Abstract
The immediate-early gene Arc is transcribed in neurons that are part of stable neural networks activated during spatial exploratory behaviors. Arc protein has been demonstrated to regulate AMPA-type glutamate receptor trafficking by recruiting endosomal pathways, suggesting a direct role in synaptic plasticity. The purpose of the present study is to examine the fidelity of Arc mRNA translation and the temporal dynamics of behaviorally induced Arc protein expression after rats explore a novel environment. These experiments reveal two waves of Arc protein expression after a single exploration session. In the initial wave, virtually all cells that express Arc mRNA in the hippocampus and parietal cortex also express Arc protein, indicating, at a cellular level, that mRNA transcription and translation are closely correlated from 30 min to 2 h in hippocampal CA and parietal neurons. A second wave of protein expression spans the interval from 8 to 24 h and is also remarkably specific to cells active in the original behavior-induced network. This second wave is detected in a subset of the original active network and displays the novel property that the proportions of Arc-positive neurons become correlated among regions at 24 h. This suggests that the second expression wave is driven by network activity, and the stabilization of circuits reflecting behavioral experience may occur in temporally discrete phases, as memories become consolidated. This is the first demonstration of network-selective translational events consequent to spatial behavior and suggests a role for immediate-early genes in circuit-specific, late-phase synaptic biology.
Keywords: Arg 3.1 protein, time course, biphasic expression, immediate early gene, place cells, imaging, hippocampal function, learning, memory consolidation
Introduction
Spatial exploration, during which rats create a representation of the explored environment (Tolman, 1948; O'Keefe and Nadel, 1978), induces location-specific firing patterns in ∼40% of hippocampal CA1 pyramidal cells in standard environments (e.g., O'Keefe, 1976; Wilson and McNaughton, 1993) and motion-specific firing patterns in ∼50% of posterior parietal cortical cells of behaving rats (McNaughton et al., 1994). The stabilization of such spatial representations in networks presumably requires the expression of genes and their translation into proteins, such as Arc (Lyford et al., 1995) or Arg3.1 (Link et al., 1995), which is an effector immediate-early gene (Lanahan and Worley, 1998). When Arc protein expression is selectively blocked in hippocampus, long-term potentiation has been shown to decay faster over days, and there is reduced behavioral retention of the spatial location of a hidden platform in the Morris swim task (Guzowski et al., 2000). Additionally, Arc mRNA travels to activated regions of dendrites (Steward et al., 1998), and its localization is NMDA receptor-dependent. Thus, Arc may play a role in facilitating plastic changes in specific dendritic domains (Steward and Worley, 2001), most likely through its interaction with dynamin and endophilin in the postsynaptic compartment, thereby modulating endocytosis and AMPA receptor trafficking (S. Chowdhury, J. D. Shepherd, H. Okuno, G. Lyford, R. S. Petralia, R. L. Huganir, and P. F. Worley, unpublished data).
Using a method that visualizes the neuronal ensembles activated during two discrete behavioral experiences [cellular compartment analysis of temporal activity by fluorescence in situ hybridization (catFISH)] (Guzowski et al., 1999), it has been shown that the same types of behaviors that induce activity in single hippocampal and parietal cortical cells recorded from freely behaving rats also selectively induce Arc mRNA in similar proportions of neurons (Guzowski et al., 1999; Vazdarjanova et al., 2002). The paradox that arises from this observation is that such close correspondence between cell activity and gene induction might suggest that the network involved in information storage might rapidly saturate. Although Arc protein is induced in most cells in the hippocampus by treatments such as maximal electroconvulsive shock, this induction method does not necessarily reflect the translation that would be initiated by behavior. In fact, Arc mRNA translation can be highly regulated (Steward and Schuman; 2003; Zalfa et al., 2003), and thus it is possible that protein expression would be modulated in a way that might avoid such potential network saturation.
As an initial test of this hypothesis, Arc mRNA and protein expression were examined using the catFISH method, which is based on the observation that Arc mRNA is present in neuronal nuclei within 2 min after behaviorally driven cell activation, after which it translocates to the cytoplasm where protein translation may occur (Guzowski et al., 1999; Vazdarjanova et al., 2002). The experimental design in the present study included two exploration epochs in the same environment. Animals were killed immediately after the second exposure to the same environment, so that Arc mRNA would be present in the nucleus, and the degree to which the same ensembles of cells expressed the protein could be determined over the interval of 30 min to 24 h.
Materials and Methods
Behavioral procedure
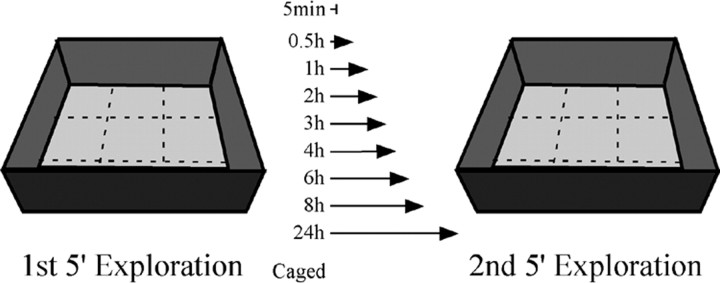
The subjects were 72 9-month-old male Fisher 344 rats (Harlan Sprague Dawley, Indianapolis, IN) that were individually caged and given water and food ad libitum. The light/dark cycle was 12 h (lights on at 9:30 A.M.). The animals were handled twice daily for 7-9 consecutive days until the day before the experiment, when the animals remained undisturbed. The exploration environment was a square open box, 61 × 61 cm with 20-cm-high walls (Fig. 1), partitioned into nine grids, each 412.09 cm2. The box was located in a 250 × 150 cm dimly lit room with several landmarks on the walls and ceiling. Each rat was transported to the exploration room and placed in the center of one of the grids. The rat was moved to the center of a different grid every 15 s on a semirandom schedule, so that each of the grids was visited two or three times during the 5 min exploration session. This procedure in which animals are assisted in traversing all areas of the box results in comparable proportions of Arc-expressing cells as those obtained from animals free to explore the box (Vazdarjanova and Guzowski, 2004), although the variability between animals in a given treatment condition is reduced somewhat in animals given assisted exploration. The two sessions were separated with one of eight intervals: 0.5, 1, 2, 3, 4, 6, 8, and 24 h (Fig. 1). During these intervals, the animals remained undisturbed. The second exploration session was performed identically to the first. Immediately after, the rat was placed into a chamber containing isofluorane, and, when deeply sedated, it was killed by decapitation. The brains were carefully removed and quick-frozen in isopentane equilibrated in an ethanol-dry ice slurry. The average time between the end of the second exploration period to freezing the brain was 160 s and always <200 s, ensuring that the killing procedure did not induce detectable Arc transcription. The brains were stored at -70°C. All animals were killed before the onset of the lights in the colony room.
