Abstract
Recent studies suggest that specific neural basic helix-loop-helix (HLH; i.e., Olig1 and Olig2, Mash1), associated inhibitory HLH (i.e., Id2 and Id4), high-mobility group domain (i.e., Sox10), and homeodomain (i.e., Nkx2.2) transcription factors are involved in oligodendrocyte (OL) lineage specification and progressive stages of maturation including myelination. However, the developmental interplay among these lineage-selective determinants, in a cell- and maturational stage-specific context, has not yet been defined. We show here in vivo and in vitro developmental expression profiles for these distinct classes of transcriptional regulators of OLs. We show that progressive stages of OL lineage maturation are characterized by dynamic changes in the subcellular distribution of these transcription factors and by different permutations of combinatorial transcriptional codes. Transient transfections of these precise combinatorial codes with a luciferase reporter gene driven by the myelin basic protein promoter define how changes in the molecular composition of these transcriptional complexes modulate myelin gene expression. Our overall findings suggest that the dynamic interplay between developmental stage-specific classes of transcriptional activators and associated inhibitory factors orchestrate myelin gene expression during terminal maturation of the mammalian CNS.
Keywords: neuronal–glial coupling, oligodendrocytes, lineage maturation, transcriptional regulation, forebrain, proneural genes
Introduction
The expression of patterning genes, predominantly homeodomain (HD) transcription factors, initially subdivides the neuraxis into distinct progenitor domains from which specific neuronal and glial subtypes are sequentially elaborated (Gunhaga et al., 2000; Novitch et al., 2001). Later, in concert with basic helix-loop-helix (bHLH) factors, these patterning genes orchestrate the timely elaboration of developmentally coupled neurons and glia. Olig2 coordinates the acquisition of pan-neuronal traits and motor neuron subtype specification within the spinal cord and promotes the expression of another bHLH factor, Neurogenin2 (Ngn2), which in turn reinforces the neuronal subtype specific developmental program (Mizuguchi et al., 2001; Novitch et al., 2001; Sun et al., 2001; Zhou and Anderson, 2002). Spinal-cord oligodendrocytes (OLs) are also elaborated within this domain after the extinction of Ngn2 (Tanagaki et al., 2001; Zhou and Anderson, 2002) and a dorsal shift of the HD protein Nkx2.2 into the Olig2+/Nkx2.2–domain (Mizuguchi et al., 2001; Novitch et al., 2001; Sun et al., 2001; Zhou and Anderson, 2002). Genetic analysis has shown that Nkx2.2 is required for OL terminal differentiation (Qi et al., 2001), whereas Olig1 is important for OL specification throughout the neuraxis (Alberta et al., 2001; Lu et al., 2002; Zhou and Anderson, 2002). Cerebral cortical OLs also arise from ventral forebrain progenitors that are initially specified by sonic hedgehog (Shh) (He et al., 2001; Tekki-Kessaris et al., 2001; Yung et al., 2002; Ivanova et al., 2003). Recently, we provided evidence for Olig2 and Mash1 being involved in ventral forebrain stem cell lineage restriction through the elaboration of Shh-responsive intermediate progenitors that sequentially give rise to GABAergic neurons and later to OLs after tangential cortical migration under the influence of developmentally modulated bone morphogenetic protein (BMP) signals (Yung et al., 2002). Furthermore, inhibitory HLH transcription factors of the inhibitor of differentiation (Id) family, particularly Id2 and Id4, modulate the OL developmental timer that couples OL progenitor cell proliferation to cell-cycle exit and early OL terminal differentiation (Kondo and Raff, 2000a; Wang et al., 2001). Id proteins are known to bind and to sequester ubiquitous class “A” bHLH factors, thereby preventing heterodimerization with neural (class “B”) bHLH factors that modulate the activation of OL-specific downstream developmental genes (Ross et al., 2003).
The myelin basic protein (MBP) promoter contains consensus sequences for Nkx2.2, for class A (E2A) bHLH proteins, and for high-mobility group (HMG) factors such as Sox10 (Stolt et al., 2002). Overexpression of Nkx2.2 induces expression of myelin genes (Qi et al., 2001). In addition, E2A proteins can also recruit appropriate class B bHLH proteins to form active transcriptional complexes with specific HD proteins at promoter sites of appropriate target genes (Poulin et al., 2000). Furthermore, Sox10 activates expression from the MBP promoter by forming both homodimers and heterodimers with Sox8 at Sox10 response elements (Stolt et al., 2004). Together, these data suggest that Nkx2.2, specific classes of HLH proteins, and HMG factors such as Sox10 have the potential to differentially modulate the activation of the MBP promoter.
Materials and Methods
Neural progenitor cell cultures. Early embryonic [embryonic day 12 (E12)] multipotent and more lineage-restricted progenitor species derived from the dorsal forebrain were plated at clonal density (15–40 cells/cm2), propagated in serum-free media containing a specified factor for various time intervals, and subsequently examined by immunofluorescence microscopy to define neural lineage profiles and expression of selected transcription factors and to detect cellular proliferation [10 μm bromodeoxyuridine (BrdU), added 3 h before fixation] as described previously (Zhu et al., 1999; Mehler et al., 2000). Multipotent progenitor clones were generated by application of basic fibroblast growth factor (bFGF; 10 ng/ml), and lineage-restricted neuronal–OL clones were generated by application of bFGF and the N-terminal active form of Shh (N-Shh; 50 ng/ml) (Yung et al., 2002). Cortical neuroblasts were generated from neuronal–OL progenitors by the application of BMP2 (10 ng/ml) for 2 d in vitro (DIV) (Yung et al., 2002). OL precursor cells were generated from post-migratory stage (E17) cortical progenitors using NG2 immunoselection as described previously (Marmur et al., 1998a,b). Early-stage OL progenitors were generated from OL precursor species by the application of platelet-derived growth factor (PDGF; 10 ng/ml) for 2 DIV. Late-stage OL progenitors were generated by application of PDGF to OL precursor species for 4 DIV. Postmitotic OLs were generated from late-stage OL progenitors by withdrawal of PDGF for an additional 2 DIV (Kondo and Raff, 2000c). We followed Institutional Animal Care and Use Committee guidelines for experiments in which primary mouse tissue specimens were used.
Cell line cultures. NIH3T3 cells were grown in DMEM supplemented with 10% fetal bovine serum, 2 mm l-glutamine, 1 mm sodium pyruvate, 100 U/ml penicillin, and 100 g/ml streptomycin. The mouse OL precursor cell line Oli-neu (Jung et al., 1995) was a kind gift from Dr. J. Trotter (University of Mainz, Mainz, Germany). Cells were grown on poly-l-lysine-coated culture dishes. The immature Oli-neu cells were maintained in growth medium consisting of DMEM supplemented with 2 mm l-glutamine, 1 mm sodium pyruvate, 10 ng/ml biotin, 100 μg/ml apotransferrin, 100 μm putrescine, 20 nm progesterone, 30 nm sodium selenite, 5 μg/ml insulin, 1% horse serum, 100 U/ml penicillin, and 100 μg/ml streptomycin. Differentiation was induced by switching the cells to a medium containing 1 mm dibutyril-cAMP (Sigma, St. Louis, MO), as described previously (Jung et al., 1995). NIH3T3 and Oli-neu cells were maintained at 37°C in 5% CO2 in a humid atmosphere.
Immunofluorescence microscopic analysis was performed as described previously (Marmur et al., 1998a,b).
