Abstract
High-content imaging of larval zebrafish behavior can be used as a screening approach to rapidly evaluate the relative potential for chemicals to cause toxicity. However, most statistical methods applied to these data transform movement values to incidence-based “hits” and calculate lowest effect levels (LELs), which loses individual fish resolution of behavior and defies hazard ranking due to reliance on applied dose levels. We developed a parallelizable workflow to calculate benchmark dose (BMD) values from dynamic, high-content zebrafish behavior data that scales for high-throughput chemical screening. To capture the zebrafish movement response from light to dark stimulus, we summarized time-dependent data using both area under the curve and the immediate change at the transition point into two novel metrics that characterized abnormal behavior as a function of chemical concentration. The BMD workflow was applied to calculate BMD10 values of 1,060 ToxCast chemicals for 24 zebrafish endpoints, including behavior, mortality and morphology. The BMD10 values provided better precision and separation than LELs for clustering chemicals since they were derived from models that best-fit their concentration-response curves. Analysis of BMD10 values revealed behavioral signatures as the most sensitive endpoints. High concordance in chemical activity was observed between ToxCast in vitro data and zebrafish in vivo behavioral data, however ToxPi analysis indicated that rankings based on in vitro data were not a reliable predictor of in vivo rankings for lower potency chemicals. This analysis method will enable the use of high-content zebrafish behavioral screening data for BMD analysis in toxicological hazard assessment.
Keywords: Benchmark dose, high-content imaging, biological modeling, zebrafish
1. Introduction
Traditional approaches to determine human exposure limits depend on overt responses, extrapolation, and safety factors. These approaches are becoming prohibitive because of the need to test large number of chemicals, and the high cost of assay standardization and testing 1–3. Adding to the growing complexity of risk assessment is the need to include mechanistic models that link molecular processes to toxicity and adverse health outcomes 4 Therefore, high- throughput biological assays have been developed for building predictive models of chemical exposure and adverse health outcomes and for the identification of potentially hazardous chemicals 5–10. The primary goal of these efforts is to enable more informed risk assessments and regulatory decisions using modern data collection approaches. High-throughput screening data are so large and complex that data-driven frameworks are essential to identify the threshold concentrations for an adverse health outcome, and to interpret the data from a regulatory perspective 11.
Most available high-throughput data are from in vitro assays because they are time- and cost- effective - compared to in vivo assays- and require only small sample sizes. However, while in vitro assays provide evidence for the potential biological activity of chemical compounds, the interpretation of assays results in the context of hazard assessment is limited compared to a whole animal physiological response. Among animal models, zebrafish (Danio rerio) is the most versatile and relatively inexpensive, and amenable for high-throughput toxicity screening 7,10,12–15. Zebrafish development is external and optically transparent 14 such that it allows for non-invasive and simultaneous evaluation of numerous development and behavioral endpoints - from individual cells to full organ development - using simple microscopic techniques. Zebrafish early development bears similarities (in cellular structure, signaling processes, anatomy, and physiology) with other vertebrates, and nearly 84% of the genes related to human diseases are present in the zebrafish genome 16. However, to be useful for future data-driven frameworks of toxicological risk assessments that set human and environmental exposure limits, a point of departure (POD) approach is essential to utilize zebrafish screening data for chemical ranking and prioritization.
Until now, the application of POD approaches for formulating a reference dose using zebrafish data have been limited to the calculation of No Observed Adverse Effect or Lowest Effect Levels (NOAELs or LELs) from the morbidity and mortality incidence data 10,17 Other POD methods, especially, the benchmark dose (BMD) approach 18–23, involves fitting dose-response models to concentration-response curves, provide more precise POD values than LEL by way of interpolation, but have not been applied to Zebrafish screening data. Since LEL values are restricted to tested concentrations, it is not possible to rank chemicals with the same LEL values. However, the multidimensional (> 20 endpoints) and high-throughput (> 1000 chemicals) nature of the data makes it challenging to apply the BMD approach, as it would involve fitting thousands of concentration-response curves. Furthermore, movement data capturing fish behavior over multiple time points is not easily amenable to dose-response analysis. Reducing high-content imaging data down to “hits” to allow comparison of treatment groups loses the resolution of individual fish movements over time.
Our work addresses the gaps and challenges of using high-content zebrafish screening data for POD analysis. First, we developed a novel method to characterize the abnormal behavior of zebrafish from the time-dependent movement response to light/dark data, which is essential for constructing concentration-response curves, maintaining the resolution of a single fish over time. Second, we established an automated workflow for computing curve fits for each chemical and zebrafish endpoint. The workflow was applied to calculate BMD10 values- benchmark concentrations (the fish were exposed to) that result in 10% extra risk compared to the background (control) response - for the 1,060 ToxCast chemicals and 24 zebrafish endpoints, including behavior, mortality and morphology. Consequently, we identified two novel and highly sensitive signatures of abnormal behavior at 120 hours post-fertilization (hpf), which provided better precision for ranking chemicals than LEL. Finally, we determined the extent of overlap in the number of active / inactive chemicals between ToxCast in vitro and zebrafish in vivo data, and analyzed the influence of zebrafish behavioral data to change hazard ranking of chemicals when integrated with ToxCast in vitro data.
2. Materials and Methods
2.1. Chemical and plate preparation for exposure studies
Stock solutions of the 1,060 unique ToxCast Phase 1 and 2 chemicals were provided by the US EPA National Center for Computational Toxicology (US EPA-NCCT), in 100% DMSO at a concentration of 20 mM in multiple 96 well plates. Information about the chemicals and their structure data files are available at http://www.epa.gov/NCCT/toxcast/chemicals.html. Details on the chemical dilutions and plate preparations can be found in Truong et al 10. The dataset used for the BMD analysis comprised of data for 1,078 chemical samples (which included some replicates) provided by the US EPA-NCCT.
2.2. Zebrafish behavioral data
Tropical 5D adult zebrafish were raised at Oregon State University Sinnhuber Aquatic Research Laboratory, Corvallis, Oregon. The zebrafish were housed in 100 gallon tanks, and spawned in location by placing spawning funnels in the tank the night prior, and embryos were collected and staged. The chorions of 4 hours post fertilization (hpf) embryos were removed using pronase as described in 24 Six hpf embryos were placed 1 per well in a 96 well plate. The embryos on each plate were exposed to 5 concentrations of the chemical, such that, 16 embryos in individual wells were exposed to a single concentration at a time. Two replicate plates for each chemical were evaluated for its effect on morphology and the movement of zebrafish embryos after 24 and 120 hpf. Each chemical was tested at concentrations with a dilution factor of 10, typically 0.0064 μΜ to 64 μΜ, but in some cases starting at lower concentrations (e.g., 0.002 μΜ) and ending at concentrations below 64 μΜ. The remaining 16 wells were used as the negative control. Thus, there were a total of 32 embryos (N = 32) exposed to each concentration and to the control solution from both plates.
The 120 hpf (5 day) behavior was collected using the larval photomotor response assay (LPR) assay for a period of 17 minutes at 28°C. The data were collected in the morning (between 9 and 11 AM) using Viewpoint Zebrabox (www.viewpoint.fr) where the plates were exposed to light (525 LUX) from 0 to 9 minutes and then to dark for the next 8 minutes. The Viewpoint system measures the distance moved (in mm) by the larval fish every 60 seconds, for the whole assay. Data from 0–3 minutes was trimmed for the analysis to account for fish acclimation.
The 5-day movement-time data have to be represented in terms of concentration-response curves for BMD and LEL analyses. Unlike with incidence (binary response) data, where the concentration-response is defined as the proportion of individual embryos that showed a response, it is not straightforward to construct a concentration-response plot from the continuous movement time data. In the case of the movement-time data, appropriate metrics that distinguish between abnormal and normal behavior have to be defined and derived from the data. Subsequently, concentration-response curves have to be constructed for each metric. Therefore, to construct a concentration-response curve for the movement-time data, we summarized the data and derived two endpoints that characterized the abnormal behavior (response) of the fish. The two endpoints were labelled as MOV21 and AUC21.
