Abstract
This study addresses one genetic regulatory mechanism that establishes the distinct identities of rod and cone photoreceptors. Previous work has shown that mutations in either humans or mice in the gene coding for photoreceptor-specific nuclear receptor Nr2e3 cause a progressive retinal degeneration characterized by increased numbers of short-wave cones. In the present work, we have examined the cellular and developmental pattern of Nr2e3 protein localization in mammals and fish, identified an optimal Nr2e3 DNA-binding site using cycles of binding to recombinant Nr2e3, characterized the transcriptional activity of wild type and one of the disease-associated point mutations in Nr2e3 in transfected cells, and characterized the transcriptional defects in the naturally occurring Nr2e3 mutant (rd7) mouse. These experiments indicate that in the mature vertebrate retina Nr2e3 is expressed exclusively in rods, that expression of Nr2e3 is one of the earliest events in the pathway of rod-specific photoreceptor development, and that Nr2e3 functions, either directly or indirectly, as a repressor of cone-specific genes in rod photoreceptor cells.
Keywords: retina, rod photoreceptor, gene expression, nuclear receptor, retinal disease, vision, development
Introduction
The rod and cone photoreceptors of the vertebrate retina differ in a variety of respects (Rodieck, 1998). Rods are specialized for vision at low light levels, whereas cones are specialized for vision at high light levels, and they also mediate color vision. Although rods and cones share a common signaling pathway for phototransduction, most of the signaling proteins are coded by distinct rod- and cone-specific genes. Mammalian rods exit their terminal mitosis and develop later than cones, and they contact a distinct type of bipolar cell, the rod bipolar. In most mammals, rods greatly outnumber cones, and cones are generally confined to only the outermost layer of photoreceptor cells. In some vertebrates, including primates and birds, cones are found at high density in a specialized region of the retina, the fovea or visual pit, which mediates high-acuity vision. Finally, most vertebrate retinas contain only a single homogeneous class of rods but multiple classes of cones that differ with respect to their visual pigment content, spectral sensitivity, synaptic contacts, and abundance and distribution across the retina.
How do rod and cone photoreceptors develop the distinct programs of gene expression that determine their separate identities? The first steps in answering this question have come from the identification of several transcription factors that control photoreceptor-specific gene expression. Crx is a homeobox gene that is expressed in both rods and cones and is required for the expression of a variety of photoreceptor-specific genes (Chen et al., 1997; Furukawa et al., 1997). Crx mutations cause photoreceptor degeneration in both mice and humans (Freund et al., 1997; Swain et al., 1997; Furukawa et al., 1999). Nrl is expressed in rods and encodes a leucine zipper transcription factor. Targeted deletion of mouse Nrl causes rod photoreceptors to acquire many of the molecular characteristics of short-wave cones (also referred to as UV or S-cones) (Mears et al., 2001). In humans, Nrl point mutations are associated with retinal dystrophy (Bessant et al., 1999). In the mouse, thyroid hormone receptor β2 is required for the development of middle wave cones (also referred to as green or M-cones); in its absence, S-cones replace the M-cones (Ng et al., 2001).
Nuclear receptor Nr2e3/photoreceptor cell-specific nuclear receptor, the focus of this study, was initially identified as an orphan nuclear receptor expressed specifically in ocular tissues (Chen et al., 1999; Kobayashi et al., 1999). Although the cellular localization of Nr2e3 transcripts was initially a matter of dispute, it is now generally accepted that these transcripts are confined to photoreceptor cells (Kobayashi et al., 1999; Akhmedov et al., 2000; Haider et al., 2000, 2001). In humans, NR2E3 mutations cause enhanced S-cone syndrome, an autosomal recessive disorder in which a progressive degeneration of rod photoreceptors is accompanied by an increase in the relative numbers of S-cones and a markedly enhanced sensitivity to short-wave light (Haider et al., 2000). Mutations in NR2E3 have also been found in patients with Goldmann-Favre syndrome and clumped pigmentary retinal degeneration (Gerber et al., 2000; Sharon et al., 2003). A naturally occurring recessive Nr2e3 mutation was identified in the rd7 mouse as the cause of progressive photoreceptor degeneration associated with retinal folds and an ∼1.5-2 fold increase in the number of S-cones (Akhmedov et al., 2000; Haider et al., 2001; Yanagi et al., 2002). Among the major unanswered questions are the identities of the genes that are regulated by Nr2e3 and the developmental context of that regulation.
In the present work, we have (1) examined the cellular and developmental pattern of Nr2e3 protein localization in mammals and fish, (2) identified an optimal Nr2e3 DNA-binding site using cycles of binding to recombinant Nr2e3, (3) characterized the transcriptional activity of wild type and one of the disease-associated point mutations in Nr2e3 in transfected cells, and (4) characterized the transcriptional defects in the rd7 mouse. These experiments indicate that Nr2e3 is expressed exclusively in rods in the mature retina, that expression of Nr2e3 is one of the earliest events in the pathway of rod photoreceptor development, and that Nr2e3 functions as a repressor of cone-specific genes in rod photoreceptor cells.
Materials and Methods
Plasmids. The human and mouse Nr2e3 coding regions were PCR-amplified from human retina and mouse eye cDNA libraries, respectively. The GAL4 DNA-binding domain (amino acids 1-147) from pCMX/GAL-L/mRXRa (a gift from Dr. David Mangelsdorf, University of Texas Southwestern Medical Center, Dallas, TX) was cloned into the SmaI and EcoRI sites of pRK5-SK to obtain pRK5-SK-GAL. The ligand-binding domain (amino acids 126-410) of human Nr2e3 (hNr2e3) was cloned into the EcoRI and HindIII sites of pRK5-SK-GAL4. PCR mutagenesis was performed according to standard protocols. Fourteen copies of the GAL4 upstream activating sequence (UAS) site from pBUASE1BlynGFP (green fluorescent protein) (a gift from Dr. Scott Fraser, California Institute of Technology, Pasadena, CA), and either two or four copies of the optimal Nr2e3-binding site (AAGGTCAAAGATCA) (this study) in either orientation were cloned into the HindIII site of pTK-Luc (a gift from Dr. David Mangelsdorf).
Cloning of the zebrafish Nr2e3 ortholog. One zebrafish expressed sequence tag (EST; wz2916.2) with significant homology to hNr2e3 was identified by a BLAST search of the zebrafish EST database at Washington University (www.zfish.wustl.edu/) and used as a query to search the zebrafish genome database from the Sanger Center sequencing project (danio.mgh.harvard.edu/blast/blast_grp.html). One bacterial artificial chromosome clone (zKp91C1) was retrieved and analyzed using Genescan (genes.mit.edu/GEN-SCAN.html) and Spidey (www.ncbi.nlm.nih.gov/spidey/index.html). Based on this analysis, the predicted coding region of zebrafish Nr2e3 (zNr2e3) was PCR-amplified from an adult zebrafish retina cDNA library (a gift from Dr. James Hurley, University of Washington, St. Louis, MO). The complete 5′ end of zNr2e3 cDNA was obtained by rapid amplification of cDNA ends (RACE)-PCR using the Smart RACE cDNA kit (BD Biosciences) and total RNA from 55 hr postfertilization (hpf) zebrafish embryos.
Antibodies. Rabbit polyclonal antibodies against the mouse Nr2e3 (mNr2e3) N-terminal (amino acids 1-39) and linker (amino acids 119-208) regions and the zNr2e3 N-terminal (amino acids 1-45) and linker (amino acids 125-239) regions were generated by immunization with T7 gene 10 fusion proteins expressed in Escherichia coli (Studier et al., 1990) and affinity-purified using immobilized glutathione S-transferase (GST) fusion proteins carrying the same Nr2e3 sequences. Sera from rabbits immunized with the T7 gene 10 protein fused to amino acids 126-410 of hNr2e3 were affinity-purified using immobilized maltose-binding protein fused to either amino acids 126-221 or 337-410 of hNr2e3 to give antibodies against the linker region or the C-terminal region, respectively. Other antibodies used were anti-β-galactosidase and anti-synaptic vesicle protein 2 (SV2) monoclonal antibodies (mAbs) (Developmental Studies Hybridoma Bank, Iowa City, IA); anti-primate cone arrestin mAb 7G6 (Dr. Peter Macleish, Morehouse School of Medicine, Atlanta, GA); anti-zebrafish rhodopsin mAb 1D1 (Dr. James M. Fadool, Florida State University, Tallahassee, FL); anti-rhodopsin mAb 1D4 (Dr. Robert Molday, University of British Columbia, Vancouver, British Columbia, Canada); and rabbit anti-cone transducin α (I-20; Santa Cruz Biotechnology, Santa Cruz, CA). Rhodamine-conjugated peanut agglutinin was purchased from Vector Laboratories (Burlingame, CA).
Immunostaining. Enucleated mouse eyes were punctured at the cornea and fixed with 1% paraformaldehyde and PBS at room temperature for 1 hr. The fixed eyes were cryoprotected in PBS with 10% sucrose at 4°C overnight and then embedded in Optimal Cutting Temperature Compound (OCT; Tissue-Tek, Tokyo, Japan). For immunostaining of zebrafish, whole embryonic fish or dissected adult fish eyes were fresh-frozen in OCT. Immunostaining of frozen sections was performed essentially as described (Rattner et al., 2001). Whole-mount antibody staining of embryonic zebrafish eyes was performed as described by Luo et al. (2004). Images were captured on a confocal microscope (LSM510; Zeiss, Oberkochen, Germany).
Electrophoretic mobility shift assay. Double-stranded DNA probes containing either of the Nr2e3 binding sites AAGCTTAAGGTCAAAGATCAGGATCC (this study) or AAGCTTTTTAAAAGTCAAAAGTCAACTTCGGATCC (Kobayashi et al., 1999) were 32P-end-labeled with the Klenow fragment of DNA polymerase I. Whole-cell extracts were prepared from dissected mouse retinas or 293 cells (mock-transfected or transfected with pRK5/mNr2e3) in extraction buffer (20 mm HEPES pH 7.8, 300 mm KCl, 1% NP-40, 20% glycerol, 0.5 mm DTT, and a protease inhibitor mixture) at 4°C for 2 hr. DNA-binding reactions were performed in 20 μl of binding buffer [20 mm HEPES pH 7.8, 0.3 mg/ml BSA, 120 mm KCl, 10% glycerol, 0.1% NP-40, 0.5 mm DTT, and 0.1 mg/ml poly(dI-dC)] with a 30,000 cpm double-stranded DNA probe and 10-20 μg of protein from the whole-cell extract. For competition experiments, the indicated amount of unlabeled double-stranded competitor DNA was included in the DNA-binding reactions. After incubation at room temperature for 30 min, the protein-DNA complex was resolved on a 5% polyacrylamide gel in 1× Tris-borate-EDTA. The gels were subsequently dried and subjected to autoradiography.
