Abstract
PURPOSE
A circulating tumor DNA (ctDNA) test to detect plasma Epstein-Barr viral DNA can be used to screen for early nasopharyngeal cancers; however, the reported sensitivity of viral ctDNA tests to detect human papillomavirus (HPV)–associated cancers is modest. We assessed the utility of droplet digital polymerase chain reaction (ddPCR) to detect early-stage HPV-associated cancers using sequential HPV16 and HPV33 assays that account for HPV subtype distribution and subtype sequence variants.
PATIENTS AND METHODS
We collected plasma specimens from 97 HPV-positive patients with oropharyngeal squamous cell carcinoma and eight patients with HPV-positive anal squamous cell carcinoma, each with locoregionally confined disease. Negative controls included samples from seven patients with HPV-negative head and neck cancers and 20 individuals without cancer.
RESULTS
Of 97 patients with nonmetastatic, locoregionally confined oropharyngeal squamous cell carcinoma, 90 patients had detectable HPV16 ctDNA and three patients had HPV33 ctDNA, indicating an overall sensitivity of 95.6%. Seven of eight patients with early anal cancer were HPV16 ctDNA positive. No HPV ctDNA was detected in 27 negative controls, indicating 100% specificity. HPV16 ctDNA was detected in 19 of 19 patients with low-volume disease, defined as patients with a single, asymptomatic positive lymph node (N1) or an isolated T1-2 asymptomatic primary tumor. HPV16 ctDNA levels directly corresponded to tumor responses to chemoradiation and surgery.
CONCLUSION
With an updated understanding of HPV subtypes and sequence variation, HPV ctDNA by ddPCR is highly sensitive and specific, identifying HPV16 and HPV33 subtypes in a similar distribution as reported in major genomic profiling studies. The detection of small tumors indicates that HPV16 and HPV33 ctDNA ddPCR could be readily used in early detection screening trials and in disease response monitoring, analogous to Epstein-Barr virus DNA.
INTRODUCTION
Detection of tumor-derived plasma Epstein-Barr virus DNA has been successfully used in a primary screening study for nasopharyngeal cancer (NPC),1 which demonstrated that NPCs could be detected in asymptomatic patients with an earlier disease stage distribution than in the unscreened population. Viral circulating tumor DNA (ctDNA) as a substrate affords unique technical advantages over single nucleotide variant detection in liquid biopsy assays because of the size of the viral genome and differentness from the human genome and because of the multiple copies of viral genome per tumor genome. These features elevate the sensitivity of detection of viral tumor DNA, although it is unknown whether viral-associated malignancies other than NPC could be similarly detected by liquid biopsy.
Cancers etiologically driven by human papillomaviruses (HPVs) include squamous cell carcinomas of the oropharynx, cervix, vulva, vagina, anal canal, and penis. Pelvic exams and Papanicolaou smears are widely adopted screening tools for early detection of early HPV-associated lesions in the cervix; however, effective screening approaches for other disease sites are lacking. Among these, HPV-associated oropharyngeal squamous cell carcinomas (OPSCCs) constitute an emerging epidemic with rapidly increasing incidence.2 However, HPV plasma ctDNA studies in head and neck cancer using low-cost polymerase chain reaction (PCR)–based methods to date have shown only modest sensitivity in patients with gross disease (19% to 79%),3-8 and in some reports, only a combination of saliva and plasma tests is recommended to boost sensitivity.
CONTEXT
The human papillomavirus (HPV) is a causative agent in cancers of the oropharynx, cervix, vulva, vagina, anal canal, and penis. Tumor-derived HPV DNA can be detected in the plasma, but the sensitivity of detection in most reports has been modest, and its optimal use as a biomarker for screening and treatment monitoring of these malignancies is uncharacterized. Here, we used droplet digital polymerase chain reaction technology to detect the most common HPV strains in HPV-associated oropharyngeal cancer, HPV types 16 and 33, and achieved near-universal detection of HPV in circulating tumor DNA derived from the plasma samples of these patients. We also demonstrated a clear relationship between HPV copies in plasma and tumor volume as tumors respond to chemoradiation. Droplet digital polymerase chain reaction of HPV types 16 and 33 was sensitive enough to detect patients with low disease burden, such as an isolated neck node or T1-2 primary tumors. These data suggest that this technology could be well suited for screening and disease monitoring.
In the study, we sought to determine the clinical utility of HPV ctDNA by measuring the sensitivity and specificity of the most sensitive ctDNA technology available, droplet digital PCR (ddPCR),9 in patients with macroscopic gross disease and in various clinical settings after initial presentation. We designed a ddPCR amplicon that makes use of an updated understanding of HPV subtype distribution and HPV subtype sequence variants identified through recent tumor genomic profiling studies. To determine the utility of ddPCR in a screening setting, we examine 19 patients in low-volume disease states that are analogous to desired subclinical, screen-detected settings. These include patients in whom a surgeon has already removed the primary tumor in the oropharynx with transoral robotic surgery, leaving only a single, asymptomatic lymph node in the neck (N1 disease). We then examine the specificity of HPV ctDNA by ddPCR to tumor as opposed to HPV infection of normal tissue by examining the response of HPV ctDNA signal to tumor-directed chemoradiation.
PATIENTS AND METHODS
This study was conducted at Memorial Sloan Kettering Cancer Center using banked plasma samples from patients from our institution who consented to institutional review board–approved biospecimen protocols. Clinical information was retrieved from the medical record under authorization through an institutional review board–approved retrospective research protocol. The clinical characteristics of patients in the series are similar to those of previously published cohorts from our institution,10-12 with stage IVA, T2, and N2b disease as the most common American Joint Committee on Cancer seventh edition cancer stage, T stage, and N stage, respectively (Table 1).
TABLE 1.
Patient Characteristics
Study Patients
Plasma specimens from 97 HPV-positive patients with OPSCC with locoregionally confined disease and eight HPV-positive patients with anal squamous cell carcinoma were accrued (Table 1). HPV positivity was defined as p16 overexpression by immunohistochemistry with greater than 70% diffuse nuclear or cytoplasmic staining (n = 95) or a clinically reported positive DNA- or RNA-based in situ hybridization test for HPV (n = 53).13 Samples from seven patients with HPV-negative head and neck cancers, eight patients with HPV-positive OPSCCs who had already undergone definitive surgery, and 20 patients without cancer were included as negative controls. Information regarding demographics, clinicopathologic features, and outcomes was obtained from the medical record and is presented in Table 1. Gross tumor volume was determined from the used radiation plans for which the treating radiation oncologist contoured gross disease and the plan was technically accessible (n = 84).
Sample Collection and Preparation
Ten milliliters of whole blood were collected from each patient into cell-free DNA (cfDNA) BCT tubes (Streck, La Vista, NE) or BD Vacutainer K2 EDTA tubes (BD Biosciences, San Jose, CA). Plasma was separated first though centrifugation at 800 × g for 10 minutes, followed by an additional centrifugation at 16,500 × g for 10 minutes. When samples were collected in BD Vacutainer K2 EDTA tubes, plasma was separated within 1 hour of blood collection and stored at −80°C in Eppendorf LoBind tubes (Eppendorf, Hamburg, Germany). When samples were collected into cfDNA BCT tubes, plasma was separated and stored identically at −80°C within 48 hours. At a later date, samples were thawed and cfDNA was extracted from 4 to 5 mL of plasma using Qiagen circulating nucleic acid kits (Qiagen, Germantown, MD) into a 50-μL final elution volume. Pretreatment plasma samples were collected a median of 2.4 weeks before the start of chemoradiation.
ctDNA Analysis
Primers and probes for ddPCR assays and quantitative PCR assay are listed in Appendix Table A1. HPV16 and HPV33 ddPCR assays were designed using Primer3Plus and ordered through Bio-Rad (Hercules, CA). All reactions were performed on a QX200 ddPCR system (Bio-Rad) in technical duplicates. EIF2C1 copy number variation primers and probes were used as internal controls for each ddPCR reaction (Bio-Rad). Reactions were partitioned into a median of approximately 15,000 droplets per well using the QX200 droplet generator. Emulsified reactions were amplified on a 96-well thermal cycler using cycling conditions identified during the optimization step. Plates were read and analyzed using QuantaSoft software (Bio-Rad) to assess the number of droplets positive for the target gene, reference gene, both, or neither. The assay threshold sensitivity was set at two mutant droplets.
