Abstract
The Pet-1 [pheochromocytoma 12 ETS (E26 transformation-specific)] gene plays a critical role in the development of serotonin (5-HT)-modulated behaviors via its control of embryonic 5-HT neuron differentiation. Pet-1 transcription is induced exclusively in 5-HT neuron postmitotic precursors before the appearance of transmitter, and its restricted expression is maintained in the adult. However, the mechanisms that direct Pet-1 expression to this single CNS neuronal cell type are unknown. Here, we show, using transgenic methods, that genomic sequences upstream, but not downstream or within the Pet-1-coding region, are sufficient for 5-HT neuron-specific transgene expression. Enhancer sequences within a 40 kb upstream fragment directed position-independent lacZ (β-d-galactosidase) transgene expression to the developing hindbrain before the appearance of 5-HT. Moreover, virtually all of the 5-HT neurons in the adult were lacZ positive in all of the lines examined. Transgene expression in 5-HT neurons was maintained when the 40 kb fragment was truncated on its 5′ end to either 12 or 1.8 kb, although position independence was then lost. Analysis of transgene expression in Pet-1 null mice indicated that Pet-1 was required to maintain the activity of the Pet-1 enhancer region in a subset of 5-HT neurons. These findings suggest that a conserved 1.8 kb region immediately flanking the Pet-1-coding region is a critical genomic target of the transcriptional cascade that governs 5-HT neuron development and provide additional evidence for 5-HT neuron heterogeneity at the genetic level. We discuss the potential application of the Pet-1 transgenes reported here to the selective genetic manipulation of 5-HT neurons.
Keywords: Pet-1, 5-HT, raphe, transcription, ETS, genetic tools
Introduction
Serotonin (5-HT) is an important modulator of neural circuitry that gives rise to and maintains numerous behaviors and homeostatic functions (Jacobs and Azmitia, 1992). A large amount of literature also suggests that 5-HT signaling is important for the proper development of neural circuitry (Whitaker-Azmitia et al., 1996; Gaspar et al., 2003; Sodhi and Sanders-Bush, 2004). It is widely hypothesized that abnormal 5-HT neuron development contributes to the pathogenesis of certain neurological and psychiatric disorders, such as autism and anxiety, and that unknown genetic factors are involved (Belmonte et al., 2004; Gross and Hen, 2004; Sodhi and Sanders-Bush, 2004). The identification of these genetic factors and the determination of their importance in pathogenesis would be greatly facilitated by elucidating the transcriptional mechanisms that control 5-HT neuron development.
The genes governing human 5-HT neuron development are unknown, but several key genes have been identified recently in rodents and chickens (for review, see Scott and Deneris, 2005). A sonic hedgehog-induced transcriptional cascade involving Nkx2.2 (NK2 transcription factor related, locus 2), Mash1 (achaete-scute homolog 1), Gata-2 (Gata binding-factor 2), Gata-3, Lmx1b (LIM homeobox transcription factor 1β), and Pet-1 [pheochromocytoma 12 (PC12) E26 transformation-specific (ETS)] transcription factors has been implicated in mouse and chicken 5-HT neuron development (van Doorninck et al., 1999; Cheng et al., 2003; Ding et al., 2003; Hendricks et al., 2003; Pattyn et al., 2003, 2004; Craven et al., 2004). Although the epistatic relationships among these factors are beginning to be elucidated (Cheng et al., 2003; Ding et al., 2003; Pattyn et al., 2003, 2004; Craven et al., 2004), nothing is known about the transcriptional mechanisms that control their spatiotemporal expression in developing and adult 5-HT neurons.
Several experimental observations indicate that the Pet-1 ETS factor plays an important role in integrating the upstream transcriptional information required for specification of 5-HT neuron phenotype. First, Pet-1 expression in the brain is restricted to developing and adult 5-HT neurons (Hendricks et al., 1999; Pattyn et al., 2003; Craven et al., 2004). Second, combined misexpression of Pet-1, Lmx1b, and Nkx2.2 is necessary and sufficient for the generation of ectopic 5-HT neurons in the dorsal chick spinal cord (Cheng et al., 2003). Third, the majority of 5-HT neuron precursors fail to develop detectable serotonergic transmitter traits in Pet-1 null mice (Hendricks et al., 2003). In the minor population that does develop a serotonergic phenotype, the levels of tryptophan hydroxylase (TPH), 5-HT transporter (SERT), and vesicular monoamine transporter 2 are decreased, suggesting that Pet-1 is required for proper gene expression in this precursor pool. These differentiation defects result in the severely reduced levels of 5-HT in the developing and adult brain that are associated with increased aggression and anxiety-like behaviors (Hendricks et al., 2003). These findings support the existence of a Pet-1-dependent genetic program that impacts adult behavior through its control of 5-HT neuron differentiation.
The pivotal role of Pet-1 in 5-HT neuron development suggests that the elucidation of its transcriptional control will be an important step in determining the upstream regulatory cascade that specifies 5-HT phenotype. Hence, we used transgenic methods to identify a conserved 1.8 kb upstream enhancer region that recapitulates the spatial and temporal expression pattern of endogenous Pet-1 in developing and adult 5-HT neurons. Analysis of transgene expression in Pet-1-deficient mice indicates that the enhancer region is subject to positive autoregulation in a subset of 5-HT neurons. These findings define an important genomic target of the transcriptional cascade that governs 5-HT neuron development and offer potential genetic-based tools to facilitate in vivo and in vitro studies of 5-HT neurons.
Materials and Methods
Transgenes
A bacterial artificial chromosome (BAC) (RP23-165D11) containing the Pet-1 locus was isolated from the Roswell Park Cancer Institute (Buffalo, NY), library 23 C57BL/6 mouse BAC library (Research Genomics, Huntsville, AL), using a probe synthesized from a λ genomic clone that had been shown previously to span the Pet-1 protein-coding region (Fyodorov et al., 1998). BAC RP23-165D11 was then mapped using NotI, MluI, SwaI, and SpeI restriction endonucleases. Three oligonucleotide probes (5′ probe, 5′-GGG GTG GGC AAA GAT AAA G-3′; 3′ probe, 5′-ATC TGG GCC GAG GAT TTC-3′; and coding region probe, 5′-GCT ACG CCT ACC GCT TTG AC-3′) were used to isolate genomic fragments for preparation of six transgenes: 52Z, 512Z, 540Z, r540Z, 2PZ, and 323Z (see Figs. 1, 6). 52Z was prepared with a SpeI/NotI fragment starting from 65 bp 5′ of Pet-1 exon 1 and extending to 1.8 kb 5′ of Pet-1. 512Z was prepared with a SwaI/NotI fragment starting from 65 bp 5′ of Pet-1 exon 1 and extending to 12 kb 5′ of Pet-1. 540Z and r540Z were prepared with an MluI/NotI fragment starting from 65 bp 5′ of Pet-1 exon 1 and extending to 40 kb 5′ of Pet-1. 2PZ was prepared with a 1.8 kb NotI/NotI Pet-1 gene fragment starting from 65 bp 5′ of Pet-1 exon 1 and ending within exon 3. 323Z was prepared with a NotI/NotI fragment starting within exon 3 and extending to 23 kb 3′ of the gene. The genomic fragments for 2PZ, 512Z, and 52Z were subcloned into a lacZ (β-d-galactosidase) reporter plasmid (BGZA) (Yee and Rigby, 1993; Helms et al., 2000), immediately upstream of its β-globin minimal promoter in their natural orientation. Also included in this plasmid was a simian virus 40 (SV40) polyadenylation sequence. The 540Z, r540Z, and 323Z transgenes were built in the pBACe3.6 vector, a plasmid capable of supporting inserted fragments of large size with low levels of chimerism (Frengen et al., 1999). Therefore, these transgenes were propagated as BACs. The polylinker of pBACe3.6 was modified by destroying its NotI site, removing the stuffer fragment, and adding the polylinker 5′-MluI, NotI, AscI-3′. The 40 kb fragment was then cloned into the MluI/NotI sites of the modified pBACe3.6. The BGZA 3′ polylinker was modified to contain KpnI, EagI, HindIII sites to allow an EagI digest to release β-globin promoter, lacZ gene, and SV40 poly(A) sequences for subsequent subcloning into the pBACe3.6 NotI site. The orientation of the 40 kb fragment relative to lacZ in pBACe3.6 was determined by PCR using the primers 5′-CTG CAG GCT AGA AGC AAA TG-3′ and 5′-GGG GTG GGC AAA GAT AAA G-3′, which annealed to the 3′ end of the 40 kb fragment and 5′ end of the β-globin minimal promoter to produce a 600 bp fragment. The r540Z transgene carrying the 40 kb fragment in reverse orientation was also isolated from the same ligation reaction by using the primers 5′-CGC TGT TTG GCT TGC TTT CTG AC-3′ and 5′-GGG ACT TGA GGC TGT GGC TTC-3′, which annealed to the 5′ end of the 40 kb fragment and 5′ end of the β-globin promoter. Cycling conditions for both primer sets were 94°C for 180 s, 35 cycles of 94°C for 30 s, 58°C for 30 s, 72°C for 60 s, and 72°C for 300 s. The 323Z transgene was prepared by removing the β-globin, lacZ, and poly(A) sequences from BGZA with EagI and then subcloning this fragment into the modified pBACe3.6 vector, followed by the subcloning of the NotI/NotI 3′ 23 kb genomic fragment into the regenerated NotI site of pBACe3.6. No attempt was made to determine its orientation relative to the lacZ gene.
