Abstract
Phosphorylation plays a key role in the precise timing of circadian clocks. Daily rhythms of phosphorylation of the Drosophila circadian clock component PERIOD (PER) were first described more than a decade ago, yet little is known about their phosphorylation sites and their function in circadian behavior. Here we show that serines 151 and 153 in PER are required for robust in vitro phosphorylation by the casein kinase 2 (CK2) holoenzyme, a cytoplasmic kinase shown to be involved in circadian rhythms. Mutation of these sites in transgenic flies results in significant period lengthening of behavioral rhythms, altered PER rhythms, and delayed PER nuclear localization in circadian pacemaker neurons. In many respects, mutation of these phosphorylation sites phenocopies mutation of the catalytic subunit of CK2. We propose that CK2 phosphorylation at these sites triggers PER nuclear localization.
Keywords: Drosophila, phosphorylation, CK2, PERIOD, circadian clock, holoenzyme, pacemaker neuron
Introduction
Circadian clocks consist mainly of transcriptional feedback loops whose components are modified by phosphorylation (Hardin, 2004; Harms et al., 2004). In Drosophila, CLOCK (CLK) and CYCLE (CYC) transcriptionally activate period (per) and timeless (tim). Around midnight, cytoplasmic PERIOD (PER) relocalizes to the nucleus of pacemaker neurons (Curtin et al., 1995) and subsequently represses CLK-CYC activation. CLK and CYC also activate members of a second interdependent feedback loop, vrille and Pdp1. These core transcriptional feedback loops bear a striking resemblance in overall architecture and individual components to those in mammals.
Clock components are modified by phosphorylation. The first clock component shown to be phosphorylated rhythmically was Drosophila PER (Edery et al., 1994). Since then, additional components from many systems have shown rhythmic phosphorylation (Hunter-Ensor et al., 1996; Lee et al., 1996; Zeng et al., 1996; Garceau et al., 1997; Lee et al., 1998, 2001; Lowrey et al., 2000; Tomita et al., 2005). Indeed, rhythmic phosphorylation may actually drive transcriptional rhythms (Nakajima et al., 2005; Tomita et al., 2005). Mutations that result in human familial advanced sleep phase syndrome appear to disrupt clock-relevant kinases, casein kinase (CK) 1δ, or phosphorylation of clock protein substrates, such as hPER2 (Toh et al., 2001; Xu et al., 2005), suggesting that fly phosphorylation studies may lead to a better understanding of human circadian clock dysfunction.
In flies, many of the strongest period phenotypes are caused by mutations in kinases (Rothenfluh et al., 2000; Suri et al., 2000; Lin et al., 2002). The most well studied clock kinase in Drosophila is Doubletime (Dbt), the fly ortholog of CK1ϵ. DOUBLETIME (DBT) is thought to promote PER phosphorylation, ubiquitination, and degradation (Kloss et al., 1998; Price et al., 1998). The Drosophila ortholog of glycogen synthase kinase 3 (GSK3), shaggy (sgg), is thought to be involved in TIM phosphorylation (Martinek et al., 2001).
We have previously described a central role for the tetrameric protein kinase CK2 in the Drosophila clock (Lin et al., 2002). CK2 consists of two catalytic α subunits linked by two regulatory β subunits (Meggio and Pinna, 2003). Studies of CK2 substrates have identified consensus sites that consist of a serine or threonine with an acidic residue at the +3 position (S/T-X-X-D/E). Mutants of either subunit result in a lengthened circadian period and altered PER rhythms (Lin et al., 2002; Akten et al., 2003). In addition, CK2 facilitates PER repression in cultured S2 cells (Nawathean and Rosbash, 2004).
In Drosophila, PER phosphorylation tightly controls its subcellular distribution within pacemaker neurons (Bao et al., 2001; Kloss et al., 2001; Martinek et al., 2001). PER nuclear localization is altered in CK2 mutants (Lin et al., 2002; Akten et al., 2003). Studies in cell culture suggest that CK2 phosphorylation promotes PER activity independent of its localization (Nawathean and Rosbash, 2004). We had shown previously that CK2α can specifically phosphorylate PER in vitro (Lin et al., 2002). Here we identify serines critical for this in vitro phenomenon and show that loss of these serines in vivo mimics the PER phenotype in CK2 mutants. These results begin to define the phosphorylation sites in a core clock protein essential for circadian behavior.
Materials and Methods
Drosophila genetics and locomotor behavior. per0; perG is the wild-type per transgenic rescue strain (Cheng et al., 1998). All transgenic DNA constructs were injected in yw embryos at the CBRC Transgenic Drosophila Core. The balanced fly lines were crossed to per0 for locomotor activity assays and Western blot analysis. For circadian locomotor activity analysis, flies were entrained for 2-5 d in 12 h light/dark conditions, and locomotor activity behavior was monitored subsequently in constant darkness for 6-8 d with TriKinetics (Waltham, MA) locomotor activity monitors, as described previously (Lin et al., 2002).
Generation of transgenic per mutants and UASCK2α. perG construct was kindly provided by P. Hardin (Texas A&M University, College Station, TX) and used for per mutation transgenic line transformation. UASCK2α was constructed by cloning a CK2α cDNA fragment into pUAST. pET-CK2α (Lin et al., 2002) was digested by BglII and XhoI. This fragment containing CK2α was cloned to pUAST vector. Transformant lines with inserts on the second or third chromosome were balanced with yw;Bl/Cy0 or yw;;TM2/TM6B, respectively.
Site-directed mutagenesis. For per cDNAs constructs, we used the wild-type pET-30(a)-per clone as a template for PCR. Overlapping PCR primers were designed according to the mutated sites and ordered from INTDNA. Platinum Pfx polymerase (Invitrogen, San Diego, CA) was used to generate all PCR fragments. To generate pET-perS59A, we generated the two fragments (the 5′ half and the 3′ half) with the overlapping region at the S59 position. The 5′ half fragment was generated from the start codon with a KpnI site to the S59 region (5′-GATCATGTCGCTGCTCGCGGCCTGCGTCGAGGG-3′). The 3′ half fragment was generated from the S59 region (5′-CTTCTGCTGCGTCATCGCGATGCACGACGGCATC-3′) to the SacI site in the per cDNAs. The two 5′ and 3′ half fragments were used to generate the overlapping PCR KpnI-to-SacI fragment for a replacing cloning to the wild-type per. The mutation clones were confirmed by sequencing (Applied Biosystems, Foster City, CA). Using the same method, we cloned the pET-perS149A (5′-CTCCGACTCGGAGCCGGCTTGATCCTCCTCCTC-3′ and 5′-GAGGAGGAGGATCAAGCCGGCTCCGAGTCGGAG-3′), pET-perS151A (5′-GTCCGCCTCCGACTCGGCGCCGGATTGATCCTC-3′ and 5′-GAGGATCAATCCGGCGCCGAGTCGGAGGCGGAC-3′), pET-perS153A (5′-GACTCGGTCCGCCTCCGCCTCGGAGCCGGATTG-3′ and 5′-CAATCCGGCTCCGAGGCGGAGGCGGACCGAGTC-3′), pET-perS149-151-153A (5′-TCGGTCCGCCTCCGCCTCGGCGCCGGCTTGATCCTCCTC-3′ and 5′-GAGGAGGATCAAGCCGGCGCCGAGGCGGAGGCGGACCGA-3′), pET-perS151-153A (5′-GGAGGAGGATCAGTCCGGCGCCGAGGCGGAGGCGGACCG-3′ and 5′-CGGTCCGCCTCCGCCTCGGCGCCGGACTGATCCTCCTCC-3′), pET-perS151-153D (5′-GAGGAGGAGGATCAATCCGGCGATGAGGATGAGGCGGACCGAGTCGAGGG-3′ and 5′-GCCCTCGACTCGGTCCGCCTCATCCTCATCGCCGGATTGATCCTCCTCCTC-3′), pET-perS245A (5′-GATGCCGTCGTGCATCGCGATGACGCAGCAGAAG-3′ and 5′-CTTCTGCTGCGTCATCGCGATGCACGACGGCATC-3′), pET-perS273A (5′-GTGCACAAAGTCGATGAACGCCCTGCCCAGCCACATG-3′ and 5′-CATGTGGCTGGGCAGGGCGTTCATCGACTTTGTGCAC-3′). For genomic per constructs, we subcloned the XbaI-to-SacI fragment in perG to pGEM7Z (Promega, Madison, WI) and the SacI-to-BamHI fragment in perG to pGEM7Z. We cut the SalI-to-AflII fragment from pET-perS149A to replace the fragment in pGEM7Z bearing the XbaI-to-SacI fragment. We used the same subcloning method for perS151A, perS153A, perS149-151-153A, perS245A, and perS273A. To generate perG bearing the site-directed mutations, we cut the XbaI-to-BamHI fragment from perG and ligated with the XbaI-to-SacI fragment and the SacI-to-BamHI fragment from pGEM7Z. For perG bearing S59A, we used an overlapping PCR to generate the XbaI-to-BamHI fragment with the same primers described previously to replace the fragment in perG.
