Abstract
Neurons modulate gene expression in response to extrinsic signals to enable brain development, cognition, and learning and to process stimuli that regulate systemic physiological functions. This signal-to-gene communication is facilitated by posttranslational modifications such as S-nitrosylation, the covalent attachment of a nitric oxide (NO) moiety to cysteine thiols. In the cerebral cortex, S-nitrosylation of histone deacetylase 2 (HDAC2) is required for gene transcription during neuronal development, but few other nuclear targets of S-nitrosylation have been identified to date. We used S-nitrosothiol resin-assisted capture on NO donor-treated nuclear extracts from rat cortical neurons and identified 614 S-nitrosylated nuclear proteins. Of these, 131 proteins have not previously been shown to be S-nitrosylated in any system, and 555 are previously unidentified targets of S-nitrosylation in neurons. The sites of S-nitrosylation were identified for 59% of the targets, and motifs containing single lysines were found at 33% of these sites. In addition, lysine motifs were necessary for promoting the S-nitrosylation of HDAC2 and methyl-CpG binding protein 3 (MBD3). Moreover, S-nitrosylation of the histone-binding protein RBBP7 was necessary for dendritogenesis of cortical neurons in culture. Together, our findings characterize S-nitrosylated nuclear proteins in neurons and identify S-nitrosylation motifs that may be shared with other targets of NO signaling.
Introduction
S-nitrosylation is a nitric oxide (NO)–dependent posttranslational modification that affects protein functions by regulating enzymatic activity, subcellular localization, or protein-protein interactions (1–3). Although more than 3000 S-nitrosylated proteins (SNO-Ps) have been identified to date, these include few nuclear proteins (4–6). In the rodent brain, expression of the enzyme responsible for neuronal NO production, the neuronal NO synthase (nNOS), is developmentally regulated and peaks during the mid-to-late embryonic stages during which the layered structure of the cortex is established (7). We previously showed that S-nitrosylation of histone deacetylase 2 (HDAC2) during this period results in dissociation of HDAC2 from chromatin, promoting histone acetylation and expression of cyclic adenosine 3′,5′-monophosphate response element–binding protein (CREB)–dependent genes that are critical for neuronal migration (4, 8). S-nitrosylation of HDAC2 is also important in the adult brain; in the hippocampus, for example, S-nitrosylation of HDAC2 is required for the updating of fear memory (9). Other examples of nuclear S-nitrosylation include glyceraldehyde-3-phosphate dehydrogenase (GAPDH), which is S-nitrosylated in the cytoplasm and translocates into the nucleus where it transnitrosylates several proteins, including HDAC2 (5, 10). Moreover, the transcription factor myocyte enhancer factor 2C (MEF2C) is S-nitrosylated in neurons in response to excitotoxic insults, resulting in reduced transcriptional activity and increased cell death (6, 11).
Previous proteomic screens report few nuclear targets of S-nitrosylation in mammalian cells (12–14). To identify nuclear proteins that are potential targets of S-nitrosylation in neurons, we performed S-nitrosothiol resin-assisted capture (SNORAC) (15) on nuclear extracts of rat cortical neurons treated with the NO donor S-nitrosocysteine (CysNO). Mass spectrometry identified 614 nuclear targets of S-nitrosylation and the site(s) of S-nitrosylation for most hits. In addition, amino acid motif analysis revealed four lysine-containing motifs that promote S-nitrosylation of nearby cysteines. We found that proteins involved in metabolism, development, and regulation of gene expression were modified by S-nitrosylation. Most subunits of the nucleosome remodeling and deacetylase (NuRD) complex were found to be S-nitrosylated. Further analysis of the histone-binding protein retinoblastoma-binding protein 7 (RBBP7) revealed that S-nitrosylation at Cys166 was required for activity-induced dendritogenesis and regulated its interaction with the chromodomain helicase DNA binding protein 4 (CHD4) subunit. The transcription factor CREB was also found to be S-nitrosylated in response to CysNO and after neuronal depolarization. Our study provides a SNO proteome of neuronal proteins that may launch further investigations into nuclear S-nitrosylation in neurons.
Results
Identification of S-nitrosylated nuclear proteins in cortical neurons
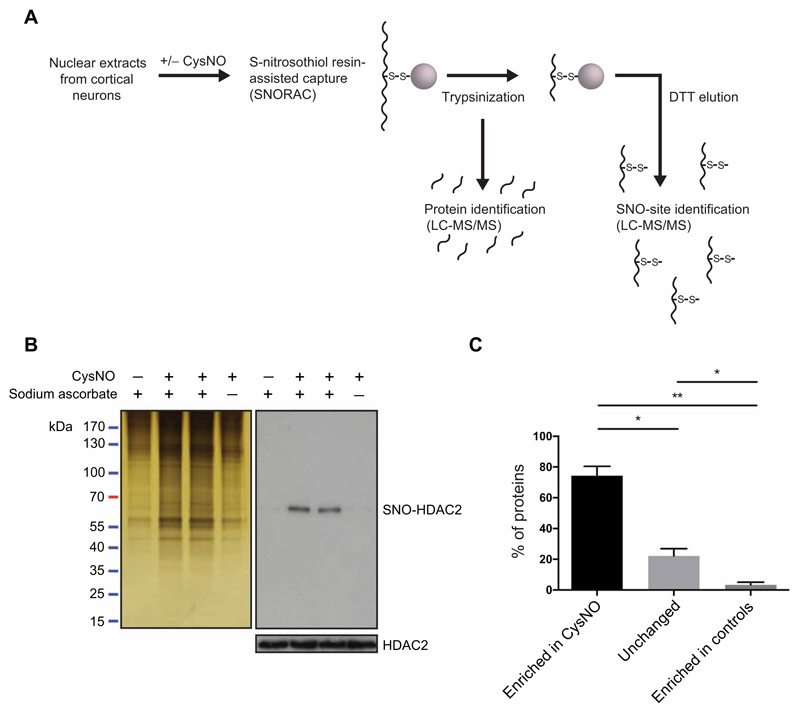
To obtain a comprehensive analysis of nuclear S-nitrosylation in neurons, we treated nuclear extracts from E17 (embryonic day 17) rat cortical neurons with the NO donor CysNO and isolated SNO-Ps using SNORAC (Fig. 1A) (15). Eluted proteins were subjected to SDS–polyacrylamide gel electrophoresis (PAGE) and silver staining to assess total SNO-Ps (Fig. 1B). As a control, we used HDAC2 (Fig. 1B and fig. S1, A and B), a known nuclear target of S-nitrosylation (4, 5, 8, 9). The corresponding samples were analyzed using quantitative mass spectrometry [liquid chromatography–tandem mass spectrometry (LC-MS/MS); see Materials and Methods] to obtain the identification of SNO-Ps and the respective sites of S-nitrosylation. Of the identified proteins, 71.75% were detected as increased in CysNO-treated samples (Fig. 1C), indicating that the SNORAC technique allows a significant enrichment of SNO-Ps. The remaining proteins were either unchanged across conditions (24.37%) or enriched in controls (3.88%) and were excluded from further analysis. Five experiments were carried out in total: two using CysNO at a 1 mM concentration and three using 200 μM CysNO (table S1 and fig. S1, C to E). Despite variability in the number of SNO-Ps identified in each experiment (table S1 and fig. S1C), 88.2% of the proteins identified using 1 mM CysNO were also S-nitrosylated using 200 μM CysNO (fig. S1D), indicating that, at both concentrations, CysNO induced robust S-nitrosylation of a similar set of proteins. Proteins were designated as S-nitrosylated if they were enriched ≥2-fold in CysNO relative to control samples in at least three independent experiments (detailed in Materials and Methods). 614 SNO-Ps met these criteria (table S2), of which 131 (21.3%) were previously unidentified targets of S-nitrosylation across all systems assayed, whereas 555 (90.4%) had not been previously identified as targets of S-nitrosylation in neurons. Additional hits were found when the SNO-P data sets were analyzed using stringency conditions of ≥1.3-fold in CysNO versus control samples in at least three independent experiments (table S3) or ≥1.5-fold in CysNO versus control samples in at least two independent experiments (table S4). We also probed experiments 1 to 4 for proteins that are basally S-nitrosylated (table S5) and proteins unchanged in binding across all conditions (table S6).
Fig. 1. Identification of S-nitrosylated nuclear proteins using SNORAC.
(A) Nuclear extracts from E17 cortical neurons were treated with CysNO and SNO-Ps captured using SNORAC. Resin-bound proteins were trypsinized, and eluted fragments were analyzed by quantitative MS to identify the proteins enriched in CysNO-treated groups. A second elution step was carried out using dithiothreitol (DTT) to release fragments containing the previously S-nitrosylated cysteines and to identify S-nitrosylation sites (SNO sites). (B) Silver stain of proteins isolated using SNORAC and HDAC2 Western blotting on SNORAC eluates and total inputs. Sodium ascorbate is required for isolation of SNO-Ps using SNORAC. Blots are representative of five independent experiments. (C) Average percentage of proteins detected by MS as enriched in CysNO (≥2-fold versus control), unchanged (<2-fold, >0.5-fold; CysNO versus control), or enriched in controls (≤0.5-fold; CysNO versus control). Control groups constitute Cys only and CysNO without ascorbate. Data are means ± SEM from five independent experiments. *P < 0.05 and **P < 0.01 by row means one-way analysis of variance (ANOVA) and Tukey post hoc test.
Gene ontology analysis of nuclear SNO-Ps
To determine whether certain types of proteins were enriched in our data set, we carried out gene ontology (GO) overrepresentation analysis (detailed in Materials and Methods) on the SNO-P data set. First, untreated nuclear extracts from E17 cortical neurons were analyzed by MS to generate a reference nuclear proteome of 3078 proteins (table S7). Using this data set as background, we found that four pathways were significantly enriched in the SNO-P data set: Huntington disease (3.48-fold–enriched), dopamine receptor–mediated signaling (4.09-fold–enriched), Wnt signaling (2.61-fold–enriched), and nicotine pharmacodynamics pathway (5.41-fold–enriched; fig. S2A). Annotations for molecular function (fig. S2B) indicated that chromatin-binding proteins were significantly enriched in the SNO-P data set (1.99-fold). Whereas several terms associated with regulation of protein binding were overrepresented in SNO-Ps (fig. S2B), hydrolase activity was the only term linked to regulation of protein activity (1.85-fold). Overrepresentation analysis for biological process revealed that 4 of 29 assigned terms (13.8%) related to metabolic processes (fig. S2C), whereas 4 of 29 (13.8%) concerned development. Two terms were associated with regulation of gene expression (gene expression, 2.5-fold; positive regulation of gene expression, 1.47-fold; fig. S2C). Nucleic acid–binding proteins were amongst the terms enriched in regards to protein class (3.11-fold; fig. S2D). We also considered standard GO classifications (fig. S3A). Overall, our data indicate that S-nitrosylation of nuclear proteins may represent a mechanism that regulates metabolic, developmental, and genetic processes.