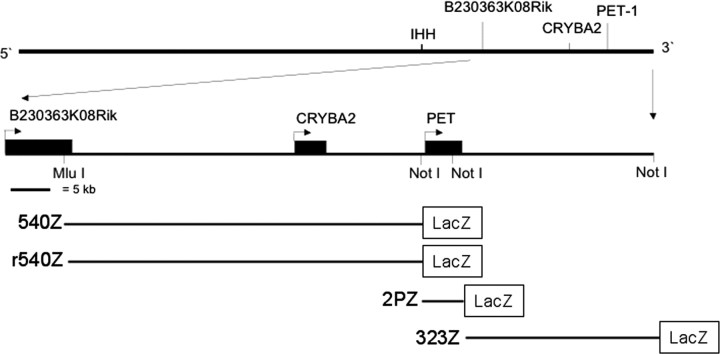
Figure 1.
Pet-1 transgene structures. Genomic organization of a bacterial artificial chromosome used to prepare transgenes. A 40 kb MluI/NotI upstream fragment was used to prepare 540Z and r540Z. A 1.8 kb NotI/NotI coding-region fragment was used to prepare 2PZ, and a 23 kb NotI/NotI downstream fragment was used to prepare 323Z. Each genomic fragment was placed immediately upstream of a β-globin promoter/lacZ/poly(A) cassette. 540Z and r540Z are identical, except that the orientation of the 40 kb insert relative to the β-globin promoter is reversed in r540Z. For details, see Materials and Methods. IHH, Indian hedgehog.
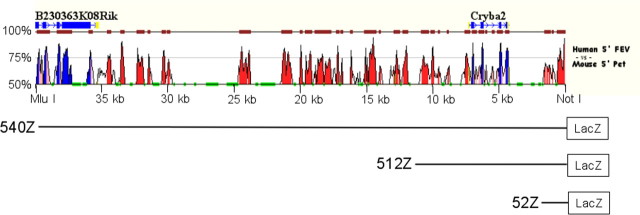
Figure 6.
Genomic sequence comparison upstream of mouse Pet-1 and human FEV. Genes are annotated as follows: boxes indicate exons (blue) or untranslated regions (yellow), and arrows show transcriptional orientation. Peaks show the degree of conservation (scale at left) between human and mouse sequences. Level of sequence identity is shown in exons (blue), introns (pink), and intergenic sequences (red). Repetitive elements are indicated in green. The zPicture sequence alignment and visualization tool was used for these comparisons (Ovcharenko et al., 2004). Based on these data, two truncations (512Z and 52Z) of 540Z were made to further localize Pet-1 enhancer sequences. The NotI site is 63 bp 5′ of Pet-1 exon 1.
Transgenic mice
Transgenes were isolated from the pBACe3.6 backbone by AscI digest and from the BGZA vector with KpnI and SacII for 2PZ or AscI/KpnI for 52Z. Constructs were purified for pronuclear injection according to Monani et al. (2000). The Case Transgenic and Targeting Facility performed pronuclear injections into hybrid C57B6/SJL F2 zygotes. Founders for each construct were genotyped by PCR. Founders 540Z, 512Z, and 52Z were identified using the same primer pairs and cycling conditions as those used to determine the 40 kb 540Z insert orientation. The PCR primers used to identify 2PZ founders were 5′-GCT ACG CCT ACC GCT TTG AC-3′ and 5′-CTG CAG GCT AGA AGC AAA TG-3′, product size 189 bp. Primers used for identification of 323Z founders were 5′-ATC TGG GCC GAG GAT TTC-3′ and 5′-CTG CAG GCT AGA AGC AAA TG-3′, product size 451 bp. Cycling parameters were the same as those used for 540Z. r540Z founders were identified using the same PCR primers and cycling conditions as those used to determine fragment orientation. C57B6/SJL founders were used for all analyses of 2PZ and 323Z transgenes and F1 C57B6/SJL offspring for 540Z, r540Z, 512Z, and 52Z transgenes.
Histology
Mice (4-6 weeks of age) were transcardially perfused with 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA), in 1× PBS, pH 7.4, followed by 3 h postfixation in 4% paraformaldehyde and PBS, pH 7.4, at 4°C. Tissue was then placed in 30% sucrose w/v, 1× PBS, pH 7.4, overnight at 4°C. Tissue sections (20 μm) were obtained using either a freezing microtome or a cryostat. Embryos were fixed by immersion for 30 min at 4°C in 4% paraformaldehyde, 1× PBS, pH 7.4, and then placed overnight in 30% sucrose, 1× PBS, pH 7.4. Fixed tissue was mounted in OCT medium (Tissue-Tek, Miles, Elkhart, IN) and stored at -80°C. Sections (10 μm) were obtained using a cryostat.
5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside staining. 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) (Alexis Biochemicals, San Diego, CA) was dissolved in 5 mm K3Fe(CN)6, 5 mm K4Fe(CN)6, and 2 mm MgCl2 in 1× PBS, pH 7.4, at 1 mg/ml. Fixed whole embryos or tissue sections were incubated overnight at 37°C in X-gal solution. Tissue sections were counterstained with 1% neutral red, dehydrated in ethanol/xylene, and mounted. Embryos were rinsed in 1× PBS, pH 7.4, and analyzed for lacZ expression.
Immunohistochemistry. The following antisera were used: rabbit polyclonal anti-5-HT (1:10,000; DiaSorin, Stillwater, MN), mouse monoclonal anti-tryptophan hydroxylase, which recognizes tryptophan hydroxylase 2 present in 5-HT neurons (Walther and Bader, 2003) (1:200; Sigma, St. Louis, MO), rabbit polyclonal anti-β-galactosidase (β-gal) (1:5000; MP Biomedicals, Irvine, CA), and goat polyclonal anti-β-galactosidase (1:2500; Biogenesis, Sandown, NH). The following secondary antisera were used: FITC (1:200), Texas Red (1:400), and cyanine 3 (1:800; all from Jackson ImmunoResearch, West Grove, PA). Fixed tissue sections were dried after mounting onto SuperFrost Plus slides (Fisher Scientific, Houston, TX) for 1 h. Slides were washed in PBST (1× PBS, pH 7.4, 0.1% Triton X-100) for 15 min and subsequently blocked for 2 h in PBST, 2% normal serum. Slides were then incubated in PBST, 2% normal serum, and primary antiserum overnight at 4°C. Slides were washed four times for 5 min each at room temperature in PBST, followed by incubation with secondary antisera for 2 h in the dark. Slides were washed for a final time in PBST for 5 min at room temperature and mounted using ProLong antifade reagent (Molecular Probes, Eugene, OR). Fluorescent images were collected on an Olympus Optical (Tokyo, Japan) BX51 microscope with a SPOT RT color digital camera (Diagnostic Instruments, Sterling Heights, MI). Postprocessing of images was performed using Adobe Photoshop 6.0 (Adobe Systems, San Jose, CA).