Pathologic Data
In a subset of patients, targeted next-generation sequencing (NGS) data of pathologic samples were available, obtained through the US Food and Drug Administration–approved MSK-IMPACT (Memorial Sloan Kettering Cancer Center, New York, NY) sequencing platform.14 To evaluate HPV16 copy number in 10 patients, we extracted genomic DNA from formalin-fixed paraffin-embedded–preserved pathologic samples. One corresponding hematoxylin and eosin–stained slide was reviewed by a pathologist (N.K.) and used as a template to scrape, collect, and extract genomic DNA from regions of other unstained slides with greater than 75% tumor cellularity. Genomic DNA was then used for HPV16 ddPCR, and the HPV16 copy number was considered as the ratio of HPV16-positive droplets to droplets containing the reference gene EIF2C1.
Statistical Analysis
Sensitivity was defined as the number of pretreatment cfDNA samples that were positive for HPV ctDNA divided by the total number of patients (N = 97). Specificity was defined as the number of negative control samples that had zero HPV-positive droplets divided by the total number of negative controls (N = 27). To evaluate for any correlation between gross tumor volume or copy number with the initial ctDNA levels, Spearman correlation coefficients were calculated using Prism software by GraphPad (San Diego, CA). For comparisons between groups, nonparametric Mann-Whitney U tests of significance were calculated using GraphPad Prism software.
RESULTS
Assay Design
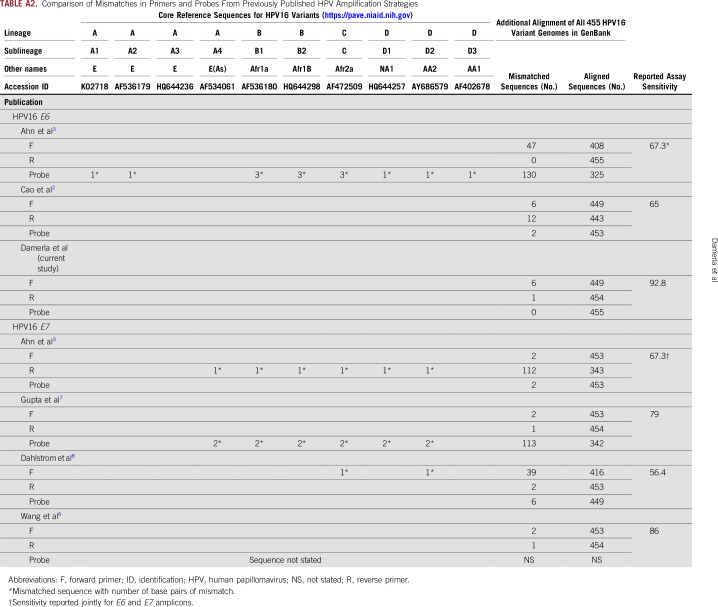
We first sought to develop the best possible primer-probe set for ddPCR using the following three criteria: location within the E6 or E7 oncogenes because these sequences are the most highly amplified sequences in tumor genomes15; smaller amplicon size, to increase probability of amplification in view of the highly fragmented nature of cfDNA; and primers or probes that perfectly match both European and non-European HPV16 isolates, maximizing universality of the test (Appendix Fig A1). Briefly, we used the Papillomavirus Episteme resource (pave.niaid.nih.gov)16 and identified the 10 sequences representative of HPV16 sublineages (A1 to A4, B1 and B2, C1, and D1 to D3), spanning European, Asian, and African isolates of HPV16 (Appendix Fig A1 and Appendix Table A2). We chose primers and probes within E6 and E7 that match these 10 most common sequences exactly and then subsequently compared these sequences with an additional 455 HPV16 isolates found in GenBank. A primer-probe set in E6 with a length of 97 base pairs met all criteria and demonstrated high PCR efficiency and sensitivity down to a single molecule of template HPV16 DNA in a ddPCR assay (Appendix Fig A2). We then compared the performance of ddPCR to two additional HPV DNA detection platforms, the Roche Cobas HPV Test (Roche, Indianapolis, IN) and quantitative PCR, and found 15.9- and 5.2-fold higher ctDNA signals with HPV16 ddPCR compared with these two platforms, respectively (Appendix Fig A2).
Sensitivity and Specificity of HPV16 and HPV33 ddPCR
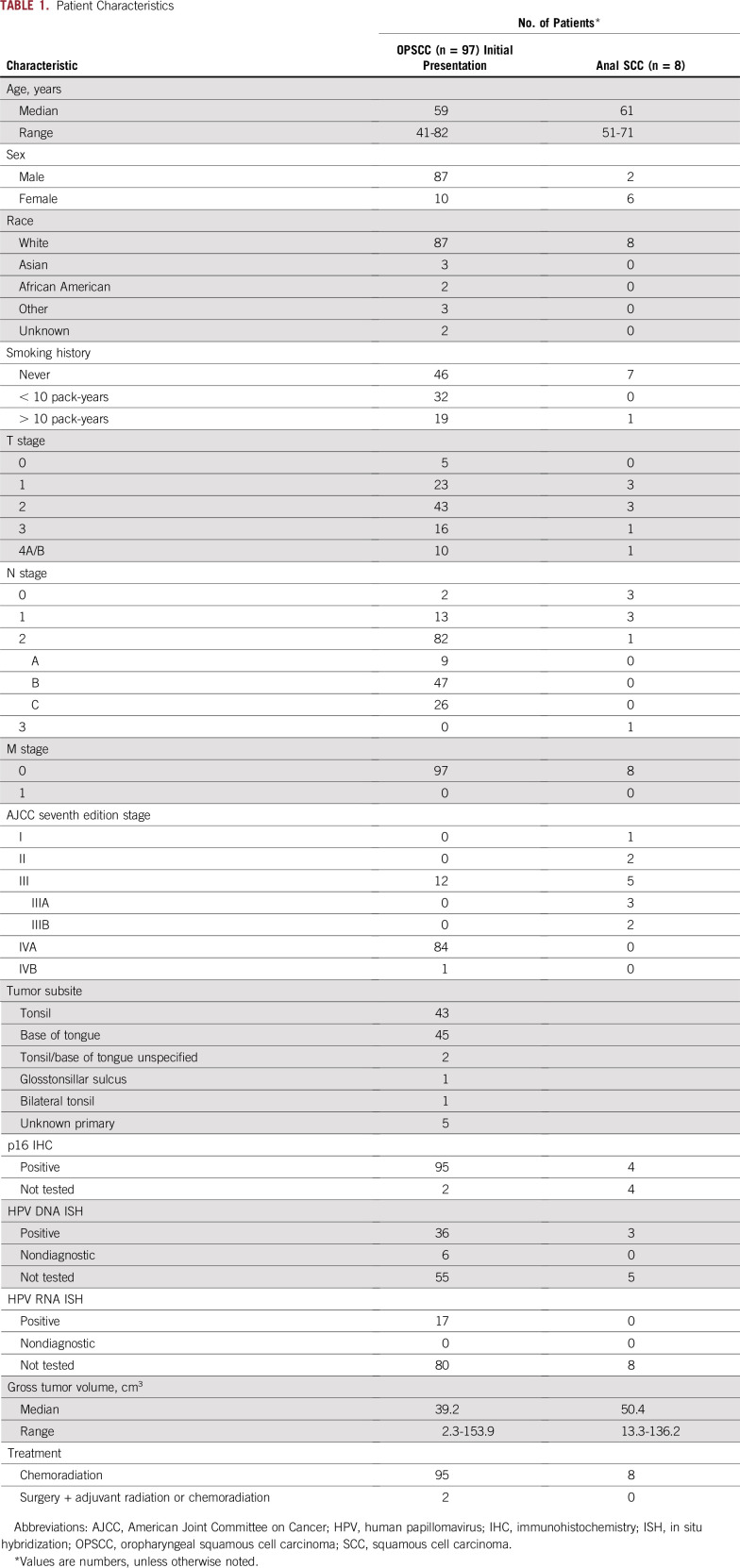
