Abstract
Lupus nephritis is a potentially fatal autoimmune disease for which the current treatment is ineffective and often toxic. To develop mechanistic hypotheses of disease, we analyzed kidney samples from patients with lupus nephritis and from healthy control subjects using single-cell RNA sequencing. Our analysis revealed 21 subsets of leukocytes active in disease, including multiple populations of myeloid cells, T cells, natural killer cells and B cells that demonstrated both pro-inflammatory responses and inflammation-resolving responses. We found evidence of local activation of B cells correlated with an age-associated B-cell signature and evidence of progressive stages of monocyte differentiation within the kidney. A clear interferon response was observed in most cells. Two chemokine receptors, CXCR4 and CX3CR1, were broadly expressed, implying a potentially central role in cell trafficking. Gene expression of immune cells in urine and kidney was highly correlated, which would suggest that urine might serve as a surrogate for kidney biopsies.
Lupus nephritis (LN) is a frequent complication of systemic lupus erythematosus (SLE)1, 2, for which current therapies are both toxic and insufficiently effective2, 3. Despite the rapid pace of immunologic discovery, most clinical trials of rationally designed therapies have failed in both general SLE and LN, with only one new drug approved for the treatment of SLE in the last five decades2, 4. Thus, there is a pressing need to decipher the immune mechanisms that drive LN.
Current knowledge of the molecular pathways dysregulated in SLE comes mainly from the unbiased analysis of blood cells5; however, the extent to which blood reflects the inflamed tissue is unclear. Immunohistochemistry and flow cytometry studies of kidney biopsies have indicated the presence of infiltrating subpopulations of immune cells6, 7, 8 but cannot reveal previously-unidentified cell types or activation states. Mouse models of LN provide detailed knowledge of the molecular pathways and cell types active in their kidney9 but vary in key aspects such as the degree of immune cell infiltration, the role of interferon and Fc receptors and responses to therapeutic interventions.
It is thus clear that the study of LN can greatly benefit from a resource allowing the generation and preliminary testing of new hypotheses. Single-cell transcriptomics is a powerful tool capable of producing a complete catalogue of cell types and states present in a given sample. Here, we employed it in the analysis of kidney, urine and blood samples from patients with LN and healthy individuals, while utilizing a standardized protocol to process patient samples acquired across a distributed clinical and research network. Our findings delineate the complex array of leukocytes active in human LN kidneys. Analysis of blood reveals both similarities to and differences from the molecular signatures detected in kidneys, highlighting the limitations of blood samples for deciphering renal disease processes. We further show that urine cells have the potential to serve as surrogates for kidney biopsies in assessing the molecular activation state of subsets of infiltrating leukocytes.
RESULTS
Isolation and processing of kidney cells for single-cell transcriptomics
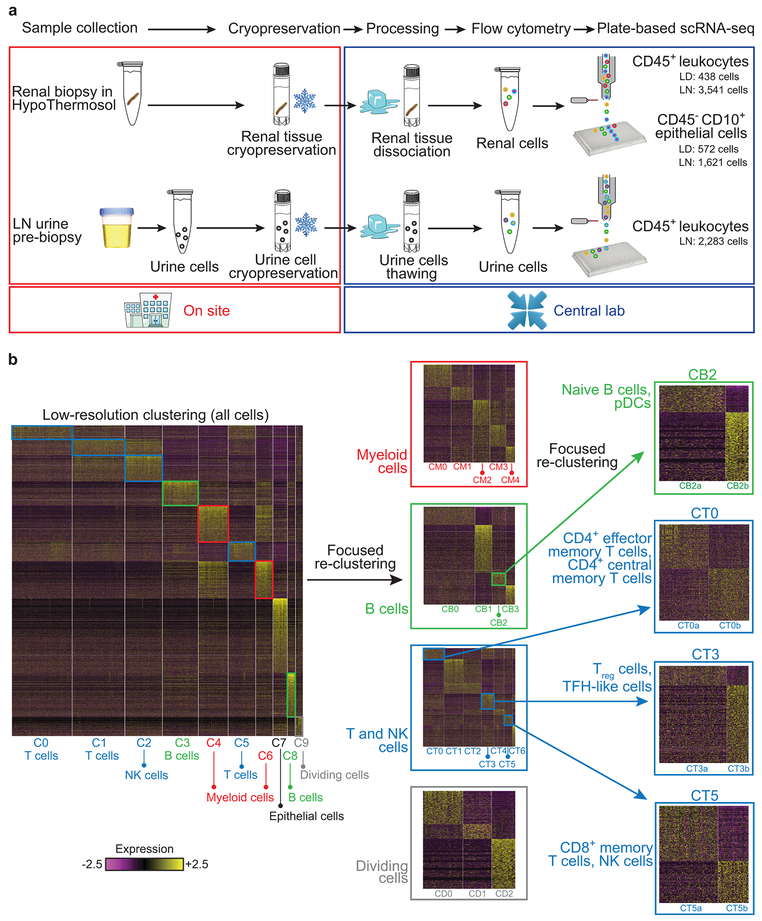
To establish a uniform pipeline to analyze kidney biopsy samples from multiple institutions, we evaluated several strategies for their preservation and transport. Cryopreservation of intact kidney tissue immediately after acquisition, followed by batched processing at a central site, offered robust leukocyte yields, intact staining for lineage markers and high quality transcriptomes (Supplementary Fig. 1a-e). We employed this pipeline to analyze kidney biopsies from 24 patients with LN and 10 control samples, acquired from living donor kidney biopsies (Fig. 1a and Supplementary Table 1). Approximately half of the LN samples, independent of histologic classification, provided leukocyte yields well above those obtained from control samples (Supplementary Fig. 2a,b), including B cells, T cells, macrophages and other leukocytes based on flow cytometry (Supplementary Fig. 2c,d).
Figure 1.
An overview of the approach used for analyzing the cellular contents and molecular states of kidney and urine samples. a, Pipeline for collecting and processing kidney and urine samples. Both types of samples were frozen on collection, then shipped to a central processing site to minimize batch effects. b, Stepwise clustering of kidney cells. Initially, all cells were analyzed together (left heatmap), and the identified clusters were labeled as containing either myeloid cells (red), B cells (green), T cells or NK cells (blue), dividing cells (gray) or epithelial cells. Each lineage, with the exception of epithelial cells, was then analyzed separately (middle heatmaps), to identify finer clusters. One B-cell cluster and three T-cell clusters were further re-clustered separately, to generate an even finer description of cell subsets (right). LD, living donor.
Viable cells were sorted into 384-well plates for single-cell RNA sequencing (scRNA-seq) using a modified CEL-Seq2 protocol10. Since our focus was on characterizing the immune cells within LN kidneys, 90% of the cells sequenced from each sample were CD45+ cells, and the rest were CD45−CD10+ cells. The quality of the collected sequencing data was comparable across plates, and higher in leukocytes compared with epithelial cells, reflecting the lower viability of the latter in the processed samples (Supplementary Fig. 1f,g). Principal component analysis performed on the gene expression data from 2,736 leukocytes and 145 epithelial cells indicated that the main sources of variability in the data corresponded to cell types, rather than batch or technical factors (Supplementary Fig. 2e,f).
Stepwise cell clustering identifies cells of the myeloid, T, natural killer (NK), B and epithelial lineages
To identify the lineage and activation state of the cells extracted from kidney samples, we clustered them based on their gene expression data, taking a stepwise approach (Fig. 1b). Low-resolution clustering of all kidney cells identified 10 clusters (Supplementary Fig. 2g), which we labeled as myeloid cells (clusters C4 and C6), T/NK cells (C0, C1, C2 and C5), B cells (C3, C8), dividing cells (C9) and kidney epithelial cells (C7), based on the expression of canonical lineage markers and other genes specifically upregulated in each cluster. Sensitivity analysis demonstrated that this labeling of cells was highly robust (Supplementary Table 2).
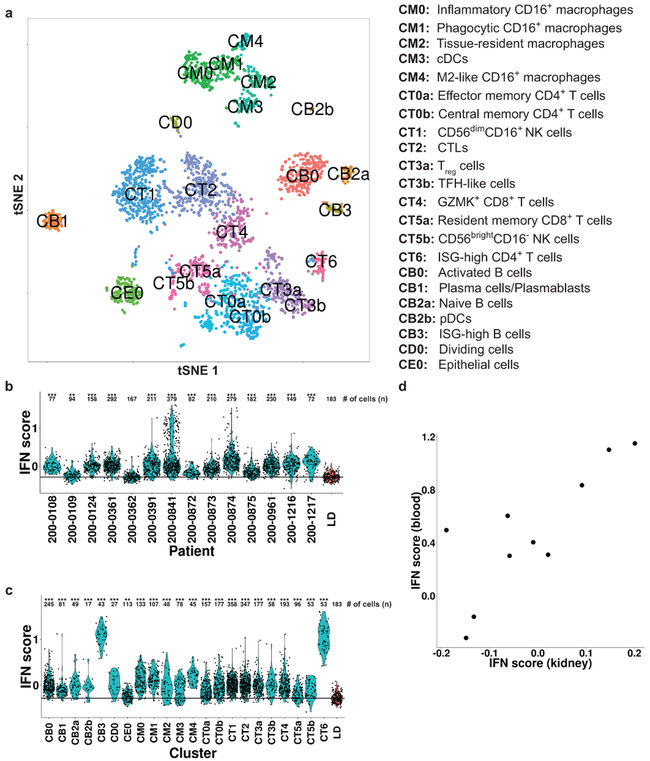
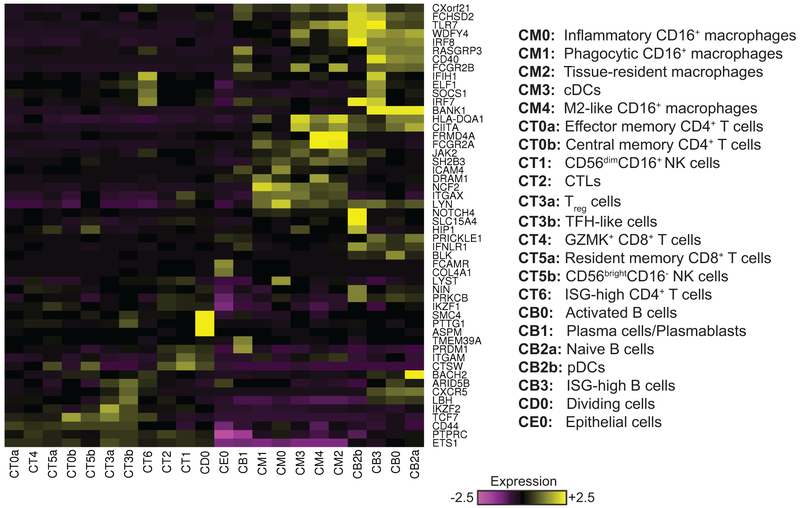
We next clustered the cells of each lineage separately, and identified 21 immune cell clusters and a single epithelial cell cluster (Fig. 2a), each containing cells from multiple patients and plates (Supplementary Tables 3 and 4). Saturation analysis indicated that the size of the present cohort is adequate to reveal most of the major clusters in LN kidneys (Supplementary Fig. 2h). Only three clusters were substantially represented in living donor control samples (Supplementary Fig. 2i and Supplementary Table 3), as verified by analyzing two additional living donor control samples, using a droplet-based approach to maximize the number of processed cells; this increased the number of living donor high-quality cells from 183 to 305, yielding largely the same results (Supplementary Table 3). For clusters that were present in sufficient numbers in both patients with LN and living donor controls, we performed a cluster-based differential expression analysis, comparing the two patient populations; the results of this analysis (Supplementary Table 5) are reported below.
