Abstract
Primary early-onset generalized dystonia is an autosomal dominant disorder caused by a deletion (ΔGAG) in the DYT1 gene encoding torsinA. The gene defect has incomplete penetrance, with ∼30% of carriers developing clinically evident dystonia. We describe lines of transgenic mice that express either human mutant torsinA (hMT) or human wild-type (hWT) torsinA. All mice demonstrated moderately increased levels of torsinA in the brain by Western blot analysis and normal subcellular distribution of torsinA in neurons by confocal microscopy. No animals had dystonic features. However, mice overexpressing hMT, but not hWT, torsinA displayed a reduced ability to learn motor skills in an accelerating rotarod paradigm. This pattern resembles the impaired motor sequence learning demonstrated in human nonmanifesting carriers of the ΔGAG mutation. Open-field testing showed no differences in spontaneous activity between transgenic mice and their nontransgenic littermates, indicating that mice overexpressing hMT torsinA did not develop overtly abnormal motor behavior. Together, these data suggest that these transgenic mice provide a useful model of the ΔGAG carrier state that can be used to probe genetic and environmental factors that can trigger the dystonic state.
Keywords: DYT1, transgenic mouse, motor learning, dystonia, TorsinA, nuclear envelope
Introduction
Dystonia is characterized by sustained muscle contractions, frequently causing twisting and repetitive movements, or abnormal postures. The most common type of primary early-onset generalized dystonia is an autosomal dominant disorder caused by a 3 bp deletion (ΔGAG) in the DYT1 gene that encodes torsinA (Ozelius et al., 1997). Symptoms of this disorder include involuntary posturing of a foot or limb that frequently generalizes to other body regions. The lifespan of affected individuals is not shortened.
The mechanism by which the ΔGAG mutation causes dystonia is unclear. There is no evidence for neuronal loss in the limited number of postmortem examinations of DYT1 patients that have been conducted (Hedreen et al., 1988), although enlargement of dopaminergic neurons (Rostasy et al., 2003) and ubiquitin-immunoreactive inclusions (McNaught et al., 2004) have been noted. TorsinA shares homology with members of the AAA+ ATPase gene family (Neuwald et al., 1999). Members of this family participate in numerous functions, including protein folding and degradation, membrane trafficking, and vesicle fusion (Ogura and Wilkinson, 2001).
An unusual feature of DYT1 dystonia is that there is a striking effect of age, such that if clinical symptoms appear, they occur almost always before 28 years of age. Thus, the ΔGAG mutation creates a susceptibility to the development of dystonia. A “second-hit” model has been proposed, in which exposure to environmental factors or modulation by other gene(s) is required to trigger the appearance of dystonic movements (Edwards et al., 2003).
Recent studies suggest that asymptomatic individuals carrying the ΔGAG mutation (“nonmanifesting carriers”) exhibit subtle abnormalities of brain function. Adult nonmanifesting carriers displayed a slower rate of acquisition of sequential motor tasks and demonstrated an altered pattern of cerebral activation at rest and when performing motor tasks (Eidelberg et al., 1998; Trost et al., 2002; Ghilardi et al., 2003). A recent epidemiological study demonstrated that early-onset recurrent major depression was also associated with the ΔGAG mutation independent of motor manifestations of dystonia (Heiman et al., 2004).
We have generated mouse models of human DYT1 dystonia expressing a transgene encoding either the human ΔGAG mutant (hMT) or human wild-type (hWT) protein. These mice did not exhibit any overt behavioral abnormalities. However, the lines expressing hMT protein demonstrated an age-dependent abnormality of motor learning, reminiscent of human nonmanifesting carriers of the ΔGAG mutation.
Materials and Methods
Mice generation and genotyping. hWT or hMT torsinA cDNAs were inserted into pcDNA3.1 under the human cytomegalovirus (CMV) immediate early promoter (Invitrogen, Carlsbad, CA) (Hewett et al., 2000). Constructs were linearized with BglII and introduced into B6C3Fq fertilized eggs by microinjection (Mangiarini et al., 1996). Three founder mice were identified and backcrossed to the C57BL/6 strain (The Jackson Laboratory, Bar Harbor, ME), and three lines of transgenic mice were established: hWT, hMT1, and hMT2. A total of 49 males, a combination of F5 and F6 progeny, were used for behavioral studies and housed with food and water ad libitum under controlled conditions (12 h light/dark cycle). Offspring were genotyped via PCR amplification of tail DNA isolated using the TissueAmp DNA kit (Qiagen, Chatsworth, CA). Two primers 5′-GCGTCTCTACTGCCTCTTCG-3′ and 5′-CGGGACTGCATTTCCACTC-3′ amplified a 740 bp segment (30 cycles; annealing temperature, 62°C) within the human torsinA coding sequence that was identified via electrophoresis on 1% agarose gels.
