Abstract
The evolution of genome editing technology based on CRISPR (clustered regularly interspaced short palindromic repeats) system has led to a paradigm shift in biological research. CRISPR/Cas9-guide RNA complexes enable rapid and efficient genome editing in mammalian cells. This system induces double-stranded DNA breaks (DSBs) at target sites and most DNA breakages induce mutations as small insertions or deletions (indels) by non-homologous end joining (NHEJ) repair pathway. However, for more precise correction as knock-in or replacement of DNA base pairs, using the homology-directed repair (HDR) pathway is essential. Until now, many trials have greatly enhanced knock-in or substitution efficiency by increasing HDR efficiency, or newly developed methods such as Base Editors (BEs). However, accuracy remains unsatisfactory. In this review, we summarize studies to overcome the limitations of HDR using the CRISPR system and discuss future direction.
Keywords: CRISPR, DNA double-strand break, Genome editing, HDR, NHEJ
INTRODUCTION
Genetically engineered mice are valuable subjects for developmental and pathomechanism studies. However, the traditional gene targeting method through embryonic stem cells (ESCs) has been time-consuming and costly. In 2013, the Jaenisch group introduced conducting gene modified mice in a one-step generation using clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR associated protein 9 (Cas9) genome engineering technology (1, 2). Since the CRISPR/Cas9-mediated system originated from the prokaryotic immune system (3–6), it enabled rapid and efficient genome editing in mammalian cells (7–11).
This system opened a new era in genome biology fields including animal, plants, and human genetic disease (12–15). Programmable endonuclease Cas9 with guide RNA (gRNA) induce DNA double-strand breaks (DSBs) on the target DNA sequences, and DSBs are repaired by non-homologous end-joining (NHEJ) or homology-directed repair (HDR) pathway, mainly (16–18). Among them, NHEJ is a predominant repair mechanism in higher eukaryotic cells or organisms. Therefore, after DSBs, NHEJ works dominantly and generates small insertions or deletions (indels), resulting in frame shifts at target genes eventually (19–21). Taking advantage of these characteristics, the efficient knock-out study through NHEJ pathway has been developed extensively in the genome editing field. However, since the NHEJ repair mechanism induces uncontrollable random mutations on target loci, NHEJ conjugated technologies showed limitations for precise genome editing, such as designated insertions and single-nucleotide substitutions (2, 22).
To overcome these limitations, many scientists made an effort to develop methods to insert donor template DNA using the HDR pathway, to perform precise gene editing. However, it was difficult to use HDR mechanism in gene editing unrestricted because of its extremely low efficiency. In mammalian cells, NHEJ is the major source of the DNA repair mechanism competing with the HDR pathway. Therefore, for more efficient HDR-mediated precise genome editing, numerous researchers have attempted to enhance HDR pathway or/and suppress NHEJ pathway by targeting key factors (23–25).
Recently, a new technology called base editors (BEs) has been introduced to overcome low accuracy of NHEJ and low efficiency of HDR. These powerful editing tools can change single nucleotide without DNA DSBs in cells (26, 27). BEs are composed of catalytically impaired Cas9 variant with deaminase classified as cytosine base editors (CBEs) and adenine base editors (ABEs), allowing direct conversion from C to T or A to G (28–30). Recent reports showed that various applications using base editors enable single nucleotide substitutions in mammalian genome successfully (31–35). Although it is clear that base-editing technique is an innovative development, limitations remain in the case of single base substitution, as well as insufficient accuracy/efficacy in vivo.
In this review, we will report recently developed methods for precise gene editing as enhanced HDR-mediated gene engineering and direct base editing in mammal species. Diverse strategies to increase HDR efficiency are introduced. One is optimization of the HDR pathway by controlling the length of homology arms of template donor DNA. Another is the inhibition of NHEJ pathway which competes with HDR. Additionally, we also introduce BEs, a method for tailored single nucleotide substitution.
