Abstract
The role of glucocorticoids and the repressor isoform of cAMP response element (CRE) modulator (CREM), inducible cAMP early repressor (ICER), in limiting corticotropin-releasing hormone (CRH) transcription during restraint stress were examined in both intact and adrenalectomized rats receiving glucocorticoid replacement. CRH primary transcript, measured by intronic in situ hybridization, increased after 30 min of restraint and returned to basal levels by 90 min, despite the persistent stressor. The decline was independent of circulating glucocorticoids, because adrenalectomized rats displayed an identical pattern. ICER mRNA in the hypothalamic paraventricular nucleus (PVN) increased after 30 min and remained elevated for up to 4 h in a glucocorticoid-independent manner. Western blot and electrophoretic mobility shift assay analyses showed increases in endogenous ICER in the PVN of rats subjected to restraint stress for 3 h. Chromatin immunoprecipitation assays showed the recruitment of CREM by the CRH CRE in conjunction with decreases in RNA polymerase II (Pol II) binding in the PVN region of rats restrained for 3 h. These data show that stress-induced glucocorticoids do not mediate the limitation of CRH transcription. Furthermore, the ability of CREM to bind the CRH CRE and the time relationship between elevated CREM and reduced Pol II recruitment by the CRH promoter suggest that inhibitory isoforms of CREM induced during stress contribute to the decline in CRH gene transcription during persistent stimulation.
Keywords: corticotropin-releasing hormone, transcription, glucocorticoids, CREM, ICER, phospho-CREB
Introduction
Precise regulation of circulating glucocorticoids, the end product of the hypothalamic-pituitary-adrenal (HPA) axis, is critical for preserving homeostasis under stress conditions. The secretion of adrenal glucocorticoids depends on the levels of circulating pituitary ACTH, which in turn are regulated by the stimulatory action of hypothalamic corticotropin-releasing hormone (CRH) and vasopressin and inhibitory effects of glucocorticoid feedback (Aguilera, 1994; Dallman et al., 1994). In addition to ACTH regulation, CRH mediates behavioral and autonomic adaptation to stress (Vale et al., 1983; Reul et al., 2000). Although these responses are essential for homeostasis, excessive CRH production leads to HPA axis dysfunction and psychiatric, immune, and metabolic disturbances; therefore, limitation of the magnitude and duration of CRH responses is essential (McEwen, 1998; Schulkin et al., 1998). Intronic in situ hybridization studies in rats have shown rapid but transient increases in CRH transcription during stress (Ma and Aguilera, 1999b; Kovacs et al., 2000), and this time pattern may be important in preventing the deleterious effects of CRH overproduction.
Glucocorticoids reduce basal levels of CRH transcription and CRH mRNA levels in the paraventricular nucleus (PVN) (Itoi et al., 1998; Ma and Aguilera, 1999a,b; Ginsberg et al., 2003) and are a likely candidate for limiting CRH transcription during stress. However, variations in plasma glucocorticoids cannot always explain changes in HPA axis activity. For example, glucocorticoid administration in adrenalectomized rats does not prevent the increase in stress-induced CRH heteronuclear RNA (hnRNA) (Ma and Aguilera, 1999b), and adrenalectomy cannot reverse the inhibitory action of dehydration on CRH expression (Aguilera et al., 1983; Watts and Sanchez-Watts, 2002). These findings suggest the involvement of factors other than glucocorticoids in limiting CRH transcription during stress.
In addition to the activation of inhibitory neural pathways afferent to the PVN, it is possible that intracellular feedback contributes to the transient CRH responses. It has been shown that the induction by cAMP of repressor isoforms of cAMP response element (CRE) modulator (CREM), such as inducible cAMP early repressor (ICER), inhibit cAMP response element-binding protein (CREB)-stimulated transcription in neuroendocrine systems (Lalli and Sassone-Corsi, 1995; Luckman and Cox, 1995; Monaco et al., 1995; Della Fazia et al., 1998; Kovacs, 1998; Borsook et al., 1999; Mazzucchelli and Sassone-Corsi, 1999). ICER is produced via activation of the second promoter of the CREM gene (Foulkes et al., 1991; Molina et al., 1993). ICER contains the DNA-binding domain, but not the transactivation domain, of CREM, thereby acting as a competitive inhibitor of transcriptional activators that bind to the CRE (Foulkes et al., 1991). Transcriptional regulation of the CRH gene involves cAMP/CREB-dependent mechanisms and a functional CRE in the CRH promoter (Seasholtz et al., 1988). Although it has been shown that CREB phosphorylation coincides with elevations in CRH hnRNA in the PVN during ether exposure (Kovacs and Sawchenko, 1996; Kovacs, 1998), a lack of correlation between the decline in phospho-CREB and reduced CRH hnRNA levels suggests that the termination of CRH transcription involves not only decreases in phospho-CREB levels but also a transcriptional repressor. The purpose of these studies is to investigate the contribution of glucocorticoid feedback and transcriptional repression by ICER in limiting CRH transcription during restraint stress.
Materials and Methods
Animals and in vivo procedures. Adult male Sprague Dawley rats weighing 275-325 g were housed three per cage for at least 1 week before experimentation, with a 14/10 h light/dark cycle and food and water available ad libitum. To determine the time course of the changes in CREM mRNA and CRH hnRNA, rats were subjected to restraint stress by placing them into plastic restrainers (2.5 × 6 inches). Groups of four rats were killed by decapitation at 0.5, 1, 2, or 3 h after initiation of restraint. Control rats were rapidly removed from the cages and decapitated. Brains were immediately removed, frozen in isopentane at -40°C, and stored at -80°C until being sectioned in a cryostat. Trunk blood was collected in plastic tubes, and serum was separated by centrifugation and stored at -80°C for corticosterone determination. To determine the effect of stress-induced elevations in plasma glucocorticoids on the termination of the CRH transcriptional response to stress, 5 d adrenalectomized rats were subjected to 3 h of restraint and killed by decapitation, as described above. Bilateral adrenalectomy was performed via a dorsal approach under ketamine/xylazine anesthesia. Adrenalectomized rats received subcutaneous slow-release corticosterone pellets to maintain steady-state levels of circulating corticosterone. Pellets were constructed by dipping a 5 × 10 mm piece of Whatman (Maidstone, UK) GF/C glass-fiber filter paper into a molten mixture of corticosterone (66%) and cholesterol (33%). Sham-operated rats were subjected to the same surgical procedure without removing the adrenals. All experiments were performed in the morning, and rats were killed between 9:00 and 11:00 A.M. For in situ hybridization, brains were frozen by immersion for 10 s in isopentane at -40°C and stored at -80°C until processing. For electrophoretic mobility shift assays (EMSAs), Western blots, and chromatin immunoprecipitation (ChIP) assays, hypothalamic sections containing the PVN were obtained by microdissection of a brain coronal section (between the mammillary bodies and optic chiasm). Coronal slices were placed on an ice-cold Petri dish, and the region between the top and 0.5 mm at each side of the third ventricle was sectioned with a microblade, placed in microtubes on dry ice, and stored at -80 C until processing. All procedures were performed according to National Institutes of Health guidelines, and experimental protocols were approved by the National Institute of Child Health and Human Development Animal Care and Use Committee.
Corticosterone assay. Corticosterone concentration was determined in serum using a commercial radioimmunoassay kit (Diagnostic Products, Los Angeles, CA). The sensitivity of this assay is 6 ng/ml, and the intraassay and interassay coefficients of variation were 3.1 and 8.5%, respectively.
In situ hybridization histochemistry. Serial coronal sections (12 μm) were cryosectioned (-15°C), thaw mounted onto poly-l-lysine-coated slides, and stored at -80°C until being processed for in situ hybridization. One in six series of sections through the PVN were collected from each animal. Levels of CRH hnRNA and CREM mRNA in the PVN were measured by in situ hybridization. An intronic CRH probe, containing a 530 bp PvuII cDNA fragment consisting of the rat CRH intron subcloned into pGEM-3 used for in situ hybridization, was kindly provided by Dr. Robert Thompson (University of Michigan, Ann Arbor, MI). A 601 bp fragment of the 3′ end of the CREM gene consisting of the DNA-binding domain II subcloned into pBluescript SK was used for generation of in situ hybridization probes (Foulkes et al., 1991).