Western blot analysis. After being killed via CO2 inhalation, brains were removed, flash-frozen in chilled isopentane, and sonicated in 2% SDS in PBS (0.01 m, pH 7.4; Protease Inhibitors Complete; Roche, Indianapolis, IN). Samples were incubated on ice for 30 min and centrifuged at 10,000 × g for 10 min at 4°C, and protein concentration was determined using the Bio-Rad (Hercules, CA) Dc reagent with a bovine serum albumin standard. Equal amounts of protein (200 μg) were resolved on 12% polyacrylamide gels and transferred to nitrocellulose (Osmonics, Minnetonka, MN), and Western blot analysis was performed (Bragg et al., 2004b) using the torsinA-specific monoclonal antibody D-M2A8 (Hewett et al., 2004). Blots were stripped and reprobed with an anti-α-tubulin antibody (1:9000; Sigma, St. Louis, MO). Densitometry was performed using a Kodak 1-D System (Eastman Kodak, Rochester, NY). TorsinA levels were normalized to α-tubulin using the average pixel intensity for each immunoreactive band in each lane. Average torsinA levels in transgenic and nontransgenic littermates were obtained by using eight mice (four transgenic, four nontransgenic) from each of the hMT lines and six mice (three transgenic, three nontransgenic) from the hWT line. The data are presented as the average fold increase of normalized torsinA expression in transgenic mice above that in their corresponding nontransgenic littermates.
Immunohistochemistry. Mice were anesthetized with pentobarbital and perfused with 4% paraformaldehyde. The brain was removed, post-fixed for 8 h, and cryoprotected in 20% sucrose overnight, and 40 μm coronal sections were prepared on a sliding microtome. Sections were placed in Antigen Retrieval Citra solution (BioGenex, Sam Ramon, CA), microwaved for 15 s, and probed with D-M2A8 (1:30) using the M.O.M. kit (Vector Laboratories, Burlingame, CA) and diaminobenzene. Sections were examined via light microscopy under a 20× objective. For immunofluorescent staining, tissue was permeabilized (0.5% Triton X-100/TBS, pH 7.4; 30 min at room temperature), blocked with 1.5% normal goat serum (Jackson ImmunoResearch, West Grove, PA), and probed with D-M2A8 (1:30) and either the nuclear dye SYTOX green (0.0625 μm; Molecular Probes, Eugene, OR) or the anti-ubiquitin antibody used to detect ubiquitin-immunoreactive torsinA inclusions in humans (1:100) (McNaught et al., 2004). Cyanine 3 (Cy3)-conjugated secondary antibody (1:500; Jackson ImmunoResearch) was used to visualize torsinA immunoreactivity and bodipy-conjugated secondary antibody (1:500; Molecular Probes) to visualize ubiquitin. Sections were examined under a 63× objective using the Leica (Nussloch, Germany) TCS SL confocal microscope.
Rotarod. Mice were placed on an accelerating rotarod at the same time daily for five consecutive days. Each trial began with a 30 s acclimation period at 4 rpm followed by acceleration by 4 rpm every 30 s to a maximum of 5 min and 40 rpm. The amount of time that elapsed before the mouse fell off was recorded. Each mouse was tested at 4, 6, and 9 months of age. A total of 49 male mice were used (hMT1, 11 transgenic, four nontransgenic; hMT2, nine transgenic, seven nontransgenic; hWT, nine transgenic, nine nontransgenic). Five hMT mice (two hMT1, three hMT2) were not tested on the fifth day of the trial at 6 months of age, so their data were excluded from analyses including data from this test period.
Spontaneous locomotor activity. Spontaneous locomotor activity was quantified at 9 months by measuring the total number of photocell beam breaks over a 15 min period in a 22 × 43 cm Plexiglas arena with a 16 × 16 photocell beam grid (Hamilton-Kinder, Poway, CA). A total of 47 male mice were used (hMT1, seven transgenic, three nontransgenic; hMT2, 11 transgenic, eight nontransgenic; hWT, nine transgenic, nine nontransgenic).