Specific antibody preparations. Olig1 and Olig2 [rabbit, rIg, 1:500] were gifts from D. Rowitch (Dana-Farber Cancer Institute, Boston, MA), H. Takebayashi (Kyoto University, Kyoto, Japan), and M. Nakafuku (University of Tokyo, Tokyo, Japan), respectively. These antibodies exhibit complete immunoreactivity for mouse cells and tissue sections, and each antibody exhibits a complete absence of cross-reactivity as judged by immunoassay techniques using individual Olig protein-producing cell lines. The additional antibodies used included the forebrain bHLH factor Mash1 (mouse, mIgG1, 1:250; PharMingen, San Diego, CA), the inhibitory HLH transcription factors Id2 and Id4 (rabbit rIg, 1:100; Santa Cruz Biotechnology, Santa Cruz, CA), the HD transcription factor Nkx2.2 (1:100, mouse, IgG2b; Developmental Studies Hybridoma Bank, Iowa City, IA), the HMG transcription factor Sox10 (guinea pig Ig, 1:3000; a gift from M. Wegner, University of Erlangen, Erlangen, Germany), the class A bHLH factor E47 (rabbit, rIg, 1:100; Santa Cruz Biotechnology), the neuroepithelial marker nestin (mIgG1, 1:200; PharMingen), the neural progenitor factor Musashi1 (rat Ig, 1:500; provided by H. Okano, Keio University, Tokyo, Japan), the OL precursor marker NG2 (rIg, 1:3500 or mIg, 1:500; provided by W. B. Stallcup, Burnham Institute, La Jolla, CA), the OL progenitor marker O4 (mIgM, 1:4), the postmitotic OL markers GC/O1 (mIgM, 1:50; provided by M. Noble, University of Rochester, Rochester, NY) and MBP (mIgG2b, 1:500; Sternberger Monoclonals, Berkeley, CA), the OL marker anti-adenomatous polyposis coli (APC) mouse monoclonal antibody clone CC-1 (mIgG2b, 1:200; Calbiochem, La Jolla, CA), the early neuronal marker type III β-tubulin (mIgG2b, 1:400; Sigma), and nuclear and cell-cycle-associated reagents BrdU (mIgG1, 1:400; Novocastra, Newcastle upon Tyne, UK) and Hoechst 33342 (2 μg/ml). All antibody preparations were tested for specificity as outlined above.
Growth factor preparations. To generate the various stem, progenitor, and more-differentiated neuronal and OL lineage species for immunocytochemical analysis, the following growth factor preparations were used: recombinant bFGF (Collaborative Biomedical Products, Bedford, MA), PDGF-AA (R & D Systems, Minneapolis, MN), BMP2 (a gift from Genetics Institute, Cambridge, MA), noggin (a gift from Regeneron, Tarrytown, NY), and N-Shh (a gift from Curis, Cambridge, MA).
Transfection and luciferase reporter assays. Cells were plated at a density of 1.4 × 105 cells/well in 6-well dishes the day before transfection. Cultures were transfected with Flag-tagged Id2 and Id4 constructs inserted into the pCE4F-EBNA vector (provided by Dr. B. Christy, University of Texas, San Antonio, TX), HA-tagged Olig1, Flag-tagged Olig2 constructs inserted into pcDNA3.1 (provided by Dr. H. Takebayashi), c-myc-tagged Nkx2.2 construct inserted into pcDNA3.1B (provided by Dr. D. Rowitch), Sox10, Mash1, and E47 full-length coding sequences that were inserted into pcDNA3.1. The MBP promoter (Miskimins et al., 2002), from –1323 to +30 upstream of the luciferase coding region in pGL3Basic was provided by Dr. R. Miskimins (University of South Dakota, Vermillion, SD). The NeuroD promoter was provided by Dr. K. Nave (Max Planck Institute, Goettingen, Germany). Transfections were performed using FuGene 6 (Roche, Welwyn Garden City, UK) according to the manufacturer protocol. Briefly, 3 μl of FuGene 6 reagent was used for every 2 μg of transfected DNA in 100 μl of DMEM. Transfections were performed using a total of 3.5 μg of cDNA, 0.5 μg of reporter DNA, and the remaining 3.0 μg varied depending on the combination tested. When a single factor was transfected, 0.5 μg corresponded to the cDNA encoding for that specific molecule, and the remaining 2.5 μg was pCDNA3. When a combination of six transcription factors was tested, each cDNA was 0.5 μg. The relative ratio between the cDNA/reporter DNA was 6:1. For pcDNA3, the total amount of transfected DNA was 3.0 μg to match the total amount of DNA transfected when multiple combinations of transcription factors were tested. The FuGene 6–DNA mixture was incubated for 30 min at room temperature and added to the cells. Cells remained in transfection medium for 48 h, after which the cells were processed for luciferase reporter assays. Assays were performed using a luciferase reporter assay system (Promega, Madison, WI) according to the manufacturer's protocol. Activity was measured in a TD20/20 luminometer (Turner Designs, Sunnyvale, CA) and normalized for protein content. The activity of the promoter was also normalized for transfection efficiency using a Renilla luciferase and showed high reproducibility of the results. No difference was found in normalizations performed using counts for cotransfected Renilla luciferase.
Western blot analysis was performed as described previously (Liu et al., 2005) using Nkx2.2 antibodies (Ericson et al., 1997) and appropriate antibody preabsorption with antigen and additional controls.
Results
Regional and cellular expression of neural bHLH, inhibitory HLH, HD, and HMG transcription factors within the embryonic forebrain suggest distinct roles for these factors in progressive OL lineage maturation from ventral forebrain stem cells
To begin to examine the role of bHLH and HD transcription factors in mediating cerebral cortical OL lineage elaboration from ventral forebrain stem and progenitor cells, we initially performed immunocytochemical analysis on embryonic forebrain tissue sections using Olig1, Olig2, Mash1, and Nkx2.2 antibodies. Olig1 is not expressed during early embryonic forebrain stages (Fig.1A1,A2, E10 and E13) and exhibits initial expression only within the cortical subventricular zone (SVZ) and cerebral cortex after tangential cortical migration (Yung et al., 2002) (Fig. 1A3, E17). In contrast, Olig2 is expressed initially within the medial (MGE) and the lateral (LGE) ganglionic eminences of the ventral forebrain at the initiation of tangential cortical migration (Fig. 1B1,B2, E10 versus E13) and within the post-migratory stage cortical SVZ and cerebral cortex (Fig. 1B3, E17). Furthermore, Mash1 is expressed initially before Olig2 in the early embryonic ventral forebrain (Fig. 1C1, E10) and in a similar pattern to Olig2 in the MGE and LGE at the time of tangential cortical migration (Fig. 1C2, E13). In contrast to the expression profiles of Olig2, Mash1 is only present within the post-migratory stage cortical SVZ (Fig. 1C3, E17). Finally, Nkx2.2 is transiently expressed within the posterolateral region of the early embryonic ventral forebrain (Fig. 1D1, E10), and its expression is negligible within the mid-embryonic forebrain (Fig.1D2,D3, E13 and E17). These distinctive regional and developmental expression profiles suggest that Olig2 plays an earlier and broader role than Olig1 in initial ventral forebrain stem cell lineage restriction and in progressive neural lineage specification within pre-migratory stage MGE and LGE progenitor cells and additional, more-advanced developmental functions within post-migratory cortical SVZ progenitors derived from the ventral forebrain in concert with Olig1. Mash1 seems to play a complementary role to that of Olig2 and an additional earlier role in ventral forebrain patterning and a more-delimited role in later stages of OL lineage specification. Furthermore, the expression profiles of Nkx2.2 suggest an early role in regional ventral forebrain patterning and a distinctive later role in cortical OL lineage maturation. Finally, the embryonic forebrain expression profiles of Id2, Id4, and Sox10 suggest early roles for these inhibitory HLH proteins in ventral forebrain stem cell lineage progression and OL progenitor lineage elaboration and for Sox10 in coupling these initial specification events with later OL maturational processes (Woodruff et al., 2001; Ross et al., 2003).
Figure 1.