Sequence comparison
Phylogenetic sequence comparisons of the mouse Pet-1 upstream region were performed using the zPicture sequence alignment and visualization tool (Ovcharenko et al., 2004). DNA sequences were obtained from the National Center for Biotechnology Information [GenBank accession numbers AC104542 (mouse) and AC097468 (human)].
Results
Strategy for identifying serotonin neuron-specific elements controlling Pet-1 transcription
Our expectation was that one or more enhancer elements direct Pet-1 expression to developing and adult 5-HT neurons. Cell type-specific enhancers are often located several kilobases away from genes under their control and can be either upstream or downstream of the promoter of the gene. Therefore, to explore each of these possibilities, we isolated several DNA fragments from a mouse bacterial artificial chromosome: a 40 kb 5′ flanking fragment whose 3′ end is 63 bp upstream of the known 5′ end of Pet-1 exon 1, a 23 kb 3′ flanking fragment whose 5′ end is within the last Pet-1 exon, and a 2 kb gene fragment. Each fragment was placed upstream of a β-globin minimal promoter/lacZ gene/poly(A) signal sequence cassette to prepare transgenes 540Z (5′ 40 kb BAC fragment), 323Z (3′ 23 kb BAC fragment), and 2PZ (1.8 kb Pet-1-transcribed region BAC fragment), respectively (Fig. 1). Although computer analysis (PromoterInspector; Genomatix Software, München, Germany) suggested that a promoter region is present immediately upstream of the 5′-untranslated region, several attempts to confirm this with 5′ rapid amplification of cDNA ends using either PC12 cells or raphe total RNA failed. We suspect that the inability to map the 5′ end of the Pet-1-transcribed region was because of the high G and C content at the 5′ end of the transcript. Because we could not determine the transcription start site region of the Pet-1 gene, it is not clear whether the Pet-1 promoter is present in the 540Z and 2PZ transgenes. Several transgenic founders were identified for each of the reporters, and these founders were then bred to generate individual lines.
Upstream enhancer elements direct 5-HT neuron-specific expression of Pet-1
The majority of 5-HT-synthesizing neurons are clustered in the midbrain and medullary raphe nuclei. However, significant numbers of them are scattered in the reticular formation lateral to each of the raphe and within the ventrolateral medulla (Steinbusch, 1981). Nine neuroanatomical clusters of raphe and reticular 5-HT neurons were originally designated by Dahlstrom and Fuxe (1964). Clusters B1-B3 are located in the medulla and provide descending serotonergic innervation to the spinal cord and other parts of the medulla. Clusters B4-B9 are located in the pons and midbrain and provide ascending innervation to forebrain regions.
Variable transgene expression was detected among the several 2PZ and 323Z founders in different forebrain and midbrain structures, but none showed expression in any of the B nuclei (Table 1 and data not shown). Therefore, 5-HT neuron-specific regulatory elements are not likely to be positioned within or downstream of the gene. In contrast, 540Z expression was detected in cells located along the midline and in the reticular formation in a pattern that corresponded to the distribution of 5-HT neuron cell bodies in the nine serotonergic B nuclei (Fig. 2). The pattern of 540Z expression in the raphe and surrounding reticular formation was highly reproducible because lacZ staining was detected in each of the B nuclei in all 10 of the lines examined (Table 1). Moreover, 540Z expression was excluded from other regions of the brain in the majority of lines (Table 1). Pet-1 transcripts are expressed in the adrenal medulla (Fyodorov et al., 1998) and in dissociated retinal cultures (Francis and Deneris, 2002). However, transgene expression was not detected in the adrenal medulla or retina in any of the 10 540Z lines (Table 1), suggesting that regulatory elements for expression in these tissues are not present in the 40 kb upstream region. Enteric neurons and enterochromaffin cells of the gut synthesize large amounts of 5-HT, and Pet-1 transcripts have been detected in intestinal tissue with an RNase protection assay (Fyodorov et al., 1998). However, although 540Z expression was clearly detectable in a very small subset of enterocytes, it was not detected in enteric neurons or enterochromaffin cells (data not shown).
Table 1.
Transgene expression patterns
|
Transgenic lines |
Number of founders |
Embryonic 5-HT |
Adult 5-HT |
Adrenal medulla |
RGC |
Ectopic |
|---|---|---|---|---|---|---|
| 540Z | 10 | +++ | +++ | − | − | + |
| (10/10) | (10/10) | (5/5) | (5/5) | (3/10) | ||
| r540Z | 6 | +++ | +++ | ND | ND | + |
| (5/5) | (6/6) | (3/6) | ||||
| 512Z | 6 | +++/+ | +++/+ | ND | ND | +++ |
| (3/3) | (4/5) | (1/5) | ||||
| 52Z | 9 | +++ | +++ | ND | ND | +++ |
| (1/1) | (7/9) | (7/9) | ||||
| 2PZ | 5 | − | − | − | − | +++ |
| (5/5) | (5/5) | (5/5) | (5/5) | (5/5) | ||
| 323Z | 5 | − | − | − | − | +++ |
|
|
|
(5/5) |
(5/5) |
(5/5) |
(5/5) |
(5/5) |
Numbers in parentheses indicate the fraction of transgene-positive lines expressed in the indicated site. +++, Strong lacZ expression; +, weak lacZ expression; +++/+, variable lacZ expression among lines; −, no lacZ expression detected; ND, not determined; RGC, retinal ganglion cell layer; Ectopic, lacZ expression in forebrain or in extra-B nucleus midbrain and medullary structures.
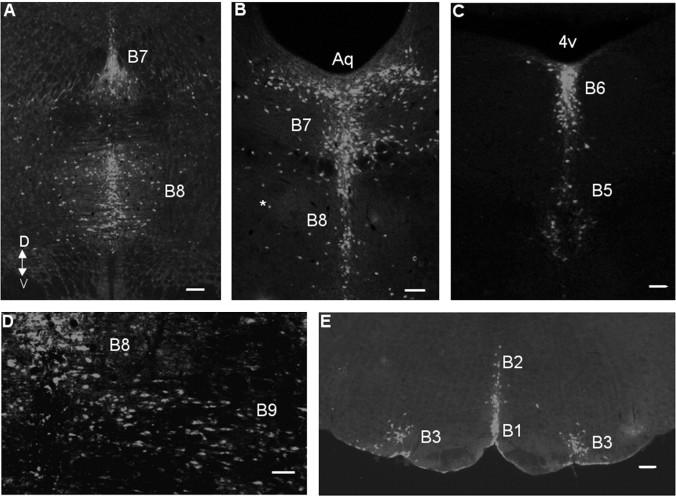
Figure 2.
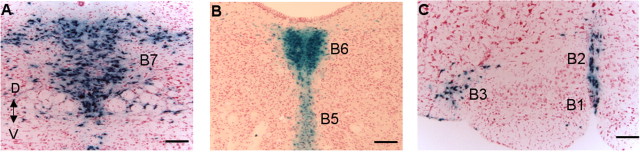
540Z is expressed specifically in midbrain and medullary B nuclei. Coronal sections (20 μm) were stained with rabbit anti-β-galactosidase antibody. A, B, Midbrain B7 dorsal (D) raphe and B8 median raphe nuclei. V, Ventral. B, The asterisk marks a representative reticular 5-HT neuron that expresses the transgene. Aq, Aqueduct. C, Pontine B6 dorsal raphe and B5 median raphe nuclei. 4v, Fourth ventricle. D, Midbrain B8 median raphe and B9 5-HT neurons. E, B1 raphe pallidus, B2 raphe obscurus, and B3 medial and ventrolateral medullary 5-HT neurons. Orientation of the section as identified for A applies to all of the panels. Scale bars: A, E, 200 μm; B-D, 100 μm.