In vitro selection of optimal Nr2e3-binding sites. A fusion protein consisting of GST and full-length mNr2e3 was expressed in transfected 293 cells (Tsai and Reed, 1997) and purified on glutathione-Sepharose 4B beads (Amersham Biosciences, Piscataway, NJ). A pool of double-stranded 56 mer oligonucleotides containing a central 20 mer of random sequence flanked by two invariant regions [ATCGATGTCGACAAGCTT(N)20 GGATCCGTCGACTCTAGA] was incubated with purified GST-mNr2e3, and the bound species was resolved by electrophoretic mobility shift assay (EMSA) as described above. The gel region corresponding to the protein-DNA complex was excised, and the DNA was eluted in Tris-EDTA. The eluted DNA was PCR-amplified with flanking primers to the invariant regions (upstream, ATCGATGTCGACAAGCTT; downstream, TCTAGAGTCGACGGATCC) for 15 cycles and then used for the next round of EMSA (Thiesen and Bach, 1990; Xiang et al., 1995). After three rounds of selection, the PCR products were digested with HindIII and BamH I and cloned, and individual clones were sequenced.
Cell culture and luciferase assay. A population of 293 cells, maintained in DMEM F-12 supplemented with 10% charcoal-treated fetal bovine sera (HyClone, Logan, UT), were transfected with 50 ng of expression plasmid, 500 ng of firefly luciferase reporter plasmid, and 10 ng of Renilla luciferase control plasmid (pRL-TK; Promega, San Luis Obispo, CA) using Fugene 6 reagent (Roche Molecular Biochemicals, Indianapolis, IN). Reporter activity was measured using a dual-luciferase report assay system (Promega) 44 hr after transfection.
In situ hybridization. In situ hybridization was performed essentially as described (Schaeren-Wiemers and Gerfin-Moser, 1993). Digoxigenin-labeled riboprobes were transcribed with T7 RNA polymerase from the indicated cDNAs, which were isolated either as cDNA clones or as cloned PCR products derived from mouse eye cDNA. Images were captured on an Axiophot microscope (Zeiss) using Openlab 3.1.2 software.
Microarray hybridization and data analysis. Microarray experiments were performed using mouse genome U74Av2 arrays (Affymetrix, Santa Clara, CA) with retina RNA probes obtained from postnatal day 10 (P10) rd7/+ and rd7/rd7 littermates or from P30 rd7/+ and rd7/rd7 mice. Data from two experiments (four independent RNA samples) at P10 and from three experiments (six independent RNA samples) were used for the analysis. For the triplicate P30 experiment, data were preprocessed using robust multichip averaging (Irizarry et al., 2003), converted into a log2-transformed format, and a p value was calculated for each transcript using t test statistics. Fold changes were calculated from the log2-transformed signal ratios. No scaling factor for variance stabilization or p value correction was used.
RNA preparation and blotting. Retinas from 1- to 2-month-old mice were snap-frozen in dry ice, and RNA was extracted using Trizol (Invitrogen, Carlsbad, CA) and an RNeasy kit (Qiagen, Valencia, CA) following the manufacturers' protocols. RNA blotting was performed using standard methods with 32P-labeled cDNA sequences, which were isolated as described under In situ hybridization.
Results
Nr2e3 sequences and localization among diverse vertebrates
As an initial step in analyzing Nr2e3 function, we asked whether the Nr2e3 gene is conserved across vertebrates and, if so, whether the expression patterns of the orthologous genes are also conserved. Starting with partial sequences for a presumptive zebrafish (Danio rerio) Nr2e3 ortholog, we used PCR amplification of an adult zebrafish eye cDNA library and RACE-PCR amplification of cDNA prepared from 55 hpf embryos to assemble the translated sequence shown in Figure 1 A. Comparison of the assembled zebrafish cDNA sequence and the subsequently determined zebrafish genomic sequence corresponding to it shows an intron-exon arrangement nearly identical to that observed for mammalian Nr2e3. The only potential difference is in the location of the first intron, a region where the amino acid sequences do not permit an unambiguous alignment. Further evidence that this zebrafish gene represents a bona fide Nr2e3 ortholog comes from its location close to the HexA, Stoml1, and Cyp11A1 genes, a linkage that is conserved in mouse and human despite an inversion that reverses the order of the mammalian Nr2e3 and HexA genes. The currently available zebrafish genome sequence contains no other genes with a comparable degree of sequence similarity to Nr2e3. The pufferfish (Fugu rubripes) genome also appears to encode a single Nr2e3 ortholog with an intron-exon arrangement similar to that seen among mammalian Nr2e3 genes. A dendrogram comparing presumptive Nr2e3 orthologs from chickens, various mammals, and fish with the most closely related human nuclear hormone receptors reveals a distinct branch of Nr2e3 sequences (Fig. 1B). Taken together, these data imply that the Nr2e3 gene has been maintained as a distinctive member of the nuclear receptor family throughout vertebrate evolution.
Figure 1.
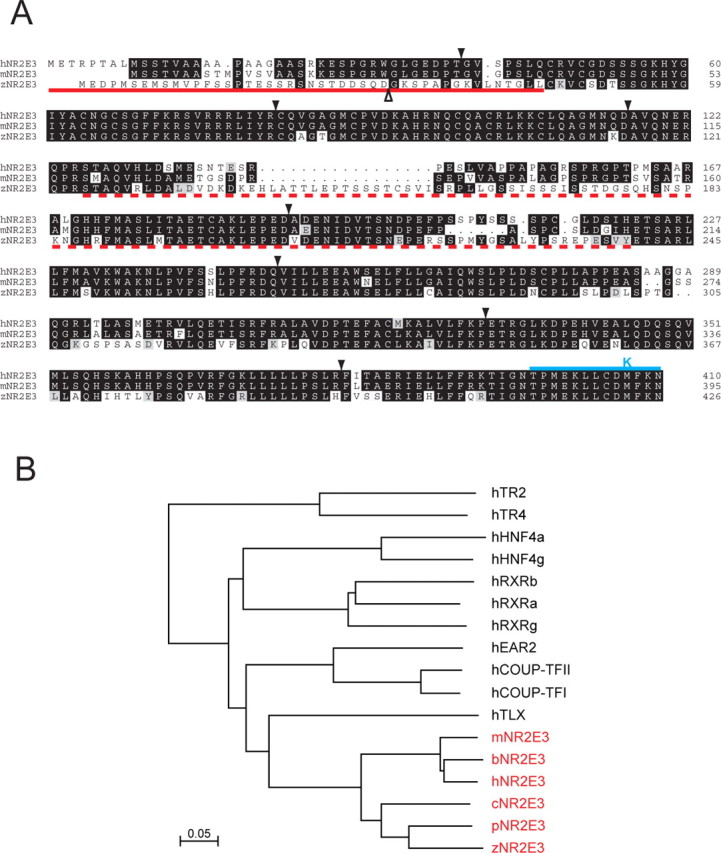
Comparison of Nr2e3 amino acid sequences among diverse vertebrates. A, Alignment of human, mouse, and zebrafish Nr2e3 sequences. Intron positions are shown with downward filled arrowheads. Except for the first intron in the zebrafish gene, shown by the upward open arrowhead, the intron locations are conserved. The solid red underline indicates immunogens used to generate anti-mouse and -zebrafish Nr2e3 N-terminal antibodies. The dashed red underline indicates immunogens used to generate anti-mouse and anti-zebrafish Nr2e3 linker region antibodies and the region included in the fusion protein used to affinity purify anti-human Nr2e3 linker region antibodies. The solid blue line and K above the C-terminal region indicate C-terminal and M407K truncation mutants, respectively, studied in human Nr2e3. B, Dendrogram showing the percent amino acid identities among Nr2e3 orthologs from various vertebrates and among the most closely related nuclear receptors from humans. Scale bar, 5% divergence. b, Bovine; c, chicken; h, human; m, mouse; p, pufferfish (Fugu rubripes); z, zebrafish.
Because mutations in NR2E3 appear to affect rod versus short-wave cone identity, a central question is whether Nr2e3 is expressed exclusively in rods, in both rods and cones, or in rods and a subset of cones. To address this question, we raised rabbit polyclonal antibodies to the linker between the DNA- and ligand-binding domains of Nr2e3, a region that is moderately conserved among mammalian Nr2e3 orthologs but has little homology with the most closely related nuclear hormone receptors such as Tlx (Fig. 1A). When macaque retinas were double-labeled for Nr2e3 and cone arrestin [the latter visualized with the primate-specific mAb 7G6, which labels the entire cell body of each cone (Zhang et al., 2003)], Nr2e3 was observed in rod but not cone nuclei (Fig. 2A,B). To address the localization of Nr2e3 in the mouse, we took advantage of a cone-specific β-galactosidase transgene (Wang et al., 1992). Like cone arrestin in the primate retina, β-galactosidase fills the entire cell body, permitting double-labeling analysis with nuclear Nr2e3. In contrast, most phototransduction proteins, including cone pigments and cone transducin, localize to the outer segment in mature retinas, limiting their utility in double-labeling analyses with nuclear antigens. Double labeling for Nr2e3 and β-galactosidase in the transgenic retina shows that Nr2e3 is confined to rod nuclei (Fig. 2D-F). In the mouse retina, Nr2e3 immunostaining is most intense near the nuclear periphery, the region where the chromatin appears to be least condensed (Carter-Dawson and LaVail, 1979). The specificity of Nr2e3 and β-galactosidase immunostaining in the mouse retina is demonstrated by their absence from a nontransgenic rd7/rd7 retina (Fig. 2C).
Figure 2.
