Abstract
Ligand-receptor complexes play a central role in mediating a range of processes in immunology and cancer biology. The ability to directly quantify the fraction of receptors occupied by a ligand in a given biospecimen, as opposed to assessing the concentration of ligand and receptor separately, could provide an additional and valuable clinical and research tool for assessing whether receptors are occupied by a ligand. To address this need, a biomarker platform was developed to quantify the fraction of receptors occupied by a ligand using pairs of RNA aptamers, where one aptamer binds preferentially to the unoccupied receptor and the other to the ligand-receptor complex. Bound aptamer was quantified using RT-qPCR colorimetric probes specific for each aptamer. The binding ratio of aptamer correlated with the fraction of receptors occupied by a ligand. This assay, termed as LIRECAP (LIgand-RECeptor complex-binding APtamer) assay, was used to determine the fraction of soluble CD25 occupied by IL2 in the serum from subjects with B-cell lymphoma. No correlation was found between the type of lymphoma and total soluble CD25 or IL2 independently. In contrast, the fraction of soluble CD25 occupied by IL2 was significantly higher in follicular lymphoma patient serum compared to diffuse large B-cell lymphoma patient serum. We conclude that this technology has the potential to serve as a high-throughput biomarker platform to quantify the fraction of receptors occupied by a ligand.
Keywords: Soluble Interleukin 2 Receptor α (CD25), Ligand-Receptor complex, RNA aptamers, T regulatory cells, B-cell lymphoma
Introduction
Multi-molecular complexes, including ligand-receptor complexes and receptor multimers, play a central role in mediating a range of processes in cancer biology. They impact cancer cell growth, differentiation, and survival, as well as the interaction between cancer cells and the microenvironment, which includes antitumor immune responses. A prominent example is the regulation of the immune response by the interaction between IL2 with its alpha receptor subunit, IL2Rα (CD25), which leads to recruitment of additional receptor subunits (IL2Rβ and γ) to mediate activation signals in lymphocytes (1). Numerous studies have illustrated the importance of IL2-CD25 complexes on the immune response in a variety of diseases, including cancer (2–7). Agents that block such interactions have been explored as antitumor immune therapeutics (8). Measurement of the receptors or the ligands involved in these interactions, e.g. IL2 or CD25, has been used as a general measure of immune cell activation (1,9–14). In the IL2-CD25 system, as well as other systems involving ligand-receptor interactions, most assays probe and measure the ligand and receptor levels separately. Determining the fraction of receptors occupied by ligand using these assays is indirect and complex. The ability to directly quantify the fraction of receptors occupied by a ligand, as opposed to assessing the concentration of ligand and receptor separately, could provide an additional and valuable tool for assessing whether receptors are occupied by a ligand in a variety of biological specimens.
Nucleic acid aptamers are short oligonucleotide molecules that recognize target antigens in a manner analogous to antibodies (15). The specificity of aptamers, including RNA aptamers, is based in large part on their nucleotide sequence that determines complementarity of secondary and tertiary structures against their targets (16). Forces like those seen with antibody-antigen interactions, including van der Waals, hydrogen bonding, and electrostatic forces, can stabilize aptamer-target interactions. In some cases, the affinity of RNA aptamers towards their targets can be similar to those of antibodies (17). RNA aptamers are most commonly generated by a process called SELEX (Systematic Evolution of Ligand by EXponential enrichment), which involves sequential enrichment of a diverse RNA library against a target (18,19). Through this iterative process, RNA aptamers that bind with high-affinity are selected (20). RNA aptamer selection is generally achieved using the native primary target and is not based on antigen processing and presentation (21). Thus, unique epitopes that would not be maintained during immunization, such as epitopes formed due to inter-molecular interactions, can be identified by aptamers, which gives them a potential advantage over antibodies. The nucleic acid nature of RNA aptamers allows them to be sequenced, synthesized, multiplied, modified, and quantified easily. Significant efforts are being made to improve the in vivo bioavailability and half-life of RNA aptamers, including PEGylation and ribose modifications, intended for use as therapeutics (22,23). Such modifications are not essential for use of RNA aptamers as in vitro diagnostic agents.
