Abstract
Neuroinflammation is associated with a variety of neurological and pathological diseases, such as Alzheimer's disease (AD), and is reliably detected by the presence of activated microglia. In early AD, the highest degree of activated microglia is observed in brain regions involved in learning and memory. To investigate whether neuroinflammation alters the pattern of rapid de novo gene expression associated with learning and memory, we studied the expression of the activity-induced immediate early gene Arc in the hippocampus of rats with experimental neuroinflammation. Rats were chronically infused with lipopolysaccharide (LPS) (0.25 μg/h) into the fourth ventricle for 28 d. On day 29, the rats explored twice a novel environment for 5 min, separated by 45 or 90 min. In the dentate gyrus and CA3 regions of LPS-infused rats, Arc and OX-6 (specific for major histocompatibility complex class II antigens) immunolabeling and Arc fluorescence in situ hybridization revealed both activated microglia (OX-6 immunoreactivity) and elevated exploration-induced Arc expression compared with control-infused rats. In contrast, in the CA1 of LPS-infused rats, where there was no OX-6 immunostaining, exploration-induced Arc mRNA and protein remained similar in both LPS- and control-infused rats. LPS-induced neuroinflammation did not affect basal levels of Arc expression. Behaviorally induced Arc expression was altered only within the regions showing activated microglia (OX-6 immunoreactivity), suggesting that neuroinflammation may alter the coupling of neural activity with macromolecular synthesis implicated in learning and plasticity. This activity-related alteration in Arc expression induced by neuroinflammation may contribute to the cognitive deficits found in diseases associated with inflammation, such as AD.
Keywords: activated microglia, Alzheimer's disease, immediate early gene, hippocampal function, cognitive impairment, spatial memory consolidation
Introduction
Chronic neuroinflammation may contribute to the impaired cognition associated with Alzheimer's disease (AD) (McGeer and McGeer, 1998; Akiyama et al., 2000). AD is characterized by a distinct pattern of neuropathological changes, including a dense pattern of highly activated astrocytes and microglia (McGeer and McGeer, 1998; Akiyama et al., 2000) and increased levels of proinflammatory cytokines (Akiyama et al., 2000). Furthermore, brain regions involved in learning and memory have the greatest degree of neuroinflammation early in the disease (Cagnin et al., 2001). Chronic neuroinflammation has been reproduced in rats by infusion of lipopolysaccharide (LPS) into the fourth ventricle. LPS activates microglia to initiate a series of inflammation-induced changes within the hippocampus and entorhinal cortex (Hauss-Wegrzyniak et al., 1998, 2002; Rosi et al., 2003, 2004). The inflammation leads to a reduction in the number of NMDA glutamate receptors (R1) within the dentate gyrus (DG) and CA3 hippocampal areas without evidence of neuronal loss (Rosi et al., 2004) and impairment in spatial, but not object recognition, memory (Hauss-Wegrzyniak et al., 1998). Furthermore, LPS-induced neuroinflammation impairs the induction of long-term potentiation (Hauss-Wegrzyniak et al., 2002).
To further define the functional consequences of chronic neuroinflammation within the hippocampus and its potential effects on cognition, we investigated whether experimentally induced neuroinflammation affects the overall hippocampal pattern of expression of the immediately early gene Arc after behavioral exploration. Arc is induced in hippocampal and parietal cortical neurons after behavioral experience, and Arc protein expression plays a critical role in the maintenance phase of long-term potentiation and spatial memory consolidation (Guzowski et al., 2000). The entry of calcium ions and activation of cAMP (Waltereit et al., 2001) after NMDA receptor activation (Steward and Worley, 2001) also leads to Arc mRNA transcription, which is protein kinase A dependent (Waltereit et al., 2001). Arc mRNA moves rapidly to the dendritic areas close to the active synaptic regions where it is locally translated and plays a fundamental role in the stabilization of behavioral experience (Steward et al., 1998; Guzowski, 2002). Alterations in the levels of Arc expression have been correlated with cognitive impairment during aging (Blalock et al., 2003) and with amyloidal deposition in mice that express multiple genes associated with AD (Dickey et al., 2004). We predicted that the cognitive impairments associated with neuroinflammation, after chronic infusion of LPS, may be associated with alterations in either Arc transcription and/or translation. The results revealed that LPS-induced chronic neuroinflammation did not significantly affect basal Arc expression but increased the number of neurons showing exploration-related Arc mRNA transcription and translation in the DG and CA3 hippocampal areas, regions that also showed the greatest number of activated microglia. Thus, the changes in Arc expression were restricted to areas with the most prominent signs of neuroinflammation. This study provides evidence of how local neuroinflammation can modify features of neural network activation that may underlie learning and memory impairments associated with several neurological diseases.
Materials and Methods
Subjects and surgical and behavioral procedures. The subjects were 37 male F-344 rats (3 months of age; Harlan Sprague Dawley, Indianapolis, IN). LPS (n = 18) or artificial CSF (aCSF; n = 19) was chronically infused (0.25 μg/h for 28 d) through a cannula implanted into the fourth ventricle of the brain that was attached to an osmotic minipump as described previously (Hauss-Wegrzyniak et al., 1998, 2000, 2002; Rosi et al., 2003, 2004). Each rat was assigned to one of six different groups: LPS-infused (n = 6) and aCSF-infused (n = 6) groups with 45 min intervals between exploration sessions, LPS-infused (n = 6) and aCSF-infused (n = 7) rats with 90 min intervals between exploration, and LPS-infused (n = 6) and aCSF-infused (n = 6) caged control rats.
The rats were handled daily for 10 d before behavioral testing began. The behavioral procedure consisted of placing the animals into the same exploration environment for 5 min on two separate occasions. The exploration environment was an open square box, 61 × 61 cm with 20-cm-high walls divided into nine grids. Each rat was moved to the center of a different grid every 15 s in a pseudorandom schedule during the 5 min exploration session, as described in detail previously (Guzowski et al., 1999). The first and second exploration sessions were separated by either 45 or 90 min. During the rest interval between the two sessions, the animals remained undisturbed in their own cage in the colony room. Immediately after the second 5 min exploration session, the rats were killed by decapitation. To ensure that any Arc transcription induced by the killing procedure would not be detectable, the brain was quickly removed (between 160 and 200 s) and frozen in -70°C isopentane. The 45 and 90 min intervals between exploration sessions were chosen so that, within the same rat, both mRNA (5 min time point) and protein (45 and 90 min) could be examined. Two “protein time points” were included to examine possible differences in protein expression over the time.
Histological procedures. The brains were divided at the midline, and half-brains from each rat were blocked together and cryosectioned (Vazdarjanova et al., 2002), such that each slide contained brain coronal sections from one rat from each of the six groups involved in this experiment (see above). All slides were stored at -70°C until processed for immunocytochemistry or in situ hybridization.