Gait dynamics. Gait dynamics was performed at 10 months on a subset of those mice that had undergone rotarod testing. A total of 41 mice were used (hMT1, nine transgenic, four nontransgenic; hMT2, eight transgenic, seven nontransgenic; hWT, seven transgenic, six nontransgenic). Digital images were collected and analyzed using a DigiGait Imaging System (Mouse Specifics, Boston, MA) (Hampton et al., 2004). Four variables were considered (stride length, paw angle, absolute paw angle, and stance/swing ratio) for each paw at two running speeds (20 and 40 cm/s). Data from all 41 mice (17 nontransgenic, 17 hMT, 7 hWT) were available at 20 cm/s; data from 25 mice (12 nontransgenic, 8 hMT, 5 hWT) were available at 40 cm/s.
Behavioral data analysis. Nontransgenic littermates for all lines were pooled to form a single control group for each age category. Because both hMT lines performed similarly, they were pooled into one group to increase statistical power. Behavioral data were analyzed with repeated-measures ANOVA with genetic status (nontransgenic, hWT, or hMT) as a between-subjects factor. For rotarod analyses, both age at testing and rotarod session were repeated-measures factors; for open-field activity, data were analyzed as activity counts at 1 min intervals. Single measures were collected for gait dynamics measures, which were analyzed with individual one-way ANOVAs with Bonferroni's correction for multiple comparisons (because 32 comparisons were made, the critical level for a family-wise error rate of 0.05 was 0.05/32 = 0.00156). The weight of each mouse was entered as a covariate in all analyses.
Results
Human hWT and hMT torsinA transgenic mice
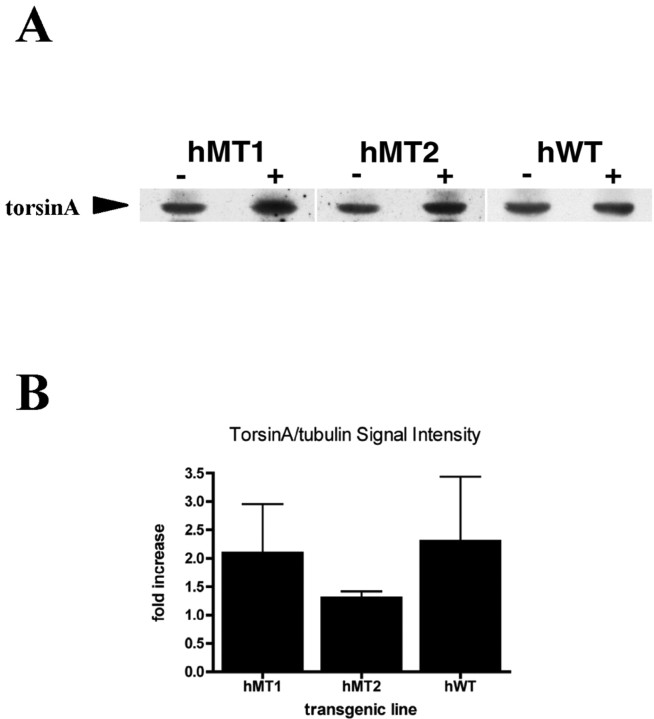
Three lines of transgenic mice were generated: hWT (hemizygous for human WT torsinA) and hMT1 and hMT2 (hemizygous for human mutant torsinA). The founders for these lines were selected from a total of 17 animals that were identified as having a strong signal for the human cDNAs on Southern blot of tail DNA (data not shown). Each founder was backcrossed to C57BL/6 mice, and offspring were analyzed by PCR. For all three lines, Western blot analysis using D-M2A8, which recognizes both human and mouse torsinA, revealed that torsinA levels in brain lysates from transgenic mice were increased relative to their nontransgenic littermate (Fig. 1A). Densitometry indicated that hMT1 and hWT transgenic mice displayed comparable increases of approximately twofold, respectively, above the levels detected in their nontransgenic littermates (Fig. 1B). In the hMT2 line, torsinA expression in transgenic mice was increased ∼1.3-fold relative to their nontransgenic littermates (hMT1, x = 2.1, SEM ± 0.9, n = 4; hMT2, x = 1.3, SEM ± 0.12, n = 4; hWT, x = 2.3, SEM ± 1.1, n = 3).
