Abstract
In a number of neuronal models of learning, signaling by the neurotransmitter nitric oxide (NO), synthesized by the enzyme neuronal NO synthase (nNOS), is essential for the formation of long-term memory (LTM). Using the molluscan model system Lymnaea, we investigate here whether LTM formation is associated with specific changes in the activity of members of the NOS gene family: Lym-nNOS1, Lym-nNOS2, and the antisense RNA-producing pseudogene (anti-NOS). We show that expression of the Lym-nNOS1 gene is transiently upregulated in cerebral ganglia after conditioning. The activation of the gene is precisely timed and occurs at the end of a critical period during which NO is required for memory consolidation. Moreover, we demonstrate that this induction of the Lym-nNOS1 gene is targeted to an identified modulatory neuron called the cerebral giant cell (CGC). This neuron gates the conditioned feeding response and is an essential part of the neural network involved in LTM formation. We also show that the expression of the anti-NOS gene, which functions as a negative regulator of nNOS expression, is downregulated in the CGC by training at 4 h after conditioning, during the critical period of NO requirement. This appears to be the first report of the timed and targeted differential regulation of the activity of a group of related genes involved in the production of a neurotransmitter that is necessary for learning, measured in an identified neuron of known function. We also provide the first example of the behavioral regulation of a pseudogene.
Keywords: long-term memory, NOS, gene expression, antisense RNA, Lymnaea, mollusk, CGC, pseudogene
Introduction
Associative long-term memory (LTM) formation depends on the ability of behavioral conditioning to evoke specific changes in patterns of gene expression in the CNS (Mayford and Kandel, 1999). This learning-evoked genomic response alters synaptic mechanisms within neuronal circuits that are required for the learned behavior. This suggests that the expression of specific neuronal genes that can affect key synaptic mechanisms must be highly responsive to associative sensory cues occurring during behavioral conditioning. Furthermore, this also means that training-induced changes in gene activity must be targeted to specific neurons that are required for the learned behavior.
A particularly favorable model system for a direct approach to the analysis of the genomic response to training is provided by the mollusc Lymnaea stagnalis. In this species, a single pairing of two chemosensory stimuli, sucrose [unconditioned stimulus (US)] and amyl acetate [conditioned stimulus (CS)], leads to the associative conditioning of feeding that can last for >2 weeks (Alexander et al., 1984). It is well established that the gaseous neurotransmitter nitric oxide (NO) is required for the chemosensory activation of the feeding central pattern generator (CPG) in Lymnaea (Elphick et al., 1995). Moreover, if signaling by NO is disrupted during a critical time window of ∼6 h after conditioning, LTM formation is impaired (Kemenes et al., 2002). This requirement for NO indicates a crucial role of this neurotransmitter in the initial stages of LTM formation and is consistent with its role in a number of other experimental models of associative learning (Müller, 1996; Lu et al., 1999; Rose, 2000; Schweighofer and Ferriol, 2000). Because NO is essential both for the chemosensory activation of feeding behavior and for the chemical conditioning of feeding, we have investigated here whether memory formation is associated with changes in the expression of the genes that encode nitric oxide synthase (NOS), the enzyme responsible for the production of NO.
The first molluscan NOS mRNA was characterized in 1998 (Korneev et al., 1998). Now we report on the cloning of another NOS-encoding transcript. We will refer to the first mRNA as Lym-nNOS1 and to the novel one as Lym-nNOS2. Here we show that after associative conditioning, the expression of Lym-nNOS1 is transiently upregulated in the CNS at 6 h after training, whereas the expression of Lym-nNOS2 is stable at all measured post-training time points. Thus, the activation of the Lym-nNOS1 gene occurs near the end of the critical period after training during which NO is required for memory consolidation. A similar result is obtained in a key modulatory neuron, the cerebral giant cell (CGC), which is essential for the conditioned feeding response (Yeoman et al., 1994). In addition to the Lym-nNOS1 mRNA, the CGC also contains an antisense RNA transcribed from a NOS pseudogene (anti-NOS gene). This anti-NOS RNA regulates NOS gene expression through the formation of an RNA-RNA duplex with Lym-nNOS1 mRNA (Korneev et al., 1999). Here we demonstrate that the expression of the anti-NOS gene is transiently downregulated in the CGC by training ∼2 h before the upregulation of Lym-nNOS1.
Materials and Methods
Experimental animals. Animals (L. stagnalis) were kept at 18-20°C on a 12 h light/dark regimen and fed on lettuce and a vegetable-based fish food.