Figure 1.
Behavioral apparatus and treatment design. A schematic drawing of the behavioral apparatus used for the exploration “treatment” and a list of the intervals intervening between the first and second exposures to the apparatus (ranging from 30 to 1440 min) are shown. Caged refers to the rats killed immediately after being taken from their home cages and is a negative control for behaviorally induced Arc. The animals in the 5 min group explored only once and were killed immediately afterward. This group was used as a negative control for Arc protein expression because at this time point, only Arc mRNA transcription is found in the nucleus.
Histological procedures
Blocking. Before sectioning, brain hemisections containing the left dorsal hippocampus from six to eight rats were molded in a block with Tissue-Tek OCT compound (Miles, Elkhart, IN), such that each block contained brains from most groups. Each block always included the caged group as a control for baseline expression of Arc and either the 0.5 or 1 h group as a positive control. The position of each group was different in each block. The blocks were cryosectioned into 20-μm-thick coronal sections at -18°C, captured on Superfrost plus slides, and stored at -70°C.
Immunohistochemistry. Six slides from each block were selected from the medial portion of the dorsal hippocampus. The tissue was fixed in 2% paraformaldehyde, 7.4 pH, for 5 min and washed in 2× SSC, pH 7.0, followed by 50:50% acetone/methanol at 4°C for 5 min. The tissue was washed in 2× SSC and 0.05% Tween 20 and quenched in 2× SSC and 1% H2O2 for 15 min. After blocking with tyramide signal amplification kit (TSA) blocking buffer (PerkinElmer Life Sciences, Emeryville, CA), the slides were incubated in polyclonal rabbit anti-Arc antibody (1:800) supplied by Dr. Worley's laboratory for 48 h at 4°C. Incubation with the anti-rabbit biotinylated secondary antibody (Vector Laboratories, Burlingame, CA) for 2 h at room temperature was followed by amplification with the avidin-biotin system (Vector Laboratories) for 45 min. The staining was visualized using the cyanine 3 (CY3) TSA fluorescence system (PerkinElmer Life Sciences), and the nuclei were conterstained with Sytox green (Molecular Probes, Eugene, OR). No staining was detected in the absence of the primary or secondary antibodies.
In situ hybridization. Six slices from each block were selected from the medial portion of the dorsal hippocampus adjacent to the ones chosen for immunohistochemistry to be stained for Arc mRNA according to the catFISH method described in detail elsewhere (Guzowski et al., 1999). Briefly, the tissue was fixed in 4% paraformaldehyde, washed in 2× SSC, and placed in an acetic-anhydride solution, followed by an acetone-methanol solution. After a prehybridization step, the tissue was hybridized with an Arc mRNA probe (100 ng/slide) tagged with digoxigenin and diluted in hybridization buffer (Sigma, St. Louis, MO) for 16-18 h at 56°C. After a series of washes, including an RNase A step, the slides were incubated overnight in an anti-digoxigenin peroxidase-conjugated antibody (Roche Products, Hertfordshire, UK) at 4°C, and the stain was visualized using the CY3 TSA fluorescence system. The nuclei were counterstained with Sytox green (Molecular Probes).
Combined in situ hybridization and immunohistochemistry. To successfully combine these two methods, the in situ hybridization procedure was conducted first using a 25% diluted solution of acetic anhydride. After the CY3 visualization step, the tissue was boiled in double-distilled H2O for 10 min, as an antigen retrieval step, and then washed in 2× SSC and quenched in 2× SSC with 1% H2O2. The rest of the steps were as described above for Arc immunohistochemistry, but the protein was visualized using the CY5 TSA fluorescence system (PerkinElmer Life Sciences). Because of the difficulty of this staining procedure, only one area was chosen for examination.
Confocal microscopy
For each rat, six sample z-stacks (1.0 μm optical thickness per plane, 25× objective) from three different slides were collected from each of three brain regions [CA1, CA3, and parietal cortex (PCx)] using a Zeiss (Thornwood, NY) SM 510NLO-meta multiphoton/confocal microscope equipped with a 488 nm argon laser and a 543 and 633 nm helium/neon laser.
Contrast and intensity parameters were set using the tissue sections from the caged and the 1 or 0.5 h (depending on the availability in each block) animals as the negative and positive controls, respectively. For consistency, these parameters were kept constant for the rest of the sections on the slide. Sample images were taken at the following coordinates: CA1 anteroposterior, -3.8 to -4.2; lateral, 1.5-2.5; and dorsoventral, 2.5; CA3 anteroposterior, 3.8; lateral, 2.8-3.8; and dorsoventral, 3.8; and PCx anteroposterior, -3.8 to -4.2; lateral, 1.5-2.5; dorsoventral, 2.0; and cortical layer V.
The staining in the dentate gyrus was so sparse that it was necessary to sample larger areas using 10× z-section stacks from the whole dentate gyrus (DG) in two sections per animal. Approximately four to eight images were taken from each DG section. To verify the number of total cells in the DG, overlapping 40× images were collected for the entire DG for the Arc mRNA analysis.
