Abstract
Understanding signaling pathways in neuroscience requires high-resolution maps of the underlying protein networks. Proximity-dependent biotinylation with engineered enzymes, in combination with mass spectrometry-based quantitative proteomics, has emerged as a powerful method to dissect molecular interactions and the localizations of endogenous proteins. Recent applications to neuroscience have provided insights into the composition of sub-synaptic structures, including the synaptic cleft and inhibitory post-synaptic density. Here we compare the different enzymes and small-molecule probes for proximity labeling in the context of cultured neurons and tissue, review existing studies, and provide technical suggestions for the in vivo application of proximity labeling.
Introduction
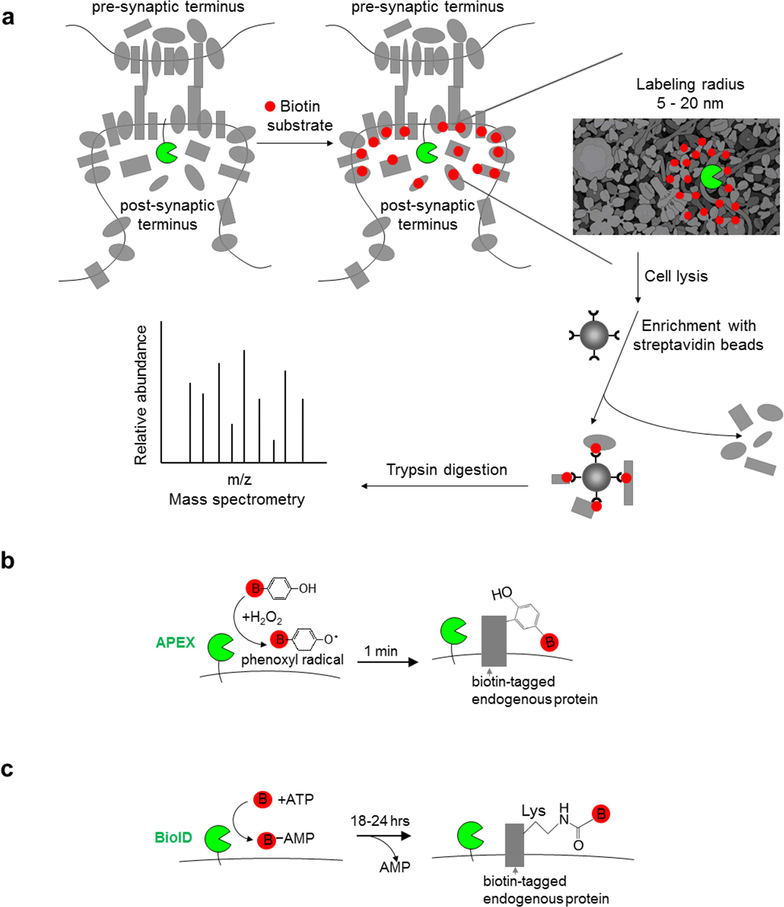
To study how spatially-compartmentalized protein networks assemble into functionally-integrated macromolecular complexes, we and others have developed a class of methods termed “proximity labeling” (PL). PL uses engineered enzymes to selectively and covalently tag neighboring proteins with biotin in living cells [1,2]. The biotinylated proteins can then be isolated after cell lysis and characterized by mass spectrometry (Figure 1a). PL has been applied to map novel components of cellular organelles [3–10] and to identify new protein-protein interaction partners with high spatial specificity [11–13]. These studies have demonstrated that PL is a powerful approach to dissect the interactions and localization patterns of molecules with nanometer spatial resolution [14,15].
Figure 1.
Workflow and mechanism of proximity labeling. (a) An engineered enzyme (green) is genetically targeted to the subcellular region of interest (e.g., the iPSD) and covalently tags proximal endogenous proteins with a biotin handle. The gray shapes are endogenous proteins residing inside and outside the region of interest. Following cell lysis, biotinylated proteins are enriched with streptavidin beads, digested to peptides on-bead, then analyzed by liquid chromatography and tandem MS. (b) For peroxidase-based labeling using APEX, H2O2 is added for 1 minute to cells preloaded with biotin-phenol (BP; red B=biotin) to initiate labeling. APEX oxidizes BP into a phenoxyl radical, which covalently tags proximal endogenous proteins at electron-rich side chains such as tyrosine. (c) For biotin ligase-based labeling using BioID, exogenously-supplied free biotin is utilized together with endogenous ATP for 18–24 hours of labeling. BioID converts biotin to reactive bioAMP, which is released from the enzyme’s active site to react with lysine residues on proximal proteins.