Of 97 patients with stage I to IVB HPV-positive OPSCC with pretreatment cfDNA, HPV16 ctDNA was detected in 90 patients, demonstrating a sensitivity of 92.8%. We also tested 20 samples from normal individuals without cancer and seven samples from patients with HPV-negative head and neck cancers, and no droplets were detected in these 27 samples, demonstrating 100% specificity. In four patients, the patient’s tumor biopsy sample had been used to specifically identify the HPV subtype. In two of those four patients, the tumor pathology was HPV16 negative, and in both patients, no HPV16 ctDNA was detected (Fig 1A, teal). In the two other patients, the tumor pathology was HPV16 positive, and in both of these patients, HPV16 ctDNA was readily detected in plasma (Fig 1A, red), demonstrating 100% concordance (four of four patients) between plasma and the tumor specimen in HPV subtyping. As an additional control, we collected plasma from eight patients who had already undergone definitive surgery, and HPV was detected in none of these patients, although in two available matched samples, HPV16 ctDNA was readily detected before surgery (Fig 1A, dark orange).
FIG 1.
Sensitivity and specificity of human papillomavirus (HPV) types 16 and 33 droplet digital polymerase chain reaction (ddPCR) assays. (A) HPV16 circulating tumor DNA (ctDNA) detected by the HPV16 ddPCR assay in samples obtained from normal individuals (n = 20) and patients with HPV-negative head and neck squamous cell carcinomas (HNSCCs; n = 7), HPV-associated oropharyngeal squamous cell carcinomas (OPSCCs; n = 97), and anal squamous cell carcinomas (n = 8). Red, teal, and blue diamonds represent HPV16 pathologic status as positive, negative, and undetermined, respectively. (B) HPV33 ddPCR assay was used for detecting HPV33 DNA in seven patient cell-free DNA samples that did not have detectable HPV16 ctDNA. (C) Combination of both HPV16 and HPV33 ddPCR assays to determine overall sensitivity of HPV detection in pretreatment samples from patients with HPV-associated OPSCC.
The mean HPV16 ctDNA level was 1,218 copies/mL (range, 0 to 13,163 copies/mL), with no clear cut points for classifications into groups. We also tested eight plasma samples from patients with anal cancers and detected HPV16 ctDNA with a mean of 2,151 copies/mL in seven samples, demonstrating that the ddPCR assay can be used in other HPV-positive malignancies (Fig 1A). We then developed a ddPCR assay for HPV33, which is the next most common HPV type associated with OPSCC.17 We observed no HPV33-positive droplets in 20 samples from normal individuals without cancer. We then tested the seven cfDNA samples that were negative for HPV16 ctDNA and found that three of these samples were HPV33 positive at 8.1, 13.6, and 10.0 copies/mL (Fig 1B). Thus, HPV16 or HPV33 ctDNA was detectable in 93 of 97 patients, with four samples remaining undetectable (overall sensitivity, 95.9%; Fig 1C). After these results, additional pathologic specimens in two of the four HPV16 and HPV33 ctDNA-negative patients were genotyped by RNA in situ hybridization and found to be negative for HPV16, suggesting other high-risk HPV subtypes. The four negative patients ranged in regard to tumor size and stage and had no distinguishing clinical characteristics (Fig 2B).
