SUMMARY:
In this study, we used ELISPOT to quantify frequencies of bronchoalveolar lavage (BAL) and peripheral blood T cells capable of producing IFNγ in response to PPD, antigen 85B, and Mtb-specific antigens CFP-10 and ESAT-6 in individuals with latent tuberculosis infection (LTBI) and Mtb-naïve controls. Compared to peripheral blood, BAL cells of LTBI subjects displayed significant enrichment for T cells responding to PPD, antigen 85B, and CFP-10, but not to ESAT-6. Baseline BAL cells of LTBI subjects displayed significant production of Mig (CXCL9) in response to PPD, antigen 85B, and CFP-10 as well. These findings suggest that enrichment for Mtb-specific T cells within BAL is not unique to active pulmonary tuberculosis and may, to the contrary, contribute to protection from re-infection in Mtb immune individuals.
INTRODUCTION
Late 20th century advances in molecular biology of the mycobacteria led to the discovery that highly antigenic Mycobacterium tuberculosis (Mtb) proteins early secreted antigen target 6kDa protein (ESAT-6) and culture filtrate protein 10 (CFP-10) were part of a gene segment present in all Mtb strains, but absent in Mycobacterium bovis BCG and most of the nontuberculous mycobacteria (1–3). This important finding was exploited in the development of IFNγ release assays (IGRAs) utilizing these antigens (such as QuantiFERON-TB Gold In-Tube [QFN-GIT] and T-Spot.TB). Unlike traditional skin testing with purified protein derivative of Mtb (PPD), IGRAs provide the ability to the distinguish immune responses to Mtb infection from those induced by BCG vaccination and most environmental mycobacteria (4). Further studies have attempted to apply this methodology to another clinical dilemma, that of distinguishing active disease in smear-negative pulmonary tuberculosis from latent tuberculosis infection (LTBI). Specifically, prior studies proposed that enrichment within bronchoalveolar lavage (BAL) for T-cell ELISPOT responses to Mtb-specific antigens could provide a means for rapid immunodiagnosis of active pulmonary TB (5–7). However, our prior studies suggested that BAL is enriched for CD4+ T cells capable of responding to Mtb protein antigens even in LTBI (8). Further, early induction of IFNγ-inducible chemokines, presumably facilitated by the presence of these local effector memory T cells (TEM), was associated with recruitment of additional antigen-specific T cells to the lung in response to bronchoscopic challenge with PPD (8,9). We therefore evaluated BAL cells of healthy individuals with LTBI and of non-vaccinated Mtb naïve subjects for ELISPOT responses to Mtb-specific antigens CFP-10 and ESAT-6, as well as to cross-reactive mycobacterial antigens Mtb Ag 85B and PPD. We also measured induction of CXCR3 chemokine ligands Mig (CXCL9) and IP-10 (CXCL10) by this same panel of antigens.
RESULTS:
Subject characteristics:
Healthy non-smokers were recruited to LTBI and Mtb-naïve subject groups. Mean age was 28 (range 20–47) for the individuals with LTBI and 34 (22–47) for control subjects. The LTBI group consisted of 5 men and 5 women; 8 were Caucasian, 1 African American, and 1 was of Indian descent. The LTBI status of 8 subjects was confirmed by QuantiFERON TB Gold In-Tube (QFN-GIT) testing, whereas 2 had positive PPD skin tests only. Lack of clinical evidence for active disease (as detailed in Methods) was confirmed by BAL fluid culture, which showed no growth in all cases. The control group was composed of 6 men and 4 women; 9 were Caucasian and 1 African American. Mtb-naïve status was determined by QFN-GIT testing for 9 subjects and by PPD skin testing for 1 subject. BAL cell differentials for LTBI subjects had 93.7 (+/−2.5)% alveolar macrophages, 3.88 (+/−1.73)% lymphocytes, and 2.27 (+/− 1.46)%; BAL of Mtb-naïve subjects displayed similar findings, with 92.38 (+/3.09%) alveolar macrophages, 4.59 (+/−2.43)% lymphocytes, and 2.36 (+/−2.71)% neutrophils. Eosinphils represented fewer than 1% of BAL cells for both subject groups.