In addition to nuclear extracts, corresponding extracts from the cytoplasmic compartment of E17 cortical neurons were exposed to CysNO and used to identify cytoplasmic SNO-Ps (detailed in Materials and Methods; fig. S1B and ftable S8). As expected, our nuclear and cytoplasmic proteins data sets were largely distinct, with 87.3% of detections unique to either data set. GO analysis also revealed marked differences in annotations (fig. S3, B to D).
S-nitrosylation of nuclear factors in cortical neurons
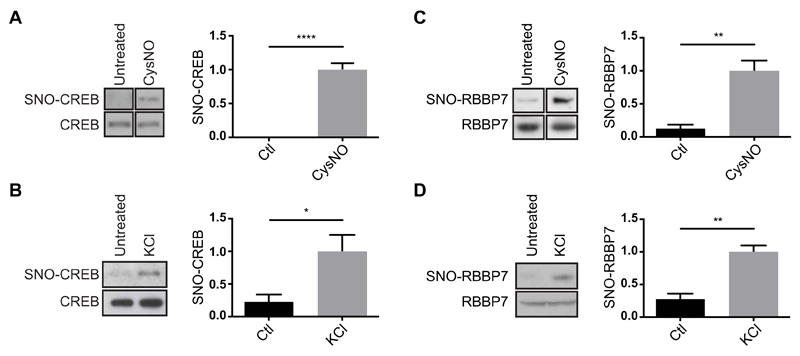
We next sought to study S-nitrosylation of nuclear proteins that are involved in the regulation of gene expression and are especially relevant for neuronal functions. We considered SNO-Ps annotated with the GO terms “transcription, DNA-dependent” (GO:0006351; table S9), and “chromatin organization” (GO:0006325; table S10). The transcription factor CREB (table S9, hit number 6) regulates the transcription of genes essential for differentiation and survival of cortical neurons (16, 17). We previously demonstrated that NO regulates CREB-dependent gene expression (17) and that S-nitrosylation of HDAC2 is necessary for CREB-activated transcription (4, 8). However, whether CREB is S-nitrosylated in neurons remained unknown. To investigate S-nitrosylation of specific nuclear proteins, we used the biotin-switch assay (18). In this assay, S-nitrosylated cysteines are labeled with biotin and subsequently isolated using streptavidin capture. Biotin switch performed on cortical neurons exposed to CysNO confirmed that CREB is S-nitrosylated (Fig. 2A). Depolarization of neurons with KCl also induced S-nitrosylation of CREB (Fig. 2B).
Fig. 2. S-nitrosylation of CREB and RBBP7 in cortical neurons.
(A) E17 rat cortical neurons were cultured for 4 days and treated with CysNO (200 μM for 20 min). SNO-Ps were isolated by SNORAC (n = 2 experiments) or biotin switch (n = 2). Noncontiguous lanes from the same experiment and blot are shown side by side. Ctl, control. (B) Cortical neurons were exposed to KCl (50 mM for 20 min), and SNO-Ps were isolated by SNORAC (n = 2) or biotin switch (n = 1). Data were combined for analysis. (C) Neurons were treated with CysNO (200 μM for 20 min), and SNO-Ps were isolated by biotin switch (n = 4 experiments). Non-contiguous lanes from the same experiment and blot are shown side by side. (D) Cortical neurons were cultured for 4 days and exposed to 50 mM KCl. SNO-Ps were isolated by biotin switch (n = 2 using KCl for 30 min, n = 1 using KCL for 20 min). Data were combined for analysis. For both CREB and RBBP7, isolated proteins were separated by SDS-PAGE, and Western blot analysis was carried out using CREB or RBBP7 antibodies. Densitometry analysis on the Western blots was performed using ImageJ. SNO-P signals were normalized to inputs. Data are means ± SEM. *P ≤ 0.05, **P ≤ 0.01, and ****P ≤ 0.0001 by unpaired t test.
RBBP7 (table S10, hit number 6) is a histone-binding protein associated with several chromatin-modifying complexes that regulate gene expression in neurons (19–21), including the NuRD complex. NuRD regulates cortical development in mice (19, 21); however, whether subunits are posttranslationally modified in response to extrinsic signals in neurons is unknown. We found that in cortical neurons, RBBP7 is S-nitrosylated in response to CysNO treatment and depolarization (Fig. 2, C and D), as well as after exposure to S-nitrosoglutathione (GSNO) (fig. S4). In addition, CHD3, CHD4, and CHD5, the adenosine triphosphatase (ATPase) subunits of the NuRD complex were all detected as S-nitrosylated in the SNO-P data set (table S2, hit numbers 372, 373, and 87) and in CysNO-treated HEK293T cell lysates (fig. S5). Together, these findings demonstrate that our SNORAC screen identified previously unknown nuclear targets of S-nitrosylation and suggest that S-nitrosylation may regulate chromatin remodeling complexes.
Identification of S-nitrosylated cysteines and predicted S-nitrosylation motifs
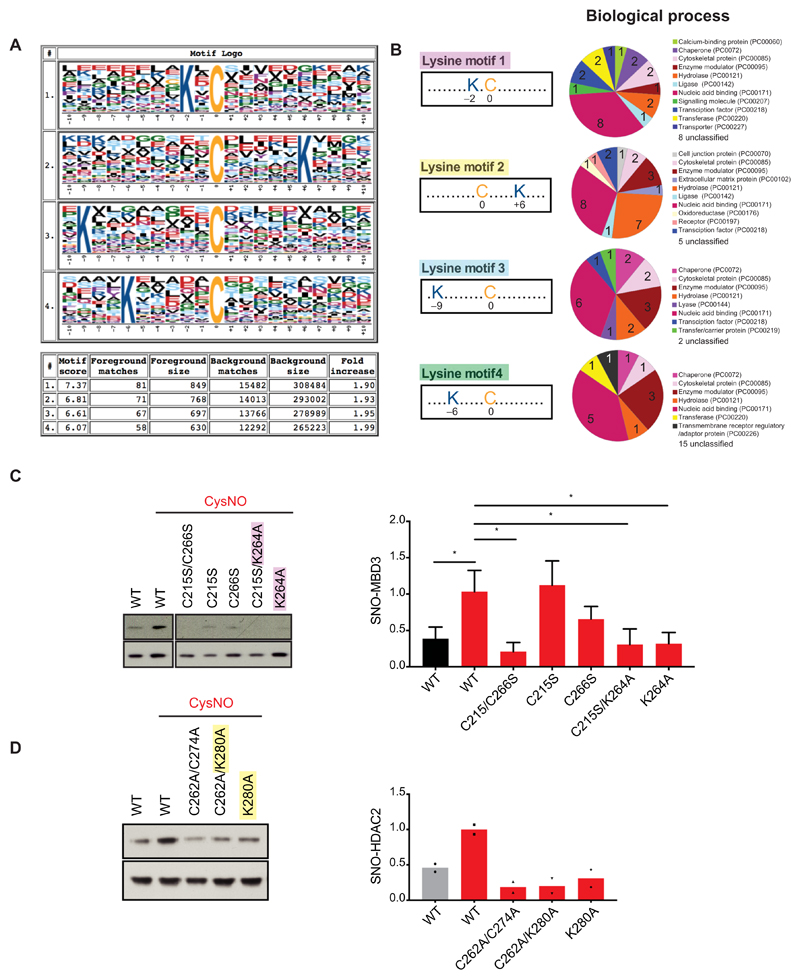
To identify the cysteines modified by nitrosylation (SNO sites), we performed a second elution step during SNORAC, which allowed the release of previously nitrosylated fragments from the resin (Fig. 1A). The fragments were then identified by LC-MS/MS. SNO-peptide analysis of experiment 5 detected 4368 cysteine-containing peptides in the CysNO sample (table S11) and 1103 in the Cys control (table S12). To identify SNO sites, we analyzed peptides detected in the CysNO data set that belonged to proteins present in the SNO-P data set (table S2). Using this approach, we assigned 942 unique SNO sites across 360 proteins (table S13). SNO sites for proteins assigned using less stringent conditions are also provided (tables S14 and S15).
Next, we searched for amino acid motifs that may render certain cysteines more prone to nitrosylation, such as sequences containing acidic or basic residues either immediately flanking (22) or distal to the SNO site (23). Analysis of the amino acid sequences surrounding SNO cysteines identified in our data set (table S13) was carried out using Motif-X (24), which revealed that lysine containing motifs were present in 277 of the 849 mappable SNO sites (32.6%; Fig. 3A). Whether specific lysine motifs are preferentially found in certain protein types is not currently known; however, GO analysis of SNO-Ps containing each lysine motif revealed different annotations for biological process (Fig. 3B). We first investigated the functional relevance of the lysine(s) by assessing the effect on S-nitrosylation of the NuRD complex protein MBD3 (table S13, hit number 271). SNO-site analysis was performed on MBD3 by generating DNA vectors expressing either wild-type (WT) or mutant forms bearing mutations of the nitrosylated cysteines to serines (C215S and C266S) or mutation of Lys264 (lysine motif 1) to Ala (K264A). CysNO-dependent S-nitrosylation of MBD3 was significantly reduced in HEK293T cells upon C215A/C266S mutation (Fig. 3C), indicating that the cysteines identified by the SNORAC were S-nitrosylated. Notably, K264A mutation alone or alongside C215S also led to a marked decrease of MBD3 S-nitrosylation. As we previously demonstrated that HDAC2 is S-nitrosylated at Cys262 and Cys274 in neurons (4), CysNO-induced S-nitrosylation of HDAC2 was assessed upon mutation of HDAC2 Lys280 to alanine (lysine motif 2; K280A; table S13, hit number 237) alone or in association with C262A (Fig. 3D). In summary, we found that a single lysine in the immediate area (−2 to +6) surrounding a cysteine promotes S-nitrosylation at that site.
Fig. 3. Lysine motifs regulate S-nitrosylation.
