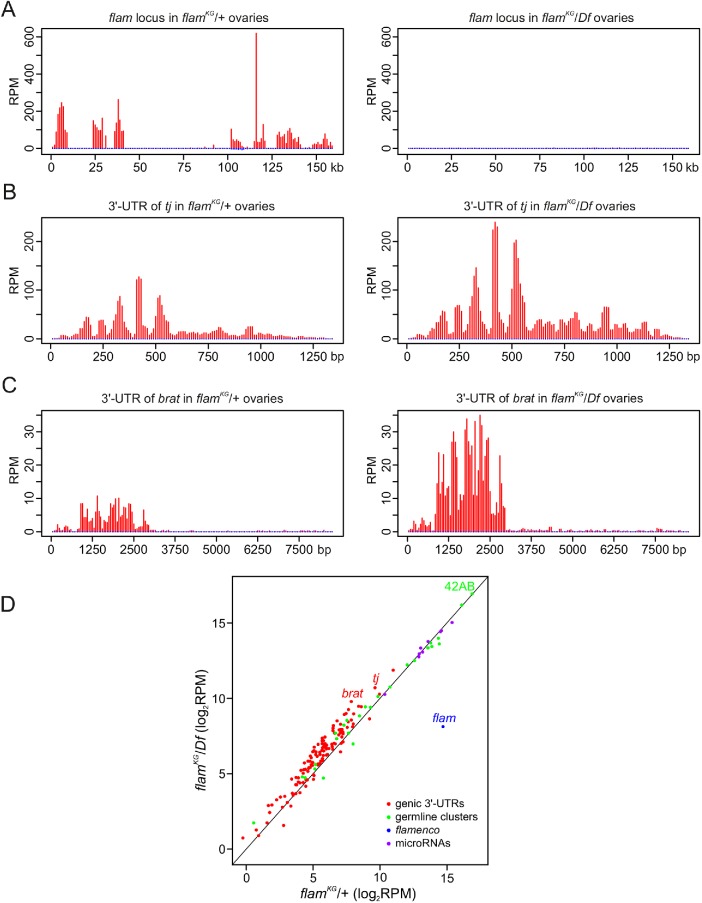
FIGURE 4:
piRNA production from the protein-coding gene transcripts is enhanced in flamKG ovaries. (A–C) piRNA density (in RPM) for uniquely mapped reads along the entire flam (A), 3′-UTR of tj (B), and 3′-UTR of brat (C) piRNA clusters. Sense and antisense piRNAs are indicated in red and blue, respectively. (D) Scatter plot showing an abundance of piRNAs (in log2 RPM) for somatic genic piRNA clusters (red dots), predominantly germline piRNA clusters (green dots), and for the top 10 microRNAs most abundant in OSCs (purple dots) in flamKG/+ (control) or flamKG/Df small RNA-seq libraries. The median fold change for somatic genic piRNA clusters and for predominantly germline piRNA clusters upon flamKG mutation is 1.6 and 1.1, respectively. The difference in piRNA production between “germline” and genic clusters is highly significant (P < 0.0001, M-W U-test).
Yb body formation depends on the flam transcripts in the follicle cells but not in the cap and escort cells
As we and others (Dennis et al., 2016) have demonstrated the disintegration of Yb bodies in flam mutant ovaries, the question arises whether the morphology of flam foci and Yb bodies is changed during normal ovarian development. The Drosophila adult ovary is composed of 12–16 ovarioles that contain a germarium and usually six to seven progressively developing egg chambers of sequentially larger sizes. The anterior end of the germarium contains germ stem cells and their daughter cystoblast cells surrounded by a somatic niche that includes terminal filament cells, cap cells, and escort cells. After mitotic divisions, germline cysts become encapsulated by a monolayer of somatic follicle cells leading to the formation of the first stage egg chamber in the germarium region 3. The egg chamber then separates from the germarium and progresses through 14 stages of maturation (Spradling et al., 2011).
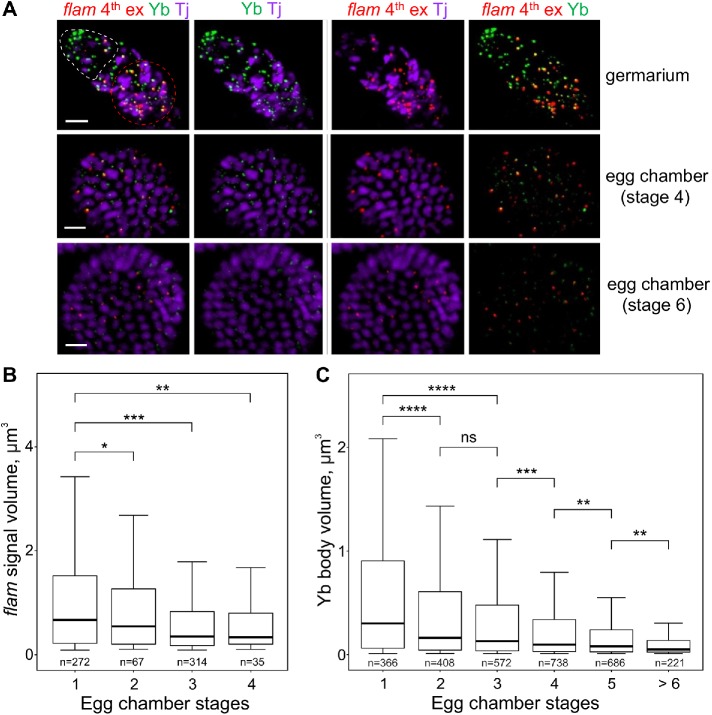
RNA FISH in WT ovaries with flam fourth or first (unpublished data) exon probes coupled with immunostaining of Yb bodies and the ovarian follicle cell marker Tj revealed that flam foci are most prominent in the germaria regions 2b and 3 (first stage egg chamber) as well as in stage 2 to 3 egg chambers, whereas at the later stages of oogenesis flam signals become weaker (Figure 5, A and B, and Supplemental Figure S7). Importantly, this flam foci reduction in the late stage follicle cells correlated with the decrease of Yb body size during the progress of oogenesis (Figure 5, A and C), thus accentuating the link between flam foci and Yb bodies in this cell type.