One-trial conditioning protocol. Reward conditioning was performed using a method based on a previously published protocol (Alexander et al., 1984). Snails were randomly assigned to experimental (paired) and control (unpaired) groups to be given a single conditioning and control trial, respectively. Experimental animals were exposed to a solution of amyl acetate (CS) and immediately after that to a sucrose solution (US). Control animals were exposed to the CS and to the US, separated by an interval of 1 h. A randomly chosen subset of 20 animals from each group was retained and tested for LTM formation at 24 h after the paired and unpaired trials, as described previously (Kemenes et al., 2002). A third group of animals was kept under the same conditions and had the same feeding regimen as experimental and unpaired control snails but was not exposed to either the CS or the US. This group is referred to as the naive control group.
Surgical procedures. At different time points (30 min, 2 h, 4 h, 6 h, 24 h) after the treatment, a randomly chosen subset of animals (usually 10 individuals per time point) was killed, and the CNS was removed. The cerebral and buccal ganglia of the CNS were separated and used for RNA extraction. For the single-cell experiments, the CNSs were placed in a buffer containing 0.1 m Tris, pH 7.2, 50% propylene glycol, and 10 mm ribonucleoside vanadyl complex, immediately after the dissection. The CNSs were kept in this solution on ice for 50 min and then at -20°C until they were used for extraction of the CGCs.
Molecular techniques and procedures. A Lymnaea CNS cDNA library was screened using a radioactively labeled fragment of Lym-nNOS1 cDNA (Korneev et al., 1998). A positive clone containing a cDNA insert of ∼6 kb was selected for further examination. Sequence analysis of the insert has shown that it was copied from a novel NOS-encoding mRNA (GenBank accession number AY769987).
Conventional multiplex reverse transcription (RT)-PCR was performed on RNAs isolated from either pooled or individual cerebral and buccal ganglia by means of the NucleoSpin RNA II kit (Macherey-Nagel, Düren, Germany). We used primers 5′-ATTGCACCGTTCAGATCAT-3′ and 5′-CATGTGTTTAGCTGTTCTG-3′ for detection of Lym-nNOS1 and Lym-nNOS2 and primers 5′-AGCTTGAGAGGATCAATG-3′ and 5′-AGTGTGTCAGTTGGAATC-3′ for detection of β-tubulin. After 25 cycles (denaturation, 94°C, 20 s; annealing, 50°C, 30 s; extension, 68°C, 1 min), PCR products were resolved on a 1% agarose gel and analyzed using the Electrophoresis Documentation and Analysis System 290 (Eastman Kodak, Rochester, NY).
Real-time RT-PCR was performed on RNAs isolated either from individual cerebral ganglia by means of the NucleoSpin RNA II kit (Macherey-Nagel) or from CGCs by means of the Absolutely RNA Nanoprep kit (Stratagene, La Jolla, CA). Both methods involve DNase treatment. cDNAs produced from RNA preparations by iScript reverse transcriptase (Bio-Rad, Richmond, CA) were amplified and analyzed on the Mx3000 real-time cycler (Stratagene) using QuantiTect SYBR Green PCR kit (Qiagen, Valencia, CA) and the following parameters: denaturation, 94°C, 30 s; annealing, 52°C, 1 min; extension, 72°C, 30 s. We used primers 5′-AGTTTGAGGGATGAGAACCT-3′ and 5′-TCCAGTGCCCATAATTACTC-3′ for detection of Lym-nNOS1, primers 5′-TTGAACAGAACACATGTAGAG-3′ and 5′-GTACTAGCAGCCGTTGG-3′ for Lym-nNOS2, primers 5′-ACTATCAGTAGTTCAAAGGTC-3′ and 5′-GTCTGATGACTAGCAAAGCT-3′ for anti-NOS, and primers 5′-AAGGGACATTACACAGAGG-3′ and 5′-GTGTCAGTTGGAATCCTTG-3′ for β-tubulin. The identity of all PCR products was confirmed by sequencing. The amount of target transcript, normalized to an endogenous reference and relative to a calibrator (CAL), was calculated as 2-ΔΔCT (Pfaffl, 2001), where ΔΔCT = ΔCT - ΔCT(CAL). ΔCT and ΔCT-(CAL) are the differences in threshold cycles for the target (Lym-nNOS1, Lym-nNOS2, anti-NOS) and reference (β-tubulin) measured in the samples and in the calibrator, respectively.