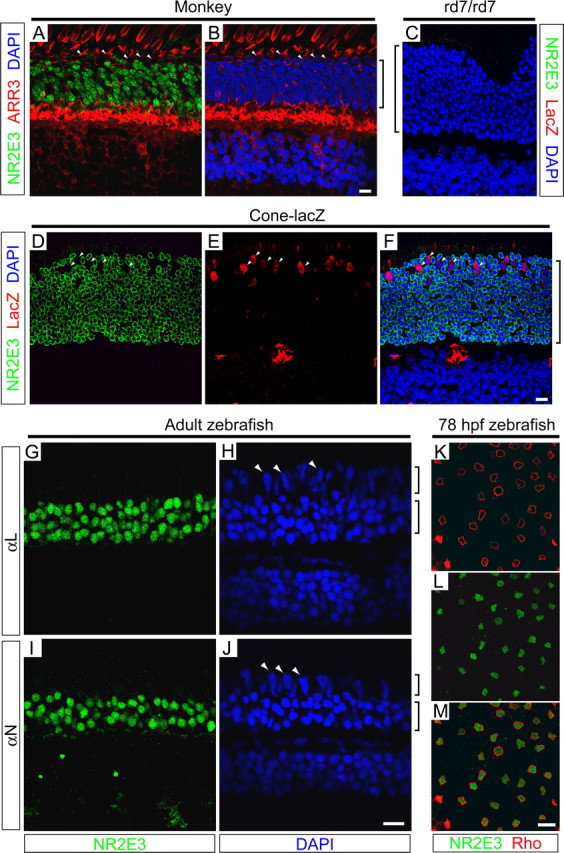
Immunolocalization of Nr2e3 to rod photoreceptors in monkey, mouse, and zebrafish retinas. A, B, Macaque retina immunostained for Nr2e3 and cone arrestin (ARR3; mAb 7G6) and counterstained with 4′, 6-diamidino-2-phenylindole (DAPI). C-F, rd7/rd7 (C) and transgenic cone-β galactosidase (D-F) mouse retinas immunostained for Nr2e3 and β-galactosidase and counterstained with DAPI. The specificities of mNr2e3 and β-galactosidase antibodies are demonstrated by the absence of immunostaining in the nontransgenic rd7/rd7 retina. The large red object in the outer plexiform layer in E and F is a blood vessel in which intravascular mouse IgG is stained with the Alexa 594-conjugated anti-mouse secondary antibody. G-J, Zebrafish retinas immunostained with antibodies to amino acids 1-45 (αN) and 125-239 (αL) of zNr2e3 and counterstained with DAPI. K-M, Flat mount of zebrafish retina at 78 hpf immunostained for rhodopsin and zNr2e3 shows that, in the central retina at this time point, Nr2e3 is observed exclusively in rods. In each of the species examined, Nr2e3 immunostaining is confined to rod nuclei; cone nuclei (A, B, D-F, H, J, white arrowheads) do not contain Nr2e3. Brackets adjacent to B, C, and F indicate the outer nuclear layer; brackets adjacent to H and J indicate the cone (top bracket) and rod (bottom bracket) nuclear layers. Scale bars, 10 μm.
To extend these observations to a more distant vertebrate, antibodies against two distinct regions of zebrafish Nr2e3 (Fig. 1A) were used to immunostain zebrafish retinas. Immunostaining of the adult retina, in which rod nuclei are spatially segregated from cone nuclei and occupy only the inner half of the outer nuclear layer, shows that Nr2e3 is expressed exclusively in rods (Fig. 2G-J). The rod specificity of Nr2e3 localization is also observed in retinal flat mounts by double labeling with anti-zebrafish rhodopsin mAb 1D1 (Fig. 2K-M). At 78 hpf, ∼2 d after the birth of most rod and cone photoreceptors, there is sufficient rhodopsin in rod cell bodies to use it as a marker for double labeling with a nuclear antigen.
Nr2e3 is expressed in rod precursors
The hypothesized role of Nr2e3 in controlling gene expression in rod photoreceptors predicts that Nr2e3 expression should precede the expression of those gene products that characterize the fully differentiated state. Rhodopsin is one of the earliest markers of rod differentiation, and in the developing mouse retina, as in the developing zebrafish retina, it initially accumulates in the cell bodies of immature rods. Double immunostaining for rhodopsin and Nr2e3 at P3 and P6 shows the expected time-dependent increase in the number of developing rods (Fig. 3A-F). However, at each of these time points, rhodopsin-expressing cells constitute only a subset of the Nr2e3-positive cells. This can be seen most clearly at P6, when recently postmitotic cells in the future inner nuclear layer commit to a rod fate and migrate outward to their final destination in the outer nuclear layer (Young, 1984). Whereas Nr2e3 accumulates in the nuclei of premigratory rod precursors, rhodopsin accumulation is generally observed only after the cells migrate to their final destination in the outer nuclear layer (Fig. 3D-H). The junction between the developing inner and outer nuclear layers is identified by the accumulation of SV2 in the developing outer plexiform layer (Fig. 3G-J). At P14, after the period of rod precursor migration, Nr2e3 immunolabeling is confined to the outer nuclear layer (Fig. 3I,J). The accumulation of Nr2e3 in presumptive photoreceptor precursors begins at least as early as embryonic day 18 (E18), when small numbers of Nr2e3-stained nuclei are seen in the outer retina (data not shown); at all developmental stages, Nr2e3 immunoreactivity localizes to the nucleus.
Figure 3.
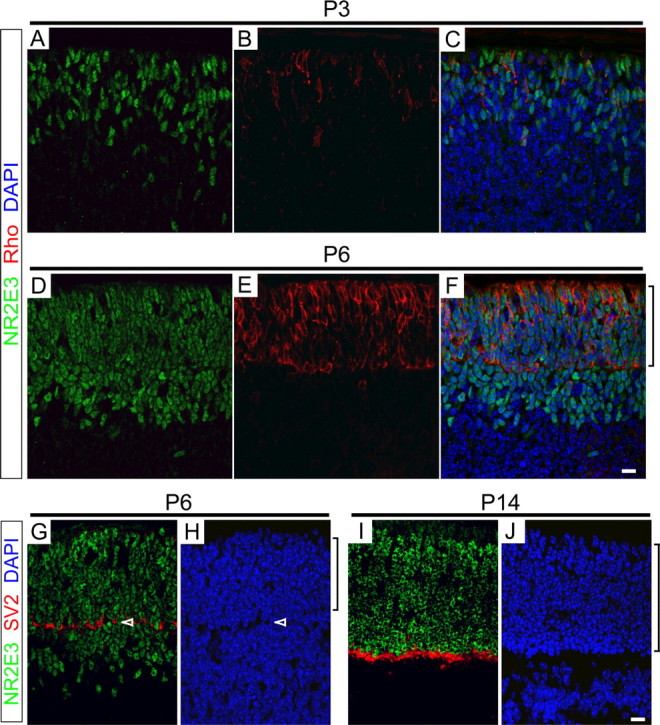
Nr2e3 accumulates in rod precursors in the developing mouse retina. A-F, P3 (A-C) and P6 (D-F) mouse retinas immunostained for Nr2e3 and rhodopsin and counterstained with DAPI. G-J, P6 (G, H) and P14 (I, J) mouse retinas immunostained for Nr2e3 and SV2 and counterstained with DAPI. Only the outer half of each retina, containing the future inner and outer nuclear layers, is shown. Brackets adjacent to F, H, and J indicate the developing outer nuclear layer. Arrowheads in G and H indicate the developing outer plexiform layer. All rhodopsin-containing cells also contain Nr2e3, but additional cells, presumably early rod precursors, contain Nr2e3 but do not contain rhodopsin. Scale bars, 10 μm.
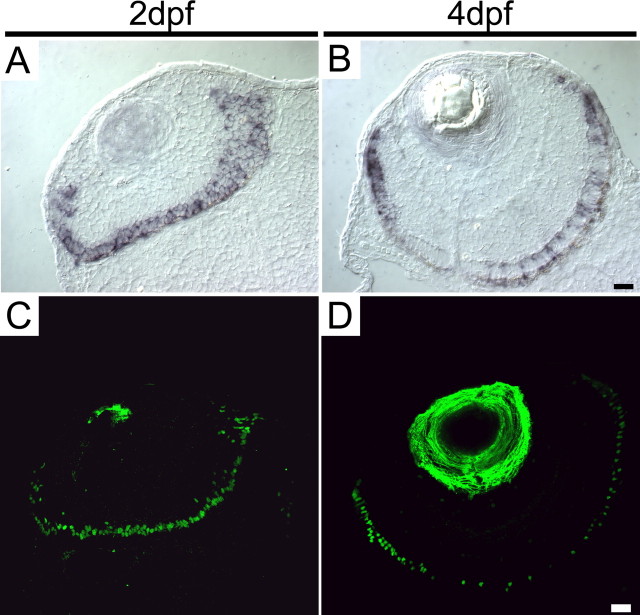
A more complex pattern of Nr2e3 expression is seen in the developing zebrafish retina. At 2 d postfertilization (dpf), a time point immediately after the birth of many cone and rod photoreceptors and before the accumulation of detectable levels of visual pigment, Nr2e3 transcripts and Nr2e3 protein accumulate in most, and perhaps all, developing photoreceptors as judged by the high density of labeled cells (Fig. 4A,C). However, by 4 dpf, Nr2e3 immunostaining in the central retina is restricted to scattered rod photoreceptors (Figs. 2K-M, 4C,D), a spatial and temporal pattern previously described for developing rods (Fadool, 2003); these cells populate the photoreceptor mosaic over the ensuing several days. In the less mature retinal periphery at 4 dpf, Nr2e3-expressing cells are present at high density and therefore are likely to include cone precursors. These data suggest that in zebrafish, Nr2e3 may play a transient role in early cone photoreceptor development.
Figure 4.
Transient Nr2e3 expression in cones in the developing zebrafish retina. In situ hybridization (A, B) and immunostaining (C, D) for Nr2e3 at 2 and 4 dpf is shown. Immunostaining of the lens is nonspecific. Cells throughout the photoreceptor layer are stained at 2 dpf, but only rods express Nr2e3 in the most mature (central) region of the retina at 4 dpf. Scale bars, 20 μm.
Optimal Nr2e3-binding sites mediate transcriptional repression in transfected cells
In earlier studies, Kobayashi et al. (1999) demonstrated the binding of expressed human Nr2e3 to a synthetic DNA target designed on the basis of a naturally occurring site for the related Drosophila protein Tailless. To define the optimal binding site(s) for Nr2e3 in an unbiased manner, we used repeated cycles of in vitro selection to isolate a family of synthetic duplex DNA targets from a library of DNA segments of random sequence (Thiesen and Bach, 1990). The synthetic DNA library was incubated with a purified GST fusion to full-length mouse Nr2e3 that was expressed in 293 cells, and the bound DNA targets were enriched by EMSA. After three rounds of binding and PCR amplification, an alignment of the sequences of 23 cloned sites yielded the consensus 5′-(A/G)AG(A/G)TCAAA(A/G)(A/G)TCA (Fig. 5A).
Figure 5.