Double immunofluorescence staining. Each of the six rats in each treatment group was represented on a series of six slides (six half-brains per slide). For each rat, three different slides were analyzed, resulting in a total of 18 slides prepared for immunofluorescence. All slides included sections from the medial portion of the dorsal hippocampus [anteroposterior (AP), approximately -3.6 mm from bregma) for the double staining for Arc protein and activated microglia (see Fig. 5A-G). The tissue was fixed in 2% paraformaldehyde, pH 7.4, for 5 min, washed in diluted 2× SSC buffer, pH 7.0 (Sigma, St. Louis, MO), and followed by washing in 50:50% acetone-methanol at 4°C for 5 min. After three washes with 2× SSC plus 0.05% Tween 20 and quenching in 2× SSC plus 3% H2O2 for 15 min, trichostatin A (TSA) blocking buffer (PerkinElmer Life Sciences, Emeryville, CA) was applied for 30 min, followed by application of polyclonal rabbit anti-Arc antibody (1:800) supplied by P.F.W.'s laboratory. After 48 h of incubation at 4°C, the sections were incubated for 2 h at room temperature with the secondary anti-rabbit biotinylated antibody (Vector Laboratories, Burlingame, CA), followed by incubation with the avidin plus biotin amplification system (Vector Laboratories) for 45 min. The staining was visualized using the TSA fluorescence system cyanine-3 (CY3) (PerkinElmer Life Sciences). After washing in TBS-Tween solution, the tissue was quenched and blocked again as described above and incubated with the monoclonal antibody OX-6 (PharMingen, San Diego, CA) in a final dilution (1:400) for 24 h. This antibody is specifically directed against class II major histocompatibility complex antigen. Before applying the biotinylated monoclonal secondary rat absorbed antibody (Vector Laboratories) for 2 h, the tissue was incubated with an avidin biotin blocking kit (Vector Laboratories) for 30 min to block cross reaction with the primary staining. After additional treatment with an avidin plus biotin amplification system (Vector Laboratories), the staining was visualized with TSA fluorescence system CY5 (PerkinElmer Life Sciences), and the nuclei were counterstained with Sytox-Green (Molecular Probes, Eugene, OR). No staining was detected in the absence of the primary or secondary antibodies.
Figure 5.
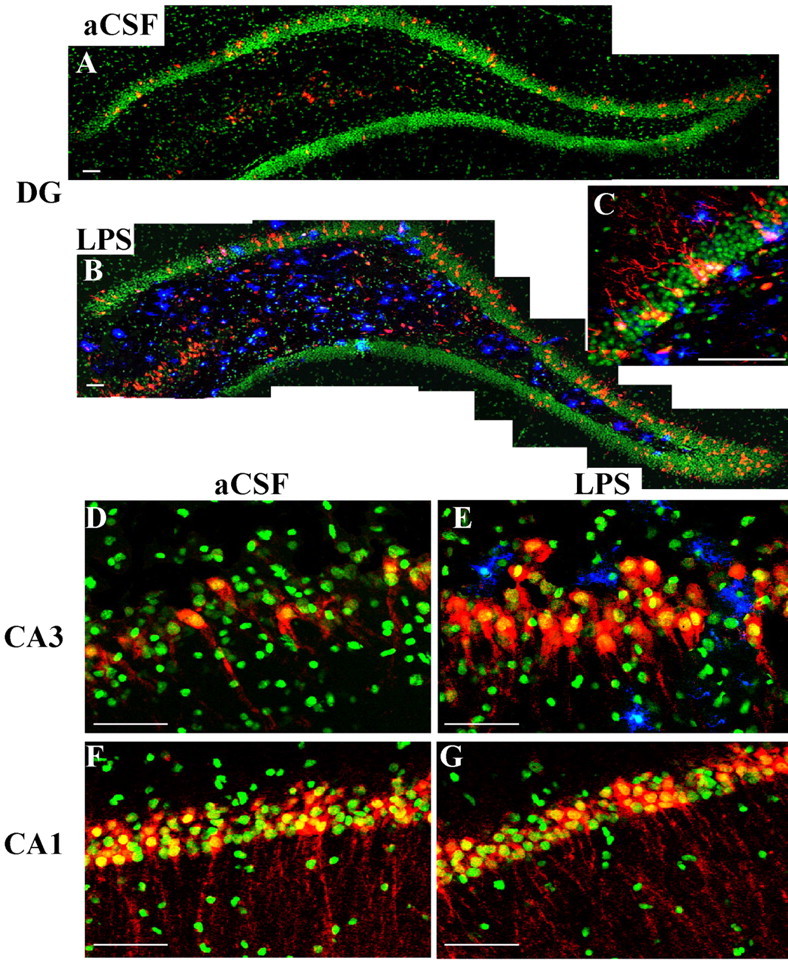
A-G, Double-immunohistochemical staining for Arc protein (red) and activated microglia (OX-6 positive; blue) within the hippocampus of aCSF rats (A, D, F) and LPS-infused rats (B, C, E, G) that explored a novel environment. Note the dramatic increase in Arc protein-positive neurons above that of aCSF controls only in regions with activated microglia. Nuclei are counterstained in green. Scale bars, 100 μm.
Fluorescence in situ hybridization. Six rats in each treatment group were represented on a series of six slides (six half-brains per slide). For each rat, three different slides were analyzed, resulting in a total of 18 slides prepared for in situ hybridization. All slides were selected from the same medial portion of the dorsal hippocampus adjacent to the ones chosen for immunohistochemistry (described above) and were processed for in situ hybridization to detect Arc mRNA (Guzowski et al., 1999; Vazdarjanova et al., 2002) (see Fig. 4 A).
Figure 4.
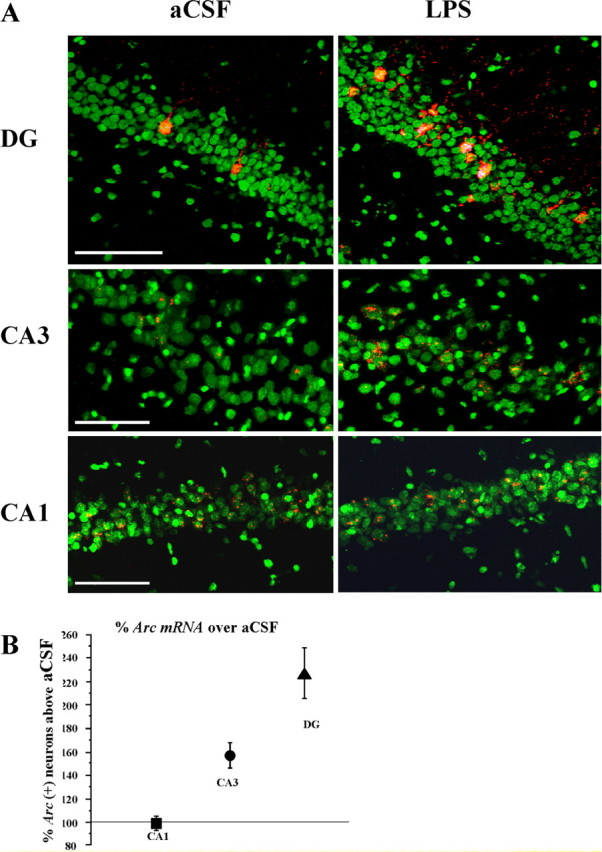
A, Examples of Arc mRNA expression (red) after behavioral exploration within the aCSF-infused (left) and LPS-infused (right) rats. Unlike aCSF-infused rats, the granule cells of LPS-infused rats contain numerous bright cytoplasmic Arc-positive neurons. Similarly, in CA3 of LPS-infused rats, there are more neurons with Arc intra nuclear foci compared with aCSF-infused rats. In CA1, both groups show similar Arc expression patterns after behavioral exploration. Nuclei are counterstained in green. B, Percentage increase in Arc-positive neurons above the mean of aCSF-infused rats for the CA3, CA1, and upper blade of the DG. Scale bars, 100 μm.