(A) Cysteine-containing peptides associated with nuclear SNO-Ps were subjected to SNO-site analysis using Motif-X (see Materials and Methods) and the IPI Rat Proteome, as background. Graphical representation of the motifs; the size of each letters refers to the probability of occurrence of that amino acid in cysteine-containing peptides. Summary table is shown for clarity. Full size letters (cysteine, C; lysine, K) indicate their recurrence in each motif. (B) GO analysis of proteins that contain either lysine motif 1, 2, 3, or 4 only around their identified S-nitrosylated cysteines. Analysis carried out using PANTHER Functional Classification for Biological Process, version 13.1 released on 03 February 2018 (www.pantherdb.org). (C) SNO-motif mutational analysis of MBD3. Western blotting and densitometry analysis of HEK293T cells transfected with vectors expressing myc-tagged WT MBD3 (WT), MBD3 Cys-to-Ser mutants (C215S and C266S), MBD3 Lys264-to-Ala mutant (K264A), and cysteine and alanine combined mutant (C215S/K264A). Twenty-four hours after transfection, cells were exposed to either Cys or CysNO (200 μM for 20 min) and subjected to the biotin-switch assay. Data are means ± SEM of n = 4 experiments. *P ≤ 0.05 by one-way ANOVA, compared to control column “WT + CysNO,” Fisher’s least significant difference (LSD). Noncontiguous lanes from the same experiment and blot are shown side by side, as indicated by the separation evident in the figure. (D) SNO-motif mutational analysis of HDAC2. HEK293T cells were transfected with vectors expressing HA-tagged WT HDAC2 (WT), HDAC2 Cys-to-Ala nitrosomutant (C264A/C272A), HDAC2 Lys280-to-Ala mutant (K280A), and cysteine and alanine combined mutant (C264A/K280A). Forty-eight hours after transfection, cells were exposed to Cys or CysNO (200 μM for 20 min) and subjected to the biotin-switch assay (n = 2 experiments).
Sites of S-nitrosylation on CREB and RBBP7
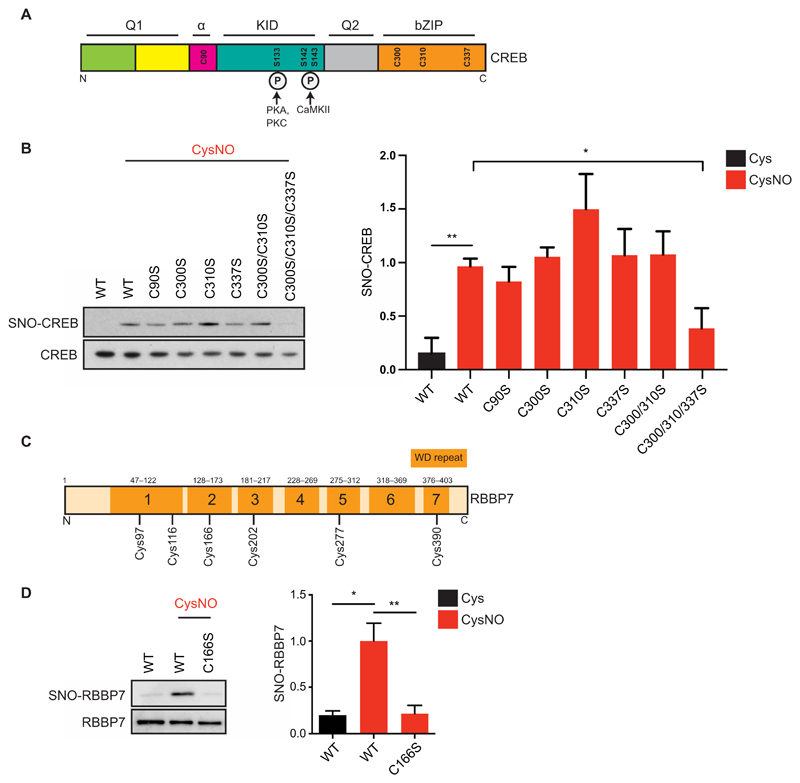
To explore the physiological implications of S-nitrosylation of selected nuclear proteins, we further investigated the site(s) of S-nitrosylation of CREB and RBBP7. SNO-peptide analysis failed to identify the cysteine(s) modified on CREB, possibly due to the small size of the predicted tryptic peptides (3, 5, and 6 amino acids) surrounding three of the four cysteines, which render these peptides unsuitable MS-based detection. Instead, CREB cysteine-to-serine mutant constructs were generated, and S-nitrosylation of these mutants was tested. CREB cysteines are located in the α domain (Cys90) and in the C-terminal Basic Leucine Zipper Domain (bZIP) domain that is important for CREB homodimerization and binding to DNA (Cys300, Cys310, and Cys337; Fig. 4A). Single mutants of Cys90, Cys300, Cys310, or Cys337 failed to reduce S-nitrosylation in CysNO-treated HEK293T cells, as did mutation of Cys300 and Cys310. However, triple mutation of Cys300, Cys310, and Cys337 led to a reduction in CREB S-nitrosylation (Fig. 4B). CREBC300/310/337S was still phosphorylated at Ser133 in response to forskolin, suggesting that, despite the mutations, the integrity of the protein and the ability to respond to intracellular signaling was preserved (fig. S6).
Fig. 4. Identification of CREB and RBBP7 SNO sites.
(A) Domain structure of CREB. Q1, glutamine-rich region 1; α, alpha domain; KID, kinase inducible domain; Q2, glutamine-rich region 2; bZIP, bZIP domain. CREB1 cysteines are annotated (Cys90, Cys300, Cys310, and Cys337). Phosphorylated serines (Ser133, Ser142, and Ser143) are shown along with their respective kinases [protein kinase A (PKA), PKC, and Ca2+/calmodulin-dependent protein kinase II]. (B) Vectors expressing myc-tagged WT CREB (WT) or CREB Cys-to-Ser mutants were transfected into HEK293T cell; 48 hours later, cell lysates were harvested and exposed to Cys or CysNO (500 μM for 20 min) and subjected to the biotin-switch assay (n = 3 experiments). (C) RBBP7 contains seven WD (tryptophan–aspartic acid)–repeat domains, composed of short motifs of about 40 amino acids and ending in a WD dipeptide. (D) Vectors expressing myc-tagged WT RBBP7 (WT) or RBBP7 Cys166-to-Ser mutant (C166S) were transfected into HEK293T cells. After 48 hours, cells were treated with Cys or CysNO (500 μM for 20 min), and lysates were subjected to the biotin-switch assay (n = 3). Isolated proteins and total inputs were separated by SDS-PAGE and then immunoblotted using an antibody against myc. Densitometry analysis was carried out using ImageJ. SNO signals were normalized to total inputs and expressed as fold change relative to WT + CysNO. All data are means ± SEM. *P ≤ 0.05 and **P < 0.01 by one-way ANOVA, compared to WT + CysNO column, and Fisher’s LSD.
SNO-peptide analysis identified Cys166 as the site of S-nitrosylation of RBBP7 (table S13, hit number 238). Cys166 was previously identified as a potential site of S-nitrosylation on RBBP7 (25); however, this finding had not been confirmed in intact cells. We generated a vector expressing a mutant form of RBBP7 carrying the C166S mutation and found that S-nitrosylation of RBBP7 was abolished in CysNO-treated HEK293T cells (Fig. 4, C and D). Because RBBP7 is closely related to RBBP4 (89.2% identical and 94.6% similar; fig. S7A), we mutated the conserved cysteine in RBBP4 and found that this event also led to decreased S-nitrosylation (fig. S7B). Because RBBPs function as scaffolding subunits for NuRD complexes, it is possible that S-nitrosylation provides a mechanism to regulate the assembly and stability of the complex.
Regulation of RBBP7 binding to CHD4 by NO
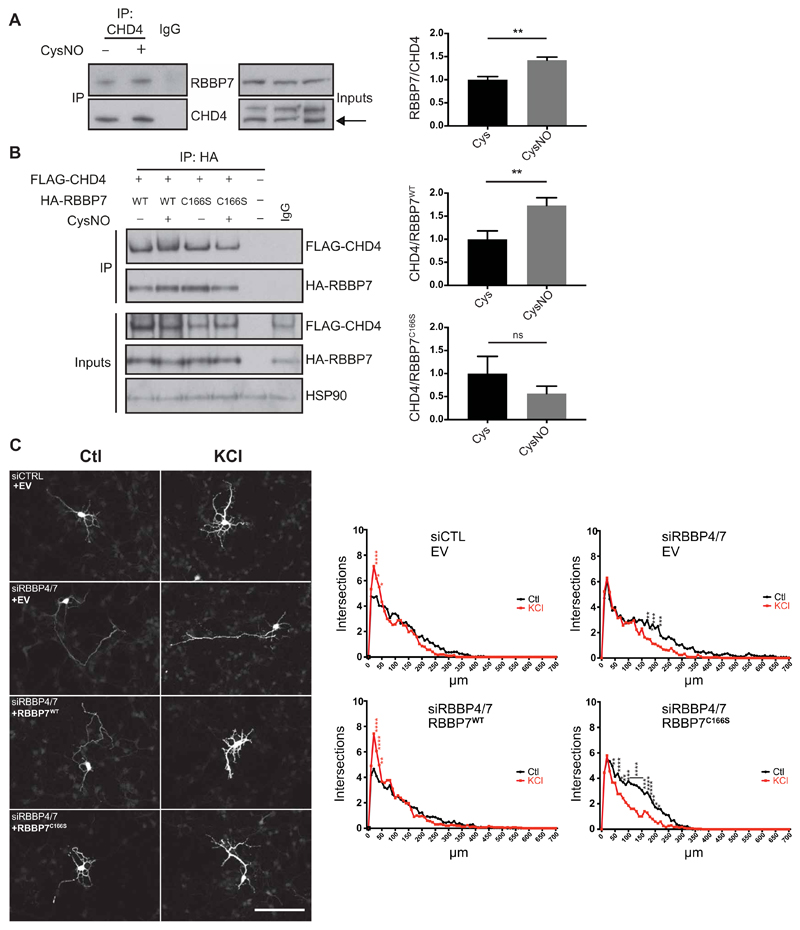
RBBP7 contains seven tryptophan–aspartic acid (WD)–repeat regions, which form a β-helical propeller structure and provide platforms for protein binding (26). As Cys166 of RBBP7 is within WD-repeat 2 (Fig. 4C), we reasoned that S-nitrosylation may regulate the interaction of RBBP7 with other subunits within chromatin remodeling complexes. We first investigated whether S-nitrosylation of RBBP7 influences its interaction with other NuRD subunits by performing pulldown experiments on E17 cortical neurons. Notably, treatment with CysNO resulted in increased binding of RBBP7 to the ATPase subunit CHD4 (Fig. 5A). To test whether the interaction depends on nitrosylation of RBBP7 at Cys166, we transfected HEK293T cells with FLAG-CHD4 alongside either HA-RBBP7WT or HA-RBBP7C166S and studied interaction using a pulldown assay. CysNO increased CHD4 binding to RBBP7 (Fig. 5B), an effect dependent on Cys166 of RBBP7. Thus, S-nitrosylation of RBBP7 regulates the interaction between at least two subunits within the NuRD complex, perhaps determining its assembly on chromatin.
