Abstract
Chronic wounds are characterized by impaired healing and uncontrolled inflammation, which compromise the protective role of the immune system and may lead to bacterial infection. Upregulation of miR-223 microRNAs (miRNAs) has been shown to drive the polarization of macrophages towards the anti-inflammatory (M2) phenotype, which could aid in the acceleration of the wound healing. However, local-targeted delivery of microRNAs is still challenging, due to their low stability. Here, we developed adhesive hydrogels containing miR-223 5p mimic (miR-223*) loaded hyaluronic acid nanoparticles to control tissue macrophages polarization during wound healing process. In vitro upregulation of miR-223* in J774A.1 macrophages demonstrated increased expression of the anti-inflammatory gene Arg-1 and a decrease in pro-inflammatory markers, including TNF-α, IL-1β, and IL-6. We also evaluated the therapeutic potential of miR-223* loaded adhesive hydrogels in vivo. Adhesive hydrogel could adhere to and cover the wounds during the healing process in an acute excisional wound model. Histological evaluation and qPCR analysis showed that local delivery of miR-223* efficiently promoted the formation of uniform vascularized skin at the wound site, which was mainly due to the polarization of macrophages to the M2 phenotype. Overall, this study demonstrates the potential of nanoparticle-laden hydrogels conveying miRNA-223* to accelerate wound healing.
Keywords: Wound healing, Local immunomodulation, Macrophage polarization, CD44 targeting hyaluronic acid nanoparticles, miRNA-223, Localized delivery, Adhesive hydrogel
1. Introduction
Chronic diseases such as cardiovascular disorders and diabetes affect approximately 133 million people in the US, which represents more than 40% of the total population.[1] From these patients, an estimated 2.4–4.5 million suffers from chronic wounds, including venous stasis ulcers, arterial ulcers, and diabetic wounds.[2] Chronic wounds fail to progress consistently through one or more stages of the normal wound healing cascade (i.e., hemostasis, local necrosis and inflammation, proliferation, and remodeling). In turn, these wounds are often associated with prolonged or excessive inflammation, persistent infections, and impaired vascularization and re-epithelization.[2] Early inflammation is triggered by infiltration of neutrophils and macrophages into the affected area, which mediate wound debridement by phagocytosis.[3, 4] In addition, macrophages mediate fundamental innate immune processes and play multiple critical roles during wound healing.[5] This process is complemented with the release of cytokines, such as transforming growth factor-beta (TGF-β), vascular endothelial growth factor (VEGF), and fibroblast growth factor (FGF), which further recruits fibroblasts, endothelial cells, and keratinocytes to aid in angiogenesis, collagen deposition and wound contraction.[4, 6–8] Classically-activated macrophages (M1) release pro-inflammatory cytokines during early stages of wound healing [8, 9], while the alternatively-activated macrophages (M2) pave the way for the resolution of the inflammatory stage by secreting anti-inflammatory cytokines that promote wound healing.[8]
Macrophage reprogramming has been explored as a therapeutic strategy to promote wound healing due to their paramount role in tissue regeneration.[10, 11] Therapies that regulate the induction of specific phenotype in macrophages have been used for the treatment of cardiovascular and inflammatory diseases and cancer.[12–15] However, there are limited studies on the effects of macrophage repolarization in wound healing in vivo. It has been demonstrated that prolonged inflammation in chronic wounds occurs due to the persistence of M1 macrophages during the later stages of tissue repair.[16–18] Therefore, therapies that efficiently induce the polarization of macrophages towards the M2 phenotype could significantly improve the healing process. miRNAs, which are short single-stranded RNAs of 21–25 nucleotides, have gained significant interest for the treatment of variety of diseases.[19] This is mainly due to their ability to target multiple genes in a particular pathway, leading to long-lasting effects at the molecular level.[20, 21] Various miRNAs have been reported to be involved in different phases of wound healing, such as inflammation, angiogenesis, and granulation.[20, 22, 23] For instance, miR-223 has been shown to act as a potent regulator of the inflammatory response by driving the polarization of inflammatory macrophages towards the tissue healing phenotype.[24] In addition, miR-223 expression has been shown to increase during the differentiation of granulocytes [25], neutrophils [26], monocytes [27], megakaryocytes [28] and eosinophils [29], and decrease during erythrocyte and macrophage differentiation [30]. Therefore, efficient delivery of miR-223 to tissue macrophages may could aid in the resolution of the inflammatory phase and promote angiogenesis and granulation. However, the development of gene therapies for wound healing based on this strategy has been hindered by the lack of efficient delivery systems.
Localized nucleic acid delivery, through the combination of a wound dressing and a nanocarrier, could enable convenient self-administration for patients, while avoiding issues with gastrointestinal tract absorption and hepatic first-pass metabolism. Therefore, the use of localized drug delivery platforms can result in improving the bioavailability and maintenance of drug concentration within the therapeutic window. Besides, local delivery enables transmission of the most significant fraction of drug molecules to the target area, maximizing therapeutic potential, and reducing systemic drug toxicity. In addition, polymeric hydrogel dressings loaded with different therapeutic agents, have been widely explored for the treatment of cutaneous wounds.[31–33] This is mainly because hydrogel-based dressings can absorb the wound exudate, which in turn promotes infiltration of dermal and epidermal cells and the eventual re-epithelialization of the wound.[34] Our groups has recently developed biodegradable adhesive hydrogels that can provide a suitable environment for tissue regeneration.[35] Gelatin methacryloyl (GelMA)-based hydrogels have been shown to support the attachment, infiltration, and proliferation of cells that mediate wound re-epithelialization and healing.[35–37] Furthermore, the adhesive properties of these visible light crosslinkable hydrogels enable rapid and robust attachment to wounds of various shapes and sizes without the need for sutures or staples.
Although hydrogel-based dressings have been extensively used for the delivery of various therapeutic molecules, the efficient delivery of miRNAs via bioadhesives for macrophage polarization remains technically challenging. This can be primarily attributed to the specific characteristics of miRNAs, including their negative charge, rapid degradation by RNases, and a short half-life of approximately 10 min in plasma.[38] Therefore, we aimed to develop a GelMA-based adhesive hydrogel for the local delivery of miR-223 5p mimic (miR-223*) encapsulated in hyaluronic acid (HA)-based nanoparticles (NPs). Two types of HA conjugates (i.e., HA-polyethyleneimine (HA-PEI) and HA-polyethylene glycol (HA-PEG)) were used to encapsulate miR-223* based on the electrostatic interaction between poly(ethylene imine) (PEI) and miRNA.[39] In particular, we used HA-PEG for the synthesis of the NPs to increase the residence time of these nanocomplexes in the circulation. HA, an anionic biopolymer that is found in the native extracellular matrix (ECM), was used because of its non-immunogenic and biocompatible nature, and because of its role in various stages of tissue regeneration/wound healing. [40, 41] Moreover, previous reports have shown that HA can be easily cleared from the body through different excretion processes with a total turnover of about 5 g of HA per day in humans.[42] HA-based NPs also hold great potential for the specific targeting of inflammatory macrophages during sustained inflammation because of the specific interaction between HA and membrane receptors such as CD44, which are highly expressed on macrophages.[43, 44] Upon the formation of the hydrogels containing miR-223* loaded HA NPs, we optimized the mechanical, physical and adhesive properties of the hydrogels to ensure that they can adhere to the native skin and cover the wound during the healing process. Next, the concentration of miR-223* loaded HA NPs was optimized to efficiently drive the polarization of macrophages in vitro. Lastly, we evaluated the feasibility of using these adhesive hydrogels for local delivery of immunomodulatory agents in vivo using a murine model of acute excisional wounds. This animal model is generally used to assess the wound healing by evaluating granulation and vascularization at different time points.[45, 46]
2. Results
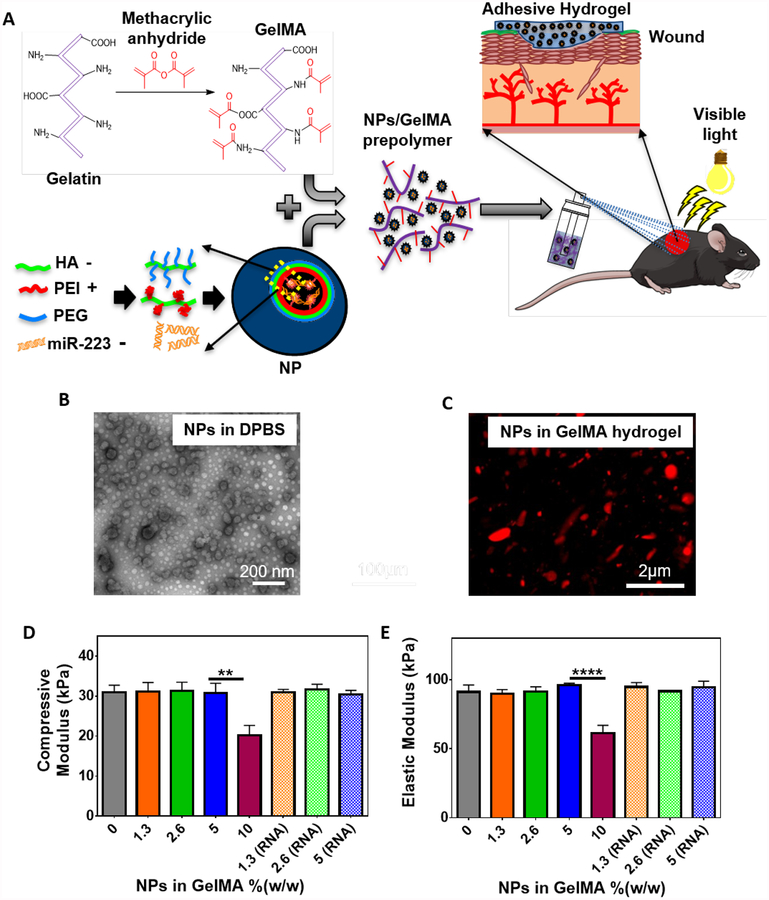
In our previous studies, we demonstrated that GelMA-based adhesives possess optimal mechanical and adhesive properties to be used as a tissue sealant or bioadhesive.[47] In this study, GelMA was synthesized by direct reaction of methacrylic anhydride with gelatin as described before (Figure 1A).[37] Visible light crosslinkable GelMA hydrogels were then formed using Eosin Y as an initiator, the triethanolamine (TEA) as a co-initiator and, N-vinylcaprolactam (VC) as a co-monomer. This visible light activated photo-initiator system helps circumvent the biosafety concerns of using UV light. We verified the chemical modification of gelatin with methacrylic anhydride via 1H-NMR (500 MHz; DMSO-D6) analysis of gelatin and GelMA (Figure S1). The appearance of peaks at δ=5.3 ppm (peak a) and δ=5.7 (peak b) ppm, corresponding to C ═ C in the structure of methacrylate and methacrylamide groups, indicated conversion of gelatin to GelMA. We then characterized the mechanical properties of GelMA hydrogels formed using different light exposure times (80 to 240 sec). Our results indicated that the mechanical properties of hydrogels did not change for exposure times longer than 240 sec, which could be due to the complete crosslinking of the pre-polymer solution at 240 sec (Figure S2). The degree of crosslinking was calculated to be 87.8 ± 1.0 % after 240 sec of light exposure as measured by integration of the peaks at δ=5.3 ppm (peak a) and 5.7 (peak b) ppm. This value did not show significant change at longer exposure times. Therefore, we selected 240 sec of exposure time to photopolymerize the hydrogels throughout our study.