Due to the complexity and heterogeneity of neurons in both spatial and temporal dimensions, PL also has the potential to benefit neuroscience research. For example, there is tremendous interest in mapping the molecular composition of synaptic subdomains (e.g., active zone, post-synaptic density, cleft, synaptic vesicles) and understanding how these change during plasticity and in disease [16]. At the level of the organism, PL could potentially be used to map the proteomic signatures of different brain regions, cell types, and synapse types [17]. For specific proteins of interest, such as ion channels and receptors, PL could yield novel interaction partners that play important functional roles in regulation or signaling [18].
Yet there have been very few examples of PL applied to neuroscience to date. In this review, we summarize the few existing studies, analyze the technical challenges to the broader application of PL to neuroscience, and envision future applications.
Enzymes used for proximity labeling
Proximity labeling methods can be divided into two categories based on the enzyme used to carry out the catalysis: peroxidase-based PL and biotin ligase-based PL. Peroxidase-based PL relies on expression of an engineered ascorbate peroxidase (APEX or APEX2) [19,20] or horseradish peroxidase (HRP) in the cells/tissues of interest. Alternatively, HRP can be targeted to specific cell surface antigens via conjugation to an antibody [21,22]. To initiate labeling, H2O2 is added for 1 minute to cells/tissues pre-loaded with the substrate biotin-phenol (BP) [1] (or its variants, such as BxxP [23], alkyne-phenol [1], and desthiobiotin phenol [24]). The peroxidase oxidizes BP into a phenoxyl radical that reacts with nearby proteins at electron-rich side chains (Figure 1b), a mechanism similar to the commonly-used tyramide signal amplification (TSA) kit. As the phenoxyl radical has a half-life of less than 1 ms [25], the labeling intensity dramatically falls off within nanometers from the peroxidase active site, generating a biotinylation contour map that is then read out by quantitative proteomics to give a ranked protein list based on proximity to the enzyme. For example, using APEX targeted to the outer mitochondrial membrane (OMM), proteins residing on the OMM could be readily distinguished from immediately adjacent cytosolic proteins [20].
The rapid kinetics of the peroxidase reaction (<1 minute labeling time) can be harnessed to interrogate dynamically evolving protein interaction networks. Two recent studies used APEX fusions to G-protein coupled receptors (GPCRs) to map protein interaction partners at various timepoints after ligand stimulation [26,27]. Because HRP/APEX can also survive cell fixation and oxidatively polymerize diaminobenzidine, these enzymes can also be used to generate contrast in fixed cells for electron microscopy (EM), enabling users to examine peroxidase fusion constructs for proper subcellular localization before initiation of a proteomic study [19].
The major distinction between APEX and HRP is that HRP is only active in the secretory pathway and extracellular environment due to its need for disulfide bond formation [19]. However, HRP is more active than APEX2, which makes it a superior choice in the compartments in which it is active [23].
The enzyme BioID, which is an R118G mutant of E. coli biotin ligase (BirA), is used in biotin ligase-based PL [2]. BirA requires both biotin and ATP as substrates to generate reactive biotinoyl-5′-AMP (bioAMP), which it then transfers onto specific lysine residues on bacterial carboxylase proteins. The R118G mutation in BioID reduces BirA’s affinity for bioAMP, resulting in release of the reactive intermediate to promiscuously tag lysine residues on proximal proteins [28] (Figure 1c). Although the half-life of bioAMP is many minutes in water [29], BioID has been shown to have a labeling radius of ~10 nm from mapping of the nuclear pore complex [11], indicating that the half-life is likely to be much less inside cells, perhaps due to the high density of intracellular nucleophiles. A major limitation of BioID compared to peroxidase-based PL is that, due to its slow kinetics, BioID requires 18–24 hours of reaction time to obtain sufficient labeled material for proteomics. The long labeling timescale makes BioID non-optimal for the study of dynamic processes. In addition, BioID fusion constructs may mistarget more than APEX fusion constructs, due to its larger size (35 kD compared to 28 kD for APEX); mistargeting has previously been observed for BirA fusions [30]. Recently, a smaller version (27 kD) of BioID, BioID2, was reported [31].
Applications of proximity labeling to neuroscience
Recent studies have demonstrated the value of PL in elucidating the molecular components of synaptic clefts and the inhibitory postsynaptic density (iPSD). Despite long-standing interest in defining the molecular components that mediate information flow between communicating neurons, biochemical purification of many synaptic subregions remains intractable.
