Abstract
The molecular mechanisms that determine the size and complexity of the neuronal dendritic tree are unclear. Here, we show that the phosphoinositide-3′ kinase (PI3K)–Akt–mammalian target of rapamycin (mTOR) signaling pathway promotes the growth and branching of dendrites in cultured hippocampal neurons. Constitutively active mutants of Ras, PI3K, and Akt, or RNA interference (RNAi) knockdown of lipid phosphatase PTEN (phosphatase and tensin homolog deleted on chromosome Ten), induced growth and elaboration of dendrites that was blocked by mTOR inhibitor rapamycin and/or by overexpression of eIF-4E binding protein 1 (4E-BP1), which inhibits translation of 5′ capped mRNAs. The effect of PI3K on dendrites was lost in more mature neurons (>14 d in vitro). Dendritic complexity was reduced by inhibition of PI3K and by RNAi knockdown of mTOR or p70 ribosomal S6 kinase (p70S6K, an effector of mTOR). A rapamycin-resistant mutant of mTOR “rescued” the morphogenetic effects of PI3K in the presence of rapamycin. By regulating global and/or local protein translation, and as a convergence point for multiple signaling pathways, mTOR could play a central role in the control of dendrite growth and branching during development and in response to activity.
Keywords: phosphoinositide, PTEN, rapamycin, dendrite development, dendrite branching, Ras
Introduction
Dendrites are the main site of information input onto neurons, and different neurons have distinctive and characteristic dendrite branching patterns. The multistep process of dendritic arbor development is controlled by both external signals and intrinsic genetic programs (for review, see McAllister, 2000; Cline, 2001; Wong and Ghosh, 2002; Jan and Jan, 2003). Diffusible cues such as neurotrophins, semaphorins or Slits (Polleux et al., 2000; Horch and Katz, 2002; Whitford et al., 2002), cell–cell interactions involving proteins such as Notch1 and β-catenin (Redmond et al., 2000; Yu and Malenka, 2003) and neuronal activity (McAllister, 2000; Wong and Gosh; 2002), have been shown to regulate various steps of dendrite development. Among the proteins that transduce these signals into changes in dendritic shape are protein kinases, such as calcium calmodulin-dependent protein (CaM) kinases II and IV (Wu and Cline, 1998; Redmond et al., 2002; Fink et al., 2003), mitogen-activated protein (MAP) kinases (Fukuda et al., 1995), Rho-associated coiled-coil-forming protein kinase (Nakayama et al., 2000), Pak1 (Hayashi et al., 2002), and phosphoinositide-3′ kinase (PI3K) (Dijkhuizen and Ghosh, 2005).
PI3Ks convert phosphatidylinositol 4,5-bisphosphate into phosphatidylinositol 3,4,5-trisphosphate (PIP3). The production of PIP3 by PI3K induces a myriad of cellular responses in neurons, such as transcription (Brunet et al., 2001), translation (Akama and McEwen, 2003), protein trafficking (Lhuillier and Dryer, 2002; Man et al., 2003; Wang et al., 2003), and cytoskeletal rearrangements (Atwal et al., 2003). These responses are mediated by a variety of downstream effectors, including protein kinases phosphoinositide-dependent kinase-1 and Akt, and small G-proteins of the Rho and Arf families (for review, see Rodgers and Theibert, 2002). The PI3K–Akt pathway has been implicated in neuronal growth, survival, neurite outgrowth, and synaptic plasticity (Atwal et al., 2000; Kuruvilla et al., 2000; Markus et al., 2002; Sanna et al., 2002).
One of the Akt-regulated proteins [the serine/threonine protein kinase mammalian target of rapamycin (mTOR)] has recently gained attention in neuroscience. mTOR controls neuronal cell size (Kwon et al., 2003), local protein synthesis in dendrites (Takei et al., 2004), and synaptic plasticity (Tang et al., 2002; Cammalleri et al., 2003, Hou and Klann, 2004). In this regard, mTOR is thought to act primarily by phosphorylating eIF-4E binding protein 1 (4E-BP1) and p70S6K, which are important regulators of protein translation.
Here, we show that PI3K and Akt, as well as their upstream activators BDNF and Ras, regulate the complexity of dendritic arborization in neurons in an mTOR-dependent manner. Activation of the PI3K–Akt pathway increases dendrite branching and alters the shape of the dendritic arbor. Furthermore, inhibition of mTOR by rapamycin or RNAi is sufficient to decrease both the number of dendrites and the complexity of dendritic branching, similar to the effect of PI3K/Akt antagonism. In addition, knockdown of p70S6K or overexpression of 4E-BP1 mimics the effect of mTOR inhibition. Thus, in addition to cell soma size and synaptic plasticity, mTOR regulates the development and elaboration of the dendritic arbor of neurons.
Materials and Methods
Antibodies and drugs. Guinea pig postsynaptic density-95 (PSD-95) antiserum was described previously (Kim et al., 1995). The following antibodies and drugs were obtained from commercial sources: rabbit anti-PTEN (phosphatase and tensin homolog deleted on chromosome Ten) (Cell Signaling Technology, Beverly, MA), mouse anti-FRAP (FKBP and rapamycin-associated protein; BD Transduction Laboratories, Woburn, MA), rat anti-hemagglutinin (HA) (Roche Diagnostics, Indianapolis, IN), mouse anti-MAP2 (Sigma, St. Louis, MO), mouse anti-Myc (Oncogene, Bedford, MA), mouse anti-Bassoon (StressGen Biotechnologies, Victoria, British Columbia, Canada), rabbit anti-green fluorescent protein (GFP) (Medical and Biological Laboratories, Woburn, MA), mouse anti-GFP (Quantum, Irvine, CA), mouse anti-β-galactosidase (Promega, Madison, WI), rabbit anti-β-galactosidase (ICN, Irvine, CA), mouse anti-FLAG (Sigma) Alexa488-, Alexa568-conjugated secondary antibodies (Invitrogen, Eugene, OR), FITC-, cyanine 3 (Cy3)-, and Cy5-conjugated secondary antibodies (Jackson ImmunoResearch, West Grove, PA), rapamycin and 2-(4-morpholinyl)-8-phenyl-1(4 H)-benzopyran-4-one LY294002 (Cell Signaling Technology), Akt inhibitor, and Akt inhibitor III (Calbiochem, La Jolla, CA).
DNA constructs. The following constructs have been described: p110-WT (wild type), p110* (Hetman et al., 1999) and FLAG-BclII (Hetman et al., 1999; Figueroa-Masot et al., 2001); Akt-WT, Akt-K179D (Manning et al., 2002); Myr-Akt (Datta et al., 1997); mTORS2035R (Hara et al., 1997); Myc-tagged Rac1 (Nakayama et al., 2000); HA-tagged Arf6T27N (Bose et al., 2001); pSUPER vector (Brummelkamp et al., 2002); pSUPERcort300 (Hering and Sheng, 2003); and pGEX-PTEN (Myers et al., 1998). 4EB-P1 cDNA was purchased from I.M.A.G.E consortium (GenBank accession number BG867440)
To generate p110CAAX, an oligonucleotide encoding the H-Ras CAAX box (TGT GTG CTC TCC) was inserted in frame at the C terminus of p110-WT, using QuickChange site-directed mutagenesis (Stratagene, La Jolla, CA). The QuickChange system was used to generate the following: HA-RasV12C40, -S35, and -G37 from pGW1-HA-RasV12; Arf6-WT by changing asparagine 27 of Arf6T27N back to threonine; Arf6-Q67L by substituting glutamine-67 in Arf6-WT into leucine; and 4EB-P1-AA by changing threonines 37 and 46 to alanines (Fingar et al., 2002). Myc-tagged PTEN was generated by inserting the PTEN cDNA (cloned by PCR) into the BglII and EcoRI sites of pGW1-Myc. For Myc-BDNF, the BDNF cDNA was cloned by PCR from rat SMART cDNA library (Clontech, Palo Alto, CA) with added restriction sites for HindIII and BamHI, and subcloned in pEGFP-N1 (Clontech). The GFP was replaced with an N-terminal myc-tag to create Myc-BDNF. β-Actin-GFP was generated by ligating GFP in the SalI and NotI sites in a modified pβ-actin-16 pl vector (Kaech et al., 1996). PTEN small interfering RNA (siRNA) sequence (gatccccagacaaggccaaccgatacttcaagagagtatcggttggc-cttgtcttttttggaa) was designed against rat PTEN mRNA (AF455569) corresponding to nucleotides 990–1008 (nucleotide 1 is the A of the start codon). mTOR siRNA sequences (gatccccgtgaagcgagccttggagtttcaaga-gaactccaaggctcgcttcactt tttggaaa, gatccccgagccacatccgtccctacttcaagagagt-agggacggatgtggctctttttggaaa, and gatccccgcgacatctcatgagaaccttcaa-gagaggttctcatgagatgtcgctttttggaaa) were targeted against rat mTOR mRNA (L37085) nucleotides 444–462, 3071–3089, and 7513–7531, respectively. p70S6K siRNA1315 targeted the rat mRNA (M58340) sequence atccgatcgcctcgaagat (1293–1312). The complementary oligonucleotides were annealed and inserted into pSUPER vector (Brummelkamp et al., 2002).
Neuron culture and transfection. Primary hippocampal cultures were prepared from embryonic day 19 (E19) rat brains (Banker and Goslin, 1991). Cells were plated on coverslips coated with poly-d-lysine (30 μg/ml) and laminin (2 μg/ml) at a density of 75,000/well. Hippocampal cultures were grown in Neurobasal medium (Invitrogen, Carlsbad, CA) supplemented with B27 (Invitrogen), 0.5 mm glutamine, 12.5 μm glutamate, and penicillin/streptomycin mix. At 7 d in vitro (DIV), 10 or 14 hippocampal neurons were transfected using the calcium phosphate method (Passafaro et al., 2001) for 3–7 d. In case of transfection with two plasmids, the DNA of interest was mixed with pEGFP-N1 in a 1:1 ratio. When three plasmids were cotransfected, pEGFP-N1 was mixed with two other plasmids in the ratio 1:2:2. When possible, cells were stained for the appropriate epitope tags and GFP to confirm cotransfection. In RNAi experiments, the siRNA-encoding plasmid and β-actin-GFP were cotransfected in 5:1 ratio. Rapamycin was added 1 d, or 1 d and 4 d after transfection to a final concentration of 10 nm. When cells were transfected with siRNA constructs, rapamycin was added 4–6 h after transfection.
Hippocampal slice culture and transfection. Organotypic hippocampal slice cultures were prepared from postnatal 7-d-old rats as described previously (Stoppini et al., 1991; Sala et al., 2003). Slices were grown in 5% CO2 at 35°C and transfected after 36–48 h in vitro using a biolistic gene gun (Bio-Rad, Hercules, CA). Rapamycin (20 nm) was added the day after transfection. Slices were fixed after 7–8 d in vitro in 4% PFA and 4% sucrose in PBS overnight.
Immunocytochemistry. For staining of endogenous (with the exception of PSD-95) and transfected proteins, neurons were fixed with ice-cold 4% paraformaldehyde and 4% sucrose in PBS for 10–20 min. For PSD-95, cells were fixed for 10 min with ice-cold 100% methanol. After fixation, cells were washed three times with PBS for 5 min at room temperature and incubated with primary antibodies in GDB buffer (0.2% gelatin, 0.8 m NaCl, 0.5% Triton X-100, 30 mm phosphate buffer, pH 7.4) overnight at 4°C. Cells were then washed three times in PBS for 10 min at room temperature. Secondary antibodies were applied in GDB for 1 h at room temperature and washed three times in PBS for 10 min. Secondary antibodies conjugated to Alexa488 and Alexa568 were used for double labeling; FITC-, Cy3-, and Cy5-secondary antibodies were used for triple labeling.