Figure 1. Synthesis and mechanical characterization of NP-laden GelMA hydrogels.
(A) Schematic illustration of the process for the formation of NP/miR-223*-laden GelMA hydrogels and the use of these adhesive hydrogels for wound healing. (B) Representative TEM image of HA/miR-223* NPs made at 325:1 (w/w) ratio in DPBS. (C) A representative confocal microscope image of Cy5.5-labeled NPs distribution in GelMA hydrogel. Red color represents HA NPs. (D) Elastic modulus and (E) compressive modulus of GelMA hydrogels containing varying concentrations of NPs in GelMA polymer %(w/w). Mechanical properties were measured after incorporation of both NP and NP/miR-223* (RNA), respectively. Data are represented as mean ± SD (**p < 0.01, and ****p < 0.0001, n ≥ 3)
To prepare HA NPs, we conjugated 20 kDa HA with poly(ethylene imine) (PEI) or poly(ethylene glycol) (PEG), and used both HA conjugates at 50:50 (w/w) ratio to form miR-223* loaded NPs as described before.[39] The addition of HA-PEI increased the efficiency of encapsulation of miRNA (~98%, as quantified by Ribogreen® RNA assay) owing to the strong interaction between PEI and miRNA (Figure S3A). Self-assembled NPs were formed at a ratio of 1:325 (w/w) miR-223* to HA conjugates in Dulbecco’s phosphate-buffered saline (DPBS) with an average size of 160.0 ± 15.0 nm, a poly disparity index of 0.22 ± 0.02, and a surface charge of −13.0 ± 2.4 mV as measured by Malvern Zetasizer. The morphology of the NPs was further characterized by TEM, which showed the presence of spherical shaped NPs, with the dark core representing uranyl acetate stained miRNA conjugated to PEI (Figure 1B). The negative surface charge of the HA NPs is consistent with a core-shell structure, where the miRNA-complexed PEI constitutes the core of the nanocomplexes, while HA-PEG constitutes the outer shell. To evaluate the miR-223* NP formation, agarose gel electrophoresis was performed, and naked miR-223* was used as a control (Figure S3B). The NPs loaded with miR-223* showed absence of miR-223* band, indicating the robustness of the nanocomplex. However, upon treatment with polyacrylic acid (PAA), the miRNA was released because of the decomplexation of the NPs. The NPs were also tested for nuclease stability, where naked miR-223* treated with RNase A and the PAA released miRNA showed complete degradation of miRNA (Figure S3B). However, the miRNA condensed inside the HA NPs was protected from nucleases, as demonstrated by the absence of an miR-223* band. Overall, the modification of the HA polymer backbone with cationic branched PEI helped to decrease the overall negative charge of the HA biopolymer and promote nucleic acid encapsulation (HA−PEI/ miRNA nanoparticles) via electrostatic interactions.[39, 40, 48] The high counter-ion density of these nanoparticles is also known to promote endosomal escape following cellular internalization via the “proton-sponge effect”.[49] Furthermore, the PEG-modified HA polymer backbones are able to self-assemble with miRNA-loaded HA−PEI, yielding multifunctional nanoparticle complexes that could help increase the residence time of these nanocomplexes in the circulation. Taken together, these results confirmed that the conjugation of miRNA to PEI led to increased half-lives and lower degradation, while also allowing controlled release of the nucleic acids upon degradation of the nanocomplexes.
We next evaluated the effect of the addition of different concentrations of NPs on the mechanical properties of GelMA hydrogels. Our results revealed no significant changes in the mechanical properties of GelMA hydrogels after the incorporation of NPs up to 5.0 %(w/w) with a compressive modulus of 30.8 ± 2.5 kPa, an elastic modulus of 95.9 ± 1.4 kPa, and an extensibility of 18.9 ± 1.8 % (Figure 1D, E). The formulation of the precursor was optimized to yield hydrogels with an elastic modulus in the range of the native dermis (≈ 88 to 300 kPa) and a compressive modulus in the range of physiological soft tissues (10–200 kPa).[50, 51] We further characterized the structure of the hydrogels containing NPs using scanning electron microscopy (SEM), which showed no significant differences between the porous structure of the hydrogel with and without NPs (Figure S4). Due to the small size of NPs (~160.0 nm), the microarchitecture and consequently the physical properties of the NP-laden hydrogels (GelMA/NP) remained unchanged.
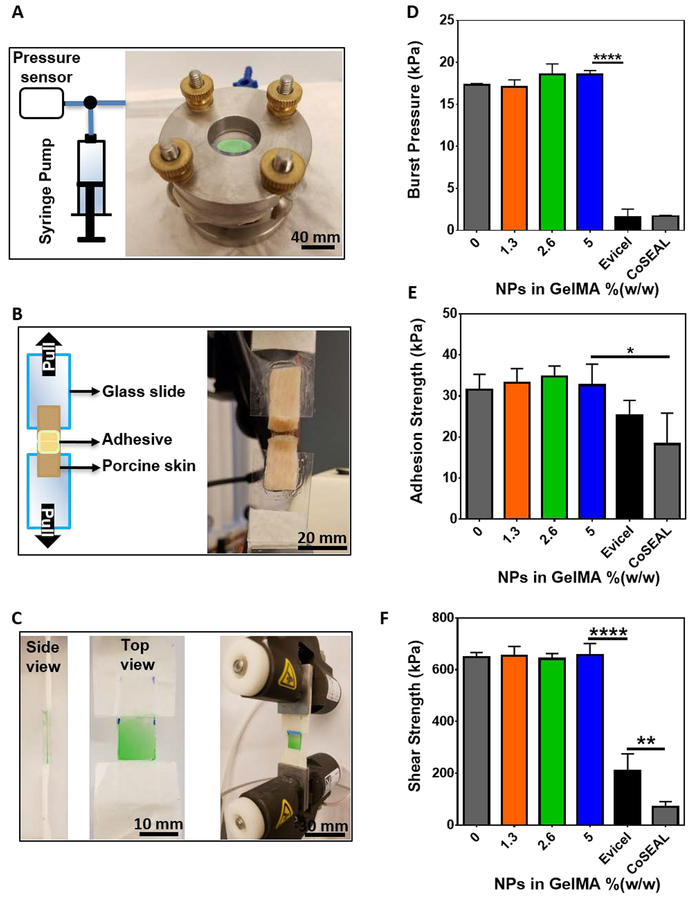
Conventional methods for wound closure such as sutures, mechanical fasteners, and staples are often associated with increased localized stress and trauma, as well as a higher risk of infection. In contrast, a hydrogel-based adhesive can strongly attach to the wound to act as a physical barrier to protect it from infection and as a depot to support the migration of cells to accelerate the rate of healing. An ideal adhesive should withstand the pressure exerted by the underlying tissue and fluids, seal the boundaries of a wound, and resist shear stresses. To evaluate the adhesive properties of the hydrogels, we performed burst pressure, wound closure, and lap shear tests based on modified American Society for Testing and Materials (ASTM) standard tests (Figure 2). Burst pressure tests are carried out to evaluate the resistivity of hydrogels against the pressure exerted by the underlying tissue and fluids. For this, air was continuously pumped into a custom-designed burst pressure apparatus. The hydrogels and commercial adhesives were applied to cover defects formed on porcine intestine sheets as a biological substrate (Figure 2A). Wound closure tests were conducted to investigate the ability of the hydrogels to seal the boundaries of a wound upon tensile stress. The hydrogels and commercial adhesives were applied on top of a 1 mm defect formed between two pieces of porcine skin as a biological substrate (Figure 2B). Lastly, lap shear tests were performed to evaluate the ability of the hydrogels to withstand shear stress (Figure 2C). Our results demonstrated that the adhesion of GelMA hydrogels containing NPs to physiological tissues was stronger than those observed for commercially available adhesives such as Evicel®, and Coseal™ (Figure 2D, E, F). In this regard, previous works have shown that GelMA-based adhesives exhibit high adhesion to different tissue surfaces due to the mechanical interlocking between GelMA and the native tissue, and covalent bonding triggered by radicals generated during photocrosslinking.[35]In addition, the presence of NPs in the hydrogel network does not affect the adhesion properties of the hydrogel.
Figure 2. Adhesive properties of NP-laden GelMA hydrogels.
(A) A representative image of burst pressure measurement setup. (B) A representative image of wound closure measurement setup, and (C) a representative image of burst lap shear setup. (D) In vitro burst pressure measurement of hydrogels with varying concentrations of NPs and two commercially available adhesives. (E) In vitro adhesion strength measurement of hydrogels with varying concentrations of NPs and two commercially available adhesives. (F) In vitro shear strength of hydrogels with varying concentrations of NPs and two commercially available adhesives. Data are represented as mean ± SD (*p < 0.05, **p < 0.01, and ****p < 0.0001, n ≥ 3).
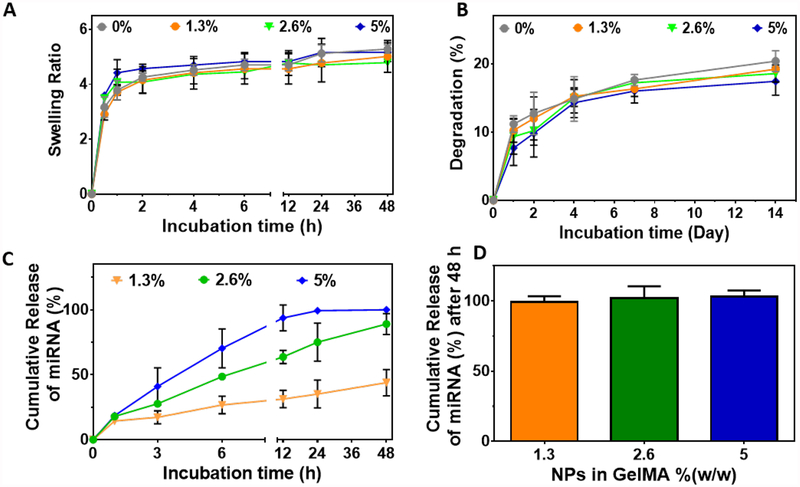
Multiple factors can affect the diffusion rate of molecules or NPs from polymeric networks, such as the type of polymer, pore size, the geometry of the particles (size and shape), preparation technique, particle concentration and physiological conditions. In our system, the diffusion rate of NPs is highly dependent on the infiltration of water into the polymeric structure (swelling) and the dissociation of polymeric bonds (degradation), which aids in the release of NPs from the hydrogels. Therefore, we first assessed the swellability and degradation rate of the GelMA/NP hydrogels in vitro. Our results showed that the hydrogels reached their maximum swelling ratios after 24 h, which corresponds to 5.17 ± 0.32 for GelMA/NP hydrogel containing 5.0 %(w/w) NPs. These values were in the range of GelMA hydrogels, which indicated that the incorporation of NPs did not alter the swellability of GelMA/NP hydrogels (Figure 3A). In particular, the high water-uptake capacity of GelMA-based hydrogels could mediate the absorption of wound exudates and induce the deposition of ECM components that promote cellular function and tissue regeneration in vivo. On the other hand, GelMA/NP hydrogels exhibited only 20% weight loss after 14 days of incubation in DPBS in vitro (Figure 3B). This observation suggested that GelMA/NP can retain structural stability throughout the different stages of wound healing.