5-HT neurons are intermingled with large numbers of nonserotonergic neurons and glia in each of the B nuclei (Steinbusch, 1981). To determine whether 540Z expression was restricted to 5-HT neurons, adult mid-hindbrain sections were coimmunostained for β-galactosidase and the 5-HT neuron-specific marker TPH. Virtually all of the TPH+ cells in each of the B nuclei were found to express lacZ, including those in the reticular formation. Similarly, nearly all of the lacZ+ cells were TPH+ (Fig. 3A-F and data not shown). We estimate that 1-2% of lacZ+ cells showed no anti-TPH immunoreactivity, and a similar number of 5-HT+ cells showed no anti-β-gal immunoreactivity. These lacZ+/TPH- and lacZ-/TPH+ cells were always intermingled with lacZ+/TPH+ cells, which suggests that variation in lacZ and TPH expression level or differential subcellular localization of β-galactosidase and TPH may be responsible for this low level of discordance. The findings presented in Figures 2 and 3 strongly suggest, therefore, that the transcriptional control elements directing 5-HT neuron-specific expression of Pet-1 in the adult brain are present within a 40 kb region upstream of Pet-1.
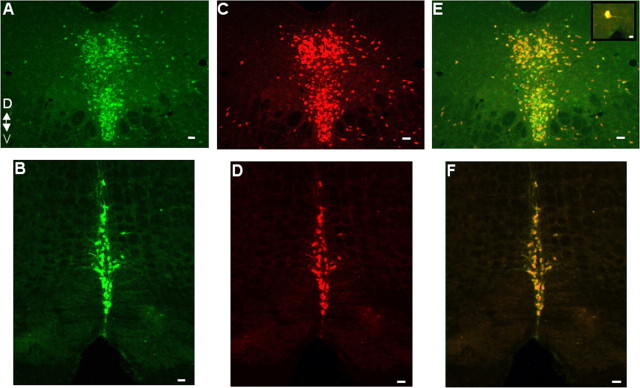
Figure 3.
5-HT neuron-specific transgene expression. A-D, Coronal adult mid-hindbrain sections (20 μm) were costained with anti-TPH antibody (A, B) and anti-β-galactosidase antibody (C, D). D, Dorsal; V, ventral. Similar results were obtained in all other B nuclei. E, F, Merged images. A, C, E, B7 dorsal raphe nuclei. B, D, F, B2 raphe obscurus. Orientation of the section as identified for A applies to all of the panels. Scale bars: A, C, E, 100 μm; B, D, F, 50 μm; E, Inset, 25 μm.
To determine whether enhancer elements within the 40 kb upstream region are responsible for the 5-HT neuron-specific expression pattern, the orientation of the 5′ 40 kb genomic fragment was reversed relative to the β-globin promoter to produce the r540Z transgene (Fig. 1). Offspring from several independent r540Z founder lines showed lacZ expression in all of the B nuclei (Fig. 4, Table 1). This finding indicates that enhancer sequences upstream of Pet-1 direct 5-HT neuron-specific expression.
Figure 4.
5-HT neuron-specific transgene expression is directed by enhancers. Coronal adult mid-hindbrain sections (20 μm) from r540Z animals stained for X-gal at the level of the indicated B nuclei are shown. Scale bars, 100 μm. Orientation of the section as identified for A applies to all of the panels. D, Dorsal; V, ventral.
540Z expression recapitulates the temporal expression pattern of Pet-1
The onset of Pet-1 expression in the mouse brain occurs at approximately embryonic day 11 (E11) in postmitotic precursors within the mantle layer (Pfaar et al., 2002; Pattyn et al., 2003). Expression is restricted to two bilateral longitudinal hindbrain domains at the level of rhombomeres r1-r3 and r5-r8. These two domains, classically referred to as rostral and caudal domains, eventually give rise to all of the 5-HT-synthesizing neurons in the rodent brain (Levitt and Moore, 1978; Lidov and Molliver, 1982; Wallace and Lauder, 1983; Pattyn et al., 2003). Expression of Pet-1 is first seen in the rostral domain, followed 1-2 d later by expression in the caudal domain. In each domain, Pet-1 expression precedes the appearance of 5-HT by <1 d (Hendricks et al., 1999).
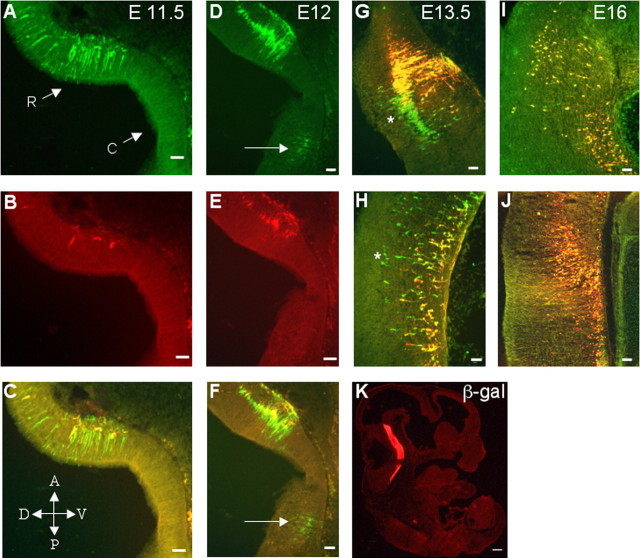
To determine whether or not the 40 kb upstream region was able to recapitulate the developmental timing of endogenous Pet-1 expression, 540Z expression was investigated in the embryonic hindbrain of several independent founder lines. As shown in Figure 5A-C, significant numbers of lacZ+ cells were detected at E11.5 in the mantle layer of the rostral, but not caudal, hindbrain. At this developmental stage, only a few lacZ+ cells located well within the mantle layer were 5-HT positive, whereas those positioned closer to the ventricular zone did not yet express the transmitter. A half-day later (Fig. 5D-F), greater numbers of double-labeled cells could be seen in the mantle layer of the rostral hindbrain. A few lacZ+ cells and no 5-HT+ cells were seen in the caudal domain at E12.0 (Fig. 5D,F, arrow). At E13.5, however (Fig. 5G,H), extensive coexpression of 540Z and 5-HT was observed in both the rostral and caudal hindbrain. Neurons that remained lacZ+/5-HT- were located more dorsally, proximal to the ventricular zone, consistent with their recent specification (Fig. 5G,H, asterisk). By E16 (Fig. 5I,J), a complete concordance between 5-HT and lacZ expression was evident. These findings show that the 40 kb upstream region in 540Z precisely recapitulated the temporal expression characteristics of endogenous Pet-1 with respect to rostral versus caudal expression and appearance of transmitter. The specificity of 540Z expression was also evident at E13.5 (Fig. 5K), when lacZ was detected exclusively in the developing hindbrain but not anteriorly, posteriorly, or in non-neural tissues.
Figure 5.
540Z expression recapitulates the temporal expression of the endogenous Pet-1 gene. Embryonic sagittal sections (10 μm) were used in all of the panels. A-K, Colocalization (C, F-J) of 5-HT (B, E) with β-galactosidase (A, D) at embryonic ages E11.5 (A-C), E12 (D-F), E13.5 (G, H, K), and E16 (I, J). K, β-galactosidase expression in whole-embryo view. R, Rostral domain; C, caudal domain; A, anterior; V, ventral; P, posterior; D, dorsal. Scale bars: A-F, I, J, 100 μm; G, H, 50 μm; K, 250 μm. Arrows (D, F) and asterisks (G, H) identify lacZ+/5-HT- neurons. Orientation of the section as identified for C applies to all of the panels.