Image acquisition (confocal microscopy). To define the anatomical boundaries and degree of microglial activation within the hippocampus, two 20 μm coronal sections of the dorsal hippocampus at approximately -3.6 mm from bregma were reconstructed for each animal from each treatment group, as shown in Figure 1 A. The collage was done by overlapping 40-45 10× flat images from the middle of Z-stacks (1.0 μm optical thickness/plane) from the same 20 μm slice. The Z-stacks were collected with a Zeiss (Thornwood, NY) LSM 510 NLO-meta multiphoton/confocal microscope equipped with a 488 nm argon laser and a 543 and 633 nm helium/neon laser. Contrast and intensity parameters were set using the tissue sections from aCSF-infused rats. The parameters were kept constant across sections on a given slide using the 488 nm (for Sytox-Green) and 543 nm (for CY3) lasers. The images were collected with a small overlap, and the shape of cell groups was used as a landmark for alignment.
Figure 1.
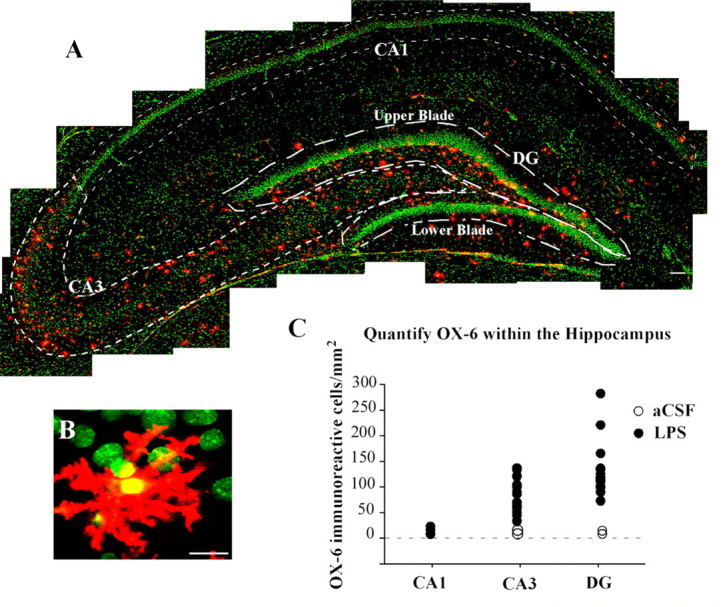
Reconstruction of a 20 μm coronal section from the dorsal hippocampus (at approximately -3.6 mm from bregma) from a young rat with chronic LPS infusion into the fourth ventricle. A, Single immunofluorescence staining revealed activated microglia (red, OX-6-immunoreactive cells) particularly elevated within the granule cell layer and the hilar region of the DG as well as inside and immediately around the CA3 molecular layer. No immunopositive staining was found in CA1. B, Activated microglia showed the characteristic bushy morphology with contracted and ramified processes. Nuclei are counterstained in green. Scale bars: A, 100 μm; B, 2.5 μm. C, Bivariate scattergram quantifying the number of OX-6-immunoreactive cells per square millimeter within each of the three areas of interest: DG, CA3, and CA1 indicated in A by dashed lines. Open circles, aCSF-infused rats; filled circles, LPS-infused rats.
Two of the three slides prepared for each animal (from immunofluorescence and in situ hybridization methods) were selected for confocal analysis. For CA1 and CA3, three Z-stacks (1.0 μm optical thickness/plane) were imaged with a 25× water-immersion lens (see Figs. 4 A, 5D-G). Contrast and intensity parameters were set using the tissue sections from the caged rats and the rats from the 45 min interval group. For consistency, these parameters were kept constant for the rest of the sections on the slide. The images were taken using the following coordinates: CA1: AP, -3.8 to -4.2; lateral (L), 1.5-2.5; dorsoventral (DV), 2.5; and CA3: AP, 3.8; L, 2.8-3.8; DV, 3.8. To study the expression of Arc mRNA and Arc protein in the DG, the entire DG from two coronal sections per rat was reconstructed as described above (see Fig. 5 A, B).
Image analysis. To quantify the activated microglia, the reconstructed hippocampal coronal sections (see Fig. 1C) were analyzed with MetaMorph imaging software (Universal Imaging Corporation, West Chester, PA). The hippocampus was divided into three areas of interest (CA1, CA3, and the granule cell layer and hilar region of the DG) as illustrated in Figure 1 A. After drawing these regions in each reconstructed image, a threshold tool was used to detect all of the OX-6 staining (see Fig. 1, red), and the area of each object was measured. All images were analyzed using the same threshold settings. The detected objects ranged from 5 to 2000 μm2. After creating a distribution curve, only those objects of sizes >65 μm2 were included in the analysis (the average size was 100 μm2). This object size was chosen to match more accurately the size of activated microglial cells and therefore significantly reduce sampling errors. The resulting number of objects was then corrected using the total area of each region of interest, and the number of objects per region (in square millimeters) was reported (see Fig. 1C).
Image analysis for the CA areas was done as described previously (Guzowski et al., 1999; Vazdarjanova et al., 2002). Each Z-stack was analyzed using MetaMorph imaging software. Briefly, Sytox nuclear counterstaining revealed two distinct morphologies. Cells with large and diffusely stained nuclei were considered neurons. The remainder of the cells (glia) had smaller nuclei (∼5 μm in diameter) and were bright and uniformly stained. Thus, only neuron-like cells present in the median planes of each Z-stack were counted. This approach is essentially an optical dissector technique minimizing sampling errors and stereological concerns, because minor variation in cell volumes does not influence sampling frequencies (Guzowski, 2002).
To analyze Arc mRNA in the CA areas, nuclei were segmented according to the criteria described above. The segmented neurons were then classified as containing fluorescently tagged Arc mRNA in the nucleus (see Fig. 4, CA areas) or in the cytoplasm (see Fig. 4, DG). Cytoplasmic Arc mRNA staining was detected only in the granule cells from the dentate gyrus, because these cells appear to show sustained transcription. The kinetics is clearly different from that observed in the CA areas in which Arc mRNA was exclusively induced by the second exploration. Because both the 45 and 90 min groups were killed immediately after the second 5 min exploration, the results from the Arc mRNA analysis in the CA areas were collapsed. In the DG, both Arc mRNA foci and cytoplasmic Arc mRNA were observed.
The analysis of Arc mRNA expression in the DG was done in reconstructed 20 μm coronal sections obtained as described above, and neurons were segmented with the same criteria used for the CA areas. The area of the granule cell layer and the total number of Arc-positive neurons was assessed in each reconstructed flat image, and this information, along with an estimate of the total number of neurons, was used to estimate the percentage of total Arc-positive neurons. The total number of neurons per dentate was estimated using a correction factor representing the number of neurons per square micrometer. This factor was derived from 92 Z-stacks from 10 different rats collected at 40× magnification. The total number of neurons per stack was counted, and the area of the granule cell layer (in square micrometers) from the middle plane was calculated. Using this factor, we calculated the percentage of neurons with Arc protein and/or Arc mRNA in the DG of each rat according to the following formula: 100 × p/[Ap × (N/A)], where p is the number of Arc+ neurons in a given reconstructed flat image, Ap is the area (in square micrometers) of the DG as measured from the reconstructed flat image, N is the total number of cells from all 40× Z-stacks, and A is the total area (in square micrometers) of the DG from the middle planes of all 40× Z-stacks. Because the behavior-induced Arc expression occurs primarily in the upper blade of the DG (Temple et al., 2003), the upper and lower blades were analyzed independently, as shown in Figure 1 A, by separating them in the middle of the genu.