FIGURE 5:
In ovarian follicle cells flam foci as well as the Yb bodies become progressively smaller during egg chamber maturation. (A) RNA FISH with flam fourth exon probe (red) in WT (flamKG/+) ovaries coupled with the immunostaining of Yb bodies by anti-Yb antibody (green) and anti-Tj antibody (violet), the nuclear marker of OSCs. Egg chambers at different stages in the same ovariole are shown. The apical region of a germarium is populated by cap and escort cells (outlined by a white dotted line), and the first stage follicle cells are located distally (outlined by a red dotted line). flam foci are most prominent in the first stage follicle cells of germaria (top panel) as well as in stage 2–3 egg chambers, whereas at the later stages of oogenesis (middle and bottom panels) flam as well as Yb body signals become weaker. Scale bars are 10 μm in the top panel and 7 μm in the middle and bottom panels. (B, C) Box plots showing a distribution of volumes of the flam fourth exon (B) or the Yb body (C) signals at different stages of egg chamber maturation in WT (flamKG/+) ovaries. Volumes of flam foci or Yb bodies from the indicated stages were measured with IMARIS software. The number of counted signals is indicated for each stage. M-W U-test was used for the pairwise comparison of distributions. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
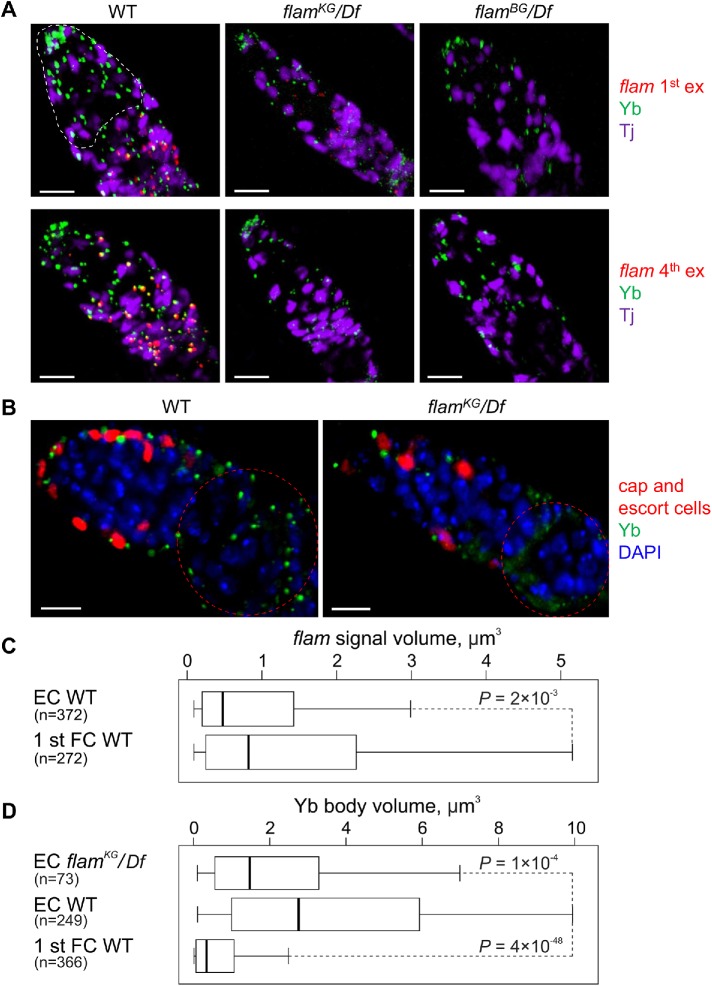
However, in the cap and escort cells, located at the anterior end of germaria, flam fourth or first exon RNA FISH signals were weaker than in the follicle cells of the first egg chamber, while the Yb bodies stained more strongly (Figure 6, A–D). Many cap and escort cells contained large Yb bodies and completely lacked flam foci. Furthermore, immunostaining of flamBG/Df and flamKG/Df ovaries with anti-Yb antibody revealed only a slight decrease of Yb body sizes in the cap and escort cells in both mutants (Figure 6, A–D). Therefore, even upon the complete absence of flam transcripts, the Yb bodies do not disintegrate, thus indicating a different mechanism of Yb body formation/maintenance in cap and escort cells.