ENHANCING KNOCK-IN EFFICACY BY CONTROLLING DONOR DNA
The most precise genome editing method is utilizing HDR mechanism to insert artificial DNA sequences to target locus or to induce single-nucleotide substitutions. However, the efficiency of HDR pathway in nature is extremely low (2, 36–38). Recently, several studies reported new methods to overcome low efficiency by optimizing template donor DNA. Researchers modulated the length of homology arms and types of donor DNA, such as single strand DNA (ssDNA) or double strand DNA (dsDNA) (Table 1). Renaud et al. explained that using single-stranded oligo DNA nucleotides (ssODNs) as template donors with chemical modifications such as phosphorothioate or LNA could improve precise knock-in efficiency, rather than using double-stranded oligo DNA nucleotides (dsODNs) (39). Paquet et al. delivered ssODN donor templates which comprise silent mutations. These mutations prevented re-cleavage of inserted sequences by CRISPR/Cas9 and increased precise knock-in efficiency (40). Easi-CRISPR was reported as a new method to generate mutant mice efficiently with insertion of exogenous artificial DNA sequences. DNA donors were prepared as ssODNs approximately 1 kb long. They delivered directly components such as ssODN donor templates, gRNAs, and Cas9 mRNA, into mouse zygotes using microinjection. They also successfully generated knock-in mice using CRISPR ribonucleoproteins (RNPs) (41). Some research groups attempted to modify Cas9 protein and gRNAs to increase HDR efficiency. Most recently, the Rossant group has shown that combining two-cell homologous recombination (2C-HR)-CRISPR with a modified biotin-streptavidin approach in mice, can increase knock-in efficiency over standard methods by more than 10-fold (up to 95 %) (42). The Gordon group demonstrated that HDR efficiency could be increased up to 30-fold using Cas9 and Porcine Circovirus 2 (PCV) Rep fusion protein delivered with ssODNs containing 13 bp PCV recognition sequences at 5'-end (43). Other groups attempted NHEJ or microhomology-mediated end-joining (MMEJ) mediated knock-in, to insert exogenous DNA sequences more efficiently to the target loci, instead of HDR pathway requiring shorter homology arms compared with HDR-mediated. A new knock-in method using MMEJ pathway, termed the precise integration into target chromosome (PITCh), was reported. They generated vectors exquisitely, which contain short micro-homology sequences approximately 5–25 bp, and enabled insertion of large DNA fragments to the target sites of various cell lines and organisms (44, 45). Also, Yao et al. successfully knocked in tagging sequences in-vivo and ex-vivo by MMEJ-mediated manner. Donor DNA sequences contain short homology arms including microhomology sequences (46). Also, they reported a new method, called homology-mediated end-joining (HMEJ) strategy. The vector for HMEJ based knock-in contains CRISPR-Cas9 cleavage sites, identical to target sequences on the genome, and approximately 800 bp-long homology arms. These methods were tested in mouse and monkey embryos and showed greater results than HDR, NHEJ, and MMEJ mediated knock-in efficiency (47). Most recently, Yao et al. demonstrated Tild-CRISPR (targeted integration with linearized dsDNA-CRISPR). They provided donor DNA with 800 bp-long homology arms by PCR-amplification. This method is based on HMEJ strategy and has advantages in preparing template donor DNA by PCR, efficiently. They claimed that it shows high integration efficiency for both, in vitro (mouse/human embryo cell) and in vivo (mouse brain) scale (48). Representative studies are summarized at Table 1.
Table 1.
Regulation of homology arm of donor DNA to enhance knock-in efficiency
| Species | Methods | Donor DNA | Insertion size | HA size | Reference | |
|---|---|---|---|---|---|---|
| Rat, Mouse | Zygote | Microinjection Cas9mRNA/gRNA |
ssODN (chemical modifications: phosphorothioate or LNA) | ~100 bp | ~100 bp | 39 |
| Human | HEK293, iPSC | Transfection, electroporation Plamid |
ssODN (silent mutations) | 100 bp/400 bp | 50 bp | 40 |
| Mouse | Zygote | Microinjection Cas9 mRNA or protein/gRNA (crRNA + tracrRNA) (Easi-CRISPR method) |
ssODN | 527 bp/893 bp | 55 bp/103 bp | 41 |
| Human | HEK293T, U2-OS | Transfection Cas9 protein/gRNA (PCV-Cas9 fusion) |
ssODN (13 bp PCV recognition sequences at 5’-end) | 50 bp | 75 bp | 43 |
| Mouse | Zygote, ESC | Microinjection Cas9 mRNA/gRNA |
ssODN dsDNA (Linearization) |
~42 bp ~2.9 bp |
60 bp ~4.5 kb |
2 |
| Human | HEK293T | Transfection Plasmid (PITCh method) |
dsDNA (Linearization) | ~1.5 kb | ~25 bp | 44, 45 |
| Mouse, monkey | Zygote E14.5 embryo Adult mouse |
Microinjection, mRNA/gRNA In utero electroporation, Cas9 mRNA/gRNA Hydrodynamic injection, Cas9 mRNA/gRNA |
dsDNA (Linearization) | 700 bp/6.1 kb | 800 bp | 46, 47 |
| Mouse, Human | Zygote E14.5 embryo |
Microinjection Cas9 mRNA/gRNA (Tild method) In utero electroporation Cas9 mRNA/gRNA |
dsDNA (Linearization or PCR amplification) | ~2 kb | 800 bp | 48 |
| Mouse | 2-cell stage embryo | Microinjection, Cas9 mRNA/gRNA (2C-HR-CRISPR with a biotin-Streptavidin approach) | dsDNA (PCR amplification) | 717 bp/1.4 kb | 100 bp/3 kb | 42 |
HA: Homology arm, iPSC: induced Pluripotent Stem Cell, ESCs: embryonic stem cells, gRNA: guide RNA, ssODNs: single-stranded oligo DNA nucleotides, dsDNA: double-strand DNA, Easi: Efficient additions with ssDNA inserts, PVC: Porcine Circovirus 2, PITCh: Precise Integration into Target Chromosome, Tild: targeted integration with linearized dsDNA, 2C-HR: two-cell homologous recombination.