Transcription of antisense 35S-labeled probes was performed as described previously after linearization of the intronic CRH plasmid with XbaI and the plasmid containing the 601 bp CREM cDNA fragment with NcoI. One microgram of linearized cDNA template was transcribed with the T7 RNA polymerase transcription kit (Promega, Madison, WI) in 1× transcription buffer containing 10 mm dithiothreitol (DTT), 20 U of RNase inhibitor, 100 μCi of [35S]UTP, 100 μCi of [35S]CTP (specific activity, 1250 Ci/mm; PerkinElmer Life and Analytical Sciences, Boston, MA), 0.5 mmol each of rATP and rGTP, with 24 μm rCTP and rUTP in a volume of 10 μl. Transcription reactions were performed at 37°C for 60 min. At this time, 20 U of DNase I were added with 10 U of RNase inhibitor, and the cDNA template was digested for 10 min at 37°C. cRNA probes were purified by gel-filtration column chromatography (Roche Diagnostics, Indianapolis, IN).
Prehybridization and hybridization procedures were performed as described previously (Luo et al., 1994). Briefly, before hybridization, sections were thawed at room temperature, fixed in 4% formaldehyde for 5 min at room temperature, washed three times each with PBS, and acetylated for 10 min at room temperature in 0.25% acetic anhydride in 0.1 m triethanolamine/0.9% NaCl, pH 8.0. Sections were dehydrated and delipidated by sequential transfers through ethanol and chloroform and air dried before hybridization. Sections were hybridized at 55°C overnight with 2 × 106 cpm of 35S-labeled probe, and then nonspecifically bound probe was removed by washing in 50% formamide/250 mm NaCl at 60°C for 10-15 min, followed by RNase A treatment for 30 min at 37°C, and three washes in 0.1× SSC at 50°C for 15 min. After the final wash, sections were dried overnight at room temperature, opposed to BioMax MR film (Eastman Kodak, Rochester, NY) for 3 weeks, developed, and fixed. Slides for each probe in each experiment were processed in a single in situ hybridization reaction.
To confirm that the induction of CREM mRNA involves increases in the cAMP-inducible isoform ICER, sets of sections were subjected to in situ hybridization using a 48-mer directed against the ICER-specific region located downstream of promoter 2 in the intron. The sequence of the oligonucleotide was CAGTTTCATC TCCAGTTACA GCCATGTTGG GCTTTTGCAT ACAGAGTG (Spencer and Houpt, 2001). Oligonucleotides were labeled at the 3′ end using [35S]deoxyATP (PerkinElmer Life and Analytical Sciences) and terminal DNA transferase (Invitrogen, Carlsbad, CA). In situ hybridization was performed as described previously for oligoprobes (Aguilera and Kiss, 1993). For quantification of CRH hnRNA and CRH mRNA in the PVN, sections were exposed to Kodak BioMax film together with 14C-labeled standards (American Radiolabeled Chemicals, St. Louis, MO) for 15 d for CRH hnRNA, 11 d for CREM mRNA, and 21 d for the ICER oligoprobe. The optical density of film autoradiographic images was measured in a computerized image-analysis system (Imaging Research, St. Catharines, Ontario, Canada), using the public-domain NIH Image program (developed at the National Institutes of Health and available at http://rsb.info.nih.gov/nih-image). Optical densities obtained in two consecutive sections per rat were averaged and used to calculate group means. The results are presented as mean ± SE of the percentage of change from basal levels in sham-operated rats.
Colocalization of CREM and CRH mRNA in parvocellular neurons was determined by double-label in situ hybridization histochemistry, as described previously (Morales and Bloom, 1997). Two radiolabeled CREM riboprobes were generated using either [35S]UTP or [33P]UTP, and a digoxigenin-labeled CRH riboprobe was prepared using kit reagents from Roche Diagnostics. Sections were incubated in 0.1 m phosphate buffer (PB), pH 7.4, containing 0.5% Triton X-100 for 10 min, rinsed three times for 10 min each in PB, treated with 0.2N HCl for 15 min, and washed three times for 10 min each in PB, followed by acetylation with 0.25% acetic anhydride in 0.1 m triethanolamine, pH 8.0, for 10 min. After three washes for 10 min in PB, sections were postfixed in 4% paraformaldehyde for 10 min, rinsed three times for 10 min each in PB, and prehybridized for 2 h at 55°C in a buffer containing 50% formamide, 10% dextran sulfate, 5× Denhart's solution, 0.62 m NaCl, 50 mm dithiothreitol, 10 mm EDTA, 20 mm PIPES, 0.2% SDS, 250 μg/ml salmon-sperm DNA, and 250 μg/ml tRNA. This was followed by incubation for 18 h at 55°C in hybridization buffer containing 107 cpm/ml of each radiolabeled CREM probe and 5 μl/ml of the digoxigenin-labeled CRH riboprobe. After hybridization, sections were washed in 2× SSC containing 10 mm β-mercaptoethanol for 30 min at room temperature, followed by RNase A treatment (5 μl/ml) for 60 min at 37°C and consecutive washes with 0.5× SSC with 10 mm β-mercaptoethanol and 0.5% Sarkosyl at 55°C for 2 h, 0.1× SSC containing 10 mm β-mercaptoethanol and 0.5% Sarkosyl at 60°C for 1 h, and 6× PB for 10 min. After 1 h of incubation in blocking buffer (2% BSA, 0.1 m Tris, 0.3% Triton X-100, and 2 mm NaN3), sections were incubated with alkaline phosphatase-conjugated antidigoxigenin antibody (1:3000; Roche Diagnostics) in blocking buffer for 18 h at 4°C. To visualize digoxigenin staining, sections were washed three times for 10 min each with Tris buffer (0.1 m Tris, 0.5 mm MgCl, and 0.15 mm NaCl, pH 9.5) and developed in the dark for 7 h with nitroblue tetrazolium salt and 0.18 mg/ml 5-bromo-4-chloro-3-indolyl phosphate toluidinium salt (Roche Diagnostics). Development was stopped by four washes for 10 min each in PB. Sections were then dehydrated with ethanol (30, 60, 90, 95, and 100%), dipped in Ilford K.5D nuclear emulsion (Polysciences, Warrenton, PA), developed after a 4 week exposure, and examined by light microscopy. CRH cells were readily identified by the dark purple staining. CREM staining was identified by clusters of grains using a Micro Video Instruments Darklite illumination system (Image Systems, North Potomac, MD) or epipol superimposed with transmitted light in a bright field (Zeiss, Thornwood, NY). Analysis was performed at the level of the medial parvocellular PVN selected from serial hypothalamic sections for each rat. The number of cells stained with digoxigenin CRH, with or without CREM mRNA, was counted bilaterally in the PVN in each rat using computer-captured images from the emulsion-dipped slides in the Zeiss microscope and averaged to obtain the values for each experimental group. The background for CREM mRNA hybridization outside the brain section was 7.3 ± 1.5 grains/mm2, and CRH-labeled neuronal cross sections containing more than threefold the background density were considered positive for CREM.
Electrophoretic mobility shift assay. Nuclear extracts from pools of hypothalamic fragments containing the PVN from 10 controls or stressed rats were prepared using a NE-PER Nuclear and Cytoplasmic Extraction Reagents kit (Pierce, Rockford, IL) according to the protocol of the manufacturer. Protein concentration was quantified by spectrophotometry using the BCA Protein Assay (Pierce) before being used for EMSA or Western blots. Double-stranded oligonucleotide probes containing the native CRH CRE (5′-TCGTTGACGTCACCAA-3′) and mutant CRH CRE (5′-TCGTAGAGAGCTCCAA-3′) were synthesized as complementary single strands and annealed at 80°C for 20 min, followed by slow cooling to room temperature overnight. Aliquots of 10 pmol of CRE double-stranded oligonucleotides were end labeled using T4 polynucleotide kinase (New England BioLabs, Beverly, MA) and 10 pmol of [γ - 33P]ATP (PerkinElmer Life and Analytical Sciences) for 20 min at 37°C in a total volume of 20 μl. Labeled CRE probe was purified by MicroSpin G-25 columns (Amersham Biosciences, Piscataway, NJ). Three to 5 μg of nuclear extract were incubated in 10 mm Tris-HCl, pH 7.5, containing 0.5 mm EDTA, 5 mm MgCl2, 50 mm NaCl, 4% glycerol, 0.5 mm DTT, 0.05 μg/μl poly (deoxyinosinic-deoxycytidylic) acid, and 25,000 cpm of radiolabeled oligonucleotide in a total volume of 20 μl for 20 min at room temperature. For super shift, 1 μl of the appropriate antibody was added before probe to the reaction. For competition, un-labeled oligonucleotides (30× and 100×) were added before incubation with the probe. Samples were electrophoresed on an 8% polyacrylamide gel (29:1 acrylamide/bis-acrylamide) in 0.5× Tris-borate EDTA (TBE) for 50 min at room temperature at 250 V. Gels were vacuum-heat dried, and bands were visualized and quantified using a Molecular Dynamics (Sunnyvale, CA) PhoshorImager after overnight exposure at room temperature.