Frequency of IFNγ-producing cells in PBMC and BAL:
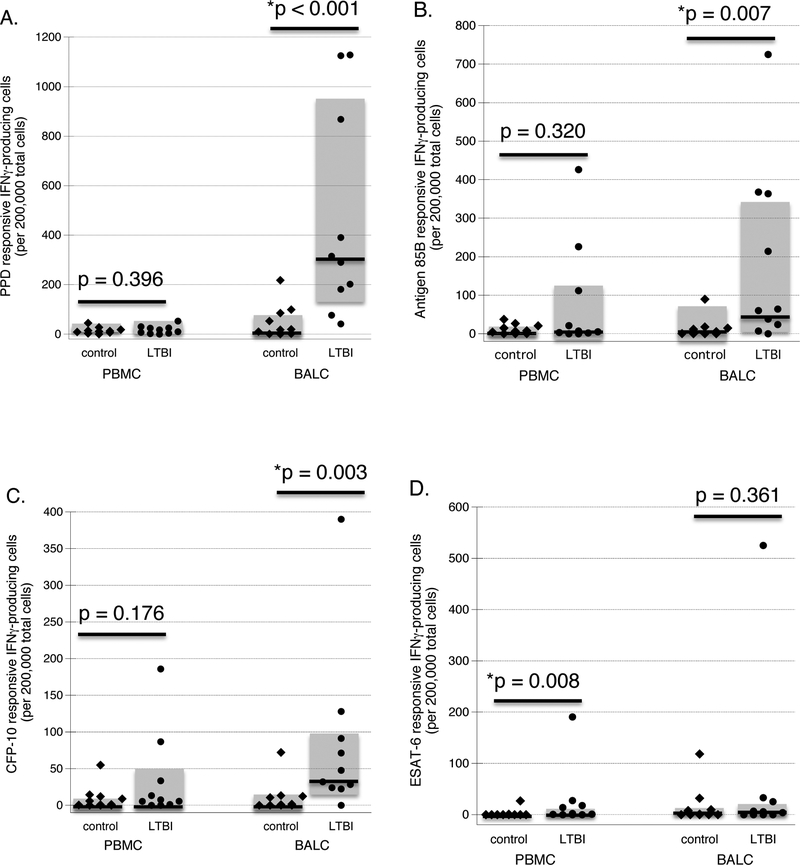
ELISPOT results for paired studies of PBMC and BAL are displayed in Figure 1. As illustrated, the frequency of IFNγ-producing cells with responsiveness to PPD, antigen 85B, and CFP-10 within PBMC did not differ between individuals with LTBI and control subjects (p=0.396, p=0.320, p=0.176, respectively, by Mann-Whitney test). However, significantly more ESAT-6 responsive IFNγ-producing PBMC were observed in LTBI subjects as compared to controls (p=0.008), although the absolute numbers of such cells were relatively infrequent for both subject groups. In contrast, in comparison to those of control subjects, BAL cells of LTBI subjects displayed markedly higher frequencies of IFNγ-producing cells with responsiveness to PPD, antigen 85B, and CFP-10 (p<0.001, p=0.007, and p=0.003, respectively), but not to ESAT-6 (p=0.361).
Figure 1: Frequency of antigen-responsive IFNγ-producing cells in peripheral blood and baseline BAL of LTBI subjects and naïve controls.
The figures illustrate frequency of cells in PBMC and baseline BAL that produce IFNγ in response to in vitro stimulation with PPD (1A), CFP-10 (1B), ESAT-6 (1C) and antigen 85B (1D), as determined by ELISPOT. Diamonds represent findings for individual control subjects, whereas results for subjects with LTBI are represented by circles. In all cases, dark horizontal lines indicate median values and shaded boxes indicate the range of 25th percentile to 75th percentile results. As detailed in the text, individuals with LTBI display higher frequencies of IFNγ-producing PBMC responsive to ESAT-6, but not to PPD, CFP-10, or antigen 85B. In contrast, BAL cells of subjects with LTBI display significantly higher frequencies of IFNγ-producing cells responsive to PPD, CFP-10, and antigen 85B than do Mtb-naïve control subjects; however, BAL cell IFNγ responses to in vitro stimulation with ESAT-6 do not differ between the two subject groups.