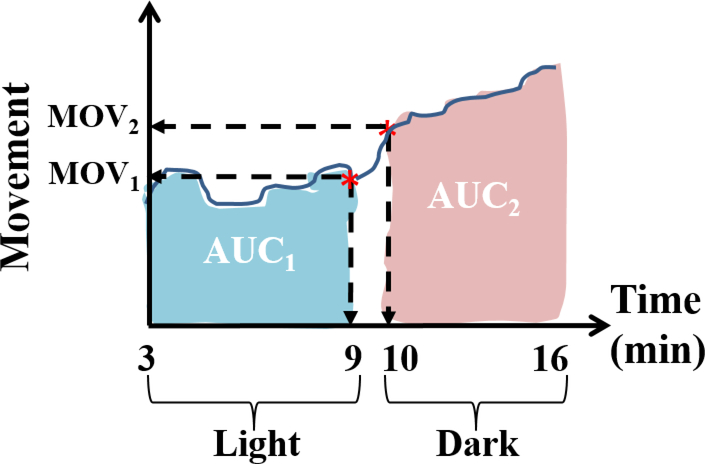
Figure 1 depicts how the movement versus time data for each zebrafish embryo was summarized to derive the MOV21 and AUC21 endpoint values. The MOV21 endpoint represents the change in movement at the light-to-dark transition time point, i.e., MOV21 = MOV2 - MOV1, where MOV1 and MOV2 are the movement values at 9 and 10 minutes, respectively. The AUC21 endpoint represents the change in the area under the movement versus time curve between the dark and light periods, i.e., AUC21 = AUC2 – AUC1, where AUC1 is the area under curve from 3 to 9 minutes (light) and AUC2 is the area under the curve from 10 to 16 minutes (dark). While the MOV21 endpoint characterizes the instantaneous change in the behavior of the fish as it goes from light to dark, the AUC21 endpoint characterizes a more sustained / global change in its behavior. A normal responding fish is expected to show an increase in activity as it goes from light to dark. Hence, a positive value in either of the endpoints would indicate that the fish is behaving normal for that endpoint. Thus, as discussed later, the ability to characterize the abnormal behavior of zebrafish embryos as a binary response (normal/abnormal) based on the sign of MOV21 and AUC21, allowed the conversion of the continuous movement values into a dichotomous data for constructing a concentration-response curve.
Figure 1.
Summarization of the movement versus time data collected at 120 hpf of the zebrafish embryos, using movement values at the light-to-dark transition point and the area under the curve. MOV1 and MOV2 are the movement values at 9 and 10 minutes, AUC1 is the area under the curve obtained during the light period, and AUC2 is the area under the curve obtained during the dark period.
2.3. Incidence data of zebrafish developmental endpoints
As reported in previous works 10, a custom program called the Zebrafish Acquisition and Analysis Program (ZAAP) was used to collect data for four 24 hpf and eighteen 120 hpf developmental endpoints as binary responses (presence or absence of an effect). Their names, labels, and observed chemical effects are listed in Table 1. At 24 hpf, each embryo on a plate was evaluated for the incidence of mortality (MO24), absence of spontaneous movement (SM24), delayed development (DP24), and notochord malformation (NC24). The four 24 hpf endpoints were assessed after subjecting the plates to a photomotor response assay at 24 hpf using the Photomotor Response Analysis Tool (PRAT) 10,12,25. At 120 hpf, the larvae were evaluated for the incidence of morphological effects and mortality after subjecting the plates to a locomotor response assay using Viewpoint Zebralab 26. A total of 18 developmental endpoints were assessed at 120 hpf: the 120 hr mortality or MORT and the 17 morphological endpoints (numbered from 6 to 22 in Table 1). If mortality occurred for an embryo at 24 hpf, the rest of the non-mortality endpoints were not recorded. Similarly, if mortality occurred at 120 hpf, the 120 hpf non-mortality endpoints were also not recorded.
Table 1.
Names and descriptions of the zebrafish endpoints.
| No. | Endpoints | Observed effect | Endpoint labels |
|---|---|---|---|
| Mortality endpoints | |||
| 1. | 120 hr mortality | Cumulative mortality by 120 hours post fertilization (hpf) | MORT |
| 2. | 24 hr mortality | Mortality before 24 hpf | MO24 |
| 24 hr morphology endpoints | |||
| 3. | 24 hr spontaneous movement | Absence of spontaneous movement | SM24 |
| 4. | 24 hr development progression | Delayed development at 24 hpf | DP24 |
| 5. | 24 hr notochord | Notochord malformation (wavy notochord) | NC24 |
| 120 hpf morphology endpoints | |||
| 6. | Axis | Curved or bent axis in either direction | AXIS |
| 7. | Brain | Brain malformations or necrosis | BRAI |
| 8. | Caudal fin | Malformed or missing | CFIN |
| 9. | Circulation | No blood circulation or flow | CIRC |
| 10. | Eye | Eyes malformed, missing or smaller/larger than normal | EYE |
| 11. | Heart | Heart malformation, pericardial edema (fluid around the heart) | PE |
| 12. | Jaw | Malformed | JAW |
| 13. | Otic | Malformed or missing | OTIC |
| 14. | Pectoral fin | Malformed or missing | PFIN |
| 15. | Pigmentation | Lack of pigmentation, overpigmentation | PIG |
| 16. | Snout | Shortened or malformed | SNOU |
| 17. | Somite | Malformed or disorganized, missing somites | SOMI |
| 18. | Swim bladder inflate | Failure of swim bladder to inflate | SWIM |
| 19. | Touch response | Not responsive to touch at 120 hpf | TR |
| 20. | Trunk | Short trunk, malformed or missing | TRUN |
| 21. | Yolk sac | Yolk sac edema, swelling around the yolk sac | YSE |
| 22. | 120 hr notochord | Notochord malformation (wavy notochord) at 120 hpf | NC |
| 120 hr movement (behavioral) endpoints | |||
| 23. | Change in movement at light-to-dark transition time point | Decrease in movement or abnormally high movement during light-to-dark transition | MOV21 |
| 24. | Change in area under movement versus time curve between dark and light periods | Decrease in activity or abnormally high activity from light to dark. | AUC21 |
3. Theory and Calculation
3.1. Construction of concentration-response curves from incidence data
Concentration-response curves were extracted from the incidence (binary response) data of the morphology, mortality and spontaneous movement endpoints (numbered from 1 to 22 in Table 1) for each chemical, as the probability for an embryo or larva to positively respond to the chemical for an endpoint at each tested concentration, d. The response probability, P, was calculated by counting and dividing the number of positively responding embryos, Npos, by the total number of embryos, Ntot, at each concentration; i.e.,
| (1) |
The response probability was calculated based on counts from both plates combined. Given that the mortality at 24 and 120 hpf do not contribute to any value to the other morphology and spontaneous movement endpoints observed at 24 and 120 hpf, data instances of embryos/larvae that died by 24 hpf or 120 hpf were ignored, respectively. Accordingly the total number, Ntot, was corrected for the observed endpoints. Specifically, Ntot was reduced by the number of embryos that died within 24 hpf (MO24 had positive hits) for the four endpoints, DP24, SM24, NC24, and MORT. Similarly, Ntot was reduced by the number of embryos/larvae that died between 24 hpf and 120 hpf (MORT had positive hits) for all other 120 hpf endpoints, except for MORT. The final dataset, comprising the concentration-response values (d, P(d)) for each chemical-endpoint pair, was used in the BMD calculations.