To measure the proportion of Arc protein-expressing cells in the CA areas and the DG, the procedures used to measure the number of neurons were performed as described above for counting Arc mRNA-positive cells. Cells were classified as either positive or negative for Arc protein according to the following criteria. Positive neurons had perinuclear/cytoplasmic staining (see Fig. 5) surrounding at least 60% of the cell and visible in at least three plains together with the cell nucleus across the Z-Stack. To avoid classification errors, we carefully verified that the staining belonged to the cell of interest and not to a dendrite or the cell body of an adjacent cell. In all hippocampal regions, Arc protein expression was not observed after 5 min of exploration but was clearly present after 30 min (data not shown). Apparently, protein translation occurred soon after Arc mRNA reached the cytoplasm. Therefore, the protein expression observed in the CA areas and the DG was induced primarily by the first exploration session.
The results for both Arc mRNA and Arc protein analysis were expressed as a percentage of total neuronal nuclei analyzed per stack. The total number of neurons analyzed for each rat ranged from 450 to 550 for the CA1 region and from 250 to 310 for the CA3 region. These counts were obtained from three images per two slides in the same dorsal hippocampal region (anteroposterior, approximately -3.6 mm from bregma). Within the DG, the number of segmented neurons ranged from 460 to 600. This number refers to single 20 μm Z-stack images used to reconstruct the coronal section. For each rat, coronal sections of the reconstructed DG were analyzed, two for Arc mRNA and two for Arc protein.
Statistical analysis. StatView software (Cary, NC) was used to perform one-way ANOVA tests for each region of interest. The control (LPS and aCSF) and experimental (LPS and aCSF) groups were the independent variables, and the percentages of total neurons, from the various categories described above, were the dependent variables. When an overall ANOVA was significant (p < 0.0001), individual between-group comparisons were performed with Bonferroni post hoc tests to correct for multiple comparisons. The percentage of Arc mRNA increase was calculated in each region using the following formula: (percentage of Arc mRNA-expressing cells in LPS/percentage of Arc mRNA-expressing cells in aCSF) × 100. A correlation using Pearson's analysis, provided in the StatView software between the number of activated microglial objects and the percentage of Arc mRNA increase, was calculated across regions.
Results
Chronic infusion of LPS was well tolerated by all rats in that they gained weight normally for the duration of the study. Quantitative counts of neurons in the DG and CA3 and CA1 hippocampal regions reveled no significant changes in the total number of neurons per region between the aCSF- and LPS-infused groups: DG (mean/stack in aCSF = 247, SD = 66.4; mean LPS = 230, SD = 84.5); CA3 (mean aCSF = 126, SD = 80; mean LPS = 144, SD = 70.9); and CA1 (mean aCSF = 419, SD = 100; mean LPS = 408, SD = 90.5). This confirms that the consequences of the neuroinflammatory processes initiated by the chronic infusion of LPS do not result in loss of neurons.
LPS-induced activated microglia is region specific within the hippocampus
Immunofluorescence staining for OX-6 (Fig. 1A) revealed numerous highly activated microglia differentially distributed throughout the hippocampus in LPS-infused rats. The activated microglia had a characteristic bushy morphology with increased cell body size and contracted and ramified processes (Fig. 1B). Within the hippocampus, the activated microglia was consistently distributed only within the hilar region of the DG and around the CA3 but not in the CA1 area (Fig. 1A,C). Rats infused with aCSF had a few mildly activated microglia evenly scattered throughout the brain (data not shown), consistent with results from previous studies (Hauss-Wegrzyniak et al., 1998, 2000; Rosi et al., 2003, 2004). The quantitative cell count analysis (Fig. 1C) revealed a statistically significant difference in the number of OX-6 immunopositive cells per square millimeter in LPS versus aCSF rats in the DG (F(1,17) = 14.739; p < 0.01) and CA3 (F(1,17) = 16.940; p < 0.001) but not in CA1 (F(1,2) = 0.30; p = 0.62). Furthermore, analysis within the LPS-infused group revealed that the DG had the highest number of OX-6-positive cells per square millimeter (mean, 144), followed by the CA3 (mean, 105) and CA1 (mean, 2) (F(2,35) = 14.63; p < 0.001; p < 0.01 for both comparisons with CA1).
Exploration-induced Arc expression in the hippocampus of aCSF- and LPS-treated rats
After spatial exploration, the DG upper blade (Fig. 2, open bars) and CA1 and CA3 (Fig. 3, open bars) of aCSF-infused rats that had explored an environment showed an increase in the percentage of neurons expressing Arc mRNA and Arc protein compared with their respective caged control groups (ANOVA: DG, F(2,31) = 24.55 for Arc mRNA and F(2,31) = 60.29 for Arc protein; CA3, F(1,33) = 129.16 for Arc mRNA and F(2,31) = 40.80 for Arc protein; CA1, F(1,33) = 112.75 for Arc mRNA and F(2,31) = 38.76 for Arc protein; p < 0.0001 in all cases). The proportion of Arc-expressing cells after exploration varies among brain regions as reported previously (e.g., ∼35% for CA1 and ∼17% for CA3) (Vazdarjanova et al., 2002). In the DG, only ∼1.9% of neurons expressed Arc after exploration (Fig. 5A), which is consistent with electrophysiological recordings showing sparse activity in the DG during exploration (Jung and McNaughton, 1993). As reported in Materials and Methods, Arc mRNA induced by the second exploration experience was observed as foci in the nuclei of CA3 and CA1 neurons, whereas no mRNA was detectable in the cytoplasm at 45 and 90 min after the first exploration session. In contrast, in the DG granule cells, Arc mRNA was observed in both the cytoplasm and the nuclei (Fig. 4). Because there were no significant differences between the percentage of DG neurons with Arc foci and with Arc cytoplasmic staining (F = 0.88; p < 0.36) (Fig. 2A,B) and because the colocalization of these two cell populations was 95%, we use Arc mRNA to mean both Arc foci and Arc cytoplasmic staining only in the DG. The percentage of Arc protein-positive neurons increased at 45 min remained elevated at 90 min postexploration within the upper blade of the dentate gyrus (Fig. 2C, open bars), whereas Arc expression was reduced in CA1 and CA3 by 90 min (comparisons of the 45 min vs 90 min exploration groups: p < 0.001 for CA1, and p = 0.016 for CA3) (Fig. 3C,D, open bars).
Figure 2.
