Abstract
The inappropriate expression/activation of cell-cycle-related molecules is associated with neuron death in many experimental paradigms and human neuropathologic conditions. However, the means whereby this links to the core apoptotic machinery in neurons have been unclear. Here, we show that the pro-apoptotic Bcl-2 homology 3 domain-only molecule Bcl-2 interacting mediator of cell death (Bim) is a target of a cell-cycle-related apoptotic pathway in neuronal cells. Induction of Bim in NGF-deprived cells requires expression and activity of cyclin-dependent kinase 4 (cdk4) and consequent de-repression of E2 promoter binding factor (E2F)-regulated genes including members of the myb transcription factor family. The Bim promoter contains two myb binding sites, mutation of which abolishes induction of a Bim promoter-driven reporter by NGF deprivation or E2F-dependent gene de-repression. NGF deprivation significantly increases endogenous levels of C-myb and its occupancy of the endogenous Bim promoter. These findings support a model in which apoptotic stimuli lead to cdk4 activation, consequent de-repression of E2F-regulated mybs, and induction of pro-apoptotic Bim.
Keywords: NGF, neuronal apoptosis, BH3-only, cell cycle, E2F, myb
Introduction
A growing body of in vitro and in vivo evidence indicates that components of the cell-cycle machinery become activated in neurons subjected to apoptotic stimuli and play required roles in their death (Husseman et al., 2000; Raina et al., 2000; Liu and Greene, 2001a; Herrup and Arendt, 2002; Greene et al., 2004). However, the downstream effectors that mediate neuron death in response to cell-cycle activation are unknown.
E2 promoter binding factor (E2F) transcription factors are cell-cycle regulatory molecules with key roles in neuron survival and death (Liu and Greene, 2001a; Greene et al., 2004). In viable neurons, E2Fs complex with retinoblastoma (Rb) family members, leading to silencing of genes with E2F binding sites (Zhang et al., 1999; Boutillier et al., 2002; Stevaux and Dyson, 2002; Liu et al., 2005). In response to apoptotic stimuli, neuronal levels of the cell-cycle molecules cyclin D and cyclin-dependent kinase 4 (cdk4) rise, and as a consequence, cdk4 activity markedly increases (Freeman et al., 1994; Kranenburg et al., 1996; Park et al., 1998; Copani et al., 1999; Padmanabhan et al., 1999). Activated cdk4 then phosphorylates Rb proteins, causing dissociation of E2F complexes and, consequently, loss of gene repression (Copani et al., 1999; Padmanabhan et al., 1999; Park et al., 2000; Stevaux and Dyson, 2002; Rideout et al., 2003; Liu et al., 2005). De-repression of E2F-responsive genes by this mechanism triggers neuron death (Liu and Greene, 2001b; Boutillier et al., 2003; Liu et al., 2005). In support of this scheme, blocking cdk activity or E2F-dependent gene de-repression suppresses neuron death (Park et al., 1997, 1998; Liu and Greene, 2001b; Rideout et al., 2003), whereas promotion of E2F-dependent gene de-repression causes neuron death (Liu and Greene, 2001b; Boutillier et al., 2003). Among E2F-regulated genes that are de-repressed in neurons by apoptotic stimuli are the transcription factors B- and C-myb (Liu and Greene, 2001b). myb overexpression induces death (Liu and Greene, 2001b), whereas downregulation of mybs protects neurons from death (Liu et al., 2004). However, transcriptional targets of mybs that mediate neuron death have been unknown.
The search for transcriptionally regulated molecules that mediate neuron death induced by trophic factor deprivation has pointed to Bcl-2 interacting mediator of cell death (Bim) (Strasser et al., 2000; Bouillet et al., 2002; Puthalakath and Strasser, 2002). Bcl-2 proteins are gatekeepers of the apoptotic machinery and possess up to four conserved Bcl-2 homology (BH) domains (Strasser et al., 2000). Family members such as Bcl-2 are anti-apoptotic, whereas others such as Bim (with a single BH3 domain) are pro-apoptotic. Trophic factor deprivation induces Bim expression in populations including sympathetic, sensory, and cerebellar granule neurons (Putcha et al., 2001; Whitfield et al., 2001; Linseman et al., 2002) and neuronal pheochromocytoma 12 (PC12) cells (Biswas and Greene, 2002). Bim deletion or downregulation reduces or delays such neuron death (Putcha et al., 2001; Biswas and Greene, 2002; Linseman et al., 2002).
Here, we identify Bim as a transcriptional target of a neuronal apoptotic cell-cycle pathway and show that this pathway is required for Bim induction in response to NGF deprivation. Bim thus represents an important link between the cell cycle and apoptotic machineries.
Materials and Methods
Materials. Platinum TaqDNA polymerase and Lipofectamine 2000 were from Invitrogen (San Diego, CA), t-butoxycarbonyl-aspartyl(OMe)-fluoromethyl ketone was from Enzyme Systems Products (Livermore, CA), anti-human NGF antiserum and anti-green fluorescent protein (GFP) antibody were from Sigma (St. Louis, MO), roscovitine and olomoucine were from Calbiochem (La Jolla, CA), anti-C-myb and anti-ERK1 (extracellular signal-regulated kinase 1) antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA), and the Bim antibody was from StressGen (Victoria, British Columbia, Canada). Human recombinant NGF was a kind gift from Genentech (South San Francisco, CA), CEP-1347 was a kind gift from Cephalon (West Chester, PA), and flavopiridol was a kind gift from Dr. P. Worland (National Cancer Institute, Bethesda, MD).
Cell culture. PC12 cells were cultured as described previously in collagen-coated dishes with RPMI 1640 medium (Mediatech, Herndon, VA) supplemented with 10% heat-inactivated horse serum and 5% fetal bovine serum (Greene and Tischler, 1976). Neuronal differentiation was induced with NGF (100 ng/ml) in medium with 1% horse serum. For NGF deprivation, on day 7 of treatment, the cultures were washed with NGF-free medium twice, and anti-NGF antibody (1:100) was added. Control cells were washed with serum-free medium and maintained in medium supplied with NGF without serum. Embryonic rat cortical neurons and neonatal rat superior cervical ganglion (SCG) sympathetic neurons were cultured as described previously (Park et al., 1998). Human embryonic kidney (HEK) 293 cells were cultured in DMEM with 10% fetal bovine serum.
Bim reporter constructs. A portion of the Bim gene that contains ∼3kb of DNA extending 5′ from exon 1 was amplified from rat genomic DNA by PCR using Platinum Taq (Invitrogen) according to the manufacturer's protocol. The primers for the amplification were 5′-GAGCTCGTGAGCCAGGCGAGAAATTTAGTG-3′ and 5′-AAGCTTCAACCAGCTGGTGACCCAGTGCCTGCG-3′ (SacI and HindIII linker sequences are underlined). The amplified products were digested with SacI and HindIII, gel purified, and subcloned into pGL3 (Promega, Madison, WI). The mutant reporters were generated by deletion of consensus sequences for binding sites or by incorporating mutation in the binding sites in the Bim promoter by PCR-based site-directed mutagenesis using PfuTurbo DNA polymerase (Stratagene, La Jolla, CA) according to the manufacturer's protocol and were verified by sequencing. The Bim–luc-(Mybm) was generated using oligonucleotides 5′-GCTTCTCTTGTCTCTTCCTTCCCACCACC-3′ and 5′-GGTGGTGGGAAGGAAGAGACAAGAGAAGC-3′; 5′-CTCCTGAGGCTTCGCGGCCGCGGCC-3′ and 5′-GGCCGCGGCCGCGAAGCCTCAGGAG-3′.
Plasmids. Constructs of B-myb, C-myb, E2F1, E2F1 (1–374), E2F1–Rb, and dominant-negative (d/n) cdk4 were described previously (Liu and Greene, 2001b). Flag-tagged rat cdk4 was generated by inserting desired mutations in the human sequence by overlapping PCR. The primers for the amplification were (1) 5′-GCTAGCAACCATGGACTACAAGGACGATGATGACAAAATGGCTACCTCTCGATATGA-3′; (2) 5′-TAAGGTGACCTTGATCTCCCGGTCAGT-3′; (3) 5′-GAGATCAAGGTCACCTTAGTGTTTGAGCATGTAGACCA; and (4) 5′-TCTAGATTACCTTCATCCTTATGTAG. PCR was performed to produce a 300 bp 5′ sequence using primers 1 and 2 and to produce 700 bp 3′ sequence using primers 3 and 4. Equal amounts of A and B were mixed and denatured. An aliquot of this mixture was used for PCR using primers 1 and 4, producing a 1 kb fragment that resolved as a single band by electrophoresis analysis. The 1 kb fragment was gel purified and TA cloned and sequenced. The correct clones were digested with NheI and XbaI, gel purified, and subcloned into pCMS–enhanced GFP (EGFP) (Clontech, Cambridge, UK).
