Abstract
Cyanobacteria produce numerous invaluable bioactive secondary metabolites (natural products) including alkaloids, isoprenoids, non-ribosomal peptides, and polyketides. However, the genomic organization of the biosynthetic gene clusters, complex gene expression patterns, and low compound yields synthesized by the native producers currently limits access to the vast majority of these valuable molecules for detailed studies. Molecular cloning and expression of such clusters in heterotrophic hosts is often precarious owing to genetic and biochemical incompatibilities. Production of such biomolecules in photoautotrophic hosts analogous to the native producers is an attractive alternative that has been under-explored. Here, we describe engineering of the fast-growing cyanobacterium Synechococcus elongatus UTEX 2973 to produce key compounds of the hapalindole family of indole-isonitrile alkaloids. Engineering of the 42-kbp “fam” hapalindole pathway from the cyanobacterium Fischerella ambigua UTEX 1903 into S2973 was accomplished by rationally re-constructing six to seven core biosynthetic genes into synthetic operons. The resulting Synechococcus strains afforded controllable production of indole-isonitrile biosynthetic intermediates and hapalindoles H and 12-epi-hapalindole U at a titer of 0.75 – 3 mg/L. Exchanging genes encoding fam cyclase enzymes in the synthetic operons was employed to control the stereochemistry of the resulting product. Establishing a robust expression system provides a facile route to scalable levels of similar natural and new forms of bioactive hapalindole derivatives and its structural relatives (e. g. fischerindoles, welwitindolinones). Moreover, this versatile expression system represents a promising tool for exploring other functional characteristics of orphan gene products that mediate the remarkable biosynthesis of this important family of natural products.
Keywords: cyanobacteria, natural product, isoprenoid, alkaloid, isonitrile
Cyanobacteria are prolific producers of more than 1,000 natural products with potent bioactivities.1, 2 While a subset of these compounds, including microcystins, is responsible for the harmful effects of algal blooms, many cyanobacterial natural products (cNPs) have promising bioactivities valuable to human health. Genome sequencing of diverse genera of cyanobacteria has revealed a tremendous diversity in the types of encoded biosynthetic gene clusters (BGCs) and identified unique enzymology involved in their production.3, 4 Genomic studies also highlight that we have still only minimally explored the true metabolic potential encoded by these organisms.2 Given that NPs and NP-inspired derivatives constitute roughly half of new pharmaceutical lead compounds,5, 6 cyanobacteria hold tremendous potential as sources of new bioactive molecules (or core scaffolds) useful to medicine and agriculture. However, ensuring a stable and sufficient production of NPs for characterization is a key limiting step hindering further development.5, 7–9 These limitations depend on the type of NP but are primarily caused by the slow growth and often challenging culture conditions for environmental strains along with the inherently low amounts produced in native organisms. Coupling these challenges with difficult purification methods and complex molecular structures can hinder or entirely preclude large-scale chemical synthesis. Accordingly, there have been several reports of expressing cNP genes and gene clusters in heterotrophic10–14 and more recently in cyanobacterial hosts15, 16 in order to gain access to these molecules. In some cases, limitations in non-native substrate pools and cofactors required by specialized NP biosynthetic enzymes can lead to low production yields and/or production of incompletely processed variants in the non-native hosts.15, 17 Expression of these pathways in related species could overcome some of these limitations owing to more closely matched cofactor availability (e.g. higher NADPH to NADH ratio for redox protein partners), compatibility of genetic elements such as promoters, and similar codon usage.18–21 Although traditionally considered slow-growing, some cyanobacterial strains exhibit rapid photoautotrophic growth rates (on par with yeast)22, 23 and thus represent attractive, sustainable production chassis for cNPs.
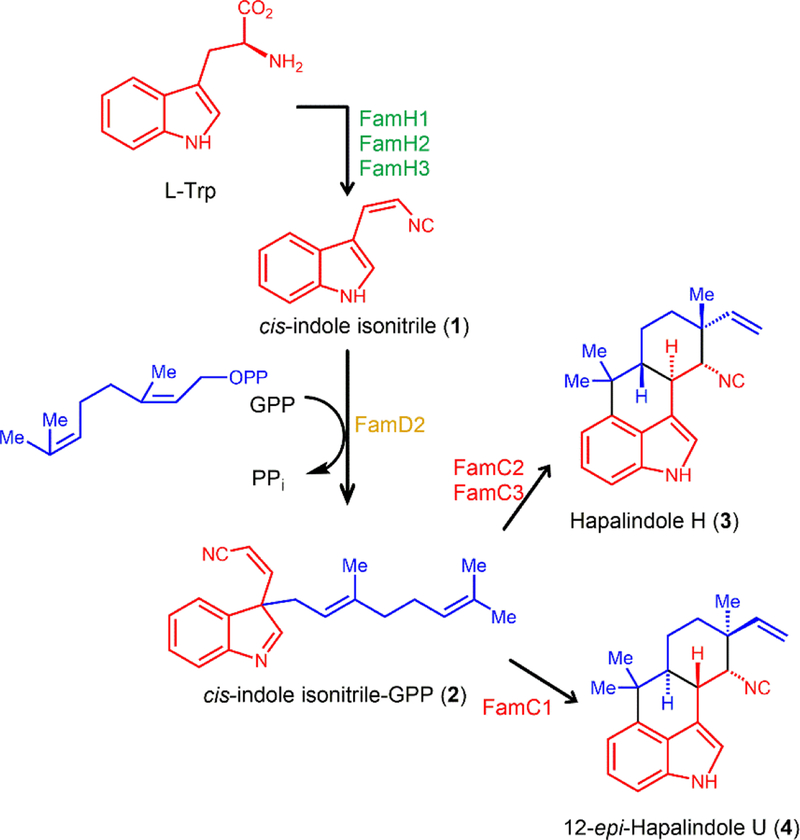
Hapalindoles are a large family of 80+ polycyclic indole-isonitrile alkaloids produced by branching cyanobacteria of the order Stigonematales, obtained from fresh water and terrestrial habitats.24 Many hapalindoles exhibit antibacterial, antimycotic, insecticidal and cytotoxic activities, sometimes with a potency exceeding that of current clinical therapeutics.24 For instance, hapalindole H is a potent inhibitor of the Nf-κB pathway in prostate cancer cells,25 is cytotoxic towards several other malignant lines26 and is active against several important fungal and bacterial pathogens.27 The hapalindole family also includes several related cNP compounds including ambiguines, fischerindoles, hapalindolinones and welwitindolinones that can have different numbers of rings and post-assembly structural modifications.24, 28 BGCs have been mined from over a dozen hapalindole-type cyanobacterial species and the early steps in the biosynthesis of their core ring systems were recently elucidated.28–30 Biosynthesis is initiated from L-tryptophan, which is converted into cis indole-isonitrile and subsequently condensed with geranyl-pyrophosphate (GPP) and cyclized to produce core hapalindoles, fischerindoles and welwitindolinones (Figure 1).29–35 However, much still remains to be understood about the subsequent alkaloid ring expansion mechanism and stereo- and site-specific late-stage tailoring reactions.36–38 It is these modifications of the core scaffold that lead to the notable structural diversity observed in hapalindole-type cNPs and modulation of the compound bioactivities.
Figure 1.
Biosynthesis of cis indole-isonitrile (1), prenylated cis indole-isonitrile-GPP intermediate (2), and hapalindoles (3, 4) from L-tryptophan. Enzymes from the hapalindole BGC in Fischerella ambigua UTEX 1903 that catalyze each step are indicated.
Here, we report the application of synthetic biology to produce hapalindole-type cNPs in engineered strains of the fast-growing unicellular cyanobacterium Synechococcus elongatus UTEX 2973 (S2973).22, 39 Cloning and rational re-factoring of key genes from the 42-kilobase hapalindole fam BGC from Fischerella ambigua UTEX 1903 afforded controllable photoautotrophic production of cis indole-isonitrile, the central geranylated indole-isonitrile intermediate, hapalindole H, and 12-epi hapalindole U in S2973 from carbon dioxide (CO2). Establishment of these foundational strains provides a broad platform to further dissect the unique steps involved in hapalindole biosynthesis and to generate more complex natural and synthetic derivatives using in vivo pathway expansion and/or in vitro semisynthesis.
Results and Discussion
Establishment of a gene promoter tool set for use in S2973.
