Abstract
Pax6 is a transcription factor with key functional roles in the developing brain. Pax6 promotes neuronal differentiation via transcriptional regulation of the Neurogenin2 (Ngn2) gene, although Pax6 expression appears in proliferating neuroepithelial cells before the onset of neurogenesis. Here, we identified Fabp7 (BLBP/B-FABP), a member of the fatty acid-binding protein (FABP) family, as a downregulated gene in the embryonic brain of Pax6 mutant rat (rSey2/rSey2) by microarray analysis. Marked reduction of Fabp7 expression was confirmed by quantitative PCR. Spatiotemporal expression patterns of Fabp7 in the wild-type rat embryos from embryonic day 10.5 (E10.5) to E14.5 were similar to those of Pax6, and expression of Fabp7 was undetectable in the rSey2/rSey2 cortex. The expression pattern of Fabp7 in the wild-type mouse embryo at E10.5 (corresponding to E12.5 rat) was different from that in the rat embryo, and no change of expression was observed in the Sey/Sey mouse embryo. Overexpression of exogenous Pax6 mainly induced ectopic expression of Fabp7, rather than of Ngn2, in the early cortical primordium. Interestingly, knocking-down FABP7 function by electroporation of Fabp7 small interfering RNA severely curtailed cell proliferation but promoted neuronal differentiation. We conclude that Fabp7 is a downstream gene of Pax6 transcription factor in the developing rat cortex and essential for maintenance of neuroepithelial cells during early cortical development.
Keywords: Pax6, Fabp7, microarray, Small eye rat, siRNA, neurogenesis
Introduction
Superb brain functions are played by a large number of neurons mostly born during embryonic development. In the initial stages of brain development, enormous proliferation of neuroepithelial (NEp) cells occurs by symmetrical cell division at the apical (ventricular) surface of the ventricular zone. Some of the NEp cells then start to make neurons, simultaneously self-renewing to keep undifferentiated NEp cells themselves. These cells are thus called “apical progenitors” of neurons (Miyata et al., 2001; Haubensak et al., 2004). With the advancement of the developmental stages, there appears to be symmetrically dividing “basal progenitors,” producing two neurons in the subventricular zone (Haubensak et al., 2004; Miyata et al., 2004; Englund et al., 2005). Therefore, both proliferation and neuronal differentiation occur in balance within the neuroepithelium, although the former dominates in the early cortical development.
Various factors coordinately regulate the proliferation and differentiation of NEp cells. Growth factors such as fibroblast growth factor and epidermal growth factor, as well as activation of Notch receptor, inhibit neuronal differentiation and promote proliferation of neural progenitor cells (Temple and Qian, 1996; Panchision and McKay, 2002). Conversely, neuronal differentiation is promoted by proneural basic helix-loop-helix transcription factors such as neurogenin (Ngn) 1 and Ngn2 (Schuurmans and Guillemot, 2002) and by a canonical Wnt signal (Hirabayashi et al., 2004). However, most of these studies focused on the midgestation stage when production of neurons is at its peak, and therefore, little is known about proliferation of NEp cells in the early embryonic stage.
Expression of the transcription factor Pax6 in NEp cells appears in the initial stage of brain development (Stoykova and Gruss, 1994; Inoue et al., 2000). The region-specific expression of Pax6 plays pivotal roles in the developing CNS, including brain patterning, neuronal specification, neuronal migration, and axonal projection (for review, see Osumi, 2001; Simpson and Price, 2002). With regard to neurogenesis, downregulation of Pax6 expression occurs in the transition from NEp cells (including apical progenitors) to basal progenitors (Englund et al., 2005). Gain and loss of function studies have shown that Pax6 promotes neuronal differentiation in the developing cerebral cortex; cells taken from the cortical primordia of Pax6 mutant Small eye mice (Sey/Sey) exhibit less neuronal differentiation compared with those taken from the wild type (WT), which is rescued by being transfected with exogenous Pax6 (Gotz et al., 1998; Heins et al., 2002). However, other studies reported that cell proliferation was decreased in Sey/Sey brain (Warren and Price, 1997; Warren et al., 1999; Estivill-Torrus et al., 2002). In the Pax6 mutant rat telencephalon, the thickness of both the ventricular zone and cortical plate is reduced (Fukuda et al., 2000). Thus, Pax6 seems to promote proliferation or differentiation in a highly context-dependent manner.
It is reasonable to assume that multiple functions of Pax6 are performed by transcriptional regulation of different target genes. Various genes are reported to be upregulated or downregulated in Pax6 mutant tissues, and some have been confirmed to be target genes of Pax6 in the in vitro studies (for review, see Simpson and Price, 2002). In brain development, the cell adhesion molecule L1 is a gene reported to be a direct target of Pax6 (Meech et al., 1999). Furthermore, expression of R-cadherin, a cell adhesion molecule, is decreased in Pax6 mutant brain (Stoykova et al., 1997; Andrews and Mastick, 2003). Wnt7b and SFRP-2, genes involved in Wnt signaling, are also downregulated (Osumi et al., 1997; Kim et al., 2001), and the former is induced by Pax6 overexpression (Takahashi et al., 2002). Furthermore, the above-mentioned neuronal differentiation-promoting effects of Pax6 are considered to be mediated by transcriptional activation of the Ngn2 gene (Scardigli et al., 2003). At present, however, no Pax6 downstream gene has been identified to promote proliferation and/or inhibit differentiation of NEp cells.
The present study was designed to identify genes that are differentially expressed in the early stage of wild-type and Pax6 mutant rat (rSey2/rSey2) brains. For this purpose, we conducted a comparative analysis using oligonucleotide microarrays. The results showed that among those genes that are upregulated or downregulated in rSey2/rSey2 brains, the expression of brain-type fatty acid binding protein (FABP) Fabp7 (B-FABP/BLBP) (Feng et al., 1994; Kurtz et al., 1994) was markedly reduced both in the forebrain (FB) and hindbrain (HB) regions. Fabp7 was expressed in Pax6-positive proliferating NEp cells, and the introduction of exogenous Pax6 by electroporation induced strong Fabp7 expression, rather than Ngn2 in the developing cortex. Furthermore, knocking-down of FABP7 function by RNA interference in NEp cells reduced their proliferation and conversely promoted neuronal differentiation. Notably, the cells in the early cortical primordium electroporated with Fabp7 small interfering RNA (siRNA) became round and failed to exhibit the morphological features of NEp with long processes. The results indicate that Fabp7 is a downstream gene of Pax6 in the early cortical development and has essential roles in the maintenance of proliferation of NEp cells.
Materials and Methods
Animals. Pax6 homozygous rat embryos were obtained by intercrossing male and female heterozygotes of Small eye rats (rSey2) (Osumi et al., 1997) that were maintained at the Tohoku University School of Medicine (Sendai, Japan). Wild-type Sprague Dawley rats were purchased from Charles River (Kanagawa, Japan). The midday of identifying the vaginal plug was designated as embryonic day 0.5 (E0.5). The Committee for Animal Experiment in Tohoku University Graduate School of Medicine approved all of the experimental procedures described in this report.
Microarray analysis. GeneChip Rat Genome U34A Array (Affymetrix, Santa Clara, CA) was used for microarray analysis. The details of the procedure are described in the supplemental material (available at www.jneurosci.org).
Real-time quantitative PCR. To analyze the validity of the microarray results, real-time quantitative PCR was performed using the LightCycler instrument and the protocol recommended by the manufacturer (Roche Diagnostics, Indianapolis, IN). The nucleotide sequences of Fabp7 primers were 5′-GATGCTTTCTGTGCCACCTG-3′ and 5′-CTGCCTCCACACCAAAGACA-3 (Nihon Gene Research Laboratories, Miyagi, Japan). Template double-stranded DNA (dsDNA) was synthesized from 2.5 μg of cDNA prepared from both WT and rSey2/rSey2 samples. Detailed conditions of the real-time PCR will be supplied on request. Real-time PCR was performed after optimizing MgCl2 concentration.
In situ hybridization. In situ hybridization on whole-mount brains and frozen sections was performed as described previously (Osumi et al., 1997). cDNA for rat Fabp7 was amplified by PCR with the same dsDNA templates and primers used for quantitative PCR, and the obtained cDNA fragment was cloned into pBluescriptIISK(-) (Stratagene, La Jolla, CA). Plasmid DNA for rat Ngn2 was a kind gift from Dr. Nakafuku (University of Tokyo, Tokyo, Japan) (Mizuguchi et al., 2001). Bright-field images were obtained using a CCD camera (Fuji, Tokyo, Japan).
