Abstract
Rho-family GTPases play key roles in regulating cytoskeletal reorganization, contributing to many aspects of nervous system development. Their activities are known to be regulated by guanine nucleotide exchange factors (GEFs), in response to various extracellular cues. P-Rex1, a GEF for Rac, has been mainly investigated in neutrophils, in which this molecule contributes to reactive oxygen species formation. However, its role in the nervous system is essentially unknown. Here we describe the expression profile and a physiological function of P-Rex1 in nervous system development. In situ hybridization revealed that P-Rex1 is dynamically expressed in a variety of cells in the developing mouse brain, including some cortical and DRG neurons. In migrating neurons in the intermediate zone, P-Rex1 protein was found to localize in the leading process and adjacent cytoplasmic region. When transfected in pheochromocytoma PC12 cells, P-Rex1 can be activated by NGF, causing an increase in GTP-bound Rac1 and cell motility. Deletion analyses suggested roles for distinct domains of this molecule. Experiments using a P-Rex1 mutant lacking the Dbl-homology domain, a dominant-negative-like form, and small interfering RNA showed that endogenous P-Rex1 was involved in cell migration of PC12 cells and primary cultured neurons from the embryonic day 14 cerebral cortices, induced by extracellular stimuli (NGF, BDNF, and epidermal growth factor). Furthermore, in utero electroporation of the mutant protein into the embryonic cerebral cortex perturbed radial neuronal migration. These findings suggest that P-Rex1, which is expressed in a variety of cell types, is activated by extracellular cues such as neurotrophins and contributes to neuronal migration in the developing nervous system.
Keywords: Rho, ruffling, neurotrophin, neocortex, PIP3, PI3-kinase
Introduction
Rho-family GTPases (i.e., Rho, Rac, and Cdc42) have been suggested to be involved in various neuronal developmental steps, such as outgrowth, extension and pathfinding of neurites, neuronal migration, and synaptogenesis (Hall, 1998; Luo, 2000; Guan and Rao, 2003). Many Rho-family GTPases are ubiquitously expressed throughout development and act as molecular switches, cycling between the GDP-bound inactive and GTP-bound active forms. However, it has not been fully understood how their activities are appropriately regulated temporally and spatially in response to various extracellular cues during the nervous system formation.
To solve this problem, a good approach would be to investigate their activators, Dbl-family guanine nucleotide exchange factors (GEFs), which catalyze GDP-GTP exchange reactions (Whitehead et al., 1997; Zheng, 2001; Schmidt and Hall, 2002). Dbl-family GEFs often show tissue-, developmental stage-, and subcellular-specific distribution and possess multiple protein motifs involved in subcellular localization or intracellular signal transduction. These features and their diversity suggest that Dbl-GEFs may integrate various extracellular signals to appropriately activate specific Rho-family GTPases at specific subcellular sites at distinct developmental stages. Among Dbl-GEFs, several GEFs for Rac have been suggested to participate in nervous system development. Still life (SIF) and Trio in Drosophila, Unc-73 in Caenorhabditis elegans, and Kalirins in mammals were shown to take part in synaptogenesis, neuritogenesis, axonal pathfinding, or dendritic morphogenesis (Sone et al., 1997, 2000; Steven et al., 1998; Awasaki et al., 2000; Lin and Greenberg, 2000; Penzes et al., 2003). Observations from our group and others also suggested that STEF (SIF and Tiam1-like exchange factor) and Tiam1 (T-cell lymphoma invasion and metastasis 1), both orthologs of Drosophila SIF, are involved in neuritogenesis as well as neuronal migration (van Leeuwen et al., 1997; Kunda et al., 2001; Matsuo et al., 2002; Kawauchi et al., 2003; Matsuo et al., 2003; Tanaka et al., 2004).
Through our extensive expression analyses of many Dbl-GEFs (Yoshizawa et al., 2003) in the developing nervous system, we found that P-Rex1, one of the Rac-specific GEFs, is expressed in the developing nervous system in a unique pattern (see Fig. 1). P-Rex1 was originally purified from human neutrophil cytosol as a phosphatidylinositol(3,4,5)P3 (PIP3)-sensitive activator of Rac (Welch et al., 2002). Welch and colleagues demonstrated that this molecule is synergistically activated by PIP3 and Gβγ and contributes to reactive oxygen species formation in a neutrophil-like cell line.
Figure 1.
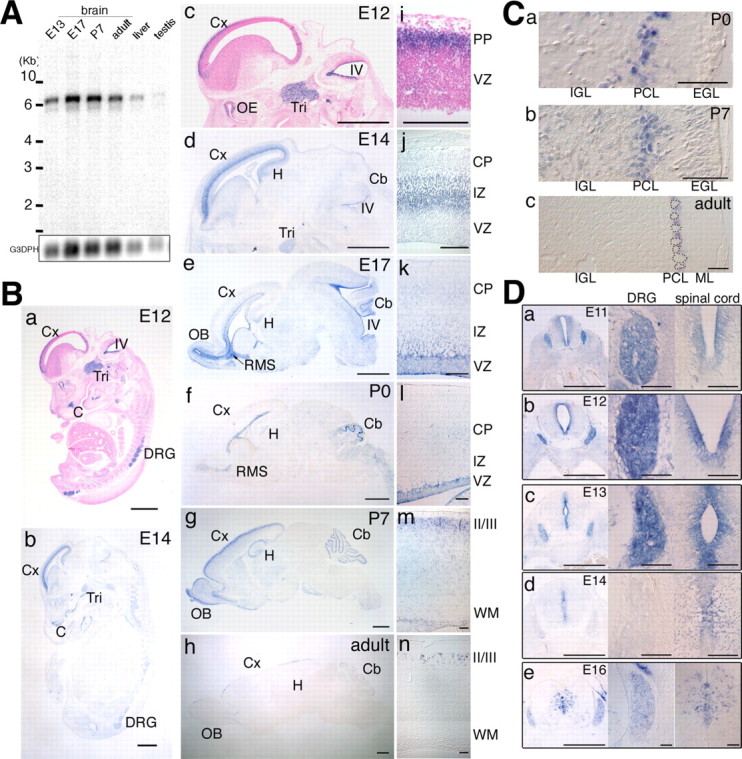
Expression of the P-Rex1 gene in the developing mouse brain. A, Northern blot analysis of murine P-Rex1 expression. Each lane contains 2 μg of poly(A+) RNA from mouse brains (E13, E17, P7, and adult), adult liver, and adult testis. Bars located to the left of the panel indicate 10, 6, 4, 3, and 2 kb, respectively. The same blot was reprobed with glyceraldehyde-3-phosphate dehydrogenase (G3PDH) cDNA as a control (bottom). B, Sagittal sections of E12 and E14 embryos (a, b) or of brains from E12 to adult (c-h) were hybridized with the mouse P-Rex1 cRNA probe. Samples in a and c were counterstained with nuclear fast red. Cx, Cerebral cortex; IV, fourth ventricle; Tri, trigeminal ganglion; C, cartilaginous tissue; DRG, dorsal root ganglion; OE, olfactory epithelium; Cb, cerebellum; H, hippocampal formation; RMS, rostral migratory stream; OB, olfactory bulb. Scale bars, 1.0 mm. i-n, Higher-magnification images of c-h around the cerebral cortices. PP, Preplate; VZ, ventricular zone; CP, cortical plate; IZ, intermediate zone; WM, white matter; II/III, the layers of the cerebral cortex. Scale bars, 100 μm. C, Distribution of P-Rex1 transcripts in the cerebellum. a-c, Higher-magnification pictures of the cerebellum in Bf-Bh. P-Rex1-positive signals were restricted in the Purkinje cell layer. Cells demarcated with dotted lines in c are Purkinje cells. Scale bars, 100 μm. D, Distribution of P-Rex1 transcripts in the spinal cord and DRG. a-e, Coronal sections of E11-E16 embryos at the lumbar level in the spinal cord were hybridized with the P-Rex1 cRNA probe (left panels). Middle and right panels are enlarged images of DRGs and central regions of spinal cords, respectively. Scale bars: left panels, 500 μm; middle and right panels, 100 μm.
In this study, we performed extensive expression analyses of P-Rex1 in the developing nervous system. We also demonstrate that P-Rex1 can be activated by extracellular NGF in pheochromocytoma PC12 cells, resulting in increased cell movement. Experiments using a dominant-negative-like form and small interfering RNA (siRNA) suggest that endogenous P-Rex1 is involved in neurotrophin-induced cell movement of PC12 cells and primary cultured neurons. Immunohistochemistry revealed that P-Rex1 protein localized in the leading process and adjacent cytoplasmic regions of migrating neurons in the cortex. Furthermore, in utero electroporation experiments showed that P-Rex1 participates in neuronal migration in the developing cerebral cortex. Our data thus suggest a role for P-Rex1 in the nervous system development, especially in neuronal migration.
Materials and Methods
Human P-Rex1 cDNA. The 3′ region of the human P-Rex1 cDNA fragment was a gift from Dr. T. Nagase (KIAA1415; Kazusa DNA Research Institute, Kisarazu, Japan), which contained an error sequence. After correcting the error sequence with appropriate oligonucleotides, the 5′ end of P-Rex1 cDNA was isolated by 5′ rapid amplification of cDNA ends to human brain poly(A+) RNA (Origene Technologies, Rockville, MD) and ligated with the corrected C-terminal cDNA, obtaining a full-length (FL) human P-Rex1 cDNA with a full-length open reading frame (ORF). Nucleotide sequence of the full-length cDNA was confirmed to be identical to that of previously reported P-Rex1 (Welch et al., 2002). In addition, we found an in-frame termination codon preceding the putative initiation codon, which was not reported in the previous study.
Mouse P-Rex1 cDNA. Although the complete sequence of mouse P-Rex1 cDNA has not been reported, the putative nucleotide and amino acid sequences are recorded in the National Center for Biotechnology Information database as XM_485102 and XP_485102, respectively. The nucleotide and amino acid sequences of XM_485102 and XP_485102 are quite homologous to those of human P-Rex1 and map to a locus on the second chromosome corresponding to the locus of P-Rex1 on chromosome 20 in humans. Referencing the nucleotide sequence of XM_485102, we designed appropriate oligonucleotides for reverse transcription (RT)-PCR to obtain fragments of mouse P-Rex1 cDNA (described below).
Northern blot hybridization. Northern blot analysis was performed as described previously (Hoshino et al., 1993; Yoshizawa et al., 2003) using the EcoRI-NotI fragment of the mouse expressed sequence tagged clone aa796530 (corresponding to amino acids 38-233 of human P-Rex1 protein) as a probe. The nucleotide sequence of this probe fragment was confirmed by sequencing.
In situ hybridization. We performed RT-PCR with poly(A+) RNA prepared from brains of embryonic day 17 (E17) ICR mouse embryos, obtaining a mouse P-Rex1 cDNA fragment that corresponds to amino acids 977-1452 of the human P-Rex1. The primer sequences are as follows: forward primer, 5′-catggaagtgtcctaccccaagac-3′; reverse primer, 5′-agggcagcctggagaagtggaagc-3′. After confirming that nucleotide sequence of the obtained DNA was identical to a part of XM_485102, we used this cDNA fragment as a probe for the in situ hybridization analyses.
Embryos and brains (neonatal and adult) of ICR strain mice were fixed in 4% paraformaldehyde (PFA) in PBS, dehydrated with methanol, embedded in paraffin, and sectioned. In situ hybridization was performed as described previously (Hoshino et al., 1999) using digoxigenin-labeled sense and antisense cRNA probes. Images were acquired with a Progres 3012 CCD camera (JENOPTIK Laser; Optik Systeme, Jena, Germany) using a 10×/0.30 numerical aperture (NA), a 20×/0.60 NA, or a 40×/0.75 NA objective lens mounted on a Leica (Heidelberg, Germany) DMRBE microscope or an APOZOOM 1:6 objective lens mounted on a Leica M420 microscope.
Generation of anti-mouse P-Rex1 antibodies. Anti-P-Rex1 rat monoclonal antibodies (3A11, 4A3) were raised against a glutathione S-transferase (GST) fusion mouse P-Rex1 polypeptide, which corresponds to amino acids 544-661 of human P-Rex1. The cDNA fragment of mouse P-Rex1 was obtained by RT-PCR to poly(A+) RNA extracted from brains of E17 ICR mouse embryos by using appropriate primers. Antibodies were purified and concentrated by protein G affinity chromatography.
