Abstract
The olfactory epithelium of fish contains two major types of olfactory sensory neurons (OSNs) that are distinct morphologically (ciliated vs microvillous) and possibly functionally. Here, we found that these OSNs express different sets of signal transduction machineries: the ciliated OSNs express OR-type odorant receptors, cyclic nucleotide-gated channel A2 subunit, and olfactory marker protein (OMP), whereas the microvillous OSNs express V2R-type receptors and transient receptor potential channel C2 (TRPC2). To visualize patterns of axonal projection from the two types of OSNs to the olfactory bulb (OB), we generated transgenic zebrafish in which spectrally distinct fluorescent proteins are expressed in the ciliated and microvillous OSNs under the control of OMP and TRPC2 gene promoters, respectively. An observation of whole-mount OB in adult double-transgenic zebrafish revealed that the ciliated OSNs project axons mostly to the dorsal and medial regions of the OB, whereas the microvillous OSNs project axons to the lateral region of the OB. A careful histological examination of OB sections clarified that the axons from the two distinct types of OSNs target different glomeruli in a mutually exclusive manner. This segregation is already established at very early developmental stages in zebrafish embryos. These findings clearly demonstrate the relationships among cell morphology, molecular signatures, and axonal terminations of the two distinct types of OSNs and suggest that the two segregated neural pathways are responsible for coding and processing of different types of odor information in the zebrafish olfactory system.
Keywords: odorant receptor, transient receptor potential channel C2, olfactory marker protein, fluorescent proteins, teleost olfactory system, odor map
Introduction
The olfactory system furnishes elaborate molecular and cellular machineries for detection and discrimination of a vast number of chemical compounds in the environment (Axel, 1995). In rodents, two functionally distinct classes of chemicals, odorants and pheromones, are detected and processed through anatomically segregated neural pathways: the main olfactory system and the vomeronasal (accessory olfactory) system (Buck, 2000; Mombaerts, 2004). Volatile odorants are received by a large repertoire of odorant receptors (ORs) expressed on ciliated olfactory sensory neurons (OSNs) in the olfactory epithelium (OE), and the information is transmitted to the main olfactory bulb (OB). In contrast, pheromones are received mostly by two families of vomeronasal receptors (VRs) on microvillous sensory neurons in the vomeronasal organ that project their axons to the accessory OB.
However, the situation is completely different in the olfactory system of fish. Fish are equipped with only one olfactory organ containing both ciliated and microvillous OSNs that project their axons to the OB. The two types of OSNs show several different properties with respect to their morphology, relative position in the OE, and molecular expression. The ciliated OSNs with long dendrites are situated in the deep layer of the OE, whereas microvillous OSNs with short dendrites are located in the superficial layer (Morita and Finger, 1998; Hansen et al., 2003, 2004). The ciliated and microvillous OSNs are reported to express OR-type and V2R-type receptors, respectively (Cao et al., 1998; Speca et al., 1999; Hansen et al., 2004). In addition, several retrograde tracing experiments have indicated that the two types of OSNs project axons to different regions in the OB. In catfish, the medial and ventral regions of the OB are innervated mostly by the ciliated OSNs, whereas the dorsal region of the OB tends to be innervated by the microvillous OSNs (Morita and Finger, 1998; Hansen et al., 2003). Similarly, in carp, the medial and lateral part of the OB are innervated by ciliated and microvillous OSNs, respectively (Hamdani et al., 2001b; Hamdani and Døving, 2002). However, detailed patterns of axonal projection from different types of OSNs to individual glomeruli in the OB remain to be elucidated.
To address this issue, we took experimental advantages of zebrafish that are feasible to genetic manipulations such as transgenesis. We analyzed detailed expression patterns of receptors and ion channels in distinct populations of zebrafish OSNs and generated lines of transgenic zebrafish in which fluorescent proteins are expressed specifically in either ciliated or microvillous OSNs by using cell type-specific promoter elements. The use of spectrally distinct fluorescent proteins enabled us to observe glomerular innervation from the two types of OSNs in single individuals simultaneously. Furthermore, the transparency of zebrafish embryos permitted us to visualize the dynamics of axonal projections in living animals.
Materials and Methods
Animals
Zebrafish, Danio rerio, were maintained at 28.5°C in our laboratory as described previously (Westerfield, 1995).
Isolation of a zebrafish transient receptor potential channel C2 gene
We searched the zebrafish genomic DNA database [National Center for Biotechnology Information (NCBI)] with the TBLASTN program using the amino acid sequence of mouse transient receptor potential channel C2 (TRPC2) (Vannier et al., 1999) and found a candidate sequence for zebrafish TRPC2 (zTRPC2). Based on a putative sequence for the N-terminal cytoplasmic region of zTRPC2, we designed a pair of oligonucleotide primers (5′-GAGATTGTGAACAAGAAGTTCCGCTTCC-3′ and 5′-GCAGAGGAAAGGCATAGTCAGAAAGATC-3′) to perform reverse transcription (RT)-PCR with adult zebrafish olfactory epithelial total RNA and obtained a 1 kb fragment of zTRPC2. We performed 5′-rapid amplification of cDNA ends (RACE) analysis with adult zebrafish olfactory epithelial total RNA using a 5′RACE kit (Invitrogen, Carlsbad, CA) and determined a putative translation initiation site of zTRPC2. To confirm that zTRPC2 is a proper ortholog of mouse vomeronasal TRPC2 (Hofmann et al., 2000; Yildirim et al., 2003), we obtained the full-length coding sequence of zTRPC2 (NCBI GenBank accession number AY974804). The predicted protein contained the conserved motifs among the TRPC subfamily and showed highest homology to mouse TRPC2 (supplemental Fig. S2, available at www.jneurosci.org as supplemental material).
Isolation of DNA sequences for in situ hybridization probes
To obtain DNA fragments as templates for cRNA probes, RT-PCR was performed with zebrafish olfactory epithelial total RNA using primers that were designed as described below.
Zebrafish olfactory marker protein. PCR primers were designed based on the cDNA sequence of the zebrafish olfactory marker protein (zOMP) (Çelik et al., 2002; Yoshida et al., 2002): zOMP-cd-F (5′-ATGTCTCTGGAGTTGACGTTCAATCCTG-3′) and zOMP-3′-R1 (5′-ATTATAACTTAAATTTAATTTTATACATATTTAGGAACAC-3′). In this case, PCR was performed with zebrafish genomic DNA as a template.
Zebrafish OR-type receptors. The pairs of primers were designed based on sequences of zebrafish odorant receptors OR2.1, OR2.2, and OR13.1 (Barth et al., 1997): zOR2.1-F (5′-GGCATTAACTGCTGGGATATTTCTTATCTG-3′) and zOR2.1-R (5′-GGTAGATAATTAGTATATTTCTACTGTACAGTTGC-3′); zOR2.2-F (5′-CTGCTCGTGAGCATCTTTACAATCATTGTC-3′) and zOR2.2-R (5′-GTTTCATCAGGACCAGAATCCCATTCATG-3′); zOR13.1-F (5′-TCATCTCATCATGTCAGCCGGGAACAT-3′) and zOR13.1-R (5′-GAACCTAATGCACAAGCAACGAAGCAAAC-3′). For the zOR13.1 cRNA probe, a partial fragment between zOR13.1-F and the PstI site in the zOR13.1 coding region was used as a template.