ENHANCING KNOCK-IN EFFICIENCY BY SMALL MOLECULES
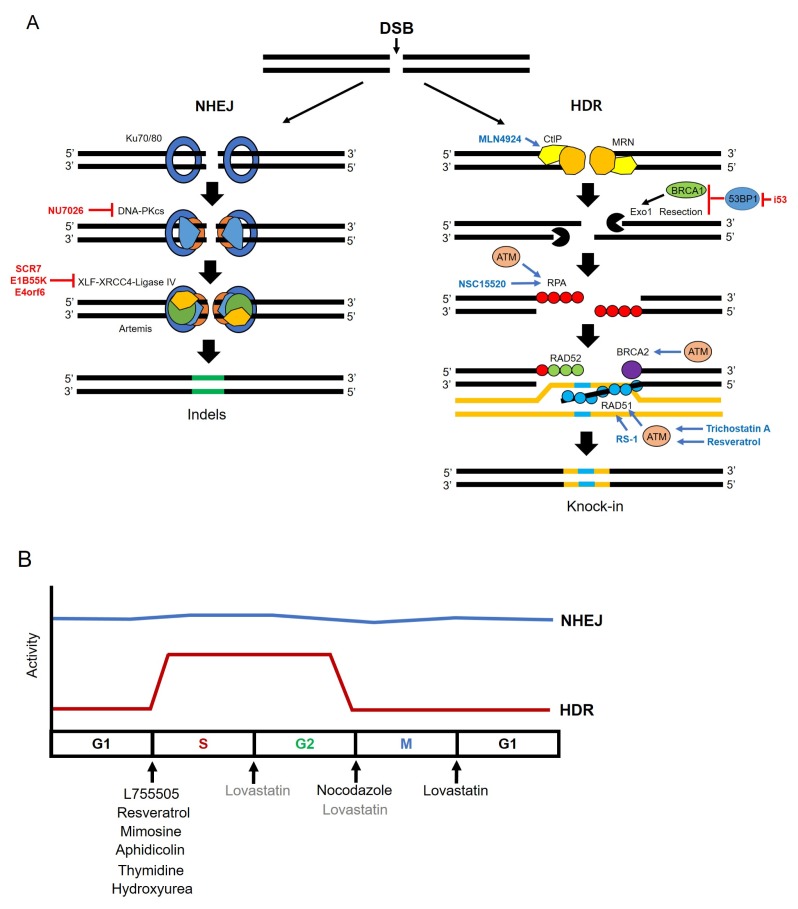
NHEJ mediated genome editing induces random mutations such as small indels on target sites. Therefore, these kinds of mutations led the frame shift on targeted genes and is proper for knock-out studies but not for inducing precise mutations, such as point mutations or knock-in studies. Conversely, HDR repair system is good in generating precise point mutations and for inserting external artificial DNA sequences. However, low efficiency has always been a major obstacle to broad use. A number of studies have attempted to increase HDR efficiency by regulating DSBs repair mechanisms (Fig. 1A). It is well known that NHEJ and HDR pathways are in competition (49–51). Several studies have shown that suppression of key molecules involved in the NHEJ pathway could increase efficiency of HDR. Many proteins are known to be relevant with NHEJ pathway including Ku heterodimers (Ku70/80), DNA-dependent protein kinase catalytic subunits (DNA-PKcs), DNA ligase IV, the X-ray repair cross-complementing protein 4 (XRCC4), and the XRCC4-like factor (XLF) as core complexes (52–54). Among these related proteins Chu et al. suppressed DNA ligase IV by Scr7, a DNA ligase IV inhibitor, and adenovirus 4 E1B55K and E4orf6 proteins, inducing proteasomal degradation of DNA ligase IV. HDR efficiency increased 4–5-fold or 8-fold, respectively (55). Also, Maruyama et al. showed that treatment of Scr7 in a mammalian cell line and mouse zygotes increase HDR efficiency approximately 19-fold (56). Yu et al. identified small molecules, L755505 and Brefeldin A. The function of these molecules in NHEJ pathway has not been clarified. However, both small molecules enhanced HDR efficiency approximately 2–3-fold for large fragment knock-in and 9-fold increase for inducing point mutation, respectively (57). Risenberg et al. identified effective small molecules to increase HDR efficiency in human induced pluripotent stem cells (hiPSCs) by screening of small molecules related with DNA repair mechanisms. The combination of small molecules termed CRISPY mix containing NU7026, Trichostatin A, MLN4924, and NSC 15520 showed the most effective HDR efficiency. Also, the related small molecules affected key molecules of major DNA repair mechanisms (58).
Fig. 1.
Small molecules enhance knock-in efficiency. (A) Small molecules related to the NHEJ or HDR repair pathway. Inhibitors are labeled in red, activators are labeled in blue. NU7026 inhibits DNA-PK, and SCR7, E1B55K, and E4orf6 inhibit DNA ligase IV. MLN4924, NSC15520, RS-1, Trichostatin A, or Resveratrol enhance CtIP, RPA, RAD51, or ATM, respectively. ATM protein also induces activation of RPA, BRCA2, and RAD51. The i53 is