Image Analysis
Image analysis was done as described earlier (Vazdarjanova et al., 2002). Briefly, neurons were segmented and classified using MetaMorph imaging software. On the basis of the nuclear counterstain, neurons and glia were discriminated. Glial nuclei were small, with intense and uniform DNA staining. Only neuron-like cells found in the middle 20% of each stack were included in the analyses. For Arc protein analysis, the segmented cells were classified as either positive or negative. Positive neurons had perinuclear or cytoplasmic staining surrounding at least 60% of the cell, visible in at least three plains together with the cell nucleus. Special attention was paid to ensure that the staining belonged to the cell of interest and not to a dendrite of a different cell.
When images with combined Arc mRNA and Arc protein staining were analyzed, cells were classified as (1) negative, neither Arc mRNA nor Arc protein; (2) Arc mRNA only, one or two intense internuclear foci present in at least three plains; (3) Arc protein only, perinuclear or cytoplasmic staining surrounding at least 60% of the cell and visible in at least three plains together with the cell nucleus; and (4) Arc mRNA and Arc protein double-stained, cells that fulfilled both criteria 2 and 3. Image analysis was performed by an experimenter blind to the experimental conditions.
Two reconstructed DG sections per rat from the middle planes of overlapping 10× z-stacks were used for the DG analysis. The area of the granule cell layer and the total number of Arc-positive neurons was assessed in each reconstructed flat image. The area was used to estimate the total number of neurons using a correction factor that represented the total neurons per square micrometer. This factor was derived from 92 z-stacks from 10 different rats collected at 40× magnification. The total number of neurons per stack was counted, and the area of the granule cell layer (in square micrometers) from the middle plane was calculated. Using this factor, we calculated the percentage of neurons with Arc protein and/or Arc mRNA in the DG of each rat according to the following formula: 100 * p/(Ap * (N/A)), where p was the number of Arc(+) neurons in a given reconstructed flat image; Ap, the area (in square micrometers) of the DG, as measured from the reconstructed flat image; N, the total number of cells from all 40× z-stacks; and A, the total area (in square micrometers) of the DG from the middle planes of all 40× z-stacks. The number of Arc-positive cells was corroborated using the 40× image stacks from the same DG.
Statistics
Statview software was used to perform one-way ANOVA tests for each studied region where the behavioral condition was the independent variable and the percentages of neurons, from the various categories described above, of the total number of neurons were used as the dependent variables. When an overall ANOVA was significant, we did individual between-group comparisons with Bonferroni post hoc tests to correct for multiple comparisons. When correlations between structures were examined, they were done split by group, and list-wise deletion was used.
Results
Spatial exploration of a novel environment (n = 6-8 rats per group) induced robust translation of Arc protein in the cytoplasm of a subpopulation of CA1, CA3, PCx, and DG neurons compared with the control group (Fig. 2). Because the caged (n = 10) and 5 min control (n = 8) groups were not statistically different (p = 0.64-0.91), they were combined to form the control group for statistical analyses. In the graphs, the caged and 5 min groups are kept separate for illustrative purposes.
Figure 2.

Exploration induces Arc protein translation. Sample confocal images of cells expressing Arc protein (red) taken at 25× magnification from CA1 (A, B), CA3 (C, D), DG (E, F), and PCx (G, H) from caged control rats (left column) and from rats that were killed 1 h after exploration (right column) are shown. Nuclei are counterstained with Sytox green. In all regions examined, the exploration rats showed greater numbers of cells with Arc protein than did the caged controls. Scale bar, 100 μm.
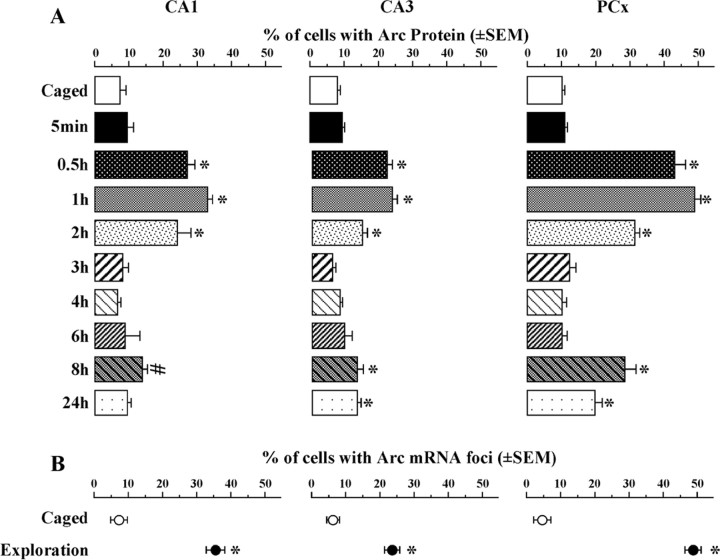
The kinetics of Arc protein expression in the cytoplasm were similar in CA1, CA3, and PCx (Fig. 3A). In all three regions, Arc protein expression was elevated between 30 min and 2 h (overall ANOVAs revealed a statistically significant change over time of the size of the neuronal ensembles expressing Arc protein: FCA1(8,63) = 20.76; p < 0.0001; FCA3(8,63) = 26.61; p < 0.0001; FPCx(8,62) = 81.64; p < 0.0001). The average numbers of neurons counted per rat were 452 for CA1, 240 for CA3, and 685 for PCx.
Figure 3.
Kinetics of exploration-induced Arc protein expression in CA1, CA3, and PCx. A, Percentage of total cells showing Arc protein (induced by the first exploration) in CA1, CA3, and PCx at all time points studied. All groups were exposed twice to the same environment, except for the caged and 5 min control groups. Caged, n = 10; 5 min, 1, 2, 4 h, n = 8; 0.5, 3, 6, 8, 24 h, n = 6. *p < 0.0014 relative to controls; #p = 0.04 (not statistically significant after correction for multiple comparisons). B, Percentage of total cells with Arc mRNA foci (induced by the second exploration) in CA1, CA3, and PCx for the caged controls (open circles) and exploration (filled circles) groups. *p < 0.001 compared with controls. Cytoplasmic Arc mRNA is not shown in these regions because after 30 min, the Arc mRNA signal decreases to baseline.
