Abstract
In mammals, olfactory signals received by odorant receptors (ORs) in the olfactory epithelium (OE) are converted to a topographical map of activated glomeruli in the olfactory bulb (OB). It has been reported that the OE can be divided into four topographically distinct zones and that olfactory sensory neurons (OSNs) expressing a particular OR gene are randomly distributed within one zone. Here, we analyzed 80 different class II OR genes for their expression patterns in the OE by in situ hybridization. It was found that the expression area in the OE does not always fit into one of the four conventional zones. Expression areas are specific to each OR gene and are arranged in an overlapping and continuous manner in the OE. We also analyzed a spatial relationship between the OE and the OB for OSN projection. Our transgenic as well as DiI retrograde staining experiments demonstrated that the dorsal/ventral arrangement of glomeruli in the OB is correlated with the expression areas of corresponding ORs along the dorsomedial/ventrolateral axis in the OE. The present study indicates that the OR gene choice may be more restricted by the OSN location in the OE than what has been thought.
Keywords: olfactory epithelium, olfactory bulb, olfactory sensory neuron, axonal projection, odorant receptor gene, positioning of glomeruli
Introduction
In the mouse olfactory system, there are ∼1200 odorant receptor (OR) genes clustered at >50 different loci, scattered among most of the chromosomes (Buck and Axel, 1991; Young et al., 2001; Zhang and Firestein, 2002; Godfrey et al., 2004). Each olfactory sensory neuron (OSN) expresses only one functional OR gene in a monoallelic manner (Chess et al., 1994; Malnic et al., 1999; Ishii et al., 2001; Serizawa et al., 2003), and expression of each OR gene is confined to a restricted area in the olfactory epithelium (OE) (Ressler et al., 1993; Vassar et al., 1993; Strotmann et al., 1994; Sullivan et al., 1996). Furthermore, OSNs expressing a given OR gene project their axons to a specific set of glomeruli in the olfactory bulb (OB) (Ressler et al., 1994; Vassar et al., 1994; Mombaerts et al., 1996). Thus, the olfactory signal received in the OE is converted to a topographic odor map of activated glomeruli in the OB.
It has been reported that the OE can be divided into four distinct zones based on the expression patterns of ORs (Ressler et al., 1993; Vassar et al., 1993; Sullivan et al., 1996), although an overlapping distribution of the OR expression zones have been implicated in recent studies (Norlin et al., 2001; Iwema et al., 2004). For some OR genes, their projection sites have been mapped in the OB by in situ hybridization (Ressler et al., 1994; Vassar et al., 1994; Tsuboi et al., 1999) or by tagging with marker genes, for example, GFP (green fluorescent protein) and lacZ (Mombaerts et al., 1996; Wang et al., 1998; Finestein et al., 2004). The zone-to-zone correlation between the OE and OB has also been proposed in previous studies (Alenius and Bohm, 1997; Yoshihara and Mori, 1997; Yoshihara et al., 1997). To study the spatial organization of the olfactory system, DiI and horseradish peroxidase (HRP) have widely been used (Astic and Saucier, 1986; Pedersen et al., 1986; Schoenfeld et al., 1994; Levai et al., 2003). Retrograde labeling with HRP in rat revealed the dorsal/ventral correlation (Astic and Saucier, 1986; Saucier and Astic, 1986). Because zone-specific expression of the OR genes appears to play an important role in determining the projection sites for OSNs, we studied the zonal structure of the OE by in situ hybridization with various OR gene probes and DiI retrograde staining of OSN axons. It was found that the expression area is unique to each OR gene and does not always fit into one of the four conventional zones. Here, we report how the OE is subdivided in terms of OR gene expression and how the OR expression areas in the OE are correlated with the OSN projection sites in the OB.
Materials and Methods
In situ hybridization. To make the OR gene probes, DNA fragments of 400-600 bp containing the coding or 3′ untranslated region (UTR) sequences were amplified by PCR from the C57BL/6 mouse genomic DNA and subcloned into pGEM-T (Promega, Madison, WI). Primer sets used in the present study are listed in supplemental Table S1 (available at www.jneurosci.org as supplemental material). The OCAM (olfactory cell adhesion molecule) coding region (a generous gift from Dr. Y. Yoshihara, RIKEN, Wako, Japan), OMACS (olfactory specific medium-chain acyl CoA synthetase) (Oka et al., 2003), and EGFP (enhanced GFP) (Clontech, Palo Alto, CA) DNAs were amplified by PCR and subcloned into pGEM-T. Digoxigenin (DIG)-labeled RNA probes were prepared with a DIG RNA labeling kit (Hoffmann-La Roche, Basel, Switzerland).
OE coronal sections were prepared from 3-week-old C57BL/6 mice as described previously (Tsuboi et al., 1999). After drying, the samples were fixed for 15 min in 4% paraformaldehyde in PBS at room temperature. The sections were rinsed with PBS and incubated with 7 μg/ml Proteinase K (in 10 mm Tris-Cl, pH 7.4, 1 mm EDTA) for 10 min at 37°C. After fixing again with 4% paraformaldehyde in PBS for 10 min and rinsing with PBS, the sections were incubated with 0.25% acetic anhydride and 0.1 m triethanolamine, pH 8.0, washed with PBS, and air dried. Probes were diluted (1:100) with the hybridization buffer (Hirota et al., 1992), and 200 μl of each sample was applied to a slide. After 16-20 h of incubation at 50°C, the sections were washed, first with 2× SSC-50% formamide, then with 2× SSC, and finally with 0.2× SSC twice for 20 min at 65°C. After blocking with the blocking reagent (Hoffmann-La Roche), slides were incubated with alkaline phosphatase-conjugated anti-DIG antibody (Hoffmann-La Roche). Probe-positive cells were stained purple with nitroblue tetrazolium salt and 5-bromo-4-chloro-3-indolyl phosphate toludinium salt. The sections were photographed with an Olympus Optical (Tokyo, Japan) AX70 microscope.