Loh et al. used HRP peroxidase in living neurons to map the proteomes of both the excitatory and inhibitory synaptic clefts, which are impossible to purify biochemically, and identified the glycosylphosphatidylinositol anchor protein Mdga2 as a potential specificity factor influencing Neuroligin-2′s recruitment of presynaptic neurotransmitters at inhibitory synapses [23]. To adapt previous peroxidase labeling protocols used for cancer cells to cultured neurons, a new lysis and enrichment protocol was developed to disassemble the detergent-insoluble and tightly crosslinked post-synaptic density (PSD), by adding a high percentage SDS lysis step with 10 minutes of boiling. This removed cytosolic contaminants co-purifying with biotinylated cleft-exposed proteins. Another methodological advance in this study was the application of an intersectional labeling strategy, using two independent peroxidase fusion constructs (HRP-Lrrtm1 and HRP-Lrrtm2 for excitatory cleft, and HRP-Nlgn2A and HRP-Slitrk3 for inhibitory cleft) targeting the same cellular locale, in order to improve the specificity of protein identifications. Finally, because non-dividing cells such as neurons are not amenable to SILAC (stable isotope labeling with amino acids in cell culture) labeling, which requires cell division and/or high protein turnover for metabolic incorporation of labels, protein quantitation was instead achieved through iTRAQ (isobaric tags for relative and absolute quantitation) chemical labeling. TMT (tandem mass tags) chemical labeling can also be used for quantitation in non-dividing cells. A similar workflow was applied to map the alpha-synuclein interactome in living neurons using APEX [13].
The iPSD is the only subcellular structure that has been investigated by PL in the brain of a living animal [32]. The application of BioID in the living mouse brain represents a major milestone for PL in neuroscience, as probe delivery and labeling procedures significantly differ from cell culture. To express BioID-fused proteins in vivo, adeno-associated viruses encoding biotin ligase fusion proteins were injected into the cortex and hippocampus of postnatal mice. The exact virus injection times, biotin dosage and labeling times were carefully optimized to achieve maximum number of synapses labeled. As biotin delivery into mouse brain is more challenging than in cell culture, the authors opted for intraperitoneal administration of biotin, which can penetrate various tissue types including the blood-brain barrier [33]. The mice were then labeled for 7 days before harvesting biotinylated material. Hence, temporal specificity for BioID in vivo is still lacking. Nevertheless, the study identified and validated many previously unknown components of the iPSD, including InSyn1 and InSyn2. Knockout of InSyn1 led to decreased postsynaptic inhibitory sites, reduced the frequency of miniature inhibitory currents, and increased excitability in the hippocampus, highlighting the power of PL to reveal new neurobiology.
In vivo application of PL requires probe delivery to tissue
Since implementation of PL requires delivery of both a genetic component (DNA encoding the enzyme) and a chemical component (the small-molecule substrates) to cells, an important consideration when performing PL in vivo is how to deliver the substrate molecules to the relevant organs or tissue. For the iPSD study described above [32], biotin was delivered to the brain via intraperitoneal injection. Mammals do not synthesize their own biotin, but it is an essential vitamin required for fatty acid biosynthesis. Import of biotin at low concentrations (less than 5 μM) into cells is mediated primarily by the Na+-dependent multivitamin transporter (SMVT1), which is ubiquitously expressed in various tissues including intestine, liver, brain, heart, lung and kidney. Passive diffusion across membranes occurs when biotin concentration exceeds 25 μM [34]. Taking advantage of these entry routes, biotin for BioID can be supplied through animal food in principle. However, this method of delivery may lead to substantial variation in labeling among individual animals as the food intake behavior may vary. The iPSD study used daily intraperitoneal injection to supply exogenous biotin, providing a simple and efficient way to control biotin dosage. Importantly, biotin is not known to be toxic even at high doses; when administered to people without biotin metabolism disorder at up to 5 mg/day for two years, adverse effects were not observed [35].
Apart from this study, one other in vivo BioID example has been reported. Here, the authors investigated c-MYC interaction partners in a tumor xenograft model, where tumor cells expressing BioID were injected into mice [36]. Again, exogenous biotin was supplied via intraperitoneal injection.
In some cases, users may opt for peroxidase-based PL in vivo, to better control the time window of labeling. In vivo substrate delivery for APEX-based PL is more challenging than for BioID, but a few recent studies have succeeded [37–39]. In insects and worms, the tightly-sealed and hydrophobic cuticle acts as a protective outer layer and is highly impermeable to small molecules in the environment. Two recent studies applying APEX proteomics to the intestine of living C. elegans [38,39] used RNAi knockdown of a glycosyltransferase gene, bus-8, to compromise cuticle integrity and enable the delivery of BP and H2O2 to the worm interior. However, disrupting a gene with known developmental functions is a concern for both the proteomic experiment and data interpretation. An alternative strategy is to dissect the tissue of interest from the intact animal body, as employed in an APEX study in Drosophila muscle mitochondria [37]. Dissection and labeling in proper tissue culture media maintains the tissue in a physiological environment for the short duration of the H2O2 reaction, providing a potentially generalizable strategy for in vivo labeling with high temporal resolution. The downside of this approach is that the dissection process may be labor-intensive and difficult for certain tissue types.