Figure 1.
Western blot analysis. A, Western blot analysis of brain homogenates from a nontransgenic and transgenic mouse from each of the three lines: torsinA immunoreactivity using the D-M2A8 antibody. B, Densitometry of Western blot analysis, expressed as fold increase of normalized torsinA signal intensity in the transgenic mouse compared with its nontransgenic littermate. hMT1, x=2.1, SEM ± 0.9, n = 4; hMT2, x = 1.3, SEM ± 0.12, n = 4; hWT, x = 2.3, SEM ± 1.1, n = 3.
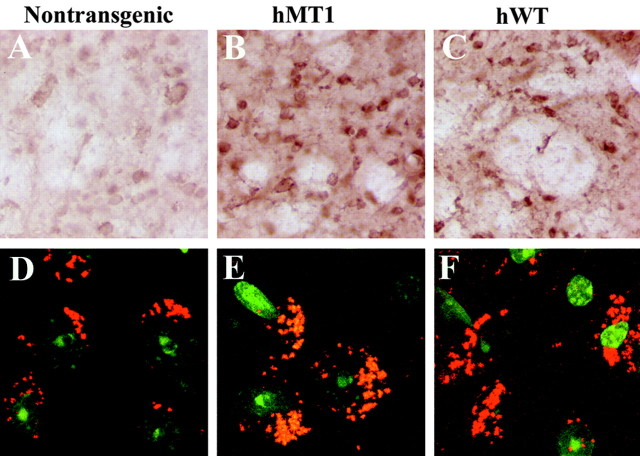
Examination of brain sections by immunocytochemistry and light microscopy indicated that nontransgenic mice displayed low-level, diffuse cytoplasmic staining in neurons throughout the brain (Fig. 2A). In hMT and hWT transgenic mice, torsinA immunoreactivity appeared more intense, although similar in distribution to nontransgenic mice (Fig. 2B, C). To determine intracellular localization, sections were examined by confocal microscopy. In nontransgenic mice (Fig. 2D), punctate cytoplasmic staining, consistent with localization in the endoplasmic reticulum (ER), was observed primarily in neurons throughout the brain. In hMT transgenic mice, the pattern was similar with no evidence of torsinA-positive inclusions or increased staining around the nuclear envelope (Fig. 2E,F). Staining for ubiquitin showed no evidence of ubiquitin-immunoreactive inclusions in the hMT transgenic mice (data not shown).
Figure 2.
Immunohistochemistry. Sections from the striatum of nontransgenic mice (A, D) and transgenic mice expressing hMT torsinA (B, E) and hWT torsinA (C, F) were examined for torsinA expression. DAB immunostaining (A-C) revealed diffuse cytoplasmic staining that was more intense in the transgenic mice but with no significant change in subcellular localization. Immunofluorescent staining, using SYTOX green to visualize the nuclei and Cy3 to visualize torsinA, did not reveal a significant difference in subcellular localization of torsinA in the different lines (D-F).