Preparation of small interfering RNA. Cdk4 small interfering RNA (siRNA) was prepared in the pU6 vector by using the GeneSuppressor System according to the manufacturer's instructions (Imgenex, San Diego, CA) based on the sequence 5′-GGTCACCTTAGTGTTTGAG-3′.A second cdk4 siRNA (cdk4 siRNA2) was prepared in the pSIREN vector by using BD Knockout RNAi systems according to the manufacturer's instructions (BD Biosciences, Franklin Lakes, NJ) based on the sequence 5′-TCTACAGCTACCAGATGGC-3′. Bim siRNA was prepared in the pSIREN vector by using BD Knockout RNAi systems according to the manufacturer's instructions (BD Biosciences) based on the sequence 5′-GACAGAGAAGGTGGACAATTG-3′. siRNA constructs for B-myb and C-myb were as described previously (Liu et al., 2004). siRNAs were transfected using Lipofectamine 2000 at a final level of 0.4 μg per 24-well culture.
Transfections. DNA was prepared with Plasmid Maxi kits (Qiagen, Hilden, Germany). Neuronal PC12 cells were cotransfected with 0.5 μg of reporter plasmid, 0.1 μgof Renilla vector, and 0.5 μg of control plasmid or plasmid expressing C-myb, E2F1, E2F1 (1–374), E2F1 (E-132), E2F–Rb, siRNA–B-myb, siRNA–C-myb, d/n cdk4, or siRNA–cdk4 in 500 μl of serum-free medium per well in 24-well dishes using Lipofectamine 2000. Six hours later, medium with Lipofectamine 2000 was replaced with fresh complete medium. Similarly, SCG neurons were transfected with various constructs at day 3 of culture with Lipofectamine 2000. HEK 293 cells were transfected as described previously (Xu et al., 2001).
Western immunoblotting. Neuronal PC12 cells were lysed, and protein was analyzed by Western immunoblotting as described previously (Biswas and Greene, 2002). Briefly, samples were incubated at 70°C for 10 min in NuPage (Invitrogen) sample buffer and electrophoretically resolved on NuPage 12% Bis-Tris polyacrylamide gels before blotting on Hybond polyvinylidene difluoride membranes (Amersham Biosciences, Arlington Heights, IL). Detection was performed using either enhanced chemiluminescence (Amersham Biosciences) or SuperSignal West Dura extended duration substrate (Pierce, Rockford, IL) according to the instructions of the manufacturers.
Immunostaining. Neuronal PC12 cells were transfected as described above, with appropriate constructs of siRNA and pCMS–EGFP. Forty-eight hours later, cells were subjected to NGF withdrawal for 18 h and then immunostained as described by Angelastro et al. (2003). Briefly, PC12 cells were fixed with 4% paraformaldehyde for 10 min. After three washes in PBS, cells were permeabilized with 0.5% Triton X-100 for 15 min. The cells were then blocked in 10% nonimmune goat serum for 2 h. The cultures were immunolabeled with rabbit anti-Bim (1:100; Stressgen) and mouse anti-GFP (1:1000; Sigma) antibody in 10% nonimmune goat serum overnight at 4°C, followed by secondary labeling with goat FITC-conjugated anti-mouse (1:1000; Alexa) and rhodamine-conjugated anti-rabbit (1:4000; Alexa) antibodies for 2 h.
Luciferase assay. SCG neurons or neuronal PC12 cells were transfected as described above, with appropriate luciferase reporter and pcDNA–LacZ/pCMS–EGFP with or without other DNAs. Twenty-four or 48 h later, cells were washed with cold PBS, triturated off the plates, and pelleted in microcentrifuge tubes. Cell pellets were lysed in buffer provided in the Promega Luciferase System. Dual luciferase assays were performed according to the manufacturer's instructions, using a TD-20/20 luminometer (Turner Designs, Sunnyvale, CA). Relative luciferase activities were obtained by normalizing the luciferase activity against Renilla luciferase activity. Data are presented in the text and figures as mean ± SEM (n = 3–6).
Survival assay. Neuronal PC12 cells were cotransfected with pCMS–EGFP and empty pU6, pU6–cdk4–siRNA, pSIREN–Luc–siRNA, or pSIREN–Bim–siRNA and 24 h later were subjected to NGF withdrawal. For E2F de-repression-induced death, cells were cotransfected with E2F1 (1–374) with pSIREN–Luc–siRNA or pSIREN–Bim–siRNA and pCMS–EGFP. The numbers of intact, EGFP-positive cells per well were assessed as described previously (Liu and Greene, 2001b). Data represent means ± SEM of three experiments performed in triplicate.
Reverse transcription-PCR. Each sample of total RNA was isolated by using TRI reagent (Molecular Research Center, Cincinnati, OH). cDNA was transcribed from total RNA with Superscript RT II (Invitrogen). The primers used for PCR amplification of Bim are 5′-GCCCCTACCTCCCTACAGAC-3′ and 5′-CCTTATGGAAGCCATTGCAC-3′. An equal amount of cDNA template was used for each PCR of actin or Bim. PCR products were resolved on an agarose gel and photographed.
Chromatin immunoprecipitation. Chromatin immunoprecipitation (ChIP) assays were performed using a kit (Upstate Biotechnology, Lake Placid, NY) according to the manufacturer's protocol, with the following exceptions. Five to 10 × 106 PC12 cells were used after treatment with or without NGF for each sample. Rabbit polyclonal C-myb antibody or E2F1 antibody was used at a final concentration of 1 μg/ml. A monoclonal Flag antibody was used as a negative control. For immunoblot analysis, the immunoprecipitated protein–DNA complexes were boiled in sample buffer for 10 min and analyzed by Western immunoblotting as described before probing with C-myb antiserum.
Quantitative PCR was performed using a Cepheid (Sunnyvale, CA) SmartCycler following the manufacturer's specifications. Purified DNA was added to a 25 μl volume reaction mix containing ready-to-go beads (Amersham Biosciences) and SYBR Green I (Molecular Probes, Eugene, OR) together with appropriate primers at 0.2 μm each. The primers used for PCR amplification of Bim promoter are 5′-GAAGGGCAAGCATCTTTCTG-3′ and 5′-CCGGCGTGTTTACCCTAGT-3′ (for C-myb response element) and 5′-CAGGCCAAGTCACTAGGGTA-3′ and 5′-CCAGCCACTCTGCTCTTACT-3′ (for E2F1 response element). Analyses of growth curves of real-time fluorescence and of melting curves were performed as described previously (Troy et al., 2001). Immunoprecipitation signals were normalized to the corresponding “input” signals (representing 1% of the DNA used in immunoprecipitations). Because the immunoprecipitation efficiencies of different experiments were often very different, each value for NGF-treated cells was arbitrarily set at 1.
Gel-shift assay. Electrophoretic mobility shift assay was performed as described previously (Liu and Greene 2001b). The following sequence of Bim promoter was used to prepare wild-type (wt) and mutant probe (myb or mutant myb site underlined): wt, 5′-TGAGGCTTCCAACTGGCGGC-3′; mutant, 5′-TGAGGCTTCTTCTCGTCAGCGGC-3′. Where appropriate, a 50-fold excess of unlabeled probe was added to the reactions 10 min before the 32P-labeled probe was added. Rabbit anti-C-myb (Santa Cruz Biotechnology) antibody was incubated after binding reaction at 4°C for 1 h as required.
Results
Induction of Bim by NGF deprivation is completely blocked by cdk inhibitors
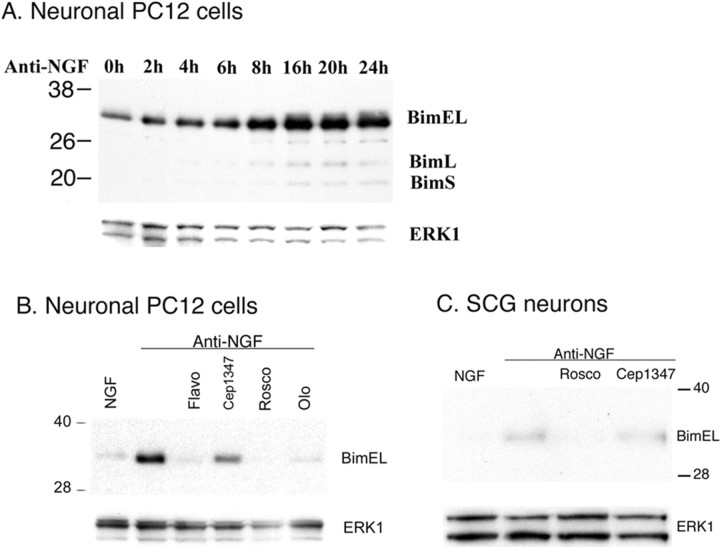
To investigate the mechanism by which Bim expression is regulated in neurons exposed to apoptotic stimuli, we first turned to neuronally differentiated PC12 cells. As in the case of their sympathetic neuron counterparts, NGF withdrawal from such cells in the absence of serum results in their apoptotic death (Greene, 1978; Batistatou and Greene, 1991). In a previous study, we established that, as for sympathetic neurons (Putcha et al., 2001; Whitfield et al., 2001), Bim is upregulated in neuronal PC12 cells in response to NGF deprivation (Biswas and Greene, 2002). A time course of NGF withdrawal from neuronal PC12 cells revealed that the expression of Bim is increased as early as 2 h and maximally by 16–24 h (Fig. 1A). The major induced form was Bim extra long (BimEL), with much lesser expression of Bim long and Bim short. As we have reported (Biswas and Greene, 2002), the MLK (mixed lineage kinase) inhibitor CEP-1347, which blocks activation of the apoptotic JNK (c-Jun N-terminal protein kinase)/c-Jun pathway in neurons (Xu et al., 2001) and fully protects neurons from trophic factor deprivation-induced death at a concentration of 100–200 nm (Maroney et al., 1999), only partially repressed Bim induction in neuronal PC12 cells when applied at a concentration of 200 nm (Fig. 1B). Increasing the concentration to 1 μm [which is more that 30-fold the IC50 and ED50 for MLK inhibition and for protection from death, respectively (Maroney et al., 1999, 2001)] did not have a substantially greater effect (Fig. S1, available at www.jneurosci.org as supplemental material). These observations are consistent with previous studies in which inhibition of the JNK/c-Jun pathway fractionally reduced Bim induction in cultured sympathetic neurons (Harris and Johnson, 2001; Whitfield et al., 2001). In contrast, we found that Bim induction was nearly completely blocked by the cdk inhibitors flavopiridol, roscovitine, and olomoucine (Fig. 1B). Past work has shown that these inhibitors protect neurons from apoptotic death evoked by NGF deprivation and DNA damage (Park et al., 1996, 1997, 1998; Courtney and Coffey, 1999) and suppress de-repression of E2F-responsive genes (Liu and Greene, 2001b).