Immunostaining. Immunostaining of whole-mount and frozen sections was performed as described previously (Osumi et al., 1997). Antibodies against FABP7 (Feng et al., 1994; Owada et al., 1996) [rabbit polyclonal; a kind gift from Drs. Y. Owada (Tohoku University, Miyagi, Japan) and M. Watanabe (Hokkaido University, Hokkaido, Japan)], Pax6 (Engelkamp et al., 1999) (mouse monoclonal; a generous gift from Dr. van Heyningen, Western General Hospital, Edinburgh, UK), Pax6 (Inoue et al., 2000) (rabbit polyclonal), nestin (Miyata and Ogawa, 1994) (mouse monoclonal; a kind gift from Dr. Ogawa), Ki-67 (mouse monoclonal; Dako Cytomation, Carpinteria, CA), p27 (mouse IgG1; BD Transduction Laboratories, San Jose, CA), class III β-tubulin (Tuj1; mouse IgG2a; BAbCO), bromodeoxyuridine (BrdU; mouse monoclonal; BD, Franklin Lakes, NJ), Ngn2 (5C6) (Lo et al., 2002) (mouse IgG1; a kind gift from Dr. Anderson, California Institute of Technology, Pasadena, CA), and green fluorescent protein (GFP; rabbit polyclonal; Chemicon, Temecula, CA) were used. As secondary antibodies, we used cyanine 3-conjugated anti-rabbit or anti-mouse IgG, FITC-conjugated anti-mouse or anti-rabbit IgG, and horseradish peroxidase-conjugated anti-rabbit IgG (Jackson Immuno-Research, West Grove, PA). Fluorescent images were obtained by a cooled CCD camera (Roper, Duluth, GA).
Electroporation of cultured rat embryos. The method used for electroporation of cultured rat embryos was described previously (Takahashi and Osumi, 2002). The transferred plasmid vector (pCAX/mPax6, pCAX/GFP, and pSUPER siRNAs) solutions were prepared at concentrations of 5 mg/ml and microinjected into telencephalic vesicles. Square pulses (70 V; 5 Hz) were delivered into the embryos using an electroporator (CUY21; Neppa Gene, Tokyo, Japan).
Gene silencing of Fabp7 with siRNA. Based on the RNA interference method for mammalian cells (Brummelkamp et al., 2002), we used the pSUPER basic vector (Oligoengine, Seattle, WA). Five types of small hairpin RNAs (shRNAs) (i-148, i-149, i-246, i-247, and i-274) were prepared (sequence data are shown in supplemental Fig. 2, available at www.jneurosci.org as supplemental material) and cloned into pSUPER vector according to the protocol provided by the manufacturer. The pSUPER/shRNA plasmid was transcribed into 19 oligonucleotide siRNAs in the cells. E11.5 WT embryos were electroporated with each of the five constructs to select the most effective construct as i-247. The mutant form of i-247 (i-247mt) was prepared by calculating the internal stability (Khvorova et al., 2003) of i-247 target sequence using Oligo 4.0 software (Molecular Biology Insights, Cascade, CO).
Cell proliferation assay. For cell proliferation analysis, rat embryos electroporated with either pSUPER/i-247, pSUPER/i-247mt, or control pSUPER vector were cultured for 24 h. After whole embryo culture, embryos were further cultured for 20 min in a medium containing 40 μm BrdU. The method used for BrdU pulse-labeling was performed as described previously (Takahashi and Osumi, 2002). The BrdU incorporation index was calculated as the percentage of BrdU-labeled cells relative to all GFP-positive cells.
Results
Identification of Fabp7 as a downregulated gene in rSey2/rSey2 brain
To search for genes downstream of Pax6 and determine their importance in the maintenance of proliferation and/or inhibition of differentiation of NEp cells, we took advantage of transcriptome analyses using WT and Pax6 mutant rat brains at E12.5 (corresponding to E10.5 in the mouse). At this developmental stage, neurogenesis has just started, and the majority of NEp cells are still proliferating. rSey2 has a mutation in the Pax6 gene (Osumi et al., 1997), resulting in production of putatively truncated Pax6 protein. In the homozygous Pax6 mutant (rSey2/rSey2), however, such truncated Pax6 protein could not be detected by immunohistochemistry and Western blot analysis (data not shown) using a monoclonal antibody that recognizes the N-terminal domain of Pax6 protein (Engelkamp et al., 1999). Therefore, we concluded that rSey2/rSey2 is a null condition for Pax6 function.
We next compared the transcriptional profiles of the WT and rSey2/rSey2 embryos using Affymetrix rat U34A oligonucleotide microarray containing 8799 genes [5382 known genes and 3417 expressed sequence tag (EST) clusters]. The mRNAs were obtained from the rostral and caudal parts of the brain tissue (here termed the forebrain and hindbrain regions, respectively) to determine the transcriptional profiles of different brain regions. We performed hybridization twice to confirm the screening results (supplemental Table 1, available at www.jneurosci.org as supplemental material). The intensity and fold changes of hybridization signals were drawn by GeneSpring 4.2 software as scatter plots (supplemental Fig. 1A, available at www.jneurosci.org as supplemental material). Among the genes that were differentially expressed in the WT and rSey2/rSey2, we found several genes that were previously reported to be downstream of Pax6 such as β-B3-2-crystallin, α-A-crystallin, and aldehyde dehydrogenase (Duncan et al., 1998; Suzuki et al., 2000) (Table 1). Thus, our strategy seemed to work for screening Pax6 downstream genes. The rat Ngn2 was neither among the known genes plotted on Affymetrix rat U34A nor identified in EST clusters that were downregulated in rSey2/rSey2 according to our criteria (supplemental Table 2A,B, available at www.jneurosci.org as supplemental material). This is because the expression of Ngn2 in WT rat brain is still very weak at E12.5 (data not shown).
Table 1.
Downregulated and upregulated genes in FB
|
Accession number |
Gene name |
Fold change |
nAdv WT |
nAdv −/− |
|---|---|---|---|---|
| U02096a | Brain-type fatty acid binding protein (Fabp 7) | −7.0 | 1.58 | 0.23 |
| M15901ab | β-B3-2-crystallin (Cvekl et al., 1995) | −6.6 | 0.90 | 0.14 |
| U47921ab | αA (insert)-crystallin (Cvekl et al., 1995) | −5.1 | 2.27 | 0.44 |
| L15556 | Phospholipase C (β4) | −3.9 | 0.33 | 0.09 |
| AF001898ab | Aldehyde dehydrogenase (ALDH) (Suzuki et al., 2000) | −3.0 | 2.14 | 0.71 |
| X13905 | Ras-related rab 1B | −2.9 | 0.55 | 0.19 |
| X54467 | Preprocathepsin D | −2.6 | 0.70 | 0.27 |
| X13412a | Flk protein | −2.6 | 0.24 | 0.09 |
| U06713 | SM-20 | −2.5 | 0.74 | 0.30 |
| S49491a | Proenkephalin | −2.4 | 0.69 | 0.29 |
| U36482 | Endoplasmic reticulum protein ERp29 | −2.0 | 4.06 | 2.00 |
| U59245a | Menkes protein (MNK) | 6.6 | 0.03 | 0.22 |
| M85299 | Sodium/hydrogen exchange protein-isoform 1 (NHE-1) | 5.6 | 0.05 | 0.30 |
| X60328 | Cytosolic epoxide hydrolase | 4.2 | 0.08 | 0.33 |
| U75358 | Myeloma protein kinase (PAK-2) | 3.4 | 0.06 | 0.21 |
| U34932 | Fos-related antigen | 3.3 | 0.09 | 0.29 |
| AF022729ab | HNK-1 sulfotransferase (Nagase et al., 2001) | 2.8 | 0.17 | 0.47 |
| S80345 | von Hippel-Lindau tumor suppressor gene homolog (VHL) | 2.6 | 0.10 | 0.26 |
| L20823a | Syntaxin 2 | 2.5 | 0.11 | 0.26 |
| AF025671a | Caspase 2 (Ich1) | 2.4 | 0.36 | 0.84 |
| AF035955a | Kinesin-related protein 6 (KRP6) | 2.4 | 0.20 | 0.52 |
| M60617a | CCAAT binding transcription factor-B subunit (CBF-A11) | 2.4 | 0.15 | 0.35 |
| AA875509a | cDNA clone similar to Mus musculus Mdm2 | 2.4 | 0.11 | 0.27 |
| Z35138 | Fibroblast growth factor receptor 2b | 2.4 | 0.22 | 0.52 |
| X61295 | L1 retroposon, ORF2 | 2.4 | 0.70 | 1.71 |
| AF061947 | Cain | 2.4 | 0.30 | 0.72 |
| S74265 | High-molecular-weight microtubule-associated protein 2 (HMW MAP2) | 2.3 | 0.11 | 0.27 |
| AF056324a | Scaffold attachment factor B | 2.3 | 0.19 | 0.45 |
| S69329b | Isl-1 (homeobox) (Stoykova et al., 2000) | 2.3 | 0.17 | 0.40 |
| D14013 | Cyclin C | 2.2 | 0.10 | 0.22 |
| AF077354a | Ischemia responsive 94 kDa protein (irp94) | 2.2 | 0.45 | 0.98 |
| AB008521 | Dynein light intermediate chain 53/55 | 2.1 | 0.12 | 0.25 |
| X70706a | T-plastin | 2.1 | 0.43 | 0.91 |
| U51583 | Zinc finger homeodomain enhancer-binding protein-1 (Zfhep-1) | 2.1 | 0.15 | 0.33 |
| X62951a | pBUS19 with repetitive elements | 2.1 | 0.51 | 1.04 |
| U33472 | Type 1 astrocyte and olfactory-limbic-associated protein AT1-46 | 2.1 | 0.37 | 0.80 |
| M90514 | DNA fragment homologous to Drosophila pecanex locus | 2.1 | 0.11 | 0.23 |
| U83666 | Cell-cycle checkpoint protein kinase Bub1 (rbub1) | 2.0 | 0.15 | 0.31 |
| L01115a
|
Adenylyl cyclase type VI |
2.0 |
0.26 |
0.53 |
List of known genes downregulated and upregulated in an rSey2/rSey2 rat FB sample based on the following criteria: a normalized average difference (nAdv) of>0.02 and a fold change of less than −2.0 or >2.0. The genes in italics and bold are those commonly downregulated and upregulated in rSey2/rSey2 rat FB and HB samples, respectively.
