Abstract
Active zone proteins play a fundamental role in regulating neurotransmitter release and defining release sites. The functional roles of active zone components are beginning to be elucidated; however, the mechanisms of active zone protein localization are unknown. Studies have shown that glutamine, leucine, lysine, and serine-rich protein (ELKS), a recently defined member of the active zone complex, acts to localize the active zone protein Rab3a-interacting molecule (RIM) and regulates synaptic transmission in cultured neurons. Here, we test the function of ELKS in vivo. Like mammalian ELKS, Caenorhabditis elegans ELKS is an active zone protein that directly interacts with the postsynaptic density-25/Discs large/zona occludens (PDZ) domain of RIM. However, RIM protein localizes in the absence of ELKS and vice versa. In addition, elks mutants exhibit neither the behavioral nor the physiological defects associated with unc-10 RIM mutants, indicating that ELKS is not a critical component of the C. elegans release machinery. Interestingly, expression of the soluble PDZ domain of RIM disrupts ELKS active zone targeting, suggesting a tight association between the two proteins in vivo. RIM truncations containing only the PDZ and C2A domains target to release sites in an ELKS-dependent manner. Together, these data identify ELKS as a new member of the C. elegans active zone complex, define the role of ELKS in synaptic transmission, and characterize the relationship between ELKS and RIM in vivo. Furthermore, they demonstrate that multiple different protein-protein interactions redundantly anchor both ELKS and RIM to active zones and implicate novel proteins in the formation of the active zone.
Keywords: synapse, active zone, RIM, ELKS, CAST, ERC
Introduction
The presynaptic active zone is an electron-dense region composed of a highly interactive protein-rich web juxtaposed to the postsynaptic density containing neurotransmitter receptors (Harlow et al., 2001; Phillips et al., 2001). Although characterization of the active zone is still incomplete, a few proteins, notably Rab3a-interacting molecule (RIM), glutamine, leucine, lysine, and serine-rich protein (ELKS) [also known as Rab6-interacting protein, CAST (CAZ-associated structural protein), ERC (ELKS, Rab6-interacting protein 2, and CAST)], mammalian UNC-13, bassoon, and piccolo, are active zone-specific proteins in the vertebrate nervous system (Dresbach et al., 2001; Rosenmund et al., 2003; Zhai and Bellen, 2004). Proteins that make up the active zone play a fundamental role in regulating neurotransmitter release and defining release sites (Richmond and Broadie, 2002; Rosenmund et al., 2003; Sudhof, 2004). Although the functional roles of these proteins are beginning to be elucidated, the mechanisms used by active zone proteins to localize are unknown.
Active zone proteins are thought to form a highly interactive protein complex at release sites; however, no mutant animal containing a gene deletion in an active zone protein has resulted in the mislocalization of other active zone proteins (Zhen and Jin, 2004; Ziv and Garner, 2004). For example, knock-out mice that lack RIM1α protein expression have defects in synaptic physiology and in learning and memory tasks but contain normal active zone structures (Wang et al., 1997; Castillo et al., 2002; Schoch et al., 2002; Calakos et al., 2004; Powell et al., 2004). However, the role RIM plays in the localization of other active zone components may be masked by cellular compensation mechanisms arising from one of three other mouse RIM genes or from one of >60 different splicing products of the mammalian RIM1α gene (Johnson et al., 2003; Wang and Sudhof, 2003). Deletion of the one rim (unc-10) gene in Caenorhabditis elegans disrupts the animals' behavior and synaptic physiology, and, like the mouse RIM1α knock-out animals, they contain normal active zone ultrastructures and active zone protein immunostaining patterns (Koushika et al., 2001). Together, these studies provide proof that RIM is critically involved in neurotransmitter release but is unnecessary for the localization of other active zone components.
Although genetic deletion studies have provided little data on how active zone proteins are localized, overexpression studies in cultured neurons have suggested a role for ELKS in the localization of RIM to the synapse (Ohtsuka et al., 2002). In the cultured neuron system, RIM localization requires an interaction between the postsynaptic density-25/Discs large/zona occludens (PDZ)-binding motif at the C-terminal tail of ELKS and the RIM PDZ domain (Ohtsuka et al., 2002). However, these studies are difficult to interpret, because the ELKS and RIM transgenes are being overexpressed in a system that already contains native ELKS and RIM, both of which are expressed from more than one gene and as multiple different isoforms (Wang et al., 2002; Johnson et al., 2003; Wang and Sudhof, 2003; Deguchi-Tawarada et al., 2004). Using C. elegans genetics, we generated animals that lack ELKS protein expression, examined ELKS involvement in synaptic transmission, and determined its relationship with RIM in vivo. Through these studies, we also implicated novel proteins involved in ELKS and RIM localization.
Materials and Methods
Growth and culture of C. elegans. C. elegans animals were grown at 22.5°C on solid medium as described previously (Sulston and Hodgkin, 1988). Aldicarb (2-methyl-2-[methylthio]proprionaldehyde O-[methylcarbamoyl] oxime; Chem Service, West Chester, PA) was prepared as a 100 mm stock solution in 70% ethanol and added to the agar growth medium after autoclaving.
Identification of C. elegans elks. Rat ELKS-1 was used to BLAST both the C. elegans genome and wormpep databases using the Washington University BLAST (basic local alignment search tool) server. F42A6.9 was the only significant homolog identified. To confirm that only one homolog was present in C. elegans, we also searched the databases specifically with the regions most conserved between ELKS-1 and ELKS-2 and C. elegans ELKS using BLAST (supplemental Fig. 1, available at www.jneurosci.org as supplemental material). These BLAST searches also only identified the F42A6.9 gene in C. elegans. Similarly, BLAST searches using these sequences also only identified a single homolog in the Drosophila genome (gene CG12933 on scaffold AE003833). This predicted gene is likely incorrectly annotated.
Cloning and minigene constructs. Molecular biology experiments were performed essentially as described previously (Sambrook et al., 1989). Plasmid pELKS04 contains the 12.3 kb NcoI/SphI genomic fragment from cosmid F42A6 inserted into pGEM T Easy (Promega, Madison, WI). Plasmid pELKS05 (full-length ELKS) removes 1889 bases before the SphI site. Plasmid pELKS09 (ELKSΔIWA) codes for an isoleucine to stop change at 835 and was constructed by site-directed mutagenesis of pELKS05 changing ATT to TGA. Construction of pRIM6 (full-length RIM) was described previously (Koushika et al., 2001). Plasmid pRIM1 [green fluorescent protein (GFP)-PDZRIM] is driven by the neuronal sng-1 promoter and contains amino acids 534-732 of the RIM protein with GFP fused the C terminus. Plasmid pRIM27 (RIMΔPDZ) is a variant of pRIM6 removing amino acids 590-752. Plasmids pRIM45 (GFP-C2A) and pRIM47 (GFP-PDZC2ARIM) are each driven by the neuronal rab-3 promoter and contain amino acids 826-974 and 627-974 of the RIM protein, respectively, with GFP fused to the C terminus.
Isolation and characterization of elks mutant animals. elks-1 mutant animals were identified and isolated by a PCR screen using a knock-out deletion library prepared with ethyl methanesulfonate as described previously (Liu et al., 1999). For simplicity, we refer to the elks-1 gene as elks throughout this manuscript. The PCR screen that identified the js805 mutant animals consisted of two rounds with the following primers to the F42A6.9 gene of C. elegans:5′ outer first round, cgaataaatttgttgccaatgctc; 3′ outer first round, gtggtcctacaacgaacacgta; 5′ inner second round, cgactttagaagctgtcgacct; and 3′ inner second round, tacaatatggcacctggtgagt. Wild-type (wt) animals showed a PCR product of ∼1100 bp, whereas the js805 animals showed a PCR product of ∼550 bp (see Fig. 2 and supplemental Fig. 3, available at www.jneurosci.org as supplemental material). For the PCR screening that identified the js816 mutant animals, we screened the C. elegans mutant library using the poison primer method described previously (Edgley et al., 2002). The js816 animals were identified with two rounds of PCR using the following primers to the F42A6.9 gene: 5′ outer first round, gcgctacaactccgaatggaac; 3′ outer first round, cataactcggctccctgctaac; 5′ inner second round, ctcaccacagctttagaatccg; 3′ inner second round, ctcaccgcttctttgcttgcac; 3′ poison second round, ctgtagcaaagtgttatgttgc; and 5′ poison second round, gggtgatgtggatgcgttaagg. Wild-type animals showed a PCR product of ∼1050 base pairs, whereas the js816 animals showed a PCR product of ∼500 base pairs (see Fig. 2 and supplemental Fig. 3, available at www.jneurosci.org as supplemental material). The homozygous js805 and js816 mutant animals were isolated and backcrossed as described previously (Liu et al., 1999). DNA sequencing was performed using a dye-terminator sequencing system (PerkinElmer, Wellesley, MA).