Figure 1.
Induction of Bim by NGF withdrawal is completely blocked by cdk inhibitors. A, Time course of the effect of NGF withdrawal on Bim induction. PC12 cells were treated with NGF for 7 d, deprived of NGF for the indicated times, and cell proteins were subjected to Western immunoblotting using enhanced chemiluminescence for the expression of Bim and ERK1. BimL, Bim long; BimS, Bim short. B, The induction of the Bim protein after NGF deprivation is completely blocked by cdk inhibitors and incompletely blocked by inhibition of JNK signaling. PC12 cells were treated with NGF for 7 d and then deprived of NGF for 24 h in presence or absence of 1 μm flavopiridol (Flavo), 200 nm CEP-1347, 20 μm roscovitine (Rosco), or 200 μm olomoucine (Olo). Western immunoblotting was as in A. C, The cdk inhibitor roscovitine blocks Bim upregulation in cultured SCG neurons. Sympathetic neurons, after 3 d of culture, were deprived of NGF for 24 h in presence or absence of 20 μm roscovitine (Rosco) or 200 nm CEP-1347. Western immunoblotting was as in A.
We next repeated these experiments with cultured sympathetic neurons. In this case also, blockade of the JNK pathway with 200 nm CEP-1347 only partially reduced Bim induction, whereas the cdk inhibitor roscovitine completely suppressed this response (Fig. 1C). Thus, in both neuronal PC12 cells and sympathetic neurons, Bim induction is fully blocked by cdk inhibitors.
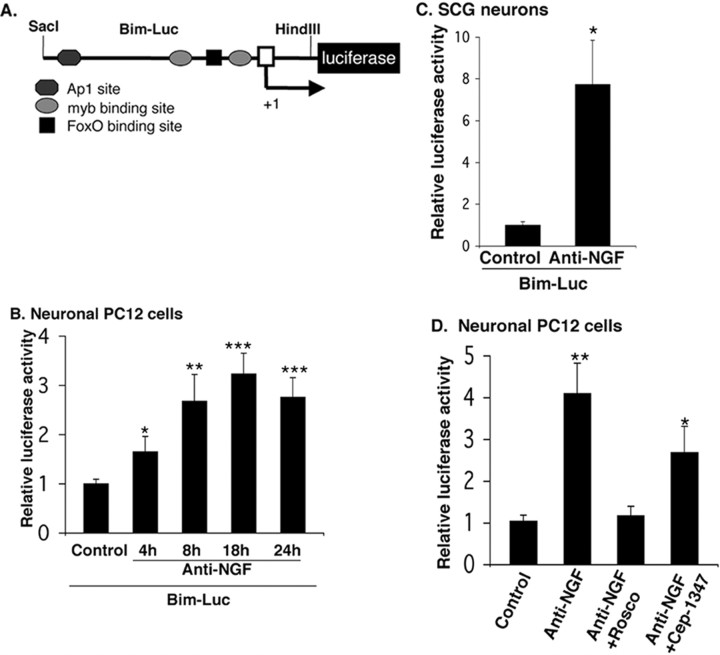
Bim induction caused by NGF withdrawal is at least in part regulated at the level of transcription (Putcha et al., 2001; Whitfield et al., 2001). To further explore the mechanism of Bim induction, we constructed a luciferase reporter driven by the Bim promoter that contains ∼3 kb of DNA extending 5′ from exon 1 of the rat Bim gene (Fig. 2A). A time course in neuronal PC12 cells transiently transfected with this reporter revealed that the Bim promoterdriven luciferase activity was enhanced by 4 h of NGF deprivation and was approximately threefold to fourfold higher than that of control after 18 h of NGF withdrawal (Fig. 2B). Withdrawal of NGF from sympathetic neurons for 18 h also induced Bim promoter-driven luciferase activity by approximately sevenfold compared with controls (Fig. 2C). As in the case of Bim protein elevation, the induction of Bim promoter-driven luciferase activity after NGF withdrawal was completely blocked by the cdk inhibitor roscovitine and only partially blocked by a JNK pathway inhibitor (Fig. 2D).
Figure 2.
The Bim promoter is activated by NGF withdrawal in neuronal PC12 cells and sympathetic neurons. A, Schematic representation of rat Bim promoter-driven luciferase reporter constructs. The Bim–lucreporter consists of ∼3 kb of the Bim rat gene extending 5′ from exon 1 in the pGL3-Basic vector. An arrow indicates the transcription start site (+1). B, Bim promoter-driven luciferase activity is induced after NGF withdrawal in neuronal PC12 cells. Neuronal PC12 cells were transfected with 0.5 μg of Bim–luc reporter and 0.1 μg of the Renilla luciferase expression construct pRL–CMV (to control for transfection efficiency) and were maintained with or without NGF for the indicated times, after which luciferase assays were performed. The data are reported as relative firefly luciferase activity normalized to Renilla luciferase activity and represent means ± SEM of four experiments. Asterisks denote statistically significant differences from control: *p < 0.05; **p < 0.005; ***p < 0.001. C, Bim promoter-driven luciferase activity is increased after NGF withdrawal from sympathetic neurons. Sympathetic neurons were transfected and assayed for luciferase activity as described in B, except that they were maintained with or without NGF for 18 h. The data are means ± SEM of five experiments. The asterisk denotes statistically significant differences from control: *p < 0.004. D, Induction of Bim promoter-driven luciferase activity after NGF withdrawal is completely blocked by a cdk inhibitor and incompletely blocked by inhibition of the JNK pathway. Neuronal PC12 cells were transfected and assayed for luciferase activity as described in B, except that they were treated with 20 μm roscovitine (Rosco) or 200 nm CEP-1347 during 18 h of NGF deprivation. The data are means ± SEM of three experiments. Asterisks denote statistically significant differences from control: *p < 0.04; **p < 0.008.
Cdk4 is required for Bim induction
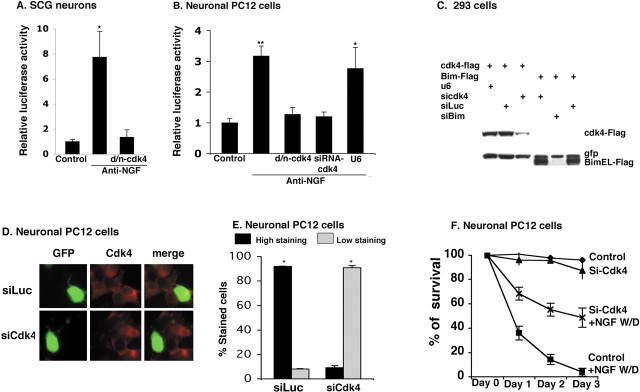
Although cdk inhibitors block induction of Bim protein and Bim reporter activity, it is possible that such actions are attributable to nonspecific actions of these molecules (Bain et al., 2003). We have reported that neuronal cyclin D-associated cdk activity (presumably because of cdk4 and/or cdk6) is elevated in response to DNA damage and that neuron death caused by apoptotic stimuli such as DNA damage and NGF deprivation is suppressed by d/n cdk4 (Park et al., 1997, 1998). Therefore, to extend our observations that cdk inhibitors block Bim induction, we transfected neuronal PC12 cells and sympathetic neurons with the Bim promoter-driven reporter and d/n cdk4. In both neuronal PC12 cells and sympathetic neurons, the d/n construct almost totally suppressed induction of reporter activity by NGF deprivation (Fig. 3A,B). As an additional test, we prepared a siRNA construct specifically directed to cdk4. The efficacy of the siRNA was shown by its ability to substantially reduce expression of Flag-cdk4 when transiently coexpressed in HEK 293 cells or of endogenous cdk4 in neuronal PC12 cells (Fig. 3C–E). Like cdk inhibitors and d/n cdk4, the cdk4 siRNA also suppressed neuron death promoted by NGF deprivation (Fig. 3F) or by the DNA-damaging agent camptothecin (data not shown). A second cdk4 siRNA targeted against a different sequence also downregulated cdk4 and protected cells from NGF deprivation (Fig. S2, available at www.jneurosci.org as supplemental material). When cotransfected in the PC12 cell Bim reporter assay, the cdk4 siRNA, but not an empty vector control or irrelevant siRNA (data not shown), blocked induction of reporter activity by NGF withdrawal (Fig. 3B). Moreover, cdk4 siRNA substantially blocked the induction of endogenous Bim in neuronal PC12 cells after NGF deprivation (Fig. 4A,B). These findings support the conclusion that cdk4 is required for Bim regulation in response to apoptotic stimuli.