Although H2O2 is used at a low concentration in the APEX PL reaction (1 mM) and only for a short period of time (<1 minute), H2O2 delivery could lead to undesired cytotoxicity arising from oxidative stress signaling. 1 mM of H2O2 has been shown to induce apoptosis in 24 hours through activation of p53-related pro-oxidant gene expression such as BAX, PIG3 and PUMA [40]. Sub-millimolar concentrations of H2O2 for just 15 minutes can cause significant aldehydic DNA lesions in HeLa cells [41]. Therefore, restricting H2O2 labeling to low concentrations and short time windows is crucial for minimizing side effects. For tissues with minimal thickness and size, such as whole worms or dissected Drosophila muscles, H2O2 can quickly penetrate and react throughout the entire sample, just as it does in cell culture [37–39]. In contrast, immersing the whole mouse brain in H2O2 solution is less likely to be effective, because by the time H2O2 penetrates the center of the tissue mass, too much damage may have been caused to the cortex by oxidation. Acute slice preparations in artificial cerebrospinal fluid, such as for slice electrophysiology, could be considered for vertebrate brain samples to avoid long incubation times in H2O2.
The challenge of high background in vivo
Another technical challenge to applying PL in vivo is that tissue contains more substantial sources of background than cultured cells. The background can come from two sources: non-specific binding of material to streptavidin-coated beads, and endogenous biotinylated proteins. The former problem arises when the experimentalist is infecting only a small fraction of the total cells in a tissue region or organism, or when the proteome of interest is very small and localized. Then the ratio of biotinylated (desired) material to non-biotinylated (undesired) material is very small, and it is difficult to implement a successful enrichment. To overcome this, crude fractionation of the sample prior to streptavidin enrichment may help to increase the ratio of desired to undesired material. For example, if conducting a PL experiment targeting a sub-synaptic region, a crude synaptosome preparation [42] prior to streptavidin enrichment could help remove background proteins from unrelated regions. Dissecting out organs of interest prior to homogenization and streptavidin enrichment could also be beneficial.
The second source of background, endogenous biotinylated proteins, also becomes a serious problem when working with small fractions of transfected cells, or small and localized proteomes. In the Drosophila brain, for example, endogenous biotinylated proteins are highly abundant, especially in glia cells [43]. These endogenous biotinylated species can vastly exceed the BioID- or APEX-biotinylated proteomes in mass, and compete for binding to streptavidin beads. Again, fractionation or dissection may help in these cases by removing background from unrelated regions. It would also be valuable in the future to develop non-biotin-based strategies for enrichment of tagged proteomes. In Rhee et al. [1], for instance, it was shown that APEX can use an alkyne-phenol substrate instead of biotin-phenol. Alkyne can be “clicked” to various azide reagents [44], potentially enabling enrichment without the use of streptavidin beads.
Tissue presents other sources of background as well. For peroxidase-based PL, background can arise from the activity of endogenous peroxidases. For example, in the C. elegans intestine, it was found that endogenous peroxidase activity is higher than that typically observed in cultured cells [39]. Therefore, designing experiments with both unlabeled and untransfected controls is essential for minimizing interference from non-specific binders, endogenous biotinylated proteins, and endogenous peroxidase activity.
Another strategy to overcome background in tissue is to increase the desired signal. For example, one could extend the reaction time to label more material, as in the iPSD in vivo study [32] and in a C. elegans study that used 2 minutes instead of the typical 1 minute for APEX tagging [39]. For peroxidase-based PL in particular, sometimes low signal results from high expression of catalases, which quench H2O2, or from low availability of the peroxidase co-factor heme, which can be improved by heme supplementation [45].
Outlook for the application of proximity labeling in neurobiology
In the last decade, proteomics in combination with biochemical approaches (e.g. fractionation, immunoprecipitation, and chemical crosslinking) has been instrumental in characterizing the molecular components of many neuronal structures, including entire synaptic terminals [42], synaptic vesicles [46], the PSD [47] and the active zone [48]. Advanced imaging approaches, such as array tomography [49], have offered neuroscientists high-resolution maps of molecular architecture in the brain. However, array tomography is limited to only ~20 targets at once, and antibody availability, while purification-based proteomics suffers from high false-positive rates due to contamination. For instance, intracellular contaminants such as mitochondrial and nuclear proteins account for 20–40% of identifications in synaptosome preparations [42]. Therefore, PL-based proteomic maps with high spatial and temporal resolution would greatly complement these existing techniques.