Zebrafish cyclic nucleotide-gated channel A2 subunit and zebrafish V2R-type receptors. We searched the zebrafish genomic DNA database as described above using amino acid sequences of rat cyclic nucleotide-gated channel A2 subunit (CNGA2) (Dhallan et al., 1990) and several goldfish V2R-type receptors (Speca et al., 1999) and found candidate sequences for zebrafish CNGA2 (zCNGA2), zVR5.3, zVR3.13a, and zVR3.13b. Based on the sequences, the following pairs of primers were designed: zCNGA2-F1 (5′-CAAGGACTGCTAGTGAAGGACCTAG-3′) and zCNGA2-R1 (5′-TCGATTACGGCCTCCATTAGGTCGT-3′); zVR5.3-F1 (5′-CGAGGATGACTATGGGAAATACGGC-3′) and zVR5.3-R1 (5′-AGCACATGTGTCGTTCTCAAAAGGCC-3′); zVR3.13a-F1 (5′-GAAGTGATAAAGAAGGGCACTGCGAG-3′) and zVR3.13a-R1 (5′-GTTATCTGCCCATAGCACATGTTCCTG-3′); zVR3.13b-F1 (5′-GTGGAAGTGATCAAAAAGGGCACATCCAA-3′) and zVR3.13b-R1 (5′-CCTTTCTAGTTCCTGGGAGACAGC-3′). The sequence of the zCNGA2 gene we obtained was identical to the zebrafish olfactory cyclic nucleotide-gated channel that was described previously by Barth et al. (1996). The partial cDNA sequences of zVR5.3, zVR3.13a, and zVR3.13b have been deposited in the NCBI GenBank database (accession numbers AY974801, AY974802, and AY974803).
RNA in situ hybridization
Fourteen-month-old wild-type fish were anesthetized with 0.016% ethyl-m-aminobenzoate methanesulfonate (tricaine; Nacalai Tesque, Kyoto, Japan). The olfactory rosettes were dissected out, fixed in 4% paraformaldehyde in PBS overnight, cryoprotected in 30% sucrose in PBS, and embedded in O.C.T. compound (Sakura, Tokyo, Japan). RNA in situ hybridization was performed on 14 μm cryostat sections, essentially as described previously (Inaki et al., 2004) with some modifications. Briefly, sections were pretreated with proteinase K (10 μg/ml) for 5 min, acetylated, and hybridized with digoxigenin-labeled cRNA probes at 60°C overnight. After hybridization, sections were washed without RNase A treatment and blocked with 1.5% blocking reagent (Roche Diagnostics, Mannheim, Germany) in 100 mm Tris-HCl, pH 7.5, containing 150 mm NaCl (NT). Digoxigenin was detected with an alkaline phosphatase-conjugated anti-digoxigenin antibody (1:1000; Roche Diagnostics) and chromogenic development in 4-nitro blue tetrazolium chloride/5-bromo-4-chloro-3-indolyl-phosphate (Roche Diagnostics). For fluorescent detection, the HNPP Fluorescent Detection Set (Roche Diagnostics) was used according to the manufacturer's instructions.
Transgenes
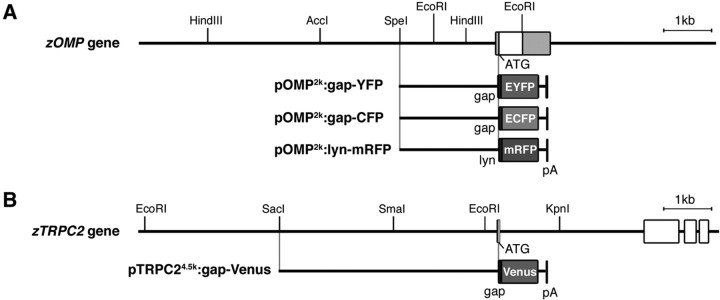
pOMP6k:gap-YFP and pOMP2k:gap-YFP. The 6 and 2 kb fragments upstream of the zOMP translation initiation site were used to drive the expression of EYFP-Mem (BD Biosciences Clontech, Palo Alto, CA), a fusion protein consisting of the N-terminal 20 aa of GAP-43 (termed gap) and enhanced yellow fluorescent protein (EYFP), as described previously (Miyasaka et al., 2005).
pOMP2k:gap-CFP-S. The EYFP-Mem cDNA in pOMP2k:gap-YFP was replaced with ECFP-Mem cDNA (BD Biosciences Clontech). The transgene construct was flanked at both ends by two I-SceI meganuclease recognition sites for the application of efficient transgenesis mediated by I-SceI (Thermes et al., 2002).
pOMP2k:lyn-mRFP-S. A monomeric red fluorescent protein (RFP), mRFP1 (Campbell et al., 2002), was fused with the N-terminal 21 aa of Lyn tyrosine kinase and substituted with the EYFP-Mem cDNA in pOMP2k:gap-YFP. Two I-SceI sites were introduced as described above.
pTRPC29k:gap-Venus-S and pTRPC24.5k:gap-Venus-S. To obtain a promoter element of the zTRPC2 gene, we screened a CHORI-211 zebrafish bacterial artificial chromosome library (BACPAC Resources, Oakland, CA). Using the obtained clone as a template, a 9 kb fragment upstream of the translation initiation site was amplified by PCR with a pair of primers [zTRPC2-F6-HindIII (5′-ACAAGCTTTGAACTGGTTAAGACCCCTCAAGAGC-3′) and zTRPC2-R10-SacII (5′-CTCCGCGGCTTCAGATATGCTGGTGATATAGATATTTCAG-3′)] and subcloned into the pGEM-T Easy vector (Promega, Madison, WI). The 9 kb zTRPC2 promoter element was excised and fused to gap-Venus, in which Venus is a variant of YFP with fast and efficient maturation (Nagai et al., 2002). Two I-SceI sites were introduced as described above. To generate a shorter promoter construct, pTRPC24.5k:gap-Venus-S, the distal 4.5 kb of the zTRPC2 promoter element in pTRPC29k:gap-Venus-S, was removed.
Generation of transgenic zebrafish
The generation of transgenic zebrafish with pOMP6k:gap-YFP and pOMP2k:gap-YFP were described previously (Miyasaka et al., 2005), and one of the established lines, Tg(pOMP2k:gap-YFP)rw032a, was used in this study and abbreviated as OMP:YFP. Plasmid constructs for transgenesis with I-SceI were purified using the QIAfilter Plasmid Maxi kit (Qiagen, Hilden, Germany) and prepared at 10 ng/μl in 0.5× commercial I-SceI buffer containing 1 U/μl I-SceI (New England Biolabs, Beverly, MA) and 0.05% Phenol Red. The DNA solution was injected into cytoplasm of the one-cell stage embryos, and the injected embryos with fluorescence in OSNs were raised to sexual maturity. Germ-line-transmitting founders were identified by the fluorescence expression in their progeny to generate transgenic lines. One transgenic line was generated from each construct and termed as follows: Tg(pOMP2k:gap-CFP)rw034a (abbreviated as OMP:CFP), Tg(pOMP2k:lyn-mRFP)rw035a (OMP:RFP), Tg(pTRPC29k:gap-Venus)rw036a, and Tg(pTRPC24.5k:gap-Venus)rw037a. Because there is no difference in the patterns of Venus expression between the two TRPC2 lines, Tg(pTRPC4.5k:gap-Venus)rw037a with strong fluorescence was used in this study and abbreviated as TRPC2:Venus. Double-transgenic zebrafish were obtained by crossing heterozygous TRPC2:Venus with heterozygous OMP:CFP and OMP:RFP, respectively.
Observation of whole-mount OB
Six- to 8-month-old fish were anesthetized as described above. Brains were exposed, and entire animals were immersed in 4% paraformaldehyde in 0.1 m phosphate buffer, pH 7.4, for 2 h at 4°C. Then the brains were removed and observed under a fluorescence stereomicroscope (MZ FLIII; Leica Microsystems, Wetzlar, Germany).