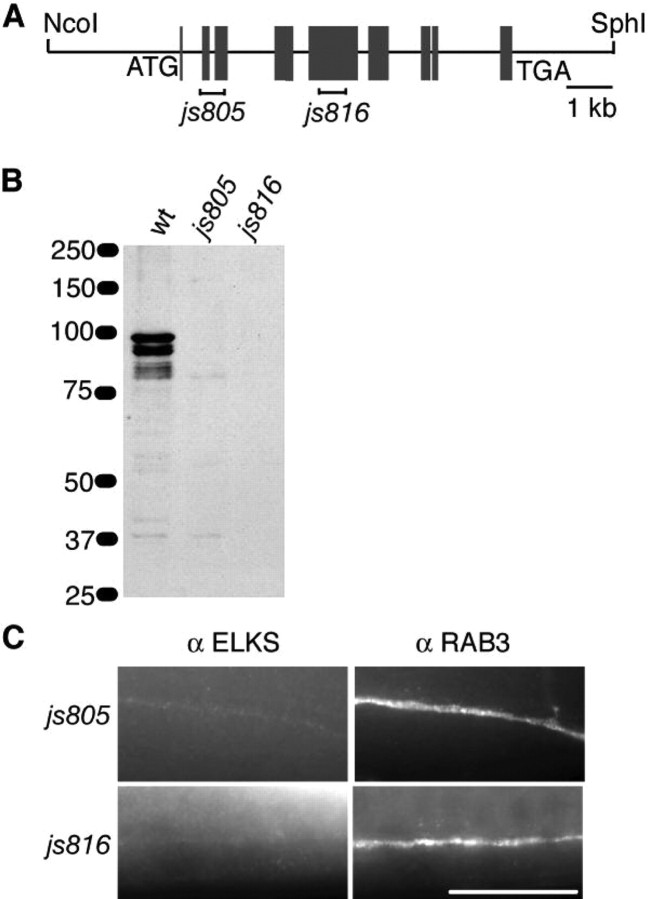
Figure 2.
The js816 allele is an elks null mutation. A, A depiction of the genomic region of the elks gene, including selected restriction sites. Exons are shown as boxes with deletions (I—I) depicted below the genomic map. B, Western blots reveal that ELKS is found in multiple isoforms in wt animals, is found only in the small isoform in js805 animals, and is absent in js816 animals. Equal volumes of worm extracts from wt, js805, and js816 animals were run over a SDS-PAGE gel, and ELKS protein was visualized using anti-ELKS antibody. Data are representative of three experiments. C, elks mutant animals js805 and js816 display minimal and no ELKS immunostaining, respectively. Whole mounts of C. elegans fixed and prepared for immunohistochemical detection of ELKS and RAB-3 in the dorsal nerve cord are shown. Scale bar, 20 μm.
Transformation of C. elegans. Microinjection was performed as described previously (Mello et al., 1991). Cosmid or plasmid DNAs were purified using Qiagen (Valencia, CA) columns following the protocol of the manufacturer. Test DNA and pJM23 (lin-15 marker) were coinjected into lin-15(n765ts), unc-10(md1117)lin-15(n765ts), or elks(js816);lin-15(n765ts), as indicated. Transformed animals were selected as non-Lin progeny from injected animals; the progeny were grown at 22.5°C.
Immunocytochemistry. Antibodies to ELKS were raised against a His6-tagged fusion protein to the last 126 amino acids of the C terminus of ELKS (His6-ELKS). His6-ELKS was purified on Ni-nitrilotriacetic acid agarose (Qiagen) in 8 m urea and renatured by dialysis against PBS. The fusion protein was injected into two rabbits by Covance (Princeton, NJ) to produce rabbit antisera Rb233 and Rb237.
Animals were fixed using Bouin's fixative for whole-mount immunocytochemistry and stained with antibodies against RIM (Koushika et al., 2001), RAB-3 (Nonet et al., 1997), UNC-13 (Kohn et al., 2000), GFP (Molecular Probes, Eugene, OR), or ELKS (Rb237). For RIM and ELKS double-labeling experiments, we used a chicken antibody against an RIM His6-tagged fusion protein described previously (Koushika et al., 2001).
Biochemical assays. A glutathione S-transferase (GST)-fusion protein containing the PDZ domain of RIM was prepared using amino acids 627-974 (GST-PDZRIM). The GST-PDZRIM fusion protein was purified with glutathione-agarose beads (Sigma, St. Louis, MO) in PBS, eluted with reduced glutathione, and dialysed in PBS.
Qualitative in vitro bindings assays were done as described previously (Deken et al., 2000). Briefly, 2 μg of purified GST-PDZRIM or GST protein alone and 2 μg of His6-ELKS (see above) were incubated at 4°C in PBS-Triton X-100 (0.2%) for 1 h. The resulting complex was “pulled down” using glutathione-agarose beads for 1 h at 4°C and precipitated at 3000 × g. The supernatant was collected as the “nonbound” fraction. The GST-bound beads were washed twice in PBS-Triton X-100 and collected in PBS-Triton X-100 plus reduced glutathione as the “bound” fraction. Equivalent percentages of each of the nonbound and bound fractions were analyzed by Western blotting using antibodies raised against ELKS (Rb237).
Western blot analysis of elks mutant animals were performed essentially as described previously (Nonet et al., 1999). Briefly, stage 1 larva of each strain were collected (∼100 μl of packed worms) in 500 μl of sucrose: HEPES buffer (0.36 m sucrose and 12 mm HEPES, with protease inhibitors), followed by four rounds of sonication. The lysates were spun at 500 × g at room temperature to discard the cuticle and nuclear contaminates. The supernatants were diluted in Laemmli's buffer with protease inhibitors, loaded onto an SDS-PAGE gel, and transferred to nitrocellulose membranes (Osmonics, Minnetonka, MN). Blocking and antibody incubations were done according to standard methods. Primary α-ELKS antibody (Rb237) was used at a 1:2000 final dilution. Secondary sheep α-rabbit IgG HRP (Promega) was used at a 1:10,000 dilution. Immunoreactivity was detected with an ECL kit (Amersham Biosciences, Piscataway, NJ).
Aldicarb assays. Animals were assayed for acute exposure to aldicarb. Aldicarb sensitivity was measured by transferring 25 animals to plates containing aldicarb and then assaying the time course of paralysis. Animals were considered paralyzed once they became hypercontracted and no longer moved even when prodded with a platinum wire.
Electrophysiological assays. The technique for recording postsynaptic currents at the C. elegans neuromuscular junction has been described previously (Wang et al., 2001), based on a technique originally developed by Richmond et al. (1999). Briefly, an adult animal was immobilized on glass coverslips by gluing along the dorsal side. A longitudinal incision was made in the dorsolateral region. After clearing the viscera, the cuticle flap was folded back and glued to the coverslip, exposing the ventral nerve cord and the two adjacent muscle quadrants. One electrode was used to voltage clamp (-60 mV) a body-wall muscle cell to monitor miniature postsynaptic currents (mPSCs) and EPSCs. A second electrode was placed at the ventral nerve cord to evoke EPSCs by applying a 0.5 ms square wave current pulse at supramaximal voltage. Postsynaptic currents were amplified with a Multiclamp 700A amplifier (Molecular Devices, Union City, CA), and acquired with Clampex software (Molecular Devices). Data were sampled at a rate of 10 kHz after filtering at 2 kHz. The recording pipette solution contained (in mm) 120 KCl, 20 KOH, 5 Tris, 0.25 CaCl2, 4 MgCl2, 36 sucrose, 5 EGTA, and 4 Na2ATP, pH adjusted to 7.2 with HCl. The external solution included (in mm) 140 NaCl, 5 KCl, 5 CaCl2, 5 MgCl2, 11 dextrose, and 5 HEPES, pH adjusted to 7.2 with NaOH.