FIGURE 6:
In cap and escort cells the assembly of Yb bodies does not depend on flam transcription. (A) RNA FISH with flam first (top panel) or fourth (bottom panel) exon probes (red) coupled with Yb (green) and Tj (violet) immunostaining. In cap and escort cells (outlined by a white dotted line in germarium) of WT (flamKG/+) ovaries (left panel) flam RNA FISH signals are weaker than in the distally located first egg chamber follicle cells, while the Yb bodies are more strongly stained. In cap and escort cells of flam mutants (middle and right panels) Yb bodies are assembled. Scale bars are 10 μm. (B) Germaria of PZ1444 (WT, left panel) or flamKG/Df; PZ1444 (right panel) ovaries (PZ1444 is a lacZ enhancer trap line that is expressed in the cap and escort cells; Margolis and Spradling, 1995; Xie and Spradling, 2000) immunostained with anti-Yb (green) and anti–β-galactosidase (red) antibodies, counterstained with DAPI (blue). Yb bodies are larger in the cap and escort cells (their nuclei are marked by red) than in the follicle cells (outlined by a red dotted line). In flamKG/Df ovaries Yb bodies are disintegrated in follicle cells, but not in the cap and escort cells. Scale bars are 10 μm. (C, D) Box plots showing a distribution of volumes of flam fourth exon (C) or Yb body (D) signals in the cap and escort cells (EC) or in the first stage egg chamber follicle cells (1 st FC) of WT (flamKG/+) or flamKG/Df ovaries. The number of counted signals is indicated for each cell type. M-W U-test was used for the pairwise comparison of distributions.
DISCUSSION
It was recently shown that in ovarian follicle cells flam piRNA precursor transcripts were assembled into foci, located close to the nuclear envelope and juxtaposed with Yb bodies, their cytoplasmic processing centers (Dennis et al., 2013; Murota et al., 2014). Our data (Supplemental Figure S1) and the results of Dennis et al. (2016) demonstrate that both in OSCs and in ovarian follicle cells flam transcripts are first gathered into foci within the nucleus and then are exported to the cytoplasm where they, with the exception of the 5′-end, become mainly localized in Yb bodies (Figure 1).
We found that the degree of colocalization of exported to the cytoplasm flam transcripts with the Yb body was progressively increased for the downstream flam regions (Figure 1C), likely reflecting the location of 5′-termini of flam transcripts outside the Yb body, and the rest of the transcript parts—inside the Yb body. Moreover, the absence of flam transcripts in the flamKG and flamBG mutants (Figure 6A and Supplemental Figure S3; Dennis et al., 2016) or their decline at the later stages of oogenesis (Figure 5, A and B) is accompanied by the disintegration of Yb bodies (Figures 3, A and B, and 5, A and C). Collectively, these results strongly support the model that flam transcripts serve not only as a substrate for piRNA production but also as a scaffold for Yb body assembly from Yb and other piRNA machinery proteins. This finding indicates the functional similarity of flam transcripts with architectural lncRNAs, which are responsible for the assembly of different protein bodies, such as paraspeckles, omega speckles, and others (for a review, see Ip and Nakagawa, 2012; Chujo and Hirose, 2017).
The 5′-end of flam transcripts producing few piRNAs (Murota et al., 2014) and only weakly colocalizing with the Yb bodies (Figure 1C) likely performs additional, but currently unknown, functions. First, the first and the second flam exons almost obligatorily present in all flam transcripts (Goriaux et al., 2014). Second, flam 5′-end contains the short cis-elements that bind Yb protein and are responsible for the targeting of downstream transcripts for processing into piRNAs (Homolka et al., 2015; Ishizu et al., 2015). Third, our results in flamKG mutant ovaries indicate that flam transcripts containing only the first and, partially, the second flam exon can still be gathered into foci near the nuclear envelope (Figure 1D). However, flam 5′-regions are not sufficient (or may not even be required) for Yb body formation (Figure 3, A and B), likely because the long flam transcripts carrying multiple sites for Yb protein binding (Murota et al., 2014) are necessary to assemble enough Yb molecules. Moreover, the peculiarities of behavior of the 5′-end of the flam transcripts imply that this region may be either cut off from the rest of the flam transcripts thus remaining in the nucleus, or shielded by an unknown complex from piRNA processing after export from the nucleus. The low abundance of continuous flam transcripts carrying both first and third exons in WT ovaries (Supplemental Figure S2) supports the former possibility.
What might be the biological function of Yb body assembly? Compartmentalization is an important feature of biological systems that increases the efficiency and specificity of different cellular activities. We propose that the compartmentalization of piRNA biogenesis machinery in the Yb body reduces the concentration of the dissolved Yb body components and thus diminishes the piRNA production from protein-coding gene transcripts, which may by chance possess cis-elements for Yb binding. Indeed, even higher levels of piRNAs from tj and other genic transcripts were generated in somatic cells of flamKG mutant ovaries (Figure 4 and Supplemental Data S1). Therefore, if flam transcripts are absent (as in flamBG ovaries) or too short (as in flamKG ovaries), the dispersed in the cytoplasm Yb body components can be gathered on the subset of protein-coding gene transcripts and induce their processing into piRNAs more efficiently than in WT ovaries.