FIG 2.
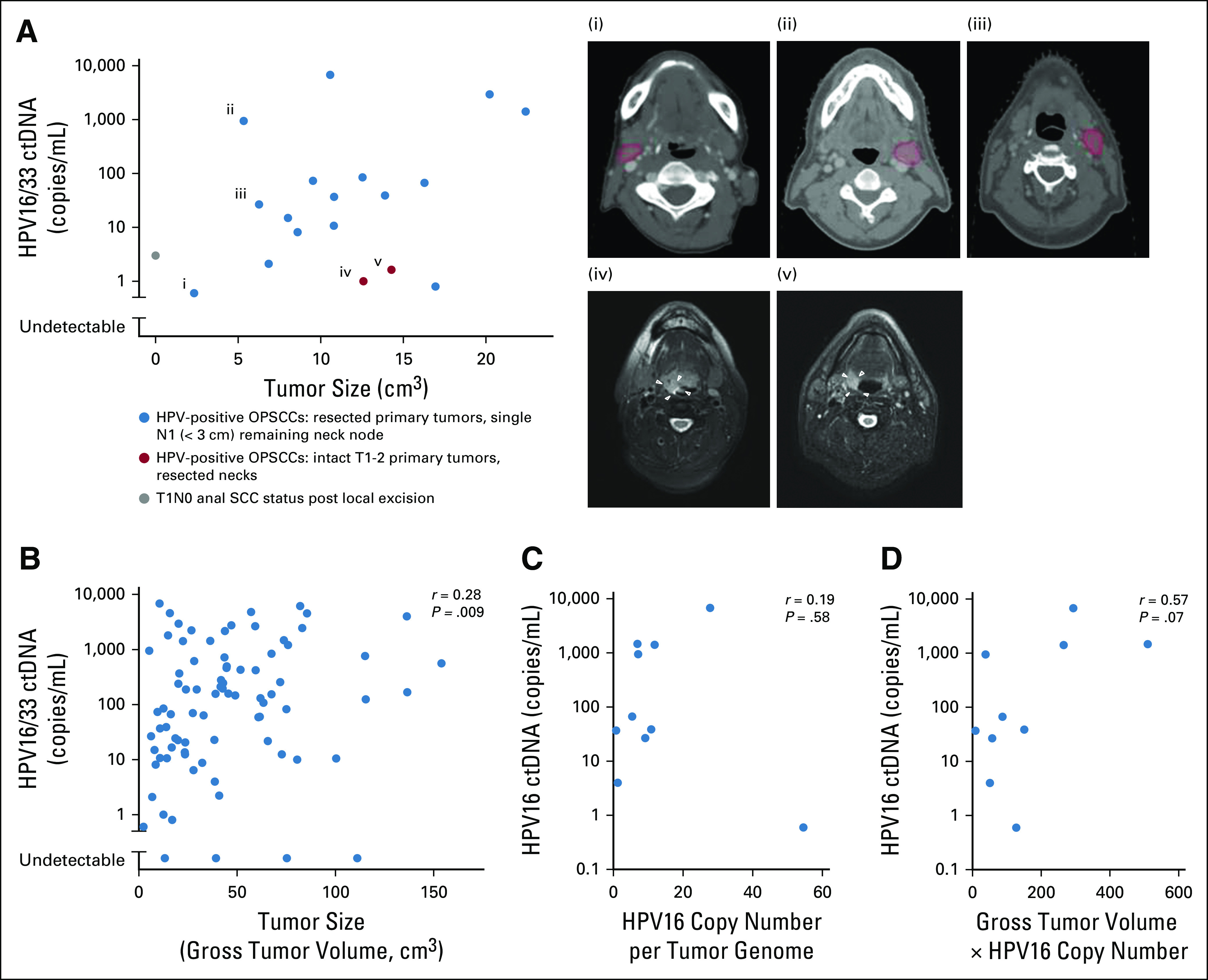
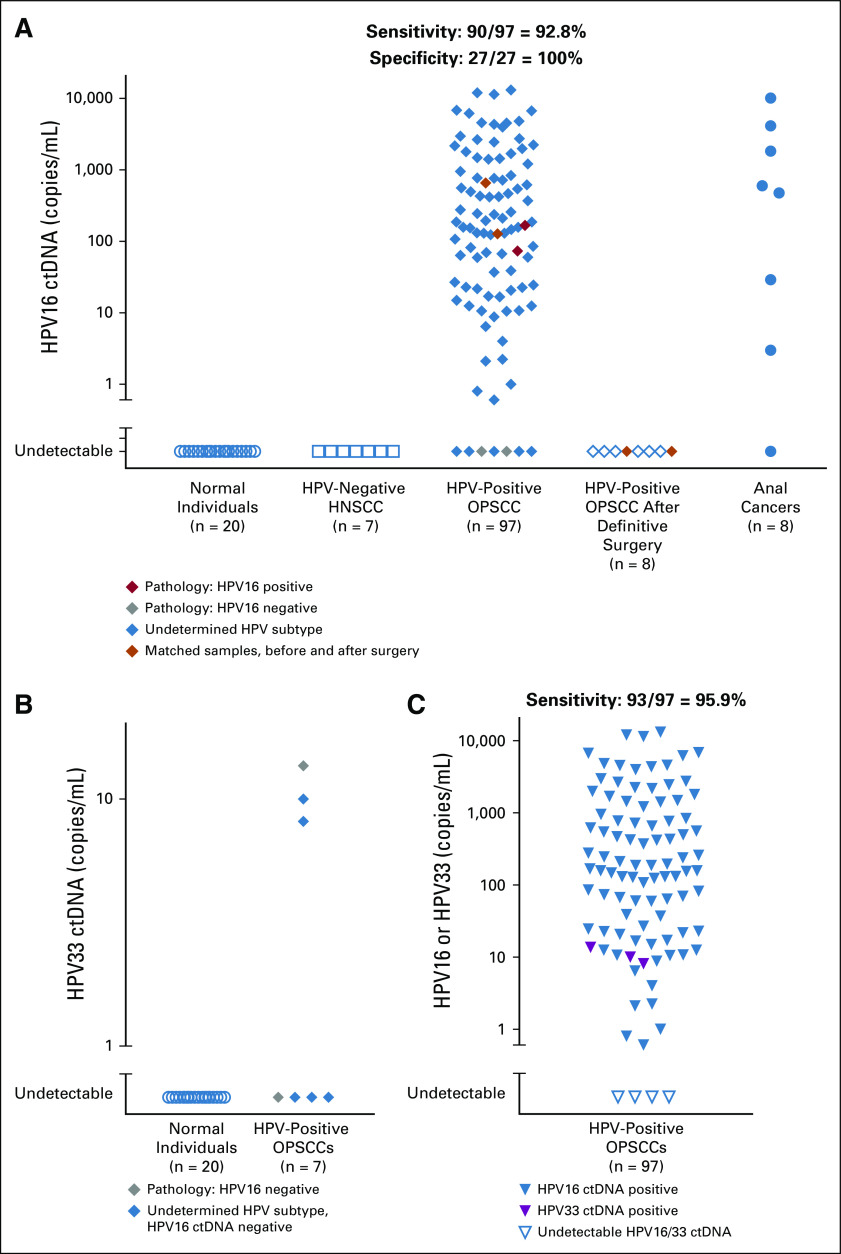
Factors influencing human papillomavirus (HPV) circulating tumor DNA (ctDNA) levels. (A) Analysis of HPV16 and HPV33 ctDNA levels in patients with low tumor burden. Panels i to iii indicate remaining lymph nodes (N1) delineated in red. White arrows in panels iv and v indicate low-volume T1 and T2 primary tumors, respectively. (B) Relationship between HPV16 and HPV33 ctDNA in pretreatment samples and gross tumor volume. (C) Correlation between pretreatment levels of HPV measured by HPV16 droplet digital polymerase chain reaction (ddPCR) and HPV copy number per tumor genome. (D) Correlation between pretreatment levels of HPV measured by HPV16 ddPCR and product of HPV copy number and gross tumor volume. OPSCC, oropharyngeal squamous cell carcinoma; SCC, squamous cell carcinoma.
HPV ctDNA Detection in Patients With Low Disease Burden
The purpose of liquid biopsy in a screening setting would be to identify subclinical tumors at early disease stages. We determined HPV ctDNA levels in 19 patients with low-volume tumor burden, and HPV16 ctDNA was readily detectable in all 19 patients by ddPCR (Fig 2A). In 16 patients with HPV-positive OPSCC, the primary tumor site in the base of tongue and tonsil had been removed with definitive transoral robotic surgery, and thus, there was no remaining gross disease in the oropharynx at the time of plasma collection. In each of these 16 patients, the only remaining gross tumor was a single, asymptomatic lymph node (N1 disease). In three patients, the remaining single lymph node is delineated in red on computed tomography images in Figure 2A. In two additional patients, a neck dissection with negative margins had been performed to remove all gross disease, leaving only T1-2 primary tumors in the oropharynx. In these two patients, low-volume T1 base of tongue and T2 tonsillar primary tumors were imperceptible to the patient, and all gross disease in the neck had been removed before plasma collection. In one patient with anal squamous cell carcinoma, a T1N0 tumor had been removed with a local excision, leaving no gross disease before plasma was collected, although the surgical margin was less than 1 mm.
Although low-volume disease was readily detectable, we observed that, across patients, the volume of gross disease was only minimally related to initial HPV16 ctDNA levels overall (Fig 2B; r = 0.28, P = .009). Because the number of HPV genomic copies per tumor genome can be widely variable,17 we determined the HPV copy number directly in 10 tumor pathologic samples, which ranged from 0.8 to 57 copies per tumor genome (Fig 2C). We then asked whether the HPV16 copy number correlates with HPV16 ctDNA level as a single covariable and again found little correlation (Fig 2C). Because tumor volume is proportional to the number of cancer cells and the HPV16 copy number per cancer cell was known in 10 patients, we postulated that the product of gross tumor volume and HPV16 copy number would be proportional to HPV16 ctDNA and found a modest, nonsignificant correlation (r = 0.57, P = .07), indicating that other unspecified factors are also important (Fig 2D).