Besides the subcellular regions already mapped with PL-based proteomics, neurons harbor many other functionally specialized subcellular structures, such as the growth cone for axon guidance [50] and the axon initial segment for signal integration [51], as well as highly distinct structures executing unique functions in certain cell types. For example, electrophysiological and structural evidence has indicated that stereocilia tips of hair cells, which are the primary mechanical sensors for hearing in mammalian ears, house the elusive mechano-to-electric transduction machinery with unknown molecular identity [52]. PL-based proteomic profiling provides an unprecedented opportunity to systematically map the protein composition of these functionally specialized regions. Notably, growing evidence suggests that glia are more than supportive cells, but are extensively involved in regulating the development and function of the nervous system as well [53]. Proteomic mapping of the interface between glia and neurons would further our understanding of the molecular basis of glial-neuronal interaction, such as the signal controlling myelination. Furthermore, glial pathogenesis has been implicated in many neurological disorders such as Alzheimer’s [54,55]. In vivo PL-based proteomics of different cellular compartments (e.g. endosomes, lysosomes, mitochondria, cell surface) in different types of glia (astrocytes, microglia, oligodendrocytes) under healthy and pathological conditions would provide a means to gain molecular insights into the role of glial cells in diseases.
Proteomic mapping of dynamic interactomes offers another exciting avenue to advance our understanding of key molecules in neurobiology. Although a huge collection of molecules essential for neural development [56–58] and signal transmission [59] has been identified through genetic analysis, most are still “orphan” genes, for which we have limited knowledge of interaction partners and function. Applying PL-based proteomics to these molecules could capture both stable and transient partners and yield a comprehensive interaction network. Moreover, the high temporal resolution of peroxidase-based PL would allow us to profile interaction dynamics under different conditions, as demonstrated in the GPCR pathways [26,27]. As many of these molecules are linked with human diseases [60], systematically profiling their interacting partners under disease conditions would not only shed light on the pathological mechanisms but also potentially identify novel disease-associated genes.
Highlights.
Proximity labeling enables proteomic interrogation of the molecular components of subcellular regions as well as protein interaction networks.
Proximity labeling has been used to uncover novel components of the synaptic cleft and inhibitory post-synaptic density.
Different strategies have been developed to delivery small molecule probes for proximity labeling in vivo.
Proximity labeling, in combination with high-resolution imaging and genetic analysis, will advance our understanding key molecules and pathways in neurobiology.
Acknowledgements
We thank members of the Ting lab for helpful comments on the manuscript. S.H. is a Stanford Bio-X Bowes graduate fellow. J. L. is a Genentech Foundation graduate fellow. Funding was provided by the NIH (R01-CA186568 to A.Y.T.).
Footnotes
Conflict of interest statement
A.Y.T. is an author of a patent application on the peroxidase technology.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Rhee H, Zou P, Udeshi ND, Martell JD, Mootha VK, Carr SA, Ting AY: Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 2013, 339:1328–31.•• This paper establishes the use of an engineered ascorbate peroxidase, APEX, and its biotin-phenol substrate for proteomic mapping of membrane-bound organelles. APEX is targeted to the mitochondria in HEK 293T cells to identify 495 proteins in the mitochondria matrix with high specificity (>94%) and high coverage (>85%).
- 2.Roux KJ, Kim DI, Raida M, Burke B: A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol 2012, 196:801–810.•• This paper utilizes the promiscuous biotin ligase mutant for proximity-dependent labeling and developed the BioID method. The authors apply BioID to study the nuclear lamina by fusing the biotin ligase mutant to the protein lamin-A. A novel nuclear lamina-associated protein, SLAP75, is discovered and validated.
- 3.Hung V, Zou P, Rhee HW, Udeshi ND, Cracan V, Svinkina T, Carr SA, Mootha VK, Ting AY: Proteomic Mapping of the Human Mitochondrial Intermembrane Space in Live Cells via Ratiometric APEX Tagging. Mol. Cell 2014, 55:332–341.• The paper describes a ratiometric approach based on quantitative proteomics to reduce background and achieve nanometer resolution for APEX labeling.
