Abstract
The fetal basis of adult disease (FeBAD) hypothesis states that many adult diseases have a fetal origin. According to FeBAD, injury or environmental influences occurring at critical periods of organ development could result in “programmatic” changes via alterations in gene expression or gene imprinting that may result in functional deficits that become apparent later in life. Alzheimer's disease (AD) is a progressive neurodegenerative disorder that is characterized by excessive deposits of aggregated β-amyloid (Aβ) peptides, which are snippets of the β-amyloid precursor protein (APP). The predominately sporadic nature of AD suggests that the environment must play a role in neurodegeneration. To examine latent responses to an environmental agent, we exposed rodents to lead and monitored the lifetime expression of the APP gene. We observed that APP mRNA expression was transiently induced in neonates, but exhibited a delayed overexpression 20 months after exposure to Pb had ceased. This upregulation in APP mRNA expression was commensurate with a rise in activity of the transcription factor Sp1, one of the regulators of the APP gene. Furthermore, the increase in APP gene expression in old age was accompanied by an elevation in APP and its amyloidogenic Aβ product. In contrast, APP expression, Sp1 activity, as well as APP and Aβ protein levels were unresponsive to Pb exposure during old age. These data suggested that environmental influences occurring during brain development predetermined the expression and regulation of APP later in life, potentially altering the course of amyloidogenesis.
Keywords: Alzheimer, β-peptide, CNS, dementia, development, environment, latency, transcription
Introduction
Epidemiological studies have shown that fetal undernutrition, lower birth weights, and obesity in humans are associated with an increased risk of death from diabetes, congestive heart failure, and stroke (Barker et al., 1989; Valdez et al., 1994; Yarbrough et al., 1998; Ozanne, 2001). The fetal basis of adult disease (FeBAD) hypothesis states that many adult diseases have a developmental origin. Thus, the promotion of amyloidogenesis may begin very early in life. Although it can be speculated that environmental insults occurring during early growth could impact the ability of an organ to respond to stress or disease later in life, the molecular targets that could mediate such delayed or latent effects have not yet been characterized. It is known that tissues and organs go through “critical periods” of development, which coincides with stages of rapid cell growth in which differential gene expression plays a major role. It has also been suggested that disturbances during these critical periods could force the body to adapt and pool resources and alter programming of the expression of genes important for normal lifelong physiology (Barker et al., 1989; Barker, 2002).
The contribution of the environment to disease processes is increasingly becoming more appreciated. Among the many chemicals one is exposed to, a few chemicals are of widespread public concern. The heavy metal Pb is among such chemicals. The total consumption of Pb in the United States is ∼1.3 × 106 metric tons (Goyer, 1996). Humans can be exposed to Pb through paint, glazed earthenware, lead piping solder in food containers, moonshine whiskey, and automobile battery casing (Goyer, 1996). Although organic forms of Pb have been removed from gasoline, inorganic Pb still remains the number one environmental hazard facing human populations today. Furthermore, the aging populations today had been exposed in the past to much higher levels of environmental Pb.
Developmental exposure to low levels of Pb has long been known to produce impairments in intellectual functioning and disturbances in a number of molecular, biochemical, and morphological events (Needleman et al., 1990; Silbergeld, 1992; Zawia, 2003; Basha et al., 2004). The cognitive impairments by Pb are believed to be permanent; however, it is not known whether early exposure to Pb can manifest latent effects that could contribute to neurodegenerative events in old age. Expression of certain genes and the transcriptional apparatus could be involved in this latent induction. To test this hypothesis, we exposed rodents during the neonatal period to Pb and monitored the lifetime expression of the APP gene and the activity of specificity protein 1 (Sp1), a transcription factor involved in the regulation of the APP gene. APP is a transmembrane protein and its proteolytic processing generates a 39-42 residue peptides referred to as β-amyloid (Aβ). Aβ peptides are the primary constituents of amyloid deposits found in the aging brain and have been implicated in the pathogenesis of Alzheimer's disease (AD) and Down syndrome by genetic, pathologic, and biochemical evidence (Selkoe, 2001; Nishimura et al., 2002; Lahiri et al., 2003). Therefore, we also examined APP and Aβ levels in old rat brains developmentally exposed to Pb.
Materials and Methods
Animal exposure. Timed-pregnant Long-Evans hooded rats were obtained from Charles River Laboratory (Bar Harbor, ME) in two batches over a 2 year period. Twenty-four hours after birth on postnatal day 1 (P1), all the pups were pooled, and new litters consisting of eight or nine males were randomly selected and placed with each dam. The dams were divided into two groups, namely, Con and Pb-E. The first group (Con) received deionized water and was considered the control group. The second group (Pb-E) was exposed to Pb from P1 through P20. The third group, Pb-L was exposed to Pb only during senescence (i.e., from 18 to 20 months of age). In each exposure scenario, 200 ppm Pb-acetate (Sigma, St. Louis, MO) was added to the deionized drinking water of the dams or the aging animals. We found that tissue Pb levels were comparatively low at this dose, and no gross morphological or nutritional disturbances were found in the pups or the aging animals (Zawia et al., 1998; Basha et al., 2003). Food and water were freely available for all the groups throughout the study. The animals were housed at constant temperature (21 ± 2°C) and relative humidity (50 ± 10%) with a 12 h light/dark cycle (7:00 A.M. to 7:00 P.M.). On selected time points (P5, P50, P350, and P600) the animals (pups/adults/senescent) were decapitated after exposure to CO2, and hypothermia and various brain regions were isolated and stored at -80°C for use in the molecular and analytical studies reported here. The animal exposure protocol and the maintenance of the animal facilities were regularly monitored by the Institutional Animal Care and Use Committee.
Nuclear protein extraction. Nuclear proteins were extracted from both cortical tissue and cultured cells according to the method given by Dignam et al. (1983), with slight modifications. Cells/tissue samples were homogenized with 1 ml of PBS, pH 7.4, and centrifuged at 2500 × g for 10 min. The pellets obtained were suspended in 5 vol of buffer A (in mm:10 HEPES, pH 7.9, 1.5 MgCl2, 0.5 DTT, 0.5 EDTA, and 0.2 PMSF) and centrifuged at 6000 × g for 2 min at 4°C. The pellets were resuspended in 3 vol of buffer A and centrifuged at 6000 × g for 2 min at 4°C. The resulting pellets were then resuspended in 5 vol of buffer C (20 mm HEPES, pH 7.9, 1.5 mm MgCl2, 0.5 mm DTT, 0.5 mm EDTA, 420 mm NaCl, 20% glycerol, 0.2 mm PMSF, 2 μg/ml aprotinin, and 0.5 μg/ml leupeptin) and homogenized with a Polytron homogenizer. The final suspensions were centrifuged at 12,000 × g for 10 min at 4°C. The supernatants were transferred to 1.5 ml tubes, snap frozen in an ethanol dry-ice bath, and then stored at -80°C (Zawia et al., 1998; Basha et al., 2003).