Fig. 5. S-nitrosylation of RBBP7 regulates dendritogenesis.
(A) E17 rat cortical neurons were cultured for 4 days, treated with 200 μM CysNO for 20 min, and subjected to coimmunoprecipitation (co-IP) using CHD4 antibody. As a control, pooled samples of Cys and CysNO were subjected to IP using an immunoglobulin G (IgG) antibody. Western blot analysis was carried out on eluates and inputs using the indicated antibodies. Densitometry analysis of coimmunoprecipitated RBBP7 was shown in relation to coimmunoprecipitated CHD4 (n = 4, **P < 0.01 by unpaired t test). Densitometry was carried out by the normalizing RBBP7 pulldown signal to RBBP7 inputs and dividing this value by the pulldown signal for CHD4. (B) S-nitrosylation of RBBP7 at Cys166 promotes the interaction with CHD4. HEK293T cells were transfected with vectors expressing Flag-mCHD4 and HA-RBBP7WT or HA-RBBP7C166S and treated with 200 μM CysNO for 20 min, and pulldown was carried out on eluates using an HA antibody. Western blot analysis was carried out on eluates and inputs using the indicated antibodies. Densitometry analysis of FLAG-CHD4 was shown in relation to HA-RBBP7 and normalized to FLAG-CHD4 inputs (n = 3, **P < 0.01 unpaired t test). Densitometry was carried out by normalizing FLAG-CHD4 pulldown signal to FLAG-CHD4 inputs and dividing this value by the pulldown signal for HA-RBBP7. ns, not significant. (C) S-nitrosylation of RBBP7 promotes dentritogenesis in cortical neurons. Control small interfering RNA (siRNA) (Ctl) or siRNAs targeting RBBP4 and RBBP7 (siRBBP4/7) were transfected into E15 mouse cortical neurons alongside a green fluorescent protein (GFP) expression vector, and either an empty vector (EV), a vector expressing siRNA-resistant myc-RBBP7WT or myc-RBBP7C166S. Neurons were maintained in control conditions (Ctl) or exposed to 50 mM KCl for 2 days and immunostained for GFP. Resulting images were analyzed using a Fiji Sholl plugin. Maximal projections of representative neurons for each condition. Scale bar, 100 μm. Summary of Sholl data analysis for each condition is shown. n = 3 biological replicates from which 10 neurons each were analyzed (n = 30 neurons in total). Data are mean values for number of intersections (intersections) against distance from the soma (in micrometers). Readings were taken every 10 μm. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Red asterisks indicate that the KCl value is significantly greater than that of Ctl, and black asterisks indicate that the Ctl value is significantly greater than that of KCl. Row means two-way ANOVA with Sidak’s test for multiple comparisons.
Role of S-nitrosylated RBBP7 in promoting dendritic growth
In cortical neurons, NO signaling enhances dendritogenesis (27) by S-nitrosylating the RBBP7-interactor HDAC2 on Cys262 and Cys274 (4). We therefore investigated whether S-nitrosylation of RBBP7 may contribute to this process. Mouse embryonic cortical neurons were maintained under depolarizing conditions with KCl (50 mM) for 2 days, and dendritic complexity assayed using Sholl analysis. Knockdown of RBBP7 alone did not inhibit KCl-induced dendritogenesis (fig. S8, A and B), possibly since the RBBP7 paralog RBBP4 is S-nitrosylated at its conserved cysteine (fig. S7B) and may therefore functionally compensate for RBBP7. Knockdown of both RBBP7 and RBBP4 concomitantly markedly reduced dendritic growth (Fig. 5C and fig. S8C). This effect was rescued by cotransfection of a construct expressing RBBP7WT but not RBBP7C166S, indicating that S-nitrosylation of RBBP7 is required for dendritic growth in vitro (Fig. 5C and fig. S8D).
Discussion
Here, we identified a nuclear SNO proteome of rat cortical neurons. To our knowledge, this is a first comprehensive screen of nuclear targets of S-nitrosylation performed in mammalian cells. Identification of nuclear proteins in previous studies (12–14) may have been limited by the low abundance of nuclear proteins relative to cytoplasmic, which renders nuclear targets of S-nitrosylation less amenable for detection. To circumvent this limitation, we assayed nuclear extracts of E17 rat cortical neurons exposed to the NO-donor CysNO. This technical approach yielded 614 predicted targets of S-nitrosylation that are suitable for further investigation. Known intracellular targets of S-nitrosylation in neurons were identified in our study, including GAPDH (10), HDAC2 (4, 9), and nucleophosmin (28). However, it should be noted that our CysNO-responsive data set may have missed some intracellular targets of S-nitrosylation, given that similar to other biotin switch–related methodologies, SNORAC only allows the identification of a subset of SNO-Ps (29).
Proteins involved in the regulation of gene expression and neuronal development were overrepresented in S-nitrosylated nuclear samples. We previously showed that S-nitrosylation of HDAC2 promotes the expression of genes necessary for neuronal radial migration during mouse embryonic development (4, 8). nNOS levels increase markedly during embryonic cortical development (7, 8); therefore, additional nuclear proteins may be targeted by S-nitrosylation at these time points to regulate corticogenesis. As an initial step toward exploring this possibility, proteins involved in the regulation of gene expression were subjected to further investigation. Both the transcription factor CREB and the histone-binding protein RBBP7 were S-nitrosylated in cortical neurons after depolarization. How S-nitrosylation affects CREB function is unknown; however, mutation of the cysteines within bZIP domain (Cys300, Cys310, and Cys337) has been shown to affect binding of CREB-regulated transcription coactivator 2 (CRTC2) (30). We found that S-nitrosylation of RBBP7 at Cys166 is required for activity-dependent dendritogenesis in vitro. The implications of this finding may be far-reaching, given that disruption of dendritogenesis at early developmental stages affects adult dendritic morphology and causes an aberrant response to fear conditioning (31). Our data also indicate that S-nitrosylation of RBBP7 may play a role in regulating subunit composition of the NuRD complex. S-nitrosylation of RBBP7 promotes the interaction with the NuRD ATPase subunit CHD4, an event that is especially relevant during corticogenesis when NuRD complexes undergo a developmentally regulated subunit switch (21). We previously showed that the mutual inclusion of CHD subunits within the NuRD complex determines the expression of specific sets of genes during mouse cortical development (21). Thus, S-nitrosylation could represent a mechanism that regulates NuRD subunit composition and interaction of the complex with chromatin. How S-nitrosylation affects protein conformation of NuRD subunits is unknown; however, recent reports suggest S-nitrosylation may promote disulfide bond formation within and between proteins, thereby inducing structural changes, which can outlast that of the initial modification (32, 33). It is possible that transient S-nitrosylation of the NuRD complex may induce disulfide bond formation that will have long-term effects on the complex subunit composition and, as a result, on gene expression.
GO analysis of S-nitrosylated nuclear proteins revealed that metabolic proteins are especially modified by S-nitrosylation. This finding is in agreement with a previous S-nitrosoproteome analysis of adult mouse brain that found that several metabolic pathways were targeted by S-nitrosylation (13). Although the role of S-nitrosylation on nuclear metabolism in mammalian cells is unknown, in plants, many nuclear proteins are S-nitrosylated in response to pathogen signaling (34). Moreover, during plant stress responses, S-nitrosylation of the protein arginine N-methyltransferase 5 promotes dimethylation of the spliceosome and pre-mRNA splicing of stress-response genes (35). The function of chromatin remodeling complexes, including NuRD, is affected by the availability of metabolically generated cofactors (36). Thus, our data suggest that in cortical neurons, S-nitrosylation of proteins involved in both nuclear metabolism and gene expression may be functionally linked.
We identified four lysine-containing motifs that may render cysteines more susceptible to S-nitrosylation. Previous work suggested that acidic and basic residues located near cysteines render nitrosylation more chemically favorable (22, 23). In macrophages, specific S-nitrosylation motifs guide the interaction between donor and acceptor proteins during transnitrosylation (37), yet similar transnitrosylation motifs have yet to be identified in neurons. We found that lysine motifs are essential for S-nitrosylation of both MBD3 and HDAC2. Lysine motifs for S-nitrosylation have been reported previously; lysine motif 1 (Lys at position −2) was found in a screen for endogenously SNO-Ps in macrophages (38), and lysine motif 2 (Lys at position +6) was identified in a screen on GSNO-treated heart homogenates (25). The mechanisms through which lysine motifs regulate S-nitrosylation of MBD3 and HDAC2 are not currently known; however, a recent bioinformatic analysis suggested that cysteines in close proximity to ubiquitylated lysines are more likely to be modified by S-nitrosylation (39), hinting at a possible interplay between cysteine S-nitrosylation and posttranslational modifications at neighboring residues. Our screen provides targets of nuclear S-nitrosylation in cortical neurons, the site(s) of S-nitrosylation for most identified proteins, and characterization of lysine-containing motifs that regulate cysteine S-nitrosylation. Further investigation of the SNO-Ps identified in this screen is needed to elucidate the full impact of NO signaling on nuclear functions in developing cortical neurons.
Materials and Methods
Animal procedures
All animal experiments were approved by the University College London Animal Welfare and Ethical Review Body and carried out in accordance to appropriate UK Home Office licenses. Timed pregnant Sprague-Dawley rats and C57BL/6J WT mice were used to prepare embryonic cortical neurons.