Notably, there was a second wave of elevated Arc expression at 8 h after exploration in CA1, CA3, and PCx. However, this elevation only reached statistical significance in CA3 (p = 0.0005) and PCx (p < 0.0001), with CA1 approaching significance (p = 0.047, not significant with Bonferroni correction). At 24 h after spatial exploration, the percentage of Arc-expressing neurons was also significantly elevated for CA3 (p = 0.0007) and PCx (p < 0.0001). Further studies with time points between 8 and 24 h may reveal whether the elevated percentage of Arc protein-positive neurons at 24 h results from a sustained second wave initiated at ∼8 h or is part of a subsequent wave of Arc expression.
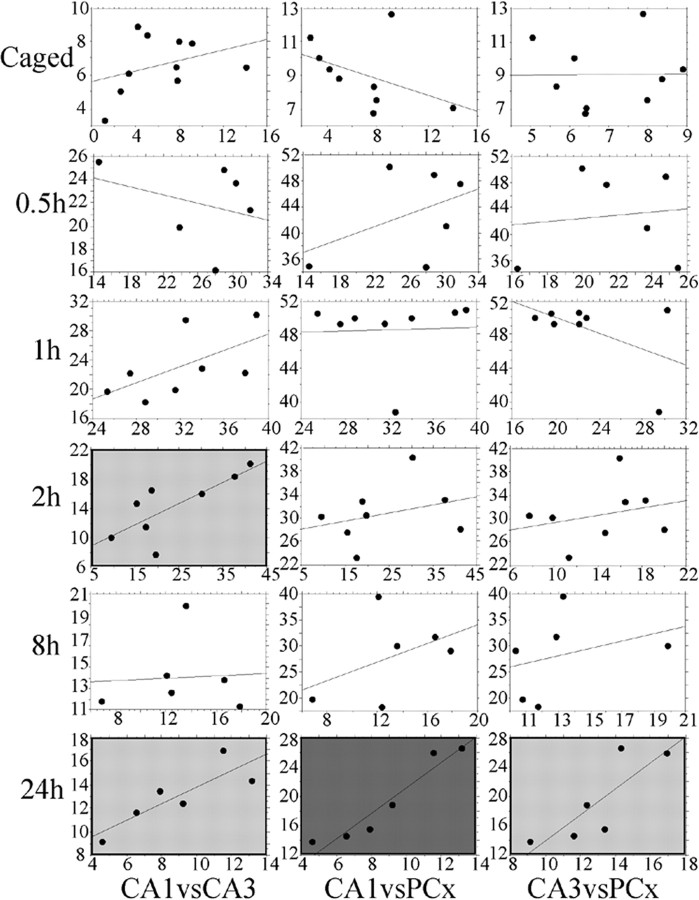
The similar kinetics of Arc protein expression in CA1, CA3, and PCx suggest that these three brain regions may be part of a coordinated system acquiring and/or consolidating spatially related information. In support of this idea, at 2 h after spatial exploration, but not earlier, there was a significant positive correlation (r = 0.772; p = 0.024) between the proportions of activated CA1 and CA3 cells (Fig. 4). Furthermore, at 24 h after exploration, there was again a significant positive correlation between all three regions in the proportions of cells with Arc protein (CA1 vs CA3, r = 0.846; p < 0.05; CA1 vs PCx, r = 0.958; p < 0.01; CA3 vs PCx, r = 0.822; p < 0.05).
Figure 4.
Regression plots comparing the proportions of cells containing Arc protein over the three regions (CA1, CA3, and PCx) and four time points studied. Regression plots between CA1 (x-axis) and CA3 (y-axis) (left column), CA1 and PCx (middle column), and CA3 and PCx (right column) are shown. For simplicity, the time points shown here (other than for caged control) reflect the times at which the expression of Arc protein was statistically above control levels. There were no significant correlations at the 3, 4, and 6 h time points. The correlations in the plots with light gray background are significant at the p < 0.05 level, whereas the correlations in the plot with dark gray background are significant at the p < 0.01 level.
In contrast to the CA regions and PCx, in the upper blade of the DG (Fig. 5A), Arc protein expression in the cytoplasm was consistently elevated from 30 min to at least 8 h after exploration (p < 0.0001 for the comparisons of the control group vs the 0.5-8 h groups), but was not different from baseline by 24 h (Fig. 5B). Interestingly, in the lower blade of the DG, there was a modest but significant increase (p < 0.001) in Arc protein levels only at 6 and 8 h after spatial exploration (Fig. 5B). This kinetics of Arc in the DG was not exclusive for the protein but was also observed for the cytoplasmic Arc mRNA (Fig. 5C), suggesting that in the DG, either the half-life of Arc mRNA is unusually long (>8 h), or Arc mRNA transcription is sustained for at least 8 h.
Figure 5.

Kinetics of exploration-induced Arc protein expression in the DG. A, Example of a reconstructed 20 μm flat image, as was used for analysis of Arc expression in granule cells of the DG. Note the sparsity of Arc staining (red) across the entire DG, with the sparsest expression observed in the lower blade. B, Percentage of cells stained for Arc protein in the upper and lower blades of the dentate gyrus at all time points studied. C, Percentage of cells stained for Arc cytoplasmic mRNA in the upper and lower blades of the dentate gyrus at all time points studied. *p < 0.0014 compared with controls. Scale bar, 100 μm.