Western blotting. Cell and tissue lysates in Laemmli's SDS sample buffer were separated with SDS-PAGE and electrophoretically transferred onto polyvinylidene difluoride membranes (Millipore, Billerica, MA). Membranes were blocked with 5% skim milk in TBST (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 0.05% Tween 20) for 1 h and probed with primary antibodies, followed by treatment with horseradish peroxidase-conjugated secondary antibodies and ECL Plus Western blotting detection reagents (Amersham Biosciences, Arlington Heights, IL). Signals were visualized by exposure to Hyperfilm (Amersham Biosciences) or detected with a cooled CCD camera (LAS-3000mini; Fujifilm, Kanagawa, Japan).
Plasmids. The cDNA encoding human FL-P-Rex1 was engineered to contain an artificial Kozak initiation sequence with a Myc epitope tag sequence on the 5′ end and a hemagglutinin (HA) epitope tag sequence with a termination codon on the 3′ end and subsequently cloned into a pcDNA3 expression vector (Invitrogen, Gaithersburg, MD), generating pMyc-FL-P-Rex1-HA. pMyc-ΔDH, -ΔPH, -ΔDEP, -ΔPDZ, and -ΔC-P-Rex1-HA constructs were made by removing nucleotide sequences, which correspond to amino acids 53-240, 273-391, 438-577, 666-772, and 892-1659, respectively, from pMyc-FL-P-Rex1-HA. pMyc-FL and -ΔDH-P-Rex1 were generated by removing the HA tag from pMyc-FL and -ΔDH-P-Rex1-HA, respectively. pEGFP-FL-P-Rex1 was generated by inserting the full-length ORF of P-Rex1 into pEGFP-C1 (Clontech, Palo Alto, CA). pEGFP-ΔDH-P-Rex1 was obtained by deleting the nucleotide sequence corresponding to amino acids 53-240 from pEGFP-FL-P-Rex1. Myc- and HA-tagged ΔDH-P-Rex1 sequence excised from pMyc-ΔDH-P-Rex1-HA was subcloned in pcCAG (Kawauchi et al., 2003), producing pcCAG-Myc-ΔDH-P-Rex1-HA. Green fluorescent protein (GFP)-tagged ΔDH-P-Rex1 sequence excised from pEGFP-ΔDH-P-Rex1 was subcloned into pcCAG, producing pcCAG-EGFP-ΔDH-P-Rex1. pΔN-STEF, pΔN-Tiam1, pcDNA3-Myc-V12Rac1, and pcDNA3-Myc-N17Rac1 were described previously (Matsuo et al., 2002). pEGFP used in electroporation experiments was described previously (Kawauchi et al., 2003). pEF-BOS-HA plasmid encoding wild-type Rac1 was a generous gift from K. Kaibuchi (Nagoya University, Nagoya, Japan). Elmo2-ΔN and Dock180-ISP were gifts from H. Katoh (Kyoto University, Kyoto, Japan). pEGFP-N2 was purchased from Clontech. All plasmids were prepared using a EndoFree plasmid purification kit (Qiagen, Hilden, Germany).
Transient transfection to KB, PC12 cells, and primary cortical neurons. KB cells (human epidermal carcinoma cell line) (Nishiyama et al., 1994) were maintained in DMEM (Sigma, St. Louis, MO) with 10% fetal bovine serum (FBS) and antibiotics. PC12 cells (rat origin) were maintained in DMEM with 10% FBS and 5% horse serum. Primary cortical neurons were cultured in Earle's balanced salt solution (EBSS) (Sigma).
Transient transfections were performed by two strategies. For less efficient gene transfer, we used the conventional lipofection method. Plasmids were transfected to KB cells and PC12 cells by Lipofectamine Plus Reagent (Invitrogen) and Lipofectamine 2000 Reagent (Invitrogen), respectively, according to the instructions of the manufacturer. For the ruffling formation assay in PC12 cells, after treatment with the Lipofectamine 2000 solution for 20 min, cells were incubated with the serum-containing media for 24 h and cultured with serum-free media for 16 h, and then 2.5S NGF (Sigma) was added for 6 min at 50 ng/ml (final concentration). For the inhibition assay for phosphatidylinositol 3 (PI3) kinase and TrkA, 50 μm [2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one hydrochloride (LY294002)] (Sigma) and 0.2 μm K-252a (isolated from the broth of Nocardiopsis sp., Calbiochem, Darmstadt, Germany) were administered for 30 min before NGF stimulation, respectively. Before migration assays, PC12 cells were treated with the Lipofectamine 2000 solution for 1 h, cultured in serum-containing media for 24 h, and starved in serum-free media for 16 h.
For high-efficiency gene transfection, PC12 cells and primary cortical neurons were electroporated using a Cell Line Nucleofector Kit V and a Mouse Neuron Nucleofector kit, respectively (Amaxa, Koeln, Germany) according to the instructions of the manufacturer.
Immunocytochemistry. PC12 and KB cells were fixed with 4% PFA in PBS for 15 min, permeabilized with 0.15% Triton X-100 in 5% bovine serum albumin (BSA)/PBS for 5 min, and blocked with 5% BSA/PBS for 30 min. After incubation with primary antibodies at 4°C for 16 h, cells were rinsed with PBS and incubated with Alexa488- or Alexa594-labeled secondary antibodies (1:400; Molecular Probes, Eugene, OR) for 1 h. Indodicarbocyanine-conjugated antibody (1:200; Jackson ImmunoResearch, West Grove, PA) was used for triple-staining experiments. Rhodamine-phalloidin (1:800; Molecular Probes) was used to visualize actin filaments. Fluorescent images were acquired with a Leica TCS SP laser scanning confocal microscope using a 63×/1.32 NA or a 100×/1.40 NA objective lens mounted on a Leica DMRE microscope. Primary antibodies used were anti-Myc polyclonal (1:200; Santa Cruz Biotechnology, Santa Cruz, CA), anti-Myc monoclonal (1:200; Santa Cruz Biotechnology), anti-HA monoclonal (1:400, 12CA5; Roche, Indianapolis, IN), anti-FLAG monoclonal (1:300, M5; Sigma), anti-GFP polyclonal (1:1500; Molecular Probes), and anti-GFP monoclonal (1:1000; provided by Dr. Imura, Kyoto University, Kyoto, Japan) antibodies.
For primary cultured neurons, cells derived from E14 mouse cerebral cortices were dissociated and seeded on poly-d-lysine-coated cover glasses in EBSS. After 3 h of incubation at 37°C, cells were fixed and stained with anti-P-Rex1 monoclonal antibodies (1:100, 3A11 or 1:10, 4A3) plus rhodamine-phalloidin or an anti-TrkB antibody (1:200; Santa Cruz Biotechnology).
Time-lapse video cell migration assay. Cell movements of transfected PC12 cells were recorded in the absence or presence of 50 ng/ml 2.5S NGF (Sigma) in a CO2 incubator chamber by time-lapse confocal microscopy (Leica TCS SL) with a 10×/0.40 NA objective lens mounted on a Leica DMIRE2 microscope. Images were captured for 12 h at 10 min intervals and analyzed with Image WM 2.12r3x software (O'Hara & Co. Ltd., Tokyo, Japan), a modified version of NIH Image software.
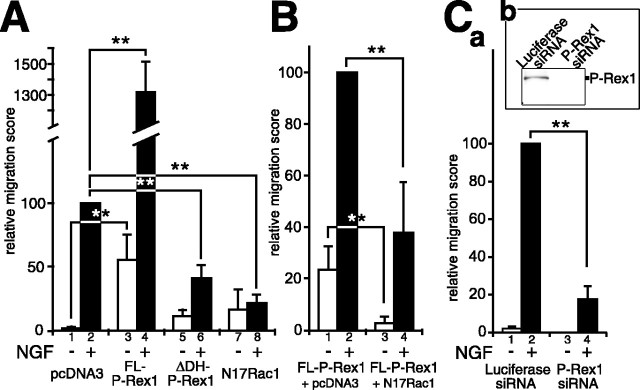
Trans-well cell migration assay. PC12 cell suspension (150 μl) containing 2 × 105 Lipofectamine-treated cells was applied to each upper well of the migration chamber (Chemotaxicell; Kurabo, Osaka, Japan). Cells were allowed to migrate across filters (5 μm pore size) coated with type I collagen (10 μg/ml) on both sides, in the presence or absence of 2.5S NGF at 50 ng/ml in media in the bottom well. The trans-well migration assay was performed at 37°C for 16-19 h. After removal of the remaining cells on the top side of filters by cotton swabs, filters were fixed with 4% PFA/PBS for 15 min, treated with 0.15% Triton X-100 in 5% BSA/PBS for 5 min, and blocked with 5% BSA/PBS for 30 min. After incubation with the Alexa488-conjugated anti-GFP antibody (Molecular Probes) at room temperature for 2 h, filters were rinsed with PBS. GFP-positive cells on the bottom side of the filters were counted with a fluorescent Axiovert S100TV microscope with a 20×/0.40 NA objective lens (Zeiss, Oberkochen, Germany) using a gridded objective. The number of counted cells was compensated with the transfection efficiency. The relative percentages of the compensated cell number to that of standard experiments (see Fig. 6 A, lane 2, B, lane 2, Ca, lane 2) were plotted as a relative migration score in the graph.
Figure 6.
Trans-well cell migration assay. A, Effects of plasmid transfection on trans-well migration of PC12 cells. PC12 cells were transfected with a mock vector plus pEGFP-N2 (lanes 1, 2), pEGFP-FL-P-Rex1 (lanes 3, 4), pEGFP-ΔDH-P-Rex1 (lane 5, 6), or pcDNA3-Myc-N17Rac1 plus pEGFP-N2 (lanes 7, 8). Cell suspensions were applied to the upper wells of the trans-well chambers (modified Boyden chambers) (Ho et al., 2001), and GFP-positive cells that migrated to the bottom side of the filter were counted after 16-19 h of incubation with or without NGF in the lower wells. The relative migration score represents the relative percentage of cell migration compared with that of mock-transfected PC12 cells with NGF treatment (lane 2) (see Materials and Methods). B, The ability of P-Rex1 to enhance PC12 cell motility with NGF treatment is dependent on Rac activity. pEGFP-FL-P-Rex1 was cotransfected with a mock vector (lanes 1, 2) or pcDNA3-Myc-N17Rac1 (lanes 3, 4) in PC12 cells, which were subsequently subjected to the trans-well migration assay with or without NGF in the bottom chambers. The relative migration score represents the relative percentage of the extent of cell migration compared with that of the control experiment (lane 2). The amounts of pEGFP-FL-P-Rex1 plasmid used were one-half of that used in A, resulting in a slightly lower response in FL-P-Rex1-expressing cells in B, lanes 1 and 2, than in A, lanes 3 and 4. C, RNA interference for P-Rex1 resulted in suppression of NGF-induced trans-well migration of PC12 cells. a, pEGFP-N2 was cotransfected with a luciferase siRNA (lanes 1,2) or P-Rex1 siRNA (lanes 3, 4) into PC12 cells. After 4 d of incubation at 37°C, trans-well migration assays were performed for 4-5 h. The relative migration score represents the relative percentage of the extent of cell migration compared with that of the control experiment (lane 2). b, Western blotting analysis showed downregulation of endogenous P-Rex1 protein. All assays were repeated at least three times. Data were evaluated by using Student's t test. **p < 0.01.
For RNA interference experiments, pEGFP-N2 plasmid was cotransfected with luciferase GL2 siRNA (Greiner Bio-one, Frickenhausen, Germany) or P-Rex1 siRNA (target sequence, 5′-gcaacgacttcaagctagtggagaa-3′; iGENE, Tsukuba, Japan) into PC12 cells with a Cell Line Nucleofector Kit V (Amaxa), and transfected PC12 cells were seeded on type I collagen-coated 6 cm dishes. After 4 d of incubation at 37°C, cells were harvested and subjected to the trans-well migration assay for 4-5 h. GFP-positive cells on the bottom side of the filters were counted.
For primary cultured neurons, cells derived from E14 mouse cerebral cortices were dissociated and suspended in EBSS (Sigma). Primary cell suspension (150 μl) containing 2 × 105 cells was applied to each upper well of the migration chamber (Chemotaxicell). Cells were allowed to migrate across filters (5 μm pore size) coated with poly-d-lysine (20 μg/ml) on both sides, in the presence or absence of 10 ng/ml BDNF (R & D Systems, Minneapolis, MN) or 100 ng/ml epidermal growth factor (EGF) (Invitrogen) in EBSS with or without 50 μm LY294002 (Sigma) in the bottom well. After 4-5 h of trans-well migration assay, cells on the top side of the filter were removed, and filters were fixed. Migrated neurons were visualized with an anti-βIII tubulin antibody (Chemicon, Temecula, CA). For transfection experiments, plasmids were transfected into dissociated primary cells using the Mouse Neuron Nucleofector kit (Amaxa). Transfected cells were seeded on poly-d-lysine-coated dishes, maintained in EBSS (Sigma) for 16 h, and applied to each migration chamber. Cell migration index was the compensated cell number estimated at 30% transfection efficiency. Relative migration score was calculated as the relative percentages of the compensated cell number to that of the standard experiment (see Fig. 8 Ab, lane 2).