Western blot. For Western blot, 50 μg of nuclear extract was loaded and separated in a 10-20% gradient SDS-PAGE. Proteins were transferred to a polyvinylidene difluoride membrane (PerkinElmer Life and Analytical Sciences), incubated with 5% blocking agent in 1× TBST (TBS plus 0.05% Tween 20) for 1 h, and incubated overnight with anti-CREM antibody (at a 1:1000 dilution; Santa Cruz Biotechnology, Santa Cruz, CA), which recognizes all CREM isoforms including ICER, or with anti-phospho-CREB antibody (at a 1:1000 dilution; Upstate Biotechnology, Lake Placid, NY). After washing in 1× TBST, membranes were incubated for 1 h at room temperature with horseradish peroxidase-conjugated donkey anti-rabbit IgG at a 1:20,000 dilution. Detection of immunoreactive CREM isoform bands was performed by using ECL Plus TM reagents (Amersham Biosciences), followed by exposure to BioMax MR film (Eastman Kodak). After film exposure, blots were stripped and assayed for tubulin using a monoclonal antibody (Sigma, St. Louis, MO) to correct for protein loading in the gel. The intensity of the bands was quantified using the computer image-analysis system (Imaging Research) and the public-domain NIH Image program described for the analysis of in situ hybridization films.
Chromatin immunoprecipitation assay. To determine whether CREM produced 3 h after stimulation can bind the CRH promoter in vivo, we performed ChIP assays in pooled dissected hypothalamic tissue of three controls and rats subjected to restraint stress for 15 min or 3 h. ChIP assays were performed according to the protocol of the manufacturer (Upstate Biotechnology) with modifications as indicated below. Microdissected hypothalamic tissue, containing the PVN, was cross-linked by the addition of 1% formaldehyde in PBS and incubation for 10 min at room temperature. After washing three times with ice-cold PBS containing 1× protease inhibitor mixture (Roche Diagnostics) and 1× phosphatase inhibitor mixture (Sigma), each PVN was resuspended in 250 μl of SDS lysis buffer containing protease and phosphatase inhibitors. After 20 min of incubation on ice, tissues were sonicated using a Sonicator 3000 (Misonix, Farmingdale, NY) for 10 s at output level 2, and sonicated four additional times for 10 s at output level 1.5 to generate 0.5-2 kb DNA fragments. Homogenized tissue was centrifuged for 10 min at 16,000 × g to remove debris, and supernatants were diluted 10-fold in a ChIP dilution buffer containing protease and phosphatase inhibitors. After removing 50 μl to quantify the DNA input, chromatin solutions were immunocleared by the addition of 75 μl of salmon-sperm DNA/protein A-agarose beads and incubation for 1 h at 4°C with rotation. Immunoprecipitation was performed in 500 μl of precleared sample by incubation at 4°C with affinity-purified anti-CREM antibody (Santa Cruz Biotechnology), anti-RNA polymerase II (Pol II) antibody (Santa Cruz Biotechnology), or anti-phospho CREB antibody (Upstate Biotechnology) at a concentration of 1:100 under rotation. After overnight incubation, 65 μl of salmon-sperm DNA/protein A-agarose slurry was added and incubated for 1 h at 4°C under rotation, and DNA-protein complexes were separated by centrifugation at 100 × g for 3 min. Pellets were washed sequentially in low-salt immune-complex wash buffer, high-salt immune-complex wash buffer, and LiCl immune-complex wash buffer (Upstate Biotechnology) for 3-5 min, followed by two washes in Tris-EDTA buffer and elution twice with 250 μl of 1% SDS in 0.1 m NaHCO3. Aliquots of eluate pools and input chromatin were incubated at 65°C for 4-18 h in 0.2 m NaCl to reverse cross-linking and then incubated at 45°C for 1 h in 40 μm Tris-HCl, pH 6.5, containing 10 μm EDTA and 20 μg of proteinase K to remove proteins. DNA fragments were purified with phenol/chloroform/isoamylalcohol (25:24:1) and ethanol precipitation and reconstituted in H2O and subjected to PCR using the following primers designed to amplify the CRH promoter region encompassing the 230 bp containing the CRE (forward, 5′-CTACCTTCTTGGCAGCTCTGC-3′; reverse, 5′-GAATCTCACATCCAATTATATC-3′). For negative controls, cross-linked DNA was subjected to immunoprecipitation with an affinity-purified antibody produced in rabbit against the C terminus of the rat V1b receptor (C. Rabadan-Diehl and G. Aguilera, unpublished observations). PCR conditions for the CRH promoter region consisted of 32 cycles each for 30 s at 95°C, 30 s at 55°C, and 45 s at 72°C, followed by a 7 min extension at 72°C. To test the specificity of the PCR product for the CRH promoter, DNA samples from the PVN immunoprecipitated with phospho-CREB, CREM, and Pol II antibodies were subjected to PCR using primers for the CRH coding region (forward, 5′-CTCTCTGGATCTCACCTTCCAC-3′; reverse, 5′-CTAAATGCAGAATCGTTTTGGC-3′). As an additional specificity control, DNA subjected to immunoprecipitation with CREM antibody was subjected to PCR using the following primers directed against the proximal vasopressin V1b receptor promoter: forward, 5′-CCTTTAGGGCATGCTTCTCAG-3′; reverse, 5′-TAGGAAACCCAGTCGGAGAA-3′. These primers yield a 430 bp PCR product of the proximal region of the V1b receptor promoter, which does not contain any CRE or CRE-like motifs (Rabadan-Diehl et al., 2000). PCR conditions for the CRH coding region were 35 cycles of 30 s at 95°C, 30 s at 55°C, and 45 s at 72°C, followed by a 7 min extension at 72°C. Similar conditions were used for the V1b receptor promoter, with the exception that the annealing temperature was 65°C instead of 55°C. PCR conditions for the input DNA were the same but used 20 cycles.
Statistical analysis. Statistical significance of the differences between groups was calculated by one-way ANOVA, followed by Bonferroni's adjusted pairwise comparisons. Statistical significance was set at p < 0.05. Data are presented as means ± SEM from the values in the number of observations indicated in Results or in Figures 1, 2, 3, 4, 5, 6, 7.
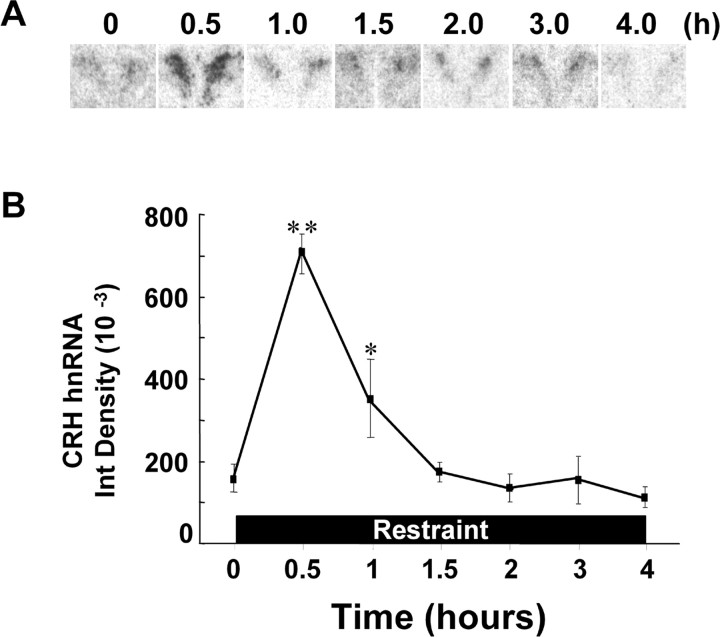
Figure 1.