The frequency and mean amplitude of mPSCs of each experiment were obtained by automatic analysis of ∼60 s continuous recordings using MiniAnalysis (Synaptosoft, Decatur, GA) with the amplitude detection threshold set at 10 pA. Amplitudes of EPSCs were measured with Clampfit (Molecular Devices). The averaged amplitude of the two largest EPSCs from each experiment was used for statistical analysis. p < 0.05 is considered statistically significant.
Results
RIM and ELKS are interacting active zone proteins in C. elegans
The C. elegans elks (F42A6.9) gene encodes a protein with homology to mammalian ELKS, sharing 20% identity to its two rat homologs with the C terminus exhibiting the most similarity (for an alignment, see supplemental Fig. 1, available at www.jneurosci.org as supplemental material). Mammalian ELKS isoforms directly interact with and colocalize with mammalian RIM at the active zone (Wang et al., 1997, 2002; Ohtsuka et al., 2002; Deguchi-Tawarada et al., 2004). The localization of RIM at the active zone is conserved in C. elegans (Koushika et al., 2001). To determine the localization of C. elegans ELKS, we raised antibodies against the ELKS C-terminal domain. Immunocytochemistry revealed that ELKS staining is found in discrete puncta in the nerve ring and along both the ventral and dorsal cords. In the nervous system, ELKS staining was restricted to synapse-rich regions and was more punctate than the staining pattern observed with the synaptic vesicle marker RAB-3 (Fig. 1A). These data suggest that ELKS is an additional member of the C. elegans active zone with both RIM and UNC-13, which share the more punctate staining pattern in relation to synaptic vesicle proteins in synapse-rich regions of the nervous system (Kohn et al., 2000; Koushika et al., 2001). Furthermore, we found that RIM and ELKS are strongly colocalized throughout the C. elegans nervous system (Fig. 1B). In addition, ELKS localization is independent of the kinesin motor protein UNC-104 (supplemental Fig. 2, available at www.jneurosci.org as supplemental material), a property shared by other active zone proteins (Kohn et al., 2000; Koushika et al., 2001) but not by synaptic vesicle proteins (Nonet et al., 1997, 1998).
Figure 1.
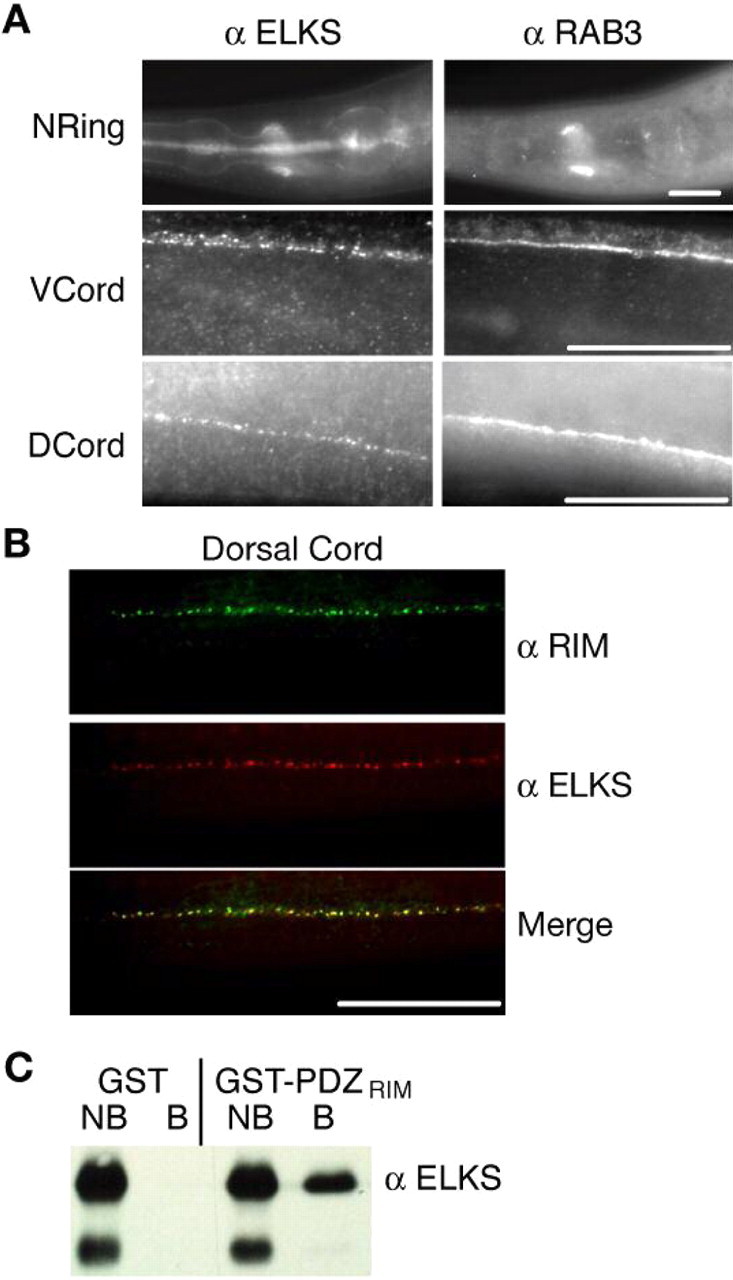
ELKS is an active zone protein in C. elegans. A, ELKS and RAB3 immunostaining in C. elegans. Whole mounts of wild-type C. elegans fixed and prepared for immunohistochemical detection of ELKS and RAB-3 proteins in the nerve ring (NRing), ventral nerve cord (VCord), and dorsal nerve cord (DCord). ELKS immunostaining is found in synapse-rich regions of the C. elegans nervous system and along the pharyngeal basal lamina. Autofluorescent background of bacteria within the lumen can also be seen. B, ELKS and RIM colocalize in C. elegans neurons. Whole mounts of wild-type C. elegans fixed and prepared for immunohistochemical detection of ELKS (red) and RIM (green) proteins in the dorsal nerve cord. C, ELKS C-terminal tail binds the PDZ domain of RIM. Western blots using ELKS antibodies revealed that recombinant ELKS C-terminal tail (last 126 amino acids) tagged to His6 bound to recombinant GST-PDZRIM (containing amino acids 627-974 of RIM) but not to GST alone. NB, Nonbound fractions; B, bound fractions. Data are representative of three experiments. Scale bars, 20 μm.
Mammalian RIM and ELKS proteins directly interact through the PDZ domain of RIM and the last three amino acids of ELKS, which contains a conserved three amino acid (Ile-Trp-Ala) PDZ-binding motif (Ohtsuka et al., 2002; Wang et al., 2002). We tested whether the C. elegans homologs also physically interact. We purified bacterial expressed His6-tagged fusion protein to the last 126 amino acids of ELKS (the same fusion protein used for antibody production) and a GST fusion protein containing the PDZ domain of RIM (GST-PDZRIM). In in vitro binding studies, the ELKS protein bound to GST-PDZRIM but not to GST alone (Fig. 1C). Together, these data strongly suggest that C. elegans ELKS, like its mammalian homolog, directly interacts with RIM and is an active zone protein.
elks mutants have no defects in behavior or synaptic physiology
Determining the neuronal function of ELKS using ELKS knockout mice will most likely be difficult because ELKS is found in multiple different tissue types and is a critical component in nuclear factor κB (NF-κB) signaling (Ducut Sigala et al., 2004). Knock-out mice containing gene deletions of core components involved in NF-κB signaling all die as late embryos (Li and Verma, 2002). To assess ELKS function in the nervous system, we set out to obtain C. elegans animals lacking the ELKS protein. We used a PCR-based strategy to screen a knock-out library for deletions in the elks (F42A6.9) gene (described in Materials and Methods). We uncovered two deletions and isolated the animals as homozygous elks mutants (Fig. 2A and supplemental Figs. 1, 3, available at www.jneurosci.org as supplemental material). elks(js805) results in a premature stop at amino acid 7 and elks(js816) results in a premature stop at amino acid 292. Homozygous js805 and js816 animals display no obvious morphological or behavioral abnormalities.