Figure 2.
A summary of the stepwise clustering of kidney cells. a, Twenty-two clusters were identified; their putative identities are specified on the right. b, The distribution of the interferon response score in all patients with LN (blue), compared with the cells of the LD controls (red). c, The distribution of the interferon response score in all cells of patients with LN, separated into clusters (blue), compared with cells of the LD controls (red). In both (b) and (c), *** - FDR-corrected p-value < 0.001; ** - FDR-corrected p-value < 0.01 (two-tailed Mann-Whitney U-test). The number of cells (n) used in each comparison is specified above the plot. The horizontal line designates the median interferon response score over the cells of the LD controls. d, A comparison of the interferon response score in kidney and in blood in 10 patients with LN for whom corresponding blood and kidney samples were available. The kidney score was calculated as the average over all kidney cells per compared patient; the blood score was calculated based on bulk RNA-seq data of total PBMCs. IFN, interferon.
The renal and systemic interferon responses are highly correlated
Type I interferons are elevated in the peripheral blood of patients with lupus11. To assess the extent of this phenomenon in kidney, we calculated for each cell an interferon response score, defined as the average expression of several known interferon-stimulated genes (ISGs; Supplementary Table 6). We found that in all patients but one there was a significant upregulation of this score in kidney cells compared with living donor controls (Fig. 2b). Furthermore, this upregulation was observed in all clusters, although it was less pronounced in the kidney epithelial cells (Fig. 2c). Two clusters, one containing B cells and the other CD4+ T cells (CB3 and CT6, respectively; see below), demonstrated particularly high values of the interferon response score; the majority of these cells were extracted from two patients (patient IDs 200–0841 and 200–0874; Supplementary Table 3). These two patients also featured B cells and CD4+ T cells with a substantially lower interferon score, suggesting that the secretion of this cytokine may be spatially localized, either in the kidney or outside of it. When we compared the interferon response score in matched blood and kidney samples from 10 patients with LN we found a significant correlation (Spearman’s ρ = 0.733, P = 0.016; Fig. 2d), indicating that the interferon response is mainly an extrarenal process.
Classification and annotation of myeloid cell clusters reveal resident and infiltrating populations
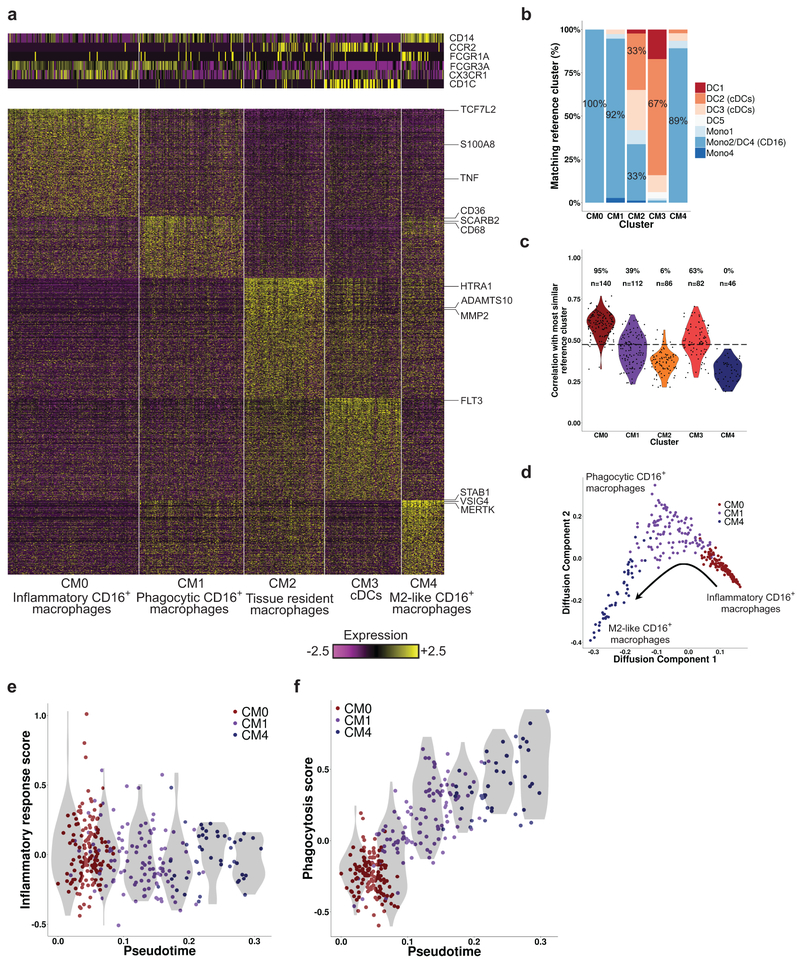
Focused analysis of the 466 cells in myeloid clusters C4 and C6 revealed 5 finer clusters (clusters CM0–CM4, Fig. 3a and Supplementary Fig. 3a–c). We determined their putative identity by comparing their global gene expression patterns with those of published reference monocyte/dendritic cell (DC) clusters identified in blood samples of healthy individuals using scRNA-seq12 (Fig. 3b,c), and by the expression of canonical lineage markers. Cluster CM3 was closest to CD1C+ DCs (reference clusters DC2 and DC3) or CLEC9A+ DCs (reference cluster DC1), in accordance with the expression of the canonical DC markers CD1C and FLT3 (Supplementary Fig. 3a), and the lack of expression of monocyte markers CD14 and CD16. Cluster CM0 cells were most similar to CD16+ patrolling monocytes (reference clusters Mono2 and DC4), with very high expression of CD16 (FCGR3A) and CX3CR1 and low expression of CD14 and CCR2. Similar results were found for clusters CM1 and CM4, though their correlation scores with the reference clusters were notably lower and, in the case of CM4, below a data-derived “assignability threshold”. CM1 cells expressed lower levels of CX3CR1 and CD16 than CM0, while CM4 cells expressed even lower levels of these two genes and higher levels of CD14 and CD64 (FCGR1A), despite being dissimilar to classical CD14+ monocytes. These three clusters likely represent infiltrating kidney monocyte/macrophage subsets as they constitute a minority of myeloid cells in normal kidneys (Supplementary Table 3).
Figure 3.
Focused analysis of kidney myeloid cells. a, Five clusters of myeloid cells were identified. The heatmaps show the expression of either canonical lineage markers (top) or genes differentially upregulated in each cluster (bottom). b, The results of classifying the kidney myeloid cells by correlating their gene expression to a set of ten reference clusters (Mono1-Mono4, DC1-DC6), taken from Villani et al.12 For each of the Five clusters identified in our data, the bars denote the percentage of cells most similar to each of the reference clusters. The percentage of cells mapped to the most frequent reference cluster in each case is specified on the corresponding bar. c, The distribution of highest Pearson correlation values per kidney myeloid cell, when compared with the reference clusters in Villani et al. The percentage of cells in each cluster for which the correlation score was above the assignability threshold is specified above the plot, followed by the number of cells in the cluster (n); the assignability threshold itself is denoted by the horizontal dashed line. d, The cells of clusters CM0 (red), CM1 (purple) and CM4 (blue), presented in two dimensions using diffusion maps. The arrow represents the direction of the putative transition between these three clusters, as explained in the text. e, The change in the inflammatory response score, calculated as the average scaled expression of several pro-inflammatory genes, along the trajectory shown in d; “pseudotime” represents the ordering of the cells along this trajectory. The violin plots (shades) show the distribution of expression levels in equally-spaced intervals along the pseudotime axis (and do not directly correspond to cell clusters). f, Same as e, but with regard to a set of genes associated with phagocytosis.
We next determined whether the pattern of gene expression in each cluster could indicate functional capabilities (Supplementary Fig. 3a). Cluster CM1 expressed upregulated levels of phagocytic receptors CD36 (SCARB3), SCARB2, CD68, CD163, NR1H3 (LXR) and GPNMB, and cluster CM4 expressed VSIG4, MSR1, CD163, MERTK, STAB1 and CD209. Cells in CM1 and especially CM4 had upregulated expression of C1Q, which acts as an opsonin for phagocytosis and promotes apoptotic cell clearance by enhancing the expression of MERTK and its soluble ligand GAS613, 14. They also demonstrated upregulated levels of CD169 (SIGLEC1), an endocytic receptor that is associated with a phagocytic and reparative phenotype15. Cluster CM0 had the highest level of expression of inflammatory genes including TNF, S100A8, S100A9, NFKB1 and the WNT pathway activator TCF7L2. By contrast, CM4 expressed many genes associated with alternatively activated macrophages, including CD163 and SLC40A1 (ferroportin), which control iron homeostasis16; IGF1 and DAB2, both drivers of the alternatively activated phenotype17, 18; and folate receptor beta (FOLR2), a receptor expressed on alternatively activated CD14+ macrophages that are found in inflammatory and malignant tissues19.
Finally, since CM2 was the main cluster found in normal kidneys (Supplementary Table 3), it probably corresponds to steady-state kidney macrophages. This cluster demonstrated low expression of CD14, CD16, CX3CR1 and CCR2, and no clear similarity to the published reference clusters of peripheral myeloid subsets (Fig. 3b,c). In comparison to the other macrophage subsets, CM2 upregulated several genes associated with tissue remodeling including MMP2, ADAMTS10 and HTRA1. These cells also upregulated BHLHE41, a gene expressed in microglia and lung resident macrophage populations20, consistent with CM2 representing resident cells. Compared with CM2 cells from living donor controls, lupus CM2 cells expressed higher levels of ISGs, as well as anti-inflammatory genes (GRN, TMSB4X, CREB5) and inhibitors of TLR signaling (GIT2, TNFAIP8L2), and lower levels of pro-inflammatory genes (ALOX15B, WNT5A) (Supplementary Table 5)20.