Image analysis and quantification. Confocal images of single- and double-transfected neurons were obtained with sequential acquisition settings at the maximal resolution of the microscope (1024 × 1024 pixels). Each image was a z-series of 7–12 images, each averaged two times with a Kalman filter and taken at 7 μm, 1 μm, or 0.75 μm depth intervals when 10×, 40×, or 63× objective were used, respectively. The resultant stack was “flattened” into a single image using maximum projection. The confocal settings were kept the same for all scans when fluorescence intensity was compared. Morphometric analysis and quantification were performed using MetaMorph image analysis software (Universal Imaging Corporation, Downingtown, PA). For measurement of total dendrite length, scans were acquired with 10× objective and 2× electronic zoom. All dendrites of individual neurons were traced, and the number of pixels was automatically counted and converted to micrometers by MetaMorph. For organotypic cultures, the entire dendritic arbor of each cell was put together in Adobe Photoshop (Adobe Systems, San Jose, CA) from images acquired with 40× objective. To measure the length of secondary and tertiary dendrites, each branch was traced from branching point to the tip, and the number of pixels was automatically counted and converted to micrometers by MetaMorph software. Data are presented as average from >12 cells of the mean length of higher-order branches per cell. Quantification of the number of primary dendrites, number of dendritic tips, and cell soma size as well as Sholl analysis was done on images acquired with 40× objective (0.7× electronic zoom) in dissociated and slice cultures. All nonaxonal protrusions initiating from the cell soma longer than 10 μm were defined as primary dendrites. For dendrite tip number, tips of all nonaxonal protrusions >10 μm were counted. For Sholl analysis, concentric circles with 15 μm differences in diameter were drawn around the cell body, and the number of dendrites crossing each circle was manually counted. The cell soma size was obtained by outlining the cell soma, followed by automatic calculation of pixel area in squared micrometers. Spine analysis was performed as described previously (Sala et al., 2001). Acquisition of the images and morphometric quantification were performed by investigators “blind” to the experimental condition. Cells were excluded from analysis that showed obvious features of toxicity such as dendrite fragmentation/blebbing, vacuoles in the cell body, and lamelopodia structures on dendrites. Data were obtained from three to four independent batches of neurons, and two coverslips (six randomly chosen cells per coverslip) were examined for each experimental condition (e.g., each transfected construct) for each culture batch. In the case of organotypic cultures, slices from two to three independent preparations were used, and a minimum of six slices per experimental condition per preparation were analyzed. For quantification of PTEN and mTOR siRNA effects on the level of endogenous proteins in transfected neurons, cell images were collected using identical intensity settings. Fluorescence intensity of transfected cells (GFP-positive) and nontransfected cells in the same field was measured by MetaMorph software. Data are presented as PTEN or mTOR fluorescence of transfected cells normalized to fluorescence of nontransfected cells.
Immunoblotting. pSUPER or pSUPER-S6K1293 were cotransfected with Myc–p70S6K and GFP in COS7 cells using Lipofectamine (Invitrogen) in a ratio of 10:1:1. Thirty six hours later, cells were immunoblotted for the transfected proteins.
Results
Activation of PI3K promotes dendrite morphogenesis
To study the role of PI3K in later stages of dendritic arbor development (after initiation of primary neurites), we transfected various forms of PI3Kα in dissociated hippocampal neurons in culture at 7, 10, or 14 DIV and expressed for 3 or 7 d. To increase PI3K activity, we used wild-type p110α subunit of PI3K or constitutively active forms of PI3Kα (p110*, p110CAAX). p110* is a fusion protein of p110 catalytic subunit with the “inter-Src homology 2” domain of p85 that was shown previously to increase PIP3 levels (Hu et al., 1995). p110CAAX consists of the p110 catalytic subunit fused to a prenylation motif, which promotes association with the plasma membrane (Egawa et al., 1999). GFP was cotransfected to visualize neuronal morphology.
Both constitutively active and wild-type p110 increased dendrite branching, resulting in increased total number of dendrites (defined as number of dendrite tips), branching complexity, and total dendrite length (Fig. 1) (supplemental Table S1, available at www.jneurosci.org as supplemental material). The degree of effect was greatest with p110CAAX > p110* > p110 WT. The effect of p110 on dendrite elaboration was more prominent when overexpressed for 1 week from 7 DIV than from 10 DIV (Fig. 1A,C,D) and virtually absent when they were transfected at 14 DIV (Fig. 1A,E). Shorter (3 d) overexpression of the p110 variants in 7 DIV neurons was insufficient to affect dendritic branching (Fig. 1A,B).
Figure 1.
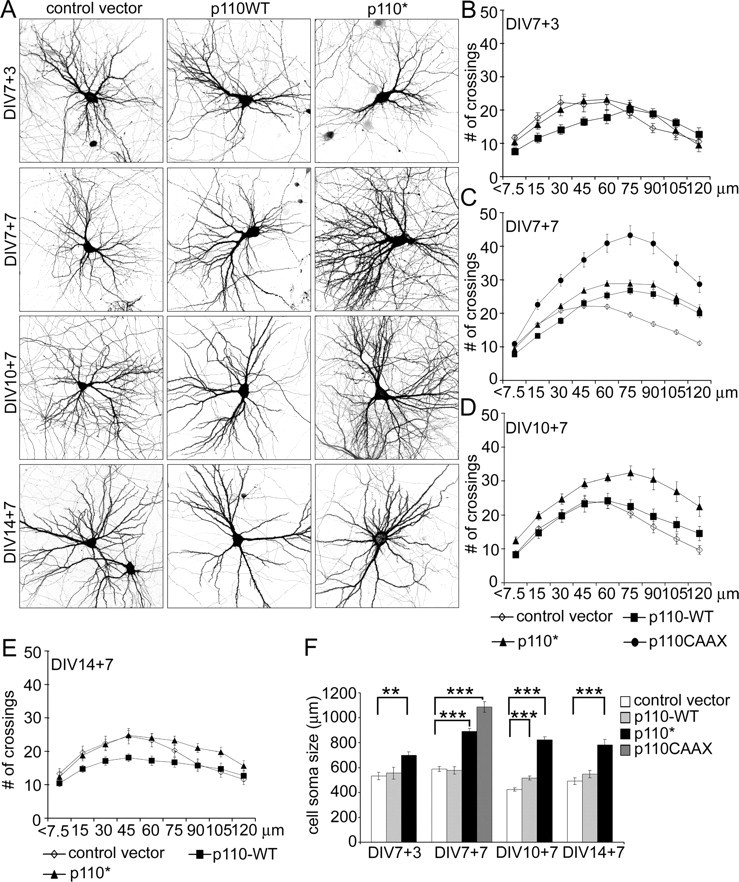
PI3K promotes dendrite branching in hippocampal neurons. A, Representative micrographs of hippocampal neurons transfected with control vector or wild-type p110α or constitutively active p110*. Neuron morphology was visualized by cotransfected GFP. Age and duration of transfection are indicated (e.g., DIV7 + 3 indicates transfected at 7 DIV and expressed for 3 d). B–E, Sholl analysis of neurons transfected with wild-type or active p110 mutants or control vector at various DIV as indicated. The horizontal axis of the Sholl plot indicates the distance from the cell soma. F, Effect of overexpression of PI3K variants on cell soma size. Age and duration of transfection are indicated. ***p < 0.001; **p < 0.01.
In neurons overexpressing p110* or p110CAAX for 7 d from 7 DIV (Fig. 1A), which showed the largest changes in dendritic arbor complexity, total number of dendrites increased ∼36% (in p110*-transfected neurons) and ∼65% (p110CAAX) relative to control (supplemental Table S1, available at www.jneurosci.org as supplemental material). The dendrite branching index (defined as the ratio of number of dendritic tips/number of primary dendrites) increased by ∼26% (p110*) and ∼38% (p110CAAX). Also, total dendrite length increased in neurons overexpressing p110* or p110CAAX by 45 and 110% of control, respectively (supplemental Table S1, available at www.jneurosci.org as supplemental material).
To analyze quantitatively the pattern of branching of the dendritic tree, we used Sholl analysis (Sholl, 1953), which measures the number of dendrites crossing circles at various radial distances from the cell soma. In normal neurons in culture at 14 DIV, the number of crossings increases with distance from the cell soma, reaching a maximum at ∼45 μm. Beyond that distance, the number of crossings decreases, until at the last measured point (120 μm from cell body), the number of crossings is approximately the same as the number of branches crossing the first circle (7.5 μm) (Fig. 1C, black diamonds) (supplemental Table S1, available at www.jneurosci.org as supplemental material). In neurons transfected with constitutively active p110, the number of crossings reached a peak at 60–75 μm from the cell body, and the number of crossings at 120 μm was much higher than the number of primary dendrites (Fig. 1C) (supplemental Table S1, available at www.jneurosci.org as supplemental material). Moreover, the number of crossings at the peak of the Sholl plot and at 120 μm was higher than in control neurons (Fig. 1C) (supplemental Table S1, available at www.jneurosci.org as supplemental material). The upward and rightward shift of the Sholl plot embodied by these numbers reflects an expansion of the “field” covered by the dendritic arbor. Notably, the number of primary dendrites (defined as number of dendrites directly emanating from the soma) was relatively unchanged by constitutively active PI3K, suggesting no impact on dendrite initiation.
Like p110* and p110CAAX, wild-type p110 overexpression also increased the complexity of branching at distances >45 μm from the soma. Curiously, however, the number of dendrite crossings was significantly lower than control in regions more proximal than 45 μm (Fig. 1C) (supplemental Table S1, available at www.jneurosci.org as supplemental material). Neurons transfected at 10 DIV with p110* for 1 week showed similar effects on dendrite branching as neurons transfected at 7 DIV (Fig. 1D) (supplemental Table S1, available at www.jneurosci.org as supplemental material); however, overexpression of p110 wild-type had little effect at this more mature stage. In neurons transfected at 14 DIV, Sholl analysis revealed minimal effect of even p110* on dendrite crossings (Fig. 1E) (supplemental Table S1, available at www.jneurosci.org as supplemental material).
PI3K activity in neurons also caused robust enlargement of the cell soma, as measured by area of the cell body in confocal projections (Fig. 1A,F) (supplemental Table S1, available at www.jneurosci.org as supplemental material). Overexpression of p110* for 7 d resulted in a significant increase in cell soma size, whether transfected at 7, 10, or 14 DIV (Fig. 1F) (supplemental Table S1, available at www.jneurosci.org as supplemental material). Wild-type p110 overexpression was much less effective, and a significant enlargement was only seen when transfected at 10 DIV (Fig. 1F) (supplemental Table S1, available at www.jneuro-sci.org as supplemental material).
Finally, we checked whether overexpression of constitutively active mutants of PI3K affects the number of synapses. Immuno-staining for postsynaptic density protein PSD-95 or presynaptic protein Bassoon revealed no significant differences in the density of puncta between cells transfected at 7 DIV for 1 week with p110* or control vector (control vector: PSD-95, 9.5 ± 0.4 clusters per 10 μm; Bassoon, 10.67 ± 0.4; p110*: PSD-95, 9.33 ± 0.27; Bassoon, 10.09 ± 0.32). We conclude that increased PI3K activity causes enlargement of the cell body and increased branching and elaboration of dendrites in cultured hippocampal neurons. Linear density of synapses is unchanged, despite an increase in total dendrite length, thus implying an increase in total synapse number. The effect on dendrite branching is lost after 14 DIV, suggesting a possible developmental “critical period” for the promoting influence of PI3K on dendrite morphogenesis in culture.