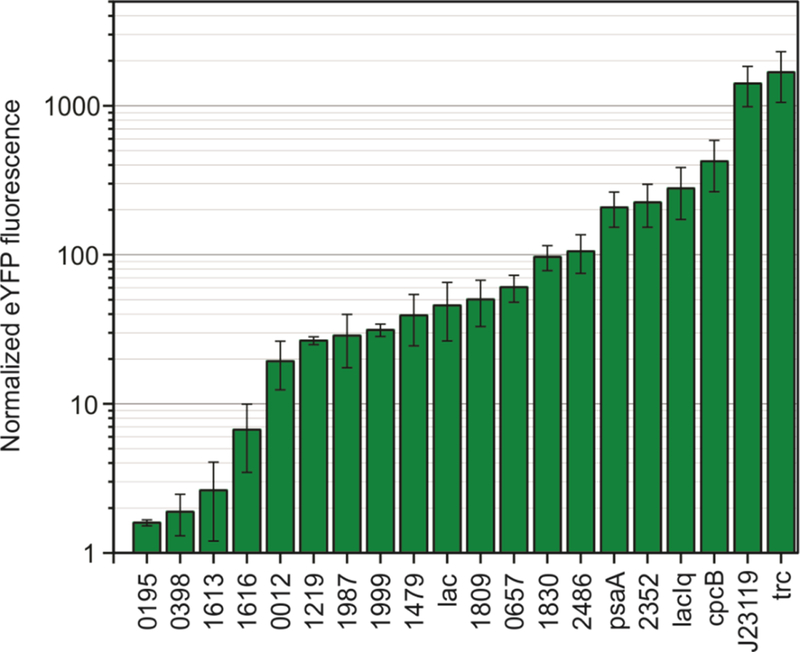
Before re-factoring the fam cluster, we worked to identify a set of endogenous and synthetic promoters that we could use to construct multiple synthetic operons, as needed. Re-using genetic elements in a design can result in instability of the construct40 and we aimed to avoid this issue by identifying a set of different promoter sequences that are functional in S2973 and that together would provide access to a wide expression range. Furthermore, there have been few reports relating to gene promoter characterization in S2973 and we sought to determine the relative strengths of different promoter elements before including them in our constructs. Thus, we screened a total of 20 native S2973, Escherichia coli (E. coli), and synthetic promoters for expression in S2973 using enhanced yellow fluorescence protein (eYFP). The promoters tested were chosen based on transcriptomic data for the closely related strain Synechococcus elongatus PCC 794241 and from a collection of promoters previously used in other cyanobacteria42, 43 (Table S1). The majority of the native S2973 promoters we tested have not been previously characterized using a reporter gene. Promoter sequences were screened by inserting them upstream of a promoter-less eYFP coding sequence on plasmid pSL3068,44 conjugating the vectors into S2973, and measuring the fluorescence of the strains cultured at 38 °C. We included the phycocyanin cpcB promoter among the native elements tested but truncated the promoter sequence to remove 218 basepairs from the 5’UTR sequence while retaining the native ribosome binding site (Table S1). This promoter set afforded a roughly 1000-fold eYFP expression range (Figure 2). The synthetic promoters trc and J23119 (BioBrick BBa_J23119) resulted in the highest eYFP production, which was roughly five-fold higher than the truncated PcpcB, the strongest native element tested. From this set of promoters, we used Ptrc, PcpcB and PJ23119 to express the regulatory and hapalindole biosynthetic genes in S2973.
Figure 2.
Enhanced yellow fluorescence protein (eYFP) data for twenty promoters characterized in S2973. Fluorescence data was normalized to optical density at 730 nm and is presented as the ratio of this normalized fluorescence to that of a strain carrying the eYFP plasmid without a promoter. Cells were cultured in air under 50 μE m−2 s−1 fluorescent illumination at 38 °C in 250 ml Erlenmeyer flasks. Errors bars are 1 SD from the mean derived from at least three biological replicates. Endogenous promoter numbers are named after the corresponding JGI gene locus tag in the closely related cyanobacterium Synechococcus elongatus PCC 7942.
Rational re-structuring of the fam BGC and production of cis indole-isonitrile in S2973.
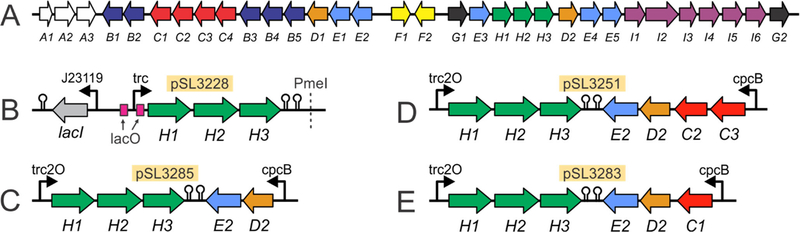
A subset of hapalindole biosynthetic genes from Fischerella ambigua UTEX 1903 (“fam” genes) were cloned for expression in S2973. In addition to biosynthetic genes that synthesize the core hapalindole scaffold, the fam cluster (Figure 3A) also encodes genes for late-stage tailoring (famB genes and famD1), tryptophan biosynthesis (famI genes) and some genes from the methylerythritol phosphate and shikimate pathways (famE genes). Putative transporter (famA1 – 3) and regulatory genes (famF1 and F2) as well as the two famG genes remain to be characterized. Hapalindole biosynthesis is initiated with L-tryptophan and is converted into the key cis indole-isonitrile (1) by the combined action of FamH1, FamH2 and FamH3 enzymes29, 34 (Figure 1). FamH1 and FamH2 share 62% sequence identity and are homologues of the isonitrile synthase enzyme PvcA from Pseudomonas aeruginosa45 while FamH3 is homologous to PvcB and has a fold similar to that of α-ketoglutarate-dependent non-heme iron oxygenases.34 FamH1 – 3 catalyze the reaction between L-tryptophan and ribulose-5-phosphate to generate indole-isonitrile 129, but little is known about the enzyme mechanisms or protein structures. Compound 1 is subsequently prenylated with geranyl pyrophosphate (GPP) by the FamD2 aromatic prenyltransferase to form a second common intermediate.29, 30 This common prenylated indole-isonitrile-GPP precursor (2) is converted into different hapalindoles by cyclase enzymes encoded in the BGCs.30, 32, 33 The synthetic fam operons were designed to express the requisite genes in S2973 on autonomously replicating broad host-range RSF1010 plasmids46 (Figure S1). Plasmid assembly is described in the Methods section; primers are listed in Table S2 and strain genotypes in Table S3. We chose to use plasmids to express the fam genes instead of integrating the pathway into the S2973 genome. The primary benefit to using plasmids is that it allows more rapid generation and testing of new constructs and genetic designs because it avoids the necessity to fully segregate strains. The fam pathway has not been targeted previously for engineering, and plasmid constructs allowed us to more efficiently screen the different genetic components needed to express and regulate the operons. A secondary reason that we chose an RSF1010-type plasmid is that it can be replicated in a variety of cyanobacterial strains.46 This versatility enables future testing of our constructs in other cyanobacteria and comparing them to S2973. However, genome integration typically provides higher strain stability43 and this option is possible in S2973 using several tools including knock-in suicide vectors with selection markers43 or markerless CRISPR genome editing.47
Figure 3.
Diagrams of the hapalindole-producing ‘fam’ BGC from Fischerella ambigua UTEX 1903 and the synthetic fam operons constructed on RSF1010 expression plasmids. (A) Diagram of the 42 kilobase pair gene cluster with the 32 fam genes. (B) Gene structure of plasmid pSL3228 expressing the famH genes and the lac repressor mutant W220F (‘lacI’). The famH genes were placed under control of the trc promoter surrounded by two lacO operator sequences (“trc2O”). The PmeI (MssI) restriction site was used to insert additional fam genes in subsequent constructs. (C) Operons on plasmid pSL3285. (D) Operons on plasmid pSL3251. (E) Operons on plasmid pSL3283. The plasmid backbone and the lacI cassette is omitted in all panels B – E. Terminators are indicated by black hairpins. Genes under control of the cpcB or J23119 promoter were constitutively expressed.
In order to control production in S2973, the famH1, famH2 and famH3 genes were restructured into a single operon under control of the trc promoter flanked by two symmetric lacO operator sequences (“trc2O”, Figure 3B) to enable IPTG induction of gene expression. We initially co-expressed the wild-type E. coli lacI repressor protein on the plasmid but this resulted in slow-growing, impaired strains that were not tractable for further experiments. We reasoned that binding of the wild-type lacI was insufficient to control transcription from Ptrc2O and that the strains were compromised owing to product toxicity and/or over-consumption of key metabolic intermediates that stunted growth. Therefore, we used the more tightly binding lacI W220F mutant repressor protein48 to control expression of the famH gene operon in all constructs reported herein. The coding sequence for lacI W220F was cloned upstream of the Ptrc2O-famH1 – 3 operon and the mutant lacI gene constitutively co-expressed from PJ23119 for the hapalindole pathway to be IPTG-inducible (Figure 3B). We only placed famH1 – famH3 under an IPTG-inducible promoter, reasoning that controlled induction of the first step in hapalindole biosynthesis would allow us to regulate flux through the downstream steps of the engineered pathway. This operon and regulatory system were cloned and assembled to form plasmid pSL3228. The E. coli rrnB T1 terminator sequence downstream of the famH operon and a T7 terminator oriented in the reverse direction downstream of rrnB T1 is also included in pSL3228. A unique PmeI restriction site introduced downstream of the T7 terminator was used to later expand the plasmid with additional fam genes (Figure 3B). Plasmid pSL3228 was introduced into S2973 by tri-parental mating to generate strain SL3228.