In the Drosophila ovarian germline, transcription of the dual-strand piRNA clusters is defined by the Rhino-Deadlock-Cutoff (RDC) complex (Mohn et al., 2014; Zhang et al., 2014). Transcripts of germline clusters recruit the TREX complex (including RNA helicase UAP56) for their transport to the cytoplasmic piRNA biogenesis centers localized in the nuage (Zhang et al., 2012; Hur et al., 2016). In the ovaries, mutant for genes of RDC factors or UAP56, the cluster transcripts are not exported from the nucleus, and nuage components become dispersed in the cytoplasm (Klattenhoff et al., 2009; Pane et al., 2011; Zhang et al., 2012). Strikingly, upon these conditions, genic piRNAs become more abundant (Zhang et al., 2012). Therefore, as in the case of flam, the transcripts from dual-strand clusters are required for the assembly of nuage structure and this compartmentalization can reduce nonselective piRNA production.
We also show that the assembly of Yb bodies on the flam transcripts operates only in the follicle cells of ovaries. Although the flam locus is required for the normal functioning of somatic escort cells (Upadhyay et al., 2016), heavily stained Yb bodies may be assembled in the cap and escort cells in the absence of flam transcripts (Figure 6, A–D). This might indicate that in these cell types the transcripts from other piRNA clusters provide the RNA backbone for Yb body formation, or that other protein constituents of the Yb body are abundantly expressed thus aggregating the Yb bodies without flam RNA. Interestingly, when visualized by electron microscopy, Yb bodies appear to be located adjacent to an unknown RNA-rich structure in the cap and escort cells, whereas in the follicle cells such neighborhood was not reported (Szakmary et al., 2009).
In conclusion, our results uncover new features of the compartmentalization of piRNA biogenesis machinery in the follicle cells of ovaries, necessary for the preferential formation of piRNAs against TEs.
MATERIALS AND METHODS
Fly stocks
Drosophila melanogaster stocks were maintained under standard conditions at 25°C. w1118 P{GT1}flamBG02658 (#13912; herein flamBG), y1 P{SUPor-P}flamKG00476/FM4 (#16453; herein flamKG), and Df(1)Exel6255, w1118 P{XP-U}Exel6255/FM7c (#7723; herein Df) mutants were obtained from the Bloomington Drosophila Stock Center. For the analysis of flam mutations, we obtained transheterozygotes by crossing Df females with flamKG or flamBG males. For the analysis of piwi mutation, piwi2 and piwi3 stocks (Lin and Spradling, 1997) were crossed. To visualize escort cells, we combined flam mutations with a PZ1444 lacZ enhancer trap line that is expressed in cap and escort cells (Margolis and Spradling, 1995; Xie and Spradling, 2000). Fly stock bearing Armi-GFP transgene (Olivieri et al., 2010) was kindly provided by J. Brennecke (Institute of Molecular Biotechnology of the Austrian Academy of Sciences).
OSC maintenance
OSCs (Niki et al., 2006), kindly provided by M. Siomi (The University of Tokyo), were cultured in Shields and Sang M3 insect medium (Sigma-Aldrich) supplemented with 10% heat-inactivated fetal bovine serum (Life Technologies), 10% fly extract (http://biology.st-andrews.ac.uk/sites/flycell/flyextract.html), 10 µg/ml insulin (Sigma-Aldrich), 0.6 mg/ml glutathione (Sigma-Aldrich), 50 U/ml penicillin, and 50 µg/ml streptomycin.
RNA isolation and RT-qPCR analysis
Total RNA was isolated from manually dissected ovaries using Trizol reagent (Invitrogen) and purified from genomic DNA by DNA-free kit (Ambion). Total RNA (1 μg) was used for the reverse transcription (RT) reaction with random primer and Superscript II reverse transcriptase (Invitrogen). The resulting cDNAs in at least three biological replicates were analyzed by RT-qPCR performed in a MJ Mini instrument (Bio-Rad) using SYBR Green chemistry (Applied Biosystems). For detection of flam transcripts containing first and third joint exons, either 1 μg of total RNA with random primer or 15 μg of total RNA with the specific for the third exon reverse primer was used for the RT reactions. Then, in the case of third exon-specific primer reaction, cDNAs were treated with RNase H (Invitrogen; 10 U, 15 min) and purified by Wizard Gel and PCR Clean-Up System (Promega). Primer pairs for RT-qPCR are listed in Supplemental Table S1.