We analyzed ELKS protein expression levels in js805 and js816 animals by Western blotting and by immunostaining. Western blots revealed that, in wild-type animals, ELKS is expressed as a full-length protein at ∼94 kDa and as two smaller protein products with molecular weights of ∼85 and ∼78 kDa (Fig. 2B). The multiple ELKS bands present on the Western blot, suggesting different ELKS isoforms produced by alternative splicing, is consistent with data showing that mammalian ELKS is present as multiple different splice variants (Wang et al., 2002; Deguchi-Tawarada et al., 2004). js805 animals contain only the 78 kDa form of ELKS and display only minimal immunostaining in synapse-rich regions of the nervous system (Fig. 2B,C). js816 animals show no staining for ELKS by Western blotting or by immunostaining, suggesting that this allele is a probable null mutation (Fig. 2B,C).
We next tested the role of ELKS in synaptic transmission by quantifying the resistance of elks mutant animals to the acetylcholinesterase inhibitor aldicarb and by electrophysiological experiments. Reduced synaptic transmission in mutant animals leads to aldicarb resistance (Miller et al., 1996). Aldicarb assays reveal that elks mutant animals expressing the short form of ELKS (js805) or no ELKS (js816) have no gross defects in synaptic transmission. The elks mutant animals and wt animals are similarly sensitive to aldicarb, whereas animals that lack RIM (md1117) are completely aldicarb resistant (Fig. 3A). Because aldicarb assays only reveal gross defects in synaptic transmission, we analyzed ELKS function more closely by examining js816 animals for changes in synaptic transmission by electrophysiology. We found no statistical difference in miniature PSC amplitude (js816, 36.19 ± 3.93 pA; wt, 35.34 ± 2.09 pA) or in miniature PSC frequency (js816, 65.54 ± 6.49 Hz; wt, 57.04 ± 9.02 Hz) between js816 (n = 8) and wild-type animals (n = 6) (Fig. 3B). There was also no statistical difference in excitatory postsynaptic amplitudes (js816, 1811.29 ± 218.83 pA; wt, 1682.09 ± 133.95 pA) between js816 (n = 6) and wild-type animals (n = 6) (Fig. 3C). The lack of a synaptic phenotype in animals lacking ELKS indicates that ELKS is a nonessential player in synaptic transmission.
Figure 3.
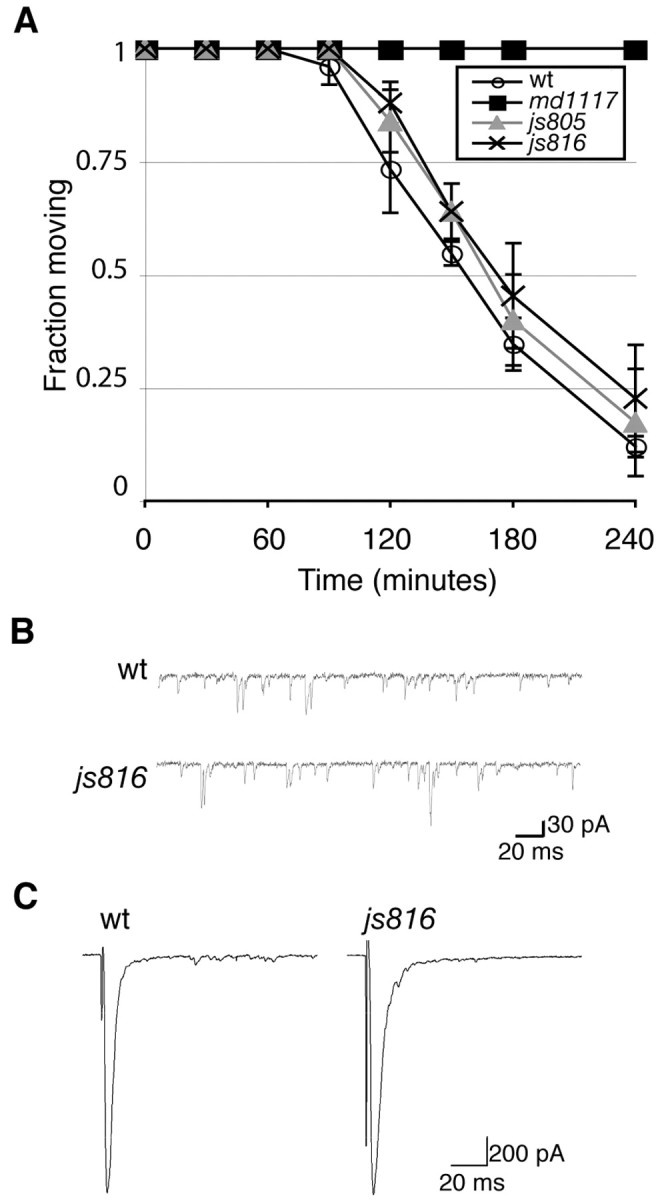
elks mutant animals have normal synaptic transmission. A, Behavior of wt, unc-10(md1117), elks(js805), and elks(js816) animals on 0.5 mm aldicarb at the indicated exposure time. Data are from three separate experiments using 25 animals per experiment. Error bars are expressed as ± SEM. B, Representative traces of spontaneous mEPSCs from wt and elks(js816) animals. C, Representative EPSCs from wt and elks(js816) animals.
ELKS is not necessary for RIM localization or function
ELKS is argued to play a role in localizing RIM to the synapse in a cultured neuron system (Ohtsuka et al., 2002). We directly tested the hypothesis that ELKS is required to localize RIM to the synapse in our js816 animals. We found that js816 animals have normal RIM staining patterns (Fig. 4A). Similarly, ELKS is not required for the localization of UNC-13, another C. elegans active zone protein (Fig. 4B). These data demonstrate that ELKS is not required for localizing RIM or UNC-13 to the active zone in vivo. Previous work has shown that RIM and UNC-13 localization is independent of one another (Koushika et al., 2001). In addition, we found that RIM localization was independent of the presynaptic calcium channel (UNC-2 in C. elegans) and α-liprin (SYD-2 in C. elegans), two other active zone components in C. elegans neurons (Fig. 4A).
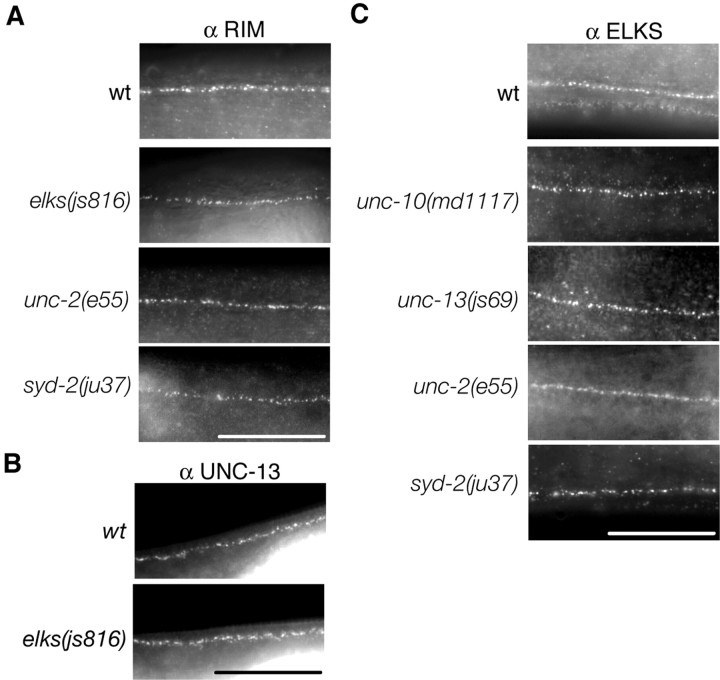
Figure 4.