Figure 3.
Cdk4 is required for Bim expression. A, B, Induction of Bim promoter-driven luciferase activity after NGF deprivation in cultures of sympathetic neurons (A) and neuronal PC12 cells (B) is blocked by inhibition/loss of cdk4 activity. Cultures were cotransfected with 0.5 μg of Bim–luc and 0.1 μgofthe Renilla luciferase expression construct pRL–CMV with 0.5 μg of empty pCMV vector (Control) or indicated construct, maintained for 24 h, and then deprived of NGF for 18 h, after which luciferase assays were performed. Data were normalized as in Figure 2 and are means ± SEM of three or four experiments. Asterisks denote statistically significant differences from control: A,*p < 0.004; B,*p < 0.001, ** p < 0.0003. C, Cdk4 and Bim siRNAs suppress cdk4 and Bim expression, respectively. HEK 293 cells were cotransfected with pCMS–EGFP expressing Flag–cdk4 or Flag–BimEL and pU6–cdk4–siRNA, pSIREN–Luc–siRNA, pSIREN–Bim–siRNA, or empty pU6 vector. After 48 h, cells were lysed and subjected to Western immunoblotting as described in Figure 1 A with anti-Flag and anti-EGFP as probes. D, E, Cdk4 siRNA reduces endogenous cdk4 expression in neuronal PC12 cells after NGF withdrawal. Neuronal PC12 cells were cotransfected with pSIREN–cdk4–siRNA (siCdk4) or pSIREN–Luc–siRNA (siLuc) and pCMS–EGFP, maintained for 48 h, and then deprived of NGF for 18 h, after which immunostaining with antibodies against cdk4 (red) and GFP (green) was performed. Percentage of stained cells pertains to the proportions of transfected cells (as indicated by staining for GFP) that show cdk4 staining either high (more than that of NGF-maintained cells) or low (equal or less than that of NGF-maintained cells). Data represent means ± SEM of three experiments. Approximately 50 cells were evaluated perculture. Asterisks denote statistically significant differences between low-staining cells and high-staining cells: *p < 0.0001. F, Cdk4 siRNA protects neuronal PC12 cells from death evoked by NGF deprivation. Cells were cotransfected with pCMS–EGFP and either empty pU6 (Control) or pU6–cdk4–siRNA and 24 h later subjected to NGF withdrawal (W/D). Numbers of transfected (green) cells were determined at indicated times, and percentage survival was calculated relative to the number of transfected cells present before exposure to apoptotic conditions. Data represent means ± SEM of three experiments performed in triplicate.
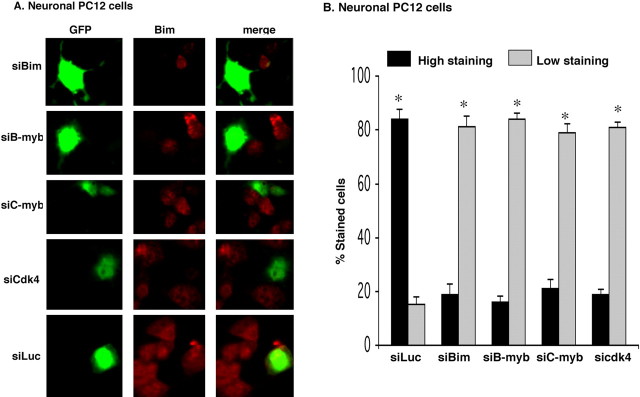
Figure 4.
siRNAs targeted to Bim, B-myb, C-myb, and cdk4 repress upregulation of endogenous Bim in neuronal PC12 cells subjected to NGF withdrawal. A, B, Neuronal PC12 cells were cotransfected with the indicated constructs and pCMS–EGFP, maintained for 48 h, and then deprived of NGF for 18 h, after which they were immunostained with antibodies against Bim (red) and GFP (green). Percentage of stained cells pertains to the proportions of transfected cells (as indicated by staining for GFP) that show Bim staining either high (more than that of NGF-maintained cells) or low (equal or less than that of NGF-maintained cells). Data represent means ± SEM of three experiments. Approximately 50 cells were evaluated per culture. Asterisks denote statistically significant differences between low-staining cells and high-staining cells: *p < 0.0001.
Bim is a target of the cdk–E2F pathway
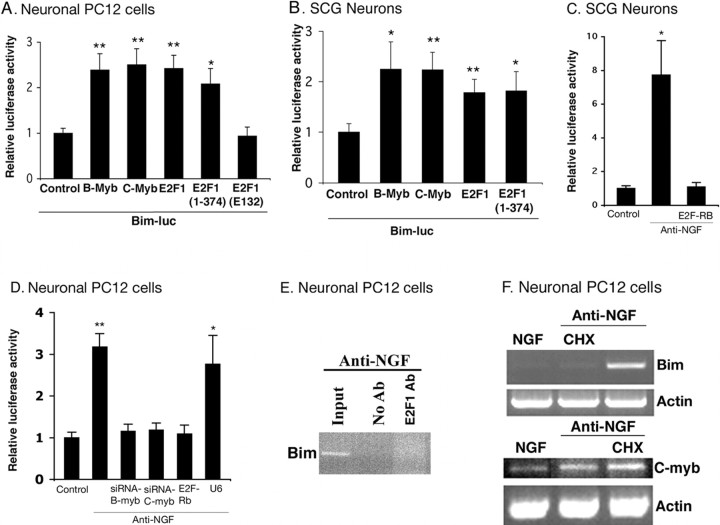
Past studies indicate that activation of neuronal cdk4 by apoptotic stimuli leads to hyperphosphorylation of Rb family members (Padmanabhan et al., 1999; Park et al., 2000; Trinh et al., 2001) and that this in turn permits de-repression of pro-apoptotic genes that are otherwise repressed by E2F–Rb family complexes in viable neurons (Liu and Greene, 2001b; Boutillier et al., 2003). We therefore next tested whether Bim can be induced by E2F-dependent de-repression by transfecting cultured neuronal PC12 cells and sympathetic neurons with E2F1(1–374), a truncated form of E2F lacking both transactivation and Rb binding domains (Sellers et al., 1995). This mutant causes de-repression (but not activation) of E2F-regulated genes as well as neuron death (Liu and Greene, 2001b). As shown in Figure 5, A and B, cotransfection of this construct caused significant induction of the Bim promoter-driven reporter. Cotransfection with wt E2F1 gave similar results (Fig. 5A,B); whereas, E2F1(E132), a point mutant of E2F1(1–374) that is unable to bind to DNA (Johnson et al., 1993), had no effect on Bim promoter-driven reporter activity (Fig. 5A). The latter observation is consistent with the past suggestion that overexpressed wt E2F1 can induce gene derepression by displacing repressive E2F–Rb complexes from E2F binding sites (Liu and Greene, 2001b).
Figure 5.
E2F-dependent de-repression of myb is required for induction of Bim promoter activity. Neuronal PC12 cells (A) and sympathetic neurons (B) were cotransfected with 0.5 μg of Bim–luc reporter and 0.1 μg of the Renilla luciferase expression construct pRL–CMV with 0.5 μg of empty pCMV vector (Control), B-myb, C-myb, E2F1, E2F1 (1–374), or E2F1 (E132) and maintained for 24 h, after which luciferase assays were performed. Data were normalized as in Figure 2 and represent means ± SEM for three or four experiments. Asterisks denote statistically significant differences from control: A, *p < 0.006, **p < 0.0001; B, *p < 0.0001, **p < 0.00004. C, D, Induction of Bim promoter-driven luciferase activity after NGF deprivation in cultures of sympathetic neurons (C) and neuronal PC12 cells (D) is blocked by interference with B- and C-myb-promoted gene regulation (D) and repression of E2F responsive genes (C, D). Cultures were cotransfected with 0.5 μg of Bim–luc and 0.1 μg of the Renilla luciferase expression construct pRL–CMV with 0.5μg of empty pCMV vector (Control) or indicated construct, maintained for 24 h, and then deprived of NGF for 18 h, after which luciferase assays were performed. Data were normalized as in Figure 2 and are means ± SEM of three or four experiments. Asterisks denote statistically significant differences from Control: C,*p < 0.004; D, *p < 0.001, **p < 0.0003. E, E2F-responsive elements of the Bim promoter are not occupied in response to NGF deprivation in PC12 cells. PC12 cells were treated with NGF for 6 d and then maintained for an additional day without NGF. Cells were then subjected to ChIP as described in Materials and Methods using anti-E2F1 (Santa Cruz Biotechnology). The immunoprecipitated material was subjected to PCR using Bim primers that flanked one E2F1 binding site. Agarose gel electrophoresis was performed on products of PCR. Templates were DNA derived from cells before ChIP (input), and DNA in immunoprecipitates were derived by ChIP with anti-E2F1. Comparable results were achieved in three independent experiments. F, The protein synthesis inhibitor cycloheximide (CHX) blocks the induction of Bim mRNA expression but not C-myb mRNA expression after NGF deprivation. PC12 cells were treated with NGF for 7 d and then deprived of NGF for 4 h in presence or absence of 10 μg/ml CHX, and total RNA was isolated, reverse transcribed, and amplified by PCR using rat Bim-, rat C-myb-, and actin-specific primers.