- 4.Hung V, Lam SS, Udeshi ND, Svinkina T, Guzman G, Mootha VK, Carr SA, Ting AY: Proteomic mapping of cytosol-facing outer mitochondrial and ER membranes in living human cells by proximity biotinylation. Elife 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han S, Udeshi ND, Deerinck TJ, Svinkina T, Ellisman MH, Carr SA, Ting AY: Proximity Biotinylation as a Method for Mapping Proteins Associated with mtDNA in Living Cells. Cell Chem. Biol 2017, 24:404–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mick DU, Rodrigues RB, Leib RD, Adams CM, Chien AS, Gygi SP, Nachury MV.: Proteomics of Primary Cilia by Proximity Labeling. Dev. Cell 2015, 35:497–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jing J, He L, Sun A, Quintana A, Ding Y, Ma G, Tan P, Liang X, Zheng X, Chen L, et al. : Proteomic mapping of ER-PM junctions identifies STIMATE as a regulator of Ca2+ influx. Nat. Cell Biol 2015, 17:1339–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Firat-Karalar EN, Rauniyar N, Yates JR, Stearns T: Proximity Interactions among Centrosome Components Identify Regulators of Centriole Duplication. Curr. Biol 2014, 24:664–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong J-M, Tay FP-L, Swa HL-F, Gunaratne J, Leung T, Burke B, Manser E: Proximity biotinylation provides insight into the molecular composition of focal adhesions at the nanometer scale. Sci. Signal 2016, 9:rs4–rs4. [DOI] [PubMed] [Google Scholar]
- 10.Gupta GD, Coyaud É, Gonçalves J, Mojarad BA, Liu Y, Wu Q, Gheiratmand L, Comartin D, Tkach JM, Cheung SWT, et al. : A Dynamic Protein Interaction Landscape of the Human Centrosome-Cilium Interface. Cell 2015, 163:1484–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim DI, KC B, Zhu W, Motamedchaboki K, Doye V, Roux KJ: Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc. Natl. Acad. Sci 2014, 111:E2453–E2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schweingruber C, Soffientini P, Ruepp M-D, Bachi A, Mühlemann O: Identification of Interactions in the NMD Complex Using Proximity-Dependent Biotinylation (BioID). PLoS One 2016, 11:e0150239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung CY, Khurana V, Yi S, Sahni N, Loh KH, Auluck PK, Baru V, Udeshi ND, Freyzon Y, Carr SA, et al. : In Situ Peroxidase Labeling and Mass-Spectrometry Connects Alpha-Synuclein Directly to Endocytic Trafficking and mRNA Metabolism in Neurons. Cell Syst. 2017, 4:242–250.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Varnaitė R, MacNeill SA: Meet the neighbors: Mapping local protein interactomes by proximity-dependent labeling with BioID. Proteomics 2016, 16:2503–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rees JS, Li X-W, Perrett S, Lilley KS, Jackson AP: Protein Neighbors and Proximity Proteomics. Mol. Cell. Proteomics 2015, 14:2848–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choquet D, Triller A: The Dynamic Synapse. Neuron 2013, 80:691–703. [DOI] [PubMed] [Google Scholar]
- 17.Buosi AS, Matias I, Araujo APB, Batista C, Gomes FCA: Heterogeneity in Synaptogenic Profile of Astrocytes from Different Brain Regions. Mol. Neurobiol 2017, doi: 10.1007/s12035-016-0343-z. [DOI] [PubMed] [Google Scholar]
- 18.Ho VM, Lee J-A, Martin KC: The cell biology of synaptic plasticity. Science 2011, 334:623–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martell JD, Deerinck TJ, Sancak Y, Poulos TL, Mootha VK, Sosinsky GE, Ellisman MH, Ting AY: Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat. Biotechnol 2012, 30:1143–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lam SS, Martell JD, Kamer KJ, Deerinck TJ, Ellisman MH, Mootha VK, Ting AY: Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods 2014, 12:51–54.• The paper describes the development of a catalytically more active version of APEX, APEX2, for proximity labeling.
- 21.Honke K, Kotani N: The enzyme-mediated activation of radical source reaction: a new approach to identify partners of a given molecule in membrane microdomains. J. Neurochem 2011, 116:690–695. [DOI] [PubMed] [Google Scholar]
- 22.Li X-W, Rees JS, Xue P, Zhang H, Hamaia SW, Sanderson B, Funk PE, Farndale RW, Lilley KS, Perrett S, et al. : New Insights into the DT40 B Cell Receptor Cluster Using a Proteomic Proximity Labeling Assay. J. Biol. Chem 2014, 289:14434–14447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loh KH, Stawski PS, Draycott AS, Udeshi ND, Lehrman EK, Wilton DK, Svinkina T, Deerinck TJ, Ellisman MH, Stevens B, et al. : Proteomic Analysis of Unbounded Cellular Compartments: Synaptic Clefts. Cell 2016, 166:1295–1307.e21.•• This paper applies HRP to probe the composition of both the inhibitory and excitatory synaptic clefts in living neurons. The two proteomes discover dozens of novel synaptic candidates, which are validated by either fluorescence microscopy or western blot. Many known synaptic proteins are also assigned to a specific cleft type in the dataset.