Figure 8.
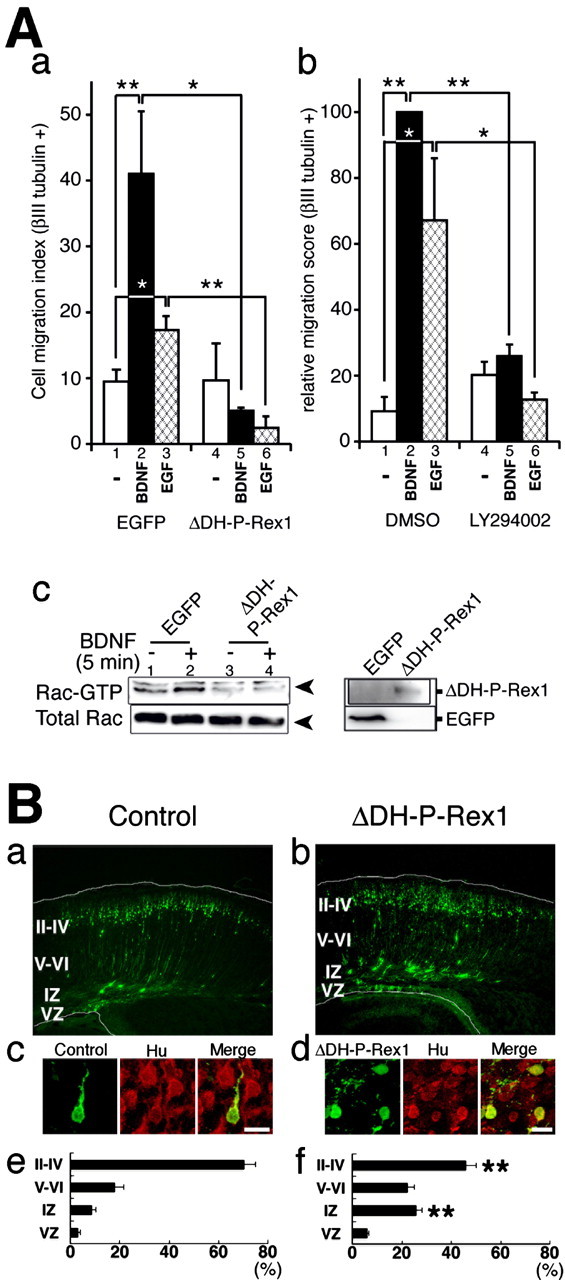
A potential role of P-Rex1 in the migrating neurons. A, P-Rex1 is involved in BDNF- and EGF-induced trans-well migration of primary cortical neurons. a, pEGFP or pcCAG-EGFP-ΔDH-P-Rex1 plasmid was transfected into primary cultured cells derived from E14 mouse cerebral cortices. After incubation at 37°C for 16 h, trans-well migration assays were performed for 4-5 h with or without 10 ng/ml BDNF or 100 ng/ml EGF stimuli. Cells that migrated out on the bottom side of the filter were stained with the neuron-specific anti-βIII-tubulin antibody, and positive-cells were counted. Cell migration indices were calculated as the compensated cell number estimated at 30% transfection efficiency. b, A PI3-kinase inhibitor blocked BDNF- and EGF-induced trans-well migration of primary cortical neurons. Primary cultured cells derived from E14 mouse cerebral cortices were applied to each trans-well migration chamber. Trans-well migration to 10 ng/ml BDNF or 100 ng/ml EGF was assayed with or without 50 μm LY294002. Relative migration score was calculated as the relative percentages of the compensated cell number to that of the standard experiment (Ab, lane 2). All assays were repeated at least three times. Data were evaluated by using Student's t test. *p < 0.05; **p < 0.01. c, GTP-bound Rac1 in primary cultured cells transfected with an EGFP or ΔDH-P-Rex1 with or without administration of BDNF. pEGFP (lanes 1, 2) or pcCAG-EGFP-ΔDH-P-Rex1 (lanes 3, 4) was transfected into dissociated cells derived from E14 mouse cerebral cortices. After transfection, cultured cells were maintained in EBSS for 16 h and treated with or without BDNF for 5 min. The top left panel indicates GTP-bound Rac1. The bottom left panel shows total Rac1. The right box depicts EGFP and EGFP-ΔDH-P-Rex1 proteins. B, Effects of ΔDH-P-Rex1 on development of the cerebral cortex. E14 embryonic brains were transfected with a control vector (a, c, e) or a ΔDH-P-Rex1-expressing vector (b, d, f) by in utero electroporation, together with pEGFP to visualize the transfected cells. a, b, At P0, frozen brain sections were examined for EGFP fluorescence. White lines represent pial and ventricular surfaces. By immunostaining with an anti-tag antibody, expression of ΔDH-P-Rex1 was confirmed in GFP-positive cells (data not shown). c, d, Expression of a neuronal marker, Hu. E14 embryos were electroporated and killed at P0. Frozen sections were immunostained with anti-GFP (green) and anti-Hu (red) antibodies (c in the cortical plate; d in the intermediate zone). Scale bars, 10 μm. e, f, Estimation of cell migration by recording fluorescence intensities of EGFP in distinct parts of the cerebral cortex (Kawauchi et al., 2003). Each bar represents the mean ± SE percentage of relative intensities; n = 11. Asterisks indicate significant differences between the scores of the intermediate zone or layers II-IV of the cortical plate in control and ΔDH-P-Rex1-transfected brains (p < 0.01; t test). II-IV, Layers II-IV of the cortical plate; V-VI, layers V-VI of the cortical plate; IZ, intermediate zone; VZ, ventricular zone.
In utero electroporation. In utero electroporation was performed as described previously (Kawauchi et al., 2003). pcCAG control vector or pcCAG-Myc-ΔDH-P-Rex1-HA was cotransfected with pEGFP (Kawauchi et al., 2003) at a ratio of 10:1 at E14, and electroporated animals were killed at postnatal day 0 (P0).
Pull-down GTPase assay. Rac pull-down GTPase assay for exogenously transfected Rac1 was performed as described previously (Matsuo et al., 2002), and the bound GTP-associated form of HA-Rac was detected by Western blotting using an antibody against the HA epitope.
As for pull-down GTPase assays for endogenously expressed Rac1, Cdc42, and RhoA, PC12 cells were electroporated with plasmids using Cell Line Nucleofector Kit V and then subjected to the detection of GFP-bound forms of small GTPases, by using Rho and Rac activation kits (Cytoskeleton, Denver, CO). GTP-bound Rho-family GTPases were detected with anti-RhoA (Cytoskeleton), anti-Rac1 (Cytoskeleton), and anti-Cdc42 (monoclonal; BD Transduction Laboratories, Franklin Lakes, NJ) antibodies, respectively. Densitometric analyses were performed using Multi Gauge (Fujifilm), and the amounts of GTP-bound Rac1 were normalized to the total amounts of Rac1 in cell lysates.
Immunohistochemistry. Frozen sections of fixed embryonic brains were treated with 10% goat serum in PBS containing 0.2% Triton X-100 (blocking buffer) for 30 min and subsequently incubated with anti-mouse P-Rex1 monoclonal antibodies (3A11, 1:50 and 4A3, 1:10) or anti-Hu monoclonal antibody (1:100; Molecular Probes) in blocking buffer at 4°C for 24 h. Specimens were subsequently rinsed with PBS and incubated with the Alexa488-labeled anti-rat IgG secondary antibody (1:400; Molecular Probes) for 2 h. Fluorescent images were obtained with a laser scanning confocal microscope (Leica TCS SP with a 20×/0.70 NA or a 100×/1.40 NA objective lens).
Results
Expression of the P-Rex1 gene in the nervous system during development
To examine the temporal expression of P-Rex1, we performed Northern blot hybridization analysis with a mouse P-Rex1-specific probe using poly(A+) RNAs extracted from brains of E13, E17, P7, and adult mice, as well as adult liver and testis (Fig. 1A). In adults, mouse P-Rex1 transcripts (6.7 kb) were strongly detected in brain and detected at low levels in liver and very weakly in testis. During development, the 6.7 kb transcript was detected in brains of E13, E17, P7, and adult mice, with the strongest expression observed at E17 and P7. In addition, a minor band (5.7 kb) was also detected in brains throughout the developmental stages, although it can be a partially degraded product of 6.7 kb transcript.
To investigate the spatial and temporal expression of P-Rex1 in the nervous system during development, in situ hybridization experiments were performed. In embryos at E12 and E14, significant signals were detected in the cerebral cortex, trigeminal ganglion, and dorsal root ganglia, as well as some cartilaginous tissues (Fig. 1Ba,Bb). P-Rex1 transcripts were observed in subregions of the CNS, in a stage-dependent manner (Fig. 1Bc-Bh). Expression was present in the cerebral cortex at all stages, ventricular zones of the fourth ventricle at E12, E14, and E17, hippocampal formation (E14, E17, P7, and adult), olfactory bulbs (E17 to adult), the rostral migratory stream (E17 and P0), and the cerebellum from E12 to adult.
We then focused on the developing cerebral cortex. The laminar structure of the adult cerebral cortex is established by sequential migration of neurons. Postmitotic neurons in the cerebral cortex can first be observed at E11 and E12 in the preplate, which would give rise to the marginal zone and subplate at later stages. Neurons, generated in the ventricular zone, move orthogonally through the intermediate zone to reach superficial layers of the cortical plate, just beneath the marginal zone in an inside-out manner. Radial neuronal migration starts at approximately E13 and continues until after birth. The final pattern of the cortical layers is established at approximately P6. At E12, P-Rex1 transcripts were observed in the preplate (Fig. 1Bi) and, at E14, was strongly detected in cells in the intermediate zone (Fig. 1Bj), which may correspond to migrating neurons, but was barely observed in the ventricular zone or the cortical plate. At E17, the signal was still found in cells in the intermediate zone (Fig. 1Bk), although the expression level decreased with the procession of the developmental stage toward P0 (Fig. 1Bl). However, in cells in the ventricular side of the intermediate zone or in the subventricular zone, considerable expression was still observed at E17 and P0. Although the transcripts of this gene were only slightly detectable in a restricted region facing the ventricle at E17 and P0 (Fig. 1Bk,Bl), gene expression could be observed in layers II/III of the cortex at P7 and adult stages (Fig. 1Bm,Bn).
P-Rex1 expression was also observed in the cerebellar primordium or cerebellum from E12 to adult. At E12 and E14, P-Rex1 transcripts were detected in the ventricular zone of the fourth ventricle, part of which would give rise to cells in the cerebellum (Fig. 1Bc,Bd). At later stages, the expression was detected in restricted parts of the cerebellum (Fig. 1Be-Bh), which correspond to the Purkinje cell layer (PCL) (Fig. 1Ca-Cc). In adults, P-Rex1 was expressed in small cells adjacent to Purkinje cells, suggesting that this gene may be expressed in Bergmann glia (Fig. 1Cc), although we do not currently know whether the P-Rex1- expressing cells in the PCL at P0 and P7 are Bergmann glia.
We then carefully examined the expression of P-Rex1 in the spinal cord and the dorsal root ganglion (DRG) during development (Fig. 1Da-De). The expression was strongly detected in most of the cells in the DRG from E11 to E13 (Fig. 1Da-Dc), whereas its expression was barely observed at E14 (Fig. 1Dd). At E16, P-Rex1 expression was observed in dispersed small cells (Fig. 1De). This transient disappearance of P-Rex1 expression at E14 was confirmed repeatedly (data not shown). In the spinal cord, P-Rex1 transcripts were strongly detected in the ventricular zone from E11 to E13 and gradually decreased after E14 (Fig. 1Da-Dd). At E16, the expression was detected in small dispersed cells in the spinal cord (Fig. 1De).