If Bim induction is caused by E2F-dependent gene derepression, then its elevation by apoptotic stimuli should be inhibited if such de-repression is blocked. To test this supposition, neuronal PC12 cells and sympathetic neurons were cotransfected with constructs encoding the Bim promoter-driven reporter and an E2F–Rb fusion protein, E2F(1–368)–Rb(379–792). The fusion protein binds to E2F sites on DNA and causes gene repression and is not subject to dissociation by cdk-dependent phosphorylation (Sellers et al., 1995). It also rescues sympathetic neurons from death evoked by NGF deprivation (Liu and Greene, 2001b)). As shown in Figure 5, C and D, the fusion protein fully blocks induction of the Bim reporter caused by NGF deprivation. Together, these findings support a mechanism in which Bim induction is downstream of cdk-promoted, E2F-dependent gene de-repression.
myb mediates induction of Bim by E2F-dependent gene de-repression
Induction of Bim by a mechanism requiring E2F-dependent gene de-repression raises the issue as to whether E2F is directly or indirectly responsible for regulating Bim transcription. It was reported previously that the mouse Bim gene promoter contains an E2F-responsive element that binds endogenous E2F1 (Hershko and Ginsberg, 2004). However, when we used ChIP assays, we were unable to detect any significant interaction of E2F1 with the Bim gene promoter in NGF-deprived neuronal PC12 cells (Fig. 5E). Moreover, in the fibroblast system, E2F1 acted to transcriptionally activate Bim; this contrasts with the present system in which Bim induction is driven by E2F-dependent gene de-repression. Finally, if Bim induction requires an intermediary, then this effect should be require de novo protein synthesis. To determine whether this is the case, neuronal PC12 cell cultures were deprived of NGF for 4 h in the presence or absence of cycloheximide and then assessed for levels of Bim transcripts. As shown in Figure 5F, cycloheximide fully blocked the induction. In contrast, elevation of C-myb transcript expression evoked by NGF deprivation, which our model indicates occurs by direct gene de-repression and therefore by a mechanism not dependent on protein synthesis (Liu and Greene, 2001b) (see below) was not blocked by cycloheximide (Fig. 5F, bottom). Together, these findings indicate that regulation of Bim expression by NGF withdrawal does not involve direct interaction of the Bim gene with E2F and that an intermediary gene product is required for Bim induction in this case.
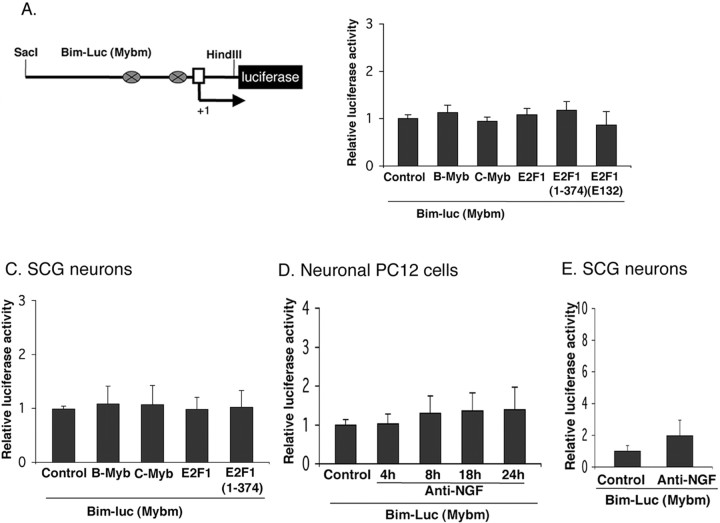
We next considered potential candidates that are subject to regulation by E2F-dependent gene de-repression and that might mediate Bim induction in response to NGF deprivation. Examination of the rat Bim promoter revealed two potential binding sites for members of the myb transcription factor family (Bouillet et al., 2001) and that these sites are conserved in mouse. We have reported that B- and C-myb are among the E2F-regulated genes that are de-repressed by apoptotic stimuli in neurons and that this occurs by a mechanism that is sensitive to cdk inhibitors (Liu and Greene, 2001b). Moreover, overexpression of B- or C-myb causes neuronal death (Liu and Greene, 2001b), whereas B- and C-myb antisense and siRNA constructs protect neurons from apoptotic death induced by NGF withdrawal or DNA damage (Liu et al., 2004). Therefore, to test the possibility that mybs mediate the actions of E2F on Bim, we evaluated the effect of E2F1 and E2F1(1–374) on a cotransfected Bim promoter-driven reporter from which the myb binding sites were deleted (Fig. 6A). In contrast with results for the intact reporter which, as noted above, was responsive to E2F1 and E2F1(1–374) (Fig. 5A,B), there was little or no induction of the mutant reporter by any E2F construct (Fig. 6B,C).
Figure 6.
The activation of Bim promoter by NGF withdrawal or B-myb, C-myb, E2F1, and an E2F truncation mutant lacking a transactivation domain in neuronal PC12 cells and sympathetic neurons requires myb binding sites. A, Schematic representation of rat mutant Bim promoter-driven luciferase reporter constructs. The mutant lacks two myb binding sites shown as ovals. B, C, The activation of Bim promoter by B-myb, C-myb, E2F1, and E2F1(1–374) requires myb binding sites. Neuronal PC12 cells (B) and sympathetic neurons (C) were cotransfected with 0.5 μg of mutant Bim–luc reporter and 0.1 μg of the Renilla luciferase expression construct pRL–CMV with 0.5 μg of empty pCMV vector (Control), B-myb, C-myb, E2F1, E2F1(1–374), or E2F1(E132) and maintained for 24 h, after which luciferase assays were performed. Data were normalized as in Figure 2 and represent means ± SEM for three or four experiments. D, Induction of Bim promoter-driven luciferase activity after NGF withdrawal in neuronal PC12 cells requires myb binding sites. Neuronal PC12 cells were transfected with 0.5 μg of mutant Bim–luc reporter and 0.1 μg of the Renilla luciferase expression construct pRL–CMV and were maintained with or without NGF for the indicated times, after which luciferase assays were performed. The data are reported as relative firefly luciferase activity normalized to Renilla luciferase activity and represent means ± SEM of four experiments. E, Induction of Bim promoter-driven luciferase activity after NGF withdrawal from sympathetic neurons requires myb binding sites. Sympathetic neurons were transfected and assayed for luciferase activity as described in D, except that they were maintained with or without NGF for 18 h. The data are means ± SEM of three experiments.
To assess directly whether mybs can regulate Bim expression, we transiently cotransfected neuronal PC12 cells and sympathetic neurons with the Bim promoter-driven reporter plasmid and a B-myb or C-myb expression vector. Both B-myb and C-myb enhanced Bim promoter-driven luciferase activity by ∼2.5-fold in both cell types (Fig. 5A,B). To verify that regulation of the reporter was indeed via the myb binding sites in the Bim promoter, we performed parallel experiments with the mutant reporter lacking both myb sites. In this case, there was little or no elevation of reporter activity by cotransfected B-myb or C-myb (Fig. 6B,C). We found similar results with cultured cortical neurons as well (Fig. S3, available at www.jneurosci.org as supplemental material).
We next determined whether mybs mediate Bim induction caused by NGF deprivation. As one approach, we used siRNA constructs targeted to either B-myb or C-myb. As shown previously (Liu et al., 2004), these siRNAs specifically disrupt expression of the corresponding endogenously and exogenously expressed genes and protect sympathetic neurons and neuronal PC12 cells from death induced by NGF withdrawal or expression of E2F(1–374). As shown in Figure 5D, cotransfection with the siRNAs abolished Bim promoter-driven reporter activity induced by NGF withdrawal. Additionally, the siRNAs significantly blocked induction of endogenous Bim in neuronal PC12 cells evoked by removal of NGF (Fig. 4A,B). Finally, withdrawal of NGF from neuronal PC12 cells or sympathetic neurons failed to significantly elevate expression of the mutant Bim promoter-driven reporter lacking myb sites (Fig. 6D,E). These findings indicate that mybs mediate the induction of Bim evoked by NGF deprivation and do so by a mechanism that is downstream of E2F de-repression.