3.2. Construction of concentration-response curves from the 5-day movement versus time data
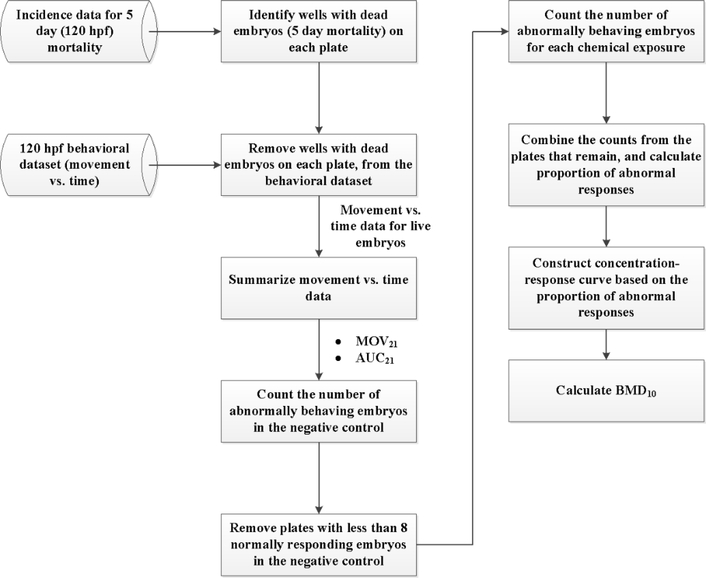
A novel workflow was developed for constructing a concentration-response curve for each chemical from the 5-day movement versus time data, summarized in Figure 2. First, wells with dead fish at 120 hpf were removed from the behavioral dataset, and only the movement data of the live fish were used for constructing the concentration-response curve. The response to chemical treatment (abnormal behavior) was defined based on two metrics, MOV21 and AUC21, which summarized the movement-over-time data (as described before based on Figure 1) compared to the normal behavior of the live larvae in the controls. For each metric (behavioral endpoint), concentration-response curves were constructed based on the number of larvae that responded abnormally in the negative control and with each concentration of the chemical. Control larvae are more active in the dark than in the light, which is signified by a positive value for each endpoint. Thus, the abnormal responses in the control were characterized by negative values of the endpoint (hypoactive fish). For the chemical-exposed larvae, the abnormal responses at each tested concentration were not only characterized by the negative endpoint values (hypoactive fish), but also by positive endpoint values that were outliers with respect to the distribution of the endpoint values of the normally responding embryos in the negative control (hyperactive fish compared to the control). The outliers were detected using the Tukey box-plot algorithm 27: the positive endpoint values of the embryos exposed to the chemicals were regarded as outliers, if they were larger than q3 + w(q3 − q1) or smaller than q1 − w(q3 – q1), where q1 and q3are the 25th and 75th percentiles of the endpoint values of the normally responding control fish, respectively, and w is 1.5.
Figure 2.
Workflow for constructing concentration-response curves from the 120 hpf movement versus time data.
After counting the number of abnormally responding larvae in the control (background response) and at the tested concentrations (chemical effect) on each plate, plates with less than 8 normally responding control fish (< 50%) were removed from the BMD analysis to increase the statistical power for separating the background effects from the chemical effects. The counts from the plates that passed the test (Ntot and Npos − as in equation 1) were added up to calculate the proportion of abnormal responses in the control fish and at each tested concentration of the chemical. Thus, a dichotomous concentration-response data (similar to the morphology/mortality concentration-response) was extracted from the continuous movement time distribution data.
3.3. Calculation of BMD10 from concentration-response data
BMD10 values were calculated by first fitting the concentration-response data of each chemical-endpoint pair, using models recommended in EPA’s technical guidance document for benchmark dose calculations 19. The concentration-response curves, derived from the 24 and 120 hpf morphology/mortality/spontaneous movement endpoint data and from the 5-day continuous movement data, represent the proportion of abnormal fish versus concentration (zero for the negative control), and are therefore, dichotomous in nature. Thus, as per the EPA guidelines for fitting dose-response models to dichotomous data, we fit 10 distinct models to the concentration-response data of each chemical-endpoint pair. These models were logistic, logistic_bgr, probit, probit_bgr, log_logistic, log_probit, gamma, Weibull, quantal linear, and multistage 2. Curve fits were done with actual concentration as well as log-transformed concentrations (see Supporting Information for calculation method) as the predictor. Thus a total of 20 different models were fit to each concentration-response curve, and the best-fit model was selected to compute the BMD10 value.
3.4. Model fitting, evaluation, and selection
Several statistical criteria were applied to evaluate the concentration-response curves and to select the best fit model. The workflow used for assessing the quality of each model’s fit to the concentration-response data of each chemical-endpoint pair is given in Figure S1. The fits were attempted only for chemical-endpoint pairs with concentration-response data that had at least 3 good concentration points (including the control). A good concentration point was one that had at least 8 embryos (Ntot) from both plates combined for that concentration, and each plate should have had at least 8 embryos with normal development in the negative control. Thus, plates that did not meet the requirement for the minimum number of normal embryos for the negative control (for each chemical-endpoint pair) were not considered for concentration-response modeling. This minimum requirement was applied to increase the statistical power for distinguishing the chemicals effects from the background effects − a threshold of 50% was considered as a reasonable choice. For example, there were 7 chemicals (cyanazine, fenarimol, D-mannitol, tert-butylbenzene, didecyldimethylammonium chloride, triisononyl trimellitate, and sodium xylenesulfonate) that had at least one morphology/mortality endpoint for which the number of normally responding embryos in the negative control on both plates were < 50%; this resulted in excluding 82 chemical-endpoint pairs (curves) from the BMD analysis. Additionally, there were chemicals did not have enough data points in at least one morphology/mortality endpoint for fitting, even though the minimum requirement for the negative control was satisfied; this resulted in excluding another 36 chemical-endpoint pairs from the analysis.
The quality of the fit for each model was evaluated based on Chi-square test: the fit was considered good if the p-value > 0.1 (Good-Fit); ok if the p-value ≤ 0.1 and R2 > 0.7 (OK-Fit), or bad if p-value ≤ 0.1 and R2 < 0.7 (Bad-Fit). The test returns a p-value that is calculated based on the difference between the log-likelihood of the fitted model and that of a perfectly fitted model (which is the data itself). A model with a p-value > 0.1 is therefore interpreted as one having a (significantly) high likelihood to fit the responses perfectly. Thus, when there are more than one model with a p-value > 0.1, the one with the highest log-likelihood value was selected as the best-fit model for the BMD calculations, and the corresponding fit is considered a Good-Fit. If no models were found to have a p-value > 0.1, then the maximum log-likelihood criterion was not used as the model-selection criterion because none of the models are likely to give a good fit. Hence, a second selection criterion was applied to select a model (that is “not the best model with a good fit” but gives an “ok fit”) based on the goodness-of-fit measure, R2 value > 0.7, and the corresponding fit was considered an OK-Fit. If none of the models fit the data (all were Bad-Fit models), then a BMD value could not be calculated for that chemical-endpoint pair. Afterwards, using the Chi-square test, each concentration-response data was evaluated for the presence of a significant concentration-response relationship compared to the background response.
3.5. BMD10 calculations
The BMD10 value is the concentration that corresponds to a 10% extra risk in response compared to the negative control. For dichotomous data, such as those analyzed in this work, the extra risk or benchmark response (BMR), is defined as
| (2) |
where P(BMD) is the proportion of affected embryos at the BMD, and P(0) is the proportion in the negative control group. Equation 1 can be rearranged to yield:
| (3) |
An extra risk of 10% corresponds to a BMR of 0.1, and the BMD10 value corresponding to the probability of response, P(BMD10), is numerically determined from the model that best fits the concentration-response curve. It is noted that the models and the definition of risk associated with BMR vary depending on whether the responses are binary or continuous. For binary response data (a.k.a. quantal / dichotomous data), there are two definitions of risk: additional risk and extra risk. Both definitions are based on the proportion (P) of individuals affected at each concentration (d) compared to the background response. The BMR for additional risk is defined as BMR = P(BMD) - P(0), whereas, the BMR for extra risk is defined as shown in Equation 3. For making comparisons across chemicals or endpoints, an extra 10% risk (BMR = 0.1) has been recommended as a standard reporting level for quantal data. The 10% response level has been traditionally used because it is close to the limit of sensitivity in most cancer bioassays and in non-cancer bioassays of comparable size 19. Therefore, we selected extra 10% risk as the BMR to calculate the BMD values (BMD10). Detailed guidance on data evaluation, endpoint and BMR level selection, model selection and fitting, and BMD computation, can be found in the EPA BMD Guidance Document 19.