Figure 3. Physical properties of NP-laden GelMA hydrogels and in vitro release profile of NP/miRNA from hydrogels.
(A) Swelling ratio, and (B) degradation rate of GelMA hydrogels loaded with varying concentrations of NPs in GelMA polymer in DPBS buffer (pH 7.4). (C) In vitro release profile of miRNA released from GelMA/NP/miRNA hydrogels in DPBS, and (D) enzyme induced release of miRNA from GelMA/NP/miRNA in the presence of 100 μg/ml collagenase type II in DPBS, 48 h post-incubation.
Kinetics of in vivo degradation for GelMA hydrogels may differ due to the presence of proteases in the wound area. In particular, collagenase type II (MMP-8) is the predominant proteolytic enzyme involved in the healing of normal and non-healing wounds.[52] Therefore, we evaluated the release rate of miRNA loaded NPs (NP/miRNA) from GelMA hydrogels in the presence and absence of collagenase type II. Our results 100 % release of NP/miRNA, as measured by Ribogreen® RNA assay, from hydrogels containing 5 %(w/w) NPs after 48 h incubation in DPBS at 37 °C and in the absence of collagenase type II (Figure 3C). However, 89 ± 8 and 43.9 ± 10.1 % release rates were observed for hydrogels loaded with 2.6 %(w/w) and 1.3 %(w/w) NPs, respectively. These results demonstrated that different doses of miRNA could be obtained by varying the concentrations of NPs that are loaded into the GelMA/NP hydrogels. Our results also showed that the diffusion rate of NPs released from the porous hydrogel network is highly dependent on the initial concentration of NPs used to synthesize the scaffolds. The burst release observed during the first 10 h could be related to the higher concentration of NPs that existed closer to the surface of the hydrogels and were in contact with the fluid. Moreover, a sustained release pattern was observed after 10 h of incubation for almost all concentrations tested, which corresponded to the NPs that were located further away from the surface of hydrogels. Overall, the results showed that the release rate for 5 %(w/w) NPs in GelMA hydrogel can potentially be used to control the macrophage polarization in the wound microenvironment during early stages of wound healing. Previous works have shown that inflammation takes place immediately after scab formation during the early stages of the healing process (1–3 days). In addition, our results demonstrated that hydrogels with 5 %(w/w) NPs/miR-223* exhibited 100% cumulative release after 48 hours (Figure 3C). These observations indicated that 5 %(w/w) scaffolds could be used to induce macrophage polarization during relevant clinical time frames in vivo and thus, this formulation was selected for further in vitro and in vivo studies. We then evaluated the effect of enzymatic degradation on the release profile of NPs/miR-223* from GelMA hydrogels. Our results showed that hydrogels incubated with DPBS containing 2.5 μg/ml of the enzyme exhibited 32.8 ± 5.3 % weight loss on day 14 (Figure S5). To assess the integrity of miRNAs during the release process, we studied the release of miRNA from the hydrogels in the elevated concentration of collagenase type II at 100 μg/ml (Figure 3D). Our results indicated 100 % release of miRNA for all concentrations of NPs in GelMA on at 48 h post-incubation, owing to the complete degradation of the hydrogels.
To assess the cytotoxicity of GelMA/NP hydrogels, we performed in vitro tests by 3D encapsulation of NIH3T3 fibroblasts within the hydrogels (Figure S6) and by exposing J774A.1 macrophages to the hydrogels using a trans-well system (Figure S7). Quantification of live/dead stained images revealed > 97% viability for both cell lines (Figure S6D, S7C). In addition, metabolic activity increased consistently during 5 days of culture (Figure S6E, S7D). These results revealed that GelMA/NP hydrogels can efficiently support the growth and proliferation of cells without eliciting any cytotoxic responses in vitro. Previous works have also shown that the administration of HA-PEI NPs is characterized by minimal systemic toxicity and a high clearance rate in vivo.[53]
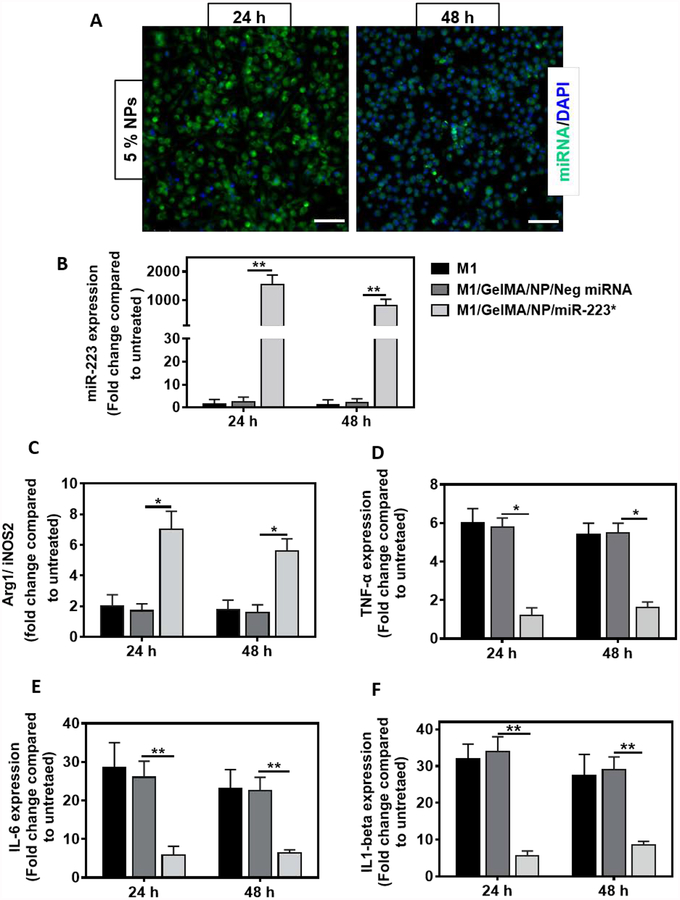
We next evaluated the ability of NP/miR-223* released from GelMA/NP/miR-223* hydrogels to be internalized by macrophages in vitro (Figure 4). First, we investigated the internalization of NP/Cy3-labeled miRNA released from GelMA hydrogels into J774A.1 macrophages. We have previously demonstrated that HA NPs can transfect J774A.1 macrophages via CD44 mediated internalization and its interaction with HA.[43] The intracellular transfection of Cy3-labeled miRNA released from 5 %(w/w) NPs in GelMA hydrogels led to a very strong signal after 24 h of incubation, followed by a gradual decrease at 48 h post-incubation (Figure 4A). Next, we performed miR-223 specific Taqman™ assay for quantification of transfected miR-223*. First, the macrophages were induced to the M1 phenotype by incubation with lipopolysaccharide (LPS) and interferon gamma (IFN-γ). The M1 induced macrophages were then incubated with GelMA/NP/miR-223* and GelMA/NP/negative miRNA hydrogels, formed in trans-wells, and were cultured for 24, and 48 h. Basal expression levels of miR-223* in J774A.1 macrophages were found to be very low with no significant increase upon LPS/IFN-γ induction. In contrast, miR-223* levels in M1 macrophages incubated with GelMA/NP/miR-223* were significantly higher, with a 1541 ± 340-fold increase after 24 h of incubation as compared to the control (Figure 4B). This was then followed by a gradual decrease to 861 ± 216-fold after 48 h, respectively, which was indicative of a higher release of miR-223* 24 h after incubation. Moreover, we observed 200 ± 51 -fold increase in the miR-223 expression of M1 macrophages transfected with NP/miR-223* released from 1.3 %(w/w) GelMA/NP/miR-223* compared to the GelMA/NP/negative miRNA group after 48 h post incubations (Supporting information, Figure S8B). These results demonstrated the efficient transfection and specific expression of miR-223 in J77A4.1 macrophages using NP/miR-223* released from GelMA/NP/miR-223*. However, 5 %(w/w) GelMA/NP/miR-223* hydrogels were used for gene expression and in vivo studies, since we achieved 100% cumulative release of NPs/miR-223* after 48 h, which coincided with the onset of the inflammatory stage of the healing process. Using 5 %(w/w) GelMA/NP/miR-223* hydrogels also enabled us to study the immunomodulatory potential of the scaffolds following a single administration of each experimental treatment, with no subsequent doses.
Figure 4. miRNA transfection and macrophage re-polarization studies in J774A.1 macrophages using GelMA/NP/miR-223* hydrogels in a trans-well set up.
(A) Representative fluorescent microscope images for transfection of Cy3-labeled miRNA released from GelMA/NP/ miRNA hydrogel, at the concentration of 5.0 %(w/w), in J774A.1 macrophages on 24, and 48 h post-incubation. Green color represents Cy3-labeled miRNA and blue color represents cell nuclei, respectively (Scale bar = 200 μm). (B) miR-223 expression was quantified by miR-223 specific Taqman™ assay performed in J774A.1 macrophages on 24, and 48 h post-incubation with GelMA/NP/Neg miRNA, and GelMA/NP/miR-223* hydrogels. The M1 represents cells that received LPS+IFN-Ƴ treatment for 16 h before incubation with either groups. Neg miRNA represents negative control miRNA and miR-223* represents miR-223 5p mimic. The gene expression level was normalized to untreated cells that did not receive any treatment. U6 snRNA was used as endogenous housekeeping gene. (C) In vitro macrophage re-polarization. qPCR analysis of Arg-1 (M2 marker)/iNOS-2 (M1 marker) gene expression was conducted on 24, and 48 h post-incubation with GelMA/NP/Neg miRNA, or GelMA/NP/miR-223* hydrogels. β-actin was used as a house keeping gene. (D-F) Anti-inflammatory effects of miR-223* transfection in J774A.1 macrophages. Decreased expression of pro-inflammatory cytokines; TNF-α, IL-6, and IL1-β mRNA levels was observed upon 24 and 48 h post-incubation with GelMA/NP/Neg miRNA and GelMA/NP/miR-223* hydrogels. The gene expression level was normalized to untreated cells that did not receive any treatment. β-actin was used as a house keeping gene. *p<0.05. Data are represented as mean ± SD (. *p<0.05, and **p < 0.01, compared to GelMA/NP/Neg miRNA group with n =4 individual experiments per group).