Immunohistochemistry
For immunohistological analysis of OB sections, 6- to 8-month-old fish were anesthetized as described above. The OBs were dissected out, immersed in Bouin's fixative for 2 h at 4°C, washed with 80% ethanol, cryoprotected in a series of sucrose solutions (10, 20, and 30% in 0.1 m phosphate buffer at pH 7.4), and embedded in O.C.T compound. Sections (20 μm thickness) were cut on a cryostat and thaw-mounted onto silane-coated glass slides (Matsunami, Osaka, Japan). The sections were rinsed in PBS, treated with 5% normal goat serum (NGS) in PBS containing 0.2% Triton X-100 (PBST), and incubated overnight with a mixture of the following primary antibodies diluted in PBST/NGS: rat anti-green fluorescent protein (GFP) monoclonal antibody (GF090R; rat IgG2a; 1:1000; Nacalai Tesque, Kyoto, Japan), rabbit anti-DsRed polyclonal antibody (1:10,000; BD Biosciences Clontech), and mouse anti-SV2 monoclonal antibody (SP2/0; mouse IgG1; 1:50; supernatant; Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA). The sections were then washed and incubated with a mixture of the following secondary antibodies in PBST/NGS: Alexa488-conjugated goat anti-rat IgG (1:300; Molecular Probes, Eugene, OR), Cy3-conjugated goat anti-rabbit IgG (1:300; Jackson ImmunoResearch, West Grove, PA), and Cy5-conjugated goat anti-mouse IgG (1:300; Jackson ImmunoResearch). The sections were washed, coverslipped with 50% glycerol in PBS, and examined with a confocal laser-scanning microscope (FV1000; Olympus, Tokyo, Japan).
For double-immunofluorescence labeling of OE sections, tissues were prepared and sectioned in the same way for RNA in situ hybridization. The sections were incubated with a mixture of rabbit anti-S100 polyclonal antibody (1:1000; Dako, Glostrup, Denmark) and rat anti-GFP monoclonal antibody (GF090R; rat IgG2a; 1:1000; Nacalai Tesque) and subsequently incubated with a mixture of Cy3-conjugated goat anti-rabbit IgG (1:300; Jackson ImmunoResearch) and Alexa488-conjugated goat anti-rat IgG (1:300; Molecular Probes).
For immunostaining of OE sections after fluorescent in situ hybridization, the sections were fixed with 4% paraformaldehyde in PBS for 10 min, washed in NT, treated with 1.5% blocking reagent (Roche Diagnostics) in NT, and incubated with rabbit anti-GFP polyclonal antibody (1:1000; BD Biosciences Clontech). After washing in NT containing 0.05% Tween 20, the sections were incubated with Alexa488-conjugated goat anti-rabbit IgG (1:300; Molecular Probes). The OE sections were examined with a confocal laser-scanning microscope (FV500; Olympus).
Time-lapse imaging of OSN projection in zebrafish embryos
Zebrafish embryos were maintained at 28.5°C in 1/3 Ringer's solution (in mm: 39 NaCl, 0.97 KCl, 1.8 CaCl2, and 1.7 HEPES, pH 7.2) supplemented with 100 U/ml penicillin and 100 μg/ml streptomycin. To prevent pigmentation, 0.002% phenylthiourea (Nacalai Tesque) was added to the solution 12 h after fertilization. At each time point for observation, zebrafish embryos were anesthetized with 0.016% tricaine, mounted in 2% low-melting point agarose in 1/3 Ringer's solution, and examined with a confocal laser-scanning microscope (FV500; Olympus). Between time points for observations, the embryos were removed from the agarose and incubated at 28.5°C.
Results
Differential expression of signaling molecules in ciliated and microvillous OSNs
We first cloned cDNAs encoding zebrafish orthologs of ORs (Barth et al., 1997), CNGA2 (Barth et al., 1996), VRs (supplemental Fig. S1, available at www.jneurosci.org as supplemental material), TRPC2 (supplemental Fig. S2, available at www.jneurosci.org as supplemental material), and OMP (Çelik et al., 2002; Yoshida et al., 2002) and examined, by in situ hybridization, what types of cells express these molecules in the zebrafish olfactory organ.
On horizontal sections of the olfactory rosette, all of the cRNA probes (zOMP, zTRPC2, zCNGA2, zORs, zVRs) labeled the medial portion of the lamella (Fig. 1A,F) (data not shown), where OSNs are located (Ngai et al., 1993a,b; Baier et al., 1994). High-power views (Fig. 1B-E,G-J) revealed that they were expressed in two distinct populations of OSNs, the cell bodies of which are located at different positions along the apical-basal axis within the OE. Although rodent OMP is expressed in both the olfactory and vomeronasal sensory neurons, zOMP mRNA was found only in OSNs with a long dendrite, the cell bodies of which are located in the deep layer of the zebrafish OE (Fig. 1B). The deep OSNs also expressed zCNGA2 (Fig. 1C) and OR-type receptors such as zOR2.1 (Fig. 1D), zOR13.1 (Fig. 1E), and zOR2.2 (data not shown), similar to the ciliated OSNs in the rodent OE. In contrast, zTRPC2-positive cells were situated in the superficial layer of the OE and bore a short dendrite (Fig. 1G). These superficial OSNs also expressed zVR5.3 (Fig. 1H), zVR3.13a (Fig. 1I), and zVR3.13b (Fig. 1J), reminiscent of the vomeronasal sensory neurons in rodents.
Figure 1.
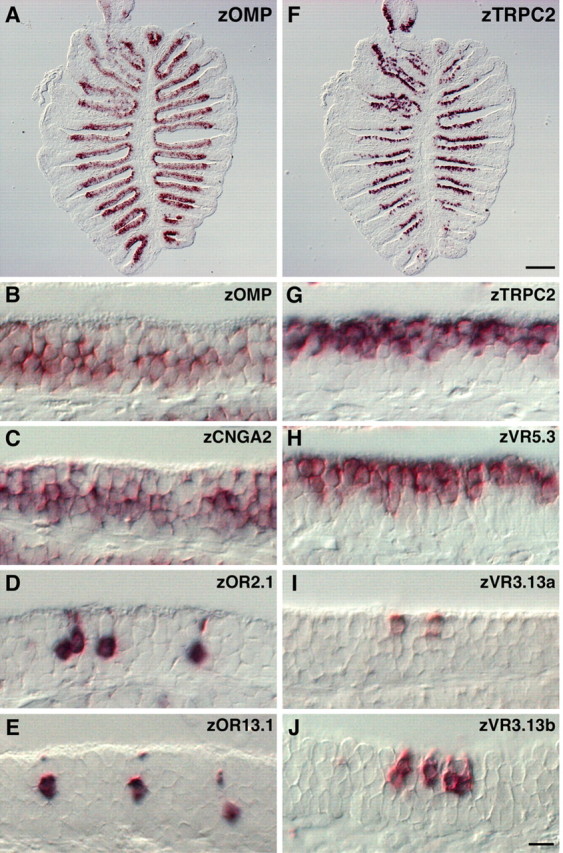
Two distinct types of OSNs with different molecular signatures in zebrafish OE. A-J, RNA in situ hybridization was performed on horizontal sections of the zebrafish olfactory rosette using digoxigenin-labeled probes specific for the following molecules: zOMP (A, B), zCNGA2 (C), zOR2.1 (D), zOR13.1 (E), zTRPC2 (F, G), zVR5.3 (H), zVR3.13a (I), and zVR3.13b (J). Note that zOMP (B), zCNGA2 (C), and OR-type receptors (D, E) were expressed in the deep layer of the OE, whereas zTRPC2 (G) and VR-type receptors (H-J) were expressed in the superficial layer. In contrast to the zVR5.3 expression in a large fraction of OSNs (H), other VR- and OR-type receptors were expressed in a small subset of OSNs (D, E, I, J). B-E, G-J, The apical surface of the OE is at the top of each panel. Scale bars: (in F) A, F, 100 μm; (in J) B-E, G-J, 20 μm.
Thus, the zebrafish OE contains two distinct populations of OSNs with different molecular signatures that probably correspond to the ciliated and microvillous OSNs identified by cellular morphology and location in the OE (Hansen et al., 2003). Together, these results support the idea that the ciliated and microvillous OSNs respond to different classes of odorants through zORs and zVRs, respectively, and use different signal transduction machineries for odor information processing.
Transgenic zebrafish lines expressing fluorescent proteins in two distinct types of OSNs
To visualize detailed morphology and axonal projection patterns of the two types of OSNs, transgenic zebrafish lines were established in which membrane-targeted fluorescent proteins are expressed under the control of individual cell type-specific promoter elements. We used the 2 kb upstream region of the zOMP gene and the 4.5 kb upstream region of the zTRPC2 gene for the transgene expression in ciliated and microvillous OSNs, respectively (Fig. 2).