an inhibitor of 53BP1. The i53 activates DNA end resection and recruitment, of BRCA1 to DSBs. (B) HDR activity is increased at S/G2 phase. NHEJ activity is labeled in blue, HDR activity is labeled in red. Small molecules are used to arrest the cell cycle at specific phase, to improve HDR efficiency. L755505, Resveratrol, Mimosine, Aphidicolin, Thymidine and Hydroxyurea block cells at the G1 to S phase before DNA replication, and Nocodazole arrests cell cycle at G2/M phase. Lovastatin also inhibits at early G1, and partially at G2/M phase.
Major DNA repair pathways, NHEJ and HDR are not always activated during all cell cycle stages. NHEJ dominates over all M, G1, S, and G2 phases, while HDR can only compete with NHEJ, during S and G2 phases. HDR is down regulated during M phase and G1 phase (59–61). Various small molecules exert their effects by controlling such stages in part (Fig. 1B). Li et al. re-tested the function of Scr7 and L755505, in porcine fetal fibroblast. Additionally, resveratrol, a novel small molecule in this field was also tested. Scr7 and L755505 in porcine fetal fibroblast led a 2-fold increase similar as tested in other cell lines, and the resveratrol could raise approximately 3-fold in porcine fetal fibroblast. It is also reported that L755505 and resveratrol could arrest cells at S phage, wherein the HDR mechanism is activated. Treatment of three molecules such as Scr7, L755505, and resveratrol, up-regulated mRNA expression level of HDR key factors, such as BRCA1, BRCA2, RPA3, SPIDR, NBN, RAD50, RAD51, and RAD52, and down-regulated key molecules of NHEJ pathway such as LIG4, MRE11, DCLRE1C, and XRCC4 (62). Also, multiple researchers identified small molecules that affect cell cycle arrest to increase HDR. Nocodazole and Lovastatin synchronize the cell cycle in G2/M phase and early G1 phase, respectively. Lovastatin also inhibits at G2/M phase, partially. Mimosine, aphidicolin, thymidine, and hydroxyurea arrest cells at between G1 phase and S phase, before DNA replication (61, 63). Recently, Canny et al. regulated another key factor: 53BP1. It is significant at the beginning of the repair mechanism between NHEJ and HDR pathways on the DSBs loci. The 53BP1 blocks DNA end resection and recruitment of BRCA1 to DSBs. This study has shown that the 53BP1 inhibitor, i53, can increase HDR efficiency (64). Song et al. reported applying RS-1 could increase HDR efficiency by stimulating Rad51. Unlike previously reported studies, in which small molecules were used to inhibit the NHEJ pathway, this study used a small molecule, RS-1, to promote the HDR pathway (65). Most of the cases of treatment of small molecules are focused on suppression of NHEJ pathways since both repair mechanisms are in competition.