Macrophage polarization following transfection of miR-223* was also evaluated by measuring the relative levels of expression of Arg1/iNOS2 (M2 or anti-inflammatory/M1 or pro-inflammatory) after 24 h and 48 h of incubation with 5 %(w/w) GelMA/NP/miR-223* hydrogels. Our results revealed a 7- and 5-fold increase in the Arg1/iNOS2 ratios at 24 and 48 h post-incubation with GelMA/NP/miR-223* hydrogels, respectively, compared to the control. These observations were indicative of the polarization of M1 induced macrophages towards the anti-inflammatory phenotype via miR-223* transfection (Figure 4C). These results were further confirmed by evaluating the expression levels of pro-inflammatory cytokines up to 48 h (Figure 4D–F). These results showed that the levels of expression of TNF-α, IL-6, and IL1-β were decreased significantly at 24 h post-incubation with GelMA/NP/miR-223* hydrogels, compared to the control (Figure 4D–F). TNF-α, IL-6, and IL1-β mediate the inflammatory phase and have been reported to suppress endothelialization, which could then impair the wound healing process. Taken together, these results confirmed the successful polarization of M1 macrophages towards the M2 state using GelMA/NP/miR-223* hydrogels in vitro.
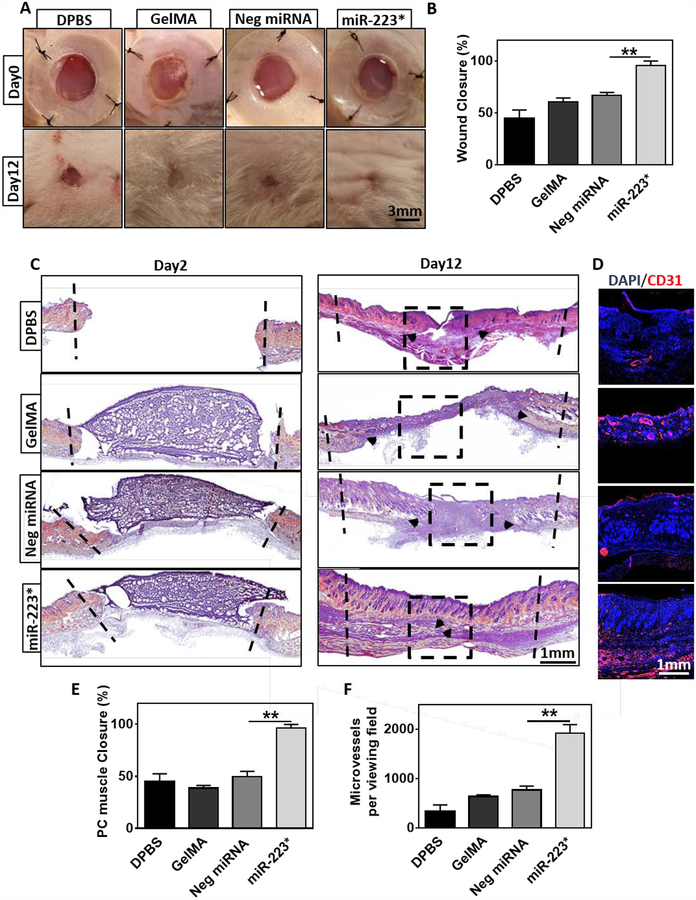
After we confirmed the ability of GelMA/NP/miR-223* hydrogels to drive macrophage polarization in vitro, we evaluated their efficacy to modulate inflammatory responses and wound healing in vivo, using a murine model of excisional wounds. Hydrogels were photopolymerized on top of wounds formed on the skin on the back of the animals on day 0. For our study, the borders of the wounds were immobilized with silicon splints to prevent the normal contraction of the wound and to better evaluate the degree of re-epithelization and granulation. Animals were divided into four treatment groups: 1) DPBS treated (DPBS), 2) pristine GelMA hydrogels (GelMA), 3) NP/negative miRNA-laden GelMA hydrogels (Neg miRNA) and 4) NP/miR-223*-laden GelMA hydrogels (miR-223*). GelMA hydrogels containing 5 %(w/w) NPs/miR-223* were chosen for in vivo studies due to their ability to induce macrophage polarization in vitro within 2 days after incubation. Visual inspection of the wounds revealed that the percentage of wound closure for miR-223* (i.e., 95.8 ± 4.3 %) treatment group was significantly higher than Neg miRNA (66.7 ± 2.9 %), GelMA (60.8 ± 3.5 %), and DPBS (44.9 ± 7.9 %) treated wounds (Figure 5A, B). These results were further confirmed by histological analysis, which shown that miR-223 5p mimic effectively promoted wound closure and healing, as demonstrated by the formation of new skin on day 12 post-surgery (Figure 5C). Quantification of panniculus carouses (PC) muscle closure under the fatty tissue layer based on Hematoxylin and Eosin Y (H&E) staining showed 96.3 ± 3.5 % closure for miR-223*, which was significantly higher than wounds treated with Neg miRNA (49.7 ± 5.1 %), GelMA (38.7 ± 2.5 %), and DPBS (45.4 ± 7.1 %) (Figure 5E).
Figure 5. Wound area and histological analysis of wounds on 2- and 12-days after GelMA/NP/miR-223* hydrogel treatment.
(A) Representative images of the wounds treated with DPBS, GelMA, Neg miRNA, and miR-223* after 0 day and 12 days of treatment. Black dashed lines demarcate the boundaries of wound. *Images for animals at day 12 were aquired after removal of the silicon splint to highlight the extent of wound healing. (B) Quantitative evaluation of wound area after 12 days of treatment. (C) H&E-stained sections of a full-thickness wound after 12 days of treatment with DPBS, GelMA, Neg miRNA, miR-223*. Black arrows indicate the edges of the panniculus carnosus (PC) muscle. (D) Representative immuno-fluorescent stained images were taken from the center of wound after 12 days of treatment (Black square indicates the area for vascularization evaluation). Endothelial cells are shown in red (CD31) and nuclei are shown in blue (DAPI). (E) Quantification of tissue regeneration evaluated by PC muscle closure. (F) Quantification of microvessel formation per viewing field (=2.5mm) for the different treatment groups. Data are represented as mean ± SD (*p < 0.05, **p < 0.01, n ≥ 3).
The formation of new blood vessels (i.e., angiogenesis) occurs during the proliferative phase of wound healing, and approximately 4–7 days after tissue disruption. During this process, new vascular sprouts extend to form capillary networks that mature into fully-formed blood vessels. In order to visualize the migration and organization of vascular endothelial cells at the center of the wounds, we performed immunofluorescent staining (IFS) against the angiogenesis-related marker CD31 (Figure 5D). Our results demonstrated that miR-223* treated group showed a >2-fold increase (p < 0.01) in the expression of CD31, as compared to the Neg miRNA group (Figure 5F). Quantification of CD31/DAPI fluorescent images revealed 1917 ± 175 microvessels per viewing field (=2.5mm) for miR-223*, which was significantly higher than the wounds treated with Neg miRNA (769 ± 80), GelMA (638 ± 34) and DPBS (329 ± 138) (Figure 5F). These results are in accordance with previous studies that have shown that the upregulation of miR-223 in the bone marrow promotes the proliferation and infiltration of hematopoietic cells to the wound site.[54] Furthermore, M2 macrophages have been shown to produce proangiogenic molecules (i.e., FGF and VEGF) that can induce the formation of new blood vessels.[55–57]
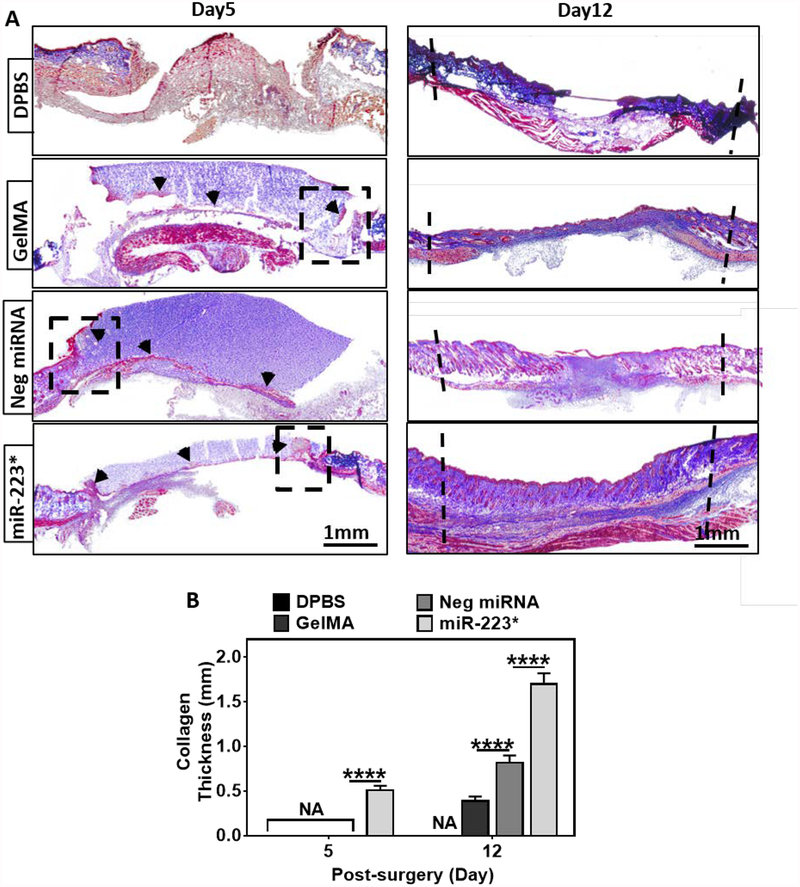
Collagen performs a key role during wound healing by enabling cell migration and acting as a foundation for ECM deposition during the proliferative phase of wound healing. Therefore, the formation of collagen-rich ECM can be an indicator of the transition from the inflammatory stage to the subsequent phases of the healing process. To characterize this phenomenon, we used Masson’s trichrome (MTC), a histological staining technique in which collagen is stained in blue, while cell bodies, muscle, and keratin are stained in red (Figure 6A). Histological assessment via MTC revealed a 2-fold increase in the thickness of the collagen layer in wounds treated with miR-223* (1.7 ± 0.1 mm), as compared to the wounds treated with Neg miRNA (0.8 ± 0.1 mm), GelMA (0.4 ± 0.0 mm) and DPBS (0.0 ± 0.0 mm) on day 12 (Figure 6B). Thicker collagen layer at day 12 in Neg miRNA treated wounds as compared to GelMA treated wounds is most likely due to non-specific target effects of Neg miRNA. However, we did not observe any other sign of better wound healing in Neg miRNA treated wounds throughout the assessment of wound healing markers. Collagen deposition was observed as early as day 5 for wounds treated with miR-223* as opposed to the control treatments, including Neg miRNA treated wounds (Figure 6A). Our results showed that GelMA/NP/miR-223* hydrogels exhibit a remarkably high-water uptake capacity and could support cell-attachment and growth in vitro. In turn, these characteristics could allow the drainage of the wound exudate and cellularization of the scaffold in vivo, as confirmed by both MTC and H&E stained images taken at day 5 post-surgery (Figure 6A and Figure S9). The formation of granulation tissue was also evaluated on H&E stained slides for GelMA, Neg miRNA, and miR-223* hydrogel-treated wounds (Figure S9A, B). Similarly, MTC stained samples taken at day 5 post-surgery revealed significant tissue ingrowth (red stain) within the polymeric scaffolds for GelMA, Neg miRNA, and miR-223*-treated wounds (Figure S9C).