Confirmed by quantitative PCR.
Reported in previous papers.
Using this approach, we detected several genes that were otherwise not reported previously as downstream to Pax6 (Table 1) (supplemental Table 2A,B, available at www.jneurosci.org as supplemental material). Among these genes, the expression of Fabp7 (B-FABP/BLBP), a brain type fatty acid binding protein, was markedly downregulated both in the FB and HB regions (-7.0-fold and -13.0-fold in FB and HB, respectively) (Table 1) (supplemental Fig. 1A,B, supplemental Table 2A, available at www.jneurosci.org as supplemental material). The reduced expression of Fabp7 in the rSey2/rSey2 brain was also confirmed by quantitative real-time PCR (supplemental Fig. 1B, available at www.jneurosci.org as supplemental material). Therefore, Fabp7 is considered a good candidate as a Pax6 target.
Fabp7 belongs to a conserved multigene family of intracellular lipid-binding proteins, which are ubiquitously expressed in vertebrate tissues with distinct expression patterns for the individual FABPs (Haunerland and Spener, 2004). In the field of developmental neurobiology, FABP7 (more often referred as BLBP) is widely used as a marker for so-called “radial glial cells” in the CNS (Feng et al., 1994; Kurtz et al., 1994; Gotz et al., 1998; Anthony et al., 2004), yet the role of FABP7 is poorly understood. Because other members of FABPs participate in cell proliferation and differentiation (for review, see Haunerland and Spener, 2004), we concentrated on analyses of this molecule.
Expression patterns of Fabp7 in WT and rSey2/rSey2 embryos
To investigate in detail the spatiotemporal expression pattern of Fabp7, we performed whole-mount in situ hybridization and compared Fabp7 and Pax6 expression levels at several embryonic stages of the developing brain (Fig. 1A). Expression of Pax6 appeared at approximately E10 in the WT rat embryo (data not shown). At E10.5, Pax6 was expressed specifically in the forebrain, hindbrain, and spinal cord. After E11.5, Pax6 expression was confined to the dorsal part in the telencephalon and diencephalon and to the ventral part of the hindbrain and spinal cord. Fabp7 expression was first detected in the hindbrain at E10.5. In the forebrain region, Fabp7 expression became evident at E11.0-E11.5. After E12.5, Fabp7 was strongly expressed in the dorsal part of the telencephalon and diencephalon and in the ventral part of the hindbrain (Fig. 1A). These results indicate that the onset of Fabp7 expression was at the stage of NEp cell proliferation and that the expression pattern of Fabp7 after E12.5 was quite similar to that of Pax6 in the rat embryonic CNS.
Figure 1.
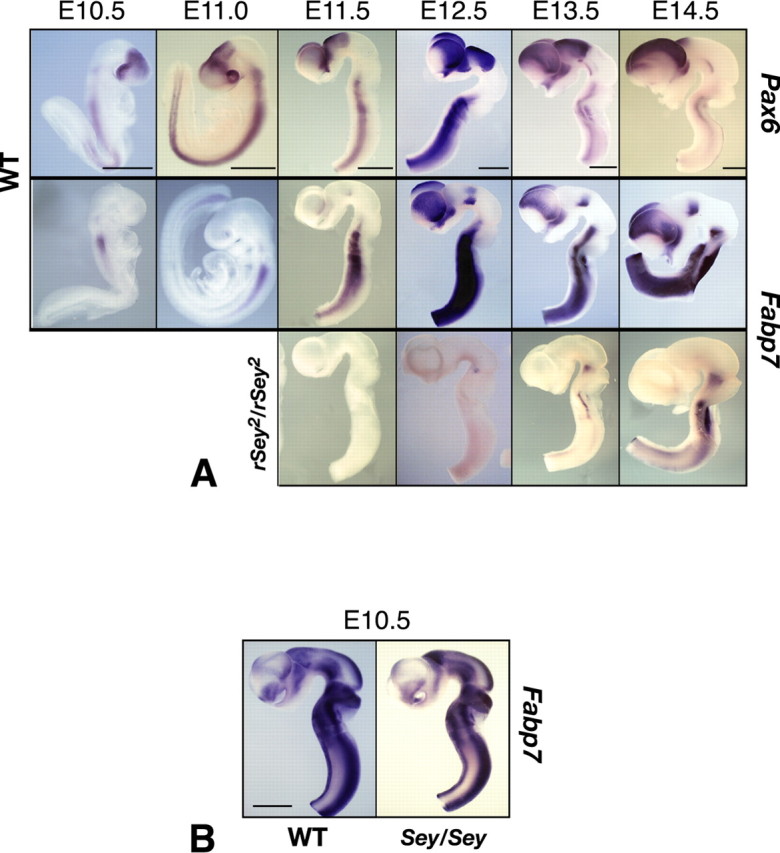
Expression patterns of Pax6 and Fabp7 in the developing rat brain. A, Whole-mount in situ hybridization of the embryo at E10.5 and E11.0 and brain tubes taken from E11.5 to E14.5. At E10.5-E11.0, Pax6 mRNA expression is positive in the forebrain and hindbrain. Fabp7 mRNA expression is positive in the hindbrain. From E12.5 and onwards, expression of Fabp7 becomes evident in the forebrain. Note that the expression pattern of Fabp7 is quite similar to that of Pax6. In rSey2/rSey2 brain, expression of Fabp7 is severely downregulated in the forebrain until E14.5. From E13.5 to E14.5, Fabp7 expression is seen in the rSey2/rSey2 hindbrain. Scale bar, 1 mm. B, Expression patterns of Fabp7 in the developing mouse brain. Whole-mount in situ hybridization of Fabp7 in the brain tube of WT and Sey/Sey at E10.5 (equivalent to rat E12.5). Contrary to the expression in rat embryos, in the mouse, Fabp7 is not specifically expressed in the dorsal telencephalon (Pax6-positive region) but intensely expressed in the ventral telencephalon and in the midbrain (Pax6-negative regions). Also note that Fabp7 expression is almost similar in WT and Sey/Sey brains. Scale bar, 1 mm.
Next, we compared the expression patterns of Fabp7 in WT and rSey2/rSey2 embryos (Fig. 1A). As expected from the microarray and quantitative PCR analyses, Fabp7 expression was undetectable in the telencephalon, diencephalon, and hindbrain in rSey2/rSey2 embryos until E12.5. After E12.5, Fabp7 was faintly expressed at the ventral part of the isthmus and hindbrain in rSey2/rSey2. These results indicate that Fabp7 expression was generally absent or severely reduced in rSey2/rSey2 brain.
The region-specific expression patterns of Fabp7 in the rat embryo are different from those in the mouse embryo; Fabp7 is more intensely expressed in the ventral telencephalon in E10.5 mouse embryo (Fig. 1B), whereas it is specifically expressed in the cortical primordium in the equivalent E12.5 rat embryo (Fig. 1A). Moreover, downregulation of Blbp/Fabp7 is not detected in the Pax6 mutant mouse (Sey/Sey) embryo (Fig. 1B) as reported previously (Gotz et al., 1998). It is possible that other members of the FABP family are expressed in the cortical primordium in the mouse embryo. Thus, Fabp7 is a downstream gene of Pax6 in the rat developing cortex.