P-Rex1 protein is expressed in the developing mouse brain and in PC12 cells
Monoclonal antibodies against the central region of the mouse P-Rex1 protein were generated, and one of the antibodies was used for Western blotting (Fig. 2A). Transfection of a full-length human P-Rex1 expression construct revealed that this antibody recognized exogenously expressed P-Rex1 (∼180 kDa) in KB cells (Nishiyama et al., 1994). Strong and moderate expression was detected in the cerebral cortex and midbrain of E14 mice, respectively. PC12 cells were also found to express this protein endogenously. Similar results were obtained with another antibody against a different region of this protein (data not shown).
Figure 2.
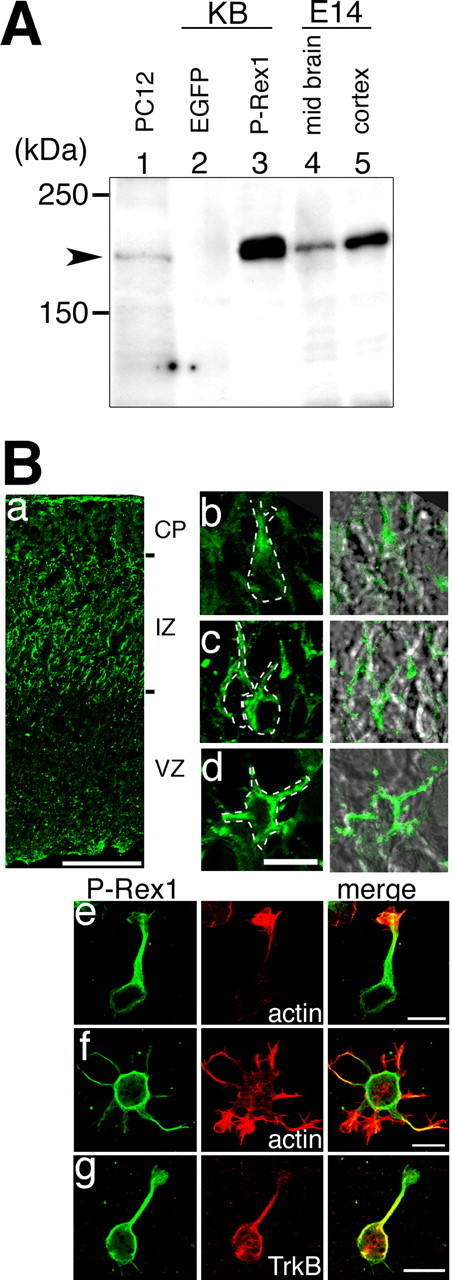
Expression of P-Rex1 protein in PC12 cells and mouse brain at E14. A, Extracts from PC12 cells (lane 1; 20 μg), KB cells transfected with pEGFP-N2 (lane 2; 10 μg) or pMyc-FL-P-Rex1-HA (lane 3; 10 μg), and embryonic day 14 mouse midbrain (lane 4; 5 μg) and cerebral cortex (lane 5; 5 μg) were subjected to SDS-PAGE. Western blot analysis was performed with an anti-mouse P-Rex1 monoclonal antibody (3A11). B, P-Rex1 localization visualized with specific monoclonal antibodies (3A11 for a, c-g; 4A3 for b). a, Frozen section of E14 mouse cerebral cortex. b-d, Higher magnification of Ba. The right panels are merged images of the left panels with Nomarski differential interference images. Cell shapes are highlighted with dotted lines. e-g, Cultured neurons derived from the cerebral cortex of an E14 mouse embryo. E14 dissociated cortical cells were plated on poly-d-lysine-coated cover glasses and fixed after incubation at 37°C for 3 h. Cells were stained with an anti-P-Rex1 monoclonal antibody 3A11 (left panels) and phalloidin (middle panels, e, f) or with an anti-TrkB antibody (middle panel, g). All neurite-bearing cells are immunopositive for neuron-specific βIII-tubulin (data not shown). Scale bars: a, 100 μm; d-g, 10 μm. CP, Cortical plate; IZ, intermediate zone; VZ, ventricular zone.
To investigate the localization of the P-Rex1 protein in the developing cerebral cortex, we performed immunohistochemistry to frozen sections of E14 cerebral cortices with anti-P-Rex1 antibodies (Fig. 2Ba-Bd). P-Rex1 protein was preferentially detected in the intermediate zone consistent with the distribution pattern of its transcript, although lower signals were also observed in the cortical plate (Fig. 2Ba). In higher-magnification pictures within the intermediate zone, this protein was detected in cells with a vertical process toward cortical plate, which are putatively radially migrating neurons. Within these cells, P-Rex1 proteins seemed to be accumulated within the presumed leading process and adjacent cytoplasmic region, both thought to be important subcellular sites for neuronal migration or nuclear translocation (Fig. 2Bb,Bc) (Rivas and Hatten, 1995; Rakic et al., 1996; Lambert de Rouvroit and Goffinet, 2001; Xie et al., 2003). In addition to these unipolar-shaped cells, we observed P-Rex1 signals in some cells with multiple processes in the intermediate zone (Fig. 2Bd). From their shape and location, those cells are thought to be multipolar cells, reported recently by Tabata and Nakajima (2003). In these cells, P-Rex1 was detected in multiple processes and adjacent cytoplasmic regions. Signals in the cortical plate may reflect the P-Rex1 localization in processes of migrating neurons extending from the intermediate zone.
Similar intracellular distribution patterns of P-Rex1 were observed in primary cultured neurons from E14 embryonic cortices. P-Rex1 protein was highly accumulated in the shafts of neurites and adjacent cytoplasmic regions of neurons with single (Fig. 2Be,Bg) or multiple (Fig. 2Bf) processes.
Exogenous expression of P-Rex1 can induce strong membrane ruffling in response to NGF in PC12 cells via Rac activation
It is known that administration of NGF to cultured PC12 cells induces membrane ruffling, which can be recognized by the accumulation of filamentous actin (F-actin) at one or more sites in the cell periphery within minutes (p < 0.01) (Fig. 3Aa,Ab, B, lanes 1, 2) (Posern et al., 2000). This phenomenon has been suggested to be Rac1 dependent because it can be suppressed by introduction of a dominant-negative form for Rac1, N17Rac1 (Posern et al., 2000) (p < 0.01) (Fig. 3B, lanes 2, 4). Conversely, V12Rac1 (a constitutively active form) transfection induced extremely strong and abnormal accumulation of filamentous actin along the cell periphery in PC12 cells (Fig. 3Ac, B, lanes 9, 10). We refer to this type of abnormal and extraordinary ruffling as “MOQ (more than one quarter) ruffling,” which in this report is defined by the criterion that the membrane ruffle covered more than one-fourth of the cell periphery. This type of ruffling was never observed in physiological conditions, such as in nontransfected PC12 cells with or without treatment of NGF (Fig. 3, compare Aa,Ab with B, lanes 1, 2). The presence of MOQ ruffling stands as a good indicator for nonphysiological strong Rac1 activity in the cell.
Figure 3.
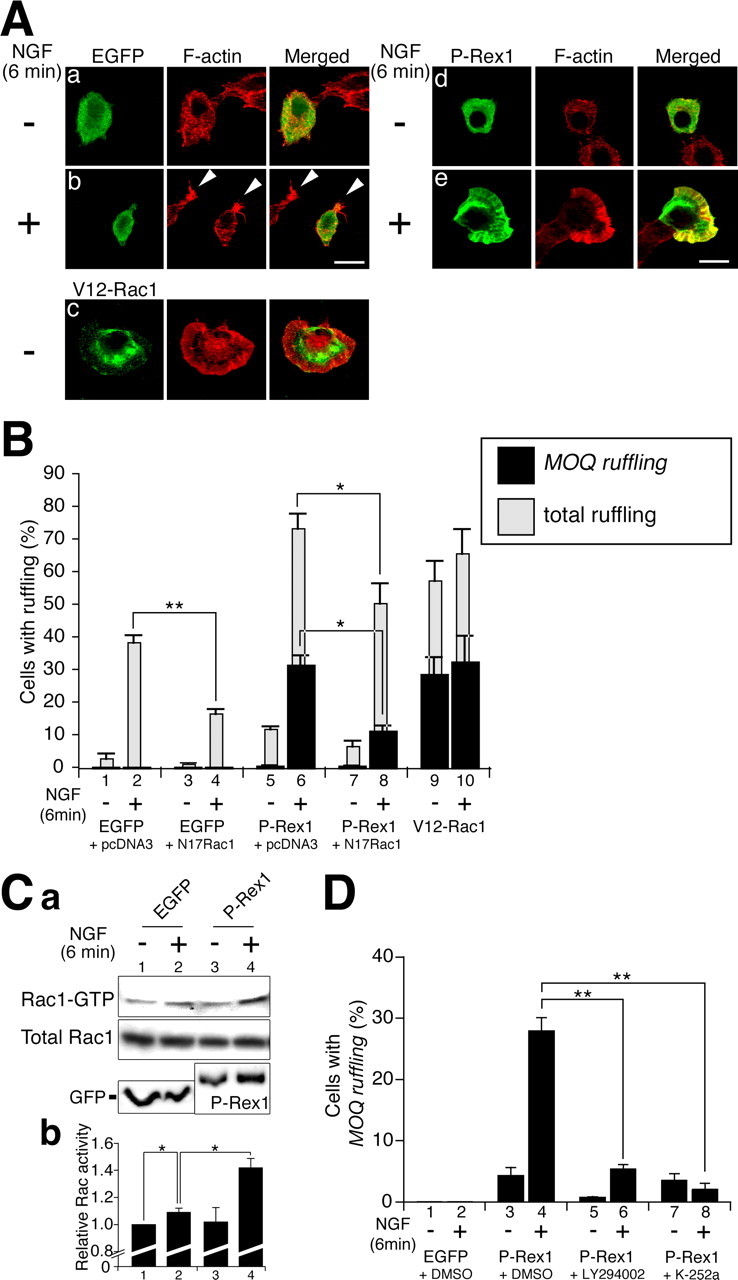
Exogenous expression of P-Rex1 in PC12 cells. A, Rapid ruffling membrane formation in PC12 cells after treatment with NGF. Cells were transfected with indicated plasmids, serum starved for 16 h, and then treated with (b, e) or without (a, c, d) NGF (50 ng/ml) for 6 min. Transfected molecules were visualized with anti-tag antibodies (green). F-actin was detected with rhodamine-phalloidin (red). Arrow heads in b indicate small ruffling membrane formation. Transfected plasmids: a, b, pEGFP-N2; c, pcDNA3-Myc-V12Rac1; d, e, pMyc-FL-P-Rex1-HA. Scale bars, 10 μm. B, Quantification of ruffling membrane formation in PC12 cells. PC12 cells were transfected with expression plasmids (below), treated with or without NGF for 6 min, fixed, and stained with anti-tag antibodies and rhodamine-phalloidin. Ratios of transfected cells exhibiting total ruffling (gray bars) and MOQ ruffling (black bars) were presented. More than 100 transfected cells were observed in each experiment in the blind-test manner. Each bar represents the mean ± SE percentage from three independent experiments. Transfected plasmids: lanes 1, 2, pEGFP-N2 plus pcDNA3; lanes 3, 4, pEFGF-N2 plus pcDNA3-Myc-N17Rac1; lanes 5, 6, pMyc-FL-P-Rex1-HA plus pcDNA3; lanes 7, 8, pMyc-FL-P-Rex1-HA plus pcDNA3-Myc-N17Rac1; lanes 9, 10, pcDNA3-Myc-V12Rac1. *p < 0.05; **p < 0.01; t test. C, GTP-bound Rac1 in PC12 cells transfected with an EGFP or FL-P-Rex1 with or without administration of NGF. a, Plasmids encoding an HA-tagged wild-type Rac1 were cotransfected with pEGFP-N2 (lanes 1, 2) or pMyc-FL-P-Rex1-HA (lanes 3, 4) into PC12 cells. After transfection, PC12 cells were serum starved for 16 h and treated with or without NGF for 6 min. The top panel indicates GTP-bound HA-Rac1. The middle panel highlights total HA-Rac1. Bottom panels depict EGFP and FL-P-Rex1, respectively. b, Quantification of the Rac1 activity. Rac1 activity is indicated by the amount of GST-PAK1 CRIB-bound Rac1, which was normalized to the amount of total Rac1 in whole-cell lysates. The values of Rac1 activity are expressed as fold increase over the value of EGFP-transfected cells that were not treated with NGF. Data are means ± SE. *p < 0.05; t test. D, Pretreatment with inhibitors for PI3-kinase and the Trk receptor family blocks NGF-induced ruffling membrane formation in PC12 cells. PC12 cells were transfected with pEGFP-N2 or pMyc-FL-P-Rex1-HA. Transfected cells were pretreated for 30 min with dimethylsulfoxide (DMSO), 50 μm LY294002, or 0.2 μm K-252a. After NGF stimulation for 6 min, cells were fixed and stained with anti-tag antibodies and rhodamine-phalloidin. Ratios of transfected cells exhibiting MOQ ruffling (black bars) were presented. More than 100 transfected cells were observed in each experiment in the blind-test manner. Each bar represents the mean ± SE percentage of at least three independent experiments. **p < 0.01; t test. Amounts of plasmid DNA used were twofold higher (3 μg) than those used in B (1.5 μg), causing a slightly higher score in FL-P-Rex1-expressing cells in D (lane 3) than in B (lane 5).