Impaired motor learning in ΔGAG transgenic mice
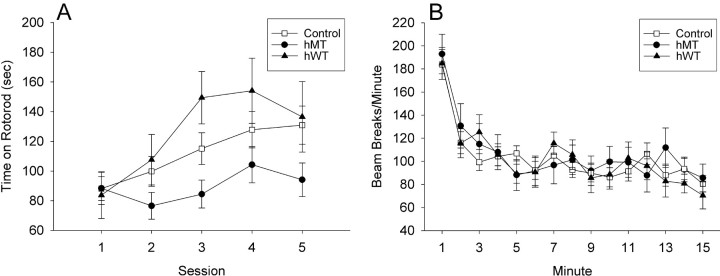
A slower learning rate was detected in hMT transgenic mice at 9 months of age (the oldest age tested) compared with nontransgenic littermates, whereas 4- and 6-month-old mice performed comparably regardless of genotype. Repeated-measures ANOVA on time to fall from the rotarod (with age and test day as repeated-measures factors) revealed main effects of genotype (F(2,38) = 3.39; p = 0.044) and age (F(2,76) = 3.14; p = 0.049) and, critically, an interaction of age, genotype, and test day (F(16,304) = 2.01; p = 0.013), indicating a difference in learning rate that varied with both age and genotype. Follow-up ANOVAs performed separately on mice at each age revealed a significant interaction of genotype and test day for 9-month-old mice (F(8,180) = 2.23; p = 0.027) but not 4-month-old mice (F(8,180) = 0.302; p = 0.97) or 6-month-old mice (F(8,160) = 1.50; p = 0.16). Data from the 9-month-old mice are shown (Fig. 3A). All mice began at the same level of performance on day 1 at all three ages tested (effect of age on session 1, F(2,38) = 1.16; p = 0.32), suggesting that there were no differences in retention from previous rotarod sessions done at 4 and 6 months of age. hMT mice showed the slowest learning across the five sessions of testing, whereas hWT mice showed a trend toward superior performance to controls. There was no indication of an “impaired” subpopulation of mice; histograms of rotarod performance at all ages appeared flat or unimodal, never bimodal. Hartley's Fmax test of homogeneity of variance revealed no differences in variability in performance between the groups (Fmax < 2.35; critical value at 0.05 level 3.54), suggesting that transgenic mice (hMT or hWT) did not demonstrate greater variability than nontransgenic mice, consistent with the lack of an impaired subpopulation.
Figure 3.
Behavioral analysis. Transgene-positive mice from the two lines expressing hMT, the line expressing hWT, and their nontransgenic littermates (Control) were systematically examined for both skill on a rotarod paradigm and spontaneous locomotor activity. A, Rotarod: transgene-positive hMT mice fell off the rotarod sooner than both their nontransgenic littermates and the transgene-positive hWT mice over repeated sessions (p < 0.05). These results were not dependent on the weight of the mice. B, Spontaneous locomotor activity. All groups of mice at 9 months of age decreased their locomotor activity during exposure to the open field, but levels of locomotor activity and rate of habituation did not differ among groups.
To determine whether the impairment in hMT transgenic mice reflected broad defects in motor function, mice were tested in an open field. All groups of mice, tested at 9 months, decreased their activity during exposure to the open field, but levels of activity and rate of habituation did not differ according to genotype (Fig. 3B). Repeated-measures ANOVA revealed a main effect of interval (F(14,602) = 2.33; p = 0.004) but no main effect of genotype (F(2,43) = 1.01; p = 0.37) or genotype by interval interaction (F(28,602) = 0.88; p = 0.65). These results indicate that all mice habituated to the novel environment; there were no overall group differences nor did they habituate at different rates. We found no correlation between open-field activity at 9 months with rotarod performance at any age (r < 0.216; p > 0.20).
Gait dynamics in mice from all three lines were characterized using the DigiGait Imaging System (supplemental Table 1, available at www.jneurosci.org as supplemental material). Only two of the 32 comparisons of gait revealed a significant effect of genotype that survived Bonferroni's correction (p > 0.05/32): stride length at 40 cm/s for left front paw, F(1,21) = 10.04 (p = 0.0009), right front paw, F(1,21) = 9.54; p = 0.0011. For both measures, hMT mice had shorter stride length than nontransgenic mice, and hWT mice had longer stride length than nontransgenic mice. Scheffé's post hoc tests revealed that hWT mice differed significantly from nontransgenic and hMT mice on these measures, but hMT and nontransgenic mice did not differ from one another. This is consistent with the hMT mice serving as a model of the human carrier state rather than an overtly dystonic state. The functional significance of differences between the hMT and hWT mice remains unclear.
Discussion
To study the pathophysiology of DYT1 dystonia, we generated three lines of transgenic mice that expressed human MT or WT torsinA at levels that were approximately twofold higher than that of the endogenous mouse protein. In all three lines, the intracellular distribution of torsinA appeared similar to that in nontransgenic littermates and was consistent with a primary localization within the ER (Hewett et al., 2000, 2003; Kustedjo et al., 2003). The most prominent defect observed in our hMT mice was a failure of older animals to improve performance in the rotarod task. Although in an initial evaluation at 9 months of age, all mice performed equally well; the hWT and nontransgenic mice showed significant improvement with repeated testing, whereas the hMT mice did not. This is clearly not simple ataxia. Ataxic animals, such as the arylsulfatase A (ASA)-deficient mouse model of metachromatic leukodystrophy (D'Hooge et al., 2001), exhibit baseline impairment in coordination. In contrast, our mice had normal exploratory behavior in the open field and normal performance on day 1 of each rotarod trial, whereas the ASA-deficient mice were impaired at the onset of a rotarod trial. Abnormalities were revealed only on repeated testing and were evident as a failure to improve performance (as normal mice do) rather than a loss of coordination.