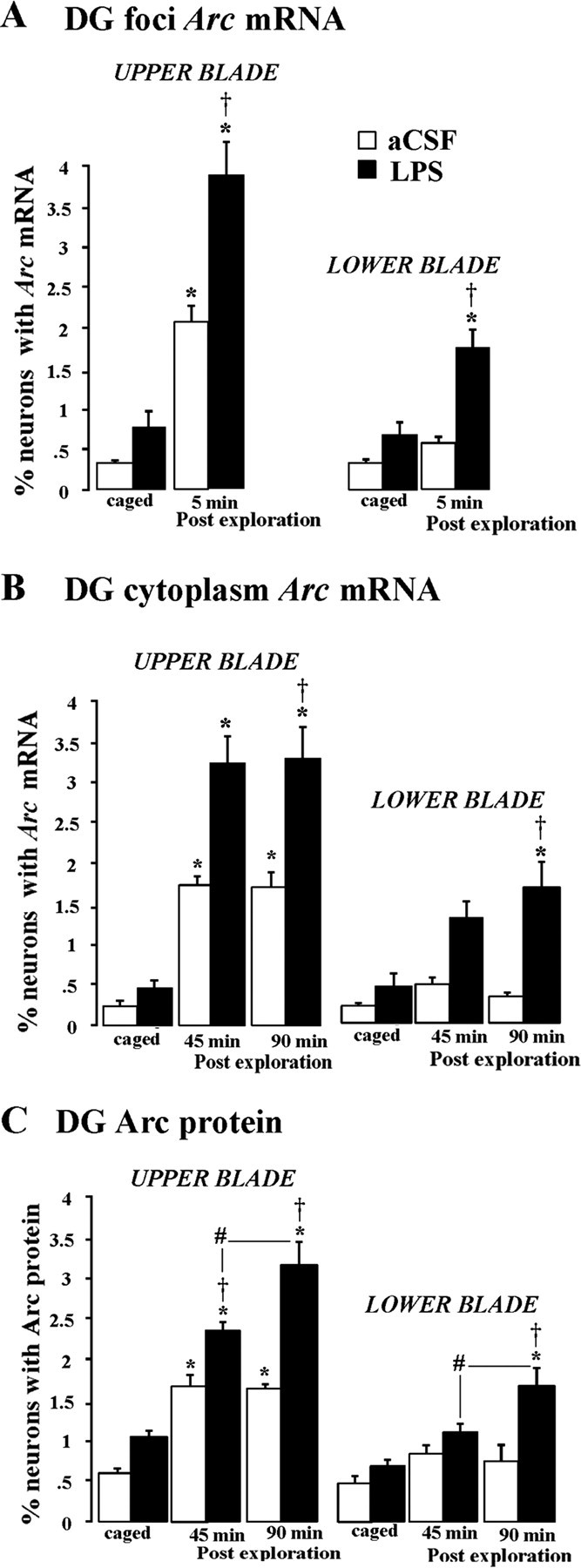
Percentage of DG neurons expressing Arc mRNA and Arc protein after spatial exploration of a novel environment in groups of aCSF or LPS chronically infused rats: A, Arc mRNA intranuclear foci; B, cytoplasmic Arc mRNA; C, Arc cytoplasmic protein. Each rat (n = 6/group) was exposed to a novel environment twice for 5 min and kept in its home cage between exposures from either 45 or 90 min. The percentage of neurons expressing Arc mRNA in the nuclear compartment and cytoplasm (A, B) was significantly increased after exploration in the upper but not the lower blade of aCSF-infused rats compared with their caged controls (open bars). LPS-infused rats (filled bars) that underwent the same behavioral treatment also showed an increased percentage of neurons expressing Arc mRNA. This increase, however, was significantly greater compared with the respective aCSF-infused rats in both the upper and lower blades. C, The pattern of Arc protein induction was similar to those of the Arc mRNA in aCSF-infused rats. In contrast, in LPS-infused rats, the 90 min time point showed greater expression than the 45 min time point. *p < 0.0001, significant versus the respective cage control group; †p < 0.0001, significant versus the aCSF group in the same behavioral exposure condition; #p < 0.0001, significant versus the LPS 45 min group.
Figure 3.
A, B, Percentage of neurons with Arc mRNA intranuclear foci (Arc+ neurons) after spatial exploration of a novel environment in hippocampal CA3 and CA1 of aCSF- or LPS-infused rats. After behavioral exploration of a novel environment, the percentage of Arc+ neurons was significantly higher in both CA3 (A) and CA1 (B) of aCSF-infused rats (open bars) compared with their caged controls. In LPS-infused rats (filled bars), the percentage of Arc+ neurons in both CA3 and CA1 was also higher than that of control rats. In addition, only in CA3 but not in CA1, the exploration-induced increase in Arc+ neurons was significantly higher than that of aCSF rats. C, D, Percentage of neurons with Arc protein across the CA3 and CA1 regions of the same rats. Similar to the results for Arc mRNA, the percentage of neurons that were positive for Arc protein was higher in the aCSF (open bars) exploration groups (both 45 and 90 min) compared with that of the caged controls in CA3 (C) and in CA1 (D). Also similar to the Arc mRNA pattern, in both CA3 and CA1 of LPS-infused rats (filled bars) from the exploration groups, the percentage of neurons positive for Arc protein was significantly higher compared with the LPS caged controls. Again, only in CA3 was this increase above caged controls and also significantly higher than the respective aCSF groups. In both CA3 and CA1 of aCSF and LPS exploration groups, the percentage of neurons positive for Arc protein was highest in the 45 min group. *p < 0.0001, significant versus the respective cage control group; †p < 0.0001, significant versus the aCSF group in the same behavioral exposure condition.
Similar to the aCSF exploration groups, the LPS groups showed a significant increase in the percentage of Arc mRNA- and Arc protein-positive neurons in the DG, CA3, and CA1 above those of caged controls (p < 0.0025 for all comparisons with the respective caged controls) (Figs. 2, 3, filled bars).
LPS treatment induced a region-specific increase in exploration-related Arc expression above that of aCSF exploration controls
In the CA3 and DG upper blade of the rats in the LPS exploration group, there was a significant increase in the number of neurons expressing Arc mRNA and protein above that of the aCSF exploration group (ANOVA: DG, F(1,31) = 15.23 for Arc mRNA, p < 0.001; DG, F(1,31) = 23.15, p < 0.001, and F(1,31) = 53.65, p < 0.0001, for Arc protein; CA3, F(1,33) = 8.30, p < 0.01, for Arc mRNA and F(1,31) = 31.30, p < 0.0001, for Arc protein). Bonferroni post hoc tests revealed a significant increase in the percentage of DG and CA3 Arc mRNA-positive neurons of the LPS exploration groups over that of the aCSF exploration groups (p < 0.005 for the DG comparisons; p < 0.0001 for the CA3 comparisons) that was paralleled by that of Arc protein at 45 and 90 min postexploration (p < 0.001) (Figs. 2, 3, filled vs open bars). Overall, this increase in the exploration-induced Arc mRNA by LPS infusion, above that of the aCSF exploration groups, was 125% in the DG (∼1.9% Arc-expressing neurons in aCSF vs ∼3.7% in LPS) and 57% in the CA3 (∼17% Arc-expressing neurons in aCSF vs ∼25% in LPS) (Fig. 4B). Importantly, no similar increases were observed in CA1 (Figs. 3B,D, filled vs open bars, 4A), suggesting that the increase in Arc-expressing neurons in LPS-infused rats is region specific. Because there were no significant differences between the aCSF and LPS caged control groups, this increase was specifically associated with the exploration behavior.
Unlike the respective aCSF groups, where exploration did not induce Arc expression in the DG lower blade, the LPS exploration groups showed an increase in the percentage of neurons with Arc mRNA and protein in the lower blade. This increase was significantly greater than caged controls and the respective aCSF exploration groups at 90 min postexploration (p < 0.001 for all four comparisons) (Fig. 2A-C, filled bars). This suggests that in the DG, LPS-induced neuroinflammation also disrupts the anatomical pattern of exploration-induced Arc expression.