When an FL-P-Rex1-expressing plasmid was transfected into PC12 cells, 11.6 ± 0.9% of cells exhibited a ruffling membrane, whereas 0.4 ± 0.4% showed MOQ ruffling (Fig. 3Ad, B, lane 5). Treatment of these cells with NGF for 6 min caused a significant increase of cells with ruffling (73.1 ± 4.7%), as well as those with MOQ ruffling (31.3 ± 3.1%; p < 0.01) (Fig. 3Ad,Ae, B, lanes 5, 6). These P-Rex1-induced events were suppressed by cotransfection of N17Rac1 (Fig. 3B, lane 8), suggesting Rac dependency. Similar phenomena were observed with EGF stimulus; administration of EGF dramatically increased both total and MOQ ruffling in P-Rex1-transfected PC12 cells (data not shown).
To monitor the GTP-bound Rac1 in the transfected PC12 cells, we performed pull-down assays using the PAK1 (p21-activated kinase 1) CRIB (Cdc42/Rac-interactive binding) domain that specifically binds to the GTP-bound active form of Rac1 (Sander et al., 1998; Matsuo et al., 2002). PC12 cells were cotransfected with HA-tagged-wild-type Rac1 plus EGFP or FL-P-Rex1 with or without treatment of NGF, and subsequently GTP-bound Rac1 was detected by pull-down assay. In the control EGFP-transfected cells, NGF treatment resulted in a slight increase of GTP-bound Rac1. However, in FL-P-Rex1-introduced cells, NGF treatment caused considerable increase of GTP-bound Rac1 (Fig. 3C).
It has been reported that the GEF activity of P-Rex1 is increased by PIP3 in vitro (Welch et al., 2002). In addition, in PC12 cells, PIP3 is known to be produced by PI3-kinase, which can be activated in response to NGF and EGF stimuli (Carter and Downes, 1992; Raffioni and Bradshaw, 1992). To test whether the MOQ ruffling-inducing ability of exogenous P-Rex1 is dependent on the NGF-PI3-kinase pathway, a PI3-kinase inhibitor, LY294002, was administered to the cultured PC12 cells. This resulted in significant suppression of the MOQ ruffling-inducing ability of the exogenous P-Rex1 in the presence of NGF (p < 0.01) (Fig. 3D, lanes 4, 6). These findings suggest that P-Rex1 can be activated by NGF in PC12 cells, probably through the activation of PI3-kinase.
The NGF signal is known to be transmitted via TrkA and p75NTR (Huang and Reichardt, 2003). However, it is likely that NGF-Rac1 link via P-Rex1 is transmitted by TrkA rather than p75NTR, because addition of an inhibitor for the Trk receptor family, K-252a, resulted in suppression of NGF-induced MOQ ruffling formation in PC12 cells, which were transfected with P-Rex1 (p < 0.01) (Fig. 3D, lanes 4, 8).
Deletion analysis of P-Rex1
P-Rex1 contains several protein motifs; Dbl-homology (DH) and Pleckstrin-homology (PH) domains, DEP (Dishevelled, Egl-10, and Pleckstrin homology) domain, and PDZ (postsynaptic density-95, Discs large and zona occludens-1 homology) domain, and an inositol polyphosphate 4 (InsP4)-phosphatase-like region. To investigate the role of each P-Rex1 domain, we transfected a series of deletion mutant constructs containing the N-terminal Myc-epitope tag (Fig. 4A). F-actin was visualized with rhodamine-phalloidin. The resulting phenotypes were assessed by percentages of cells bearing MOQ ruffling with or without administration of NGF (Fig. 4B).
Figure 4.
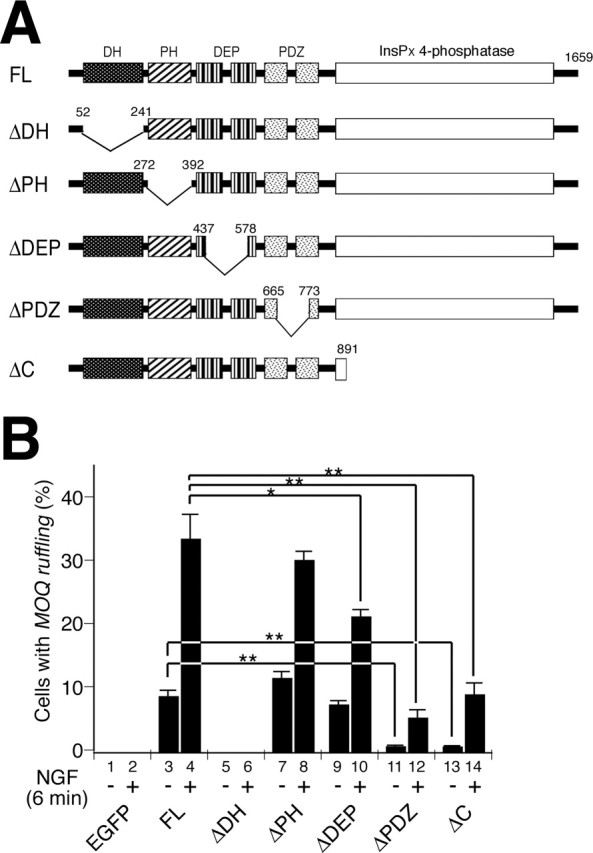
Deletion analysis of P-Rex1. A, Schematic structures of mutant P-Rex1 proteins. Numbers denote amino acid positions. B, Cells were transfected with 2.5 μg of indicated constructs. Amounts of plasmid DNA used were 1.7-fold higher than those used in Figure 3B, causing a slightly higher score in FL-P-Rex1-expressing cells in B than in Figure 3B. At least 100 cells positively stained with anti-tag antibodies were observed in each experiment in the blind-test manner. Each bar represents the mean ± SE from three independent experiments. *p < 0.05; **p < 0.01; t test.
Deletion of the DH domain (ΔDH-P-Rex1) resulted in the complete loss of MOQ ruffling-inducing ability either with or without NGF (lanes 5, 6), consistent with the consensus that the DH domain is a catalytic domain for the exchange reaction (Schmidt and Hall, 2002). The ΔPH-P-Rex1 mutant was able to induce MOQ ruffling to an extent similar to FL-P-Rex1 in either NGF-containing or absent conditions (lanes 7, 8), suggesting that the PH domain in P-Rex1 is neither essential for the exchange activity nor receiving the NGF-derived signal in PC12 cells. ΔDEP-P-Rex1 was able to induce MOQ ruffling that was indistinguishable from FL-P-Rex1 when no trophic factor was administered (lane 9). However, when cells were treated with NGF, ΔDEP-P-Rex1 showed significantly less MOQ ruffling-inducing ability than FL-P-Rex1, although prominent enhancement of the inducing ability could still be observed with addition of NGF (lane 10). This suggests that the DEP domains play a role in receiving the NGF-derived signal in PC12 cells. Despite the presence of the catalytic DH domain, both ΔPDZ-P-Rex1 and ΔC-P-Rex1 mutants exhibited drastically reduced inducing abilities with or without NGF, indicating that both PDZ and InsPx 4-phosphatase-like domains are required for optimal activity of P-Rex1 in PC12 cells.
Exogenous expression of P-Rex1 enhances migration of PC12 cells when treated with NGF
Using time-lapse confocal microscopy, we recorded the movement of PC12 cells that were transfected with N-terminal-EGFP-tagged P-Rex1 (pEGFP-FL-P-Rex1) or control EGFP-expressing plasmid in either NGF absent or containing media. pEGFP-FL-P-Rex1 showed basically identical characteristics to pMyc-FL-P-Rex1 in PC12 cells, with regard to its ability to induce membrane ruffling as well as its NGF dependency (data not shown). In our experimental condition, control EGFP-transfected PC12 cells did not show remarkable cell movement, even in the presence of NGF (Fig. 5Aa,Ab, B, lanes a, b). In contrast, FL-P-Rex1-expressing cells exhibited vigorous and rapid cell motility when treated with NGF (Fig. 5Ad, B, lane d), whereas they did not show significant increase in cell movement without administration of NGF (Fig. 5Ac, B, lane c). This NGF-dependent enhancement of cell motility by P-Rex1 was suggested to be dependent on Rac activity, because coexpression of N17Rac1 suppressed this effect (Fig. 5Ae, B, lane e).
Figure 5.
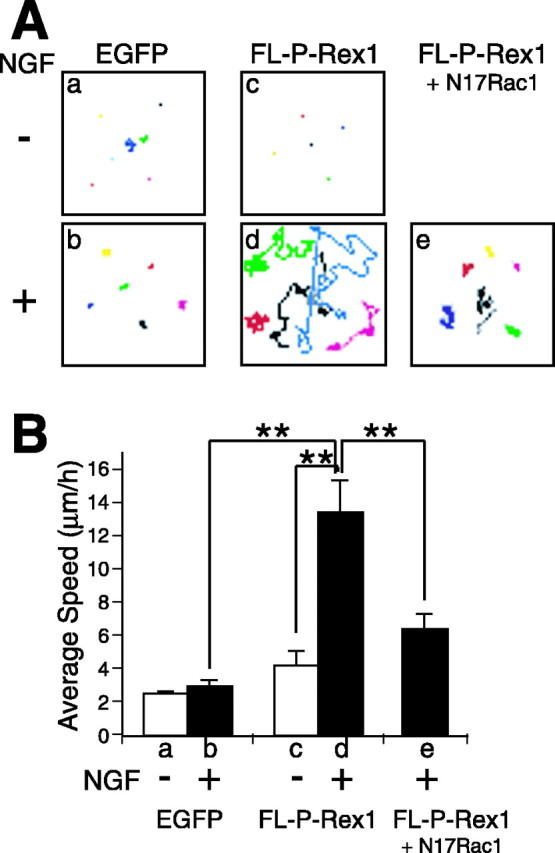
Time-lapse cell motility recording of PC12 cells transfected with P-Rex1 in NGF-containing and absent conditions. A, B, PC12 cells were transiently transfected with control pEGFP-N2 (a, b), pEGFP-FL-P-Rex1 (c, d), or pEGFP-FL-P-Rex1 plus pcDNA3-Myc-N17Rac1 (e). Movement of GFP-positive cells was recorded for 12 h with or without NGF treatment by time-lapse microscopy. A, Traces of each GFP-positive cell are depicted in a distinct color. B, Bars represent estimated average ± SE speed of distinct experiments (a-e). Data were evaluated by Student's t test. **p < 0.01.
Next, we performed the trans-well migration assay (Ho et al., 2001). In this experiment, cells that had migrated from the upper well to the lower well were counted. PC12 cells were transfected with either control EGFP or FL-P-Rex1 expression vector. Consistent with the previous study (Ho et al., 2001), addition of NGF to the lower well resulted in considerable enhancement of control PC12 cell migration (p < 0.01) (Fig. 6A, lanes 1, 2). Whereas introduction of FL-P-Rex1 also increased the trans-well cells even in the NGF-free condition (p < 0.01) (Fig. 6A, lanes 1, 3), administration of NGF to those cells resulted in extremely drastic increases in cell migration (p < 0.01) (Fig. 6A, lanes 3, 4). These phenomena were suppressed by coexpression of N17Rac1 with FL-P-Rex1, suggesting Rac dependency (p < 0.01) (Fig. 6B, lanes 1,3 and 2,4).
The findings obtained from both the time-lapse observation and the trans-well migration assay suggest that exogenously expressed P-Rex1 can be activated by NGF in PC12 cells to enhance cell migration, probably through activating Rac1.