Preparation of cortical neurons
E17 rat cortices were isolated in cold dissection buffer [1× Hanks’ balanced salt solution, 2.5 mM Hepes (pH 7.4), 30 mM d-glucose, 1 mM CaCl2, 1 mM MgSO4, and 4 mM NaHCO3] and digested in digestion buffer [1 mM Hepes (pH 7.4), 20 mM glucose, 82 mM Na2SO4, 30 mM K2SO4, 6 mM MgCl2, 0.25 mM CaCl2, 0.001% phenol red, and 0.126 mM NaOH] plus 200 U of papain activated with cysteine-HCl and neutralized to pH 7. Cortices were washed, dissociated, and plated in plating media [minimum essential medium with 10% fetal bovine serum (FBS), 5% horse serum, and 1 mM glutamine]. Cells were plated on 10 cm Nunc plates coated with poly-d-lysine (20 μg/ml) and laminin (2 μg/ml) at 12.5 million per dish and kept at 37°C and 5% CO2. After 1 day in culture, media were replaced with supplemented neurobasal [neurobasal with 1× B27, 1 mM glutamine, 1× penicillin-streptomycin, and 10 μM 5-flouro-2′-deoxyuridine (FdU)]. Before stimulation, cells were starved for 16 hours by replacing twothirds of media with NB without B27 (1 mM glutamine, 1× penicillin-streptomycin, and 10 μM FdU). E15.5 mouse cortices were isolated and digested as for rat cortices. Cells were plated on coverslips in 24-well plates coated with poly-d-lysine (40 μg/ml) and laminin (2 μg/ml) at 0.35 million per well and kept at 37°C and 5% CO2.
Cell culture and transfections
HEK293T cells [American Type Culture Collection (ATCC) CRL-11268] and Neuro2a cells (ATCC CCL-131) were cultured according to standard conditions. Cells were transfected using Lipofectamine 2000 in OptiMEM according to the manufacturer’s guidelines.
Biotin-switch technique
S-nitrosylation is light-sensitive, and light can also react with ascorbate to induce artifactual signals; therefore, all procedures involving detection or stimulation of S-nitrosylation were carried out under minimal light conditions using brown Eppendorf tubes and foil-covered polypropylene falcon tubes. No glassware/metalware was used to avoid contaminating metal species that could interfere with the reaction (40, 41). CysNO was prepared as previously described (42). Cys only was prepared as for CysNO using Bio Perfomance Certified (BPC)–grade water (W3513, Sigma-Aldrich) in place of NaNO2. Buffers were prepared as in (15) with additional buffers used as specified.
For neuronal samples, 3 × 10 cm plates of E17 cortical neurons per sample were treated as appropriate. For HEK experiments, a transfected 6 cm plate of HEK293T cells was treated per condition. Cells were washed twice with ice-cold phosphate-buffered saline (PBS) and harvested in cold HEN lysis buffer [100 mM Hepes, 1 mM EDTA, and 0.1 mM neocuproine (pH 8.0) with 0.2% NP-40]. Samples were then homogenized five times with a 25-gauge needle and centrifuged for 10 min at 10,000g at 4°C, and the supernatant was collected (neuronal samples contained ~1.2 mg of protein per three plates, and HEK293T cells contained 650 μg of protein per sample). Except for experiments that were lysed and blocked immediately in Fig. 2, low–molecular weight S-nitrosothiols were removed by acetone precipitation at −20°C for >1 hour, followed by centrifugation of protein pellets for 20 min at 2000g at 4°C, two washes with 70% acetone, and resuspension in 200 μl of HENS [100 mM Hepes, 1 mM EDTA, and 0.1 mM neocuproine (pH 8.0) with 1% SDS]. Samples were blocked by adding an equal volume of 2× methyl thiocysteine (MMTS) blocking buffer (2% MMTS and 5% SDS in HEN buffer) and incubated for 30 min at 50°C with intermittent vortexing. Protein pellets were precipitated and, as before, then washed three times with 70% acetone and resuspended in 220 μl of HEN lysis buffer. Sodium ascorbate was added to a final concentration of 50 mM, and samples were incubated for 5 min at room temperature (RT) with gentle mixing. HPDP-biotin (catalog no. 21341, Thermo Fisher Scientific) was added to 1 mM, and samples were rotated for 45 min, followed by acetone precipitation as above. After three washes with 70% acetone, samples were resuspended in 220 μl of HENS, and 400 μl of neutralization buffer (25 mM Hepes, 100 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, pH 7.5) was added. Samples were precleared by incubation with protein A beads for 1 hour at RT and then centrifuged at 500g for 5 min at RT, and the supernatant was removed. Total inputs (10%) were taken, and samples were incubated with 30 μl of bead volume of streptavidin agarose beads overnight, rotating at 4°C. Samples were then centrifuged at 500g for 5 min at 4°C and washed four times in wash buffer (neutralization buffer, 600 mM NaCl). Beads were dried using a 29-gauge needle, and 40 μl of 2× Western loading buffer [2% SDS, 2% beta mercaptoethanol, 20% glycerol, 100 mM tris (pH 6.8), and 0.008% bromophenal blue] was added. Samples were then boiled for 5 min and then centrifuged at 10,000g for 2 min at RT, and the supernatant was run on homemade 10% polyacrylamide gels or precast NuPAGE 4-12% bis-tris gels (NP0335). For biotin switch on treated cellular lysates, the above protocol was followed, except that the treatment took place after harvesting in samples made up to 500 μl with HEN buffer. In experiments using N-ethylmaleimide-cysteine (NEM) blocking (140 mM in HEN buffer with SDS at 2.44%), blocking was carried out for 45 min at 50°C.
S-nitrosothiol resin-assisted capture
Nuclear and cytoplasmic protein extractions of E17 neurons were carried out using an NE-PER kit (catalog no. 78833, Thermo Fisher Scientific) as per the manufacturer’s instructions, with an extra wash with cytoplasmic extraction reagent (CER1) before addition of buffer nuclear extraction reagent (NER1). Four hundred micrograms of protein was used per sample. CysNO treatment for 20 min was carried out in the CER1 (cytoplasmic) or NER1 (nuclear) buffer with intermittent mixing. CysNO was removed after treatment either by acetone precipitation or using P6 spin columns prewashed in HEN buffer. Protein amounts were equalized by bicinchoninic acid (BCA) assay, and then, the volume was made up to 500 μl with HEN buffer. SNORAC was carried out according to (15) with the following modifications. Thiopropyl sepharose 6B resin (catalog no. 17-0420-01, GE Healthcare) was used, after three washes in BPC water and one wash in HEN buffer. Beads were centrifuged at 500g for 10 min at 4°C in between washes. Beads were stored (no longer than 1 day) in HEN buffer at 4°C in a 1:1 ratio. MS samples were frozen with HEN buffer diluted 10-fold in water and stored at −20°C.
Silver staining
Standard silver staining protocol was followed. Briefly, the polyacrylamide gel was soaked in 50% methanol two times for 15 min and then in 5% methanol for 10 min before rinsing in water. Gel was then soaked in 10 μM DTT for 20 min and then in 0.1% silver nitrate for 20 min. After three washes with water, gel was washed two times in developer solution (500 ml of ~285 mM sodium carbonate solution plus 250 μl of 37% formaldehyde) and soaked until bands became visible. The reaction was stopped with addition of citric acid, and gel was washed in water before imaging.
Coimmunoprecipitation
Co-IP experiments were performed in low-light conditions [see sample guidelines for working with S-nitrosylation (41)]. For neuronal co-IPs, cells were seeded onto 10 cm plates, treated with 200 μM CysNO donor or Cys control for 20 min, then washed with cold PBS, and harvested in cold IP lysis buffer [50 mM tris (pH 7.4), 150 mM NaCl, 1% Triton X-100, and 1% sodium deoxycholate (DOC)] containing a protease inhibitor cocktail, phosphatase inhibitors 2 and 3, and phenylmethylsulfonyl fluoride (all at 1:100). Samples were lysed on ice for 30 min, homogenized, and then cleared by centrifugation at 1000g for 10 min at 4°C. Lysates were precleared with protein G Sepharose beads (catalog no. 17-0618-01, GE Healthcare) for 2 hours at 4°C. Total inputs (10%) were taken. The remaining sample was incubated overnight 4°C with rabbit anti-CHD4 (ab72418) or rabbit IgG (X0903, Dako). Lysates were then incubated with protein G Sepharose beads for 2 hours at 4°C. Beads were washed once with wash buffer [50 mM tris (pH 7.5), 150 mM NaCl, 0.1% Triton X-100, 5% glycerol], with protease, and phosphatase inhibitors at 1:1000 twice with wash buffer, and twice with PBS. Excess PBS was removed, and proteins were eluted by boiling the beads with 2× Western loading buffer. Total inputs and samples were then run on precast 3 to 8% tris-acetate gels.
For HEK293T cell co-IPs, cells were seeded onto 6 cm plates and cotransfected with Flag-tagged mCHD4 and either HA-RBBP7WT or HA-RBBP7C166S. Cells were then treated as described for E17 neurons, then washed with cold PBS, and harvested in cold radioimmunoprecipitation assay buffer [50 mM tris (pH 7.5), 150 mM NaCl, 1% NP40, and 0.5% DOC] containing inhibitors as described above at 1:100. Samples were lysed and cleared, and then, protein concentration was determined using BCA Protein Assay; 250 to 400 μg of protein was used per experiment. Lysates were precleared, and total inputs were taken. The remaining sample was incubated overnight in the dark at 4°C with rabbit anti-HA (3724S, Cell Signaling Technology) or rabbit IgG (X0903, Dako). Lysates were then incubated with protein G–Sepharose beads for 2 hours at 4°C. Beads were washed, and proteins were eluted as for E17 neuron samples. Samples were then run on precast NuPAGE 4-12% bis-tris gels.
Western blotting
Western blotting was carried out according to standard procedures. Transfer conditions were 100 V for 3 hours at 4°C in 10% methanol for experiments involving detection of high–molecular weight proteins and were 330 mA for 1 hour and 30 min in 20% methanol for routine transfers. Membranes were blocked in 5% milk/TBST (tris buffered saline with 0.1% Tween20) before overnight incubation at 4°C with primary antibodies at a 1:1000 dilution. The following primary antibodies were used: mouse anti–c-Myc (A7; sc-56634, Santa Cruz Biotechnology), mouse anti-Flag (M2; F3165, Sigma-Aldrich), mouse anti-HA (901502, BioLegend), rabbit anti-HA (3724, Cell Signaling Technology), mouse anti-HDAC2 (05-814, Merck Millipore), mouse anti-HSP90 (Ab13492, Abcam), rabbit anti-RBBP4 (79416, Abcam), rabbit-RBBP7 (R4279, Sigma-Aldrich), goat anti-HSP90 (sc-1055, Santa Cruz Biotechnology), rabbit anti-CREB (9197, Cell Signaling Technology), and phospho-CREB ser133 (9191S, Cell Signaling Technology). After TBST washes, membranes were incubated with the appropriate secondary antibody at a 1:20,000 dilution. The following secondary antibodies were used: anti-mouse horseradish peroxidase (HRP; NXA931, GE Healthcare), anti-goat HRP (A5420, Sigma-Aldrich), and anti-rabbit HRP (NA93AV, GE Healthcare). Signal was detected by using enhanced chemiluminesence (ECL) or ECL Prime detecting reagents (GE Healthcare Life Sciences) and by exposing the immunoblot to the Amersham Hyperfilm (GE Healthcare Life Sciences).