Interestingly, in the DG upper and lower blade, the percentage of Arc protein-positive neurons between 45 and 90 min after exploration showed significant differences (p < 0.001) (Fig. 2C, filled bars). Although in aCSF-treated animals, the percentage of Arc protein-expressing neurons was similar between 45 and 90 min after exploration (Fig. 2C, open bars). In contrast, CA1 and CA3 areas showed a similar pattern of change over time (Fig. 3C, open vs filled bars).
The increase in exploration-induced Arc in LPS-treated rats is evident only in hippocampal regions with activated microglia
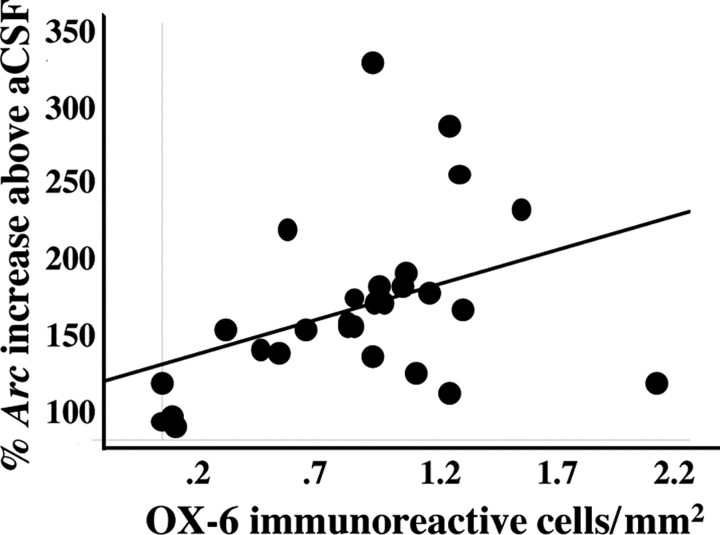
The spatial pattern of OX-6 immunoreactivity overlapped with that of LPS-induced increases in exploration-related Arc expression (Fig. 5). Therefore, we examined directly the relationship between these two variables. We found a significant correlation between the number of OX-6-immunoreactive cells per square millimeter and LPS-specific Arc increase across all three regions examined (r = 0.391; p < 0.05) (Fig. 6).
Figure 6.
Significant correlation between the number of activated microglia and the percentage increase in exploration-induced Arc in LPS-treated rats, above that of aCSF-infused rats, for all three regions.
Discussion
The present findings are the first to demonstrate that neuroinflammation disrupts the activity patterns associated with neuroplasticity and memory. Consistent with previous studies (Hauss-Wegrzyniak et al., 1998, 2000, 2002; Rosi et al., 2003, 2004), chronic LPS infusion into the fourth ventricle resulted in selective microglia activation within DG and CA3 but not in CA1 hippocampal areas. Although gene expression was not significantly altered in LPS-treated controls, exploration-induced Arc mRNA and protein were significantly elevated above those of aCSF exploration groups. Additionally, chronic neuroinflammation disrupted the spatial pattern of Arc expression in the DG by increasing the behavior-induced percentage of lower blade granule cells that expressed Arc mRNA and protein, above that of caged controls. This region of the dentate gyrus does not normally show exploration-induced Arc expression (Temple et al., 2003). Thus, chronic neuroinflammation does not appear to affect gene expression globally; the increase in cells expressing Arc occurs selectively after activation of hippocampal networks engaged in exploratory behavior. This relationship suggests that such a mechanism of disrupted activity patterns could contribute to the dysfunction in cognition observed during neurodegenerative diseases.
Activated microglia are selectively distributed within the hippocampus
Although it was known that chronic infusion of LPS results in the appearance of numerous OX-6-immunoreactive cells in the dentate gyrus and CA3 (Hauss-Wegrzyniak et al., 1998; Rosi et al., 2003, 2004), the present study quantified the distribution of OX-6 immunoreactivity over the entire hippocampus, showing that there was a 73-fold increase in activated microglia in the dentate gyrus and a 53-fold increase in CA3, compared with CA1 where no activated microglia were found. The distribution of LPS-induced activated microglia in this model develops over time (Wenk et al., 1999) and may result from either a differential susceptibility of the local microglia normally present in the affected regions and/or to a selective migration of activated microglia from neighboring regions. Alternatively, LPS may preferentially accumulate in the DG and thus activate a higher proportion of the locally present microglia. Neuroinflammation associated with a distinct distribution of activated microglia is also seen in AD (McGeer and McGeer, 1998; Akiyama et al., 2000). In early AD, activated microglia are found in those brain regions, such as the hippocampus (Cagnin et al., 2001), that later show the greatest degree of neuropathological changes and neurodegeneration (Griffin et al., 1998).
Neuroinflammation selectively increased behaviorally induced Arc in the DG and CA3
The results presented here demonstrate that rats with significant neuroinflammation exhibit exaggerated Arc mRNA and Arc protein expression after behavioral exploration. Furthermore, this increased expression is specific to the regions of the hippocampus with the greatest numbers of activated microglia (CA3 and dentate gyrus). These data demonstrate for the first time a close correlation between the presence of activated microglia and an altered pattern of hippocampal ensemble activation that can be induced by spatial exploration. The significant correlation between the presence of activated microglia and altered behaviorally induced Arc expression in hippocampal ensembles suggests that substantial disruption of network activity would be expected in response to such an inflammatory insult. Because Arc mRNA specifically travels to dendrites and accumulates at sites of intense synaptic activity, and Arc protein appears to be locally synthesized, it is in a critical position to play a fundamental role in the stabilization of behavioral experience. In fact, recent evidence suggests that Arc may selectively traffic glutamate receptors in spines of hippocampal neurons (Chowdhury et al., 2005). Thus, neuroinflammation may lead to altered synaptic plasticity in affected cells through an alteration of this trafficking function.
Neuroinflammation does not alter Arc translation
In the CA regions of the hippocampus, Arc transcription and translation appear to be tightly coupled. The proportion of cells expressing Arc protein was similar to that of cells expressing Arc mRNA in both the aCSF- and LPS-infused groups. Moreover, the decay across time (from 45 to 90 min) in the percentage of Arc-expressing cells was similar in both the LPS- and aCSF-treated groups. Combined, these findings suggest that in CA3 and CA1, neuroinflammation alters neither Arc translation nor the kinetics of Arc protein expression. Similarly, in the DG of aCSF-infused rats, Arc protein and cytoplasmic Arc mRNA were also tightly coupled and remained similar at 45 and 90 min. The percentage of neurons expressing Arc mRNA and Arc protein in the DG of the LPS group was also not significantly different; however, the kinetics of Arc protein expression appeared to be slightly delayed compared with that of the aCSF animals.
Although the present results suggest that during neuroinflammation the observed neuronal dysfunction is not attributable to gross disruption of translational processes, we cannot be confident with these methods that specific translational processes in the dendrites are unaltered. Additional research should address the possibility that neuroinflammation might disrupt Arc translation in the dendrites where Arc mRNA is regulated by synaptic signals such as BDNF and reelin (Yin et al., 2002; Dong et al., 2003).