Figure 2.
DNA constructs for generating transgenic zebrafish lines expressing fluorescent proteins under the control of OMP and TRPC2 promoters. A, Genomic organization of the zOMP gene (top). The white box indicates the coding region of the zOMP gene. Gray boxes indicate noncoding regions of the cDNA. The 2 kb upstream region of the zOMP translation initiation site was used to express gap-YFP, gap-CFP, and lyn-mRFP (pOMP2k:gap-YFP, pOMP2k:gap-CFP, and pOMP2k:lyn-mRFP). B, Genomic organization of the zTRPC2 gene (top). White boxes indicate the coding region of the zTRPC2 gene. The 4.5 kb upstream region of the zTRPC2 translation initiation site was used to express gap-Venus (pTRPC24.5k:gap-Venus). Each construct contains the SV40 polyadenylation signal (in picoamperes).
To confirm that the expression of transgenes recapitulates that of endogenous molecules, in situ hybridization and immunohistochemistry were performed on the same sections of the adult OE (Fig. 3). In OMP:YFP transgenic fish, YFP was detected in long dendrite-bearing OSNs, the cell bodies of which are located in the deep layer of the OE, faithfully to the expression of endogenous zOMP mRNA (Fig. 3A-C). In contrast, Venus expression in TRPC2:Venus transgenic fish was observed in short dendrite-bearing OSNs in the superficial layer of the OE that expressed endogenous zTRPC2 mRNA (Fig. 3D-F). Similar results were obtained in transgenic lines with longer promoter elements (OMP, 6 kb; TRPC2, 9 kb) (data not shown). These findings suggest that the ciliated and microvillous OSNs were selectively labeled with fluorescent proteins by using zOMP and zTRPC2 promoters, respectively.
Figure 3.
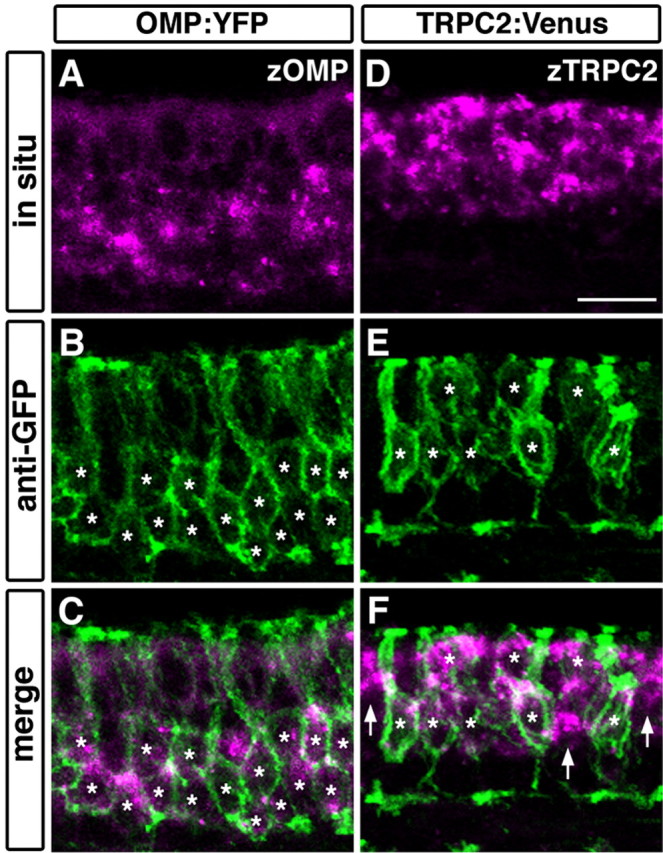
Double fluorescent labeling of transgene products and endogenous mRNA in the OE of OMP:YFP and TRPC2:Venus transgenic fish.A-C, The expression of zOMP mRNA (magenta; A, C) and YFP protein (green; B, C) in the OMP:YFP line. D-F, The expression of zTRPC2 mRNA (magenta; D, F) and Venus protein (green; E, F) in the TRPC2:Venus line. All photographs represent stacked images of optical sections. Asterisks denote the position of somata labeled with anti-GFP. In the OMP:YFP line, YFP was expressed in cells that express zOMP mRNA in the deep layer of the OE (asterisks; B, C). In the TRPC2:Venus line, Venus was expressed in zTRPC2-positive cells in the superficial layer (asterisks; E, F). However, some zTRPC2-positive cells appeared to be unlabeled (arrows; F). The apical surface of the OE is at the top. Scale bar, 50 μm.
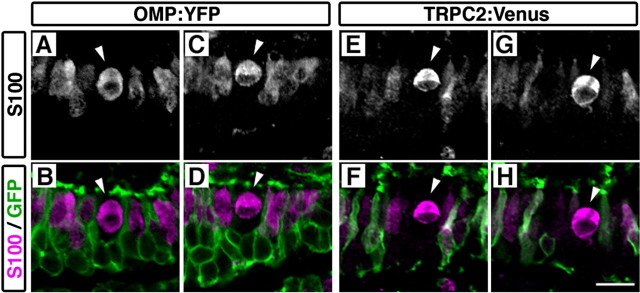
In fish, there is a third type of OSN: crypt cells, which comprise only a minor population in the OE. The crypt OSNs are located in the superficial layer of the OE and have unique characters such as typical ovoid-shaped cell bodies and strong S100 protein-like immunoreactivity (Hansen and Finger, 2000; Hansen et al., 2003, 2004; Germanà et al., 2004). We examined whether crypt OSNs are labeled with fluorescent proteins in the OMP:YFP and TRPC2:Venus transgenic fish. Double fluorescence immunohistochemical analysis showed that strongly S100-immunoreactive cells with ovoid somata (Fig. 4, arrowheads) were devoid of YFP or Venus expression (Fig. 4), indicating that crypt OSNs were not labeled in either of the transgenic lines.
Figure 4.
S100-positive crypt OSNs were not labeled in either OMP:YFP or TRPC2:Venus transgenic fish. OE sections of two transgenic fish, OMP:YFP (A-D) and TRPC2:Venus (E-H), were immunostained with antibodies against S100 (A, C, E, G; magenta in B, D, F, H) and GFP (green in B, D, F, H). Two representative crypt OSNs are shown for each transgenic line (arrowheads; A-H). All photographs represent single optical sections. The apical surface of the OE is at the top. Scale bar, 50 μm.
Mutually exclusive innervation of OB glomeruli by ciliated and microvillous OSNs
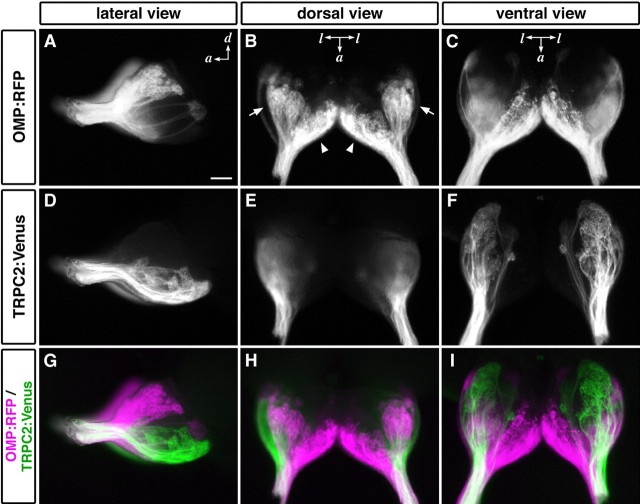
To visualize axonal projections from ciliated and microvillous OSNs simultaneously and separately, we crossed the OMP:RFP transgenic line with the TRPC2:Venus line and obtained double-transgenic zebrafish OMP:RFP;TRPC2:Venus. A whole-mount observation revealed differential distributions of RFP and Venus fluorescence on the surface of the OB (Fig. 5). Lateral views of the OB clearly demonstrated segregated targeting of RFP- and Venus-positive axons to the dorsal and ventral regions on the lateral surface of the OB (Fig. 5A,D,G). Dorsal views showed that RFP-expressing axons targeted two major glomerular subgroups called dorsal cluster (Fig. 5B, arrows) and anterior plexus (Fig. 5B, arrowheads), whereas Venus-positive glomeruli were not present on the dorsal surface (Fig. 5E,H). On the ventral surface of the OB, RFP- and Venus-expressing axons target the medial and lateral regions, respectively (Fig. 5C,F,I). Thus, the ciliated and microvillous OSNs exhibit differential patterns of axonal innervation onto the OB.