Bacterial expression and recombinant protein purification. Wild-type CK2α has been described previously (Lin et al., 2002). The wild-type CK2β coding region was amplified (platinum Pfx polymerase; Invitrogen) from clone RE31047 (Invitrogen) and subcloned into pET-30a (Novagen, Madison, WI). The wild-type PER coding region was subcloned into pET-30(a). We also cloned CK2α, CK2β, wild-type PER, and PER mutations into pQE-80L (Qiagen, Hilden, Germany). 6xHis and S tag purifications followed the manufacturers' instructions (Novagen and Qiagen). Expression of glutathione S-transferase (GST)-TIM1-1159 and hamster CK1ϵ were as described previously (Lowrey et al., 2000; Martinek et al., 2001). For copurification of CK2 holoenzyme, we loaded bacterial expressed lysates with CK2α (6xHis tag) into a His-tag affinity purification column and then loaded bacterial expressed lysates with CK2β (6xHis tag). The copurified CK2 holoenzyme was eluted and checked by SDS-PAGE.
Kinase assay. All in vitro kinase reactions were performed at room temperature. The amount of PER, PER mutation, and PER fragments varied from 0.1 to 1 μg (see details in figure legends). We used 0.2 μg of CK2α, 0.2 μg of CK2 holoenzyme, and 0.1 μg of CK1 for kinase experiments for 10, 2, and 30 min, respectively. For all phosphorylation reactions, 100 μl of CK2 kinase assay buffer (50 mm Tris, pH 8.5, 100 mm NaCl, 10 mm MgCl2, 0.5 μl of ATP; 10 mCi/ml) containing recombinant protein was incubated. After the kinase reaction, the proteins were precipitated by trichloroacetic acid to remove free nucleotide, and boiled in 2× SDS gel loading buffer.
For priming phosphorylation assays, 40 μl of S-protein agarose (Novagen) was washed with 500 μl of 1× S-tag buffer (Novagen). Agarose was spun down at 3000 rpm for 30 s to remove washed buffer, and 0.2 μg of 6xHis and S-tagged PER (H-S-Per) was incubated with washed S-protein agarose containing 100 μl of 1 × S-tag buffer end over end for 10 min. Then agarose was spun down, and buffer was removed. Either CK2α (0.2 μg) or CK2 holoenzyme with 1 μl of 10 mm ATP in 100 μl of 1× S-tag and CK2 assay buffer was added to the S-protein agarose for 10 or 2 min, respectively. After this, S-protein agarose was spun down and washed three times with 500 μl of 1× S-tag buffer. One microliter of GSK3 (New England Biolabs, Beverly, MA; 250,000 U/ml) and [γ-32P]ATP (PerkinElmer, Boston, MA; 10 mCi/ml) in 100 μl of 1× GSK3 and CK2 assay buffer was added to S-protein agarose for 20 min. S-protein agarose was spun down and washed three times with 1× S-tag buffer to removed free nucleotide. Then 25 μl of 2× SDS gel loading buffer was added to S-protein agarose and boiled for 5 min, and 25 μl was applied to SDS-PAGE. The gel was stained with Coomassie for autoradiography. GST-TIM1-1159 (0.1 μg) was incubated with same amount of GSK3 for a positive control.
For the in vitro PER and CK2 holoenzyme interaction, 40 μl of S-protein agarose was washed with 500 μl of 1× S-tag buffer. Agarose was spun down at 3000 rpm for 30 s to remove washed buffer. Then 0.1 μg of H-S-PER was incubated with washed S-protein agarose containing 100 μl of 1 × S-tag buffer end over end for 20 min. S-protein agarose was spun down and washed once with 1× S-tag buffer, and 100 μl of 1 × S-tag and CK2 assay buffer containing 1 μl of 10 mm ATP was added to the beads with 0.2 μg of H-CK2 holoenzyme (6xHis-tagged CK2). S-protein agarose was incubated end over end for 2 min. S-protein agarose was spun down and washed three times with 500 μl of 1 × S-tag buffer, and 30 μl of 2× SDS gel loading buffer was added to S-protein agarose and boiled for 5 min. Then 20 μl was applied to SDS-PAGE, and gels were transferred to membranes for Western blotting with anti-CK2 holoenzyme (Dahmus et al., 1984).
Western blotting. Quantification of PER Western blotting was performed as described previously with NIH Image (Lin et al., 2002). Equal loading and transfer were confirmed with Ponceau S staining of membranes. The dilutions for the primary and secondary antibodies were 1:10,000 and 1:2000 in TBS-T buffer (ECL protocol; Amersham Biosciences, Arlington Heights, IL). Five TBS-T buffer washes lasting 5 min each were performed to block the primary and the secondary antibody incubations (5% milk; Bio-Rad, Hercules, CA). The incubation times for blocking the primary and the secondary antibodies were as follows: 1 h at room temperature, overnight at 4°C, and 1 h at room temperature, respectively. ECL reagents were used for immunoassay signals.
Immunofluorescence. Immunostaining was performed as described previously with the exception that images were collected on a Nikon (Tokyo, Japan) C1 confocal microscope, and PER signal was amplified with TSA Kit #15 (Invitrogen, Eugene, OR) (Lin et al., 2002).
Results
Serines 149, 151, and 153 of PERIOD are required for robust phosphorylation by CK2 in vitro
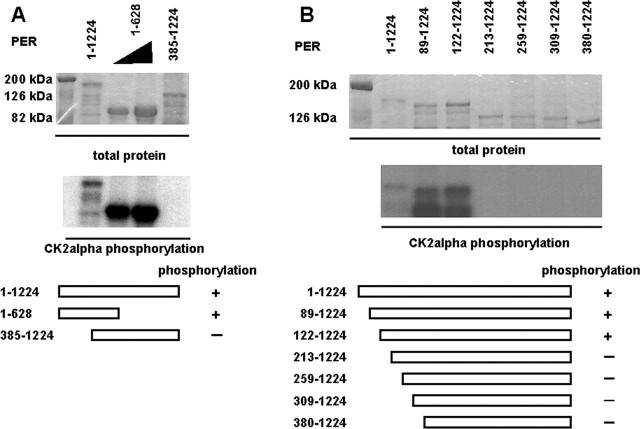
To determine the molecular mechanisms by which CK2 influences PER action, we assayed CK2 phosphorylation of various PER deletion constructs in vitro (Fig. 1). Our findings showed that the regulatory β subunit was dispensable for specific PER phosphorylation in vitro (Lin et al., 2002). As a result, we initially assayed PER phosphorylation by the catalytic α subunit alone. We observed that N-terminal fragments of PER (1-628) were robustly phosphorylated, whereas we could not detect significant PER phosphorylation of fragments containing the C-terminal portion of PER (385-1224) (Fig. 1A). We then generated and tested a series of N-terminal deletions that removed increasing parts of PER (Fig. 1B). These studies showed robust phosphorylation of N-terminal deletions including 122-1224 but lack of phosphorylation of deletions from 213 to 1224 and beyond (Fig. 1B). These results suggested that the relevant in vitro CK2 sites might be between amino acids 122 and 213.