Results
Two NOS-encoding RNAs and one noncoding anti-NOS RNA are present in Lymnaea CNS
In our search for new neuronal NOS (nNOS)-related transcripts in Lymnaea, we screened a cDNA library prepared from the CNS using a fragment of the cDNA derived from a previously characterized nNOS-encoding mRNA referred to here as Lym-nNOS1 (Korneev et al., 1998). One of the selected clones contained a cDNA insert of 6313 nt with an open reading frame (ORF) of 3654 nt and 5′ and 3′ untranslated regions (UTRs) of 244 and 2415 nt, respectively. Sequencing shows that the new cDNA is related to Lym-nNOS1 but represents a copy from an independent mRNA. The identity of this novel mRNA to the published Lym-nNOS1 sequence is 89% within the ORF, and there is no homology in the UTRs (Fig. 1A). The deduced new protein consists of 1218 aa and is ∼90% identical to the Lym-nNOS1 protein. Clearly therefore, the novel protein is a member of the Lymnaea NOS family, and we will refer to it here as Lym-nNOS2. Both Lym-nNOS1 and Lym-nNOS2 contain a tandem repeat region located near the COOH terminal. In Lym-nNOS2, this region is composed of 20 copies of the 7 aa elementary motif and is 56 aa longer than the repetitive region in Lym-nNOS1. We exploited this difference to detect simultaneously the expression of Lym-nNOS1 and Lym-nNOS2 in our multiplex RT-PCR experiments (Fig. 1B).
Figure 1.
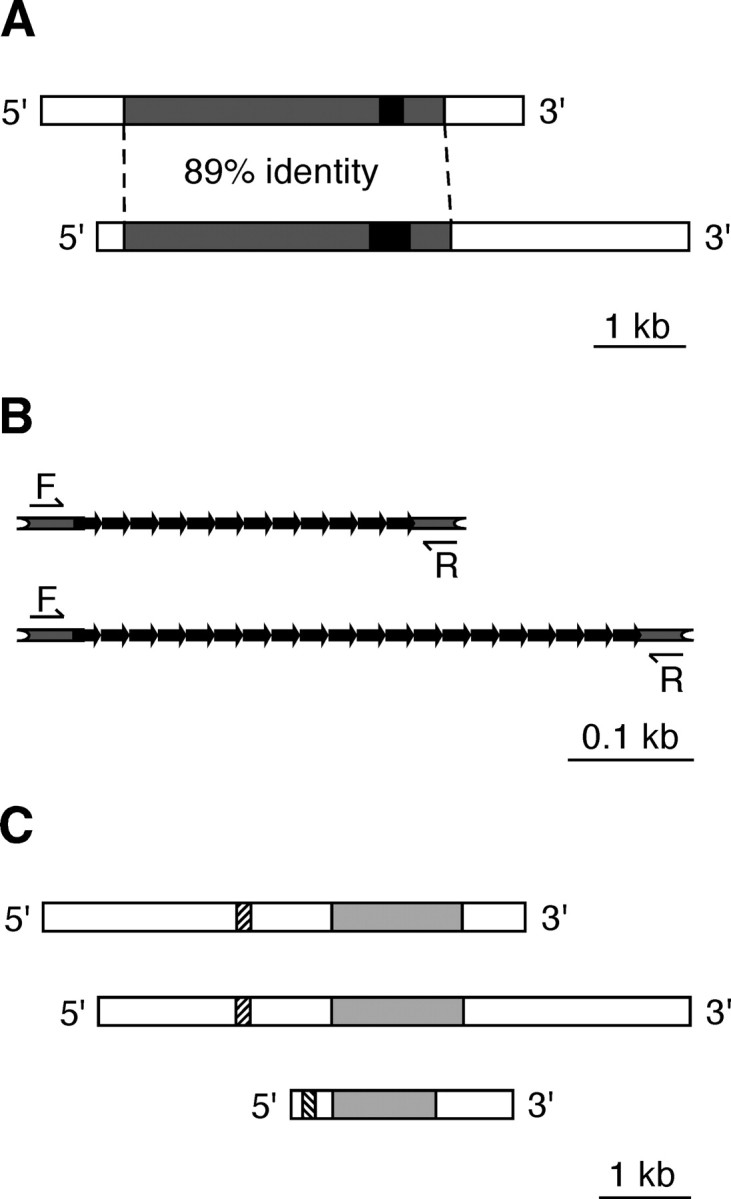
Two types of NOS-encoding mRNAs are present in the Lymnaea CNS. A, Schematic representation of the Lym-nNOS1 (top) and the novel Lym-nNOS2 (bottom) transcripts. Gray boxes indicate the protein-coding regions. Black boxes indicate tandem repeats in the coding regions. The unfilled areas correspond to the 5′ and 3′ UTRs. Note that there is no homology between UTRs in Lym-nNOS1 and Lym-nNOS2. B, Organization of tandem repeats in the coding regions of Lym-nNOS1 (top) and Lym-nNOS2 (bottom). Bold black arrows indicate elementary repeats. Half arrows indicate the positions of primers used in conventional multiplex RT-PCR experiments (primers F and R). C, Schematic organization of the Lym-nNOS1 (top), Lym-nNOS2 (middle), and anti-NOS (bottom) transcripts. Regions of homology are gray. Hatched boxes indicate the antisense region in the anti-NOS and its complementary counterparts in Lym-nNOS1 and Lym-nNOS2.