Figure 5.
Differential axonal projections from two types of OSNs. Whole-mount OB from OMP:RFP;TRPC2:Venus adult zebrafish were observed under a fluorescence stereomicroscope: A, D, G, lateral views; B, E, H, dorsal views; C, F, I, ventral views. Ciliated OSNs labeled with RFP projected their axons to almost all over the dorsal region and the ventromedial portion of the OB (A-C; magenta in G-I). In contrast, microvillous OSNs labeled with Venus projected their axons exclusively to the ventrolateral region (D-F; green in G-I). The arrows and arrowheads indicate dorsal cluster and the anterior plexus, respectively. a, Anterior; d, dorsal; l, lateral. Scale bar, 100 μm.
A detailed glomerular map established by differential innervation from ciliated and microvillous OSNs was examined on OB horizontal sections of the OMP:RFP;TRPC2:Venus double-transgenic fish by immunohistochemistry. Counterstaining for a synaptic vesicle protein, SV2, could help us to distinguish the glomerular layer from the olfactory nerve layer and to identify individual glomeruli that were previously designated by Baier and Korsching (1994).
Most of the glomeruli in the dorsal part of the OB, including dorsal cluster and its associated glomeruli, were innervated by RFP-expressing ciliated OSNs (Fig. 6Q-S, numbers 1 and 2). In mediodorsal posterior glomeruli, faint RFP expression was detected in only a small portion (Fig. 6A-C, Q-S, number 3). In the dorsal part, no labeled glomerulus was detected for Venus signals originating from microvillous OSNs (Fig. 6I,J,Q,R).
Figure 6.
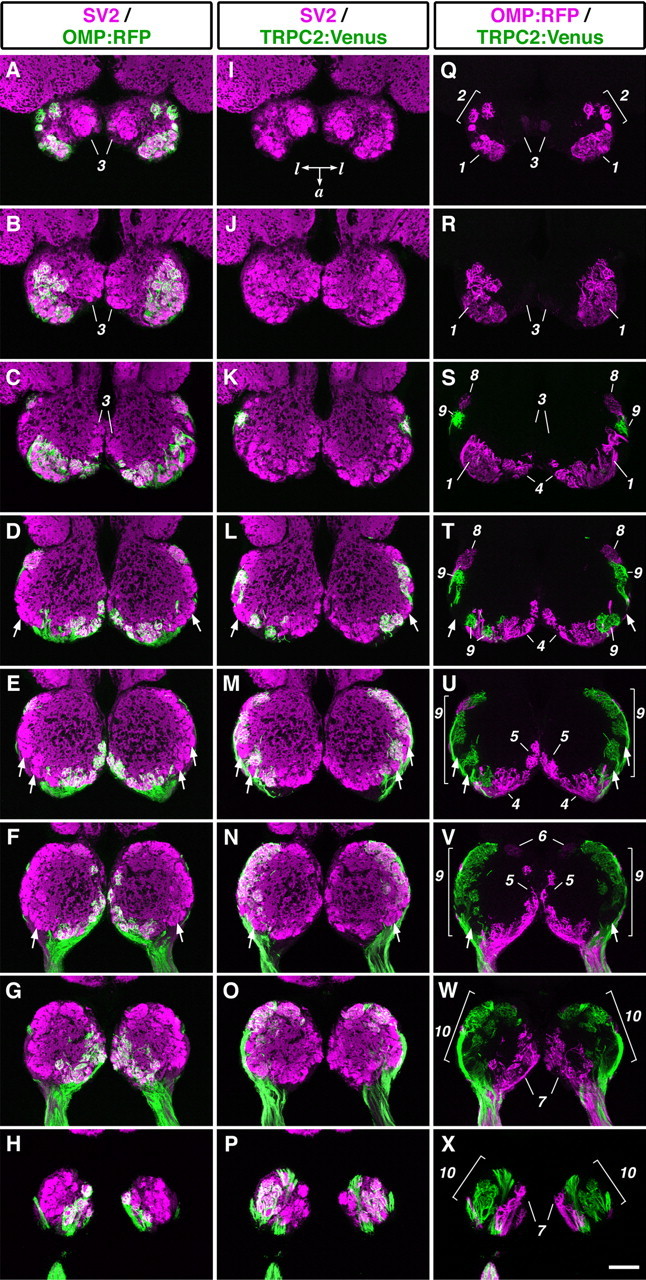
Mutually exclusive glomerular innervation by two types of OSNs. Horizontal sections through the OB of OMP:RFP; TRPC2:Venus adult zebrafish were stained with anti-RFP, anti-GFP, and anti-SV2 antibodies. A-H, Merged images of staining for RFP (green) and SV2 (magenta). I-P, Merged images of staining for Venus (green) and SV2 (magenta). Q-X, Merged images of staining for RFP (magenta) and Venus (green). The alternate eight sections from a series of OB sections are ordered from the dorsalmost (top) to the ventralmost (bottom) section. All photographs represent stacked images of optical sections. Arrows indicate glomeruli that were not labeled with either RFP or Venus. Numbers for single glomeruli or glomerular subgroup: 1, dorsal cluster; 2, dorsal cluster-associated glomeruli; 3, mediodorsal posterior glomeruli; 4, anterior plexus; 5, medial glomeruli; 6, medioventral posterior glomerulus; 7, ventromedial glomeruli; 8, lateroposterior glomerulus; 9, lateral chain; 10, ventrolateral glomeruli. Posterior is at the top of each panel. a, Anterior; l, lateral. Scale bar, 100 μm.
In the middle and ventral parts of the OB, clearly segregated terminations of RFP- and Venus-expressing axons were observed. RFP-positive ciliated OSNs projected axons mostly to the medial region, including the anterior plexus, its neighboring medial glomeruli, a medioventral posterior glomerulus, and ventromedial glomeruli (Fig. 6S-X, numbers 4-7). There was only one lateral glomerulus innervated by RFP-positive ciliated OSNs, at the most posterior position of the lateral chain (Fig. 6S,T, number 8). In contrast, Venus-positive microvillous OSNs innervated only the lateral region of the OB but not the medial region. The Venus-positive glomeruli included most glomeruli in the lateral chain and ventrolateral glomeruli (Fig. 6S-X, numbers 9 and 10). The patterns of glomerular targeting were stereotyped in all individuals, regardless of sex (data not shown).
Importantly, we could not find any double-positive glomeruli that were targeted by both ciliated and microvillous OSNs, suggesting that the two distinct types of OSNs project their axons in a mutually exclusive manner. In contrast, several glomeruli were double negative for RFP and Venus (Fig. 6D-F,L-N,T-V, arrows), raising a possibility that these glomeruli may be innervated by the third type of OSNs, crypt cells.