Figure 6. Collagen deposition and granulation tissue formation at 5- and 12-days post-surgery.
(A) Representative images of Masson’s trichrome (MTC) stained sections of a full-thickness skin wound after 5 and 12 days of treatment with DPBS, GelMA, Neg miRNA, and miR-223* hydrogels (black dashed lines indicate the boundaries of wound). Cell migration is observed on day 5 post-surgery inside the hydrogels using MTC staining at the wound and hydrogel intersection (black dashed boxes). (B) Quantification of the collagen thickness at the center of the wound (blue stain). miR-223* treated wounds showed higher collagen deposition as compared to the control groups after 5 and 12 days of treatment. Data are represented as mean ± SD (*p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001, n ≥ 3).
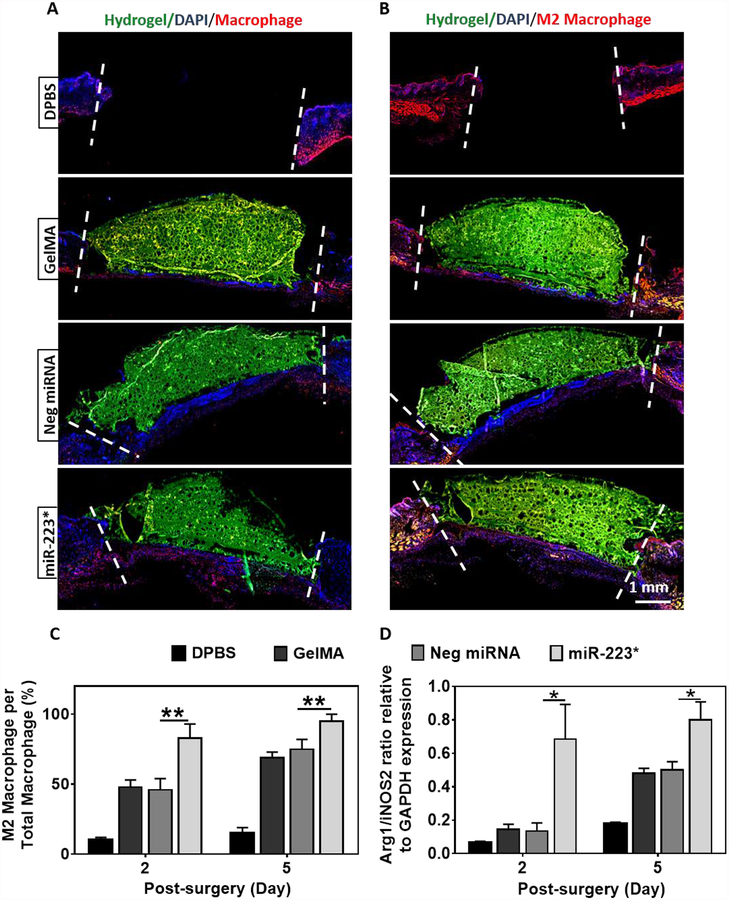
Inflammation is triggered during the early stages of wound healing via the infiltration of neutrophils and macrophages, approximately 48 h after tissue disruption. It is mainly mediated by pro-inflammatory cytokines secreted by M1 macrophages in response to exogenous or endogenous signals. Therefore, we performed immunofluorescence staining (IFS) against the pan-macrophage marker F4/80, as well as the M2 macrophage marker CD206 (C-Type Mannose Receptor 1) to assess the ability of GelMA/NP/miR-223* to selectively direct macrophage polarization towards the M2 phenotype (Figure 7A–C). For this, the ratio of M2 macrophages over total macrophages was calculated based on IFS stained tissue sections. IFS against CD206 revealed a significant increase in the ratio of M2 macrophages over total macrophages (Figure 7C). These results confirmed that local polarization of macrophages was directed toward the M2 phenotype for miR-223* treated wounds. We did not observe any positive fluorescent staining against both F4/80 and CD206 on day 12, which was indicative of macrophage withdrawal during the latter stages of wound healing. In parallel, we further quantified the change in the relative levels of expression of M1 (i.e., iNOS2) and M2 (i.e., Arg1) markers via quantitative PCR (qPCR) from tissue samples retrieved on days 2 and 5 post-surgery (Figure 7D). Our results revealed a significant increase in the expression of M2-specific genes on days 2 and 5 post-surgery, as demonstrated by the higher ratio of Arg1/iNOS2 in wounds treated with miR-223* compared to the controls (Figure 7D). Furthermore, the slight increase observed in the Arg1/iNOS2 ratio for GelMA and Neg miRNA treated groups could be explained due to the natural repolarization of macrophages from the M1 toward the M2 phenotype during later stages of wound healing. Taken together, these observations suggest that NP/miR-233 5p mimic loaded hydrogels not only increased the level of macrophage infiltration but also effectively mediated the local polarization of macrophages toward the M2 phenotype in vivo. This in turn led to comparatively better healing for wounds treated with NP/miR-233 5p mimic (miR-223*), as demonstrated by increased re-epithelization, formation of granulation tissue, and vascularization at the wound area.
Figure 7. Immuno-histological and quantitative analysis of wounds for macrophage evaluation after 2- and 5-days of treatment.
(A, B) Representative immunofluorescent stained images for total macrophage and M2 phenotype expression from the center of wound after 2 days of treatment with DPBS, GelMA, Neg miRNA, and miR-223* hydrogels. Total macrophage marker (F4/80) and M2 macrophage (anti-mannose receptor antibody) are shown in red color. Blue color represents nuclei (DAPI), and green color represents GelMA. (C) Quantitative analysis of M2 macrophages/total number of macrophages, per viewing field after 2 and 5 days of treatment (Only red stained cells in the wound area are counted, that is the area between 2 white dashed lines) (viewing field=2.5mm). (D) qPCR analysis of Arg1/iNOS2 mRNA expression for wound biopsies after 2 and 5 days of treatment relative to GAPDH expression. Data are represented as mean ± SD (*p < 0.05, **p < 0.01, n ≥ 3).
3. Conclusion
miRNAs have emerged as a potential therapy for the treatment of non-healing wounds, due to their pivotal role in the modulation of cellular processes involved in wound healing. Among all reported miRNAs, miR-223 has been shown to be a potent regulator of bone marrow-derived cells. However, its role in tissue regeneration/wound healing has not been fully explored mainly due to the limitations of current delivery systems. Here, we developed a delivery system using a macrophage-targeting HA NP-laden adhesive hydrogel. Both the adhesive hydrogel and the miR-223* loaded NPs were synthesized from highly biocompatible and naturally-derived biopolymers (i.e., GelMA and HA, respectively). The main advantage of the hydrogel precursor is its ability to be readily delivered to wounds with varying shapes and sizes and be rapidly photocrosslinked in situ via exposure to visible light. The high encapsulation and transfection efficiency of the HA NPs inside the adhesive hydrogels allowed the use of relatively small concentrations of miR-223*. HA-based NPs loaded with miR-223* were able to efficiently induce the polarization of M1 macrophages towards the anti-inflammatory M2 state both in vitro and in vivo, accompanied by accelerated tissue regeneration rate. Histological evaluation of explanted tissue samples showed that the GelMA/NP/miR-223* hydrogels could drive wound healing by not only triggering the resolution of the inflammatory phase but also by promoting the formation of new vascularized skin tissue. Our adhesive hydrogel system provides a suitable platform to deliver different nucleic acids for gene therapy.
Future work will focus on evaluating the ability of this system to elicit macrophage polarization and wound healing using a more physiologically relevant model of chronic non-healing wounds. This is mainly because there are significant differences at the cellular and molecular level between the wound microenvironment in normal subjects when compared to patients exhibiting non-healing wounds. Furthermore, we will explore the incorporation of additional miRNAs that can modulate the function of other inflammatory cells involved in wound healing. We also aim to further characterize global changes in gene expression occurring in macrophages during in vivo polarization towards the anti-inflammatory phenotype. Lastly, chronic wounds are highly susceptible to microbial infection, and the incorporation of an antimicrobial strategy based on different biocidal agents such as ZnO NPs or antimicrobial peptides will be explored. Moreover, diabetic wounds create a fertile environment for biofilm formation because the necrotic tissue and debris found in them are easily susceptible to infection and bacterial colonization. The presence of biofilm creates a self-perpetuating cycle, prolonging the existence of macrophages and neutrophils in the wound bed. Therefore, the adhesive wound dressing can be used as a reservoir to deliver novel antibacterial agents that can prevent the growth of drug-resistant microbial infections. Taken together, our results showed the remarkable potential of NP/miR-233*-laden adhesive hydrogels to be used as sutureless, regenerative, and immunomodulatory wound dressings to promote the healing of chronic wounds.
4. Materials and Methods
4.1. Materials
Sodium hyaluronate (HA, 20 kDa) was purchased from Lifecore Biomedical Co. (Chaska, MN). Branched poly (ethylene mine) (bPEI, 10 kDa) and Masson’s Trichrome (MTC) Stain Kit were purchased from Polysciences Inc. (Warrington, PA). Gelatin from cold water fish skin, methacrylic anhydride (MA), Eosin Y disodium salt, Triethanolamine (TEA), N-Vinylcaprolactam (VC), hematoxylin-eosin y staining (H&E) solutions, N-(3-dimethylaminopropyl)-N’-ethylcarbodiimide hydrochloride (EDC), N-hydroxysuccinimide (NHS), lipopolysaccharide (LPS), 3-(Trimethoxysilyl)propyl methacrylate (TMSPMA) and 4-morpholineethanesulfonate (MES) were purchased from Sigma-Aldrich. Collagenase type II was purchased from Worthington Biochemical Co. Mono-functional polyethyleneglycol amine (PEG-amine, 2kDa) was purchased from Creative PEG Works (Winston Salem, NC). Dulbecco’s modified Eagle medium (DMEM) was purchased from Cellgro (Manassas, VA) and Fetal Bovine Serum (FBS) was obtained from HyClone (Logan, UT). Murine IFNγ was obtained from PeproTech (Rocky Hill, NJ). 4’,6-diamidino-2-phenylindole (DAPI), Alexa-Fluor 488-labeled rhodamine-phalloidin, and live/dead viability kit were purchased from Invitrogen (San Diego, CA). mmu-mirVana™ miR-223 5p mimic (Thermo Fischer scientific, catalogue no. 4464066; Assay ID: MC14755), mirVana™ miRNA mimic negative control #1 (Thermo Fischer scientific, catalog no. 4464058), TaqMan™ probes specific for miR-223 (Thermo Fischer scientific, catalog no. 4427975), nuclease free water, and penicillin/streptomycin antibiotics were purchased from Life Technologies (Woburn, MA). Primers specific for iNOS-2, TNF-α, Arg-1, IL-1β, IL-6, and β-actin were purchased from Thermo Fisher Scientific (Waltham, MA). Quant-iT™ RiboGreen® RNA assay kit was purchased from Thermo Fischer scientific. Quick-RNA™ isolation kit was purchased from Zymoresearch (Irvine, CA). The iScript™ Reverse Transcription Supermix and iTaq™ Universal SYBR® Green Supermix were purchased from Bio-Rad (Hercules, CA).