Immunostaining
Immunostaining was performed basically as described in Shpiz et al. (2014) with some modifications. OSCs in the growth phase were collected and rinsed two times in phosphate-buffered saline (PBS). Ovaries from 1- to 5-d-old females were manually isolated in PBT (PBS containing 0.01% Tween-20) at 4°C and then rinsed in PBS. Cells or ovaries were fixed in 4% formaldehyde (on PBT) for 25 min at room temperature. Fixation was stopped by incubation with 0.25 M glycine (Sigma-Aldrich) for 5 min. Then cells or ovaries were washed in PBS three times for 10 min at room temperature, permeabilized with PBTX (PBS with 0.1% Tween-20, 0.3% Triton X-100) for 10 min, blocked with PBTX containing 3% normal goat serum (NGS; Invitrogen) at room temperature for 1 h (cells) or for 3 h (ovaries), incubated with primary antibody in PBTX containing 3% NGS for 3 h (cells) or for 7 h (ovaries) at room temperature, or overnight at 4°C, washed in PBTX three times for 10 min at room temperature, incubated with secondary antibodies (1:1000) in PBTX containing 3% NGS for 3 h (cells) or for 7 h (ovaries) at room temperature, or overnight at 4°C, and then washed in PBTX three times for 10 min at room temperature in a dark chamber. Coverslips were mounted with a drop of SlowFade Gold Antifade reagent (Invitrogen) containing DAPI. As the primary, rabbit polyclonal anti-lamin Dm0 (1:500; Osouda et al., 2005), mouse monoclonal anti-Yb (1:100; Murota et al., 2014), anti-Armi (1:50; Saito et al., 2010), anti-Piwi (1:500; Saito et al., 2006), chicken anti–β-galactosidase (1:500; Abcam; ab9361), and guinea pig anti-Tj (1:5000; Li et al., 2003) antibodies were used. As the secondary, Alexa Fluor 546–conjugated goat anti-rabbit immunoglobulin G (IgG) (Invitrogen) or Alexa Fluor 488–conjugated goat anti-mouse IgG (Invitrogen) antibodies were used.
RNA FISH coupled with immunostaining
RNA FISH was performed basically as described in Shpiz et al. (2014) with some modifications. DNA fragments corresponding to flam RNA FISH probes were PCR-amplified on the Drosophila genomic DNA using primer pairs (Supplemental Table S1) carrying the T7 RNA polymerase promoter sequence at their 5′-ends. Digoxigenin (DIG)-11-UTP (Roche) labeling of transcripts generated from these fragments by T7 RNA polymerase was carried out according to manufacturer’s instructions. RNA probes were dissolved in 100 µl of hybridization buffer (HB; 50% formamide, 4× SSC [1× SSC: 150 mM NaCl, 15 mM Na3 citrate, pH 7.0], 100 mM Na3PO4, pH 7.0) at a concentration of ∼10 µg/ml.
OSCs in the growth phase or ovaries, manually isolated from 1- to 5-d-old females, were rinsed twice with PBS and then fixed with 4% formaldehyde (on PBT) for 25 min. Fixation was stopped by incubation with 0.25 M glycine (Sigma-Aldrich) for 5 min. Cells or ovaries were washed in PBT three times for 10 min, permeabilized in PBTX for 10 min, blocked with PBTX containing 3% NGS (Invitrogen) for 1 h at room temperature, and washed in PBTX three times for 10 min. Cells or ovaries were prepared for hybridization by sequential incubation for 15 min at 42°C each in 20, 50, 80, and 100% HB on PBT. RNA FISH probes (in HB) were heated at 80°C for 5 min and then cooled on ice. Herring sperm DNA (Promega) and yeast tRNA (Invitrogen) were then added to the probes (both to 0.5 µg/µl), and RNA hybridization was performed overnight at 42°C. Cells or ovaries were washed sequentially for 15 min at 42°C in 50% formamide, 2× SSC; 40% formamide, 2× SSC; 30, 20, and 10% formamide on PBT. Further, cells or ovaries were washed several times in PBT at room temperature, blocked in PBT containing 3% NGS at room temperature for 2 h, incubated with primary antibody in PBT containing 3% NGS for 7 h at room temperature, or overnight at 4°C, and then washed in PBT three times for 10 min. RNA FISH signals in ovaries were subjected to a tyramide amplification procedure according to manufacturer’s instructions (Perkin Elmer). Cells or ovaries were incubated with the secondary antibody (1:1000) in PBT containing 3% NGS for 7 h at room temperature, or overnight at 4°C, and then washed in PBT three times for 10 min in a dark chamber. Coverslips were mounted with a drop of SlowFade Gold Antifade reagent (Invitrogen) containing DAPI. As the primary, rabbit polyclonal anti-lamin Dm0 (1:500; Osouda et al., 2005), mouse monoclonal anti-Yb (1:100; Murota et al., 2014), and sheep polyclonal anti–DIG-FITC (1:200; Roche) antibodies were used. As the secondary, Alexa Fluor 546–conjugated goat anti-rabbit IgG (Invitrogen), Alexa Fluor 633–conjugated goat anti-mouse IgG (Invitrogen), and Alexa Fluor 488–conjugated goat anti-Fluorescein/Oregon Green IgG (Invitrogen) antibodies were used.
Western blot analysis
Ovarian lysates were fractionated by SDS–PAGE (10% acrylamide gel) and transferred to a polyvinylidene difluoride membrane (Immobilon-P; Millipore). Blots were developed using alkaline phosphatase–conjugated secondary antibody (Sigma) and the Immun-Star AP detection system (Bio-Rad). The following primary antibodies were used: mouse monoclonal anti-Yb (1:500; Murota et al., 2014) and mouse monoclonal anti-beta actin (1:3000; Abcam; ab8224).