Establishment of the segregated axonal terminations at early stages of development
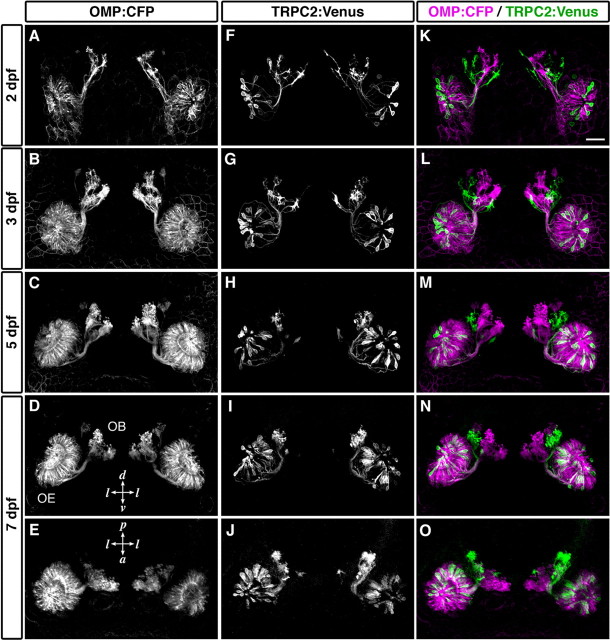
Next, we examined the developmental process of axonal projection from the two types of OSNs in double-transgenic embryos that express cyan fluorescent protein (CFP) in ciliated OSNs and Venus in microvillous OSNs. Time-lapse imaging of dynamic behavior of fluorescently labeled OSNs was performed in living embryos. At 2 d postfertilization (dpf), CFP and Venus signals were detected in different populations of OSNs in the OE (Fig. 7A,F,K). Axons from the two types of OSNs projected toward distinct regions in the presumptive OB even at this very early stage: the medial region of the OB was targeted by CFP-expressing ciliated OSNs, whereas the lateral and ventral regions were targeted by Venus-expressing microvillous OSNs (Fig. 7A,F,K). At later stages (3 and 5 dpf) when the number of OSNs increased and the OB glomeruli differentiated, the axonal terminations became more apparent (Fig. 7B,C,G,H,L,M). At 7 dpf, the overall pattern of segregated innervation was almost similar to that in adult fish (Fig. 7D,E,I,J,N,O). These results suggest that the segregated axonal terminations of ciliated and microvillous OSNs are approximately established at very early stages of development and refined with glomerular maturation as the development proceeds further.
Figure 7.
Establishment of the segregated axonal terminations at early stages of development. Time-lapse imaging of labeled OSNs in living OMP:CFP;TRPC2:Venus zebrafish embryos was performed. CFP-labeled (A-E; magenta in K-O) and Venus-labeled OSNs (F-J; green in K-O) in a single representative embryo are shown. All photographs represent stacked images of optical sections. Axonal terminations from CFP-labeled and Venus-labeled OSNs were already segregated at 2 dpf (K) and maintained through development (L, 3 dpf; M, 5 dpf; N, O, 7 dpf). A-D, F-I, K-N, Frontal views; E, J, O, dorsal views. l, Lateral; d, dorsal; v, ventral; p, posterior; a, anterior. Scale bar, 50 μm.
Discussion
The key points of this study are (1) ciliated and microvillous OSNs in the zebrafish OE express different sets of receptors and ion channels, (2) transgenic zebrafish lines were established that express fluorescent proteins either in the ciliated or microvillous OSNs under the control of zOMP or zTRPC2 promoters, (3) the two types of OSNs project their axons to different regions of the OB in a mutually exclusive manner, and (4) the segregated axonal terminations occur already at early developmental stages. From these results, we propose that the two segregated neural pathways are the basis for odor information coding and processing in the zebrafish olfactory system.
A genetic approach to dissection of neural circuitry in the zebrafish olfactory system
Previously, our knowledge on the anatomy of olfactory pathways in fish was mainly provided by retrograde tracing experiments in which DiI (1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate) was injected into various regions of the OB to trace back the cell bodies of OSNs in the OE (Morita and Finger, 1998; Hansen et al., 2003). These studies showed a tendency of differential localization of labeled OSNs in the superficial and deep layers of the OE corresponding to the injection sites in the OB. Conversely, anterograde tracing of OSN axonal trajectory by DiI injection into the nasal cavities resulted in uniform labeling of OSN axons and OB glomeruli (Baier and Korsching, 1994). Thus, these classical approaches were not adequate for identification of target glomeruli innervated by distinct types of OSNs.
Our genetic strategy for visualization of axonal projections from the OE to the OB has several advantages over the dye-tracing method. First, distinct functional types of OSNs can be selectively labeled by using different promoter elements. Therefore, we can track overall axonal projection patterns from transgene-expressing OSNs in whole-mount preparations (Fig. 5) and distinguish in detail the target glomeruli on histological sections with high resolution (Fig. 6). Second, the transparency of zebrafish embryos permits us to monitor the dynamics of axon outgrowth and targeting from the OE to the OB during in vivo development. In particular, by expressing two spectrally distinct fluorescent proteins in ciliated and microvillous OSNs, the segregation of distinct neural pathways can be visualized simultaneously in single individuals (Fig. 7). Third, once the transgenic lines are established, we can observe the specific labeling of distinct neural pathways with great accuracy and reproducibility. This feature will enable us to efficiently screen and analyze mutant zebrafish that show impairment in the projection of OSN axons. For instance, we have identified Robo2 as a crucial axon guidance molecule for the establishment of a precise glomerular map in the OB by crossing the OMP:YFP transgenic fish with the robo2 mutant fish astray (Miyasaka et al., 2005).
Cellular morphology and molecular signatures of different types of OSNs
It has been suggested that OSN morphology (microvillous or ciliated) is related to the receptor gene expression in fish as well as in rodents (Cao et al., 1998; Speca et al., 1999; Hansen et al., 2004). In the present study, we cloned the zebrafish ortholog of TRPC2 for the first time and demonstrated that zTRPC2, together with VR-type receptors, were expressed exclusively by microvillous OSNs in zebrafish (Fig. 1). Additionally, we confirmed previous reports (Speca et al., 1999; Hansen et al., 2004) that ciliated OSNs express zCNGA2, OR-type receptors (Fig. 1), and GTP-binding protein Gαolf/s (data not shown). Thus, the combinatorial usage of receptors, channels, and possibly G-proteins in the microvillous and ciliated OSNs is evolutionarily conserved between fish and rodents. These results implicate common molecular and cellular mechanisms among vertebrates with which structurally different classes of odorants activate the microvillous and ciliated OSNs through the distinct signal transduction cascades after binding to the VR-type and OR-type receptors, respectively.
Odorant response specificities of ciliated and microvillous OSNs
Teleost fish can detect and discriminate several structurally different classes of naturally occurring odorants: amino acids, nucleotides, bile acids, gonadal steroids, and prostaglandins (Sorensen and Caprio, 1998). How does the fish olfactory system accomplish this feat? Previous molecular biological, physiological, and imaging studies have provided important clues to addressing this question. One of the goldfish V2R-type receptors and its zebrafish ortholog are activated by amino acids with preferences for arginine and glutamate, respectively (Speca et al., 1999; Kuang et al., 2003; Luu et al., 2004). Activity-dependent labeling of OSNs using a cation channel-permeant probe, agmatine, has shown that amino acid odorants stimulate at least microvillous OSNs in zebrafish (Lipschitz and Michel, 2002). Functional imaging of OB glomeruli in zebrafish using calcium- and voltage-sensitive dyes has revealed that various amino acids and nucleotides elicit activation of glomeruli in the lateral region of the OB (Friedrich and Korsching, 1997, 1998). On the contrary, neural activities elicited by bile acids, a fish gonadal steroid (17α, 20β-dihydroxy-4-pregnene-3-one-20-sulfate), and prostaglandin F2α are observed in the medial region of the OB (Friedrich and Korsching, 1998). By combining these previous findings with our present data, a presumption is made that the microvillous OSNs are activated by amino acids and nucleotides through V2R-type receptors and project their axons mostly to the lateral region of the OB, whereas the ciliated OSNs are stimulated by bile acids, steroids, and prostaglandins through OR-type receptors and innervate the medial region of the OB.