Trajectory analysis identifies a continuum of intermediate states spanning patrolling, phagocytic and alternatively activated monocytes
Dimensionality reduction using either diffusion maps21 (Fig. 3d) or t-Distributed Stochastic Neighbor Embedding22 (tSNE; Fig. 2a) indicated possible transitions between the three clusters of infiltrating monocytes/macrophages, with CM1 linking CM0 and CM4. Furthermore, since the cells in cluster CM0 tended to be the most similar to peripheral blood CD16+ monocytes, while the cells in cluster CM4 were the least similar (Fig. 3c), the suggested progression is from an inflammatory blood monocyte (CM0) to a phagocytic (CM1) and then an alternatively activated (CM4) phenotype. Indeed, we found a gradual reduction along the trajectory from CM0 to CM4 in the expression of NFKB1, an inflammatory gene; a transient increase in CD36, an important phagocytic receptor; and a continuous increase in MERTK, a key signaling receptor induced by CD36 (Supplementary Fig. 3d-f)23. Overall, a general downregulation of inflammatory genes and a concurrent upregulation of genes associated with phagocytosis (Supplementary Table 6) was observed along this trajectory (Fig. 3e,f).
To further investigate this hypothesized within-kidney transition, we analyzed blood samples from two of the patients who had high numbers of CM1 and CM4 cells in their kidneys (patient IDs 200–0873 and 200–0874; Supplementary Table 3). We used droplet-based scRNA-seq, yielding 1,411 sorted high-quality myeloid blood cells that included a subpopulation of CD16+ monocytes (Supplementary Fig. 3g). We next compared the gene expression data of each cell in this subpopulation with that of the myeloid kidney clusters. As expected, the vast majority of peripheral blood CD16+ cells were most similar to the CM0 cluster, with a few cells mapped to either CM1 or CM3 and no cell mapped to CM4 or CM2 (Supplementary Fig. 3h). This held true when considering all sorted blood myeloid cells, not just those identified as CD16+ monocytes.
To determine whether the hypothesized differentiation begins before entering the kidney, we examined the relative upregulation of phagocytosis-associated genes in cluster CM1 compared with CM0, in both blood and kidney (Supplementary Fig. 3i-j). We found that while there was a significant increase in these genes in kidneys (P < 0.001; Mann-Whitney U-test), no such increase could be observed in blood.
These analyses are consistent with differentiation of CD16+ monocytes into CM1 and CM4 cells within the kidney, but do not rule out differentiation of a small number of blood cells coupled with selective migration into the kidney. Furthermore, other schemes of transitions (or their absence) between these clusters are possible, and further investigation is required.
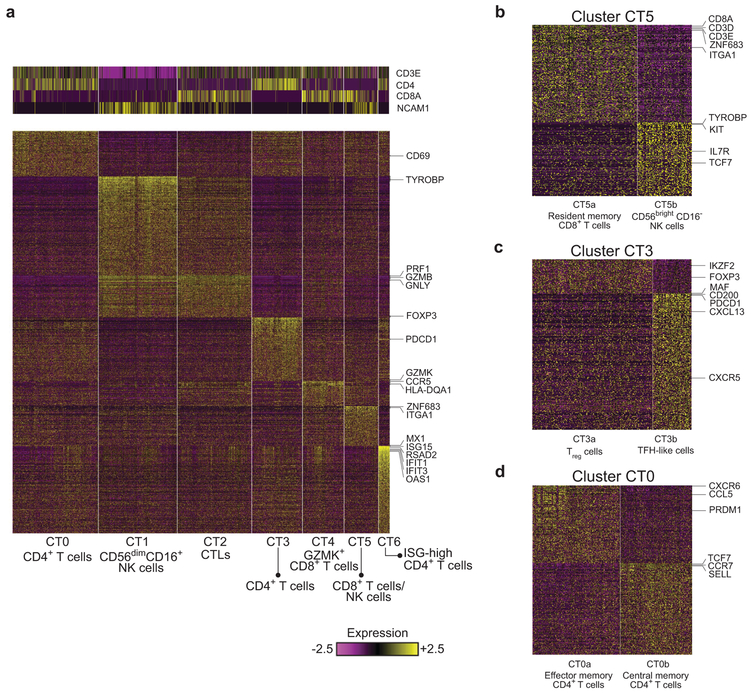
LN kidneys contain two clusters of NK cells and three clusters of CD8+ T cells
Clusters C0, C1, C2 and C5, comprising 1,764 cells, contained T cells and NK cells. A focused clustering of these cells separated them into seven finer clusters of NK, CD8+ T and CD4+ T cells (clusters CT0–CT6, Fig. 4a and Supplementary Fig. 4a). Cluster CT1 contained NK cells, which could be identified by the lack of CD3E and CD3D combined with expression of CD56 (NCAM1) and DAP12 (TYROBP), as well as high expression of cytotoxic genes including PRF1, GZMB, and GNLY. A similar cytotoxic program was observed in the CD8+ T cell cluster CT2, pointing to a cytotoxic T lymphocyte (CTL) identity. A second population of CD8+ T cells, demonstrating high levels of the granzyme GZMK24 rather than GZMB and GNLY, populated cluster CT4. These cells expressed relatively low levels of PRF1 compared with cluster CT2 and also showed high expression of HLA-DR/DP/DQ molecules and CCR5, consistent with an earlier report25. Cluster CT5 could be further split into two subclusters (Fig. 4b and Supplementary Fig. 4b): a third CD8+ T cell population (cluster CT5a), and a small population of NK cells (CT5b). The cells in cluster CT5a had features of resident memory cells, including expression of ZNF683 (HOBIT), ITGAE, ITGA1 and XCL1, and lack of KLF226, 27, and accordingly were relatively abundant in normal kidney biopsies (Supplementary Table 3). Cluster CT5b cells expressed TYROBP and CD56, suggesting an NK cell identity, but differed from CT1 NK cells by higher expression of KIT, TCF7, IL7R, and RUNX2, and lower expression of PRF1, GZMB, FCGR3A, TBX21, and S1PR5, consistent with the identification of these cells as tissue-resident CD56brightCD16− NK cells, in contrast to the CD56dimCD16+ NK cell features observed in CT128.
Figure 4.
Focused analysis of kidney T cells and NK cells. a, Preliminary analysis identified seven clusters. The heatmaps show the expression of either canonical markers defining T-cell and NK cell subsets (top) or genes differentially upregulated in each cluster specifically (bottom). b, The splitting of cluster CT5 into two subclusters, representing resident memory CD8+ T cells (CT5a) and CD56brightCD16− NK cells (CT5b). c, Cluster CT3 can be split into two subclusters, putatively corresponding to CD4+ Treg cells (CT3a) and TFH-like cells (CT3b). d, Analyzing the cells of cluster CT0 reveals two populations of cells, one putatively identified as early effector memory CD4+ T cells (CT0a), the other late central memory CD4+ T cells (CT0b).
It was previously reported that an exhaustion signature in peripheral blood CD8+ T cells of patients with SLE associates with lower flare rates29 and that such exhausted T cells are also detected in the kidneys of lupus mice30. In our data, however, all three CD8+ T cell clusters (CT2, CT4, CT5a) expressed only low levels of canonical exhaustion markers (Supplementary Fig. 4c); this was probably not due to technical limitations, as PD-1 (PDCD1), CTLA4 and BTLA were highly expressed in the T follicular helper (TFH)-like cells (cluster CT3b – see the following section). We sorted CD8+ T cells from matched blood samples obtained at the time of kidney biopsy along with ten healthy control samples, and used data from bulk RNA-seq to measure an “exhaustion score” defined as the average expression of a comprehensive, published list of exhaustion markers31 (Supplementary Table 6). We found that this exhaustion score was significantly higher in blood CD8+ T cells of patients with LN compared with those of healthy controls (P < 0.01, Mann-Whitney U-test), but not in kidney CD8+ T cells (Supplementary Fig. 4d-f), suggesting that the CD8+ T cell exhaustion seen in blood does not occur in the affected organ.
Analysis of CD4+ T-cell subsets identifies five clusters, including TFH-like cells
Clusters CT0, CT3 and CT6 contained CD4+ T cells. CT3 could be divided into two subclusters, with one (CT3a) containing cells expressing genes associated with T regulatory (Treg) cells, including FOXP3 and IKZF2 (HELIOS), and the other (CT3b) consisting of cells with low FOXP3 and features consistent with TFH cells, including the expression of CXCL13, CXCR5, PDCD1, MAF, and CD200 (Fig. 4c and Supplementary Fig. 4g).
Cluster CT0 could be further split into two subclusters, the first containing primarily effector memory CD4+ T cells (CT0a), with more frequent expression of PRDM1, CCL5 and CXCR6, and the second consisting of mostly CCR7+SELL+TCF7+ central memory T cells (CT0b; Fig 4d and Supplementary Fig. 4h). The similar expression of CD69 in both clusters suggests that CT0b cells are more likely to be central memory than naive CD4+ T cells. Expression of TCF7, KLF2 and LEF1 may indicate an early central memory T-cell (Tcm) phenotype of CT0b cells, in contrast to the late effector phenotype of CT0a cells32. Of note, CT0a was the only CD4+ T cell cluster found with substantial frequency in living donor samples (Supplementary Table 3). Differential expression analysis of this cluster indicated a dysregulation of ISGs in the LN samples (Supplementary Table 5).
While some LN kidney T cells have been previously annotated as TH1 and TH17 cells, in our data CD4+ T cells did not segregate into distinct clusters with characteristic effector lineage features. IFNG and CXCR3 could be identified in few CT0 cells, primarily within CT0a (Supplementary Fig. 4h). In contrast, IL17A, IL17F and CCR6 were very rarely detected, and no IL4, IL5 or IL13 expression was observed. TBX21 and RORC were found in a minority of CT0 cells, with TBX21 much more frequently expressed in CD56dimCD16+ NK cells (cluster CT1) and CTLs (CT2; Supplementary Fig. 4a).