Specificity of HPV ctDNA Signal to Tumor-Directed Therapy
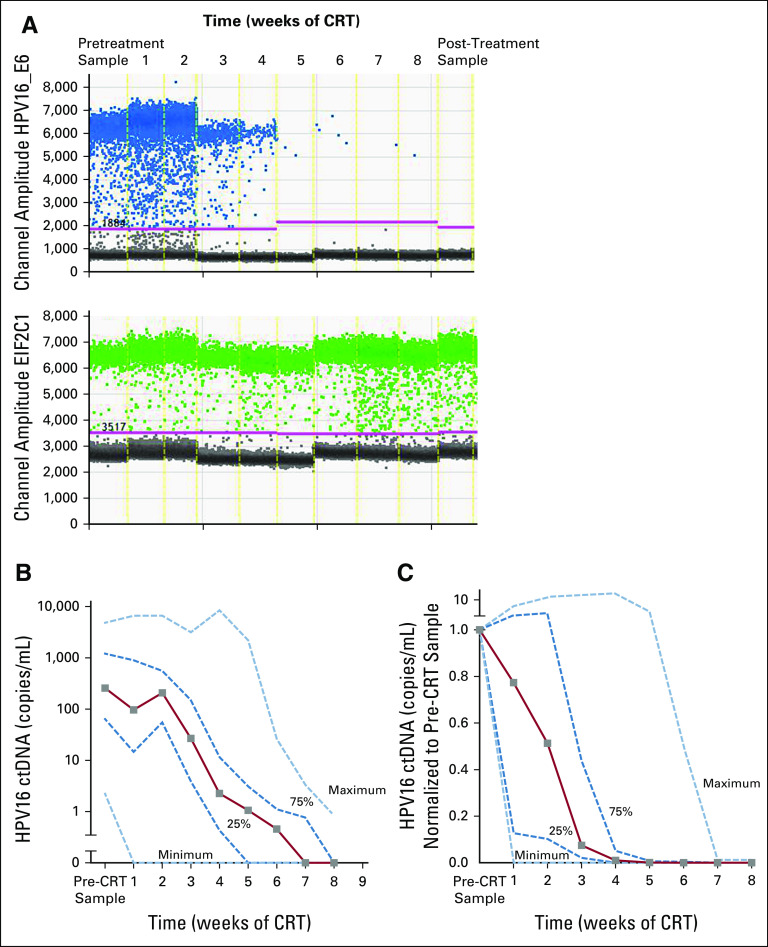
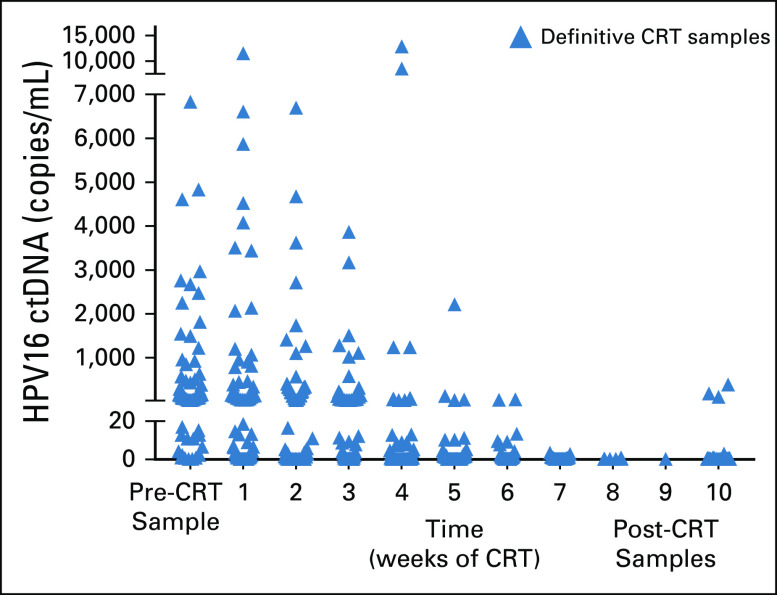
A major potential challenge of HPV screening for early cancers is the specificity of the test because the natural prevalence of oral, penile, cervical, and anal HPV16 infection is 1% to 4% in the general population.18-21 It is unknown whether or how often an HPV infection without cancer results in detectable plasma HPV DNA. We demonstrated the specificity of the test to tumor HPV DNA by examining the responsiveness of the signal to tumor-directed therapies. In a subset of patients with HPV-positive OPSCC, we obtained samples every week during and after definitive chemoradiation, and in all patients, we observed rapid declines. Figure 3A shows the raw data of one patient during the course of radiotherapy, and the number of droplets positive for HPV16 (blue) decreases rapidly, whereas the number of droplets positive for a reference gene (EIF2C1) remains unchanged. In 68 patients with multiple samples, HPV16 ctDNA was generally cleared by week 7 after the start of chemoradiation, with the exception of three patients in whom HPV16 ctDNA levels remained detectable at 10 weeks (Appendix Fig A3). In 26 patients in whom a complete set of samples were available every week, we found a high degree of heterogeneity in response kinetics but universal responses to chemoradiation (Figs 3B and 3C).
FIG 3.
Human papillomavirus (HPV) droplet digital polymerase chain reaction (ddPCR) assay in monitoring treatment response to chemoradiation (CRT). (A) Amplitude plots of the ddPCR raw data are shown for CRT per week of one patient with oropharyngeal squamous cell carcinoma, starting from pretreatment sample on the left, samples collected per week through 8 weeks of CRT, and post-treatment sample on the right. (B) Graph representing median HPV copies per milliliter at each treatment time point from B along with interquartile range (dark blue dashed lines) and minimum and maximum values (light blue dashed lines) in patients with complete sets of plasma samples (28 patients) every week. (C) Median HPV copies per milliliter measures in patient samples after every week of CRT are represented as a fraction of the same-patient pretreatment sample (28 patients). Light blue dashed lines indicate minimum and maximum values, and dark blue dashed lines show the interquartile range. ctDNA, circulating tumor DNA.
DISCUSSION
HPV-associated malignancies constitute 4.5% of all cancers worldwide,22 but there is an effective primary screening paradigm available only for cervical cancers. Head and neck squamous cell carcinomas of the oropharynx constitute the next most frequent HPV-associated cancer, but most patients present with advanced locoregional disease. In this study, we demonstrated that HPV ctDNA can be detected in nearly all patients with HPV-positive OPSCC, a marked improvement over previously reported low-cost PCR-based technologies and likely a result of the following four primary improvements: use of the ddPCR technique, which we found to be superior to quantitative PCR; design of HPV16 primers and probes that match all HPV16 variants; use of a combination of HPV16 and HPV33 tests; and plasma collection and storage procedures optimized for cfDNA. The overall sensitivity was 95.9% in our cohort, a figure similar to the HPV subtype distribution for HPV16 and HPV33 in HPV-positive OPSCC. We demonstrated that the assay was portable to other HPV-associated malignancies because we also detected HPV ctDNA in seven of eight anal squamous cell carcinomas.
Because plasma Epstein-Barr virus DNA can be used to detect subclinical NPCs in early stages, we examined whether HPV ctDNA can be used to detect tumors with low disease burden. In 19 patients, plasma was collected after the symptomatic site of disease had already been resected, and yet HPV16 ctDNA remained detectable. Early detection by liquid biopsy of patients with T1N0 or T1N1 HPV-positive OPSCCs would be beneficial to patients because these patients may be candidates for curative transoral robotic resections alone, obviating the need for postoperative radiation. Likewise, patients with early T1N0 anal squamous cell carcinoma are candidates for curative local excisions without any radiotherapy.
In addition, the specificity of the test, which is of paramount importance in any screening application, was 100% in 27 negative control samples. The plasma HPV16 ctDNA signal completely responded to tumor-directed therapies and paralleled plasma levels of driver mutations, indicating that HPV ctDNA is highly tumor specific.