Gel mobility shift assays. Changes in the profiles of the Sp1 transcription factor were studied through electrophoretic mobility shift assay. Nuclear extracts were derived from the cortical tissues as described above. Consensus oligonucleotide sequence for Sp1 was obtained from commercial sources (Promega, Madison, WI) and radiolabeled by 5′ labeling procedures. The oligos (1.75 pmol) were incubated with 1 μl of [γ-32P]ATP (3000 Ci/mmol; PerkinElmer, Boston, MA) and 10 U of T4 polynucleotide kinase (Promega) in a final volume of 10 μl for 15 min at 37°C. The reaction was terminated, and the DNA probe was isolated through purification with a Sephadex G-25 column. A binding reaction was set up containing: 5000-20,000 cpm of the DNA probe (0.1-0.5 ng), 0.5-2 μg of poly dI-dC, 5-20 μg of the nuclear extract and binding buffer [2 mm HEPES, 2.5 mm MgCl2, 2.5 mm KCl, 0.5 mm DTT, 2.5% glycerol, 0.001% NP-40, 50 μg/ml BSA, 0.2% protease inhibitor mixture (Sigma), 0.2% ficoll, and 20 ng/ml poly dI-dC]. The binding reaction was performed at room temperature for 15 min. The reaction mix was separated by 6% polyacrylamide gel electrophoresis. The gel was dried and exposed to a phosphor screen for 2 h and scanned in the Typhoon Variable Mode Imager 9410. The resulting scanned gels were analyzed for shifted bands and quantitated using the Image Quant 5.0 software (Zawia et al., 1998; Basha et al., 2003).
Macroarray analysis. The Message Hunter macroarray screening system (Genotech, St. Louis, MO) was used to study the simultaneous expression of multiple genes. Blots containing fragments of 15 genes from various families of transcription factors in variable concentrations (2 and 20 ng), prearranged on nylon membranes, along with actin, GAPDH, and pUC19 DNA as positive or negative controls, were used. Samples of brain mRNA from control and Pb-exposed animals were isolated and reverse-transcribed in the presence of [α-32P]deoxy-CTP and hybridized to the gene array organized on the nylon membrane. Each gene on the membrane was 200-bp-long and contained equimolar amount of DNA. Sequences corresponding to ∼35 genes of representative transcription factor families were immobilized on several filters. The filters were then stringently washed and exposed to x-ray films. The resultant bands were scanned and quantitated using UVP Epi chemi II Darkroom software. The array data were collected from two sets of experiments, and each set consisted of tissue pooled from three animals (Bakheet and Zawia, 2004).
Total RNA isolation, synthesis of cDNA, and PCR. RNA from various control and exposed cortical tissues was isolated according to the TRIzol method (Invitrogen, Carlsbad, CA). The RNA was reverse-transcribed to obtain cDNAs, catalyzed by SuperScript II RNase H reverse transcriptase (RT). The RNA/primer mixture containing 50 ng of total RNA, 1 μl of 10 mm deoxy-NTP mix, and 1 μl of oligo(dT) were incubated at 65°C for 5 min. A reaction mixture containing 10× RT buffer (200 mm Tris-HCl, pH 8.4, 500 mm KCl), 25 mm MgCl2, 0.1 m DTT, and RNaseOUT recombinant RNase inhibitor was added and incubated at 42°C for 2 min. One microliter of SuperScript II RT was then added and incubated at 42°C for 50 min. The reaction was terminated at 70°C for 15 min. One microliter of RNase H was added, and the reaction was incubated for 20 min at 37°C. The final product was stored at -20°C and used in the PCR step. Primers were designed from the APP gene sequences so that their amplified PCR products include all spliced forms of the messages [sense, GAG GAG GAG GCC GAG GAG CCC TA (base pairs 788-810); antisense, CTC GGT GCT TGG CTT CCA ACC TC (base pairs 1213-1235)] modified from the method of Smith et al. (1991). Control primers for housekeeping genes such as G3PDH and β-actin were obtained commercially. The general PCR step consisted of the reaction of 1 μl of each of the primers, 2 μl of the cDNA strand, and 46 μl of PCR Supermix obtained from Invitrogen. Using a MiniCycler, the reaction was denatured at 94°C for 2 min and subsequently underwent 28-30 temperature cycles (94°C denaturing for 30 s, 55°C annealing for 30 s, and 72°C elongation for 30 s). The reaction was held at 72°C for 5-7 min for a final elongation period. PCR conditions were varied depending on the primers used. The final PCR product was analyzed onto a 1% agarose/ethidium bromide gel by running the gel at 80 V in 0.5× Tris-borate EDTA buffer for ∼90 min and scanned in the Typhoon Variable Mode Imager 9410. The resulting scanned gels were quantitated and analyzed using the Image Quant 5.0 software (Zawia et al., 1998; Basha et al., 2003).
Western blot analysis. Polyclonal antibodies for rabbit APP (Sigma) were used to probe Western blots. Western blotting and quantitation procedures were performed as described previously (Atwood et al., 1998; Nunomura et al., 1999) with slight modifications. The protein samples (2-4 μg) were subjected to SDS-PAGE, transferred to nitrocellulose membranes, and probed with a polyclonal antibody that recognizes rodent APP [N-terminal antibody (Sigma) developed in rabbit using a synthetic peptide NVQNGKWDSDPSGTK corresponding to the N-terminal region of human APP695 (46-60 aa), conjugated to KLH as the epitope]. The blots were incubated with anti-rabbit horseradish peroxidase, the chemiluminescent signal was captured using Kodak Image Station, and the images were analyzed and quantified.
Metal analysis. On P20 (completion of early exposure) and at 20 months of age (completion of late exposure), the animals from all the three groups were used to determine the levels of Pb in the blood and brain. The metal analysis was performed following the method described previously (Smith et al., 1998; Basha et al., 2003). Animals were deeply anesthetized, and blood was collected using a sterile syringe through cardiac puncture. For the collection of brain tissue, animals were perfused with sterile saline, the brains were excised, and cortical tissue from the brains was isolated. Samples were pooled from three or four animals, and three to five sample determinations were made for each exposure condition. Tissue and blood samples were analyzed by Rhode Island Analytical Laboratory (Warwick, RI), using atomic absorption spectroscopy following EPA approved methodology.
