Abstract
Somatic mutation of the PP2A Aα-subunit gene PPP2R1A is highly prevalent in high-grade endometrial carcinoma (EMCA). The structural, molecular, and biological basis by which the most recurrent EMCA-specific mutation site P179 facilitates features of EMCA malignancy have yet to be fully determined. Here we used a series of structural, biochemical, and biological approaches to investigate the impact of the P179R missense mutation on PP2A function. Enhanced sampling molecular dynamics simulations showed that arginine-to-proline substitution at the P179 residue changes the protein’s stable conformation profile. A crystal structure of the tumor-derived PP2A mutant revealed marked changes in A-subunit conformation. Binding to the PP2A catalytic subunit was significantly impaired, disrupting holoenzyme formation and enzymatic activity. Cancer cells were dependent on PP2A disruption for sustained tumorigenic potential, and restoration of wildtype Aα in a patient-derived P179R mutant cell line restored enzyme function and significantly attenuated tumorigenesis and metastasis in vivo. Furthermore, small molecule-mediated therapeutic reactivation of PP2A significantly inhibited tumorigenicity in vivo. These outcomes implicate PP2A functional inactivation as a critical component of high-grade EMCA disease pathogenesis. Moreover, they highlight PP2A reactivation as a potential therapeutic strategy for patients who harbor P179R PPP2R1A mutations.
Keywords: Endometrial Cancer, PP2A, PPP2R1A, P179R, Tumorigenesis
Introduction
Uterine cancer is the most common gynecologic malignancy in the United States with approximately 60,000 women diagnosed each year [1]. While most cases of uterine endometrial carcinoma (EMCA) have favorable outcomes with recurrence-free long-term survival, outcomes for the high-grade, treatment-refractory histological subtypes remain an important clinical problem [2, 3]. Serous EMCA (USC) accounts for only 10% of uterine cancer cases but a disproportionate 39% of deaths, with a 5-year survival of 55% [2]. Endometrial carcinosarcoma (UCS), a more aggressive mixed histological subtype, is rare, <5%, but accounts for >15% of deaths, with a 5-year survival around 35% [4–6]. Importantly, despite an overall decline in cancer-related deaths in the US, mortality rates for uterine cancers continue to rise [1]. A lack of established disease driving mechanisms for high-grade subtypes has limited the use of targeted treatment strategies, an approach that has had some success in the treatment of other cancers. New insight and validation of biological drivers of disease progression are therefore needed to improve disease management. Large-scale genomic profiling efforts have made substantial progress in identifying alterations characteristic for USC and UCS [7–10]. Foremost, both are typified by TP53 mutation, which is present in 80–90% of cases. In addition to TP53, PPP2R1A mutation was also more frequent in high-grade subtypes, occurring in ~30% of USC or UCS patients, versus ~5% in patients with the endometrioid subtype (UEC). PPP2R1A encodes the scaffold subunit of the PP2A tumor suppressive phosphatase.
Protein phosphatase 2A (PP2A) is a serine/threonine phosphatase that is involved in the physiologic regulation of diverse signaling pathways. PP2A is also a tumor suppressor whose diminished activity contributes to transformation and tumor development [11–13]. While a number of mechanisms can underlie diminished PP2A activity in cancer, notable is the occurrence of ‘hotspot’ mutations to PPP2R1A, the gene encoding the PP2A Aα subunit. The PP2A holoenzyme is a heterotrimer composed of a scaffolding A-subunit (PP2A-A), catalytic C-subunit (PP2A-C), and regulatory B-subunit (PP2A-B). Each is a subunit family composed of multiple isoforms; for PP2A-A this includes α and β isoforms.
The biogenesis of canonical PP2A heterotrimers is sequential: first a C-subunit binds to an A-subunit, and is followed by incorporation of one of 15 identified B-subunits [14, 15]. Several activation steps must also take place, facilitated by PP2A regulatory proteins, to convert the C-subunit into its catalytically active form and to modulate the binding of B-subunits [15]. Critically, throughout this process the A-subunit serves as a highly flexible scaffolding protein to facilitate protein interactions and trimerization. Cancer-derived hotspot mutations of PPP2R1A cluster at the structural interface between A- and B-subunits, and have been shown to disrupt subunit binding to varying degrees [11, 16–18]. A recent report characterizing several hotspot mutations found that these mutant proteins have increased binding to the PP2A inhibitor TIPRL, resulting in a dominant negative phenotype [18]. Our data suggests this mechanism may be true for some hotspot mutations but may not hold true for others that are most specific and recurrent in high-grade EMCA. Specifically, while PPP2R1A mutations are found in cancers of multiple origins, they are striking for USC and UCS not only for their high prevalence, but also for the nearly exclusive occurrence of mutation at two hotspot sites: P179 and S256.
Our work focuses on P179R, the most common of these recurrent, EMCA-enriched mutations, for which we have developed crystallography, modeling, and biochemical data to describe its impact on PP2A function. The P179R missense mutation induces global changes in A-subunit protein structure and its preferred conformational dynamics, which results in loss of interaction with the catalytic subunit and diminished holoenzyme stability. When wildtype protein is restored in the P179R mutant context, tumorigenesis is significantly inhibited. Moreover, pharmacologic activation of PP2A with a Small Molecule Activator of PP2A (SMAP) phenocopied this outcome of tumor growth inhibition [19]. Altogether these works demonstrate that PP2A is a key tumor suppressor in EMCA and is functionally disrupted by recurrent P179R PPP2R1A mutation to drive tumorigenesis.
Materials and Methods
Analysis of primary human endometrial and ovarian cancer samples:
De-identified tissue specimens were obtained from the Case CCC Gynecologic Tumor Biobank (Dr. Analisa DiFeo). DNA and RNA were isolated from tissue pieces using the AllPrep DNA/RNA Mini Kit (Qiagen) in combination with a QIAshredder (Qiagen) for initial tissue homogenization. The DNA region of interest was amplified by PCR (PCR Master Mix, Promega) and submitted for Sanger sequencing. Results were viewed using the software program 4Peaks. DNA and RNA were stored at −20°C and −80°C respectively.
Cell lines and culture conditions:
The UT42, UT89, UT150, and UT185 cell lines were generated by Dr. Analisa DiFeo. These stable lines were derived from primary human endometrial cancer tissue following protocols described previously [20]. HEK293T cells were purchased from the ATCC. All lines were cultured in DMEM (Corning, 10–013) supplemented with 10% fetal bovine serum (GE Healthcare, SH30070.03) and 1% penicillin-streptomycin (GE Healthcare, SV30010). Cells were maintained in humidified sterile incubator conditions with 5% CO2 at 37°C. Cells are passaged once 70–90% confluency is reached, and are maintained in culture for no more than 20 passages. Freshly thawed cells are passaged 2–3 times before use in experiments. Mycoplasma testing of cell lines is performed once annually using a detection kit (Lonza, LT07–710) and positive cell lines are discarded. Authentication of cell lines was performed via short tandem repeat (STR) profiling after the cell line was established (typically after two passages in culture), and compared to that of the primary tumor.
Generation of stable cell lines:
standard methodologies were used; details are provided in Supplementary Methods.
Immunoblotting:
standard methodologies were used; details and the primary antibodies used are provided in Supplementary Methods.
Real-time PCR:
standard methodologies were used; details and primer sequences are provided in Supplementary Methods.
Co-immunoprecipitation and phosphatase activity assays:
UT89 and UT42 isogenic cell lines were plated to achieve 70% confluency at 24 (UT89) or 48 (UT42) hours. Cells were harvested and co-immunoprecipitation (IP) of V5-tagged protein was performed according to the Dynabeads Co-immunoprecipitation kit (Invitrogen) protocol. 1.5 mg Dynabeads per IP reaction were coupled to V5 antibody (Abcam ab27671) at a concentration of 8 ug Ab/mg beads. Cells were lysed in a 1:9 ratio of mg cells to ml lysis buffer and equal volumes of this pre-IP lysate were incubated with antibody-conjugated beads. Pre-IP lysates, and IP isolates obtained after elution from beads, were stored at −20°C for immunoblotting.
The phosphatase activity assay was performed using the IP protocol with modifications: cells were lysed in a phosphatase assay buffer (25 mM HEPES, 1 mM MgCl, 0.1 mM MnCl, 1% Triton-X, and protease inhibitor); 5 mg of protein lysate was incubated with beads; beads were triple-washed with a wash buffer (25 mM HEPES, 1 mM MgCl, 0.1 mM MnCl) and aliquots portioned out for immunoblotting or activity assay. For assessment of phosphatase activity, beads with bound proteins were suspended in phosphatase assay buffer supplemented with 1 mM DTT. Samples were incubated with increasing concentrations of DiFMUP (ThermoFisher, D6567) for 15 min on a shaker at 37°C, at which point fluorescence was measured at 365/455nm. As a control for specificity of activity, 50 nM of Okadeic Acid (OA) was also added to select wells of V5-WT IP isolate. Measurements from V5-EGFP IP isolates were considered background and subtracted out. Phosphatase activity rate was then calculated for V5-WT and V5-P179R as fluorescent units per minute incubation time. Final values were normalized to total C-subunit protein in IP isolates as determined by immunoblotting.
Cell-free GST pull-down assay:
Approximately 0.6 μM of purified GST-B56α, 6 μM of His8-Cα, and 6 μM of Aα wildtype (WT) or mutant protein were incubated with 50 μL of GSH Sepharose 4B resin (GE Healthcare) for 1 hr at 4°C in binding buffer (20 mM Tris pH8.0, 150 mM NaCl, 5% Glycerol, 0.05% Tween 20, 2mM DTT; total volume of 250 μL). The beads were then washed with binding buffer three times. The bound proteins were resolved on SDS-PAGE and stained with Coomassie Brilliant Blue. The intensities of the bands were quantified by Image Lab Software (Bio-Rad) and values were normalized to the GST-B56α of WT. Each experiment was repeated three times.
Proteasome inhibition and half-life studies:
Cycloheximide solution was obtained from Sigma-Aldrich as a stock solution of 100 mg/ml in DMSO, and used at a concentration of 100 μg/ml for experiments. Bortezomib (Velcade) was obtained from Selleck Chemicals; a stock solution was prepared in DMSO and diluted to 1 μM for experiments. Treatments were added 24 hours after cell plating. At the reported timepoints, cells were harvested and lysed for protein immunoblotting. To calculate protein half-life, cycloheximide protein quantification data was Ln-transformed and fit with a regression line using GraphPad Prism 7 software. Details of the statistical analysis of these data are presented in Supplementary Methods. The slope of the best-fit line was used to calculate half-life with the formula T(1/2) = ln(2) / slope.