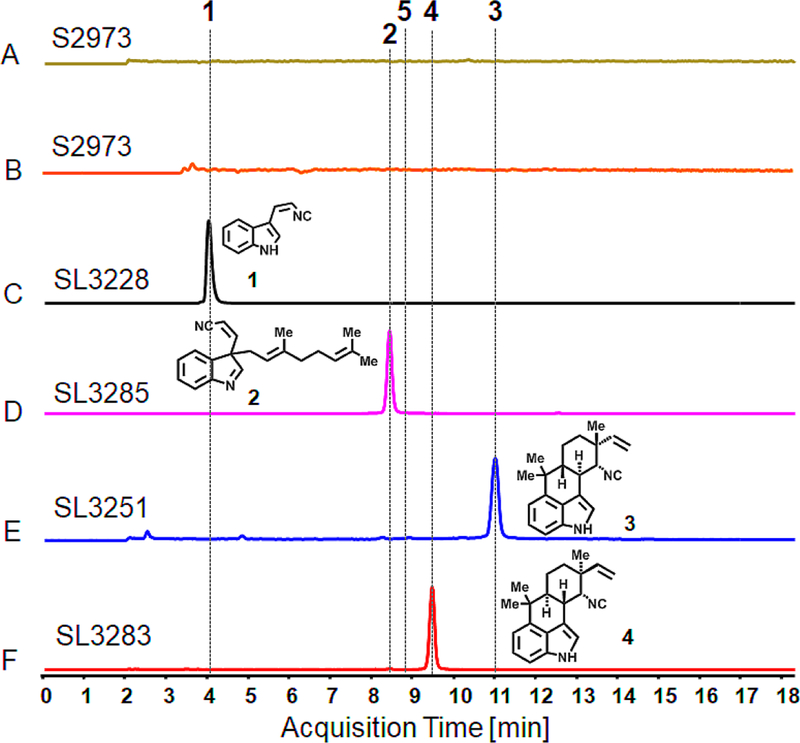
Organic extracts of freeze-dried SL3228 cell biomass cultured under photoautotrophic conditions revealed production of cis indole-isonitrile 1 (Figure 4A, 4B and 4C and Figures S2 and S3). We also detected production of 1 by SL3228 under uninduced conditions owing to some trc promoter leakiness, but this was apparently not sufficient to compromise the strain. Notably, we determined that all three FamH enzymes were required for biosynthesis of 1 in S2973. Strains expressing famH1 and famH3 or famH2 and famH3 did not produce detectable quantities of 1 (Figure S2). This was surprising as FamH1 and FamH3 alone were shown to be sufficient for in vitro production of this molecule.29 We did not detect production of the trans 1 isomer in SL3228 under any growth conditions. The trans isomer is not converted into hapalindoles according to previous in vitro studies.29 Based on a comparison to a cis 1 HPLC standard curve, we estimate that SL3228 cultured in 60 ml volume in a bioreactor produced 10 μg/g dry cell weight (DCW) when uninduced and 200 μg/g DCW when induced with 75 μM IPTG and cultured under 500 μmol photons (μE) m−2 s−1. Growth of SL3228 was negatively impacted by expression of the fam genes and the exponential- phase growth rate was reduced by roughly 50% when uninduced compared to a control strain without fam genes (Figure S4). Induction with IPTG further reduced the growth rate of the strain to a fraction of the uninduced conditions and the cells were a deep yellow color distinct from the green color typical of S2973 (Figure S4). We considered that cells induced with IPTG may be deprived of L-tryptophan due to its consumption by FamH1 – 3. However, we found that L-tryptophan content of SL3228 was comparable under induced and uninduced conditions (Figure S5), suggesting that consumption by the fam enzymes is compensated by increased biosynthesis of this amino acid.
Figure 4.
Analysis of isolated hapalindole products generated by engineered strain S2973 using LCMS. Extracted ion chromatograms (EICs) are shown in all panels. Extracts of wild-type S2973 (A) m/z of 168 for cis indole-isonitrile (1) and (B) m/z of 305 for indole-isonitrile-GPP intermediate (2) and other hapalindole related compounds. (C) Strain SL3228 engineered to produce 1 (m/z of 168). (D) Strain SL3285 produces 2 (m/z of 305). (E) SL3251 produces hapalindole H (m/z of 305) (3). (F) SL3283 produces 12-epi hapalindole U (m/z of 305) (4).
Production of cis indole-isonitrile-GPP and hapalindole H in S2973.
Having established that S2973 could generate the precursor cis indole-isonitrile, we expanded the biosynthetic pathway by adding the famD2 prenyltransferase and fam cyclase genes into a second operon under control of the S2973 cpcB promoter (Figure 3C and 3D). We chose cpcB from the promoter set (Figure 2) because it is the strongest of the native promoter elements tested and we reasoned it would result in comparable expression levels to the IPTG-titratable trc2O promoter. The fam cluster encodes four cyclases (FamC1-FamC4, Figure 3A) that form homo- and heterodimers to stereo-specifically catalyze hapalindole cyclization via a Cope rearrangement.32, 33 The type of cyclase dimer that catalyzes conversion of the indole-isonitrile-GPP intermediate 2 determines the stereochemistry of the resulting hapalindole.30, 32, 33, 35 These cyclases belong to a unique protein family and the first crystal structures of hapalindole cyclases were recently reported by the Sherman group and others.33,49 In vitro characterization showed that Ca2+ ions are required for the activity of some cyclases and that calcium also controls the relative ratio of the hapalindole products generated by their reactions.33,35 The enzymes are not known to require other cofactors.
In order to explore the versatility of our expression system, we constructed two versions of the expanded fam pathway on vectors. In the first, we added the famC2 and famC3 cyclase genes, famD2 prenyltransferase gene, and the famE2 GPP synthase gene (plasmid pSL3251, Figure 3D). In the second we included famD2 and famE2, but omitted the cyclase genes (pSL3285, Figure 3C). The cyclases FamC2 and FamC3 form a heterodimer that converts compound 2 into hapalindole H (3, Figure 1).32 FamD2 is an aromatic prenyltransferase that requires Mg2+ for activity50 and FamE2 is a dedicated GPP synthase that catalyzes the condensation of dimethylallyl pyrophosphate and isopentenyl pyrophosphate to form GPP.29 These plasmids were conjugated into S2973 to generate strains SL3285 and SL3251.
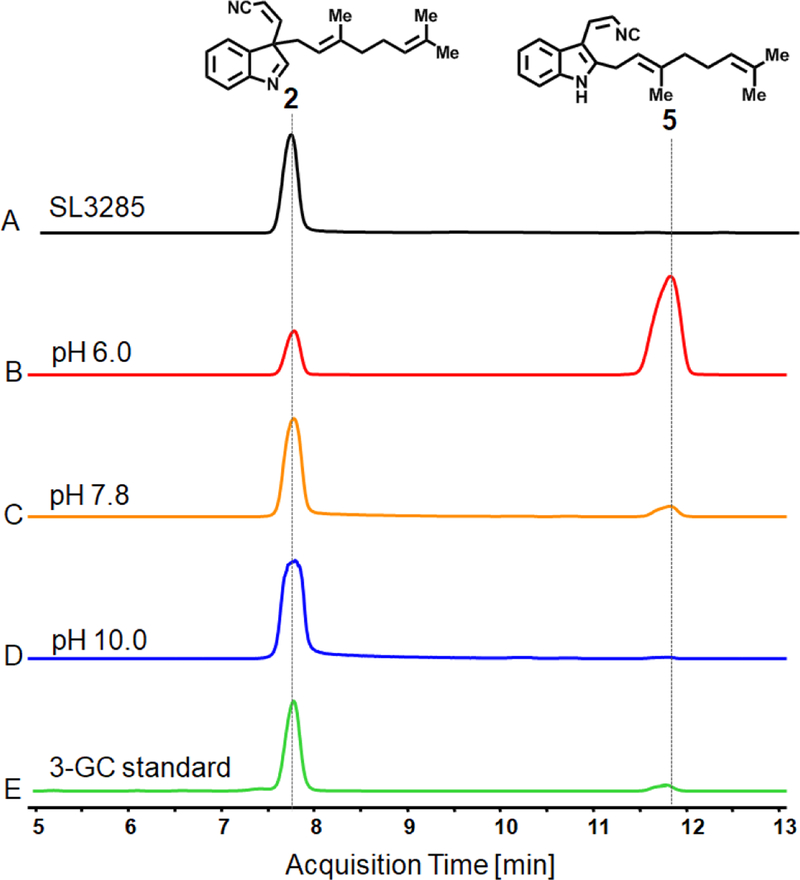
Organic extracts of SL3285 showed conversion of cis indole-isonitrile 1 into a new product (2, Figure 4C) that possessed the exact retention time of authentic indole-isonitrile-GPP. Product 2 also matched the results obtained by an in vitro chemoenzymatic product of synthesized 1 and GPP by purified FamD2 enzyme at various pHs (pH 6.0, 7.8 and 10, Figure 5), as shown before.18 In addition, the UV-spectrum of product 2 also showed the lack of distinct absorption features at 250–350 nm consistent with de-aromatization of the indole ring and loss of intact indolyl vinyl isonitrile group (Figure S6). At mildly acidic pH in the in vitro assays, 2 was converted into the C2-prenylated isomer (5, Figure 5) that was previously shown to be a terminal shunt metabolite unable to be cyclized into hapalindoles or converted back into compound 2.30
Figure 5.
Determination of the indole isonitrile-GPP intermediate isomer produced by engineered SL3285 using LCMS. EIC trace (168 m/z) showing the comparison of SL3285 product (A) with the in vitro aromatic prenyltransferase assay of purified FamD2 enzyme at different pH 6.0 (B), 7.8 (C) and 10 (D) with the substrate 1 and GPP. The authentic indole-isonitrile-GPP (2, peak 7.75 min) standard showed that the intermediate produced by SL3285 (and SL3283) is exclusively the C3-geranylated isomer. The C2-geranylated isomer (5, 11.8 min) is formed under low-pH conditions but is not generated in S2973.