All OR gene probes used in this study were <80% similar in sequence with any other related subfamily gene. We found that, when sequence similarities were >84%, the probes detected the related genes. By BLAST (basic local alignment search tool) search of the mouse genomic (C57BL/6) sequence, we identified 80 OR sequences that have no related OR genes with similarities >80% in the coding or 3′ UTR region. Consecutive coronal sections (8 μm thick) of the OE from 3-week-old C57BL/6 mice were analyzed by in situ hybridization using the OR gene probes listed in supplemental Table S1 (available at www.jneurosci.org as supplemental material).
Transgenic mice. The BAC (bacteria artificial chromosome)-A16 construct was generated by inserting the IRES-gap-EGFP (internal ribosomal entry site-growth-associated protein-EGFP)-containing SacII-NotI fragment from p29IG (Nakatani et al., 2003) immediately downstream of the stop codon of the A16 gene in the BAC clone RPCI-24-151H7 (Children's Hospital Oakland Research Institute, Oakland, CA). The flanking regions of IRES-gap-EGFP, the A16 coding region, and the 3′ UTR were obtained by recombinant PCR, using RPCI-24-151H7 as a template. The IRES-gap-EGFP with flanking regions was subcloned in pKOV-KanF and was used to tag the A16 gene in Escherichia coli by homologous recombination (Lalioti and Heath, 2001). For BAC-HA16, a 2.1 kb ScaI-ScaI fragment, containing the H region (homology region) of the MOR28 (murine odorant receptor) cluster (Serizawa et al., 2003), was subcloned into the EcoRV site in pBluescript II (Stratagene, La Jolla, CA) to generate pHr. Flanking sequences for the H region were amplified by two pairs of primers (GGTACCATTGAGACCCAGGGCTC: GTCGACTTATTTCAGTGCAGACACT and GTCTAGAACAGGTGATTGACAG: GCGGCCGCTACTTGCATATTCCTGTTT) and were inserted into pHr to generate pHupA16. The insert of pHupA16 was cloned into pKOV-KanF and used for homologous recombination in E. coli that contains BAC-A16, generating BAC-HA16. For BAC-T, IRES-tau-lacZ, IRES-gap-EGFP, and IRES-WGA (wheat germ agglutinin), each with its flanking regions, were isolated from YAC-290 (Serizawa et al., 2003), cloned into pKOV-KanF, and used to tag the MOR28, MOR10, and MOR83 genes in RPCI-23-339P24 (Children's Hospital Oakland Research Institute) by homologous recombination in E. coli (Lalioti and Heath, 2001). The BAC constructs were linearized by AscI digestion, isolated by preparative pulse-field gel electrophoresis as described previously (Serizawa et al., 2000), and injected into fertilized mouse eggs.
The EGFP fluorescence on the whole-mount OB was imaged by a fluorescence microscope (model SZX12; Olympus Optical) and photographed with a cooled CCD camera (model C4742-95-12ERG; Hamamatsu Photonics, Hamamatsu, Japan).
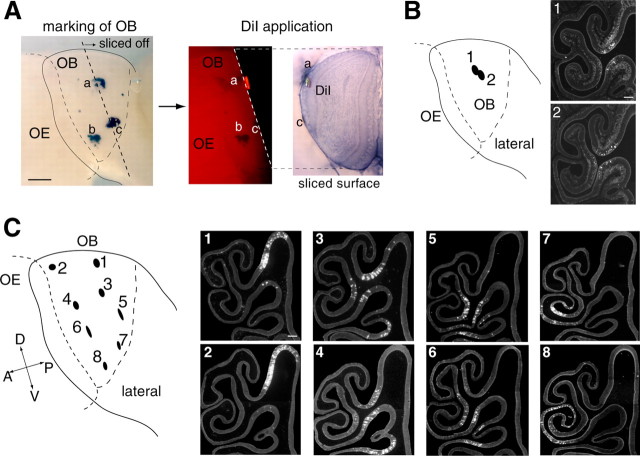
DiI retrograde labeling. Three-week-old mice (C57BL/6) were anesthetized with sodium pentobarbital (2.5 mg/animal) and perfused intracardially with 4% paraformaldehyde in PBS. The olfactory tissues (OE and OB) were dissected out and fixed overnight with 4% paraformaldehyde in PBS. The skull was removed, and the surface of the OB was marked with a felt-tip pen. The tissues were then embedded in 4% agarose gel in PBS, and the lateral view of the OB was photographed. The posterior portion of the OB was sliced off with a microslicer (model PRO7; Douhan EM, Kyoto, Japan) until the sliced surface reached the target spot for labeling. The sliced surface was stained with trypan blue to visualize the glomerular structure. DiI crystal (Molecular Probes, Eugene, OR) was precisely placed on the glomerular layer, and samples were covered with 4% agarose gel. The lateral view of the OB was photographed to identify the DiI location on the previously photographed OB picture. Using the previously marked spots with a felt-tip pen as positional markers, locations of DiI crystals were plotted on the schematic diagram of the OB. Samples were then incubated for 2-3 weeks at 37°C to allow the diffusion of DiI through nicked axon termini to the OSN cell bodies in the OE. Tissues were decalcified by incubating in PBS with 0.5 m EDTA at 4°C for 24 h and placed in PBS containing 30% sucrose for 24 h. The tissues were removed from the gel, and the OB was detached from the OE. Keeping the OE-OB boundary horizontal, the OE was embedded in OCT compound (Tissue-Tek, Miles, Torrance, CA). OE sections (16 μm thick) were prepared with a cryostat (model CM3050S; Leica, Nussloch, Germany) and analyzed with a fluorescence microscope (model IX70; Olympus Optical), coupled with a cooled CCD camera [C4742-95-12ERG (Hamamatsu Photonics) or VB-6010 (Keyence, Osaka, Japan)]. The DiI staining in the OE for each experiment, along with endoturbinate II and ectoturbinate 2′, is represented by one of the different colors in the color gradation diagram (see Fig. 4A). This OE region includes all zones from 1 to 4 and is reproducibly stained with DiI from any point on the lateral OB.