Clonogenicity assay:
standard methodologies were used; details are provided in Supplementary Methods.
SMAP dose-response assay:
Evaluation of SMAP treatment-induced growth inhibition was performed to determine the IC50 of four endometrial carcinoma cell lines. Cells were plated into 96-well plates at optimized cell densities such that approximately 90% confluency is achieved at 72 hrs. 24 hrs after plating, wells received media containing DMSO or SMAP-061, with triplicate wells per treatment condition. The IncuCyte live-cell imaging system (Essen Biosciences) was utilized to monitor cell number in each well for 72 hrs. The fold-change in cell number was determined for DMSO or SMAP, at each dose, and used to construct the dose-response curves. In GraphPad Prism, data was log-transformed and fit with a nonlinear curve.
Xenograft tumor assays:
All animal work was conducted in accordance with CWRU Animal Resource Center approved protocols and ethical guidelines. For the sub-cutaneous xenograft studies, 1×106 (UT89) or 5 ×106 (UT42) cells suspended in a 1:1 mixture of matrigel and cell media were injected sub-cutaneous into the right flank of 6–8 week-old female Balb/c nu/nu mice. Tumor growth was assessed by caliper measurement and volume (V) was calculated using the formula V = 0.5 × (L × W2) where L = length and W = width. At study end, tumor tissue was harvested, formalin-fixed, and paraffin-embedded for histologic evaluation, or snap-frozen in liquid nitrogen for protein isolation and immunoblotting.
For the UT42 intra-uterine xenograft study, procedures were modeled after a published study [21]. In brief, 6–8 week-old female athymic NSG mice (NOD.Cg-Prkdcscid Il2rgtm1Wjl / SzJ) were put under general anesthesia, a small abdominal incision was made, and the left uterine horn was identified and ligated at either end using 4–0 Vicryl suture (Ethicon). 5×106 cells suspended in 50 μl sterile PBS were then injected into the left uterine horn; uterine enlargement and absence of visible leakage was confirmed. Injections were performed across three days for an equal number of animals per experimental group. The study was ended at 8 weeks from the date of injection. Animals were euthanized and the left and right uterine horns were isolated, evaluated for tumor presence, and weighed intact for all animals. Metastatic nodules throughout the abdominal cavity were collected, counted, and weighed separately from the uterus as the ‘metastatic tumor burden’. Collected tissue specimens were formalin-fixed and paraffin-embedded for H&E staining. Upon microscopic evaluation for tumor presence, the largest linear dimension (LLD) was calculated as the sum of the lengths (in mm) of all tumor cell foci observed in a single section. Metastases were also confirmed microscopically.
The UT42 SMAP treatment study was performed using patient-derived xenograft (PDX). Tumor pieces were surgically implanted into the flank of female NSG mice. Successful outgrowth was monitored, and upon reaching a tumor volume of 100 mm3 animals were enrolled and randomly assigned to a treatment group. Treatment was delivered by oral gavage once daily (QD): vehicle control (N,N-Dimethylacetamide, DMA), 50 mg/kg SMAP-1154, or 100 mg/kg SMAP-1154. Animal body weight and tumor volume were measured every 2–3 days for the study duration. Animals were observed for signs of toxicity (i.e. debilitating diarrhea, abdominal stiffness, jaundice, hunched posture, rapid weight loss). Animals were euthanized once morbidity criteria were met as established in the protocol; in all instances this was due to a tumor size that accounted for >10% of body weight. Terminal sacrifice was considered the survival endpoint for construction of Kaplan-Meier curves. Tumor tissue was harvested 2 hours after a final treatment dose and was formalin-fixed for IHC evaluation or snap-frozen in liquid nitrogen for immunoblotting.
H&E staining was performed following standard protocols. H&E stained sections were imaged using a Leica SCN400 slide scanner at 2× magnification. Images were exported through the SCN400 Digital Imaging Hub and cropped where appropriate for publication (no other image adjustments were performed).
Protein expression, purification, crystallization and structure determination:
A mouse PP2A Aα P179R mutant construct was cloned using site-directed mutagenesis based on a mouse wildtype PP2A Aα pGEX-4T1 construct with an N-terminal GST tag and a TEV cleavage site in-between [22]. The GST fusion protein was over-expressed in E. coli BL21 (DE3) cells grown in Luria broth media. Bacteria cell pellets were lysed by sonication. The GST fusion proteins were eluted from Glutathione Sepharose 4B beads. GST tag was removed by TEV at 4°C overnight. Then the proteins were further purified by an anion exchange column, and finally purified by gel filtration on a Superdex 200 10/300 GL column (GE Healthcare). The peak fractions were pooled, and concentrated to ~5 mg/ml in a buffer containing 10 mM Tris HCl pH 80, 100 mM NaCl, 2 mM DTT.
The hanging-drop vapor diffusion method for crystallization was used to prepare crystals of mouse PP2A Aα P179R. To obtain protein crystals for structural study, 1 μL of protein sample (5 mg/mL) was mixed with 1 μL of well solution containing 0.2M Sodium malonate pH 7.0, 20% w/v PEG 3,350. The crystals were flash frozen by liquid nitrogen in a cryogenic solution containing 0.2 M Sodium malonate pH 7.0, 10% glycerol and 20% w/v PEG 3,350.
Screening and data collection were performed at the Advanced Light Source (ALS), beamline 8.2.1 at wavelength 1.00 Å. Details of the data collection and refinement are provided in Supplementary Methods. All diffraction data were processed by HKL2000 [23]. The initial phase was determined by molecular replacement in Phaser with model from PDB: 2IAE [22, 24]. All models were improved using iterative cycles of manual rebuilding with the program COOT and refinement with Refmac5 of the CCP4 6.1.2 program suite [25, 26].
Molecular dynamics simulations:
Structural coordinates for PP2A Aα were retrieved from the Protein Data Bank (PDB: 2IAE) [22]. The mutants were constructed in silico using the ICM mutagenesis program [27]. The Amber ff14SB force field was used to describe the systems, with explicit TIP3P water molecules [28–30]. PROPKA 3.1 and PDB2PQR 2.1, as implemented in the proteinPrepare() functionality of HTMD was used to determine the protonation state of the residues [31]. 1,000 steps of steepest descent were used to minimize all systems. Energy minimization and equilibrium for volume relaxation were carried out under NPT conditions at 1 atm, with initial velocities sampled from the Boltzmann distribution at 300 K. The temperature was kept at 300 K by a Langevin thermostat. The PME algorithm was used for electrostatic interactions with a cut-off of 1.2 nm. A 1.05 μs production run was carried out for all the systems in the NVT ensemble with hydrogen mass repartitioning that allowed for a time step of 4 fs. All simulations were performed using the ACEMD program using an identical protocol [32]. (Continued in Supplementary Methods).
Statistics:
Results are displayed as mean ± standard deviation (SD), unless otherwise indicated. In each figure legend, n indicates the number of independent experimental replicates completed, or, when in reference to an in vivo study, the number of animals per group. These were distinct samples whose data points were averaged to obtain group means. The statistical tests used are identified in the figure legends and/or text. All Student t-tests performed were two-sided. GraphPad Prism 7 software was utilized for graph construction and statistical analysis.
Data Availability:
The atomic coordinate and structure factor of mouse PP2A Aα P179R is deposited in the Protein Data Bank (PDB) with the accession code 6EF4. Other data that support the findings of this study are available from the corresponding author upon reasonable request.
Code Availability:
Details regarding the application and use of previously published algorithms available upon request
Results
Mutation of PPP2R1A at P179 and S256 is specific to high-grade subtypes of endometrial carcinoma.
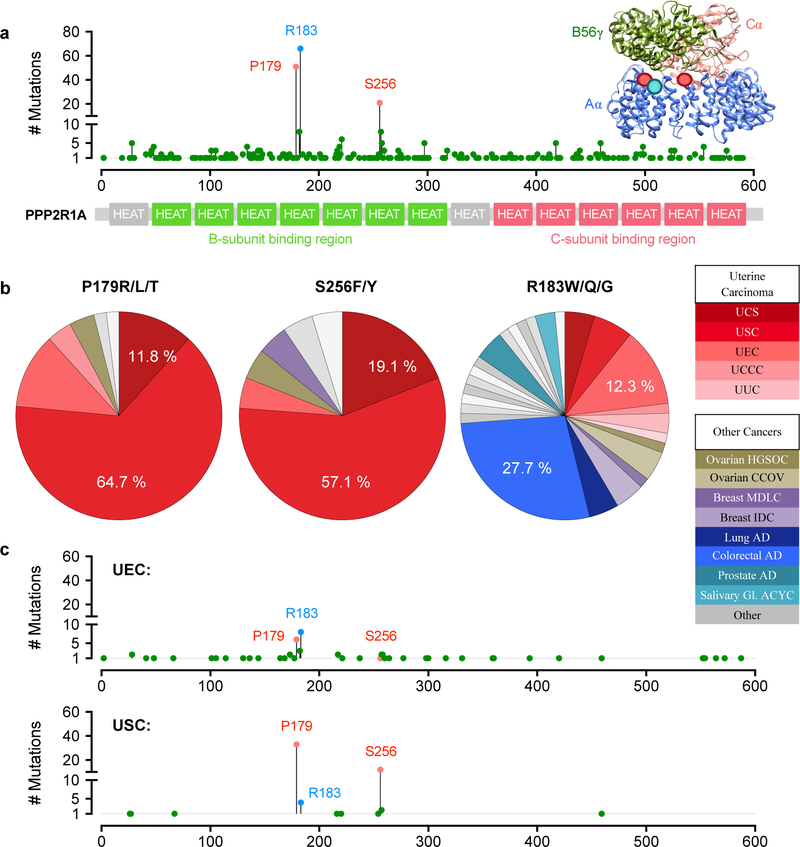
While three amino acid residues are notable PPP2R1A hotspot mutation sites in cancer (Figure 1a), two of these residues, P179 and S256, display a striking specificity in occurrence that is in contrast to the third, most frequently mutated site, R183 (Figure 1b). Specifically, P179 and S256 mutations are highly enriched in endometrial carcinoma (EMCA), occurring nearly exclusively in uterine serous carcinoma (USC) and carcinosarcoma (UCS), and also accounting for nearly all PPP2R1A mutations in these EMCA subtypes (Figure 1b–c). This is in stark contrast to the R183 mutation, which was identified in cancers of >20 different organ systems, with colorectal carcinoma being the most common. Although R183 mutations do occur in EMCA, they are much more prevalent in the less aggressive endometrioid carcinoma (UEC) subtype. Critically, this differential pattern of occurrence may indicate differing mechanistic roles through which these mutations contribute to cancer. In addition, mutation at these hotspot sites is also largely mutually exclusive. Across cancer, only one tumor was found to harbor two PPP2R1A hotspot mutations: a mixed ductal and lobular breast carcinoma was identified with both R183W and S256F mutations. In EMCA, occasional tumors were found to harbor more than one PPP2R1A mutation but in none of these cases were there two co-occurring hotspot site mutations (Supplementary Figure 1a).