This idea may be also supported by ligand selectivities of receptors. The V2R-type receptors are structurally related to the calcium-sensing receptor and metabotropic glutamate receptor families (Tanabe et al., 1992; Hebert and Brown, 1995), all of which bind hydrophilic ligands (calcium ion and glutamate) in their long extracellular N-terminal regions (Pin et al., 2003). In contrast, the OR-type receptors interact with hydrophobic compounds in the ligand-binding pocket of the transmembrane region (Floriano et al., 2000; Singer, 2000; Katada et al., 2005). Actually, a cell surface receptor for bile acids in mammals shows significant structural homology to the OR-type receptors (Maruyama et al., 2002; Kawamata et al., 2003). Thus, these selective binding profiles of receptors to hydrophilic or hydrophobic ligands are consistent with our notion that amino acids and nucleotides are detected by V2R-type receptors expressed on microvillous OSNs, whereas bile acids, steroids, and prostaglandins are detected by OR-type receptors expressed on ciliated OSNs. However, there are several studies reporting that amino acids are detected not only by microvillous OSNs but also by ciliated OSNs (Sato and Suzuki, 2001; Hansen et al., 2003). The transgenic zebrafish lines established in the present study will be useful in future experiments to elucidate odorant response specificities of ciliated versus microvillous OSNs.
Two segregated olfactory pathways in fish: functional relevance
Different classes of odorants exert different behavioral and physiological responses in fish: amino acids and nucleotides trigger arousal and feeding behaviors, whereas, steroids, prostaglandins, and possibly bile acids elicit unique social behaviors and/or endocrine responses (Sorensen and Caprio, 1998). Several lines of evidence suggest that such distinct types of behaviors are mediated through two major axonal trajectories of OB projection neurons. Behavioral and electrophysiological studies in crucian carp and goldfish have shown that the lateral olfactory tract (LOT) mediates feeding behaviors (von Rekowski and Zippel, 1993; Hamdani et al., 2001a), whereas the medial olfactory tract (MOT) mediates reproductive and alarm behaviors (Demski and Dulka, 1984; Sorensen et al., 1991; Hamdani et al., 2000; Weltzien et al., 2003). Furthermore, DiI injection into the lateral part of the carp OB results in the retrograde labeling of microvillous OSNs and the concomitant anterograde labeling of the LOT (Hamdani et al., 2001b), whereas ciliated OSNs and the MOT are labeled by DiI injection into the medial part of the OB (Hamdani and Døving, 2002). Together with our present findings, we speculate that the two segregated neural pathways are responsible for coding and processing of different types of odor information in the fish olfactory system. A genetic transneuronal tracing method to express plant lectin transgene under the control of cell type-specific promoters (Yoshihara et al., 1999) would satisfactorily meet the verification of this hypothesis and help to understand how the odor information is coded and processed in the telencephalon of fish.
The microvillous OSNs in fish appear to respond feeding cues, whereas those in rodents respond preferentially to pheromones. Interestingly, in snakes, the vomeronasal organ that has only microvillous OSNs mediates both social and nonsocial behaviors (Halpern and Martínez-Marcos, 2003). Although two segregated olfactory pathways with distinct molecular and cellular mechanisms are quite likely conserved among most vertebrates, how to use the two olfactory pathways in light of distinct functions seems to differ among animal species, depending on their living environments.
Footnotes
This work was supported in part by the Special Coordination Funds for Promoting Science and Technology from the Japan Science and Technology Corporation to Y.Y., by a Grant-in-Aid for Scientific Research on Priority Areas (C)-Advance Brain Science Project to Y.Y., and by a Grant-in-Aid for Young Scientists to N.M. from the Ministry of Education, Culture, Sports, Science, and Technology of Japan. We thank A. Miyawaki (RIKEN Brain Science Institute, Saitama, Japan) for Venus cDNA, R.Y. Tsien (University of California San Diego, La Jolla, CA) for mRFP1 cDNA, Y. Emori and A. Yasuoka (University of Tokyo, Tokyo, Japan) for valuable advice, A. Funaki for the initial part of this study, S. Yamashita and N. Kobayashi for maintenance of our zebrafish facility, and members of the Yoshihara laboratory for helpful discussions.
Correspondence should be addressed to Dr. Yoshihiro Yoshihara, Laboratory for Neurobiology of Synapse, RIKEN Brain Science Institute, 2-1 Hirosawa, Wako, Saitama 351-0198, Japan. E-mail: yoshihara@brain.riken.jp.
Copyright © 2005 Society for Neuroscience 0270-6474/05/254889-09$15.00/0
References
- Axel R (1995) The molecular logic of smell. Sci Am 273: 154-159. [DOI] [PubMed] [Google Scholar]
- Baier H, Korsching S (1994) Olfactory glomeruli in the zebrafish form an invariant pattern and are identifiable across animals. J Neurosci 14: 219-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier H, Rotter S, Korsching S (1994) Connectional topography in the zebrafish olfactory system: random positions but regular spacing of sensory neurons projecting to an individual glomerulus. Proc Natl Acad Sci USA 91: 11646-11650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth AL, Justice NJ, Ngai J (1996) Asynchronous onset of odorant receptor expression in the developing zebrafish olfactory system. Neuron 16: 23-34. [DOI] [PubMed] [Google Scholar]
- Barth AL, Dugas JC, Ngai J (1997) Noncoordinate expression of odorant receptor genes tightly linked in the zebrafish genome. Neuron 19: 359-369. [DOI] [PubMed] [Google Scholar]
- Buck LB (2000) The molecular architecture of odor and pheromone sensing in mammals. Cell 100: 611-618. [DOI] [PubMed] [Google Scholar]
- Cao Y, Oh BC, Stryer L (1998) Cloning and localization of two multigene receptor families in goldfish olfactory epithelium. Proc Natl Acad Sci USA 95: 11987-11992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY (2002) A monomeric red fluorescent protein. Proc Natl Acad Sci USA 99: 7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Çelik A, Fuss SH, Korsching SI (2002) Selective targeting of zebrafish olfactory receptor neurons by the endogenous OMP promoter. Eur J Neurosci 15: 798-806. [DOI] [PubMed] [Google Scholar]
- Demski LS, Dulka JG (1984) Functional-anatomical studies on sperm release evoked by electrical stimulation of the olfactory tract in goldfish. Brain Res 291: 241-247. [DOI] [PubMed] [Google Scholar]
- Dhallan RS, Yau KW, Schrader KA, Reed RR (1990) Primary structure and functional expression of a cyclic nucleotide-activated channel from olfactory neurons. Nature 347: 184-187. [DOI] [PubMed] [Google Scholar]
- Floriano WB, Vaidehi N, Goddard III WA, Singer MS, Shepherd GM (2000) Molecular mechanisms underlying differential odor responses of a mouse olfactory receptor. Proc Natl Acad Sci USA 97: 10712-10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich RW, Korsching SI (1997) Combinatorial and chemotopic odorant coding in the zebrafish olfactory bulb visualized by optical imaging. Neuron 18: 737-752. [DOI] [PubMed] [Google Scholar]
- Friedrich RW, Korsching SI (1998) Chemotopic, combinatorial, and non-combinatorial odorant representations in the olfactory bulb revealed using a voltage-sensitive axon tracer. J Neurosci 18: 9977-9988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germanà A, Montalbano G, Laurà R, Ciriaco E, del Valle ME, Vega JA (2004) S100 protein-like immunoreactivity in the crypt olfactory neurons of the adult zebrafish. Neurosci Lett 371: 196-198. [DOI] [PubMed] [Google Scholar]