Figure 4.
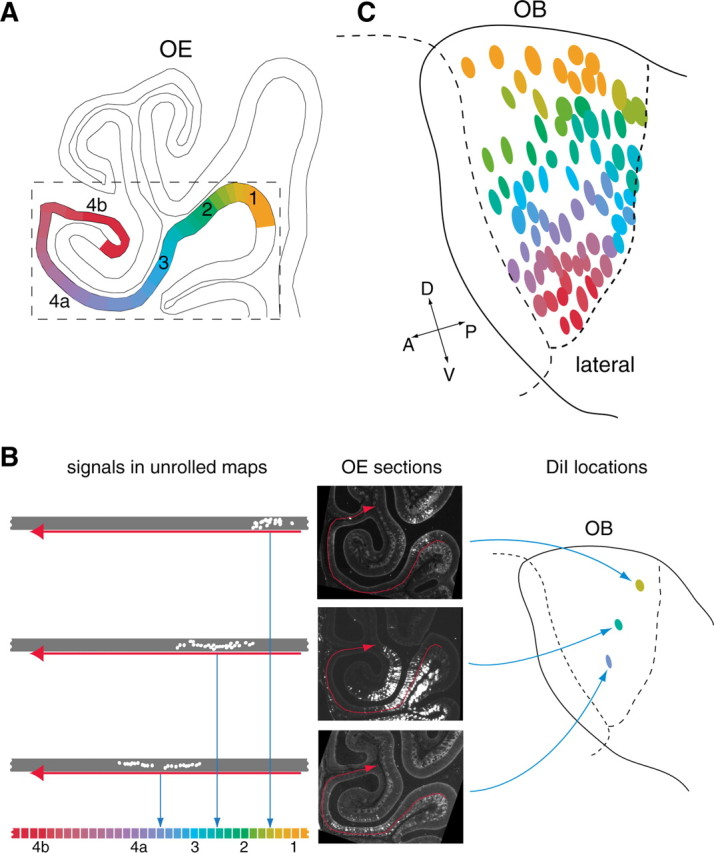
Correlation between the DiI-stained areas in the OE and locations of DiI spots in the OB. A, The retrograde staining of OSN cell bodies was analyzed along the endoturbinate II and ectoturbinate 2′ of the OE. This region includes all zones from 1 to 4 and is reproducibly stained with DiI from any point on the lateral side of the OB. B, Staining signals were plotted on the unrolled maps of the OE region along endoturbinate II and ectoturbinate 2′. The midpoint of the signal distribution on the map was determined, and its location was represented by one of the different colors as defined by a color gradation diagram. C, Eighty different DiI spots analyzed in the present study are colored and located in the schematic diagram of the OB. A, Anterior; D, dorsal; P, posterior; V, ventral.
Results
In situ hybridization of OE sections
We analyzed 80 different OR genes for their expression patterns in the OE. Probes used in the hybridization study were unique, specific, and would not cross-hybridize with other related OR genes. In our hybridization conditions, the probes detected related genes when sequence similarities were >84% (see supplemental Fig. S1, Table S1, available at www.jneurosci.org as supplemental material). Thus, probes chosen for this study were <80% similar in sequence to any other related subfamily gene.
The mouse ORs have been classified into two classes: the class I receptors, which resemble the OR family originally identified in fish (Ngai et al., 1993) and appear to recognize relatively hydrophilic ligands (Malnic et al., 1999), and class II receptors, which seem to have evolved in amphibians for detection of volatile ligands (Zhang and Firestein, 2002). In mouse, the class I OR genes are mostly expressed in the OCAM-negative region (Zhang et al., 2004) or zone 1 of the OE according to the definition by Sullivan et al. (1996). Some class II OR genes, for example, 265-1, are also expressed in zone 1 (Fig. 1A). For the class II genes expressed in the OCAM-positive (non-zone 1) region, hybridization signals were confined to specific areas, as it has been reported previously (Ressler et al., 1993; Vassar et al., 1993; Strotmann et al., 1994) (Fig. 1A). For example, the murine OR genes 277-1 and 103-1 were found to be expressed in the conventional zones 2 and 3, respectively. Both 103-15 (mouse I7) and 244-1 (MOR28) were classified as conventional zone 4 receptor genes (Tsuboi et al., 1999; Bozza et al., 2002), although we noticed that expression area of 244-1 was confined to the ventral half of zone 4 and was complementary to that of 103-15, which was confined to the dorsal part of zone 4. It should be noted that, for each OR gene, its hybridization pattern is symmetrical between the left and right OEs and conserved across individuals.