The expression level and distribution of hMT torsinA in the transgenic mice described here differ from those observed in previously reported transgenic mouse models of DYT1 dystonia (Goodchild and Dauer, 2004; Shashidharan et al., 2005). Goodchild and Dauer (2004) generated mice in which hMT torsinA expression is driven by a tetracycline-regulated minimal CMV promoter. In these mouse brains, mutant torsinA is concentrated and colocalized with laminB in association with the nuclear envelope (NE). Although Western blot analysis was not shown for these mice, strong staining around the NE is consistent with a high level of expression of the mutant protein, as shown in cultured cells (Hewett et al., 2000; Kustedjo et al., 2000; Bragg et al., 2004a; Gonzalez-Alegre and Paulson, 2004; Naismith et al., 2004). Similarly, the transgenic mice established by Shashidharan et al. (2005) incorporate a neuronal-specific enolase promoter to drive high levels of hMT torsinA expression. Although not quantified, on Western blot analysis, hMT torsinA levels appear to be ∼10-fold higher than endogenous levels of mouse torsinA. These mice exhibited torsinA-immunoreactive perinuclear inclusions that also stained for ubiquitin.
Our mice also differ in behavior from those described by Shashidharan et al. (2005), the only lines for which behavioral characteristics have been reported, in that our mice did not display hyperkinetic activity. Behavioral defects were only uncovered in our hMT lines using a motor-learning paradigm. This difference in phenotype may be attributable to a combination of differences in promoter strength and genetic background. Although our transgenic lines and those described by Shashidharan et al. (2005) were generated by crossing to C57BL/6 mice, we used a combination of F5 and F6 mice, whereas Shashidhdaran et al. (2005) used F1 mice for behavioral analysis.
The hMT mice described here provide a model for the human DYT1 carrier state. They express mutant torsinA at near-physiologic levels and display delayed motor learning on the rotarod task, reminiscent of motor learning deficits described in human “nonmanifesting DYT1 carriers” (Ghilardi et al., 2003). In a motor sequence-learning task, nonmanifesting DYT1 carriers demonstrate normal movement speed and accuracy but have marked impairment in learning new motor sequences and abnormal patterns of cerebral activation while learning the task. Specifically, they exhibit enhanced responses in the left prefrontal cortex, right presupplementary motor area, occipital association cortex, and lateral cerebellum. These differences are thought to represent compensatory mechanisms arising as a result of underlying striatal dysfunction. This motor learning deficit has not been studied longitudinally, and it is not known at what age it appears in humans. As a result, correlating the timing of these events with the features of our mice is difficult. In addition, the timing of gene expression driven by the CMV transgene promoter used in our mice likely differs from the endogenous promoter. Specifically, in rats (the only rodent species carefully studied), endogenous torsinA expression levels are highest between embryonic day 20 and postnatal day 1 (Xiao et al., 2004). Determining whether temporal differences in expression contribute to the delayed onset of the motor learning deficit and the lack of overt dystonia in our mice will require additional study. It will be of interest to determine whether our transgenic mice have distinct patterns of cerebral activation during motor learning and whether their impairments extend to other assessments, such as sequence learning tasks.
The possibility of environmental factors triggering dystonia has a parallel in a recently described Drosophila model of DYT1 dystonia (Koh et al., 2004). Expression of the human ΔE302/303 protein, but not wild-type torsinA, in either neurons or muscles did not produce any motor abnormality but did result in morphologically aberrant synapses lacking synaptic vesicles and having abnormal membrane structures. Interestingly a behavioral phenotype (impaired locomotion) was unmasked in the flies expressing the mutant protein when they were exposed to high environmental temperature. This raises the interesting question of whether it will be possible to identify a stimulus that can provoke a dystonic phenotype in the transgenic mice described in this study and whether this would provide insight into the precipitants of dystonic symptoms in human carriers of the ΔGAG mutation. Thus, our transgenic mice offer a paradigm for testing environmental factors that have been implicated in the onset of DYT1 dystonia in humans, including drugs, stress, and injury. Understanding the nature of these triggers is important, because if the onset of symptoms could be delayed beyond 28 years of age, it might be possible to avert the disease entirely.