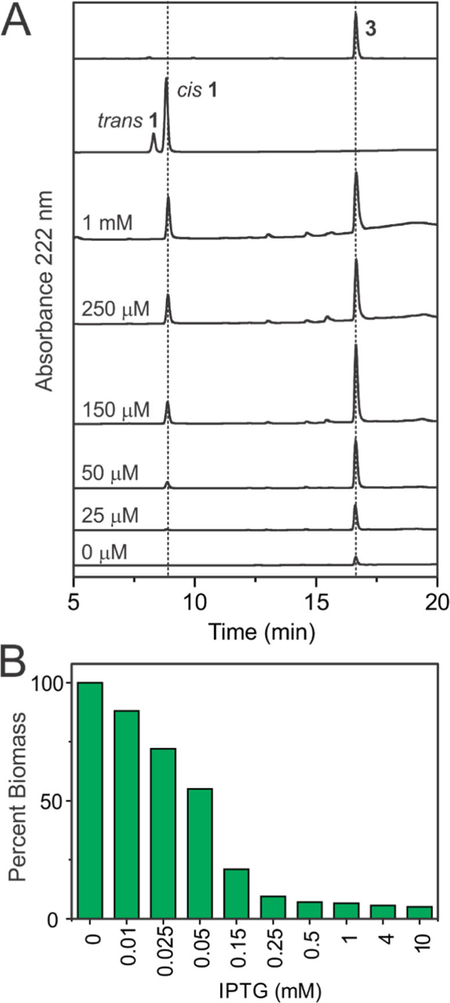
Extracts of SL3251 biomass also expressing the famC2 and famC3 cyclase genes contained a new metabolite that was identified as hapalindole H (3) using LC/MS, 1H NMR and by comparison to an authentic standard (Figure 4D, S3 and S7 and Table S4). Uninduced SL3251 produced some hapalindole 3 but did not contain detectable amounts of cis indole-isonitrile 1 likely due to conversion by the constitutively expressed prenyltransferase (famD2) and cyclase (famC2/famC3) genes (Figure 6A). Titration with IPTG using strain SL3251 indicated that increasing the concentration above 100 μM results in a >75% reduction in S2973 biomass production and accumulation of unreacted 1 in cells (Figure 6B). These data suggest that the Ptrc2O/lacI-W220F regulatory system is titratable and that over-induction of the fam pathway incurs a heavy penalty in S2973 biomass production. This may be due to toxicity of the metabolites and/or over-consumption of isoprenoid precursors by FamE2 that are required for pigment and quinone biosynthesis in cyanobacteria.51 Growth of SL3251 was again reduced compared to a control strain but improved relative to SL3228, particularly under induced conditions with 75 μM IPTG (Figure S4). When induced with 75 μM IPTG and cultured in a bioreactor at 250 μE m−2 s−1, SL3251 produced hapalindole 3 at a titer of 1.6 – 2 mg/g DCW or roughly 2.5 – 3.1 mg/L. We assessed whether 3 was being secreted from the cells but did not detect this compound or the two intermediates in the culture medium.
Figure 6.
Production of indole-isonitrile (1) and hapalindole H (3) and dry biomass yield in strain SL3251 as a function of IPTG concentration. (A) HPLC traces of extracts of SL3251 at different IPTG concentrations (labeled). The amount of extract loaded for HPLC was normalized to dry cell biomass. Standards for hapalindole H and indoleisonitrile are shown in the top two traces. (B) Dry biomass yield of SL3251 as a function of IPTG concentration normalized to uninduced biomass yield. Cells were cultured under 400 μE m−2 s−1 in a bioreactor bubbling with 5% CO2 in air. IPTG was added at the start of culturing.
We ascertained whether E. coli was able to produce hapalindole 3 by screening extracts of cells grown in LB media using HPLC. E. coli hosting plasmid pSL3251 induced with 200 μM IPTG contained cis indole-isonitrile 1, but no detectable hapalindole 3 was observed (Figure S8). To test whether cpcB gene promoter incompatibility was responsible for failed production in E. coli, we generated another plasmid pSL3325 where we replaced the cyanobacterial cpcB promoter on plasmid pSL3251 with the native E. coli lac promoter to drive expression of famC3, famC2, famD2, and famE2. IPTG-induced E. coli bearing this plasmid remained unable to produce 3, but again contained unreacted 1 (Figure S8). We did not ascertain whether E. coli was able to generate indole-isonitrile-GPP 2, but previous reports have shown that E. coli can produce active FamD2 prenyltransferase.30, 31 We also used E. coli to produce this enzyme for in vitro assays (Figure 5). The inability of E. coli to synthesize hapalindole H is thus likely due to failed production of active FamC2 and/or FamC3 cyclases.
Production of 12-epi-hapalindole U by exchanging the fam cyclase genes.
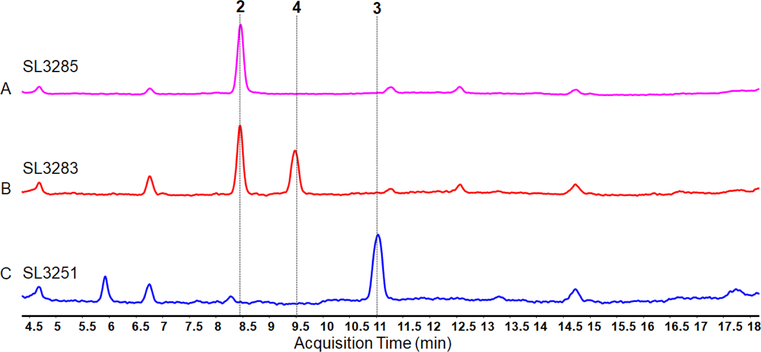
We next aimed to establish production of other hapalindole metabolites in S2973 by exchanging FamC2 and FamC3 with another cyclase from the fam cluster. Homodimers of the FamC1 cyclase generate 12-epi hapalindole U (Figure 1).30 We exchanged the famC2 and famC3 cyclases with the famC1 gene under control of the cpcB promoter to generate strain SL3283 (Figures 3E). Organic extracts of SL3283 induced with IPTG contained 12-epi hapalindole U (4, Figure 4E). Production of hapalindole 4 was confirmed using LC/MS, 1H NMR, and by comparison to an authentic standard (Figures S3, S9 and Table S5). In contrast to SL3251, SL3283 also contained unreacted indole-isonitrile-GPP 2 indicating its incomplete conversion into cyclized hapalindole (Figure 7). Overall, yields for hapalindole 4 were lower than those for hapalindole 3 in strain SL3251. In a bioreactor, SL3283 produced 0.7 mg/g DCW 12-epi hapalindole U (0.75 mg/L). Growth of SL3283 in a bioreactor were, like SL3251, improved relative to SL3228 under IPTG-induced conditions (Figure S3). We also tested whether E. coli could produce 4 by expressing the FamC1 cyclase alongside FamH1 – H3. However, IPTG-induced E. coli with plasmid pSL3283 failed to produce detectable levels of hapalindole 4, but rather contained unreacted cis indole-isonitrile 1 (Figure S8).
Figure 7.
LCMS traces showing the total ion chromatograms (TICs) of SL3285 (A), SL3283 (B), and SL3251 (C) evidencing unutilized indole-isonitrile-GPP intermediate 2 during the production of 12-epi-hapalindole U (4) but not hapalindole H (3).
S2973 hapalindole yields compared to native producers and other engineered microbes.
The S2973 strains reported herein produced roughly 0.7 – 2 mg hapalindole/g DCW under photoautotrophic conditions. Compared to previous publications, our yields for hapalindole H are slightly below those of the native producer Fischerella ambigua or related strains that are reported to yield 4 – 7 mg/g DCW.26, 27 The yield of hapalindole U has been reported at near 0.7 mg/g DCW from Westiellopsis sp.26 which is on par with our titer for 12-epi hapalindole U. Although current yields are comparable to the native producers, S2973 offers distinct advantages over these strains. Depending on the culturing conditions used, we were able to generate dense cultures of S2973 within 3 – 4 days in a bioreactor, significantly faster than the 6 – 8 weeks for the native producers.26, 27 Furthermore, S2973 is genetically tractable with a burgeoning synthetic biology toolset22, 47, 52, 53 enabling higher yield potential. For instance, metabolic engineering of the substrate-supplying shikimate54 and methylerythritol phosphate pathways51 alongside central photoautotrophic metabolism55, 56 are viable strategies to increase titers of the hapalindoles in future S2973 strains. Such genetic tools are currently unavailable for the Stigonematales and many other NP-producing cyanobacteria. We compared the current yields of hapalindoles produced by S2973 to those in other cNPs and some plant NPs produced by engineered microbes. Hapalindole yields were comparable (low mg/g DCW or mg/L range) to those of other NPs in engineered cyanobacteria including shinorine in Synechocystis,16 the non-ribosomal peptide lyngbyatoxin A in Anabaena15 and the plant NPs dhurrin19 and manoyl oxide18 in Synechocystis. However, S2973 exhibits a faster optimal photoautotrophic doubling time (1.6 – 2 hours) than both Anabaena (14 hours)15 and Synechocystis (6 – 7 hours).22 The yields in S2973 were 100- to 1000-fold higher compared to some engineered heterotrophic systems including 4-O-demethylbarbamide in Streptomyces venezuelae10 and microcystin-LR in E. coli12 (both in the low μg/L range) and comparable to those of microviridins and cyanobactins in E. coli (~0.2 – 0.7 mg/L).11, 13 However, significant further strain optimization and metabolic engineering is required to achieve yields commensurate with lyngbyatoxin A produced by engineered E. coli (~26 mg/L).14
The yield-limiting step is different between the hapalindole-producing S2973 strains.