PTEN knockdown mimics PI3K effect on dendrite branching
To confirm that the observed p110 effect on dendrite branching is mediated by elevated levels of PIP3, we tested the effect of RNAi knockdown of PTEN, the PIP3 phosphatase that counteracts PI3K. An RNAi-plasmid targeted against PTEN (pSUPER-PTEN990) inhibited expression of myc-tagged PTEN, but not p110*, when cotransfected in COS7 cells; empty vector (pSUPER) did not affect any of these proteins (data not shown). pSUPER-PTEN990 was also effective in suppressing expression of endogenous PTEN in neurons, resulting in >80% reduction in PTEN immunofluorescence in transfected neurons (Fig. 2A) (p < 0.001 when PTEN990 was compared with controls). Controls pSUPER or pSUPERcort300 (which knocks down cortactin expression) (Hering and Sheng, 2003) had no effect on PTEN.
Figure 2.
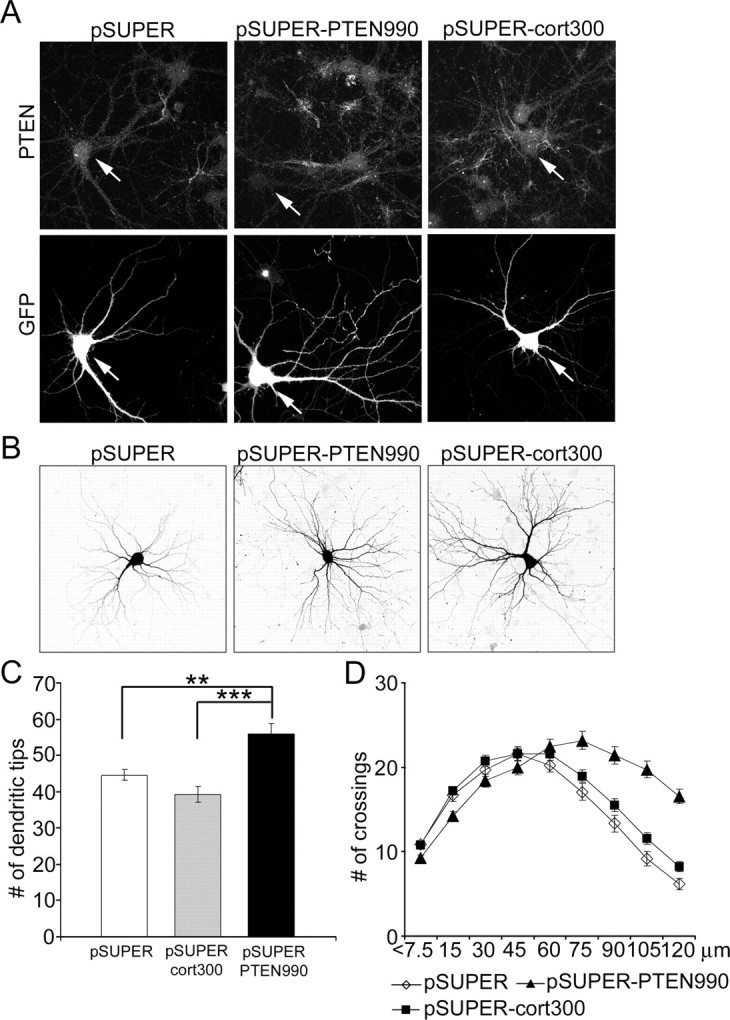
Knockdown of PTEN by RNAi increases dendritic branching of hippocampal neurons. A, PTEN immunostaining of hippocampal neurons transfected at 7 DIV for 4 d with pSUPER vector control, pSUPERcort300 (siRNA directed against cortactin), or pSUPER-PTEN990 (siRNA directed against PTEN). GFP was cotransfected to identify transfected cells and to visualize neuronal morphology. Arrows indicate transfected cells. B, Representative images of hippocampal neurons transfected at 7 DIV with pSUPER, pSUPERcort300, or pSUPER-PTEN990 for 5 d. Morphology of transfected cells was visualized by cotransfected GFP. C, Number of dendritic tips in hippocampal neurons transfected at 7 DIV for 5 d with pSUPER, pSUPERcort300, or pSUPER-PTEN990. ***p < 0.001; **p < 0.01. D, Sholl analysis of neurons transfected with pSUPER, pSUPERcort300, or pSUPER-PTEN990.
Four to 5 d after transfection with pSUPER-PTEN990, neurons showed ∼25% increase in total number of dendrite tips, compared with control cells transfected with pSUPER or pSUPERcort300 (Fig. 2B,C) (supplemental Table S1, available at www.jneurosci.org as supplemental material). Sholl analysis showed that although the number of crossings at the maximum was not significantly different, the “peak” of branching was shifted rightward (to more distal from the soma) in PTEN-RNAi cells (Fig. 2 D) (supplemental Table S1, available at www.jneurosci.org as supplemental material), similar to overexpression of constitutively active p110 (Fig. 1B,C). At 120 μm, the number of dendrite crossings reached 201 and 269% of pSUPER and pSUPERcort300 controls, respectively (supplemental Table S1, available at www.jneurosci.org as supplemental material). Knockdown of PTEN also resulted in a slight but significant increase in cell soma size (supplemental Table S1, available at www.jneurosci.org as supplemental material). Overall, RNAi knockdown of PTEN resulted in a similar dendritic phenotype as overexpression of p110, arguing that the more elaborate dendritic branching is attributable to elevated PIP3 in the cell.
Akt increases complexity of dendritic tree
Which downstream mechanism is responsible for the effect of PI3K on dendrite morphology? We transfected neurons at 7 or 10 DIV with known effectors of PI3K: Akt, Rac1, or Arf6. Only Akt increased the complexity of dendritic tree and cell soma size (Fig. 3). As with PI3K, the greatest effect was observed in neurons transfected at 7 DIV with a constitutively active form of Akt (myr-Akt; in which the catalytic domain of Akt is fused to a myristoylation-signal) (Fig. 3A) (supplemental Table S2, available at www.jneurosci.org as supplemental material). Indeed, myr-Akt had a greater effect on dendrite complexity than p110* when transfected at 7 DIV, increasing the total number of dendrite tips by 78% and branching index by 54% (Fig. 3A) (supplemental Table S2, available at www.jneurosci.org as supplemental material). The number of primary dendrites was only modestly increased by myr-Akt (15%). In addition to increasing the number of branches, myr-Akt also pushed the peak of branch crossings to greater distances from the cell body (Fig. 3B) (supplemental Table S2, available at www.jneurosci.org as supplemental material). The number of dendritic branches at the peak of the Sholl plot and, at 120 μm, were much higher than in control.
Figure 3.
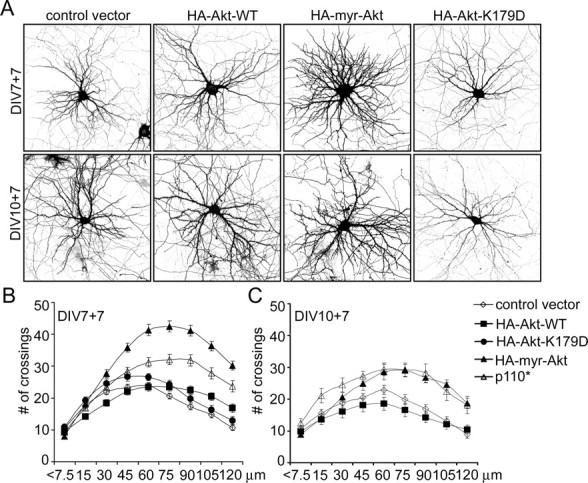
Constitutively active Akt promotes dendritic branching similar to PI3K. A, Representative micrographs of hippocampal neurons transfected with control vector, wild-type Akt (WT), constitutively active Akt (myr-Akt), or kinase-dead Akt (Akt-K179D). Neuronal morphology was visualized by cotransfected GFP. Age and duration of transfection are indicated. B, C, Sholl analysis of neurons transfected with control vector or Akt or PI3K constructs at various DIV as indicated.
Myr-Akt had similar effects on dendrites when transfected at 10 DIV, albeit less dramatic than when transfected at 7 DIV (Fig. 3A,C). At this age of culture, the effects of myr-Akt were quantitatively similar to p110*. Cell soma size was enhanced by myr-Akt when transfected at either 7 or 10 DIV (supplemental Table S2, available at www.jneurosci.org as supplemental material).
Myr-Akt and p110* caused subtle changes in number and morphology of dendritic spines (supplemental Fig. S1, available at www.jneurosci.org as supplemental material). When transfected at 10 DIV for 1 week, both constructs slightly decreased spine density (number of spines per 10 μm: control, 4.7 ± 0.2; p110*, 4.0 ± 0.2; myr-Akt, 3.9 ± 0.3; p < 0.05). Average spine length increased in cells transfected with p110* and myr-Akt, respectively (in μm: control, 1.7 ± 0.04; p110*, 2.0 ± 0.06; myr-Akt, 2.2 ± 0.08; p < 0.001). Spine width was increased slightly by myr-Akt but not by p110* (in μm: control, 0.96 ± 0.01; p110*, 0.98 ± 0.02; myr-Akt, 1.08 ± 0.02; p < 0.001).
In parallel experiments, wild-type Akt and a kinase-dead mutant of Akt (Akt-K179D, which has no dominant-negative activity) did not increase dendrite arborization or cell soma size (Fig. 3A–C) and had no effect on spine characteristics (data not shown). None of the tested Rac1 constructs (Rac1-N17 dominant negative, Rac1 wild-type, Rac1-L61 constitutively active) or Arf6 constructs (Arf6-T27N dominant negative, Arf6-WT, and Arf6-Q71L constitutively active) mimicked the effects of p110* or p110CAAX (data not shown). Thus, these results suggest that Akt is the main PI3K effector involved in regulating dendritic tree shape.
Inhibition of PI3K and Akt causes loss of dendrites
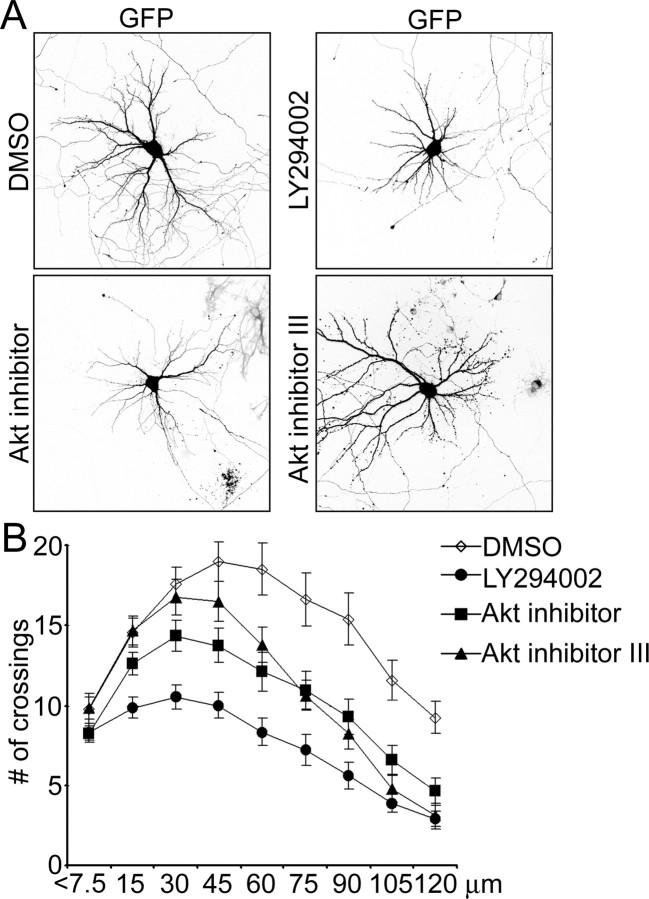
To investigate whether endogenous PI3K and Akt activities are important for dendrite morphogenesis, we used pharmacological inhibitors. Because activity of both these kinases is important for neuronal survival, we transfected neurons with the anti-apoptotic protein Bcl-2 (tagged with FLAG epitope) before drug application to prevent cell death (Markus et al., 2002; Gaudilliere et al., 2004). In these “protected” cells, the PI3K inhibitor LY294002 (50 μm) reduced dendrite number as well as soma size (Fig. 4A,B) (supplemental Table S1, available at www.jneurosci.org as supplemental material). Sholl analysis revealed that the point of maximum crossings shifted closer to the cell soma in LY294002-treated cells, which is opposite to the effect of increasing PI3K or Akt activity. The number of dendrite crossings was reduced by LY294002 at all measured points (Fig. 4B) (supplemental Table S1, available at www.jneurosci.org as supplemental material).