To calculate the BMD10 values of 1,078 (1,060 unique) chemicals for each of the 24 endpoints, 25,872 concentration-response curves (1,078 chemicals × 24 endpoints) were constructed and 20 models were fit to each curve (1,078 chemicals × 24 endpoints × 20 endpoints = 517,440 fits). In order to perform the calculations efficiently in a high-throughput manner, we implemented and automated the various steps of the BMD10 calculation workflow in Matlab. The steps included processing the plate datasets of the 22 endpoints with binary response values (morphology and mortality at 24 and 120 hpf; spontaneous movement at 24 hpf) and of the two 120 hpf behavioral endpoints (MOV21 and AUC21); summarizing the movement-time data; constructing the concentration-response curves from plate data; fitting the models; and, evaluating and selecting the best models for computing the BMD10 values. The BMD calculations were performed in Matlab installed on a 64-bit Windows 7 desktop that had 16 GB RAM and two 1.60 GHz Intel Xeon® 6-Core processors. To speed up the calculations, we designed the Matlab code to run the calculations for multiple endpoints in parallel across multiple processors.
3.6. Imputation of BMD10 values for clustering and ranking of chemicals
BMD10 values between 10−5 and 103, derived from the good and OK fits, were categorized as good BMD10 values. Values below 10−5 and above 103, derived from good and OK fits, cannot be ascertained without visual inspection and re-fitting of the data, and/or without repeating the assay. But these values indicate whether a chemical is active or inactive for a given endpoint, respectively. Therefore, for clustering and ranking analysis, values below 10−5 and above 103, were imputed the values, 10−6 and 104, respectively. Values for chemical-endpoint pairs with bad or no fits, or with any negative values, were imputed the value, 105. Finally, chemicals that had at least one endpoint with a good BMD10 value (i.e., between 10−5 and 103), were considered for clustering and ranking analysis, and for identifying the most sensitive endpoints.
3.7. Automation and manual iteration of the BMD analysis workflow
In the fully automated workflow, an attempt was made to fit each curve and to calculate a BMD10, without manual / visual inspection of the slope of the curve. But there were chemicals for which a BMD10 value could not be computed because of a bad fit. The bad fits can be improved by visualizing the concentration-response plots to see if they can be re-fit well after removing one or two data points, especially in the concentration dose range. Since it is not practically possible to do a manual inspection for all the endpoints, we considered to only re-fit the data with bad fits for the two 120 hpf behavioral endpoints, MOV21 and AUC21 − after visually inspecting the fits and removing one or two data points in the high concentration range. If the re-fit was good or OK and if the corresponding BMD10 value was in the range between 10−5 and 103, then the fit and the BMD10 value was accepted for the clustering and ranking analysis.
3.8. LEL calculations
As done for the BMD analysis, the incidence data from each plate of a chemical was required to have at least 8 normally responding control fish for a given endpoint, in order to include the data in the LEL calculations. Additionally, only those concentration points with at least 8 total embryos (from both plates combined) were selected for the calculating a LEL value. The LEL values were estimated using the same statistical approach described in Truong et al10. Since the concentration-response data comprised of binary responses from Ntot(d) embryos for each concentration d, the LEL value, x, was estimated for each chemical-endpoint pair from the binomial distribution function as
| (4) |
where nc is the number of controls and pc is the observed incidence (positive hits) in the controls, and P(X > x) is the probability of finding a significant response above the threshold concentration x (LEL). Thus the estimated LEL value, x, was the lowest concentration at which the incidence exceeded the background incidence with a p-value ≤ 0.05. Some chemical-endpoint pairs did not have an LEL value in the tested concentration range, if none of the tested concentration groups showed an effect significantly different from the control group − these were recorded as ‘NA’. LEL values for chemical-endpoint pairs with zero incidence values for all concentration groups, including the control were recorded as ‘No Response’, while of those with no concentration points with minimum 8 total embryos were recorded as ‘No Data’.
3.9. ToxCast in Vitro Assay Data
The in vitro assay data (AC50 values) of the 1,060 ToxCast chemicals, and information about the assay study design and the biological process targets of each assay endpoint were obtained from files downloaded from the website, http://epa.gov/ncct/toxcast/data.html. These files were ‘Dashboard_Export_04–19-2015_08–22-27.csv’, ‘ToxCast Assay Annotation Study_Design_info_20141021.csv’, and ‘ToxCast Assay Annotation Assay_Target_Info_20141021.csv’, respectively. The assay data reports AC50 values of chemicals for 821 ToxCast in vitro assay endpoints. In this work, the ToxCast in vitro assay endpoints were grouped according to their biological process targets (if recorded in the dataset). Consequently, 788 endpoints were grouped based on 11 biological process targets: cell proliferation (30 endpoints), cell cycle (24 endpoints), cell death (31 endpoints), protein stabilization (10 endpoints), mitochondrial depolarization (7 endpoints), oxidative phosphorylation (12 endpoints), regulation of transcription factor activity (104 endpoints), regulation of gene expression (149 endpoints), cell morphology (2 endpoints), regulation of catalytic activity (284 endpoints), and receptor binding (135 endpoints).
3.10. Ranking chemicals based on their activity in ToxCast in vitro assays and zebrafish in vivo assays
The Toxicological Priority Index (ToxPi) software 28 was used to rank chemicals based on their AC50 values for the 788 ToxCast in vitro assay endpoints and based on their BMD10 values for the most sensitive zebrafish endpoints (MOV21 and AUC21). For the ranking, the AC50 and BMD10 values, were scaled as − log10(AC50) + 6 and −log10(BMD10) +6, respectively. Only chemicals with 10−5 < BMD10 < 103 from Good/OK-Fit models and active in ToxCast in vitro assays were considered in the ToxPi ranking analysis. The 788 ToxCast in vitro assay endpoints were grouped according to their biological process targets and each group was considered as one ToxPi slice. The MOV21 and AUC21 zebrafish endpoints were considered as a separate ToxPi slice. In this work, all the slices were equally weighted, irrespective of the number of endpoints that belonged to each slice.
4. Results
4.1. Benchmark dose analysis of the 24 and 120 hpf morbidity and mortality endpoints
The BMD10 values and the best-fit models of each chemical-endpoint pair, corresponding to the 1,078 chemicals and the 22 endpoints (mortality and morbidity), are listed in Tables S1 and S2, respectively. A total of 23,716 concentration-response (C-R) curves (1078 chemicals × 22 endpoints) were analyzed through the BMD workflow (Figure S1). Some of the curves were not fit to the data due to insufficient number of concentration points that satisfied the minimum number of embryos requirement (8 negative controls per plate and 8 for a chemical concentration effect from both plates combined). But for the ones that could be fit, the quality of the fit (whether it was good, ok, or a bad fit) was evaluated based on the statistical criteria shown in Figure S1 (as described in Methods). An example concentration-response plot for each fit type for the chemical cymoxanil is illustrated in Figure S2. After calculating the BMD10 values, the 23,716 C-R data sets were grouped into 10 categories according to the various outcomes of the BMD analysis, such as the quality of the fits (good, ok, bad, or not enough points to fit), range of BMD10 values (good, uncertain, inactive), and presence of significant concentration-response. The categories, the C-R counts, and the classification rules used for defining each category are listed in Table S3. Among the rest of the chemical-endpoint pairs, there were 1,120 (4.7%) C-R curves that gave a bad fit; out of which, 39 were found to be ones that did show any significant concentration-response. And there were only 25 C-R curves that gave a good/ok fit but the corresponding BMD10 values were uncertain as they were in the range 0 to 10-5. By automating the workflow and running the analysis in parallel on 10 processors across the 22 endpoints, it took nearly 2 days to complete the fitting of the 23,716 concentration-response plots to 20 different models (total 474,320 fits). These results demonstrate that the BMD analysis workflow was applied in a high-throughput manner to quickly evaluate and fit the concentration-response data of the 24 and 120 hpf morbidity and mortality endpoints.
4.2. Benchmark dose analysis of the 120 hpf behavioral data
The 120 hpf behavioral metrics, MOV21 and AUC21, were analyzed separately from the morbidity and mortality data, to determine if the fish behaved abnormally or did not change in lighting conditions (as explained in the Methods). Subsequently, a dichotomous concentration- response curve was constructed for each endpoint, based on the number of abnormal fish in the control (background response) and for each chemical exposure concentration (as shown in the workflow, in Figure 2). Since the concentration-response curves were dichotomous, the same models used for the fitting the concentration-response curves of the morbidity and mortality endpoints, were also used for fitting the MOV21 and AUC21 C-R curves. Figure S3 shows an example comparing the concentration-response plots and BMD10 values of the MOV21 and AUC21 metrics for the chemical flusilazole.