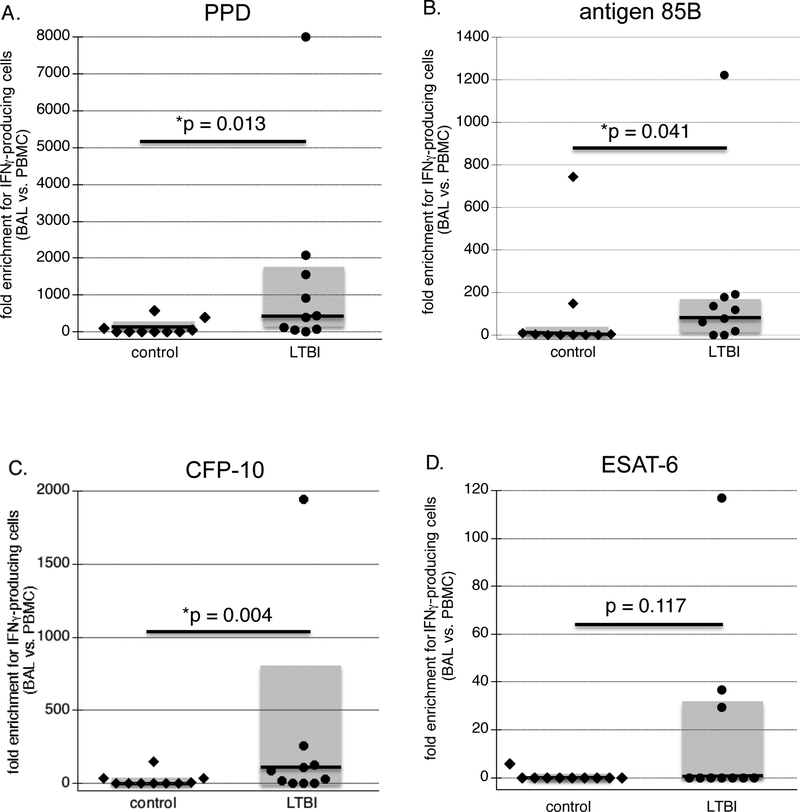
Calculation of enrichment of antigen-specific lymphocytes with BAL as compared to PBMC was based on the conservative estimation that PBMC are generally composed of approximately 85% lymphocytes, and on the observed percentage of lymphocytes in BAL differential cell counts of each subject (Figure 2). As illustrated, compared to Mtb-naïve control subjects, individuals with LTBI displayed significant enrichment for BAL lymphocytes producing IFNγ in response to PPD, antigen 85B, and CFP-10 (p=0.013, 0.041, and 0.003, respectively), but not for ESAT-6 responsive lymphocytes (p=0.117). These findings thus indicate that enrichment within BAL for Mtb-specific IFNγ-producing lymphocytes for these antigens is not specific to active TB, but is observed in healthy individuals with LTBI as well.
Figure 2: Enrichment for lymphocytes responsive to Mtb antigens in BAL of healthy subjects with LTBI:
Enrichment was calculated from the data displayed in Figure 1 using BAL cell differentials of each subject and the conservative assumption that lymphocytes accounted for 85% of PBMC samples in each case. Figures display the fold-differences in frequencies of antigen-responsive IFNγ-producing lymphocytes in these samples for each subject, again displayed as individual data (diamonds and circles), median values (dark lines) and range of 25th to 75th percentiles (shaded boxes). As described in the text, compared to results for control subjects, BAL of LTBI subjects displayed significant enrichment for IFNγ-producing cells in response to PPD, antigen 85B, and CFP-10, but not to ESAT-6.
Induction of BAL cell production of IFNγ-inducible chemokines CXCL9 (Mig) and CXCL10 (IP-10) by M. tuberculosis antigens:
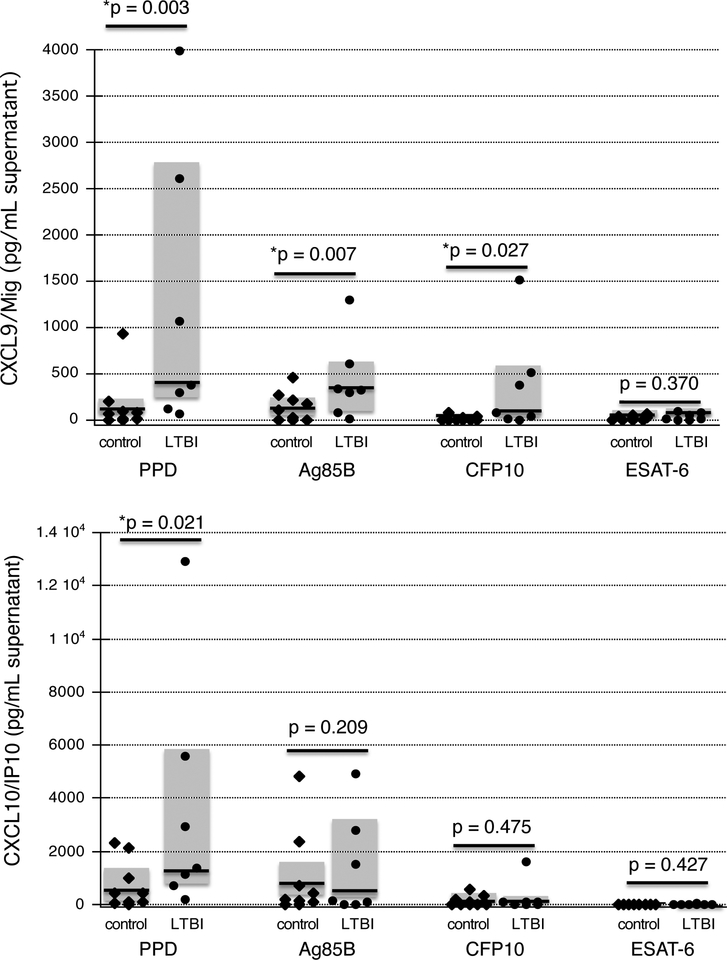
Our prior studies have indicated that individuals with LTBI produce the IFNγ-inducible chemokines CXCL9 (Mig) and CXCL10 (IP-10) in response to in vivo bronchoscopic challenge with PPD, and that this response is associated with subsequent recruitment of additional PPD-responsive Th1 cells to the lung (8,9). Further, these chemokines were also induced by in vivo PPD stimulation of baseline BAL cells for LTBI subjects (8). In order to evaluate the potential functional consequences of CFP-10 and ESAT-6 responsive cells in baseline BAL, we therefore assessed the ability of these antigens to stimulate in vitro production of these chemokines. As illustrated in Figure 3A, BAL cells of individuals with LTBI produced significantly more Mig (CXCL9) than did those of naïve controls following incubation with PPD, CFP10, and antigen 85B (p=0.003, p=0.027, and 0.007, respectively). No differences were observed in ESAT-6 induced production of Mig, which was minimal in both subject groups (p=0.370). However, these results were not echoed by the findings observed for production of IP-10 (CXCL10), which