Searching the rat genomic sequence with TFBIND software based on TRANSFAC R.3.4 database (Tsunoda and Takagi, 1999), we found four prospective Pax6 binding sites (with matching scores >0.80 against Pax6 binding consensus sequence matrix: M00097 V$PAX6_01) on the 5 kb upstream to Fabp7 (-4571/-4558, -3268/-3255, -2503/-2490, -1927/-1940 bp; the last site was in opposite direction). Therefore, it is very likely that the expression of Fabp7 gene is regulated by Pax6 transcription factor in the developing rat brain.
Induction of Fabp7 expression by Pax6 overexpression
The marked reduction of Fabp7 expression in rSey2/rSey2 embryos led us to test whether exogenous Pax6 can induce Fabp7 expression in the developing brain. Pax6-expression vector was electroporated into E11.5 telencephalon before the appearance of endogenous Fabp7 expression. GFP-expression vector was cotransfected with Pax6-expression vector for conveniently monitoring exogenous gene expression. Exogenous Pax6 protein was detected at 3 h after electroporation, and it was maintained for at least 18 h after electroporation (Fig. 2).
Figure 2.
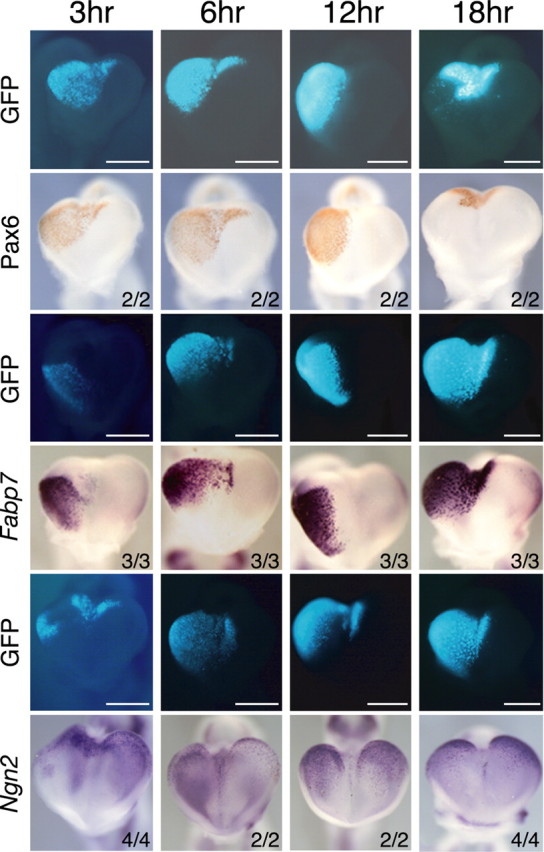
Induction of Fabp7 and Ngn2 mRNAs by Pax6 at the early stage of cortical development. pCAX/mPax6 expression vector was coelectroporated with pCAX/GFP into the telencephalic vesicle of E11.5 WT embryos, which were cultured for 3, 6, 12, and 18 h. GFP fluorescence, immunostaining of Pax6 protein, and Fabp7 mRNA are shown in whole mount. At 3 h after electroporation when Pax6 protein was just synthesized, Fabp7 mRNA was immediately induced by Pax6 overexpression. Strong and persistent expression of Fabp7 was observed at least 18 h after electroporation. In contrast, Ngn2 expression was weakly induced at 3 h but not well maintained until 18 h. Numbers represent the number of embryos analyzed. Scale bar, 0.5 mm.
Ectopic Fabp7 expression was observed at the region where Pax6 was overexpressed at 3 h after electroporation, and it continued at a robust level for at least 18 h (Fig. 2). Induction of Fabp7 gene was also confirmed at a protein level by staining with anti-FABP7 antibody (data not shown). Such effect was not observed after control electroporation using GFP-expression vector only or GFP-expression vector plus pCAX mock vector (data not shown). These results indicate that introduction of Pax6-expression vector can immediately induce strong expression of Fabp7 in the early cortex primordium.
Ngn2 was downregulated in rSey2/rSey2 embryos (data not shown). In this regard, a previous report described the induction of Ngn expression by Pax6 in vitro (Scardigli et al., 2003). Next, we investigated whether Ngn2 gene is induced by overexpression of Pax6 in the cortical primordium at the same stage. Ngn2 expression was induced by transfection of exogenous Pax6, but this induction was very weak and rather transient compared with that of Fabp7 (Fig. 2). Thus, in the early cortex, Pax6 positively regulates the expression of Fabp7 more strongly than that of Ngn2.
FABP7 expression in proliferating NEp cells
The interesting expression patterns of Fabp7 gene prompted us to investigate its functions in the developing brain. In the first step, we performed immunostaining using a FABP7-specific antibody and other markers and examined the morphological features of FABP7-positive cells (Fig. 3). At E12.5, FABP7 protein was specifically expressed in NEp cells with a strong level in the nucleus and a weak level in the cytoplasm. We also noticed different levels of FABP7 expression in NEp cells (see below). Double staining for FABP7 and Pax6 revealed that nearly all FABP7-positive cells coexpressed Pax6. This was also confirmed by immunostaining of dissociated cells taken from E12.5 telencephalon; 88.6% of Pax6-expressing cells were positive for FABP7. Expression of nestin, which labels immature NEp cells and that of Ki-67, labeling actively dividing precursors in all phases of the cell cycle, were also observed in FABP7-positive cells. In contrast, the expression of p27, which labels nonproliferating cells and of acetylated β-tubulin, a specific marker for neurons, never overlapped with that of FABP7. These results indicate that the proliferating NEp cells, but not mature neurons, expressed FABP7 during early cortical development.
Figure 3.
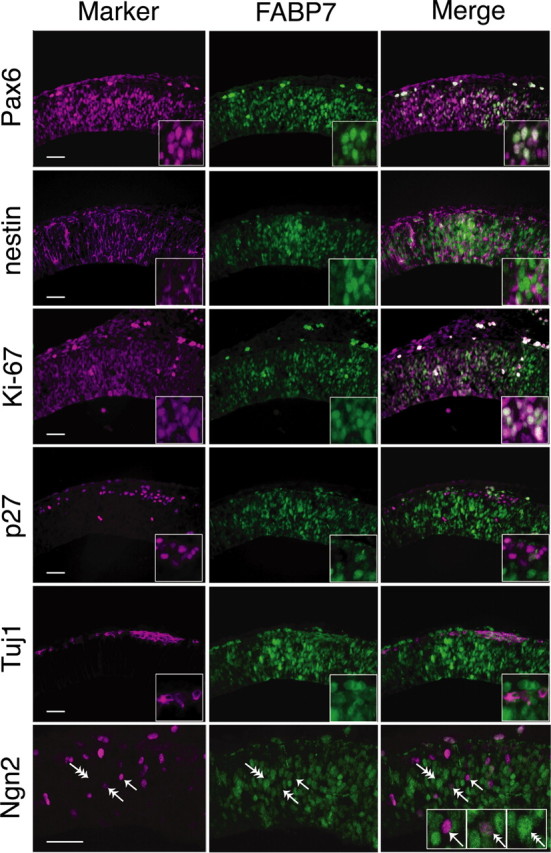
FABP7 expression in proliferating NEp cells. Adjacent coronal sections of the WT telencephalon at E12.5. FABP7 expression overlaps with that of Pax6. FABP7-positive cells coexpress progenitor markers, nestin, and Ki-67 but do not express p27 (a nonproliferating marker) or Tuj1 (a neuronal marker). Double immunostaining for FABP7 and Ngn2 showed three types of NEp cells: FABP7 positive/Ngn2 negative (triple arrow), FABP7 moderately positive/Ngn2 moderately positive (double arrow), and FABP7 negative/Ngn2 positive (single arrow). Scale bar, 100 μm.
The presence of FABP7-strongly positive and -weakly positive NEp cells prompted us to examine the expression of FABP7 and Ngn2 by double immunostaining (Fig. 3). Interestingly, FABP7-strongly expressing NEp cells were negative for Ngn2 (Fig. 3, triple arrow), whereas those strongly expressing Ngn2 were negative for FABP7 (Fig. 3, single arrow). Other cells were moderately double-positive for FABP7 and Ngn2 (Fig. 3, double arrow). These findings suggest a possible transition of FABP7 to Ngn2 in the neuroepithelium of the developing cortex.