Cell culture. Pheochromocytoma (PC12) cells (American Type Culture Collection, Manassas, VA) were maintained under conditions of 5% CO2 and 37°C. Cells were grown in DMEM (Hyclone, Logan, UT) supplemented to a final concentration with 10% fetal bovine serum (Hyclone), 5% horse serum (Hyclone), and 1% penicillin-streptomycin solution (Sigma).
Transfection and responsiveness of APP promoters. PC12 cells were plated for 48 h, and the transfection mixture (100 μl) containing vectors suspended in Lipofectamine 2000 reagent (Invitrogen) was added in the wells. Cells were transfected with plasmid vectors (pGL3) containing the human APP promoter. pRL-TK, a plasmid construct driven by the herpes simplex virus thymidine kinase promoter, was cotransfected as a negative control. In some experiments basic pGL3 that lacks any promoter was transfected to serve as a background control. Twenty-four hours after transfection, a 400 μl medium containing 0.1 μm of Pb was added. Cells were harvested using Passive Lysis Buffer (Promega) at 48 h after exposure.
Silencing of the Sp1 gene using small interfering RNA. For small interfering RNA (siRNA), sense, antisense, and T7 promoter oiligonucleotides were synthesized by Invitrogen. The synthetic oligos had the following sequences: T7 promoter, 5′-TAATACGACTCACTATAG-3′; sense, 5′-ATGATCGTGCCTGCCTGAGAGCTATAGTGAGTCGTATTA-3′; antisense, 5′-CAGCTCTCAGGCAGGCACGATCTATAGTGAGTCGTATTA-3′. One nanomole of T7 and sense/antisense were annealed in 50 μl of Tris EDTA buffer, heated at 95°C for 5 min to obtain double-stranded DNA (dsDNA). In vitro transcription was performed using the AmpliScribe T7 High Yield Transcription kit (Epicenter Technologies, Madison, WI). Both transcription reactions of sense and antisense were mixed and heated at 95°C for 5 min and then cooled down. After purification of siRNA with 0.2 m sodium acetate and 70% ethanol, the siRNA was reconstituted with water that it is ready for transfection into cells. The extent of silencing the Sp1 gene was validated using the luciferase assay described below.
Measurement of reporter gene activity. For the luciferase assay, 10 μl of homogenized lysates were analyzed using Dual-Luciferase Reporter Assay System (Promega) in an LB 96 V microplate luminometer. The APP promoter constructed on pGL3 [APP(+)], pGL3-Basic [APP(-)], and pRL-TK vectors were used. Their reporter genes following translation into enzymes result in bioluminescent products during the analysis. The APP(+) and APP(-) vectors contain firefly luciferase luc+ as the reporter gene, which is quantifiable by the luciferase assay reagent. The pRL-TK has the renilla luciferase reporter gene (rluc) to serve as a control for transfection efficiency. The renilla luciferase gene product has different substrate specificity from firefly luciferase. Applying specific substrates as described by the manufacturer can distinguish between the activities of both enzymes. Thus, the same assay records both firefly and renilla luciferase activities. The ratio of firefly luminescence signal for APP(+) and APP(-) over renilla luminescence signal (pRL-TK) in samples gives APP(+) and APP(-) relative activities.
Amyloid β assay. The levels of amyloid β-40 were measured using the IBL assay kit (IBL, Gunma, Japan). This kit is a solid phase sandwich ELISA that uses two kinds of highly specific antibodies. This kit measures both soluble and insoluble Aβ levels in tissue and serum according to the information provided by the suppliers. The assay conditions were followed according to the method described by Morishima-Kawashima et al. (2000), with slight modifications as suggested by the manufacturers. Brain tissue extracts (test samples) were prepared following the guanidine-HCl extraction method. Brain tissue was homogenized in Tris-saline (TS) (50 mm Tris-HCl buffer, pH 7.4, 150 mm NaCl, 1 μg/ml N-α-p-tosyl-l-lysine chloromethyl ketone, 1 μg/ml antipain, 0.5 mm diisopropyl fluorophosphates, 0.5 mm PMSF, and 0.1% protease inhibitor mixture) and centrifuged at 100,000 × g for 20 min at 4°C. The pellet was resuspended in 4 vol of TS and centrifuged at 70,000 × g for 20 min at 4°C. The resultant pellet was dissolved in 500 μl of 6 m guanidine-HCl (in 50 mm Tris buffer, pH 7.6), incubated at room temperature for 30 min and centrifuged at 70,000 × g for 20 min at 4°C. The resultant supernatant was collected and diluted by EIA buffer (supplied with the kit) to 12 times to reduce the guanidine-HCl concentration in the samples. The test samples were aliquoted and stored in a freezer at -80°C. The protein content was estimated using BCA reagent, and the test samples were prepared with 200 μg of protein in 100 μl using EIA buffer to adjust the volume. The test samples and the standards were added to the 96 well plate [precoated with anti-human Aβ (35-40) (1A10) mouse IgG monoclonal antibody] and incubated overnight at 4°C. After several washes with EIA wash buffer, 100 μl of labeled antibody was added to each well (except the wells corresponding to reagent blanks) and incubated at 4°C for 1 h. After a thorough wash, 100 μl of TMB buffer was added to each well and incubated in the dark (shielded) for 30 min at room temperature. The reaction was stopped by adding 100 μl of 1N H2SO4, and the readings were taken at 450 nm. The levels of Aβ in the test samples were calculated by incorporating the unknown values into the standard curve obtained in the assay.
Statistical treatment. Data were analyzed by two-way ANOVA followed by a Duncan's post hoc test to compare the effects among various treatments; values marked with an asterisk are significantly different from their corresponding controls (p < 0.05). The relationship between APP mRNA expression and Sp1 DNA-binding was analyzed using Spearman and Pearson correlation analysis using SAS 9.1 (Raleigh, NC).
Results
The promotion of amyloidogenesis may begin very early in life by interference in the processes that determine the level of responsiveness of the APP gene. The data reported below monitored the expression of the APP mRNA, the activity of Sp1, APP, and Aβ levels in developmentally exposed animals as well as in animals exposed during old age.