Finally, cluster CT6 contained CD4+ T cells demonstrating exceptionally higher levels of ISGs, including ISG15, MX1, RSAD2, OAS3, IFIT1 and IFIT2, compared with other T cells (Supplementary Fig. 4a).
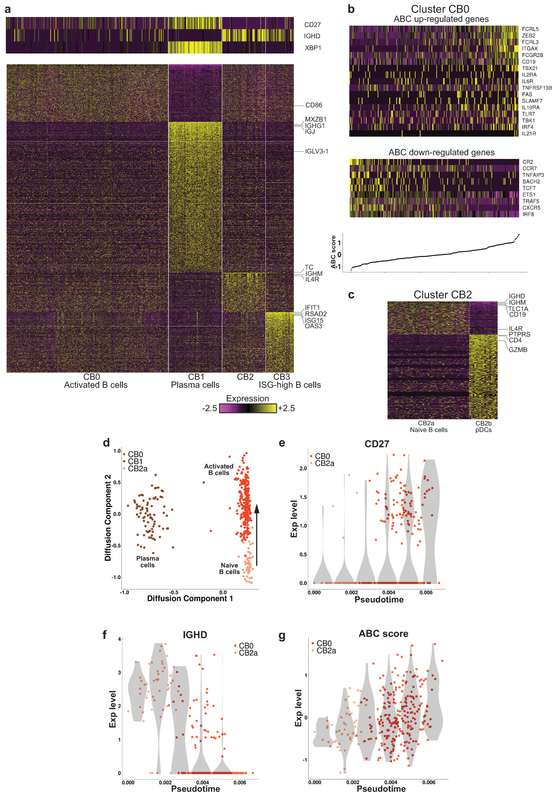
Analysis of B-cell clusters reveals age-associated B cells (ABCs)
Analysis of the 435 cells mapped to clusters C3 and C8 identified 4 different B-cell clusters in LN samples, but almost no B cells in healthy kidneys (clusters CB0–CB3; Fig. 5a, Supplementary Figs. 5a and 2d and Supplementary Table 3). Cluster CB1 contained plasmablasts/plasma cells, expressing high levels of XBP1 and MZB1, as well as immunoglobulin genes. The cells in cluster CB3 demonstrated high levels of several ISGs, including IFIT1, IFIT2, IFIT3, ISG15, OAS3 and RSAD2. Expression of these genes was also detected in the other B-cell clusters, but at substantially lower levels.
Figure 5.
Focused analysis of kidney B cells. a, Preliminary analysis identified four clusters. The heatmaps show the expression of either canonical markers defining B-cell subsets (top) or genes differentially upregulated in each cluster specifically (bottom). b, The expression of genes previously found to be differentially expressed in ABCs. The top heatmap pertains to genes known to be upregulated in ABCs, the bottom heatmap to genes downregulated in this subset. Columns are sorted by the ABC score, defined as the difference between the average expression of these two sets of genes. The bottom panel shows the ABC score per cell, such that each point on the line corresponds to the heatmap column directly above it. c, Cluster CB2 split into two subclusters, one corresponding to naive B cells (CB2a), the other to pDCs (CB2b). d, Projection of the cells in clusters CB0, CB1 and CB2a onto a two-dimensional plane, using diffusion maps. The arrow represents the hypothesized direction of transition along the trajectory from naive to activated B cells. e-f, The changes in the expression of CD27 and IGHD along the trajectory shown in d. g, The change in the ABC score along this trajectory. In (e)-(g), the violin plots (shades) show the distribution of expression levels in equally spaced intervals along the pseudotime axis (and do not directly correspond to cell clusters).
Cluster CB0 cells had upregulated expression of activation markers such as CD27, CD86, IGJ and IGHG1, and low levels of IGHD and IGHM, suggesting an activated B-cell identity. Furthermore, we could detect in this cluster a gene expression signature consistent with ABCs (Fig. 5b) that are implicated in both aging and autoimmunity33. Based on a panel of genes reported to be differentially expressed in ABCs34, we computed for each cell in cluster CB0 a score representing the extent to which its gene expression pattern matched that expected by an ABC (“ABC score”; Supplementary Table 6). A continuous range of values of this score could be observed in cluster CB0, without a clear separation into distinct subpopulations (Fig. 5b). The ABC score per patient, calculated as the average of the ABC score over the CB0 cells of each patient, did not positively correlate with age (Spearman’s ρ = −0.255), suggesting that the presence of these cells indeed reflected disease rather than age.
The tSNE plot for the B cells suggested that cluster CB2 may contain multiple subsets (Supplementary Fig. 5b). Accordingly, we were able to split the cells in cluster CB2 into two subclusters (Fig. 5c and Supplementary Fig. 5c): the first of these (CB2a), expressing the B cell markers CD19 and CD20 (MS4A1), demonstrated upregulation of genes typical of naive B cells, including high levels of IGHD, IGHM, TCL1A and IL4R, and had nearly undetectable expression of CD27; the other cluster (CB2b) expressed genes known to be upregulated in plasmacytoid DCs (pDCs) (Fig. 5c and Supplementary Fig. 5c), including PTPRS, GZMB, CLEC4C, CD123 (IL3RA) and CD317 (BST2). To further validate this hypothesized identification, we calculated the Pearson correlation in gene expression between each cell in cluster CB2 and 3 independent sets of reference samples: FANTOM535,36, containing bulk RNA-seq data from 360 cell types, 17 of which are immune cell subsets; bulk RNA-seq data from 13 immune cell populations sorted from healthy individuals (Browne et al., manuscript in preparation); and an scRNA-seq data set, which includes data from 10 different clusters of DCs and monocytes from healthy blood12. This analysis classified all CB2b cells as pDCs, using any of the three reference data sets (Supplementary Fig. 5d-f). Furthermore, as predicted almost all of the CB2a cells were classified as naive B cells, when compared with the data from Browne et al. (the only data set of the three tested that contained multiple B-cell populations).
Trajectory analysis reveals intermediate states between naive B cells and ABCs
Projecting the gene expression data of the B cells onto two dimensions using diffusion maps21, we found that the naive (CB2a) and activated (CB0) B cells formed a continuum of states, demonstrating a gradual increase in CD27 expression and a parallel decrease in IGHD expression, reflecting activation (Fig. 5d-f). Furthermore, traversing the trajectory from CB2a to CB0 coincided with a continuous increase in the ABC score (Fig. 5g), indicating that activation and differentiation into ABCs are highly correlated processes in our data. In contrast, very few cells occupied intermediate states between plasma and naive or activated B cells, consistent with a lack of differentiation into plasma cells in the inflamed kidney. However, as it was previously suggested37 that ABCs are preplasma cells, this question requires additional investigation, in particular employing B-cell receptor (BCR) repertoire analysis.
The dividing cell cluster includes T and NK cells
Cluster C9 contained three subclusters. Two of them (CD1 and CD2) demonstrated upregulated levels of mitochondrial genes and genes associated with a stress response (Supplementary Fig. 6a,b), indicating lower viability and/or quality, and were excluded from subsequent analyses. Cluster CD0 demonstrated elevated levels of genes participating in cell division. Classification of its cells by comparison with FANTOM5 indicated CD8+ T cells, NK cells and CD4+ Treg cells (Supplementary Fig. 6c).
Cluster-specific expression of genes associated with disease risk
Genome-wide association studies (GWASs) have identified numerous risk alleles and their susceptibility genes in SLE and LN38, 39. We analyzed the expression of these genes across the 22 clusters identified in kidneys, and found both expected and surprising cluster-specific expression patterns (Fig. 6). For example, TLR7, whose role in nucleic acid sensing, B cell activation and differentiation is well established40, 41 is expressed here in pDCs, myeloid cells and B cells. We found HIP1, suggested to regulate DC activity42, to be expressed in resident memory CD8+ T cells (cluster CT5a), CD56brightCD16− NK cells (CT5b), conventional dendritic cells (cDCs) (CM3) and pDCs (CB2b). LBH, implied to modulate synovial hyperplasia43, was expressed here in T- and B-cell subsets, raising the possibility that the LBH risk locus impacts both fibroblasts and lymphocyte subsets. We also observed cluster-specific expression of several poorly annotated SLE susceptibility genes, including WDFY4, CXorf21 and TMEM39A. Finally, our analysis identified both innate and adaptive immune cell subsets expressing several transcription factors associated with SLE, including ARID5B, CIITA, ETS, IKZF1, IKZF2, IRF7, IRF8 and PRDM1.
Figure 6.
The expression of GWAS genes in LN kidneys. The heatmap shows, for each gene, the scaled average expression over all cells in each cluster. Included are genes previously indicated in lupus by GWAS, considering only genes that demonstrated high variability across clusters in our data. Both rows and columns are clustered based on Euclidean distance.
Expression patterns of chemokines and cytokines
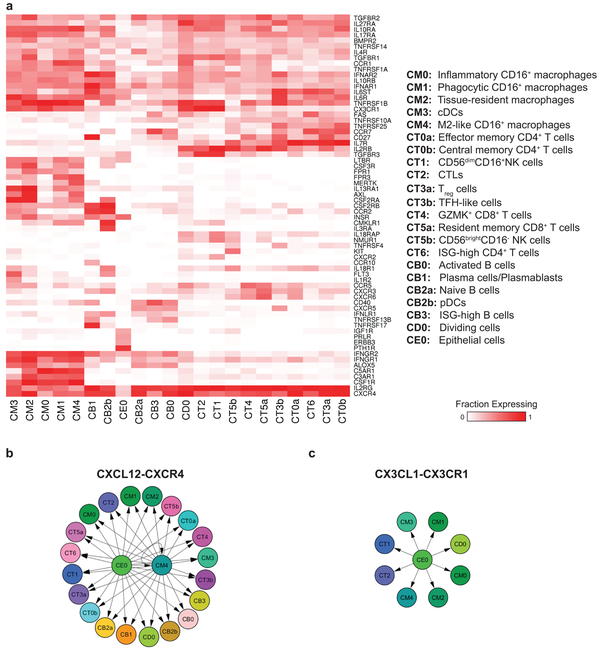
We next analyzed the expression patterns of chemokine and cytokine receptors (Fig. 7a), focusing on receptors that were expressed by a relatively large fraction (> 30%) of the cells in at least 1 cluster (this threshold was set based on the observed distribution of expression frequency, considering all receptors; Supplementary Fig. 7a). We found that a single chemokine receptor, CXCR4, was expressed in the majority of infiltrating cells in nearly all clusters (Supplementary Fig. 7b). A second chemokine receptor, CX3CR1, was expressed in most myeloid cells, as well as CD56dimCD16+ NK cells (cluster CT1) and CTLs (CT2) (Supplementary Fig. 7c). Of note, the expression frequency of other chemokine receptors previously implicated in LN, such as CCR5, CXCR3 and CCR2, was found to be much lower (Supplementary Fig. 7d-f). For cytokine receptors, we observed that IL2RG, encoding the common gamma chain and important for signaling of several cytokines, was frequently expressed in almost all clusters. TGFBR2, a subunit of the receptor for the cytokine TGFβ, was also expressed on the majority of cells. IL10RA, IL27RA, IL17RA and TNFRSF1B were expressed by a large fraction of cells in all clusters, with the exception of the B cells.