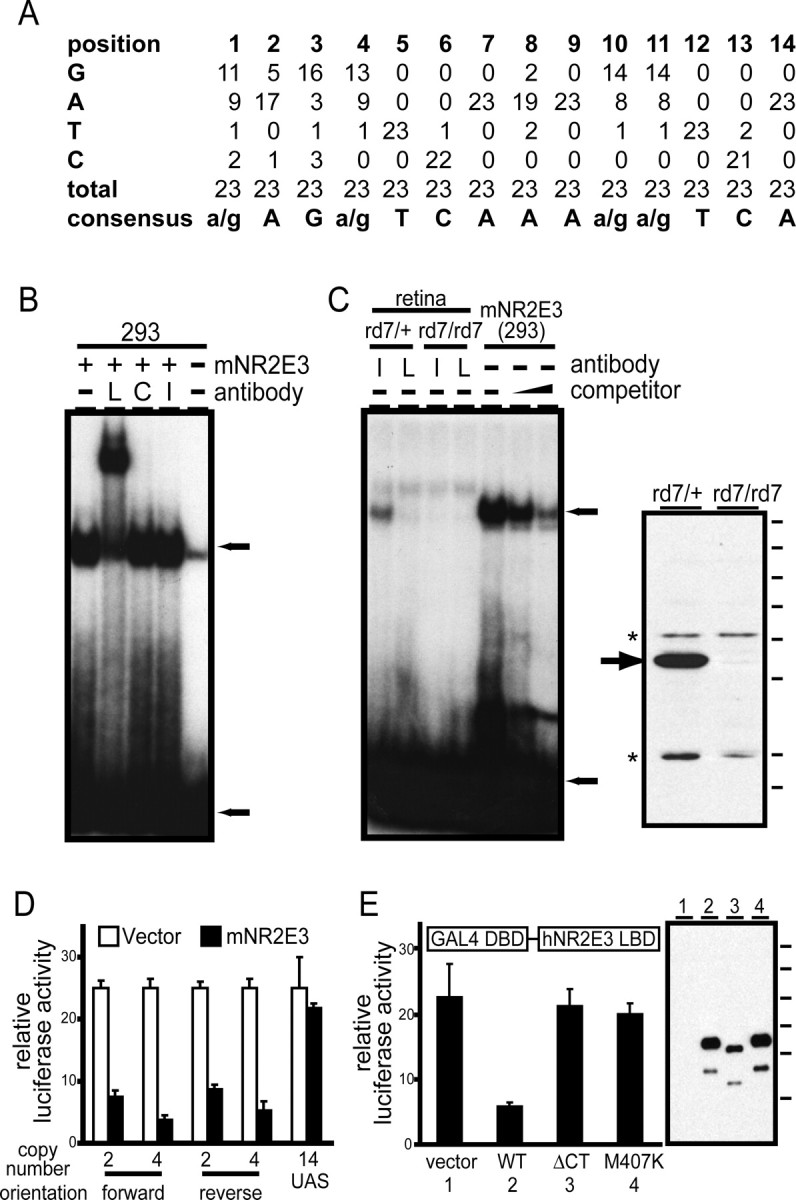
An optimal Nr2e3-binding site mediates Nr2e3-dependent transcriptional repression in transfected cells. A, Compilation of 23 cloned oligonucleotide sequences after three rounds of selection with a GST fusion to full-length mNr2e3. B, EMSA with mNr2e3 produced in transfected 293 cells using the DNA duplex probe defined by Kobayashi et al. (1999) (see Materials and Methods). Antibodies directed against the hNr2e3 linker region (L) produce a supershift; antibodies directed against the C-terminal 73 amino acids of hNr2e3 (C) or against an irrelevant protein (photoreceptor cadherin; I) do not produce a mobility shift. Extracts from untransfected 293 cells (-) produce a weak EMSA band that has a slightly higher electrophoretic mobility than that produced by mNr2e3. C, Left panel, EMSA using an optimal DNA duplex probe determined by in vitro selection (see Materials and Methods). Left four lanes, EMSA using retina extracts from rd7/+ or rd7/rd7 littermates incubated with anti-hNr2e3 linker region or irrelevant antibodies. Right three lanes, EMSA with mNr2e3 produced in transfected 293 cells in the presence of increasing quantities (0.5 and 2.5 μm) of the same optimal DNA duplex as competitor. Top and bottom arrows, Bound and free probe, respectively. Right panel, Specificity of the anti-hNr2e3 linker region antibodies used for super shifting experiments determined by immunoblotting of retina proteins from rd7/+ or rd7/rd7 littermates. Arrow, Nr2e3; asterisks, cross-reacting proteins. Molecular mass standards (from top to bottom): 183, 114, 81, 64, 50, 37, 26, and 20 kDa. D, Cotransfection of 293 cells with a vector (pRK5) directing the expression of full-length mNr2e3 or with pRK5 alone, together with a luciferase reporter under the control of a minimal herpes simplex virus thymidine kinase promoter 3′ of the indicated number and orientation of optimal Nr2e3-binding sites determined by in vitro selection. A control reporter carries 14 Gal4 UAS sites in place of the Nr2e3 sites. Nr2e3 represses expression in a target-site-dependent manner. E, Cotransfection of 293 cells with pRK5 directing the expression of a Gal4DBD-hNr2e3LBD fusion protein together with a luciferase reporter under the control of a minimal herpes simplex virus thymidine kinase promoter 3′ of 14 tandem Gal4 UAS sites. The hNr2e3 LBD corresponds to amino acids 126-410. Repression of the reporter is seen with the WT Nr2e3 LBD sequence but not with a C-terminal truncation of 13 amino acids (ΔCT), substitution mutation M407K, or empty pRK5 vector. Right panel, Immunoblot of the different Gal4-Nr2e3LBD constructs expressed in 293 cells with anti-hNr2e3 antibodies. Lanes 1-4 refer to the four numbered samples in the graph. Each fusion protein shows similar levels of cleavage between the Gal4 and Nr2e3 domains, producing a lower-molecular weight Nr2e3LBD polypeptide. Molecular mass standards (from top to bottom): 183, 114, 81, 64, 50, and 37 kDa. Error bars in D and E indicate SDs of at least three experiments.
The core binding site for nonsteroid nuclear hormone receptors is the hexamer AG(G/T)TCA, and the binding specificity of different receptor dimers is determined by the orientation and spacing of pairs of hexamers (for review, see Arannda and Pascual, 2001). Based on these observations, we presume that Nr2e3 binds to the optimal site as a dimer. The optimal Nr2e3 site appears to include sequence constraints outside the core hexamers because the first and eighth bases (i.e., the bases preceding the first and second hexamers) were strongly biased to A/G and A, respectively. Substitutions of either C or T in the first position of the optimal site decreased the efficiency of EMSA binding by >10-fold (data not shown).
Binding to a consensus DNA duplex site (AAAGTCAAAAGTCA) was demonstrated by EMSA using mNr2e3 expressed in 293 cells (Fig. 5B). The shifted species is presumed to represent an mNr2e3-DNA complex because it is absent from untransfected 293 cells, and it can be supershifted by antibodies against the linker region of hNr2e3 but not by irrelevant antibodies. Antibodies to the C-terminal 73 amino acids also fail to supershift, presumably because these antibodies were generated against denatured protein and fail to bind native mNr2e3. Expressed human and zebrafish Nr2e3 exhibit similar binding and supershifting behavior in the EMSA (data not shown).
A protein-DNA complex between the Nr2e3 consensus site AAGGTCAAAGATCA and mNr2e3 was also observed using extracts from mouse retina (Fig. 5C). The authenticity of this complex is inferred from (1) its absence when extracts were prepared from rd7/rd7 retinas, (2) its disruption (or conversion to an electrophoretically heterogeneous species) on addition of anti-hNr2e3 linker region antibodies but not irrelevant antibodies, and (3) its indistinguishable electrophoretic mobility relative to the complex formed by mNr2e3 expressed in 293 cells. The specificity of mNr2e3 for the consensus DNA target was further confirmed by competition with unlabeled DNA (Fig. 5C). The specificity of the antilinker antibodies used in these experiments for mNr2e3 is shown in the immunoblot in Figure 5C, which also demonstrates the expected absence of Nr2e3 protein in rd7/rd7 retina extracts.
The effect of mNr2e3 on the transcription of a minimal herpes simplex virus thymidine kinase promoter carrying two, four, or no copies of the Nr2e3 consensus binding site was determined by transient cotransfection in 293 cells (Fig. 5D). This experiment shows that mNr2e3 represses the basal activity of the luciferase reporter and that repression is more effective in the presence of additional Nr2e3 consensus binding sites, independent of their orientation. If this transcriptional repression is mediated by the binding of corepressors to the Nr2e3 ligand-binding domain (LBD), as seen for other nuclear hormone receptors, then it should be observed when this domain alone is localized to the target promoter. Chen et al. (1999) demonstrated this property of Nr2e3 by cotransfecting a plasmid coding for the Gal4 DNA-binding domain (DBD) fused to the Nr2e3 LBD and a plasmid carrying a reporter controlled by a promoter containing Gal4-binding sites. We have confirmed this finding and additionally observed that mutations in the hNr2e3 LBD that either eliminate the predicted C-terminal helix or introduce an M407K substitution, a mutation associated with the enhanced S-cone syndrome (Haider et al., 2000), mostly abolish transcriptional repression (Fig. 5E). These data are consistent with structural data from other nuclear hormone receptors showing that the C-terminal helix of the LBD plays a critical role in the corepressor complex (Perissi et al., 1999; Ghosh et al., 2002; Xu et al., 2002).
Accumulation of cone-specific transcripts in rd7/rd7 retinas
As described in the Introduction, electrophysiologic and anatomic analyses of Nr2e3-deficient retinas in humans and mice suggest that Nr2e3 normally acts to repress the development of S-cones. To define the action of Nr2e3 at the level of gene regulation, we used in situ hybridization to analyze the expression patterns of a series of cone- and rod-specific genes in the retinas of rd7/+ and rd7/rd7 littermates before significant photoreceptor cell loss (Fig. 6). [With respect to retinal histology, the rd7 mutation appears to be fully recessive, and therefore we have used wild-type (WT) and rd7/+ retinas as equivalent representatives of the normal retina.]
Figure 6.