Exposure to Pb and latent expression of the APP mRNA
To determine whether developmental exposure to Pb resulted in long-term alterations in the expression of APP, we examined the lifelong expression of the APP gene. We also monitored the concurrent expression of housekeeping genes. We found that exposure to Pb during the postnatal period resulted in a transient increase in APP mRNA expression during the first month after birth followed by a return to basal levels by 1 year, and a subsequent rise in APP expression again at 20 months of age (Fig. 1). β-actin mRNA levels remained unchanged by age or Pb exposure during the same period (Fig. 1), as well as those of G3PDH (data not shown). Whereas both blood (46.43 ± 1.95 μg/dl) and tissue (0.41 ± 0.04 μg/g) levels of Pb were increased during developmental exposure to Pb, both blood and tissue levels of Pb were at background levels in the neonatally exposed old animals. There were also no significant changes in the body weights of Pb-exposed animals (control, 701.67 ± 16.02 g; Pb-E, 684 ± 25.46 g), and the survival rate of the exposed group was identical to that of the control group to 20 months of age.
Figure 1.
Lifetime changes in APP695 and β-actin mRNA expression in the cortex. Data shown are derived from two cohorts of animals separated in time by 1 year. The bar in the figure indicates the neonatal period of lactational exposure to Pb. Each data point represents the mean ± SEM for four to six animals. Data were analyzed by two-way ANOVA followed by a Duncan's post hoc test to compare the effects among various treatments; values marked with an asterisk are significantly different from their corresponding controls (p < 0.05).
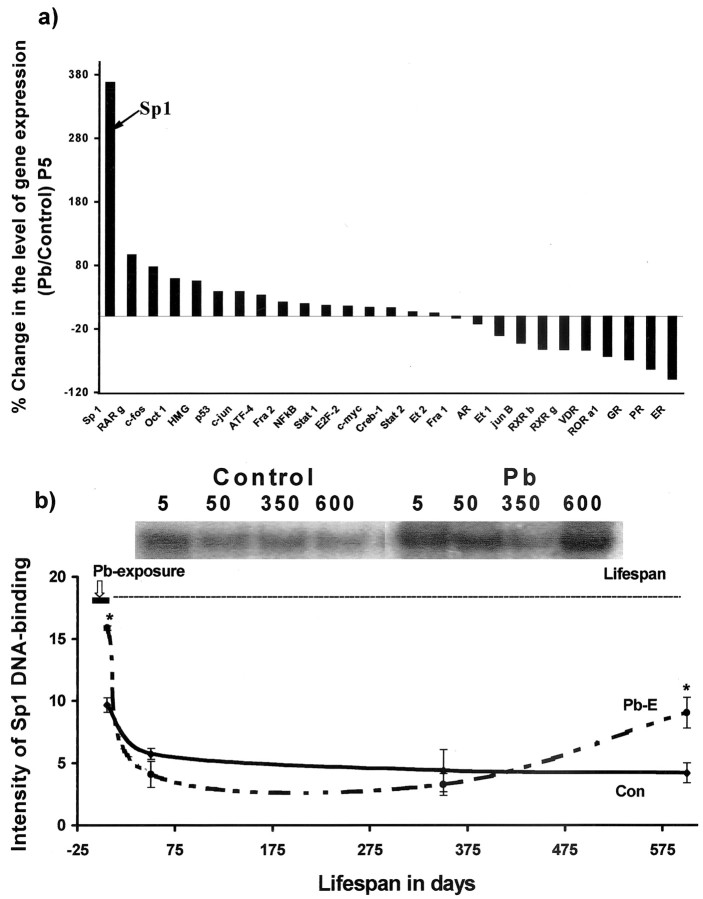
mRNA profiling of transcription factors and Sp1 DNA-binding
The regulatory portion of the APP gene is a GC-rich region, which lacks a TATA box and contains consensus sites for several transcription factors (Salbuam et al., 1988; La Fauci et al., 1989). To determine whether any of these transcription factors was prominently altered by Pb exposure, we conducted a macroarray screen of a series of transcription factors on postnatal day 5, the age of highest changes in APP expression. We found that Sp1 expression exhibited a high level of induction after Pb exposure (Fig. 2a). Consequently, we monitored the lifelong profiles of Sp1 DNA-binding in the cortex of control and Pb-exposed animals and found that exposure to Pb resulted in a premature peak of Sp1 DNA-binding in the early postnatal period and returned to basal levels weeks later. Commensurate with APP expression, Sp1 DNA-binding also exhibited a startling rise in activity, 20 months after Pb exposure had ceased (Fig. 2b).
Figure 2.
mRNA profiling of transcription factors after developmental exposure to Pb and lifetime changes in Sp1 DNA-binding in the cortices of control and Pb-exposed animals. a, mRNA profiling. Cortical total RNA was isolated from P5 rats and used to probe cDNA arrays of transcription factors: AR, androgen receptor; ATF-4, activating transcription factor-4; c-fos, c-jun, c-myc, oncogenes; Creb-1, cAMP-responsive element-binding protein-1; E2F-2, ubiquitin-conjugating enzyme F-2; ER, estrogen receptor; Et, erythroblastosis virus oncogene homology; GR, glucocorticoid receptor; HMG, high-mobility group protein; NFκB, nuclearfactor κB; Oct 1, octamer transcription factor 1; p53, phosphoprotein 53; PR, progesterone receptor; RAR, retinoic acid receptor [related factors: retinoid orphan receptor (ROR), retinoid X receptor (RXR)]; Stat, signal transducer and activator of transcription; VDR, vitamin D3 receptor. The array data shown were derived from two sets of experiments, and each set consisted of tissue pooled from three animals. b, Sp1 DNA-binding. Nuclear extracts were prepared from controls and animals exposed as indicated by the bar in the figure. Sp1 DNA-binding was analyzed using gel mobility shift assays. Sp1 DNA-binding data shown are derived from two cohorts of animals separated in time by 1 year; each data point represents the mean ± SEM for four to six animals. Data were analyzed by two-way ANOVA followed by a Duncan's post hoc test to compare the effects among various treatments. Values marked with an asterisk are significantly different from their corresponding controls (p < 0.05).
We performed correlation analysis to determine whether a relationship existed between APP mRNA expression and Sp1 DNA-binding at various time points. We found an overall statistical correlation between APP expression and Sp1 activity (Spearman correlation coefficient, 0.8); this statistical correlation became higher when one of the time points (P5) was dropped from the analysis (Pearson correlation coefficient, 0.98). To establish a direct biological link between APP and Sp1, we conducted cell culture studies involving the APP promoter and Sp1 presented below.