Figure 1.
Mapping CK2 in vitro phosphorylation sites in PERIOD by deletion analysis. The top panels show Coomassie-stained recombinant PER fragments as a control for protein amount. The molecular mass of markers is indicated. The middle panels indicate in vitro phosphorylation of PER fragments by CK2α, and the bottom panel is a schematic of summarized results. A, Two fragments of recombinant PER (shown in the diagram at the bottom) were assayed for in vitro phosphorylation by CK2α. Only PER1-628 is phosphorylated. The full-length PER1-1224 and PER 385-1224 are used at 0.2 μg. PER1-628 varies from 0.5 to 1 μg. B, N-terminal PER deletions were assayed for phosphorylation: only the first two deletion fragments become phosphorylated. Equivalent amounts (0.2 μg) were used for all fragments.
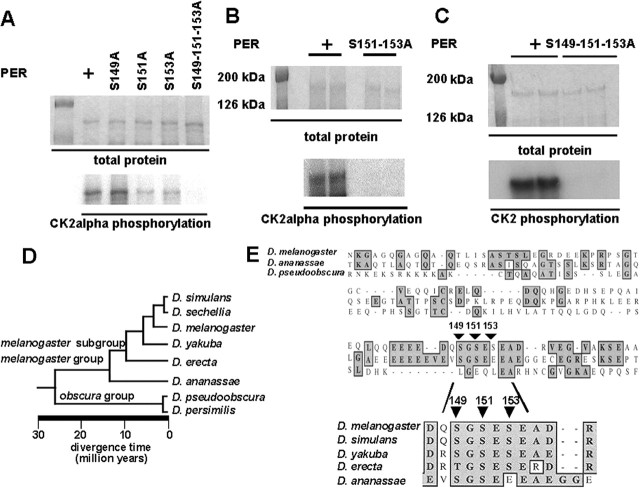
Using a web-based phosphorylation site program, Phosphobase (Blom et al., 1998), we searched this region (PER122-213) for CK2 consensus sites. We identified two consensus CK2α sites at Ser149 and Ser153. Between these two serines was a third serine (Ser151) that also contained an acidic residue at the +3 position, suggesting that it may also represent a CK2 site. We mutated this serine cluster to alanines and found that it significantly blocked phosphorylation by CK2α (Fig. 2A). We then mutated them individually but observed reductions only in phosphorylation with single Ser151 and 153 mutations and nearly complete loss with the double Ser151-153 double mutant (Fig. 2A,B).
Figure 2.
Serines 149, 151, and 153 are required for in vitro phosphorylation by CK2. A, Each of three Ser (S149A, S151A, and S153A) and all three Ser residues (S149-151-153A) were mutagenized to alanine in full-length PER. Each PER mutant was assayed for CK2α phosphorylation. The top panel shows Coomassie-stained PER as a protein amount control. The bottom panel shows that the phosphorylation of PERS149A by CK2α compared with PER is similar, and the phosphorylation of PERS151A and PERS153A by CK2α compared with PER is reduced. Phosphorylation is not observed for PERS149-151-153A by CK2α compared with PER; 0.2 μg of PER were used. B, PERS151-153A was assayed for phosphorylation against CK2α. The top panel shows Coomassie-stained PER and PERS151-153A as a protein amount control. The bottom panel shows that PER phosphorylation by CK2α and PERS151-153A phosphorylation by CK2α is not observed. Duplicate experiments were performed; 0.1 μg of PER was used. C, PERS149-151-153A was assayed for phosphorylation against CK2 holoenzyme. The top panel shows Coomassie-stained PER and PERS149-151-153A as a protein amount control. The bottom panel shows that PER phosphorylation by CK2 holoenzyme and PERS149-151-153A phosphorylation by CK2 are dramatically reduced. Duplicate experiments were performed; 0.1 μg of PER was used. D, The phylogenetic relationships of divergence of eight Drosophila species separated by >30 million years (adapted from http://bugbane.bio.indiana.edu:7151). E, The PERS149, -S151, and -S153 of D. melanogaster were aligned with another five Drosophila species with the CLUSTAL W algorithm of the MacVector 7.2 Package. The top panel shows a low similarity between D. melanogaster and D. pseudoobscura in the region close to S149, S151, and S153 indicated by arrows. D. melanogaster and D. ananassae are similar in this cluster but less so in the surrounding region. The bottom panel shows an inset from the top panel in which S149, S151, and S153 are highly conserved in D. melanogaster, D. simulans, D. yakuba, D. erecta (T149 instead of S149), and D. ananassae (S153E change). Of note, the CK2 potential phosphorylation consensus sequence is also highly conserved.
These preliminary studies relied on phosphorylation by the α subunit alone. Previous studies have established an in vivo function for the β subunit in circadian clock function (Akten et al., 2003), suggesting that the CK2 holoenzyme represents a more physiological state for the kinase. Thus, we wanted to determine whether these serine mutations also affected phosphorylation by the CK2 holoenzyme. We prepared holoenzyme by copurification of the α and β subunits and found that it robustly phosphorylated PER (Fig. 2B). Indeed, we observed substantial increases in phosphorylation over that observed with the α subunit alone (data not shown), as has been observed for other CK2 substrates; however, this holoenzyme phosphorylation was also dramatically reduced in the Ser149-151-153 triple mutant (Fig. 2C). Thus, these serines are also required for phosphorylation by the CK2 holoenzyme.
We reasoned that important functional phosphorylation sites might be highly conserved. To assess the potential in vivo role of these in vitro sites, we performed an alignment of the Drosophila melanogaster PER sequence including Ser149-151-153 with other closely related Drosophila species, some of which are thought to have been separated from D. melanogaster by ∼25 million years (Fig. 2D). First, we noted that these serines (CK2 consensus site = S/T-X-X-D/E) do not appear to be present in D. pseudoobscura (Fig. 2E) or in mammals, consistent with a previous designation that this domain is part of a poorly conserved region (Colot et al., 1988); however, this cluster is unaltered in the most closely related species, D. yakuba and D. simulans. It should be noted that there is a discrepancy between a reported yakuba sequence, which is identical at serine 153 (Piccin et al., 2000), and that indicated in the DroSpeGe database that shows a serine 153 to leucine change, perhaps because of a sequencing error or polymorphism. In the species that are intermediate in distance between these two groups, substitutions of these sequences are suggestive of a function in phosphorylation. In D. erecta, melanogaster serine 149 is changed to another potential phosphorylation substrate, threonine, retaining its CK2 site. In D. ananassae, serine 149 is intact but serine 153 is changed to glutamate E, a residue that mimics a phosphorylated amino acid. Moreover, if one compares the melanogaster and ananassae over a wider sequence, one can observe that this serine cluster appears to be an island of conservation in a less well conserved region. On the basis of these sequence comparisons and in vitro data, we suggest that this conservation reflects an underlying role as an in vivo kinase substrate.