was produced by BAL cells of LTBI subjects to a greater extent than those of controls in response to PPD (p-0.021), but not in response to CFP10, ESAT-6, or antigen 85B. These chemokine findings thus indicate that CFP10 responsive IFNγ-producing cells in baseline BAL of individuals with LTBI can contribute to recruitment of additional cells to the lung as part of the pulmonary recall response to Mtb via production of CXCL9, but not of CXCL10, whereas ESAT-6 responsive cells did not stimulate production of either chemokine.
Figure 3: Production of IFNγ-inducible chemokines by baseline BAL cells of individuals with LTBI and naïve controls.
Figures display production of CXCL9 (Mig), 2A, and CXCL10 (IP-10), 2B as determined by ELISA of culture supernatants. Again, findings for individual control subjects are indicated by diamonds, whereas circles indicate results for individual LTBI subjects. Dark horizontal lines indicate median values and shaded boxes represent the 25th percentile to 75th percentile ranges of study results. Baseline BAL cells of subjects with LTBI displayed significantly more production of CXCL9 (Mig) than did those of naïve controls in response to in vivo stimulation with PPD, CFP-10, and antigen 85B, but not in response to ESAT-6 (2A). In contrast, production of CXCL10 (IP-10) by BAL cells from subjects with LTBI was higher than that of control subjects only following stimulation with PPD, and not in response to CFP-10, ESAT-6, or antigen 85B (2B).
DISCUSSION:
Identification of a means for definitive diagnosis of active pulmonary TB in smear-negative patients is a longstanding and important goal of TB control programs, but one that has remained elusive (10). To this end, the development of Mtb-specific immunodiagnostics provided a new means to approach the appealing concept that pulmonary TB may be most accurately diagnosed by evaluating samples from the lung. This application of T-cell IFNγ release assays for Mtb-specific antigens to this approach was first described by Jafari and colleagues, and has been re-addressed by them and others in additional publications (5–7). For the most part, these studies have indicated that T cells capable of responding to Mtb-specific antigens are enriched within the lungs of individuals with active tuberculosis, as assessed by comparison of T-cell responses in PBMC and BAL. Our findings, however, suggest that equivalent local enrichment for T cells capable of responding to at least some Mtb antigens is observed in BAL of healthy individuals with LTBI as well. Further, the rapid production of CXCL9 (Mig) by BAL cells from these individuals in response to PPD, antigen 85B, and CFP-10 suggests that localization of memory cells to the lung may serve a protective role in facilitating recruitment of additional Mtb-responsive cells to the lung at the time of re-exposure to the organism.