Regional and temporal expression profiles of specific bHLH (Olig1, Olig2, and Mash1) and HD (Nkx2.2) transcription factors within the embryonic forebrain suggest complementary roles for Olig2 and Mash1 in ventral forebrain stem-cell lineage restriction and for Olig1 in OL lineage specification. A–D, Comparative immunofluorescence microscopy of Olig1 (A), Olig2 (B), Mash1 (C), and Nkx2.2 (D) protein expression in transverse (A1, B1, C1, D1) and coronal (A2, A3, B2, B3, C2, C3, D2, D3) sections of embryonic (E10, E13, and E17) mouse forebrain using Olig1, Olig2, Mash1, and Nkx2.2 antibodies. Olig1 is not expressed at early embryonic forebrain stages (A1, A2) and exhibits initial expression only at a time consistent with the developmental stage after tangential cortical migration within the cortical SVZ and cerebral cortex (CTX) (A3). In contrast, Olig2 is initially expressed within the MGE and LGE of the ventral forebrain at the time of tangential cortical migration (B2 vs B1) and within the cortical SVZ and CTX after tangential migration (B3). Mash1 is initially expressed before Olig2 in the early embryonic ventral forebrain (C1) and in a similar pattern to Olig2 in the MGE and LGE at the time of tangential cortical migration (C2). Like Olig2, Mash1 is expressed at a time consistent with the developmental stage after tangential cortical migration within the cortical SVZ and CTX (C3). In contrast, Nkx2.2 is transiently expressed in the early embryonic posterolateral region of the ventral forebrain (D1) and exhibits negligible expression in later stages of embryonic forebrain development (D2, D3).
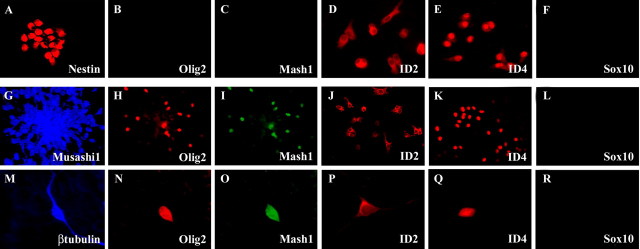
To further define the roles of these selective classes of transcription factors in OL lineage elaboration from ventral forebrain progenitor species, we performed immunocytochemical analysis on dissociated cellular preparations derived from the ventral forebrain before tangential cortical migration using Olig1, Olig2, Mash1, Id2, Id4, Sox10, and Nkx2.2 antibodies. Early embryonic (E12.5) ventral forebrain FGF-responsive neural stem cells express high levels of the neuroepithelial marker nestin (Fig. 2A) and of the inhibitory HLH factors Id2 and Id4 (Fig. 2D,E), but they do not express Olig2, Mash1, or Sox10 proteins (Fig. 2B,C,F). Coapplication of bFGF and Shh to these cells results in the continued expression of nestin (data not shown) and the RNA-binding protein Musashi1 (Fig. 2G) (Kaneko et al., 2000; Imai et al., 2001) but the absence of expression of neuronal, OL, or astroglial lineage markers (data not shown). Under these early differentiating conditions but before the overt expression of neuronal and glial lineage markers, these neuronal–OL lineage-restricted progenitors (Yung et al., 2002) now display selective nuclear expression of Olig2, Mash1 (Fig. 2H,I), and Id4 (Fig. 2K), whereas Id2 remains predominantly cytosolic (Fig. 2J). Our previously published observations confirm that Shh-mediated lineage restriction of early embryonic ventral forebrain stem cells to GABAergic neuronal–OL progenitors is mediated in part by the induction of Olig2 with Mash1 (Yung et al., 2002). Putative neuronal precursors of cortical GABAergic neurons derived from post-migratory cortical SVZ progenitors continue to express Olig2 and Mash1 (Fig. 2N,O) but have undetectable levels of Sox10 (Fig. 2R) and Nkx2.2 (data not shown). These cumulative observations suggest that specific combinations of bHLH, inhibitory HLH, HD, and HMG transcription factors orchestrate a distinct spectrum of cellular events associated with ventral forebrain patterning, neuronal–OL lineage restriction of embryonic ventral forebrain stem cells, and later in neuronal and glial lineage elaboration.
Figure 2.
Cellular expression profiles of Olig2 and Mash1 support complementary roles for these transcription factors in orchestrating neuronal–OL lineage restriction of ventral forebrain stem cells. A–R, Immunofluorescence microscopy analysis of transcription factor expression within neural progenitors derived from early embryonic ventral forebrain self-renewing multipotent progenitors (Nestin+; A) reveals the absence of Olig2 (B) and Mash1 (C) expression in these cells. However, Shh-mediated neuronal–OL lineage restriction in vitro is accompanied by the induction of nuclear expression of Olig2 (H) and Mash1 (I). These neuronal–OL progenitors (Musashi1+; G) sequentially give rise to βtubulin+ neuroblasts (M) that are the likely precursors of cortical GABAergic neurons and later to OL precursor species under the influence of developmentally modulated BMP signals (Yung et al., 2002). It is important to note that βtubulin+ neuroblasts exhibit nuclear expression of both Olig2 (N) and Mash1 (O). During these progressive stages of neuronal–OL lineage elaboration, inhibitory HLH proteins display consistent expression profiles in cytosolic (Id2; D, J, P) and nuclear (Id4; E, K, Q) compartments, respectively. In contrast, the expression of HMG protein Sox 10 was noted to be absent in all of these developmental stages (F, L, R).
The regional and temporal forebrain expression profiles of neural bHLH, inhibitory HLH, HD, and HMG transcription factors within the early postnatal cerebral cortex suggest distinctive roles for these factors in OL lineage elaboration and OL maturation
To further examine the in vivo expression profiles of specific neural bHLH, inhibitory HLH, HD, and HMG factors during early postnatal cortical development, we performed immunofluorescence microscopic analysis of coronal sections obtained from early postnatal stage [postnatal day 0 (P0) and P6] mouse brains. At P0, Olig1 and Olig2, Id2 and Id4, and Sox10 are expressed within both the cortical SVZ and cerebral cortex (Fig. 3A). Interestingly, only Id2 and Id4 display predominantly cytosolic expression profiles (Fig. 3A). In contrast, Mash1 and Nkx2.2 are only observed within cerebral cortical regions (Fig. 3A). At P6, Mash1 and Nkx2.2 are also detected in the subcortical white-matter tracts, although Nkx2.2 expression is predominantly cytosolic (Fig. 3B) (supplemental Fig. 1, available at www.jneurosci.org as supplemental material). These additional in vivo expression profiles suggest that Olig1and Olig2 together with Id2 and Id4 are involved in OL lineage specification, whereas Nkx2.2, Sox10, and Mash1 have distinct roles in cortical OL maturation. We also performed colocalization studies during more-advanced stages of cortical development by using the OL cell somal marker APC in conjunction with suitable antibody isotypes for different HLH factors (Fig. 4). Our studies demonstrate the complete overlap of APC with Id2 (Fig. 4C′), Id4 (F′), and Mash1 (I′).
Figure 3.
Regional and temporal expression profiles of Olig1, Olig2, Mash1, Nkx2.2, and Sox10 during early postnatal cerebral cortical development suggest distinct roles for these transcription factors in OL lineage commitment and progressive OL maturation. A, B, Comparative immunofluorescence microscopy of Olig1, Olig2, Mash1, Nkx2.2, and Sox10 protein expression in coronal sections of early postnatal [P0 (A) and P6 (B)] mouse cerebral cortex (CTX) using Olig1, Olig2, Mash1, Nkx2.2, and Sox 10 antibodies reveals that although Olig1, Olig2, and Sox 10 are expressed in both the SVZ and CTX (A), Mash1 and Nkx2.2 expression is only detected in the developing CTX but not in the SVZ (A). However, during later stages of postnatal cortical development, both Mash1 and Nkx2.2 expression are detected within the subcortical white matter (SCWM) in parallel with the initiation of OL lineage maturation (B).
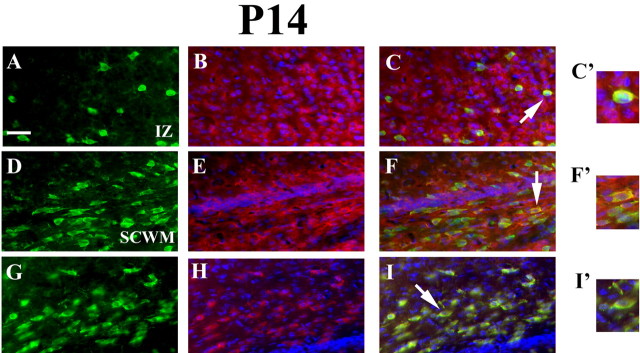
Figure 4.