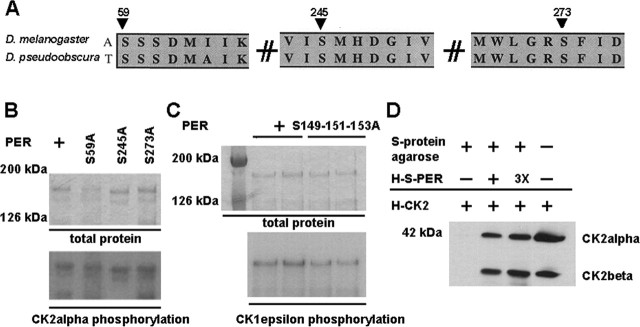
In the N-terminal region of PER, we did identify other well conserved CK2 consensus sites in the N terminus, including Ser59, Ser245, and Ser259 (Fig. 3A). Given their conservation, we thought that they might be functional. To determine whether these sites were also essential for CK2 phosphorylation, we mutated them to alanine and assayed the ability of CK2 to phosphorylate. Unlike Ser149-151-153, we found that mutation of these residues did not affect phosphorylation by CK2 in vitro (Fig. 3B). These data suggest that they are not CK2 substrates, although their conservation may nonetheless reflect some function in PER regulation.
Figure 3.
Conserved PER serines are not required for CK2 phosphorylation and serines 149, 151, and 153 are not required for phosphorylation by CK1ϵ or interactions with CK2 holoenzyme. A, Conserved S59, S245, and S273 are indicated by arrows for D. melanogaster and D. pseudoobscura. B, Ser59, Ser245, or Ser273 was mutagenized to Ala and assayed for phosphorylation by CK2α. The top panel shows Coomassie-stained PER and mutagenized PER as a protein amount control. The bottom panel shows that all CK2α phosphorylation of PER is similar; 0.1 μg of PER was used. C, The top panel shows Coomassie-stained PER and PERS149-151-153A as a protein amount control. The bottom panel shows that there is no significant change for the phosphorylation of PER and PERS149-151-153A by CK1ϵ; 0.1 μg of PER was used. D, Western blot of pull down of CK2 holoenzyme (α and β) by PER shows that the interaction of PER and PER S149-151-153A with CK2 holoenzyme is similar. PER and PERS149-151-153A (3X) were fused to 6xHis and S tags indicated as H-S-PER. The tag-fused PER was bound to S-protein agarose and tested for pull down of 6xHis-tagged CK2 holoenzyme (α and β subunits). Bands indicate CK2α and -β subunits recognized by anti-CK2 holoenzyme. The molecular mass of markers is indicated.
We were concerned that S149-151-153A mutations might only secondarily affect phosphorylation by CK2 by primarily affecting protein folding, rendering PER a poor kinase substrate. To test this hypothesis, we assayed wild-type and S149-151-153A phosphorylation by CK1ϵ. Previous studies have shown a function for CK1ϵ, the closely related CK1δ, and their fly ortholog, DBT, in PER phosphorylation (Kloss et al., 1998; Price et al., 1998; Lowrey et al., 2000; Lee et al., 2001); however, previous studies have failed to show kinase activity with recombinant fly DBT (Kloss et al., 1998; Suri et al., 2000) and thus have resorted to vertebrate orthologs (Preuss et al., 2004). Here we assayed PER phosphorylation with the hamster ortholog mutated in the short-period tau mutant (Lowrey et al., 2000). We found that hamster CK1ϵ robustly labeled Drosophila PER as well as the triple mutant, indicating that it remains a good kinase substrate and that the effects of the serine mutations are specific to CK2 (Fig. 3C).
Another possibility is that this serine cluster is required for PER-CK2α interactions that in turn are required for phosphorylation elsewhere in the protein. Using an in vitro pull-down assay, we detected stable CK2 holoenzyme-PER interactions in vitro, consistent with the proposed role of PER as a CK2 substrate (Fig. 3D). Moreover, the strength of this interaction was not significantly affected by the Ser149-151-153 triple mutation. Together, these results are most consistent with in vitro phosphorylation of Ser149-151-153 by CK2.
Serines 149, 151, and 153 are required for wild-type circadian period
To assess the in vivo role of serines 149, 151, and 153, we mutated them to alanine and assayed their circadian function. Previous studies have shown full rescue of arrhythmic per01 mutants with a 13.2 kb genomic per transgene (Zehring et al., 1984; Hamblen et al., 1986; Cheng et al., 1998). We mutated this transgene using site-directed mutagenesis singly at Ser149 (S149A), Ser151 (S151A), and Ser153 (S153A), and all three serines mutated to alanine (S149-151-153A) and assayed behavioral rescue of per01. To compare the in vivo function of in vitro and evolutionarily defined CK2 sites in PER, we also mutated the conserved Ser59 to alanine and assayed its function in vivo. Ser59 appears to be widely conserved from flies to mammals (Fig. 3A) (data not shown). For each mutation, multiple independent lines were assayed as heterozygotes (single transgene copy). The average of multiple independent inserts for each line was then used to obtain an average for each transgene to control for positional effect. These mutant lines were compared with two strains of per01 flies harboring two independent rescuing genomic per fragments (perG). In most cases, robust rescue of rhythmicity was observed. In exceptional lines in which rhythmicity was poor, period assessments were deemed unreliable and therefore not used for overall period calculations. We found that the S149-151-153A mutations resulted in periods that were ∼2 h longer than perG controls (Fig. 4, Table 1). Interestingly, single mutations of Ser149, -151, and -153 as well as the double mutation S151-153A also resulted in similarly long periods. Paralleling the in vitro results, mutation of Ser59 did not result in a significant circadian rhythm phenotype (Table 1). We also examined the behavioral phenotypes of these mutants in CK2αTik (Tik) heterozygous mutants, in which catalytic activity is reduced by ∼50% (Lin et al., 2002). That period is still additionally lengthened in these mutants (Table 1), suggesting that there may be additional CK2 phosphorylation sites in PER or other clock genes. Nonetheless, these data suggest that these potential phosphorylation sites, Ser149-151-153, act as a unit, wherein mutation of any site within the cluster effectively knocks out its function.
Figure 4.
Circadian locomotor activity of transgenic per and per mutant flies. A, The locomotor activity for transgenic wild-type per (perG) fly in a per0 background. For visual continuity, 2 d are plotted per row. Flies were entrained for 2-5 d in a 12 h light/dark cycle and subsequently maintained in constant darkness for the locomotor analysis. Open bar indicates 12 h light/dark cycle; closed bar indicates constant darkness. B, The locomotor activity for transgenic PERS153A (perS153A) fly in per0 background. C, The locomotor activity for transgenic PERS149-151-153A (perS149-151-153A) fly in per0 background.
Table 1.