ΔDH-P-Rex1 can suppress the function of P-Rex1
Previous studies showed that deletion mutants that lacked the DH domain of the Dbl-family proteins often acted as specific dominant-negative forms (Patel et al., 2002; Kawasaki et al., 2003). Therefore, we investigated whether ΔDH-P-Rex1 can suppress the function of P-Rex1. To test this, we used KB cells in which endogenous P-Rex1 expression was not detected (Fig. 2). Introduction of FL-P-Rex1 caused prominent ruffling membrane formation in KB cells in serum-containing media (Fig. 7Aa, B, lane 2). However, cotransfection of ΔDH-P-Rex1 (ΔDH-P-Rex1) with FL-P-Rex1 resulted in significant reduction of ruffling-bearing cells (p < 0.01) (Fig. 7Ab, B, lanes 2, 3). A similar effect was also observed by cotransfection of N17Rac1 with FL-P-Rex1 (p < 0.01) (Fig. 7Ac, B, lanes 2, 4). In contrast, ΔDH-P-Rex1 did not suppress V12Rac1-induced ruffling (Fig. 7Ad, B, lanes 5, 6), implying that ΔDH-P-Rex1 affects the function of P-Rex1 rather than that of Rac or molecules in its downstream pathway.
Figure 7.
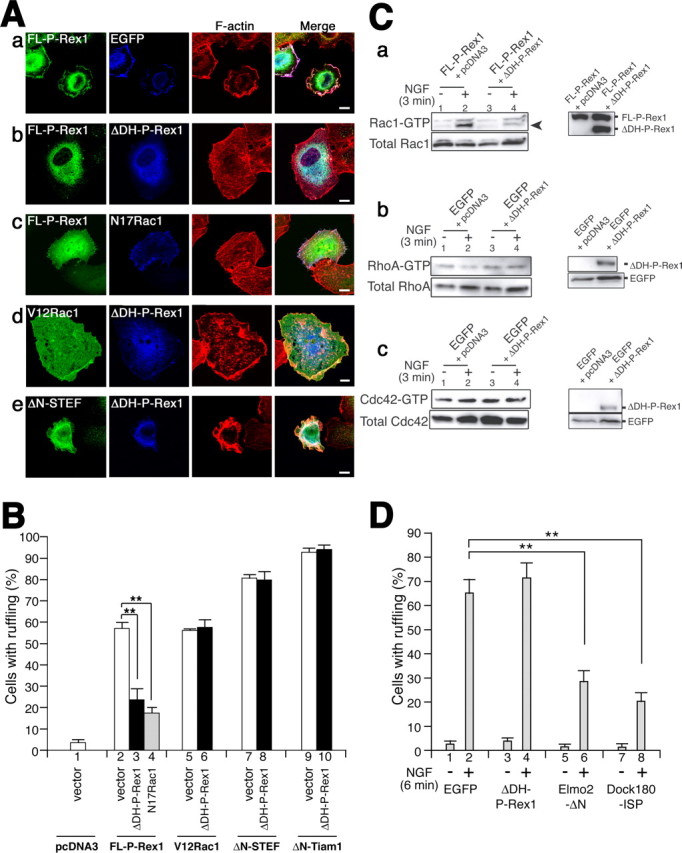
ΔDH-P-Rex1 suppresses the function of P-Rex1. A, KB cells were transfected with certain combinations of expression plasmids (below) in serum-containing media. Transfected molecules were visualized with their own tags (green and blue) as described in white letters in the panels. F-actin was visualized with rhodamine-phalloidin (red). Merged pictures are also represented. Scale bars, 10 μm. B, Quantification of KB cells with ruffling membrane formation, which were transfected with certain combinations of constructs (below). Transfected molecules are presented in bold black letters. At least 100 cells positively stained with anti-tag antibodies were scored as to whether they exhibit membrane ruffling. Each bar represents the mean ± SE from three independent experiments. **p < 0.01; t test. Transfected constructs: Aa, B, lane 2, pMyc-FL-P-Rex1 plus pEGFP-N2; Ab, B, lane 3, pMyc-FL-P-Rex1-HA plus pEGFP-ΔDH-P-Rex1; Ac, B, lane 4, pEGFP-FL-P-Rex1 plus pcDNA3-Myc-N17Rac1; Ad, B, lane 6, pcDNA3-Myc-V12Rac1 plus pEGFP-ΔDH-P-Rex1; Ae, B, lane 8, pΔN-STEF plus pEGFP-ΔDH-P-Rex1; B, lane 1, pcDNA3 plus pEGFP-N2; B, lane 5, pcDNA3-Myc-V12Rac1 plus pEGFP-N2; B, lane 7, pΔN-STEF plus pEGFP-N2; B, lanes 9, 10, pΔN-Tiam1 plus pcDNA3 or pMyc-ΔDH-P-Rex1, respectively. C, a, ΔDH-P-Rex1 can suppress the function of P-Rex1 to activate Rac1 in response to NGF. pEGFP-FL-P-Rex1 was cotransfected with pcDNA3 mock vector (lanes 1, 2) or pMyc-ΔDH-P-Rex1 (lanes 3, 4) into PC12 cells with a Cell Line Nucleofector Kit V (Amaxa). After transfection, cells were incubated in culture medium at 37°C for 24 h and serum starved for 16 h. After administration of 50 ng/ml NGF for 3 min, cells were lysed, and Rac pull-down GTPase assays were performed. The top left panel indicates GTP-bound endogenous Rac1. The bottom left panel highlights total endogenous Rac1. The right panel depicts proteins derived from transfected constructs. Arrowheads indicate endogenous Rac1 proteins. b, c, ΔDH-P-Rex1 does not affect RhoA and Cdc42 activities. Endogenous GTP-bound RhoA (b) or Cdc42 (c) in PC12 cells transfected with indicated plasmids with or without NGF is shown. Cells were lysed, and pull-down GTPase assays were performed. Right panels indicate proteins derived from transfected constructs. D, Quantification of ruffling membrane formation in PC12 cells. PC12 cells were transfected with indicated constructs, treated with or without 50 ng/ml NGF for 6 min, fixed, and stained with anti-tag antibodies and rhodamine-phalloidin. Ratios of transfected cells exhibiting total ruffling (gray bars) were shown. More than 50 transfected cells were observed in each experiment in the blind-test manner. Each bar represents the mean ± SE percentage from three independent experiments. Specific inhibitor constructs for Elmo/Dock180 pathway, Elmo2-ΔN and Dock180-ISP, were described previously (Katoh and Negishi, 2003). **p < 0.01; t test.
To evaluate the specificity of this dominant-negative-like effect of ΔDH-P-Rex1 on Rac-GEFs, we tested whether ΔDH-P-Rex1 could suppress the activity of other Rac-GEFs, STEF and Tiam1. Cotransfection of ΔDH-P-Rex1 with ΔN-STEF or ΔN-Tiam1, which has a strong Rac-GEF activity, could not suppress the ruffling membrane-inducing ability (Fig. 7Ae, B, lanes 7,8 and 9,10), in contrast to our previous finding that the function of both ΔN-STEF and ΔN-Tiam1 could be suppressed by the specific dominant-negative form (PHnTSS STEF) in KB cells (Matsuo et al., 2002).
To test whether ΔDH-P-Rex1 can also suppress the function of P-Rex1 in PC12 cells, ΔDH-P-Rex1 was cotransfected with FL-P-Rex1 in PC12 cells, and the endogenous Rac1 activity was evaluated by pull-down GTPase assays. The results showed that ΔDH-P-Rex1 suppressed the induction of Rac1 activity evoked by the exogenous P-Rex1 in the presence of NGF (Fig. 7Ca, lanes 2, 4). This suggests that ΔDH-P-Rex1 can suppress the function of P-Rex1 in PC12 cells.
Next, we investigated whether ΔDH-P-Rex1 affects other small GTPase activities, such as RhoA and Cdc42. Pull-down GTPase assays using a PAK CRIB domain and a Rhotekin RBD (Rho-GTP-binding domain), the latter of which is known to specifically bind to GTP-bound RhoA, showed that transfection of ΔDH-P-Rex1 did not significantly affect the activities of endogenous RhoA (Fig. 7Cb) nor Cdc42 (Fig. 7Cc).
These results suggest that ΔDH-P-Rex1 can specifically interfere with the function of P-Rex1. Indistinguishable effects were observed with transfection of EGFP-tagged-ΔDH-P-Rex1 (EGFP-ΔDH-P-Rex1) and Myc-tagged-ΔDH-P-Rex1 (Myc-ΔDH-P-Rex1) in our experimental conditions (data not shown).
ΔDH-P-Rex1 inhibits NGF-induced trans-well migration of PC12 cells
As shown in the trans-well migration assay, the motility of PC12 cells was increased by NGF. This phenomenon was suggested to be Rac dependent, because it was suppressed by introduction of N17Rac1 (p < 0.01) (Fig. 6A, lanes 2, 8). Because P-Rex1 is endogenously expressed in PC12 cells (Fig. 2) and because exogenously expressed P-Rex1 could enhance the NGF-dependent trans-well migration of PC12 cells, it is likely that endogenously expressed P-Rex1 might be involved in this event. To test this, we introduced ΔDH-P-Rex1 to PC12 cells to suppress the function of endogenous P-Rex1, which resulted in significant reduction of NGF-induced trans-well cell migration (p < 0.01) (Fig. 6A, lanes 2, 6). To further confirm the involvement of P-Rex1 in trans-well migration of PC12 cells, we designed siRNA for P-Rex1, which could effectively suppress the expression of P-Rex1 protein (Fig. 6Cb). Introduction of this siRNA to PC12 cells resulted in efficient suppression of NGF-induced migration of PC12 cells (Fig. 6Ca, lanes 2, 4), confirming that endogenous P-Rex1 is involved in this event. Pull-down GTPase assays were performed to assess the Rac1 activity in PC12 cells used in these experiments (supplemental Fig. S1a,b, available at www.jneurosci.org as supplemental material).
Interestingly, introduction of ΔDH-P-Rex1 could not suppress NGF-induced ruffling membrane formation of PC12 cells (Fig. 7D, lanes 2, 4), indicating that another endogenous Rac-GEF may be involved in this event. This is also supported by the result that Rac1 activity was not suppressed by ΔDH-P-Rex1 in NGF-treated PC12 cells (supplemental Fig. S2, lanes 2, 4, available at www.jneurosci.org as supplemental material). One of the candidates for the Rac-GEF was the Elmo/Dock180 complex, which was reported to be required for NGF-dependent neurite extension of PC12 cells (Katoh and Negishi, 2003). To test this, we introduced mutant forms of Elmo and Dock180, which can interfere with the functions of the wild-type Elmo/Dock180 complex, and succeeded in suppressing the NGF-induced ruffling membrane formation in PC12 cells (Fig. 7D, lanes 2,6 and 2,8).
ΔDH-P-Rex1 inhibits BDNF- and EGF-induced trans-well migration of cultured cortical neurons
From the localization pattern of P-Rex1 (Fig. 2B), we suspected that P-Rex1 may be involved in cell migration of cortical neurons. It has been reported that cortical neurons express TrkB (Fig. 2Bg) (Behar et al., 1997), a major receptor for BDNF, and that their motility can be enhanced by BDNF stimuli (Behar et al., 1997). EGF was also implied to accelerate cell motility of cortical neurons (Threadgill et al., 1995; Caric et al., 2001). Accordingly, our trans-well migration assay showed that cell migration of primary cultured neurons from E14 embryonic cerebral cortices was significantly enhanced by BDNF and EGF (p < 0.01 and p < 0.05, respectively) (Fig. 8A, lanes 1,2 and 1,3). This phenomenon was suggested to be PI3-kinase dependent, because it was suppressed by a PI3-kinase inhibitor, LY294002 (p < 0.01 and p < 0.05, respectively) (Fig. 8Ab, lanes 2,5 and 3,6). Introduction of ΔDH-P-Rex1 effectively suppressed both BDNF- and EGF-induced migration of these primary cultured neurons (p < 0.05 and p < 0.01, respectively) (Fig. 8A, lanes 2,5 or 3,6). Pull-down GTPase assay using primary cultured cells from E14 embryonic cerebral cortices demonstrated that Rac1 activity was enhanced by administration of BDNF in control vector transfected cells, whereas no enhancement of the Rac1 activity was observed in ΔDH-P-Rex1-transfected cells (Fig. 8Ac, lanes 1,2 and 2,4). These findings indicate that activities of endogenous P-Rex1 and PI3-kinase are required in BDNF- and EGF-induced cell migration of primary cultured cortical neurons.