Changes in CRH hnRNA in the PVN during 4 h of restraint stress, measured by intronic in situ hybridization. Rats were placed in plastic restrainers and killed by decapitation at the times indicated. Images are representative of five rats per experimental group. Data points are the mean ± SE of integrated density (Int Density) values in five rats per group. *p < 0.01 compared with basal; **p < 0.001 compared with basal. Error bars represent SEM.
Figure 2.
Glucocorticoids do not mediate the rapid decline in CRH transcription during stress. A, B, Plasma corticosterone (Cort; A) and CRH hnRNA levels in the PVN (B) during the course of 3 h of restraint stress in sham-operated (Sham) and adrenalectomized (ADX) rats receiving constant levels of corticosterone via subcutaneous implants. Rats were killed by decapitation at the indicated time points. Data points are the mean ± SE of the values obtained in five rats per experimental group. *p < 0.001 compared with basal; #p < 0.05 compared with basal sham-operated rats. Int Density, Integrated density. Error bars represent SEM.
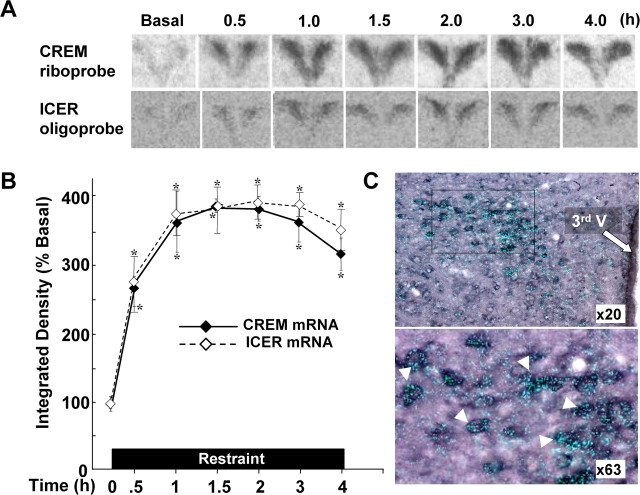
Figure 3.
Changes in CREM/ICER mRNA measured by in situ hybridization using either a riboprobe recognizing all forms of CREM (CREM mRNA) or a 48-mer oligoprobe specific for ICER (ICER mRNA). A, Representative images from film autoradiography of one of three rats per experimental group for CREM mRNA and five rats for ICER mRNA. B, Data points are the mean ± SE of the pooled data, expressed as percentage of basal values. Basal integrated density values were 7983 and 1019 arbitrary units for CREM mRNA and ICER mRNA, respectively. *p < 0.001 compared with basal. C, Cellular localization of CREM mRNA in PVN sections of rats subjected to restraint stress for 3 h. Double-labeling in situ hybridization using 35S-labeled CREM cRNA probes (bright green grains) and digoxigenin-labeled CRH cRNA probes shows 35S-CREM staining (bright grains) overlaying digoxigenin-labeled parvicellular CRH neurons. 3rd V, Third ventricle. C, Bottom, Higher magnification of the area enclosed in the square. The white arrowheads indicate some of the CRH neurons labeled with CREM. CREM staining in control nonstressed rats was no different from background staining [12.3 ± 1.8 and 9.6 ± 1.4 grains/cell (data not shown)]. Error bars represent SEM.
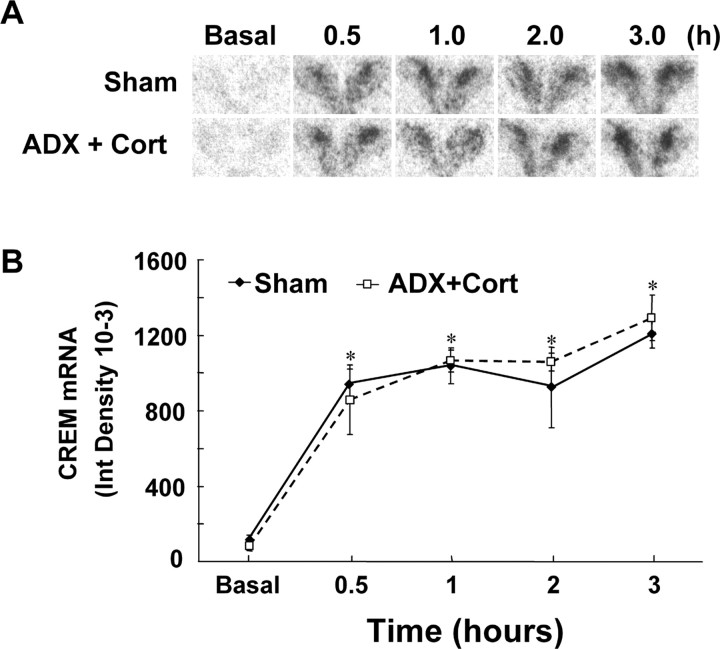
Figure 4.
Increases in CREM expression during stress are independent of glucocorticoids. Changes in CREM mRNA during restraint stress in sham-operated (Sham) or adrenalectomized (ADX) rats receiving constant low levels of corticosterone (Cort) replacement via subcutaneous implants are shown. A, Representative autoradiographic images of three rats per experimental group. B, Data points are the mean ± SE of the integrated density (Int Density) values in three rats per experimental group. *p < 0.001 versus basal in sham-operated and adrenalectomized rats; treatment had no effect (adrenalectomy). Error bars represent SEM.
Figure 5.
Restraint stress leads to sequential increases in phospho-CREB and CREM protein in the hypothalamic PVN region. A, Western blot analysis of phospho-CREB and CREM in nuclear extracts of microdissected PVN of rats under basal conditions and after 15 min or 3 h of restraint stress. Data were obtained from pooled hypothalamic tissue from 10 rats per experimental group. Protein loading in the gel was verified by tubulin levels, as shown in the bottom row. B, Semiquantitative analysis of the ICER bands, expressed as percentage change from basal values. Error bars represent SEM.
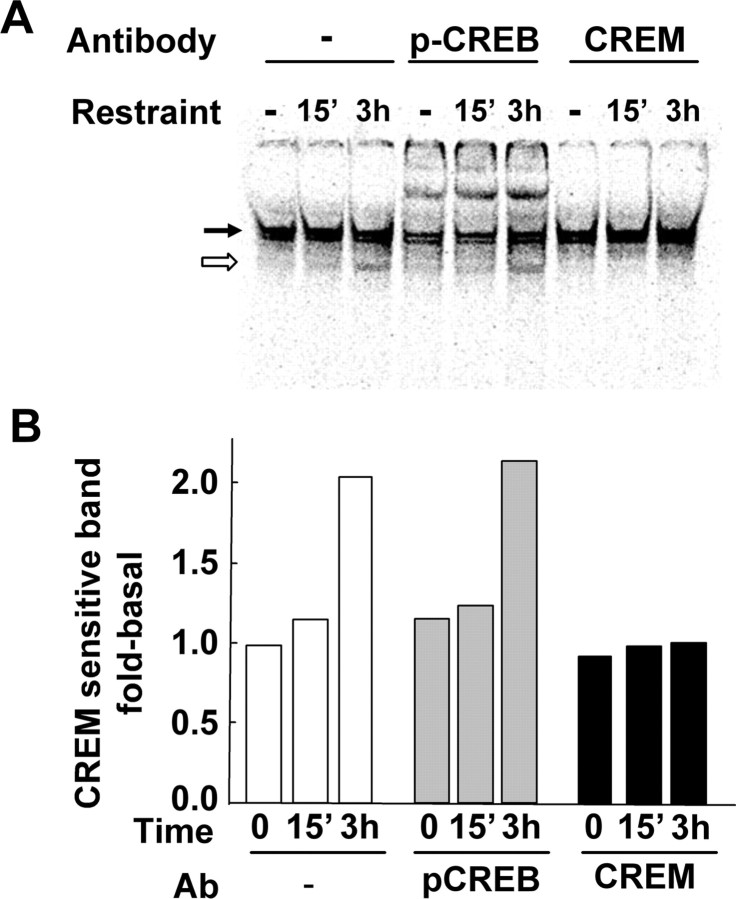
Figure 6.