Bim mediates death induced by de-repression of E2F-regulated genes
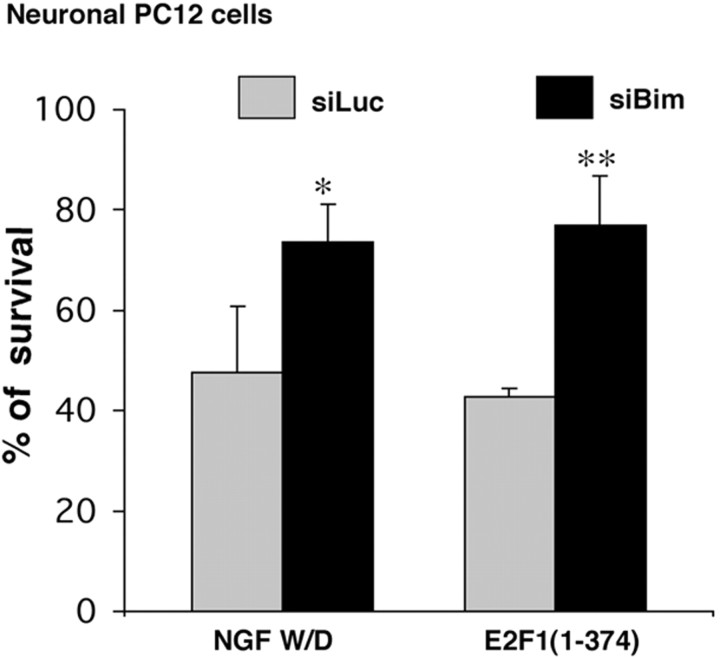
If, as our model predicts, NGF causes de-repression of E2F-regulated genes, which in turn promote death by inducing Bim, then Bim should mediate neuron death evoked by overexpression of E2F1(1–374). To approach this experimentally, we generated a siRNA construct specifically targeted to Bim. The efficacy of the siRNA was shown by its ability to substantially reduce expression of Flag–Bim when cotransfected in HEK 293 cells (Fig. 3C) or of endogenous Bim when transiently expressed in neuronal PC12 cells (Fig. 4A,B) as well as to suppress death promoted by NGF deprivation in neuronal PC12 cell cultures (Fig. 7). However, it does not affect the induction of the Bim reporter (Fig. S4, available at www.jneurosci.org as supplemental material). As shown in Figure 7, Bim siRNA provided significant protection from death evoked by overexpression of E2F1(1–374). These findings thus support the model that Bim lies downstream of E2F-dependent de-repression in neuronal apoptosis.
Figure 7.
Bim siRNA protects neuronal PC12 cells from death evoked by NGF withdrawal (W/D) and E2F derepression. Neuronal PC12 cells were cotransfected with pCMS–EGFP and pSIREN–Luc–siRNA (siLuc) or pSIREN–Bim–siRNA (siBim) and E2F1 (1–374) as indicated, maintained for 48 h, and then deprived of NGF for 18 h as indicated, after which time numbers of surviving transfected (EGFP+) cells were counted. Data represent means ± SEM of three experiments performed in triplicates. Asterisks denote statistically significant differences from Control (siLuc): *p < 0.05; **p < 0.01.
C-myb binds the Bim promoter in situ and apoptotic stimuli increase occupancy of the promoter by C-myb
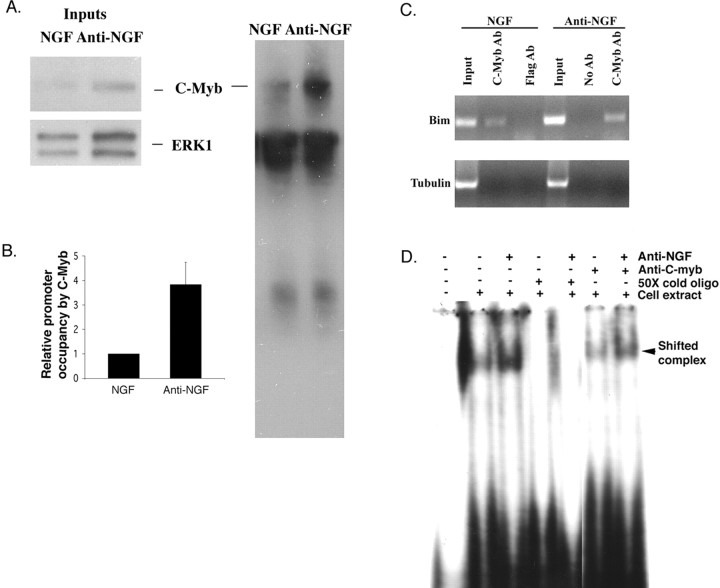
To determine whether C-myb actually occupies the Bim promoter in living cells and whether such occupancy is enhanced in response to NGF withdrawal, we next turned to ChIP assays. Accordingly, neuronal PC12 cells were maintained with or deprived of NGF for 24 h and subjected to ChIP. Figure 8A shows that endogenous C-myb protein is immunoprecipated in this procedure and that it is substantially elevated in response to NGF deprivation. Quantitative real-time PCR of DNA in the immunoprecipitates (see Materials and Methods for details) using primers that flank one C-myb binding site in the Bim promoter revealed that there is a corresponding fourfold to fivefold increase in C-myb–Bim promoter complexes in the NGF-deprived cells (Fig. 8B). Analysis of the PCR products on a 2% agarose gel confirmed that the C-myb antiserum, but not an irrelevant antibody (anti-Flag), immunoprecipitated a portion of the Bim promoter containing the predicted 200 bp fragment flanked by the primers (Fig. 8C). The identity of the PCR product as the predicted portion of the Bim promoter was further verified by sequencing. Finally, a no-antiserum control as well as PCR with α-tubulin primers confirms that the immunoprecipitation was specific (Fig. 8C). Additionally, an electrophoretic mobility gel-shift assay was performed to confirm that C-myb binds to the putative C-myb site of the Bim promoter. Figure 8D shows that endogenous C-myb protein binds to a Bim probe that contains a C-myb binding site and that the Bim–C-myb complex is elevated after NGF deprivation. In conformation that the complex contains endogenous C-myb, it was super-shifted by C-myb antibody. No complex was formed with the Bim probe containing a scrambled C-myb site (data not shown). Together, these findings show that NGF deprivation increases endogenous cellular C-myb levels as well as occupancy of the endogenous Bim promoter by C-myb.
Figure 8.
NGF deprivation induces occupation of the endogenous Bim promoter by endogenous C-myb. A, NGF deprivation increases the amount of endogenous C-myb recovered in ChIP assays. PC12 cells were treated with NGF for 6 d and then maintained for an additional day with or without NGF. Equal numbers of cells from each condition were subjected to ChIP as described in Materials and Methods using anti-C-myb. The immunoprecipitated protein–DNA complexes were boiled in sample buffer and analyzed by Western immunoblotting using C-myb antiserum. Comparable results were achieved in an independent experiment. B, NGF withdrawal enhances occupancy of the Bim promoter by C-myb. Neuronal PC12 cells treated as in A were subjected to ChIP with anti-C-myb. The immunoprecipitated material was subjected to real-time quantitative PCR using Bim primers that flanked one C-myb binding site. Data were normalized to the amount of Bim DNA present in the sample before immunoprecipitation (input) as determined by real-time quantitative PCR using the same primers. Comparable results were achieved in four independent experiments. Error bars represent SEM. C, Verification that C-myb occupies the endogenous Bim promoter. Agarose gel electrophoresis was performed on products of quantitative real-time PCR using material derived from neuronal PC12 cells treated as in A as template. Templates were DNA derived from cells before ChIP (input) and DNA in immunoprecipitates derived by ChIP with anti-C-myb or anti-Flag. Primers corresponded to a portion of the Bim promoter as in B (top) or to a portion of theα-tubulin gene (bottom). D, Endogenous C-myb complexes with the Bim promoter in neuronal PC12 cells, and such complexes are increased in response to NGF deprivation. PC12 cells were treated with NGF for 7 d and then maintained for 8 h with or without NGF. Equal amounts of cell extracts from each condition were subjected to electrophoretic mobility gel-shift assays using a rat Bim promoter sequence(lanes 2–7). A50-fold excess of unlabeled probe was included where indicated. Samples in lanes 6 and 7 were incubated with anti-C-myb antibody (Santa Cruz Biotechnology). The solid arrowhead marks the Bim–C-myb complex that is selectively supershifted by anti-C-myb.
Discussion
Bim regulation by the apoptotic cdk4–E2F–myb pathway
In this study, we investigated the mechanisms by which NGF deprivation induces the pro-apoptotic BH3-only protein Bim. We found that Bim is a target of a previously described pathway that includes cdk4, E2F, and myb. In viable nonmitotic cells such as neurons, E2F is bound by Rb family members that in turn bind chromatin modifiers. Such complexes repress genes such as myb that have E2F binding sites. Apoptotic stimuli such as NGF deprivation lead to activation of cdk4, which in turn phosphorylates members of the Rb family, leading to dissociation of Rb family members and chromatin modifiers from the E2F complex and to gene derepression. This de-repression of genes such as B- and C-myb appears to participate in and to be required for neuron death. Our findings now link this pathway to the cellular apoptotic machinery by identifying Bim as one of its regulated components.
A number of experimental observations tie Bim to the cdk4–E2F–myb pathway. First, Bim induction evoked by NGF deprivation is completely blocked by small molecule cdk inhibitors. Such inhibitors suppress other apoptotic-stimulus-promoted events in the pathway including phosphorylation of Rb family members and de-repression of E2F-dependent genes, as well as death itself (Park et al., 1996, 1997, 1998; Padmanabhan et al., 1999; Liu and Greene, 2001b). Second, because of the possibility that the observed effect of the small molecule cdk inhibitors might arise from nonspecific actions, we also used a d/n cdk4 and a cdk4 siRNA. Both reagents inhibited neuron death evoked by apoptotic stimuli, and both effectively blocked the capacity of NGF withdrawal to activate a Bim promoter-driven reporter. Third, E2F1, E2F1(1–374), and C-myb, all of which promote neuron death, activated the Bim promoter-driven reporter and did so by a mechanism dependent on the myb binding sites in the promoter construct. The effectiveness of E2F1(1–374), a mutant that retains the capacity to bind DNA but that lacks transactivation and Rb-binding domains, supports a de-repression mechanism. Fourth, a Bim siRNA inhibited neuron death induced by E2F1(1–374). Fifth, NGF deprivation activated the promoter-driven reporter but did not activate one lacking myb binding sites. Moreover, both B-myb and C-myb siRNAs effectively blocked the capacity of NGF withdrawal to induce endogenous Bim protein as well as to activate a Bim promoter-driven reporter. Finally, ChIP and electrophoretic mobility gel-shift assay experiments reveal that NGF deprivation leads to enhanced occupancy of the endogenous Bim promoter by endogenous C-myb.