Figure 1.
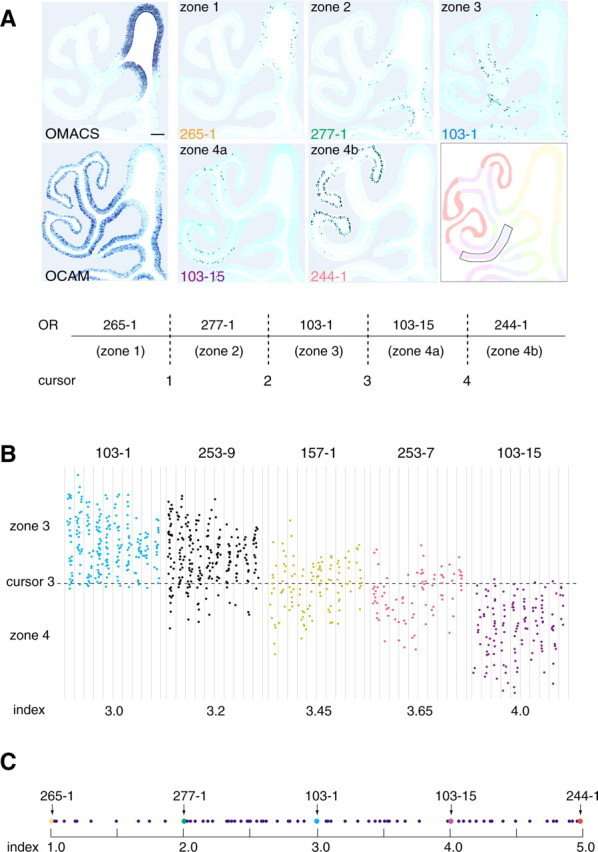
In situ hybridization of the mouse OE sections. A, Conventional OE zones are represented by the following standard OR genes: MOR 265-1 (zone 1), 277-1 (zone 2), 103-1 (zone 3), 103-15 (zone 4a), and 244-1 (zone 4b). Zone 4 was subdivided into two subzones, 4a and 4b, to calculate zone indices for the zone 4 OR genes. Four cursor lines, 1-4, were placed between the two neighboring zones or subzones as shown. The expression of the OMACS gene that codes for the olfactory-specific medium-chain acyl-CoA synthetase (Oka et al., 2003) was used as a marker for zone 1. Zone 1 is also defined as an OCAM-negative zone by in situ hybridization. Scale bar, 100 μm. B, Distribution of hybridization signals. Five OR genes, MOR 103-1, 253-9, 157-1, 253-7, and 103-15, expressed in zone 3 and/or zone 4 are analyzed. The zone 3-4 regions (boxed in the diagram of A) in every sixth coronal section (8 μm thick) were examined from the anterior-to-posterior direction (left to right). Hybridization signals were plotted on the unrolled maps of the OE region along endoturbinate II and ectoturbinate 2′. The zone index of each OR gene was calculated as a ratio of hybridization signals in two neighboring zones. For example, MOR 253-9, for which the zone index was 3.2, showed 20% of total signals in zone 4a and 80% in zone 3. C, Distribution of zone indices. A total of 80 different OR genes were analyzed. Zone indices for the standard OR genes are indicated by arrows. The scale does not reflect the physical distances between the neighboring zone boundaries.
During the process of analyzing the hybridization patterns of other OR genes, we found that the signals were not always restricted to one of the four zones, with the exception of the zone 1 OR genes. Many OR genes gave hybridization signals across the boundaries of the OE zones (Fig. 1B). For example, the 253-9 gene gave 80% of the total signal in zone 3 and 20% in zone 4 (the zone index, as defined in the legend of Fig. 1B, was 3.2). Some receptors gave signals almost equally in the both zones: 157-1 gave 55% of the total signal in zone 3 and 45% in zone 4 (zone index, 3.45). The 253-7 gene was distributed 65% in zone 4 and 35% in zone 3 (zone index, 3.65). As summarized in Figure 1C, the OR genes expressed in the OCAM-positive region of the OE appear to possess unique and distinct zone indices that are arranged in a continuous and overlapping manner along the dorsomedial/ventrolateral axis of the OE (for primer sets, see supplemental Table S1, available at www.jneurosci.org as supplemental material).
Retrograde DiI staining of OSN axons
For some OR genes, their projection sites (glomeruli) have been mapped in the OB by in situ hybridization (Ressler et al., 1994; Vassar et al., 1994; Tsuboi et al., 1999) or by tagging with marker genes, for example, GFP and lacZ (Mombaerts et al., 1996; Wang et al., 1998; Finestein et al., 2004). In our previous studies, we determined the projection sites for four OR genes: 235-1 (A16), 236-1 (MOR18), 244-3 (MOR83), and 244-1 (MOR28) (Tsuboi et al., 1999). These glomerular locations are schematically shown in Figure 2A, along with their zone index values. There appears to be a strong correlation between their expression areas in the OE and corresponding glomeruli in the OB along the dorsal/ventral axis.
Figure 2.