Figure 1. P179R/L/T and S256F/Y Aα mutations are highly specific to high-grade endometrial cancer subtypes.
a, The distribution of PPP2R1A mutations identified in human cancers across the Aα-subunit protein length (adapted from cBioportal.org) [64, 65]. Inset image of a PP2A heterotrimer (PDB: 2IAE) indicating the location of mutated amino acids at the A-/B-subunit interface. b, The cancers in which point mutation of the P179, S256, or R183 residue was identified. c, Occurrence and distribution of PPP2R1A mutations in endometrioid (UEC) and serous (USC) subtypes of endometrial cancer. Acronyms: UCS, uterine carcinosarcoma; USC, uterine serous carcinoma; UEC, uterine endometrioid carcinoma; UCCC, uterine clear cell carcinoma; UUC, uterine undifferentiated carcinoma; HGSOC, high-grade serous ovarian carcinoma; CCOV, clear cell ovarian carcinoma; MDLC, mixed ductal and lobular carcinoma; IDC, invasive ductal carcinoma; AD, adenocarcinoma; ACYC, adenoid cystic carcinoma.
The P179 and S256 mutation sites reside in HEAT domains 5 and 7, respectively, of the PP2A Aα subunit. The P179 mutation is the more common of the two, and is substituted with arginine (P179R, 48 cases), lysine (P179L, 2 cases), or threonine (P179T, 1 case). The S256 is mutated to phenylalanine (S256F, 15 cases) or tyrosine (S256Y, 6 cases). Importantly, large-scale sequencing studies have revealed that 28–32% of USC patients and 8–12% of UCS patients harbor one of these mutations (TCGA, MSK-IMPACT; Supplementary Figure 1a). These patient cases display features characteristic of the high-grade USC and UCS subtypes in which they occur, and which are distinct from, and often in contrast to, the characteristic features of UEC tumors. First, P179 mutant tumors are frequently high-stage, with greater than 50% of patients diagnosed with stage III or IV disease (Supplementary Figure 1b). A stage III or IV diagnosis indicates cancer progression involving extra-uterine dissemination. This advanced disease is typical of USC and UCS, whereas 80% or more of UEC tumors are low-stage. Correspondingly, P179 tumors, similar to all USC tumors, display poor disease-free survival (Supplementary Figure 1c). At the genome-level, P179 tumors present with other defining features of USC and UCS, such as a low overall mutational burden, high copy number alteration, TP53 mutant, and MYC, CCNE1, or ERBB2 amplification (Supplementary Figure 1d–f). Hallmarks that distinguish UEC, including microsatellite instability (MSI) or PTEN, ARID1A, or CTNNB1 mutation, are rare in P179 tumors.
We sought to identify patient tumor specimens harboring recurrent PPP2R1A mutations in the Case Comprehensive Cancer Center (Case CCC) Gynecologic Tumor Biobank. 74 EMCA tumors and 18 ovarian carcinoma (OVCA) tumors underwent targeted Sanger sequencing at the HEAT 5 and 7 hotspot foci. 16% (15 tumors) were found to harbor PPP2R1A mutations and all were at residues P179, R183, or S256 (Supplementary Figure 2a–b). In addition to the previously reported P179R and P179L variants, we also identified a new variant, P179H, in a mixed serous-/endometrioid-subtype tumor (Supplementary Figure 2b–c). In this cohort, 38% of USC tumors were found to harbor P179 / S256 site mutations (Supplementary Figure 2a). Consistent with previous studies, our Sanger sequencing results demonstrated that these tumors are frequently heterozygous for mutation, with a wildtype allele remaining (Supplementary Figure 2c) [39–41]. However, for a subset of tumors there was no wildtype allele detected, which suggests that mutations can be either homozygous or co-occur with allelic deletion.
P179R-Aα disrupts PP2A holoenzyme assembly.
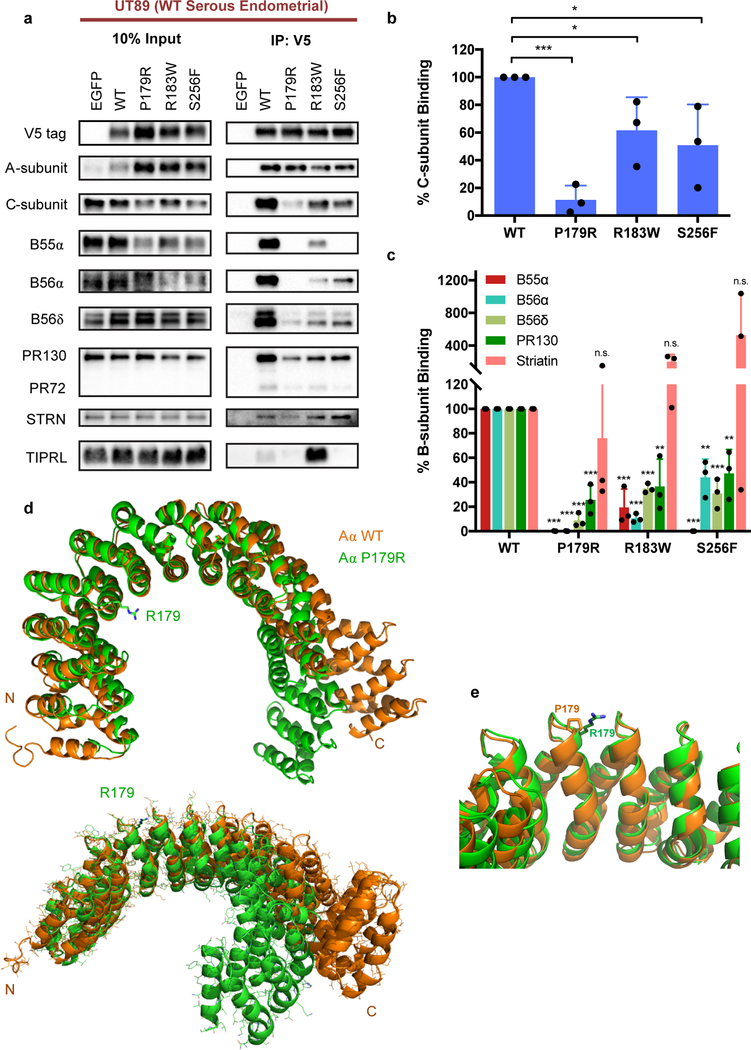
Studies have previously reported altered interactome and holoenzyme complex formation with cancer-associated mutant isoforms of the PP2A Aα-subunit [16–18]. However, these studies were not performed in USC or UCS EMCA cells, which is the most relevant disease context for studying the P179 and S256 mutations. Thus, we generated a primary serous EMCA cell line, UT89, to characterize the interactome of P179R, which represents the most common missense mutation in high-grade EMCA. For comparison, we also evaluated S256F and R183W. Cell lines with stable expression of V5-tagged wildtype (WT) or mutant Aα-subunit were generated using a lentivirus approach and tagged Aα protein was co-immunoprecipitated with its binding partners from whole cell lysates. Binding to PP2A family members was evaluated by western blotting of co-immunoprecipitants. Overall, the P179R mutation induced significant disruption of PP2A holoenzyme formation, presenting with a significant reduction in binding to members of each B-subunit family except the Striatins, for which binding remained intact, as well as a significant loss of catalytic C-subunit binding (Figure 2a–c). By comparison, S256F and R183W mutants also demonstrated losses in B-subunit interactions, but in many cases these losses were less severe. Interestingly, P179R and S256F shared a near-complete loss of binding to the B55α-subunit, whereas R183W retained some binding. Importantly, P179R was the most severely impaired in its interaction with the PP2A catalytic C-subunit. Finally, a published report identifies increased TIPRL binding to mutant Aα as a dominant-negative mechanism through which PP2A is inactivated in cancer [18]. We therefore also assessed mutant protein binding to TIPRL. From our results, the P179R and S256F mutants do not display increased TIPRL binding in a serous endometrial carcinoma cell line (Figure 2a). Meanwhile, R183W did display this gain of binding.
Figure 2. P179R-Aα displays impaired binding to both PP2A B- and C-subunits, which is supported by a global protein conformation change in the resolved crystal structure.
a, Western blots of co-immunoprecipitation (IP) isolates reveals altered interactome with mutant P179R-Aα protein. b–c, Quantification of the percent C- or B-subunit binding relative to WT. Band intensity measurements for subunit protein in the co-IP samples has been normalized to the amount of subunit protein present in the pre-IP input. Data is the average of three independent co-IP experiments (n=3), and represents the mean ± SD. Statistical significance was determined by Student’s t-test (P179R vs. WT). * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001, n.s. = not significant. d–e, Overlay of the resolved P179R-Aα crystal structure (green; PDB: 6EF4) with a published WT-Aα crystal structure (orange; PDB: 1B3U) [42]. Crystallographic data collection and refinement details are provided in Supplementary Methods.
To complement the co-immunoprecipitation approach, we additionally employed a cell-free binding assay to evaluate the impact of P179 mutation on holoenzyme assembly (Supplementary Figure 3a–b). GST-tagged B56α-subunit was incubated with purified C-subunit and purified wildtype (WT) or mutant Aα-subunit proteins, and then isolated and assessed for binding. When in the presence of P179 mutant Aα isoforms, assembly of the complete Aα-B56α-C holoenzyme heterotrimer was impaired. This is notable, as even the sole loss of B56α holoenzymes has been identified as sufficient for cell transformation when substituted into an established transformation model involving PP2A inactivation [13].