Figure 4.
Inhibition of PI3K and Akt reduces dendritic tree complexity. A, Representative micrographs of hippocampal neurons transfected with Bcl-2 at 7 DIV and treated for 3 d from 9 DIV with either 50 μm LY294002 or 10 μm Akt inhibitor or 25 μm Akt inhibitor III. Neuron morphology was visualized by cotransfected GFP. B, Sholl analysis of hippocampal neurons treated with PI3K or Akt inhibitors or DMSO vehicle as in A.
Application of either 10 μm Akt inhibitor (Hu et al., 2000) or 25 μm Akt inhibitor III (Kozikowski et al., 2003) for 3 d resulted in reduced dendritic tip number as well as a downward and leftward shift of the Sholl plot (Fig. 4A,B) (supplemental Table S2, available at www.jneurosci.org as supplemental material). The concentrations of drugs used in these experiments blocked BDNF-induced autophosphorylation of Akt (data not shown). These results indicate that endogenous PI3K and Akt activity are required for proper growth and branching of the dendritic tree.
Rapamycin blocks PI3K and Akt effects on dendrite branching
mTOR is an important downstream node in the PI3K–Akt pathway. To test whether PI3K and Akt act through mTOR to regulate dendrite complexity, we used a specific inhibitor of mTOR, rapamycin. Neurons were transfected at 7 DIV and treated with 10 nm rapamycin (one or two doses) over the following 6 d. This treatment was enough to decrease phosphorylation of p70S6K on Thr389 (data not shown), which depends on mTOR activity (Burnett et al., 1998). Rapamycin decreased the total number of dendrite tips in vector-transfected cells by ∼20% (single dose) and ∼25% (two doses) (Fig. 5A) (supplemental Table S3, available at www.jneurosci.org as supplemental material). Rapamycin, even in a single dose, abolished the increase of total branch number induced by p110*, p110CAAX, or myr-Akt (Figs. 5A, 7A) (supplemental Table S3, available at www.jneurosci.org as supplemental material). Rapamycin shifted the Sholl plots of neurons overexpressing constitutively active PI3K and Akt back to the control size and shape (Fig. 5B). In control vector-transfected cells, rapamycin decreased the number of crossings at all measured points beyond 30 μm (Fig. 5B) (supplemental Table S3, available at www.jneurosci.org as supplemental material). Rapamycin also decreased the soma size of vector-transfected neurons and prevented the increase of soma size in neurons expressing constitutively active PI3K (supplemental Table S3, available at www.jneurosci.org as supplemental material).
Figure 5.
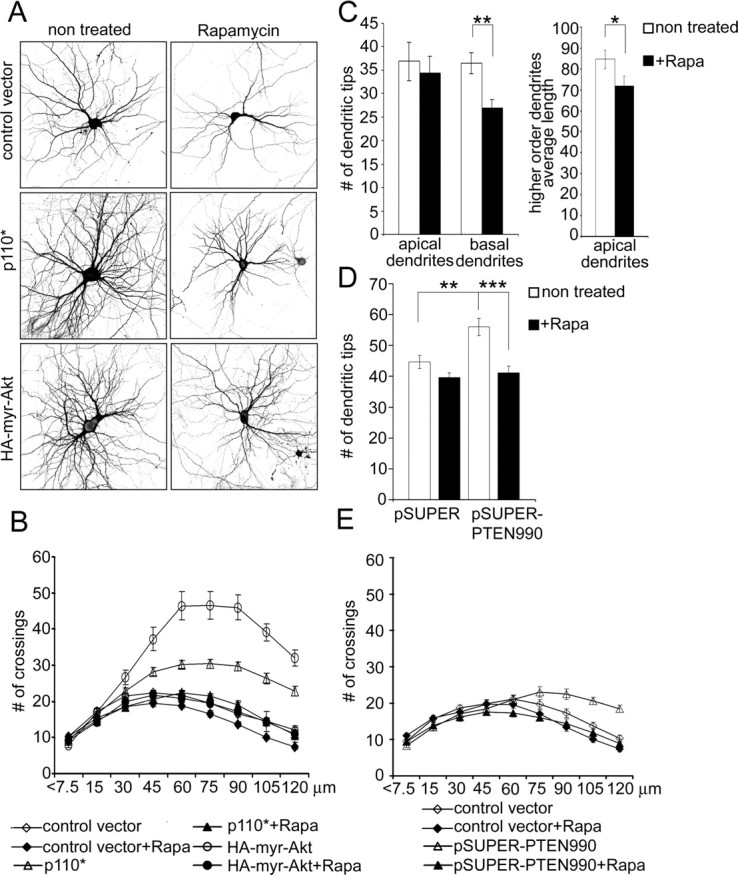
Rapamycin inhibits dendrite branching and the effects of PI3K and Akt. A, Representative micrographs of hippocampal neurons treated with vehicle or 10 nm rapamycin for 6 d. Neurons were transfected with control vector, constitutively active PI3K (p110*), or constitutively active Akt (myr-Akt) 1 d before rapamycin treatment. Neuron morphology was visualized by cotransfected GFP. B, Sholl analysis of neurons transfected with constructs and treated with rapamycin as in A. C, Effect of rapamycin on total number of apical and basal dendritic branches and average length of secondary and tertiary (higher-order) apical branches in organotypic cultures of hippocampus. Dendrites of CA1 neurons were visualized by transfection with GFP. Rapamycin (20 nm) was added 18 h after transfection for 5 d. D, Effect of rapamycin on number of dendrite tips in hippocampal neurons in dissociated culture, transfected at 7 DIV for 5 d with pSUPER or pSUPER-PTEN990. Rapamycin was added 8 h after transfection. ***p < 0.001; **p < 0.01. E, Sholl analysis of neurons transfected with pSUPER or pSUPER-PTEN990, cultured with or without rapamycin.
Figure 7.
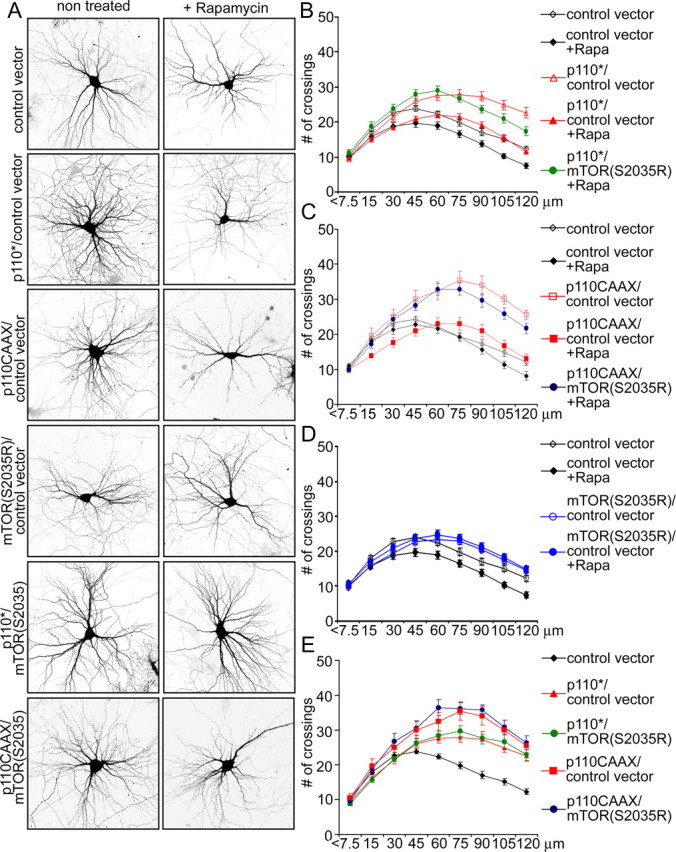
Rapamycin inhibition of PI3K effect can be circumvented by rapamycin-insensitive mutant of mTOR. A, Representative micrographs of hippocampal neurons transfected at 7 DIV with the indicated constructs and treated with vehicle or rapamycin (10 nm) for 6 d. Neuronal morphology was visualized by cotransfected GFP. B–E, Sholl analysis of neurons transfected with the indicated constructs in the absence or presence of rapamycin (+Rapa).
Given that it can inhibit translation, a trivial explanation for the blocking effect of rapamycin is that it prevents the expression of transfected PI3K and myr-Akt. To test this possibility, we verified that rapamycin also antagonizes the dendritic branching induced by RNAi of PTEN (which should not depend on translation of transfected construct). Indeed, rapamycin prevented the enhancing effect of PTEN RNAi on total number of dendritic tips (Fig. 5D) (supplemental Table S3, available at www.jneurosci.org as supplemental material). By Sholl analysis, rapamycin reverted the dendrite branching pattern of PTEN-RNAi neurons back to the normal profile (Fig. 5E). Thus, mTOR activity is required for dendrite elaboration induced by PI3K, Akt, and RNAi of PTEN.
mTOR is important for dendrite arborization
To investigate the function of endogenous mTOR in dendrite development, we targeted mTOR with three different siRNAs: mTOR-444, mTOR-3071, and mTOR-7513. Four days after transfection of any of these constructs into 1-week-old neurons, staining for endogenous mTOR was reduced by ∼90% (p < 0.001; Mann–Whitney test) (Fig. 6A, arrows), whereas transfection of pSUPER or pSUPERcort300 had no effect. All three mTOR siRNAs reduced the total number of dendrite tips (by 35–52%) (Fig. 6B) (supplemental Table S3, available at www.jneurosci.org as supplemental material). Of the three, only mTOR-3071 siRNA caused a small but significant decrease in the number of primary dendrites when compared with control pSUPER- or pSUPERcort300-transfected cells (supplemental Table S3, available at www.jneurosci.org as supplemental material). These results indicate a relatively selective effect of mTOR RNAi on dendrite branching rather than primary dendrite formation.
Figure 6.