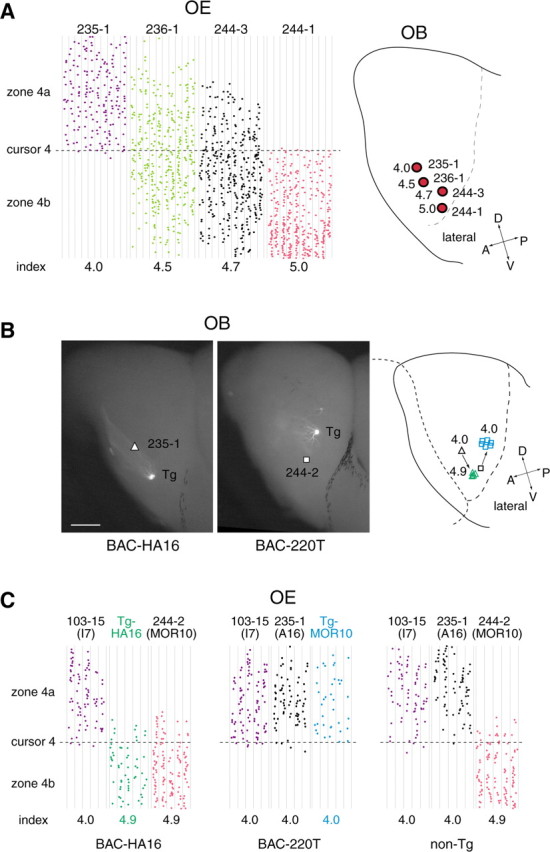
Identification of OSN projection sites in the OB. Glomerular locations are compared for four OR genes (right). Distribution of hybridization signals in the OE was analyzed for each OR gene (left), and the zone index was calculated as described in Figure 1 B. Projection sites for 235-1 (A16), 236-1 (MOR18), 244-3 (MOR83), and 244-1 (MOR28) were determined by in situ hybridization of OB sections (Tsuboi et al., 1999) and by fluorescent staining of the OB with the differently tagged OR transgenes (Serizawa et al., 2003) (K. Miyamichi and S. Serizawa, unpublished result). B, Projection sites for the transgenic OR genes are shown. To visualize the glomeruli for the GFP-tagged HA16 and MOR10 (244-2) transgenes, the whole-mount OBs were observed for GFP by fluorescence microscopy. Projection sites for the endogenous A16 (235-1) and MOR10 (244-2) genes (indicated by a triangle and a square, respectively) were determined by in situ hybridization of OB sections using the OR gene sequences as probes (Tsuboi et al., 1999). Projection sites for the transgenes (green triangles for the Tg-HA16 and blue squares for the Tg-MOR10) are shifted compared with those for the endogenous genes, as shown in the schematic diagram. Scale bar, 500 μm. C, OE sections of the BAC-HA16, BAC-220T, and non-Tg mice were analyzed by in situ hybridization with OR gene probes. Hybridization signals were plotted schematically, and the zone indices were calculated as described for Figure 1 B. Expression of the GFP-tagged transgenes was detected with the GFP coding sequence as a probe. Distributions of hybridization signals across the boundary of zones 4a and 4b are schematically shown with the zone indices. A, Anterior; D, dorsal; P, posterior; V, ventral.
To further examine the correlation between the OE and OB in the OSN axonal projection, we performed retrograde DiI staining of glomeruli (Fig. 3). DiI and HRP have been used to study the spatial organization of the olfactory system (Astic and Saucier, 1986; Pedersen et al., 1986; Schoenfeld et al., 1994; Levai et al., 2003). The olfactory tissues (OE and OB) were dissected out and embedded in agarose gel after fixation in paraformaldehyde. To ensure that DiI crystals directly contacted the axons within the glomeruli, we sliced off the posterior portion of the OB with a microslicer until the sliced surface reached the target glomeruli marked previously with a felt-tip pen (Fig. 3A). This method was highly advantageous in preventing unwanted dye diffusion. In the present study, ∼80 different locations on the OB were examined. Only one spot, usually containing three to five glomeruli, was stained and analyzed in each experiment. To allow the diffusion of DiI through nicked axon termini to the OSN cell bodies, samples were incubated at 37°C for 2-3 weeks after DiI crystal application. The OE was then sliced in parallel with the cribriform plate and analyzed for the distribution of DiI-stained OSN cell bodies with a fluorescence microscope. Stained OSNs were confined to a restricted area of the OE, and staining patterns varied depending on the locations of DiI crystals on the OB. When two neighboring spots in the OB were compared (Fig. 3B), staining patterns of OSNs in the OE were quite similar, if not identical, whereas distant pairs usually gave different staining patterns in the OE. Interestingly, DiI spots located on the same level along the dorsal/ventral axis showed similar retrograde staining patterns in the OE, even when they were far apart along the anterior/posterior axis of the OB. Four sets of such examples are shown in Figure 3C. There appears to be a strong correlation between the DiI-stained OE areas and DiI spotted glomerular locations along the dorsal/ventral axis of the OB.
Figure 3.
Retrograde DiI staining of OSN axons. A, The OB was marked with a felt-tip pen to determine the location of the target spot for DiI labeling (a-c). The posterior portion of the OB was sliced off with a microslicer until the target glomerulus appeared on the sliced surface of the remaining OB. DiI crystal was placed on the target spot, which usually covered three to five glomeruli. The sample was then embedded in agarose gel and incubated for 2-3 weeks at 37°C to allow the DiI to diffuse from the glomeruli to the OSN cell bodies through their axons. B, Coronal sections of the OE were analyzed for the distribution of DiI fluorescence. Two neighboring DiI spots on the OB gave similar retrograde staining patterns in the OE, whereas spots that were far apart usually yielded different patterns. C, A pair of DiI spots (1:2), located on the same level along the dorsal/ventral axis, were analyzed for the staining patterns in the OE. Note that their patterns are quite similar, despite the large separation of the DiI spots on the OB along the anterior/posterior axis. Similar examples are shown for three other sets of DiI spots (3:4, 5:6, and 7:8). Scale bars: A, 500 μm; B, C, 100 μm. A, Anterior; D, dorsal; P, posterior; V, ventral.