Completing the complement of transcripts discussed in this work is the previously reported noncoding RNA transcribed from a pseudogene, which is also a member of the Lymnaea NOS gene family (Korneev et al., 1999). This RNA contains a region of significant antisense homology to the nNOS-encoding mRNAs. We refer to the noncoding transcript as anti-NOS RNA. Figure 1C provides an overview comparison of each of the three NOS transcripts referred to in this paper.
Training selectively targets Lym-nNOS1 expression in the CNS
In all experiments in which the consequences of conditioning on gene expression were measured, a randomly chosen subset of each group of animals was retained and tested for LTM formation at 24 h after training. This was to confirm that LTM would have occurred in the animals that were killed at earlier time points to measure training-induced changes in NOS gene expression. Importantly, the mean feeding response to amyl acetate (CS) of the trained snails was always significantly higher than the response of the control animals. The results of one such experiment are shown in Figure 2A. The mean feeding response to the CS of the trained snails was 8.06 ± 2.8 rasps (feeding movements) per 2 min; this was significantly higher (p < 0.005) than the response of the control animals (-1.4 ± 1.4 rasps per 2 min).
Figure 2.
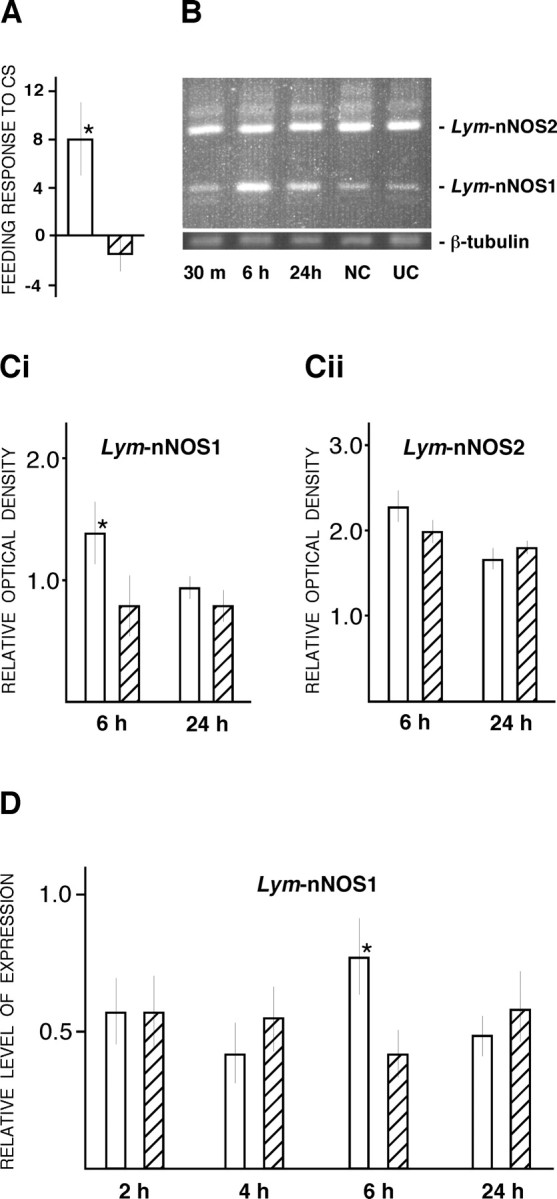
Lym-nNOS1 gene is upregulated in cerebral ganglia at 6 h after training. A, A typical result of the experimental test of LTM formation at 24 h after training. Note that the mean feeding response to amyl acetate (the CS) of the trained snails (white bar) is significantly higher than the response of the control animals (hatched bar). B, Results of conventional multiplex RT-PCR on pooled cerebral ganglia dissected from conditioned [30 min (m), 6 h, and 24 h after training] and control (NC, naive control; UC, 6 h unpaired control) animals. Primers specific for Lym-nNOS1, Lym-nNOS2, and β-tubulin mRNAs were used. C, Statistical analysis of multiplex RT-PCR experiments performed on individual cerebral ganglia dissected from trained (white bars) and unpaired control (hatched bars) animals. PCR products were resolved on a 1% agarose gel, and the levels of Lym-nNOS1 (Ci) and Lym-nNOS2 (Cii) gene expression were calculated relative to β-tubulin (internal control) using the Electrophoresis Documentation and Analysis System 290 (Eastman Kodak). D, Results of real-time RT-PCR analysis of Lym-nNOS1 gene expression performed on individual cerebral ganglia at 2, 4, 6, and 24 h after training. The level of Lym-nNOS1 gene expression, normalized to an endogenous control (β-tubulin) and relative to a calibrator, is indicated by white bars (conditioned groups) and hatched bars (unpaired control groups). Asterisks in A, Ci, and D indicate significant differences from controls of at least p < 0.05.