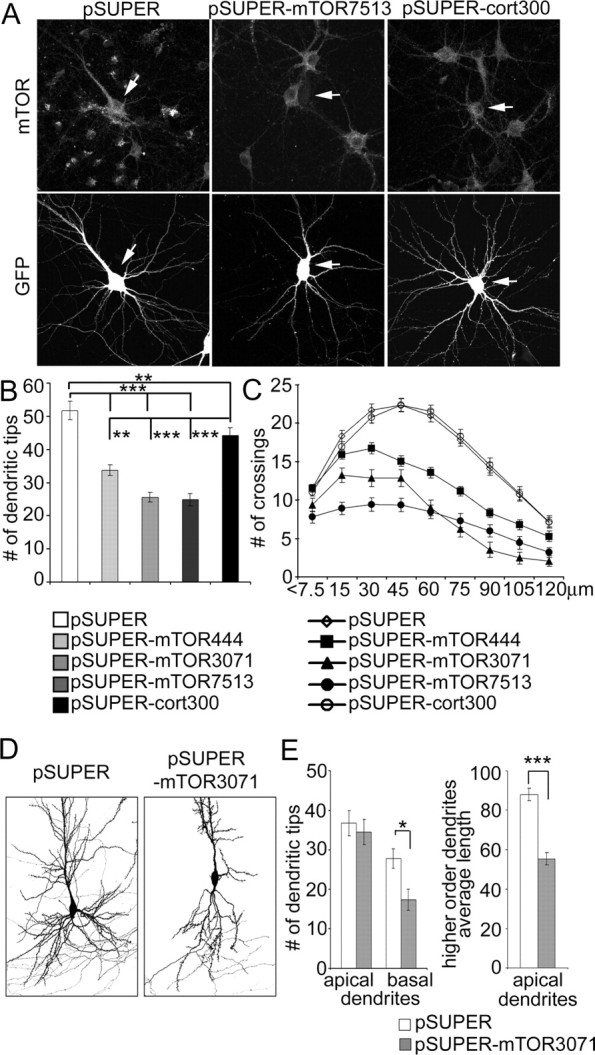
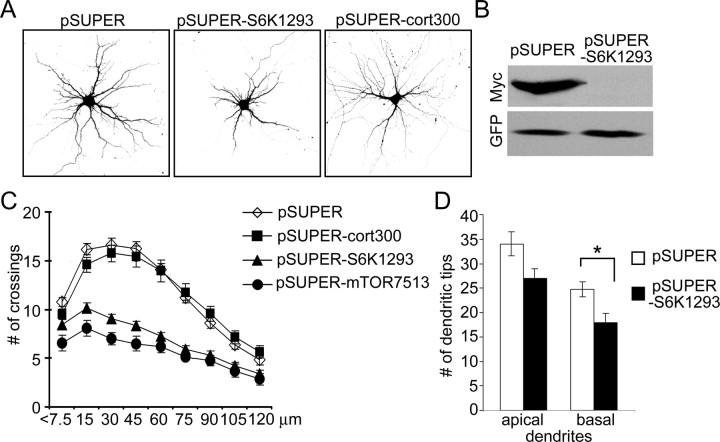
RNAi knockdown of mTOR decreases dendrite branching. A, Representative images of GFP and mTOR staining of hippocampal neurons transfected at 7 DIV for 4 d with pSUPER, pSUPERcort300, or pSUPER-mTOR7513 (RNAi directed against mTOR). GFP was cotransfected to visualize transfected cells (indicated by arrows). B, Number of dendrite tips in hippocampal neurons transfected at 7 DIV for 4 d with pSUPER, pSUPERcort300, or one of pSUPER-mTOR siRNA plasmids (444, 3071, or 7513). C, Sholl analysis of neurons transfected with control or mTOR siRNA plasmids. D, Images of CA1 neurons in organotypic culture transfected for 5 d with pSUPER vector or pSUPER-mTOR3071 siRNA plasmid (only basal dendrites shown in full). GFP was cotransfected for visualization of cell. E, Effect of mTOR3071 siRNA on total number of basal dendrites and average length of secondary and tertiary branches (referred to as higher-order dendrites) of apical dendrites in CA1 neurons of hippocampal organotypic cultures. ***p < 0.001; **p < 0.01; *p < 0.05.
In addition to confirming the reduction of dendrite branches, Sholl analysis showed that all three mTOR-RNAi constructs shifted the peak of branching closer to the cell soma, whereas RNAi of cortactin had a minimal effect on branching pattern (Fig. 6C) (supplemental Table S3, available at www.jneurosci.org as supplemental material). In these RNAi experiments, mTOR-444 consistently had the weakest effect on dendrite branching, which correlated with the fact that only mTOR-3071 and mTOR-7513 siRNAs were able to produce a significant decrease in soma size (supplemental Table S3, available at www.jneurosci.org as supplemental material).
Finally, we tested whether mTOR is sufficient to mediate the effects of PI3K on dendrite branching by using a rapamycin-resistant mutant of mTOR (mTOR-S2035R). Cotransfection of mTOR-S2035R prevented the decrease in dendrite number and complexity induced by rapamycin in control vector-transfected cells (Fig. 7A,D) (supplemental Table S3, available at www.jneurosci.org as supplemental material). In the presence of rapamycin, mTOR-S2035R also rescued the increase of total number of dendrite tips and of branching complexity induced by p110* or p110CAAX (Fig. 7A,B,C) (supplemental Table S3, available at www.jneurosci.org as supplemental material). Coexpression of mTOR-S2035R in the absence of rapamycin did not further enhance the elaboration of dendrites attributable to p110* or p110CAAX (Fig. 7A,E) (supplemental Table S3, available at www.jneurosci.org as supplemental material).
Role of p70S6K and 4EB-P1 in dendrite development
mTOR stimulates translation by activating p70S6K, a kinase that increases synthesis of several components of the translation machinery (Jefferies et al., 1997). In addition, mTOR phosphorylates 4E-BP1 and thereby inhibits binding of 4E-BP1 to initiation factor eif4E, which leads to enhanced formation of the initiation complex for translation of 5′ guanine cap-containing mRNAs (Beretta et al., 1996). We tested the role of these pathways in dendrite arborization by RNAi knockdown of p70S6K and overexpression of 4E-BP1 in hippocampal neurons.
One-week-old neurons were transfected with RNAi construct pSUPER-S6K1293, which effectively suppressed expression of p70S6K protein in COS7 cells (Fig. 8B). Three days after transfection with pSUPER-S6K1293, neurons displayed ∼25% reduction in total dendritic tip number, compared with neurons transfected with pSUPER or pSUPER-cort300 (Fig. 8A) (supplemental Table S4, available at www.jneurosci.org as supplemental material). The number of primary dendrites also fell by 11% (supplemental Table S4, available at www.jneurosci.org as supplemental material). p70S6K-RNAi neurons showed a downward and leftward shift of the Sholl plot, similar to that observed with mTOR-RNAi, indicating simplification of dendritic tree (Fig. 8C) (supplemental Table S4, available at www.jneurosci.org as supplemental material).
Figure 8.
RNAi knockdown of p70S6K decreases dendrite branching. A, Representative micrographs of hippocampal neurons transfected at 7 DIV for 4 d with pSUPER, pSUPERcort300, or pSUPER-S6K1293 (RNAi directed against p70S6K). GFP was cotransfected for visualization of transfected cells. B, Western blot analysis of effect of pSUPER-S6K1293 on expression of cotransfected Myc-p70S6K protein in COS7 cells. C, Sholl analysis of neurons transfected at 7 DIV for 4 d with pSUPER, pSUPERcort300, or pSUPER-S6K1293. D, Effect of pSUPER-S6K1293 transfection on total number of apical and basal dendrites in CA1 neurons of hippocampal organotypic cultures. *p < 0.05
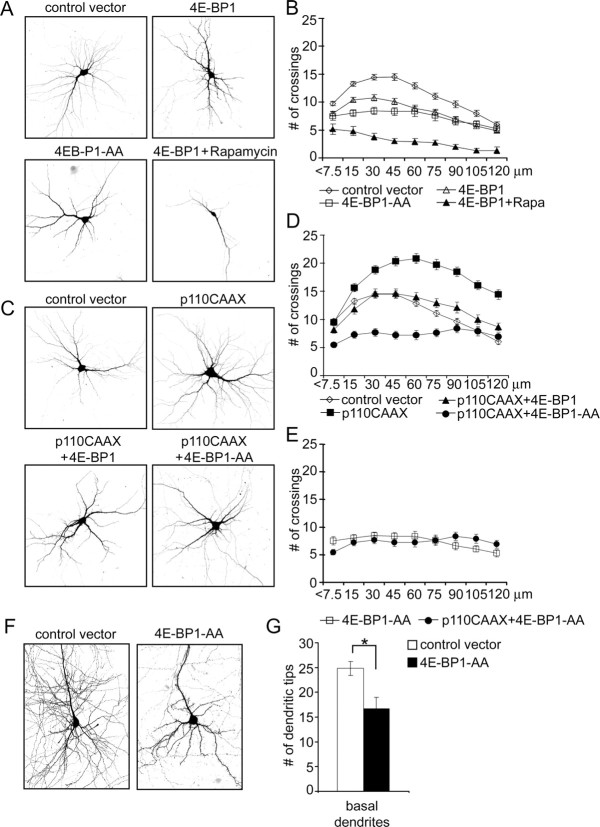
In complementary experiments, 7 DIV neurons were transfected for 1 week with wild-type 4E-BP1 or a mutant 4E-BP1 that cannot be phosphorylated by mTOR (4E-BP1-AA) (Fingar et al., 2002). Expression of either of these proteins reduced the total number of dendrite tips without affecting the number of primary dendrites (Fig. 9A) (supplemental Table S4, available at www.jneurosci.org as supplemental material). The inhibitory effect was smaller for wild-type 4E-BP1 (16% decrease) than 4E-BP1-AA (33% decrease), presumably because the former can still be phosphorylated by endogenous mTOR. Chronic application of 10 nm rapamycin to cells transfected with wild-type 4E-BP1 reduced the number of dendritic branches even further (Fig. 9A) (supplemental Table S4, available at www.jneurosci.org as supplemental material) and additionally decreased the number of primary dendrites. Overexpression of 4E-BP1 and 4E-BP1-AA also reduced complexity of branching as revealed by Sholl analysis (Fig. 9B).
Figure 9.
Overexpression of 4E-BP1 inhibits dendritic branching. Representative micrographs of hippocampal neurons transfected at 7 DIV for 1 week with control vector, wild-type 4E-BP1, or 4E-BP1-AA mutant. Rapamycin (10 nm) was added 1 d after transfection, as indicated. Neuron morphology was visualized by cotransfected GFP. B, Sholl analysis of neurons transfected and/or treated as in A. C, Representative micrographs of hippocampal neurons transfected at 7 DIV for 7 d with p110CAAX and 4E-BP1 constructs, as indicated. Neuronal morphology was visualized by cotransfected GFP. D, E, Sholl analysis of neurons transfected with the indicated constructs as described in A and C. F, G, Effect of 4E-BP1-AA mutant over expression on total number of basal dendrites in CA1 neurons of hippocampal organotypic cultures. *p < 0.05.
If the PI3K–Akt–mTOR pathway regulates dendrite arborization through mTOR-dependent protein synthesis, then one might expect that overexpression of 4E-BP1 would block the increase in dendritic branching induced by active PI3K. Indeed, the increase of total dendrite tip number induced by p110CAAX was reduced (58%) by cotransfection of wild-type 4E-BP1 and completely blocked by the nonphosphorylatable 4E-BP1-AA, which reduced the total number of dendrite tips to that seen with 4E-BP1-AA alone (Fig. 9C) (supplemental Table S4, available at www.jneurosci.org as supplemental material). Sholl analysis confirmed that coexpression of 4E-BP1 with p110CAAX shifted the branching pattern toward control (Fig. 9D) (supplemental Table S4, available at www.jneurosci.org as supplemental material). The Sholl plot for coexpression of 4E-BP1-AA with p110CAAX was flat (implying much reduced branching) and similar to overexpression of 4E-BP1-AA alone (Fig. 9C–E).