NUCLEOTIDE REPLACEMENT WITH BASE EDITORS
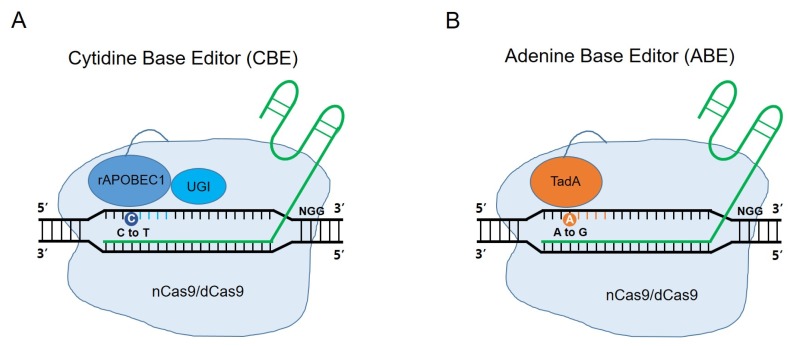
More than 50% of human pathogenic mutations are point mutations or single nucleotide polymorphisms (SNPs) (26). As the importance of precise medicine arises, accurate single nucleotide substitutions in the genome have been required for pathology or mechanistic studies. However, in the beginning of the CRISPR technology, specific nucleotide substitutions at desired target sites could only be induced by an HDR-based CRISPR system, despite its low efficiency. To overcome such limitation, new tools called Base Editors (BEs) were developed to induce single-nucleotide substitution, which do not need a template donor DNA (Fig. 2A and 2B) (28–30). Because these techniques do not introduce DSBs, they never use DNA repair mechanisms as NHEJ, MMEJ, or HDR pathways. BEs were composed of nuclease activity deficient Cas9, nickase Cas9 (nCas9) or dead Cas9 (dCas9), and cytidine deaminase or adenine deaminase. They enable conversion of C to T, or A to G, and vice versa. They are newly-developed methods not affected by HDR efficiency in case of inducing substitutions. These tools were verified through various research groups and applied to many other organisms, including mice and rabbits (31, 32, 66, 67). The substitution efficiency was higher than the HDR mechanism. However, the unique characteristic of BEs, such as base editing window which indicates the specific region occurring substitution, could be a limitation to inducing single-nucleotide substitution to the exact target base pair. So, some researchers attempted to change the base editing window. One study induced some mutations at cytidine deaminase domains to narrow the base editing window for more specific substitutions (68). Conversely, to extend coverage of BE systems, some researchers demonstrated that using the extended guided RNA could extend coverage of BEs and using Cas9 variants with different protospacer adjacent motif (PAM) sequences, such as xCas9 and VQR variants (32, 69, 70). There remain several improvements in the BE system. Accuracy and efficacy have not been satisfied for clinical demands and knock-in of external DNA sequences are impossible.
Fig. 2.
Schematics of base editors (BEs). (A) The cytidine base editor (CBE) consists of cytidine deaminase rAPOBEC1 (blue), uracil glycosylase inhibitor (UGI) and nickase Cas9 (nCas9) or dead Cas9 (dCas9). CBE can induce targeted nucleotide substitutions, such as C to T, or G to A conversion. (B) The adenine base editor (ABE) consists of adenine deaminase TadA (orange,) and nCas9 or dCas9. ABE can induce targeted nucleotide substitutions, such as A to G, or T to C conversion. The active window of CBE and ABE is 4–8 nucleotides, in the distal region of the guide RNA.