A, Gel-shift assay and supershift of CRH CRE-radiolabeled oligonucleotides and nuclear proteins from microdissected PVNs from 10 rats per group of control rats or rats subjected to restraint stress for 15 min or 3 h. The black arrow indicates the major shifted band that is supershifted with phospho-CREB antibody. The white arrow shows a faster-migrating band that was supershifted by the CREM antibody. The image is representative of three similar experiments. The semiquantitative analysis of this band in the presence or absence of phospho-CREB and CREM antibodies is shown in B. Ab, Antibody. Data are expressed as fold-change from basal without antibody.
Figure 7.
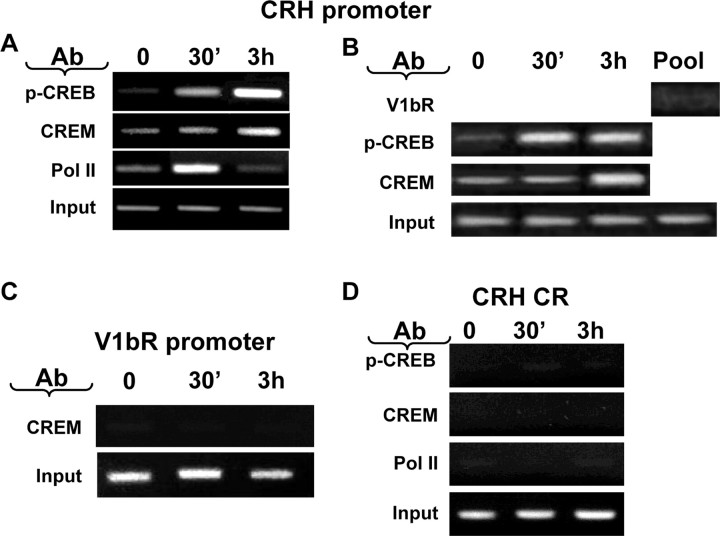
Restraint stress induces recruitment of phospho-CREB and CREM by the CRH promoter in the hypothalamic PVN region. A, ChIP assays using phospho-CREB, CREM, and Pol II antibodies and cross-linked DNA from the microdissected hypothalamic PVN region of control rats and rats subjected to restraint stress for 30 min or 3 h. Similar increases in CREM recruitment by 3 h are shown in B. B, PCR for the CRH promoter in the pooled remaining cross-linked DNA for this experiment (Pool), immunoprecipitated with an unrelated antibody against a polypeptide corresponding to the C terminus of the vasopressin V1b receptor, yielded no PCR band. C, No PCR product was found using primers against the V1b receptor on DNA immunoprecipitated with CREM antibody. D, PCR for the CRH coding region (CR) performed on the immunoprecipitants in A showed nonspecific bands, which did not change with stress. Gel images are representative of the results in three experiments. Ab, Antibody; p-CREB, phospho-CREB; V1bR, vasopressin V1b receptor.
Results
Effects of prolonged stress and glucocorticoids on CRH transcription
Levels of CRH hnRNA were low under basal conditions, increased by 4.3-fold after 30 min of restraint stress (p < 0.0001 compared with basal), declined by 60 min, and returned to near basal levels by 90 min, despite the continuous presence of the restraint stress for 4 h (Fig. 1). To determine the involvement of the stress-induced glucocorticoid surge on the decline in the CRH transcriptional response, we measured CRH hnRNA responses to prolonged restraint in adrenalectomized rats receiving glucocorticoid replacement. Sham-operated rats showed marked and sustained increases in plasma corticosterone in response to restraint (p < 0.0005), and, as expected, adrenalectomy prevented these increases. Adrenalectomized rats with glucocorticoid replacement showed constant serum corticosterone levels, which were slightly but significantly elevated compared with the resting levels in controls (p < 0.02) (Fig. 2A).
Despite the lack of glucocorticoid surge in adrenalectomized rats, CRH hnRNA responses in the PVN showed a pattern similar to that of sham-operated rats by 30 min (5.2 ± 1.2 and 7.2 ± 0.9 for sham-operated and adrenalectomized rats, respectively) and returned to near basal levels by 1 h (Fig. 2B). These results indicate that the elevation in circulating glucocorticoids induced by restraint stress is not responsible for the rapid decline in CRH transcription.
Effect of stress and glucocorticoids on CREM mRNA expression
To assess the involvement of repressor isoforms of CREM limiting CRH transcription responses to stress, levels of CREM mRNA were measured in the PVN of rats subjected to prolonged restraint stress. As shown in Figure 3, A and B, CREM mRNA was low under basal conditions and increased markedly by 30 min, with a distribution pattern consistent with CRH neurons; levels remained elevated at similar values for the 4 h duration of the stress (p < 0.0001). Similar results were obtained using an ICER-specific oligoprobe for the in situ hybridization (Fig. 3A, B). To confirm that the increases in CREM mRNA expression during stress occur in CRH neurons, we performed double-staining in situ hybridization using digoxigenin-labeled CRH and 35S-labeled CREM riboprobes in hypothalamic sections of rats subjected for 3 h to restraint stress. As shown in Figure 3C, the concentration of bright grains corresponding to CREM mRNA silver staining was much higher over cells labeled for CRH mRNA than in the surrounding areas. Of the CRH mRNA-stained cells, 95.3 ± 2.4% were also labeled for CREM mRNA. The average of the number of grains overlaying CRH cells was 33.5 ± 0.8 per cell (n = 50 cells), whereas the number in equivalent neighboring areas was 9.5 ± 0.78. As in the film analysis, silver grain density overlaying CRH mRNA-stained neurons of control nonstress rats was low and not significantly different from the background [12.3 ± 1.8 and 9.6 ± 1.4 grains per cell or equivalent area (data not shown in Fig. 3)]. CREM mRNA expression was identical in sham-operated and adrenalectomized rats receiving replacement levels of corticosterone (Fig. 4A, B).
Effects of stress on CREM protein levels in the PVN
Western blot analysis of PVN nuclear extracts revealed a marked increase in a 43 kDa band corresponding to phosphorylated CREB by 15 min after restraint of stress. Phosphorylated CREB remained elevated for the duration of the experiment, up to 3 h (Fig. 5A). Western blot using the CREM antibody revealed the expected bands corresponding to the different isoforms of CREM, of which three bands with molecular weights of ∼18, 15, and 13.5 kDa were consistent with the migration pattern of ICER (Molina et al., 1993). These bands increased markedly after 3 h of restraint (Fig. 5A, B). The molecular size of the higher ICER band was identical to that observed in protein extracts from the hypothalamic cell line (4B) transfected with ICER I (N. Kalitchenko, Y. Liu, and Aguilera, unpublished observations).
The ability of phospho-CREB and CREM to interact with the CRH CRE was studied by EMSA using hypothalamic nuclear extracts from control and stressed rats and 33P-labeled double-stranded oligonucleotide corresponding to the rat CRE sequence. EMSA revealed two adjacent major shifted complexes of high density. These bands remained similar in intensity during stress and were supershifted with the phospho-CREB antibody (Fig. 6A). An additional faster-migrating band of lower intensity became evident after 15 min of restraint stress. The intensity of this band increased by approximately twofold at 3 h of restraint stress, and it was abolished by an anti-CREM antibody that recognizes ICER and other CREM isoforms but not by the phospho-CREB antibody (Fig. 6A, B). Similar to observations in cells incubated with forskolin for 3 h (Kalitchenko et al., 2003), overexposure of the film revealed an additional faster-migrating band, which was supershifted with CREM antibody. It was not possible to quantify this band because of interference from the high background of the film after overexposure (data not shown).
Recruitment of CREM by the CRH promoter
To determine whether CREM is recruited by the CRH promoter during stress, we performed ChIP assays using cross-linked DNA from the PVN region of rats subjected to restraint stress for 15 min or 3 h. As shown in Figure 7A, CRH promoter immunoprecipitation with the phospho-CREB antibody increased by 15 min of stress in correspondence with the increase in Pol II, but the recruitment of Pol II decreased by 3 h, despite a progressive increase in phospho-CREB. However, the amount of CRH promoter immunoprecipitated with the CREM antibody increased by 3 h in association with the decrease in Pol II recruitment. Similar results were observed in an additional experiment using phospho-CREB and CREM antibodies (Fig. 7B). Confirming the specificity of the protein binding to the CRH promoter, only a negligible PCR band was observed in the pooled remaining cross-linked DNA from the different groups after immunoprecipitation with a nonrelated affinity-purified antibody (Fig. 7B). No significant PCR product was observed in DNA immunoprecipitated with CREM antibody using PCR primers against a region of the vasopressin V1b receptor promoter lacking a CRE (Fig. 7C). Similarly, ChIP assay using primers against the CRH coding region in DNA immunoprecipitated with phospho-CREB, CREM, or Pol II antibodies yielded almost undetectable PCR bands that remained unchanged during stress (Fig. 7D).