Active zone proteins RIM, UNC-13, and ELKS localize independently of one another. A, RIM immunostaining in the dorsal nerve cord of wt and animals lacking ELKS elks(js816), the N-type calcium channel unc-2(e55), and SYD-2/α-liprin syd-2(ju37) animals. B, UNC-13 immunostaining in the dorsal nerve cord of wt and elks(js816) animals. C, ELKS immunostaining in the dorsal nerve cord of wt and animals lacking RIM unc-10(md1117), UNC-13 unc-13(js69), the N-type calcium channel unc-2(e55), and SYD-2/α-liprin syd-2(ju37) animals. Scale bars, 20 μm.
We next tested the reverse hypothesis that ELKS is localized to the active zone through single interactions with other known active zone components. We stained animals devoid of RIM, UNC-13, SYD-2, and UNC-2 for ELKS. ELKS immunostaining is unaltered in each of these mutant animals, demonstrating that these proteins are not essential for the localization of ELKS (Fig. 4C).
The PDZ domain of RIM was shown to be necessary for RIM localization in cultured mammalian neurons (Ohtsuka et al., 2002). We showed that an RIM interaction with ELKS is not essential for RIM localization (Fig. 4A), but it remains a possibility that some other RIM PDZ-binding protein is essential to localize RIM. We tested this by making a RIM construct lacking the PDZ domain (RIMΔPDZ) and expressing this construct in C. elegans animals that lack RIM. Full-length RIM and RIMΔPDZ transgenic expression in unc-10(md1117) mutant animals display immunostaining patterns similar to native RIM immunostaining in wild-type animals (Fig. 5A). These data support our findings that RIM can localize independently of ELKS and show that the PDZ domain is not necessary for RIM localization.
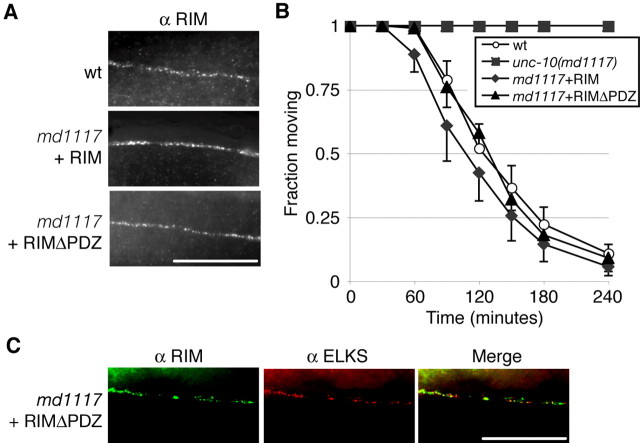
Figure 5.
The PDZ domain of RIM is not required for RIM localization or function. A, The PDZ domain of RIM is not required for RIM localization. RIM immunostaining in the dorsal nerve cord in wt and in transgenic unc-10(md1117) animals expressing full-length RIM or RIMΔPDZ (deletion of amino acids 590-752). B, The PDZ domain of RIM is not required for RIM function. Behavior of wt, unc-10(md1117), and unc-10(md1117) transgenic animals expressing full-length RIM or RIMΔPDZ on 0.5 mm aldicarb at the indicated exposure time. Data are from three separate experiments using 25 animals per experiment. Error bars are expressed as ± SEM. C, ELKS is localized without the PDZ domain of RIM. RIM (green) and ELKS (red) immunostaining in the dorsal nerve cord in unc-10(md1117) transgenic animals expressing RIMΔPDZ. Scale bars, 20 μm.
The interaction between ELKS and RIM has also been proposed to have functional relevance in synaptic transmission of cultured neurons (Takao-Rikitsu et al., 2004). To check the hypothesis that the ELKS-RIM interaction is important for RIM function, we tested the behavior of our RIMΔPDZ-expressing animals. RIMΔPDZ-expressing animals rescue unc-10 locomotory defects (data not shown) as well as pharmacological defects to acetylcholinesterase inhibitors. We found that the unc-10(md1117) animals expressing RIMΔPDZ are sensitive to aldicarb, indicating that the ELKS-interacting PDZ domain of RIM is not required for normal RIM function (Fig. 5B). Consistent with our data that RIM is not required for ELKS localization (Fig. 4C), we found that ELKS staining in md1117 animals expressing RIMΔPDZ is normal (Fig. 5C). These data demonstrate that RIM function and localization do not require its PDZ domain or an interaction with ELKS.
RIM and ELKS are tightly associated in vivo
The PDZ domain of RIM alone is required and is sufficient for localizing to the synapse in transfected hippocampal neuron cultures (Ohtsuka et al., 2002). However, we found that ELKS, which binds to the PDZ domain of RIM (Fig. 1C), and the PDZ domain of RIM are not required for RIM localization in vivo (Figs. 4A, 5A). To test whether the PDZ domain of RIM is sufficient for RIM localization through some other unknown interaction, we expressed a GFP-tagged RIM construct containing only the PDZ domain (GFP-PDZRIM) in C. elegans neurons. We found that the PDZ domain of RIM is not sufficient to localize RIM to the synapse in vivo in either the presence or absence of native RIM. Unexpectedly, ELKS localization in animals expressing GFP-PDZRIM is totally disrupted (Fig. 6A). These data are surprising because animals lacking RIM have normal ELKS immunostaining patterns (Fig. 4C).
Figure 6.
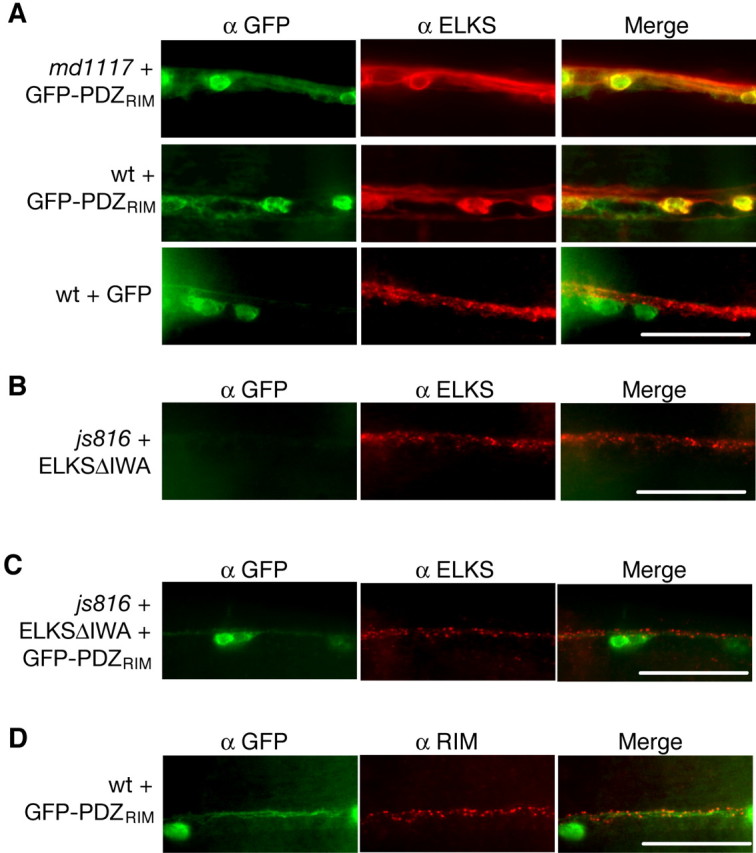
RIM and ELKS strongly interact in vivo. A, ELKS is mislocalized in transgenic animals expressing a RIM construct containing only the PDZ domain of RIM fused to GFP (GFP-PDZRIM). GFP (green) and ELKS (red) immunostaining in the ventral nerve cord of transgenic unc-10(md1117) and wt animals expressing GFP-PDZRIM or GFP alone is shown. B, ELKS localizes independently of its C-terminal IWA sequence. GFP (green) and ELKS (red) immunostaining in the ventral nerve cord of transgenic elks(js816) animals expressing an elks construct that deletes the final three amino acids of ELKS (ELKSΔIWA) is shown. C, ELKS localization is restored in transgenic animals expressing both GFP-PDZRIM and ELKSΔIWA. GFP (green) and ELKS (red) immunostaining in the ventral nerve cord of transgenic elks(js816) animals expressing GFP-PDZRIM and an elks construct that deletes the final three amino acids of ELKS (ELKSΔIWA). D, RIM is localized when ELKS is mislocalized. GFP (green) and RIM (red) immunostaining in the ventral nerve cord of animals expressing GFP-PDZRIM. Scale bars, 20 μm.