Altogether, these data support the previous literature describing loss of PP2A subunit interactions upon Aα mutation [16–18]. Moreover, it highlights a near complete loss of catalytic subunit binding to P179R-Aα, which ultimately suggests a marked change in protein structure that influences C-subunit contact points distant from the site of mutation.
The P179R-Aα protein displays altered conformational dynamics.
To understand how the overall protein structure may be altered by the P179R mutation, we determined the crystal structure of the P179R mutant protein at 3.4Å resolution (Figure 2d–e). Comparison of this PP2A P179R-Aα structure with the WT-Aα crystal structure (PDB: 1B3U) revealed an obvious conformational difference between the mutant and wildtype proteins (Figure 2d–e, overlays) [42]. Although we cannot rule out a contribution from crystal packing, our crystal structure indicates a change of the conformation-energy landscape of the PP2A P179R-Aα protein.
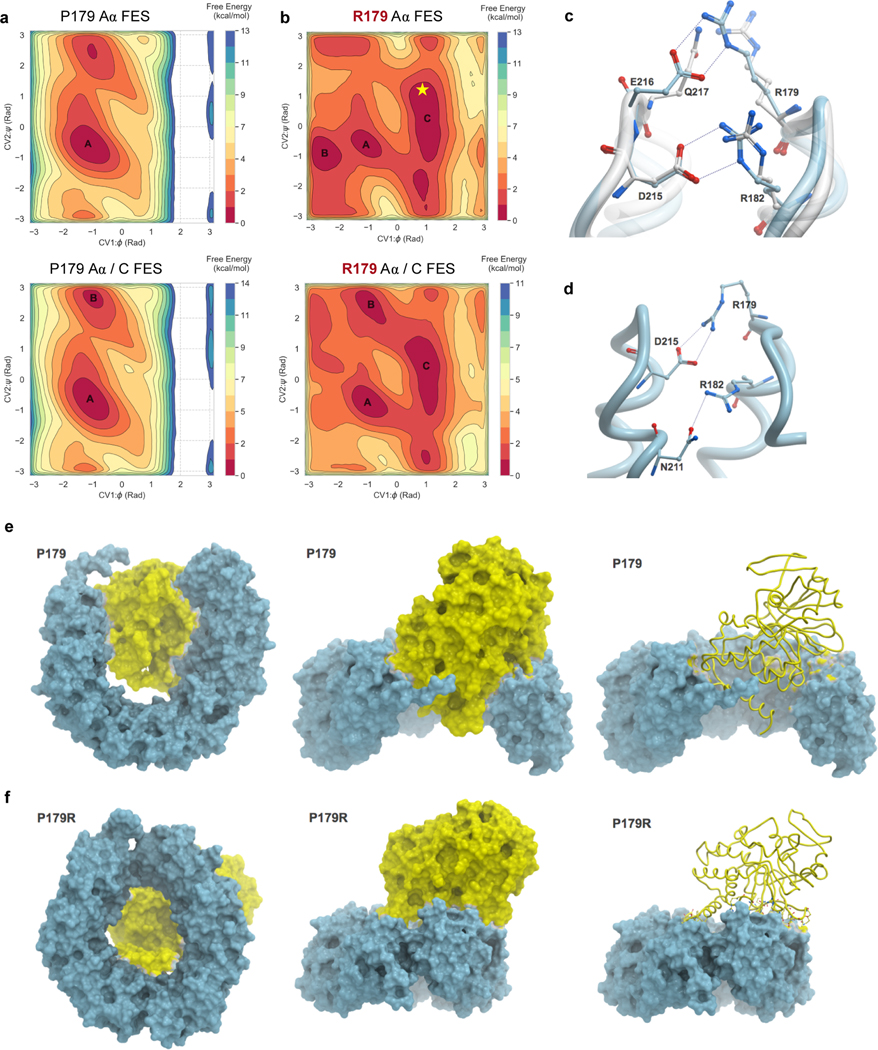
Therefore, we next performed classical and enhanced sampling molecular dynamics simulations of WT-Aα and P179R-Aα to identify stable protein conformations. Free energy landscape (FES) plots were generated as a function of φ and ψ dihedral angles of the P179 and mutant R179 residues, for both apo Aα and Aα in complex with the C-subunit (Figure 3a–b, Supplementary Figure 4a–p). These landscapes provide evidence that the R179 residue of the Aα mutant is capable of exploring an altered free energy landscape to adopt multiple metastable conformations of the Aα subunit protein, which may alter interactions with other PP2A subunits and proteins. We further extracted and analyzed the most stable conformations from the free energy minima of the WT (basin A) and mutant (basin C) FES (Figure 3a–b). Importantly, when we extrapolate the obtained P179R-Aα crystal structure on the FES of the simulated P179R-Aα, we found that the conformation adopted by the crystal structure is indeed one of the stable conformations and can be found in the largest minimum of the R179 apo Aα FES (Figure 3b, indicated by star).
Figure 3. Molecular modeling reveals distinct conformational dynamics of the P179R-Aα mutant protein.
a–b, Free energy surfaces (FES) plots of the apo Aα-subunit (top) and Aα/C complex (bottom) generated as a function of φ and ψ dihedral angles of residues P179 and R179. a, The P179 wildtype residue explores a similar conformational landscape in both apo and complex states. b, By comparison, the R179 mutant FES is modified, indicating sampling of additional conformations. The landscape between apo and complex states of R179 is also altered, highlighting additional sampling of interactions when in the Aα/C complex. Labelled basins represent energy minima, with basins A and C representing the most populated minima for P179 and R179, respectively. The resolved crystal structure (Figure 2d–e) extrapolates onto basin C of R179, indicated by the star. c–d, A comparison of inter-residue interactions made by R179. c, Overlay of the P179R-Aα crystal structure (white), with the representative conformation extracted from the largest populated basin of the apo R179 Aα FES (cyan). In the crystal structure, R179 makes ion pair interactions with Q217 while R182 interacts with D215. In the simulated apo P179R-Aα, R179 interacts with E216 while the interaction between R182 and D215 remains unaltered. d, In the representative conformation of P179R-Aα in the Aα/C complex, R179 makes an ion pair with D215 while R182 interacts with the side chain of N211. e–f, Conformational changes observed in the representative structures extracted from the most populated minima for the WT Aα/C complex (e) and the P179R mutant Aα/C complex (f).
Due to a cyclized side chain, P179 does not make inter-residue interactions in the wildtype crystalline conformation or throughout the course of any of our molecular dynamics simulations. By contrast, the R179 residue of the mutant protein makes direct interactions with nearby residues. In the resolved crystal structure, the guanidinium side chain of R179 orients towards the solvent, but still interacts with the side chain of Q217 of the adjacent HEAT repeat (Figure 3c, white structure). Comparably, extracting the most stable, representative conformation from the largest basin on the FES identifies an ion pair interaction between R179 and E216 (Figure 3c, cyan structure). In both, the interaction of R179 is facilitated by an additional ion pair interaction between R182 and D215.
The cyclic side chain of proline is effective in imparting conformational rigidity to the secondary structure of proteins [43]. Proline is also exceptional in the cis-trans isomerization of the peptidyl-prolyl backbone dihedral angle (ω; defined as Cα-C-N-Cα) preceding itself (0° for cis and ±180° for trans). The cis-trans configuration is isoenergetic, with the difference being ~1 kcal/mol between the two states [44–46]. This isomerization is an important structural mechanism through which proteins achieve large conformational changes and reach various macrostates of multidomain proteins without affecting the covalent structures. Therefore, mutation of proline can have an influence not only on the conformation of the protein but also on these biological processes [45–48]. To study the structural influence of P179R mutation on isomerization of the Aα P179 residue, including its dynamics in binding to the catalytic C-subunit, we carried out additional simulations of the wildtype and mutant residues. First we analyzed cis-trans state as a function of ω dihedral of P179 (Cα178-C178-N179-Cα179) in both Aα/C complex and apo states of Aα. The simulations identified that the ω dihedral of the wildtype P179 residue exists predominantly in the trans configuration (Supplementary Figure 4q, r). This is more pronounced in the apo state when compared with the complex. By contrast, in the P179R mutant, the ω dihedral of R179 preferentially adopts a cis configuration in apo and complex states (Supplementary Figure 4s, t). In this cis conformation, R179 makes a strong ion pair with D215, while R182 interacts with N211 (Figure 3d).
Next, we analyzed the global conformational changes and the effect of mutation on C-subunit binding to the Aα subunit. The FES plots indicated a complete change in conformational landscape between the wildtype and mutant complex (Figure 3a–b). We extracted the representative conformations from the most populated minimum of the Aα/C complex FES (basin A in P179 wildtype and C in R179 mutant). The WT-Aα subunit adopts a ‘clam-like’ conformation, with the catalytic subunit sandwiched between the two ends of the Aα subunit (Figure 3e). The catalytic subunit is deeply embedded within the ends, thereby maximizing its interactions with the Aα subunit. In the P179R mutant, the Aα subunit prefers to adopt a near closed ‘donut-shaped’ conformation (Figure 3f). The result is that the C-subunit appears to be squeezed out of the complex and now makes peripheral interactions with the Aα subunit, which is indicative of a destabilized complex. We calculated the binding energy for the C-subunit to either the WT or P179R mutant Aα-subunit, and determined them to be 236 kJ/mol and 347 kJ/mol, respectively. The greater binding energy between P179R-Aα and C, versus WT-Aα and C, is indicative of a greater C-subunit binding affinity for the WT isoform than the mutant isoform. This is again consistent with a destabilized complex, and in support of observations from the extracted structures. Altogether, these data show that a P179R mutation of the Aα protein leads to conformational disruption and a destabilized interaction with the C-subunit, which in turn lends context to the altered P179R mutant interactome described previously.
Impaired binding to P179R-Aα results in C-subunit destabilization.