We were able to control the production levels of cis indole-isonitrile 1 using the titratable Ptrc2O/lacI-W220F regulator system (Figure 6). The observation that all of the hapalindole-producing strains grew more quickly than SL3228 upon induction with 50 – 75 μM IPTG (Figure S4) suggests that 1 is toxic to S2973 when it accumulates in cells. Metabolic burden alone is unlikely to explain the slower growth of the induced strain, as the hapalindole-producing strains SL3251 and SL3283 are also expressing the famH genes, cyclase(s), prenyltransferase, and GPP synthase that would be expected to result in an even higher burden. The efficient turnover of cis indole-isonitrile is of key importance in improving strain productivity and titer in S2973. Yields of cis indole-isonitrile in strain SL3228 were three- or more fold lower than those of the hapalindoles and we reason that the toxicity of this metabolite prevents accumulation of higher amounts in S2973. In contrast, consumption of this intermediate by the constitutively expressed FamD2 and cyclases in strains SL3251 and SL3283 prevents this accumulation, and thus leads to higher yields of the presumably less toxic hapalindoles. Our observations also suggest that the rate-limiting step in the engineered hapalindole pathway is different between the strains. SL3283 accumulated indole-isonitrile-GPP 2 (Figure 7) suggesting that the activities and/or the expression levels of the cyclases were limiting in this construct. Incomplete conversion of 2 in SL3283 is likely to be responsible for the lower overall yield of 12-epi hapalindole U compared to hapalindole H. In contrast, in SL3251, we did not detect any accumulation of 2 (Figure 7), indicating that FamC2 and FamC3 activity or expression level is higher relative to FamC1 in SL3283 and that turnover of 1 by the prenyltransferase FamD2 may be limiting in this strain. Alternatively, this could be due to limitations in the cellular availability of dimethylallyl pyrophosphate in SL3251 that is needed by FamE2 to generate GPP.51
Rational re-factoring of the fam pathway to direct biosynthesis and a means to study hapalindole tailoring.
Several reports have shown successful targeting of cNP clusters for heterologous expression in cyanobacteria15, 16 with the 42-kilobase fam cluster, to our knowledge, representing the largest pathway reported so far. Typically, one of the major challenges in working with NP BGCs is that a biosynthetic pathway often results in production of a mixture of possibly dozens of different structural variants. For instance, the fam BGC in Fischerella ambigua produces primarily hapalindole H but also about 20 derivatives.28 Complex mixtures of these isomers demand arduous fractionation of extracts to purify individual molecules for structural analysis. We used biochemical knowledge of the hapalindole biosynthesis to re-factor six to seven genes from the fam BGC and thereby direct production toward individual regio- and stereo-chemically pure hapalindole molecules out of the 20 potential variants.28 Furthermore, the stepwise expansion of the core hapalindole pathway that we established in S2973 provides a powerful approach to determine the role of NP biosynthetic enzymes by inserting single genes with as-of-yet unknown function and analyzing the chemical result. The fam BGC and related clusters from other Stigonematales encode a variety of tailoring enzymes including cytochrome P450s.28 These monooxygenases can be challenging to express at full activity in E. coli or study in vitro, but are often active in engineered cyanobacteria due to the presence of compatible reductase partners.19–21 Importantly, bio-production in S2973 did not result in mixtures of pathway intermediates such as the trans indole-isonitrile29 or the C2-geranylated shunt product 5 that is produced in chemoenzymatic reactions under pH conditions below 8 (see Figure 5 and reference30). Neither of these compounds are substrates for conversion into hapalindoles. The stereo- and regio-specificity of the in vivo S2973 production system provides distinct advantages for scale-up and gene-function studies compared to in vitro or synthetic chemical production methods for hapalindoles. We observed that E. coli accumulated cis indole-isonitrile but was unable to produce the hapalindoles. This is likely due to its inability to process the N-terminal transmembrane region of fam cyclases.32
Conclusions
We established a photoautotrophic production system for key members of the hapalindole cNP family as well as critical biosynthetic intermediates in a fast-growing heterologous cyanobacterial host. Historically, bioengineering of cyanobacteria has focused on the production of ‘low-cost, high-volume’ commodity chemicals and biofuels.56 Although improvements in engineering strategies and a better understanding of photoautrotrophic metabolism have yielded significant increases in cyanobacterial titer and productivity,53 the need to compete with cheap petrochemical-derived fuels and chemicals currently prevents these strains from being economically viable. Photosynthetic production of complex ‘high-cost, low-volume’ compounds such as NPs could overcome this type of direct economic competition. Our S2973 strains demonstrate that fast-growing unicellular cyanobacteria are a promising production chassis for CO2-derived NP compounds that are challenging to produce in E. coli or via chemical synthesis. The two hapalindole variants that we produced have useful bioactivities including antibacterial and antifungal properties and toxicity against malignant cell lines.24, 27 Moreover, these strains serve as a foundation for future engineering to improve hapalindole titers and as a means to dissect the fascinating biochemical transformations involved in the biosynthesis of this family of compounds. Once the tailoring steps of the fam pathway (and other hapalindole pathways) are more clearly understood, S2973 strains can be engineered to produce more complex hapalindoles or generate entirely novel derivatives by combinatorially expressing tailoring genes from other Stigonematales species. The inability of E. coli to produce the hapalindoles illustrates that heterotrophs are poor expression hosts for the critical Fam cyclase enzymes. This provides an impetus to further develop cyanobacteria as expression platforms for these types of pathways. Improvements in cyanobacterial synthetic biology provides a means to study both known cNP clusters as well as the hundreds of cryptic cNP biosynthetic pathways that remain to be explored.2, 4 This could provide access to entirely new classes of cNPs and a means to explore the rich diversity of fascinating chemical transformations involved in their assembly and structural diversification.
Methods
Cyanobacterial culturing and conjugation.
S2973 was maintained on BG11 plates or liquid media at 38 °C. Kanamycin was added to a concentration of 10 μg/ml on plates and in liquid when required. Strains were routinely cultured in 250 ml shaker flask under 50 – 150 μmol photons (μE) m−2 s−1 or in a MC1000 Multicultivator bioreactor (Photon Systems Instruments, psi.cz) under 200 – 900 μE m−2 s−1 LEDs while bubbling with 5% CO2-supplemented air. Expression plasmids were introduced into S2973 via tri-parental mating with an Escherichia coli strain hosting the IncP helper plasmid pRL443 and with another hosting the plasmid to be introduced into S2973. For mating, S2973 was cultured overnight to an O.D. 730 nm of 0.3 – 0.6 and E. coli were cultured overnight in LB media. Cells were collected by centrifugation, washed twice with deionized water, mixed, and spread on nitrocellulose membranes (Millipore Immobilon-NC) then placed on BG11 agar plates with 5 % (v/v) LB. After outgrowth for 1 day at 38 °C in air, membranes were moved to BG11 plates with 50 μg/ml kanamycin. Ex-conjugants were individually picked and moved to BG11 plates with 10 μg/ml kanamycin and PCR genotyped to confirm transfer of the plasmid. In order to mitigate issues with strain instability, we resurrected strains from −80 °C frozen stocks (with 10% v/v DMSO as cryoprotectant) roughly every two months. We did not add additional calcium to the media when culturing the hapalindole-producing strains (unmodified BG11 contains 250 μM Ca2+).
Construction of eYFP and fam expression vectors.
All plasmids were constructed using Gibson Assembly.57 PCR primers used to amplify the promoters, fam genes and other genetic elements are listed in Supplementary Table S2. All vectors described herein were derived from pVZ321, a RSF1010 broad host-range plasmid46 and maps for these constructs are shown in Figure S1. To screen the promoter activity using eYFP, we used plasmid pSL3068 that was described previously.44 Promoters were amplified from S2973 or E. coli genomic DNA (gDNA) isolated using phenol/choloroform extraction and ethanol precipitation. To assemble the promoter-eYFP constructs, pSL3068 was linearized using AarI restriction enzyme, gel-purified using a Purelink Quick Gel Extract kit (Thermo Fisher), and mixed with the PCR-amplified promoters for Gibson Assembly. The following primers were used to PCR-amplify the promoter regions for Gibson assembly: 0012, P0012-F/P0012-R; 0195, P0195-F/P0195-R; 0398, P0398-F/P0398-R; 0657, P0657-F/P0657-R; 1219, P1219-R/P1219-R; 1479, P1479-F/P1479-R; 1613, P1613-F/P1613-R; 1616, P1616-F/P1616-R; 1809, P1809-F/P1809-R; 1830, P1830-F/P1830-R; 1987, P1987-F/P1987-R; 1999, P1999-F/P1999-R; 2352, P2352-F/P2352-R; 2486, P2486-F/P2486-R; cpcB, PcpcB-F/PcpcB-R; J23119, PJ23119-F/PJ23119-R; lac, Plac-F/Plac-R; lacIq, PlacIq-F/PlacIq-R; psaA, PpsaA-F/PpsaA-R; trc, Ptrc-F/Ptrc-R. These primers amplified the native ribosome binding site along with the promoter region. The assembly reactions were transformed into XL1-Blue E. coli and plated on LB with 50 μg/ml kanamycin overnight. Resistant colonies were selected for culturing in LB liquid media. Plasmids were isolated using a Genejet Plasmid Miniprep kit (Thermo Scientific) and correct sequences verified using Sanger sequencing (Genewiz, www.genewiz.com). Correctly assembled plasmids were introduced into S2973 using tri-parental mating.
To construct the fam expression plasmids, we generated a derivative vector pSL3084 from the RSF1010 plasmid pSL3067.44 pSL3084 has the direct repeat region around the aphA kanamycin resistance cassette removed in order to improve vector stability. pSL3084 was used as the backbone/parent plasmid to generate the fam plasmids. The pSL3084 fragments were amplified using the two primer pairs 3084–1F/3084–1R and 3084–2F/3084–2R from pSL3067 template and the two purified fragments joined using Gibson assembly. To assemble the fam plasmids, pSL3084 was digested with XbaI restriction enzyme (Thermo), purified, and this linearized vector used in Gibson Assembly reactions with PCR products amplified from Fischerella ambigua UTEX 1903 gDNA or the promoter-eYFP plasmids unless otherwise noted.