To examine the effect of conditioning on the expression of the nNOS-encoding genes, we first used comparative multiplex RT-PCR to monitor simultaneously the activity of the Lym-nNOS1 and Lym-nNOS2 genes. Two regions of the CNS containing the feeding circuits (the cerebral and buccal ganglia) were analyzed separately, and the activity of the genes was determined at 30 min, 6 h, and 24 h after a single conditioning trial. Because the length of the repetitive regions in Lym-nNOS1 and Lym-nNOS2 is different, we could detect both nNOS-encoding transcripts simultaneously using a single pair of primers (Fig. 1B). The results of the analysis performed on pooled (n = 10) cerebral ganglia (Fig. 2B) indicate upregulation of the Lym-nNOS1 gene in the 6 h experimental group compared with the naive and unpaired controls. Note that the level of Lym-nNOS2 gene expression remains unchanged in all tested groups of snails. No significant trainingrelated changes in nNOS gene expression have been detected in the buccal ganglia. Thus, our data suggest that the regulation of Lym-nNOS1 but not Lym-nNOS2 is sensitive to the effects of training and that training-induced changes in Lym-nNOS1 gene activity are not generalized but are targeted in particular to the cerebral ganglia.
The statistical significance of the results obtained on pooled ganglia was examined by additional experiments in which cerebral ganglia (n = 10) dissected at 6 and 24 h after training were analyzed individually by conventional RT-PCR (Fig. 2C). The data were subjected to a two-way ANOVA, and it was confirmed that there is a significant stimulation of Lym-nNOS1 gene expression at 6 h after training (p < 0.05) and that Lym-nNOS2 expression is unaffected by conditioning.
To quantify more precisely the effects of conditioning on Lym-nNOS1 gene expression in the cerebral ganglia with a finer time resolution, we exploited the advantages of real-time RT-PCR. The cerebral ganglia were removed from groups of snails (n = 10) at 2, 4, 6, and 24 h after conditioning and were individually subjected to real-time RT-PCR in which the expression of the Lym-nNOS1 gene was analyzed using a calibrator-normalized relative quantification method. Results of the analysis, shown in Figure 2D, demonstrate a statistically significant upregulation of the Lym-nNOS1 gene in the 6 h post-training group only (p < 0.05). This confirms the transient and highly selective nature of the training-associated Lym-nNOS1 gene activation.
Training differentially regulates the expression of Lym-nNOS1 and anti-NOS genes in a single identified neuron
It is known that CGC activity is required for the feeding response and that CGC has widespread synaptic connections with the rest of the feeding circuit, including neurons of the CPG and motoneurons (Fig. 3A) (Yeoman et al., 1996). Also, the CGC is one of the key neurons involved in the modulation of conditioned feeding behavior, for which it has a crucial gating function (Yeoman et al., 1994; Straub and Benjamin, 2001). Based on this, we selected the CGCs for an investigation of training-induced changes in Lym-nNOS1 gene expression.
Figure 3.