The finding that PBMC ELISPOT responses of LTBI subjects were not dramatically different from those of Mtb naïve controls at first appears surprising in that CFP-10 and ESAT-6 are both components of the QuantiFERON TB Gold blood test used to confirm the Mtb-infection in the LTBI group. However, unlike the ELISPOT tests in which responses to each antigen are assessed separately, QuantiFERON results examine INFγ responses to a mixture of Mtb-specific antigens that includes TB7.7 (aka Rv2654c, ref. 11) in addition to ESAT-6 and CFP-10. Further, QuantiFERON positivity is based on levels of IFNγ induced by this combination of antigens (as compared to whole blood incubated in the absence of antigen), rather than the frequency of cells that produce IFNγ as determined by ELISPOT. Thus, both the stimuli and the read-outs of these two approaches are quite distinct (12).
An additional unexpected finding was that BAL cells from LTBI subjects showed significant induction of CXCL9 (Mig) but not CXCL10 (IP-10) in response to specific Mtb antigens. As Mig and IP-10 levels were evaluated from the same supernatant samples for each stimulus, differences in the responses of these chemokines cannot be attributed to variability in IFNγ levels. The findings may instead be consistent with prior observations that, whereas phagocyte production of Mig is uniquely induced by IFNγ, IP-10 can be induced by multiple other stimuli as well (13). Thus, these differing results may suggest that a higher IFNγ threshold must be surpassed to stimulate production of CXCL10, or, alternatively, that IFNγ alone is not sufficient for optimal induction of CXCL10 within this system.
The original studies that applied Mtb-specific ELISPOT to evaluation of BAL samples compared individuals in whom BAL cultures ultimately confirmed the diagnosis of pulmonary TB to those of individuals in whom other lung diseases were ultimately diagnosed. These studies utilized the commercially available T-spot. TB kit (Oxford Immunotec, Marlborough MA). Although our studies did not make use of this same product, we did use commercially-available IFNγ ELISPOT kits as well as antigens obtained from national reference laboratories; further, BAL cells were stimulated with the same concentrations of ESAT-6 and CFP-10 (5μg/mL) reported earlier. Accordingly, it seems unlikely that differences in our conclusions from those of prior studies can be attributed to differences in ELISPOT technique alone. Instead, our findings may simply reflect our focus on evaluating these findings in LTBI specifically. In the prior studies, subsets of the “non TB” groups in each case had diagnostic evaluation that could be consistent with LTBI (eg, PPD skin-test positivity); however, comparison of findings from patients with culture-positive TB to those with LTBI was not directly performed. In addition, the detection of Mtb-specific T cells within BAL of patients without active TB could have been further impaired by the fact that positive PPD skin-testing results were not always evaluated further using Mtb-specific IGRAs. Accordingly, positive PPD results could have resulted from prior BCG vaccination in these European and African patient populations, reducing the percentage of “non TB” patients studied who actually had LTBI (5–7). In contrast, a study of immunodiagnosis based on sputum sample IFNγ responses to PPD reported a lack of such responses in BCG vaccinated individuals, and hypothesized that this reflected a lack of localization of the cross-reactive mycobacterial cells to the lung following standard intradermal vaccination (14). Our approach differs from these prior investigations in presenting a comparison of responses of BAL and peripheral blood T cells in healthy nonsmokers in a (non-vaccinated) US population with those of individuals with confirmed LTBI.
The challenges of distinguishing LTBI from active tuberculosis in patients with culture-negative disease have been emphasized by studies involving more systems-biology based approaches to immunodiagnosis. Several studies have investigated the use of possible latency-associated antigens as identified by their upregulation in states of stress designed to simulate immune containment. In peripheral blood, responses to antigens comprising the “dormancy” regulon DosR did not individually distinguish responses of individuals with LTBI from those of patients with active TB. Responses to the panel as a whole, however, indicated that T cells from LTBI subjects recognized more latency-associated antigens with stronger cumulative IFNγ response than did those of individuals with active TB (15). BAL cell responses to a specific DosR-associated protein (Rv2628) were enriched compared to those of peripheral blood in both LTBI subjects and patients with active TB, however (16). Other studies suggest that loss of T-cell polyfunctionality may also be a distinguishing feature of the immune response in active TB as compared to LTBI (17). Another potential variable, whether or not treatment for LTBI affects the resulting immune profile of these individuals, has generally not been addressed. In our subject group, no clear differences were observed between the responses of individuals who had or had not completed a course of INH (not shown); however, it remains possible that such distinctions could be identified with more extensive evaluations involving larger subject cohorts.