To assemble pSL3228, the PCR fragments were amplified with 28−30 bp terminal overhangs or directly synthesized prior to Gibson assembly with XbaI-digested pSL3084: the J23119 promoter and lacI W220F cassette synthesized with terminal overlaps for Gibson assembly (see Supplementary Information); the trc promoter sequence was also synthesized with overlaps (see Supplementary Information); famH1 was amplified using CK142/CK065 primers; famH2 and famH3 were amplified using CK066/CK112 primers; and the bi-directional terminator was amplified using CK113/CK143 primers and cloning vector pSR26–258 as a template. These five fragments were mixed with XbaI-digested pSL3084 for Gibson assembly. CK143 also introduced a unique PmeI (MssI) enzyme restriction site downstream of the terminator useful for future cloning. To make plasmids pSL3251, pSL3283, and pSL3285, pSL3228 was digested with PmeI, gel-purified, and used as a linearized backbone for Gibson assembly. The PCR fragments for pSL3251 were amplified using the following primers: famE2, CK213/CK170; famD2, CK171/CK073; famC2, CK074/CK075; famC3, CK076/CK041; cpcB promoter, CK037/CK077. pSL3283 PCR fragments for assembly were amplified using these primers: famE2 and famD2, CK213/CK214 using pSL3251 DNA template; famC1, CK215/CK216; cpcB promoter, CK217/CK172. Primers for generating pSL3285 PCR fragments were as follows: famE2 and famD2, CK213/CK188; cpcB promoter, CK189/CK172. Primers amplifying the start codon of the fam genes were designed to engineer the RBS for higher translation rates using RBS calculator v2.0 (https://salislab.net/software/forward).59 The input target translation initiation rate (TIR) using RBS calculator was 5,000. To generate plasmid pSL3325 replacing the cpcB with the lac promoter the following primers were used: the famE2, D2, C2, and famC3 fragment were amplified from pSL3251 using CK213/CK270 primers and the lac promoter was amplified from the Plac-eYFP plasmid using primers CK271/CK272. After Gibson assembly, plasmids were propagated in E. coli, isolated, and Sanger sequenced to ensure correct assembly as described for the eYFP plasmids above.
Fluorescence measurements.
Fluorescence from S2973 strains was measured when the culture optical density at 730 nm was between 0.2 – 0.6. For these experiments, cells were cultured at 38 °C in air in 250 ml glass shaker flasks under 50 μE m−2 s−1 fluorescent lights. O.D.s and fluorescence were measured for 150 μl of culture in 96-well plastic trays using a Bio-Tek μQuant Plate Reader. Fluorescence measurements were made using an excitation wavelength of 514 nm and emission was monitored at 527 nm.
Analysis and purification of hapalindoles from S2973 biomass.
The biosynthesized compounds in S2973 were isolated by macerating 20 mg of the corresponding biomasses in 5 ml of extraction solvent (1:1 mixture of dichloromethane and methanol) using Tissue Tearor (BioSpec Products Inc.) for 10 min. The extract was centrifuged at 8,200 rpm for 15 min, and the pellet was re-extracted once. The pooled organic extract was dried in a nitrogen stream and dissolved in 400 μl of methanol, and 1 μl of the extract was analyzed either in Q-TOFF or TOFF as described below in the in vitro chemoenzymatic method. For the structural elucidation, 500 to 700 mg of the corresponding biomass was macerated in 10 ml of the extraction solvent, and extracted twice. The extracted products were dried and concentrated as before and purified by HPLC (Column: XBridgeTM Prep Phenyl 5 μM, 10 × 250 mm), using a 60–100% gradient of acetonitrile in water over 28 min. The purified compounds were concentrated, dissolved in C6D6, and analyzed using a Varian 600 MHz NMR spectrometer. Proton and carbon signals are reported in parts per million (δ) using residual solvent signals as an internal standard.
Expression and purification of FamD2.
The expression and purification of FamD2 was performed as described.30 Briefly, a single BL21(DE3) colony of FamD2 transformant was inoculated in LB medium containing 50 μg/mL kanamycin and grown overnight at 37 °C shaking at 200 rpm. The main culture (1 L) was prepared at the dilution of 1:100 in 2.8 L Fernbach flask containing LB medium and the same concentration of antibiotic. The cells were grown (37 °C, 200 rpm) to an optical density (A600 nm) of 0.6. The culture flasks were chilled in ice and was induced with IPTG (0.2 mM) and was further incubated (16 °C, 200 rpm) for 16 h. The cells were harvested (5000 rpm, 4 °C, 15 min), flash frozen and stored at −80°C until purification. The cell pellets were re-suspended at 4°C in the lysis buffer (10 mM HEPES, 50 mM NaCl, 0.2 mM TCEP, 10% glycerol), containing 0.5 mg/mL of lysozyme and 1 μl of benzonase. The mixture was stirred for 30 min and sonicated on ice for 100 s total time using 10 s pulses followed by a 50 s pause. The cellular debris was removed by centrifugation (35,000 rpm, 4 °C, 35 min). Imidazole (10 mM) was added in the clarified lysate and loaded onto Ni-NTA agarose column equilibrated with lysis buffer. The column was washed with five column volume of wash buffer (10 mM HEPES, 300 mM NaCl, 0.2 mM TCEP, 10% glycerol, 20 mM imidazole) and the His-tagged protein was eluted with elution buffer (10 mM HEPES, 50 mM NaCl, 0.2 mM TCEP, 10% glycerol, 300 mM imidazole). The fractions were pooled and dialyzed using a PD10-desalting column (GE Healthcare) using lysis buffer (10 mM HEPES, 50 mM NaCl, 0.2 mM TCEP, 10% glycerol). The purified protein was analyzed by SDS-PAGE gel for purity, measured by Nanodrop for concentration, and flash-frozen in liquid nitrogen to store at −80 °C.
Synthesis of geranyl diphosphate triammonium salt.
Synthesized as previously described.60 Briefly, in a 10 ml round-bottom flask purged with nitrogen, tris (tetrabutylammonium) hydrogen pyrophosphate (1.33 g, 1.39 mmol) was dissolved in CH3CN (1.50 ml). Geranyl chloride (0.12 ml, 0.63 mmol) was added and the reaction mixture was stirred at room temperature for 2 h.
Synthesis of (Z)-2-(3-Indolyl)vinyl isocyanide (cis indole-isonitrile).
Synthesized as previously described.60 Briefly, to a 50 ml two-neck round-bottom flask purged with nitrogen at −78°C (dry ice/acetone), diethyl (isocyanomethyl) phosphonate (0.20 ml, 1.248 mmol) was diluted with THF (5 ml). KHMDS (1 M THF, 2.60 ml, 2.60 mmol) was added dropwise and the reaction was stirred at −78°C for 30 minutes. To a separate 4 ml vial, indole-3-carboxaldehyde (164 mg, 1.13 mmol) was dissolved in THF (5 ml) and the resulting solution was added dropwise to the KHMDS solution at −78°C. The resulting mixture was stirred at 0°C (cryocool) for 21 h. The resulting red solution was quenched by the addition of AcOH (0.15 ml, 2.6 mmol) and concentrated. The dark red residue was diluted with EtOAc (20 ml), washed with 1 M aqueous potassium phosphate buffer (20 ml, pH 7), washed with brine, dried with Na2SO4 and concentrated to a dark red oil. The residue was dissolved in Et2O and purified by flash chromatography (24%–100% pentane/Et2O, SiO2) to afford the titled compound as a red crystalline solid (20.7mg, 10.8%). Spectral data were in accord with those reported.60
1H NMR (599 MHz, CDCl3) δ 5.75 (d, J = 8.8 Hz, 1H), 6.80 (dt, J = 9.1, 4.6 Hz, 1H), 7.23 (t, J = 7.5 Hz, 1H), 7.27 – 7.30 (m, 1H), 7.44 (d, J = 8.1 Hz, 1H), 7.68 (d, J = 7.9 Hz, 1H), 8.14 (d, J = 2.8 Hz, 1H), 8.61 (s, 1H).
13C NMR (151 MHz, CDCl3) δ 168.45, 135.03, 126.76, 126.41, 124.16, 123.19, 120.93, 118.00, 111.50, 110.08, 104.4 (t).
In vitro aromatic prenyltransferase FamD2 assay.
The cis indole-isonitrile and GPP were synthesized as described above and the aromatic prenyltransferase FamD2 was used to generate the indole-isonitrile-GPP intermediate in vitro. The in vitro assay was performed in a 50 μl reaction containing FamD2 (10 μM), indole isonitrile (1 mM), GPP (1 mM) and MgCl2 (20 mM), either in 50 mM PIPES buffer, pH 6.0, or 50 mM Tris buffer, pH 7.8 or 50 mM Glycine buffer, pH 10.0. The reaction was incubated at 30 °C for 2 h. and quenched and extracted twice with double volume of ethyl acetate. The organic layer was combined, dried and re-constituted in 50 μl of acetonitrile for LCMS analysis. The analytical scale liquid chromatography-mass spectrometry (LC-MS) run was performed on an Agilent G6545A quadrupole-time of flight (Q-TOF) or Agilent 6230 time of flight (TOF) mass spectrometer equipped with a dual AJS ESI source and an Agilent 1290 Infinity series diode array detector, auto-sampler and binary pump. A XBridgeR Shield RP18 3.5 μm, 3.0 × 150 mm from Waters was used for all separations. The chromatographic method for all substrates gradient was from 60% to 100% of 95% acetonitrile water containing 0.1% formic acid over 27 min. The volume of 1 μl injections were made for each sample.
Dowex 50WX8 resin preparation and GPP purification.