4.2. Preparation and characterization of NPs/miRNA
HA-PEI and HA-PEG conjugates were synthesized by coupling reaction between HA (20 kDa) and bPEI (10 kDa) or PEG-amine (2 kDa), respectively, in the presence of EDC/NHS, as described previously.[40] Briefly, for the synthesis of HA−PEI conjugate, HA (100 mg) was dissolved in 1 M NaCl. EDC (50 mg) and NHS (50 mg) were added, and the solution was stirred for 30 min at room temperature. PEI (15 mg) was added, and the pH of the final solution was adjusted to 7. After stirring overnight at room temperature, the solution was dialyzed first against 1 M NaCl for 8 h and then against deionized water for 24 h using a cellulose dialysis membrane (MW cut off ~12−14 kDa). Alternatively, for the synthesis of HA−PEG conjugate, 100 mg of HA was dissolved in 20 mM MES buffer and 500 mM NaCl, pH 6. 50 mg of PEG-amine was added, and the solution was stirred for 15 min. Next, EDC (50 mg)/NHS (50 mg) solution (dissolved in 20 mM MES buffer and 500 mM NaCl, pH 6) was added to the HA/PEG-amine solution and stirred overnight at room temperature. The solution was then dialyzed against deionized water for 24 h using a cellulose dialysis membrane (MW cut off ~12−14 kDa). Both dialyzed solutions of HA-PEI and HA-EG were frozen at – 80 °C overnight, lyophilized, and stored at −20 °C until used for experiments.
HA-PEI/HA-PEG NPs were prepared by mixing HA-PEI and HA-PEG at a 1:1 ratio in DPBS. miR-223 5p mimic (miR-223*) was added to the HA-PEI/HA-PEG solution at a 1:325 (w/w) ratio, vortexed, and incubated for 20 min at room temperature. The miRNA encapsulation efficiency was measured by an indirect method. Briefly, the NPs were centrifuged for 20 min at 13,000 rpm, and the un-encapsulated miRNA was measured in the supernatant using Ribogreen® RNA assay (Thermoscientific). The average particle size, polydispersity index, and zeta potential were measured using Malvern Zetasizer (Westborough, MA). The morphology of the NPs was also analyzed using transmission electron microscopy (TEM) (JEOL, Peabody, MA). For TEM, the samples were prepared by adding a drop of NPs in DPBS on a Formvar/Carbon film grid followed by air-drying and 2 % uranyl acetate staining.
4.3. Synthesis and mechanical characterization of NP/miRNA-laden GelMA adhesives
GelMA was synthesized as described previously.[58] Briefly, gelatin from cold water fish skin (10 g) was dissolved in DPBS at 50 °C. 8 %(v/v) methacrylic anhydride (MA) was added dropwise under continuous stirring at 50 °C. After 3 h, the solution was diluted with DPBS to stop the reaction. The diluted solution was dialyzed against distilled water at 50 °C for 5 days. The resulting solution was then filtered under sterile conditions, frozen at – 80 °C overnight, and lyophilized for 4 days. The lyophilized GelMA was dissolved in a solution containing TEA (3.75 %(w/v)) and VC (2.5 %(w/v)) in DPBS. NPs were formed and kept at 4 °C prior for crosslinking. NP solutions with varying concentrations were mixed with the GelMA precursor solution at a 1:1 (v/v) ratio. Eosin Y disodium salt (1 mM) was dissolved separately in DPBS and was then added to the GelMA/NP/TEA/VC solution to form the final precursor. To form the hydrogels, 70 μl of the prepolymer solution containing NPs was pipetted into polydimethylsiloxane (PDMS) cylindrical molds (diameter: 6 mm; height: 2.5 mm) for compressive tests or rectangular molds (12.5 × 4.5 × 1.25 mm) for tensile test. The solutions were then photopolymerized via exposure to visible light (450–520 nm) for 240 s, using an LS1000 FocalSeal Xenon Light Source (Genzyme). Hydrogels were synthesized using different ratios of NPs to GelMA (i.e., 0, 1.3, 2.6, and 5 %(w/w)). The compressive tests and tensile tests were performed according to the previously published methods.[35]
4.4. Evaluation of in vitro swelling ratio and degradation
The swellability of GelMA/NP hydrogels was defined by calculating the ratio of weight change of the swollen hydrogel to the initial weight of the hydrogel in the dry state. For this, GelMA/NP hydrogels were incubated in DPBS at 37 °C and weighed at different time points for up to 48 h. GelMA/NPs hydrogels were prepared as described in the previous section. Next, samples were lyophilized and their initial (dry) weight was measured. After incubation for each time point, the excess buffer was gently removed using a disposable wipe, and the wet weight was measured.
To evaluate the percentage of in vitro degradation, GelMA/NP hydrogels were lyophilized, weighed, and placed in Eppendorf tubes containing 1 ml of DPBS at 37 °C for 2 weeks. The DPBS solutions were changed every 2 days. The samples were then freeze-dried and weighed on days 1, 4, 7, and 14 post-incubation (n = 4). Similarly, for in vitro enzymatic degradation tests, hydrogels were placed in 1 ml of DPBS containing 0, 2.5, 5, 10, 40, 70, and 100 μg/ml collagenase type II. The enzyme-containing solutions were changed every 3 days, and samples were lyophilized on 1, 2, 4, 7, and 14 days post-incubations. The percentage of degradation of the gels was determined using Equation (1).
| (1) |
Where W0 is the initial sample dry weight, and Wt is the dry weight after time t.
4.5. Scanning electron microscope (SEM) imaging
The samples for SEM imaging were dehydrated through an ascending series of ethanol in water solutions (30, 50, 70, 90 and 100% (v/v)) and were then dried using a critical point dryer. SEM images of pristine GelMA and GelMA/NP/miR-223* hydrogels were obtained using a Hitachi S-4800 scanning electron microscope.
4.6. In vitro release study
GelMA/NP containing different concentrations of NPs (1.3, 2.6, and 5 %(w/w)) were formed on the upper layer of the trans-well membranes. NPs were formed at different miRNA to polymer ratio (1:84, 1:169, 1:325 respectively for 1.3, 2.6, and 5 %(w/w)). Next, trans-well membranes containing GelMA/NP hydrogels were submerged into 1 ml of DPBS in 12-well plates. The concentration of miRNA in the solution was then assessed using a Quant-iT™ RiboGreen™ RNA Assay Kit at 1, 3, 6, 12, 24, 48, 72 h post-incubation at 37 °C.
The release test was also performed under enzymatic degradation conditions for GelMA/NP/negative miRNA at different concentrations of NPs (1.3, 2.6, 5 %(w/w)) in GelMA. The hydrogels were formed on the upper layer of trans-well membrane and were then submerged into a 12-well plate filled with 1 ml collagenase type II solution in DPBS (100 μg/ml). Next, the concentration of miRNA in solution was measured at 3, 6, 12, 24, 48, 72 h post-incubation at 37 °C using a Quant-iT™ RiboGreen™ RNA Assay Kit.
4.7. NP/miRNA stability and nuclease protection Assay:
To test the stability of miR-223* in HA NPs, intact miR-223*, miR-223* loaded NPs and de-complexed miR-223* (released from NPs using polyacrylic acid (PAA)) were run on 4% EX E-gels (Invitrogen). Briefly, the miRNA was de-complexed from NPs by using an equal volume of 4% PAA (strongly negatively charged) and vortex mixing.[59] The mixture was then kept at room temperature for 30 min and was later centrifuged at 11,000 g for 2 min. The supernatant containing released miRNA was loaded on 4% EX E-gels (Invitrogen). Ultra-low range DNA ladder was loaded as a marker. For nuclease protection assay, naked miR-223*, miR-223*/NP, and PAA released miR-223* from NPs were incubated with 20 μl of RNase A (0.03 U per ml) for 1 h at 37 °C, and the samples were run on 4% EX E-gels for evaluation of miRNA protection.
4.8. In vitro burst pressure
The burst pressure of GelMA/NP and commercially available surgical sealants Evicel® (Ethicon, Somerville, NJ, USA) and Coseal™ (Baxter, Deerfield, IL, USA) were measured based on the standard ASTM F2392–04 testing methodology. Porcine intestine (Savenor’s Market) was cut to a dimension of 40 mm× 40 mm. Next, the porcine intestine was placed between two stainless steel plates (35 mm× 35 mm), in which the upper piece had a 10-mm-diameter hole in its center and a circular defect (2 mm in diameter) was created on the center of the porcine intestine (Figure S10A). 20 μl of the adhesive was injected on the defect and, in the case of GelMA, the precursor was exposed to visible light. Next, the porcine intestine was placed into the burst pressure testing system, consisting of a pressure sensor, a recording unit. Air was then applied using a syringe pump at a rate of 5 ml/s to the sample until bursting (n ≥ 5).
4.9. In vitro wound closure
The adhesion strength of GelMA/NP, Evicel® and Coseal™ was evaluated according to a modified ASTM testing methodology (ASTM F2458–05). Briefly, fresh porcine skin was obtained from a local slaughterhouse and after removing the adipose tissue layer, the skin samples were cut into rectangular sections (10 mm × 15 mm). The porcine skin was kept moist with DPBS prior to use. The tissues were fixed onto two precut glass slides (30 mm × 60 mm) using Krazy Glue. The tissue was then cut from the middle with a single edge cutter blade. Afterward, 40 μl of the precursor solution or the commercial adhesives was pipetted onto the tissue interface and subsequently crosslinked with visible light (Figure S10B). The commercial sealants were applied according to instructions from the manufacturer.
4.10. In vitro lap shear
The shear strength of GelMA/NP, Evicel® and Coseal™ was evaluated according to the standard testing methodology (ASTM F2255–05). Two precut glass slides (10 mm × 50 mm) were used to hold the adhesives. One end of each glass slides (10 mm× 15 mm) was coated with gelatin from porcine skin (20 %(w/v) in distilled water) that was dried at room temperature before adding the adhesive precursors. Afterward, 10 μl of the desired solution was added on the gelatin-coated region of a precut glass slide, and then another gelatin-coated glass slide was placed over the precursor solution (Figure S10C). After assembly, the GelMA/NP hydrogel between the two gelatin-coated regions of the two glass slides was photo-crosslinked by visible light. The two glass slides were placed into the grips of an Instron mechanical tester for shear testing. For this, tensile loading with a strain rate of 10 mm/min was applied to the samples. The point of detachment of the two glass slides was measured and reported as the sealant shear strength (n ≥ 5).
4.11. In vitro cell studies
J774A.1 murine macrophages were cultured at 37 °C and 5 % CO2 in DMEM culture media supplemented with 10 %(v/v) FBS and 1 %(v/v) penicillin/streptomycin. For toxicity test, 200,000 cells were seeded in 12-well plates and then allowed to attach for 24 h before applying the hydrogels. 150 μl of the precursor solution containing GelMA/NP/miR-223* were photo-crosslinked on the membrane of trans-well inserts (75 mm polycarbonate inserts with a 1.0 μm pore size). After photo-crosslinking, the inserts were placed into the 12-well plates containing J774A.1 macrophages. Each well was filled with 850 μl of DMEM, and another 150 μl of media was added to the upper layer of the trans-well insert.