Despite the potential of local immune responses to contribute to both the understanding of protective immunity to Mtb and the ability to distinguish between LTBI and active disease, our studies and others suggest that focus on enrichment for IFNγ responses to a limited number of antigens within the lung may remain premature. Historically, use of more comprehensive approaches to defining pulmonary immune responses in human subjects has been restricted by the low numbers of lymphocytes obtainable using BAL; perhaps new approaches to defining T-cell epitopes using limited numbers of cells (18) may now provide a means to facilitate greater progress in these studies.
MATERIALS AND METHODS
Subjects:
Eligibility for research bronchoscopy was based on age 18–50, non-smoking status, and lack of other significant medical problems including asthma or other chronic respiratory disease, cardiac disease, or ongoing use of systemic immunosuppressive agents for any reason. LTBI subjects were self-identified on the basis of prior positive tuberculosis skin test (ranging from 1 to 10 years prior to their participation) and confirmed by either repeat skin testing or blood testing using QFN-GIT. For the purposes of this study, skin-test responses of 10 mm or more of induration were considered positive. One subject who had received BCG vaccination was confirmed to have LTBI by QFN-GIT testing. No LTBI subjects had a prior history of active TB or of any symptoms suggestive of current disease, such as cough, night sweats, fevers, or weight loss. All underwent chest-rays that showed no evidence for active TB. In addition, BAL fluid obtained during bronchoscopy procedures of each LTBI subject was sent for culture; as detailed below, no Mtb growth was observed for any of these samples. Six of the subjects had completed INH therapy, three had not, and one (who was first noted to have a positive skin test as a young child) was uncertain if she had ever received treatment. The status of control subjects was also confirmed by negative responses to either PPD skin testing or QFN-GIT blood testing.
All protocols involving human subjects were approved by the Institutional Board of Review of Case Western Reserve University and University Hospitals of Cleveland and of the Louis Stokes Cleveland Department of Veterans’ Affairs Medical Center.
Collection and processing of BAL cells:
All bronchoscopies were performed in the Dahms Clinical Research Unit (DCRU) at University Hospitals Case Medical Center as previously described (9). BAL samples were obtained by instillation and subsequent withdrawal of up to eight 30 mL aliquots of pre-warmed buffered saline. Recovered BAL fluid was placed on ice for transport to the laboratory. Samples were aliquoted into 50 ml polypropylene tubes and immediately centrifuged at 2000 RPM (480 × g) for 10 minutes. BAL cells were stained and counted using a hemocytometer. Cytospin preparations were made using approximately 25–50,000 cells from each BAL sample and cell differentials determined by light microscopy as detailed previously.
BAL fluid cultures for M. tuberculosis
BAL fluid from all LTBI subjects was stored at −80°C and ultimately sent for culture in the clinical microbiology laboratory of University Hospitals Case Medical Center. As per standard clinical practice, 500μL of BAL fluid was inoculated into MGITs (Mycobacterial Growth Indicator Tubes, Becton Dickinson Co, Sparks, MD) that contain 7 mL of modified Middlebrook 7H9 broth to which is added a 800μL of OADC/PANTA, a reconstituted mixture of PANTA (lyophilized antibiotics polymyxin B, amphotericin B, naldixic acid, trimethoprim, azlocillin) and OADC supplement (oleic acid, bovine albumin, dextrose, catalase). Culture bottles were incubated continuously at 37°C in a growth-detecting incubator (MGIT 960, Becton Dickinson) with protocol length of 42 days for final interpretation of “no growth”.
Isolation of peripheral blood mononuclear cells (PBMC):
Peripheral blood was obtained by venipuncture and mononuclear cells isolated by density sedimentation using Ficoll-Hypaque (Ficoll-Paque PLUS, Amersham Bioscience AB, Uppsala, Sweden) as previously described (8). PBMC were collected from the Ficoll-serum interface using transfer pipettes, washed three times in RPMI 1640 (BioWhittaker) and counted using a hemocytometer.