Application of the BMD analysis workflow (Figure S1) resulted in 78 and 104 chemicals with bad fits for the MOV21 and AUC21 endpoints, respectively. To know if some of these chemicals could be redeemed with a good/ok fit and a good BMD10 value, a second iteration of the BMD workflow was performed for these chemicals after visualizing their concentration-response plots and removing some data points in the high concentration range. Consequently, 27 and 32 chemicals were redeemed with good/ok fits and good BMD10 for the MOV21 and AUC21 metrics, respectively. This iterative procedure resulted in a final number of 596 and 591 chemicals with good/ok fits and good BMD10 values for the MOV21 and AUC21 metrics, respectively. The final computed BMD10 values and the best-fit models of the 1,078 chemicals for both metrics are provided in Tables S1 and S2. Based on the C-R fit classification rules (defined in Table S3), there were 772 chemicals with good/ok fits for the MOV21 metric; out of which, 596 chemicals had good BMD10 values, 62 chemicals were inactive, and 114 active chemicals had uncertain BMD10 values. For the AUC21 metric, there were 762 chemicals with good/ok fits; among which there were 591 chemicals with good BMD10 values, 65 inactive chemicals, and 106 active chemicals with uncertain BMD10 values. The number of curves for each C-R fit classification are listed in Table S3. These results demonstrate for the first time how time-series data of 120 hpf fish movements can be converted into dichotomous concentration response curves for POD analysis.
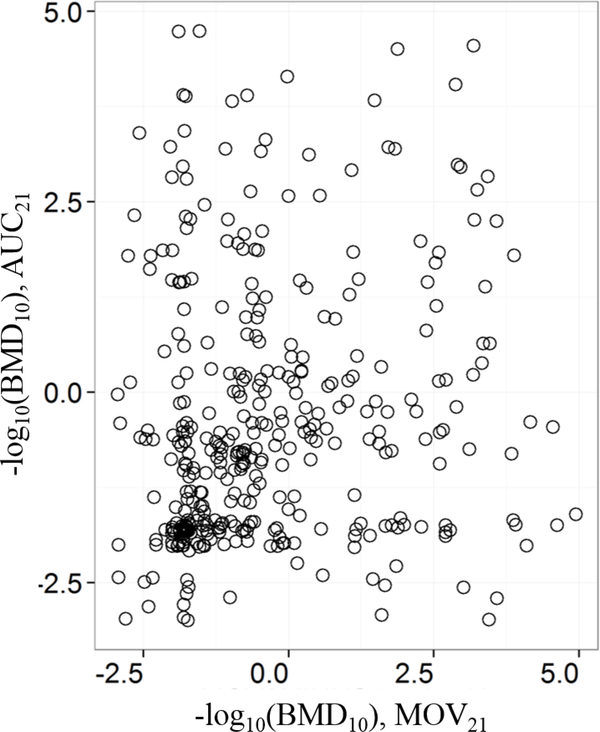
Next, the concordance between the MOV21 and AUC21 metrics was analyzed by counting the number of chemicals common to both endpoints for each C-R fit classification. The contingency table is shown in Table S4. Between the two metrics, there were 379 chemicals that had a good BMD10 value, but there was no correlation between them (Figure 3). The Pearson’s correlation coefficient was 0.165. This lack of correlation suggests that the two metrics − one representing the instantaneous abnormal behavior at light-to-dark transition (MOV21) and the other representing a more sustained abnormal behavior (AUC21) − are two distinct signatures (endpoints) of zebrafish behavior and possibly associated with two different mechanisms of developmental toxicity.
Figure 3.
Comparison between the MOV21 and AUC21 BMD10 values of 379 chemicals that had good BMD10 values for both endpoints.
4.3. Differences between 120 hpf behavioral endpoints and the other endpoints
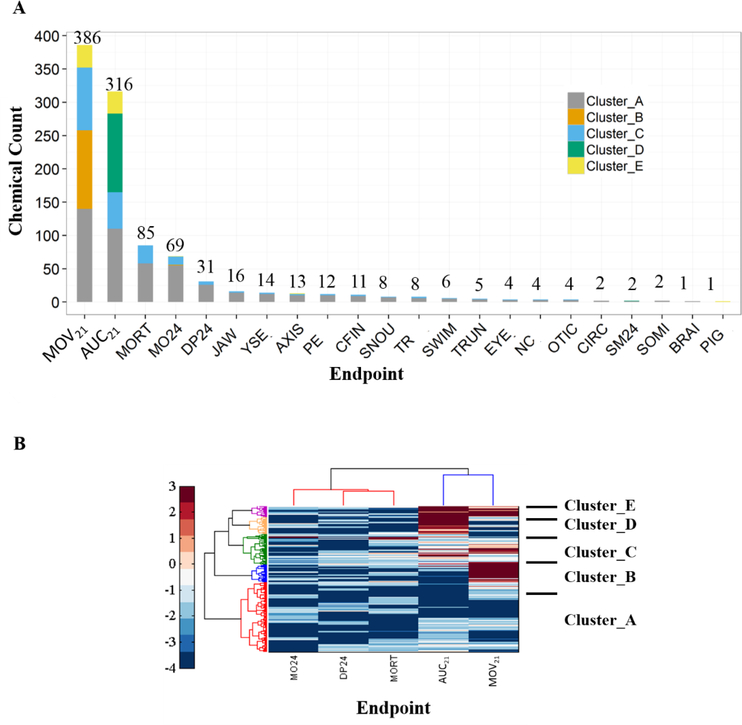
Although no correlation was found between the MOV21 and AUC21 endpoints, they clustered together but separately from the 24 hpf and 120 hpf endpoints of morbidity and mortality (Figure S4), signifying that the behavioral endpoints contain novel information about the adverse effects of each chemical. Furthermore, the 120 hpf behavioral signatures turned out to be the most sensitive (i.e., they have higher likelihood of a hit) of all the endpoints, since the BMD10 values of more than 60% of the chemicals were lower for these endpoints than for the morphology/mortality endpoints (Figure 4A). Specifically, 724 chemicals were found to be the most active through the behavioral endpoints (397 for MOV21 and 327 for AUC21).
Figure 4.
Chemical clustering and ranking based on the morphology / mortality and 120 hpf behavioral endpoints, and identification of the most sensitive endpoints: (A) Number of chemicals with the minimum BMD10 value for each endpoint and their cluster membership (4B). The number is shown above each bar. (B) Hierarchical clustering based on −log10 (BMD10) values of 1,006 chemicals that had at least one endpoint with a good BMD10 value (10−5 ≤ BMD10 ≤ 103), among the top 5 sensitive endpoints. Chemical clusters are labelled from A to G as shown. Heat map color bar represents −log10 (BMD10) values.
The top five sensitive endpoints were MOV21, AUC21, MORT, MO24, and DP24 (Figure 4A). There were 1,000 chemicals that had a good BMD10 value for at least one of these endpoints. Clustering analysis based on these five endpoints revealed 5 chemical clusters, which are labeled from A to E in Figure 4B. The distribution of the members of each chemical cluster by the endpoint with the minimum BMD10 value is indicated on Figure 4A. Cluster D chemicals (e.g. mesotrione and busulfan) have higher potency for causing a sustained abnormal behavior in zebrafish at 120 hpf (AUC21); whereas, Cluster B chemicals (e.g. butanoic acid and hydroquinone) have higher potency for affecting the normal ability of zebrafish to instantly respond to change in lighting conditions (MOV21). While Cluster E chemicals (e.g. thiram and etoxazole) are more potent in affecting the two behavioral endpoints, Cluster A (e.g. anthracene and oryzalin) and Cluster C chemicals (e.g. amiodarone hydrochloride and fenamiphos) are potent in affecting all endpoints.