One possible explanation of these results is that the PDZ-binding motif (IWA) of ELKS is required for ELKS localization, and the PDZ domain of RIM competes with the PDZ domain of an unknown protein that normally acts to localize ELKS. If this is true, deletion of the ELKS IWA sequence should result in ELKS mislocalization. Consistent with mammalian data (Ohtsuka et al., 2002), deletion of the PDZ-binding domain of ELKS (ELKSΔIWA) had no effect on ELKS localization, suggesting that ELKS localizes independently of its PDZ-binding domain (Fig. 6 B).
Another possible explanation is that soluble GFP-PDZRIM, through a tight, direct interaction with ELKS, disrupts ELKS localization by sequestering ELKS away from the synapse. Consistent with this, the localization of ELKS is rescued in the presence of the GFP-PDZRIM by the deletion of the ELKS IWA sequence (Fig. 6C). This suggests that ELKS and RIM are very tightly associated with one another in vivo and that RIM promotes ELKS localization under normal, wild-type conditions but is not an essential component in ELKS localization (Fig. 4C). Together, these data implicate the role of a novel protein involved in ELKS localization at the active zone.
We next tested whether native RIM is localized in the presence of GFP-PDZRIM. RIM is normally localized in wild-type animals expressing soluble GFP-PDZRIM, suggesting that RIM is localized to the synapse even when ELKS is mislocalized (Fig. 6D).
ELKS helps localize RIM at the synapse
We next tested whether we could make a minimal RIM construct sufficient to localize to discrete puncta in the C. elegans nervous system. Addition of the RIM C2A domain to the PDZ domain of RIM (GFP-PDZC2ARIM), but not the C2A domain of RIM alone (GFP-C2ARIM), was sufficient to localize RIM and restore ELKS localization (Fig. 7A). Interestingly, the localization of GFP-PDZC2ARIM is dependent on ELKS expression (Fig. 7B). These data suggest that ELKS plays a role, but not an essential role (Fig. 4A), in localizing RIM to the synapse. These data implicate another RIM-binding protein that interacts outside of the PDZ and C2A domains to aid in RIM localization to the synapse.
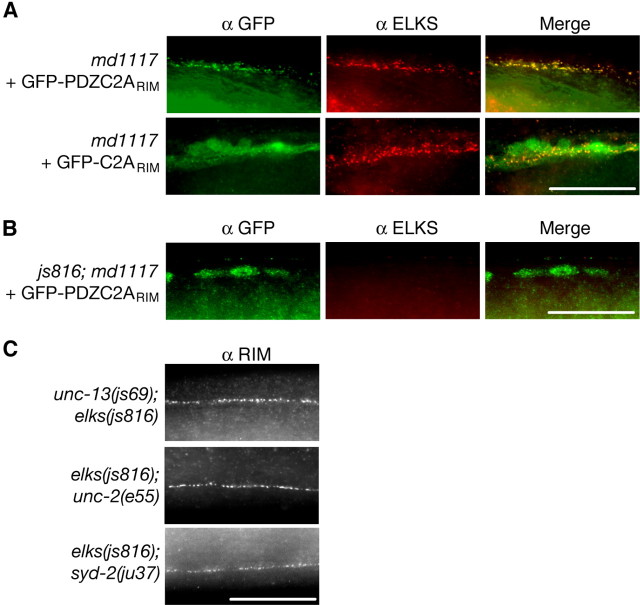
Figure 7.
RIM localization is partially dependent on ELKS. A, The PDZ and C2A domains of RIM are sufficient for RIM localization. GFP (green) and ELKS (red) immunostaining in the ventral nerve cord of transgenic unc-10(md1117) animals expressing GFP-PDZC2ARIM (GFP-tagged RIM containing amino acids 627-974) or GFP-C2ARIM (GFP-tagged RIM containing amino acids 826-974) is shown. B, The localization of GFP-PDZC2ARIM is ELKS dependent. GFP (green) and ELKS (red) immunostaining in the ventral nerve cord of transgenic elks(js816); unc-10(md1117) expressing GFP-PDZC2ARIM. C, RIM localizes in the absence of ELKS and other active zone components. RIM immunostaining in the dorsal nerve cord of unc-13(s69); elks(js816), elks(js816); unc-2(e55), and elks(js816); syd-2(ju37) double-mutant animal is shown. Scale bars, 20 μm.
In an attempt to identify this partner, we constructed double mutants between elks and other mutants lacking active zone proteins known to interact with RIM and tested for RIM localization. UNC-13, like ELKS, directly interacts with RIM (Betz et al., 2001). To determine whether RIM localizes to the active zone through joint interactions with ELKS and UNC-13, we made double-mutant animals that lack both ELKS and UNC-13 protein expression. The unc-13(s69); elks(js816) animals display normal RIM immunostaining, suggesting that UNC-13 does not act with ELKS to localize RIM (Fig. 7C). We also tested whether ELKS acts with UNC-2 or SYD-2 to localize RIM. Double-mutant animals of ELKS and UNC-2 [elks(js816); unc-2(e55)] or SYD-2 [elks(js816); syd-2(ju37)] did not result in RIM mislocalization. Together, these data suggest a joint RIM interaction with ELKS, and a novel protein acts to localize RIM.
Discussion
Active zone proteins are specifically located and spatially restrict sites of neurotransmitter release and function by conferring regulatory interactions with proteins involved in the release process (Dresbach et al., 2001; Rosenmund et al., 2003; Zhai and Bellen, 2004). However, the composition and localization mechanisms of proteins specific to the active zone region are essentially unknown. We characterized ELKS as a member of the C. elegans active zone complex and generated mutant animals devoid of the ELKS protein to determine its role in neurotransmitter release and its involvement in the localization of other known active zone proteins in vivo.
ELKS was first identified as a ubiquitously expressed protein and part of a gene rearrangement with the receptor-type tyrosine kinase RET (Nakata et al., 1999). N-terminal dimerization of ELKS causes the ELKS-RET chimeric protein to oligomerize and act as an oncoprotein, leading to papillary thyroid carcinoma (Nakata et al., 2002). ELKS has since been identified as a Rab6A-interacting and -regulating protein (Monier et al., 2002), as a member of the IκB kinase (IKK) complex involved in NF-κB signaling (Ducut Sigala et al., 2004), and as a member of the active zone complex in the mammalian nervous system (Ohtsuka et al., 2002; Wang et al., 2002; Deguchi-Tawarada et al., 2004). The role of ELKS in the nervous system is unclear.
Our data reveal that ELKS is a nonessential player in neurotransmitter release. This is surprising, because genetic knockouts of all other active zone specific proteins tested thus far have rather severe defects in neurotransmission and in animal behavior (Aravamudan et al., 1999; Augustin et al., 1999; Richmond et al., 1999; Koushika et al., 2001; Castillo et al., 2002; Altrock et al., 2003; Powell et al., 2004). It is also surprising, in light of published reports showing microinjection of ELKS peptides that interact with RIM disrupt synaptic transmission in cultured neurons (Takao-Rikitsu et al., 2004). We found that animals devoid of ELKS protein display normal behaviors and normal synaptic physiology. ELKS may play a different role in synaptic transmission in C. elegans neurons than in mammalian neurons. Indeed, there is substantial morphological variation in active zone structure in different types of synapses in different organisms (Zhai and Bellen, 2004). Analysis of ELKS mutants in other organisms will be required to address this issue.
We found that the direct interaction between ELKS and RIM is conserved in C. elegans but, in contrast to other reports (Ohtsuka et al., 2002), this interaction is not necessary for RIM to localize to the synapse. Our data demonstrate that the ELKS-RIM interaction plays a redundant role in conjunction with a novel protein to localize RIM in vivo. Our data strongly suggest that RIM localizes to the synapse independently of ELKS through an interaction with a novel protein that can localize RIM even in the absence of ELKS. Our data indicate that ELKS acts as an auxiliary scaffolding protein at the active zone.