Cellular expression profiles of Id2, Id4, and Mash1 in OLs localized to the intermediate zone and the subcortical white matter of P14 mouse brain. A–I, Comparative immunofluorescence microscopy of Id2 (A–C), Id4 (D–F), and Mash1 (G, H) expression in coronal sections of P14 mouse cerebral cortex using Id2, Id4, and Mash1 antibodies in combination with the OL marker anti-APC mouse monoclonal antibody clone CC-1 reveals complete overlap of Id2 (C), Id4 (F), and Mash1 (I) at OL cell bodies. C′, F′, and I′ are high-resolution images of OL cell bodies displaying cytoplasmic staining for Id2, Id4, and Mash1. APC (FITC; A, D, G), Id2 (B), Id4 (E), and Mash1 (H; tetramethylrhodamine isothiocyanate) with Hoechst 33342 (UV) are shown. IZ, Intermediate zone; SCWM, subcortical white matter.
Distinct combinatorial transcriptional complexes are identified in proliferating and differentiated Oli-neu cells, thus validating this cell line as a suitable model system for characterization of myelin gene expression
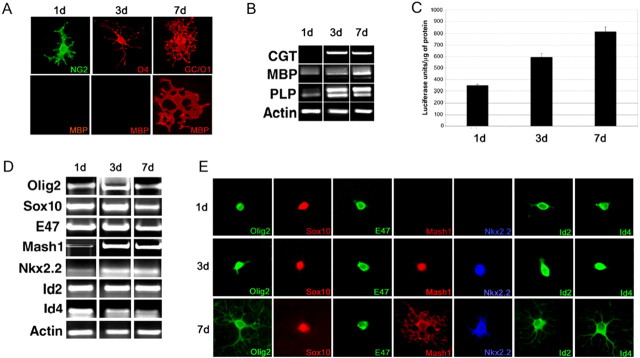
Myelin gene expression is characteristic of differentiating OL and is likely affected by the presence of specific transcription factors. To validate the use of the Oli-neu progenitor cell line (Jung et al., 1995) as a suitable model system for the functional studies of transcriptional codes, we have adopted a multidisciplinary experimental approach. First, we show that the antigenic profile of Oli-neu cells at sequential stages of maturation resembles that of cells of the OL lineage. Undifferentiated Oli-neu progenitors express NG2 but do not express MBP or other late differentiation markers (Fig. 5A,B). However, as these cells are allowed to differentiate, they begin to express myelin genes, including ceramide galactosyl-transferase (CGT), MBP, and proteolipid protein (PLP) (Fig. 5B). We then demonstrate a progressive increase in expression and activity of the luciferase reporter gene driven by the 1.3 kb MBP promoter (MBP-Luc) sequence (Miskimins et al., 2002). As expected, transfection of progenitor cells with the MBP-Luc results in very low basal activity that progressively increases as the cells differentiate (Fig. 5C). Finally, we observe that the progressive increase in myelin gene promoter activity is accompanied by corresponding changes in RNA levels (Fig. 5D) and protein expression patterns (Fig. 5E) of specific families of transcription factors, including bHLH, homeobox, and HMG-containing proteins. Together, these data suggest that specific transcriptional codes identify sequential stages of OL differentiation.
Figure 5.
Characterization of the Oli-neu cells as a model system for studying myelin gene expression. Antigenic characterization of Oli-neu cells maintained in differentiating conditions for 1, 3, or 7 d. Immunocytochemistry with antibodies against NG2, O4, GalC, and MBP reveals the progressive acquisition of late OL differentiation markers. Note that the MBP can be detected only after 7 d in differentiation medium (A). Reverse transcription (RT)-PCR of RNA samples isolated from Oli-neu cells at the same stages of differentiation is shown. The cDNA is amplified with primers for CGT, MBP, and PLP and actin is used as an internal control. Note the very low levels of MBP and of the DM20 isoform of PLP detected at day 1. The levels of MBP expression increase after 3 d and 7 d in differentiation medium (B). The progressive expression of differentiation markers is also reflected in a progressive increase in the basal activity of a luciferase reporter gene driven by the MBP promoter (C). Immature Oli-neu cells were transfected with the reporter construct, and the cells were later induced to differentiate for 1, 3, and 7 d. Note the progressive increase in luciferase activity, reflecting the increase in basal promoter activity as the cells differentiated (C). The progressive increase in myelin gene expression was associated with a stage-specific pattern of expression of distinct families of transcription factors as detected by RT-PCR (D) and by immunocytochemistry (E).
Distinct combinatorial transcriptional complexes differentially regulate MBP promoter activity during OL differentiation
To further define the roles of these neural bHLH, inhibitory HLH, HD, and HMG transcription factors in mediating OL lineage elaboration and maturation, we obtained early proliferative OL precursors from the SVZ (NG2+/O4–) and performed a detailed immunocytochemical analysis on dissociated cellular preparations after the implementation of subsequent OL maturational paradigms (Kondo and Raff, 2000c). At the OL precursor (NG2+/O4–) stage, cells express Olig2, Id2, Id4, and Sox10 but have undetectable levels of Olig1, Nkx2.2, and Mash1 (Fig. 6A–J). To address the role of these transcription factors for myelin gene expression, we expressed them both individually and in stage-specific combinations in immature Oli-neu cells and assayed the activity of the luciferase reporter gene. Cultures transfected with the same amount of pcDNA3 were used as control for the endogenous levels of MBP promoter activity, and the fidelity of gene introduction was verified by Western blot analysis using antibodies for the appropriate primary factor or epitope-tagged gene construct (data not shown). The effect of the different transcription factors was then expressed as fold changes over the activity detected in pcDNA3-transfected immature Olineu cells (Fig. 6K). When single factors present at the NG2+/O4– precursor stage were cotransfected in immature Oli-neu cells with the MBP-promoter, only Sox 10 was able to activate the MBP promoter sevenfold over the baseline (x = 7.2 ± 0.5; n = 6), whereas the class B bHLH protein Olig2 (x =–0.4 ± 0.02; n = 6) or the class A bHLH protein E47 (x = –0.23 ± 0.008; n = 6) alone or in combination were unable to activate the promoter (Fig. 6K). In contrast, when Olig2 and E47 were cotransfected with Sox10, there was a synergistic and statistically significant increase in the activity of the MBP promoter (x = 19.9 ± 3.2; n = 6). Because at the NG2 precursor stage the presence of the inhibitory HLH factors Id2 and Id4 was quite prominent, we asked whether their inhibitory effects prevailed over the activation of the promoter observed. Indeed, the expression of Id2 and Id4 with Olig2/Sox10/E47 prevented the synergistic activation of the promoter (x = 6.7 ± 0.24-fold; n = 3) and reduced the MBP promoter activity to that observed in the presence of Sox10 alone (Fig. 6K). These data suggest that at the NG2+/O4– stage, Id2 and Id4 exert an inhibitory control over the function of Olig2 and E47.
Figure 6.
The combinatorial transcriptional code expressed at the OL precursor stage of OL lineage elaboration is characterized by the presence of Olig2 and Sox10 and by the inhibitory activity of Id2 and Id4. Immunofluorescence microscopy of transcription factor expression in OL precursors derived from E17 cortical SVZ progenitors (A–J) reveals that NG2+/O4– OL precursors (A, F) express Olig2 (C), Id2 (G), Id4 (H), and Sox10 (I) but not Olig1 (B), Nkx2.2 (D), Mash1 (E), or MBP (J). When this combination of transcription factors is transiently transfected in the immature Oli-neu cells together with the MBP promoter driving a luciferase reporter gene, the result is a net activation of the promoter that is equivalent to that observed in cells transfected with Sox10 alone (K). In the absence of the inhibitory proteins Id2 and Id4, Olig2, and Sox10 are able to activate the promoter, thus suggesting that the NG2 developmental stage is characterized by the predominant inhibitory effect of the Ids. Each bar graph represents the mean of three to six experiments (*p < 0.0001).