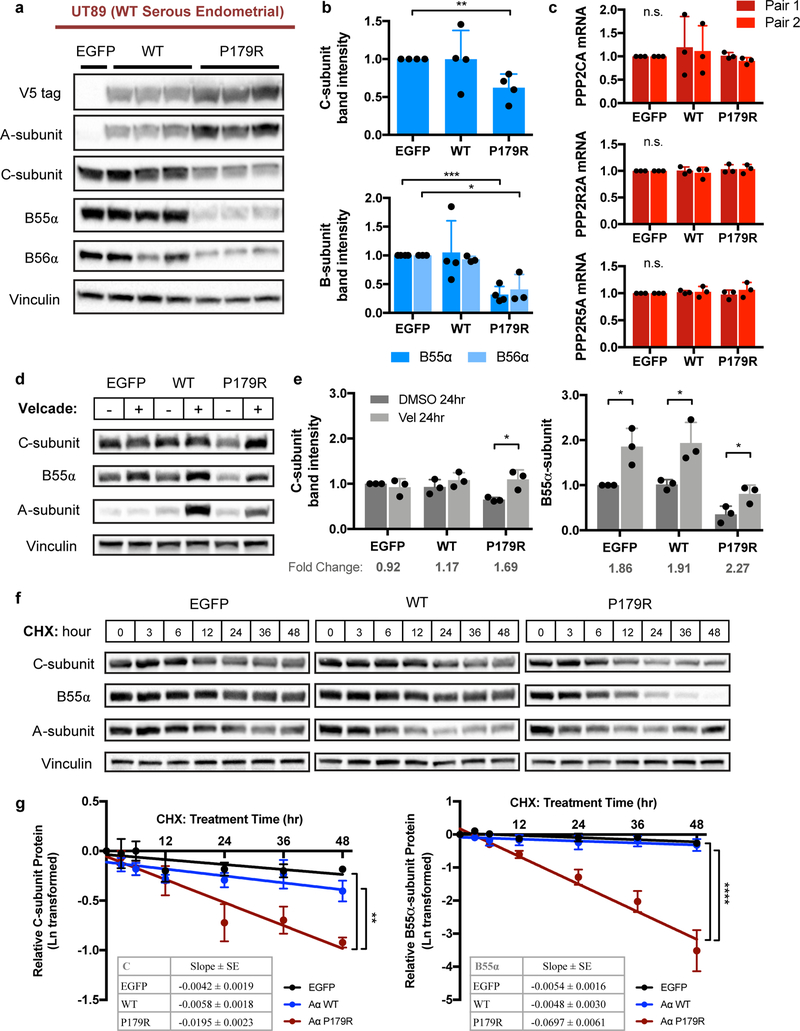
Expression of the P179R mutant isoform in UT89 cells was found to reduce total protein levels of C- and B-subunits to a degree that reflected their respective loss of binding in co-IP experiments (Figure 4a). Indeed, the loss of catalytic C-subunit total protein, as well as the regulatory B55α- and B56α-subunit proteins, was significant for the P179R mutant, which had demonstrated marked impairment of binding and conformation-related destabilization of the Aα/C complex (Figure 4b). This led us to hypothesize that impaired subunit incorporation into the trimeric holoenzyme results in increased degradation of the free subunit monomers. Real-time PCR verified that there was no change in PPP2CA (C-subunit), PPP2R2A (B55α), or PPP2R5A (B56α) mRNA, confirming that the decreased protein resulted from a post-translational mechanism (Figure 4c). We then asked whether this protein loss could be reversed upon treatment with the proteasome inhibitor Velcade. After a 24-hour treatment, total C-subunit protein was restored to a level comparable to that of EGFP control lines (Figure 4d–e) implicating proteasome-mediated degradation of C-subunit in P179R-expressing lines. Evaluation of a B-subunit isoform, B55α, revealed that its abundance was also increased with proteasome blockade in P179R-expressing cells, as well as in the EGFP and WT lines (Figure 4d–e). This could suggest that at least some B-subunits are present in excess and continuously turned over. To further verify decreased subunit stability in the presence of P179R-Aα, we investigated protein half-life using the translation inhibitor cycloheximide. P179R Aα-expressing cells demonstrated an increased rate of C-subunit degradation, represented by the slope of protein reduction over time, which was significantly different from both EGFP and WT-Aα transduced cells (Figure 4f–g). While EGFP and WT demonstrated modest C-subunit degradation within the experiment timeframe (~20–30% reduced relative to time=0 hrs), the higher rate of reduction in P179R allowed for calculation of a half-life of 36 hrs, at which point C-subunit total protein was 50% reduced. The protein degradation rate of B55α was also significantly increased in P179R mutant cells relative to EGFP or WT cells (Figure 4f–g). Finally, we evaluated a panel of PPP2R1A wildtype and P179R mutant patient tumor specimens from the Case CCC tumor biobank for C- and B-subunit protein levels; wildtype samples representative of both serous and endometrioid EMCA tumors were included (Supplementary Figure 5a). We calculated the ratio of C:A (C-subunit to A-subunit) and B:A (B55α-subunit to A-subunit) to account for variation in the basal level of A-subunit protein present in individual tumor specimens. Overall, P179R mutant tumors had lower levels of C- and B-subunit protein when compared to wildtype serous or endometrioid tumors (Supplementary Figure 5a–c). The differences were statistically significant for B55α (Mut vs Serous p=0.0012; Mut vs. Endometrioid p=0.0028), and trended toward significant for the C-subunit (Mut vs. Endometrioid p=0.0688; Mut vs. Serous p=0.1838).
Figure 4. P179R-Aα expression induced a loss of PP2A C- and B-subunit stability.
a, Western blots demonstrating loss of total C, B55α, and B56α protein upon expression of the P179R mutant Aα isoform that has impaired subunit binding. Triplicate lanes represent independent clonal lines generated from parental cells transduced in parallel wells. b, Quantification of total C-, B55α-, and B56α-subunits for selected clones used in subsequent experiments; presented as fold change relative to EGFP (n=3). c, mRNA transcript levels of PPP2CA (Cα), PPP2R2A (B55α), and PPP2R5A (B56α) from real-time PCR; normalized to β-Actin (n=3). Statistical significance for b and c was determined by Student’s t-test (WT or P179R vs. EGFP) with data graphed as mean ± SD. d–e, Representative westerns and quantification of total C and B55α protein present following treatment with DMSO or Velcade (1.0 μM) for 24 hours (hr) (n=3). Statistical significance was determined by Student’s t-test (Treated vs. Untreated), with data graphed as mean ± SD. f, Representative westerns for change in total C and B55α protein following treatment with Cycloheximide (CHX; 100 μg/ml) for the indicated time. g, Linear regression analysis was performed on Ln-transformed western blot densitometries to represent the change in protein abundance across time-points. Graphs represent the mean ± SEM; tables provide the slope ± SE of the best-fit line (n=3). Details of the statistical analysis of CHX data are provided in Supplementary Methods. * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001, n.s. = not significant.
Given the PP2A Aα mutant-driven disruption of PP2A holoenzyme assembly and reduction of catalytic subunit protein, and that PP2A has prominent tumor suppressive functions, we investigated whether expression of P179R-Aα in the PPP2R1A wildtype UT89 cell line led to an alteration of in vitro or in vivo tumor cell growth. No significant difference in clonogenesis or tumorigenesis was observed by colony formation assay or sub-cutaneous xenograft assay, respectively, when P179R-Aα was introduced into a complete wildtype background (Supplementary Figure 6a–b).
Restoration of wildtype PP2A function suppresses malignant features of P179R-Aα mutant cells.
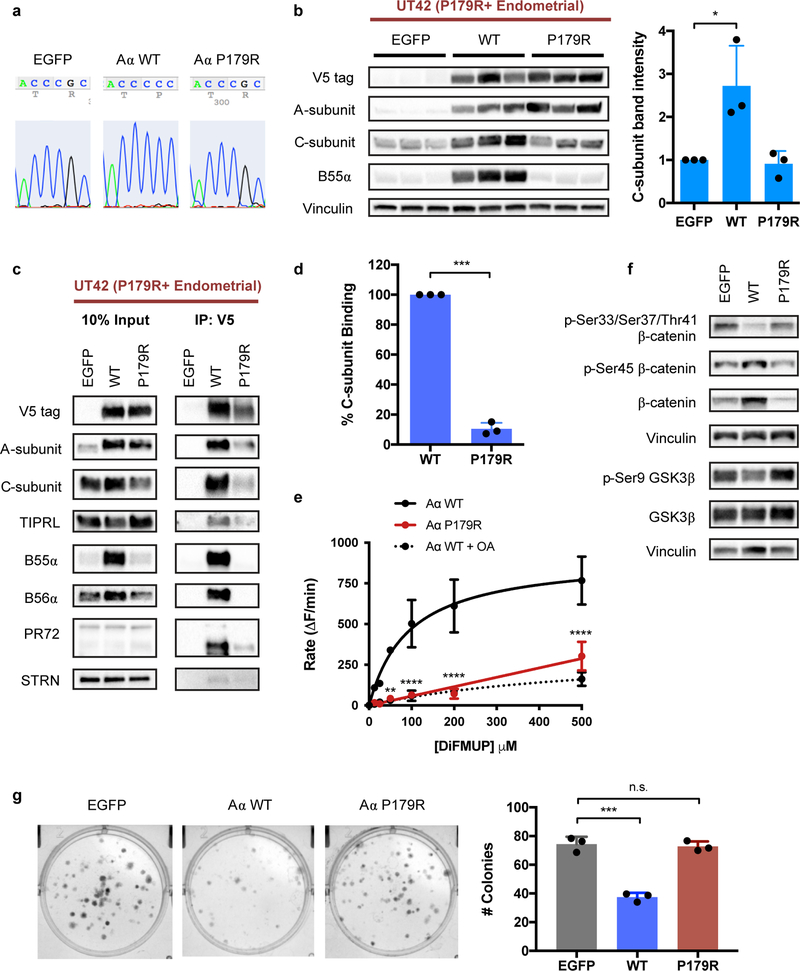
We derived another primary patient cell line, UT42, from a patient tumor that harbors the P179R mutation endogenously, in order to directly characterize mutant biology (Supplementary Figure 2). This model system provides the environmental, cellular, and genetic context native to a cell that acquired the P179R mutation as it underwent transformation. The UT42 cell line can be stably maintained in cell culture for greater than 30 passages, and was verified to retain the P179R mutation by Sanger sequencing. UT42 cells were transduced with V5-tagged WT-Aα or P179R-Aα to evaluate the impact of reconstitution with wildtype Aα protein on tumorigenic potential (Figure 5a). Expression of WT-Aα protein increased C- and B-subunit protein while expression of additional P179R-Aα did not, suggesting that these subunits are stabilized when in the presence of the binding-competent wildtype protein (Figure 5b, c). In this model system, co-immunoprecipitation of the V5-tagged P179R-Aα displayed an 80% reduction in C-subunit binding, which is consistent with data from UT89 (Figure 5c, d). To assess the catalytic activity of reconstituted WT-Aα and compare to that of mutant P179R-Aα protein, a phosphatase activity assay was performed on the co-immunoprecipitants of WT- or P179R-Aα V5-tagged protein from the UT42 isogenic lines. Robust dephosphorylation of the substrate DiFMUP was observed for extracted WT-Aα protein bound to C- and B-subunits (Figure 5e); this activity could be reversed by treatment with the serine/threonine phosphatase inhibitor OA [49]. By contrast, significantly less dephosphorylation activity was recorded for extracted P179R-Aα, and was comparable to that seen with OA-mediated PP2A inhibition, suggesting a near complete loss of catalytic activity with this recurrent, patient-derived Aα mutant protein. Phosphatase activity has been normalized to the relative amount of C-subunit in the pull-down isolate, confirmed by western blotting, to account for the decreased catalytic C-subunit binding to P179R-Aα. Finally, expression of WT-Aα led to dephosphorylation of the established PP2A substrates β-catenin and GSK3β at the specific residues against which the PP2A-B55α and PP2A-B56δ holoenzymes, respectively, have previously demonstrated phosphatase activity (Figure 5f) [50–52]. Consistent with the literature, dephosphorylation of β-catenin led to increased total protein as this event protects it from degradation.