This study describes an RNA aptamer-based assay platform designated as the “LIgand-REceptor Complex-binding APtamers” or “LIRECAP” assay that allows for quantification of the fraction of receptors occupied by a ligand. The LIRECAP assay reported here was developed and used to measure the fraction of soluble CD25 occupied by IL2 in the serum of subjects with B-cell lymphoma. Although these studies focused on the IL2-CD25 ligand-receptor complex in lymphoma, the LIRECAP assay platform could be applied to measure a broad range of multi-molecular complexes that could serve as novel biomarkers for cancer progression or response to therapy.
Methods
Cells and reagents
The DNA template, primers, and fluorochrome-labeled DNA probes (Integrated DNA technologies) were chemically synthesized and described in detail in their respective sections. The template, primer, and probe sequences were resuspended in nuclease-free water at 100 μM concentration upon receipt. Peripheral blood mononuclear cells (PBMCs) were obtained from healthy donors (without any exclusion criteria) in the form of leukocyte retention system cones (LRS) from the DeGowin Blood Center, University of Iowa, according to the institutional Review Board guidelines and after obtaining informed consent from subjects. Primary human T cell subsets, such as CD4+ T cells, CD8+ T cells, and CD4+CD25+ regulatory T cells (Tregs), were immediately isolated from the PBMCs using commercially-available magnetic cell separation kits (Miltenyi Biotech). Untagged recombinant human soluble CD25 (Cat # 200–02RC) and IL2 (Cat # 200–02) were from Peprotech. His-tag binding Dynabeads (Cat # 10103D), Protein G-coated Dynabeads (Cat # 10003D), 6X His- and Fc-tagged recombinant human CD25 (Cat # 10165-H08H and 10165-H02H) were from Thermo Fisher. TaqMan™ Universal PCR, SYBR Green PCR Master Mixes, Superscript™ III RT, and Platinum Taq DNA polymerase were obtained from Thermo Fisher or Promega. Buffers, common chemicals and salts, including HEPES, sodium chloride, and magnesium chloride, were obtained from Sigma Aldrich and Fisher Scientific.
Human subjects and serum
Serum samples were obtained from subjects with B-cell follicular lymphoma (FL; n=4) and diffuse large B-cell lymphoma (DLBCL; n=6) after obtaining informed consent according to the guidelines from the Declaration of Helsinki and Institutional Review Board. Serum was stored frozen at –80°C until use. CD25 and IL2 levels in these samples have been reported previously (14) (Supplementary Table S1).
Generation of CD25-binding RNA aptamers using Treg cell-based SELEX
CD25-binding RNA aptamers were generated using cell-based SELEX (summarized in Figure 1A and Supplementary Table S2)(24). Briefly, the initial RNA library was transcribed from DNA template using a recombinant Y639F mutant T7 RNA polymerase, as described previously (24), and 2′-fluoro-modified pyrimidines (TriLink Biotechnologies). The RNA library was pre-cleared by incubating with primary human CD4+CD25neg Teff cells (to remove aptamers that bound to common T-cell antigens) followed by binding with primary human Tregs (CD4+CD25high T cells) from the same donor. After washing the Tregs thrice with pre-warmed AIM V™ medium with AlbuMax™ supplement [AIM-V (BSA) medium] (Thermo Fisher), the Treg-bound aptamer fraction was extracted using TRIzol, amplified by RT-PCR using 1 μM primers (Supplementary Table S2), from which the RNA pool for the next SELEX round was transcribed. SELEX was repeated for eight rounds, with each round using T cells from a different donor to ensure that the aptamers recognized antigens across diverse subjects.