LPS-induced molecular cascades can alter exploration-related Arc expression
The LPS-induced increase in exploration-related Arc expression could result from a sequence of biochemical processes initiated by activation of microglial cells. The chronic elevation of proinflammatory cytokines and nitric oxide (Quan et al., 1994; Brown and Bal-Price, 2003; Bodles and Barger, 2004) leads to a cascade of self-propagating cellular events (Griffin et al., 1998; Emerit et al., 2004), including a blockade of glutamate uptake by glia (Robinson et al., 1993), increased synthesis and release of prostaglandins (Katsuura et al., 1989), enhanced release of glutamate from astrocytes (Bezzi et al., 1998; Brezzi et al., 2001), and disruption of normal physiological activity within the hippocampus (Angulo et al., 2004). Elevated levels of glutamate may act on non-NMDA-type glutamate receptors to cause chronic membrane depolarization that would partially relieve the voltage-dependent Mg2+ block at NMDA receptors. Subsequent activation of NMDA receptors by ordinary glutamatergic synaptic activity may thus permit a continuous influx of calcium ions into neurons, theoretically overwhelming the endogenous mechanisms that regulate calcium ion homeostasis. This scenario has become known as the “weak excitotoxicity” model (Albin and Greenamyre, 1992). Because Arc expression is NMDA receptor dependent (Steward and Worley, 2001), these elevated intracellular levels of calcium may contribute to Arc induction in a larger than usual number of neurons. Inflammation can also relieve the Mg2+ blockade of voltage-gated NMDA channels by increasing nitric oxide levels, leading to impaired mitochondrial respiration, oxidative stress, a decline in energy production, and continued membrane depolarization (Willard et al., 2000; Beal, 2003; Brown and Bal-Price, 2003; Emerit et al., 2004). The inflammation-induced threshold change for Arc induction may be influenced also by elevated levels of cytokines that enhance NMDA-dependent intracellular calcium levels (Viviani et al., 2003; Bodles and Barger, 2004).
We have documented previously a decrease in the number of NMDA receptor channels within the DG and CA3 using the chronic LPS infusion model (Rosi et al., 2004). The apparent discrepancy of a decreased number of DG and CA3 NMDA receptors, and elevated Arc expression in these regions (which is NMDA receptor dependant), can be reconciled by hypothesizing that a significant proportion of the remaining NMDA channels are chronically depolarized attributable to the consequences of neuroinflammation. By itself, the elevated extracellular glutamate level resulting from proinflammatory processes appears in-sufficient to induce Arc expression, because there was no significant increase in Arc expression in LPS-infused caged controls. Thus, it is the combined effect of elevated glutamate levels and normal synaptic glutamate release that induces Arc in a larger than usual proportion of neurons in the neuroinflammation model. A similar series of molecular processes likely occurs in other chronic degenerative conditions that are associated with brain inflammation, mitochondrial impairment, and dementia such as AD (Akiyama et al., 2000), Parkinson's disease, Huntington's disease, and autoimmunodeficiency syndrome (Beal, 2000, 2003).
Exploration-induced Arc expression and cognitive impairment
The increase in the percentage of neurons expressing Arc after chronic LPS infusion may contribute to the cognitive impairment associated with neuroinflammation. In aCSF-infused rats, the low proportion of Arc-expressing neurons in the upper blade of the DG after behavioral exploration is consistent with the sparse activity of DG cells observed in electrophysiological recordings (Jung and McNaughton, 1993) and with the principle of sparse distributed coding (Marr, 1971; McNaughton and Morris, 1987). This principle suggests that to achieve maximally efficient storage, only a small fraction of the total population of cells ultimately should represent an episode (McNaughton et al., 1996; Sakurai, 1999). The dramatic increase in the number of neurons expressing Arc within the upper and lower blades of the DG during chronic inflammation may disrupt sparse coding, thereby decreasing the memory capacity of the system. Thus, the altered pattern of Arc expression within the DG may account, at least in part, for cognitive impairments observed in neurodegenerative diseases associated with neuroinflammation.
Footnotes
This work was supported by National Institutes of Health Grants AG009219, AG100546, and MH064357 and by Human Frontiers Science Program Grant LTF000112-2002-C. We thank Dalia Mikhael and Aldrich Sy for their technical assistance and Michelle Carroll for editorial assistance.
Correspondence should be addressed to Dr. Gary Wenk, University of Arizona, Arizona Research Laboratories, Division of Neural Systems, Memory, and Aging, Life Sciences, North Building, Room 384, P.O. Box 245115, Tucson, AZ 85724-5115. E-mail: Gary@nsma.arizona.edu.
Copyright © 2005 Society for Neuroscience 0270-6474/05/250723-09$15.00/0
References
- Akiyama H, Barger S, Barnum S, Bradt B, Bauer J, Cooper NR, Eikelenboom P, Emmerling M, Fiebich B, Finch CE, Frautschy S, Griffin WST, Hampel H, Landreth G, McGeer PL, Mrak R, MacKenzie I, O'Banion K, Pachter J, Pasinetti G, et al. (2000) Inflammation in Alzheimer's disease. Neurobiol Aging 21: 383-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albin RL, Greenamyre JT (1992) Alternative excitotoxic hypotheses. Neurology 42: 733-738. [DOI] [PubMed] [Google Scholar]
- Angulo MC, Kozlov AS, Charpak S, Audinat E (2004) Glutamate released from glial cells syncronizes neuronal activity in the hippocampus. J Neurosci 24: 6920-6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal MF (2000) Energetics in the pathogenesis of neurodegenerative diseases. Trends Neurosci 23: 298-304. [DOI] [PubMed] [Google Scholar]
- Beal MF (2003) Mitochondria, oxidative damage, and inflammation in Parkinson's disease. Ann NY Acad Sci 991: 120-131. [DOI] [PubMed] [Google Scholar]
- Bezzi P, Carmignoto G, Pasti L, Vesce S, Rossi D, Rizzini BL, Pozzan T, Volterra A (1998) Prostaglandins stimulate calcium-dependent glutamate release in astrocytes. Nature 391: 281-285. [DOI] [PubMed] [Google Scholar]
- Blalock EM, Chen K-C, Herman JP, Porter NM, Foster TC, Landfield PW (2003) Gene microarrays in hippocampal aging: statistical profilino identifies novel processes correlated with cognitive impairment. J Neurosci 23: 3807-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodles AM, Barger SW (2004) Cytokines and the aging brain—what we don't know might help us. Trends Neurosci 27: 621-626. [DOI] [PubMed] [Google Scholar]
- Brezzi P, Domercq M, Brambilla L, Galli R, Schols D, De Clercq E, Vescovi A, Bagetta G, Kollias G, Meldolesi J, Volterra A (2001) CXCR4-activated astrocyte glutamate release via TNFα: amplification by microglia triggers neurotoxicity. Nat Neurosci 4: 702-710. [DOI] [PubMed] [Google Scholar]
- Brown GC, Bal-Price A (2003) Inflammatory neurodegeneration mediated by nitric oxide, glutamate, and mitochondria. Mol Neurobiol 27: 325-355. [DOI] [PubMed] [Google Scholar]