The scaffolding function of ELKS is not unique to the nervous system. Recently, ELKS was shown to be part of the IKK complex and acts as an essential player in recruiting IκBα to the protein complex, allowing for the activation of the transcription factor NF-κB (Ducut Sigala et al., 2004). NF-κB signaling is essential for the expression of a number of genes, including those involved in the immune response, oncogenesis, apoptosis, and development (Li and Verma, 2002). Interestingly, ELKS, through Rab6A, is also involved in endosome-to-Golgi trafficking (Monier et al., 2002). This suggests that ELKS may function as a link for signals arising at the synapse to reach the cell body. In light of this and the fact that NF-κB is known to function at the synapse, ELKS may act in nervous system development or neuronal survival by regulating gene expression from the synapse (Meffert et al., 2003). Although we did not directly test this hypothesis, ELKS likely does not play a major role in nervous system development or apoptosis, because the C. elegans genome contains no counterparts of NF-κB coding genes (Ruvkun and Hobert, 1998). Our data are consistent with this hypothesis, because elks mutant animals have no gross abnormalities in nervous system structure. In fact, the number of synapses along the dorsal nerve cord in elks mutant (js816 animals, 1.10 ± 0.05 RIM puncta/μm) and wild-type animals (N2 animals, 1.12 ± 0.03 RIM puncta/μm) are indistinguishable. These data, however, do not rule out the possibility that ELKS functions in other signaling pathways in the C. elegans nervous system.
Our data demonstrate that ELKS localization is capable of transitioning between the active zone and a ubiquitously distributed state. This is indirect evidence that ELKS is in a more peripheral position within the active zone complex. Thus, ELKS could be appropriately positioned to shuttle or regulate the shuttling of signaling components to and from the active zone, similar to its role in regulating the NF-κB signaling complex. However, this remains a hypothesis, because we have no direct evidence that ELKS distribution is dynamic in vivo.
There is mounting evidence that active zone components traffic to the synapse differently than synaptic vesicles. During synaptogenesis, it has been suggested that active zone proteins are packaged and transported to the synapse together on an active zone precursor vesicle distinct from synaptic vesicles (Ahmari et al., 2000; Zhai et al., 2001). RIM, UNC-13, and ELKS have been shown to be associated with this vesicle (Zhai et al., 2001; Ohtsuka et al., 2002). Our work demonstrates that ELKS and UNC-13 are not required for RIM localization at the synapse, suggesting that RIM can interact with the active zone precursor vesicle and/or traffics to the synapse independently of its association with ELKS and UNC-13. In addition, ELKS must also associate with this trafficking vesicle independent of a single association with RIM, UNC-13, SYD-2/α-liprin, or the presynaptic calcium channel. Our studies and others have shown that active zone proteins traffic to the synapse independently of the kinesin motor protein UNC-104, whereas the trafficking of synaptic vesicle proteins like Rab3 and synaptotagmin depend on UNC-104 (Kohn et al., 2000; Koushika et al., 2001). Through genetic approaches, we are trying to identify the motor proteins used to traffic active zone proteins to the synapse.
Because active zone proteins do not span the membrane, they must be anchored to the plasma membrane at the site of neurotransmitter release. Our data suggest that ELKS acts as an auxiliary scaffolding protein at the active zone. Overexpression of ELKS in cultured neurons enhances the synaptic localization of SYD-2/α-liprin (Ko et al., 2003), and our work shows that the localization of a minimal RIM protein containing the PDZ and C2A domains requires ELKS expression (Fig. 7B). However, ELKS alone is not required for the synaptic localization of RIM (Fig. 4A), which suggests that a novel protein is also involved in RIM maintenance at the active zone. In addition, the maintenance of ELKS at the active zone is through the actions of a novel protein.
Although a specific description of ELKS function is still unknown, this study clearly demonstrates that ELKS is a C. elegans active zone protein, is a nonessential player in neurotransmitter release, directly interacts with RIM, and plays a supplemental role in RIM localization. Importantly, we also implicated the role of novel proteins involved in ELKS and RIM localization to the active zone.
Footnotes
This work was supported by National Institutes of Health Grants 5 F32 NS047000 (S.L.D.) and R01 NS033535 (M.L.N.). We thank J. Rand for providing UNC-13 antisera and A. Wei and the Nonet laboratory for providing comments on this manuscript.
Correspondence should be addressed to Michael L. Nonet, 660 South Euclid Avenue, St. Louis, MO 63110. E-mail: nonetm@pcg.wustl.edu.
Copyright © 2005 Society for Neuroscience 0270-6474/05/255975-09$15.00/0
References
- Ahmari SE, Buchanan J, Smith SJ (2000) Assembly of presynaptic active zones from cytoplasmic transport packets. Nat Neurosci 3: 445-451. [DOI] [PubMed] [Google Scholar]
- Altrock WD, tom Dieck S, Sokolov M, Meyer AC, Sigler A, Brakebusch C, Fassler R, Richter K, Boeckers TM, Potschka H, Brandt C, Loscher W, Grimberg D, Dresbach T, Hempelmann A, Hassan H, Balschun D, Frey JU, Brandstatter JH, Garner CC, et al. (2003) Functional inactivation of a fraction of excitatory synapses in mice deficient for the active zone protein bassoon. Neuron 37: 787-800. [DOI] [PubMed] [Google Scholar]
- Aravamudan B, Fergestad T, Davis WS, Rodesch CK, Broadie K (1999) Drosophila UNC-13 is essential for synaptic transmission. Nat Neurosci 2: 965-971. [DOI] [PubMed] [Google Scholar]
- Augustin I, Rosenmund C, Sudhof TC, Brose N (1999) Munc13-1 is essential for fusion competence of glutamatergic synaptic vesicles. Nature 400: 457-461. [DOI] [PubMed] [Google Scholar]
- Betz A, Thakur P, Junge HJ, Ashery U, Rhee JS, Scheuss V, Rosenmund C, Rettig J, Brose N (2001) Functional interaction of the active zone proteins Munc13-1 and RIM1 in synaptic vesicle priming. Neuron 30: 183-196. [DOI] [PubMed] [Google Scholar]
- Calakos N, Schoch S, Sudhof TC, Malenka RC (2004) Multiple roles for the active zone protein RIM1alpha in late stages of neurotransmitter release. Neuron 42: 889-896. [DOI] [PubMed] [Google Scholar]
- Castillo PE, Schoch S, Schmitz F, Sudhof TC, Malenka RC (2002) RIM1alpha is required for presynaptic long-term potentiation. Nature 415: 327-330. [DOI] [PubMed] [Google Scholar]
- Deguchi-Tawarada M, Inoue E, Takao-Rikitsu E, Inoue M, Ohtsuka T, Takai Y (2004) CAST2: identification and characterization of a protein structurally related to the presynaptic cytomatrix protein CAST. Genes Cells 9: 15-23. [DOI] [PubMed] [Google Scholar]
- Deken SL, Beckman ML, Boos L, Quick MW (2000) Transport rates of GABA transporters: regulation by the N-terminal domain and syntaxin 1A. Nat Neurosci 3: 998-1003. [DOI] [PubMed] [Google Scholar]
- Dresbach T, Qualmann B, Kessels MM, Garner CC, Gundelfinger ED (2001) The presynaptic cytomatrix of brain synapses. Cell Mol Life Sci 58: 94-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducut Sigala JL, Bottero V, Young DB, Shevchenko A, Mercurio F, Verma IM (2004) Activation of transcription factor NF-kappaB requires ELKS, an IkappaB kinase regulatory subunit. Science 304: 1963-1967. [DOI] [PubMed] [Google Scholar]