Downregulation of FABP7 expression by RNA interference in cultured embryos
The robust expression of FABP7 in proliferating NEp cells suggests that this fatty acid-binding protein plays an important role in proliferation or fate of NEp cells. To investigate this issue, we first performed knocking-down experiments using Fabp7-specific siRNA in the cortex primordium of rat embryos. We designed five types of double-stranded siRNAs that recognized different sequences of Fabp7 mRNA (i-148, i-149, i-246, i-247, and i-274) (supplemental Fig. 2A,B, available at www.jneurosci.org as supplemental material), which were cloned into the pSUPER vector. We found a marked reduction of Fabp7 mRNA with pSUPER/i-247 (Fig. 4B, red boxes) and with pSUPER/i-149 (supplemental Fig. 2C, available at www.jneurosci.org as supplemental material) when electroporated into the telencephalon of E11.5 WT rats. Because pSUPER/i-247 most effectively downregulated Fabp7 mRNA, we used this construct in subsequent experiments. The reduction of FABP7 protein was also confirmed with anti-FABP7 antibody (Fig. 4C). Neither the control mock vector nor a mutant form of siRNA (pSUPER/i-247mt) (Fig. 4A) reduced the expression of Fabp7 at mRNA and protein levels (Fig. 4B,C). These results indicate that Fabp7 expression is downregulated by electroporation with Fabp7-specific siRNA.
Figure 4.
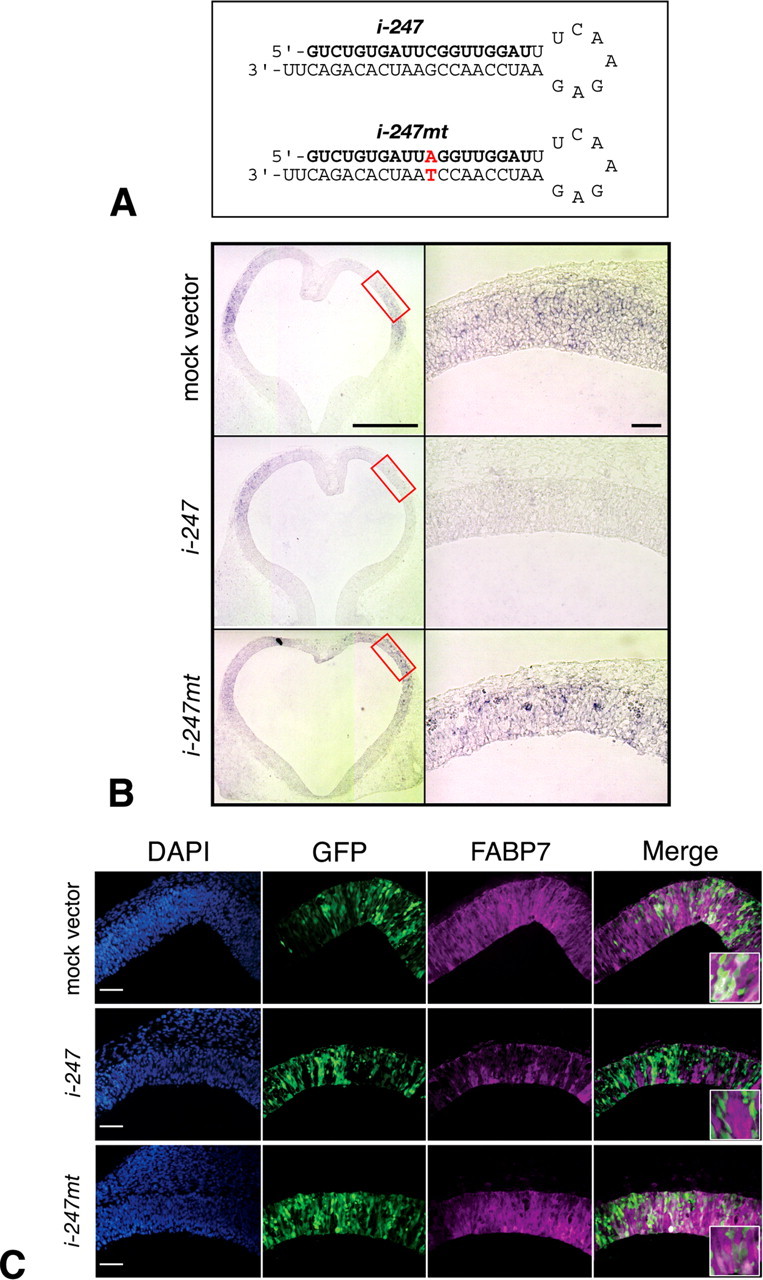
Downregulation of FABP7 expression by siRNA. A, The sequence of the most effective shRNA (i-247) and its mutant (i-247mt) sequences. B, C, pSUPER control mock vector, pSUPER/i-247,or pSUPER/i-247 mt was coelectroporated with pCAX/GFP in the telencephalic vesicle of E11.5 WT rat embryos, which were cultured for 24 h. Reduced expression of Fabp7 mRNA was specifically observed at the region electroporated with pSUPER/i-247 (B, boxes in the left panel), which was not observed in cases electroporated with pSUPER or pSUPER/i-247 mt. Downregulation of FABP7 protein expression was confirmed at the protein level (GFP-positive NEp cells in C). Scale bar, 100 μm.
Misexpression of Fabp7-specific siRNA reduces cell proliferation and promotes neuronal differentiation
Using the above-mentioned Fabp7-specific siRNA, we next examined the role of Fabp7 on proliferation and differentiation of NEp cells. After siRNA electroporation, the proliferating cells were pulse-labeled by BrdU and then the number of BrdU-incorporating cells at 24 h after electroporation was counted. The number of these cells was extremely reduced in embryos electroporated with pSUPER/i-247, compared with those electroporated with the mock vector or pSUPER/i-247mt (Fig. 5A). Quantitative analysis showed a 77.1% reduction in the percentage of BrdU-incorporating cells in embryos transfected with pSUPER/i-247 (Fig. 5B). Cell death was not specifically induced by siRNA electroporation at this stage (data not shown). Therefore, functional knock-down of FABP7 decreased proliferation of telencephalic NEp cells at early embryonic development.
Figure 5.
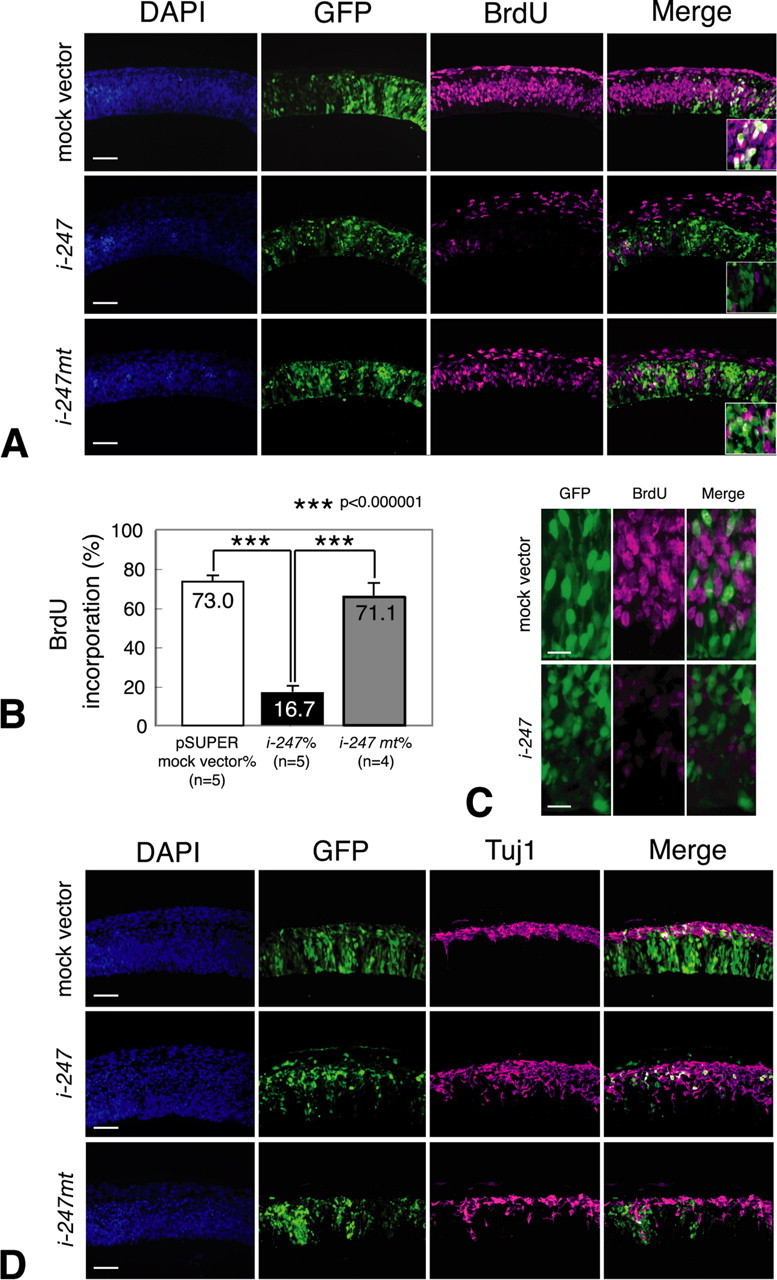
Reduction of FABP7 in neuroepithelial cells is associated with diminished cell proliferation and enhanced neuronal differentiation. A, E11.5 WT rat embryos were electroporated with either control mock vector, pSUPER/i-247,or pSUPER/i-247 mt together with pCAX/GFP in the telencephalic vesicle and cultured for 24 h. Note that immunoreactivity of BrdU is severely reduced in neuroepithelial cells when pSUPER/i-247 was electroporated. B, Quantitative analysis of neuroepithelial cell proliferation. The ordinate shows the percentage of BrdU incorporated cells relative to all GFP-positive cells (ANOVA and Student's t test). C, Morphological changes in pSUPER/i-247 electroporated GFP-positive cells. Cells treated with Fabp7 siRNA become round in shape. Scale bar, 30 μm. D, E11.5 WT rat embryos were electroporated with each expression vector and pCAX/GFP in the telencephalic vesicle and cultured for 48 h. Immunoreactivity of Tuj1 is increased and ectopically observed in the ventricular zone of embryos electroporated with pSUPER/i-247 but not with pSUPER or pSUPER/i-247 mt. Scale bar, 100 μm.