Figure 5. Restoration of C-subunit protein and PP2A catalytic activity upon expression of WT-Aα in a P179R+ tumor-derived cell line, UT42.
a, Sanger sequencing of UT42 cell lines transduced with expression vectors for EGFP, WT-Aα, or P179R-Aα. b, Representative western blots demonstrate significantly increased C and B55α protein with expression of WT-Aα. Triplicate lanes are independent clonal lines generated from parental cells transduced in parallel wells. Graphed is the total C-subunit protein for clones selected for subsequent experiments (n=3). Data is presented as the mean ± SD fold change relative to EGFP, and statistical significance was determined by Student’s t-test (WT or P179R vs. EGFP). c, Western blots of co-IP isolates from the UT42 isogenic lines show that the expressed WT-Aα protein is capable of binding to other PP2A C- and B-subunits; expressed P179R-Aα protein does not. d, Percent C-subunit binding of P179R-Aα relative to WT from co-IP in the UT42 cell lines (n=3). Data presented as mean ± SD with significance determined by Student’s t-test. e, Co-IP isolates of WT or P179R Aα-containing complexes were assessed for phosphatase activity using a DiFMUP-based fluorescence assay (n=3), wherein WT-Aα isolates displayed robust dephosphorylation activity. This phosphatase activity could be blocked by Okadeic Acid treatment (OA, 50 nM). Data was analyzed by two-way ANOVA (p<0.0001, dF=28) with Tukey’s post-hoc t-tests. f, Expression of WT-Aα results in dephosphorylation of the PP2A substrates β-catenin and GSK3β. Images are representative of western blotting results from three independent biological replicates (n=3). g, Representative images and quantification of colony formation by UT42 cells expressing EGFP, WT-Aα, or P179R-Aα (n=3). Data presented as mean ± SD. Statistical significance was determined by Student’s t-test (WT or P179R vs. EGFP). * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001, **** p ≤ 0.0001, n.s. = not significant.
A colony formation assay was used to assess the impact of WT-Aα expression on in vitro clonogenic growth. UT42 cells reconstituted with WT-Aα protein were significantly impaired in their ability to form colonies, whereas expression of additional mutant P179R-Aα protein did not impact clonogenesis compared to control (Figure 5g).
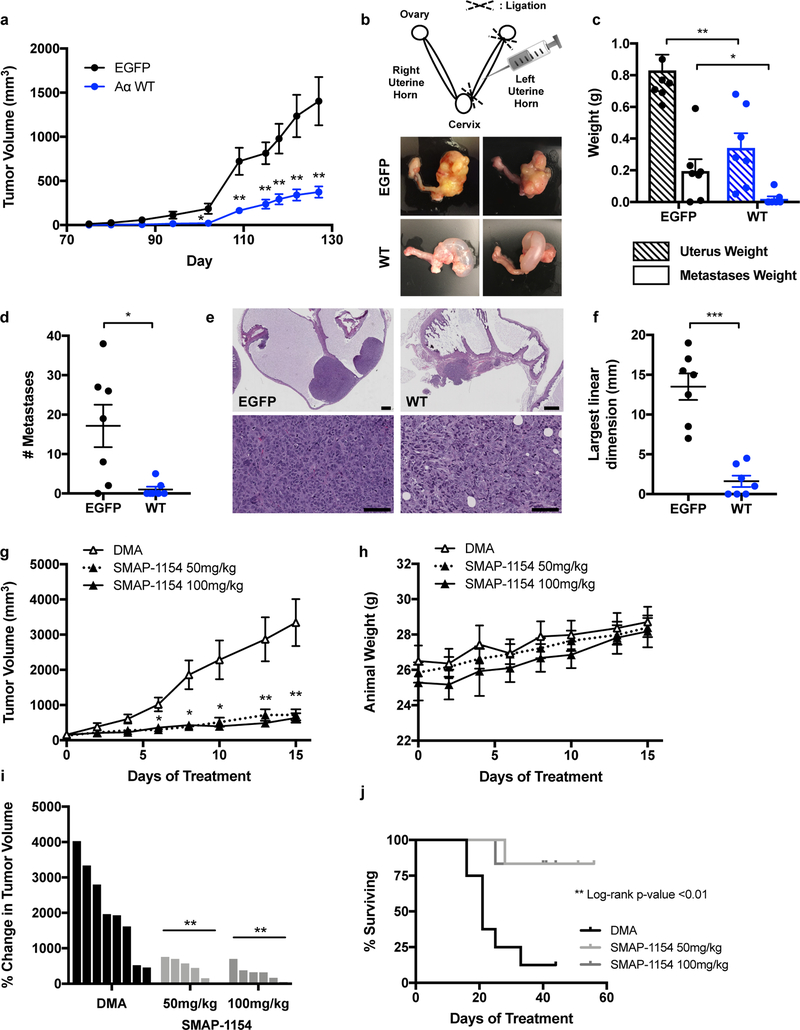
Using both genetic and pharmacologic approaches, we sought to determine the sensitivity of UT42 tumor growth to restoration of PP2A activity. First, UT42 cells that were reconstituted with catalytically active, wildtype (WT) Aα protein were injected sub-cutaneous in the flank of female mice. Tumor growth was compared to that of control EGFP-expressing cells (Figure 6a). Expression of WT-Aα protein significantly inhibited sub-cutaneous tumor growth.
Figure 6. PP2A activation in UT42 suppressed tumorigenesis and metastasis in vivo.
a, Tumor growth following sub-cutaneous injection of EGFP- or WT-Aα-expressing UT42 cells into the animal’s hind flank (n=11). b, Schematic of the intra-uterine cell injection method (adapted from ref. 21) with representative images of primary tumor formation in the extracted gynecologic tract (images for all animals presented in Supplementary Figure 6c). c, Weight of the injected left uterine horn, and of collected metastatic nodules, isolated from mice at 8 weeks post-injection (n=7). d, Number of metastatic nodules per animal and group. e, H&E stained sections of representative EGFP and WT primary uterine tumors at low magnification (top, scale bar = 1 mm) and high magnification (bottom, scale bar = 100 μm). f, The largest linear dimension was calculated as the sum of the lengths of all tumor foci observed within the sampled H&E stained tissue section. g, Tumor growth of sub-cutaneous UT42 PDX implants treated with vehicle (DMA; n=8), 50 mg/kg SMAP (n=6), or 100 mg/kg SMAP (n=6). h, Animal weights recorded across the duration of treatment. i, Waterfall plot presenting the percent change in tumor volume between the start of treatment and treatment day 15. j, Kaplan-Meier curves for animal survival within each treatment group. Data for all in vivo experiments are presented as the mean ± SEM with statistical significance determined by Student’s t-test, except for survival curves which were evaluated for significance by Log-rank (Mantel-Cox) test. * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001, n.s. = not significant.
We next employed an orthotopic xenograft model with tumor cell injection into the uterus (Figure 6b). In a pilot study with parental UT42 cells, all animals developed primary tumors within the uterus by 8 weeks, as well as intra-peritoneal metastatic nodules; no lymph node enlargement was discerned. This model thus allows for assessment of both primary tumor growth within the native environment of the uterus corpus, as well as cell metastatic potential. We carried out the full experiment under the same experimental conditions, injecting UT42 EGFP or WT-Aα cells into the left uterine horn of female NSG mice. At study end, tumor burden was evaluated macroscopically through gross inspection of the isolated gynecologic tract, as well as inspection of the pelvic cavity, abdominal cavity organs, and lungs for metastatic nodules. Tissue was collected for further microscopic evaluation and verification of tumor cell presence. At 8 weeks, four of seven EGFP animals had tumor filling the entirety of the injected uterine horn, and the remaining three animals had partial tumor presence visible macroscopically as a mass along the uterine wall (Figure 6b, Supplementary Figure 6c–d). By comparison, no WT animals had tumor filling the uterine cavity; four had partial tumor presence along the uterine wall, and the remaining three had no macroscopically discernable tumor presence. Reduced tumor formation resulted in a significantly reduced uterus weight, despite apparent hydrometra (Figure 6c). WT animals also demonstrated a significant decrease in metastatic burden, as determined by metastasis nodule weight and nodule count (Figure 6c–d). In both groups, metastatic nodules were predominantly peritoneal and surrounded by sub-peritoneal fat. No organ-invasive metastases were identified in WT animals. By contrast, two EGFP animals displayed metastatic nodules that were invasive into the liver parenchyma, and one animal had a retro-peritoneal metastasis attached to the kidney capsule (Supplementary Figure 6d–e). It should be noted that it is not possible to separate the decreased occurrence of metastasis from the decreased primary tumor burden of WT animals as an independent phenotype in this study. We did however perform a scratch wound assay to assess whether the WT-Aα cells may be impaired in migratory function and observed no significant difference from EGFP cells (Supplementary Figure 6f).