At the early OL progenitor (NG2+/O4+) stage, cells continue to express Olig2, Id2, Id4, and Sox10 but now also display nuclear expression of Olig1 and Mash1 (Fig. 7A–J). Therefore, single factors expressed at this stage, such as Mash1 or Olig1, were expressed in immature Oli-neu cells alone or in combination (Fig. 7K). Mash1 overexpression increased the MBP promoter activity sixfold over baseline (x = 5.8 ± 0.3; n = 6), whereas Olig1 did not activate the promoter (x = 1.0 ± 0.1-fold; n = 6) (Fig. 7K). The combination of Olig1/Olig2/Mash1 reversed the inductive effect of Mash1 alone (x = –0.9 ± 0.04; n = 6) (Fig. 7K). However, when these same factors were expressed together with the relevant heterodimeric binding partner E47, a synergistic activation of the promoter was observed (x = 10.6 ± 2.9-fold over baseline; n = 6). Cotransfection of Sox10 enhanced the synergism to a 19 ± 1.8-fold activation over baseline in the absence of E47 (n = 6) and 30.5 ± 4.3-fold in the presence of E47 (n = 9). Interestingly, at this later OL developmental stage, the inhibitory proteins Id2 and Id4, when expressed together with this activating combination, only modestly inhibited the promoter activity (Fig. 7K). Together, these data suggest that at the early OL progenitor stage, the additional expression of Olig1 and Mash1 together with Olig2 and Sox10 allows near optimal myelin gene expression even in the presence of the inhibitory HLH factors Id2 and Id4.
Figure 7.

The combinatorial transcriptional code expressed at the early OL progenitor stage of OL lineage elaboration is characterized by the presence of Olig1, Olig2, and Mash1 that together with Sox10 and E47 activate the MBP promoter even in the presence of Id2 and Id4. Immunofluorescence microscopy of transcription factor expression at the early O4+ OL progenitor stage (A, J) reveals the presence of Olig1 (B) and Olig2 (C) together with Mash1 (E), Id2 (G), Id4 (H), and Sox10 (I) in the absence of MBP staining(J). These cells continue to express NG2 (F). When the same combination of transcription factors is transiently transfected in the immature Oli-neu cells together with the MBP promoter driving a luciferase reporter gene (K), the result is a statistically significant (*p < 0.0001) activation of the promoter. In the absence of Id2 and Id4, the activity was even higher, thus suggesting that the presence of Olig1 and Mash1 renders the promoter more resilient to the inhibitory activity of the Id proteins. Each bar graph represents the average of six to nine independent determinations (K).
At later stages of development, OL progenitors (NG2–/O4+) acquire Nkx2.2 nuclear expression while exhibiting decreasing levels of expression of nuclear-localized Id2 and Id4 (Fig. 8A–J). Therefore, we first transfected Nkx2.2. alone in immature Olineu cultures and assessed the consequences for MBP promoter activity (Fig. 8K). The difference in luciferase activity between Nkx2.2 overexpressors and pcDNA3 controls was not statistically significant (x =–0.64 ± 0.07; n = 6). In contrast, a statistically significant decrease in promoter activity was observed in cells coexpressing Nkx2.2 with Sox10, because the activity decreases from x = 7.2 ± 0.5 in the absence of Nkx2.2. to x =–0.9 ± 0.01 in the presence of Nkx2.2 (Fig. 8K). The inhibitory effect of Nkx2.2 was even more evident when it was cotransfected with the combinatorial codes Olig1/Olig2/Mash1/E47 or Olig1/Olig2/Mash1/Sox10 (Fig. 8K). Intriguingly, however, no inhibition of promoter activity was observed when Nkx2.2 was coexpressed with the combinatorial transcription code detected at the late NG2–/O4+ stage. When the combination Olig1/Olig2/Mash1/Sox10/E47 was transfected in the immature Oli-neu cells, the activity of the MBP promoter increased 30-fold compared with controls, regardless of the presence of Nkx2.2 (Fig. 8K). Together, these data suggest that although Nkx2.2 has the ability to repress the MBP promoter, its functional role is defined by the combinatorial transcriptional code present at specific stages of OL development.
Figure 8.
The HD transcription factor Nkx2.2 inhibits the activity of several combinations of transcription factors but only partially affects the activation induced by Olig1, Olig2, and Mash1 together with Sox10 and E47. At the later stages of differentiation, OL progenitors (NG2–/O4+) (A, F) begin to express Nkx2.2 (D), whereas they continue to express Olig1 (B), Olig2 (C), Mash1 (E), and Sox10 (I) in the nucleus. Please note the absence of MBP staining (J). Transfection of Nkx2.2 in immature Oli-neu cells together with Sox10 in the absence or presence of Olig1/Olig2/Mash1 or E47 with Olig1/Olig2/Mash1 results in a statistically significant (*p < 0.0001) repression of the MBP promoter activity, whereas together with Olig1, Olig2, Mash1, Sox10, and E47 it increases the MBP promoter activity (K).
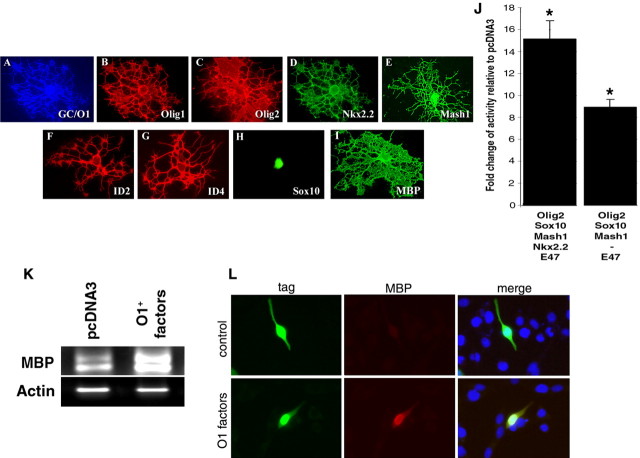
As OL cells mature and acquire GalC/O1 expression, they continue to display predominantly nuclear expression of Sox10, and they exhibit both nuclear and cytoplasmic localization of Olig2, Mash1, and Nkx2.2 and predominantly cytoplasmic expression of Olig1, Id2, and Id4 (Fig. 9A–I). If the combination of transcription factors observed at this developmental stage (i.e., Olig2/Mash1/Sox10/E47) is transfected in immature Oli-neu cells, the activity of the MBP promoter is significantly increased in the absence (x = 9.0 ± 0.7; n = 6) or presence (x = 15.2 ± 1.7; n = 6) of Nkx2.2. To further assess the functional significance of these data, we also expressed the Olig2/Mash1/Sox10/E47 combination in immature Oli-neu cells and demonstrate that this combinatorial transcription factor code is sufficient to activate the endogenous promoter (Fig. 9J) and results in MBP expression (Fig. 9K,L) in cells that would normally not express this protein at the immature stage of development.
Figure 9.
Ectopic expression of factors detected at the GalC/O1+ stage is sufficient to induce MBP expression. A–I, At the GalC/O1+ postmitotic OL stage (A), the most dramatic changes observed are the cytoplasmic localization of Olig1 (B), Nkx2.2 (D), Id2 (F), and Id4 (G). Only Olig2 (C), Mash1 (E), and Sox10 (H) remain in the nucleus. The transfection of these combinations in immature Oli-neu cells result in a statistically significant increase in promoter activity (J) (*p < 0.001). The effect of the transfected combination of transcription factors on the endogenous promoter is indicated by the increased levels of MBP mRNA detected by semiquantitative reverse transcription-PCR (K). The increased MBP transcripts result also in increased expression of the MBP protein (L) as documented by immunocytochemistry. Note the presence of MBP immunoreactivity (red immunofluorescence) in immature Oli-neu cells transfected with the plasmids encoding for the factors expressed at the GalC/O1+ stage (O1+ factors) and not in cultures transfected with pcDNA3 encoding enhanced green fluorescent protein (EGFP; control). Transfected cells (green immunofluorescence) were identified by antibodies against GFP (for pcDNA3-EGFP-transfected cells) or against FLAG tag (for Olig2-FLAG-transfected cells).