CONCLUSION
CRISPR/Cas9 mediated genome engineering applicable to a variety of organisms is crucial as a tool for research and clinical applications. In this review, we showed efforts to increase efficiency of HDR, one of the genetic manipulation strategies, for accurate and specific targeted knock-in. Recent efforts to improve HDR efficiency have focused on controlling the homology arm length, or suppressing the NHEJ pathway using small molecules. In particular, the Tild-CRISPR method, a method of controlling donor DNA homology arm length, is expected to greatly improve the efficiency of HDR. Based on these results, HDR efficiency is expected to be enhanced by combining NHEJ pathway inhibition with small molecules and the control of homology arm length. Additionally, the BEs (nucleotide substitution methods for specific target sites) are expected to be applied to studies of clinical pathology mechanism by allowing tailored point mutation. Recently, development of gene editing technology has suggested the possibility of clinical application as a genetic disease therapeutic agent. However, accuracy of gene correction fails to meet clinical demands and additionally, the stable in vivo delivery system is lacking. To overcome these problems and to apply clinical applications for therapeutic purposes, it is necessary to improve gene editing accuracy/efficiency and develop in vivo delivery systems delivery systems, simultaneously.
ACKNOWLEDGEMENTS
This study was supported by the Chung Yang, Cha Young Sun & Jang Hi Joo Memorial fund, Korea university grant (K1804351), and the Bio & Medical Technology Development Program of the National Research Foundation (NRF) of Korea (NRF-2018M3A9H3021707, NRF-2018R1D1A1B07048434, and NRF-2014M3A9D5A01075128).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Wang H, Yang H, Shivalila CS, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 4.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 5.Terns MP, Terns RM. CRISPR-based adaptive immune systems. Curr Opin Microbiol. 2011;14:321–327. doi: 10.1016/j.mib.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhaya D, Davison M, Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu Rev Genet. 2011;45:273–297. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- 7.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 8.Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woo JW, Kim J, Kwon SI, et al. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat Biotechnol. 2015;33:1162–1164. doi: 10.1038/nbt.3389. [DOI] [PubMed] [Google Scholar]
- 14.Amoasii L, Hildyard JCW, Li H, et al. Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science. 2018;362:86–91. doi: 10.1126/science.aau1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niu D, Wei HJ, Lin L, et al. Inactivation of porcine endogenous retrovirus in pigs using CRISPR-Cas9. Science. 2017;357:1303–1307. doi: 10.1126/science.aan4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang F, Han M, Romanienko PJ, Jasin M. Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc Natl Acad Sci U S A. 1998;95:5172–5177. doi: 10.1073/pnas.95.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kakarougkas A, Jeggo PA. DNA DSB repair pathway choice: an orchestrated handover mechanism. Br J Radiol. 2014;87 doi: 10.1259/bjr.20130685. 20130685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindahl T. DNA repair enzymes. Annu Rev Biochem. 1982;51:61–87. doi: 10.1146/annurev.bi.51.070182.000425. [DOI] [PubMed] [Google Scholar]
- 19.Steentoft C, Vakhrushev SY, Vester-Christensen MB, et al. Mining the O-glycoproteome using zinc-finger nuclease-glycoengineered SimpleCell lines. Nat Methods. 2011;8:977–982. doi: 10.1038/nmeth.1731. [DOI] [PubMed] [Google Scholar]
- 20.Kim Y, Kweon J, Kim A, et al. A library of TAL effector nucleases spanning the human genome. Nat Biotechnol. 2013;31:251–258. doi: 10.1038/nbt.2517. [DOI] [PubMed] [Google Scholar]
- 21.Lehner K, Mudrak SV, Minesinger BK, Jinks-Robertson S. Frameshift mutagenesis: the roles of primer-template misalignment and the nonhomologous end-joining pathway in Saccharomyces cerevisiae. Genetics. 2012;190:501–510. doi: 10.1534/genetics.111.134890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smithies O, Gregg RG, Boggs SS, Koralewski MA, Kucherlapati RS. Insertion of DNA sequences into the human chromosomal beta-globin locus by homologous recombination. Nature. 1985;317:230–234. doi: 10.1038/317230a0. [DOI] [PubMed] [Google Scholar]