- Edgley M, D'Souza A, Moulder G, McKay S, Shen B, Gilchrist E, Moerman D, Barstead R (2002) Improved detection of small deletions in complex pools of DNA. Nucleic Acids Res 30: e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow ML, Ress D, Stoschek A, Marshall RM, McMahan UJ (2001) The architecture of active zone material at the frog's neuromuscular junction. Nature 409: 479-484. [DOI] [PubMed] [Google Scholar]
- Johnson S, Halford S, Morris AG, Patel RJ, Wilkie SE, Hardcastle AJ, Moore AT, Zhang K, Hunt DM (2003) Genomic organisation and alternative splicing of human RIM1, a gene implicated in autosomal dominant cone-rod dystrophy (CORD7). Genomics 81: 304-314. [DOI] [PubMed] [Google Scholar]
- Ko J, Na M, Kim S, Lee JR, Kim E (2003) Interaction of the ERC family of RIM-binding proteins with the liprin-alpha family of multidomain proteins. J Biol Chem 278: 42377-42385. [DOI] [PubMed] [Google Scholar]
- Kohn RE, Duerr JS, McManus JR, Duke A, Rakow TL, Maruyama H, Moulder G, Maruyama IN, Barstead RJ, Rand JB (2000) Expression of multiple UNC-13 proteins in the Caenorhabditis elegans nervous system. Mol Biol Cell 11: 3441-3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koushika SP, Richmond JE, Hadwiger G, Weimer RM, Jorgensen EM, Nonet ML (2001) A post-docking role for active zone protein Rim. Nat Neurosci 4: 997-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Verma IM (2002) NF-kappaB regulation in the immune system. Nat Rev Immunol 2: 725-734. [DOI] [PubMed] [Google Scholar]
- Liu LX, Spoerke JM, Mulligan EL, Chen J, Reardon B, Westlund B, Sun L, Abel K, Armstrong B, Hardiman G, King J, McCague L, Basson M, Clover R, Johnson CD (1999) High-throughput isolation of Caenorhabditis elegans deletion mutants. Genome Res 9: 859-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meffert MK, Chang JM, Wiltgen BJ, Fanselow MS, Baltimore D (2003) NF-kappa B functions in synaptic signaling and behavior. Nat Neurosci 6: 1072-1078. [DOI] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V (1991) Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J 10: 3959-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KG, Alfonso A, Nguyen M, Crowell JA, Johnson CD, Rand JB (1996) A genetic selection for Caenorhabditis elegans synaptic transmission mutants. Proc Natl Acad Sci USA 93: 12593-12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier S, Jollivet F, Janoueix-Lerosey I, Johannes L, Goud B (2002) Characterization of novel Rab6-interacting proteins involved in endosome-to-TGN transport. Traffic 3: 289-297. [DOI] [PubMed] [Google Scholar]
- Nakata T, Kitamura Y, Shimizu K, Tanaka S, Fujimori M, Yokoyama S, Ito K, Emi M (1999) Fusion of a novel gene, ELKS, to RET due to translocation t(10;12)(q11;p13) in a papillary thyroid carcinoma. Genes Chromosomes Cancer 25: 97-103. [DOI] [PubMed] [Google Scholar]
- Nakata T, Yokota T, Emi M, Minami S (2002) Differential expression of multiple isoforms of the ELKS mRNAs involved in a papillary thyroid carcinoma. Genes Chromosomes Cancer 35: 30-37. [DOI] [PubMed] [Google Scholar]
- Nonet ML, Staunton JE, Kilgard MP, Fergestad T, Hartwieg E, Horvitz HR, Jorgensen EM, Meyer BJ (1997) Caenorhabditis elegans rab-3 mutant synapses exhibit impaired function and are partially depleted of vesicles. J Neurosci 17: 8061-8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet ML, Saifee O, Zhao H, Rand JB, Wei L (1998) Synaptic transmission deficits in Caenorhabditis elegans synaptobrevin mutants. J Neurosci 18: 70-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet ML, Holgado AM, Brewer F, Serpe CJ, Norbeck BA, Holleran J, Wei L, Hartwieg E, Jorgensen EM, Alfonso A (1999) UNC-11, a Caenorhabditis elegans AP180 homologue, regulates the size and protein composition of synaptic vesicles. Mol Biol Cell 10: 2343-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka T, Takao-Rikitsu E, Inoue E, Inoue M, Takeuchi M, Matsubara K, Deguchi-Tawarada M, Satoh K, Morimoto K, Nakanishi H, Takai Y (2002) Cast: a novel protein of the cytomatrix at the active zone of synapses that forms a ternary complex with RIM1 and munc13-1. J Cell Biol 158: 577-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips GR, Huang JK, Wang Y, Tanaka H, Shapiro L, Zhang W, Shan WS, Arndt K, Frank M, Gordon RE, Gawinowicz MA, Zhao Y, Colman DR (2001) The presynaptic particle web: ultrastructure, composition, dissolution, and reconstitution. Neuron 32: 63-77. [DOI] [PubMed] [Google Scholar]
- Powell CM, Schoch S, Monteggia L, Barrot M, Matos MF, Feldmann N, Sudhof TC, Nestler EJ (2004) The presynaptic active zone protein RIM1alpha is critical for normal learning and memory. Neuron 42: 143-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond JE, Broadie KS (2002) The synaptic vesicle cycle: exocytosis and endocytosis in Drosophila and C. elegans Curr Opin Neurobiol 12: 499-507. [DOI] [PubMed] [Google Scholar]
- Richmond JE, Davis WS, Jorgensen EM (1999) UNC-13 is required for synaptic vesicle fusion in C. elegans Nat Neurosci 2: 959-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenmund C, Rettig J, Brose N (2003) Molecular mechanisms of active zone function. Curr Opin Neurobiol 13: 509-519. [DOI] [PubMed] [Google Scholar]
- Ruvkun G, Hobert O (1998) The taxonomy of developmental control in Caenorhabditis elegans Science 282: 2033-2041. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning, a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
- Schoch S, Castillo PE, Jo T, Mukherjee K, Geppert M, Wang Y, Schmitz F, Malenka RC, Sudhof TC (2002) RIM1alpha forms a protein scaffold for regulating neurotransmitter release at the active zone. Nature 415: 321-326. [DOI] [PubMed] [Google Scholar]
- Sudhof TC (2004) The synaptic vesicle cycle. Annu Rev Neurosci 27: 509-547. [DOI] [PubMed] [Google Scholar]
- Sulston J, Hodgkin J (1988) Methods. In: The nematode Caenorhabditis elegans (Wood WB, ed), pp 587-606. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
- Takao-Rikitsu E, Mochida S, Inoue E, Deguchi-Tawarada M, Inoue M, Ohtsuka T, Takai Y (2004) Physical and functional interaction of the active zone proteins, CAST, RIM1, and Bassoon, in neurotransmitter release. J Cell Biol 164: 301-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Sudhof TC (2003) Genomic definition of RIM proteins: evolutionary amplification of a family of synaptic regulatory proteins. Genomics 81: 126-137. [DOI] [PubMed] [Google Scholar]
- Wang Y, Okamoto M, Schmitz F, Hofmann K, Sudhof TC (1997) Rim is a putative Rab3 effector in regulating synaptic-vesicle fusion. Nature 388: 593-598. [DOI] [PubMed] [Google Scholar]
- Wang Y, Liu X, Biederer T, Sudhof TC (2002) A family of RIM-binding proteins regulated by alternative splicing: implications for the genesis of synaptic active zones. Proc Natl Acad Sci USA 99: 14464-14469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZW, Saifee O, Nonet ML, Salkoff L (2001) SLO-1 potassium channels control quantal content of neurotransmitter release at the C. elegans neuromuscular junction. Neuron 32: 867-881. [DOI] [PubMed] [Google Scholar]
- Zhai RG, Bellen HJ (2004) The architecture of the active zone in the presynaptic nerve terminal. Physiology (Bethesda) 19: 262-270. [DOI] [PubMed] [Google Scholar]
- Zhai RG, Vardinon-Friedman H, Cases-Langhoff C, Becker B, Gundelfinger ED, Ziv NE, Garner CC (2001) Assembling the presynaptic active zone: a characterization of an active one precursor vesicle. Neuron 29: 131-143. [DOI] [PubMed] [Google Scholar]
- Zhen M, Jin Y (2004) Presynaptic terminal differentiation: transport and assembly. Curr Opin Neurobiol 14: 280-287. [DOI] [PubMed] [Google Scholar]
- Ziv NE, Garner CC (2004) Cellular and molecular mechanisms of presynaptic assembly. Nat Rev Neurosci 5: 385-399. [DOI] [PubMed] [Google Scholar]