The cdk4–E2F pathway and neuron death
Although a substantial body of circumstantial and experimental evidence supports a model for involvement of cell-cycle-related molecules in a number of paradigms of neuron death, there are several areas in which this model has been unclear. For one, much of the evidence for participation of cdk4/6 has come from studies with small molecule inhibitors that, although very effective in blocking neuron death, may exert nonspecific actions. A limited number of studies with d/n cdk4 and cdk6 have supported the small molecule findings by showing interference with neuron death in several models (Park et al., 1997, 1998; Rideout et al., 2003). Our observation that d/n cdk4 interferes with activation of a Bim promoter-driven reporter evoked by NGF deprivation provides additional support. Because it is conceivable the d/n construct acts nonspecifically, it is significant that Bim reporter activation and endogenous Bim induction after NGF withdrawal were also inhibited by a cdk4 siRNA and that the latter suppressed death induced by DNA damage and NGF deprivation. A second issue with the model was the role of mybs in neuron death. Although we provided evidence that neuronal myb levels increase in response to apoptotic stimuli via relief of E2F-mediated repression and that myb overexpression promotes neuron death (Liu and Greene, 2001b), it was unclear how mybs might participate in the death mechanism. The present findings corroborate elevation of myb protein in response to NGF deprivation, demonstrate increased C-myb occupation of the endogenous Bim promoter under the same conditions, and provide evidence that C-myb binding sites and mybs are required for NGF-withdrawal-induced Bim promoter activation as well as endogenous Bim induction after NGF withdrawal. Thus, the data provide a mechanism by which myb de-repression promotes neuron death. Similarly, the present findings fill in an important gap in the cdk4–E2F–myb model by identifying for the first time that this pathway induces the synthesis of a pro-apoptotic effector (Bim) that is directly linked to the core cell death machinery.
Although the present work has focused on the NGF withdrawal and DNA damage paradigms, activation of the cdk4–E2F pathway has been documented in a variety of neuropathologic conditions including spinal cord injury, stroke, Alzheimer's disease, and amyotrophic lateral sclerosis, and experimental studies in a number of model systems support a causal role for the pathway in neuron death associated with these maladies (Greene et al., 2004). This raises the possibility that Bim will also be elevated and contributes to neuron death in such disorders. In this regard, it is of interest that Bim levels are elevated in frontal cortex of Alzheimer's disease brain (Engidawork et al., 2001).
Bim regulation by multiple apoptotic pathways
In addition to the cdk–E2F pathway, other pathways have been described that participate in neuron death by regulating transcription of apoptosis-related genes such as Bim. One is the JNK/c-Jun pathway. As shown previously with a d/n form of c-Jun and confirmed here by the use of a small molecule inhibitor, interference with this pathway partially blocks Bim induction (Harris and Johnson, 2001; Whitfield et al., 2001; Biswas and Greene, 2002). Such observations raise the question as to whether the two pathways might interact to regulate Bim. There are several ways in which this is possible. Apoptotic stimuli such as NGF deprivation lead to induction of c-Jun in neurons (Ham et al., 2000), and this induction presumably contributes to the efficacy with which the JNK/c-Jun pathway promotes death. Studies with small molecule cdk inhibitors and d/n cdk4 indicate that induction (but not activation) of c-Jun is dependent on the cdk4–E2F pathway (Ghahremani et al., 2002). Inhibition of the cdk–E2F pathway would thus also reduce regulation of Bim by blocking induction of c-Jun. Another way in which the two pathways might interact is at the level of the Bim promoter. The latter contains one AP1 site, raising the possibility of cooperative actions of the two pathways on Bim transcription (S. C. Biswas and L. A. Greene, unpublished observations).
An additional transcriptional pathway reported to regulate neuronal Bim expression is one that involves the FOXO family of Forkhead transcription factors (Linseman et al., 2002; Gilley et al., 2003). Apoptotic stimuli lead to dephosphorylation of FOXO transcription factors, their consequent nuclear translocation, and participation in neuron death (Brunet et al., 1999). Examination of the Bim gene revealed two FOXO binding elements in the 5′ region of the gene; a reporter driven by a Bim promoter construct was activated in response to NGF withdrawal, whereas there was no activation of a reporter mutated at both FOXO binding sites (Gilley et al., 2003). Moreover, either FOXO site was sufficient to mediate such a response. The promoter construct used in our study contains one of the two FOXO binding sites identified by Gilley et al. (2003), and the reporter used for the FOXO studies includes the myb sites identified here. Mutation of the binding sites for either FOXO (Gilley et al., 2003) or mybs (present study) completely suppressed reporter activation induced by NGF deprivation. This raises the possibility that each of the pathways and their target transcription factors plays a necessary, but not sufficient, role in inducing Bim and subsequent death. This is supported by the observation that the increase of Bim expression evoked by inhibition of phosphatidylinositol 3-kinase (which results in FOXO activation) is only ∼60% of that seen after NGF withdrawal (Gilley et al., 2003). Similarly, induction of Bim reporter activity by overexpression of mybs or E2F is significantly less than that obtained after NGF withdrawal, at least in case of sympathetic neurons (compare Figs. 3A, 5B). The contribution of JNK/c-Jun pathway to Bim induction also appears to be partial in that specific JNK/c-Jun pathway blockers only incompletely suppress Bim induction after NGF withdrawal (Harris and Johnson, 2001; Whitfield et al., 2001; Biswas and Greene, 2002). Although overexpressing components of each pathway can be sufficient to induce Bim and death, this does not appear to be so under physiologic conditions such as represented by NGF withdrawal.
Footnotes
This work was supported by grants from the National Institutes of Health–National Institute of Neurological Disorders and Stroke.
Correspondence should be addressed to Subhas C. Biswas, Department of Pathology, Columbia University College of Physicians and Surgeons, P & S 15-401, 630 West 168th Street, New York, NY 10032. E-mail: scb34@columbia.edu.