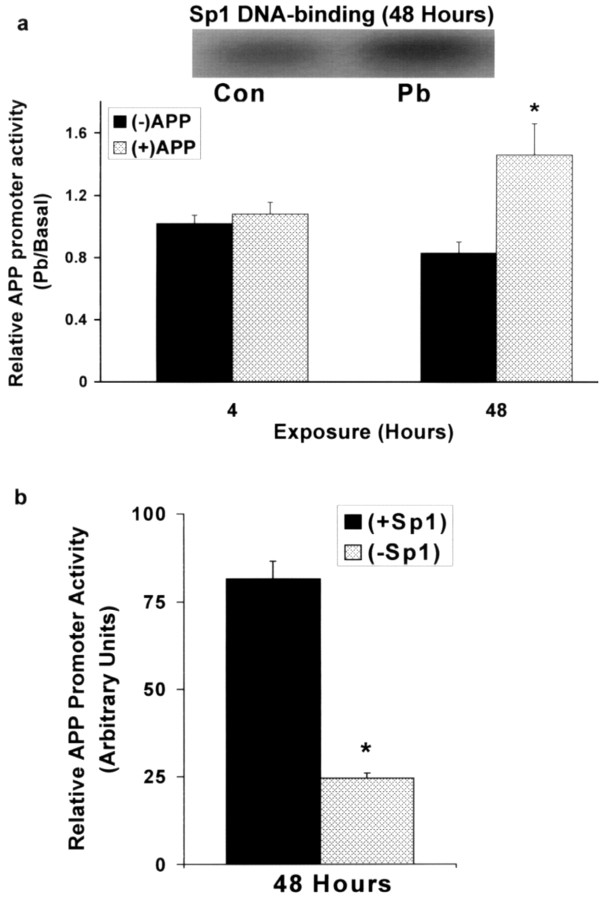
The APP promoter, Pb, and Sp1 siRNA
In vivo experiments have shown that exposure to Pb increases the expression of the APP gene and the activity of Sp1; however, it is uncertain whether Sp1 was responsible for the changes in APP expression and whether Pb disturbed the expression of the APP gene through alterations in Sp1 activity. Therefore, we conducted independent cell culture experiments to test the responsiveness of the human APP promoter to Pb exposure and the dependence of the APP promoter on Sp1 activity. We transfected PC12 cells with a 1.2 kb human APP promoter (-1100/+104, +1 being the transcription start site) (Lahiri and Robakis, 1991) linked to a luciferase reporter gene. We found that the APP promoter was active in these cells and that Pb (0.1 μm, determined from a range of doses) stimulated promoter activity in a delayed manner (Fig. 3a). Pb-induced promoter activity was also accompanied by an increase in Sp1-DNA binding (Fig. 3a, see inset), and these responses were selective for Pb (Ba or Hg had no effect; data not shown). To further confirm the connection between Sp1 and APP gene expression, we knocked out the expression of the Sp1 gene in an effort to deplete Sp1 levels using siRNA technology. Silencing the Sp1 gene resulted in a 75% decrease in the responsiveness of the APP promoter (Fig. 3b).
Figure 3.
Human APP promoter responsiveness after exposure to Pb and silencing of the Sp1 gene using siRNA in PC12 cells. a, Responsiveness of the APP promoter and Sp1 DNA-binding to Pb exposure. Con, Control. b, Silencing of the Sp1 gene. Cells were incubated in the presence or absence of Pb (0.1 μm) or siRNA (to knock out the Sp1 gene) and harvested to determine reporter gene activity or Sp1 DNA-binding. APP promoter activity plotted above is the ratio of activity of cells that contain pGL3 with the APP promoter versus cells transfected with pGL3 minus the APP promoter. Both cells contain the negative pRLTK vector. Promoter values shown are the means ± SEM of five to seven determinations. Data were analyzed by two-way ANOVA followed by a Duncan's post hoc test to compare the effects among various treatments; values marked with an asterisk are significantly different from their corresponding controls (p < 0.05).
Effect of Pb exposure during old age on APP and Aβ Levels
To examine whether APP mRNA expression and Sp1 DNA-binding are responsive to Pb exposure during old age, we compared the levels of APP mRNA, Sp1 DNA-binding, APP, and Aβ levels in aged animals exposed to Pb from 18 to 20 months of age (Fig. 4). Exposure of naive animals during old age had no significant effect on APP mRNA levels, Sp1 DNA-binding, APP, and Aβ levels (Fig. 4), although Pb concentrations in the blood and cortex of these animals were significantly elevated (blood Pb = 60.1 ± 15.4 μg/dl, and tissue Pb = 0.32 ± 0.03 μg/g). In contrast, animals developmentally exposed to Pb exhibited an elevation in APP mRNA, Sp1 DNA-binding, APP and Aβ levels.
Figure 4.
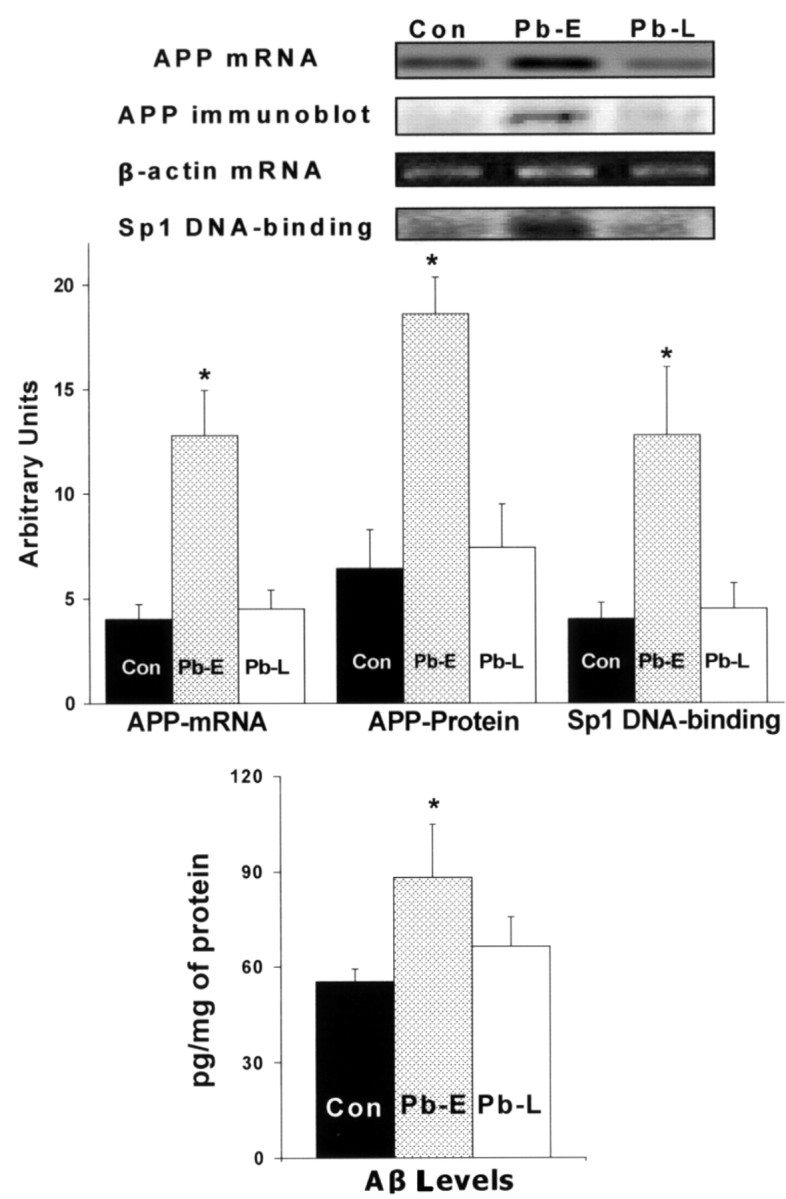
Unresponsiveness of APP expression, Sp1 DNA-binding, APP and Aβ formation to Pb exposure in old age. Brain cortices were obtained from control 20-month-old animals and those directly exposed to 200 ppm of Pb-acetate as follows: control (Con, unexposed), Pb-E (exposed from P1 to P20), and Pb-L (exposed from 18 to 20 months of age). These tissues were used to analyze the mRNA expression of APP, β-actin (RT-PCR), Sp1 DNA-binding (gel shift), APP protein levels (Western blot), and Aβ activity (ELISA). Data shown are derived from two cohorts of animals separated in time by 1 year; each data point represents the mean ± SEM for four to six animals. Data were analyzed by two-way ANOVA followed by a Duncan's post hoc test to compare the effects among various treatments; values marked with an asterisk are significantly different from their corresponding controls (p < 0.05).
Discussion
Developmental exposure to some environmental agents has been shown to remain “silent” for a long period of time without any overt manifestations. Behavioral studies after developmental exposure to environmental agents such as methylmercury (Rice, 1989), methylazoxymethanol (Lee and Rabe, 1992), and triethyltin (Barone et al., 1995) have provided evidence that latent neurotoxicity occurs in adult animals. More recently, developmental exposure to pesticides has been shown to result in a loss of nigral dopamine neurons and a decrease in dopamine content, 9 months later (Thiruchelvam et al., 2002; Reeves et al., 2003). These data suggest that behavioral and neurodegenerative outcomes later in life can be influenced by developmental exposure to environmental agents.