Footnotes
This work was supported by the Jack Fasciana Fund for Dystonia Research (X.O.B.) and by National Institute of Neurological Disorders and Stroke Grants K08 NS 044272 (N.S.), R01 NS28384 (X.O.B.), and NS37409 (X.O.B., D.G.S.). We thank Laurie Ozelius for advice on mouse models.
Correspondence should be addressed to Dr. Nutan Sharma, Department of Neurology, Massachusetts General Hospital, 114 16th Street, Charlestown, MA 02129. E-mail: Nsharma@partners.org.
Copyright © 2005 Society for Neuroscience 0270-6474/05/255351-05$15.00/0
References
- Bragg DC, Camp SM, Kaufman CA, Wilbur JD, Boston H, Schuback DE, Hanson PI, Sena-Esteves M, Breakefield XO (2004a) Perinuclear biogenesis of mutant torsin-A inclusions in cultured cells infected with tetracycline-regulated herpes simplex virus type 1 amplicon vectors. Neuroscience 125: 651-661. [DOI] [PubMed] [Google Scholar]
- Bragg DC, Kaufman CA, Kock N, Breakefield XO (2004b) Inhibition of N-linked glycosylation prevents inclusion formation by the dystonia-related mutant form of torsinA. Mol Cell Neurosci 27: 417-426. [DOI] [PubMed] [Google Scholar]
- D'Hooge R, Van Dam D, Franck F, Gieselmann V, De Deyn PP (2001) Hyperactivity, neuromotor defects, and impaired learning and memory in a mouse model for metachromatic leukodystrophy. Brain Res 907: 35-43. [DOI] [PubMed] [Google Scholar]
- Edwards M, Huang Y-Z, Wood NW, Rothwell JC, Bhatia KP (2003) Different patterns of electrophysiological deficits in manifesting and nonmanifesting carriers of the DYT1 gene mutation. Brain 126: 2074-2080. [DOI] [PubMed] [Google Scholar]
- Eidelberg D, Moeller JR, Antonini A, Kazumata K, Nakamura T, Dhawan V, Spetsieris P, deLeon D, Bressman SB, Fahn S (1998) Functional brain networks in DYT1 dystonia. Ann Neurol 44: 299-300. [DOI] [PubMed] [Google Scholar]
- Ghilardi MR, Carbon M, Silvestri G, Dhawan V, Tagliati M, Bressman S, Ghez C, Eidelberg D (2003) Impaired sequence learning in carriers of the DYT1 dystonia mutation. Ann Neurol 54: 102-109. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Alegre P, Paulson HL (2004) Aberrant cellular behavior of mutant torsinA implicates nuclear envelope dysfunction in DYT1 dystonia. J Neurosci 24: 2593-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodchild RE, Dauer WT (2004) Mislocalization of the nuclear envelope: an effect of the dystonia-causing torsinA mutation. Proc Natl Acad Sci USA 1001: 847-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton TG, Stasko MR, Kale A, Amende I, Costa AC (2004) Gait dynamics in trisomic mice: quantitative neurological traits of Down syndrome. Physiol Behav 82: 381-389. [DOI] [PubMed] [Google Scholar]
- Hedreen JC, Zweig RM, DeLong MR, Whitehouse PJ, Price DL (1988) Primary dystonias: a review of the pathology and suggestions for new directions of study. Adv Neurol 50: 123-132. [PubMed] [Google Scholar]
- Heiman GA, Ottman R, Saunders-Pullman RJ, Ozelius LJ, Risch NJ, Bressman SB (2004) Increased risk for recurrent major depression in DYT1 dystonia mutation carriers. Neurology 63: 631-637. [DOI] [PubMed] [Google Scholar]
- Hewett J, Gonzalez-Agosti C, Slater D, Li S, Ziefer P, Bergeron D, Jacoby DJ, Ozelius LJ, Ramesh V, Breakefield XO (2000) Mutant torsinA, responsible for early onset torsion dystonia, forms membrane inclusions in cultured neural cells. Hum Mol Genet 22: 1403-1413. [DOI] [PubMed] [Google Scholar]