Circadian locomotor activity for transgenic PER mutant flies
|
Genotype |
Period (h) ± SEM |
Rhythmic% (n) |
|---|---|---|
| per0;;perG(27)/+ | 23.8 ± 0.1 | 92 (48) |
| per0;perG(32)/+ | 23.6 ± 0.1 | 93 (46) |
| Average (2) | 23.7 ± 0.1 | 93 (47) |
| per0;;perG(27)/Tik | 25.8 ± 0.1 | 98 (60) |
| per0;perG(32)/+;Tik/+ | 26.2 ± 0.1 | 97 (60) |
| Average (2) | 26.0 ± 0.1 | 98 (60) |
| per0;perS59A(f37)/+ | 23.6 ± 0.1 | 100 (37) |
| per0;perS59A(m10)/+ | 23.8 ± 0.1 | 100 (36) |
| per0;perS59A(m16)/+ | 23.8 ± 0.1 | 91 (35) |
| per0;;perS59A(m17)/+ | 24.5 ± 0.1 | 97 (36) |
| Average (4) | 23.9 ± 0.1 | 97 (36) |
| per0;;perS149A(6)/+ | 25.2 ± 0.1 | 97 (36) |
| per0;perS149A(40)/+ | 25.0 ± 0.1 | 100 (36) |
| per0;;perS149A(53)/+ | 25.3 ± 0.1 | 92 (36) |
| per0;;perS149A(54)/+ | 24.9 ± 0.1 | 90 (31) |
| per0;perS149A(70)/+ | 24.9 ± 0.1 | 94 (36) |
| per0;perS149A(72)/+ | 25.4 ± 0.1 | 100 (36) |
| per0;;perS149A(73)/+ | 25.2 ± 0.1 | 83 (24) |
| Average (7) | 25.1 ± 0.1 | 94 (34) |
| per0;perS151A(11)/+ | 25.4 ± 0.1 | 98 (48) |
| per0;perS151A(15)/+ | 25.1 ± 0.1 | 96 (48) |
| per0;perS151A(18)/+ | 25.4 ± 0.1 | 100 (36) |
| Average (3) | 25.3 ± 0.1 | 98 (44) |
| per0;perS153A(33)/+ | 25.3 ± 0.1 | 93 (60) |
| per0;perS153A(75)/+ | 25.1 ± 0.1 | 100 (35) |
| per0;perS153A(77)/+ | 26.9 ± 0.1 | 97 (35) |
| average (3) | 25.7 ± 0.1 | 97 (43) |
| per0;perS151-153A(f2)/+ | 25.3 ± 0.1 | 96 (24) |
| per0;perS151-153A(f4)/+ | 26.7 ± 0.2 | 92 (24) |
| per0;;perS151-153A(f28)/+ | 25.4 ± 0.1 | 100 (12) |
| Average (3) | 25.8 ± 0.1 | 96 (20) |
| per0;perS151-153A(f2)/+;Tik/+ | 27.8 ± 0.1 | 100 (12) |
| per0;perS151-153A(f4)/+;Tik/+ | 29.1 ± 0.1 | 100 (12) |
| Average (2) | 28.5 ± 0.1 | 100 (12) |
| per0;;perS151-153D(f15)/+ | 25.1 ± 0.1 | 91 (11) |
| per0;;perS151-153D(f33)/+ | 25.1 ± 0.1 | 90 (20) |
| per0;;perS151-153D(f36)/+ | 24.6 ± 0.2 | 83 (12) |
| per0;perS151-153D(f39)/+ | 25.0 ± 0.1 | 90 (21) |
| per0;;perS151-153D(m8)/+ | 25.2 ± 0.1 | 100 (13) |
| per0;perS151-153D(m12)/+ | 25.3 ± 0.1 | 92 (24) |
| per0;perS151-153D(m48)/+ | 25.3 ± 0.2 | 75 (12) |
| Average (7) | 25.1 ± 0.1 | 89 (16) |
| per0;perS149-153A(40)/+ | 25.3 ± 0.0 | 98 (48) |
| per0;perS149-151-153A(A)/+ | 25.5 ± 0.1 | 88 (48) |
| per0;;perS149-151-153A(B)/+ | 25.3 ± 0.1 | 96 (48) |
| Average (3) | 25.4 ± 0.1 | 94 (48) |
| per0;perS149-151-153A(40)/+;Tik/+ | 27.7 ± 0.1 | 100 (29) |
| per0;perS149-151-153A(A)/+;Tik/+ | 27.8 ± 0.1 | 100 (48) |
| per0;;perS149-151-153A(B)/Tik | 27.7 ± 0.1 | 100 (36) |
| Average (3) |
27.7 ± 0.1 |
100 (38) |
The number of flies analyzed is indicated in parentheses in the final column. For each genotype, the number in the parentheses indicates an independent transgenic fly line. For each genotype average, the number in the parentheses indicates the total independent fly lines that were tested. Rhythmic% (n) refers to the percentage of flies that were scored as rhythmic (see Materials and Methods).
Given the strong in vitro effects of S151-153A (Fig. 2B), we mutated these residues to the phosphomimetic aspartate (S151-153D) and assayed the consequences on behavior. We found that mutation to aspartate also results in period lengthening (Table 1). The interpretation of these results is further complicated by CK2α overexpression studies. To assay the consequences of CK2α expression on the circadian clock, we used the GAL4/UAS system (Brand and Perrimon, 1993). By using different pacemaker neuron drivers, we found that CK2α overexpression results in long periods (Table 2). These results suggest that both increases and decreases in CK2 activity may result in longer periods. Similar results have been observed with gain- and loss-of-function of per (Yang and Sehgal, 2001). In addition, Dbt mutants that result in reduced catalytic activity can have both short and long period phenotypes (Preuss et al., 2004). If phosphomimetic (e.g., Ser to Asp) and phosphoinactive (e.g., Ser to Ala) mutations mimic gain- and loss-of-function CK2 mutants, then they both may have long periods, as we have observed. Nonetheless, the aspartate mutants generally have shorter periods than the corresponding alanine mutants (average 25.1 vs 25.8 h) (Table 1), perhaps abrogating the alanine mutant effects, although these effects do not reach statistical significance (p = 0.25).
Table 2.
Circadian locomotor Activity for CK2α-overexpressing flies
|
Genotype |
Period (h) ± SEM |
Rhythmic% (n) |
|---|---|---|
| pdfGAL4/+ | 24.4 ± 0.1 | 100 (8) |
| timGAL4/+ | 24.5 ± 0.2 | 88 (8) |
| timUASGAL4/+ | 24.2 ± 0.1 | 88 (8) |
| UASCK2α(33)/+ | 24.1 ± 0.1 | 88 (16) |
| pdfGAL4/+;UASCK2α(33)/+ | 25.2 ± 0.1* | 91 (22) |
| timGAL4/+;UASCK2α(33)/+ | 25.4 ± 0.1* | 75 (16) |
| timUASGAL4/+;UASCK2α(33)/+ | 24.9 ± 0.1* | 94 (16) |
| UASCK2α(35)/+ | 24.1 ± 0.1 | 100 (24) |
| pdfGAL4/UASCK2α(35) | 25.7 ± 0.2* | 81 (16) |
| timGAL4/UASCK2α(35) | 25.9 ± 0.2* | 87 (15) |
| timUASGAL4/UASCK2α(35) | 25.5 ± 0.2* | 75 (16) |
| UASCK2α(47)/+ | 24.1 ± 0.1 | 88 (16) |
| pdfGAL4/+;UASCK2α(47)/+ | 25.5 ± 0.1* | 79 (24) |
|
timGAL4/+;UASCK2α(47)/+ |
25.4 ± 0.2*
|
96 (24) |
p < 0.005 versus GAL4 and UAS controls.
Our in vitro data suggest that mutation Ser149, unlike Ser151 or Ser153, does not affect CK2 phosphorylation (Fig. 2A), yet mutation to alanine does result in a circadian behavioral phenotype. To determine whether this serine might be the target of other kinases, we used Phosphobase to determine whether other kinases might phosphorylate this residue. We found that this site also represents a consensus site for GSK3, a kinase previously reported to phosphorylate TIM (Martinek et al., 2001). GSK3 often recognizes sites that have been previously phosphorylated at a position four amino acids C-terminal to the GSK3 site. In this case, that position (Ser153) represents a potential CK2 phosphorylation site, suggesting that CK2 phosphorylation of Ser153 might prime phosphorylation of Ser149. Indeed, previous studies have shown that CK2 can prime phosphorylation by GSK3 (Picton et al., 1982); however, we were unable to show priming, at least in vitro (data not shown). It will be of interest to examine potential in vivo interactions between CK2 and GSK3 mutants.