Figure 7.
Chemokine- and cytokine-mediated cellular networks. a, The pattern of chemokine receptor expression over the cell clusters. The color codes for fraction of cells expressing each receptor. Shown are receptors that are expressed in at least 30% of the cells of at least one cluster. Both rows and columns are clustered based on Euclidean distance. b, The producers-consumers cellular network corresponding to the chemokine CXCL12 and its receptor CXCR4. c, The producers-consumers cellular network of the chemokine CX3CL1 and its receptor CX3CR1.
To identify potential interactions between the cells acting in the inflamed kidney, we analyzed the expression patterns of the corresponding ligands. We found that the CXCR4 ligand, CXCL12, was expressed mainly in the cells in cluster CM4, as well as in the epithelial cells (Fig. 7b and Supplementary Fig. 7g). The latter were also the main source of the CX3CR1 ligand, CX3CL1 (Fig. 7c and Supplementary Fig. 7h). Of note, CM4 cells were in addition the top producers of CCL2 and CCL8 (Supplementary Fig. 7i); these are the ligands of CCR2, which is expressed in a large fraction of plasma cells (cluster CB1) and pDCs (cluster CB2b). These findings imply that kidney epithelial cells and M2-like macrophages may be coordinating traffic of immune cells infiltrating the kidney. It should be noted though that this analysis does not cover other cell types, such as endothelial cells, which were not profiled here.
Comparison of urine and kidney leukocytes
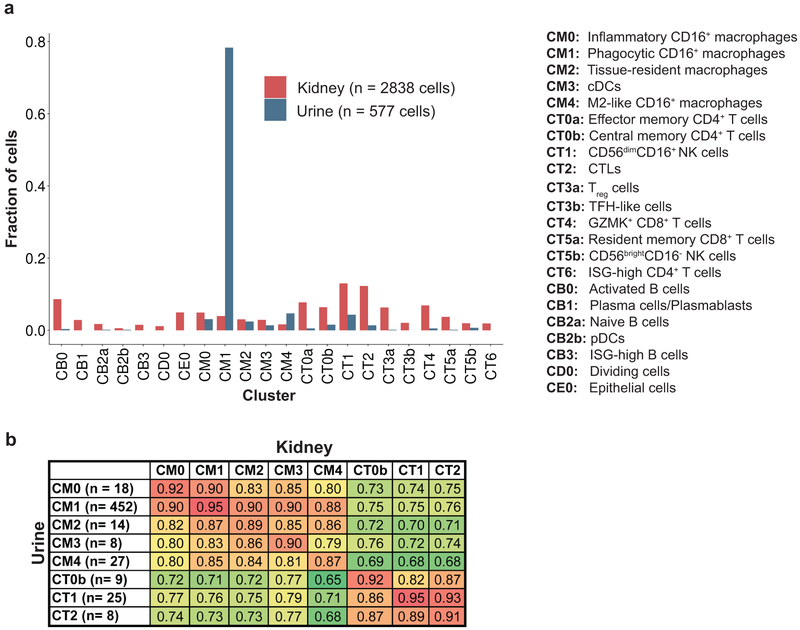
Leukocytes isolated from urine samples of patients with LN were processed in the same way as kidney cells (Fig. 1a and Supplementary Fig. 1h). Following filtering, 577 high-quality cells, collected from 8 patients, were included in subsequent analyses.
We first assigned each urine cell to the kidney cluster most similar in its gene expression data. Urine samples had a higher frequency of myeloid cells (in particular cluster CM1) and fewer T cells than kidneys (Fig. 8a). We next compared gene expression across corresponding urine and kidney clusters, restricting the comparison to clusters with at least five urine cells. High correlations were observed, typically ranging from 0.85 to 0.95 (Fig. 8b and Supplementary Fig. 8), suggesting that urine cells can serve to estimate gene expression in their kidney counterparts.
Figure 8.
Comparison of immune cells extracted from urine samples and from kidney samples. a, The relative frequency of each cluster in urine and in kidney. b, Pearson correlation values between gene expression data of urine and kidney clusters, computed using the average gene expression taken over the cells in each cluster, and considering only clusters that had at least five urine cells.
DISCUSSION
Using single-cell transcriptomics to study kidney samples obtained from patients with LN and living donor controls, we reveal the complexity of immune populations in LN kidneys, identifying multiple disease-specific subsets of myeloid, NK, T and B cells, and giving rise to several observations. We found: (1) evidence for within-tissue differentiation of inflammatory CD16+ macrophages into M2-like cells, which may orchestrate the renal infiltration and retention of other leukocyte subsets; (2) an abundance of dividing CTLs and NK cells, indicated to be a major source of IFNγ and cytolytic molecules, and lack of expression of exhaustion markers in CD8+ T cells, suggesting a role for cytotoxic activity in LN; (3) two additional populations of CD8+ T cells, which could not be easily identified by cell surface markers; (4) a range of B-cell activation states from naive cells to ABCs in the kidney; (5) an interferon response signature in infiltrating leukocytes, correlated with the same signature in blood; (6) frequent expression by kidney immune cells of the chemokine receptors CXCR4 and CX3CR1, suggesting they may serve as potential therapeutic targets; (7) cell subset-specific expression of genes associated with lupus in GWAS; and (8) a high correlation of gene expression in urine immune cells and corresponding kidney leukocytes.
Our transcriptomic analysis offered a detailed view of the T-cell populations in LN kidneys. The co-clustering of CD4+ TFH cells and FoxP3+Helios+ regulatory T cells raises the possibility that T follicular regulatory cells are also present44, 45. CD4+ T cell clusters were not clearly associated with TH1 or TH17 signatures, suggesting that T cell polarization may not be a major feature in LN. The identification of three distinct CD8+ T-cell subsets raises the question whether the previously reported localization pattern of CD8+ T cells in the kidney46 is subset-specific.
B cells are found in more than half of lupus biopsies but not in healthy samples47. Our finding of B cells spanning a spectrum of states between naive and activated cells, together with the presence of TFH-like cells, is consistent with the view that immune responses to tissue damage are being driven in situ48. Of note, activation was correlated with an ABC signature previously suggested to be driven by BCR/TLR ligands49. Understanding whether these ABCs are clonally expanded or enhance inflammation locally in patients with LN, and determining the clonal relatedness of naive, activated and antibody-secreting B cells will require a larger data set, combined with analysis of BCR sequences.
Peripheral monocytes can enter injured tissues and differentiate into inflammatory and reparative/resolving macrophages50. If the cells are chronically exposed to damage-associated molecular pattern molecules (DAMPs) and endosomal TLR ligands, resolution may fail, and macrophages with mixed functions may emerge51. Here, the three subpopulations of CD16+ macrophages are suggested to transition through an inflammatory to a resolution phase; such a functional switch was previously identified in a mouse model of acute inflammation52. Of note, we did not identify a cluster of infiltrating cells with high similarity to CD14+ monocytes. It is still unclear why some types of tissue injury recruit CD14+ macrophages while others recruit CD16+ macrophages; influences may include the types of expressed DAMPS and/or other microenvironmental cues such as cytokines and chemokines.
Our study demonstrates the feasibility of profiling kidney samples using single-cell transcriptomics, employing a freezing strategy to minimize batch effects that could mask subtle gene expression signatures. While this strategy may result in the loss of neutrophils and can alter the relative frequency of other cell subsets, we did not observe a major effect on gene expression due to freezing.
Despite our cohort’s relatively high diversity with respect to histologic appearance and intercurrent therapies, we observed a surprising number of commonalities. Furthermore, saturation analysis indicated that increasing the cohort size is not expected to drastically change the presented cell subset catalogue. Rather, such an increase can enable investigating how the presence and transcription profiles of particular cell infiltrates are related to disease manifestations and treatment responses. A study currently in progress, performed as part of the AMP RA/SLE consortium, will utilize the sample processing strategy developed here to analyze a much larger cohort, addressing these questions. Furthermore, profiling stromal renal cells together with leukocytes will elucidate their interactions. The results discovered in such studies will be further validated using tissue staining, functional studies in cell lines or primary human cells, and animal models of disease.
METHODS
Human kidney tissue, urine and blood acquisition
Renal tissue, urine and blood specimens from patients with LN were acquired at ten clinical sites in the United States. Institutional review board approval was received at each site. Research biopsy cores were collected from consented subjects either as an additional biopsy pass obtained specifically for research during a clinically indicated biopsy procedure (nine sites), or as a portion of a biopsy specimen acquired for diagnostic pathology during a clinically-indicated biopsy procedure (one site). Control kidney samples were obtained at a single site by biopsy of a living donor kidney after removal from the donor and before implantation in the recipient.
After acquisition, kidney biopsy samples were placed into HypoThermosol FRS preservation solution for 10–30min on ice and then transferred to a cryovial containing 1ml CryoStor CS10 cryopreservation medium (BioLife Solutions). The cryovial was incubated on ice for 20–30min and was then placed in a Mr. Frosty freezing container (Nalgene, catalog no. 5100–0001) and transferred to a −80°C freezer overnight. Cryopreserved samples were then stored in liquid nitrogen and shipped on dry ice to the central processing site, where they were stored in liquid nitrogen until processing.