Dendritogenesis assay
E15.5 mouse neurons were transfected using Lipofectamine 2000 2 hours after plating, with 220 ng of Pbird GFP with 500 ng of mycEV, mycRBBP7WT siRNA-resistant, or mycRBBP7C166S siRNA-resistant plasmids. Cells were cotransfected with 400 nM of each siRNA. Media were changed 3 hours after transfection to NB media supplemented with 0.33% B27, 1 mM glutamine, 1× penicillin-streptomycin, and 10 μM FdU, and coverslips were fixed in 4% paraformaldehyde/PBS 48 hours after transfection. Coverslips were washed once in PBS, permeabilized in 0.3% Triton X-100/PBS, blocked in a 5% normal goat serum/5% FBS/PBS solution for 1 hour at RT, and incubated with chicken anti-GFP (ab13970, Abcam) overnight. After three PBS washes, samples were incubated with 4′,6-diamidino-2-phenylindole and Alexa Fluor goat anti-chicken 488 (A11039, Life Technologies) at 1:1000 for 1 hour at RT. Coverslips were washed and mounted. Slides were then blinded before imaging and analysis. Images were acquired using an SPE or SPE3 confocal microscope (Leica) with LAS AF software and processed using ImageJ/Fiji. Sholl analysis was performed using ImageJ Simple Neurite Tracer (fig. S8) or a Fiji Sholl Analysis plugin (Fig. 4), and samples were deblinded after final processing.
Plasmids
The cytomegalovirus (CMV) myc-CREB plasmid was created by B. Lonze (Ginty Lab). msMBD3, ratRBBP4, and ratRBBP7 were subcloned into the CMV-myc expression vector. Mutagenesis to create RBBP7, RBBP4 and MBD3 cystine-to-serine mutants, MBD3 lysine-to-alanine mutants, and mycRBBP7WT and mycRBBP7C166S siRNA-resistant plasmids was carried out using a QuikChange multisite-directed mutagenesis kit (Agilent). HA-tagged ratRBBP7 and HA-tagged msHDAC2 were each cloned into AdEasy pShuttle-CMV vectors, and cysteine and lysine mutants were created using QuikChange multisite-directed mutagenesis kit (Agilent). Flag-mCHD4, hCHD5, and hCHD3 were generated as previously described (21). siRNAs were generated by Invitrogen. The following sequences were used: siRBBP7, 5′-CCACAUAAUGAAACUAUUCUGGCUU-3′ (forward) and 5′-AAGCCAGAAUAGUUUCAUUAUGUGG-3′ (reverse); siRBBP4, 5′-AAAUCUUUCCCUUCAGGCCUGGUCA-3′ (forward) and 5′-UGACCAGGCCUGAAGGGAAA-GAUUU-3′ (reverse).
GO analysis
The GO shown in tables S2 to S4, S8, and S13 was assigned from UniProt by Proteome Discoverer 1.4 (Thermo Fisher Scientific). All other GO analyses were carried out using PANTHERdb (www.pantherdb.org), and the version number is listed in the respective figure legend. For overrepresentation analysis, PANTHER Overrepresentation Test (13th April 2017 release) was used with Bonferroni correction for multiple testing.
Mass spectrometry
For the experiments performed on neuronal nuclear extracts treated with either 1 mM Cys or CysNO (table S16), beads were thawed at 4°C, centrifuged at 4000 rpm for 1 min at RT. The supernatant was digested at 37°C with 800 ng of trypsin proteomics grade (Sigma-Aldrich) in 100 μl of 100 mM tris buffer (pH 8.5; buffer A). After tryptic digestion, supernatants were recovered. Beads were washed three times with 20 mM ammonium bicarbonate in 50% MeOH (v/v) (buffer B). Ammonium bicarbonate washes were pooled, dried, and then combined with the first supernatant. Digested proteins samples were subjected to 18O labeling as in (43). Thirty microliters of aliquots of A2, A3, B1, and B2 (table S16) was dried and resuspended in 50 μl of H2O18 98% pure (Sigma-Aldrich)/methanol 4:1 (v/v). The solution was then buffered by tris buffer. A fresh trypsin aliquot (200 ng in 0.5 μl) was added to catalyze the oxygen exchange reaction, which was allowed to proceed overnight at 37°C. Thirty microliters of aliquots of A1, A2, B2, and B3 was dried, resuspended in 30 μl of normal high-performance LC water, and subjected to a parallel trypsin-catalyzed oxygen exchange reaction. Although no change in the molecular weight of control peptides [“light” (L) peptides] was produced, the reaction was allowed to proceed in parallel to the “heavy” (H) labeling reaction to avoid any possible source of bias. After labeling, to inactivate trypsin and avoid the phenomenon of back exchange, samples were heated for 1 hour at 56°C and boiled for 10 min at 100°C. Labeled samples were mixed in a 1:1 ratio as follows: A2(H)/A1(L), A3(H)/A2(L), B2(H)/B3(L), and B1(H)/B2(L). This arrangement allowed the relative comparison of proteomes isolated from CysNO-treated cells versus untreated cells and CysNO-treated cells versus CysNO minus ascorbate-treated cells, in duplicate analysis with forward/reverse labeling. Mixed samples were purified by strong cation exchange (SCX) StageTips (44). Peptide mixtures were diluted with 1.5 ml of 80% acetonitrile/0.5% formic acid (solution SCX-a) and loaded onto a 200-μl micropipette tip stacked with one layer of a SCX resin (Empore extraction disks, Sigma-Aldrich) previously conditioned with 20 μl of solution SCX-b (20% acetonitrile/0.5% formic acid) and 20 μl of solution SCX-a. After two washing steps (20 μl of solution SCX-a and 20 μl of solution SCX-b), elution of tryptic peptides was achieved by adding 7 μl of 500 mM ammonium acetate/20% acetonitrile (solution E). The eluate was dried and resuspended in 15 μl of 0.1% formic acid/2% acetonitrile. Five microliters of aliquot of each sample was subjected to nanoscale liquid chromatography coupled to tandem mass spectrometry (nanoLC-MS/MS) analysis.
For the experiments on neuronal nuclear extracts treated with either 200 μM Cys or CysNO (table S17), beads were thawed at 4°C, centrifuged at 4000 rpm for 1 min at RT. The supernatant was removed and digested at 37°C with 300 ng of trypsin proteomics grade (Sigma-Aldrich) in 100 μl of 100 mM triethylammonium hydrogen carbonate buffer (TEAB) (pH 8.5; buffer C). After tryptic digestion, the supernatants were recovered, and the beads were washed three times with 20 mM TEAB in 50% (v/v) MeOH (buffer D). The washes and the first supernatant of each sample were pooled, dried, and resuspended in 100 μl of buffer C (samples: SNO-Ps). Proteins were reduced further by adding 10 μl of 100 mM DTT (1 hour at 37°C), and residual free cysteines were alkylated by adding 12 μl of 200 mM iodoacetamide. After quenching the reaction with additional 2 μl of 100 mM DTT, complete overnight digestion was achieved by adding a new 300 ng of aliquot of trypsin. Samples were labeled according to the standard dimethyl labeling procedure (45). Briefly, samples were subjected to reductive amination by adding 4 μl of 0.6 M sodium cyanoborohydride and 4 μl of either 4% (w/v) formaldehyde (“light” labeling) or 4% (w/v) formaldehyde-d2 (“medium” labeling). Samples were labeled as light (L) or medium (M) and mixed in pairs (table S18). Digest pairs were diluted to 2.4 ml with SCX-a solution and purified by SCX StageTips, as described above. The six eluates were evaporated, and peptides were resuspended in 12 μl of 0.1% formic acid/2% acetonitrile. Four microliters of aliquot of each sample was subjected to nanoLC-MS/MS analysis with technical duplicates (two injections per sample).
For the experiment on nuclear and cytoplasmic extracts treated with either 200 μM Cys or CysNO (table S19), SNO-Ps were isolated, and digested proteins were prepared and labeled by 18O as described above, with minor modifications in starting amounts of samples subjected to isotopic labeling. The volume of the supernatant and evaporated washes recovered from on-bead digestion was 100 μl. Starting amounts for the heavy labeling reaction were as follows: 45 μl for samples 1 and 2, 25 μl for samples 3 and 5, and 2 × 25 μl for sample 4. The same amount for each of the five samples was taken for the light labeling reaction.
Labeled samples (table S18) were mixed in a 1:1 ratio as follows: 1(H)/2(L), 1(L)/2(H), 4(H)/3(L), and 3(H)/4(L). Mixed samples were purified by SCX StageTips, as described in experiment 1. Because of the higher amount of protein sample recovered in this experiment, sample fractionation at the peptide level could be achieved (four fractions). After tip conditioning, sample loading, and washing, stepwise elution of tryptic peptides was achieved by sequential addition of 7 μl of four solutions, all containing 20% acetonitrile, of increasing ionic strength and pH: (i) 60 mM ammonium acetate and 0.5% formic acid, (ii) 120 mM ammonium acetate and 0.5% formic acid, (iii) 250 mM ammonium acetate and 0.5% formic acid, and (iv) 500 mM ammonium acetate. The four SCX fractions were evaporated by vacuum centrifugation and resuspended in 8 μl of 0.1% formic acid/2% acetonitrile. Six microliters of aliquot of each fraction (four fractions × six H/L pairs) was subjected to nanoLC-MS/MS analysis.
SNO-site identification
To elute cys-nitrosylated peptides that remained covalently attached to the beads after on-bead digestion of SNO-Ps, 100 μl of buffer B were added to each sample, followed by the addition of 10 μl of 100 mM DTT (incubation for 1 hour at 37°C). Alkylation of released cysteines was achieved by adding 10 μl of 200 mM iodoacetamide (for 1 hour at 37°C in the dark). The supernatants were recovered, and the beads were washed with 50 μl of buffer B. Supernatants and their respective washes were pooled and evaporated in a vacuum centrifuge (samples: SNO peptides). For a qualitative analysis of putative nitrosylation sites, SNO peptides from the evaporated extracts were dissolved in 200 μl of solution SCX-a, purified by SCX StageTips as described, evaporated to dryness, and reconstituted in 10 μl of 0.1% formic acid/2% acetonitrile. Five microliters of aliquot of the pooled sample was subjected to nanoLC-MS/MS analysis. Peptides detected in the CysNO data set of experiment 5 were assigned to proteins present in the SNO-P data set (biological n of ≥3). Notably, shared peptides that were detected under both Cys and CysNO conditions were also included in our combined data set (table S13).