Previous research has shown various degrees of sensitivities for HPV detection for patients with head and neck cancer3-6,8 and anogenital cancer23-25 using PCR-based methods. A key advantage of ddPCR in a screening setting is the low cost. A disadvantage of ddPCR would be the requirement to use limited material for independent HPV16 and HPV33 tests. These two HPV subtypes constitute more than 95% of all oropharyngeal26,27 and anal HPV-associated squamous cell carcinomas.28 If a single test were to be used to detect cervical cancer as well, an HPV18 ddPCR test would need to be added. An alternative approach is NGS, which, although much more costly, can capture multiple subtypes in one test. A recent report using NGS for determining circulating HPV DNA in 55 patients with locally advanced head and neck squamous carcinoma showed 100% sensitivity.29 In practice, liquid biopsy screening for HPV-associated tumors would most likely involve an initial low-cost test, such as ddPCR, followed by additional tests such as physical exams, Papanicolaou smears, and NGS of additional plasma specimens.
There are some limitations to our study. Although our ctDNA results match the distribution of HPV16 and HPV33 frequencies in HPV-associated OPSCC, we were not able to specifically HPV genotype all pathologic specimens that were diagnosed as HPV positive by p16 positivity and/or DNA or RNA in situ hybridization by standard clinical definitions.13 In addition, it should be noted that although HPV ctDNA was detected in all 19 patients with low tumor burden, these patients may not be representative of patients with early subclinical disease because micrometastatic lesions in these patients with locoregional disease could artificially boost sensitivity of the assay. Our results demonstrate that HPV ctDNA can be effectively used with high sensitivity and specificity for intact tumors, even in patients with low disease burden, indicating clinical utility for screening and systemic treatment response monitoring.
ACKNOWLEDGMENT
We thank Erik Anderson, Luis Diaz, Jonathan Leeman, Jeremy Setton, and Dana Tsui for helpful discussions.
APPENDIX
FIG A1.
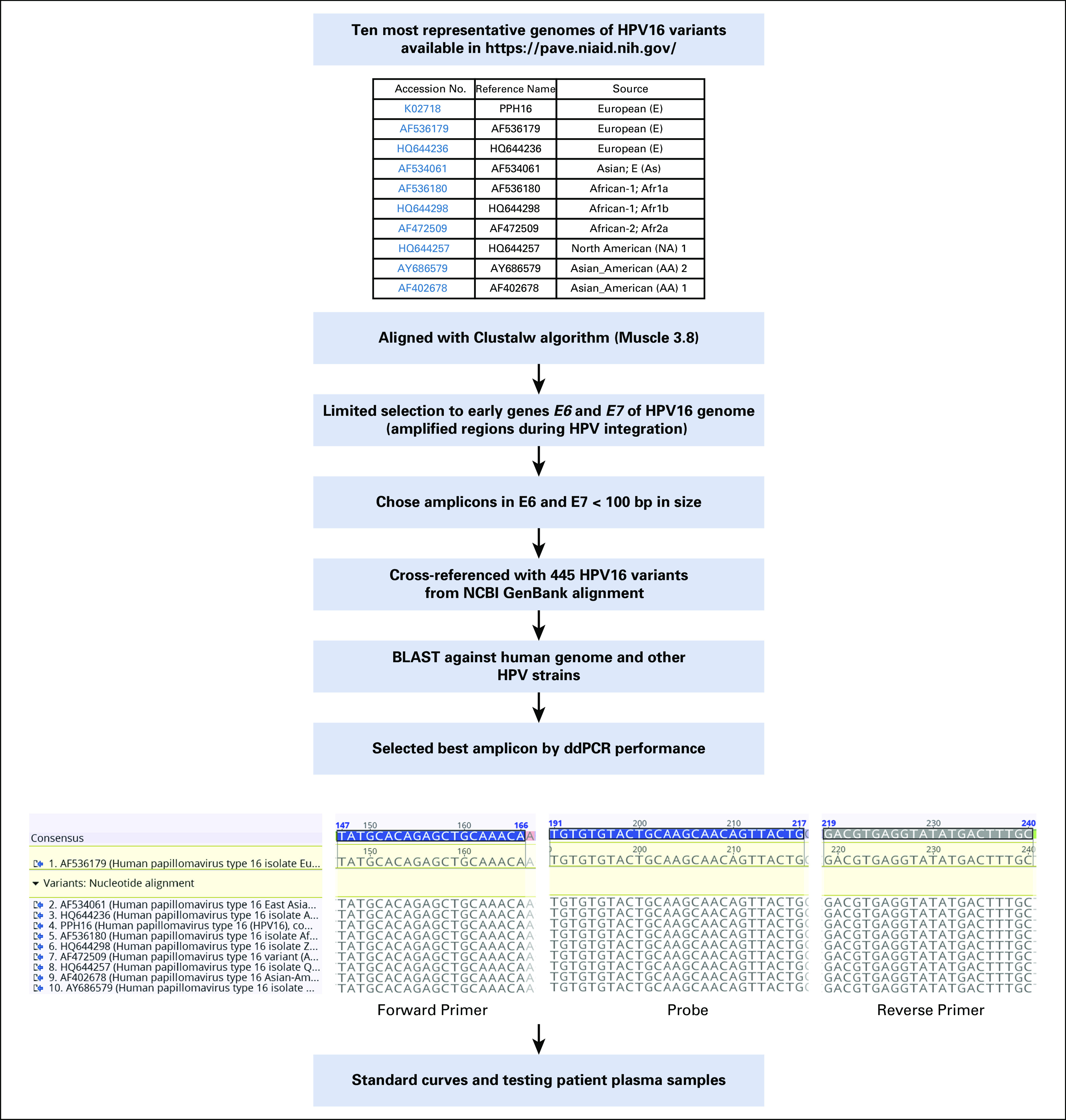
Schema for optimal primer and probe design. BLAST, Basic Local Alignment Search Tool; bp, base pairs; ddPCR, droplet digital polymerase chain reaction; HPV, human papillomavirus; NCBI, National Center for Biotechnology Information.
FIG A2.