In H&E stained sections, tumor growth was found to expand within the endometrial layer of the uterus and through the myometrium, frequently present within parametrial soft tissue and occasionally encasing adnexal structures (Figure 6e). Growth in the outer uterine wall, subserosa, and parametrial soft tissue was observed in animals from both experimental groups. This growth pattern mirrors typical tumor growth observed in human tumor specimens. One additional WT animal was found to have microscopic tumor foci that were not apparent during gross evaluation. The longest linear dimension (LLD) was calculated for each animal as an aggregate measure of tumor foci size. Expression of WT-Aα led to a marked reduction in tumor foci size relative to the tumors formed by EGFP-expressing cells (Figure 6f).
Finally, we sought to evaluate the sensitivity of the UT42 P179R mutant tumor to a pharmacologic method of PP2A reactivation. We have previously reported on a novel series of Small Molecule Activators of PP2A (SMAP) that demonstrate in vivo suppression of tumor growth via activation of PP2A [19, 53, 54]. SMAP-061 and SMAP-1154 are two lead compounds of the SMAP series, which was generated through reverse engineering of tricyclic neuroleptics to remove anti-CNS toxicity and enhance anti-proliferative properties (structures published in ref. 19) [55]. Through in vitro characterization, the UT42 cell line demonstrated robust growth inhibition with SMAP treatment and displayed an IC50 that was significantly lower than three other PPP2R1A wildtype EMCA lines, including UT89 (Supplementary Figure 7a). Relative to UT89, the UT42 cell line also demonstrated greater sensitivity and marked growth impairment in the face of SMAP treatment during a two-week colony formation assay (Supplementary Figure 7b).
To investigate SMAP response in vivo, patient-derived xenografts (PDX) of the UT42 tumor were implanted sub-cutaneous in the flank of female mice and treated with vehicle (DMA), 50 mg/kg SMAP-1154, or 100 mg/kg SMAP-1154 QD. Tumor growth was markedly reduced by SMAP treatment at both doses (Figure 6g). Animals receiving SMAP treatment did not display signs of weight loss (Figure 6h) or other side effects during monitoring. From the initiation of treatment to study end, vehicle-treated tumors demonstrated robust increases in tumor volume, whereas minimal change in tumor volume was observed with SMAP treatment (Figure 6i). Critically, animals in the SMAP treatment groups exhibited a greater rate of survival (Figure 6j), as tumors receiving vehicle treatment grew rapidly to meet morbidity criteria for animal euthanasia. Evaluation of protein isolates collected from vehicle- or SMAP-treated tumors confirmed dephosphorylation of the PP2A substrates Akt, GSK3β, and c-Myc (Supplementary Figure 7c–d). Alteration of these targets is consistent with our previous works investigating the treatment of tumors with SMAPs [19, 53, 54]. Overall, the UT42 tumor harboring a P179R mutation was highly responsive to SMAP treatment, which significantly reduced its growth in vivo.
Discussion
PP2A provides critical regulatory activity that counter-balances kinase-mediated phosphorylation to maintain appropriate cell growth, regulate cell division, and prevent tumor development. A recurrent mutation of the PP2A Aα scaffolding subunit (PPP2R1A) is present in ~30% of high-grade subtypes of endometrial carcinoma (EMCA), with mutation at the P179 site accounting for ~20–25%. The UT42 patient-derived cell model harbors an endogenous P179R mutation and thus provides a unique system in which to investigate mutant biology as this tumor developed in the immediate context of somatic PPP2R1A mutation. The striking impairment of tumor formation and metastasis that occurred when wildtype Aα protein was restored provides evidence that functional disruption of PP2A is critical to this tumor and its malignant features. In a similar manner, tumor growth was significantly reduced when treated with a pharmacologic PP2A activator, again highlighting sensitivity of the P179R mutant UT42 tumor to PP2A activity. Overall, PP2A’s robust roles as a suppressor of transformation and tumor development have been well-documented and reviewed in the literature, to which our outcomes add further emphasis on its importance as a tumor suppressor in high-grade EMCA [56–58].
P179 site mutations are notable for displaying marked disease specificity, occurring almost exclusively in EMCA and the serous carcinoma and carcinosarcoma subtypes. This pattern suggests a role for PP2A as a disease driver, in which the biochemical consequences of mutating this residue is specifically advantageous to endometrial tumorigenesis. Consistent with this perspective, a large-scale sequencing study has shown that PPP2R1A mutations are somatic and truncal, and so are presumed to be acquired during the early processes of malignant cell transformation [9]. Several research groups have investigated PP2A as a tumor suppressor whose functional loss contributes to cell transformation. Interestingly, a majority of developed models rely on co-inactivation of p53 with PP2A for complete transformation, meanwhile, TP53 and PPP2R1A P179 mutations highly co-occur and are both truncal in these otherwise low mutational burden EMCA tumors [8–10, 59]. P179 mutation may therefore be a mechanism through which models of PP2A in transformation bear out in EMCA.
To understand how the P179R mutant alters PP2A function we began with investigation of PP2A subunit binding. Our findings are in agreement with previous literature that identified disrupted PP2A subunit interactions due to P179R PPP2R1A mutation [17, 18]. The consequence for unbound B- and C-subunits, an increased rate of protein degradation, is a novel finding for this cancer-associated mutation but is in support of a standing view that monomeric PP2A B- and C-subunits are less stable than their trimeric counterparts [11, 60]. Together these findings suggest that acquisition of a P179R mutation will impede assembly of holoenzymes containing catalytic and regulatory subunits and thereby induce a corresponding loss of canonical PP2A function within the cell.
The P179 residue resides at the A/B subunit interface and alteration of its side chain chemistry could reasonably be predicted to directly impair B-subunit binding. On the other hand, significant loss of C-subunit binding suggested to us that this mutation had a larger, global effect on the scaffolding protein’s conformation. By utilizing crystallography and molecular dynamics modeling, we provide context to the interactome dataset by detailing the structural alterations induced by the P179R substitution. Simulations of the wildtype protein reveal proline residue 179 as a site of cis-trans isomerization that can confer dynamic flexibility to the A-subunit scaffolding protein. The substituted arginine changed the dominant isomerization state and introduced new inter-residue interactions within the protein tertiary structure. The P179R substitution modifies the A-subunit’s most stable, and preferentially adapted, conformation in a manner that is unfavorable to C-subunit binding, as evidenced by Aα/C complex simulations for the mutant isoform as well as its increased binding energy requirement. Additionally, we were able to resolve the crystal structure of the mutant protein at a 3.4Å resolution. While published crystallographic structures have resolutions ranging from 1–4Å, with an average resolution of ~2.2Å, many structures, especially those with all-helical secondary structures such as the PP2A Aα protein, can be clearly resolved in 3–4Å without ambiguity. This is because alpha-helices have more characteristic electron densities and have well-defined main-chain structural restrictions. P179R-Aα is the first resolved crystal structure for a cancer-derived mutant PP2A protein and it underscores that mutation is a pathogenic mechanism through which cancer cells can disrupt the PP2A structure.
Importantly, our work suggests that a previously described mechanism of PP2A inactivation by PPP2R1A hotspot mutation may not hold true for P179R [18]. While some PPP2R1A mutations display increased binding to the PP2A inhibitor TIPRL and may be inactivated through a dominant negative mechanism, P179R did not display an increase in TIPRL binding in our model systems. This included protein co-immunoprecipitation in both a wildtype serous EMCA cell line (Figure 2a), as well as within a cell line that itself harbors the P179R mutation endogenously (Figure 5c). It is possible that the conflicting observations in P179R interactome reflect on the context-specificity of this mutation, however, a targeted investigation would be needed to lend more conclusive clarity to this discrepancy.
There is a strong clinical need for new therapeutic options in EMCA, and in particular for high-grade subtypes like serous EMCA that portend a poor prognosis due to tumor recurrence. Recently, immunotherapy has become an exciting prospective therapeutic opportunity for microsatellite instable (MSI) EMCA tumors due to their abundance of neoantigens [61]. However, tumors that harbor P179 mutations are unlikely to be amenable to the immunotherapy approach as we show this mutation occurs exclusively in low mutational burden, MSI-negative tumor types. The outcomes of the in vivo work presented here highlight a path forward for use of a targeted therapeutic approach that capitalizes on PP2A reactivation.
Finally, in addition to cancer, instances of germline PPP2R1A mutation have been reported in association with developmental and intellectual disabilities [62, 63]. A P179L mutation was one of several missense mutations identified. Whether this confers increased risk for cancer is unknown. From our results, one could reasonably predict these mutations lead to significant disruption of PP2A physiologic activity within neuronal cells.
Overall, this body of work suggests a loss-of-function of PP2A tumor suppressive activity due to P179R mutation of the Aα subunit. ‘Rescue’ through expression of holoenzyme-forming and catalytically competent wildtype Aα protein, or through pharmacologic PP2A activation, suppressed tumorigenesis. These findings support PPP2R1A P179R mutation as key driver of disease in the 20–30% of high-grade EMCA tumors that harbor one, and present pharmacologic targeting of PP2A as a potential therapeutic direction for this patient population.
Supplementary Material
Significance.
This study characterizes a highly recurrent, disease-specific PP2A PPP2R1A mutation as a driver of endometrial carcinoma and a target for novel therapeutic development.
Acknowledgments
The authors would like to thank the Case Comprehensive Cancer Center, which supports the Gynecologic Tumor Biobank, for the invaluable resources provided by this work; Kristen Weber Bonk and the CWRU Athymic Animal and Xenograft Core for their continued support with the in vivo studies; Daniela Schlatzer and the CWRU Proteomics Core for support with proteomics work; and finally, Richard Lee and the CWRU Light Microscopy Imaging Core. The Imaging Core’s Leica SCN400 slide scanner utilized for this work is made available through the NIH Office of Research Infrastructure (NIH-ORIP) Shared Instrumentation Grant (S10 RR031845). Funding for this work was provided by grants from the National Cancer Institute (NIH-NCI) to G. Narla (R01 CA181654), A. DiFeo (R01 CA197780), and S.E. Taylor (F30 CA224979); the Department of Defense to A. DiFeo (OC150553); and The Young Scientist Foundation to A. DiFeo. S.E. Taylor and D. Leonard are additionally supported by T32 GM007250 (NIH-NIGMS), and C.M. O’Connor by T32 GM008803 (NIH-NIGMS). S. Avril is supported by a Clinical and Translational Science Award KL2 TR0002547 (NIH-NCATS).