Protein identification from neuronal extracts
Neuronal nuclear protein extracts (50 μg) were precipitated by adding cold acetone [4:1 (v/v)] and incubating the solution for 1 hour at −20°C. The pellet was solubilized in 50 μl of 5% sodium DOC containing 60 mM tris buffer (pH 8.8). Cysteine alkylation was achieved by adding sequential aliquots of, respectively, 5 μl of 100 mM DTT (1-hour incubation at 37°C), 6 μl of 200 mM iodoacetamide (1-hour incubation at 37°C), and 1 μl of 100 mM DTT (20-min incubation at 37°C). Solution volume was brought to 500 μl by adding 20 mM tris buffer. Overnight protein digestion was carried out at 37°C after adding sequencing-grade trypsin [1:100 (w/w); Sigma-Aldrich]. The enzymatic reaction was quenched by the addition of 50 μl of 5% trifluroacetic acid, which served also for precipitating the DOC detergent. After spinning down the precipitate, 200 μl of the supernatant was removed and purified on RP StageTips, as described in previous paragraphs. The purified peptide mixture was fractionated on SCX StageTips, as described above. Stepwise elution of tryptic peptides from SCX StageTips was achieved by sequential addition of 20 μl of seven solutions, all containing 20% acetonitrile, of increasing ionic strength and pH: (i) 50 mM ammonium acetate and 0.5% acetic acid, (ii) 75 mM ammonium acetate and 0.5% acetic acid, (iii) 100 mM ammonium acetate and 0.5% acetic acid, (iv) 150 mM ammonium acetate and 0.5% acetic acid, (v) 250 mM ammonium acetate and 0.5% acetic acid, (vi) 350 mM ammonium acetate and 0.5% acetic acid, and (vii) 500 mM ammonium acetate. Fractions were evaporated by vacuum centrifugation and resuspended in 15 μl of 0.1% formic acid/2% acetonitrile. Two microliters of aliquot of each fraction was subjected to nanoLCMS/MS analysis using a long elution gradient (see below).
NanoLC-MS/MS analysis of neuronal extracts
Chromatography was performed on an Easy LC 1000 nanoLC system (Thermo Fisher Scientific). The analytical nanoLC column was a pulled fused silica capillary (75 μm, intradermally), in-house packed to a length of 10 cm with 3 μm of C18 silica particles from Dr. Maisch. The peptide mixtures were loaded at 500 nl/min directly onto the analytical column. A binary gradient was used for peptide elution. Mobile phase A was 0.1% formic acid and 2% acetonitrile, whereas mobile phase B was 0.1% formic acid and 80% acetonitrile. Peptides were separated by a gradient elution at a flow rate of 300 nl/min, ramping from 5% B to 35% B in 60 min (50 min for SNO peptides and 120 min for nuclear extracts) and from 35% B to 100% B in additional 15 min; after 5 min at 100% B, the column was re-equilibrated at 2% B for 10 min before the following injection. MS detection was performed on a quadrupole-orbitrap MS Q Exactive (Thermo Fisher Scientific) operating in positive ion mode, with nanoelectrospray (nESI) potential at 1800 V applied on the column front end via a tee piece. Data-dependent acquisition was performed by using a top-12 method (top 8 for SNO peptide), where the 12 most abundant ions were automatically selected for higher-energy collisional dissociation fragmentation at normalized collision energy of 25%. Resolution (full width at half maximum), automatic gain control (AGC) target, and maximum injection time for full MS and MS/MS were 70,000/17,500, 1 × 106/1 × 105, and 20 ms/60 ms, respectively. MS/MS parameters for detection of SNO-peptides were as follows: resolution, 35,000; AGC target, 2 × 105; maximum injection time, 120 ms. MS full scan range was 350 to 1800 mass/charge ratio (m/z). Mass window for precursor ion isolation was 1.6 m/z. Ion threshold for triggering MS/MS events was 5 × 104. Dynamic exclusion was 30 s.
Analysis of neuronal extract data
Protein identification and quantification were achieved in Proteome Discoverer 1.4 (Thermo Fisher Scientific) using Sequest as the search engine and the Rattus proteome sequence database (www.ebi.ac.uk/uniprot; accessed on March 2015). The database was merged with a list of common contaminants named “Common Repository of Adventitious Proteins” retrieved from the Global Proteome Machine website (www.thegpm.org/crap/index.html). In total, 27,927 entries were searched. The following general search parameters were used: MS tolerance, 15 parts per million; MS/MS tolerance, 0.02 Da; enzyme, trypsin; maximum missed cleavages, 2; variable modification, oxidized methionine. In addition, the following search parameters were used depending on the experiment: experiments 1 and 2: variable modification, C-terminal 18O labeling; fixed modification, MMTS; experiments 3 and 4: variable modifications, NEM and carbamidomethyl-cysteine (CAM); fixed modifications (in two parallel searches), (i) N-terminal dimethyl (light) and dimethyl-lysine (light) and (ii) N-terminal dimethyl (medium) and dimethyl-lysine (medium); experiment 5: variable modification, C-terminal 18O labeling; fixed modification, NEM. For SNO-peptide searches, the following additional parameters were used: experiments 1 and 5: fixed modification, CAM. False discovery rate estimation was performed in Proteome Discoverer using the Target-decoy validator node. Minimum requirement for protein ID for every experiment was two peptides at 95% confidence, whereas for SNO-peptide identification, the peptide confidence threshold was set to 99%.
Hit criteria for SNO-Ps
To be considered as a target for S-nitrosylation, proteins detected from MS had to be enriched ≥2-fold in CysNO samples versus control in three experiments and not enriched ≥2-fold in any control. Sodium ascorbate is required as a reducing agent for S-nitrosothiols; therefore, samples in which sodium ascorbate were omitted were used as negative controls (see table S2).
Motif-X analysis
Analysis of the animo acid sequences surrounding SNO sites identified in table S13 was carried out using the web-based tool Motif-X (http://motif-x.med.harvard.edu/) (24, 46). The following settings were used: foreground data set (extended from IPI Rat Proteome): central character, C; width, 21; significance, 0.000001; background data set (IPI Rat Proteome): background central character, C.
Statistical analysis
Statistical analysis was performed as indicated in figure legends using GraphPad Prism software (version 7.0a).
Supplementary Material
Acknowledgments
We thank E. Brookes, H. Crerar [Medical Research Council (MRC) Laboratory for Molecular Cell Biology University College London (LMCB), UK], and P. Salomoni (German Center for Neurodegenerative Disease, Bonn Germany) for insightful comments on the manuscript and all members of the Riccio laboratory for helpful suggestions.
Funding: This work was supported by an MRC Senior Non-Clinical Fellowship (SNCF G0802010 to A.R.), a Wellcome Trust Investigator Award (103717/Z/14/Z to A.R.), the MRC LMCB Core Grant (MC_U12266B), and a Biomedpark University Magna Graecia Award (PONa3_00435 to G.C).
Footnotes
Author contributions: A.R. and J.G.S. designed the research. M.G. and G.C. performed the MS analysis. J.G.S., S.G.A., and C.A. performed all other experiments. J.G.S., S.G.A., M.G., and A.R. analyzed data. J.G.S. and A.R. wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The MS proteomics data have been deposited to the ProteomeXchange Consortium (47) via the PRoteomics IDEntifications (PRIDE) (48) partner repository with the data set identifier PXD009071. All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References
- 1.Anand P, Stamler JS. Enzymatic mechanisms regulating protein S-nitrosylation: Implications in health and disease. J Mol Med (Berl) 2012;90:233–244. doi: 10.1007/s00109-012-0878-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hess DT, Matsumoto A, Kim S-O, Marshall HE, Stamler JS. Protein S-nitrosylation: Purview and parameters. Nat Rev Mol Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 3.Gould N, Doulias P-T, Tenopoulou M, Raju K, Ischiropoulos H. Regulation of protein function and signaling by reversible cysteine S-nitrosylation. J Biol Chem. 2013;288:26473–26479. doi: 10.1074/jbc.R113.460261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nott A, Watson PM, Robinson JD, Crepaldi L, Riccio A. S-nitrosylation of histone deacetylase 2 induces chromatin remodelling in neurons. Nature. 2008;455:411–415. doi: 10.1038/nature07238. [DOI] [PubMed] [Google Scholar]
- 5.Kornberg MD, Sen N, Hara MR, Juluri KR, Nguyen JVK, Snowman AM, Law L, Hester LD, Snyder SH. GAPDH mediates nitrosylation of nuclear proteins. Nat Cell Biol. 2010;12:1094–1100. doi: 10.1038/ncb2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okamoto S-i, Nakamura T, Cieplak P, Chan SF, Kalashnikova E, Liao L, Saleem S, Han X, Clemente A, Nutter A, Sances S, et al. S-nitrosylation-mediated redox transcriptional switch modulates neurogenesis and neuronal cell death. Cell Rep. 2014;8:217–228. doi: 10.1016/j.celrep.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bredt DS, Snyder SH. Transient nitric oxide synthase neurons in embryonic cerebral cortical plate, sensory ganglia, and olfactory epithelium. Neuron. 1994;13:301–313. doi: 10.1016/0896-6273(94)90348-4. [DOI] [PubMed] [Google Scholar]