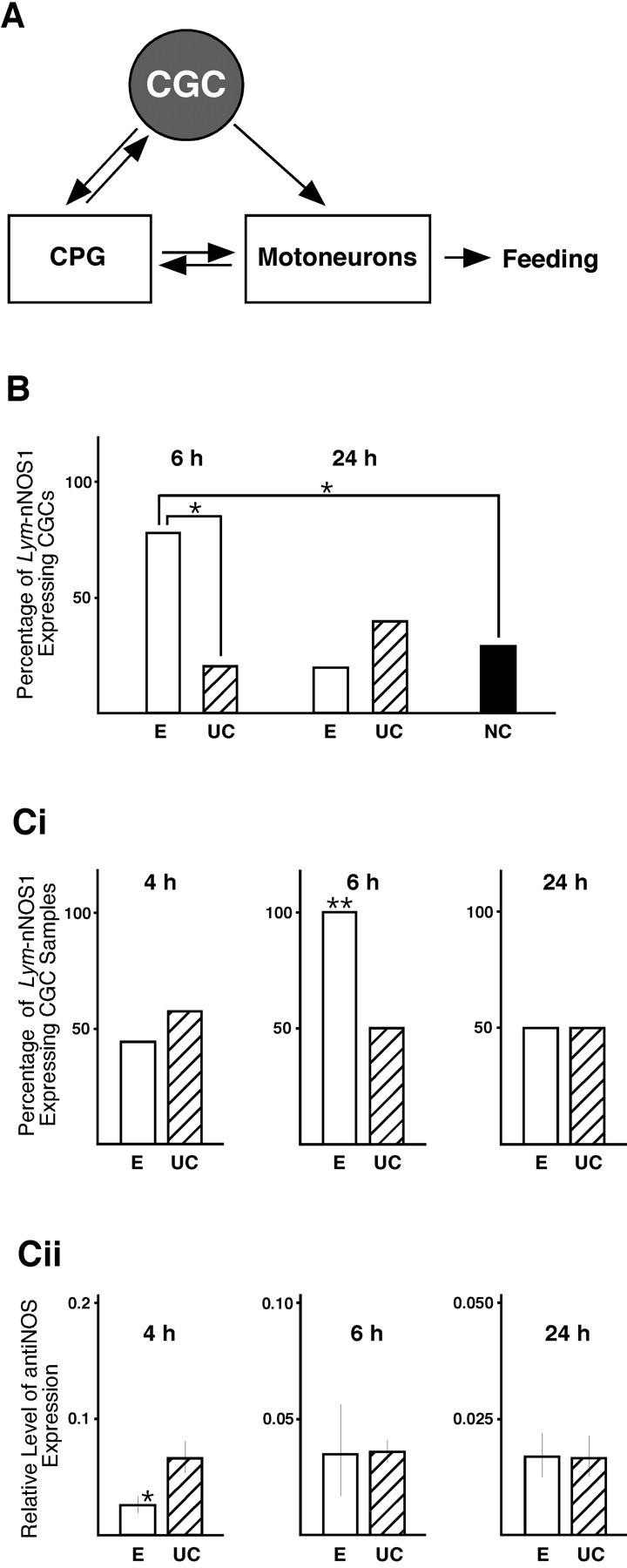
Training-induced differential regulation of the Lym-nNOS1 and anti-NOS genes in CGCs. A, Schematic diagram showing the neural circuit underlying feeding behavior. The CGCs play an important gating role in feeding behavior through their modulatory actions on interneurons of the CPG and feeding motoneurons. B, Results of real-time RT-PCR performed on individual CGCs. The percentage of Lym-nNOS1-expressing CGCs dissected from conditioned (E), unpaired control (UC), and naive control (NC) animals is indicated by white, hatched, and black bars, respectively. Single asterisks indicate a significant difference between the 6 h conditioned group and both control groups (p < 0.05). C, Results of simultaneous analysis of Lym-nNOS1 (Ci) and anti-NOS (Cii) expression in CGCs at 4, 6, and 24 h after training by real-time RT-PCR. Ci, Induction of the Lym-nNOS1 gene at 6 h after training. The double asterisk in the 6 h conditioned group (white bar) indicates a significant difference from the unpaired control (hatched bar) of at least p < 0.005. Cii, Downregulation of the anti-NOS gene at 4 h after training. The level of anti-NOS gene expression normalized to an endogenous control (β-tubulin) and relative to a calibrator is indicated by white bars (conditioned groups) and hatched bars (unpaired control groups). The single asterisk in the 4 h conditioned group indicates a significant difference from controls of p = 0.05.
The CGCs dissected from conditioned, unpaired control and naive snails were subjected individually to quantitative real-time RT-PCR analysis using the Qiagen QuantiTect SYBR Green PCR kit. Very interesting temporal dynamics of training-induced changes in Lym-nNOS1 gene expression were revealed (Fig. 3B). Specifically, we found that in the majority of CGCs dissected from either unpaired or naive control snails, Lym-nNOS1 mRNA cannot be detected. At the same time, almost 80% of the CGCs isolated from the 6 h conditioned group contain Lym-nNOS1 mRNA. Fisher's exact test confirmed that this difference between controls and the 6 h conditioned group is statistically significant (p < 0.05). Thus the results of the experiment clearly demonstrate that there is a training-associated induction of Lym-nNOS1 gene expression in CGCs occurring at 6 h after training.