ΔDH-P-Rex1 introduction affects normal development of the cerebral cortex
To investigate the function of P-Rex1 in the developing cortex, we introduced ΔDH-P-Rex1 or a mock vector plasmid into cells in the ventricular zone of E14 cortices by in utero electroporation (Kawauchi et al., 2003) and killed the animals at P0. An EGFP-expressing plasmid was coelectroporated to visualize transfected cells. In the control animals, many transfected cells that had been born in the ventricular zone reached the pial side of the cortical plate at P0, 5 d after electroporation, although a smaller number of cells were observed in the deep layers of the cortical plate or in the intermediate zone (Fig. 8Ba). In ΔDH-P-Rex1-introduced animals, a considerable number of cells that were stalled in the intermediate zone were also observed, although a fair number of cells reached the superficial layers of the cortical plate (Fig. 8Bb). EGFP-positive transfected cells that stalled in the intermediate zone were labeled with a neuronal marker, Hu (Fig. 8Bd) and β-III tubulin (data not shown), suggesting that those cells were neurons. The extent of cell migration was statistically estimated by fluorescent intensities in distinct layers of the cerebral cortex (Fig. 8Be,Bf) and revealed that the relative signals detected in the intermediate zone and the cortical plate were significantly higher and lower, respectively, in ΔDH-P-Rex1 electroporated brains than in the control brains. Together with our findings that P-Rex1 is expressed in migrating neurons in the developing cortex and is required for BDNF- and EGF-induced migration of cultured cortical neurons, the results from the electroporation experiment suggest the involvement of P-Rex1 in cortical neuronal migration, at least to some extent.
Discussion
Expression of P-Rex1 during nervous system development
Our experiments showed that the P-Rex1 gene was expressed in restricted areas during the development of the nervous system. However, this gene was found to be expressed in a variety of cells: neurons, neural precursor cells, and glial cells. Regarding neurons, they include transiently emerging neurons in preplate, migrating neurons in the intermediate zone and rostral migratory stream and in immature/mature neurons in layers II/III in the neonatal and adult cerebral cortex. P-Rex1 expression was also detected in the presumed neural precursor cells in the ventricular zone of the fourth ventricle and of the spinal cord. Moreover, some glial cells were found to express this gene (Bergmann glia in cerebellum, etc.). Together with our previous reports that the Rac-GEFs STEF and Tiam1 were involved in several developmental processes such as neurite extension and neuronal migration (Matsuo et al., 2002, 2003; Kawauchi et al., 2003), P-Rex1 may also have multiple roles in many types of cells during the development of the nervous system, although we show its involvement only in cell migration in this report.
Activation of P-Rex1 with NGF
Although ectopic expression of P-Rex1 in PC12 cells had weak effects on ruffling membrane formation without the addition of NGF, it exhibited a marked ability to induce extraordinarily strong ruffling, namely, MOQ ruffling, when treated with NGF in a Rac1-dependent manner. We also observed that EGF induced MOQ ruffling in P-Rex1-transfected PC12 cells (data not shown). These results imply that the GEF activity of P-Rex1 can be activated by extracellular cues, such as NGF and EGF, in PC12 cells. This notion was also supported by the result that NGF treatment significantly increased GTP-bound Rac1 in P-Rex1-transfected PC12 cells.
In a neutrophil-like cell line, P-Rex1 was shown to be activated by PIP3 and Gβγ, acting as a coincidence detector in the two distinct signaling pathways (Welch et al., 2002). In PC12 cells, NGF stimulus has been known to activate several intracellular signaling pathways via TrkA and p75NTR (Huang and Reichardt, 2003). We showed that NGF-Rac1 link via P-Rex1 is mediated by TrkA rather than p75NTR. PIP3 is produced by NGF stimulus in PC12 cells, because PI3-kinase is known to be activated by NGF through the TrkA-Shc-Grb2 pathway (Carter and Downes, 1992; Raffioni and Bradshaw, 1992; Holgado-Madruga et al., 1997). We showed the involvement of PI3-kinase in NGF-induced MOQ ruffling formation in P-Rex1-transfected PC12 cells.
Very recently, a paradoxical in vitro biochemical study has been reported (Hill et al., 2005). Hill et al. showed that a P-Rex1 mutant lacking the PH domain could not be stimulated by PIP3, a substrate of PI3-kinase, suggesting that the PH domain confers PIP3 regulation of P-Rex1 Rac-GEF activity, in contrast to our result that ΔPH-P-Rex1 could induce NGF-dependent MOQ ruffling formation. Although we do not have a good explanation for this contradiction, the behavior of P-Rex1 may be different between in vitro and in vivo studies.
Although our experiments showed that exogenously expressed P-Rex1 could induce membrane ruffling in PC12 cells in an NGF-dependent manner, introduction of ΔDH-P-Rex1 failed to suppress NGF-induced ruffling membrane formation in PC12 cells. Furthermore, introduction of ΔDH-P-Rex1 did not suppress the neurite extension or differentiation of PC12 cells evoked by NGF treatment (data not shown). Consistent with the report of Kato and Negishi (2003) that the Rac-GEF complex Elmo/Dock180 is required for NGF-induced neurite extension and differentiation of PC12 cells, we observed the involvement of Elmo/Dock180 in NGF-induced ruffling membrane formation in PC12 cells. Conversely, our experiments with ΔDH-P-Rex1 demonstrated that endogenously expressed P-Rex1 is involved in NGF-induced cell migration of PC12 cells. These results suggest that multiple Rac-GEFs in the same cell may be differentially used for distinct cellular events, responding to the same extracellular signal, NGF. One possible mechanism underlying this plural usage of Rac-GEFs may be their distinct subcellular localization, although this has not been resolved in PC12 cells. However, as discussed below, subcellular accumulation of P-Rex1 in spindle-shaped migrating neurons may contribute to the specific subcellular function of this protein.
A potential role of P-Rex1 in neuronal migration
It is known that long-term NGF treatment (>48 h) forces PC12 cells to differentiate into neuronal cells with extended neurites (Greene et al., 1998). However, NGF administration for <24 h did not induce neurite extension in PC12 cells but increased the cell motility (Altun-Gultekin and Wagner, 1996). Our time-lapse study and trans-well migration assay showed that exogenously expressed P-Rex1 significantly enhanced the cell migration of PC12 cells in a Rac-dependent manner during addition of NGF. Experiments using ΔDH-P-Rex1 and siRNA for P-Rex1 further suggested that endogenous P-Rex1 is involved in the NGF-induced trans-well migration of PC12 cells. We further showed that ΔDH-P-Rex1 could suppress BDNF- and EGF-induced trans-well migration of primary cultured neurons from embryonic cortices, in which P-Rex1 accumulation is found in neuronal processes and adjacent cytoplasmic regions. Moreover, electroporation of ΔDH-P-Rex1 into the developing cerebral cortex resulted in perturbed cell migration of cortical neurons, indicating that P-Rex1 is involved in neuronal migration in the developing cerebral cortex. Neuronal migration is thought to consist of many complex processes, such as extension of the leading process, translocation of the nucleus, etc. In addition, many in vitro studies have suggested that cell migration is usually accompanied by ruffling membrane formation at the leading edge or leading process. However, we showed that endogenously expressed P-Rex1 was not required for ruffling membrane formation in PC12 cells. Therefore, other Rac-GEFs, which are involved in ruffling membrane formation, may also be cooperatively required for neuronal migration.
Our previous study using in utero electroporation showed that Rac plays an indispensable role in cortical neuronal migration and that two Rac-GEFs, namely STEF and Tiam1, also participate in that event (Kawauchi et al., 2003). Whereas stef and Tiam1 are expressed in both the intermediate zone and cortical plate during embryonic stages (Ehler et al., 1997; Yoshizawa et al., 2002; Kawauchi et al., 2003), P-Rex1 expression is more restricted. At E14, P-Rex1 transcripts were detected in the intermediate zone but not in the cortical plate in which migrating neurons reside. At E17, P-Rex1 expression was observed only in the ventricular side of the intermediate zone. This restricted expression pattern may imply a more specified function of P-Rex1 in neuronal migration.
Immunohistochemistry with anti-P-Rex1 antibodies revealed that P-Rex1 protein was localized in restricted subcellular regions in presumably radially migrating neurons in the intermediate zone; protein accumulation was observed in the putative leading process and adjacent cytoplasmic region of each cell. Previous reports suggested that those regions, situated at the migratory front, are important intracellular sites for neuronal migration, especially for nuclear translocation (Rivas and Hatten, 1995; Rakic et al., 1996; Lambert de Rouvroit and Goffinet, 2001; Xie et al., 2003). Furthermore, experiments using fluorescence resonance energy transfer showed that Rac is activated in the restricted cytoplasm at the migratory front in fibroblasts (Kraynov et al., 2000). Together with the fact that P-Rex1 is a GEF for Rac, the restricted localization of P-Rex1 in migrating neurons suggests a role in neuronal migration.
Previous studies suggested the involvement of some neurotrophins or growth factors in cortical neuronal migration. TrkB and TrkC, neurotrophin tyrosine kinase receptors, are expressed in migrating neurons in the cortical plate, although TrkA was not detected in the developing cortex (Ernfors et al., 1992). In addition, BDNF and neurotrophin-3 (NT-3) were shown to induce tyrosine phosphorylation of Trk receptors in cells of the developing neocortex, whereas NGF did not have that effect (Knüsel et al., 1994). Furthermore, BDNF and NT-4 stimulated the motility of cultured embryonic cortical cells, and additional application of BDNF or NT-4 to the developing neocortex caused abnormal cell migration (Behar et al., 1997; Brunstrom et al., 1997; Ringstedt et al., 1998). Accordingly, TrkB conditional knock-out mice showed a delay in neuronal migration in the developing cerebral cortex (Medina et al., 2004). Another tyrosine kinase receptor, EGFR, is also implicated in the regulation of radial migration in the developing cerebral cortex. EGFR and its ligands, including heparin-binding EGF (HB-EGF) and TGFα, are expressed in the developing cortex (Kornblum et al., 1997; Nakagawa et al., 1998). EGFR-targeted disruption and EGFR overexpression studies showed that EGFR is involved in normal radial migration (Threadgill et al., 1995; Caric et al., 2001). In PC12 cells, P-Rex1 was shown to be activated by NGF as well as EGF. Furthermore, P-Rex1 was shown to participate in both BDNF- and EGF-induced cell migration of cultured cortical neurons. These facts suggest that, in migrating neurons, P-Rex1 may be activated by neurotrophins or EGF to catalyze GDP-GTP exchange reactions of Rac in restricted subcellular regions, contributing to proper migration.
Footnotes
This work was supported by Grants-in-Aid for Scientific Research on Priority Areas [“Elucidation of Glia-Neuron Network-Mediated Information-Processing Systems” (#16047220) and “Membrane Traffic” (#16044225)] and by a Center of Excellence Grant, all from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and by grants from the Japan Brain Foundation and Takeda Science Foundation. We thank T. Nagase for providing KIAA1415 plasmid, K. Kaibuchi for pEF-BOS-HA wt-Rac1, H. Katoh for Elmo2-ΔN and Dock180-ISP plasmid, and A. Imura for anti-GFP monoclonal antibody. We are also grateful to N. Matsuo and R. Yu for helpful comments and T. Obata for technical assistance.
Correspondence should be addressed to Mikio Hoshino, Department of Pathology and Tumor Biology, Kyoto University Graduate School of Medicine, Yoshida-Konoe-cho, Sakyo-ku, Kyoto 606-8501, Japan. E-mail: mikio@lmls.med.kyoto-u.ac.jp.
Copyright © 2005 Society for Neuroscience 0270-6474/05/254406-14$15.00/0
T.K. and M.S. contributed equally to this work.
References
- Altun-Gultekin ZF, Wagner JA (1996) Src, ras, and rac mediate the migratory response elicited by NGF and PMA in PC12 cells. J Neurosci Res 44: 308-327. [DOI] [PubMed] [Google Scholar]
- Awasaki T, Saito M, Sone M, Suzuki E, Sakai R, Ito K, Hama C (2000) The Drosophila trio plays an essential role in patterning of axons by regulating their directional extension. Neuron 26: 119-131. [DOI] [PubMed] [Google Scholar]
- Behar TN, Dugich-Djordjevic MM, Li YX, Ma W, Somogyi R, Wen X, Brown E, Scott C, McKay RD, Barker JL (1997) Neurotrophins stimulate chemotaxis of embryonic cortical neurons. Eur J Neurosci 9: 2561-2570. [DOI] [PubMed] [Google Scholar]
- Brunstrom JE, Gray-Swain MR, Osborne PA, Pearlman AL (1997) Neuronal heterotopias in the developing cerebral cortex produced by neurotrophin-4. Neuron 18: 505-517. [DOI] [PubMed] [Google Scholar]
- Caric D, Raphael H, Viti J, Feathers A, Wancio D, Lillien L (2001) EGFRs mediate chemotactic migration in the developing telencephalon. Development 128: 4203-4216. [DOI] [PubMed] [Google Scholar]
- Carter AN, Downes CP (1992) Phosphatidylinositol 3-kinase is activated by nerve growth factor and epidermal growth factor in PC12 cells. J Biol Chem 267: 14563-14567. [PubMed] [Google Scholar]
- Ehler E, van Leeuwen F, Collard JG, Salinas PC (1997) Expression of Tiam-1 in the developing brain suggests a role for the Tiam-1-Rac signaling pathway in cell migration and neurite outgrowth. Mol Cell Neurosci 9: 1-12. [DOI] [PubMed] [Google Scholar]
- Ernfors P, Merlio JP, Persson H (1992) Cells expressing mRNA for neurotrophins and their receptors during embryonic rat development. Eur J Neurosci 4: 1140-1158. [DOI] [PubMed] [Google Scholar]
- Greene LA, Farinelli SE, Cunningham ME, Park DS (1998) Culture and experimental use of the PC12 rat pheochromocytoma cell line. In: Culturing nerve cells, Ed 2 (Banker G, Goslin K, eds), pp 161-188. Cambridge, MA: MIT.