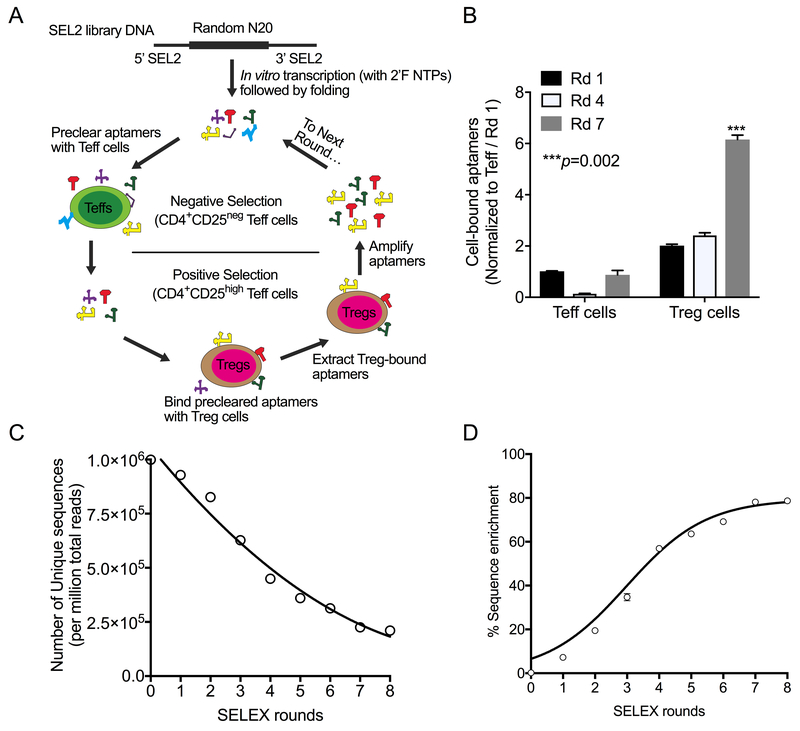
Figure 1: Selection and identification of Treg-binding RNA aptamers.
(A) Treg-binding RNA aptamers were selected using a cell-based SELEX approach starting with a SEL2 generation-based library (with a 20-mer Random region). Each round of SELEX consisted of a negative selection step with CD4+CD25− Teff cells to preclear aptamers that bound to common T-cell antigens and a positive selection step with CD4+CD25high Tregs to select for Treg-binding aptamers. Treg-bound aptamers were extracted, amplified by RT-PCR, transcribed, and used for the next round of SELEX. A total of eight rounds of SELEX was done, with each round involving cells from a different normal donor. (B) Evaluation of binding to CD4+CD25− Teff cells and CD4+CD25high Tregs at the 3 rounds (Rd) indicated. A representative plot of two independent sets is shown (Mean±SEM). P values shown were calculated using the unpaired Student’s t-test. (C) High-throughput sequencing of enriched aptamers from each round of SELEX showing the number of unique sequences (per million total reads). (D) Percent of sequence enrichment during each round was calculated as 100 – [(Unique sequences/Total reads) * 100]. The data shows the linear enrichment of Treg-binding sequences from Rds 1 through 7.
High-throughput sequencing and bioinformatics analysis
DNA from each round’s RNA aptamer pool was sequenced by Illumina-based high-throughput sequencing (Hiseq paired-end sequencing, 100 cycles) using primers containing unique barcodes to amplify the pool and each amplicon. A total of 77.1 million sequences were analyzed using a standardized bioinformatics workflow to determine aptamer enrichment efficiency, cluster size, and copy numbers (23). The percentage of sequence enrichment during each round was calculated as 100 – [(Unique sequences/Total reads) * 100]. Aptamer sequences were grouped into sequence (edit and tree distance) and structure families (RNA structure). Individual sequences were ranked based on the copy numbers, fold-enrichment, and read count in the final round of SELEX. A sequence was selected for further binding analysis if it was enriched to a higher total copy number across all rounds, showed a progressive enrichment, based on log2 fold changes across all or majority of SELEX rounds, and represented a unique structure and sequence family.