DOI:10.1523/JNEUROSCI.1570-05.2005
Copyright © 2005 Society for Neuroscience 0270-6474/05/258349-10$15.00/0
References
- Angelastro JM, Ignatova TN, Kukekov VG, Steindler DA, Stengren GB, Mendelsohn C, Greene LA (2003) Regulated expression of ATF5 is required for the progression of neural progenitor cells to neurons. J Neurosci 23: 4590–4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain J, McLauchlan H, Elliott M, Cohen P (2003) The specificities of protein kinase inhibitors: an update. Biochem J 371: 199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistatou A, Greene LA (1991) Aurintricarboxylic acid rescues PC12 cells and sympathetic neurons from cell death caused by nerve growth factor deprivation: correlation with suppression of endonuclease activity. J Cell Biol 115: 461–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas SC, Greene LA (2002) Nerve growth factor (NGF) down-regulates the Bcl-2 homology 3 (BH3) domain-only protein Bim and suppresses its proapoptotic activity by phosphorylation. J Biol Chem 277: 49511–49516. [DOI] [PubMed] [Google Scholar]
- Bouillet P, Zhang LC, Huang DC, Webb GC, Bottema CD, Shore P, Eyre HJ, Sutherland GR, Adams JM (2001) Gene structure alternative splicing, and chromosomal localization of pro-apoptotic Bcl-2 relative Bim. Mamm Genome 12: 163–168. [DOI] [PubMed] [Google Scholar]
- Bouillet P, Purton JF, Godfrey DI, Zhang LC, Coultas L, Puthalakath H, Pellegrini M, Cory S, Adams JM, Strasser A (2002) BH3-only Bcl-2 family member Bim is required for apoptosis of autoreactive thymocytes. Nature 415: 922–926. [DOI] [PubMed] [Google Scholar]
- Boutillier AL, Trinh E, Loeffler JP (2002) Constitutive repression of E2F1 transcriptional activity through HDAC proteins is essential for neuronal survival. Ann NY Acad Sci 973: 438–442. [DOI] [PubMed] [Google Scholar]
- Boutillier AL, Trinh E, Loeffler JP (2003) Selective E2F-dependent gene transcription is controlled by histone deacetylase activity during neuronal apoptosis. J Neurochem 84: 814–828. [DOI] [PubMed] [Google Scholar]
- Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME (1999) Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96: 857–868. [DOI] [PubMed] [Google Scholar]
- Copani A, Condorelli F, Caruso A, Vancheri C, Sala A, Giuffrida Stella AM, Canonico PL, Nicoletti F, Sortino MA (1999) Mitotic signaling by beta-amyloid causes neuronal death. FASEB J 13: 2225–2234. [PubMed] [Google Scholar]
- Courtney MJ, Coffey ET (1999) The mechanism of Ara-C-induced apoptosis of differentiating cerebellar granule neurons. Eur J Neurosci 11: 1073–1084. [DOI] [PubMed] [Google Scholar]
- Engidawork E, Gulesserian T, Seidl R, Cairns N, Lubec G (2001) Expression of apoptosis related proteins in brains of patients with Alzheimer's disease. Neurosci Lett 303: 79–82. [DOI] [PubMed] [Google Scholar]
- Freeman RS, Estus S, Johnson Jr EM (1994) Analysis of cell cycle-related gene expression in postmitotic neurons: selective induction of Cyclin D1 during programmed cell death. Neuron 12: 343–355. [DOI] [PubMed] [Google Scholar]
- Ghahremani MH, Keramaris E, Shree T, Xia Z, Davis RJ, Flavell R, Slack RS, Park DS (2002) Interaction of the c-Jun/JNK pathway and cyclin-dependent kinases in death of embryonic cortical neurons evoked by DNA damage. J Biol Chem 277: 35586–35596. [DOI] [PubMed] [Google Scholar]
- Gilley J, Coffer PJ, Ham J (2003) FOXO transcription factors directly activate bim gene expression and promote apoptosis in sympathetic neurons. J Cell Biol 162: 613–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene LA (1978) Nerve growth factor prevents the death and stimulates the neuronal differentiation of clonal PC12 pheochromocytoma cells in serum-free medium. J Cell Biol 78: 747–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene LA, Tischler AS (1976) Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc Natl Acad Sci USA 73: 2424–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene LA, Biswas SC, Liu DX (2004) Cell cycle molecules and vertebrate neuron death: E2F at the hub. Cell Death Differ 11: 49–60. [DOI] [PubMed] [Google Scholar]
- Ham J, Eilers A, Whitfield J, Neame SJ, Shah B (2000) c-Jun and the transcriptional control of neuronal apoptosis. Biochem Pharmacol 60: 1015–1021. [DOI] [PubMed] [Google Scholar]
- Harris CA, Johnson Jr EM (2001) BH3-only Bcl-2 family members are coordinately regulated by the JNK pathway and require Bax to induce apoptosis in neurons. J Biol Chem 276: 37754–37760. [DOI] [PubMed] [Google Scholar]
- Herrup K, Arendt T (2002) Re-expression of cell cycle proteins induces neuronal cell death during Alzheimer's disease. J Alzheimers Dis 4: 243–247. [DOI] [PubMed] [Google Scholar]
- Hershko T, Ginsberg D (2004) Up-regulation of Bcl-2 homology 3 (BH3)-only proteins by E2F1 mediates apoptosis. J Biol Chem 279: 8627–8634. [DOI] [PubMed] [Google Scholar]
- Husseman JW, Nochlin D, Vincent I (2000) Mitotic activation: a convergent mechanism for a cohort of neurodegenerative diseases. Neurobiol Aging 21: 815–828. [DOI] [PubMed] [Google Scholar]
- Johnson DG, Schwarz JK, Cress WD, Nevins JR (1993) Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature 365: 349–352. [DOI] [PubMed] [Google Scholar]
- Kranenburg O, van der Eb AJ, Zantema A (1996) Cyclin D1 is an essential mediator of apoptotic neuronal cell death. EMBO J 15: 46–54. [PMC free article] [PubMed] [Google Scholar]
- Linseman DA, Phelps RA, Bouchard RJ, Le SS, Laessig TA, McClure ML, Heidenreich KA (2002) Insulin-like growth factor-I blocks Bcl-2 interacting mediator of cell death (Bim) induction and intrinsic death signaling in cerebellar granule neurons. J Neurosci 22: 9287–9297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu DX, Greene LA (2001a) Neuronal apoptosis at the G1/S cell cycle check-point. Cell Tissue Res 305: 217–228. [DOI] [PubMed] [Google Scholar]
- Liu DX, Greene LA (2001b) Regulation of neuronal survival and death by E2F-dependent gene repression and derepression. Neuron 32: 425–438. [DOI] [PubMed] [Google Scholar]
- Liu DX, Biswas SC, Greene LA (2004) B-myb and C-myb play required roles in neuronal apoptosis evoked by NGF deprivation and DNA damage. J Neurosci 24: 8720–8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu DX, Nath N, Chellappan SP, Greene LA (2005) Regulation of neuron survival and death by p130 and associated chromatin modifiers. Genes Dev 19: 719–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroney AC, Finn JP, Bozyczko-Coyne D, O'Kane TM, Neff NT, Tolkovsky AM, Park DS, Yan CY, Troy CM, Greene LA (1999) CEP-1347 (KT7515), an inhibitor of JNK activation, rescues sympathetic neurons and neuronally differentiated PC12 cells from death evoked by three distinct insults. J Neurochem 73: 1901–1912. [PubMed] [Google Scholar]
- Maroney AC, Finn JP, Connors TJ, Durkin JT, Angeles T, Gessner G, Xu Z, Meyer SL, Savage MJ, Greene LA, Scott RW, Vaught JL (2001) Cep-1347 (KT7515), a semisynthetic inhibitor of the mixed lineage kinase family. J Biol Chem 276: 25302–25308. [DOI] [PubMed] [Google Scholar]
- Padmanabhan J, Park DS, Greene LA, Shelanski ML (1999) Role of cell cycle regulatory proteins in cerebellar granule neuron apoptosis. J Neurosci 19: 8747–8756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DS, Farinelli SE, Greene LA (1996) Inhibitors of cyclin-dependent kinases promote survival of post-mitotic neuronally differentiated PC12 cells and sympathetic neurons. J Biol Chem 271: 8161–8169. [DOI] [PubMed] [Google Scholar]
- Park DS, Levine B, Ferrari G, Greene LA (1997) Cyclin dependent kinase inhibitors and dominant negative cyclin dependent kinase 4 and 6 promote survival of NGF-deprived sympathetic neurons. J Neurosci 17: 8975–8983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DS, Morris EJ, Padmanabhan J, Shelanski ML, Geller HM, Greene LA (1998) Cyclin-dependent kinases participate in death of neurons evoked by DNA-damaging agents. J Cell Biol 143: 457–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DS, Obeidat A, Giovanni A, Greene LA (2000) Cell cycle regulators in neuronal death evoked by excitotoxic stress: implications for neurodegeneration and its treatment. Neurobiol Aging 21: 771–781. [DOI] [PubMed] [Google Scholar]
- Putcha GV, Moulder KL, Golden JP, Bouillet P, Adams JA, Strasser A, Johnson EM (2001) Induction of BIM, a proapoptotic BH3-only BCL-2 family member, is critical for neuronal apoptosis. Neuron 29: 615–628. [DOI] [PubMed] [Google Scholar]
- Puthalakath H, Strasser A (2002) Keeping killers on a tight leash: transcriptional and post-translational control of the pro-apoptotic activity of BH3-only proteins. Cell Death Differ 9: 505–512. [DOI] [PubMed] [Google Scholar]
- Raina AK, Zhu X, Monteiro M, Takeda A, Smith MA (2000) Abortive oncogeny and cell cycle-mediated events in Alzheimer disease. Prog Cell Cycle Res 4: 235–242. [DOI] [PubMed] [Google Scholar]
- Rideout HJ, Wang Q, Park DS, Stefanis L (2003) Cyclin-dependent kinase activity is required for apoptotic death but not inclusion formation in cortical neurons after proteasomal inhibition. J Neurosci 23: 1237–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers WR, Rodgers JW, Kaelin Jr WG (1995) A potent transrepression domain in the retinoblastoma protein induces a cell cycle arrest when bound to E2F sites. Proc Natl Acad Sci USA 92: 11544–11548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevaux O, Dyson NJ (2002) A revised picture of the E2F transcriptional network and RB function. Curr Opin Cell Biol 14: 684–691. [DOI] [PubMed] [Google Scholar]
- Strasser A, O'Connor L, Dixit VM (2000) Apoptosis signaling. Annu Rev Biochem 69: 217–245. [DOI] [PubMed] [Google Scholar]
- Trinh E, Boutillier AL, Loeffler JP (2001) Regulation of the retinoblastoma-dependent Mdm2 and E2F-1 signaling pathways during neuronal apoptosis. Mol Cell Neurosci 17: 342–353. [DOI] [PubMed] [Google Scholar]
- Troy CM, Rabacchi SA, Hohl JB, Angelastro JM, Greene LA, Shelanski ML (2001) Death in the balance: alternative participation of the caspase-2 and -9 pathways in neuronal death induced by nerve growth factor deprivation. J Neurosci 21: 5007–5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield J, Neame SJ, Paquet L, Bernard O, Ham J (2001) Dominant-negative c-Jun promotes neuronal survival by reducing BIM expression and inhibiting mitochondrial cytochrome c release. Neuron 29: 629–643. [DOI] [PubMed] [Google Scholar]
- Xu Z, Maroney AC, Dobrzanski P, Kukekov NV, Greene LA (2001) The MLK family mediates c-Jun N-terminal kinase activation in neuronal apoptosis. Mol Cell Biol 21: 4713–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HS, Postigo AA, Dean DC (1999) Active transcriptional repression by the Rb-E2F complex mediates G1 arrest triggered by p16INK4a, TGF-beta, and contact inhibition. Cell 97: 53–61. [DOI] [PubMed] [Google Scholar]