We note that the behavioral data analysis results are based on the abnormal responses of both malformed and non-malformed fish. It is possible that a fish is behaving abnormally simply because it is malformed; therefore, excluding them from the analysis may exclude the effect of malformation on fish behavior. However, we cannot be certain that malformation is the only reason for the observed abnormal behavior in malformed fish. Further well-by-well analysis of the data for each concentration of a chemical revealed that not all malformed fishbehave abnormally and many non-malformed fish do behave abnormally. At least 90% of behavioral response based on MOV21 and AUC21, come from non-malformed fish in more than 75% and 72% of the chemical-concentration pairs, respectively. Thus, while abnormal behavior in a non-malformed fish can be due to other (neurological) effects, abnormality in a malformed fish can be due to malformation and/or neurological effects. Since, the possibility of neurological effects in malformed fish cannot be ruled out, it is safer to include abnormal malformed fish in the behavior analysis than to exclude them. Thus, with the behavioral endpoints being the most sensitive, including the malformed fish in the behavioral data analysis allows for high-throughput screening and ranking of chemicals based on abnormal fish movements without knowledge about what causes the abnormal behavior.
Nevertheless, to determine how many chemicals would be most active through abnormal behavior of non-malformed fish alone, we re-analyzed the data and calculated BMD10 values for MOV21 and AUC21 after removing the abnormally behaving malformed fish. The calculated BMD10 values were also highly correlated to those calculated earlier based on abnormal responses of both malformed and non-malformed fish (see Figure S5). Analysis show that the behavioral endpoints continue to be the most sensitive endpoints. The abnormal behavior of non-malformed fish can alone reveal 657 (386 for MOV21 and 271 for AUC21) chemicals as the most active, out of which 626 were revealed as active when abnormal malformed fish were included (see Venn diagram in Figure S6). Thus, the behavioral data can be analyzed with and without the abnormal malformed fish, but it is essential to include them to capture the full behavioral response since other (neurological) effects might also be contributing to their abnormal behavior.
4.4. Differences between BMD and LEL approaches
The LEL values for each chemical-endpoint pair were also calculated and are listed in Table S5. The BMD10 values of the behavioral endpoints were compared with the respective LEL values, and the results are shown in Figures 6A and 6B. The limitations of a minimal effect level versus the BMD point of departure is clearly illustrated by a ‘laddering’ created due to the restriction of LEL values to the experimental concentrations (Figure 5A & B), while the BMD values provide better separation, precision, and rank order. The minimum BMD10 (BMD10 of the most sensitive endpoint among the 22 morphology/mortality endpoints) for each chemical was also compared to its minimum LEL value, as shown in Figure 5C. Using the LEL approach, most of the chemicals defy ranking because their minimum LEL values are equivalent, whereas using the BMD approach, these chemicals could be ranked for hazard potential. A direct one-to-one correlation between the BMD10 and LEL values cannot be evaluated, as the LEL values (by definition) do not necessarily correspond to the extra 10% risk in the response level as the BMD10 values. Therefore, the LEL value of a chemical can be greater or less than its BMD10 value, which would correspond to an extra risk that is less or greater than 10%, respectively. Similarly, a chemical without an LEL value can have a BMD10 value in the tested concentration range, or vice-versa. Consequently, chemicals that are inactive based on LEL can be active based on BMD10 in the tested concentration range or vice-versa; and this is illustrated in Figures S7A and S7B for chemicals with BMD10 values (including the imputed values) in the range 10−6 to 104 for the MOV21 and AUC21 endpoints, respectively. The number of chemicals without LEL values was found to be higher for those with high BMD10 values. At the same time, there are also chemicals with very low LEL values that correspond to an extra risk greater than 10%. These differences between the LEL and BMD10 values result from the fact that the calculation of the BMD10 values is influenced by all the points of the fit concentration-response curve (thus the slope) rather than the potential to be influenced by a single spurious point as in the LEL approach.
Figure 6.
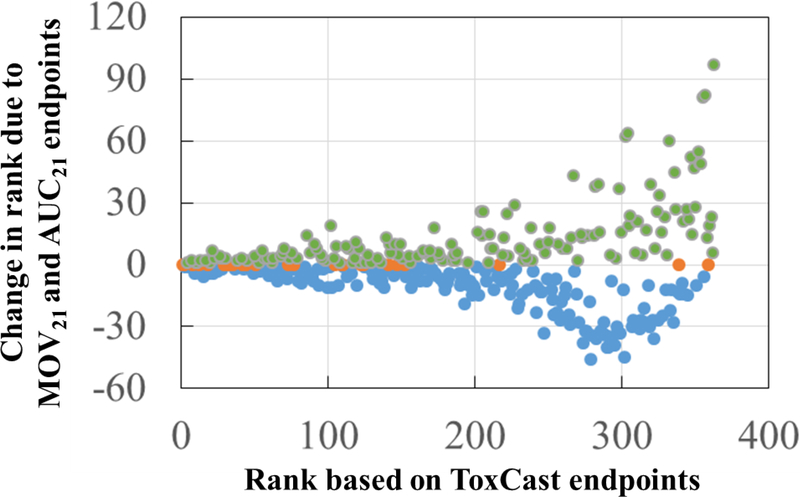
Effect of the 120 hpf zebrafish behavioral endpoints (MOV21 and AUC21) on the ToxPi ranking of 363 active chemicals using results from in vitro ToxCast assays with good BMD10 values (10−5 ≤ BMD10 ≤ 103) for the zebrafish endpoints. Positive, negative and zero change in rankings are plotted as filled green, blue, and orange circles, respectively.
Figure 5.
Comparison between LEL and BMD10 values based on: (A) MOV21; (B) AUC21; and, (C) the most sensitive endpoint among the 22 morphology/mortality endpoints.
4.5. Differences in chemical activity based on ToxCast in vitro endpoints and the 120 hpf Zebrafish behavioral endpoints
We found 865 active chemicals common between the ToxCast in vitro and zebrafish behavioral assay endpoints. A chemical was considered active towards a behavioral zebrafish endpoint (MOV21 and AUC21) based on the BMD10 values: active if BMD10 ≤ 103 and inactive if the BMD10 could be calculated but > 103. The same chemical was considered active or inactive towards a ToxCast in vitro endpoint based on the labels (Active or Inactive) listed in the ‘Activity Call’ column of the ToxCast in vitro data set (Dashboard_Export_04–19-2015_08–22-27.csv). The high overlap of active chemicals indicates a high concordance in the chemical activity observed between the ToxCast in vitro endpoints and the 120 hpf zebrafish behavioral endpoints.
However, there are 130 ToxCast-active chemicals that are inactive in Zebrafish (Table S6), and 13 ToxCast-inactive chemicals that are active in zebrafish (Table S7). The lack of correlation between ToxCast in vitro and Zebrafish behavior assays for the 130 chemicals seems to suggest that these chemicals may need to be re-evaluated at higher concentrations or perhaps they were not bioavailable in the exposure system. Alternatively, it is possible that the in vitro molecular interactions of these chemicals are not related or relevant to adverse zebrafish behavior, and not all molecular interactions necessarily lead to adverse effects at the organism level. For the 13 chemicals that were inactive in ToxCast in vitro assays, it seems that their effect on the zebrafish behavioral endpoint may be associated with a biological mechanism that is not currently captured by the ToxCast in vitro assays.