In addition to the above changes, we also noticed morphological changes in GFP-positive cells with reduced FABP7 expression and BrdU incorporation. Specifically, these cells no longer exhibited the morphological features of NEp cells, which have thin processes extending apically and basally. Instead, the cells appeared round in shape (Fig. 5C). These findings suggest that FABP7 is involved in maintaining the morphology of proliferating NEp cells.
To understand further the functional role of FABP7 in NEp cells, we examined neuronal differentiation in embryos electroporated with Fabp7 siRNA. At 24 h after electroporation, no ectopic neurons were yet detected in the ventricular zone (data not shown). After 48 h of electroporation, the number of Tuj1-immunoreactive cells was markedly increased in embryos transfected with pSUPER/i247 (Fig. 5D). Furthermore, in pSUPER/i-247 misexpressed embryos, Tuj1-positive cells were ectopically located in the ventricular zone without significant upregulation of Ngn2 mRNA and Ngn2 protein (data not shown). Such an increase of neuronal differentiation was not observed in embryos electroporated with the mock vector or pSUPER/i-247mt. Together, these experiments indicate that misexpression of Fabp7-specific siRNA was associated with a marked reduction of NEp cell proliferation, morphological changes in these cells, and induction of premature neurogenesis in the early stage of development of the cortex primordium.
Discussion
Several reports have suggested that Pax6 promotes neuronal differentiation in embryonic and adult neurogenesis (Gotz et al., 1998; Hartfuss et al., 2001; Hack et al., 2004). However, little is known about the functional role of Pax6 in proliferating NEp cells at stages before neurogenesis. In the present microarray analysis, we identified Fabp7 as a downregulated gene in the CNS of rSey2/rSey2 embryos. Importantly, we also showed in siRNA experiments that FABP7 is required for maintenance of proliferating NEp cells.
Fabp7 gene is a suitable candidate as a target of Pax6 transcription factor in the early development of the rat cortex for the following reasons. First, the marked downregulation of Fabp7 expression in rSey2/rSey2 was reproduced in quantitative PCR (supplemental Fig. 1A,B, available at www.jneurosci.org as supplemental material). In situ hybridization studies in the WT embryo also revealed spatiotemporal expression patterns of Fabp7 that were quite similar to those of Pax6 (Fig. 1A). In rSey2/rSey2, Fabp7 expression was severely reduced in various brain regions (Fig. 1A). Furthermore, Fabp7 expression was strongly induced within just a few hours when Pax6-expression vector was electroporated into the developing telencephalon (Fig. 2), whereas overexpression of a dominant-negative type Pax6 (Pax6-En) (Takahashi et al., 2002) reduced the expression of Fabp7 (our unpublished data). Regarding the cis-element of mouse Fabp7 (BLBP), Feng and Heintz (1995) reported a putative PAX consensus sequence at -548 to -565 bp, although another group did not mention Pax6 binding sites on the mouse Fabp7 promoter region (Josephson et al., 1998). In the rat, we found four prospective Pax6 binding sites on the 5 kb upstream to Fabp7. Considered together, these findings suggest that Fabp7 is a direct downstream gene of Pax6 in early brain development of the rat. Additional studies are required to elucidate the functional interactions between Pax6 protein and Fabp7 gene.
FABP7-expressing NEp cells coexpressed neural stem/progenitor markers such as nestin (Fig. 3). Interestingly, cells that showed strong expression of FABP7 did not coexpress Ngn2 and vice versa, whereas some cells expressed both FABP7 and Ngn2 at moderate levels. A model that could consistently explain these results would incorporate transition from FABP7-expressing self-renewing NEp cells to Ngn2-expressing neuronal progenitors in the developing cortex (supplemental Fig. 3, available at www.jneurosci.org as supplemental material). Support for such a model is provided by a recent real-time imaging study, in which cells that exhibited intense immunoreactivity for Ngn2 appeared as “basal progenitors” during midneurogenesis (Miyata et al., 2004). In contrast, Pax6 expression is downregulated in these basal progenitor cells (Englund et al., 2005). Furthermore, in our unpublished observation using the E10.5 Tis-21-GFP knock-in mice, in which neuron-generating apical progenitors and their descendants express GFP (Haubensak et al., 2004; Calegari et al., 2005), FABP7 never colocalized with GFP-positive cells in the telencephalic neuroepithelium. In our study, electroporation of Pax6 in the early cortical primordium resulted in weak and transient induction of Ngn2 (Fig. 2), suggesting that Pax6 predominantly transcribes Fabp7 rather than Ngn2 in the early developing cortex. Together, FABP7 seems to maintain NEp cell lineage, thereby producing numerous progenitor cells to form a large cortex primordium. Pax6 may keep a balance between proliferation and differentiation by regulating the transcription of various genes (e.g., Fabp7 and Ngn2) during cortical neurogenesis (supplemental Fig. 3, available at www.jneurosci.org as supplemental material).
Various functions have been proposed for different members of the FABP family, including promotion of cellular uptake and transport of fatty acids, targeting of fatty acids to specific metabolic pathways, and participation in the regulation of gene expression and cell growth (for review, see Haunerland and Spener, 2004). Each member of the FABP family of proteins shows a unique expression pattern distinct to each tissue, and some FABPs are expressed in “stem cells” or “transiently-amplifying cells” in certain tissues (Roth et al., 1991; O'Shaughnessy et al., 2000). It is also generally accepted that many FABPs participate in cell growth rather than differentiation. Although FABP7 (BLBP) has been used for a long time as a marker of “radial glial cells,” which also behave like stem cells in the nervous system (Feng et al., 1994; Kurtz et al., 1994; Gotz et al., 1998; Hartfuss et al., 2001; Anthony et al., 2004), very little is known about its cellular function. Genetically manipulated Fabp7-deficient mice show no obvious defects in brain development (Y. Owada, personal communications), a discrepancy with our results with Fabp7 siRNA (Fig. 5). This is probably a result of the different expression patterns of Fabp7 in the mouse embryo in which Fabp7 is not expressed in the early cortical primordium (Fig. 1B). Another possibility would be compensation by other members of the FABP family, because the expression of H-type FABP is upregulated in E-FABP knock-out mice (Owada et al., 2002). In this regard, affinity-purified anti-BLBP (FABP7) antibodies blocked glial and neuronal differentiation in primary cultures of cerebellar cells but had no effect on cell proliferation (Feng et al., 1994). In our study, Fabp7 siRNA altered NEp cell morphology and reduced cell proliferation in the ventricular zone, whereas overexpression of Fabp7 in the telencephalon of both WT and rSey2/rSey2 embryos had no effect on cell proliferation (our unpublished results). This could be a result of insufficient supply of ligand(s) for FABP7 in our experimental condition. Moreover, overexpression of Fabp7 did not alter expression of region-specific genes such as Nkx2.2 and Dbx1 (our unpublished results). Hence, we conclude that FABP7 is required, at least in part, for the proliferation of NEp cells.
An important factor in maintaining NEp cells in the proliferative undifferentiated state is the Notch-Hes signal (Ross et al., 2003). A recent report showed premature neurogenesis in Hes-1 and Hes-5 double knock-out mice (Hatakeyama et al., 2004). Interestingly, changes in the morphology of NEp cells noted in the cortical primordium of Hes-1 and Hes-5 double knock-out mice were quite similar to our results in Fabp7 RNAi studies. Feng et al. (1994) also observed changes in cell morphology when they treated granule cells with anti-BLBP (FABP7) antibody. Recently, the Notch effecter CBF1 was reported to regulate the transcription of Blbp (Fabp7) gene (Anthony et al., 2005). We did not observe upregulation of Ngn2 in cells electroporated with Fabp7 siRNA (data not shown). Thus, the primary role of FABP7 in cortical neurogenesis could be to maintain epithelial cell cytoskeleton, thereby promoting the proliferation of NEp cells (supplemental Fig. 3, available at www.jneurosci.org as supplemental material).