Measuring distances between RNA FISH signals, Yb bodies, and nuclear lamina
Three-dimensional image stacks were recorded with a confocal LSM 510 Meta laser scanning microscope (Zeiss). Optical sections with 0.5- to 1.0-μm intervals along the Z-axis were captured. Images were processed and analyzed using IMARIS 7.4.2 software (Bitplane AG) with blind experimental setup. Images were thresholded to eliminate hybridization and immunostaining background, and nuclei containing Yb body foci obviously located in the cytoplasm were taken for further analysis. The distances between the signals and the nuclear envelope were counted as was previously described (Shevelyov et al., 2009). Briefly, the nuclear rim colored by anti-lamin Dm0 antibody was manually outlined in each section by the middle to automatically reconstruct the surface of the nuclei. One “measurement point” was positioned in the optical section with the brightest FISH or Yb signal, at its visually determined center, and another one was placed on the reconstructed nuclear surface at the point of its earliest intersection with the progressively growing sphere of the first “measurement point.” The distance between two “measurement points” (the shortest distance between the RNA FISH signal or Yb signal and the nuclear rim as well as the distances between the centers of the RNA FISH and Yb signals) was measured for each nucleus (Supplemental Figure S8). Data were obtained in two independent experiments for 40–50 nuclei per experiment (Supplemental Table S2).
Measuring of Yb body volumes in WT and mutant ovaries
Confocal images containing several hundreds of flam RNA FISH signals or immunostained Yb bodies from two to three independent experiments were processed and analyzed with the same set of parameters for WT or for flam mutant ovaries using IMARIS 7.4.2 software (Bitplane AG). The surfaces of flam signals or Yb bodies were automatically reconstructed. For background subtraction, the images were thresholded to the 40% (for Yb) or 25% (for flam) of maximal intensity of the channel because at this value the Yb or flam fluorescence was not detected outside egg chambers. The volumes of flam signals or Yb bodies exceeding the background threshold (>0.09 µm3 for flam signals or >0.01 µm3 and <10 µm3 for Yb signals) were retrieved for further analysis.
Small RNA cloning
Small RNA cloning was performed as described in Muerdter et al. (2012). In brief, total RNA was isolated from flamKG/Df or flamKG/+ ovaries by Trizol reagent (Invitrogen), resolved on a denaturing 12% polyacrylamide gel, and small RNAs corresponding to 19–29 nt fraction were isolated. Libraries were barcoded according to Illumina TrueSeq Small RNA sample prep kit instructions and sequenced using the Illumina NovoSec-6000 instrument. Small RNA sequencing data are deposited in the Gene Expression Omnibus (GEO) under GSE126016 accession number.
piRNA mapping
piRNA data for the flamKG/+ and flamKG/flamKG ovaries were retrieved from the GEO NCBI database (GSM379054, GSM379055; Malone et al., 2009). Adapter sequences were removed using the “FlexBar” tool (Dodt et al., 2012). Sequenced reads of small RNA libraries obtained in our work were trimmed for adapter sequences, masked for low-complexity or low-quality sequences and filtered by size ranging from 23 to 29 nt (to obtain piRNA fraction) using “Cutadapt” (Martin, 2011). For the analysis of microRNA abundance filtering by size was omitted. Processed reads were then uniquely mapped to Release 5.41 of Drosophila genome using “Bowtie2” (Langmead and Salzberg, 2012). “R” with the additional packages including “GenomicRanges” and “GenomicAlignments” (Lawrence et al., 2013) was used to calculate reads per million (RPM) values for the various genomic regions: for the 3′-UTRs of genic piRNA clusters (their coordinates were obtained from Robine et al., 2009), for the flam locus, for the collection of the predominantly germline piRNA clusters (including 42AB cluster; their coordinates were retrieved from Rosenkranz, 2016), as well as for the microRNAs (coordinates of pre-microRNAs were retrieved from FlyBase dmel_r5.57 annotation). For the analysis of data from Malone et al. (2009), only genes and microRNAs with read counts more than 10 were used. The top 10 expressed microRNAs in OSCs were identified by mapping reads from the OSC small RNA libraries (Sienski et al., 2012; GSM1914769) to Drosophila genome and by counting reads mapped to microRNA genes using STAR (Dobin et al., 2013). The scatter plots of RPM values were generated using “R” with the “ggplot2” package (Wickham, 2016).
Statistical analysis
For P value estimation, the Mann-Whitney (M-W) U-test was used for comparison of two sample distributions.
Supplementary Material
Acknowledgments
We thank Alla I. Kalmykova and Pavel A. Komarov for assistance with the construction of small RNA-seq libraries; Mikiko C. Siomi for anti-Piwi, anti-Yb, anti-Armi, and anti-Tj antibodies as well as for the OSC culture; Paul A. Fisher for anti-lamin Dm0 antibody; and Julius Brennecke and the Bloomington Drosophila Stock Center for the fly stocks. This work was supported by grants from the Russian Science Foundation (Grant no. 14-14-01076), from the Russian Foundation for Basic Research (Grant no.16-34-01015 to O.A.S., Grant no.16-04-01524 to M.S.K., and Grant no.18-34-00140 to A.A.I.), and from the Presidium of Russian Academy of Sciences (Program for Molecular and Cellular Biology).
Abbreviations used:
- CLIP
cross-linking immunoprecipitation
- EJC
exon junction complex
- FISH
fluorescence in situ hybridization
- lncRNA
long noncoding RNA
- microRNA
micro RNA
- OSC
ovarian somatic cell
- piRNA
PIWI-interacting small RNA
- RT-qPCR
quantitative PCR after reverse transcription
- RPM
reads per million
- TE
transposable element
- 3′-UTR
3′-untranslated region.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-10-0591) on April 3, 2019.