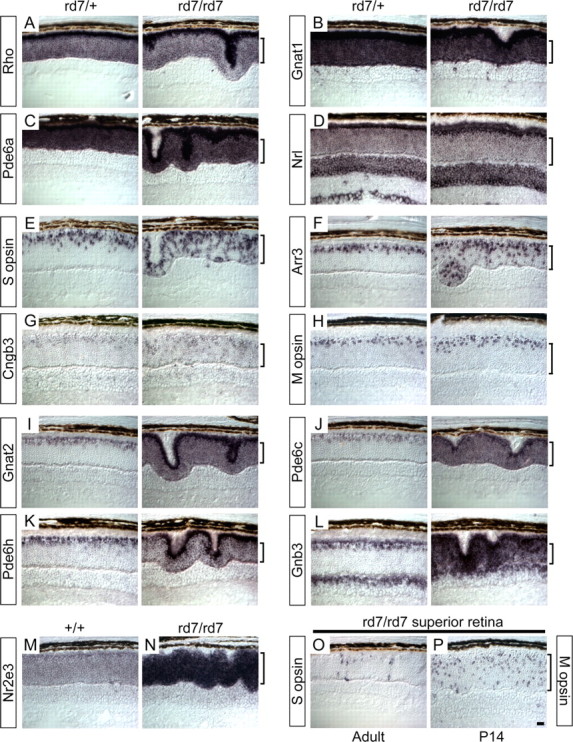
In situ hybridization reveals distinct patterns of cone-specific gene expression in rd7/rd7 retinas. A-L, In situ hybridization to retinas obtained from 6-week-old rd7/+ (left) or rd7/rd7 (right) littermates with probes for the indicated rod and cone transcripts. A-D, Rod transcripts with no detectable alteration in expression in the rd7/rd7 retina. E-H, Cone transcripts that are mostly excluded from rods in both rd7/ + and rd7/rd7 retinas. I-L, Conetranscripts that are expressed in rods in the rd7/rd7 retina. The sections with S-opsin (E) and M-opsin (H) hybridizations are derived from inferior and superior retina, respectively, the regions with the highest concentrations of S- and M-cones. M, N, Hybridization to 6-week-old +/+ and rd7/rd7 retinas with an Nr2e3 probe. O, P, Superior region of rd7/rd7 retinas at the indicated ages hybridized with S-opsin (left) or M-opsin (right) probes. As in the WT retina, S-cones are present at low density in the superior half of the rd7/rd7 retina, but in the rd7/rd7 retina, they reside in both normal and abnormal (i.e., inner) regions of the outer nuclear layer (ONL). In the rd7/rd7 retina, M-cone cell bodies are initially found through out the ONL, and this distribution progressively shifts to the outer edge of the ONL, as seen by comparing M-opsin hybridization patterns at P14 versus 6 weeks (P, H). Each panel shows the full thickness of the retina, with the retinal pigment epithelium at the top and the ganglion cell layer at the bottom. The vertical brackets indicate the extent of the ONL. Characteristic retinal folds are seen in the rd7/rd7 sections, but the relative composition of photoreceptor cell types does not appear to differ between folded and normal regions of the retina. For each probe, the pair of sections of different genotypes was processed on the same slide; therefore, relative signal intensities directly reflect differences in transcript abundance. Chromogenic substrate development times differed among probes. Scale bar, 20 μm.
This analysis shows that the distribution and abundance of three rod-specific transcripts, rhodopsin, the α subunit of rod cGMP phosphodiesterase (Pde6a), and the α subunit of rod transducin (Gnat1), are unaltered in the rd7/rd7 retina (Fig. 6A-C), as is the distribution and abundance of Nrl transcripts (Fig. 6D). Consistent with the anti-Nr2e3 immunostaining shown in Figure 2, we observe Nr2e3 transcripts confined to the outer nuclear layer in both WT and rd7/rd7 retinas (Fig. 6M,N). Interestingly, the Nr2e3 hybridization signal is ∼10-fold stronger in rd7/rd7 retinas compared with WT retinas, an observation discussed more fully below.
This analysis also reveals two distinct patterns of aberrant accumulation of cone-specific transcripts in the rd7/rd7 retina. One pattern of aberrant transcript accumulation is most evident in the inferior retina, where S-cones are most highly concentrated. Here, transcripts coding for S-opsin, cone arrestin (Arr3), and the β subunit of the cone cyclic nucleotidegated channel (Cngb3) accumulate in cells in the outer third of the outer nuclear layer, the normal location for cone cell bodies, as well as in a class of cells of approximately comparable abundance scattered throughout the inner two-thirds of the outer nuclear layer (Fig. 6E-G). A similar pattern is likely to hold for the α subunit of the cone cyclic nucleotide-gated channel (Cnga3) and the γ subunit of cone transducin (Gngt2), but the low signals observed with these transcripts make an unambiguous determination difficult (data not shown). We will refer to these additional cells as inner cones. In the superior retina, in situ hybridization with the S-opsin probe shows that inner S-cones decrease in abundance approximately in proportion to the decrease in the abundance of normally positioned S-cones (Fig. 6O). In contrast to S-opsin transcripts, M-opsin transcripts remain confined to the outer third of the outer nuclear layer in the adult (Fig. 6H); they were the only cone-specific transcripts that showed a normal pattern of localization in the adult rd7/rd7 retina. However, during late retinal development (P14), M-opsin transcripts transiently localize to both normally positioned cones and inner cones in the rd7/rd7 retina (Fig. 6P), whereas in age-matched rd7/+ littermates, they are confined to the outer aspect of the outer nuclear layer (data not shown). This analysis also shows that the retinal folds that characterize the rd7/rd7 retina reflect a general spreading out and buckling of the outer nuclear layer (Fig. 6E,F) rather than a local enrichment of excess cones, as suggested by Haider et al. (2001).
A second pattern of abnormal transcript accumulation is seen with the α and β subunits of cone transducin (Gnat2 and Gnb3) and the α and γ subunits of the cone cGMP phosphodiesterase (Pde6c and Pde6h) (Fig. 6I-L). For each of these transcripts, loss of Nr2e3 leads to a dramatic accumulation in rods, as determined by the hybridization signal across the outer nuclear layer. Figure 6L shows that the β subunit of cone transducin is also normally expressed in a subset of neurons in the inner nuclear layer. Some rod transcripts, such as those encoding rhodopsin (Fig. 6A) and the aberrantly expressed α subunit of cone transducin in the rd7/rd7 retina (Fig. 6I), are enriched in the inner segment region of rod photoreceptors. Thus, loss of Nr2e3 leads both to a derepression of many cone-specific genes in rods and the production of S-cone-like cells in the inner two-thirds of the outer nuclear layer.
Microarray and RNA blot hybridization
To obtain a global assessment of the changes in gene expression in the rd7/rd7 retina, retina RNA was prepared from P10 rd7/+ or rd7/rd7 littermates and from P30 rd7/+ or rd7/rd7 mice and hybridized to the Affymetrix U74Av2 chip, which analyzes ∼12,500 mouse transcripts. The scatterplot in Figure 7A summarizes the comparison of rd7/rd7:rd7/+ transcript ratios at P30 for three independent RNA preparations; similar changes in transcript abundance were observed with P10 retinas (data not shown). For the vast majority of transcripts with detectable hybridization signals, the absence of Nr2e3 causes little or no change in abundance. A number of transcripts encoding crystallins are present at appreciable levels in the rd7/+ RNA samples; these transcripts could be retina-derived (Xi et al., 2003), or they might reflect variable contamination with small amounts of lens epithelium. In agreement with the in situ hybridization results described above, the abundances of several cone-specific transcripts are consistently higher in the rd7/rd7 retina. An additional set of transcripts correspond to a set that were recently reported to be altered in abundance in the Nrl(-/-) retina (Yoshida et al., 2004), and these are also consistently altered in the same manner in the rd7/rd7 retina. These include transcripts for the Clca3 chloride channel, fatty acid-binding protein 7 (Fabp7), and a presumptive long-chain fatty acid synthase (Elovl2). rd7/rd7 retinas show little or no change in the levels of transcripts coding for rod phototransduction proteins, in contrast to the downregulation of these transcripts seen in Nrl(-/-) retinas (Yoshida et al., 2004). A summary of the changes in transcript abundance in the rd7/rd7 retina is given in supplemental Table 1 (available at www.jneurosci.org as supplemental material).
Figure 7.
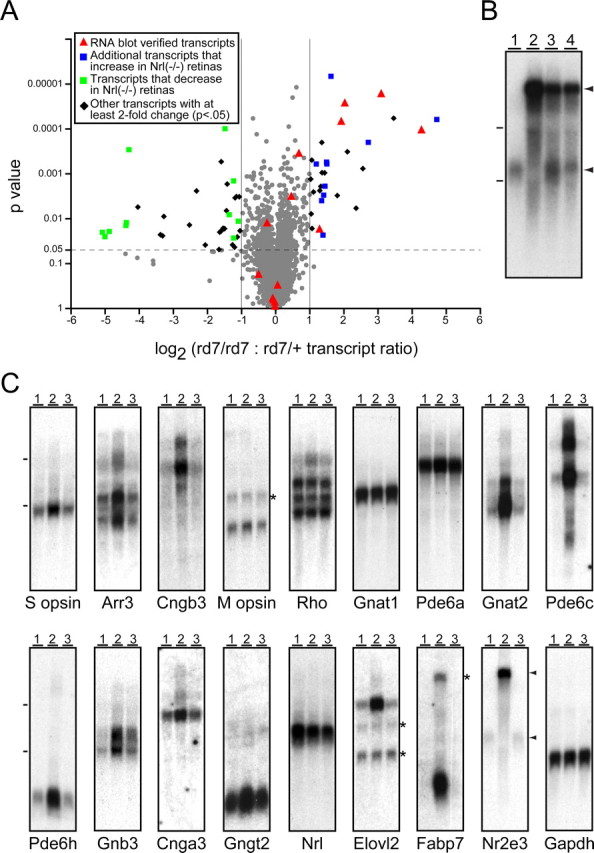
Microarray and RNA blot hybridization shows selectivity in rd7/rd7 misregulation of cone-specific transcripts. A, Scatterplot showing the rd7/rd7:rd7/ + abundance ratio for P30 retina transcripts averaged over three independent microarray experiments using the U74Av2 chip from Affymetrix. Abundance ratios are plotted on the x-axis and are stratified by p value on the y-axis. Most transcripts change by less than twofold and are represented by gray circles. Transcripts tested by RNA blotting (C) are represented by red triangles. Additional upregulated transcripts that are also upregulated in Nrl(-/-) retinas (Yoshida et al., 2004) are shown as blue squares, and down regulated transcripts that are also downregulated in Nrl(-/-) retinas (Yoshida et al., 2004) are shown as green squares. Other transcripts with at least a twofold change and a p < 0.05 are shown as black diamonds. We note that several photoreceptor transcripts (e.g., Nr2e3, Cngb3, Pde6c, Pde6h, Arr3) are not represented on the U74Av2 chip. B, Equal quantities of total RNA from 1- to 2-month-old WT (lane 1), rd7/rd7 (lane 2), rd7/+ (lane 3), and a 1:1 mixture of WT and rd7/rd7 (lane 4) retinas hybridized with an Nr2e3 probe. Arrowheads to the right of the Nr2e3 blots in B and C indicate the normal Nr2e3 transcript (bottom arrowhead) and the high-molecular weight variant Nr2e3 transcript (top arrowhead). C, Total RNA from 1- to 2-month-old WT (lane 1), rd7/rd7 (lane 2), and prCAD(-/-) (lane 3) retinas hybridized with the indicated probes. At these ages, the rd7/rd7 and prCAD(-/-) retinas show only a minimal loss of photoreceptors. Dashes to the left of the blots in B and C show the mobilities of the large and small ribosomal RNAs. The asterisks to the right of several blots show the locations of residual hybridization signals from earlier experiments. Exposure times are not matched across blots.