Transcriptional events in the brain are selectively susceptible to environmental influences, and their disturbance may result in perturbations in the regulation of gene expression. Although short-term disturbances to transcriptional processes can occur during any period of life, perturbations of the transcriptional apparatus in the brain during development may result in permanent reprogramming and alterations in the requirements for gene expression. The selective changes in APP gene expression occurring during development are transient and a direct result of concurrent Pb exposure; however, the delayed overexpression of APP in old age, long after Pb exposure has ceased, suggests that developmental exposure to Pb had reprogrammed the responsiveness of the APP gene. This is also supported by the observation that the late rise in APP mRNA expression at 20 months of age happened when both blood and tissue concentrations of Pb in these old animals were at background levels, thus ruling out the possibility that Pb is stored in bones, and leaches out, during old age, to stimulate the upregulation in APP mRNA expression. If we extrapolate the blood Pb levels we found in these animals (control and Pb-E) at 20 months of age (<2 μg/dl) to humans, these levels are well below the 10 μg/dl safety level for blood Pb set for humans by the Centers for Disease Control and Prevention (1991). When we exposed old animals to 200 ppm Pb in their drinking water for 3 months, we found 40-60 μg/dl of Pb in their blood. At least one in 20 children in the United States today have blood Pb levels >10 μg/dl (Pirkle et al., 1998).
The lifelong lag between initial exposure and the delayed elevations in gene expression suggests that the regulatory apparatus of the APP gene may have been altered by developmental Pb exposure, rendering the gene more responsive to endogenous stimuli that appear during old age. Changes in the gene expression are mediated through intercellular and/or intracellular signaling pathways that are finally transmitted through the action of transcription factors. Because the APP gene could be activated by several transcription factors, it was important to identify the factor most responsible for Pb-induced changes in the APP gene expression. We screened transcription factors (TFs) that are “constitutive” and those, whose activation is “conditional” (i.e., dependent on some signaling event).
We found changes in the signal-dependent TFs such as the steroid receptor superfamily (glucocorticoid receptor, estrogen receptor, progesterone receptor, retinoid X receptor, retinoic acid receptor, etc.), development-specific factors [specificity protein 1 (Sp1), octamer transcription factor-1], those respondent to internal signals (phosphoprotein 53), and those linked to cell-surface receptors (cAMP-responsive element-binding protein, activator protein-1, nuclear factor κB, signal transducer and activator of transcription, etc.). Although changes in expression levels of most of these TFs were small, it was interesting to observe that the steroid receptor superfamily exhibited a downregulation trend. The screening analysis revealed Sp1 to be the most highly expressed TF after Pb exposure, and the DNA-binding studies showed that the lifelong changes in Sp1 activity were similar to those of APP. Sp1 is one of the transcription factors that can promote the production of APP mRNA (Pollwein et al., 1992; Querfurth et al., 1999), and Sp1 DNA-binding has been shown to be highly responsive to the presence of heavy metals such as Pb (Zawia et al., 1998; Basha et al., 2003).
The lifelong profiles of both APP expression and Sp1 DNA-binding do not exactly mirror each other; however, their similar responses to Pb exposure during early and late life suggested that they may be linked. The responsiveness of the human APP promoter to Pb exposure and the ability of Sp1 siRNA to dampen such responsiveness strongly suggest that Pb-induced changes in APP expression are mediated via a mechanism that involves Sp1 (Fig. 3).
Although both APP gene expression and Sp1 activity were responsive to Pb exposure during brain development or in cells actively growing in vitro, the delayed changes in both APP and Sp1 DNA-binding in animals previously exposed to Pb required additional examination. Furthermore, it was important to know whether both APP and Sp1 are responsive to Pb exposure during old age. Interestingly we found that both the APP gene and the transcription factor that regulates it were not affected by Pb exposure during old age (Fig. 4). This lack of responsiveness of these molecular markers persisted in the face of brain tissue Pb concentrations that were high, and the animals were directly exposed for 3 months. This affirmed that the latent expression of APP mRNA and the concurrent rise in Sp1 DNA-binding in old animals was a programmed event that was set in motion by exposure to Pb in early life.