Role of mTOR in dendrite morphogenesis in hippocampal slice cultures
We investigated the role of mTOR in a more intact preparation: organotypic cultures of the hippocampus. CA1 pyramidal neurons of hippocampal slice cultures treated for 5 d with rapamycin showed a reduced number of basal dendrites (Fig. 5C). Rapamycin had no significant effect on the total number of apical branches, but it decreased the average length of higher-order (secondary and tertiary) branches of apical dendrites by ∼15% (Fig. 5C). Similarly, CA1 neurons transfected with RNAi construct against mTOR (pSUPER-mTOR3071) showed a reduced number of basal dendrites as well as shortening of secondary and tertiary apical branches (Fig. 6D,E). RNAi against p70S6K (pSUPER-S6K1293) and overexpression of 4E-BP1-AA also decreased the number of basal dendrites (Figs. 8D, 9F,G). Thus, endogenous mTOR as well as mTOR-regulated protein synthesis are important for normal development of dendrite arbors in organotypic slice cultures as well as dissociated cultures of hippocampus.
Ras and BDNF also promote dendrite branching
PI3K is activated directly by Ras, and Ras was shown previously to regulate dendritic branching in transgenic animals (Alpar et al., 2003). Constitutively active Ras mutants have been identified that selectively stimulate PI3K (RasV12C40), the MAP kinase (MAPK) pathway (RasV12S35), or Ral–guanine nucleotide dissociation stimulator (GDS) (RasV12G37) (Rodriguez-Viciana et al., 1997; Koh et al., 2002). When transfected into neurons at 7 DIV and overexpressed for 1 week, all three forms of active Ras led to increased neuronal branching (Fig. 10A,B) (supplemental Table S5, available at www.jneurosci.org as supplemental material). Cells transfected with any of the Ras mutants showed an elevated number of crossings at the peak of the Sholl plot (Fig. 10B) (supplemental Table S5, available at www.jneurosci.org as supplemental material), which implies greater branching. However, only RasV12C40 was able to move the Sholl peak more distal from the cell soma and increase the number of dendrite crossings at 120 μm, which is a sign of outward expansion of the “dendrite field” (Fig. 10 B) (supplemental Table S5, available at www.jneurosci.org as supplemental material). Thus, the dendritic elaboration phenotype caused by overexpression of active PI3K or Akt is most closely mimicked by RasV12C40, the Ras mutant that selectively stimulates PI3K. Interestingly, rapamycin (10 nm) prevented not only the RasV12C40 effect but also the increased dendritic branching induced by RasV12S35 and RasV12G37 (Fig. 10A), suggesting a possible convergence of these Ras pathways on mTOR for promoting dendrite elaboration.
Figure 10.
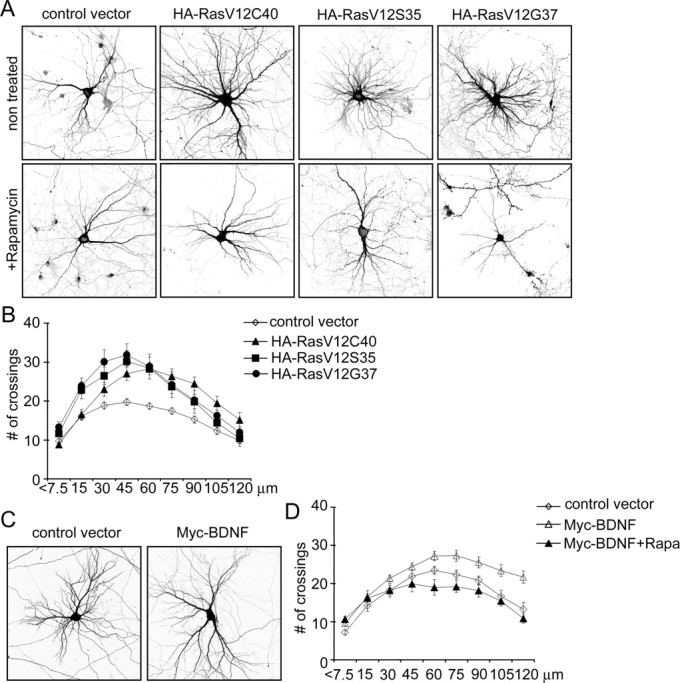
Ras and BDNF regulate dendritic branching in an mTOR-dependent manner. A, Representative micrographs of hippocampal neurons transfected at 7 DIV for 1 week with control vector, PI3K-specific (RasV12C40), MAPK-specific (RasV12S35), or Ral–GDS-specific (RasV12G37) constitutively active mutants of Ras, and treated with vehicle or single dose of 10 nm rapamycin 1 d after transfection. Neuron morphology was visualized by cotransfected GFP. B, Sholl analysis of neurons transfected with different Ras mutants, as indicated. C, Representative micrographs of hippocampal neurons transfected at 7 DIV for 1 week with control vector or myc-BDNF. D, Sholl analysis of neurons transfected with myc-BDNF grown in the absence or presence of 10 nm rapamycin for 6 d.
BDNF is a physiological activator of Ras–PI3K pathways in neurons and has been implicated in dendrite development (McAllister et al., 1995; Horch and Katz, 2002; Tolwani et al., 2002; Wirth et al., 2003). We tested whether BDNF might act via the Ras–PI3K–Akt pathway to regulate dendrite morphogenesis. Cells transfected with myc-tagged BDNF for 1 week showed a slight but significant increase in the number of dendrite branches, as evidenced by greater number of dendrite crossings at the peak of the Sholl plot (Fig. 10C,D) (supplemental Table S5, available at www.jneurosci.org as supplemental material). We presume this is attributable to an autocrine effect of overexpressed BDNF (Horch and Katz, 2002). As with activation of the PI3K–Akt pathway, BDNF shifted the point of maximum crossings distally from the cell soma and increased the number of dendrite crossings at 120 μm. The BDNF effects were blocked by 10 nm rapamycin (Fig. 10D) (supplemental Table S5, available at www.jneurosci.org as supplemental material), implying that BDNF promotion of dendrite branching depends on mTOR.
Discussion
PI3Ks are involved in multiple aspects of neuronal development and physiology, including differentiation, polarization, growth, survival, cytoskeletal reorganization, regulated secretion, and receptor trafficking (Rodgers and Theibert, 2002). PI3K has been reported to promote neurite initiation and outgrowth as well as stability of growth cones, whereas PI3K inhibitors impair neurite outgrowth induced by various stimuli (Kita et al., 1998; Atwal et al., 2000, 2003; Sanchez et al., 2001; Dijkhuizen and Ghosh, 2005). However, in most cases, the role of PI3K was assessed at very early developmental stages or in neuron-like cell lines, thus obscuring its possible role in regulating the elaboration of the dendritic tree. Indeed, studies of the function of PI3K in neurite initiation and stability usually did not specify the type of the outgrowths or focused on axons and primary dendrites only.
This study focused on dendrite morphogenesis occurring after primary neurite initiation. We found that PI3K activity promotes the branching and growth of dendrites during a stage of development of hippocampal neurons in culture (7–14 DIV) when synaptogenesis is particularly active. During this phase, both overexpression and loss-of-function experiments (LY294002, rapamycin, and/or RNAi) establish the importance of PI3K and the downstream Akt–mTOR pathway in normal dendrite elaboration. More work is needed, however, to confirm the role of this pathway in vivo.
PI3K in stages of dendrite development
Development of dendrites can be divided into several overlapping stages. After an initial phase of slow dendrite growth, rapid extension and arborization occurs, characterized by an increased rate of additions and retractions of branches (Wu et al., 1999; Williams and Truman, 2004) and high motility of filopodia on dendritic growth cones (Portera-Cailliau et al., 2003; Williams and Truman, 2004). Subsequently, the dendritic arbor becomes more stable, showing a reduced rate of branch additions and retractions (Wu et al., 1999) and disappearance of growth cone-associated filopodia (Portera-Cailliau et al., 2003). Because the main mode of dendritic branching is splitting of growth cones (Acebes and Ferrus, 2000), dendrite branching is significantly reduced in mature neurons. Thus, our finding that PI3K activation does not affect dendritic arborization beyond 14 DIV is compatible with the idea that dendrites mature from a dynamic phase to a less plastic state.
Rac1 and c-Jun N-terminal protein kinase have been implicated as PI3K effectors in control of neurite initiation (Kita et al., 1998). Another potential PI3K target, Arf6, can affect dendrite outgrowth and branching in young neurons (Hernandez-Deviez et al., 2002). However, our work highlights the importance of Akt in control of dendritic branching and reveals the essential role of mTOR activity in this regulation. At least in more mature neurons (7–14 DIV), our results point to the Akt–mTOR signaling pathway as the primary mediator of PI3K regulation of the pattern and degree of dendrite branching.
mTOR: controller of dendritic arbor “size”
A major conclusion of the study is the central importance of mTOR in the development of the dendritic tree. Inhibition of mTOR by rapamycin or RNAi was sufficient to reduce the number of branches and to “proximalize” the shape of the dendritic tree. Moreover, the effects of BDNF, Ras, PI3K, PTEN RNAi, and Akt on branching were all blocked by rapamycin. As for other cell types, mTOR seems to control neuronal soma size (Kwon et al., 2003), which we confirmed here. Our findings, however, extend the functions of mTOR beyond cell body growth to include the control of dendritic field size and complexity.
It is known that mTOR controls cell growth in response to mitogens and nutrient availability by regulating translation (for review, see Schmelzle and Hall, 2000; Harris and Lawrence, 2003). Active mTOR phosphorylates 4E-BP1 and p70S6K (Burnett et al., 1998), which stimulate translation of 5′ capped and 5′TOP (5′-terminal oligo pyrimidine tract) mRNAs, respectively (Beretta et al., 1996; Jefferies et al., 1997). mTOR and its substrates 4E-BP1 and p70S6K are present in dendrites (Tang et al., 2002), and phosphorylated p70S6K accumulates in dendrites in response to synaptic activity in a rapamycin-sensitive manner (Cammalleri et al., 2003). Thus, mTOR is in an appropriate location to regulate protein synthesis in response to synaptic activity and neurotrophins, and indeed it was recently shown to control local dendritic translation in response to BDNF and insulin (Takei et al., 2004; Lee et al., 2005).
How do we explain that multiple Ras effector pathways (PI3K, MAPK, Ral–GDS) can stimulate dendritic growth and elaboration, and that all these effects were blocked by rapamycin? The PI3K pathway to mTOR is mediated by Akt and results in inhibition of tuberin, a GAP (GTPase-activating protein) for Rheb (Ras homolog enriched in brain); elevated GTP-bound Rheb then stimulates mTOR (Manning et al., 2002; Garami et al., 2003; Tee et al., 2003b). Another activator of mTOR is phosphatidic acid (Fang et al., 2001), which is generated by phospholipase D. Phospholipase D is stimulated by Ral GT-Pases, which are activated by Ras through RalGDS (Jiang et al., 1995). Whether these mechanisms mediate any of the Ras effects on dendrites in neurons remains to be established.
More difficult to explain is how dendritic branching induced by a MAPK-specific Ras mutant can depend on mTOR activity. Rapamycin was shown to block MAP kinase-induced phosphorylation of p70S6K in cardiomyocytes (Wang and Proud, 2002), and MAP kinase activity seems to be important for regulation of Tuberin (Tee et al., 2003a). However, no such interaction has yet been shown in neurons. Experiments by Kelleher et al. (2004) show that in neurons, mTOR and MAP kinases can act in concert to regulate activity-dependent protein synthesis.