To independently confirm the results of in situ and microarray hybridization and to compare transcript abundances between the rd7/rd7 retina and a retina with a similarly slow but mechanistically unrelated degeneration, RNA blots were prepared from 1- to 2-month-old WT, rd7/rd7, and prCAD(-/-) (photoreceptor cadherin) retinas (Rattner et al., 2001) at ages before substantial photoreceptor loss. As seen in Figure 7C, RNA blotting confirms the elevation of transcripts coding for S-opsin, cone arrestin, the α and β subunits of cone transducin (Gnat2 and Gnb3), and the α and γ subunits of the cone cGMP phosphodiesterase (Pde6c and Pde6h) in rd7/rd7 retinas. The elevation of Gnat2, Pde6c, and Pde6h transcripts is especially dramatic, consistent with their accumulation in rod photoreceptors seen by in situ hybridization (Fig. 6H-J). RNA blotting also reveals a modest elevation of transcripts coding for the α subunit of the cone cGMP-gated channel (Cnga3) and the cone-specific γ subunit of transducin (Gngt2), consistent with their expression in both normal and inner cones in rd7/rd7 retinas. Importantly, the levels of these transcripts are not elevated in the prCAD(-/-) retina, implying that this pattern of transcript accumulation is not a general response to progressive photoreceptor degeneration. For each of the photoreceptor-specific transcripts, RNA blot hybridization also shows no elevation in the rds/rds retina (data not shown). Also consistent with the microarray hybridization analysis, RNA blotting shows an elevation of Fabp7 and Elovl2 transcripts in rd7/rd7 retinas (Fig. 7C). These transcripts code, respectively, for a high-affinity docosa-hexanoic acid (DHA)-binding protein (Xu et al., 1996) and an elongase that synthesizes long-chain polyunsaturated fatty acids, including DHA (Leonard et al., 2002), suggesting that Nr2e3 plays a role in controlling the metabolism of this class of lipids.
Finally, we investigated the levels of Nr2e3 transcripts in rd7/rd7 and prCAD(-/-) retinas and observed the accumulation of a novel high-molecular weight Nr2e3 transcript specifically in rd7/rd7 retinas at levels manyfold higher than that of the normal Nr2e3 transcript in WT retinas (Fig. 7B). [This observation is at odds with a previous report showing no Nr2e3 transcripts in the rd7/rd7 retina (Akhmedov et al., 2000).] The high-molecular weight Nr2e3 transcript is also abundant in rd7/+ retinas, and, interestingly, the normal Nr2e3 transcript is more abundant in the rd7/+ retina than in the WT retina (Fig. 7B). The high-molecular weight Nr2e3 transcript also hybridizes to a probe from intron 5 (data not shown). We note that the accumulation of a partially spliced mutant transcript is highly unusual because most transcripts of this type are efficiently eliminated by nonsense-mediated RNA decay (Frisch-meyer and Dietz, 1999). Taken together, these data suggest the possibility that Nr2e3 normally feedback-inhibits the production of its own transcripts, and the rd7/+ heterozygote displays haploinsufficiency, or, less likely, protein (or RNA) products of the rd7 allele may dominantly interfere with that inhibition in both rd7/+ and rd7/rd7 retinas.
Cone transducin in rd7/rd7 rod outer segments
In rd7/rd7 rods, the high level of transcripts encoding cone phototransduction proteins suggests that some or all of these proteins might be present at substantial levels in rod outer segments. To test this possibility for the α subunit of cone transducin (Gnat2), retinas from rd7/+ and rd7/rd7 littermates were immunostained with an antipeptide antibody that specifically recognizes this protein and also stained with peanut agglutinin (PNA), which labels the extracellular matrix sheath surrounding all cone outer segments (Fig. 8). In the rd7/+ retina, cone α-transducin is confined to cones, as expected (Fig. 8A-C). In contrast, in the rd7/rd7 retina, cone α-transducin is present in both rod and cone outer segments, a pattern that is most clearly seen in the superior retina where the cone density is lower than in the inferior retina (Fig. 8D-L). In both the superior and inferior retina, the intensity of rod outer segment immunostaining is somewhat lower than that of cone outer segments. These data suggest that rd7/rd7 mice, and by inference Nr2e3(-/-) humans, use a mixture of rod and cone proteins for rod phototransduction.
Figure 8.
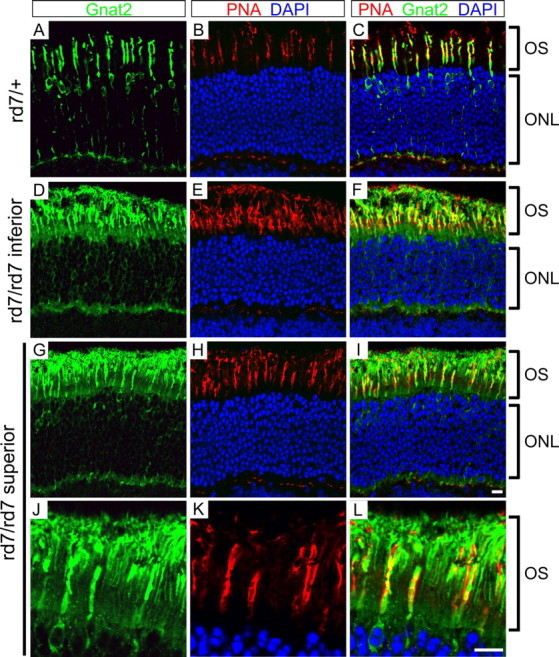
The α subunit of cone transducin localizes to both cone and rod photoreceptors in rd7/rd7 retinas. A-C, rd7/+ retinas immunostained for the α subunit of cone transducin (Gnat2; green) and costained with PNA (red) and DAPI (blue). D-L, rd7/rd7 stained as in A-C. D-F, The inferior rd7/rd7 retina has an approximately twofold higher density of S-cones than an rd7/+ retina. G-L, the superior rd7/rd7 retina has a normal concentration of M-cones. The outer segment region of G-I is enlarged in J-L, showing the localization of cone transducin α to both rod and cone outer segments (OS). Retinas were harvested from littermates at 6 weeks of age. PNA marks each cone sheath, an extracellular matrix structure that encompasses the cone OS and extends distal to it. Scale bar, 10 μm.
Discussion
Implications of Nr2e3 conservation and rod expression
In this study, we show that Nr2e3 orthologs are present in diverse vertebrates, including fish and mammals, suggesting that Nr2e3 arose in concert with the evolution of rod and cone photoreceptors. In keeping with this evolutionary history, our observations regarding Nr2e3 expression in rods and the effects of the Nr2e3 mutation in the mouse retina point to a central role for Nr2e3 in orchestrating the patterns of gene expression that distinguish rod and cone photoreceptors.
In mature primate, mouse, and zebrafish retinas, Nr2e3 is localized exclusively to the nuclei of all rods. In early development in the mouse, Nr2e3 accumulates in rod precursors before the appearance of rhodopsin, making Nr2e3 one of the earliest markers of rod precursors. Consistent with the data reported here, O'Brien et al. (2004) have recently reported that Nr2e3 expression is excluded from developing cones in the fetal human retina. In contrast, in zebrafish there appears to be a transient wave of Nr2e3 expression in developing cone photoreceptors before the onset of cone pigment gene expression.
Gene expression defects in the rd7/rd7 retina
By in situ, microarray, and RNA blot hybridization, we observe a highly selective pattern of gene expression defects in the rd7/rd7 retina. Several cone-specific transcripts, including the S-opsin message, are expressed in abnormally located cells within the outer nuclear layer, which we have termed inner cones; numerous cone-specific transcripts are aberrantly expressed in rods; and rod transcripts are unaltered in pattern and abundance, except for the high level of an aberrant Nr2e3 transcript. Thus, cone-specific genes show a fundamental division into two classes based on their responses to loss of Nr2e3. None of the above-mentioned alterations are observed in a mechanistically unrelated retinal degeneration caused by loss of photoreceptor cadherin, and microarray analysis indicates that most transcripts are unaltered in the rd7/rd7 retina. These data, together with the observation that Nr2e3 functions as a transcriptional repressor in transfected cells (Chen et al., 1999; this study), suggest that one principal function of Nr2e3 is to repress the expression of cone-specific genes in rods. The observation of modestly elevated levels of the WT Nr2e3 transcript in rd7/+ retinas further suggests the possibility that Nr2e3 may directly or indirectly act in a negative feedback loop to regulate the levels of its own transcript.
Haider et al. (2001) described a 1.5- to 2-fold increase in the number of S-cones in the rd7/rd7 retina using peanut agglutinin staining and anti-S-opsin immunostaining, and our in situ hybridization analyses suggest that this excess can be accounted for mostly or entirely by the inner S-cones. To explain this excess of S-cones in mice and the even greater excess of S-cones in humans with Nr2e3 mutations, several groups have suggested that Nr2e3 might function as a repressor of S-cone proliferation (Haider et al. 2001; Milam et al., 2002). This explanation would be most plausible if, as in zebrafish, Nr2e3 was transiently expressed in mammalian cone precursors and could regulate cone proliferation in a cell-autonomous manner. Alternatively, the observations that Nr2e3 is expressed in rods but not cones in mammalian retinas and that many cone-specific transcripts are derepressed in rd7/rd7 rods suggest that the Nr2e3(-/-) phenotype in both mice and humans might be explained strictly as a defect in gene regulation in postmitotic cells. In this conceptualization, all rods in the rd7/rd7 retina are somewhat cone-like in their gene expression patterns. The inner S-cones may represent a subset of photoreceptors that were initially destined for a rod fate but became de facto cones by activating a sufficiently large number of cone-specific genes. This discussion highlights the difficulties associated with using the terms rod and cone when, as in the rd7/rd7 retina, the patterns of gene expression, which constitute the operational distinction between cell types, are altered to produce intermediate types.