After comparing the list of active and inactive chemicals, we used the ToxPi software 28 to look at how the addition of the 120 hpf behavioral endpoint chemical activity data to the in vitro ToxCast data affects the ranking of chemicals. Only those chemicals with good BMD10 values (10−5 ≤ BMD10 ≤ 103) from Good/OK-Fit models and active in ToxCast in vitro assays were selected for the ranking. A total of 363 chemicals were ranked in descending order of their ToxPi scores, with lower ranking numbers corresponding to higher ToxPi scores (higher toxicity). Thus, a chemical with a ranking of 1 has the highest ranking and the highest toxicity. Kendall’s tau rank correlation test showed that the two ranked lists − with and without the 120 hpf behavioral endpoints − were highly correlated (correlation coefficient, tau = 0.914, p-value < 0.01), although the individual chemical rankings changed as shown in Figure 6. For the chemicals that fell in the top 2/3 of the rankings (above 242), there were minimal changes with addition of the zebrafish behavioral data (−23 to +29). However, for the chemicals that initially ranked in the bottom 1/3 (below 242) using ToxCast in vitro data alone, the rankings dramatically changed with addition of the zebrafish MOV21 and AUC21 endpoints (−46 to +97). For example, the ranking for quinoline increased from 357 to 275 (357 – 275 = 82, change is+82) when the MOV21 and AUC21 BMD10 values (0.0236 μΜ and 0.0733 μΜ) were included in the ToxPi analysis (Figure S8). Nevertheless, the highest ranking that was attained due to the behavioral endpoints by a low ranking chemical (from the bottom 1/3 chemicals) is 229, which is still lower than the 228 ToxCast-based high ranking chemicals. These results demonstrate that zebrafish behavior as an assay of bioactivity can significantly influence hazard rankings of chemicals when used in addition to ToxCast data.
5. Discussion
Establishing a reference dose for adverse outcomes is a fundamental step in developing risk management decisions 29. Typically, this is derived using the lowest value of a relevant endpoint’s point of departure (POD) from normal (e.g., LEL or BMD). This work has addressed the gaps and challenges of using high-throughput zebrafish screening data with point-of-departure (POD) methods for ranking and assessing the toxicity of chemicals.
To enable high-throughput calculation of BMD values from embryonic and larval zebrafish concentration-response data, we established and automated a BMD calculation workflow (Figure S1). A key component of the workflow is to determine which models to select for fitting the concentration-response curves and to identify the best-fit model for deriving the BMD value that corresponds to a specified benchmark response (BMR). All steps related to the data processing and the BMD calculations were implemented in a Matlab code. To speed up the calculations, the Matlab code was designed to split and run the calculations for multiple endpoints at once (in parallel) using multiple processors. Thus, the various endpoints related to zebrafish morphology, mortality, and movement (behavior) could be evaluated using the BMD workflow for all the 1,078 chemicals, in a high-throughput manner. Other software programs are available to calculate BMD values − the Benchmark Dose Software (BMDS) 18 and the BMDExpress tool30, but we couldn’t use them in a programmatic way to fit with our overall workflow, and to fulfill our need to automate the BMD workflow for high-throughput data processing and calculation of BMD values with parallel computing capabilities. Hence, we developed our own Matlab code for calculating BMD values. However, we have used the same dose-response models and model selection criteria described in EPA’s BMD technical guidance document19 for our BMD calculations. The dichotomous nature of the concentration-response curves made it possible to apply the same workflow (Figure S1) and curve-fitting models for all chemical-endpoint pairs. A standalone software program that implements the workflow is available for download at https://github.com/pnnl/ZebrafishBMD.
The BMD calculations are subject to the statistical criteria applied at various steps of the BMD analysis workflow. These include 1) setting the requirement for the minimum number of normal control fish to be available on a plate (minimum 8) for a given chemical-endpoint effect; 2)setting the requirement for the minimum number of good concentration points in the concentration-response curve so that a fit can be attempted (3 including the control group); and, 3) the significance test criteria used for determining whether the model fit was good, ok, or bad. Changing one or more of these criteria can modify the results. For instance, raising the minimum number requirement for the normal controls on each plate will result in excluding more chemicals from the BMD analysis. In the current criterion, the minimum number was set to 8 (50%) for both plates, which resulted in excluding 62 and 68 chemicals for the MOV21 and AUC21 endpoints, respectively. Similar conclusions can be arrived for morphology and mortality endpoints, where 82 chemical-endpoint pairs (of 7 chemicals) were excluded from the present BMD analysis. For fitting the concentration-response curve, at least 3 good concentration points (including the control) were required, and a good concentration point was defined as one that had a minimum of total 8 embryos. This criterion was always met for the control since only plates with a minimum of 8 normal embryos for the control were considered in the analysis. The criterion was however, not stringent compared to the minimum requirement imposed for the control; since there were only 36 chemical-endpoint pairs among the morphology and mortality endpoints (Table S3) and only 1 chemical for the MOV21 endpoint (Table S3), which did not have enough points for fitting a concentration-response curve. In fact, most of the concentration- response curves that were fit had at least 4 good concentration points.
The novel method that we developed for constructing concentration-response curves from the 120 hpf zebrafish movement time-series data enabled the use of these data for POD analysis. The method is based on the fact that a normal zebrafish would show increased activity in the dark compared to that in the light. Accordingly, we defined two novel signatures (MOV21 and AUC21) to characterize the abnormal responses from the data. The MOV21 endpoint defined difference between the fish movements at the transition time point from light to dark, and the AUC21 endpoint defined the difference between the area under the movement-time curve (AUC) of the dark period (10–16 minutes) and that of the light period (3–9 minutes). While the MOV21 endpoint characterized the instantaneous change in fish movement, the AUC21 endpoint characterized a more sustained change in movement. Lack of correlation between the AUC21 and MOV21 values seems to suggest that the biological mechanisms are different for the two behavioral endpoints. In addition to these two endpoints, we considered defining an endpoint based on the difference between the time-averaged movement values of the dark and light periods. But a visual comparison of the distribution of the movement values between the light and dark periods seemed to suggest that averaging the movement values would lead to loss of information, and the average values would lack the sensitivity to accurately distinguish between normal and abnormal responses.
To construct a concentration-response curve for each behavioral endpoint, a response has to be defined based on the endpoint values. One way to construct the curve is to average the endpoint values of all the fish exposed to a given concentration (in solution), and then use the average value as the response. Since the endpoint values can be positive or negative, averaging them can lead to loss of information, and the resulting average value wouldn’t be sensitive to chemical effects. Hence, it is not advisable to use average values. Since by definition, the positive and negative endpoint values for the control correspond to fish with increased (normal behavior) and decreased activity (abnormal behavior), respectively, we modeled the response as a binary response variable. Specifically, the negative values for the control and for each chemical concentration group signified that the zebrafish response was abnormal (hypoactive fish) to change in light conditions, whereas the positive values signified normal response. However, if the positive value for a given concentration group was an outlier with respect to the distribution of the positive values of the control, then the response was considered abnormal (hyperactive fish), else normal. By counting the number of fish with normal and abnormal behavior in the control and in each chemical concentration group, the proportion of abnormally responding fish was calculated to construct the dichotomous concentration-response curves. Thus, the same dichotomous models and BMR definition (10% extra risk), used for the concentration-response curves of the morphology/mortality endpoints, was also used for those of the behavioral endpoints.
Compared to LEL, the BMD10 values provided greater precision for clustering chemicals and identifying the most sensitive zebrafish endpoints. The BMD approach (unlike the LEL) captured the resolution of the individual fish movements over time. The LEL value for a chemical-endpoint pair can be greater or less than the BMD10 value; and, chemicals with no LEL value can have a BMD10 value in the tested concentration range, or vice-versa. These differences between BMD and LEL result from the fact that LEL values do not necessarily correspond to the 10% extra risk as the BMD10 values, and that the BMD10 calculation is influenced not only by how well the concentration-response curve is fitted but also by all the points of the fitted curve (hence the slope of the curve) rather than a single point as in the LEL approach. Consequently, the classification of active and inactive chemicals can vary between the two approaches.
The BMD approach allowed us to rank and identify the most sensitive zebrafish endpoints (MOV21, AUC21, MORT, MO24, and DP24), with MOV21 and AUC21 being the top two. These endpoints could form the minimum set of endpoints to evaluate in zebrafish toxicity screening of chemicals, thereby reducing the burden of the analysis and interpretation for 24 endpoints. The high sensitivity of the behavioral endpoints suggests that the addition of other behavior driven endpoints in the future would provide a more comprehensive profile of a chemical. The behavioral data can be analyzed with and without the abnormal malformed fish. However, it is not safe to exclude the abnormal malformed fish from the analysis by simply assuming that their abnormal behavior is due to their malformation, because it is not known if neurological effects might also be contributing to their abnormal behavior. Therefore, additional endpoints that specifically represent neurological effects could be used to confirm whether the abnormal behavior of malformed fish are also associated with neurological effects.