- Cagnin A, Brooks DJ, Kennedy AM, Gunn RN, Myers R, Turkheimer FE (2001) In vivo measurement of activated microglia in dementia. Lancet 358: 461-467. [DOI] [PubMed] [Google Scholar]
- Chowdhury S, Shepherd JD, Okuno H, Lyford G, Petralia RS, Huganir RL, Worley PF (2005) Arc interacts with endophilin to regulate AMPA receptor trafficking. Cell, in press. [DOI] [PMC free article] [PubMed]
- Dickey CA, Gordon MN, Mason JE, Wilson NJ, Diamond DM, Guzowski JF, Morgan D (2004) Amyloid suppresses induction of genes critical for memory consolidation in APP+PS1 transgenic mice. J Neurochem 88: 434-442. [DOI] [PubMed] [Google Scholar]
- Dong E, Caruncho H, Liu WS, Smalheiser NR, Grayson DR, Costa E, Guidotti A (2003) A reelin-integrin receptor interaction regulates Arc mRNA translation in synaptoneurosomes. Proc Natl Acad Sci USA 100: 5479-5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerit J, Edeas M, Bricaire F (2004) Neurodegenerative diseases and oxidative stress. Biomed Pharmacother 58: 39-46. [DOI] [PubMed] [Google Scholar]
- Griffin WS, Sheng JG, Royston MC, Gentleman SM, McKenzie JE Graham DI, Roberts GW, Mrak RE (1998) Glial-neuronal interactions in Alzheimer's disease: the potential role of a “cytokine cycle” in disease progression. Brain Pathol 8: 65-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzowski JF (2002) Insights into immediate-early gene function in hippocampal memory consolidation using antisense oligonucleotide and fluorescent imaging approaches. Hippocampus 12: 86-104. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, McNaughton BL, Barnes CA, Worley PF (1999) Environment-specific expression of the immediate-early gene Arc in hippocampal neuronal ensembles. Nat Neurosci 2: 1120-1124. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, Lyford GL, Stevenson GD, Houston FP, McGaugh JL, Worley PF, Barnes CA (2000) Inhibition of activity-dependent Arc protein expression in the rat hippocampus impairs the maintenance of long-term potentiation and the consolidation of long-term memory. J Neurosci 20: 3993-4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauss-Wegrzyniak B, Dobrzanski P, Stoehr JD, Wenk GL (1998) Chronic neuroinflammation in rats reproduces components of the neurobiology of Alzheimer's disease. Brain Res 780: 294-303. [DOI] [PubMed] [Google Scholar]
- Hauss-Wegrzyniak B, Galons JP, Wenk GL (2000) Quantitative volumetric analysis of brain magnetic resonance imaging from rat with chronic neuroinflammation and correlation with histology. Exp Neurol 16: 347-354. [DOI] [PubMed] [Google Scholar]
- Hauss-Wegrzyniak B, Lynch MA, Vraniak PD, Wenk GL (2002) Chronic brain inflammation results in cell loss in the entorhinal cortex and impaired LTP in perforant path-granule cell synapses. Exp Neurol 176: 336-341. [DOI] [PubMed] [Google Scholar]
- Jung MW, McNaughton BL (1993) Spatial selectivity of unit activity in the hippocampus granular layer. Hippocampus 3: 165-182. [DOI] [PubMed] [Google Scholar]
- Katsuura G, Gottschall PE, Dahl RR, Airmura A (1989) Interleukin-1β increases prostaglandin E2 in rat astrocyte cultures: modulatory effect of neuropeptides. Endocrinology 124: 3125-3127. [DOI] [PubMed] [Google Scholar]
- Marr D (1971) Simple memory: a theory for archicortex. Philos Trans R Soc Lond B Biol Sci 262: 23-81. [DOI] [PubMed] [Google Scholar]
- McGeer EG, McGeer PL (1998) The importance of inflammatory mechanisms in Alzheimer disease. Exp Gerontol 33: 371-378. [DOI] [PubMed] [Google Scholar]
- McNaughton BL, Morris RG (1987) Hippocampal synaptic enhancement and information storage within a distributed memory system. Trends Neurosci 10: 408-415. [Google Scholar]
- McNaughton BL, Barnes CA, Gerrard JL, Gothard K, Jung NW, Knierim JJ, Kudrimoti H, Qin Y, Skaggs WE, Suster M, Weaver KL (1996) Deciphering the hippocampal polyglot: the hippocampus as a path integration system. J Exp Biol 199: 173-185. [DOI] [PubMed] [Google Scholar]
- Quan N, Sundar SK, Weiss JM (1994) Induction of interleukin-1 in various brain regions after peripheral and central injections of lipopolysaccharide. J Neuroimmunol 49: 125-134. [DOI] [PubMed] [Google Scholar]
- Robinson MB, Djali S, Buchhalter JR (1993) Inhibition of glutamate uptake with l-transpyrrolidine-2,4-dicarboxylate potentiates glutamate neurotoxicity in primary hippocampal cultures. J Neurochem 61: 2099-2103. [DOI] [PubMed] [Google Scholar]
- Rosi S, McGann K, Hauss-Wegrzynia B, Wenk GL (2003) Potential role of A2b receptor in the reduction of microglial activation pathway in an Alzheimer's disease model. J Neurochem 86: 220-226.12807441 [Google Scholar]
- Rosi S, Ramirez-Amaya V, Hauss-Werzniak B, Wenk GL (2004) Chronic brain inflammation leads to a decline in hippocampal NMDA-R1 receptors. J Neuroinflamm 1: 12-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai Y (1999) How do cell assemblies encode information in the brain? Neurosci Biobehav Rev 23: 785-796. [DOI] [PubMed] [Google Scholar]
- Steward O, Worley PF (2001) Selective targeting of newly synthesized Arc mRNA to active synapses requires NMDA receptor activation. Neuron 30: 227-240. [DOI] [PubMed] [Google Scholar]
- Steward O, Wallace CS, Lyford GL, Worley PF (1998) Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron 21: 741-751. [DOI] [PubMed] [Google Scholar]
- Temple MD, Worley PF, Steward O (2003) Visualizing changes in circuit activity resulting from denervation and reinnervation using immediate early gene expression. J Neurosci 23: 2779-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazdarjanova A, McNaughton BL, Barnes CA, Worley PF, Guzowski JF (2002) Experience-dependent coincident expression of the effector immediate-early genes Arc and Homer 1a in hippocampal and neocortical neuronal networks. J Neurosci 22: 10067-10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viviani B, Bartesaghi S, Gardoni F, Mezzani A, Behrens A, Bartfai T, Binaglia M, Corsini E, Di Luca M, Galli CL, Marianovich M (2003) Interleukin-1β enhances NMDA receptor-mediated intracellular calcium increase through activation of Src family. J Neurosci 24: 8692-8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltereit R, Dammermann B, Wulff P, Stabuli U, Kauselmann G, Bundman M, Khul D (2001) Arc3.1/Arc mRNA induction by Ca2+ and cAMP protein kinase A and mitogen-activated protein kinase/extracellular regulated kinase activation. J Neurosci 21: 5484-5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenk GL, Hauss-Werzniak B, Willard LB (1999) Pathological and biochemical studies of chronic neuroinflammation may lead to therapies for Alzheimer's disease. In: Research prospectives in neuroscience: neuroimmune, neurodegenerative, and psychiatric disorders and neural injury (Petterson P, Kordon C, Christen Y, eds), pp 73-77. Heidelberg: Springer.
- Willard LB, Hauss-Wegrzyniak B, Danysz W, Wenk GL (2000) The cytotoxicity of chronic neuroinflammation on basal forebrain cholinergic neurons of rats can be attenuated by glutamatergic antagonism or cyclooxygenage-2 inhibition. Exp Brain Res 134: 58-65. [DOI] [PubMed] [Google Scholar]
- Yin Y, Edelman GM, Vanderklish PW (2002) The brain-derived neurotrophic factor enhances synthesis of Arc in synaptoneurosomes. Proc Natl Acad Sci USA 99: 2368-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]