REFERENCES
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. (2007). Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila . Cell , 1089–1103. [DOI] [PubMed] [Google Scholar]
- Chujo T, Hirose T. (2017). Nuclear bodies built on architectural long noncoding RNAs: unifying principles of their construction and function. Mol Cells , 889–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Hannon GJ. (2016). One loop to rule them all: the ping-pong cycle and piRNA-guided silencing. Trends Biochem Sci , 324–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis C, Brasset E, Sarkar A, Vaury C. (2016). Export of piRNA precursors by EJC triggers assembly of cytoplasmic Yb-body in Drosophila . Nat Commun , 13739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis C, Zanni V, Brasset E, Eymery A, Zhang L, Mteirek R, Jensen S, Rong YS, Vaury C. (2013). “Dot COM,” a nuclear transit center for the primary piRNA pathway in Drosophila . PLoS One , e72752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis C, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras T. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics , 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodt M, Roehr JT, Ahmed R, Dieterich C. (2012). FLEXBAR—flexible barcode and adapter processing for next-generation sequencing platforms. Biology (Basel) , 895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goriaux C, Desset S, Renaud Y, Vaury C, Brasset E. (2014). Transcriptional properties and splicing of the flamenco piRNA cluster. EMBO Rep , 411–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase AD, Fenoglio S, Muerdter F, Guzzardo PM, Czech B, Pappin DJ, Chen C, Gordon A, Hannon GJ. (2010). Probing the initiation and effector phases of the somatic piRNA pathway in Drosophila . Genes Dev , 2499–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler D, Olivieri D, Novatchkova M, Gruber FS, Meixner K, Mechtler K, Stark A, Sachidanandam R, Brennecke J. (2011). A systematic analysis of Drosophila TUDOR domain-containing proteins identifies Vreteno and the Tdrd12 family as essential primary piRNA pathway factors. EMBO J , 3977–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homolka D, Pandey RR, Goriaux C, Brasset E, Vaury C, Sachidanandam R, Fauvarque MO, Pillai RS. (2015). PIWI slicing and RNA elements in precursors instruct directional primary piRNA biogenesis. Cell Rep , 418–428. [DOI] [PubMed] [Google Scholar]
- Huang X, Fejes Tóth K, Aravin AA. (2017). piRNA biogenesis in Drosophila melanogaster . Trends Genet , 882–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur JK, Luo Y, Moon S, Ninova M, Marinov GK, Chung YD, Aravin AA. (2016). Splicing-independent loading of TREX on nascent RNA is required for efficient expression of dual-strand piRNA clusters in Drosophila . Genes Dev , 840–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip JY, Nakagawa S. (2012). Long non-coding RNAs in nuclear bodies. Dev Growth Differ , 44–54. [DOI] [PubMed] [Google Scholar]
- Ipsaro JJ, Haase AD, Knott SR, Joshua-Tor L, Hannon GL. (2012). The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature , 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizu H, Iwasaki YW, Hirakata S, Ozaki H, Iwasaki W, Siomi H, Siomi MC. (2015). Somatic primary piRNA biogenesis driven by cis-acting RNA elements and trans-acting Yb. Cell Rep , 429–440. [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Xi H, Li C, Lee S, Xu J, Khurana JS, Zhang F, Schultz N, Koppetsch BS, Nowosielska A, et al (2009). The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell , 1137–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods , 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ. (2013). Software for computing and annotating genomic ranges. PLoS Comput Biol , e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MA, Alls JD, Avancini RM, Koo K, Godt D. (2003). The large Maf factor Traffic Jam controls gonad morphogenesis in Drosophila . Nat Cell Biol , 994–1000. [DOI] [PubMed] [Google Scholar]
- Lin H, Spradling AC. (1997). A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development , 2463–2476. [DOI] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ. (2009). Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell , 522–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis J, Spradling A. (1995). Identification and behavior of epithelial stem cells in the Drosophila ovary. Development , 3797–3807. [DOI] [PubMed] [Google Scholar]
- Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal , 10–12. [Google Scholar]
- Mével-Ninio M, Pelisson A, Kinder J, Campos AR, Bucheton A. (2007). The flamenco locus controls the gypsy and ZAM retroviruses and is required for Drosophila oogenesis. Genetics , 1615–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Sienski G, Handler D, Brennecke J. (2014). The Rhino-Deadlock-Cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila . Cell , 1364–1379. [DOI] [PubMed] [Google Scholar]
- Muerdter F, Olovnikov I, Molaro A, Rozhkov NV, Czech B, Gordon A, Hannon GJ, Aravin AA. (2012). Production of artificial piRNAs in flies and mice. RNA , 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murota Y, Ishizu H, Nakagawa S, Iwasaki YW, Shibata S, Kamatani MK, Saito K, Okano H, Siomi H, Siomi MC. (2014). Yb integrates piRNA intermediates and processing factors into perinuclear bodies to enhance piRISC assembly. Cell Rep , 103–113. [DOI] [PubMed] [Google Scholar]