Serines 149, 151, and 153 are required for intact PER rhythms of expression and nuclear entry
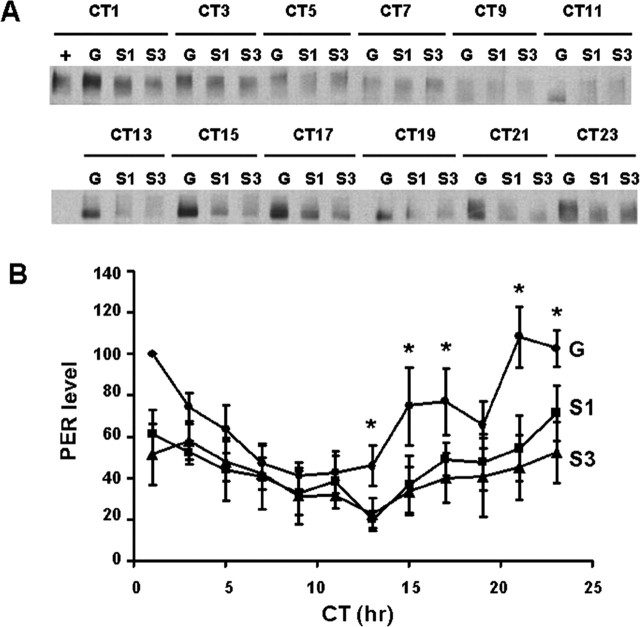
Given the effects of mutation of Ser149-151-153 on circadian behavior, we wanted to assess the effect on PER rhythms as assayed by Western blotting (Fig. 5). We assayed perG, perS153A, and perS149,151,153A mutants as heterozygotes in a per01 background under constant darkness, where CK2 mutant effects on PER were observed previously (Lin et al., 2002; Akten et al., 2003). Here we observe that these mutants result in reduced amplitude rhythms relative to wild-type perG (Fig. 5). Changes are most evident during the accumulation phase; however, if we normalize peaks to 100 for all genotypes, these differences are lost, suggesting that they can essentially be attributed to level differences among the strains (data not shown). The single (Ser153A) and triple mutants (Ser149-151-153A) showed essentially indistinguishable temporal patterns of expression from each other, consistent with their similar behavioral periods. Although rhythms of PER levels were affected, we did not observe any significant change in mobility, a reflection of gross phosphorylation state. Mobility differences in PER have been observed previously in CK2α mutants (Lin et al., 2002), suggesting that the triple mutant may affect only a subset of CK2 sites on PER. It should be mentioned that CK2β mutants that show a similar magnitude of circadian period phenotype as these serine mutants do not have a demonstrable effect on gross PER mobility (Akten et al., 2003). Nonetheless, these results indicate that these PER mutations do affect PER metabolism.
Figure 5.
Effects of per mutants on PER rhythms. A, Western blot of PER. CT indicates circadian time; CT0 is 12 h after lights off. G, PER wild-type; S1, PERS153A; S3, PERS149-151-153A. B, Quantification of PER Western blots. The PER rhythms were altered for PERS 153A (filled squares) and PERS 149-151-153A (filled triangles) compared with wild-type PER (filled circles), especially for changes during the accumulation phase. Each point is the average of three independent experiments. Error bars indicate SEM. t test; p < 0.05.
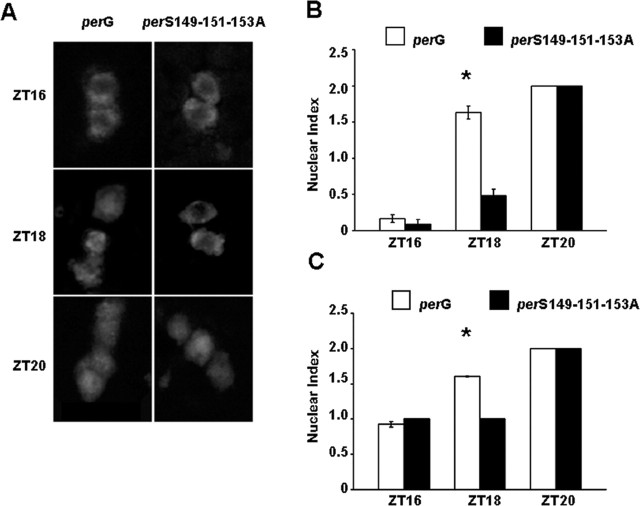
Previous studies of CK2 mutants also showed delays in the timing of PER nuclear entry. A core set of pacemaker neurons, the ventral group of lateral neurons (LNvs), are especially important for circadian behavior and express PIGMENT DISPERSING FACTOR (PDF), a key neuropeptide (Helfrich-Forster, 1995; Park and Hall, 1998; Renn et al., 1999; Park et al., 2000). We assayed PER subcellular localization in the PDF+ core pacemaker neurons in the adult brain during 12 h light/dark conditions in perG and perS149-151-153A mutants (Fig. 6). Consistent with previous reports in wild-type flies, we observed cytoplasmic PER at Zeitgeber time (ZT) 16 (ZT 0 = lights on; ZT 12 = lights off), mixed-nuclear staining at ZT 18, and nuclear staining at ZT 20. In perS149-151-153A mutants, staining remains cytoplasmic at ZT 18 and becomes nuclear by ZT 20. This ∼2 h shift relative to perG is comparable with the period difference between these two strains. perG and perS149-151-153A mutants both entrain to 12 light/dark cycles (data not shown); thus, any difference between the two strains cannot be attributed solely to differences in the free-running period. Similar to CK2α and -β mutants, the timing of PER nuclear localization is delayed (Lin et al., 2002; Akten et al., 2003).
Figure 6.
S149-151-153A nuclear translocation is delayed by ∼2 h compared with PerG control. A, Immunofluorescence. PerG and S149-151-153A flies in a per0 background were entrained to 12 h light/dark cycle. Brains were dissected at ZT 16, ZT 18, and ZT 20, fixed, and stained for PER. Small LNvs were imaged by confocal microscopy. At ZT 18, S149-151-153A remains mostly cytoplasmic, whereas PerG has predominantly translocated to the nucleus. By ZT 20, both PerG and S149-151-153A have localized to the nucleus. B, C, Quantification of immunocytochemistry experiment with a nuclear index. Cells with predominantly cytoplasmic PER localization were assigned a value of 0; cells with PER protein localized in both the cytoplasm and nucleus were assigned a value of 1; and cells with per localization in the nucleus were assigned a value of 2. Nuclear index values from two replicates were averaged, and the SE and t test value were calculated to determine significance. For perG, the number of sLNv/lLNvs analyzed = 48/54 (ZT 16), 30/33 (ZT 18), and 61/80 (ZT 20). For perS149-151-153A, the number of sLNv/lLNvs analyzed = 22/29 (ZT 16), 29/28 (ZT 18), and 42/69 (ZT 20). B, Quantification of small lateral neurons. C, Quantification of large lateral neurons. Lack of error bars indicates uniform results for the entire time point. *p < 0.05.
Discussion
Here we provide evidence for a function for PER CK2 phosphorylation sites in circadian clock function and behavior in Drosophila. We initially identified these phosphorylation sites in vitro, and site-directed mutagenesis studies focused our attention on three key serines in the N terminus required for robust in vitro phosphorylation of PER by CK2 α and the CK2 holoenzyme (Figs. 1, 2). This mutant form is still phosphorylated by another kinase, CK1ϵ, and is still able to directly interact with CK2, suggesting that these serines represent in vitro phosphorylation targets for CK2. Mutation of these sites in vivo resulted in a long circadian period, altered PER rhythms, and delayed PER nuclear entry. Overall, these phenotypes significantly overlap with those observed previously in CK2 mutants.
These data contribute significantly to the role of phosphorylation in the regulation of core clock components and in turn the effects on circadian behavior. Despite considerable progress in defining clock components and clock kinases, remarkably little is known about the target phosphorylation sites essential for their function, especially in metazoans. Recent work has defined key serines that are important for CK1ϵ regulation of mouse PER nuclear localization and phosphorylation in cultured cell lines (Takano et al., 2004). Although they defined critical serines, these studies were not performed in the context of a functioning circadian clock. Thus, it is not known whether these sites are essential for clock function. Second, because the studies were performed in cultured cells, it is not clear whether these putative phosphorylation sites will be important for circadian behavior. In contrast, we have shown functioning of critical serine targets of CK2 in an in vivo functioning circadian system and on circadian behavior.