Kidney tissue thawing and dissociation into a single-cell suspension
Kidney samples were thawed and processed in batches of four samples, with most batches containing both LN and control kidney samples. The cryovial containing the kidney tissue was rapidly warmed in a 37°C water bath until almost thawed. The sample was then poured into a well of a 24-well dish and rinsed in a second well containing warmed RPMI/10%FBS. The tissue was incubated for 10min at room temperature. Specimens were cut into 2–3 pieces and placed into a 1.5 ml centrifuge tube containing 445ul of Advanced DMEM/F-12 (ThermoFisher Scientific, catalog no.12634–028) and 5ul of DNase I (Roche, catalog no.04536282001, 100U/ml final concentration). 50ul of Liberase TL (Roche, catalog no.05401020001, 250ug/mL final concentration) was added, and the tube was placed on an orbital shaker (300–500 r.p.m.) at 37°C for 12min. At 6min into the digestion, the mixture was gently pipetted up and down several times using a cut 1 mL pipette tip. After 12min, 500ul of RPMI/10% FBS was added to stop the digestion. The resulting cell suspension was filtered through a 70-µm filter into a new 1.7 ml microfuge tube. The cells were washed with RPMI/10%FBS, centrifuged at 300g at 4°C for 10min, and resuspended in cold PBS for downstream analyses. Quantification of cell yields was performed by hemocytometer with trypan blue exclusion and by flow cytometry with propidium iodide-exclusion. Yields of cell subsets (leukocytes, epithelial cells) were quantified by acquiring the entire sample through the flow sorter and plotting the number of intact, PI-negative cell events with the appropriate surface markers. Cell yields were normalized to input tissue mass.
Urine cell pellet collection and cryopreservation protocol
Midstream urine samples were collected from patients with LN before kidney biopsy. The total urine volume (15–90 mL) was split into two 50 mL Falcon tubes. Urine cells were pelleted by centrifugation at 200g for 10min, and then resuspended in 1 ml cold X-VIVO10 medium (Lonza BE04–743Q). Cells were transferred to a microcentrifuge tube, washed once in 1ml X-VIVO10 medium, and then resuspended in 0.5 ml cold CryoStor CS10. Cells were transferred into a 1.8 mL cryovial, placed in a Mr. Frosty freezing container, stored in at −80ºC overnight, and then transferred to liquid nitrogen. For downstream analyses, cryopreserved urine cells were rapidly thawed by vigorous shaking in a 37°C water bath, transferred into warm RPMI/10%FBS, centrifuged at 300g for 10min, and resuspended in cold HBSS/1%BSA.
Flow cytometric cell sorting of kidney and urine samples
An 11-color flow cytometry panel was developed to identify epithelial cells and leukocyte populations within dissociated kidney cells. Antibodies include anti-CD45-FITC (HI30), anti-CD19-PE (HIB19), anti-CD11c-PerCP/Cy5.5 (Bu15), anti-CD10-BV421 (HI10A), anti-CD14-BV510 (M5E2), anti-CD3-BV605 (UCHT1), anti-CD4-BV650 (RPA-T4), anti-CD8-BV711 (SK1), anti-CD31-AlexaFluor700 (WM59), anti-PD-1-APC (EH12.2H7), and propidium iodide (all from BioLegend). Kidney or urine cells were incubated with antibodies in HBSS/1%BSA for 30min. Cells were washed once in HBSS/1%BSA, centrifuged, and passed through a 70 μm filter. Cells were sorted on a 3-laser BD FACSAria Fusion cell sorter. Intact cells were gated according to FSC-A and SSC-A. Doublets were excluded by serial FSC-H/FSC-W and SSC-H/SSC-W gates. Non-viable cells were excluded based on propidium iodide uptake. Cells were sorted through a 100 micron nozzle at 20 psi. For each sample, 10% of the sample was allocated to sort CD10+CD45− epithelial cells as single cells, and the remaining 90% of the sample was used to sort CD45+ leukocytes as single cells. Single cells were sorted into 384-well plates containing 6µl 1% NP-40 with index sorting, and plates were immediately frozen and stored at −80 °C. Flow cytometric quantification of cell populations was performed using FlowJo 10.0.7.
Library preparation and RNA sequencing of kidney and urine samples
scRNA-seq was performed using the CEL-Seq2 method10 with the following modifications. Single cells were sorted into 384-well plates containing 0.6µl 1% NP-40 buffer in each well. Then, 0.6µl dNTPs (10mM each; NEB) and 5nl of barcoded reverse transcription primer (1µg/µl) were added to each well along with 20nL of ERCC spike-in (diluted 1:800,000). Reactions were incubated at 65°C for 5min, and then moved immediately to ice. Reverse transcription reaction and second-strand complementary DNA (cDNA) synthesis were carried out as previously described10 and double-stranded c-DNA was purified using 0.8X volumes of AMPure XP beads (Beckman Coulter). In vitro transcription reactions were performed as described followed by treatment with ExoSAP-IT PCR Product Cleanup Reagent (ThermoFisher Scientific, catalog no. 78201.1.ML). Amplified RNA (aRNA) was fragmented at 80°C for 3min and purified using RNAClean XP beads (Beckman Coulter). The purified aRNA was converted to cDNA using an anchored random primer and Illumina adaptor sequences were added by PCR. The final cDNA library was purified using AMPure XP beads (Beckman Coulter). Paired-end sequencing of ~1 million paired-end reads per cell was performed on the HiSeq 2500 in Rapid Run Mode with a 5% PhiX spike-in using 15 bases for Read1, 6 bases for the Illumina index and 36 bases for Read2.
Frozen needle core biopsies obtained from two additional healthy donor kidneys before to reperfusion were processed as above to produce single cell suspensions. Unsorted cells in 0.04% BSA (Sigma) were used to generate single cell libraries with the Chromium Single Cell Gene Expression system using 3′ Library & Gel Bead Kit v2 (10X Genomics) and paired-end sequencing was performed on a HiSeq X.
Processing of blood samples
For profiling blood cells, blood was collected from 10 patients with LN before kidney biopsy and peripheral blood mononuclear cells (PBMCs) were isolated using Ficoll-Paque PLUS (GE Healthcare) density gradient centrifugation in 15ml SepMate tubes (Stemcell) according to manufacturer instructions and cryopreserved in CryoStor CS10 Freezing Media (STEMCELL Technologies). Sex-matched PBMCs from healthy donors isolated and cryopreserved at the University of North Carolina Kidney Center were used as controls.
For bulk RNA-seq experiments, thawed cells were stained with antibodies against CD45-PE (HI30), CD3-PE/Cy7 (UCHT1), and CD8a-APC (HIT8a). 5,000 viable (DAPI−) PBMC (CD45+) or 1,000–5,000 CD8+ T cells (CD45+CD3+CD8+) were sorted into microcentrifuge tubes containing 20μl TCL buffer (Qiagen) supplemented with 1% b-mercaptoethanol (Sigma) using a Sony SH800S cell sorter, and the lysate was frozen and stored at −80 degrees. Of ten LN patient, 8 had enough CD8+ T cells to allow sequencing. RNA was isolated with Agencourt RNAClean XP beads (Beckman Coulter) and converted to sequencing libraries using the Smart-seq2 method53. 38bp paired-end reads were generated on a Nextseq500 (Illumina).
For droplet-based scRNA-seq, thawed cells were stained with antibodies from Biolegend: CD3-FITC (HIT3a), CD19-FITC (HIB19), CD20-FITC (2H7), CD56-FITC (HCD56), HLADR-PE/Dazzle (L243), CD16-PerCP/Cy5.5 (3G8), and from BD Biosciences: CD14-APC/Cy7 (MφP9). Viable (DAPI−) monocytes (CD3−, CD19−, CD20−, CD56−, HLADR+, CD14+ or CD16+) were sorted into RPMI (Life Technologies) + 0.04% BSA (Sigma) and single cell libraries were generated using the Chromium Single Cell Gene Expression system using 3′ Library & Gel Bead Kit v2 (10X Genomics). Paired-end sequencing was performed on a Nextseq500.
RNA-seq data processing
For the cells processed using CEL-Seq2, we used a modified version of the Drop-seq pipeline developed by the McCarroll lab54, to perform all steps necessary to produce gene by cell expression matrices of reads as well as unique molecular identifiers (UMIs). These steps include demultiplexing, quality filtering, polyA and adapter trimming, aligning, and collapsing reads with unique combinations of cell+gene+UMI. We used STAR-2.5.1b to align reads to the Hg19 human genome reference. Only uniquely mapped reads were counted. UMIs with fewer than 10 reads were filtered out before creating the final expression matrices, to minimize read cross-contamination across cells. For each cell, the computed gene expression counts were then normalized for read depth and log transformed. For cells processed using 10x, sequencing output was aligned using the 10x standard pipeline.
Cell filtering and quality control
For kidney cells processed using CEL-Seq2, high-quality cells were defined as having at least 1,000 detected genes (that is, with positive count values); for urine cells, which tended to have fewer detectable genes, this threshold was set to 500 genes; for cells processed using 10x, the threshold used was 250 genes. We further required the percentage of reads mapped to mitochondrial genes per cell to be lower than 25% (8% for blood cells processed using 10x). To remove wells that were suspected to contain messenger RNA from multiple cells, we required the number of genes per cell to be smaller than 5,000 for the kidney cells processed using CEL-Seq; 4,000 for urine cells; 1,700 for blood cells processed using 10x; 3,500 for the kidney cells processed using 10x (all thresholds were set based on empirical distributions).
To minimize the effect of technical factors, we tested different regression models, taking into account such variables as the plate identifier, number of UMIs per cell, and the percentage of mitochondrial genes per cell. We found that using such models had a negligible effect on the gene by cell expression matrix, as well as the overall results of clustering. We therefore decided to avoid employing them in cleaning the data for subsequent analyses.
In the analysis of myeloid blood cells, initial comparison to the gene expression data of immune cell subsets in FANTOM5 was performed to further validate their myeloid cell identify, in addition to filtering based on flow cytometry cell sorting.
Cell clustering
Clustering of kidney cells was done using Seurat (v1.4.0.8)55, in a stepwise manner. We initially performed low-resolution clustering, analyzing all cells together, then labeled each of the resulting clusters as myeloid cells, T/NK cells, B cells, dividing cells or epithelial cells. The cells of each such general class were then analyzed separately, to identify finer clusters. In some cases, as described in the main text, the resulting clusters were further split into subclusters. In each case, clustering was done following principal component analysis, based on context-specific variable genes that were identified independently for each set of analyzed cells.
Sensitivity analysis was performed in each clustering step, with a particular focus on the low-resolution clustering stage. Briefly, all parameters in the clustering process, including the number of variable genes and principal components considered, were varied, and the robustness of the results was determined. To assess this robustness, we estimated in each case the Rand index: looking at a large number (1,000) of random pairs of cells, we counted how many pairs were either included in the same cluster in both of the compared clustering runs, or not included in the same cluster, and referred to these as consistent pairs; we then calculated the fraction of consistent pairs of all random cell pairs considered. We repeated this procedure 100 times, to calculate the mean of the Rand index estimate.