- Hewett J, Ziefer P, Bergeron D, Naismith T, Boston H, Slater D, Wilbur J, Schuback D, Kamm C, Smith N, Camp S, Ozelius LJ, Ramesh V, Hanson PI, Breakefield XO (2003) TorsinA in PC12 cells: localization in the endoplasmic reticulum and response to stress. J Neurosci Res 72: 158-168. [DOI] [PubMed] [Google Scholar]
- Hewett J, Kamm C, Boston H, Beauchamp R, Naismith T, Ozelius L, Hanson PI, Breakefield XO, Ramesh V (2004) TorsinB—perinuclear location and association with torsinA. J Neurochem 89: 1186-1194. [DOI] [PubMed] [Google Scholar]
- Koh YH, Rehfeld K, Ganetzky B (2004) A Drosophila model of early onset torsion dystonia suggests impairment in TGF-beta signaling. Hum Mol Genet 13: 2019-2030. [DOI] [PubMed] [Google Scholar]
- Kustedjo K, Bracey MH, Cravatt BF (2000) Torsin A and its torsion dystonia-associated mutant forms are lumenal glycoproteins that exhibit distinct subcellular localizations. J Biol Chem 275: 27933-27939. [DOI] [PubMed] [Google Scholar]
- Kustedjo K, Deechongkit S, Kelly JW, Cravatt BF (2003) Recombinant expression, purification, and comparative characterization of torsinA and its torsin dystonia-associated variant delta E-torsinA. Biochemistry 42: 15333-15341. [DOI] [PubMed] [Google Scholar]
- Mangiarini L, Sathawivam K, Seller M, Cozens B, Harper A, Hetherington C, Lawton M, Trottier Y, Lehrach H, Davies SW, Bates GP (1996) Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell 87: 493-506. [DOI] [PubMed] [Google Scholar]
- McNaught KS, Kapustin A, Jackson T, Jengelley TA, Jnobaptiste R, Shashidharan P, Perl DP, Pasik P, Olanow CW (2004) Brainstem pathology in DYT1 primary torsion dystonia. Ann Neurol 56: 540-547. [DOI] [PubMed] [Google Scholar]
- Naismith TV, Heuser JE, Breakefield XO, Hanson PI (2004) TorsinA in the nuclear envelope. Proc Natl Acad Sci USA 101: 7612-7617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald AF, Aravind L, Spouge JL, Koonin EV (1999) AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res 9: 27-43. [PubMed] [Google Scholar]
- Ogura T, Wilkinson AJ (2001) AAA+ superfamily ATPases: common structure-diverse function. Genes Cells 6: 575-597. [DOI] [PubMed] [Google Scholar]
- Ozelius LJ, Hewett J, Page C, Bressman S, Kramer P, Shalish C, de Leon D, Brin M, Raymond D, Corey DP, Fahn S, Risch N, Buckler A, Gusella JF, Breakefield XO (1997) The early-onset torsion dystonia gene (DYT1) encodes an ATP-binding protein. Nat Genet 17: 40-48. [DOI] [PubMed] [Google Scholar]
- Rostasy K, Augood SJ, Hewett JW, Leung JC, Sasaki H, Ozelius LJ, Ramesh V, Standaert DG, Breakefield XO, Hedreen JC (2003) TorsinA protein and neuropathology in early onset generalized dystonia with GAG deletion. Neurobiol Dis 12: 11-24. [DOI] [PubMed] [Google Scholar]
- Shashidharan P, Sandu D, Potla U, Armata IA, Walker RH, McNaught KS, Weisz D, Sreenath T, Brin MF, Olanow CW (2005) A transgenic mouse model of early-onset DYT1 dystonia. Hum Mol Genet 14: 125-133. [DOI] [PubMed] [Google Scholar]
- Trost M, Carbon M, Edwards C, Ma Y, Raymond D, Mentis MJ, Moeller JR, Bressman SB, Eidelberg D (2002) Primary dystonia: is abnormal functional brain architecture linked to genotype? Ann Neurol 52: 853-856. [DOI] [PubMed] [Google Scholar]
- Xiao J, Gong S, Zhao Y, LeDoux MS (2004) Developmental expression of rat torsinA transcript and protein. Brain Res Dev Brain Res 152: 47-60. [DOI] [PubMed] [Google Scholar]