- 8.Nott A, Nitarska J, Veenvliet JV, Schacke S, Derijck AAHA, Sirko P, Muchardt C, Pasterkamp RJ, Smidt MP, Riccio A. S-nitrosylation of HDAC2 regulates the expression of the chromatin-remodeling factor Brm during radial neuron migration. Proc Natl Acad Sci U S A. 2013;110:3113–3118. doi: 10.1073/pnas.1218126110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graff J, Joseph NF, Horn ME, Samiei A, Meng J, Seo J, Rei D, Bero AW, Phan TX, Wagner F, Holson E, et al. Epigenetic priming of memory updating during reconsolidation to attenuate remote fear memories. Cell. 2014;156:261–276. doi: 10.1016/j.cell.2013.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, Takahashi M, Cheah JH, Tankou SK, Hester LD, Ferris CD, et al. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol. 2005;7:665–674. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- 11.Ryan SD, Dolatabadi N, Chan SF, Zhang X, Akhtar MW, Parker J, Soldner F, Sunico CR, Nagar S, Talantova M, Lee B, et al. Isogenic human iPSC Parkinson’s model shows nitrosative stress-induced dysfunction in MEF2-PGC1α transcription. Cell. 2013;155:1351–1364. doi: 10.1016/j.cell.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doulias P-T, Tenopoulou M, Greene JL, Raju K, Ischiropoulos H. Nitric oxide regulates mitochondrial fatty acid metabolism through reversible protein S-nitrosylation. Sci Signal. 2013;6:rs1. doi: 10.1126/scisignal.2003252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raju K, Doulias P-T, Evans P, Krizman EN, Jackson JG, Horyn O, Daikhin Y, Nissim I, Yudkoff M, Nissim I, Sharp KA, et al. Regulation of brain glutamate metabolism by nitric oxide and S-nitrosylation. Sci Signal. 2015;8:ra68. doi: 10.1126/scisignal.aaa4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seneviratne U, Nott A, Bhat VB, Ravindra KC, Wishnok JS, Tsai L-H, Tannenbaum SR. S-nitrosation of proteins relevant to Alzheimer’s disease during early stages of neurodegeneration. Proc Natl Acad Sci U S A. 2016;113:4152–4157. doi: 10.1073/pnas.1521318113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forrester MT, Thompson JW, Foster MW, Nogueira L, Moseley MA, Stamler JS. Proteomic analysis of S-nitrosylation and denitrosylation by resin-assisted capture. Nat Biotechnol. 2009;27:557–559. doi: 10.1038/nbt.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lonze BE, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35:605–623. doi: 10.1016/s0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 17.Riccio A, Alvania RS, Lonze BE, Ramanan N, Kim T, Huang Y, Dawson TM, Snyder SH, Ginty DD. A nitric oxide signaling pathway controls CREB-mediated gene expression in neurons. Mol Cell. 2006;21:283–294. doi: 10.1016/j.molcel.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 18.Jaffrey SR, Snyder SH. The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE. 2001;2001:pl1. doi: 10.1126/stke.2001.86.pl1. [DOI] [PubMed] [Google Scholar]
- 19.Xue Y, Wong J, Moreno GT, Young MK, Côté J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 20.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nitarska J, Smith JG, Sherlock WT, Hillege MMG, Nott A, Barshop WD, Vashisht AA, Wohlschlegel JA, Mitter R, Riccio A. A functional switch of NuRD chromatin remodeling complex subunits regulates mouse cortical development. Cell Rep. 2016;17:1683–1698. doi: 10.1016/j.celrep.2016.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stamler JS, Toone EJ, Lipton SA, Sucher NJ. (S)NO signals: Translocation, regulation, and a consensus motif. Neuron. 1997;18:691–696. doi: 10.1016/s0896-6273(00)80310-4. [DOI] [PubMed] [Google Scholar]
- 23.Marino SM, Gladyshev VN. Structural analysis of cysteine S-nitrosylation: A modified acid-based motif and the emerging role of trans-nitrosylation. J Mol Biol. 2010;395:844–859. doi: 10.1016/j.jmb.2009.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chou MF, Schwartz D. Biological sequence motif discovery using motif-x . Curr Protoc Bioinformatics. 2011;31:13.15.1–13.15.24. doi: 10.1002/0471250953.bi1315s35. [DOI] [PubMed] [Google Scholar]
- 25.Kohr MJ, Aponte AM, Sun J, Wang G, Murphy E, Gucek M, Steenbergen C. Characterization of potential S-nitrosylation sites in the myocardium. Am J Physiol Heart Circ Physiol. 2011;300:H1327–H1335. doi: 10.1152/ajpheart.00997.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murzina NV, Pei X-Y, Zhang W, Sparkes M, Vicente-Garcia J, Pratap JV, McLaughlin SH, Ben-Shahar TR, Verreault A, Luisi BF, Laue ED. Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure. 2008;16:1077–1085. doi: 10.1016/j.str.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carrel D, Hernandez K, Kwon M, Mau C, Trivedi MP, Brzustowicz LM, Firestein BL. Nitric oxide synthase 1 adaptor protein, a protein implicated in schizophrenia, controls radial migration of cortical neurons. Biol Psychiatry. 2015;77:969–978. doi: 10.1016/j.biopsych.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee SB, Kim CK, Lee K-H, Ahn J-Y. S-nitrosylation of B23/nucleophosmin by GAPDH protects cells from the SIAH1–GAPDH death cascade. J Cell Biol. 2012;199:65–76. doi: 10.1083/jcb.201205015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chung HS, Murray CI, Venkatraman V, Crowgey EL, Rainer PP, Cole RN, Bomgarden RD, Rogers JC, Balkan W, Hare JM, Kass DA, Van Eyk JE. Dual labeling biotin switch assay to reduce bias derived from different cysteine subpopulations: A method to maximize S-nitrosylation detection. Circ Res. 2015;117:846–857. doi: 10.1161/CIRCRESAHA.115.307336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo Q, Viste K, Urday-Zaa JC, Senthil Kumar G, Tsai WW, Talai A, Mayo KE, Montminy M, Radhakrishnan I. Mechanism of CREB recognition and coactivation by the CREB-regulated transcriptional coactivator CRTC2. Proc Natl Acad Sci U S A. 2012;109:20865–20870. doi: 10.1073/pnas.1219028109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin Q, Ponnusamy R, Widagdo J, Choi JA, Ge W, Probst C, Buckley T, Lou M, Bredy TW, Fanselow MS, Ye K, et al. MicroRNA-mediated disruption of dendritogenesis during a critical period of development influences cognitive capacity later in life. Proc Natl Acad Sci U S A. 2017;114:9188–9193. doi: 10.1073/pnas.1706069114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolhuter K, Whitwell HJ, Switzer CH, Burgoyne JR, Timms JF, Eaton P. Evidence against stable protein S-nitrosylation as a widespread mechanism of post-translational regulation. Mol Cell. 2018;69:438–450.e5. doi: 10.1016/j.molcel.2017.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y-T, Piyankarage SC, Williams DL, Thatcher GRJ. Proteomic profiling of nitrosative stress: Protein S-oxidation accompanies S-nitrosylation. ACS Chem Biol. 2014;9:821–830. doi: 10.1021/cb400547u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaki M, Shekariesfahlan A, Ageeva A, Mengel A, von Toerne C, Durner J, Lindermayr C. Identification of nuclear target proteins for S-nitrosylation in pathogen-treated Arabidopsis thaliana cell cultures. Plant Sci. 2015;238:115–126. doi: 10.1016/j.plantsci.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 35.Hu J, Yang H, Mu J, Lu T, Peng J, Deng X, Kong Z, Bao S, Cao X, Zuo J. Nitric oxide regulates protein methylation during stress responses in plants. Mol Cell. 2017;67:702–710.e4. doi: 10.1016/j.molcel.2017.06.031. [DOI] [PubMed] [Google Scholar]
- 36.Berger SL, Sassone-Corsi P. Metabolic signaling to chromatin. Cold Spring Harb Perspect Biol. 2016;8:a019463. doi: 10.1101/cshperspect.a019463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jia J, Arif A, Terenzi F, Willard B, Plow EF, Hazen SL, Fox PL. Target-selective protein S-nitrosylation by sequence motif recognition. Cell. 2014;159:623–634. doi: 10.1016/j.cell.2014.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ibáñez-Vea M, Huang H, Martínez de Morentin X, Pérez E, Gato M, Zuazo M, Arasanz H, Fernández-Irigoyen J, Santamaría E, Fernandez-Hinojal G, Larsen MR, et al. Characterization of macrophage endogenous S-nitrosoproteome using a cysteine-specific phosphonate adaptable tag in combination with TiO2 chromatography. J Proteome Res. 2018;17:1172–1182. doi: 10.1021/acs.jproteome.7b00812. [DOI] [PubMed] [Google Scholar]
- 39.Fowler NJ, Blanford CF, de Visser SP, Warwicker J. Features of reactive cysteines discovered through computation: From kinase inhibition to enrichment around protein degrons. Sci Rep. 2017;7:16338. doi: 10.1038/s41598-017-15997-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Forrester MT, Foster MW, Stamler JS. Assessment and application of the biotin switch technique for examining protein S-nitrosylation under conditions of pharmacologically induced oxidative stress. J Biol Chem. 2007;282:13977–13983. doi: 10.1074/jbc.M609684200. [DOI] [PubMed] [Google Scholar]
- 41.Forrester MT, Foster MW, Benhar M, Stamler JS. Detection of protein S-nitrosylation with the biotin-switch technique. Free Radic Biol Med. 2009;46:119–126. doi: 10.1016/j.freeradbiomed.2008.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cook JA, Kim SY, Teague D, Krishna MC, Pacelli R, Mitchell JB, Vodovotz Y, Nims RW, Christodoulou D, Miles AM, Grisham MB, et al. Convenient colorimetric and fluorometric assays for S-nitrosothiols. Anal Biochem. 1996;238:150–158. doi: 10.1006/abio.1996.0268. [DOI] [PubMed] [Google Scholar]
- 43.Bernaudo F, Monteleone F, Mesuraca M, Krishnan S, Chiarella E, Scicchitano S, Cuda G, Morrone G, Bond HM, Gaspari M. Validation of a novel shotgun proteomic workflow for the discovery of protein–protein interactions: Focus on ZNF521. J Proteome Res. 2015;14:1888–1899. doi: 10.1021/pr501288h. [DOI] [PubMed] [Google Scholar]
- 44.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 45.Boersema PJ, Raijmakers R, Lemeer S, Mohammed S, Heck AJR. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat Protoc. 2009;4:484–494. doi: 10.1038/nprot.2009.21. [DOI] [PubMed] [Google Scholar]
- 46.Schwartz D, Gygi SP. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat Biotechnol. 2005;23:1391–1398. doi: 10.1038/nbt1146. [DOI] [PubMed] [Google Scholar]
- 47.Deutsch EW, Csordas A, Sun Z, Jarnuczak A, Perez-Riverol Y, Ternent T, Campbell DS, Bernal-Llinares M, Okuda S, Kawano S, Moritz RL, et al. The ProteomeXchange consortium in 2017: Supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 2017;45:D1100–D1106. doi: 10.1093/nar/gkw936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vizcaino JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, Xu Q-W, et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:11033. doi: 10.1093/nar/gkw880. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.