Classification by correlation
In determining their putative identity, we compared the gene expression of individual cells with external gene expression data sets of reference samples. In each comparison, we computed the Pearson correlation between the log-transformed gene expression data of the cell and the reference sample, and chose the reference sample that produced the maximal correlation value (using Spearman correlation instead of Pearson correlation did not drastically change the classification results). To assess the confidence in this classification, we computed in each case an “assignability threshold”: we generated 1,000 “random cells”, by averaging for each gene the raw counts across the classified cells using random weights, such that the sum of weights for each gene was 1; we then normalized the sum of counts to 10,000. For each random cell, we identified the most similar reference sample, and recorded the corresponding Pearson correlation. The assignability threshold was set to the 95th percentile of the distribution of these Pearson correlations. We note that this approach preserves the main aspects of the original data; in particular, highly expressed genes (such as house-keeping genes) remain high in the generated random cells.
Myeloid cells were compared with the scRNA-seq data published in Villani et al.12, such that each cluster in that study was represented by the average expression over the cells included in it, taking into account only genes showing high variability in that data set. Similar results were found if all genes, or only cluster markers, were considered. Comparison to FANTOM5 and the data from Browne et al. was done based on the median of reference sample replicates, considering all genes.
Differential expression analysis
Identification of genes differentially expressed between patients with LN and living donor controls was done using the framework proposed by McDavid et al.56, as implemented by Seurat. P values were corrected for multiple comparisons using the Benjamini-Hochberg method57. For each gene with a corrected P value smaller than 0.05 (“candidate differentially expressed genes”), a further correction for the number of patients was performed: 1,000 random permutations of patients across the two groups were generated, while keeping the number of patients in each group fixed. For each candidate differentially expressed gene and each random permutation, the McDavid test statistic was computed as above. We then calculated a new P value for each candidate differentially expressed gene, defined as the fraction of random permutations in which the value of the test statistic was more extreme than its value in the original partition of patients between groups (while adding 1 to both numerator and denominator). Finally, the new P values were corrected for multiple comparisons using the Benjamini-Hochberg method. This analysis was performed separately for each cluster with at least 20 cells in both patient groups.
Trajectory analysis
Trajectory analysis was performed based on dimensionality reduction using diffusion maps, as implemented in the Destiny software package (v2.6.2)21. In each case, only the cells relevant to the question at hand were analyzed.
Calculation of gene set-based scores
Scores based on specific gene sets (for example, interferon response score, ABC score etc.) were calculated for each cell as the average of the scaled (Z-normalized) expression of the genes in the list. To control for the variable quality and complexity of the data of different cells, the score of each cell was corrected by subtracting the average of a large set of similarly-expressed genes, as proposed by Tirosh et al58. When the list contained genes that are expected to be upregulated in a particular condition and genes that are expected to be downregulated in it (as was the case for the ABC score), the average of scaled expression was calculated separately for each set of genes, and the difference between the scores of the upregulated genes and the downregulated genes was taken as the overall score. A similar approach was used when calculating gene-set based scores in bulk RNA-seq data. The gene sets used for particular scores can be found in Supplementary Table 6.
Analysis of interferon response score
To assess the statistical significance of ISGs upregulation per patient, an interferon response score was calculated for each cell based on a given list of ISGs, as explained above. For each patient with LN, the distribution of the calculated scores was compared with that of cells collected from living donor controls, using the two-tailed Mann-Whitney U-test. The derived P values were then corrected for multiple comparisons, using the Benjamini-Hochberg method57. A similar approach was used when comparing the distribution of ISG scores per cluster in the patient with LN, as compared with the LD cells taken as a whole.
Analysis of GWAS gene expression
We analyzed the expression patterns of 180 genes previously reported in GWASs of either SLE or LN. For each such gene, we calculated its average scaled (Z-normalized) expression in each cell cluster, taking into account only cells coming from LN samples. For biclustering of GWAS genes and cell clusters, we kept only genes that had an average scaled expression value of more than 1 or less than −1 in at least one cell cluster, such that biclustering was based only on the GWAS genes that were relatively variable in our data. Biclustering was then performed, based on the average scaled expression in each cell cluster and using a Euclidean distance metric. cDCs, conventional dendritic cells.
Analysis of chemokine/cytokine receptors
Analysis of chemokine/cytokine receptors was based on a receptor-ligand pairs list downloaded from the International Union of Basic and Clinical Pharmacology (IUPHAR) and British Pharmacological Society (BPS) database59 and extended manually to incorporate a number of missing, previously published pairs. For each receptor, we calculated the percentage of cells expressing it in each cell cluster, where a receptor was said to be expressed by a cell if it had at least one mapped read (the results reported here were found to be robust to changes in this threshold). For biclustering of receptors and cell clusters, we kept only receptors that appeared in at least 30% of the cells in at least one cluster.
Assignment of urine cells to kidney clusters
For each urine cell, we computed its Pearson correlation with each kidney cluster, taking the average over the kidney cells included in the cluster. The urine cell was then assigned to the cluster that produced the highest correlation value.
CODE AVAILABILITY
All R scripts used to analyze the data reported in this publication are available from the corresponding authors upon request.
DATA AVAILABILITY
The data reported in this publication, including the clinical and serological data of the study participants, are deposited in the ImmPort repository (accession code SDY997). The raw single-cell RNA-seq data are also deposited in dbGAP (accession code phs001457.v1.p1). The processed data can be viewed using an interactive browser at https://immunogenomics.io/ampsle, https://immunogenomics.io/cellbrowser/, and https://portals.broadinstitute.org/single_cell/study/amp-phase-1.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the Accelerating Medicines Partnership (AMP) in Rheumatoid Arthritis and Lupus Network. AMP is a public-private partnership (AbbVie Inc., Arthritis Foundation, Bristol-Myers Squibb Company, Foundation for the National Institutes of Health, Janssen Pharmaceuticals, Lupus Foundation of America, Lupus Research Alliance, Merck Sharp & Dohme Corp., National Institute of Allergy and Infectious Diseases, National Institute of Arthritis and Musculoskeletal and Skin Diseases, Pfizer Inc., Rheumatology Research Foundation, Sanofi, and Takeda Pharmaceuticals International, Inc.) created to develop new ways of identifying and validating promising biological targets for diagnostics and drug development. Funding was provided through grants from the National Institutes of Health (UH2-AR067676, UH2-AR067677, UH2-AR067679, UH2-AR067681, UH2-AR067685, UH2-AR067688, UH2-AR067689, UH2-AR067690, UH2-AR067691, UH2-AR067694, and UM2-AR067678). N.H. was supported by the David P. Ryan, MD Endowed Chair in Cancer Research. We thank participating Lupus Nephritis Trials Network clinical sites and participants.
The complete list of authors (including consortium members) is as follows:
Arnon Arazi1, Deepak A. Rao2, Celine C. Berthier3, Anne Davidson4, Yanyan Liu2, Paul J. Hoover1, Adam Chicoine2, Thomas M. Eisenhaure1, A. Helena Jonsson2, Shuqiang Li1, David J. Lieb1, Fan Zhang2, Kamil Slowikowski2, Edward P. Browne5, Akiko Noma1, Danielle Sutherby6, Scott Steelman7, Dawn E. Smilek8,9, Patti Tosta8,9, William Apruzzese2, Elena Massarotti2, Maria Dall’Era10, Meyeon Park11, Diane L. Kamen12, Richard A. Furie13, Fernanda Payan-Schober14, William F. Pendergraft III15, Elizabeth A. McInnis16, Jill P. Buyon17, Michelle A. Petri18, Chaim Putterman19, Kenneth C. Kalunian20, E. Steve Woodle21, James A. Lederer22, David A. Hildeman23,24, Chad Nusbaum7, Soumya Raychaudhuri2, Matthias Kretzler3, Jennifer H. Anolik25, Michael B. Brenner2, David Wofsy10, Nir Hacohen1, Betty Diamond4, Dia Waguespack26, Sean M. Connery14, Maureen A. McMahon27, William J. McCune3, Ruba B. Kado3, Raymond Hsu11, Melissa A. Cunningham12, Paul J. Utz28, Mina Pichavant28, Holden T. Maecker29, Rohit Gupta30, Judith A. James31, Joel M. Guthridge31, Chamith Fonseka2, Evan Der19, Robert Clancy17, Thomas Tuschl32, Hemant Suryawanshi32, Andrea Fava18, H. Michael Belmont17, Peter M. Izmirly17, Pavel Morozov34, Manjunath Kustagi34 and Daniel H. Goldman18
1 Broad Institute of MIT and Harvard, Cambridge Massachusetts, USA
2 Division of Rheumatology, Immunology, Allergy, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston Massachusetts, USA
3 Internal Medicine, Department of Nephrology, University of Michigan, Ann Arbor, Michigan, USA
4 Center for Autoimmune and Musculoskeletal Diseases, The Feinstein Institute for Medical Research, Northwell Health, Manhasset, New York, USA
5 UNC HIV Cure Center and Department of Medicine, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA.
6 Celsius Therapeutics, Cambridge, Massachusetts, USA
7 Cellarity, Inc., Cambridge, Massachusetts, USA
8 Lupus Nephritis Trials Network, University of California San Francisco, San Francisco, California, USA
9 Immune Tolerance Network, University of California San Francisco, San Francisco, California, USA
10 Rheumatology Division, University of California San Francisco, San Francisco, California, USA
11 Division of Nephrology, University of California San Francisco, San Francisco, California, USA
12 Division of Rheumatology and Immunology, Medical University of South Carolina, Charleston, South Carolina, USA
13 Division of Rheumatology, Northwell Health, Great Neck, New York, USA
14 Department of Medicine, Paul L. Foster School of Medicine, Texas Tech University Health Sciences Center, El Paso, Texas, USA.