Although these data argue for a role for CK2 in the phosphorylation of Ser151-153, they cannot exclude the effects of CK2 elsewhere on the PER protein or on other proteins in the circadian clock. We have identified several potential CK2 sites in PER and in other clock proteins using Phosphobase, raising the possibility that CK2 may work through multiple sites on PER (data not shown). Our in vitro conditions (e.g., absence of TIM) may not fully replicate the in vivo situation and thus miss these other important sites. CK2 action also may not occur exclusively through PER. We reported previously in vitro phosphorylation of TIM by CK2α as well as effects on in vivo TIM rhythms (Lin et al., 2002). Thus, CK2 may affect multiple targets to regulate clock function.
Consistent with this hypothesis, we observe period lengthening of perS149-151-153A mutants in a Tik mutant background (Table 1). Under natural conditions, we propose that some fraction of these serines is phosphorylated. In the Tik mutant, in which catalytic activity is ∼50% of wild type (Lin et al., 2002), we expect that this fraction is reduced but not absent both at this cluster and at other clock-relevant CK2 sites. In the S149-151-153A mutants, these residues cannot be phosphorylated, a more severe situation than that seen in wild type or even Tik. In wild type, normal CK2 phosphorylation elsewhere in PER or in other clock components partially compensates for the loss of these phosphorylation sites. Nonetheless, when CK2 is impaired, as in Tik mutants, we still observe an additional period lengthening caused by the combination of a complete loss of phosphorylation of these key sites by alanine mutation and reduction of clock-relevant CK2 phosphorylation elsewhere.
Moreover, these data do not exclude a function for other kinases in the phosphorylation of Ser149-151-153. Serine 149 also represents a potential GSK3/SGG site. Although our in vitro data fail to show GSK3 phosphorylation at this site, it is possible that it is a true in vivo target. Given the similarity of perS149-151-153A and CK2 mutant phenotypes, we propose that Ser149-151-153 represents one cluster of these in vivo phosphorylation sites that is phosphorylated, at least, by CK2.
Based on our findings, we propose that the distinct CK2 sites present in D. melanogaster PER but not in other closely related species, such as D. pseudoobscura, may represent the basis for species-specific aspects of circadian function. Such variation has been proposed to underlie the process of allochronic speciation (Tauber et al., 2003). Temporal isolation through the use of species-specific circadian programs may serve as a barrier to gene flow and thus facilitate speciation. Indeed, species-specific differences in locomotor activity and mating behavior have been observed between D. melanogaster and other Drosophila species (Petersen et al., 1988; Sakai and Ishida, 2001; Tauber et al., 2003). By using cross-species rescue of per01 behavior, many of these changes have been attributed to variation in the per gene, i.e., the species of per determines the nature of diurnal, circadian, and/or mating behavior (Petersen et al., 1988; Tauber et al., 2003).
We propose that variation in phosphorylation sites may underlie these species differences in behavior. A comparison of different Drosophila PERs (Fig. 2D) reveals that Ser149-151-153 is part of one of the nonconserved modules of PER (Colot et al., 1988). This small region within the larger module is well conserved only within the melanogaster subgroup. Of note, this cluster is conserved in D. ananassae, in which the remainder of the module is not as well conserved, consistent with an underlying function (Fig. 2D). The single change of Ser153 to glutamate, a phosphomimetic residue, is also consistent with a function in phosphorylation.
Species specificity of clock function has also been noted at the molecular level in terms of PER nuclear localization. Of note, PER is predominantly cytoplasmic in the putative pacemaker neurons of the silkmoth (Antherea pernyi) (Levine et al., 1995), beetle (Frisch et al., 1996), and hawkmoth (Manduca sexta) (Wise et al., 2002). It is interesting that the CK2 sites described here are also not conserved with Antherea PER, and mutation of these serines alters PER nuclear entry, a process apparently present in melanogaster but not the silkmoth. We propose that nuclear entry triggered by CK2 phosphorylation of PER may be a species-specific aspect of the clock program.
Footnotes
This work was funded by National Institutes of Health Grant R01 MH067870. We thank Paul Hardin for the per01; perG flies and 13.2 kb per genomic clone; the CBRC Transgenic Drosophila Core for performing embryo injections; M. Young for GST-TIM1-1159; J. Takahashi for an expression clone for hamster CK1ϵ; DroSpeGe website (bugbane.bio.indiana.edu) for period sequences for other Drosophila species; and M. Rosbash and C. Glover for antibodies. We thank Rob Jackson for independently pointing out potential CK2 and GSK3 sites in PER.
Correspondence should be addressed to Ravi Allada, Department of Neurobiology and Physiology, Northwestern University, Evanston, IL 60208. E-mail: r-allada@northwestern.edu.
Copyright © 2005 Society for Neuroscience 0270-6474/05/2511175-09$15.00/0
References
- Akten B, Jauch E, Genova GK, Kim EY, Edery I, Raabe T, Jackson FR (2003) A role for CK2 in the Drosophila circadian oscillator. Nat Neurosci 6: 251-257. [DOI] [PubMed] [Google Scholar]
- Bao S, Rihel J, Bjes E, Fan JY, Price JL (2001) The Drosophila double-time S mutation delays the nuclear accumulation of period protein and affects the feedback regulation of period mRNA. J Neurosci 21: 7117-7126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom N, Kreegipuu A, Brunak S (1998) PhosphoBase: a database of phosphorylation sites. Nucleic Acids Res 26: 382-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401-415. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Gvakharia B, Hardin PE (1998) Two alternatively spliced transcripts from the Drosophila period gene rescue rhythms having different molecular and behavioral characteristics. Mol Cell Biol 18: 6505-6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot HV, Hall JC, Rosbash M (1988) Interspecific comparison of the period gene of Drosophila reveals large blocks of non-conserved coding DNA. EMBO J 7: 3929-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin KD, Huang ZJ, Rosbash M (1995) Temporally regulated nuclear entry of the Drosophila period protein contributes to the circadian clock. Neuron 14: 365-372. [DOI] [PubMed] [Google Scholar]
- Dahmus GK, Glover CV, Brutlag DL, Dahmus ME (1984) Similarities in structure and function of calf thymus and Drosophila casein kinase II. J Biol Chem 259: 9001-9006. [PubMed] [Google Scholar]
- Edery I, Zwiebel LJ, Dembinska ME, Rosbash M (1994) Temporal phosphorylation of the Drosophila period protein. Proc Natl Acad Sci USA 91: 2260-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch B, Fleissner G, Brandes C, Hall JC (1996) Staining in the brain of Pachymorpha sexguttata mediated by an antibody against a Drosophila clock-gene product: labeling of cells with possible importance for the beetle's circadian rhythms. Cell Tissue Res 286: 411-429. [DOI] [PubMed] [Google Scholar]
- Garceau NY, Liu Y, Loros JJ, Dunlap JC (1997) Alternative initiation of translation and time-specific phosphorylation yield multiple forms of the essential clock protein FREQUENCY. Cell 89: 469-476. [DOI] [PubMed] [Google Scholar]