Although the kinetics of Arc expression in the CA1, CA3, and the PCx was similar, the size of the respective neuronal ensembles expressing Arc was different. At 1 h after exploration, 32.3 ± 1.7% of the total neurons in the dorsal hippocampal CA1 contained detectable cytoplasmic Arc protein; in CA3, the percentage was 23.1 ± 1.6%; and in PCx, it was 48.6 ± 1.4% (Fig. 3A). These proportions of neurons expressing Arc protein closely matched the proportions of cells expressing Arc mRNA (CA1, 35.8 ± 2.1%; CA3, 23.6 ± 1.1%; PCx, 49.1 ± 1.4%) immediately after the second exploration session (Fig. 3, compare A,B). The proportions of neurons expressing Arc mRNA were also similar to those previously reported (Guzowski et al., 1999; Vazdarjanova et al., 2002). During the second wave of Arc protein expression at 8 h, the sizes of the Arc-expressing neuronal ensembles were only 40-55% of those initially activated (13% of the total neurons in CA1, 13% in CA3, and 28% in PCx) (Fig. 3). Similarly, at 24 h, the sizes of the Arc-expressing ensembles in CA3 and PCx were 13 and 19%, respectively. In the upper blade of the DG, the highest proportion of Arc protein-expressing neurons (Fig. 5B) as well as Arc mRNA-expressing neurons (Fig. 5C) was ∼2%, and in the lower blade, it was ∼ 1%.
The finding that after exploration of a novel environment the proportion of Arc protein-expressing neurons closely matches the proportion of Arc mRNA-expressing neurons (Fig. 3) suggests that, in the cytoplasm, Arc is translated into protein with high fidelity. The reliability of Arc translation into protein was studied further by combining fluorescence immunohistochemistry for Arc protein and in situ hybridization for Arc mRNA (Fig. 6A-D). Because the rats of all experimental groups explored the novel environment for a second time for 5 min immediately before they were killed (see Materials and Methods) (Fig. 1), neurons marked with intranuclear foci of transcription revealed the neuronal ensembles activated by the second exposure, whereas neurons with Arc protein staining revealed the ensembles activated during the first exposure to the environment. Indeed, as illustrated in Figure 6G, in the 30 min and 1 h groups, the percentages of PCx neurons that expressed Arc after both exploration sessions, i.e., neurons double-labeled for Arc protein and Arc mRNA, were very high: 92% (30 min group) and 83% (1 h group) of the total neurons expressing Arc protein, whereas the percentage of neurons that expressed only Arc protein (Fig. 6E-H) was low for all groups. In addition, the overall percentage of neurons with Arc mRNA (sum of the Arc mRNA-only and double-labeled values) was similar to that of Arc protein (sum of the Arc protein-only and double-labeled values) for the 1 h group (∼49% of the total number of neurons). Taken together, these results suggest that the same cells that express Arc mRNA also express Arc protein.
Figure 6.
Arc protein is expressed in the same neurons that express Arc mRNA. A-D, Example confocal images from parietal cortex (nuclei shown in green) taken from a caged control animal (A), an animal killed 5 min after a single exploration session (B), and an animal given two exploration sessions separated by 0.5 h (C) or 8 h (D). The latter two time points correspond to the first appearance of Arc protein in the first and second waves of protein expression, respectively. Arc mRNA intranuclear foci are shown in red, Arc protein is shown in purple, and the colocalization of Arc mRNA and protein is shown in pink (25× magnification; scale bar, 100 μm). E-H, Quantification of the proportions of neurons containing Arc mRNA foci only (red), Arc cytoplasmic protein only (blue), and double-stained neurons with Arc mRNA and protein (green), for the caged, 5 min, and 0.5 and 8 h conditions. To reflect the total proportions of neurons that expressed Arc mRNA and protein, the Double category is added to each of the mRNA and protein counts, indicated by the crosshatched bars. For each group, the total number of cells that expressed Arc protein and also mRNA was also determined (yellow histogram to the right, reflecting colocalization). Compared with caged controls, the 5 min and 8 h groups showed significantly increased numbers of Arc mRNA-expressing cells (*p < 0.05; **p < 0.01, with Bonferroni correction); neither the mRNA nor protein alone is significantly different from caged controls for the 0.5 h group, but the double-stained population is significantly increased from controls (**p < 0.01); for the 8 h group, the double-labeled cells are also significantly different from controls (**p < 0.01). The proportions of neurons that showed colocalization of Arc mRNA and protein are statistically greater than caged controls only in the 0.5 and 8 h groups (yellow bars). The results (data not shown) from the 3, 4, and 6 h groups are similar to those from the 5 min group shown; those from the 1 h group (data not shown) are similar to those from the 0.5 h group shown; and those from the 24 h group (data not shown) are similar to those from the 8 h group shown. Note that in the caged control animals, the colocalization is close to chance levels, whereas in the 0.5, 1, 8, and 24 h groups, the colocalization is well above chance (>80%).
Furthermore, by detecting Arc mRNA and Arc protein in the same tissue, it was possible to demonstrate that the neurons expressing Arc protein at 8 and 24 h were a subpopulation of the initially activated neuronal ensemble. At 8 h, the percentages of double-labeled neurons were 81% of the total number of neurons expressing Arc protein and 82% for the 24 h time point. In the control animals, the percentage of double-labeled neurons was at a chance level, ∼50%. These results show that the initially activated neuronal ensemble underwent “reactivation” of Arc expression at 8 and 24 h.
Discussion
Several novel observations arose from the data collected in the present study: (1) there is synchronized (30 min-2 h) Arc protein expression in pyramidal cells of CA3, CA1, and parietal cortex activated by exploratory behavior; (2) the proportions of Arc protein-expressing cells match the proportions of cells that express Arc mRNA and those defined by electrophysiological recordings; (3) there is a more sustained (∼8 h) Arc expression pattern in granule cells of the dentate gyrus; (4) there is a network-specific reactivation of Arc protein at 8 and 24 h in a subset of cells from the original behavior-induced ensemble; and (5) there is a correlation, which only emerges at 24 h, in the proportions of Arc-positive CA and parietal cells from the initially activated network.
Initial wave of Arc protein expression: Arc mRNA is translated into protein with high fidelity
The present data do not support the original hypothesis that Arc protein translation may be highly regulated in a way that might avoid saturation of information in networks. Arc protein was expressed in a neuronal ensemble similar in size to that expressing Arc mRNA. Furthermore, using a combined in situ hybridization and immunohistochemical staining procedure for Arc, a large percentage of the neurons show double labeling for Arc mRNA and protein (the degree of colocalization was >80%). This finding is consistent with a previous study reporting correspondence between expression patterns of mRNA and protein of another activity-induced immediate-early gene, zif268 (Worley et al., 1991). The present data suggest that if translational regulation of Arc does occur in the cytoplasm, as has been reported in dendrites (Yin et al., 2002; Dong et al., 2003), it may affect the amount of synthesized protein at specific regions (such as synapses) and/or its half-life, as shown for Arc in synaptoneurosome preparations (Yin et al., 2002).