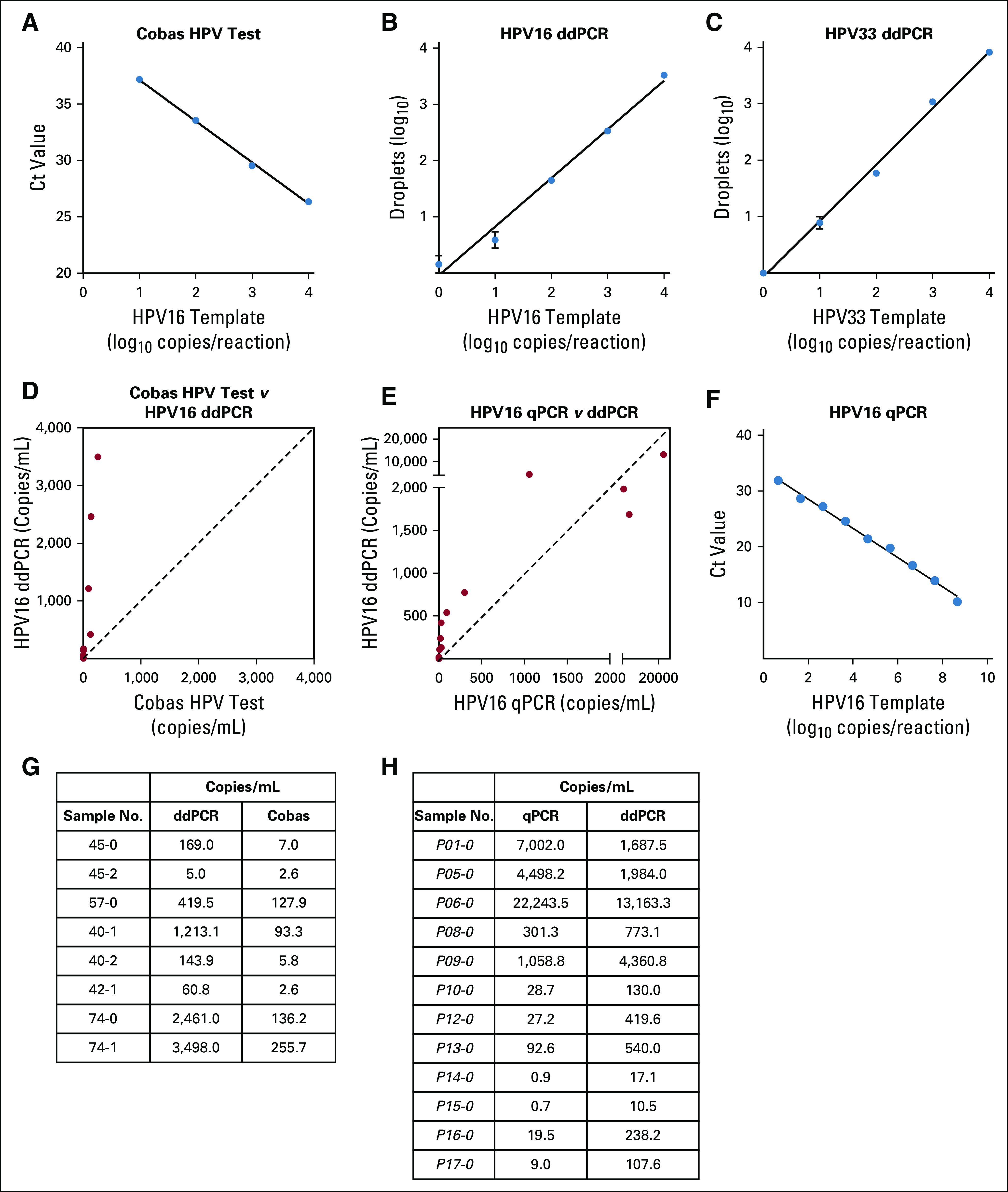
Human papillomavirus (HPV) droplet digital polymerase chain reaction (ddPCR) assay performance relative to established polymerase chain reaction (PCR)–based HPV detection techniques. (A-C and F) Standard curves of Roche Cobas HPV test, HPV16 ddPCR, HPV33 ddPCR, and HPV16 quantitative PCR (qPCR) using a serial dilution of HPV16 plasmid template on a plasmid. (D and G) Comparison of HPV16 ddPCR and the Cobas HPV test in the same eight samples. ddPCR detected a higher number of copies per milliliter of plasma (P = .008, Wilcoxon matched-pairs signed rank test), whereas circulating tumor DNA (ctDNA) levels detected by both methods are highly correlated (r2 = 0.82, P = .002). The signal in paired samples was a median of 15.9-fold higher with ddPCR. The proprietary primer and probe sequences of the Roche Cobas test result in an amplicon length of approximately 200 base pairs (bp), likely too large for fragmented ctDNA, which has a median size of 176 bp.9 (E and H) Comparison of HPV16 ddPCR and HPV16 qPCR in the same 12 samples. We found ddPCR to be more sensitive at lower ctDNA values (arbitrary cutoff, less than 1,000 HPV ctDNA copies/mL) by Wilcoxon matched-pairs signed rank test (P = .008), although each approach was highly correlated (r2 = 0.88, P < .001). Among all paired samples, HPV ctDNA was a median of 5.2-fold higher with ddPCR. At high levels of HPV16, the HPV ctDNA signal can begin to saturate, as observed in three samples because only 20,000 droplets are read on the Bio-Rad QX200 platform. The higher sensitivity of the ddPCR assay despite a larger amplicon size may be secondary to the partitioning of DNA into droplets and reduced inhibitory factors compared with qPCR29 Ct, cycle threshold.
FIG A3.
Human papillomavirus (HPV) type 16 droplet digital polymerase chain reaction (ddPCR) data before, during, and after chemoradiation (CRT). During the course of 7 weeks of CRT, plasma samples were collected every week in 68 patients. ctDNA, circulating tumor DNA.
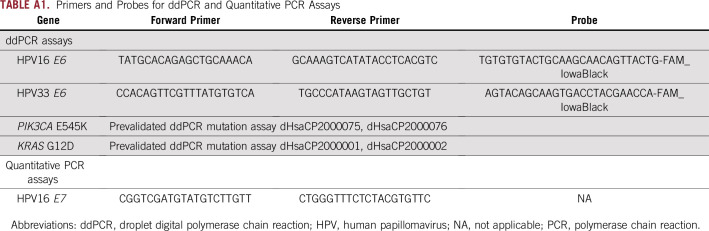
TABLE A1.
Primers and Probes for ddPCR and Quantitative PCR Assays
TABLE A2.
Comparison of Mismatches in Primers and Probes From Previously Published HPV Amplification Strategies
Footnotes
Supported by The James and Judith K. Dimon Foundation and a research grant from the Society of Memorial Sloan Kettering Cancer Center (D.S.H.). Also supported in part by a Cancer Center Support Grant No. P30CA008748 from the National Institutes of Health/National Cancer Institute, Cycle for Survival, and the Marie-Josée and Henry R. Kravis Center for Molecular Oncology.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
AUTHOR CONTRIBUTIONS
Conception and design: Rama R. Damerla, Nancy Y. Lee, Marsha Reyngold, Chiaojung Jillian Tsai, Simon N. Powell, Daniel S. Higginson
Financial support: Nancy Y. Lee, Simon N. Powell, Daniel S. Higginson
Administrative support: Simon N. Powell
Provision of study materials or patients: Nora Katabi, Agnes Viale
Collection and assembly of data: Rama R. Damerla, Daoqui You, Rekha Soni, Rachna Shah, Marsha Reyngold, Vanessa Wu, N. Esther Babady, Agnes Viale, Daniel S. Higginson
Data analysis and interpretation: Rama R. Damerla, Daoqui You, Nora Katabi, Sean M. McBride, Nadeem Riaz, Daniel S. Higginson
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/site/ifc.
Rama R. Damerla
Patents, Royalties, Other Intellectual Property: Daniel S. Higginson and Rama R. Damerla are inventors on a provisional patent application filed by their institution, Memorial Sloan Kettering Cancer Center (Inst)
Nancy Y. Lee
Stock and Other Ownership Interests: AstraZeneca (I)
Consulting or Advisory Role: Merck, Pfizer, Merck Serono, Sanofi
Research Funding: AstraZeneca
Sean M. McBride
Consulting or Advisory Role: Bristol-Myers Squibb, Janssen
Nadeem Riaz
Honoraria: PeerView
Speakers' Bureau: Illumina
Research Funding: Bristol-Myers Squibb, Pfizer
Travel, Accommodations, Expenses: Varian Medical Systems
Simon N. Powell
Consulting or Advisory Role: Varian Medical Systems
N. Esther Babady
Research Funding: GenMark, Luminex, Roche Molecular Diagnostics
Travel, Accommodations, Expenses: Roche Molecular Diagnostics
Daniel S. Higginson
Patents, Royalties, Other Intellectual Property: Daniel S. Higginson and Rama R. Damerla are inventors on a provisional patent application filed by their institution, Memorial Sloan Kettering Cancer Center (Inst)
No other potential conflicts of interest were reported.