3D encapsulation of NIH3T3 fibroblasts was carried out using a protocol described in our previous publication.[35] Briefly, precursor GelMA/NP/ miR-223* solutions were prepared in cell culture media containing TEA (1.875% w/v) and VC (1.25% w/v), and gently mixed with 3T3 cells (5 × 106 cells/ml). A 10 μl drop of this mixture was pipetted on a 150 mm spacer, and covered by a TMSPMA-coated glass slide. After photo-crosslinking, the hydrogels were washed several times with warm media to remove the unreacted photo-initiators. The cell-laden hydrogels were then placed in 24 well plates and incubated at 37 °C, 5% CO2, and humidified atmosphere.
4.12. Cell viability and proliferation assays
Cell viability was determined using a calcein AM/ethidium homodimer-1 live/dead kit (Invitrogen) according to instructions from the manufacturer. Cell viability was evaluated on days 1, 3, and 5 post-incubation with GelMA/NP/miRNA (n ≥ 6). Fluorescent images were taken using a Zeiss Axio Observer Z1 inverted microscope and were analyzed using the ImageJ software. Cell viability was calculated as the ratio of viable cells to the total number of live and dead cells. The PrestoBlue assay was used to evaluate the metabolic activity of cells according to instructions from the manufacturer on days 1, 3, and 5 post-incubation with GelMA/NP/miRNA (n ≥ 6). Fluorescence intensity of the resulting solutions was recorded at 535–560 nm excitation and 590–615 nm emission.
4.13. In vitro miRNA transfection and macrophage polarization
For transfection and polarization studies, 200,000 J774A.1 macrophages were cultured on 12 well-plates and were first polarized toward the M1 (pro-inflammatory) phenotype by overnight incubation with LPS+IFN-γ (100 ng/mL) using our previously published method.[59] The cells were then transfected with i) GelMA/blank NP, ii) GelMA/NP/negative miRNA, and iii) GelMA/NP/miR-223* (5.0 %(w/w)) crosslinked on the trans-well insert. The cells were then processed for RNA isolation on 24, and 48 h post-incubation at 37 °C, using Quick-RNA™ MiniPrep RNA isolation kit (Zymo Research Corp., Irvine, CA). The efficiency of transfection was assessed using a Taqman® miR-223 specific assay (Thermo Fischer scientific) via qPCR (LightCycler® 480, Roche, Branford, CT). U6 small RNA (snRNA) was used as a housekeeping gene. The changes in the levels of expression of iNOS2 (M1/ pro-inflammatory marker) and Arg1 (M2/ anti-inflammatory marker) were quantified using qPCR (iTaq™ Universal SYBR® Green supermix, Biorad, CA).[59] β-actin was used as a house-keeping gene. The levels of expression of the pro-inflammatory cytokine’s TNF-α, IL1β, and IL-6 were quantified by using qPCR.[59] β-actin was used as a housekeeping gene. Primers are listed in Table S1.
4.14. Full-thickness in vivo wound model
All animal protocols were reviewed and approved by the Institutional Animal Care and Use Committee (ICAUC; protocol 17–0516R) at Northeastern University (Boston, MA, USA). CD-1 male mice (7 to 8-week old, average wt. 30 g) were purchased from Charles River (MA, USA). Animals were housed for at least 48 h prior to the surgery at the animal facilities with 12 h light/dark cycles, and food and water ad libitum. 24 h before the surgery, the dorsal skin of the animals was shaved, and the remaining hair was removed using a commercial hair-removing cream. On surgery day, the animals were anesthetized via inhalation of 4% isoflurane in an induction chamber. Following induction, the animals were positioned in ventral recumbency on top of a heating blanket at 37 °C to stabilize their body temperature and connected to a breathing circuit (2% isoflurane) for maintenance of anesthesia. 5–10 mg/kg of Meloxicam in 150 μl of sterile saline was administered subcutaneously prior to the incision. Following analgesia, the surgical site was disinfected using three consecutive applications of iodine and 70% ethanol. 6-mm sterile biopsy punches were used to create bilateral full-thickness wounds on the dorsum of the mice, and silicon splints (inner diameter = 6 mm, outer diameter = 9 mm, thickness = 1 mm) were then sutured on top of each wound.[60] The animals were assigned randomly to two endpoints and four groups for analysis: i) DPBS control (DPBS), ii) pure GelMA hydrogels (GelMA), iii) GelMA/NP negative miRNA control (Neg miRNA) and iv) GelMA/NP miR-233* (5.0 %(w/w) NPs to GelMA concentration). For animals receiving the DPBS control, 50 μl of sterile DPBS was pipetted on top of each wound. For animals receiving control and treatment GelMA hydrogels, 50 μl of the hydrogel precursor was pipetted on the wound area and photo-crosslinked in situ with visible light for 4 min. The wound beds were then covered with Tegaderm films and protected using cohesive bandages. Meloxicam (5–10 mg/kg) was administered subcutaneously after the procedure for analgesia, followed by placing the mice under a warming lamp until recovered from anesthesia.
The animals were weighed and monitored daily for signs of pain or discomfort, and digital photographs were acquired for all wounds to monitor wound closure. Following administration of the treatments, the wounds were covered with a protective Tegaderm film and a cohesive bandage. No additional administrations were carried out, and the adhesive Tegaderm film was not changed to avoid additional trauma to the wound site. Changes in the wound area, relative to the corresponding initial measurement, were calculated using ImageJ to compare the rate of wound closure among the experimental groups. On days 2, 5 and 12 post-surgery, animals were euthanized via CO2 inhalation followed by cervical dislocation. Following euthanasia, the full-thickness wounds (n = 6 wounds for each group) and 1–2 mm of the adjacent skin were harvested for qRT-PCR analysis and histopathological examination.
4.15. Morphological assessment and histopathological evaluation
Wound biopsies with hydrogels were fixed in 10 % formalin for 24 h. They were then incubated in 30 % sucrose solution overnight, followed by overnight incubation in 100 % FBS. Next, wound biopsies were flash-frozen in liquid nitrogen in Optimal Cutting Temperature (OCT) Compound. 9-μm cryosections were acquired using a Leica Biosystems CM3050 S Research Cryostat and processed with hematoxylin-eosin and Masson’s trichrome staining as well as immunofluorescence staining. Tissue sections were stained with primary antibodies against CD31, F4/80 and CD206, and an Alexa Fluor 594-conjugated secondary antibody was used for detection. For evaluation of the number of microvessels (CD31), endothelial cell or endothelial-cell cluster that was clearly separated from adjacent microvessel was considered a single countable microvessel (4 mm2 of the wounds’ center).[61] All sections were counterstained with DAPI, and visualized on an Axio Observer Z1 inverted microscope (n = 3).
4.16. Total RNA isolation and qPCR analysis
The wound biopsy samples were excised and stored in RNA later at −20 °C before further processing. For RNA isolation, the tissue samples were triturated using a mortar and pestle under liquid nitrogen. Total RNA was isolated from the triturated samples using TRI reagent (Fischer scientific) following the instructions from the manufacturer. RNA (1 μg) was reversely transcribed into cDNA and was used for qPCR analysis using SYBR® green supermix reagent. The expression of Arg1 and iNOS2 was quantified using a 7300 Real-Time PCR System from Applied Biosystems. GAPDH was used as a housekeeping gene. The comparative CT method was used to analyze the expression of Arg1 and iNOS2 mRNA.
4.17. Data analysis
Data analysis was carried out using a 1- or 2-way ANOVA test with GraphPad Prism 6.0 software. Error bars represent mean ± standard deviation (SD) of measurements (*p < 0.05, **p < 0.01, ***p < 0.001, and ****p<0.0001).
Supplementary Material
Figure S1. 1H-NMR analysis of GelMA pre-polymer and gelatin.
Figure S2. Mechanical characterization and 1H-NMR analysis of GelMA hydrogels formed at varying photo-crosslinking time.
Figure S3. Synthesis and characterization of HA-PEI/HA-PEG nanoparticles.
Figure S4. SEM images of pristine GelMA and GelMA/NP/miRNA.
Figure S5. Enzymatic degradation of hydrogels.
Figure S6. In vitro 3D encapsulation of NIH3T3 fibroblasts in NP laden GelMA.
Figure S7. In vitro cytocompatibility of GelMA/NP hydrogels using J774A.1 macrophages.
Figure S8. In vitro evaluation of miRNA transfection in J77A4.1 macrophages incubated with 1.3 %(w/w) GelMA/NP/miR-223*.
Figure S9. Histological analysis of wounds on day 5 post-surgery.
Figure S10. Schematics of burst pressure, wound closure test, and shear stress measurement.
Table S1. List of primers used for qPCR.
Acknowledgments
General: Electron microscopy was performed at the Electron Microscopy Center of Northeastern University with the assistance of Mr. William Fowle. Special thanks to Professor Rebecca L. Carrier and Professor Eno E. Ebong for sharing their equipment (Light Microscope and Cryostat) for this work.
Funding: NA acknowledge the support from National Institutes of Health (NIH) (R01EB023052; R01HL140618).
Footnotes
Competing interests: Include any financial interests of the authors that could be perceived as being a conflict of interest. Also include any awarded or filed patents pertaining to the results presented in the paper. If there are no competing interests, please state so.
Data and materials availability: All relevant data are available from the authors upon reasonable request.