Together, these data indicate that specific permutations of transcription factors regulate myelin gene expression at sequential developmental stages. Olig1 nuclear expression is essential to permit maximal activation of the MBP promoter in late O4 progenitors. Nkx2.2 seems to switch developmental roles from that of a transcriptional repressor, in the early OL progenitor stage to that of a transcriptional activator at later OL developmental stages. This is indicative of the degree of developmental plasticity inherent in the OL-specific program of transcriptional regulation of myelin gene expression.
The OL developmental transcription factor code is selective for the MBP promoter and also induces gene activation in nonmyelinating cells
The Oli-neu cell line has been characterized previously as an OL precursor line (Jung et al. 1995), and therefore it could be argued that the effect of introduction of specific combinatorial transcriptional codes into this cell line may interact with endogenous components expressed in OL lineage species. To further examine this possibility, we repeated the transient cotransfection assays in a nonmyelinating cell line, the NIH3T3 line (Fig. 10). The combination of transcription factors present at the OL precursor and at the early and late OL progenitor stage were separately cotransfected with the MBP promoter driving a luciferase reporter in the fibroblast cell line. Remarkably, a similar activation of the transiently transfected exogenous promoter is detected in the NIH3T3 cells, thus suggesting that discrete combinations of neural and more ubiquitous bHLH, HD, and HMG factors are sufficient to activate the MBP promoter independent of the cellular context. Finally, to address the specificity of the different transcription factor combinations on other OL lineage-dependent (CGT, PLP) and OL lineage-independent (NeuroD) promoters, we transiently transfected the same OL-selective combinatorial transcription factor codes and assayed the effects of these factor combinations on the activation of the neuronal promoter NeuroD (Fig. 10). The activity of the reporter gene driven by the NeuroD promoter is not affected by the transfection of these distinct OL-selective combinatorial codes. Moreover, only activation of the CGT but not the PLP myelin gene promoter was significantly enhanced (data not shown). Together, these data suggest that myelin gene expression is the result of the complex interplay between HD proteins, bHLH molecules, and HMG domain proteins that may assemble in distinct permutations to differentially modulate the expression of myelin-associated genes.
Figure 10.
Comparative effects of the precise combination of transcription factors specific for the MBP promoter when transfected into myelinating and non-myelinating cells and assessed for MBP and NeuroD promoter activation. The functional role of the specific combinations of positive transcription factors detected at each developmental stage was studied in a myelinating cell line (A, B) or in a fibroblastic cell line (C, D). Transient transfection of the same positive transcription factors detected during OL differentiation in NIH3T3(3T3) cells induced a pattern of MBP promoter activation, similar to that observed in the Oli-neu cells. This effect was specific for myelin genes because it was not observed on a neuronal promoter (NeuroD).
Discussion
In vivo and in vitro expression profiles of neural bHLH (Olig1, Olig2, Mash1), inhibitory HLH (Id2, Id4), HD (Nkx2.2), and HMG (Sox10) proteins suggest that these transcription factors have specific roles at multiple stages of ventral forebrain stem cell lineage restriction, OL lineage specification, and progressive stages of OL maturation including myelination (Figs. 1, 2, 3, 4, 5, 6, 7, 8, 9). In this regard, we have demonstrated that Olig1, Olig2, and Mash1 are involved in the regulation of multiple stages of cortical OL lineage specification from ventral forebrain stem and more lineage-restricted progenitor species (S. Gokhan, S. Y. Yung, and M. F. Mehler, manuscript in preparation). However, the molecular mechanisms involved in coupling OL lineage specification and progressive stages of OL terminal differentiation to the characteristic spatiotemporal profiles of myelination have not been defined previously. After OL lineage commitment and OL progenitor cell proliferation, OL maturation requires precise coordination of the processes of cell-cycle exit and progressive stages of cellular differentiation culminating in myelination (Durand and Raff, 2000). For example, the timing of cell-cycle exit is crucial for appropriate process outgrowth during OL lineage maturation (Liu et al., 2003). Chromatin modifiers regulate this process by changing the levels and availability of protein cofactors that are involved in gene activation and repression (Marin-Husstege et al., 2002; Liu et al., 2003; Shen et al., 2005). In addition, neurogenesis and gliogenesis are sequentially elaborated, and neuronal–OL developmental coupling helps to ensure that axonal myelination is also precisely regulated during lineage maturation (Mehler, 2002; Yung et al., 2002).
In this study, we have addressed the question of how these stage-specific combinatorial transcriptional codes affect MBP gene expression. The MBP promoter has been chosen because MBP is an abundant component of myelin and because of the presence of several consensus sequences for Nkx and Sox transcriptional regulators. The MBP promoter also contains multiple binding sites for ubiquitous class A bHLH (E47, E12, HEB) E2A proteins, which represent heterodimeric binding partners for class B neural bHLH factors (Olig1, Olig2, Mash1) (Norton, 2000). Class B bHLH factors are also known to be recruited to Nkx-containing transcriptional complexes by class A bHLH proteins, thereby increasing the combinatorial profiles of these transcriptional regulators (Poulin et al., 2000). However, the bioavailability of class A and B bHLH proteins is strictly regulated by inhibitory HLH proteins (e.g., Id2, Id4) that exert their developmental actions by direct binding and sequestration of specific members of these two classes of bHLH transcription factors (Kondo and Raff, 2000a,b; Norton, 2000; Wang et al., 2001; Samanta and Kessler, 2004; Gokhan, Yung, and Mehler, manuscript in preparation). In this regard, we demonstrate that at the OL precursor stage, the transcriptional inhibitory roles of Id2 and Id4 predominate over the basal “activating” functions of Olig2/E47/Sox10 factor combinations, presumably by direct sequestration of class A and B bHLH components. In support of this interpretation, we show that the presence of Olig2/E47/Sox10 factor combinations is able to activate the MBP promoter in the absence of the Id proteins. Conversely, in the presence of Id2 and Id4, the effect of the same transcription factor combination on the MBP promoter is only equivalent to that of Sox10 alone. These observations suggest that myelin gene expression in OL precursors is maintained at very low levels by the inhibitory actions of Id2 and Id4. At the early OL progenitor stage, cells acquire the expression of two additional bHLH proteins, Olig1 and Mash1. Therefore, at this developmental stage, both Olig1 and Olig2 but not Mash1 compete with Id2 and Id4 for the E2A pool (Gokhan, Yung, and Mehler, manuscript in preparation). The presence of Olig1, Olig2, and Mash1 results in a combinatorial transcription factor code that is able to synergistically activate the MBP promoter in concert with Sox10, even in the presence of Id2 and Id4. It is tempting to speculate that at the early OL progenitor stage, the Id inhibitory effect is counteracted by competition with multiple neural bHLH proteins for the pool of E2A factors, whereas at later stages of OL progenitor maturation, the loss of inhibition is the consequence of developmentally mediated reductions in the levels and cellular expression of both Id2 and Id4 (Kondo and Raff 2000a,b; Wang, 2001).
Remarkably, at all OL developmental stages, the combination of class A (e.g., E47) and class B (e.g., Olig1, Olig2, Mash1) bHLH proteins together with the HMG box containing protein Sox10 results in a synergistic rather than merely an additive activation of the MBP promoter. These findings can be explained by the several possible ways of regulation of gene expression by Sox family members (for review, see Wegner, 1999). It is known that the Sox gene family promotes OL specification (e.g., Sox9) and OL terminal differentiation including myelination (e.g., Sox8, Sox10) with Sox10 playing a dominant role in the latter developmental events (Stolt et al., 2004). In addition to the direct binding to the minor groove of DNA, Sox proteins have also been shown to promote the assembly of multimeric protein complexes, in part by their ability to alter the conformation of DNA, and thereby enhance local chromatin remodeling (van de Wetering et al., 1993; Wolffe, 1994; Werner and Burley, 1997). We therefore propose that Sox10 may act both as a DNA-binding protein and as a molecular scaffold to recruit bHLH heterodimers to specific promoter sites resulting in activation of MBP gene expression.