Antigens:
Sterile commercially-prepared PPD (Tubersol®, Aventis Pasteur, Toronto) was used in skin-testing in standard intradermal dosing of 0.1 mL (5 tuberculin units). PPD for in vitro use was obtained from the Staten Serum Institut (Copenhagen, Denmark). CFP-10, ESAT-6, and Antigen 85B were purchased from BEIresources. Staphylococcal enterotoxin B (SEB, Sigma) was used as a positive control for IFNγ production.
IFNγ ELISPOT:
IFNγ ELISPOT was performed using a commercially available kit (BD bioscience-#551849) as detailed previously (7). Following coating of 96-well Unifilter plates with anti-human IFNγ, BAL cells and PBMC were plated at densities of 2.5 × 105, 1.25 × 105 and 0.65 × 105 per well in triplicate and incubated 37°C overnight in the presence of medium alone, PPD, CFP-10, ESAT-6, and antigen 85B (each at concentration 5 μg/ml) and the positive control SEB (1μg/ml). The next day, plates were washed prior to incubation with biotin-conjugated anti-IFNγ for 2 hours at room temperature. Following washing, Strepavidin-AP was added to the plate for 1 hour at room temperature. The plate was washed and final substrate solution (AEC/0.1M acetate solution/H2O2) was added prior to incubation in the dark for 5–60 minutes at room temperature. The plates were then rinsed with deionized water and allowed to dry. Plates were scanned with CTL Technologies ImmunoSpot Plate Scanning Services (CTL Technology, Ltd., Cleveland, OH), and images analyzed using ImmunoSpot software (CTL Technology). Results were expressed as the number of cells displaying IFNγ production in response to each antigen per 200,000 total cells in culture.
Assessment of in vitro chemokine production:
Unsorted BAL cells and PBMC were resuspended in IMDM with 2% FCS and 1% penicillin G at concentration 400K/ml, and aliquoted into 24-well plates (1 ml/well). Cultures were incubated at 37°C for 24 hours with PPD (10μg/ml), CFP-10 (10μg/ml), ESAT-6 (10μg/ml), Antigen 85B (10μg/ml), SEB (1μg/ml), or medium alone. Culture supernatants were collected and chemokine concentrations measured using commercially available ELISA kits, MIG (CXCL-10) and IP-10 (CXCL-9) (R and D Systems).
Statistical analysis:
Statistical analysis were performed using the Mann-Whitney test for non-parametric data with GraphPad Prism 5.0c software (GraphPad, LaJolla, CA). Observed p values of </= 0.05 were considered statistically significant.
ACKNOWLEDGEMENTS:
These studies were funded by the Merit Review program of the United States Department of Veterans’ Affairs Office of Research and Development and by NIH RO1-HL111523 (to RFS). Research bronchoscopies were performed in the Dahms Clinical Research Unit (DCRU), which is a clinical core of the Case Western Reserve University/University Hospitals Case Medical Center Clinical and Translational Science Collaborative, supported by NIH UL1 RR024989 from the National Center for Research Resources.
REFERENCES:
- 1.Van Pinxteren LAH, Ravn P, Agger EM, Pollock J and Andersen P. Diagnosis of tuberculosis based on the two specific antigens ESAT-6 and EFP10. 2000. Clin and Diag Lab Immunol, 7: 155–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berthet FX, Rasmussen PB, Rosenkrands I, Andersen P, Gicquel B. A Mycobacterium tuberculosis operon encoding ESAT-6 and a novel low molecular mass culture filtrate protein (CFP-10). 1998, Microbiology. 1144: 3195–203. [DOI] [PubMed] [Google Scholar]
- 3.Behr MA, Wilson MA, Gill WP, Salamon H, Schoolnik GK, Rane S and Small PM. Comparative genomics of BCG vaccines by whole-genome DNA microarray. 1999. Science, 284: 1520–1523. [DOI] [PubMed] [Google Scholar]
- 4.Moon H-W and Hur M, Interferon-gamma release assays for the diagnosis of latent tuberculosis infection: an updated review. 2013. Ann Clin and Lab Sci, 43: 221–229. [PubMed] [Google Scholar]
- 5.Jafari C, Ernst M, Strassburg A, Greinert U, Kalsdorf B, Kirsten D and Lange C. Local immunodiagnosis of pulmonary tuberculosis by enzyme-lnked immunospot. 2008, Eur Resp J, 31: 261–265. [DOI] [PubMed] [Google Scholar]