This notion became more evident when we colored the DiI-placed spots in the schematic diagram of the OB according to the areal distribution of DiI stained OSNs in the OE (Fig. 4). The stained cell bodies were analyzed along the endoturbinate II and ectoturbinate 2′ of the OE (Fig. 4A). This region includes all conventional zones and is reproducibly stained with DiI from any point on the lateral side of the OB. Staining signals were plotted on the unrolled maps of the OE region (Fig. 4B). The midpoint of the signal distribution was determined, and its location was represented by one of the different colors as defined by the color gradation diagram in Figure 4B. This color index, representing the DiI-stained areas in the OE, was used to color the origin of DiI in the OB. Gradation is evident along the dorsal/ventral axis of the OB (Fig. 4C).
Shifted projection sites for the OR transgenes
As described above, projection of OSNs appears to be affected along the dorsal/ventral axis of the OB by the expression areas of ORs in the OE. Our transgenic experiments revealed this property. The murine OR gene A16 (235-1) has been reported to be a typical zone 4 gene (Sullivan et al., 1996) with a zone index of 4.0. This gene was poorly expressed from the BAC construct, BAC-A16, in the transgenic mice. We reported previously a cis-acting DNA region named H that activates the MOR28 (244-1) cluster for transcription (Serizawa et al., 2003). When the H region was inserted into the BAC construct, the expression frequency of the transgenic A16 (Tg-HA16) was restored to normal, comparable with that of the endogenous A16 (235-1). However, its expression in the OE was found to be in a more ventrolateral position, with a zone index of 4.9 (Fig. 2C). This ventral shift in the expression area in the OE was seen for all three transgenic lines of BAC-HA16. Although the exact cause of the shift in the expression area is unknown, the H region of MOR28 may have influenced the zonal specificity of the transgenic A16 (Tg-HA16). Because the expression area was shifted ventrally in the OE, we were interested in knowing the glomerular locations for the Tg-HA16. As shown in Figure 2B (left), the projection site was shifted ventrally for the Tg-HA16 compared with that for the endogenous A16 (235-1) (Ressler et al., 1994; Tsuboi et al., 1999). This shift was seen in all three mouse lines containing the BAC-HA16 construct. It is interesting that a fraction of transgene-expressing OSNs projected their axons to the endogenous A16 glomeruli, probably attributable to a homophilic interaction among the axons expressing the same type of OR molecules, as reported previously (Vassalli et al., 2002; Feinstein and Mombaerts, 2004).
A similar shift in the axonal projection was also seen for the MOR10 (244-2) in the BAC-220T construct. In one of the mouse lines, the zone index for the transgenic MOR10 (Tg-MOR10) was 4.0, whereas the index for the endogenous MOR10 (244-2) was 4.9 (Fig. 2C). When we examined the projection sites for the Tg-MOR10, we found a dorsal shift of the 244-2 glomeruli in the BAC-220T transgenic mouse, as shown in Figure 2B (middle). Together, these results suggest that the dorsomedial/ventrolateral OE axis of OR expression strongly influences the projection target in the OB. In other transgenic lines in which the expression area of the Tg-MOR10 was normal, its projection sites in the OB were indistinguishable from those for the endogenous MOR10 (244-2).
Discussion
In the present study, we analyzed OSNs for their OR gene expression in the OE and their axonal projection in the OB. In situ hybridization with various OR gene probes demonstrated that each OR gene is expressed in a restricted area in the OE. It has been reported that OSNs expressing a given OR gene are confined to one of the four OE zones but are randomly distributed within the zone (Ressler et al., 1993; Vassar et al., 1993; Sullivan et al., 1996). This appears to be true for the class I OR genes (Zhang et al., 2004) (T. Miyazaki, A. Tsuboi, and H. Sakano, unpublished observation). However, for most class II OR genes, the expression area in the OE appears to be unique to each OR gene and does not always fit into one of the conventional zones (Fig. 1B). No clear boundaries or borders appear to be present between the neighboring zones. For ∼80 class II OR genes examined thus far, their expression areas are distinct and distributed in an overlapping and continuous manner along the dorsomedial/ventrolateral axis of the OE (Fig. 1C). There may be a gradient of a specific guidance molecule, for example, neuropilin 2 (Norlin et al., 2001), along this axis. How is the area-specific expression in the OE regulated for the OR genes? It has been assumed that zone-specific transcription factors for the OR genes are responsible for the regulation. However, if the expression areas of various ORs are arranged in a continuous and overlapping manner, we may have to consider a different mechanism, one that detects a gradient or a relative location in the OE for each OR gene along the dorsomedial/ventrolateral axis. If this is the case, the choice of the OR genes may be more restricted by the location of the OSN in the OE than what has been thought (Serizawa et al., 2004).