Discussion
Previous reports using short-lasting stressors have suggested that the activation of CRH transcription rapidly returns to basal levels after stress (Kovacs et al., 2000; Ma et al., 2001). The present studies extend this observation by showing that the activation of CRH transcription is transient, despite the continuous presence of restraint stress. The major aim of this study was to identify possible mechanisms limiting CRH transcription, using restraint stress as an experimental model. The present data provide evidence (1) that the transient nature of CRH transcription during restraint stress is not mediated by the glucocorticoid surge and (2) that the repressor isoform of CREM, ICER, is induced during stress and that CREM is capable of binding to the CRH promoter and inhibiting CRH transcription. In addition, the presence of high levels of phosphorylated CREB at 3 h of restraint indicates that the decline in CRH transcription is not solely attributable to the desensitization of signaling pathways or to reduced phospho-CREB levels. Although it is likely that presynaptic mechanisms such as desensitization of afferent pathways to the PVN and/or activation of inhibitory pathways (e.g., GABAergic) (Bartanusz et al., 2004; Herman et al., 2004) contribute to limiting CRH transcriptional responses to stress, the present data suggest that CREM-mediated repression plays a role in this process.
Studies in rats have shown that elevated levels of circulating glucocorticoids decrease basal levels of CRH transcription (Itoi et al., 1998; Ma et al., 2001; Ginsberg et al., 2003) and CRH mRNA levels in the PVN (Jingami et al., 1985; Sawchenko, 1987; Lightman and Young, 1989; Swanson and Simmons, 1989). However, although some observations suggest that glucocorticoids limit the stress response (Kovacs and Sawchenko, 1996), others show a lack of effect of glucocorticoids or even a facilitation of the CRH transcriptional response to stress (Ma and Aguilera, 1999b; Watts and Sanchez-Watts, 2002). It is evident from the present data using adrenalectomized rats that restraint stress-induced CRH hnRNA levels (and presumably CRH transcription) decline despite the lack of glucocorticoid surge. Because rats were maintained under low levels of glucocorticoid replacement, it is not possible to rule out that basal steroid levels have a permissive role in limiting CRH transcriptional responses by interacting with an additional repressor. Nevertheless, it is clear from the results that stress-induced increases in circulating glucocorticoids are not responsible for limiting CRH responses. This indicates that, although glucocorticoid feedback may have a role in limiting the sensitivity and magnitude of CRH transcription responses (Ma and Aguilera, 1999b; Helmreich et al., 2001), additional mechanisms must be involved.
Because cAMP is an important regulator of CRH transcription, we sought the possible involvement of the cAMP-inducible repressor ICER in the regulation of CRH transcription. ICER, a product of activation of the second promoter of the CREM gene, has been reported to repress cAMP-induced transcription in other neuroendocrine tissues. For example, in the pineal gland, ICER is involved in the repression of melatonin synthesis during the course of normal circadian rhythm (Foulkes et al., 1996) and is involved in the desensitization of thyroid-stimulating hormone and follicle-stimulating hormone receptors in the pituitary and Sertoli cells, respectively (Lalli and Sassone-Corsi, 1995; Monaco et al., 1995). In pheochromocytoma PC12 cells, ICER inhibits PKA-stimulated transcriptional activity of tyrosine hydroxylase (Piech-Dumas and Tank, 1999). ICER has also been implicated in the regulation of ACTH secretion from a pituitary-derived corticotroph cell line (Lamas et al., 1997). Supporting a role for ICER in limiting HPA axis activity, a recent study showed that the antidepressant desipramine blunted plasma corticosterone and hypothalamic CRH mRNA responses to swim stress in wild-type mice, but not in CREM knock-out, ICER-deficient mice (Conti et al., 2004).
The present study shows that stress induces ICER in the PVN and that CREM can interact with the CRH promoter. This was shown by the increases in ICER mRNA and ICER protein in the Western blot and the ability of CREM antibodies to supershift the fast-migrating bands observed in gel-shift assays from PVN nuclear extracts of stressed rats. Although the CREM antibody recognizes all CREM isoforms, the presence of lower-migrating bands in the EMSA at the time when ICER levels increase suggests that the CRH CRE does indeed interact with ICER. In addition, the increased CREM recruitment by the CRH promoter at 3 h of stress, associated with reduced Pol II recruitment, strongly suggests that CREM interacts with the CRH promoter and represses CRH transcription in vivo. Because ICER is the only CREM isoform increasing at 3 h of restraint stress, it is likely that the CREM antibody-sensitive protein immunoprecipitated in the ChIP assay corresponds to ICER. ICER inhibits cAMP-inducible genes by competing with the transcriptional activators CREB and CREM to CRE or CRE-like sequences in the promoter of target genes (Sassone-Corsi, 1998). Because ICER lacks the CREM transactivation domain, binding of ICER to a CRE impairs recruitment of CREB or the CREB-binding protein (p300) to activate transcription. The binding of CREM to the CRH CRE in the EMSA and the ability of the CREM antibody to immunoprecipitate the region of the CRH promoter containing the CRE suggest that CREM, and probably ICER, can modulate CRH transcription by interacting with the CRE in the promoter. In this regard, recent in vitro studies using CRH promoter-luciferase constructs show that ICER inhibits forskolin-stimulated CRH promoter activity, an effect that is abolished by mutation of the CRE (Kalitchenko et al., 2003). The migration and supershift patterns of PVN nuclear protein complexes from stressed rats are identical to those observed in gel-shift assays from nuclear extracts of hypothalamic cell lines stimulated with forskolin or cells transfected with ICER (Kalitchenko et al., 2003). The rapid migration and supershift patterns of the lower shifted band suggest that it corresponds to smaller DNA-protein complexes, which could correspond to ICER homodimers. In contrast, the bands migrating just below the major shifted complex could be CREM-ICER heterodimers. Because these bands were not supershifted by the phospho-CREB antibody, it is unlikely that the complexes contain phospho-CREB. An unexpected finding was the lack of correlation between levels of phospho-CREB and the pattern of CRH transcription. Although the increase in phospho-CREB at 30 min correlated with transcriptional activation, reflected by the levels of CRH hnRNA and Pol II recruitment by the CRH promoter, these parameters declined at 3 h of restraint stress, despite continually increasing levels of phospho-CREB. Although the significance of the elevated phospho-CREB levels at 3 h remains to be elucidated, the data indicate that the decline in CRH transcription is unlikely to result from the reduction of phospho-CREB levels or dissociation of phospho-CREB from the promoter but that it could involve active repression by another transcription factor such as ICER.
The major neural pathways mediating the activation of CRH neurons (and probably CRH transcription) during stress are norepinephrinergic, glutamatergic, and serotoninergic (Pacak and Palkovits, 2001; Herman et al., 2004). The signaling mechanism of these neurotransmitters does not involve cAMP (Lefkowitz and Caron, 1988a,b; Aarts and Tymianski, 2003). However, they could induce CREB phosphorylation through alternative pathways, such as the transactivation of the mitogen-activated protein kinase cascade, and activate CRE-dependent transcription (Luttrell, 2002; Pokorska et al., 2003) or calcium-calmodulin-dependent kinase IV (Tiraboschi et al., 2004). Although norepinephrine preferentially stimulates α-adrenergic receptors, it could also cause small increases in cAMP by activation of β-adrenergic receptors (Lefkowitz and Caron, 1988a). Studies using the hypothalamic cell line 4B have shown that minimal elevations in intracellular cAMP are sufficient to elicit full activation of a CRH promoter-luciferase reporter in the hypothalamic cell line 4B (Nikodemova et al., 2003). It has been shown that stimulation of CRH transcription by depolarization or calcium-dependent signaling involves the potentiation of cAMP-dependent transcriptional activation (Guardiola-Diaz et al., 1994). Therefore, the stimulation of calcium-dependent pathways by stress may potentiate basal levels of cAMP-dependent transcription. An additional mechanism stimulating cAMP signaling in the CRH neuron involves CRH itself. Morphological studies have shown contact of CRH-containing terminals, probably dendritic, with CRH cell bodies (Rho and Swanson, 1987; Liposits et al., 1988). In addition, stress causes marked increases in the expression of CRH-R1, and the cell content of these receptors increases markedly during stress (Luo et al., 1994). Thus, CRH release within the PVN will increase cAMP levels, and this may sequentially facilitate and then limit CRH transcription, first via CREB phosphorylation and later via induction of the repressor ICER.