References:
- 1.McGlynn EA; Asch SM; Adams J; Keesey J; Hicks J; DeCristofaro A; Kerr EA, N Engl J Med 2003, 348 (26), 2635–45. DOI 10.1056/NEJMsa022615. [DOI] [PubMed] [Google Scholar]
- 2.Frykberg RG; Banks J, Adv Wound Care (New Rochelle) 2015, 4 (9), 560–582. DOI 10.1089/wound.2015.0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koh TJ; DiPietro LA, Expert Rev Mol Med 2011, 13, e23 DOI 10.1017/S1462399411001943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Opdenakker G; Van Damme J; Vranckx JJ, Trends in immunology 2018, 39 (4), 341–354. [DOI] [PubMed] [Google Scholar]
- 5.Koh TJ; DiPietro LA, Expert reviews in molecular medicine 2011, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brancato SK; Albina JE, Am J Pathol 2011, 178 (1), 19–25. DOI 10.1016/j.ajpath.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Velnar T; Bailey T; Smrkolj V, Journal of International Medical Research 2009, 37 (5), 1528–1542. [DOI] [PubMed] [Google Scholar]
- 8.Kwon MJ; An S; Choi S; Nam K; Jung HS; Yoon CS; Ko JH; Jun HJ; Kim TK; Jung SJ, The journal of gene medicine 2012, 14 (4), 272–278. [DOI] [PubMed] [Google Scholar]
- 9.Barrientos S; Stojadinovic O; Golinko MS; Brem H; Tomic-Canic M, Wound Repair Regen 2008, 16 (5), 585–601. DOI 10.1111/j.1524-475X.2008.00410.x. [DOI] [PubMed] [Google Scholar]
- 10.Hesketh M; Sahin KB; West ZE; Murray RZ, International journal of molecular sciences 2017, 18 (7), 1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krzyszczyk P; Schloss R; Palmer A; Berthiaume F, Frontiers in physiology 2018, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schultze JL, Current opinion in pharmacology 2016, 26, 10–15. [DOI] [PubMed] [Google Scholar]
- 13.Saeed S; Quintin J; Kerstens HH; Rao NA; Aghajanirefah A; Matarese F; Cheng SC; Ratter J; Berentsen K; van der Ent MA; Sharifi N; Janssen-Megens EM; Ter Huurne M; Mandoli A; van Schaik T; Ng A; Burden F; Downes K; Frontini M; Kumar V; Giamarellos-Bourboulis EJ; Ouwehand WH; van der Meer JW; Joosten LA; Wijmenga C; Martens JH; Xavier RJ; Logie C; Netea MG; Stunnenberg HG, Science 2014, 345 (6204), 1251086 DOI 10.1126/science.1251086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoogeveen RM; Nahrendorf M; Riksen NP; Netea MG; de Winther MP; Lutgens E; Nordestgaard B; Neidhart M; Stroes ES; Catapano AL, European heart journal 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Genard G; Lucas S; Michiels C, Frontiers in immunology 2017, 8, 828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wetzler C; Kampfer H; Stallmeyer B; Pfeilschifter J; Frank S, J Invest Dermatol 2000, 115 (2), 245–53. DOI 10.1046/j.1523-1747.2000.00029.x. [DOI] [PubMed] [Google Scholar]
- 17.Shouval DS; Biswas A; Goettel JA; McCann K; Conaway E; Redhu NS; Mascanfroni ID; Al Adham Z; Lavoie S; Ibourk M, Immunity 2014, 40 (5), 706–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wynn TA; Vannella KM, Immunity 2016, 44 (3), 450–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cha W; Fan R; Miao Y; Zhou Y; Qin C; Shan X; Wan X; Cui T, MedChemComm 2018, 9 (3), 396–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banerjee J; Sen CK, microRNA and wound healing In microRNA: Medical Evidence, Springer: 2015; pp 291–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lam JK; Chow MY; Zhang Y; Leung SW, Mol Ther Nucleic Acids 2015, 4, e252 DOI 10.1038/mtna.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aunin E; Broadley D; Ahmed MI; Mardaryev AN; Botchkareva NV, Scientific reports 2017, 7 (1), 3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li D; Li X; Wang A; Meisgen F; Pivarcsi A; Sonkoly E; Ståhle M; Landén NX, Journal of Investigative Dermatology 2015, 135 (6), 1676–1685. [DOI] [PubMed] [Google Scholar]
- 24.Zhuang G; Meng C; Guo X; Cheruku PS; Shi L; Xu H; Li H; Wang G; Evans AR; Safe S, Circulation 2012, CIRCULATIONAHA. 111.087817. [DOI] [PubMed] [Google Scholar]
- 25.Johnnidis JB; Harris MH; Wheeler RT; Stehling-Sun S; Lam MH; Kirak O; Brummelkamp TR; Fleming MD; Camargo FD, Nature 2008, 451 (7182), 1125. [DOI] [PubMed] [Google Scholar]
- 26.Gentner B; Schira G; Giustacchini A; Amendola M; Brown BD; Ponzoni M; Naldini L, Nature methods 2008, 6 (1), 63. [DOI] [PubMed] [Google Scholar]
- 27.Haneklaus M; Gerlic M; Kurowska-Stolarska M; Rainey A-A; Pich D; McInnes IB; Hammerschmidt W; O’Neill LA; Masters SL, The Journal of Immunology 2012, 1200312. [DOI] [PubMed] [Google Scholar]
- 28.Yuan JY; Wang F; Yu J; Yang GH; Liu XL; Zhang JW, Journal of cellular and molecular medicine 2009, 13 (11‐12), 4551–4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu TX; Lim E-J; Besse JA; Itskovich S; Plassard AJ; Fulkerson PC; Aronow BJ; Rothenberg ME, The Journal of Immunology 2013, 1202897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haneklaus M; Gerlic M; O’Neill LA; Masters S, Journal of internal medicine 2013, 274 (3), 215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ng VW; Chan JM; Sardon H; Ono RJ; Garcia JM; Yang YY; Hedrick JL, Adv Drug Deliv Rev 2014, 78, 46–62. DOI 10.1016/j.addr.2014.10.028. [DOI] [PubMed] [Google Scholar]
- 32.Veiga AS; Schneider JP, Biopolymers 2013, 100 (6), 637–44. DOI 10.1002/bip.22412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uehara M; Li X; Sheikhi A; Zandi N; Walker B; Saleh B; Banouni N; Jiang L; Ordikhani F; Dai L, Scientific reports 2019, 9 (1), 6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao Y; Reis LA; Feric N; Knee EJ; Gu J; Cao S; Laschinger C; Londono C; Antolovich J; McGuigan AP, Proceedings of the National Academy of Sciences 2016, 113 (40), E5792–E5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Annabi N; Rana D; Shirzaei Sani E; Portillo-Lara R; Gifford JL; Fares MM; Mithieux SM; Weiss AS, Biomaterials 2017, 139, 229–243. DOI 10.1016/j.biomaterials.2017.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jang H-J; Kim Y.-m.; Yoo B-Y; Seo Y-K, Journal of biomaterials applications 2018, 32 (6), 716–724. [DOI] [PubMed] [Google Scholar]
- 37.Noshadi I; Hong S; Sullivan KE; Shirzaei Sani E; Portillo-Lara R; Tamayol A; Shin SR; Gao AE; Stoppel WL; Black LD III; Khademhosseini A; Annabi N, Biomater Sci 2017, 5 (10), 2093–2105. DOI 10.1039/c7bm00110j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ben-Shushan D; Markovsky E; Gibori H; Tiram G; Scomparin A; Satchi-Fainaro R, Drug delivery and translational research 2014, 4 (1), 38–49. [DOI] [PubMed] [Google Scholar]
- 39.Aldawsari HM; Dhaliwal HK; Aljaeid BM; Alhakamy NA; Banjar ZM; Amiji MM, Molecular pharmaceutics 2018. [DOI] [PubMed] [Google Scholar]
- 40.Ganesh S; Iyer AK; Morrissey DV; Amiji MM, Biomaterials 2013, 34 (13), 3489–502. DOI 10.1016/j.biomaterials.2013.01.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frenkel JS, International wound journal 2014, 11 (2), 159–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Platt VM; Szoka FC Jr, Molecular pharmaceutics 2008, 5 (4), 474–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tran T-H; Rastogi R; Shelke J; Amiji MM, Scientific reports 2015, 5, 16632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stefanello TF; Szarpak-Jankowska A; Appaix F; Louage B; Hamard L; De Geest BG; van der Sanden B; Nakamura CV; Auzély-Velty R, Acta biomaterialia 2014, 10 (11), 4750–4758. [DOI] [PubMed] [Google Scholar]
- 45.Wang X; Ge J; Tredget EE; Wu Y, Nature protocols 2013, 8 (2), 302. [DOI] [PubMed] [Google Scholar]
- 46.Dunn L; Prosser HC; Tan JT; Vanags LZ; Ng MK; Bursill CA, JoVE (Journal of Visualized Experiments) 2013, (75), e50265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Assmann A; Vegh A; Ghasemi-Rad M; Bagherifard S; Cheng G; Sani ES; Ruiz-Esparza GU; Noshadi I; Lassaletta AD; Gangadharan S; Tamayol A; Khademhosseini A; Annabi N, Biomaterials 2017, 140, 115–127. DOI 10.1016/j.biomaterials.2017.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Han S-E; Kang H; Shim GY; Kim SJ; Choi H-G; Kim J; Hahn SK; Oh Y-K, Journal of drug targeting 2009, 17 (2), 123–132. [DOI] [PubMed] [Google Scholar]
- 49.Akinc A; Thomas M; Klibanov AM; Langer R, The Journal of Gene Medicine: A cross‐disciplinary journal for research on the science of gene transfer and its clinical applications 2005, 7 (5), 657–663. [DOI] [PubMed] [Google Scholar]
- 50.Li C; Guan G; Reif R; Huang Z; Wang RK, Journal of The Royal Society Interface 2011, 9 (70), 831–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang C; Xu L; Zhou Y; Zhang X; Huang X; Wang M; Han Y; Zhai M; Wei S; Li J, Carbohydrate Polymers 2010, 82 (4), 1297–1305. [Google Scholar]
- 52.Nwomeh BC; Liang HX; Cohen IK; Yager DR, J Surg Res 1999, 81 (2), 189–95. DOI 10.1006/jsre.1998.5495. [DOI] [PubMed] [Google Scholar]
- 53.Parayath NN; Parikh A; Amiji MM, Nano letters 2018, 18 (6), 3571–3579. [DOI] [PubMed] [Google Scholar]
- 54.Chen C-Z; Li L; Lodish HF; Bartel DP, science 2004, 303 (5654), 83–86. [DOI] [PubMed] [Google Scholar]
- 55.Mantovani A; Sozzani S; Locati M; Allavena P; Sica A, Trends in immunology 2002, 23 (11), 549–555. [DOI] [PubMed] [Google Scholar]
- 56.Italiani P; Boraschi D, Frontiers in immunology 2014, 5, 514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jetten N; Verbruggen S; Gijbels MJ; Post MJ; De Winther MP; Donners MM, Angiogenesis 2014, 17 (1), 109–118. [DOI] [PubMed] [Google Scholar]
- 58.Sani ES; Kheirkhah A; Rana D; Sun Z; Foulsham W; Sheikhi A; Khademhosseini A; Dana R; Annabi N, Science Advances 2019, 5, eaav1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tran T-H; Krishnan S; Amiji MM, PloS one 2016, 11 (5), e0152024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yao Z; Huang Y; Luo G; Wu J; He W, Burns & trauma 2014, 2 (4), 196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weidner N; Semple JP; Welch WR; Folkman J, N Engl J Med 1991, 324 (1), 1–8. DOI 10.1056/NEJM199101033240101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. 1H-NMR analysis of GelMA pre-polymer and gelatin.
Figure S2. Mechanical characterization and 1H-NMR analysis of GelMA hydrogels formed at varying photo-crosslinking time.
Figure S3. Synthesis and characterization of HA-PEI/HA-PEG nanoparticles.
Figure S4. SEM images of pristine GelMA and GelMA/NP/miRNA.
Figure S5. Enzymatic degradation of hydrogels.
Figure S6. In vitro 3D encapsulation of NIH3T3 fibroblasts in NP laden GelMA.
Figure S7. In vitro cytocompatibility of GelMA/NP hydrogels using J774A.1 macrophages.
Figure S8. In vitro evaluation of miRNA transfection in J77A4.1 macrophages incubated with 1.3 %(w/w) GelMA/NP/miR-223*.
Figure S9. Histological analysis of wounds on day 5 post-surgery.
Figure S10. Schematics of burst pressure, wound closure test, and shear stress measurement.
Table S1. List of primers used for qPCR.