A conserved 1.8 kb region immediately upstream of Pet-1 is sufficient for 5-HT neuron-specific expression
The 40 kb region upstream of Pet-1 is located on mouse chromosome 1 and is syntenic with a region on human chromosome 2 that includes fifth Ewing variant (FEV), a gene highly related to mouse and rat Pet-1 (Peter et al., 1997). A map displaying the genetic organization and conservation of the 40 kb region is shown in Figure 6. At the 5′ end of this region is a conserved open reading frame (ORF) designated B230363K08Rik, which encodes a protein of unknown function. Downstream of this ORF is a region of noncoding DNA of ∼25 kb in which large portions are at least 70% identical between the mouse and human sequences. A β-crystallin gene, CRYBA2, which terminates 5 kb 5′ of the Pet-1/FEV genes, follows this region. Sequences extending ∼2 kb immediately upstream of Pet-1/FEV show 70-90% identity, whereas the remainder of the intergenic region between CRYBA2 and Pet-1/FEV is not well conserved (Fig. 6). Comparison of the mouse sequence to that of chicken and zebrafish indicated little or no conservation in this region (data not shown).
To determine whether the conserved sequences between B230363K08Rik and CRYBA2 were required for 5-HT neuron-specific transgene expression, we prepared a construct, 512Z, which contains 12 kb of Pet-1 5′ flanking sequences and whose 3′ end is identical to that of the 540Z genomic sequence (Fig. 6). Similar to 540Z expression, the 512Z transgene was expressed in all nine of the serotonergic B nuclei (Table 1, Fig. 7A,B, and data not shown). However, although two 512Z lines showed 5-HT neuron-specific expression, transgene expression in several other lines was either very weak in raphe or not restricted to 5-HT neurons, suggesting significant position effects. In strongly expressing 512Z lines, coimmunostaining with anti-TPH antiserum indicated that 512Z expression was restricted to 5-HT neurons (Fig. 7 A,B). 512Z expression was also present in the rostral and caudal domains of the embryonic hindbrain (Fig. 7 C,D, Table 1). These findings demonstrate that the noncoding region downstream of B230363K08Rik is dispensable for 5-HT neuron-specific transgene expression.
Figure 7.
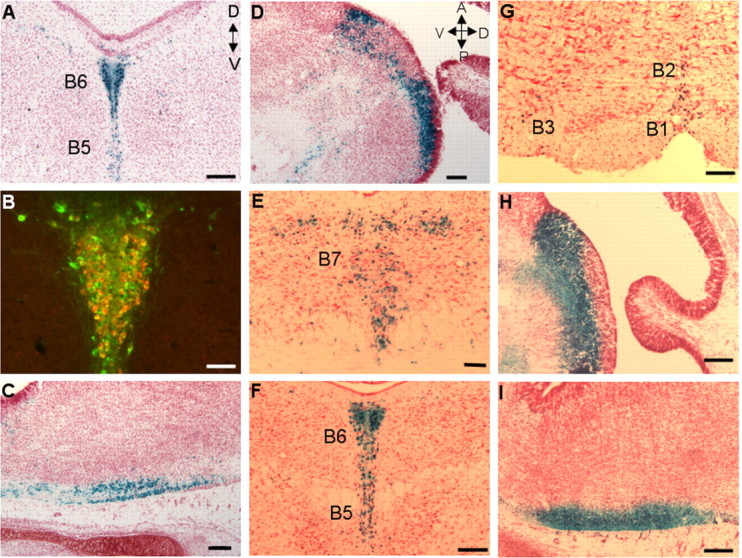
A 1.8 kb fragment is sufficient for 5-HT neuron-specific transgene expression. A, B, E-G, Coronal sections (20 μm) of adult mid-hindbrain. D, Dorsal; V, ventral. C, D, H, I, Sagittal sections (20 μm) of E16 embryonic rostral and caudal hindbrain. A, Anterior; P, posterior. A, X-gal reactivity showing 512Z expression in B6 dorsal raphe and B5 median raphe nuclei. B, Colocalization of TPH and 512Z in B5 and B6 nuclei. C, D, X-gal reactivity in E16 512Z caudal (C) and rostral (D) hindbrain. E-I, X-gal reactivity showing 52Z expression in midbrain B7 (E), B5 and B6 (F), and B1-B3 (G) nuclei, E16 rostral hindbrain (H), and E16 caudal hindbrain (I). Scale bars: A, C-I, 100 μm; B, 50 μm. Orientation of the section as identified for A also applies to B and E-G. Orientation of section as identified for D also applies to C, H, and I.
The 512Z findings suggested that conserved noncoding sequences within the CRYBA2/Pet-1 intergenic region might be sufficient for 5-HT neuron-specific expression. Thus, to further localize the transcriptional control region of the Pet-1 gene, the 512Z transgene was truncated on its 5′ end to produce a 1.8 kb transgene, 52Z, whose genomic sequences correspond to the highly conserved Pet-1 proximal portion of the CRYBA2/Pet-1 intergenic region (Fig. 6). Each founder line investigated showed reporter expression in all of the serotonergic B nuclei (Fig. 7E-G and data not shown) similar to that found with the longer transgene constructs. Moreover, 52Z expression was present in both the rostral and caudal serotonergic domains of developing hind-brain (Fig. 7H,I). Of the several adult lines examined, one showed restricted expression to B nuclei, whereas the others displayed noticeable ectopic expression in the midbrain and forebrain (Table 1, Fig. 7E-G). This result indicated that, unlike 540Z expression, but similar to 512Z, the 52Z pattern was influenced by the site of genomic integration (Table 1), which is consistent with the much smaller size of the transgene. Nevertheless, these findings demonstrate that genomic sequences sufficient for 5-HT neuron-specific expression reside within a conserved 1.8 kb region that immediately flanks the Pet-1 protein-coding region.
Pet-1 transcription is positively autoregulated in a subset of 5-HT neurons
Many transcription factors, including several bHLH (basic helix-loop-helix) and POU (Pit-Oct-Unc) domain factors, have been shown to either positively or negatively regulate their own transcription (DiMattia et al., 1997; Trieu et al., 1999; Helms et al., 2000; Meredith and Johnson, 2000), which is thought to be important for maintaining differentiated cell phenotypes. To determine whether or not Pet-1 is involved in regulating its own transcription, we introduced the 540Z transgene into Pet-1 null mice (Hendricks et al., 2003) and scored the number of transgene-positive cells that expressed 5-HT. Although normal numbers of 5-HT neuron precursors are present in Pet-1 homozygous null mice at E11.5, ∼70% fail to ever express 5-HT. These mutant precursors are likely eliminated and account for the absence of the majority of 5-HT neuron cell bodies in the adult null brain (Hendricks et al., 2003). However, it has not yet been determined whether these precursors are eliminated prenatally or postnatally. In Pet-1 heterozygotes, 540Z expression was detected in virtually all of the 5-HT+cells (Fig. 8A-C). In contrast, a complex pattern of 540Z expression was found in the homozygous null back-ground (Fig. 8D-F,H). Of all of the lacZ+ or 5-HT+ cells in the embryo, ∼30% expressed both lacZ and 5-HT, whereas ∼40% were lacZ-/5-HT+. The remainder were lacZ+/5-HT- (Fig. 8G,H). All three of the classes of cells were intermingled with one another and were not confined to any particular B nucleus. The tripartite pattern of transgene expression was maintained postnatally (Fig. 8H), demonstrating that the heterogeneity in transgene expression was not transient. These findings suggest that Pet-1 is involved either directly or indirectly in positive autoregulation in a subset of 5-HT neurons.
Figure 8.