In summary, the present data show that the activation of CRH transcription during restraint stress is transient and that transcriptional inhibition is independent of the stress-induced glucocorticoid surge or the levels of phosphorylated CREB. The increase in ICER expression and recruitment of CREM by the CRH promoter late during restraint stress, in conjunction with the demonstrated ability of ICER to reduce basal and cAMP-induced CRH promoter activation (Kalitchenko et al., 2003), suggest that ICER plays a role in the regulation of CRH transcription. The data suggest that fine modulation of CRH expression depends on a complex balance between activation by phospho-CREB and repression by ICER. Induction of ICER could serve as a protective mechanism to limit transcription of hypothalamic CRH and avoid pathological consequences of excessive expression of the neuropeptide.
Footnotes
We thank Dr. Ana da Costa for her help with the in situ hybridization.
Correspondence should be addressed to Dr. Greti Aguilera, Section on Endocrine Physiology, Developmental Endocrinology Branch, National Institute of Child Health and Human Development, National Institutes of Health, Building 10, Room 10N262, Bethesda, MD 20892-1862. E-mail: Greti_Aguilera@nih.gov.
J. D. Shepard's present address: Cellular Neurobiology Research Branch, Intramural Research Program, National Institute on Drug Abuse, National Institutes of Health, Baltimore, MD 21224.
Copyright © 2005 Society for Neuroscience 0270-6474/05/254073-09$15.00/0
J.D.S. and Y.L. contributed equally to this work.
References
- Aarts MM, Tymianski M (2003) Novel treatment of excitotoxicity: targeted disruption of intracellular signalling from glutamate receptors. Biochem Pharmacol 66: 877-886. [DOI] [PubMed] [Google Scholar]
- Aguilera G (1994) Regulation of pituitary ACTH secretion during chronic stress. Front Neuroendocrinol 15: 321-350. [DOI] [PubMed] [Google Scholar]
- Aguilera G, Kiss A (1993) Activation of magnocellular vasopressin responses to non-osmotic stress after chronic adrenal demedullation in rats. J Neuroendocrinol 5: 501-507. [DOI] [PubMed] [Google Scholar]
- Aguilera G, Harwood JP, Wilson JX, Morell J, Brown JH, Catt KJ (1983) Mechanisms of action of corticotropin-releasing factor and other regulators of corticotropin release in rat pituitary cells. J Biol Chem 258: 8039-8045. [PubMed] [Google Scholar]
- Bartanusz V, Muller D, Gaillard RC, Streit P, Vutskits L, Kiss JZ (2004) Local gamma-aminobutyric acid and glutamate circuit control of hypophyseotrophic corticotropin-releasing factor neuron activity in the paraventricular nucleus of the hypothalamus. Eur J Neurosci 19: 777-782. [DOI] [PubMed] [Google Scholar]
- Borsook D, Smirnova O, Behar O, Lewis S, Kobierski LA (1999) Phospho-CREB and CREM/ICER: positive and negative regulation of proenkephalin gene expression in the paraventricular nucleus of the hypothalamus. J Mol Neurosci 12: 35-51. [DOI] [PubMed] [Google Scholar]
- Conti AC, Kuo YC, Valentino RJ, Blendy JA (2004) Inducible cAMP early repressor regulates corticosterone suppression after tricyclic antidepressant treatment. J Neurosci 24: 1967-1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallman MF, Akana SF, Levin N, Walker C-D, Bradbury MJ, Suemaru S, Scribner KS (1994) Corticosteroids and the control of function in the hypothalamo-pituitary-adrenal (HPA) axis. Ann NY Acad Sci 746: 22-28. [DOI] [PubMed] [Google Scholar]
- Della Fazia MA, Servillo G, Foulkes NS, Sassone-Corsi P (1998) Stress-induced expression of transcriptional repressor ICER in the adrenal gland. FEBS Lett 434: 33-36. [DOI] [PubMed] [Google Scholar]
- Foulkes NS, Borrelli E, Sassone-Corsi P (1991) CREM gene: use of alternative DNA-binding domains generates multiple antagonists of cAMP-induced transcription. Cell 64: 739-749. [DOI] [PubMed] [Google Scholar]
- Foulkes NS, Borjigin J, Snyder SH, Sassone-Corsi P (1996) Transcriptional control of circadian hormone synthesis via the CREM feedback loop. Proc Natl Acad Sci USA 93: 14140-14145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg AB, Campeau S, Day HE, Spencer RL (2003) Acute glucocorticoid pretreatment suppresses stress-induced hypothalamic-pituitary-adrenal axis hormone secretion and expression of corticotropin-releasing hormone hnRNA but does not affect c-fos mRNA or fos protein expression in the paraventricular nucleus of the hypothalamus. J Neuroendocrinol 15: 1075-1083. [DOI] [PubMed] [Google Scholar]
- Guardiola-Diaz HM, Boswell C, Seasholtz AF (1994) The cAMP-responsive element in the corticotropin-releasing hormone gene mediates transcriptional regulation by depolarization. J Biol Chem 269: 14784-14791. [PubMed] [Google Scholar]
- Helmreich DL, Itoi K, Lopez-Figueroa MO, Akil H, Watson SJ (2001) Norepinephrine-induced CRH and AVP gene transcription within the hypothalamus: differential regulation by corticosterone. Brain Res Mol Brain Res 88: 62-73. [DOI] [PubMed] [Google Scholar]
- Herman JP, Mueller NK, Figueiredo H (2004) Role of GABA and glutamate circuitry in hypothalamo-pituitary-adrenocortical stress integration. Ann NY Acad Sci 1018: 35-45. [DOI] [PubMed] [Google Scholar]
- Itoi K, Seasholtz AF, Watson SJ (1998) Cellular and extracellular regulatory mechanisms of hypothalamic corticotropin-releasing hormone neurons. Endocr J 45: 13-33. [DOI] [PubMed] [Google Scholar]
- Jingami H, Matsukura S, Numa S, Imura H (1985) Effects of adrenalectomy and dexamethasone administration on the level of prepro-corticotropin-releasing factor mRNA in the hypothalamus and adrenocorticotropin/beta-lipotropin precursor mRNA in the pituitary in rats. Endocrinology 117: 1314-1320. [DOI] [PubMed] [Google Scholar]
- Kalitchenko N, Liu Y, Kasckow J, Aguilera G (2003) Self-limitation of cAMP-stimulated CRH transcription in the hypothalamic cell line 4B: role of inducible cAMP early repressor (ICER). Soc Neurosci Abstr 29: 15.11. [Google Scholar]
- Kovacs KJ (1998) Functional neuroanatomy of the parvocellular vasopressinergic system: transcriptional responses to stress and glucocorticoid feedback. Prog Brain Res 119: 31-43. [DOI] [PubMed] [Google Scholar]
- Kovacs KJ, Sawchenko PE (1996) Regulation of stress-induced transcriptional changes in the hypothalamic neurosecretory neurons. J Mol Neurosci 7: 125-133. [DOI] [PubMed] [Google Scholar]
- Kovacs KJ, Foldes A, Sawchenko PE (2000) Glucocorticoid negative feedback selectively targets vasopressin transcription in parvocellular neurosecretory neurons. J Neurosci 20: 3843-3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalli E, Sassone-Corsi P (1995) Thyroid-stimulating hormone (TSH)-directed induction of the CREM gene in the thyroid gland participates in the long-term desensitization of the TSH receptor. Proc Natl Acad Sci USA 92: 9633-9637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamas M, Molina C, Foulkes NS, Jansen E, Sassone-Corsi P (1997) Ectopic ICER expression in pituitary corticotroph AtT20 cells: effects on morphology, cell cycle, and hormonal production. Mol Endocrinol 11: 1425-1434. [DOI] [PubMed] [Google Scholar]