- Niki Y, Yamaguchi T, Mahowald AP. (2006). Establishment of stable cell lines of Drosophila germ-line stem cells. Proc Natl Acad Sci USA , 16325–16330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Ishizu H, Saito K, Fukuhara S, Kamatani MK, Bonnefond L, Matsumoto N, Nishizawa T, Nakanaga K, Aoki J, et al (2012). Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature , 284–287. [DOI] [PubMed] [Google Scholar]
- Olivieri D, Senti KA, Subramanian S, Sachidanandam R, Brennecke J. (2012). The cochaperone Shutdown defines a group of biogenesis factors essential for all piRNA populations in Drosophila . Mol Cell , 954–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri D, Sykora MM, Sachidanandam R, Mechtler K, Brennecke J. (2010). An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila . EMBO J , 3301–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osouda S, Nakamura Y, de Saint Phalle B, McConnell M, Horigome T, Sugiyama S, Fisher PA, Furukawa K. (2005). Null mutants of Drosophila B-type lamin Dm0 show aberrant tissue differentiation rather than obvious nuclear shape distortion or specific defects during cell proliferation. Dev Biol , 219–232. [DOI] [PubMed] [Google Scholar]
- Pandey RR, Homolka D, Chen KM, Sachidanandam R, Fauvarque MO, Pillai RS. (2017). Recruitment of Armitage and Yb to a transcript triggers its phased processing into primary piRNAs in Drosophila ovaries. PLoS Genet , e1006956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane A, Jiang P, Zhao DY, Singh M, Schüpbach T. (2011). The Cutoff protein regulates piRNA cluster expression and piRNA production in the Drosophila germline. EMBO J , 4601–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane A, Wehr K, Schupbach T. (2007). zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell , 851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H, Watanabe T, Ku HY, Liu N, Zhong M, Lin H. (2011). The Yb body, a major site for Piwi-associated RNA biogenesis and a gateway for Piwi expression and transport to the nucleus in somatic cells. J Biol Chem , 3789–3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robine N, Lau NC, Balla S, Jin Z, Okamura K, Kuramochi-Miyagawa S, Blower MD, Lai EC. (2009). A broadly conserved pathway generates 3’UTR-directed primary piRNAs. Curr Biol , 2066–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenkranz D. (2016). piRNA cluster database: a web resource for piRNA producing loci. Nucleic Acids Res , D223–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC. (2009). A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila . Nature , 1296–1299. [DOI] [PubMed] [Google Scholar]
- Saito K, Ishizu H, Komai M, Kotani H, Kawamura Y, Nishida KM, Siomi H, Siomi MC. (2010). Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila . Genes Dev , 2493–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC. (2006). Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev , 2214–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevelyov YY, Lavrov SA, Mikhaylova LM, Nurminsky ID, Kulathinal RJ, Egorova KS, Rozovsky YM, Nurminsky DI. (2009). The B-type lamin is required for somatic repression of testis-specific gene clusters. Proc Natl Acad Sci USA , 3282–3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpiz S, Lavrov S, Kalmykova A. (2014). Combined RNA/DNA fluorescence in situ hybridization on whole-mount Drosophila ovaries. Methods Mol Biol , 161–169. [DOI] [PubMed] [Google Scholar]
- Sienski G, Dönertas D, Brennecke J. (2012). Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell , 964–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A, Fuller MT, Braun RE, Yoshida S. (2011). Germline stem cells. Cold Spring Harb Perspect Biol , a002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szakmary A, Reedy M, Qi H, Lin H. (2009). The Yb protein defines a novel organelle and regulates male germline stem cell self-renewal in Drosophila melanogaster . J Cell Biol , 613–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay M, Martino Cortez Y, Wong-Deyrup S, Tavares L, Schowalter S, Flora P, Hill C, Nasrallah MA, Chittur S, Rangan P. (2016). Transposon dysregulation modulates dWnt4 signaling to control germline stem cell differentiation in Drosophila . PLoS Genet , e1005918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J, Mohammed J, Bortolamiol-Becet D, Tsai H, Robine N, Westholm JO, Ladewig E, Dai Q, Okamura K, Flynt AS, et al (2014). Diversity of miRNAs, siRNAs, and piRNAs across 25 Drosophila cell lines. Genome Res , 1236–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. (2016). ggplot2: Elegant Graphics for Data Analysis, New York: Springer-Verlag. [Google Scholar]
- Xie T, Spradling AC. (2000). A niche maintaining germ line stem cells in the Drosophila ovary. Science , 328–330. [DOI] [PubMed] [Google Scholar]
- Zamparini AL, Davis MY, Malone CD, Vieira E, Zavadil J, Sachidanandam R, Hannon GJ, Lehmann R. (2011). Vreteno, a gonad-specific protein, is essential for germline development and primary piRNA biogenesis in Drosophila . Development , 4039–4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wang J, Schultz N, Zhang F, Parhad SS, Tu S, Vreven T, Zamore PD, Weng Z, Theurkauf WE. (2014). The HP1 homolog Rhino anchors a nuclear complex that suppresses piRNA precursor splicing. Cell , 1353–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wang J, Xu J, Zhang Z, Koppetsch BS, Schultz N, Vreven T, Meignin C, Davis I, Zamore PD, et al (2012). UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery. Cell , 871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.