- Halpern M, Martínez-Marcos A (2003) Structure and function of the vomeronasal system: an update. Prog Neurobiol 70: 245-318. [DOI] [PubMed] [Google Scholar]
- Hamdani EH, Døving KB (2002) The alarm reaction in crucian carp is mediated by olfactory neurons with long dendrites. Chem Senses 27: 395-398. [DOI] [PubMed] [Google Scholar]
- Hamdani EH, Stabell OB, Alexander G, Døving KB (2000) Alarm reaction in the crucian carp is mediated by the medial bundle of the medial olfactory tract. Chem Senses 25: 103-109. [DOI] [PubMed] [Google Scholar]
- Hamdani EH, Kasumyan A, Døving KB (2001a) Is feeding behaviour in crucian carp mediated by the lateral olfactory tract? Chem Senses 26: 1133-1138. [DOI] [PubMed] [Google Scholar]
- Hamdani EH, Alexander G, Døving KB (2001b) Projection of sensory neurons with microvilli to the lateral olfactory tract indicates their participation in feeding behaviour in crucian carp. Chem Senses 26: 1139-1144. [DOI] [PubMed] [Google Scholar]
- Hansen A, Finger TE (2000) Phyletic distribution of crypt-type olfactory receptor neurons in fishes. Brain Behav Evol 55: 100-110. [DOI] [PubMed] [Google Scholar]
- Hansen A, Rolen SH, Anderson K, Morita Y, Caprio J, Finger TE (2003) Correlation between olfactory receptor cell type and function in the channel catfish. J Neurosci 23: 9328-9339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen A, Anderson KT, Finger TE (2004) Differential distribution of olfactory receptor neurons in goldfish: structural and molecular correlates. J Comp Neurol 477: 347-359. [DOI] [PubMed] [Google Scholar]
- Hebert SC, Brown EM (1995) The extracellular calcium receptor. Curr Opin Cell Biol 7: 484-492. [DOI] [PubMed] [Google Scholar]
- Hofmann T, Schaefer M, Schultz G, Gudermann T (2000) Cloning, expression and subcellular localization of two novel splice variants of mouse transient receptor potential channel 2. Biochem J 351: 115-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaki K, Nishimura S, Nakashiba T, Itohara S, Yoshihara Y (2004) Laminar organization of the developing lateral olfactory tract revealed by differential expression of cell recognition molecules. J Comp Neurol 479: 243-256. [DOI] [PubMed] [Google Scholar]
- Katada S, Hirokawa T, Oka Y, Suwa M, Touhara K (2005) Structural basis for a broad but selective ligand spectrum of a mouse olfactory receptor: mapping the odorant-binding site. J Neurosci 25: 1806-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata Y, Fujii R, Hosoya M, Harada M, Yoshida H, Miwa M, Fukusumi S, Habata Y, Itoh T, Shintani Y, Hinuma S, Fujisawa Y, Fujino M (2003) A G protein-coupled receptor responsive to bile acids. J Biol Chem 278: 9435-9440. [DOI] [PubMed] [Google Scholar]
- Kuang D, Yao Y, Wang M, Pattabiraman N, Kotra LP, Hampson DR (2003) Molecular similarities in the ligand binding pockets of an odorant receptor and the metabotropic glutamate receptors. J Biol Chem 278: 42551-42559. [DOI] [PubMed] [Google Scholar]
- Lipschitz DL, Michel WC (2002) Amino acid odorants stimulate microvillar sensory neurons. Chem Senses 27: 277-286. [DOI] [PubMed] [Google Scholar]
- Luu P, Acher F, Bertrand HO, Fan J, Ngai J (2004) Molecular determinants of ligand selectivity in a vertebrate odorant receptor. J Neurosci 24: 10128-10137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T, Miyamoto Y, Nakamura T, Tamai Y, Okada H, Sugiyama E, Nakamura T, Itadani H, Tanaka K (2002) Identification of membrane-type receptor for bile acids (M-BAR). Biochem Biophys Res Commun 298: 714-719. [DOI] [PubMed] [Google Scholar]
- Miyasaka N, Sato Y, Yeo SY, Hutson LD, Chien CB, Okamoto H, Yoshihara Y (2005) Robo2 is required for establishment of a precise glomerular map in the zebrafish olfactory system. Development 132: 1283-1293. [DOI] [PubMed] [Google Scholar]
- Mombaerts P (2004) Genes and ligands for odorant, vomeronasal and taste receptors. Nat Rev Neurosci 5: 263-278. [DOI] [PubMed] [Google Scholar]
- Morita Y, Finger TE (1998) Differential projections of ciliated and microvillous olfactory receptor cells in the catfish, Ictalurus punctatus J Comp Neurol 398: 539-550. [DOI] [PubMed] [Google Scholar]
- Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A (2002) A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol 20: 87-90. [DOI] [PubMed] [Google Scholar]
- Ngai J, Dowling MM, Buck L, Axel R, Chess A (1993a) The family of genes encoding odorant receptors in the channel catfish. Cell 72: 657-666. [DOI] [PubMed] [Google Scholar]
- Ngai J, Chess A, Dowling MM, Necles N, Macagno ER, Axel R (1993b) Coding of olfactory information: topography of odorant receptor expression in the catfish olfactory epithelium. Cell 72: 667-680. [DOI] [PubMed] [Google Scholar]
- Pin JP, Galvez T, Prézeau L (2003) Evolution, structure, and activation mechanism of family 3/C G-protein-coupled receptors. Pharmacol Ther 98: 325-354. [DOI] [PubMed] [Google Scholar]
- Sato K, Suzuki N (2001) Whole-cell response characteristics of ciliated and microvillous olfactory receptor neurons to amino acids, pheromone candidates and urine in rainbow trout. Chem Senses 26: 1145-1156. [DOI] [PubMed] [Google Scholar]
- Singer MS (2000) Analysis of the molecular basis for octanal interactions in the expressed rat I7 olfactory receptor. Chem Senses 5: 155-165. [DOI] [PubMed] [Google Scholar]
- Sorensen PW, Caprio J (1998) Chemoreception. In: The physiology of fishes, Ed 2 (Evans DH, ed), pp 375-405. Boca Raton, FL: CRC.
- Sorensen PW, Hara TJ, Stacey NE (1991) Sex pheromones selectively stimulate the medial olfactory tracts of male goldfish. Brain Res 558: 343-347. [DOI] [PubMed] [Google Scholar]
- Speca DJ, Lin DM, Sorensen PW, Isacoff EY, Ngai J, Dittman AH (1999) Functional identification of a goldfish odorant receptor. Neuron 23: 487-498. [DOI] [PubMed] [Google Scholar]
- Tanabe Y, Masu M, Ishii T, Shigemoto R, Nakanishi S (1992) A family of metabotropic glutamate receptors. Neuron 8: 169-179. [DOI] [PubMed] [Google Scholar]
- Thermes V, Grabher C, Ristoratore F, Bourrat F, Choulika A, Wittbrodt J, Joly JS (2002) I-SceI meganuclease mediates highly efficient transgenesis in fish. Mech Dev 118: 91-98. [DOI] [PubMed] [Google Scholar]
- Vannier B, Peyton M, Boulay G, Brown D, Qin N, Jiang M, Zhu X, Birnbaumer L (1999) Mouse trp2, the homologue of the human trpc2 pseudogene, encodes mTrp2, a store depletion-activated capacitative Ca2+ entry channel. Proc Natl Acad Sci USA 96: 2060-2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Rekowski C, Zippel HP (1993) In goldfish the qualitative discriminative ability for odors rapidly returns after bilateral nerve axotomy and lateral olfactory tract transection. Brain Res 618: 338-340. [DOI] [PubMed] [Google Scholar]
- Weltzien FA, Höglund E, Hamdani EH, Døving KB (2003) Does the lateral bundle of the medial olfactory tract mediate reproductive behavior in male crucian carp? Chem Senses 28: 293-300. [DOI] [PubMed] [Google Scholar]
- Westerfield M (1995) The zebrafish book. Eugene, OR: University of Oregon.
- Yildirim E, Dietrich A, Birnbaumer L (2003) The mouse C-type transient receptor potential 2 (TRPC2) channel: alternative splicing and calmodulin binding to its N terminus. Proc Natl Acad Sci USA 100: 2220-2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Ito A, Matsuda N, Mishina M (2002) Regulation by protein kinase A switching of axonal pathfinding of zebrafish olfactory sensory neurons through the olfactory placode-olfactory bulb boundary. J Neurosci 22: 4964-4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara Y, Mizuno T, Nakahira M, Kawasaki M, Watanabe Y, Kagamiyama H, Jishage K, Ueda O, Suzuki H, Tabuchi K, Sawamoto K, Okano H, Noda T, Mori K (1999) A genetic approach to visualization of multisynaptic neural pathways using plant lectin transgene. Neuron 22: 33-41. [DOI] [PubMed] [Google Scholar]