Changes in transcriptional events may not be quite important unless such transcriptional alterations are translated into products that play a role in physiological and pathological processes. Therefore we examined the levels of APP and its amyloidogenic Aβ product in old animals. Consistent with the changes in APP gene expression, latent elevation in both APP and Aβ levels occurred. This suggested that the “reprogramming” of gene expression after developmental Pb exposure had a significant impact on the functional and pathological events of old age. Furthermore, recent immunohistochemical data from our lab confirm that Sp1 and APP are colocalized to the same cells in the brain, and the areas surrounding these cells are also abundant in Aβ levels (N. H. Zaiwa, B. Brock, K. Dipalma, W. Wei, M. R. Basha, unpublished data). The elevations in APP and the subsequent increase in Aβ peptide, a hallmark of Alzhemier's pathology, could influence the course of neurodegeneration in these aging rodents.
The relationship between Sp1, APP, and Aβ in Pb exposed animals is strongly supported by recent molecular and cell culture studies. In addition to APP, another important protein closely involved in AD amyloidogenesis is BACE (β-APP cleaving enzyme). Indeed, the BACE gene promoter has been shown recently to contain a functional Sp1 site (Ge et al., 2004). Furthermore, overexpression of Sp1 in transfected cells elevates APP thereby generating higher Aβ levels (Christensen et al., 2004). Therefore, the promotion of amyloidogenesis may begin very early in life by interference in the processes that determine the level of responsiveness of the APP gene. It is unclear why APP mRNA expression, Sp1 DNA-binding, and de novo synthesis of APP are resistant to Pb exposure during old age; however, this lack of responsiveness cannot be attributed to the inability of Pb to enter the brain, but rather to permissive programming which is established during development.
The findings of this study raise fundamental biological and toxicological questions. Among these are: (1) How is an environmental insult maintained for the life of an organism after the damaging agent had long disappeared? (2) What are the cellular targets that sustain or retain such perturbations? (3) Why does such an alteration remain “silent” for a period of time, and what precipitates its manifestations later in life? and (4) Is this property unique to the nervous system?
To answer some of these questions, one must deal with the latency of the response to Pb exposure and the ability of these molecular markers to register the passage of time. The latent responses reported here are probably unique to the nervous system and a few other tissues. Because cells in most tissues turn over, and proteins are in a continual cycle of synthesis and degradation, the molecular targets that would store and transmit this information would have to reside in the genome of terminally differentiated cells. Furthermore, for the damage to persist on the structure of the DNA, the perturbation has to escape recognition of DNA repair enzymes. Thus, it is probable that the environmental insult interfered with epigenetic phenomena such as: DNA methylation patterns, histone deacetylation, and chromatin restructuring. Errors in some of these processes are not subject to repair because they are not viewed as damage by DNA repair pathways. Therefore, one interpretation of these data are that Pb had permanently interfered with the programmatic settings of the regulation of the APP gene. These settings are established during development and are critical for the responsiveness of the gene regulatory apparatus in the adult. Finally, these data suggest that developmental exposure to Pb exhibits latent effects on the expression of genes that are known to be involved in neurodegenerative processes during old age. These developmental events may influence the course of neurodegenerative changes later in life.
Footnotes
This work was supported by grants from the National Institutes of Health (ES08104, ES013022, and RR16457 to N.H.Z.; AG18379 and AG18884 to D.K.L.). We thank Prof. Tomas Guilarte (Environmental Health Sciences, Johns Hopkins University, Baltimore, MD), Dr. H. A. Lashuel (Center for Neurologic Diseases, Harvard Medical School, Boston, MA), and Dr. Fernando Cardozo-Pelaez (Environmental Toxicology, University of Montana, Missoula, MT) for the advice and comments on this manuscript. We are also grateful to Dr. X. Sun (Research Assistant Professor of Statistics, Cancer Prevention Research Center, University of Rhode Island, Kingston, RI) and Dr. Hui Cai (Vanderbilt University, Nashville, TN) for providing assistance in the statistical analysis.
Correspondence should be addressed to Dr. N. H. Zawia, Department of Biomedical and Pharmaceutical Sciences, University of Rhode Island, Kingston, RI 02881. E-mail: nzawia@uri.edu.
Copyright © 2005 Society for Neuroscience 0270-6474/05/250823-07$15.00/0
References
- Atwood CS, Moir RD, Huang X, Scarpa RC, Bacarra NM, Romano DM, Hartshorn MA, Tanzi RE, Bush AI (1998) Dramatic aggregation of Alzheimer abeta by Cu(II) is induced by conditions representing physiological acidosis. J Biol Chem 273: 12817-12826. [DOI] [PubMed] [Google Scholar]
- Bakheet SA, Zawia NH (2004) The role of pou domain transcription factors in lead neurotoxicity. In: Molecular neurotoxicology, environmental agents, and transcription-transduction coupling (Zawia NH, ed), pp 183-198. Boca Raton, FL: CRC.
- Barker DJ (2002) Fetal programming of coronary heart disease. Trends Endocrinol Metab 13: 364-368. [DOI] [PubMed] [Google Scholar]
- Barker DJ, Winter PD, Osmond C, Margetts B, Simmonds SJ (1989) Weight in infancy and death from ischaemic heart disease. Lancet 2: 577-580. [DOI] [PubMed] [Google Scholar]
- Barone Jr S, Stanton ME, Mundy WR (1995) Neurotoxic effects of neonatal triethyltin (TET) exposure are exacerbated with aging. Neurobiol Aging 16: 723-735. [DOI] [PubMed] [Google Scholar]
- Basha MR, Wei W, Brydie M, Razmiafshari M, Zawia NH (2003) Lead-induced developmental perturbations in hippocampal Sp1 DNA-binding are prevented by zinc supplementation: in vivo evidence for Pb and Zn competition. Int J Dev Neurosci 21: 1-12. [DOI] [PubMed] [Google Scholar]
- Basha MR, Wei W, Reddy GR, Zawia NH (2004) Zinc finger transcription factors mediates perturbations of brain gene expression elicited by heavy metals. In: Molecular neurotoxicology, environmental agents, and transcription-transduction coupling (Zawia NH, ed), pp 43-64. Boca Raton, FL: CRC.
- Centers for Disease Control and Prevention (CDC) (1991) Preventing lead poisoning in young children: a statement by the CDC, Atlanta, GA, United States Department of Health and Human Services.