In situ hybridization and DiI retrograde staining experiments demonstrated that the dorsal/ventral arrangement of glomeruli in the OB is correlated with the expression areas of corresponding ORs along the dorsomedial/ventrolateral axis in the OE (Figs. 2A, 4). Retrograde labeling with HRP in rat also revealed this dorsal/ventral correlation (Astic and Saucier, 1986; Saucier and Astic, 1986). Our transgenic experiments shown in Figure 2B indicate that this correlation is independent of the expressed OR molecules. In contrast, the anterior/posterior arrangement of glomeruli appears to be independent of the epithelial locations of OSNs and more dependent on the expressed ORs. Recently, the ephrin-A system has been considered for the determination of axonal projection sites along the anterior/posterior axis in the OB (Cutforth et al., 2003), although it is yet to be clarified how the ORs are involved in the OSN projection. In any case, the present study indicates that the spatial information within the OE plays an important role not only in the axonal projection of OSNs along the dorsal/ventral axis of the OB but also in the choice of the OR genes in the OSNs. The OR gene choice appears to be more restricted by the OSN location and not totally stochastic within the conventional zones.
Footnotes
This work was supported by grants from Japan Science and Technology Agency, the Ministry of Education, Culture, and Science of Japan, and the Mitsubishi Foundation. We thank Anthony Otsuka for critical comments and Tokiko Tsubokawa and Hiroko Nakatani for expert technical assistance. K.M. is a predoctoral fellow of Japan Society for the Promotion of Science.
Correspondence should be addressed to Hitoshi Sakano, Department of Biophysics and Biochemistry, Graduate School of Science, The University of Tokyo, Tokyo 113-0032, Japan. E-mail:sakano@mail.ecc.u-tokyo.ac.jp.
Copyright © 2005 Society for Neuroscience 0270-6474/05/253586-07$15.00/0
K.M. and S.S. contributed equally to this work.
References
- Alenius M, Bohm S (1997) Identification of a novel neural cell adhesion molecule-related gene with a potential role in selective axonal projection. J Biol Chem 17: 26083-26086. [DOI] [PubMed] [Google Scholar]
- Astic L, Saucier D (1986) Anatomical mapping of the neuroepithelial projection to the olfactory bulb in the rat. Brain Res 16: 445-454. [DOI] [PubMed] [Google Scholar]
- Bozza T, Feinstein P, Zheng C, Mombaerts P (2002) Odorant receptor expression defines functional units in the mouse olfactory system. J Neurosci 22: 3033-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck L, Axel R (1991) A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell 65: 175-187. [DOI] [PubMed] [Google Scholar]
- Chess A, Simon I, Cedar H, Axel R (1994) Allelic inactivation regulates olfactory receptor gene expression. Cell 78: 823-834. [DOI] [PubMed] [Google Scholar]
- Cutforth T, Moring L, Mendelsohn M, Nemes A, Shah NM, Kim MM, Frisen J, Axel R (2003) Axonal ephrin-As and odorant receptors: coordinate determination of the olfactory sensory map. Cell 114: 311-322. [DOI] [PubMed] [Google Scholar]
- Feinstein P, Mombaerts P (2004) A contextual model for axonal sorting into glomeruli in the mouse olfactory system. Cell 117: 817-831. [DOI] [PubMed] [Google Scholar]
- Godfrey PA, Malnic B, Buck LB (2004) The mouse olfactory receptor gene family. Proc Natl Acad Sci USA 101: 2156-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota S, Ito A, Morii E, Wanaka A, Tohyama M, Kitamura Y, Nomura S (1992) Localization of mRNA for c-kit receptor and its ligand in the brain of adult rats: an analysis using in situ hybridization histochemistry. Mol Brain Res 15: 47-54. [DOI] [PubMed] [Google Scholar]
- Ishii T, Serizawa S, Kohda A, Nakatani H, Shiroishi T, Okumura K, Iwakura Y, Nagawa F, Tsuboi A, Sakano H (2001) Monoallelic expression of the odourant receptor gene and axonal projection of olfactory sensory neurones. Genes Cells 6: 71-78. [DOI] [PubMed] [Google Scholar]
- Iwema CL, Fang H, Kurtz DB, Youngentob SL, Schwob JE (2004) Odorant receptor expression patterns are restored in lesion-recovered rat olfactory epithelium. J Neurosci 24: 356-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalioti M, Heath J (2001) A new method for generating point mutations in bacterial artificial chromosomes by homologous recombination in Escherichia coli Nucleic Acids Res 29: E14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levai O, Breer H, Strotmann J (2003) Subzonal organization of olfactory sensory neurons projecting to distinct glomeruli within the mouse olfactory bulb. J Comp Neurol 458: 209-220. [DOI] [PubMed] [Google Scholar]
- Malnic B, Hirono J, Sato T, Buck LB (1999) Combinatorial receptor codes for odors. Cell 96: 713-723. [DOI] [PubMed] [Google Scholar]
- Mombaerts P, Wang F, Dulac C, Chao SK, Nemes A, Mendelsohn M, Edmondson J, Axel R (1996) Visualizing an olfactory sensory map. Cell 87: 675-686. [DOI] [PubMed] [Google Scholar]
- Nakatani H, Serizawa S, Nakajima M, Imai T, Sakano H (2003) Developmental elimination of ectopic projection sites for the transgenic OR gene that has lost zone specificity in the olfactory epithelium. Eur J Neurosci 18: 2425-2432. [DOI] [PubMed] [Google Scholar]