Proliferation of NEp cells is a key process necessary to establish a huge brain region such as the neocortex of mammals. Production of neurons would be reduced if the size of the progenitor pool is decreased because of a low rate of NEp cell proliferation. Previous studies describing phenotypes of Pax6 mutant suggest that Pax6 contributes to both cell proliferation and neuronal differentiation (Warren et al., 1999; Fukuda et al., 2000; Philips et al., 2005). In gain-of-function studies using dissociated cell cultures in which the structure of the neuroepithelium has been lost, Pax6 is reported to promote neuronal differentiation (Gotz et al., 1998; Heins et al., 2002). In contrast, in Drosophila eye development, cell specification and cell proliferation in the eye primordium are independently controlled by eyeless and eyegone, two types of Pax6 homologs (Jang et al., 2003; Dominguez et al., 2004). The eyegone gene product has a truncated paired domain and binds to a sequence similar to Pax6(5a) binding site (Jun et al., 1998). It is reasonable to assume that differential usage of Pax6(5a) and Pax6 by alternative splicing could regulate the balance between differentiation and proliferation through the transcriptional control of different genes. Alternatively, Pax6 may differentially regulate its downstream genes by differential usage of cobinding partners to activate the target genes. For example, Pax6 and Sox2 regulate crystallin gene expression in the lens (Kamachi et al., 2001), and Pax6 and Cdx2 regulate glucagon gene expression in the pancreas (Hussain and Habener, 1999). Therefore, differential expression of partner molecules may influence the expression of different target genes. Third, a change in chromatin modification might cause differential specificity of target genes; Fabp7 promoter sequence in the rat genome might be inactivated in later cortical development so that it is inaccessible for Pax6 protein to bind. Additional studies using various molecular approaches are necessary to understand the mechanisms by which Pax6 regulates NEp cell proliferation and neuronal differentiation during cortical development.
Footnotes
This work was supported by Grants-in-Aid for Scientific Research from the Japanese Ministry of Education, Culture, Sports, Science and Technology (10220209, 16015205), by a research grant from the TORAY Foundation, and by the Core Research for Evolutional Science and Technology from Japanese Science and Technology Corporation (N.O.). Y.A. was supported by the Sugawara Foundation and the 21st Century Center of Excellence Program “Future Medical Engineering Based On Bio-Nanotechnology.” We thank Drs. Gord Fishell, Yoshio Wakamatsu, Masanori Takahashi, and Daisuke Sakai for valuable suggestions on our work and a critical reading of this manuscript; Noriko Takashima, Nao Kamata, Michi Otonari, and Hisako Yusa for maintenance of the rSey2 colony; and all other members of Keitaikeisei Laboratory for encouragement and important discussion. We also thank Drs. Yuji Owada, Veronica van Heyningen, Masaharu Ogawa, David Anderson, and Masato Nakafuku for providing reagents.
Correspondence should be addressed to Prof. Noriko Osumi, Division of Developmental Neuroscience, Department of Functional Genomics, Center for Translational and Advanced Animal Research, Tohoku University Graduate School of Medicine, 2-1, Seiryo-machi, Aoba-ku, Sendai 980-8575, Japan. E-mail: osumi@mail.tains.tohoku.ac.jp.
Y. Arai's present address: Max-Planck-Institute of Molecular Cell Biology and Genetics, Pfotenhauerstrasse 108, D-01307 Dresden, Germany.
Copyright © 2005 Society for Neuroscience 0270-6474/05/259752-10$15.00/0
References
- Andrews GL, Mastick GS (2003) R-cadherin is a Pax6-regulated, growth-promoting cue for pioneer axons. J Neurosci 23: 9873-9880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony TE, Klein C, Fishell G, Heintz N (2004) Radial glia serve as neuronal progenitors in all regions of the central nervous system. Neuron 41: 881-890. [DOI] [PubMed] [Google Scholar]
- Anthony TE, Mason HA, Gridley T, Fishell G, Heintz N (2005) Brain lipid-binding protein is a direct target of Notch signaling in radial glial cells. Genes Dev 19: 1028-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550-553. [DOI] [PubMed] [Google Scholar]
- Calegari F, Haubensak W, Haffner C, Huttner WB (2005) Selective lengthening of the cell cycle in the neurogenic subpopulation of neural progenitor cells during mouse brain development. J Neurosci 25: 6533-6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvekl A, Kashanchi F, Sax CM, Brady JN, Piatigorsky J (1995) Transcriptional regulation of the mouse alpha A-crystallin gene: activation dependent on a cyclic AMP-responsive element (DE1/CRE) and a Pax-6-binding site. Mol Cell Biol 15: 653-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez M, Ferres-Marco D, Gutierrez-Avino FJ, Speicher SA, Beneyto M (2004) Growth and specification of the eye are controlled independently by Eyegone and Eyeless in Drosophila melanogaster Nat Genet 36: 31-39. [DOI] [PubMed] [Google Scholar]
- Duncan MK, Haynes Jr JI, Cvekl A, Piatigorsky J (1998) Dual roles for Pax-6: a transcriptional repressor of lens fiber cell-specific beta-crystallin genes. Mol Cell Biol 18: 5579-5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelkamp D, Rashbass P, Seawright A, van Heyningen V (1999) Role of Pax6 in development of the cerebellar system. Development 126: 3585-3596. [DOI] [PubMed] [Google Scholar]
- Englund C, Fink A, Lau C, Pham D, Daza RA, Bulfone A, Kowalczyk T, Hevner RF (2005) Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci 25: 247-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estivill-Torrus G, Pearson H, van Heyningen V, Price DJ, Rashbass P (2002) Pax6 is required to regulate the cell cycle and the rate of progression from symmetrical to asymmetrical division in mammalian cortical progenitors. Development 129: 455-466. [DOI] [PubMed] [Google Scholar]
- Feng L, Heintz N (1995) Differentiating neurons activate transcription of the brain lipid-binding protein gene in radial glia through a novel regulatory element. Development 121: 1719-1730. [DOI] [PubMed] [Google Scholar]
- Feng L, Hatten ME, Heintz N (1994) Brain lipid-binding protein (BLBP): a novel signaling system in the developing mammalian CNS. Neuron 12: 895-908. [DOI] [PubMed] [Google Scholar]
- Fukuda T, Kawano H, Osumi N, Eto K, Kawamura K (2000) Histogenesis of the cerebral cortex in rat fetuses with a mutation in the Pax-6 gene. Brain Res Dev Brain Res 120: 65-75. [DOI] [PubMed] [Google Scholar]
- Gotz M, Stoykova A, Gruss P (1998) Pax6 controls radial glia differentiation in the cerebral cortex. Neuron 21: 1031-1044. [DOI] [PubMed] [Google Scholar]
- Hack MA, Sugimori M, Lundberg C, Nakafuku M, Gotz M (2004) Regionalization and fate specification in neurospheres: the role of Olig2 and Pax6. Mol Cell Neurosci 25: 664-678. [DOI] [PubMed] [Google Scholar]
- Hartfuss E, Galli R, Heins N, Gotz M (2001) Characterization of CNS precursor subtypes and radial glia. Dev Biol 229: 15-30. [DOI] [PubMed] [Google Scholar]
- Hatakeyama J, Bessho Y, Katoh K, Ookawara S, Fujioka M, Guillemot F, Kageyama R (2004) Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development 131: 5539-5550. [DOI] [PubMed] [Google Scholar]
- Haubensak W, Attardo A, Denk W, Huttner WB (2004) Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc Natl Acad Sci USA 101: 3196-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haunerland NH, Spener F (2004) Fatty acid-binding proteins-insights from genetic manipulations. Prog Lipid Res 43: 328-349. [DOI] [PubMed] [Google Scholar]
- Heins N, Malatesta P, Cecconi F, Nakafuku M, Tucker KL, Hack MA, Chapouton P, Barde YA, Gotz M (2002) Glial cells generate neurons: the role of the transcription factor Pax6. Nat Neurosci 5: 308-315. [DOI] [PubMed] [Google Scholar]