The role of Nr2e3 in controlling photoreceptor gene expression
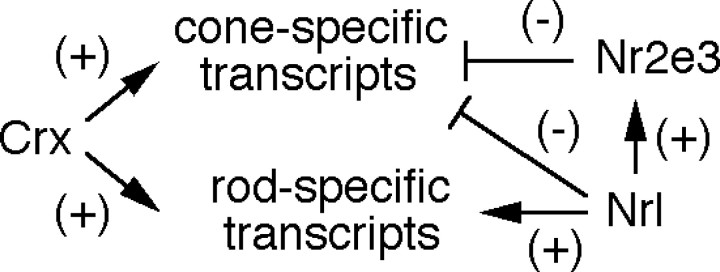
The rd7/rd7 defects reported here are reminiscent of some of the defects observed in Nrl(-/-) retinas (Mears et al., 2001). However, Nrl(-/-) retinas show a more extreme transformation of rods toward a cone-like fate, including an absence of transcripts coding for rhodopsin, rod transducin α, and rod cGMP phosphodiesterase β and a large increase in S-opsin transcripts. Because Nrl(-/-) retinas also lack Nr2e3 transcripts, it is likely that all of the molecular defects seen in the rd7/rd7 retina are included in the Nrl(-/-) phenotype. Figure 9 presents a diagram showing the relationship among Crx, Nrl, and Nr2e3 in controlling rod- and cone-specific genes in rod photoreceptor cells. Crx activates various rod- and cone-specific genes (Furukawa et al., 1999), and Nrl activates the transcription of various rod-specific genes, including Nr2e3 (Mears et al., 2001). Both Nrl (Mears et al., 2001) and Nr2e3 repress the transcription of cone-specific genes. It would be interesting to determine whether a symmetric system exists in cones for repressing rod-specific genes.
Figure 9.
Model for transcriptional control in rod photoreceptors. Crx activates the transcription of both rod- and cone-specific genes (Furukawa et al., 1999), and Nrl activates the transcription of rod-specific genes, including Nr2e3 (Mears et al., 2001). Both Nrl (Mears et al., 2001) and Nr2e3 repress the transcription of cone-specific genes. In the mouse retina, Crx transcripts appear at ∼E12.5 (Furukawa et al., 1997), Nrl transcripts appear at ∼E14.5 (Liu et al., 1996), and Nr2e3 transcripts appear at ∼E16.5 (Cheng et al., 2004), suggesting a temporal hierarchy with Crx and Nrl above Nr2e3.
Recent in vitro and cell culture experiments suggest that Nr2e3 may interact with the nuclear hormone receptor Nr1d1 (Reverb-α) to synergistically activate the rhodopsin and rod transducin α promoters (Cheng et al., 2004). In contrast, our cell culture analyses with full-length Nr2e3 and with Gal4DBD-Nr2e3LBD fusions have shown only transcriptional repression. However, our cell culture experiments should be interpreted cautiously because they are at best suggestive of Nr2e3 function on the relevant target promoters, in rod photoreceptors, and in the presence of interacting partners and/or small molecule ligands. We note, however, that our failure to see significant alterations in rod transcripts in the rd7/rd7 retina suggest that, if the transcriptional activation postulated by Cheng et al. (2004) also occurs in the retina, it is subject to compensatory regulation in the absence of Nr2e3.
Implications for Nr2e3-associated retinal diseases
NR2E3 mutations were first identified in patients with the enhanced S-cone syndrome, but more recent work has shown that NR2E3 mutations are a common cause of clumped pigmentary retinopathy in the context of enhanced S-cone syndrome, Goldman-Favre syndrome, and retinitis pigmentosa (Sharon et al., 2003). In one postmortem study, the pigmentary clumps seen ophthalmoscopically were shown to correspond to large numbers of melanin granules within retinal pigment epithelial cells (To et al., 1996), suggesting the possibility that the retinal degeneration caused by NR2E3 mutations may occur in a distinctive spatial pattern. Enhanced S-cone syndrome is characterized by reduced rod, L-cone, and M-cone responses, an early disorganization of retinal lamination, and a progressive loss of photoreceptors (Jacobson et al., 1990, 2004; Haider et al., 2000). Abnormal patterns of cone photoreceptor immunoreactivity have also been described in a postmortem study of one late-stage enhanced S-cone syndrome retina (Milam et al., 2002; Cideciyan et al., 2003).
In the rd7/rd7 retina, the increase in the total number of S-but not M-cones and the progressive loss of rod photoreceptors appear to faithfully model the human disease process. The misexpression of numerous cone transcripts in rd7/rd7 rods suggests that rod loss in patients with NR2E3 mutations may arise from the toxic consequences of gene misexpression. It would be of interest to determine whether phototransduction is altered in rd7/rd7 rods as a consequence of this misexpression. It is also intriguing that transcripts coding for two proteins implicated in long-chain polyunsaturated fatty acid metabolism, Fabp7 and Elovl2, are induced. Photoreceptor outer segments contain unusually high levels of DHA and other long-chain polyunsaturated fatty acids (Penn and Anderson, 1987), and DHA levels in rod outer segments decline in diverse models of inherited retinal degeneration, which has led several groups to suggest that decreased DHA may be a protective response to retinal degeneration (Penn and Anderson, 1987; Anderson et al., 2001; Bicknell et al., 2002). Moreover, mutations in a gene coding for a presumptive fatty acid elongase, Elovl4, cause an autosomal dominant macular degeneration (Edwards et al., 2001; Zhang et al., 2001). Thus, retinal degeneration in patients with NR2E3 mutations might also be caused, at least in part, by aberrant metabolism of long-chain fatty acids.
Footnotes
This work was supported by the National Eye Institute and the Howard Hughes Medical Institute. We thank Dr. Peter MacLeish, Dr. James Fadool, Dr. Scott Fraser, Dr. James Hurley, Wenqin Luo, Dr. David Manglesdorf, Dr. Robert Molday, Dr. Francisco Murillo, Wenyi Wang, and Dr. Yanshu Wang for advice and/or materials and Dr. Thomas Rotolo, Dr. Chunqiao Liu, and two anonymous referees for helpful comments on this manuscript.
Correspondence should be addressed to Dr. Jeremy Nathans, Johns Hopkins University School of Medicine, 805 Preclinical Teaching Building, 725 North Wolfe Street, Baltimore, MD 21205. E-mail: jnathans@jhmi.edu.
Copyright © 2005 Society for Neuroscience 0270-6474/05/250118-12$15.00/0
References
- Akhmedov NB, Piriev NI, Chang B, Rapoport AL, Hawes NL, Nishina PM, Nusinowitz S, Heckenlively JR, Roderick TH, Kozak CA, Danciger M, Davisson MT, Farber DB (2000) A deletion in a photoreceptor-specific nuclear receptor mRNA causes retinal degeneration in the rd7 mouse. Proc Natl Acad Sci USA 97: 5551-5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RE, Maude MB, Bok D (2001) Low docosahexaenoic acid levels in rod outer segment membranes of mice with rds/peripherin and P216L peripherin mutations. Invest Ophthalmol Vis Sci 42: 1715-1720. [PubMed] [Google Scholar]
- Arannda A, Pascual A (2001) Nuclear hormone receptors and gene expression. Physiol Rev 81: 1269-1304. [DOI] [PubMed] [Google Scholar]
- Bessant DA, Payne AM, Mitton KP, Wang QL, Swain PK, Plant C, Bird AC, Zack DJ, Swaroop A, Bhattacharya SS (1999) A mutation in NRL is associated with autosomal dominant retinitis pigmentosa. Nat Genet 21: 355-356. [DOI] [PubMed] [Google Scholar]
- Bicknell IR, Darrow R, Barsalou L, Fliesler SJ, Organisciak DT (2002) Alterations in retinal rod outer segment fatty acids and light-damage susceptibility in P23H rats. Mol Vis 8: 333-340. [PubMed] [Google Scholar]
- Carter-Dawson LD, LaVail MM (1979) Rods and cones in the mouse retina: structural analysis using light and electron microscopy. J Comp Neurol 188: 245-262. [DOI] [PubMed] [Google Scholar]
- Chen F, Figueroa DJ, Marmorstein AD, Zhang Q, Petrukhin K, Caskey CT, Austin CP (1999) Retina-specific nuclear receptor: a potential regulator of cellular retinaldehyde-binding protein expressed in retinal pigment epithelium and Muller glial cells. Proc Natl Acad Sci USA 96: 15149-15154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Wang QL, Nie Z, Sun H, Lennon G, Copeland NG, Gilbert DJ, Jenkins NA, Zack DJ (1997) Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron 19: 1017-1030. [DOI] [PubMed] [Google Scholar]
- Cheng H, Khanna H, Oh EC, Hicks D, Mitton KP, Swaroop A (2004) Photoreceptor-specific nuclear receptor NR2E3 functions as a transcriptional activator in rod photoreceptors. Hum Mol Genet 13: 1563-1575. [DOI] [PubMed] [Google Scholar]
- Cideciyan AV, Jacobson SG, Gupta N, Osawa S, Locke KG, Weiss ER, Wright AF, Birch DG, Milam AH (2003) Cone deactivation kinetics and GRK1/GRK7 expression in enhanced S cone syndrome caused by mutations in NR2E3. Invest Ophthalmol Vis Sci 44: 1268-1274. [DOI] [PubMed] [Google Scholar]
- Edwards AO, Donoso LA, Ritter R (2001) A novel gene for autosomal dominant Stargardt-like macular dystrophy with homology to the SUR4 protein family. Invest Ophthalmol Vis Sci 42: 2652-2663. [PubMed] [Google Scholar]
- Fadool JM (2003) Development of a rod photoreceptor mosaic revealed in transgenic zebrafish. Dev Biol 258: 277-290. [DOI] [PubMed] [Google Scholar]
- Freund CL, Gregory-Evans CY, Furukawa T, Papaioannou M, Looser J, Ploder L, Bellingham J, Ng D, Herbrick JA, Duncan A, Scherer SW, Tsui LC, Loutradis-Anagnostou A, Jacobson SG, Cepko CL, Bhattacharya SS, McInnes RR (1997) Cone-rod dystrophy due to mutations in a novel photoreceptor-specific homeobox gene (CRX) essential for maintenance of the photoreceptor. Cell 91: 543-553. [DOI] [PubMed] [Google Scholar]