The details of Ras signaling to mTOR remain to be completed. Nevertheless, because Ras transduces signals from multiple extracellular factors (via receptor tyrosine kinases, adhesion receptors, and G-protein-coupled receptors) and is also activated by calcium influx (Finkbeiner and Greenberg, 1996; Bivona and Philips, 2003), mTOR could play an integrative role in controlling dendrite morphology in response to diverse extracellular signals.
mTOR-regulated translation and dendrite morphogenesis
We found that suppression of p70S6K or overexpression of 4E-BP1 impairs dendrite branching similarly to inhibition of mTOR. These results imply that mTOR-dependent protein synthesis is involved in the regulation of dendritic arborization. In contrast, Dijkhuizen and Ghosh (2005) showed that BDNF-induced formation of primary dendrites was not blocked by the protein synthesis inhibitor cycloheximide. Differences in age of neurons, primary dendrites versus dendrite branches, and time course offer explanations for the apparent discrepancy between the two studies. Although Dijkhuizen and Ghosh (2005) observed rapid changes in primary dendrite formation in young neurons, our research focused on a longer-term function of the PI3K–Akt–mTOR pathway in shaping dendritic trees in mature neurons.
The dendritic distribution of mTOR and its downstream effectors suggest that mTOR could regulate growth and branching through modulating dendritic protein synthesis. The idea is attractive because it might allow for localized control of dendrite morphology in response to local influences, such as synaptic activity or neurotrophins; however, direct evidence is lacking. In contrast, certain proteins such as BDNF, MAP2, and CaMKIIα can be synthesized in dendrites (Mayford et al., 1996; Tongiorgi et al., 1997; Steward and Halpain, 1999), and their potential to affect dendrite morphogenesis is already established. Genetic screens in Drosophila have identified RNA binding/trafficking proteins that regulate dendrite branching, further supporting the idea that localized translation might play a role in dendrite development (Lee et al., 2003; Ye et al., 2004).
Footnotes
C.C.H. was supported by the International Human Frontier Science Program Organization. M.S. is an Investigator of the Howard Hughes Medical Institute. We thank the following for plasmids: M. Hetman for p110-WT, p110*, and pFLAG-BclII; L. Cantley for Akt-WT and Akt-K179D; H. Hering for pSUPERcort 300; S. Datta for Myr-Akt; D. Sabatini for mTORS2035R and Myc-p70S6K; L. Luo for Myc-tagged Rac1 variants; M. Czech for HA-tagged Arf6T27N; D. Pak for pGW1-HA-RasV12; and N. Tonks for pGEX-PTEN variants. We thank Z. Li and N. Hussain for comments on this manuscript.
Correspondence should be addressed to Morgan Sheng, The Picower Institute for Learning and Memory, Massachusetts Institute of Technology, 77 Massachusetts Avenue (46-4303), Cambridge, MA 02139. E-mail: msheng@mit.edu.
DOI:10.1523/JNEUROSCI.2270-05.2005
Copyright © 2005 Society for Neuroscience 0270-6474/05/2511300-13$15.00/0
References
- Acebes A, Ferrus A (2000) Cellular and molecular features of axon collaterals and dendrites. Trends Neurosci 23: 557–565. [DOI] [PubMed] [Google Scholar]
- Akama KT, McEwen BS (2003) Estrogen stimulates postsynaptic density-95 rapid protein synthesis via the Akt/protein kinase B pathway. J Neurosci 23: 2333–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpar A, Palm K, Schierwagen A, Arendt T, Gartner U (2003) Expression of constitutively active p21H-rasval12 in postmitotic pyramidal neurons results in increased dendritic size and complexity. J Comp Neurol 467: 119–133. [DOI] [PubMed] [Google Scholar]
- Atwal JK, Massie B, Miller FD, Kaplan DR (2000) The TrkB-Shc site signals neuronal survival and local axon growth via MEK and P13-kinase. Neuron 27: 265–277. [DOI] [PubMed] [Google Scholar]
- Atwal JK, Singh KK, Tessier-Lavigne M, Miller FD, Kaplan DR (2003) Semaphorin 3F antagonizes neurotrophin-induced phosphatidylinositol 3-kinase and mitogen-activated protein kinase kinase signaling: a mechanism for growth cone collapse. J Neurosci 23: 7602–7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banker G, Goslin K (1991) Culturing nerve cells. Cambridge, MA: MIT.
- Beretta L, Gingras AC, Svitkin YV, Hall MN, Sonenberg N (1996) Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J 15: 658–664. [PMC free article] [PubMed] [Google Scholar]
- Bivona TG, Philips MR (2003) Ras pathway signaling on endomembranes. Curr Opin Cell Biol 15: 136–142. [DOI] [PubMed] [Google Scholar]
- Bose A, Cherniack AD, Langille SE, Nicoloro SM, Buxton JM, Park JG, Chawla A, Czech MP (2001) G(alpha)11 signaling through ARF6 regulates F-actin mobilization and GLUT4 glucose transporter translocation to the plasma membrane. Mol Cell Biol 21: 5262–5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550–553. [DOI] [PubMed] [Google Scholar]
- Brunet A, Datta SR, Greenberg ME (2001) Transcription-dependent and -independent control of neuronal survival by the PI3K-Akt signaling pathway. Curr Opin Neurobiol 11: 297–305. [DOI] [PubMed] [Google Scholar]
- Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM (1998) RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci USA 95: 1432–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammalleri M, Lutjens R, Berton F, King AR, Simpson C, Francesconi W, Sanna PP (2003) Time-restricted role for dendritic activation of the mTOR-p70S6K pathway in the induction of late-phase long-term potentiation in the CA1. Proc Natl Acad Sci USA 100: 14368–14373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline HT (2001) Dendritic arbor development and synaptogenesis. Curr Opin Neurobiol 11: 118–126. [DOI] [PubMed] [Google Scholar]
- Datta SR, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg ME (1997) Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell 91: 231–241. [DOI] [PubMed] [Google Scholar]
- Dijkhuizen PA, Ghosh A (2005) BDNF regulates primary dendrite formation in cortical neurons via the PI3-kinase and MAP kinase signaling pathways. J Neurobiol 62: 278–288. [DOI] [PubMed] [Google Scholar]
- Egawa K, Sharma PM, Nakashima N, Huang Y, Huver E, Boss GR, Olefsky JM (1999) Membrane-targeted phosphatidylinositol 3-kinase mimics insulin actions and induces a state of cellular insulin resistance. J Biol Chem 274: 14306–14314. [DOI] [PubMed] [Google Scholar]
- Fang Y, Vilella-Bach M, Bachmann R, Flanigan A, Chen J (2001) Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science 294: 1942–1945. [DOI] [PubMed] [Google Scholar]
- Figueroa-Masot XA, Hetman M, Higgins MJ, Kokot N, Xia Z (2001) Taxol induces apoptosis in cortical neurons by a mechanism independent of Bcl-2 phosphorylation. J Neurosci 21: 4657–4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingar DC, Salama S, Tsou C, Harlow E, Blenis J (2002) Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev 16: 1472–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink CC, Bayer KU, Myers JW, Ferrell Jr JE, Schulman H, Meyer T (2003) Selective regulation of neurite extension and synapse formation by the beta but not the alpha isoform of CaMKII. Neuron 39: 283–297. [DOI] [PubMed] [Google Scholar]
- Finkbeiner S, Greenberg ME (1996) Ca(2+)-dependent routes to Ras: mechanisms for neuronal survival, differentiation, and plasticity? Neuron 16: 233–236. [DOI] [PubMed] [Google Scholar]
- Fukuda M, Gotoh Y, Tachibana T, Dell K, Hattori S, Yoneda Y, Nishida E (1995) Induction of neurite outgrowth by MAP kinase in PC12 cells. Oncogene 11: 239–244. [PubMed] [Google Scholar]
- Garami A, Zwartkruis FJ, Nobukuni T, Joaquin M, Roccio M, Stocker H, Kozma SC, Hafen E, Bos JL, Thomas G (2003) Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol Cell 11: 1457–1466. [DOI] [PubMed] [Google Scholar]
- Gaudilliere B, Konishi Y, de la Iglesia N, Yao G, Bonni A (2004) A CaMKII-NeuroD signaling pathway specifies dendritic morphogenesis. Neuron 41: 229–241. [DOI] [PubMed] [Google Scholar]
- Hara K, Yonezawa K, Kozlowski MT, Sugimoto T, Andrabi K, Weng QP, Kasuga M, Nishimoto I, Avruch J (1997) Regulation of eIF-4E BP1 phosphorylation by mTOR. J Biol Chem 272: 26457–26463. [DOI] [PubMed] [Google Scholar]
- Harris TE, Lawrence Jr JC (2003) TOR signaling. Sci STKE 2003: re15. [DOI] [PubMed]
- Hayashi K, Ohshima T, Mikoshiba K (2002) Pak1 is involved in dendrite initiation as a downstream effector of Rac1 in cortical neurons. Mol Cell Neurosci 20: 579–594. [DOI] [PubMed] [Google Scholar]
- Hering H, Sheng M (2003) Activity-dependent redistribution and essential role of cortactin in dendritic spine morphogenesis. J Neurosci 23: 11759–11769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Deviez DJ, Casanova JE, Wilson JM (2002) Regulation of dendritic development by the ARF exchange factor ARNO. Nat Neurosci 5: 623–624. [DOI] [PubMed] [Google Scholar]
- Hetman M, Kanning K, Cavanaugh JE, Xia Z (1999) Neuroprotection by brain-derived neurotrophic factor is mediated by extracellular signal-regulated kinase and phosphatidylinositol 3-kinase. J Biol Chem 274: 22569–22580. [DOI] [PubMed] [Google Scholar]
- Horch HW, Katz LC (2002) BDNF release from single cells elicits local dendritic growth in nearby neurons. Nat Neurosci 5: 1177–1184. [DOI] [PubMed] [Google Scholar]
- Hou L, Klann E (2004) Activation of the phosphoinositide 3-kinase-Akt-mammalian target of rapamycin signaling pathway is required for metabotropic glutamate receptor-dependent long-term depression. J Neurosci 24: 6352–6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Klippel A, Muslin AJ, Fantl WJ, Williams LT (1995) Ras-dependent induction of cellular responses by constitutively active phosphatidylinositol-3 kinase. Science 268: 100–102. [DOI] [PubMed] [Google Scholar]
- Hu Y, Qiao L, Wang S, Rong SB, Meuillet EJ, Berggren M, Gallegos A, Powis G, Kozikowski AP (2000) 3-(Hydroxymethyl)-bearing phosphatidylinositol ether lipid analogues and carbonate surrogates block PI3-K, Akt, and cancer cell growth. J Med Chem 43: 3045–3051. [DOI] [PubMed] [Google Scholar]
- Jan YN, Jan LY (2003) The control of dendrite development. Neuron 40: 229–242. [DOI] [PubMed] [Google Scholar]
- Jefferies HB, Fumagalli S, Dennis PB, Reinhard C, Pearson RB, Thomas G (1997) Rapamycin suppresses 5′TOP mRNA translation through inhibition of p70s6k. EMBO J 16: 3693–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Luo JQ, Urano T, Frankel P, Lu Z, Foster DA, Feig LA (1995) Involvement of Ral GTPase in v-Src-induced phospholipase D activation. Nature 378: 409–412. [DOI] [PubMed] [Google Scholar]