- Ngai J, Dowling MM, Buck L, Axel R, Chess A (1993) The family of genes encoding odorant receptors in the channel catfish. Cell 72: 657-666. [DOI] [PubMed] [Google Scholar]
- Norlin EM, Alenius M, Gussing F, Hagglund M, Vedin V, Bohm S (2001) Evidence for gradients of gene expression correlating with zonal topography of the olfactory sensory map. Mol Cell Neurosci 18: 283-295. [DOI] [PubMed] [Google Scholar]
- Oka Y, Kobayakawa K, Nishizumi H, Miyamichi K, Hirose S, Tsuboi A, Sakano H (2003) O-MACS, a novel member of the medium-chain acyl-CoA synthetase family, specifically expressed in the olfactory epithelium in a zone-specific manner. Eur J Biochem 270: 1995-2004. [DOI] [PubMed] [Google Scholar]
- Pedersen PE, Jastreboff PJ, Stewart WB, Shepherd GM (1986) Mapping of an olfactory receptor population that projects to a specific region in the rat olfactory bulb. J Comp Neurol 250: 93-108. [DOI] [PubMed] [Google Scholar]
- Ressler KJ, Sullivan SL, Buck LB (1993) A zonal organization of odorant receptor gene expression in the olfactory epithelium. Cell 73: 597-609. [DOI] [PubMed] [Google Scholar]
- Ressler KJ, Sullivan SL, Buck LB (1994) Information coding in the olfactory system: evidence for a stereotyped and highly organized epitope map in the olfactory bulb. Cell 79: 1245-1255. [DOI] [PubMed] [Google Scholar]
- Saucier D, Astic L (1986) Analysis of the topographical organization of olfactory epithelium projections in the rat. Brain Res 16: 455-462. [DOI] [PubMed] [Google Scholar]
- Schoenfeld TA, Clancy AN, Forbes WB, Macrides F (1994) The spatial organization of the peripheral olfactory system of the hamster. I. Receptor neuron projections to the main olfactory bulb. Brain Res 34: 183-210. [DOI] [PubMed] [Google Scholar]
- Serizawa S, Ishii T, Nakatani H, Tsuboi A, Nagawa F, Asano M, Sudo K, Sakagami J, Sakano H, Ijiri T, Matsuda Y, Suzuki M, Yamamori T, Iwakura Y, Sakano H (2000) Mutually exclusive expression of odorant receptor transgenes. Nat Neurosci 3: 687-693. [DOI] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Nakatani H, Suzuki M, Saito M, Yoshihara Y, Sakano H (2003) Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse. Science 302: 2088-2094. [DOI] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Sakano H (2004) One neuron-one receptor rule in the mouse olfactory system. Trends Genet 20: 648-653. [DOI] [PubMed] [Google Scholar]
- Strotmann J, Wanner I, Helfrich T, Beck A, Breer H (1994) Rostro-caudal patterning of receptor-expressing olfactory neurones in the rat nasal cavity. Cell Tissue Res 278: 11-20. [DOI] [PubMed] [Google Scholar]
- Sullivan SL, Adamson MC, Ressler KJ, Kozak CA, Buck LB (1996) The chromosomal distribution of mouse odorant receptor genes. Proc Natl Acad Sci USA 93: 884-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi A, Yoshihara S, Yamazaki N, Kasai H, Asai-Tsuboi H, Komatsu M, Serizawa S, Ishii T, Matsuda Y, Nagawa F, Sakano H (1999) Olfactory neurons expressing closely linked and homologous odorant receptor genes tend to project their axons to neighboring glomeruli on the olfactory bulb. J Neurosci 19: 8409-8418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassalli A, Rothman A, Feinstein P, Zapotocky M, Mombaerts P (2002) Minigenes impart odorant receptor-specific axon guidance in the olfactory bulb. Neuron 35: 681-696. [DOI] [PubMed] [Google Scholar]
- Vassar R, Ngai J, Axel R (1993) Spatial segregation of odorant receptor expression in the mammalian olfactory epithelium. Cell 74: 309-318. [DOI] [PubMed] [Google Scholar]
- Vassar R, Chao SK, Sitcheran R, Nunez JM, Vosshall LB, Axel R (1994) Topographic organization of sensory projections to the olfactory bulb. Cell 79: 981-991. [DOI] [PubMed] [Google Scholar]
- Wang F, Nemes A, Mendelsohn M, Axel R (1998) Odorant receptors govern the formation of a precise topographic map. Cell 93: 47-60. [DOI] [PubMed] [Google Scholar]
- Yoshihara Y, Mori K (1997) Basic principles and molecular mechanisms of olfactory axon pathfinding. Cell Tissue Res 290: 457-463. [DOI] [PubMed] [Google Scholar]
- Yoshihara Y, Kawasaki M, Tamada A, Fujita H, Hayashi H, Kagamiyama H, Mori K (1997) OCAM: a new member of the neural cell adhesion molecule family related to zone-to-zone projection of olfactory and vomeronasal axons. J Neurosci 17: 5830-5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JM, Friedman C, Williams EM, Ross JA, Tonnes-Priddy L, Trask BJ (2001) Different evolutionary processes shaped the mouse and human olfactory receptor gene families. Hum Mol Genet 11: 535-546. [DOI] [PubMed] [Google Scholar]
- Zhang X, Firestein S (2002) The olfactory receptor gene superfamily of the mouse. Nat Neurosci 5: 124-133. [DOI] [PubMed] [Google Scholar]
- Zhang X, Rogers M, Tian H, Zhang X, Zou DJ, Liu J, Ma M, Shepherd GM, Firestein SJ (2004) High-throughput microarray detection of olfactory receptor gene expression in the mouse. Proc Natl Acad Sci USA 101: 14168-14173. [DOI] [PMC free article] [PubMed] [Google Scholar]