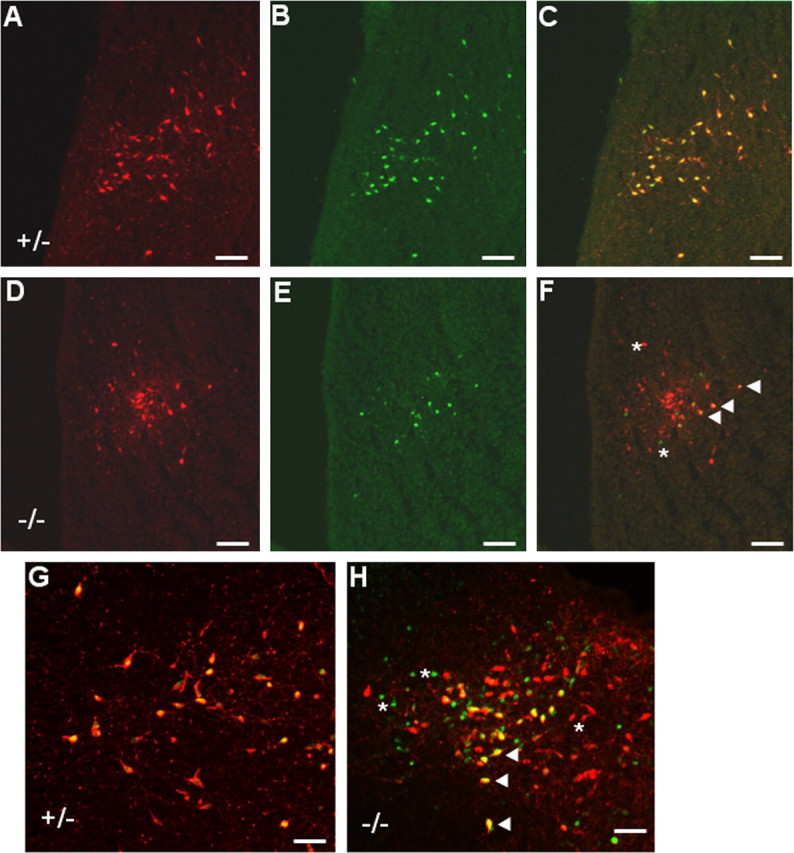
Pet-1 enhancer activity is autoregulated in a subset of 5-HT neurons. A-F, Coimmunostaining (C, F) of 5-HT (A, D) with β-galactosidase (B, E) in 20 μm sagittal sections of Pet-1+/- (A-C) and Pet-1-/- (D-F) E18 rostral hindbrain. 5-HT/β-gal coimmunostaining of Pet-1+/- (G) and Pet-1-/- (H) postnatal day 3 midbrain. F, H, Asterisks indicate representative lacZ-/5-HT+ and lacZ+/5-HT- cells in the Pet-1-/- embryo (F) and neonate (H). Arrowheads indicate cells coexpressing 540Z and 5-HT in the Pet-1-/- embryo (F) and neonate (H). Scale bars: A-F, 100 μm; G, H, 50 μm.
Discussion
Pet-1 is the only known transcription factor whose expression in the brain is restricted to developing and adult 5-HT neurons. Its expression in postmitotic 5-HT neuron precursors is a key step in the development of 5-HT-modulated behaviors, but the mechanisms that direct Pet-1 transcription to this single neuronal cell type are not known. In the present study, we used transgenic methods to identify a conserved 1.8 kb enhancer region immediately flanking the Pet-1 gene that is able to recapitulate the endogenous spatiotemporal pattern of Pet-1 transcription. We show that Pet-1 enhancer function is positively autoregulated in a subset of 5-HT neurons. These findings identify a critical genomic target of the transcriptional cascade that governs 5-HT neuron development and provide additional evidence for genetic heterogeneity within the 5-HT neuron population.
The transcriptional cascade that governs 5-HT neuron development converges on a 1.8 kb Pet-1 enhancer region
Our findings suggest that enhancer sequences controlling 5-HT neuron-specific transcription of Pet-1 are located in a conserved 1.8 kb region upstream of the gene and not within or downstream of it. First, lacZ expression in 5-HT neurons was not seen in any of the several 2PZ and 323Z lines that carry a Pet-1 gene fragment or 23 kb of 3′ noncoding sequences, respectively. Second, lacZ expression in developing and adult 5-HT neurons was evident in all 540Z and r540Z lines. In the majority of the 540Z and r540Z lines, transgene expression was not detected elsewhere in the developing or adult brain. Transgene expression was present in what appeared to be, within experimental error, all of the 5-HT neurons in all nine B nuclei. Thus, the Pet-1 upstream region was able to direct highly reproducible transgene expression specifically in developing and adult 5-HT neurons. Third, transgene expression, under the control of the upstream region, occurred first in the rostral domain and then in the caudal domain of the developing hindbrain and in each before the appearance of 5-HT. Finally, the pattern of transgene expression was reproduced with a conserved 1.8 kb upstream fragment immediately flanking the Pet-1-coding region. However, transgene expression directed by the 1.8 kb region was not restricted to 5-HT neurons in the majority of lines examined; therefore, boundary elements may be located farther upstream. As a consequence, the relatively small size of this fragment may subject it to significant position effects at the site of its genomic integration. Nevertheless, together, our findings suggest that the transcriptional cascade that controls 5-HT neuron development converges on a 1.8 kb conserved region located immediately upstream of the Pet-1 gene.
What are the transcription factors that directly interact with the Pet-1 enhancer region? Pet-1 expression is initiated after the expression of Nkx2.2 and Mash1 in progenitors and after Gata-2 and Lmx1b expression in postmitotic precursors (Hendricks et al., 1999; Ding et al., 2003; Pattyn et al., 2003, 2004; Craven et al., 2004). Mouse loss-of-function studies indicate that embryonic Pet-1 expression is dependent on Gata-2 and Mash1 function at all levels of the ventral hindbrain (Craven et al., 2004; Pattyn et al., 2004), whereas Nkx2.2 is required for the expression of Pet-1 at all rostrocaudal levels, except for the r1 rhombomere. In ovo misexpression of Gata-2 can induce the ectopic expression of Pet-1 in the r1 hindbrain (Craven et al., 2004). Lmx1b is not required for the initiation of Pet-1 expression (Cheng et al., 2003) but may be required to maintain Pet-1 expression after the appearance of serotonergic transmitter traits (Ding et al., 2003). In the absence of Phox2b (paired-like homeodomain protein 2b) function, Pet-1 is ectopically expressed in r4, in which 5-HT neurons are never normally generated, and it is prematurely expressed at all other axial levels of the hindbrain (Pattyn et al., 2003). Thus, Nkx2.2, Mash1, Gata-2, Lmx1b, and Phox2b may directly interact with the 1.8 kb region to control the proper spatiotemporal activation of Pet-1. Our analysis of 540Z expression in Pet-1-deficient mice indicated that a subset of 5-HT neurons were able to maintain transgene expression, whereas others were not. This finding suggests that Pet-1 may be required to maintain its own expression in some 5-HT neurons through direct interaction with the Pet-1 enhancer region. Alternatively, Pet-1 may autoactivate indirectly via positively acting downstream targets or via repression of a Pet-1 transcriptional repressor.
Although we have not yet addressed experimentally which, if any, of these factors directly interact with the Pet-1 enhancer region, database searches for conserved transcription factor binding sites identified several potential ETS and Gata-2 sites clustered within 500 bp of the Pet-1-coding region. Interestingly, the ETS-like sites conform to a previously determined Pet-1 consensus binding site (Hendricks et al., 1999); therefore, these sites may mediate direct autoactivation. Functional analysis of putative transcription factor binding sites in the 1.8 kb region with additional transgenic studies may begin to identify those components of the upstream transcriptional cascade that are directly responsible for Pet-1 expression in developing and adult 5-HT neurons.
The restricted expression of Pet-1 to a single neuronal cell type in the brain suggests that global transcriptional repression may also be involved. Albert and colleagues have determined that the REST (repressor element 1 silencing transcription factor), Freud-1 (5′ repressor element under dual repression binding protein-1), and NUDR (nuclear deformed epidermal autoregulatory factor-1 related homolog) repressors are all involved in transcriptional control of the 5-HT1A receptor gene (Lemonde et al., 2003, 2004; Ou et al., 2003). Sequence searches, however, failed to identify conserved REST, Freud-1, and NUDR repressor elements in the 1.8 kb Pet-1 enhancer region, although an NUDR site is present in the human sequence. Because the 1.8 kb enhancer region is, to our knowledge, the first description of enhancer elements sufficient for 5-HT neuron-specific expression in vivo, the Pet-1 enhancer region may be useful as a model for understanding the transcriptional mechanisms that direct the expression of other genes, such as tph2 (Walther and Bader, 2003), specifically to 5-HT neurons.