In pyramidal cells of CA3, CA1, and parietal cortex, the first wave of Arc protein expression occurred between 30 min and 2 h after exploration of a novel environment, returning to levels equivalent to those of caged control rats at time points ranging from 3 to 6 h. Furthermore, the proportions of cells activated in these regions are consistent with what have been observed in electrophysiological recording studies of these areas (Barnes et al., 1990; McNaughton et al., 1994; Qin et al., 1997). In contrast, the upper blade of the dentate gyrus showed a single wave of sustained Arc protein expression that lasted for at least 8 but not longer than 24 h, and the lower blade showed expression of Arc at 6 and 8 h only. The data from granule cells could indicate either persistent transcription or greater stability of mRNA in these cells. It is unlikely that the sustained Arc expression in granule cells results from sustained neuronal activity, however, because granule cell recordings have not shown similar sustained unit activity after exploration (Jung and McNaughton, 1993; Shen et al., 1998; Gothard et al., 2001). The differences in the exact time course between hippocampal granule cells and pyramidal cells in CA1, CA3, and parietal cortex may have implications for synaptic function in these regions because Arc protein has recently been shown to mediate protein synthesis-dependent endocytosis of postsynaptic AMPA receptors (Chowdhury, Shepherd, Okuno, Lyford, Petralia, Huganir, and Worley, unpublished data).
Second wave of Arc protein expression in CA1, CA3, and PCx
Because the present experimental design included time points at 8 and 24 h after the first behavioral exploration session, the serendipitous finding arose that the CA and parietal cortical regions showed a second wave of Arc expression at these times. Interestingly, the size of the Arc-expressing ensemble was only ∼50% of that initially activated. This observed multiphasic pattern of Arc expression is consistent with a previous report of biphasic Arc mRNA expression after mice were trained on an active avoidance task (Montag-Sallaz and Montag, 2003). In that study, however, it could not be determined whether the observed second wave of Arc expression occurred in the cells of the initially activated ensemble or in a different one. By using the catFISH method in the present study, Arc protein and Arc mRNA could be assessed in the same tissue, and it was possible to show that this second wave of Arc expression is a highly specific reactivation of a subset of the initial cell ensemble. The reactivation of Arc expression might occur by a cell-autonomous mechanism if, for example, the initial genomic response includes factors that reinduce Arc transcription. We do not favor such a mechanism as an explanation, however, because the percentage of Arc-positive neurons in various brain regions differ between the first and second waves. Moreover, networks are known to show activity pattern reactivation electrophysiologically, during subsequent quiet resting or sleep states after specific behavioral experiences (Wilson and McNaughton, 1994).
Could this network-specific reactivation of Arc expression be a marker of memory consolidation? Memory consolidation is not a uniform process (McGaugh, 2000) but has an early phase initiated during memory acquisition with the activation of glutamatergic receptors and a late phase triggered by the activation of either glutamate receptors (Williams et al., 1998) or receptors for modulatory neurotransmitters, such as norepinephrine, glucocorticoids, and dopamine (Bernabeu et al., 1997; Izquierdo and McGaugh, 2000; Roozendaal, 2000). Both phases appear to involve cAMP-dependent phosphorylation of the transcription factor cAMP response element-binding protein (CREB) (Bourtchuladze et al., 1994; Bernabeu et al., 1997; Stanciu et al., 2001). CREB is also thought to regulate the expression of immediate-early genes (including Arc) (Guzowski, 2002), consistent with the idea that Arc may participate, along with CREB, in both phases of memory consolidation.
The hypothesis that the second wave of Arc expression results from neural activity initiated by the original exploration, rather than some independent molecular process, is in accord with the electrophysiological observation of replay of ensemble activity patterns that reflect previous experience (Wilson and McNaughton, 1994). The idea that the consolidation process consists of a hippocampal-neocortical dialog (Marr, 1971; Squire, 1992; McNaughton et al., 2003) has gained support recently from a number of single-cell recording experiments in behaving rats. In these studies, reactivation of network states reflecting behavioral experience during quiet waking and sleep behaviors has been observed and may provide the mechanism by which an assembly of neurons can produce Arc reactivation (Pavlides and Winson, 1989; Wilson and McNaughton, 1994; Kudrimoti et al., 1999; Louie and Wilson, 2001; Ribeiro et al., 2004). In fact, there is some suggestion from these recording experiments that a persistent experience-dependent ensemble activity pattern can be detected, even after ≥20 h has elapsed (Kudrimoti et al., 1999; Louie and Wilson, 2001; Ribeiro et al., 2004). The reexpression of Arc out to 24 h, in a subset of the original network, makes an explicit prediction for such recording studies: if ensembles of hippocampal and parietal cortical cells are recorded continuously for ≥24 h after a discrete behavioral experience, then reactivation of network states reflecting that experience should be detected in the full ensemble at intervals between 30 min and 2 h and in a subset of this network at intervals from 8 to 24 h. Taken together, it appears that the catFISH method was able to reveal patterns of network activity consistent with the neural implementation of memory consolidation.
Long-term correlation among a subset of the initial, behaviorally activated ensemble
At least two possible “systems” explanations can be raised to explain the observation that the reactivation of Arc expression occurred in only half the initially activated neurons. First, a relatively large number of neurons may initially participate in representing a behavioral experience, whereas only a subset of these neurons may be necessary to elicit pattern completion or retrieval of the entire experience (McNaughton and Morris, 1987). Mechanistically, the reexpression of Arc in this subset may directly promote the synaptic changes that were initiated immediately after the behavioral experience. It is equally possible, however, that reactivation occurs only in the subpopulation of cells that requires further modification of their synaptic connections. The observation that this second wave of protein expression is driven by activity in the circuits that reflect a discrete behavioral experience suggests that network stabilization may occur in temporally discrete phases.