- 24.Lee S-Y, Kang M-G, Shin S, Kwak C, Kwon T, Seo JK, Kim J-S, Rhee H-W: Architecture Mapping of the Inner Mitochondrial Membrane Proteome by Chemical Tools in Live Cells. J. Am. Chem. Soc 2017, 139:3651–3662. [DOI] [PubMed] [Google Scholar]
- 25.Mortensen A, Skibsted LH: Importance of Carotenoid Structure in Radical-Scavenging Reactions. J. Agric. Food Chem 1997, 45:2970–2977. [Google Scholar]
- 26.Paek J, Kalocsay M, Staus DP, Wingler L, Pascolutti R, Paulo JA, Gygi SP, Kruse AC: Multidimensional Tracking of GPCR Signaling via Peroxidase-Catalyzed Proximity Labeling. Cell 2017, 169:338–349.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lobingier BT, Hüttenhain R, Eichel K, Miller KB, Ting AY, von Zastrow M, Krogan NJ: An Approach to Spatiotemporally Resolve Protein Interaction Networks in Living Cells. Cell 2017, 169:350–360.e12.• This paper describes the workflow to deconvolve APEX labeling data to reveal protein interaction dynamics.
- 28.Choi-Rhee E, Schulman H, Cronan JE: Promiscuous protein biotinylation by Escherichia coli biotin protein ligase. Protein Sci. 2008, 13:3043–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu Y, Beckett D: Kinetics of Biotinyl-5’-adenylate Synthesis Catalyzed by the Escherichia coli Repressor of Biotin Biosynthesis and the Stability of the Enzyme- Product Complex. Biochemistry 1994, 33:7354–7360. [DOI] [PubMed] [Google Scholar]
- 30.Liu DS, Loh KH, Lam SS, White KA, Ting AY: Imaging Trans-Cellular Neurexin-Neuroligin Interactions by Enzymatic Probe Ligation. PLoS One 2013, 8:e52823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim DI, Jensen SC, Noble KA, KC B, Roux KH, Motamedchaboki K, Roux KJ: An improved smaller biotin ligase for BioID proximity labeling. Mol. Biol. Cell 2016, 27:1188–1196.• This paper reports the development of a smaller, more efficient version of BioID, BioID2.
- 32.Uezu A, Kanak DJ, Bradshaw TWA, Soderblom EJ, Catavero CM, Burette AC, Weinberg RJ, Soderling SH: Identification of an elaborate complex mediating postsynaptic inhibition. Science (80-. ). 2016, 353:1123–1129.•• This paper describes the first use of BioID in vivo in mouse brains to discover inhibitory postsynaptic proteins. Proximity labeling followed by quantitative proteomics reveals a large number of previously uncharacterized proteins. The function of one candidate, InSyn1, is investigated by CRISPR-Cas9 deletion of the gene.
- 33.Spector R, Mock D: Biotin transport through the blood-brain barrier. J. Neurochem 1987, 48:400–4. [DOI] [PubMed] [Google Scholar]
- 34.Polyak SW: Mechanisms of Biotin Transport. Biochem. Anal. Biochem 2015, 4. [Google Scholar]
- 35.Koutsikos D, Agroyannis B, Tzanatos-Exarchou H: Biotin for diabetic peripheral neuropathy. Biomed. Pharmacother 1990, 44:511–4. [DOI] [PubMed] [Google Scholar]
- 36.Dingar D, Kalkat M, Chan P-K, Srikumar T, Bailey SD, Tu WB, Coyaud E, Ponzielli R, Kolyar M, Jurisica I, et al. : BioID identifies novel c-MYC interacting partners in cultured cells and xenograft tumors. J. Proteomics 2015, 118:95–111. [DOI] [PubMed] [Google Scholar]
- 37.Chen C-L, Hu Y, Udeshi ND, Lau TY, Wirtz-Peitz F, He L, Ting AY, Carr S a, Perrimon N: Proteomic mapping in live Drosophila tissues using an engineered ascorbate peroxidase. Proc. Natl. Acad. Sci. U. S. A 2015, 112:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reinke AW, Balla KM, Bennett EJ, Troemel ER: Identification of microsporidia host-exposed proteins reveals a repertoire of rapidly evolving proteins. Nat. Commun 2017, 8:14023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reinke AW, Mak R, Troemel ER, Bennett EJ: In vivo mapping of tissue- and subcellular-specific proteomes in Caenorhabditis elegans. Sci. Adv 2017, 3:e1602426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sablina AA, Budanov AV, Ilyinskaya GV, Agapova LS, Kravchenko JE, Chumakov PM: The antioxidant function of the p53 tumor suppressor. Nat. Med 2005, 11:1306–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakamura J: Micromolar concentrations of hydrogen peroxide induce oxidative