- Hamblen M, Zehring WA, Kyriacou CP, Reddy P, Yu Q, Wheeler DA, Zwiebel LJ, Konopka RJ, Rosbash M, Hall JC (1986) Germ-line transformation involving DNA from the period locus in Drosophila melanogaster: overlapping genomic fragments that restore circadian and ultradian rhythmicity to per0 and permutants. J Neurogenet 3: 249-291. [DOI] [PubMed] [Google Scholar]
- Hardin PE (2004) Transcription regulation within the circadian clock: the E-box and beyond. J Biol Rhythms 19: 348-360. [DOI] [PubMed] [Google Scholar]
- Harms E, Kivimae S, Young MW, Saez L (2004) Posttranscriptional and posttranslational regulation of clock genes. J Biol Rhythms 19: 361-373. [DOI] [PubMed] [Google Scholar]
- Helfrich-Forster C (1995) The period clock gene is expressed in central nervous system neurons which also produce a neuropeptide that reveals the projections of circadian pacemaker cells within the brain of Drosophila melanogaster Proc Natl Acad Sci USA 92: 612-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter-Ensor M, Ousley A, Sehgal A (1996) Regulation of the Drosophila protein timeless suggests a mechanism for resetting the circadian clock by light. Cell 84: 677-685. [DOI] [PubMed] [Google Scholar]
- Kloss B, Price JL, Saez L, Blau J, Rothenfluh A, Wesley CS, Young MW (1998) The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iepsilon. Cell 94: 97-107. [DOI] [PubMed] [Google Scholar]
- Kloss B, Rothenfluh A, Young MW, Saez L (2001) Phosphorylation of period is influenced by cycling physical associations of double-time, period, and timeless in the Drosophila clock. Neuron 30: 699-706. [DOI] [PubMed] [Google Scholar]
- Lee C, Parikh V, Itsukaichi T, Bae K, Edery I (1996) Resetting the Drosophila clock by photic regulation of PER and a PER-TIM complex. Science 271: 1740-1744. [DOI] [PubMed] [Google Scholar]
- Lee C, Bae K, Edery I (1998) The Drosophila CLOCK protein undergoes daily rhythms in abundance, phosphorylation, and interactions with the PER-TIM complex. Neuron 21: 857-867. [DOI] [PubMed] [Google Scholar]
- Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM (2001) Posttranslational mechanisms regulate the mammalian circadian clock. Cell 107: 855-867. [DOI] [PubMed] [Google Scholar]
- Levine JD, Sauman I, Imbalzano M, Reppert SM, Jackson FR (1995) Period protein from the giant silkmoth Antheraea pernyi functions as a circadian clock element in Drosophila melanogaster Neuron 15: 147-157. [DOI] [PubMed] [Google Scholar]
- Lin JM, Kilman VL, Keegan K, Paddock B, Emery-Le M, Rosbash M, Allada R (2002) A role for casein kinase 2a in the Drosophila circadian clock. Nature 420: 816-820. [DOI] [PubMed] [Google Scholar]
- Lowrey PL, Shimomura K, Antoch MP, Yamazaki S, Zemenides PD, Ralph MR, Menaker M, Takahashi JS (2000) Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science 288: 483-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinek S, Inonog S, Manoukian AS, Young MW (2001) A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell 105: 769-779. [DOI] [PubMed] [Google Scholar]
- Meggio F, Pinna LA (2003) One-thousand-and-one substrates of protein kinase CK2? FASEB J 17: 349-368. [DOI] [PubMed] [Google Scholar]
- Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, Oyama T, Kondo T (2005) Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science 308: 414-415. [DOI] [PubMed] [Google Scholar]
- Nawathean P, Rosbash M (2004) The doubletime and CKII kinases collaborate to potentiate Drosophila PER transcriptional repressor activity. Mol Cell 13: 213-223. [DOI] [PubMed] [Google Scholar]
- Park JH, Hall JC (1998) Isolation and chronobiological analysis of a neuropeptide pigment-dispersing factor gene in Drosophila melanogaster J Biol Rhythms 13: 219-228. [DOI] [PubMed] [Google Scholar]
- Park JH, Helfrich-Forster C, Lee G, Liu L, Rosbash M, Hall JC (2000) Differential regulation of circadian pacemaker output by separate clock genes in Drosophila Proc Natl Acad Sci USA 97: 3608-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen G, Hall JC, Rosbash M (1988) The period gene of Drosophila carries species-specific behavioral instructions. EMBO J 7: 3939-3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccin A, Couchman M, Clayton JD, Chalmers D, Costa R, Kyriacou CP (2000) The clock gene period of the housefly, Musca domestica, rescues behavioral rhythmicity in Drosophila melanogaster: evidence for intermolecular coevolution? Genetics 154: 747-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picton C, Woodgett J, Hemmings B, Cohen P (1982) Multisite phosphorylation of glycogen synthase from rabbit skeletal muscle: phosphorylation of site 5 by glycogen synthase kinase-5 (casein kinase-II) is a prerequisite for phosphorylation of sites 3 by glycogen synthase kinase-3. FEBS Lett 150: 191-196. [DOI] [PubMed] [Google Scholar]
- Preuss F, Fan JY, Kalive M, Bao S, Schuenemann E, Bjes ES, Price JL (2004) Drosophila doubletime mutations which either shorten or lengthen the period of circadian rhythms decrease the protein kinase activity of casein kinase I. Mol Cell Biol 24: 886-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JL, Blau J, Rothenfluh A, Abodeely M, Kloss B, Young MW (1998) double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell 94: 83-95. [DOI] [PubMed] [Google Scholar]
- Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH (1999) A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila Cell 99: 791-802. [DOI] [PubMed] [Google Scholar]
- Rothenfluh A, Abodeely M, Young MW (2000) Short-period mutations of per affect a double-time-dependent step in the Drosophila circadian clock. Curr Biol 10: 1399-1402. [DOI] [PubMed] [Google Scholar]
- Sakai T, Ishida N (2001) Circadian rhythms of female mating activity governed by clock genes in Drosophila Proc Natl Acad Sci USA 98: 9221-9225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suri V, Hall JC, Rosbash M (2000) Two novel doubletime mutants alter circadian properties and eliminate the delay between RNA and protein in Drosophila J Neurosci 20: 7547-7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano A, Isojima Y, Nagai K (2004) Identification of mPer1 phosphorylation sites responsible for the nuclear entry. J Biol Chem 279: 32578-32585. [DOI] [PubMed] [Google Scholar]
- Tauber E, Roe H, Costa R, Hennessy JM, Kyriacou CP (2003) Temporal mating isolation driven by a behavioral gene in Drosophila Curr Biol 13: 140-145. [DOI] [PubMed] [Google Scholar]
- Toh KL, Jones CR, He Y, Eide EJ, Hinz WA, Virshup DM, Ptacek LJ, Fu YH (2001) An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science 291: 1040-1043. [DOI] [PubMed] [Google Scholar]
- Tomita J, Nakajima M, Kondo T, Iwasaki H (2005) No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science 307: 251-254. [DOI] [PubMed] [Google Scholar]
- Wise S, Davis NT, Tyndale E, Noveral J, Folwell MG, Bedian V, Emery IF, Siwicki KK (2002) Neuroanatomical studies of period gene expression in the hawkmoth, Manduca sexta J Comp Neurol 447: 366-380. [DOI] [PubMed] [Google Scholar]
- Xu Y, Padiath QS, Shapiro RE, Jones CR, Wu SC, Saigoh N, Saigoh K, Ptacek LJ, Fu YH (2005) Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature 434: 640-644. [DOI] [PubMed] [Google Scholar]
- Yang Z, Sehgal A (2001) Role of molecular oscillations in generating behavioral rhythms in Drosophila Neuron 29: 453-467. [DOI] [PubMed] [Google Scholar]
- Zehring WA, Wheeler DA, Reddy P, Konopka RJ, Kyriacou CP, Rosbash M, Hall JC (1984) P-element transformation with period locus DNA restores rhythmicity to mutant, arrhythmic Drosophila melanogaster Cell 39: 369-376. [DOI] [PubMed] [Google Scholar]
- Zeng H, Qian Z, Myers MP, Rosbash M (1996) A light-entrainment mechanism for the Drosophila circadian clock. Nature 380: 129-135. [DOI] [PubMed] [Google Scholar]