- Frischmeyer PA, Dietz HC (1999) Nonsense-mediated mRNA decay in health and disease. Hum Mol Genet 8: 1893-1900. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Morrow EM, Cepko CL (1997) Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell 91: 531-541. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Morrow EM, Li T, Davis FC, Cepko CL (1999) Retinopathy and attenuated circadian entrainment in Crx-deficient mice. Nat Genet 23: 466-470. [DOI] [PubMed] [Google Scholar]
- Gerber S, Rozet JM, Takezawa SI, dos Santos LC, Lopes L, Gribouval O, Penet C, Perrault I, Ducroq D, Souied E, Jeanpierre M, Romana S, Frezal J, Ferraz F, Yu-Umesono R, Munnich A, Kaplan J (2000) The photoreceptor cell-specific nuclear receptor gene (PNR) accounts for retinitis pigmentosa in the Crypto-Jews from Portugal (Marranos), survivors from the Spanish Inquisition. Hum Genet 107: 276-284. [DOI] [PubMed] [Google Scholar]
- Ghosh JC, Yang X, Zhang A, Lambert MH, Li H, Xu HE, Chen JD (2002) Interactions that determine the assembly of a retinoid X receptor/corepressor complex. Proc Natl Acad Sci USA 99: 5842-5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider NB, Jacobson SG, Cideciyan AV, Swiderski R, Streb LM, Searby C, Beck G, Hockey R, Hanna DB, Gorman S, Duhl D, Carmi R, Bennett J, Weleber RG, Fishman GA, Wright AF, Stone EM, Sheffield VC (2000) Mutation of a nuclear receptor gene, NR2E3, causes enhanced S cone syndrome, a disorder of retinal cell fate. Nat Genet 24: 127-131. [DOI] [PubMed] [Google Scholar]
- Haider NB, Naggert JK, Nishina PM (2001) Excess cone cell proliferation due to lack of a functional NR2E3 causes retinal dysplasia and degeneration in rd7/rd7 mice. Hum Mol Genet 10: 1619-1626. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson SG, Marmor MF, Kemp CM, Knighton RW (1990) SWS (blue) cone hypersensitivity in a newly identified retinal degeneration. Invest Opthalmol Vis Sci 31: 827-838. [PubMed] [Google Scholar]
- Jacobson SG, Sumaroka A, Aleman TS, Cideciyan AV, Schwartz SB, Roman AJ, McInnes RR, Sheffield VC, Stone EM, Swaroop A, Wright AF (2004) Nuclear receptor NR2E3 gene mutations distort human retinal laminar architecture and cause an unusual degeneration. Hum Mol Genet 13: 1893-1902. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Takezawa S, Hara K, Yu RT, Umesono Y, Agata K, Taniwaki M, Yasuda K, Umesono K (1999) Identification of a photoreceptor cell-specific nuclear receptor. Proc Natl Acad Sci USA 96: 4814-4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard AE, Kelder B, Bobik EG, Chuang LT, Lewis CJ, Kopchick JJ, Mukerji P, Huang YS (2002) Identification and expression of mammalian long-chain PUFA elongation enzymes. Lipids 37: 733-740. [DOI] [PubMed] [Google Scholar]
- Liu Q, Ji X, Breitman ML, Hitchcock PF, Swaroop A (1996) Expression of the bZIP transcription factor gene Nrl in the developing nervous system. Oncogene 12: 207-211. [PubMed] [Google Scholar]
- Luo W, Williams J, Smallwood PM, Touchman JW, Roman LM, Nathans J (2004) Proximal and distal sequences control UV cone pigment gene expression in transgenic zebrafish. J Biol Chem 279: 19286-19293. [DOI] [PubMed] [Google Scholar]
- Mears AJ, Kondo M, Swain PK, Takada Y, Bush RA, Saunders TL, Sieving PA, Swaroop A (2001) Nrl is required for rod photoreceptor development. Nat Genet 29: 447-452. [DOI] [PubMed] [Google Scholar]
- Milam AH, Rose L, Cideciyan AV, Barakat MR, Tang WX, Gupta N, Aleman TS, Wright AF, Stone EM, Sheffield VC, Jacobson SG (2002) The nuclear receptor NR2E3 plays a role in human retinal photoreceptor differentiation and degeneration. Proc Natl Acad Sci USA 99: 473-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng L, Hurley JB, Dierks B, Srinivas M, Salto C, Vennstrom B, Reh TA, Forrest D (2001) A thyroid hormone receptor that is required for the development of green cone photoreceptors. Nat Genet 27: 94-98. [DOI] [PubMed] [Google Scholar]
- O'Brien KM, Cheng H, Jiang Y, Schulte D, Swaroop A, Hendrickson AE (2004) Expression of photoreceptor-specific nuclear receptor NR2E3 in rod photoreceptors of fetal human retina. Invest Ophthalmol Vis Sci 45: 2807-2812. [DOI] [PubMed] [Google Scholar]
- Penn JS, Anderson RE (1987) Effect of light history on rod outer-segment membrane composition in the rat. Exp Eye Res 44: 767-778. [DOI] [PubMed] [Google Scholar]
- Perissi V, Staszewski LM, McInerney EM, Kurokawa R, Krones A, Rose DW, Lambert MH, Milburn MV, Glass CK, Rosenfeld MG (1999) Molecular determinants of nuclear receptor-corepressor interaction. Genes Dev 13: 3198-3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattner A, Smallwood PM, Williams J, Cooke C, Savchenko A, Lyubarsky A, Pugh EN, Nathans J (2001) A photoreceptor-specific cadherin is essential for the structural integrity of the outer segment and for photoreceptor survival. Neuron 32: 775-786. [DOI] [PubMed] [Google Scholar]
- Rodieck RW (1998) The first steps in seeing. Sunderland, MA: Sinauer.
- Schaeren-Wiemers N, Gerfin-Moser A (1993) A single protocol to detect transcripts of various types and expression levels in neural tissue and cultured cells: in situ hybridization using digoxigenin-labelled cRNA probes. Histochemistry 100: 431-440. [DOI] [PubMed] [Google Scholar]
- Sharon D, Sandberg MA, Caruso RC, Berson EL, Dryja TP (2003) Shared mutations in NR2E3 in enhanced S-cone syndrome, Goldmann-Favre syndrome, and many cases of clumped pigmentary retinal degeneration. Arch Ophthalmol 121: 1316-1323. [DOI] [PubMed] [Google Scholar]
- Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW (1990) Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol 185: 60-89. [DOI] [PubMed] [Google Scholar]
- Swain PK, Chen S, Wang QL, Affatigato LM, Coats CL, Brady KD, Fishman GA, Jacobson SG, Swaroop A, Stone E, Sieving PA, Zack DJ (1997) Mutations in the cone-rod homeobox gene are associated with the cone-rod dystrophy photoreceptor degeneration. Neuron 19: 1329-1336. [DOI] [PubMed] [Google Scholar]
- Thiesen HJ, Bach C (1990) Target detection assay (TDA): a versatile procedure to determine DNA binding sites as demonstrated on SP1 protein. Nucleic Acids Res 18: 3203-3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai RY, Reed RR (1997) Using a eukaryotic GST fusion vector for proteins difficult to express in E. coli Biotechniques 23: 794-800. [DOI] [PubMed] [Google Scholar]
- To KW, Adamian M, Jakobiec FA, Berson EL (1996) Clinical and histopathologic findings in clumped pigmentary retinal degeneration. Arch Ophthalmol 114: 950-955. [DOI] [PubMed] [Google Scholar]
- Wang Y, Macke JP, Merbs SL, Zack DJ, Klaunberg B, Bennett J, Gearhart J, Nathans J (1992) A locus control region adjacent to the human red and green visual pigment genes. Neuron 9: 429-440. [DOI] [PubMed] [Google Scholar]
- Yanagi Y, Takezawa S, Kato S (2002) Distinct functions of photoreceptor cell-specific nuclear receptor, thyroid hormone receptor beta2 and CRX in cone photoreceptor development. Invest Ophthalmol Vis Sci 43: 3489-3494. [PubMed] [Google Scholar]
- Yoshida S, Mears AJ, Friedman JS, Carter T, He S, Oh E, Jing Y, Farjo R, Fleury G, Barlow C, Hero AO, Swaroop A (2004) Expression profiling of the developing and mature Nrl-/- mouse retina: identification of retinal disease candidates and transcriptional regulatory targets of Nrl. Hum Mol Genet 13: 1487-1503. [DOI] [PubMed] [Google Scholar]
- Young RW (1984) Cell death during differentiation of the retina in the mouse. J Comp Neurol 229: 362-373. [DOI] [PubMed] [Google Scholar]
- Xi J, Farjo R, Yoshida S, Kern TS, Swaroop A, Andley UP (2003) A comprehensive analysis of the expression of crystallins in mouse retina. Mol Vis 9: 410-419. [PubMed] [Google Scholar]
- Xiang M, Zhou L, Macke JP, Yoshioka T, Hendry SH, Eddy RL, Shows TB, Nathans J (1995) The Brn-3 family of POU-domain factors: primary structure, binding specificity, and expression in subsets of retinal ganglion cells and somatosensory neurons. J Neurosci 15: 4762-4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu HE, Stanley TB, Montana VG, Lambert MH, Shearer BG, Cobb JE, McKee DD, Galardi CM, Plunket KD, Nolte RT, Parks DJ, Moore JT, Kliewer SA, Willson TM, Stimmel JB (2002) Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARalpha. Nature 415: 813-817. [DOI] [PubMed] [Google Scholar]
- Xu LZ, Sanchez R, Sali A, Heintz N (1996) Ligand specificity of brain lipid-binding protein. J Biol Chem 271: 24711-24719. [DOI] [PubMed] [Google Scholar]
- Zhang H, Cuenca N, Ivanova T, Church-Kopish J, Frederick JM, MacLeish PR, Baehr W (2003) Identification and light-dependent translocation of a cone-specific antigen, cone arrestin, recognized by monoclonal antibody 7G6. Invest Ophthalmol Vis Sci 44: 2858-2867. [DOI] [PubMed] [Google Scholar]
- Zhang K, Kniazeva M, Han M, Li W, Yu Z, Yang Z, Li Y, Metzker ML, Allikmets R, Zack DJ, Kakuk LE, Lagali PS, Wong PW, MacDonald IM, Sieving PA, Figueroa DJ, Austin CP, Gould RJ, Ayyagari R, Petrukhin K (2001) A 5-bp deletion in ELOVL4 is associated with two related forms of autosomal dominant macular dystrophy. Nat Genet 27: 89-93. [DOI] [PubMed] [Google Scholar]