DNA lesions more efficiently than millimolar concentrations in mammalian cells. Nucleic Acids Res. 2003, 31:1790–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Biesemann C, Grønborg M, Luquet E, Wichert SP, Bernard V, Bungers SR, Cooper B, Varoqueaux F, Li L, Byrne JA, et al. : Proteomic screening of glutamatergic mouse brain synaptosomes isolated by fluorescence activated sorting. EMBO J. 2014, 33:157–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Erdmann I, Marter K, Kobler O, Niehues S, Abele J, Müller A, Bussmann J, Storkebaum E, Ziv T, Thomas U, et al. : Cell-selective labelling of proteomes in Drosophila melanogaster. Nat. Commun 2015, 6:7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uttamapinant C, Tangpeerachaikul A, Grecian S, Clarke S, Singh U, Slade P, Gee KR, Ting AY: Fast, Cell-Compatible Click Chemistry with Copper-Chelating Azides for Biomolecular Labeling. Angew. Chemie Int. Ed 2012, 51:5852–5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martell JD, Yamagata M, Deerinck TJ, Phan S, Kwa CG, Ellisman MH, Sanes JR, Ting AY: A split horseradish peroxidase for the detection of intercellular protein-protein interactions and sensitive visualization of synapses. Nat. Biotechnol 2016, 34:774–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takamori S, Holt M, Stenius K, Lemke EA, Grønborg M, Riedel D, Urlaub H, Schenck S, Brügger B, Ringler P, et al. : Molecular Anatomy of a Trafficking Organelle. Cell 2006, 127:831–846. [DOI] [PubMed] [Google Scholar]
- 47.Bayés À, Collins MO, Croning MDR, van de Lagemaat LN, Choudhary JS, Grant SGN: Comparative Study of Human and Mouse Postsynaptic Proteomes Finds High Compositional Conservation and Abundance Differences for Key Synaptic Proteins. PLoS One 2012, 7:e46683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boyken J, Grønborg M, Riedel D, Urlaub H, Jahn R, Chua JJE: Molecular Profiling of Synaptic Vesicle Docking Sites Reveals Novel Proteins but Few Differences between Glutamatergic and GABAergic Synapses. Neuron 2013, 78:285–297. [DOI] [PubMed] [Google Scholar]
- 49.Micheva KD, Smith SJ: Array Tomography: A New Tool for Imaging the Molecular Architecture and Ultrastructure of Neural Circuits. Neuron 2007, 55:25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dent EW, Gupton SL, Gertler FB: The Growth Cone Cytoskeleton in Axon Outgrowth and Guidance. Cold Spring Harb. Perspect. Biol 2011, 3:a001800–a001800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kole MHP, Stuart GJ: Signal Processing in the Axon Initial Segment. Neuron 2012, 73:235–247. [DOI] [PubMed] [Google Scholar]
- 52.Zhao B, Müller U: The elusive mechanotransduction machinery of hair cells. Curr. Opin. Neurobiol 2015, 34:172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zuchero JB, Barres BA: Glia in mammalian development and disease. Development 2015, 142:3805–3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kurosinski P, Götz J: Glial Cells Under Physiologic and Pathologic Conditions. Arch. Neurol 2002, 59:1524. [DOI] [PubMed] [Google Scholar]
- 55.Chung W-S, Welsh CA, Barres BA, Stevens B: Do glia drive synaptic and cognitive impairment in disease?. Nat. Neurosci 2015, 18:1539–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kolodkin AL, Tessier-Lavigne M: Mechanisms and Molecules of Neuronal Wiring: A Primer. Cold Spring Harb. Perspect. Biol 2011, 3:a001727–a001727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zipursky SL, Sanes JR: Chemoaffinity Revisited: Dscams, Protocadherins, and Neural Circuit Assembly. Cell 2010, 143:343–353. [DOI] [PubMed] [Google Scholar]
- 58.Hong W, Luo L: Genetic Control of Wiring Specificity in the Fly Olfactory System. Genetics 2014, 196:17–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Triller A, Sheng M: Synaptic structure and function. Curr. Opin. Neurobiol 2012, 22:363–365. [DOI] [PubMed] [Google Scholar]
- 60.Sullivan PF, Daly MJ, O’Donovan M: Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat. Rev. Genet 2012, 13:537–551. [DOI] [PMC free article] [PubMed] [Google Scholar]