- Kaech S, Ludin B, Matus A (1996) Cytoskeletal plasticity in cells expressing neuronal microtubule-associated proteins. Neuron 17: 1189–1199. [DOI] [PubMed] [Google Scholar]
- Kelleher III RJ, Govindarajan A, Jung HY, Kang H, Tonegawa S (2004) Translational control by MAPK signaling in long-term synaptic plasticity and memory. Cell 116: 467–479. [DOI] [PubMed] [Google Scholar]
- Kim E, Niethammer M, Rothschild A, Jan YN, Sheng M (1995) Clustering of Shaker-type K+ channels by interaction with a family of membrane-associated guanylate kinases. Nature 378: 85–88. [DOI] [PubMed] [Google Scholar]
- Kita Y, Kimura KD, Kobayashi M, Ihara S, Kaibuchi K, Kuroda S, Ui M, Iba H, Konishi H, Kikkawa U, Nagata S, Fukui Y (1998) Microinjection of activated phosphatidylinositol-3 kinase induces process outgrowth in rat PC12 cells through the Rac-JNK signal transduction pathway. J Cell Sci 111: 907–915. [DOI] [PubMed] [Google Scholar]
- Koh YH, Ruiz-Canada C, Gorczyca M, Budnik V (2002) The Ras1-mitogen-activated protein kinase signal transduction pathway regulates synaptic plasticity through fasciclin II-mediated cell adhesion. J Neurosci 22: 2496–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozikowski AP, Sun H, Brognard J, Dennis PA (2003) Novel PI analogues selectively block activation of the pro-survival serine/threonine kinase Akt. J Am Chem Soc 125: 1144–1145. [DOI] [PubMed] [Google Scholar]
- Kuruvilla R, Ye H, Ginty DD (2000) Spatially and functionally distinct roles of the PI3-K effector pathway during NGF signaling in sympathetic neurons. Neuron 27: 499–512. [DOI] [PubMed] [Google Scholar]
- Kwon CH, Zhu X, Zhang J, Baker SJ (2003) mTor is required for hypertrophy of Pten-deficient neuronal soma in vivo. Proc Natl Acad Sci USA 100: 12923–12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A, Li W, Xu K, Bogert BA, Su K, Gao FB (2003) Control of dendritic development by the Drosophila fragile X-related gene involves the small GTPase Rac1. Development 130: 5543–5552. [DOI] [PubMed] [Google Scholar]
- Lee CC, Huang CC, Wu MY, Hsu KS (2005) Insulin stimulates postsynaptic density-95 protein translation via the phosphoinositide 3-kinase-Akt-mammalian target of rapamycin signaling pathway. J Biol Chem 280: 18543–18550. [DOI] [PubMed] [Google Scholar]
- Lhuillier L, Dryer SE (2002) Developmental regulation of neuronal K(Ca) channels by TGFbeta1: an essential role for PI3 kinase signaling and membrane insertion. J Neurophysiol 88: 954–964. [DOI] [PubMed] [Google Scholar]
- Man HY, Wang Q, Lu WY, Ju W, Ahmadian G, Liu L, D'Souza S, Wong TP, Taghibiglou C, Lu J, Becker LE, Pei L, Liu F, Wymann MP, MacDonald JF, Wang YT (2003) Activation of PI3-kinase is required for AMPA receptor insertion during LTP of mEPSCs in cultured hippocampal neurons. Neuron 38: 611–624. [DOI] [PubMed] [Google Scholar]
- Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC (2002) Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol Cell 10: 151–162. [DOI] [PubMed] [Google Scholar]
- Markus A, Zhong J, Snider WD (2002) Raf and akt mediate distinct aspects of sensory axon growth. Neuron 35: 65–76. [DOI] [PubMed] [Google Scholar]
- Mayford M, Baranes D, Podsypanina K, Kandel ER (1996) The 3′-untranslated region of CaMKII alpha is a cis-acting signal for the localization and translation of mRNA in dendrites. Proc Natl Acad Sci USA 93: 13250–13255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister AK (2000) Cellular and molecular mechanisms of dendrite growth. Cereb Cortex 10: 963–973. [DOI] [PubMed] [Google Scholar]
- McAllister AK, Lo DC, Katz LC (1995) Neurotrophins regulate dendritic growth in developing visual cortex. Neuron 15: 791–803. [DOI] [PubMed] [Google Scholar]
- Myers MP, Pass I, Batty IH, Van der Kaay J, Stolarov JP, Hemmings BA, Wigler MH, Downes CP, Tonks NK (1998) The lipid phosphatase activity of PTEN is critical for its tumor supressor function. Proc Natl Acad Sci USA 95: 13513–13518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama AY, Harms MB, Luo L (2000) Small GTPases Rac and Rho in the maintenance of dendritic spines and branches in hippocampal pyramidal neurons. J Neurosci 20: 5329–5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passafro M, Piech V, Sheng M (2001) Subunit-specific temporal and spatial patterns of AMPA receptor exocytosis in hippocampal neurons. Nat Neurosci 4: 917–926. [DOI] [PubMed] [Google Scholar]
- Polleux F, Morrow T, Ghosh A (2000) Semaphorin 3A is a chemoattractant for cortical apical dendrites. Nature 404: 567–573. [DOI] [PubMed] [Google Scholar]
- Portera-Cailliau C, Pan DT, Yuste R (2003) Activity-regulated dynamic behavior of early dendritic protrusions: evidence for different types of dendritic filopodia. J Neurosci 23: 7129–7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond L, Oh SR, Hicks C, Weinmaster G, Ghosh A (2000) Nuclear Notch1 signaling and the regulation of dendritic development. Nat Neurosci 3: 30–40. [DOI] [PubMed] [Google Scholar]
- Redmond L, Kashani AH, Ghosh A (2002) Calcium regulation of dendritic growth via CaM kinase IV and CREB-mediated transcription. Neuron 34: 999–1010. [DOI] [PubMed] [Google Scholar]
- Rodgers EE, Theibert AB (2002) Functions of PI 3-kinase in development of the nervous system. Int J Dev Neurosci 20: 187–197. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Warne PH, Khwaja A, Marte BM, Pappin D, Das P, Waterfield MD, Ridley A, Downward J (1997) Role of phosphoinositide 3-OH kinase in cell transformation and control of the actin cytoskeleton by Ras. Cell 89: 457–467. [DOI] [PubMed] [Google Scholar]
- Sala C, Piech V, Wilson NR, Passafaro M, Liu G, Sheng M (2001) Regulation of dendritic spine morphology and synaptic function by Shank and Homer. Neuron 31: 115–130. [DOI] [PubMed] [Google Scholar]
- Sala C, Futai K, Yamamoto K, Worley PF, Hayashi Y, Sheng M (2003) Inhibition of dendritic spine morphogenesis and synaptic transmission by activity-inducible protein Homer1a. J Neurosci 23: 6327–6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez S, Sayas CL, Lim F, Diaz-Nido J, Avila J, Wandosell F (2001) The inhibition of phosphatidylinositol-3-kinase induces neurite retraction and activates GSK3. J Neurochem 78: 468–481. [DOI] [PubMed] [Google Scholar]
- Sanna PP, Cammalleri M, Berton F, Simpson C, Lutjens R, Bloom FE, Francesconi W (2002) Phosphatidylinositol 3-kinase is required for the expression but not for the induction or the maintenance of long-term potentiation in the hippocampal CA1 region. J Neurosci 22: 3359–3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelzle T, Hall MN (2000) TOR, a central controller of cell growth. Cell 103: 253–262. [DOI] [PubMed] [Google Scholar]
- Sholl DA (1953) Dendritic organization in the neurons of the visual and motor cortices of the cat. J Anat 87: 387–406. [PMC free article] [PubMed] [Google Scholar]
- Steward O, Halpain S (1999) Lamina-specific synaptic activation causes domain-specific alterations in dendritic immunostaining for MAP2 and CAM kinase II. J Neurosci 19: 7834–7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoppini L, Buchs PA, Muller D (1991) A simple method for organotypic cultures of nervous tissue. J Neurosci Methods 37: 173–182. [DOI] [PubMed] [Google Scholar]
- Takei N, Inamura N, Kawamura M, Namba H, Hara K, Yonezawa K, Nawa H (2004) Brain-derived neurotrophic factor induces mammalian target of rapamycin-dependent local activation of translation machinery and protein synthesis in neuronal dendrites. J Neurosci 24: 9760–9769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang SJ, Reis G, Kang H, Gingras AC, Sonenberg N, Schuman EM (2002) A rapamycin-sensitive signaling pathway contributes to long-term synaptic plasticity in the hippocampus. Proc Natl Acad Sci USA 99: 467–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee AR, Anjum R, Blenis J (2003a) Inactivation of the tuberous sclerosis complex-1 and -2 gene products occurs by phosphoinositide 3-kinase/Akt-dependent and -independent phosphorylation of tuberin. J Biol Chem 278: 37288–37296. [DOI] [PubMed] [Google Scholar]
- Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J (2003b) Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol 13: 1259–1268. [DOI] [PubMed] [Google Scholar]
- Tolwani RJ, Buckmaster PS, Varma S, Cosgaya JM, Wu Y, Suri C, Shooter EM (2002) BDNF overexpression increases dendrite complexity in hippocampal dentate gyrus. Neuroscience 114: 795–805. [DOI] [PubMed] [Google Scholar]
- Tongiorgi E, Righi M, Cattaneo A (1997) Activity-dependent dendritic targeting of BDNF and TrkB mRNAs in hippocampal neurons. J Neurosci 17: 9492–9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Proud CG (2002) Ras/Erk signaling is essential for activation of protein synthesis by Gq protein-coupled receptor agonists in adult cardiomyocytes. Circ Res 91: 821–829. [DOI] [PubMed] [Google Scholar]
- Wang Q, Liu L, Pei L, Ju W, Ahmadian G, Lu J, Wang Y, Liu F, Wang YT (2003) Control of synaptic strength, a novel function of Akt. Neuron 38: 915–928. [DOI] [PubMed] [Google Scholar]
- Whitford KL, Marillat V, Stein E, Goodman CS, Tessier-Lavigne M, Chedotal A, Ghosh A (2002) Regulation of cortical dendrite development by Slit-Robo interactions. Neuron 33: 47–61. [DOI] [PubMed] [Google Scholar]
- Williams DW, Truman JW (2004) Mechanisms of dendritic elaboration of sensory neurons in Drosophila: insights from in vivo time lapse. J Neurosci 24: 1541–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth MJ, Brun A, Grabert J, Patz S, Wahle P (2003) Accelerated dendritic development of rat cortical pyramidal cells and interneurons after biolistic transfection with BDNF and NT4/5. Development 130: 5827–5838. [DOI] [PubMed] [Google Scholar]
- Wong RO, Ghosh A (2002) Activity-dependent regulation of dendritic growth and patterning. Nat Rev Neurosci 3: 803–812. [DOI] [PubMed] [Google Scholar]
- Wu GY, Cline HT (1998) Stabilization of dendritic arbor structure in vivo by CaMKII. Science 279: 222–226. [DOI] [PubMed] [Google Scholar]
- Wu GY, Zou DJ, Rajan I, Cline H (1999) Dendritic dynamics in vivo change during neuronal maturation. J Neurosci 19: 4472–4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye B, Petritsch C, Clark IE, Gavis ER, Jan LY, Jan YN (2004) Nanos and Pumilio are essential for dendrite morphogenesis in Drosophila peripheral neurons. Curr Biol 14: 314–321. [DOI] [PubMed] [Google Scholar]
- Yu X, Malenka RC (2003) Beta-catenin is critical for dendritic morphogenesis. Nat Neurosci 6: 1169–1177. [DOI] [PubMed] [Google Scholar]