Our findings have uncovered an additional developmental mechanism for timely inhibition of MPB promoter activity at the late OL progenitor stage through the actions of the HD protein Nkx2.2. This result is consistent with previous reports suggesting cross-repressive interactions between Nkx2.2. and Olig2 (Sun et al., 2003) and the relief of Nkx2.2-mediated repression by the Sp1 transcription factor (Wei et al., 2005). In the spinal cord, the ablation of Nkx2.2 expression leads to an expansion of Olig1/2 expression into the Nkx2.2 domain, resulting in an increased elaboration of OL progenitors but a dramatic delay in later OL myelination (Qi et al., 2001). In this regard, it is interesting that the presence of Nkx2.2 can inhibit MBP promoter activity in concert with different combinatorial profiles of neural bHLH, E2A, and HMG factors but not with the profile of these transcriptional regulators present at the stage of OL terminal differentiation, when the actions of both Id proteins and Nkx2.2 are nullified. Interestingly, the highest degree of MBP promoter activation was observed at the late OL progenitor stage rather than at the postmitotic OL stage. These results are consistent with the highest activation of the promoter for MBP at the onset of myelination, when the cell requires the elaboration of all of the necessary molecular components to be assembled into the evolving myelin sheath. It is conceivable that after initial completion of the process of regional myelination, only a lower level of activation of myelin gene expression will be necessary during the adult maintenance phase when gradual replacement of specific myelin components may occur. This hypothesis is supported by the data for postmitotic OLs, a stage of OL terminal differentiation during which time the majority of OL-selective transcriptional activators undergo nuclear to cytoplasmic translocation.
It is important to emphasize that we have identified specific transcriptional codes that result in the selective activation of the MBP promoter (Fig. 10A,C). Conversely, no significant activation was observed for the neuronal bHLH differentiation effector factor NeuroD (Fig. 10B,D). In addition, the enhancement of MBP promoter activation by the OL-selective combinatorial codes of transcription factors identified here is not limited to OL lineage species but also occurs in non-myelinating cells, such as the fibroblast NIH3T3 cell line (Fig. 10B). Together, these data indicate that the expression of the specific combinatorial transcriptional codes detected at each stage of OL lineage maturation is sufficient to confer different degrees of activation of the MBP promoter. Because these various transcription factors exhibit distinct temporal expression profiles in evolving OL lineage species, the ability to exert different combinatorial effects on myelin gene transcription represents an extremely powerful and versatile mechanism to orchestrate the precise spatiotemporal profiles of myelination characteristic of the mammalian central nervous system. Furthermore, several of these transcription factors also promote the developmental coupling and asynchronous maturation of regional neuronal and OL subtypes. Therefore, these transcriptional regulators may play additional novel roles in coordinately regulating the timely myelination of developing axons (Mehler, 2002; Yung et al., 2002). In conclusion, this study demonstrates an elegant mechanism to couple earlier developmental events in OL lineage elaboration with later stages of OL maturation, particularly myelination, by the use of common sets of transcription factors that exert distinct cellular effects at progressive stages of OL lineage maturation.
Footnotes
This work was supported by grants from the Rosanne H. Silbermann Foundation (S.G.), the National Multiple Sclerosis Society (RG 3421-A-4 to P.C.-B., FA 1512-A-1 to M.M.-H.), the National Institutes of Health (NIH) (NS42925 to P.C.-B., NS38902 and MH66290 to M.F.M.), and the F. M. Kirby Foundation (M.F.M.). D.F. was supported by an NIH initiative for underrepresented minorities in science (GM55145). We are grateful to Dr. R. Miskimins for providing valuable reagents for the MBP promoter analysis.
Correspondence should be addressed to either of the following: Dr. Mark F. Mehler, Kennedy Center 401, Albert Einstein College of Medicine, 1410 Pelham Parkway South, Bronx, NY 10461, E-mail: mehler@aecom.yu.edu; or Dr. Patrizia Casaccia-Bonnefil, Department of Neuroscience R304, University of Medicine and Dentistry of New Jersey, Robert Wood Johnson Medical School, 675 Hoes Lane, Piscataway, NJ 08854, E-mail: casaccpa@umdnj.edu.
DOI:10.1523/JNEUROSCI.1850-05.2005
Copyright © 2005 Society for Neuroscience 0270-6474/05/258311-11$15.00/0
S.G. and M.M.-H. contributed equally to this work.
References
- Alberta JA, Song-Kyu P, Mora J, Yuk D-i, Pawlitzky I, Iannarelli P, Vartanian T, Stiles CD, Rowitch DH (2001) Sonic hedgehog is required during an early phase of oligodendrocyte development in mammalian brain. Mol Cell Neurosci 18: 434–441. [DOI] [PubMed] [Google Scholar]
- Durand B, Raff M (2000) A cell-intrinsic timer that operates during oligodendrocyte development. BioEssays 22: 64–71. [DOI] [PubMed] [Google Scholar]
- Ericson J, Rashbass P, Schedl A, Brenner-Morton S, Kawakami A, van Heyningen V, Jessell TM, Briscoe J (1997) Pax6 controls progenitor cell identity and neuronal fate in response to graded Shh signaling. Cell 90: 169–180. [DOI] [PubMed] [Google Scholar]
- Gunhaga L, Jessell TM, Edlund T (2000) Sonic hedgehog signaling at gastrula stages specifies ventral telencephalic cell in the chick embryo. Development 127: 3283–3293. [DOI] [PubMed] [Google Scholar]
- He W, Ingraham C, Rising L, Goderie S, Temple S (2001) Multipotent stem cells from the mouse basal forebrain contribute GABAergic neurons and oligodendrocytes to the cerebral cortex during embryogenesis. J Neurosci 21: 8854–8862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T, Tokunaga A, Yoshida T, Hashimoto M, Mikoshiba K, Weinmaster G, Nakafuku M, Okano H (2001) The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol Cell Biol 21: 3888–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova A, Nakahira E, Kagawa T, Oba A, Tamaki W, Takebayashi H, Spassky N, Levine J, Zalc B, Ikenaka K (2003) Evidence for a second wave of oligodendrogenesis in the postnatal cerebral cortex of the mouse. J Neurosci Res 73: 581–592. [DOI] [PubMed] [Google Scholar]
- Jung M, Kramer E, Grzenkowski M, Tang K, Blakemore W, Aguzzi A, Khazaie K, Chlichlia K, von Blankenfeld G, Kettenmann H, Trotter J (1995) Lines of murine oligodendroglial precursor cells immortalized by an activated neu tyrosine kinase show distinct degrees of interaction with axons in vitro and in vivo. Eur J Neurosci 7: 1245–1265. [DOI] [PubMed] [Google Scholar]
- Kaneko Y, Sakakibara S, Imai T, Suzuki A, Nakamura Y, Sawamoto K, Ogawa Y, Toyama Y, Miyata T, Okano H (2000) Musashi1: an evolutionally conserved marker for CNS progenitor cells including neural stem cells. Dev Neurosci 22: 139–153. [DOI] [PubMed] [Google Scholar]
- Kondo T, Raff M (2000a) The Id4 HLH protein and the timing of oligodendrocyte differentiation. EMBO J 19: 1998–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Raff M (2000b) Basic helix-loop-helix proteins and the timing of oligodendrocyte differentiation. Development 127: 2989–2998. [DOI] [PubMed] [Google Scholar]