- Hirabayashi Y, Itoh Y, Tabata H, Nakajima K, Akiyama T, Masuyama N, Gotoh Y (2004) The Wnt/beta-catenin pathway directs neuronal differentiation of cortical neural precursor cells. Development 131: 2791-2801. [DOI] [PubMed] [Google Scholar]
- Hussain MA, Habener JF (1999) Glucagon gene transcription activation mediated by synergistic interactions of pax-6 and cdx-2 with the p300 co-activator. J Biol Chem 274: 28950-28957. [DOI] [PubMed] [Google Scholar]
- Inoue T, Nakamura S, Osumi N (2000) Fate mapping of the mouse prosencephalic neural plate. Dev Biol 219: 373-383. [DOI] [PubMed] [Google Scholar]
- Jang CC, Chao JL, Jones N, Yao LC, Bessarab DA, Kuo YM, Jun S, Desplan C, Beckendorf SK, Sun YH (2003) Two Pax genes, eye gone and eyeless, act cooperatively in promoting Drosophila eye development. Development 130: 2939-2951. [DOI] [PubMed] [Google Scholar]
- Josephson R, Muller T, Pickel J, Okabe S, Reynolds K, Turner PA, Zimmer A, McKay RD (1998) POU transcription factors control expression of CNS stem cell-specific genes. Development 125: 3087-3100. [DOI] [PubMed] [Google Scholar]
- Jun S, Wallen RV, Goriely A, Kalionis B, Desplan C (1998) Lune/eye gone, a Pax-like protein, uses a partial paired domain and a homeodomain for DNA recognition. Proc Natl Acad Sci USA 95: 13720-13725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamachi Y, Uchikawa M, Tanouchi A, Sekido R, Kondoh H (2001) Pax6 and SOX2 form a co-DNA-binding partner complex that regulates initiation of lens development. Genes Dev 15: 1272-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD (2003) Functional siRNAs and miRNAs exhibit strand bias. Cell 115: 209-216. [DOI] [PubMed] [Google Scholar]
- Kim AS, Anderson SA, Rubenstein JL, Lowenstein DH, Pleasure SJ (2001) Pax-6 regulates expression of SFRP-2 and Wnt-7b in the developing CNS. J Neurosci 21: RC132(1-5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz A, Zimmer A, Schnutgen F, Bruning G, Spener F, Muller T (1994) The expression pattern of a novel gene encoding brain-fatty acid binding protein correlates with neuronal and glial cell development. Development 120: 2637-2649. [DOI] [PubMed] [Google Scholar]
- Lo L, Dormand E, Greenwood A, Anderson DJ (2002) Comparison of the generic neuronal differentiation and neuron subtype specification functions of mammalian achaete-scute and atonal homologs in cultured neural progenitor cells. Development 129: 1553-1567. [DOI] [PubMed] [Google Scholar]
- Meech R, Kallunki P, Edelman GM, Jones FS (1999) A binding site for homeodomain and Pax proteins is necessary for L1 cell adhesion molecule gene expression by Pax-6 and bone morphogenetic proteins. Proc Natl Acad Sci USA 96: 2420-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T, Ogawa M (1994) Developmental potentials of early telencephalic neuroepithelial cells: a study with microexplant culture. Dev Growth Differ 36: 319-331. [DOI] [PubMed] [Google Scholar]
- Miyata T, Kawaguchi A, Okano H, Ogawa M (2001) Asymmetric inheritance of radial glial fibers by cortical neurons. Neuron 31: 727-741. [DOI] [PubMed] [Google Scholar]
- Miyata T, Kawaguchi A, Saito K, Kawano M, Muto T, Ogawa M (2004) Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development 131: 3133-3145. [DOI] [PubMed] [Google Scholar]
- Mizuguchi R, Sugimori M, Takebayashi H, Kosako H, Nagao M, Yoshida S, Nabeshima Y, Shimamura K, Nakafuku M (2001) Combinatorial roles of olig2 and neurogenin2 in the coordinated induction of pan-neuronal and subtype-specific properties of motoneurons. Neuron 31: 757-771. [DOI] [PubMed] [Google Scholar]
- Nagase T, Nakamura S, Harii K, Osumi N (2001) Ectopically localized HNK-1 epitope perturbs migration of the midbrain neural crest cells in Pax6 mutant rat. Dev Growth Differ 43: 683-692. [DOI] [PubMed] [Google Scholar]
- O'Shaughnessy RF, Seery JP, Celis JE, Frischauf A, Watt FM (2000) PA-FABP, a novel marker of human epidermal transit amplifying cells revealed by 2D protein gel electrophoresis and cDNA array hybridisation. FEBS Lett 486: 149-154. [DOI] [PubMed] [Google Scholar]
- Osumi N (2001) The role of Pax6 in brain patterning. Tohoku J Exp Med 193: 163-174. [DOI] [PubMed] [Google Scholar]
- Osumi N, Hirota A, Ohuchi H, Nakafuku M, Iimura T, Kuratani S, Fujiwara M, Noji S, Eto K (1997) Pax-6 is involved in the specification of hindbrain motor neuron subtype. Development 124: 2961-2972. [DOI] [PubMed] [Google Scholar]
- Owada Y, Yoshimoto T, Kondo H (1996) Spatio-temporally differential expression of genes for three members of fatty acid binding proteins in developing and mature rat brains. J Chem Neuroanat 12: 113-122. [DOI] [PubMed] [Google Scholar]
- Owada Y, Suzuki I, Noda T, Kondo H (2002) Analysis on the phenotype of E-FABP-gene knockout mice. Mol Cell Biochem 239: 83-86. [PubMed] [Google Scholar]
- Panchision DM, McKay RD (2002) The control of neural stem cells by morphogenic signals. Curr Opin Genet Dev 12: 478-487. [DOI] [PubMed] [Google Scholar]
- Philips GT, Stair CN, Young Lee H, Wroblewski E, Berberoglu MA, Brown NL, Mastick GS (2005) Precocious retinal neurons: Pax6 controls timing of differentiation and determination of cell type. Dev Biol 279: 308-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross SE, Greenberg ME, Stiles CD (2003) Basic helix-loop-helix factors in cortical development. Neuron 39: 13-25. [DOI] [PubMed] [Google Scholar]
- Roth KA, Hermiston ML, Gordon JI (1991) Use of transgenic mice to infer the biological properties of small intestinal stem cells and to examine the lineage relationships of their descendants. Proc Natl Acad Sci USA 88: 9407-9411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scardigli R, Baumer N, Gruss P, Guillemot F, Le Roux I (2003) Direct and concentration-dependent regulation of the proneural gene Neurogenin2 by Pax6. Development 130: 3269-3281. [DOI] [PubMed] [Google Scholar]
- Schuurmans C, Guillemot F (2002) Molecular mechanisms underlying cell fate specification in the developing telencephalon. Curr Opin Neurobiol 12: 26-34. [DOI] [PubMed] [Google Scholar]
- Simpson TI, Price DJ (2002) Pax6; a pleiotropic player in development. BioEssays 24: 1041-1051. [DOI] [PubMed] [Google Scholar]
- Stoykova A, Gruss P (1994) Roles of Pax-genes in developing and adult brain as suggested by expression patterns. J Neurosci 14: 1395-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoykova A, Gotz M, Gruss P, Price J (1997) Pax6-dependent regulation of adhesive patterning, R-cadherin expression and boundary formation in developing forebrain. Development 124: 3765-3777. [DOI] [PubMed] [Google Scholar]
- Stoykova A, Treichel D, Hallonet M, Gruss P (2000) Pax6 modulates the dorsoventral patterning of the mammalian telencephalon. J Neurosci 20: 8042-8050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki R, Shintani T, Sakuta H, Kato A, Ohkawara T, Osumi N, Noda M (2000) Identification of RALDH-3, a novel retinaldehyde dehydrogenase, expressed in the ventral region of the retina. Mech Dev 98: 37-50. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Osumi N (2002) Pax6 regulates specification of ventral neurone subtypes in the hindbrain by establishing progenitor domains. Development 129: 1327-1338. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Sato K, Nomura T, Osumi N (2002) Manipulating gene expressions by electroporation in the developing brain of mammalian embryos. Differentiation 70: 155-162. [DOI] [PubMed] [Google Scholar]
- Temple S, Qian X (1996) Vertebrate neural progenitor cells: subtypes and regulation. Curr Opin Neurobiol 6: 11-17. [DOI] [PubMed] [Google Scholar]
- Tsunoda T, Takagi T (1999) Estimating transcription factor bindability on DNA. Bioinformatics 15: 622-630. [DOI] [PubMed] [Google Scholar]
- Warren N, Price DJ (1997) Roles of Pax-6 in murine diencephalic development. Development 124: 1573-1582. [DOI] [PubMed] [Google Scholar]
- Warren N, Caric D, Pratt T, Clausen JA, Asavaritikrai P, Mason JO, Hill RE, Price DJ (1999) The transcription factor, Pax6, is required for cell proliferation and differentiation in the developing cerebral cortex. Cereb Cortex 9: 627-635. [DOI] [PubMed] [Google Scholar]