15 The Integrative Medical Clinic of North Carolina, PLLC, Chapel Hill, North Carolina, USA
16 University of North Carolina Kidney Center, Division of Nephrology and Hypertension, Department of Medicine, UNC School of Medicine, Chapel Hill, North Carolina, USA
17 Department of Medicine, Division of Rheumatology, New York University School of Medicine, New York, New York, USA
18 Division of Rheumatology, Johns Hopkins University, Baltimore, Maryland, USA
19 Division of Rheumatology and Department of Microbiology and Immunology, Albert Einstein College of Medicine and Montefiore Medical Center, Bronx, New York, USA
20 University of California San Diego School of Medicine, La Jolla, California, USA
21 Division of Transplantation, Department of Surgery, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA
22 Department of Surgery, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA
23 Department of Pediatrics, University of Cincinnati, Cincinnati, Ohio, USA
24 Division of Immunobiology, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA
25 Department of Medicine, Division of Allergy, Immunology, and Rheumatology, University of Rochester Medical Center, Rochester, New York, USA
26 Department of Internal Medicine, University of Texas, Houston, Texas, USA
27 Department of Medicine, University of California Los Angeles, Los Angeles, California, USA
28 Department of Medicine, Division of Immunology and Rheumatology, Stanford University School of Medicine, Stanford, California, USA
29 Department of Microbiology & Immunology, Stanford University School of Medicine, Stanford, California, USA
30 Institute for Immunity, Transplantation, and Infection, Stanford University School of Medicine, Stanford, California, USA
31 Arthritis and Clinical Immunology, Oklahoma Medical Research Foundation, Oklahoma City, Oklahoma, USA
32 Laboratory for RNA Molecular Biology, The Rockefeller University, New York, New York, USA
34 Laboratory of RNA Molecular Biology, Rockefeller University, New York, New York, USA
Footnotes
COMPETING INTERESTS
M.B.B. is a consultant to Roche in the area of stromal cells.
REFERENCES
- 1.Costenbader KH et al. Trends in the incidence, demographics, and outcomes of end-stage renal disease due to lupus nephritis in the US from 1995 to 2006. Arthritis Rheum 63, 1681–1688 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Narain S & Furie R Update on clinical trials in systemic lupus erythematosus. Curr Opin Rheumatol 28, 477–487 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Tektonidou MG, Dasgupta A & Ward MM Risk of End-Stage Renal Disease in Patients With Lupus Nephritis, 1971–2015: A Systematic Review and Bayesian Meta-Analysis. Arthritis Rheumatol 68, 1432–1441 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thanou A & Merrill JT Treatment of systemic lupus erythematosus: new therapeutic avenues and blind alleys. Nat Rev Rheumatol 10, 23–34 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Banchereau R et al. Personalized Immunomonitoring Uncovers Molecular Networks that Stratify Lupus Patients. Cell 165, 1548–1550 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Liarski VM et al. Cell distance mapping identifies functional T follicular helper cells in inflamed human renal tissue. Sci Transl Med 6, 230ra246 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hutloff A et al. Involvement of inducible costimulator in the exaggerated memory B cell and plasma cell generation in systemic lupus erythematosus. Arthritis Rheum 50, 3211–3220 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Kassianos AJ et al. Increased tubulointerstitial recruitment of human CD141(hi) CLEC9A(+) and CD1c(+) myeloid dendritic cell subsets in renal fibrosis and chronic kidney disease. Am J Physiol Renal Physiol 305, F1391–1401 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Davidson A What is damaging the kidney in lupus nephritis? Nat Rev Rheumatol 12, 143–153 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hashimshony T et al. CEL-Seq2: sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol 17, 77 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hooks JJ et al. Immune interferon in the circulation of patients with autoimmune disease. N Engl J Med 301, 5–8 (1979). [DOI] [PubMed] [Google Scholar]
- 12.Villani AC et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghebrehiwet B, Hosszu KH & Peerschke EI C1q as an autocrine and paracrine regulator of cellular functions. Mol Immunol 84, 26–33 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hulsebus HJ, O’Conner SD, Smith EM, Jie C & Bohlson SS Complement Component C1q Programs a Pro-Efferocytic Phenotype while Limiting TNFalpha Production in Primary Mouse and Human Macrophages. Front Immunol 7, 230 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ikezumi Y et al. The sialoadhesin (CD169) expressing a macrophage subset in human proliferative glomerulonephritis. Nephrol Dial Transplant 20, 2704–2713 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Knutson MD Iron transport proteins: Gateways of cellular and systemic iron homeostasis. J Biol Chem 292, 12735–12743 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adamson SE et al. Disabled homolog 2 controls macrophage phenotypic polarization and adipose tissue inflammation. The Journal of clinical investigation 126, 1311–1322 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spadaro O et al. IGF1 Shapes Macrophage Activation in Response to Immunometabolic Challenge. Cell Rep 19, 225–234 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Varghese B, Paulos C & Low PS Optimization of Folate-Targeted Immunotherapy for the Treatment of Experimental Arthritis. Inflammation 39, 1345–1353 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Kreslavsky T et al. Essential role for the transcription factor Bhlhe41 in regulating the. Nat Immunol 18, 442–455 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Angerer P et al. destiny: diffusion maps for large-scale single-cell data in R. Bioinformatics 32, 1241–1243 (2016). [DOI] [PubMed] [Google Scholar]
- 22.van den Maaten LJPHG Visualizing High-Dimensional Data Using t-SNE. J Mach Learn Res 9, 2579–2605 (2008). [Google Scholar]
- 23.Dehn S & Thorp EB Myeloid receptor CD36 is required for early phagocytosis of myocardial infarcts and induction of Nr4a1-dependent mechanisms of cardiac repair. Faseb j (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bengsch B et al. Deep immune profiling by mass cytometry links human T and NK cell differentiation and cytotoxic molecule expression patterns. J Immunol Methods (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bratke K, Kuepper M, Bade B, Virchow JC Jr. & Luttmann W Differential expression of human granzymes A, B, and K in natural killer cells and during CD8+ T cell differentiation in peripheral blood. Eur J Immunol 35, 2608–2616 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Boddupalli CS et al. ABC transporters and NR4A1 identify a quiescent subset of tissue-resident memory T cells. The Journal of clinical investigation 126, 3905–3916 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mackay LK et al. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science 352, 459–463 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Allan DSJ et al. Transcriptome analysis reveals similarities between human blood CD3(−) CD56(bright) cells and mouse CD127(+) innate lymphoid cells. Sci Rep 7, 3501 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKinney EF, Lee JC, Jayne DR, Lyons PA & Smith KG T-cell exhaustion, co-stimulation and clinical outcome in autoimmunity and infection. Nature 523, 612–616 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tilstra JS et al. Kidney-infiltrating T cells in murine lupus nephritis are metabolically and functionally exhausted. The Journal of clinical investigation (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wherry EJ et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity 27, 670–684 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Nish SA et al. CD4+ T cell effector commitment coupled to self-renewal by asymmetric cell divisions. J Exp Med 214, 39–47 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karnell JL et al. Role of CD11c(+) T-bet(+) B cells in human health and disease. Cell Immunol 321, 40–45 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Jenks SA et al. Distinct Effector B Cells Induced by Unregulated Toll-like Receptor 7 Contribute to Pathogenic Responses in Systemic Lupus Erythematosus. Immunity 49, 725–739 e726 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.The FANTOM Consortium and the RIKEN PMI and CLST (DGT). A promoter-level mammalian expression atlas. Nature 507, 462–470 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lizio M et al. Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol 16, 22 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen L, Morris DL & Vyse TJ Genetic advances in systemic lupus erythematosus: an update. Curr Opin Rheumatol 29, 423–433 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Wang S et al. IL-21 drives expansion and plasma cell diferentiation of autoreactive CD11chiT-bet+ B cells in SLE. Nat. Commun 9, 1758 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chung SA et al. Lupus nephritis susceptibility loci in women with systemic lupus erythematosus. J Am Soc Nephrol 25, 2859–2870 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hua Z & Hou B TLR signaling in B-cell development and activation. Cell Mol Immunol 10, 103–106 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawasaki T & Kawai T Toll-like receptor signaling pathways. Front Immunol 5, 461 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Madan-Lala R et al. Mycobacterium tuberculosis impairs dendritic cell functions through the serine hydrolase Hip1. J Immunol 192, 4263–4272 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsuda S et al. Regulation of the Cell Cycle and Inflammatory Arthritis by the Transcription Cofactor LBH Gene. J Immunol 199, 2316–2322 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Botta D et al. Dynamic regulation of T follicular regulatory cell responses by interleukin 2 during influenza infection. Nat Immunol 18, 1249–1260 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wing JB et al. A distinct subpopulation of CD25(−) T-follicular regulatory cells localizes in the germinal centers. Proc Natl Acad Sci U S A 114, E6400–e6409 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winchester R et al. Immunologic characteristics of intrarenal T cells: trafficking of expanded CD8+ T cell beta-chain clonotypes in progressive lupus nephritis. Arthritis Rheum 64, 1589–1600 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peterson KS et al. Characterization of heterogeneity in the molecular pathogenesis of lupus nephritis from transcriptional profiles of laser-captured glomeruli. The Journal of clinical investigation 113, 1722–1733 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davidson A Editorial: autoimmunity to vimentin and lupus nephritis. Arthritis Rheumatol 66, 3251–3254 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Myles A, Gearhart PJ & Cancro MP Signals that drive T-bet expression in B cells. Cell Immunol 321, 3–7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huen SC & Cantley LG Macrophages in Renal Injury and Repair. Annual review of physiology 79, 449–469 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Tabas I & Bornfeldt KE Macrophage Phenotype and Function in Different Stages of Atherosclerosis. Circ Res 118, 653–667 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Auffray C et al. Monitoring of blood vessels and tissues by a population of monocytes with patrolling behavior. Science 317, 666–670 (2007). [DOI] [PubMed] [Google Scholar]
METHODS-ONLY REFERENCES
- 53.Picelli S et al. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc 9, 171–181 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Nemesh J Drop-seq core computational protocol. McCarroll Laboratory http://mccarrolllab.com/wp-content/uploads/2016/03/DropseqAlignmentCookbookv1.2Jan2016.pdf (2016). [Google Scholar]
- 55.Satija R, Farrell JA, Gennert D, Schier AF & Regev A Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 33, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McDavid A et al. Data exploration, quality control and testing in single-cell qPCR-based gene expression experiments. Bioinformatics 29, 461–467 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Benjamini YH, Y. Controlling the False Discovery Rate: a Practical and Powerful Approach to Multiple Testing. J R Statist Soc B 57, 289–300 (1995). [Google Scholar]
- 58.Tirosh I et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.IUPHAR/BPS. Guide to Pharmacology. http://www.guidetopharmacology.org/ download.jsp (2019).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data reported in this publication, including the clinical and serological data of the study participants, are deposited in the ImmPort repository (accession code SDY997). The raw single-cell RNA-seq data are also deposited in dbGAP (accession code phs001457.v1.p1). The processed data can be viewed using an interactive browser at https://immunogenomics.io/ampsle, https://immunogenomics.io/cellbrowser/, and https://portals.broadinstitute.org/single_cell/study/amp-phase-1.