- Kondo T, Raff M (2000c) Oligodendrocyte precursor cells reprogrammed to become multipotential CNS stem cells. Science 289: 1754–1757. [DOI] [PubMed] [Google Scholar]
- Liu A, Muggironi M, Marin-Husstege M, Casaccia-Bonnefil P (2003) Oligodendrocyte process outgrowth in vitro is modulated by epigenetic regulation of cytoskeletal severing proteins. Glia 44: 264–274. [DOI] [PubMed] [Google Scholar]
- Liu A, Stadelmann C, Moscarello M, Bruck W, Sobel A, Mastronardi FG, Casaccia-Bonnefil P (2005) Expression of stathmin, a developmentally controlled cytoskeleton-regulating molecule, in demyelinating disorders. J Neurosci 25: 737–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu QR, Sun T, Zhu Z, Ma N, Garcia M, Stiles CD, Rowitch DH (2002) Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell 109: 75–86. [DOI] [PubMed] [Google Scholar]
- Marin-Husstege M, Muggironi M, Liu A, Casaccia-Bonnefil P (2002) Histone deacetylase activity is necessary for oligodendrocyte lineage progression. J Neurosci 22: 10333–10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmur R, Kessler JA, Zhu G, Gokhan S, Mehler MF (1998a) Differentiation of oligodendroglial progenitors from cortical multipotent cells requires extrinsic signals including activation of gp130/LIFβ receptors. J Neurosci 18: 9800–9811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmur R, Mabie PC, Gokhan S, Song Q, Kessler JA, Mehler MF (1998b) Isolation and developmental characterization of cerebral cortical multipotent progenitors. Dev Biol 204: 577–591. [DOI] [PubMed] [Google Scholar]
- Mehler MF (2002) Mechanisms regulating lineage diversity during cerebral cortical neurogenesis and gliogenesis. In: Cortical development, results and problems in cell differentiation, Vol 39 (Hohmann C, ed), pp 27–52. Heidelberg: Springer. [DOI] [PubMed] [Google Scholar]
- Mehler MF, Mabie PC, Zhu G, Gokhan S, Kessler JA (2000) Developmental changes in progenitor cell responsiveness to bone morphogenetic proteins differentially modulate progressive CNS lineage fate. Dev Neurosci 22: 74–85. [DOI] [PubMed] [Google Scholar]
- Miskimins R, Srinivasan R, Marin-Husstege M, Miskimins WK, Casaccia-Bonnefil P (2002) p27Kip1 enhances myelin basic promoter activity. J Neurosci Res 67: 100–105. [DOI] [PubMed] [Google Scholar]
- Mizuguchi R, Sugimori M, Takebayashi H, Kosako H, Nagao M, Yoshida S, Nabeshima Y-i, Shimamura K, Nakafuku M (2001) Combinatorial roles of Olig2 and Neurogenin2 in the coordinated induction of pan-neuronal and subtype-specific properties of motor neurons. Neuron 31: 757–771. [DOI] [PubMed] [Google Scholar]
- Norton JD (2000) Id helix-loop-helix proteins in cell growth, differentiation and tumorigenesis. J Cell Sci 113: 3897–3905. [DOI] [PubMed] [Google Scholar]
- Novitch BG, Chen AI, Jessell TM (2001) Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron 31: 773–789. [DOI] [PubMed] [Google Scholar]
- Poulin G, Lebel M, Chamberland M, Paradis FW, Drouin J (2000) Specific protein-protein interaction between basic helix-loop-helix transcription factors and homeoproteins of the Pitx family. Mol Cell Biol 20: 4826–4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross SE, Greenberg ME, Stiles CD (2003) Basic helix-loop-helix factors in cortical development. Neuron 39: 13–25. [DOI] [PubMed] [Google Scholar]
- Qi Y, Cai J, Wu Y, Wu R, Lee J, Fu H, Rao M, Sussel L, Rubenstein J, Qiu M (2001) Control of oligodendrocyte differentiation by the Nkx2.2 homeodomain transcription factor. Development 128: 2723–2733. [DOI] [PubMed] [Google Scholar]
- Samanta J, Kessler JA (2004) Interactions between ID and OLIG proteins mediate the inhibitory effects of BMP4 on oligodendroglial differentiation. Development 131: 4131–4142. [DOI] [PubMed] [Google Scholar]
- Shen S, Li J, Casaccia-Bonnefil P (2005) Histone modifications affect timing of oligodendrocyte progenitor differentiation in the developing rat brain. J Cell Biol 169: 577–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt CC, Rehberg S, Ader M, Lommes P, Riethmacher D, Schachner M, Bartsch U, Wegner M (2002) Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev 16: 165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt CC, Lommes P, Friedrich RP, Wegner M (2004) Transcription factors Sox8 and Sox10 perform non-equivalent roles during oligodendrocyte development despite functional redundancy. Development 131: 2349–2358. [DOI] [PubMed] [Google Scholar]
- Sun T, Echelard Y, Lu R, Yuk D-i, Kaing S, Stiles CD, Rowitch DH (2001) Olig bHLH proteins interact with homeodomain proteins to regulate cell fate acquisition in progenitors of the ventral neural tube. Curr Biol 11: 1413–1420. [DOI] [PubMed] [Google Scholar]
- Sun T, Dong H, Wu L, Kane M, Rowitch DH, Stiles CD (2003) Cross-repressive interaction of the Olig2 and Nkx2.2 transcription factors in developing neural tube associated with formation of a specific physical complex. J Neurosci 23: 9547–9556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanagaki K, Nogaki F, Takahashi J, Tashiro K, Kurooka H, Honjo T (2001) Notch1 and Notch3 instructively restrict bFGF-responsive multipotent neural progenitor cells to an astroglial fate. Development 128: 689–702. [DOI] [PubMed] [Google Scholar]
- Tekki-Kessaris N, Pringle N, Richardson WD (2001) Ventral neurogenesis and the neuron-glial switch. Neuron 31: 677–680. [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Oosterwegel M, van Norren K, Clevers H (1993) Sox-4, and Sry-like HMG box protein, is a transcriptional activator in lymphocytes. EMBO J 12: 3847–3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Sdrulla A, Johnson JE, Yokota Y, Barres BA (2001) A role for the helix-loop-helix protein Id2 in the control of oligodendrocyte development. Neuron 29: 603–614. [DOI] [PubMed] [Google Scholar]
- Wegner M (1999) From head to toes: the multiple facets of Sox proteins. Nucleic Acid Res 27: 1409–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Miskimins WK, Miskimins R (2005) Stage-specific expression of myelin basic protein in oligodendrocytes involves Nkx2.2-mediated repression that is relieved by the Sp1 transcription factor. J Biol Chem 280: 16284–16294. [DOI] [PubMed] [Google Scholar]
- Werner MH, Burley SK (1997) Architectural transcription factors: proteins that remodel DNA. Cell 88: 733–736. [DOI] [PubMed] [Google Scholar]
- Wolffe AP (1994) Architectural transcription factors. Science 264: 1100–1101. [DOI] [PubMed] [Google Scholar]
- Woodruff RH, Tekki-Kessaris N, Stiles CD, Rowitch DH, Richardson WD (2001) Oligodendrocyte development in the spinal cord and telencephalon: common themes and new perspectives. Int J Dev Neurosci 19: 370–385. [DOI] [PubMed] [Google Scholar]
- Yung S-Y, Gokhan S, Jurcsak J, Molero AE, Abrajano JJ, Mehler MF (2002) Differential modulation of BMP signaling promotes the elaboration of cerebral cortical GABAergic neurons or oligodendrocytes from a common sonic hedgehog-responsive ventral progenitor species. Proc Natl Acad Sci USA 99: 16273–16278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Anderson DJ (2002) The bHLH transcription factors DLJG1 and OLIG2 couple neuronal and glial subtype specification. Cell 109: 61–73. [DOI] [PubMed] [Google Scholar]
- Zhu G, Mehler MF, Zhao J, Yung S, Kessler JA (1999) Sonic hedgehog and BMP2 exert opposing actions on proliferation and differentiation of embryonic neural progenitor cells. Dev Biol 215: 118–129. [DOI] [PubMed] [Google Scholar]