- 23.Zelensky AN, Schimmel J, Kool H, Kanaar R, Tijsterman M. Inactivation of Pol theta and C-NHEJ eliminates off-target integration of exogenous DNA. Nat Commun. 2017;8:66. doi: 10.1038/s41467-017-00124-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schimmel J, Kool H, van Schendel R, Tijsterman M. Mutational signatures of non-homologous and polymerase theta-mediated end-joining in embryonic stem cells. EMBO J. 2017;36:3634–3649. doi: 10.15252/embj.201796948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mateos-Gomez PA, Kent T, Deng SK, et al. The helicase domain of Poltheta counteracts RPA to promote alt-NHEJ. Nat Struct Mol Biol. 2017;24:1116–1123. doi: 10.1038/nsmb.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rees HA, Liu DR. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat Rev Genet. 2018;19:770–788. doi: 10.1038/s41576-018-0059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim JS. Precision genome engineering through adenine and cytosine base editing. Nat Plants. 2018;4:148–151. doi: 10.1038/s41477-018-0115-z. [DOI] [PubMed] [Google Scholar]
- 28.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishida K, Arazoe T, Yachie N, et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science. 2016;353:aaf8729. doi: 10.1126/science.aaf8729. [DOI] [PubMed] [Google Scholar]
- 30.Gaudelli NM, Komor AC, Rees HA, et al. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim K, Ryu SM, Kim ST, et al. Highly efficient RNA-guided base editing in mouse embryos. Nat Biotechnol. 2017;35:435–437. doi: 10.1038/nbt.3816. [DOI] [PubMed] [Google Scholar]
- 32.Ryu SM, Koo T, Kim K, et al. Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat Biotechnol. 2018;36:536–539. doi: 10.1038/nbt.4148. [DOI] [PubMed] [Google Scholar]
- 33.Liang P, Ding C, Sun H, et al. Correction of beta-thalassemia mutant by base editor in human embryos. Protein Cell. 2017;8:811–822. doi: 10.1007/s13238-017-0475-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Z, Chen M, Chen S, et al. Highly efficient RNA-guided base editing in rabbit. Nat Commun. 2018;9:2717. doi: 10.1038/s41467-018-05232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yeh WH, Chiang H, Rees HA, Edge ASB, Liu DR. In vivo base editing of post-mitotic sensory cells. Nat Commun. 2018;9:2184. doi: 10.1038/s41467-018-04580-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang HHY, Pannunzio NR, Adachi N, Lieber MR. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat Rev Mol Cell Biol. 2017;18:495–506. doi: 10.1038/nrm.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee SH, Kim S, Hur JK. CRISPR and Target-Specific DNA Endonucleases for Efficient DNA Knock-in in Eukaryotic Genomes. Mol Cells. 2018;41:943–952. doi: 10.14348/molcells.2018.0408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cox DB, Platt RJ, Zhang F. Therapeutic genome editing: prospects and challenges. Nat Med. 2015;21:121–131. doi: 10.1038/nm.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Renaud JB, Boix C, Charpentier M, et al. Improved Genome Editing Efficiency and Flexibility Using Modified Oligonucleotides with TALEN and CRISPR-Cas9 Nucleases. Cell Rep. 2016;14:2263–2272. doi: 10.1016/j.celrep.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 40.Paquet D, Kwart D, Chen A, et al. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature. 2016;533:125–129. doi: 10.1038/nature17664. [DOI] [PubMed] [Google Scholar]
- 41.Quadros RM, Miura H, Harms DW, et al. Easi-CRISPR: a robust method for one-step generation of mice carrying conditional and insertion alleles using long ssDNA donors and CRISPR ribonucleoproteins. Genome Biol. 2017;18:92. doi: 10.1186/s13059-017-1220-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gu B, Posfai E, Rossant J. Efficient generation of targeted large insertions by microinjection into two-cellstage mouse embryos. Nat Biotechnol. 2018;36:632–637. doi: 10.1038/nbt.4166. [DOI] [PubMed] [Google Scholar]
- 43.Aird EJ, Lovendahl KN, St Martin A, Harris RS, Gordon WR. Increasing Cas9-mediated homology-directed repair efficiency through covalent tethering of DNA repair template. Commun Biol. 2018;1:54. doi: 10.1038/s42003-018-0054-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakade S, Tsubota T, Sakane Y, et al. Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nat Commun. 2014;5:5560. doi: 10.1038/ncomms6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sakuma T, Nakade S, Sakane Y, Suzuki KT, Yamamoto T. MMEJ-assisted gene knock-in using TALENs and CRISPR-Cas9 with the PITCh systems. Nat Protoc. 2016;11:118–133. doi: 10.1038/nprot.2015.140. [DOI] [PubMed] [Google Scholar]