- Christensen MA, Zhou W, Qing H, Lehman A, Philipsen S, Song W (2004) Transcriptional regulation of BACE1, the beta-amyloid precursor protein beta-secretase, by Sp1. Mol Cell Biol 24: 865-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11: 1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge YW, Maloney B, Sambamurti K, Lahiri DK (2004) Functional characterization of the 5′ flanking region of the BACE gene: identification of a 91 bp fragment involved in basal level of BACE promoter expression. FASEB J 18: 1037-1039. [DOI] [PubMed] [Google Scholar]
- Goyer RA (1996) Toxic effects of metals. In: Casarett and Doull's toxicology: the basic science of poisons (Klaassan CD, ed), pp 691-736. New York: McGraw-Hill.
- La Fauci G, Lahiri DK, Salton SR, Robakis NK (1989) Characterization of the 5′-end region and the first two exons of the beta-protein precursor gene. Biochem Biophys Res Commun 159: 297-304. [DOI] [PubMed] [Google Scholar]
- Lahiri DK, Robakis NK (1991) The promoter activity of the gene encoding Alzheimer beta-amyloid precursor protein (APP) is regulated by two blocks of upstream sequences. Mol Brain Res 9: 253-257. [DOI] [PubMed] [Google Scholar]
- Lahiri DK, Farlow MR, Sambamurti K, Greig NH, Giacobini E, Schneider LS (2003) A critical analysis of new molecular targets and strategies for drug developments in Alzheimer's disease. Curr Drug Targets 4: 97-112. [DOI] [PubMed] [Google Scholar]
- Lee MH, Rabe A (1992) Premature decline in Morris water maze performance of aging micrencephalic rats. Neurotoxicol Teratol 14: 383-392. [DOI] [PubMed] [Google Scholar]
- Morishima-Kawashima M, Oshima N, Ogata H, Yamaguchi H, Yoshimura M, Sugihara S, Ihara Y (2000) Effect of apolipoprotein E allele epsilon4 on the initial phase of amyloid beta-protein accumulation in the human brain. Am J Pathol 157: 2093-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman HL, Schell A, Bellinger D, Leviton A, Allred EN (1990) The long-term effects of exposure to low doses of lead in childhood. An 11-year follow-up report. N Engl J Med 322: 83-88. [DOI] [PubMed] [Google Scholar]
- Nishimura I, Uetsuki T, Kuwako K, Hara T, Kawakami T, Aimoto S, Yoshikawa K (2002) Cell death induced by a caspase-cleaved transmembrane fragment of the Alzheimer amyloid precursor protein. Cell Death Differ 9: 199-208. [DOI] [PubMed] [Google Scholar]
- Nunomura A Perry G, Hirai K, Aliev G, Takeda A, Chiba S, Smith MA (1999) Neuronal RNA oxidation in Alzheimer's disease and Down's syndrome. Ann NY Acad Sci 893: 362-364. [DOI] [PubMed] [Google Scholar]
- Ozanne SE (2001) Metabolic programming in animals. Br Med Bull 60: 143-152. [DOI] [PubMed] [Google Scholar]
- Pirkle J, Kaufmann R, Brody D, Hickman T, Gunter E, Paschal D (1998) Exposure of the US population to lead, 1991-1994. Environ Health Perspect 106: 745-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollwein P, Masters C, Beyruther K (1992) The expression of the amyloid precursor protein (APP) is regulated by two GC-elements in the promoter. Nucleic Acids Res 20: 63-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querfurth HW, Jiang J, Xia W, Selkoe DJ (1999) Enhancer function and novel DNA binding protein activity in the near upstream beta APP gene promoter. Gene 232: 125-141. [DOI] [PubMed] [Google Scholar]
- Reeves R, Thiruchelvam M, Richfield EK, Cory-Slechta DA (2003) Behavioral sensitization and long-term neurochemical alterations associated with the fungicide triadimefon. Pharmacol Biochem Behav 76: 315-326. [DOI] [PubMed] [Google Scholar]
- Rice D (1989) Delayed neurotoxicity in monkeys exposed developmentally to methyl mercury. Neurotoxicology 10: 645-650. [PubMed] [Google Scholar]
- Salbuam JM, Weidemann A, Lemaire H, Masters CL, Beyreuther K (1988) The promoter of Alzheimer's disease amyloid A4 precursor gene. EMBO J 7: 2807-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe DJ (2001) Alzheimer's disease: genes, proteins, and therapy. Physiol Rev 81: 741-766. [DOI] [PubMed] [Google Scholar]
- Silbergeld EK (1992) Mechanisms of lead neurotoxicity, or looking beyond the lamppost. FASEB J 6: 3201-3206. [DOI] [PubMed] [Google Scholar]
- Smith CJ, Wion D, Brachet P (1991) Nerve growth factor-induced neuronal differentiation is accompanied by differential splicing of beta-amyloid precursor mRNAs in the PC12 cell line. Mol Brain Res 10: 351-354. [DOI] [PubMed] [Google Scholar]
- Smith D, Bayer L, Strupp BJ (1998) Efficacy of succimer chelation for reducing brain Pb levels in a rodent model. Environ Res 78: 168-176. [DOI] [PubMed] [Google Scholar]
- Thiruchelvam M, Richfield EK, Goodman BM, Baggs RB, Cory-Slechta DA (2002) Developmental exposure to the pesticides paraquat and maneb and the Parkinson's disease phenotype. Neurotoxicology 23: 621-633. [DOI] [PubMed] [Google Scholar]
- Valdez R, Athens MA, Thompson GH, Bradshaw BS, Stern MP (1994) Birth weight and adult health outcomes in a biethnic population in the USA. Diabetologia 37: 624-631. [DOI] [PubMed] [Google Scholar]
- Yarbrough DE, Barrett-Connor E, Kritz-Silverstein D, Wingard DL (1998) Birth weight, adult weight, and girth as predictors of the metabolic syndrome in postmenopausal women: the Rancho Bernardo Study. Diabetes Care 21: 1652-1658. [DOI] [PubMed] [Google Scholar]
- Zawia NH (2003) Transcriptional involvement in neurotoxicity. Toxicol Appl Pharmacol 190: 177-188. [DOI] [PubMed] [Google Scholar]
- Zawia NH, Sharan R, Brydie M, Oyama T, Crumpton T (1998) SP1 as a target site for metal-induced perturbations of transcriptional regulation of developmental brain gene expression. Brain Res Dev Brain Res 107: 291-298. [DOI] [PubMed] [Google Scholar]