It is becoming increasingly evident that certain genetic variants within the transcriptional control region of serotonergic genes, such as SERT and the 5-HT1A receptor, are associated with increased risk for some mood disorders (Lesch et al., 1996; Caspi et al., 2003; Lemonde et al., 2003). It seems likely that other genes, particularly those that control the development of 5-HT neurons, contain polymorphisms that may also contribute to increased disease risk. One such candidate is FEV, a human gene that encodes an ETS factor whose DNA binding domain is identical to that of Pet-1 (Peter et al., 1997; Fyodorov et al., 1998). However, it is not yet known whether Pet-1 and FEV perform similar functions. Furthermore, although FEV transcripts are present in human raphe regions (Maurer et al., 2004), it remains to be determined whether FEV is expressed in 5-HT neurons and, if so, whether its spatiotemporal pattern is similar to that of Pet-1. However, the substantial degree of sequence identity between the mouse and human 1.8 kb enhancer region suggests that the transcriptional control of Pet-1 and FEV may be conserved. Thus, it will be of great interest to test this idea by investigating the function of the FEV upstream region and to determine whether naturally occurring genetic variation within it impacts FEV transcriptional control. If Pet-1 and FEV are indeed functional orthologs, polymorphisms in the FEV upstream region would be prime candidates for genetic factors that increase the risk for particular neurological or psychiatric disorders in which dysfunction of the 5-HT system has been implicated.
Pet-1 autoregulation and 5-HT neuron heterogeneity
The presence of lacZ+/5-HT+ and lacZ+/5-HT- cells in Pet-1 null mice suggests that Pet-1 is not required to maintain enhancer activity in these cells. However, intermingled with these cells were lacZ-/5-HT+ cells, and, therefore, Pet-1 may be required to maintain its own expression in a subset of 5-HT neurons. lacZ+/5-HT- cells are likely those serotonergic precursors that fail to ever express the transmitter and are eventually eliminated in the Pet-1 null brain (Hendricks et al., 2003). Given that normal numbers of 5-HT neuron precursors are present at the time of serotonergic neuron differentiation in the developing Pet-1 null hindbrain (Hendricks et al., 2003), the small numbers of lacZ+/5-HT- cells at E18 suggests that the majority of them have been eliminated by this stage. However, another equally likely possibility is that a significant portion of 5-HT- precursors require Pet-1 to maintain enhancer activity.
The differential requirement for Pet-1 in autoregulation provides additional support for 5-HT neuron heterogeneity at the genetic level. For instance, previous studies showed that the number of caudal 5-HT neurons in the developing hindbrain that depend on Gata-3 function for proper differentiation is much greater than those in the rostral domain (van Doorninck et al., 1999; Craven et al., 2004; Pattyn et al., 2004). Furthermore, the Nkx2.2/Phox2b-dependent switch from visceral motoneuron production to 5-HT neuron production occurs at the level of r2-r3 in the rostral hindbrain, but not in r1, which points to a genetically distinct program for 5-HT neuron specification in the most rostral portions of the hindbrain (Briscoe et al., 1999; Pattyn et al., 2003). Finally, the presence of a remaining population of 5-HT neurons in Pet-1 null mice indicates a differential dependence on Pet-1 within the 5-HT neuron population (Hendricks et al., 2003). What might be the potential significance of this genetic heterogeneity? Although this cannot yet be answered, we speculate that it may reflect mechanisms that determine the well documented differences in axonal morphology and trajectory, synaptic status, neurotoxin sensitivity, and physiological characteristics within the 5-HT neuron population (Kosofsky and Molliver, 1987; Tork, 1990; Mamounas et al., 1991; Beck et al., 2004).
Potential applications of the Pet-1 enhancer region
To conclude, we suggest that an important application of our findings is the selective genetic manipulation of 5-HT neurons. The high reproducibility of 540Z expression among different lines and no significant position effects in the majority of lines suggest the 40 kb Pet-1 upstream fragment can be used as a reliable tool to direct 5-HT neuron-specific transgene expression. In contrast, significant position effects were seen with both the 512Z and 52Z lines (Table 1), making the 12 and 2 kb fragments much less attractive as tools for spatial control of transgene expression. Embryonic stem cell knock-in (Soriano, 1995) or traditional BAC transgenic (Gong et al., 2003) approaches could also be used to exploit the Pet-1 expression pattern. However, gene expression directed by the 40 kb fragment offers greater specificity because 540Z was not expressed in peripheral cell types that express the endogenous Pet-1 gene. Moreover, simple subcloning methods for transgene preparation will allow the relatively rapid generation of lines. Although position independence was lacking with the 52Z transgene, its reproducible expression in 5-HT neurons suggests that the 1.8 kb upstream region may be useful for directing viral gene expression selectively to this neuronal cell type. Thus, the Pet-1 transgenes described here offer approaches that will greatly increase the accessibility of 5-HT neurons for a broad range of in vivo and in vitro studies.
Footnotes
This work was supported by National Institutes of Health Grant MH62723. We thank Dr. Jane Johnson for the BGZA plasmid and Kathy Lobur for excellent technical assistance in breeding and maintenance of transgenic lines. We also thank the Case Transgenic and Targeting Facility for outstanding service.
Correspondence should be addressed to Evan Deneris, Department of Neuroscience, Case Western Reserve University School of Medicine, 2109 Adelbert Road, Cleveland, OH 44106-4975. E-mail: esd@case.edu.
Copyright © 2005 Society for Neuroscience 0270-6474/05/252628-09$15.00/0
References
- Beck SG, Pan YZ, Akanwa AC, Kirby LG (2004) Median and dorsal raphe neurons are not electrophysiologically identical. J Neurophysiol 91: 994-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmonte MK, Cook Jr EH, Anderson GM, Rubenstein JL, Greenough WT, Beckel-Mitchener A, Courchesne E, Boulanger LM, Powell SB, Levitt PR, Perry EK, Jiang YH, DeLorey TM, Tierney E (2004) Autism as a disorder of neural information processing: directions for research and targets for therapy. Mol Psychiatry 9: 646-663. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Sussel L, Serup P, Hartigan-O'Connor D, Jessell TM, Rubenstein JL, Ericson J (1999) Homeobox gene Nkx2.2 and specification of neuronal identity by graded Sonic hedgehog signalling. Nature 398: 622-627. [DOI] [PubMed] [Google Scholar]
- Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, McClay J, Mill J, Martin J, Braithwaite A, Poulton R (2003) Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science 301: 386-389. [DOI] [PubMed] [Google Scholar]
- Cheng L, Chen CL, Luo P, Tan M, Qiu M, Johnson R, Ma Q (2003) Lmx1b, Pet-1, and Nkx2.2 coordinately specify serotonergic neurotransmitter phenotype. J Neurosci 23: 9961-9967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven SE, Lim KC, Ye W, Engel JD, De Sauvage F, Rosenthal A (2004) Gata2 specifies serotonergic neurons downstream of sonic hedgehog. Development 131: 1165-1173. [DOI] [PubMed] [Google Scholar]
- Dahlstrom A, Fuxe K (1964) Localization of monoamines in the lower brain stem. Experientia 20: 398-399. [DOI] [PubMed] [Google Scholar]
- DiMattia GE, Rhodes SJ, Krones A, Carriere C, O'Connell S, Kalla K, Arias C, Sawchenko P, Rosenfeld MG (1997) The Pit-1 gene is regulated by distinct early and late pituitary-specific enhancers. Dev Biol 182: 180-190. [DOI] [PubMed] [Google Scholar]