Dowex 50WX8 resin (26 g, hydrogen form) was washed with half saturated aqueous ammonium chloride (5 × 50 ml) and water (5 × 50 ml) until the pH of the supernatant equaled 5. The slurry was rinsed twice with ion exchange buffer (2% isopropanol in 25 mM aqueous ammonium bicarbonate) and loaded into a flash column and equilibrated with ion exchange buffer. The reaction mixture was concentrated to afford an orange residue which was diluted with ion exchange buffer. The crude mixture was chromatographed with two column volumes of ion exchange buffer (75 ml) over Dowex 50WX8 resin previously prepared. The fractions were combined and concentrated by rotary evaporation, flash frozen and lyophilized for 2d. The resulting white powder was diluted with 0.1 M ammonium bicarbonate (4 mL) and 50% isopropanol/CH3CN (10 ml), vortexed for 30 seconds and centrifuged (2,000 rpm, room temperature for 5 min). The organic layer was extracted and the residual 0.5 ml of yellow liquid was diluted with 50% isopropanol/CH3CN and this process was repeated twice. The combined organic layers were concentrated to afford a while solid (360 mg). The white solid was taken up in 50% isopropanol: 25% CH3CN:25% 0.1 M aqueous ammonium bicarbonate and chromatographed over cellulose. The resulting fractions were combined and lyophilized affording the titled compound as a white powder (126.4 mg, 54.7%).
1H NMR (400 MHz, D2O/ND4OD) δ 1.92 (d, J = 1.3 Hz, 3H), 1.98 (s, 3H), 2.01 (d, J = 1.3 Hz, 3H), 2.39 (d, J = 6.5 Hz, 2H), 2.41 – 2.49 (m, 2H), 5.45 – 5.53 (m, 1H), 5.74 (dt, J = 6.1, 3.9 Hz, 1H).
13C NMR (151 MHz, D2O/ND4OD) δ 142.46, 133.67, 124.40, 120.46, 62.61, 39.10, 25.92, 25.16, 17.26, 15.89.
31P NMR (162 MHz, D2O/ND4OD) δ −9.93 (d, J = 21.6 Hz), −6.08 (d, J = 21.6 Hz).
Supplementary Material
Acknowledgements
We thank all members of the Pakrasi and Sherman groups for collegial discussions. Studies in the Pakrasi lab were supported by a Postdoctoral Fellowship from the Arnold and Mabel Beckman Foundation (Irvine, CA) to Cory Knoot. NIH (R35 GM118101), and the Hans W. Vahlteich Professorship are gratefully acknowledged for financial support (to D.H.S).
Abbreviations
- S2973
Synechococcus elongatus UTEX 2973
- NP
natural product
- cNP
cyanobacterial natural product
- GPP
geranyl pyrophosphate
- BGC
biosynthetic gene cluster
- fam
42-kbp hapalindole gene cluster from Fischerella ambigua UTEX 1903
- eYFP
enhanced yellow fluorescence protein
- DCW
dry cell weight
Footnotes
Supporting Information
Plasmid maps, NMR spectra, LC/MS data with standards, strain growth in bioreactor, analysis of L-tryptophan content, tables of promoter sequences, primers used in plasmid construction, strain genotypes, and 1H NMR shifts for hapalindoles.
The authors declare no competing financial interests.
References
- (1).Burja AM, Banaigs B, Abou-Mansour E, Grant Burgess J, and Wright PC (2001) Marine cyanobacteria - a prolific source of natural products. Tetrahedron 57, 9347–9377. [Google Scholar]
- (2).Dittmann E, Gugger M, Sivonen K, and Fewer DP (2015) Natural Product Biosynthetic Diversity and Comparative Genomics of the Cyanobacteria. Trends Microbiol. 23, 642–652. [DOI] [PubMed] [Google Scholar]
- (3).Kehr JC, Picchi DG, and Dittmann E (2011) Natural product biosyntheses in cyanobacteria: A treasure trove of unique enzymes. Beilstein J. Org. Chem 7, 1622–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Kleigrewe K, Gerwick L, Sherman DH, and Gerwick WH (2016) Unique marine derived cyanobacterial biosynthetic genes for chemical diversity. Nat. Prod. Rep 27, 1048–1065. [DOI] [PubMed] [Google Scholar]
- (5).Butler MS (2004) The Role of Natural Product Chemistry in Drug Discovery. J. Nat. Prod 67, 2141–2153. [DOI] [PubMed] [Google Scholar]
- (6).Newman DJ, and Cragg GM (2016) Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat. Prod 79, 629–661. [DOI] [PubMed] [Google Scholar]
- (7).Cragg GM, Schepartz SA, Suffness M, and Grever MR (1993) The Taxol Supply Crisis. New NCI Policies for Handling the Large-Scale Production of Novel Natural Product Anticancer and Anti-HIV Agents. J. Nat. Prod 56, 1657–1668. [DOI] [PubMed] [Google Scholar]
- (8).Paterson I, and Anderson EA (2005) The Renaissance of Natural Products as Drug Candidates. Science 310, 451. [DOI] [PubMed] [Google Scholar]
- (9).Marienhagen J, and Bott M (2013) Metabolic engineering of microorganisms for the synthesis of plant natural products. J. Biotechnol 163, 166–178. [DOI] [PubMed] [Google Scholar]
- (10).Kim EJ, Lee JH, Choi H, Pereira AR, Ban YH, Yoo YJ, Kim E, Park JW, Sherman DH, Gerwick WH, and Yoon YJ (2012) Heterologous Production of 4-O-Demethylbarbamide, a Marine Cyanobacterial Natural Product. Org. Lett 14, 5824–5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Ziemert N, Ishida K, Liaimer A, Hertweck C, and Dittmann E (2008) Ribosomal Synthesis of Tricyclic Depsipeptides in Bloom-Forming Cyanobacteria. Angew. Chem. Int. Ed 47, 7756–7759. [DOI] [PubMed] [Google Scholar]
- (12).Liu T, Mazmouz R, Ongley SE, Chau R, Pickford R, Woodhouse JN, and Neilan BA (2017) Directing the Heterologous Production of Specific Cyanobacterial Toxin Variants. ACS Chem. Biol 12, 2021–2029. [DOI] [PubMed] [Google Scholar]
- (13).Tianero MDB, Donia MS, Young TS, Schultz PG, and Schmidt EW (2012) Ribosomal Route to Small-Molecule Diversity. J. Am. Chem. Soc 134, 418–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Ongley SE, Bian X, Zhang Y, Chau R, Gerwick WH, Müller R, and Neilan BA (2013) High-Titer Heterologous Production in E. coli of Lyngbyatoxin, a Protein Kinase C Activator from an Uncultured Marine Cyanobacterium. ACS Chem. Biol 8, 1888–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Videau P, Wells KN, Singh AJ, Gerwick WH, and Philmus B (2016) Assessment of Anabaena sp. Strain PCC 7120 as a Heterologous Expression Host for Cyanobacterial Natural Products: Production of Lyngbyatoxin A. ACS Synth. Biol, acssynbio.6b00038. [DOI] [PubMed] [Google Scholar]
- (16).Yang G, Cozad MA, Holland DA, Zhang Y, Luesch H, and Ding Y (2018) Photosynthetic Production of Sunscreen Shinorine Using an Engineered Cyanobacterium. ACS Synth. Biol 7, 664–671. [DOI] [PubMed] [Google Scholar]
- (17).Lassen LM, Nielsen AZ, Ziersen B, Gnanasekaran T, Møller BL, Jensen PE, Lassen LM, Nielsen AZ, Ziersen B, Gnanasekaran T, Møller BL, and Jensen PE (2014) Redirecting Photosynthetic Electron Flow into Light-Driven Synthesis of Alternative Products Including High-Value Bioactive Natural Compounds. ACS Synth. Biol 3, 1–12. [DOI] [PubMed] [Google Scholar]
- (18).Englund E, Andersen-Ranberg J, Miao R, Hamberger B, and Lindberg P (2015) Metabolic Engineering of Synechocystis sp. PCC 6803 for Production of the Plant Diterpenoid Manoyl Oxide. ACS Synth. Biol 4, 1270–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Wlodarczyk A, Gnanasekaran T, Nielsen AZ, Zulu NN, Mellor SB, Luckner M, Thøfner JFB, Olsen CE, Mottawie MS, Burow M, Pribil M, Feussner I, Møller BL, and Jensen PE (2016) Metabolic engineering of light-driven cytochrome P450 dependent pathways into Synechocystis sp. PCC 6803. Metab. Eng 33, 1–11. [DOI] [PubMed] [Google Scholar]
- (20).Xue Y, Zhang Y, Grace S, and He Q (2014) Functional expression of an Arabidopsis p450 enzyme, p-coumarate-3-hydroxylase, in the cyanobacterium Synechocystis PCC 6803 for the biosynthesis of caffeic acid. J. Appl. Phycol 26, 219–226. [Google Scholar]