Footnotes
Conflict of Interest Statement:
The Icahn School of Medicine at Mount Sinai and Case Western Reserve University on behalf of the author G.N. have filed patents covering composition of matter on the small molecules disclosed herein for the treatment of human cancer and other diseases and for methods of use for using these small molecule PP2A activators. RAPPTA Therapeutics LLC has licensed this intellectual property for the clinical and commercial development of this series of small molecule PP2A activators. The author G.N. has an ownership interest in RAPPTA Therapeutics LLC.
References
- 1.Siegel RL, Miller KD & Jemal A Cancer statistics, 2018. CA Cancer J Clin 68, 7–30 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Hamilton CA et al. Uterine papillary serous and clear cell carcinomas predict for poorer survival compared to grade 3 endometrioid corpus cancers. Br J Cancer 94, 642–646 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Remmerie M & Janssens V Targeted Therapies in Type II Endometrial Cancers: Too Little, but Not Too Late. Int J Mol Sci 19, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cantrell LA, Blank SV & Duska LR Uterine carcinosarcoma: A review of the literature. Gynecol Oncol 137, 581–588 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Arend R, Doneza JA & Wright JD Uterine carcinosarcoma. Curr Opin Oncol 23, 531–536 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Felix AS et al. Comparison of survival outcomes between patients with malignant mixed mullerian tumors and high-grade endometrioid, clear cell, and papillary serous endometrial cancers. Int J Gynecol Cancer 21, 877–884 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao S et al. Landscape of somatic single-nucleotide and copy-number mutations in uterine serous carcinoma. Proc Natl Acad Sci U S A 110, 2916–2921 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kandoth C et al. Integrated genomic characterization of endometrial carcinoma. Nature 497, 67–73 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibson WJ et al. The genomic landscape and evolution of endometrial carcinoma progression and abdominopelvic metastasis. Nature Genetics 48, 848–855 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cherniack AD et al. Integrated Molecular Characterization of Uterine Carcinosarcoma. Cancer Cell 31, 411–423 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen W, Arroyo JD, Timmons JC, Possemato R & Hahn WC Cancer-associated PP2A Aalpha subunits induce functional haploinsufficiency and tumorigenicity. Cancer Research 65, 8183–8192 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Chen W et al. Identification of specific PP2A complexes involved in human cell transformation. Cancer Cell 5, 127–136 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Sablina AA, Hector M, Colpaert N & Hahn WC Identification of PP2A complexes and pathways involved in cell transformation. Cancer Research 70, 10474–10484 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janssens V, Longin S & Goris J PP2A holoenzyme assembly: in cauda venenum (the sting is in the tail). Trends in Biochemical Sciences 33, 113–121 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Sents W, Ivanova E, Lambrecht C, Haesen D & Janssens V The biogenesis of active protein phosphatase 2A holoenzymes: a tightly regulated process creating phosphatase specificity. FEBS J 280, 644–661 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Ruediger R, Pham HT & Walter G Disruption of protein phosphatase 2A subunit interaction in human cancers with mutations in the A alpha subunit gene. Oncogene 20, 10–15 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Jeong AL et al. Patient derived mutation W257G of PPP2R1A enhances cancer cell migration through SRC-JNK-c-Jun pathway. Sci Rep 6, 27391 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haesen D et al. Recurrent PPP2R1A Mutations in Uterine Cancer Act through a Dominant-Negative Mechanism to Promote Malignant Cell Growth. Cancer Research 76, 5719–5731 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Sangodkar J et al. Activation of tumor suppressor protein PP2A inhibits KRAS-driven tumor growth. Journal of Clinical Investigation 127, 2081–2090 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagaraj AB et al. Critical role of Wnt/beta-catenin signaling in driving epithelial ovarian cancer platinum resistance. Oncotarget 6, 23720–23734 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takahashi K et al. Development of a mouse model for lymph node metastasis with endometrial cancer. Cancer Sci 102, 2272–2277 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Cho US & Xu W Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme. Nature 445, 53–57 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Otwinowski Z & Minor W Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 24.McCoy AJ et al. Phaser crystallographic software. J Appl Crystallogr 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50, 760–763 (1994). [DOI] [PubMed] [Google Scholar]
- 27.Abagyan R, Totrov M & Kuznetsov D ICM—A new method for protein modeling and design: Applications to docking and structure prediction from the distorted native conformation. J Comput Chem 15, 488–506 (1994). [Google Scholar]
- 28.Case DA et al. The Amber biomolecular simulation programs. J Comput Chem 26, 1668–1688 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maier JA et al. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J Chem Theory Comput 11, 3696–3713 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price DJ & Brooks CL 3. A modified TIP3P water potential for simulation with Ewald summation. J Chem Phys 121, 10096–10103 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Doerr S, Harvey MJ, Noe F & De Fabritiis G HTMD: High-Throughput Molecular Dynamics for Molecular Discovery. J Chem Theory Comput 12, 1845–1852 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Harvey MJ, Giupponi G & Fabritiis GD ACEMD: Accelerating Biomolecular Dynamics in the Microsecond Time Scale. J Chem Theory Comput 5, 1632–1639 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Bonomi M, Barducci A & Parrinello M Reconstructing the equilibrium Boltzmann distribution from well-tempered metadynamics. J Comput Chem 30, 1615–1621 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Laio A & Gervasio FL Metadynamics: a method to simulate rare events and reconstruct the free energy in biophysics, chemistry and material science. Reports on Progress in Physics 71, 126601 (2008). [Google Scholar]
- 35.Skliros A et al. The importance of slow motions for protein functional loops. Phys Biol 9, 014001 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hess B, Kutzner C, van der Spoel D & Lindahl E GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J Chem Theory Comput 4, 435–447 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Dolinsky TJ, Nielsen JE, McCammon JA & Baker NA PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res 32, W665–7 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baker NA, Sept D, Joseph S, Holst MJ & McCammon JA Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A 98, 10037–10041 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McConechy MK et al. Subtype-specific mutation of PPP2R1A in endometrial and ovarian carcinomas. J Pathol 223, 567–573 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Shih I-M et al. Somatic mutations of PPP2R1A in ovarian and uterine carcinomas. Am J Pathol 178, 1442–1447 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagendra DC, Burke J 3., Maxwell GL & Risinger JI PPP2R1A mutations are common in the serous type of endometrial cancer. Mol Carcinog 51, 826–831 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Groves MR, Hanlon N, Turowski P, Hemmings BA & Barford D The structure of the protein phosphatase 2A PR65/A subunit reveals the conformation of its 15 tandemly repeated HEAT motifs. Cell 96, 99–110 (1999). [DOI] [PubMed] [Google Scholar]
- 43.Morris AL, MacArthur MW, Hutchinson EG & Thornton JM Stereochemical quality of protein structure coordinates. Proteins 12, 345–364 (1992). [DOI] [PubMed] [Google Scholar]
- 44.Velazquez HA & Hamelberg D Dynamical role of phosphorylation on serine/threonine-proline Pin1 substrates from constant force molecular dynamics simulations. J Chem Phys 142, 075102 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Dugave C & Demange L Cis-trans isomerization of organic molecules and biomolecules: implications and applications. Chem Rev 103, 2475–2532 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Lu KP, Finn G, Lee TH & Nicholson LK Prolyl cis-trans isomerization as a molecular timer. Nat Chem Biol 3, 619–629 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Wedemeyer WJ, Welker E & Scheraga HA Proline cis-trans isomerization and protein folding. Biochemistry 41, 14637–14644 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Andreotti AH Native state proline isomerization: an intrinsic molecular switch. Biochemistry 42, 9515–9524 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Xing Y et al. Structure of protein phosphatase 2A core enzyme bound to tumor-inducing toxins. Cell 127, 341–353 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Hein AL et al. PR55alpha Subunit of Protein Phosphatase 2A Supports the Tumorigenic and Metastatic Potential of Pancreatic Cancer Cells by Sustaining Hyperactive Oncogenic Signaling. Cancer Research 76, 2243–2253 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang W et al. PR55 alpha, a regulatory subunit of PP2A, specifically regulates PP2A-mediated beta-catenin dephosphorylation. J Biol Chem 284, 22649–22656 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu L & Eisenman RN Regulation of c-Myc Protein Abundance by a Protein Phosphatase 2A-Glycogen Synthase Kinase 3beta-Negative Feedback Pathway. Genes & Cancer 3, 23–36 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kauko O et al. PP2A inhibition is a druggable MEK inhibitor resistance mechanism in KRAS-mutant lung cancer cells. Sci Transl Med 10, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Allen-Petersen BL et al. Activation of PP2A and Inhibition of mTOR Synergistically Reduce MYC Signaling and Decrease Tumor Growth in Pancreatic Ductal Adenocarcinoma. Cancer Research 79, 209–219 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kastrinsky DB et al. Reengineered tricyclic anti-cancer agents. Bioorganic & Medicinal Chemistry 23, 6528–6534 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mumby M PP2A: unveiling a reluctant tumor suppressor. Cell 130, 21–24 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Janssens V, Goris J & Van Hoof C PP2A: the expected tumor suppressor. Curr Opin Genet Dev 15, 34–41 (2005). [DOI] [PubMed] [Google Scholar]
- 58.Hahn WC et al. Creation of human tumour cells with defined genetic elements. Nature 400, 464–468 (1999). [DOI] [PubMed] [Google Scholar]
- 59.Hahn WC et al. Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Molecular and Cellular Biology 22, 2111–2123 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strack S, Cribbs JT & Gomez L Critical role for protein phosphatase 2A heterotrimers in mammalian cell survival. J Biol Chem 279, 47732–47739 (2004). [DOI] [PubMed] [Google Scholar]
- 61.Dudley JC, Lin M-T, Le DT & Eshleman JR Microsatellite Instability as a Biomarker for PD-1 Blockade. Clin Cancer Res 22, 813–820 (2016). [DOI] [PubMed] [Google Scholar]
- 62.Houge G et al. B56delta-related protein phosphatase 2A dysfunction identified in patients with intellectual disability. Journal of Clinical Investigation 125, 3051–3062 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.DDDS. Large-scale discovery of novel genetic causes of developmental disorders. Nature 519, 223–228 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gao J et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science Signaling 6, pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cerami E et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discovery 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinate and structure factor of mouse PP2A Aα P179R is deposited in the Protein Data Bank (PDB) with the accession code 6EF4. Other data that support the findings of this study are available from the corresponding author upon reasonable request.