Concordance in the number of active and inactive chemicals between ToxCast assay endpoints and 120 hpf zebrafish behavioral endpoints was observed for 865 chemicals; however the ranking was not the same (but correlated) between the two. In particular, chemicals with lowest rankings based on ToxCast in vitro endpoints, such as quinolone, showed significant change in hazard ranking when the zebrafish MOV21 and AUC21 endpoints were integrated into a ToxPi analysis. Hence, the ranking of chemicals using in vitro assays may not be a reliable predictor of the ranking in vivo for chemicals with lower potency.
6. Conclusions
BMD as a POD approach, when applied to high throughput in vivo screening assays, can provide additional and relevant information for chemical risk assessment. The methods developed in this work allow for the identification of novel signatures of abnormal fish responses from behavioral time series data using high-content imaging approaches. By constructing dichotomous concentration-response curves for these signatures, this approach can be used, adapted, or extended for high-throughput zebrafish toxicity screening and hazard ranking of new chemicals. A software program that implements the BMD workflow and methods developed in this work is freely available for community use at https://github.com/pnnl/ZebrafishBMD.
Supplementary Material
Highlights.
Benchmark dose values have greater precision for ranking chemicals than LEL values.
Benchmark dose analysis allows to identify the most sensitive zebrafish endpoints.
Zebrafish movement versus time data reveal new signatures of chemical toxicity.
Behavioral signatures are the most sensitive to chemical toxicity in zebrafish.
In vitro rankings do not reliably predict in vivo rankings of low potency chemicals.
Acknowledgements
This work was supported by the National Institute of Environmental and Health Sciences at the National Institutes of Health [P42 ES016465, P30 ES000210] and the Environmental Protection Agency [R835168]. The authors gratefully acknowledge Justin Teeguarden for providing valuable comments and feedback on the manuscript. The Pacific Northwest National Laboratory is operated for DOE by Battelle Memorial Institute under contract DE-AC05–76RL01830.
Footnotes
Appendix A. Supplementary Information
Supplementary_methods_and_figures.docx
Supplementary_Tables.xlsx
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hartung T A toxicology for the 21st century--mapping the road ahead. Toxicological sciences : an official journal of the Society of Toxicology 109, 18–23, doi: 10.1093/toxsci/kfp059 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Judson R et al. The toxicity data landscape for environmental chemicals. Environmental health perspectives 117, 685–695, doi: 10.1289/ehp.0800168 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.RSC. A brief guide to REACH 2008:What you need to know., (2008).
- 4.NRC. Toxicity Testing in the 21st Century: A Vision and a Strategy., (2007).
- 5.Dix DJ et al. The ToxCast program for prioritizing toxicity testing of environmental chemicals. Toxicological Sciences 95, 5–12, doi:DOI 10.1093/toxsci/kfl103 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Kodell RL & Chen JJ On the use of hierarchical probabilistic models for characterizing and managing uncertainty in risk/safety assessment. Risk Analysis 27, 433–437, doi:DOI 10.1111/j.1539-6924.2007.00895.x (2007). [DOI] [PubMed] [Google Scholar]
- 7.Padilla S et al. Zebrafish developmental screening of the ToxCast Phase I chemical library. Reproductive toxicology 33, 174–187, doi: 10.1016/j.reprotox.2011.10.018 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Thomas RS et al. A Comprehensive Statistical Analysis of Predicting In Vivo Hazard Using High-Throughput In Vitro Screening. Toxicological Sciences 128, 398–417, doi:DOI 10.1093/toxsci/kfs159 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Tice RR, Austin CP, Kavlock RJ & Bucher JR Improving the Human Hazard Characterization of Chemicals: A Tox21 Update. Environmental health perspectives 121, 756–765, doi:Doi 10.1289/Ehp.1205784 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Truong L et al. Multidimensional in vivo hazard assessment using zebrafish. Toxicological sciences : an official journal of the Society of Toxicology 137, 212–233, doi: 10.1093/toxsci/kft235 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rowlands JC, Sander M, Bus JS & FutureTox Organizing C FutureTox: building the road for 21st century toxicology and risk assessment practices. Toxicological sciences : an official journal of the Society of Toxicology 137, 269–277, doi: 10.1093/toxsci/kft252 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Truong L, Harper SL & Tanguay RL Evaluation of embryotoxicity using the zebrafish model. Methods in molecular biology 691, 271–279, doi: 10.1007/978-1-60761-849-2_16 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sipes NS, Padilla S & Knudsen TB Zebrafish: as an integrative model for twenty-first century toxicity testing. Birth Defects Res C Embryo Today 93, 256–267, doi: 10.1002/bdrc.20214 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Hill AJ, Teraoka H, Heideman W & Peterson RE Zebrafish as a model vertebrate for investigating chemical toxicity. Toxicological Sciences 86, 6–19, doi: 10.1093/toxsci/kfi110 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Brady CA, Rennekamp AJ & Peterson RT Chemical Screening in Zebrafish. Methods in molecular biology 1451, 3–16, doi: 10.1007/978-1-4939-3771-4_1 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Howe K et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 496, 498–503, doi: 10.1038/nature12111 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang G, Truong L, Tanguay RL & Reif DM A New Statistical Approach to Characterize Chemical-Elicited Behavioral Effects in High-Throughput Studies Using Zebrafish. PloS one 12, e0169408, doi: 10.1371/journal.pone.0169408 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis JA, Gift JS & Zhao QJ Introduction to benchmark dose methods and US EPA’s benchmark dose software (BMDS) version 2.1.1. Toxicology and applied pharmacology 254, 181–191, doi:DOI 10.1016/j.taap.2010.10.016 (2011). [DOI] [PubMed] [Google Scholar]
- 19.EPA, U. S. Benchmark Dose Technical Guidance., (U.S. EPA, 2012). [Google Scholar]
- 20.EU. Technical Guidance Document (TGD) on Risk Assessment of Chemical Substances Following European Regulations and Directives, Parts I-IV., (2003).
- 21.OECD. Draft Guidance Document on the Performance of Chronic Toxicity and Carcinogenicity Studies, Supporting TG 451, 452 and 453., Organisation for Economic Co-Operation and Development, (2008). [Google Scholar]
- 22.Gaylor DW & Kodell RL A procedure for developing risk-based reference doses. Regulatory toxicology and pharmacology : RTP 35, 137–141, doi: 10.1006/rtph.2002.1533 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Izadi H, Grundy JE & Bose R Evaluation of the benchmark dose for point of departure determination for a variety of chemical classes in applied regulatory settings. Risk analysis : an official publication of the Society for Risk Analysis 32, 830–835, doi: 10.1111/j.1539-6924.2011.01732.x (2012). [DOI] [PubMed] [Google Scholar]
- 24.Mandrell D et al. Automated zebrafish chorion removal and single embryo placement: optimizing throughput of zebrafish developmental toxicity screens. Journal of laboratory automation 17, 66–74, doi: 10.1177/2211068211432197 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reif DM et al. High-throughput characterization of chemical-associated embryonic behavioral changes predicts teratogenic outcomes. Archives of toxicology 90, 1459–1470, doi: 10.1007/s00204-015-1554-1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saili KS et al. Neurodevelopmental low-dose bisphenol A exposure leads to early life-stage hyperactivity and learning deficits in adult zebrafish. Toxicology 291, 83–92, doi:DOI 10.1016/j.tox.2011.11.001 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mcgill R, Tukey JW & Larsen WA Variations of Box Plots. Am Stat 32, 12–16, doi:Doi 10.2307/2683468 (1978). [DOI] [Google Scholar]
- 28.Reif DM et al. Endocrine profiling and prioritization of environmental chemicals using ToxCast data. Environmental health perspectives 118, 1714–1720, doi: 10.1289/ehp.1002180 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.EPA US Technical Guidance Document (TGD) on Risk Assessment of Chemical Substances Following European Regulations and Directives, Parts I-IV., (U.S. EPA, 2003). [Google Scholar]
- 30.Yang L, Allen BC & Thomas RS BMDExpress: a software tool for the benchmark dose analyses of genomic data. Bmc Genomics 8, 387, doi: 10.1186/1471-2164-8-387 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.