- Lefkowitz RJ, Caron MG (1988a) Adrenergic receptors. Adv Second Messenger Phosphoprotein Res 21: 1-10. [PubMed] [Google Scholar]
- Lefkowitz RJ, Caron MG (1988b) Adrenergic receptors. Models for the study of receptors coupled to guanine nucleotide regulatory proteins. J Biol Chem 263: 4993-4996. [PubMed] [Google Scholar]
- Lightman SL, Young III WS (1989) Influence of steroids on the hypothalamic corticotropin-releasing factor and preproenkephalin mRNA responses to stress. Proc Natl Acad Sci USA 86: 4306-4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liposits Z, Sievers L, Paull WK (1988) Neuropeptide-Y and ACTH-immunoreactive innervation of corticotropin releasing factor (CRF)-synthesizing neurons in the hypothalamus of the rat. An immunocytochemical analysis at the light and electron microscopic levels. Histochemistry 88: 227-234. [DOI] [PubMed] [Google Scholar]
- Luckman SM, Cox HJ (1995) Expression of inducible cAMP early repressor (ICER) in hypothalamic magnocellular neurons. Brain Res Mol Brain Res 34: 231-238. [DOI] [PubMed] [Google Scholar]
- Luo X, Kiss A, Makara G, Lolait SJ, Aguilera G (1994) Stress-specific regulation of corticotropin releasing hormone receptor expression in the paraventricular and supraoptic nuclei of the hypothalamus in the rat. J Neuroendocrinol 6: 689-696. [DOI] [PubMed] [Google Scholar]
- Luttrell LM (2002) Activation and targeting of mitogen-activated protein kinases by G-protein-coupled receptors. Can J Physiol Pharmacol 80: 375-382. [DOI] [PubMed] [Google Scholar]
- Ma XM, Aguilera G (1999a) Transcriptional responses of the vasopressin and corticotropin-releasing hormone genes to acute and repeated intraperitoneal hypertonic saline injection in rats. Brain Res Mol Brain Res 68: 129-140. [DOI] [PubMed] [Google Scholar]
- Ma XM, Aguilera G (1999b) Differential regulation of corticotropin-releasing hormone and vasopressin transcription by glucocorticoids. Endocrinology 140: 5642-5650. [DOI] [PubMed] [Google Scholar]
- Ma XM, Camacho C, Aguilera G (2001) Regulation of corticotropin-releasing hormone (CRH) transcription and CRH mRNA stability by glucocorticoids. Cell Mol Neurobiol 21: 465-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzucchelli C, Sassone-Corsi P (1999) The inducible cyclic adenosine monophosphate early repressor (ICER) in the pituitary intermediate lobe: role in the stress response. Mol Cell Endocrinol 155: 101-113. [DOI] [PubMed] [Google Scholar]
- McEwen BS (1998) Protective and damaging effects of stress mediators. N Engl J Med 338: 171-179. [DOI] [PubMed] [Google Scholar]
- Molina CA, Foulkes NS, Lalli E, Sassone-Corsi P (1993) Inducibility and negative autoregulation of CREM: an alternative promoter directs the expression of ICER, an early response repressor. Cell 75: 875-886. [DOI] [PubMed] [Google Scholar]
- Monaco L, Foulkes NS, Sassone-Corsi P (1995) Pituitary follicle-stimulating hormone (FSH) induces CREM gene expression in Sertoli cells: involvement in long-term desensitization of the FSH receptor. Proc Natl Acad Sci USA 92: 10673-10677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales M, Bloom FE (1997) The 5-HT3 receptor is present in different subpopulations of GABAergic neurons in the rat telencephalon. J Neurosci 17: 3157-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikodemova M, Kasckow J, Liu H, Manganiello V, Aguilera G (2003) Cyclic adenosine 3′,5′-monophosphate regulation of corticotropin-releasing hormone promoter activity in AtT-20 cells and in a transformed hypothalamic cell line. Endocrinology 144: 1292-1300. [DOI] [PubMed] [Google Scholar]
- Pacak K, Palkovits M (2001) Stressor specificity of central neuroendocrine responses: implications for stress-related disorders. Endocr Rev 22: 502-548. [DOI] [PubMed] [Google Scholar]
- Piech-Dumas KM, Tank AW (1999) CREB mediates the cAMP-responsiveness of the tyrosine hydroxylase gene: use of an antisense RNA strategy to produce CREB-deficient PC12 cell lines. Brain Res Mol Brain Res 70: 219-230. [DOI] [PubMed] [Google Scholar]
- Pokorska A, Vanhoutte P, Arnold FJ, Silvagno F, Hardingham GE, Bading H (2003) Synaptic activity induces signalling to CREB without increasing global levels of cAMP in hippocampal neurons. J Neurochem 84: 447-452. [DOI] [PubMed] [Google Scholar]
- Rabadan-Diehl C, Lolait S, Aguilera G (2000) Isolation and characterization of the promoter region of the rat vasopressin V1b receptor gene. J Neuroendocrinol 12: 437-444. [DOI] [PubMed] [Google Scholar]
- Reul JM, Bilang-Bleuel A, Droste S, Linthorst AC, Holsboer F, Gesing A (2000) New mode of hypothalamic-pituitary-adrenocortical axis regulation: significance for stress-related disorders. Z Rheumatol 59 [Suppl 2]: II/22-II/25. [DOI] [PubMed] [Google Scholar]
- Rho JH, Swanson LW (1987) Neuroendocrine CRF motoneurons: intrahypothalamic axon terminals shown with a new retrograde-Lucifer-immuno method. Brain Res 436: 143-147. [DOI] [PubMed] [Google Scholar]
- Sassone-Corsi P (1998) Coupling gene expression to cAMP signalling: role of CREB and CREM. Int J Biochem Cell Biol 30: 27-38. [DOI] [PubMed] [Google Scholar]
- Sawchenko PE (1987) Evidence for a local site of action for glucocorticoids in inhibiting CRF and vasopressin expression in the paraventricular nucleus. Brain Res 403: 213-223. [DOI] [PubMed] [Google Scholar]
- Schulkin J, Gold PW, McEwen BS (1998) Induction of corticotropin-releasing hormone gene expression by glucocorticoids: implication for understanding the states of fear and anxiety and allostatic load. Psychoneuroendocrinology 23: 219-243. [DOI] [PubMed] [Google Scholar]
- Seasholtz AF, Thompson RC, Douglass JO (1988) Identification of a cyclic adenosine monophosphate-responsive element in the rat corticotropin-releasing hormone gene. Mol Endocrinol 2: 1311-1319. [DOI] [PubMed] [Google Scholar]
- Spencer CM, Houpt TA (2001) Dynamics of c-fos and ICER mRNA expression in rat forebrain following lithium chloride injection. Brain Res Mol Brain Res 93: 113-126. [DOI] [PubMed] [Google Scholar]
- Swanson LW, Simmons DM (1989) Differential steroid hormone and neural influences on peptide mRNA levels in CRH cells of the paraventricular nucleus: a hybridization histochemical study in the rat. J Comp Neurol 285: 413-435. [DOI] [PubMed] [Google Scholar]
- Tiraboschi E, Tardito D, Kasahara J, Moraschi S, Pruneri P, Gennarelli M, Racagni G, Popoli M (2004) Selective phosphorylation of nuclear CREB by fluoxetine is linked to activation of CaM kinase IV and MAP kinase cascades. Neuropsychopharmacology 29: 1831-1840. [DOI] [PubMed] [Google Scholar]
- Vale W, Rivier C, Brown MR, Spiess J, Koob G, Swanson L, Bilezikjian L, Bloom F, Rivier J (1983) Chemical and biological characterization of corticotropin releasing factor. Recent Prog Horm Res 39: 245-270. [DOI] [PubMed] [Google Scholar]
- Watts AG, Sanchez-Watts G (2002) Interactions between heterotypic stressors and corticosterone reveal integrative mechanisms for controlling corticotropin-releasing hormone gene expression in the rat paraventricular nucleus. J Neurosci 22: 6282-6289. [DOI] [PMC free article] [PubMed] [Google Scholar]