- 46.Yao X, Wang X, Liu J, et al. CRISPR/Cas9 - Mediated Precise Targeted Integration In Vivo Using a Double Cut Donor with Short Homology Arms. EBioMedicine. 2017;20:19–26. doi: 10.1016/j.ebiom.2017.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yao X, Wang X, Hu X, et al. Homology-mediated end joining-based targeted integration using CRISPR/Cas9. Cell Res. 2017;27:801–814. doi: 10.1038/cr.2017.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yao X, Zhang M, Wang X, et al. Tild-CRISPR Allows for Efficient and Precise Gene Knockin in Mouse and Human Cells. Dev Cell. 2018;45:526–536 e525. doi: 10.1016/j.devcel.2018.04.021. [DOI] [PubMed] [Google Scholar]
- 49.Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- 50.Allen C, Halbrook J, Nickoloff JA. Interactive competition between homologous recombination and non-homologous end joining. Mol Cancer Res. 2003;1:913–920. [PubMed] [Google Scholar]
- 51.Hartlerode AJ, Scully R. Mechanisms of double-strand break repair in somatic mammalian cells. Biochem J. 2009;423:157–168. doi: 10.1042/BJ20090942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ceccaldi R, Rondinelli B, D'Andrea AD. Repair Pathway Choices and Consequences at the Double-Strand Break. Trends Cell Biol. 2016;26:52–64. doi: 10.1016/j.tcb.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pannunzio NR, Watanabe G, Lieber MR. Nonhomologous DNA end-joining for repair of DNA double-strand breaks. J Biol Chem. 2018;293:10512–10523. doi: 10.1074/jbc.TM117.000374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shibata A. Regulation of repair pathway choice at two-ended DNA double-strand breaks. Mutat Res. 2017;803–805:51–55. doi: 10.1016/j.mrfmmm.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 55.Chu VT, Weber T, Wefers B, et al. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol. 2015;33:543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- 56.Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol. 2015;33:538–542. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu C, Liu Y, Ma T, et al. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell. 2015;16:142–147. doi: 10.1016/j.stem.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Riesenberg S, Maricic T. Targeting repair pathways with small molecules increases precise genome editing in pluripotent stem cells. Nat Commun. 2018;9:2164. doi: 10.1038/s41467-018-04609-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Orthwein A, Fradet-Turcotte A, Noordermeer SM, et al. Mitosis inhibits DNA double-strand break repair to guard against telomere fusions. Science. 2014;344:189–193. doi: 10.1126/science.1248024. [DOI] [PubMed] [Google Scholar]
- 60.Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annu Rev Genet. 2010;44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin S, Staahl BT, Alla RK, Doudna JA. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife. 2014;3:e04766. doi: 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li G, Zhang X, Zhong C, et al. Small molecules enhance CRISPR/Cas9-mediated homology-directed genome editing in primary cells. Sci Rep. 2017;7:8943. doi: 10.1038/s41598-017-09306-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang D, Scavuzzo MA, Chmielowiec J, et al. Enrichment of G2/M cell cycle phase in human pluripotent stem cells enhances HDR-mediated gene repair with customizable endonucleases. Sci Rep. 2016;6:21264. doi: 10.1038/srep21264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Canny MD, Moatti N, Wan LCK, et al. Inhibition of 53BP1 favors homology-dependent DNA repair and increases CRISPR-Cas9 genome-editing efficiency. Nat Biotechnol. 2018;36:95–102. doi: 10.1038/nbt.4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Song J, Yang D, Xu J, Zhu T, Chen YE, Zhang J. RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency. Nat Commun. 2016;7:10548. doi: 10.1038/ncomms10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee HK, Willi M, Miller SM, et al. Targeting fidelity of adenine and cytosine base editors in mouse embryos. Nat Commun. 2018;9:4804. doi: 10.1038/s41467-018-07322-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu Z, Lu Z, Yang G, et al. Efficient generation of mouse models of human diseases via ABE- and BE-mediated base editing. Nat Commun. 2018;9:2338. doi: 10.1038/s41467-018-04768-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim YB, Komor AC, Levy JM, Packer MS, Zhao KT, Liu DR. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat Biotechnol. 2017;35:371–376. doi: 10.1038/nbt.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Banno S, Nishida K, Arazoe T, Mitsunobu H, Kondo A. Deaminase-mediated multiplex genome editing in Escherichia coli. Nat Microbiol. 2018;3:423–429. doi: 10.1038/s41564-017-0102-6. [DOI] [PubMed] [Google Scholar]
- 70.Hu JH, Miller SM, Geurts MH, et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 2018;556:57–63. doi: 10.1038/nature26155. [DOI] [PMC free article] [PubMed] [Google Scholar]