Another interesting aspect of this selective, network-specific reactivation of Arc is the fact that the sizes of the CA3, CA1, and parietal cortical neuronal ensembles become correlated at 24 h after spatial exploration. This is consistent with the expectation that all three regions may undergo plastic changes at the same time if they are, in fact, participating in a sophisticated neural network that processes spatial information (McNaughton et al., 1996; Burgess et al., 1999; Nadel et al., 2000). This finding is also consistent with electrophysiological studies that have described ensemble reactivation of hippocampal and parietal cortical cell pairs that reflect recent spatial exploratory behavior (Qin et al., 1997). Together, the present findings suggest that two or more waves of Arc expression may be required to stabilize behaviorally induced spatial representations in hippocampal-neocortical circuits, and the reactivation of Arc may represent an anatomical signature of the synaptic activity that underlies consolidation.
Footnotes
This work was supported by Human Frontier Science Program Grants AG09219, LTF000112-2002-C, and MH064357. We thank Frank Houston, Kathy Olson, and Aldrich Sy for technical assistance, Dr. John Guzowski for critical methodological input, and Dr. Thane Plummer for helpful comments on a previous version of this manuscript.
Correspondence should be addressed to Dr. C. A. Barnes, Arizona Research Laboratories, Division of Neural Systems, Memory, and Aging, University of Arizona, Life Sciences North Building, Room 384, P.O. Box 245115, Tucson, AZ 85724-5115. E-mail: carol@nsma.arizona.edu.
Copyright © 2005 Society for Neuroscience 0270-6474/05/251761-08$15.00/0
References
- Barnes CA, McNaughton BL, Mizumori SJ, Leonard BW, Lin LH (1990) Comparison of spatial and temporal characteristics of neuronal activity in sequential stages of hippocampal processing. Prog Brain Res 83: 287-300. [DOI] [PubMed] [Google Scholar]
- Bernabeu R, Bevilaqua L, Ardenghi P, Bromberg E, Schmitz P, Bianchin M, Izquierdo I, Medina JH (1997) Involvement of hippocampal cAMP/cAMP-dependent protein kinase signaling pathways in a late memory consolidation phase of aversively motivated learning in rats. Proc Natl Acad Sci USA 94: 7041-7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourtchuladze R, Frenguelli B, Blendy J, Cioffi D, Schutz G, Silva AJ (1994) Deficient long-term memory in mice with a targeted mutation of the cAMP-responsive element-binding protein. Cell 79: 59-68. [DOI] [PubMed] [Google Scholar]
- Burgess N, Jeffery KJ, O'Keefe JO (1999) Integrating hippocampal and parietal functions: a spatial point of view. In: The hippocampus and parietal foundations of spatial cognition (Burgess N, Jeffery KJ, O'Keefe JO, eds), pp 3-29. New York: Oxford UP.
- Dong E, Caruncho H, Liu WS, Smalheiser NR, Grayson DR, Costa E, Guidotti A (2003) A reelin-integrin receptor interaction regulates Arc mRNA translation in synaptoneurosomes. Proc Natl Acad Sci USA 100: 5479-5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gothard KM, Hoffman KL, Battaglia FP, McNaughton BL (2001) Dentate gyrus and ca1 ensemble activity during spatial reference frame shifts in the presence and absence of visual input. J Neurosci 21: 7284-7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzowski JF (2002) Insights into immediate-early gene function in hippocampal memory consolidation using antisense oligonucleotide and fluorescent imaging approaches. Hippocampus 12: 86-104. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, McNaughton BL, Barnes CA, Worley PF (1999) Environment-specific expression of the immediate-early gene Arc in hippocampal neuronal ensembles. Nat Neurosci 2: 1120-1124. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, Lyford GL, Stevenson GD, Houston FP, McGaugh JL, Worley PF, Barnes CA (2000) Inhibition of activity-dependent arc protein expression in the rat hippocampus impairs the maintenance of long-term potentiation and the consolidation of long-term memory. J Neurosci 20: 3993-4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo I, McGaugh JL (2000) Behavioural pharmacology and its contribution to the molecular basis of memory consolidation. Behav Pharmacol 11: 517-534. [DOI] [PubMed] [Google Scholar]
- Jung MW, McNaughton BL (1993) Spatial selectivity of unit activity in the hippocampus granular layer. Hippocampus 3: 165-182. [DOI] [PubMed] [Google Scholar]
- Kudrimoti HS, Barnes CA, McNaughton BL (1999) Reactivation of hippocampal cell assemblies: effects of behavioral state, experience, and EEG dynamics. J Neurosci 19: 4090-4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanahan A, Worley P (1998) Immediate-early genes and synaptic function. Neurobiol Learn Mem 70: 37-43. [DOI] [PubMed] [Google Scholar]
- Link W, Konietzko U, Kauselmann G, Krug M, Schwanke B, Frey U, Kuhl D (1995) Somatodendritic expression of an immediate early gene is regulated by synaptic activity. Proc Natl Acad Sci USA 92: 5734-5738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie K, Wilson MA (2001) Temporally structured replay of awake hippocampal ensemble activity during rapid eye movement sleep. Neuron 29: 145-156. [DOI] [PubMed] [Google Scholar]
- Lyford GL, Yamagata K, Kaufmann WE, Barnes CA, Sanders LK, Copeland NG, Gilbert DJ, Jenkins NA, Lanahan AA, Worley PF (1995) Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron 14: 433-445. [DOI] [PubMed] [Google Scholar]
- Marr D (1971) Simple memory: a theory for archicortex. Philos Trans R Soc Lond B Biol Sci 262: 23-81. [DOI] [PubMed] [Google Scholar]
- McGaugh JL (2000) Memory: a century of consolidation. Science 287: 248-251. [DOI] [PubMed] [Google Scholar]
- McNaughton BL, Morris RGM (1987) Hippocampal synaptic enhancement and information storage within a distributed memory system. Trends Neurosci 10: 408-415. [Google Scholar]
- McNaughton BL, Mizumori SJ, Barnes CA, Leonard BJ, Marquis M, Green EJ (1994) Cortical representation of motion during unrestrained spatial navigation in the rat. Cereb Cortex 4: 27-39. [DOI] [PubMed] [Google Scholar]
- McNaughton BL, Barnes CA, Gerrard JL, Gothard K, Jung NW, Knierim JJ, Kudrimoti H, Qin Y, Skaggs WE, Suster M, Weaver KL (1996) Deciphering the hippocampal polyglot: the hippocampus as a path integration system. J Exp Biol 199: 173-185. [DOI] [PubMed] [Google Scholar]
- McNaughton BL, Barnes CA, Battaglia FP, Bower MR, Cowen SL, Ekstrom AD, Gerrard JL, Hoffman KL, Houston PF, Karten Y, Lipa P, Pennartz CMA, Sutherland GR (2003) Off-line reprocessing of recent memory and its role in memory consolidation: a progress report. In: Sleep and brain plasticity (Maguet P, Smith C, Stickgold B, eds), pp 225-246. Oxford: Oxford UP.