- Ding YQ, Marklund U, Yuan W, Yin J, Wegman L, Ericson J, Deneris E, Johnson RL, Chen ZF (2003) Lmx1b is essential for the development of serotonergic neurons. Nat Neurosci 6: 933-938. [DOI] [PubMed] [Google Scholar]
- Francis N, Deneris ES (2002) Retinal neuron activity of ETS domain-binding sites in a nicotinic acetylcholine receptor gene cluster enhancer. J Biol Chem 277: 6511-6519. [DOI] [PubMed] [Google Scholar]
- Frengen E, Weichenhan D, Zhao B, Osoegawa K, van Geel M, de Jong PJ (1999) A modular, positive selection bacterial artificial chromosome vector with multiple cloning sites. Genomics 58: 250-253. [DOI] [PubMed] [Google Scholar]
- Fyodorov D, Nelson T, Deneris E (1998) Pet-1, a novel ETS domain factor that can activate neuronal nAchR gene transcription. J Neurobiol 34: 151-163. [PubMed] [Google Scholar]
- Gaspar P, Cases O, Maroteaux L (2003) The developmental role of serotonin: news from mouse molecular genetics. Nat Rev Neurosci 4: 1002-1012. [DOI] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N (2003) A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 425: 917-925. [DOI] [PubMed] [Google Scholar]
- Gross C, Hen R (2004) The developmental origins of anxiety. Nat Rev Neurosci 5: 545-552. [DOI] [PubMed] [Google Scholar]
- Helms AW, Abney AL, Ben-Arie N, Zoghbi HY, Johnson JE (2000) Autoregulation and multiple enhancers control Math1 expression in the developing nervous system. Development 127: 1185-1196. [DOI] [PubMed] [Google Scholar]
- Hendricks T, Francis N, Fyodorov D, Deneris ES (1999) The ETS domain factor Pet-1 is an early and precise marker of central serotonin neurons and interacts with a conserved element in serotonergic genes. J Neurosci 19: 10348-10356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks TJ, Fyodorov DV, Wegman LJ, Lelutiu NB, Pehek EA, Yamamoto B, Silver J, Weeber EJ, Sweatt JD, Deneris ES (2003) Pet-1 ETS gene plays a critical role in 5-HT neuron development and is required for normal anxiety-like and aggressive behavior. Neuron 37: 233-247. [DOI] [PubMed] [Google Scholar]
- Jacobs BL, Azmitia EC (1992) Structure and function of the brain serotonin system. Physiol Rev 72: 165-229. [DOI] [PubMed] [Google Scholar]
- Kosofsky BE, Molliver ME (1987) The serotoninergic innervation of cerebral cortex: different classes of axon terminals arise from dorsal and median raphe nuclei. Synapse 1: 153-168. [DOI] [PubMed] [Google Scholar]
- Lemonde S, Turecki G, Bakish D, Du L, Hrdina PD, Bown CD, Sequeira A, Kushwaha N, Morris SJ, Basak A, Ou XM, Albert PR (2003) Impaired repression at a 5-hydroxytryptamine 1A receptor gene polymorphism associated with major depression and suicide. J Neurosci 23: 8788-8799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemonde S, Rogaeva A, Albert PR (2004) Cell type-dependent recruitment of trichostatin A-sensitive repression of the human 5-HT1A receptor gene. J Neurochem 88: 857-868. [DOI] [PubMed] [Google Scholar]
- Lesch KP, Bengel D, Heils A, Sabol SZ, Greenberg BD, Petri S, Benjamin J, Muller CR, Hamer DH, Murphy DL (1996) Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science 274: 1527-1531. [DOI] [PubMed] [Google Scholar]
- Levitt P, Moore RY (1978) Developmental organization of raphe serotonin neuron groups in the rat. Anat Embryol (Berl) 154: 241-251. [DOI] [PubMed] [Google Scholar]
- Lidov HGW, Molliver ME (1982) Immunohistochemical study of the development of serotonergic neurons in the rat CNS. Brain Res Bull 9: 559-604. [DOI] [PubMed] [Google Scholar]
- Mamounas LA, Mullen CA, O'Hearn E, Molliver ME (1991) Dual serotoninergic projections to forebrain in the rat: morphologically distinct 5-HT axon terminals exhibit differential vulnerability to neurotoxic amphetamine derivatives. J Comp Neurol 314: 558-586. [DOI] [PubMed] [Google Scholar]
- Maurer P, Rorive S, de Kerchove d'Exaerde A, Schiffmann SN, Salmon I, de Launoit Y (2004) The Ets transcription factor Fev is specifically expressed in the human central serotonergic neurons. Neurosci Lett 357: 215-218. [DOI] [PubMed] [Google Scholar]
- Meredith A, Johnson JE (2000) Negative autoregulation of Mash1 expression in CNS development. Dev Biol 222: 336-346. [DOI] [PubMed] [Google Scholar]
- Monani UR, Sendtner M, Coovert DD, Parsons DW, Andreassi C, Le TT, Jablonka S, Schrank B, Rossol W, Prior TW, Morris GE, Burghes AH (2000) The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in Smn(-/-) mice and results in a mouse with spinal muscular atrophy. Hum Mol Genet 9: 333-339. [DOI] [PubMed] [Google Scholar]
- Ou XM, Lemonde S, Jafar-Nejad H, Bown CD, Goto A, Rogaeva A, Albert PR (2003) Freud-1: a neuronal calcium-regulated repressor of the 5-HT1A receptor gene. J Neurosci 23: 7415-7425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovcharenko I, Loots GG, Hardison RC, Miller W, Stubbs L (2004) zPicture: dynamic alignment and visualization tool for analyzing conservation profiles. Genome Res 14: 472-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattyn A, Vallstedt A, Dias JM, Samad OA, Krumlauf R, Rijli FM, Brunet JF, Ericson J (2003) Coordinated temporal and spatial control of motor neuron and serotonergic neuron generation from a common pool of CNS progenitors. Genes Dev 17: 729-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattyn A, Simplicio N, Van Doorninck JH, Goridis C, Guillemot F, Brunet JF (2004) Ascl1/Mash1 is required for the development of central serotonergic neurons. Nat Neurosci 7: 589-595. [DOI] [PubMed] [Google Scholar]
- Peter M, Couturier J, Pacquement H, Michon J, Thomas G, Magdelenat H, Delattre O (1997) A new member of the ETS family fused to EWS in Ewing tumors. Oncogene 14: 1159-1164. [DOI] [PubMed] [Google Scholar]
- Pfaar H, von Holst A, Vogt Weisenhorn DM, Brodski C, Guimera J, Wurst W (2002) mPet-1, a mouse ETS-domain transcription factor, is expressed in central serotonergic neurons. Dev Genes Evol 212: 43-46. [DOI] [PubMed] [Google Scholar]
- Scott MM, Deneris ES (2005) Making and breaking serotonin neurons and autism. Int J Dev Neurosci, in press. [DOI] [PubMed]
- Sodhi MS, Sanders-Bush E (2004) Serotonin and brain development. Int Rev Neurobiol 59: 111-174. [DOI] [PubMed] [Google Scholar]
- Soriano P (1995) Gene targeting in ES cells. Annu Rev Neurosci 18: 1-18. [DOI] [PubMed] [Google Scholar]
- Steinbusch HWM (1981) Distribution of serotonin-immunoreactivity in the central nervous system of the rat-cell bodies and terminals. Neuroscience 6: 557-618. [DOI] [PubMed] [Google Scholar]
- Tork I (1990) Anatomy of the serotonergic system. Ann NY Acad Sci 600: 9-35. [DOI] [PubMed] [Google Scholar]
- Trieu M, Rhee JM, Fedtsova N, Turner EE (1999) Autoregulatory sequences are revealed by complex stability screening of the mouse brn-3.0 locus. J Neurosci 19: 6549-6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorninck JH, van Der Wees J, Karis A, Goedknegt E, Engel JD, Coesmans M, Rutteman M, Grosveld F, De Zeeuw CI (1999) GATA-3 is involved in the development of serotonergic neurons in the caudal raphe nuclei. J Neurosci 19: RC12(1-8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace JA, Lauder JM (1983) Development of the serotonergic system in the rat embryo: an immunocytochemical study. Brain Res Bull 10: 459-479. [DOI] [PubMed] [Google Scholar]
- Walther DJ, Bader M (2003) A unique central tryptophan hydroxylase isoform. Biochem Pharmacol 66: 1673-1680. [DOI] [PubMed] [Google Scholar]
- Whitaker-Azmitia PM, Druse M, Walker P, Lauder JM (1996) Serotonin as a developmental signal. Behav Brain Res 73: 19-29. [DOI] [PubMed] [Google Scholar]
- Yee SP, Rigby PW (1993) The regulation of myogenin gene expression during the embryonic development of the mouse. Genes Dev 7: 1277-1289. [DOI] [PubMed] [Google Scholar]