- (21).Lassen LM, Nielsen AZ, Olsen CE, Bialek W, Jensen K, Møller BL, and Jensen PE (2014) Anchoring a plant cytochrome P450 via PsaM to the thylakoids in Synechococcus sp. PCC 7002: Evidence for light-driven biosynthesis. PLoS One 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Yu J, Liberton M, Cliften PF, Head RD, Jacobs JM, Smith RD, Koppenaal DW, Brand JJ, and Pakrasi HB (2015) Synechococcus elongatus UTEX 2973, a fast growing cyanobacterial chassis for biosynthesis using light and CO2. Sci. Rep 5, 8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Jaiswal D, Sengupta A, Sohoni S, Sengupta S, Phadnavis AG, Pakrasi HB, and Wangikar PP (2018) Genome Features and Biochemical Characteristics of a Robust, Fast Growing and Naturally Transformable Cyanobacterium Synechococcus elongatus PCC 11801 Isolated from India. Sci. Rep 8, 16632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Walton K, and Berry PJ (2016) Indole Alkaloids of the Stigonematales (Cyanophyta): Chemical Diversity, Biosynthesis and Biological Activity. Mar. Drugs 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Acuña UM, Mo S, Zi J, Orjala J, and De Blanco EJC (2018) Hapalindole H Induces Apoptosis as an Inhibitor of NF-ĸB and Affects the Intrinsic Mitochondrial Pathway in PC-3 Androgen-insensitive Prostate Cancer Cells. Anticancer Res. 38, 3299–3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Kim H, Lantvit D, Hwang CH, Kroll DJ, Swanson SM, Franzblau SG, and Orjala J (2012) Indole alkaloids from two cultured cyanobacteria, Westiellopsis sp. and Fischerella muscicola. Biorg. Med. Chem 20, 5290–5295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Mo S, Krunic A, Chlipala G, and Orjala J (2009) Antimicrobial ambiguine isonitriles from the cyanobacterium Fischerella ambigua. J. Nat. Prod 72, 894–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Micallef ML, Sharma D, Bunn BM, Gerwick L, Viswanathan R, and Moffitt MC (2014) Comparative analysis of hapalindole, ambiguine and welwitindolinone gene clusters and reconstitution of indole-isonitrile biosynthesis from cyanobacteria. BMC Microbiol. 14, 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Hillwig ML, Zhu Q, and Liu X (2014) Biosynthesis of Ambiguine Indole Alkaloids in Cyanobacterium Fischerella ambigua. ACS Chem. Biol 9, 372–377. [DOI] [PubMed] [Google Scholar]
- (30).Li S, Lowell AN, Yu F, Raveh A, Newmister SA, Bair N, Schaub JM, Williams RM, and Sherman DH (2015) Hapalindole/Ambiguine Biogenesis Is Mediated by a Cope Rearrangement, C-C Bond-Forming Cascade. J. Am. Chem. Soc 137, 15366–15369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Liu X, Hillwig ML, Koharudin LMI, and Gronenborn AM (2016) Unified biogenesis of ambiguine, fischerindole, hapalindole and welwitindolinone: identification of a monogeranylated indolenine as a cryptic common biosynthetic intermediate by an unusual magnesium-dependent aromatic prenyltransferase. Chem. Commun 52, 1737–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Li S, Lowell AN, Newmister SA, Yu F, Williams RM, and Sherman DH (2017) Decoding cyclase-dependent assembly of hapalindole and fischerindole alkaloids. Nat. Chem. Biol 13, 467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Newmister SA, Li S, Garcia-Borràs M, Sanders JN, Yang S, Lowell AN, Yu F, Smith JL, Williams RM, Houk KN, and Sherman DH (2018) Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis. Nat. Chem. Biol 14, 345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Brady SF, and Clardy J (2005) Systematic Investigation of the Escherichia coli Metabolome for the Biosynthetic Origin of an Isocyanide Carbon Atom. Angew. Chem 117, 7207–7210. [DOI] [PubMed] [Google Scholar]
- (35).Zhu Q, and Liu X (2017) Discovery of a Calcium-Dependent Enzymatic Cascade for the Selective Assembly of Hapalindole-Type Alkaloids: On the Biosynthetic Origin of Hapalindole U. Angew. Chem. Int. Ed 56, 9062–9066. [DOI] [PubMed] [Google Scholar]
- (36).Hillwig ML, Zhu Q, Ittiamornkul K, and Liu X (2016) Discovery of a Promiscuous Non-Heme Iron Halogenase in Ambiguine Alkaloid Biogenesis: Implication for an Evolvable Enzyme Family for Late-Stage Halogenation of Aliphatic Carbons in Small Molecules. Angewandte Chemie - International Edition, 5780–5784. [DOI] [PubMed] [Google Scholar]
- (37).Hillwig ML, and Liu X (2014) A new family of iron-dependent halogenases acts on freestanding substrates. Nat. Chem. Biol 10, 6–10. [DOI] [PubMed] [Google Scholar]
- (38).Wong CP, Awakawa T, Nakashima Y, Mori T, Zhu Q, Liu X, and Abe I (2018) Two Distinct Substrate Binding Modes for the Normal and Reverse Prenylation of Hapalindoles by the Prenyltransferase AmbP3. Angew. Chem. Int. Ed 57, 560–563. [DOI] [PubMed] [Google Scholar]
- (39).Ungerer J, Lin P-C, Chen H-Y, and Pakrasi HB (2018) Adjustments to Photosystem Stoichiometry and Electron Transfer Proteins Are Key to the Remarkably Fast Growth of the Cyanobacterium Synechococcus elongatus UTEX 2973. mBio 9, e02327–02317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Sleight SC, Bartley BA, Lieviant JA, and Sauro HM (2010) Designing and engineering evolutionary robust genetic circuits. J. Biol. Eng 4, 12–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Choi SY, Park B, Choi I-G, Sim SJ, Lee S-M, Um Y, and Woo HM (2016) Transcriptome landscape of Synechococcus elongatus PCC 7942 for nitrogen starvation responses using RNA-seq. Sci. Rep 6, 30584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Camsund D, and Lindblad P (2014) Engineered transcriptional systems for cyanobacterial biotechnology. Front. Bioeng. Biotechnol 2, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Berla BM, Saha R, Immethun CM, Maranas CD, Moon TS, and Pakrasi HB (2013) Synthetic biology of cyanobacteria: unique challenges and opportunities. Front. Microbiol 4, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Knoot CJ, and Pakrasi HB (2019) Diverse hydrocarbon biosynthetic enzymes can substitute for olefin synthase in the cyanobacterium Synechococcus sp. PCC 7002. Sci. Rep 9, 1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Brady SF, and Clardy J (2005) Cloning and Heterologous Expression of Isocyanide Biosynthetic Genes from Environmental DNA. Angew. Chem. Int. Ed 44, 7063–7065. [DOI] [PubMed] [Google Scholar]
- (46).Taton A, Unglaub F, Wright NE, Zeng W. Y. u., Paz-Yepes J, Brahamsha B, Palenik B, Peterson TC, Haerizadeh F, Golden SS, and Golden JW (2014) Broad-host-range vector system for synthetic biology and biotechnology in cyanobacteria. Nucleic Acids Res. 42, e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Ungerer J, and Pakrasi HB (2016) Cpf1 Is A Versatile Tool for CRISPR Genome Editing Across Diverse Species of Cyanobacteria. Sci. Rep 6, 39681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Gatti-Lafranconi P, Dijkman WP, Devenish SRA, and Hollfelder F (2013) A single mutation in the core domain of the lac repressor reduces leakiness. Microb. Cell Fact 12, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Chen C-C, Hu X, Tang X, Yang Y, Ko T-P, Gao J, Zheng Y, Huang J-W, Yu Z, Li L, Han S, Cai N, Zhang Y, Liu W, and Guo R-T (2018) The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement. Angew. Chem. Int. Ed 57, 15060–15064. [DOI] [PubMed] [Google Scholar]
- (50).Awakawa T, Mori T, Nakashima Y, Zhai R, Wong CP, Hillwig ML, Liu X, and Abe I (2018) Molecular Insight into the Mg2+-Dependent Allosteric Control of Indole Prenylation by Aromatic Prenyltransferase AmbP1. Angew. Chem. Int. Ed 57, 6810–6813. [DOI] [PubMed] [Google Scholar]
- (51).Lin P-C, and Pakrasi HB (2018) Engineering cyanobacteria for production of terpenoids. Planta. [DOI] [PubMed] [Google Scholar]
- (52).Wendt KE, Ungerer J, Cobb RE, Zhao H, and Pakrasi HB (2016) CRISPR/Cas9 mediated targeted mutagenesis of the fast growing cyanobacterium Synechococcus elongatus UTEX 2973. Microb. Cell Fact 15, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Li S, Sun T, Xu C, Chen L, and Zhang W (2018) Development and optimization of genetic toolboxes for a fast-growing cyanobacterium Synechococcus elongatus UTEX 2973. Metab. Eng 48, 163–174. [DOI] [PubMed] [Google Scholar]
- (54).Averesch NJH, and Krömer JO (2018) Metabolic Engineering of the Shikimate Pathway for Production of Aromatics and Derived Compounds—Present and Future Strain Construction Strategies. Front. Bioeng. Biotechnol 6, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Oliver NJ, Rabinovitch-Deere CA, Carroll AL, Nozzi NE, Case AE, and Atsumi S (2016) Cyanobacterial metabolic engineering for biofuel and chemical production. Curr. Opin. Chem. Biol 35, 43–50. [DOI] [PubMed] [Google Scholar]
- (56).Knoot CJ, Ungerer J, Wangikar PP, and Pakrasi HB (2018) Cyanobacteria: Promising biocatalysts for sustainable chemical production. J. Biol. Chem 293, 5044–5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison Iii CA, and Smith HO (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343. [DOI] [PubMed] [Google Scholar]
- (58).Landry BP, Palanki R, Dyulgyarov N, Hartsough LA, and Tabor JJ (2018) Phosphatase activity tunes two-component system sensor detection threshold. Nature Communications 9, 1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Espah Borujeni A, Channarasappa AS, and Salis HM (2014) Translation rate is controlled by coupled trade-offs between site accessibility, selective RNA unfolding and sliding at upstream standby sites. Nucleic Acids Res. 42, 2646–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Davisson VJ, Woodside AB, Neal TR, Stremler KE, Muehlbacher M, and Poulter CD (1986) Phosphorylation of isoprenoid alcohols. The Journal of Organic Chemistry 51, 4768–4779. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.