- Montag-Sallaz M, Montag D (2003) Learning-induced arg 3.1/arc mRNA expression in the mouse brain. Learn Mem 10: 99-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadel L, Samsonovich A, Ryan L, Moscovich M (2000) Multiple trace theory of human memory: computational, neuroimaging and neurophsychological results. Hippocampus 10: 352-368. [DOI] [PubMed] [Google Scholar]
- O'Keefe J (1976) Place units in the hippocampus of the freely moving rat. Exp Neurol 51: 78-109. [DOI] [PubMed] [Google Scholar]
- O'Keefe J, Nadel L (1978) The hippocampus as a cognitive map Oxford: Clarendon.
- Pavlides C, Winson J (1989) Influences of hippocampal place cell firing in the awake state on the activity of these cells during subsequent sleep episodes. J Neurosci 9: 2907-2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin YL, McNaughton BL, Skaggs WE, Barnes CA (1997) Memory reprocessing in corticocortical and hippocampocortical neuronal ensembles. Philos Trans R Soc Lond B Biol Sci 352: 1525-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro S, Gervasoni D, Soares ES, Zhou Y, Lin SC, Pantoja J, Lavine M, Nicolelis MAL (2004) Long-lasting novelty-induced neuronal reverberation during slow-wave sleep in multiple forebrain areas. PLoS Biol 2: 0126-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roozendaal B (2000) 1999 Curt P. Richter award. Glucocorticoids and the regulation of memory consolidation. Psychoneuroendocrinology 253: 213-238. [DOI] [PubMed] [Google Scholar]
- Shen J, Kudrimoti HS, McNaughton BL, Barnes CA (1998) Reactivation of neuronal ensembles in hippocampal dentate gyrus during sleep after spatial experience. J Sleep Res 1: 6-16. [DOI] [PubMed] [Google Scholar]
- Squire LR (1992) Memory and the hippocampus: a synthesis from findings with rats, monkeys, and humans. Psychol Rev 99: 195-231. [DOI] [PubMed] [Google Scholar]
- Stanciu M, Radulovic J, Spiess J (2001) Phosphorylated cAMP response element binding protein in the mouse brain after fear conditioning: relationship to Fos production. Brain Res Mol Brain Res 94: 15-24. [DOI] [PubMed] [Google Scholar]
- Steward O, Schuman EM (2003) Compartmentalized synthesis and degradation of proteins in neurons. Neuron 40: 347-359. [DOI] [PubMed] [Google Scholar]
- Steward O, Worley PF (2001) Selective targeting of newly synthesized Arc mRNA to active synapses requires NMDA receptor activation. Neuron 2001 30: 227-240. [DOI] [PubMed] [Google Scholar]
- Steward O, Wallace CS, Lyford GL, Worley PF (1998) Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron 21: 741-751. [DOI] [PubMed] [Google Scholar]
- Tolman EC (1948) Cognitive maps in rats and men. Psychol Rev 55: 189-208. [DOI] [PubMed] [Google Scholar]
- Vazdarjanova A, Guzowski JF (2004) Differences in hippocampal neuronal population responses to modifications of an environmental context: evidence for distinct, yet complementary, functions of CA3 and CA1 ensembles. J Neurosci 24: 6489-6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazdarjanova A, McNaughton BL, Barnes CA, Worley PF, Guzowski JF (2002) Experience-dependent coincident expression of the effector immediate-early genes arc and Homer 1a in hippocampal and neocortical neuronal networks. J Neurosci 22: 10067-10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JM, Mason-Parker SE, Abraham WC, Tate WP (1998) Biphasic changes in the levels of N-methyl-d-aspartate receptor-2 subunits correlate with the induction and persistence of long-term potentiation. Brain Res Mol Brain Res 60: 21-27. [DOI] [PubMed] [Google Scholar]
- Wilson MA, McNaughton BL (1993) Dynamics of the hippocampal ensemble code for space. Science 261: 1055-1058. [DOI] [PubMed] [Google Scholar]
- Wilson MA, McNaughton BL (1994) Reactivation of hippocampal ensemble memories during sleep. Science 265: 676-679. [DOI] [PubMed] [Google Scholar]
- Worley PF, Christy BA, Nakabeppu Y, Bhat RV, Cole AJ, Baraban JM (1991) Constitutive expression of zif268 in neocortex is regulated by synaptic activity. Proc Natl Acad Sci USA 88: 5106-5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Edelman GM, Vanderklish PW (2002) The brain-derived neurotrophic factor enhances synthesis of Arc in synaptoneurosomes. Proc Natl Acad Sci USA 99: 2368-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalfa F, Giorgi M, Primerano B, Moro A, Di Penta A, Reis S, Oostra B, Bagni C (2003) The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell 112: 317-327. [DOI] [PubMed] [Google Scholar]