Next we investigated changes in Lym-nNOS gene expression with finer time resolution and simultaneously measured the expression of the anti-NOS gene, which we know acts in CGCs as a translational negative regulator of NOS gene expression (Korneev et al., 1999). Temporal dynamics of the post-training expression of both Lym-nNOS1 and anti-NOS were studied at three time points in three experimental and three unpaired control groups. Within each group, the CGCs were pooled to form eight separate samples, each containing three neurons. The samples were then subjected to real-time RT-PCR. The results of the experiment, showing statistically significant (p < 0.005) induction of Lym-nNOS1 gene expression at 6 h after conditioning (Fig. 3Ci), confirm our findings described above and reveal its transient and precisely timed nature. As for the anti-NOS gene, it also exhibits transient training-provoked changes in expression (Fig. 3Cii). However, the temporal pattern of these changes is different. The anti-NOS gene is downregulated in the 4 h experimental group but shows no changes in activity at 6 and 24 h after training compared with controls.
Discussion
In this study, we highlight the acute sensitivity to exposure to a very brief behavioral stimulus of the transcriptional activity of genes associated with the production of NO, a neurotransmitter with an established role in memory formation and in the chemosensory activation of feeding (Elphick et al., 1995; Kemenes et al., 2002).
We show that a single conditioning trial acts selectively and differentially within the NOS gene family. The expression of the Lym-nNOS1 gene is transiently upregulated at 6 h after conditioning, an anti-NOS gene is downregulated at 4 h, and the activity of the newly discovered Lym-nNOS2 gene is stable at all measured time points. Importantly, the training-induced changes in the activity of these genes are targeted to the cerebral ganglia. This is significant because chemosensory neurons that detect the US and the CS terminate in these ganglia and also because the cerebral ganglia are the central locus of plasticity underlying associative LTM formation (Straub et al., 2004). Moreover, we know that the cerebral ganglia contain modulatory neurons that are essential for the gating or initiation of feeding behavior in response to the CS. One such neuron, a giant serotonergic cell known in Lymnaea as the CGC, has been a particular focus of our interest.
We report here that the expression of Lym-nNOS1 and anti-NOS genes is differentially regulated in this neuron as the result of training. This finding is significant for a number of reasons. For example, the CGCs are essential for the activation of feeding, but they are not components of the feeding CPG, and their effects on feeding are indirect. The CGCs in fact are interconnected diffusely to many neural components of the feeding system (Fig. 3) and function in a permissive manner to modulate the probability that feeding behavior will be initiated (Yeoman et al., 1994). This modulatory function of the CGCs, coupled to their diffuse connectivity with the feeding system, suggests that these neurons have multiple and distributed effects, rather than highly local synaptic effects, within the feeding neural network. Although speculative, it is attractive to suggest that the ability of the CGC to signal broadly within the feeding neural network via NO is altered during a critical early stage of memory formation by the traininginduced differential regulation of the Lym-nNOS1 and anti-NOS genes. One particularly intriguing explanation of how exactly this might happen is based on the results of our previous experiments, in which we showed that the expression of the nNOS gene in the CGCs is suppressed by a natural antisense mechanism through the formation of duplex molecules between Lym-nNOS1 mRNA and anti-NOS RNA (Korneev et al., 1999). Consequently, we hypothesize that the decrease in the amount of anti-NOS RNA observed at 4 h after conditioning can lead to an increase in the production of NO that facilitates memory formation. Whether this change in the NO signaling capability of the CGC is required for memory formation or is one of many subtle effects of training that together are required has yet to be determined.
The existence of a second NOS-encoding mRNA (Lym-nNOS2) in the Lymnaea CNS was not unexpected, because we have shown recently that the NOS gene in Lymnaea was subjected to a duplication (Korneev and O'Shea, 2002). Therefore, it was reasonable to expect that other copies of the gene could exist in the genome. Our discovery of Lym-nNOS2 has confirmed this. Clearly, however, the Lym-nNOS1 and Lym-nNOS2 genes respond very differently to conditioning, the former being induced and the latter being stably and constitutively expressed. These observations are reminiscent of the situation in mammals in which some NOS genes (nNOS and endothelial NOS) are consistently expressed and one [inducible NOS (iNOS)] is inducible (for review, see Stuehr, 1999). So far the existence of distinct constitutive and inducible NOS genes has not been demonstrated in invertebrates, and indeed in Drosophila there is just one gene encoding NOS (Stasiv et al., 2001). However, this gene produces a number of alternatively spliced transcripts that might encode proteins with different patterns of expression and function (Stasiv et al., 2004). Molluscs as well as mammals show that there is another way to achieve heterogeneity and flexibility within the system regulating NO signaling. This involves the creation of several differentially expressed homologous NOS genes “specialized” for different functions. Because Lym-nNOS1 in Lymnaea is dynamically responsive to subtle associative sensory cues, it is likely that this gene is functionally associated with molecular mechanisms underlying adaptive behavioral plasticity. Finally, this dynamic sensitivity to a sensory cue is also shared by anti-NOS, providing a clear example of a pseudogene with expression that is regulated by a behavioral stimulus.