REFERENCES
- 1.Chan KCA, Woo JKS, King A, et al. Analysis of plasma Epstein-Barr virus DNA to screen for nasopharyngeal cancer. N Engl J Med. 2017;377:513–522. doi: 10.1056/NEJMoa1701717. [DOI] [PubMed] [Google Scholar]
- 2.Chaturvedi AK, Engels EA, Pfeiffer RM, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29:4294–4301. doi: 10.1200/JCO.2011.36.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahn SM, Chan JY, Zhang Z, et al. Saliva and plasma quantitative polymerase chain reaction-based detection and surveillance of human papillomavirus-related head and neck cancer. JAMA Otolaryngol Head Neck Surg. 2014;140:846–854. doi: 10.1001/jamaoto.2014.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao H, Banh A, Kwok S, et al. Quantitation of human papillomavirus DNA in plasma of oropharyngeal carcinoma patients. Int J Radiat Oncol Biol Phys. 2012;82:e351–e358. doi: 10.1016/j.ijrobp.2011.05.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mazurek AM, Rutkowski T, Fiszer-Kierzkowska A, et al. Assessment of the total cfDNA and HPV16/18 detection in plasma samples of head and neck squamous cell carcinoma patients. Oral Oncol. 2016;54:36–41. doi: 10.1016/j.oraloncology.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, Springer S, Mulvey CL, et al. Detection of somatic mutations and HPV in the saliva and plasma of patients with head and neck squamous cell carcinomas. Sci Transl Med. 2015;7:293ra104. doi: 10.1126/scitranslmed.aaa8507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gupta GP, Kumar S, Marron D, et al: Circulating tumor HPV16 DNA as a biomarker of tumor genomics and disease control in HPV-associated oropharyngeal squamous cell carcinoma. Int J Radiat Oncol Biol Phys 100:1310-1311, 2018. [Google Scholar]
- 8.Dahlstrom KR, Li G, Hussey CS, et al. Circulating human papillomavirus DNA as a marker for disease extent and recurrence among patients with oropharyngeal cancer. Cancer. 2015;121:3455–3464. doi: 10.1002/cncr.29538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diaz LA, Jr, Bardelli A. Liquid biopsies: Genotyping circulating tumor DNA. J Clin Oncol. 2014;32:579–586. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leeman JE, Li JG, Pei X, et al. Patterns of treatment failure and postrecurrence outcomes among patients with locally advanced head and neck squamous cell carcinoma after chemoradiotherapy using modern radiation techniques. JAMA Oncol. 2017;3:1487–1494. doi: 10.1001/jamaoncol.2017.0973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zumsteg ZS, Lok BH, Ho AS, et al. The toxicity and efficacy of concomitant chemoradiotherapy in patients aged 70 years and older with oropharyngeal carcinoma in the intensity-modulated radiotherapy era. Cancer. 2017;123:1345–1353. doi: 10.1002/cncr.30495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Setton J, Caria N, Romanyshyn J, et al. Intensity-modulated radiotherapy in the treatment of oropharyngeal cancer: An update of the Memorial Sloan-Kettering Cancer Center experience. Int J Radiat Oncol Biol Phys. 2012;82:291–298. doi: 10.1016/j.ijrobp.2010.10.041. [DOI] [PubMed] [Google Scholar]
- 13.Ang KK, Harris J, Wheeler R, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363:24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akagi K, Li J, Broutian TR, et al. Genome-wide analysis of HPV integration in human cancers reveals recurrent, focal genomic instability. Genome Res. 2014;24:185–199. doi: 10.1101/gr.164806.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Doorslaer K, Li Z, Xirasagar S, et al. The Papillomavirus Episteme: A major update to the papillomavirus sequence database. Nucleic Acids Res. 2017;45:D499–D506. doi: 10.1093/nar/gkw879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.The Cancer Genome Atlas Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deshmukh AA, Tanner RJ, Luetke MC, et al. Prevalence and risk of penile human papillomavirus infection: Evidence from the National Health and Nutrition Examination Survey 2013-2014. Clin Infect Dis. 2017;64:1360–1366. doi: 10.1093/cid/cix159. [DOI] [PubMed] [Google Scholar]
- 19.Fakhry C, Gillison ML, D’Souza G. Tobacco use and oral HPV-16 infection. JAMA. 2014;312:1465–1467. doi: 10.1001/jama.2014.13183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunne EF, Unger ER, Sternberg M, et al. Prevalence of HPV infection among females in the United States. JAMA. 2007;297:813–819. doi: 10.1001/jama.297.8.813. [DOI] [PubMed] [Google Scholar]
- 21.Gillison ML, Chaturvedi AK, Anderson WF, et al. Epidemiology of human papillomavirus-positive head and neck squamous cell carcinoma. J Clin Oncol. 2015;33:3235–3242. doi: 10.1200/JCO.2015.61.6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Martel C, Plummer M, Vignat J, et al. Worldwide burden of cancer attributable to HPV by site, country and HPV type. Int J Cancer. 2017;141:664–670. doi: 10.1002/ijc.30716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang Z, Stevanović S, Hinrichs CS, et al. Circulating cell-free DNA for metastatic cervical cancer detection, genotyping, and monitoring. Clin Cancer Res. 2017;23:6856–6862. doi: 10.1158/1078-0432.CCR-17-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cabel L, Jeannot E, Bieche I, et al. Prognostic impact of residual HPV ctDNA detection after chemoradiotherapy for anal squamous cell carcinoma. Clin Cancer Res. 2018;24:5767–5771. doi: 10.1158/1078-0432.CCR-18-0922. [DOI] [PubMed] [Google Scholar]
- 25.Jeannot E, Becette V, Campitelli M, et al. Circulating human papillomavirus DNA detected using droplet digital PCR in the serum of patients diagnosed with early stage human papillomavirus-associated invasive carcinoma. J Pathol Clin Res. 2016;2:201–209. doi: 10.1002/cjp2.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Faust H, Eldenhed Alwan E, Roslin A, et al. Prevalence of human papillomavirus types, viral load and physical status of HPV16 in head and neck squamous cell carcinoma from the South Swedish Health Care Region. J Gen Virol. 2016;97:2949–2956. doi: 10.1099/jgv.0.000611. [DOI] [PubMed] [Google Scholar]
- 27.Bratman SV, Bruce JP, O’Sullivan B, et al. Human papillomavirus genotype association with survival in head and neck squamous cell carcinoma. JAMA Oncol. 2016;2:823–826. doi: 10.1001/jamaoncol.2015.6587. [DOI] [PubMed] [Google Scholar]
- 28.Serup-Hansen E, Linnemann D, Skovrider-Ruminski W, et al. Human papillomavirus genotyping and p16 expression as prognostic factors for patients with American Joint Committee on Cancer stages I to III carcinoma of the anal canal. J Clin Oncol. 2014;32:1812–1817. doi: 10.1200/JCO.2013.52.3464. [DOI] [PubMed] [Google Scholar]
- 29.Lee JY, Garcia-Murillas I, Cutts RJ, et al. Predicting response to radical (chemo)radiotherapy with circulating HPV DNA in locally advanced head and neck squamous carcinoma. Br J Cancer. 2017;117:876–883. doi: 10.1038/bjc.2017.258. [DOI] [PMC free article] [PubMed] [Google Scholar]