- Guan KL, Rao Y (2003) Signalling mechanisms mediating neuronal responses to guidance cues. Nat Rev Neurosci 4: 941-956. [DOI] [PubMed] [Google Scholar]
- Hall A (1998) Rho GTPases and the actin cytoskeleton. Science 279: 509-514. [DOI] [PubMed] [Google Scholar]
- Hill K, Krugmann S, Andrews SR, Coadwell WJ, Finan P, Welch HC, Hawkins PT, Stephens LR (2005) Regulation of P-Rex1 by phosphatidylinositol (3,4,5)-trisphosphate and Gβγ subunits. J Biol Chem 280: 4166-4173. [DOI] [PubMed] [Google Scholar]
- Ho W, Uniyal S, Meakin SO, Morris VL, Chan BM (2001) A differential role of extracellular signal-regulated kinase in stimulated PC12 pheochromocytoma cell movement. Exp Cell Res 263: 254-264. [DOI] [PubMed] [Google Scholar]
- Holgado-Madruga M, Moscatello DK, Emlet DR, Dieterich R, Wong AJ (1997) Grb2-associated binder-1 mediates phosphatidylinositol 3-kinase activation and the promotion of cell survival by nerve growth factor. Proc Natl Acad Sci USA 94: 12419-12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino M, Matsuzaki F, Nabeshima Y, Hama C (1993) hikaru genki, a CNS-specific gene identified by abnormal locomotion in Drosophila, encodes a novel type of protein. Neuron 10: 395-407. [DOI] [PubMed] [Google Scholar]
- Hoshino M, Sone M, Fukata M, Kuroda S, Kaibuchi K, Nabeshima Y, Hama C (1999) Identification of the stef gene that encodes a novel guanine nucleotide exchange factor specific for Rac1. J Biol Chem 274: 17837-17844. [DOI] [PubMed] [Google Scholar]
- Huang EJ, Reichardt LF (2003) Trk receptors: roles in neuronal signal transduction. Annu Rev Biochem 72: 609-642. [DOI] [PubMed] [Google Scholar]
- Katoh H, Negishi M (2003) RhoG activates Rac1 by direct interaction with the Dock180-binding protein Elmo. Nature 424: 461-464. [DOI] [PubMed] [Google Scholar]
- Kawasaki Y, Sato R, Akiyama T (2003) Mutated APC and Asef are involved in the migration of colorectal tumour cells. Nat Cell Biol 5: 211-215. [DOI] [PubMed] [Google Scholar]
- Kawauchi T, Chihama K, Nabeshima Y, Hoshino M (2003) The in vivo roles of STEF/Tiam1, Rac1 and JNK in cortical neuronal migration. EMBO J 22: 4190-4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knüsel B, Rabin SJ, Hefti F, Kaplan DR (1994) Regulated neurotrophin receptor responsiveness during neuronal migration and early differentiation. J Neurosci 14: 1542-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblum HI, Hussain RJ, Bronstein JM, Gall CM, Lee DC, Seroogy KB (1997) Prenatal ontogeny of the epidermal growth factor receptor and its ligand, transforming growth factor alpha, in the rat brain. J Comp Neurol 380: 243-261. [DOI] [PubMed] [Google Scholar]
- Kraynov VS, Chamberlain C, Bokoch GM, Schwartz MA, Slabaugh S, Hahn KM (2000) Localized Rac activation dynamics visualized in living cells. Science 290: 333-337. [DOI] [PubMed] [Google Scholar]
- Kunda P, Paglini G, Quiroga S, Kosik K, Cáceres A (2001) Evidence for the involvement of Tiam1 in axon formation. J Neurosci 21: 2361-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert de Rouvroit C, Goffinet AM (2001) Neuronal migration. Mech Dev 105: 47-56. [DOI] [PubMed] [Google Scholar]
- Lin MZ, Greenberg ME (2000) Orchestral maneuvers in the axon: trio and the control of axon guidance. Cell 101: 239-242. [DOI] [PubMed] [Google Scholar]
- Luo L (2000) Rho GTPases in neuronal morphogenesis. Nat Rev Neurosci 1: 173-180. [DOI] [PubMed] [Google Scholar]
- Matsuo N, Hoshino M, Yoshizawa M, Nabeshima Y (2002) Characterization of STEF, a guanine nucleotide exchange factor for Rac1, required for neurite growth. J Biol Chem 277: 2860-2868. [DOI] [PubMed] [Google Scholar]
- Matsuo N, Terao M, Nabeshima Y, Hoshino M (2003) Roles of STEF/Tiam1, guanine nucleotide exchange factors for Rac1, in regulation of growth cone morphology. Mol Cell Neurosci 24: 69-81. [DOI] [PubMed] [Google Scholar]
- Medina DL, Sciarretta C, Calella AM, Von Bohlen Und Halbach O, Unsicker K, Minichiello L (2004) TrkB regulates neocortex formation through the Shc/PLCgamma-mediated control of neuronal migration. EMBO J 23: 3803-3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Sasahara M, Hayase Y, Haneda M, Yasuda H, Kikkawa R, Higashiyama S, Hazama F (1998) Neuronal and glial expression of heparin-binding EGF-like growth factor in central nervous system of prenatal and early-postnatal rat. Brain Res Dev Brain Res 108: 263-272. [DOI] [PubMed] [Google Scholar]
- Nishiyama T, Sasaki T, Takaishi K, Kato M, Yaku H, Araki K, Matsuura Y, Takai Y (1994) rac p21 is involved in insulin-induced membrane ruffling and rho p21 is involved in hepatocyte growth factor- and 12-O-tetradecanoylphorbol-13-acetate (TPA)-induced membrane ruffling in KB cells. Mol Cell Biol 14: 2447-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel JC, Hall A, Caron E (2002) Vav regulates activation of Rac but not Cdc42 during FcγR-mediated phagocytosis. Mol Biol Cell 13: 1215-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes P, Beeser A, Chernoff J, Schiller MR, Eipper BA, Mains RE, Huganir RL (2003) Rapid induction of dendritic spine morphogenesis by trans-synaptic ephrinB-EphB receptor activation of the Rho-GEF kalirin. Neuron 37: 263-274. [DOI] [PubMed] [Google Scholar]
- Posern G, Saffrich R, Ansorge W, Feller SM (2000) Rapid lamellipodia formation in nerve growth factor-stimulated PC12 cells is dependent on Rac and PI3K activity. J Cell Physiol 183: 416-424. [DOI] [PubMed] [Google Scholar]
- Raffioni S, Bradshaw RA (1992) Activation of phosphatidylinositol 3-kinase by epidermal growth factor, basic fibroblast growth factor, and nerve growth factor in PC12 pheochromocytoma cells. Proc Natl Acad Sci USA 89: 9121-9125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P, Knyihar-Csillik E, Csillik B (1996) Polarity of microtubule assemblies during neuronal cell migration. Proc Natl Acad Sci USA 93: 9218-9222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringstedt T, Linnarsson S, Wagner J, Lendahl U, Kokaia Z, Arenas E, Ernfors P, Ibáñez CF (1998) BDNF regulates reelin expression and Cajal-Retzius cell development in the cerebral cortex. Neuron 21: 305-315. [DOI] [PubMed] [Google Scholar]
- Rivas RJ, Hatten ME (1995) Motility and cytoskeletal organization of migrating cerebellar granule neurons. J Neurosci 15: 981-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander EE, van Delft S, ten Klooster JP, Reid T, van der Kammen RA, Michiels F, Collard JG (1998) Matrix-dependent Tiam1/Rac signaling in epithelial cells promotes either cell-cell adhesion or cell migration and is regulated by phosphatidylinositol 3-kinase. J Cell Biol 143: 1385-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Hall A (2002) Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes Dev 16: 1587-1609. [DOI] [PubMed] [Google Scholar]
- Sone M, Hoshino M, Suzuki E, Kuroda S, Kaibuchi K, Nakagoshi H, Saigo K, Nabeshima Y, Hama C (1997) Still life, a protein in synaptic terminals of Drosophila homologous to GDP-GTP exchangers. Science 275: 543-547. [DOI] [PubMed] [Google Scholar]
- Sone M, Suzuki E, Hoshino M, Hou D, Kuromi H, Fukata M, Kuroda S, Kaibuchi K, Nabeshima Y, Hama C (2000) Synaptic development is controlled in the periactive zones of Drosophila synapses. Development 127: 4157-4168. [DOI] [PubMed] [Google Scholar]
- Steven R, Kubiseski TJ, Zheng H, Kulkarni S, Mancillas J, Ruiz Morales A, Hogue CW, Pawson T, Culotti J (1998) UNC-73 activates the Rac GTPase and is required for cell and growth cone migrations in C elegans. Cell 92: 785-795. [DOI] [PubMed] [Google Scholar]
- Tabata H, Nakajima K (2003) Multipolar migration: the third mode of radial neuronal migration in the developing cerebral cortex. J Neurosci 23: 9996-10001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Ohashi R, Nakamura R, Shinmura K, Kamo T, Sakai R, Sugimura H (2004) Tiam1 mediates neurite outgrowth induced by ephrin-B1 and EphA2. EMBO J 23: 1075-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threadgill DW, Dlugosz AA, Hansen LA, Tennenbaum T, Lichti U, Yee D, LaMantia C, Mourton T, Herrup K, Harris RC, Barnard JA, Yuspa SH, Coffey RJ, Magnuson T (1995) Targeted disruption of mouse EGF receptor: effect of genetic background on mutant phenotype. Science 269: 230-234. [DOI] [PubMed] [Google Scholar]
- van Leeuwen FN, Kain HE, van der Kammen RA, Michiels F, Kranenburg OW, Collard JG (1997) The guanine nucleotide exchange factor Tiam1 affects neuronal morphology; opposing roles for the small GTPases Rac and Rho. J Cell Biol 139: 797-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch HC, Coadwell WJ, Ellson CD, Ferguson GJ, Andrews SR, Erdjument-Bromage H, Tempst P, Hawkins PT, Stephens LR (2002) P-Rex1, a PtdIns(3,4,5)P3- and Gβγ-regulated guanine-nucleotide exchange factor for Rac. Cell 108: 809-821. [DOI] [PubMed] [Google Scholar]
- Whitehead IP, Campbell S, Rossman KL, Der CJ (1997) Dbl family proteins. Biochim Biophys Acta 1332: F1-F23. [DOI] [PubMed] [Google Scholar]
- Xie Z, Sanada K, Samuels BA, Shih H, Tsai LH (2003) Serine 732 phosphorylation of FAK by Cdk5 is important for microtubule organization, nuclear movement, and neuronal migration. Cell 114: 469-482. [DOI] [PubMed] [Google Scholar]
- Yoshizawa M, Hoshino M, Sone M, Nabeshima Y (2002) Expression of stef, an activator of Rac1, correlates with the stages of neuronal morphological development in the mouse brain. Mech Dev 113: 65-68. [DOI] [PubMed] [Google Scholar]
- Yoshizawa M, Sone M, Matsuo N, Nagase T, Ohara O, Nabeshima Y, Hoshino M (2003) Dynamic and coordinated expression profile of dbl-family guanine nucleotide exchange factors in the developing mouse brain. Gene Expr Patterns 3: 375-381. [DOI] [PubMed] [Google Scholar]
- Zheng Y (2001) Dbl family guanine nucleotide exchange factors. Trends Biochem Sci 26: 724-732. [DOI] [PubMed] [Google Scholar]