Footnotes
This work was supported by a United Kingdom Biotechnology and Biological Sciences Research Council grant. S.A.K. and E.I.K. are supported by the Wellcome Trust.
Correspondence should be addressed to Sergei Korneev or Michael O'Shea at the above address. E-mail: S.Korneev@sussex.ac.uk or M-Oshea@sussex.ac.uk.
Copyright © 2005 Society for Neuroscience 0270-6474/05/251188-05$15.00/0
References
- Alexander Jr J, Audesirk TE, Audesirk GJ (1984) One-trial reward learning in the snail Lymnaea stagnalis J Neurobiol 15: 67-72. [DOI] [PubMed] [Google Scholar]
- Elphick MR, Kemenes G, Staras K, O'Shea M (1995) Behavioral role for nitric oxide in chemosensory activation of feeding in a mollusc. J Neurosci 15: 7653-7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemenes I, Kemenes G, Andrew RJ, Benjamin PR, O'Shea M (2002) Critical time-window for NO-cGMP-dependent long-term memory formation after one-trial appetitive conditioning. J Neurosci 22: 1414-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneev S, O'Shea M (2002) Evolution of nitric oxide synthase regulatory genes by DNA inversion. J Mol Biol Evol 19: 1228-1233. [DOI] [PubMed] [Google Scholar]
- Korneev SA, Piper MR, Picot J, Phillips R, Korneeva EI, O'Shea M (1998) Molecular characterization of NOS in a mollusc: expression in a giant modulatory neuron. J Neurobiol 35: 65-76. [DOI] [PubMed] [Google Scholar]
- Korneev SA, Park JH, O'Shea M (1999) Neuronal expression of neural nitric oxide synthase (nNOS) protein is suppressed by an antisense RNA transcribed from an NOS pseudogene. J Neurosci 19: 7711-7720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YF, Kandel ER, Hawkins RD (1999) Nitric oxide signaling contributes to late-phase LTP and CREB phosphorylation in the hippocampus. J Neurosci 19: 10250-10261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayford M, Kandel ER (1999) Genetic approaches to memory storage. Trends Genet 15: 463-470. [DOI] [PubMed] [Google Scholar]
- Müller U (1996) Inhibition of nitric oxide synthase impairs a distinct form of long-term memory in the honeybee, Apis mellifera Neuron 16: 541-549. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: 2002-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose SP (2000) God's organism? The chick as a model system for memory studies. Learn Mem 7: 1-17. [DOI] [PubMed] [Google Scholar]
- Schweighofer N, Ferriol G (2000) Diffusion of nitric oxide can facilitate cerebellar learning: a simulation study. Proc Natl Acad Sci USA 97: 10661-10665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasiv Y, Regulski M, Kuzin B, Tully T, Enikolopov G (2001) The Drosophila nitric-oxide synthase gene (dNOS) encodes a family of proteins that can modulate NOS activity by acting as dominant negative regulators. J Biol Chem 276: 42241-42251. [DOI] [PubMed] [Google Scholar]
- Stasiv Y, Kuzin B, Regulski M, Tully T, Enikolopov G (2004) Regulation of multimers via truncated isoforms: a novel mechanism to control nitricoxide signaling. Genes Dev 18: 1812-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub VA, Benjamin PR (2001) Extrinsic modulation and motor pattern generation in a feeding network: a cellular study. J Neurosci 21: 1767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub VA, Styles BJ, Ireland JS, O'Shea M, Benjamin PR (2004) Central localization of plasticity involved in appetitive conditioning in Lymnaea Learn Mem 11: 787-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuehr DJ (1999) Mammalian nitric oxide synthases. Biochim Biophys Acta 1411: 217-230. [DOI] [PubMed] [Google Scholar]
- Yeoman MS, Pieneman AW, Ferguson GP, Ter Maat A, Benjamin PR (1994) Modulatory role for the serotonergic cerebral giant cells in the feeding system of the snail, Lymnaea I. Fine wire recording in the intact animal and pharmacology. J Neurophysiol 72: 1357-1371. [DOI] [PubMed] [Google Scholar]
- Yeoman MS, Brierley MJ, Benjamin PR (1996) Central pattern generator interneurons are targets for the modulatory serotonergic cerebral giant cells in the feeding system of Lymnaea J Neurophysiol 72: 1372-1382. [DOI] [PubMed] [Google Scholar]