- 6.Jafari C, Thijsen S, Sotgiu G, Goletti D, Dominguez Benitez JA, Losi M, Eberhardt R, Kirsten D, Kalsdorf B, Bossink A, Latorre I, Migliori GB, Strassburg A, Winteroll S, Greinert U, Richeldi L, Ernst M, and Lange C. Bronchoalveolar lavage enzyme-linked immunospot for a rapid diagnosis of tuberculosis. Am J Resp Crit Care Med, 2009. 180: 666–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dheda K, van Zyl-Smit RN, Meldau R, Meldau S, Symons G, Khalfey H, Govender N, Rosu V, Sechi LA, Maredza A, Semple P, Whitelaw A, Wainwright H, Madri M, Dawson R, Bateman ED, and Zumla. Quantitative lung T cell responses aid the rapid diagnosis of pulmonary tuberculosis. 2009. Thorax, 64: 847–853. [DOI] [PubMed] [Google Scholar]
- 8.Walrath J, Zukowski L, Krywiak A, and Silver RF. Resident Th1-like effector-memory cells in pulmonary recall responses to Mycobacterium tuberculosis. Am. J. Resp. Cell. and Mol. Biol, 2005, 33: 48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silver RF, Li Q, Zukowski L, Kotake S, Pozuelo F, Krywiak A, and Larkin R. Recruitment of antigen-specific Th1-like responses to the human lung following segmental antigen challenge with Purified Protein Derivative of M. tuberculosis. Am. J. Resp. Cell. and Mol. Biol, 2003, 29: 117–123. [DOI] [PubMed] [Google Scholar]
- 10.World Health Organization. Global tuberculosis report, 2014 [Internet]. Geneva: World Health Organization; 2014. Accessed April 2015 at: http://apps.who.int/iris/bitstream/10665/137094/1/9789241564809_eng.pdf?ua=1. [Google Scholar]
- 11.Aagaard C, Brock I, Oslon A, Ottenhoff THM, Weldingh K, Andersen P. Mapping immune reactivity toward Rv2653 and Rv2654: tow novel low-molecular-mass antigens found specifically in the Mycobacterium tuberculosis complex. 2004. J. Infect. Dis, 189: 812–819. [DOI] [PubMed] [Google Scholar]
- 12.Mazurek GH, Jereb J, Vernon A, LoBue P, Goldberg S, Castro K; IGRA Expert Committee; Centers for Disease Control and Prevention (CDC). Updated guidelines for using interferon gamma release assays to detect Mycobacterium tuberculosis infection—United States 2010, 2010. MMWR Recomm Rep, 59:1–25. [PubMed] [Google Scholar]
- 13.Farber JM. Mig and IP-10: CXC chemokines that target lymphocytes, 1997. J. Leuk. Biol 61: 246–257. [PubMed] [Google Scholar]
- 14.Breen RAM, Hardy GAD, Perrin FMR, Leaer S, Kinloch S, Smith CJ, Cropley I, Jannossy G, and Lipman MCI. Rapid diagnosis of smear-negative tuberculosis using immunology and microbiology with induced sputum in HIV-infected and uninfected individuals, 2007. PLoS one, 12: E1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leyten EML, Lin MY, Franken LMC, Friggen AH, Prins C, van Meijgaarden KE, Voskuil MI, Weldingh K, Andersen P, Schoolnik GK, Arend AM, Ottenhoff THM, and Klein MR. Human T-cell response to 25 novel antigens encoded by genes of the dormancy regulon of Mycobacterium tuberculosis. 2006. Microbes and Infection, 8: 2052–2060. [DOI] [PubMed] [Google Scholar]
- 16.Chiacchio T, Petruccioli E, Vanini V, Butera O, Cuzzi G, Petrone L, Matteucci G, Lauria FN, Franken KL, Girardi E, Ottenhoff TH, Goletti D. Higher frequency of T-cell response to M. tuberculosis latency antigen Rv2628 at the site of active tuberculosis disease than in peripheral blood. 2011. PLoS One, 6: E27539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Day CL, Abrahams DA, Lerumo L, Janse van Rensburg E, Stone L, O’rie T, Pienaar B, Kaplan G, Mahomed H, Dheda K and Hanekom WA. 2011Functional capacity of Mycobacterium tuberculosis-specific T cell responses in humans is associated with mycobacterial load. J. Immunol, 187: 2222–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li D, Gao G, Li Z, Sun W, Li X, Chen N, Sun J Yang Y. Profiling the T-cell receptor repertoire of a patient with pleural tuberculosis by high throughput sequencing. 2014. Immunol. Letters, 162: 170–180. [DOI] [PubMed] [Google Scholar]