Aptamer binding assays
Cell-based aptamer binding assays
Aptamer binding to CD4+ T cell subsets was measured at 37°C. Briefly, T-cell subsets were blocked with 0.1 mg/mL transfer (t)RNA in AIM-V (BSA) medium for 15 minutes and incubated with folded aptamers (at 100 nM for 30 minutes). For identifying aptamers recognizing the IL2-CD25 complex, Treg cells were treated with 2 nM (equivalent to 300 IU/mL) IL2 (Peprotech) or with PBS for 30 minutes before incubating with aptamers. Cells were washed thrice with AIM-V (BSA) medium, and total RNA was extracted with TRIzol spiked with a reference control aptamer (M12–23) (TriLink Biotechnologies) to control for variations in RNA extraction (24,25). Cellular RNA was digested with RNase A. Aptamers were precipitated with ice-cold absolute ethanol, resuspended in 50 μL of water and quantified by standard SYBR green-based RT-qPCR assay (Promega). A typical qPCR reaction was performed with 2 μL of precipitated RNA and standard SEL2 primers (Supplementary Table S2). Bound aptamers were quantified using the standard curve method and the data for each treatment was first normalized to the respective reference M12–23 control aptamer, and then normalized to the control (C-248) aptamer, a sequence that was lost in SELEX Rd1 implying that it did not bind to either of the T-cell subsets used for selecting aptamers.
Bead-based aptamer binding assays
One μL of His-tagged binding Dynabeads coated with CD25 protein (Thermo Fisher) were blocked with binding buffer (HEPES-buffered saline, pH 7.5 with 2 mM CaCl2) containing BSA (0.1 mg/mL) and tRNA (0.1 mg/mL). For creating IL2-CD25 complexes, equimolar amounts of IL2 (Peprotech) was incubated with CD25-coated beads for 30 minutes at 37°C, and the unbound IL2 washed away with the binding buffer. Target-coated beads were incubated with aptamers at 100 nM for 30 minutes at 37°C, and washed thrice with pre-warmed binding buffer. Target-bound aptamers were extracted using Trizol and quantified using SyBR green-based qPCR method, as described above. Aptamer binding was normalized to M12–23 reference control. Binding of Treg aptamers was next normalized to the negative control aptamer (Supplementary Table S3).
Biolayer interferometry
Biolayer interferometry was used to measure the binding kinetics between the aptamer and target (Octet Rded96, ForteBio). Briefly, sensors loaded with synthetic biotinylated aptamers (TriLink) were dipped into Fc-tagged CD25 or IL2-CD25 complexes serially diluted two-fold in binding buffer at 30°C and analyzed using the Data Analysis software v10.0, as per the manufacturer’s recommendations (26)(Supplementary Table S4).
Enzyme-linked immunosorbent assay (ELISA) for measuring EC50 of IL2-CD25 binding
The effect of aptamers on the CD25 and IL2 interaction was determined by an ELISA-based assay. Briefly, plates coated with His-tagged CD25 (at 1mg/mL in carbonate-bicarbonate buffer, pH 9.6 overnight) were blocked with binding buffer containing 3% BSA and 0.1 mg/mL tRNA, washed in wash buffer (Binding buffer containing 0.01% Tween-20) thrice and incubated with 400 nM folded synthetic aptamers for 15 minutes at 37°C. Serially diluted human recombinant IL2 was added to the wells containing 100 nM aptamers and incubated for 30 minutes at 37°C. Plates were washed thrice and incubated with 1:2000 diluted anti-human IL2 polyclonal IgG (R&D Systems). Plates were washed and developed using HRP-conjugated anti-goat IgG antibody (Southern Biotech), incubated with 3,3’,5,5’-tetramethylbenzidine+H2O2 substrate (eBiosciences) and read at 450 nm in a 96-well plate reader (Molecular Devices) with the background subtraction at 650 nm. Absorbance values were used to calculate the percent ligand binding using the Hills equation (GraphPad Prism v8.0.1).
Measurement of IL10 from cell culture supernatants:
To determine whether the fractional occupancy of IL2-CD25 complexes correlated with IL10 secretion, primary human CD4+ T cells were cultured with IL2-CD25 complex-coated Dynabeads created at different fractional occupancies (89% to 0%) using His-CD25-coated Dynabeads (as above) and 0 nM to 160 nM of IL2. Cells were cultured in a 96-well cell culture plate (Corning) for 3 days at 37°C. Fifty μL of cell culture supernatant per well was collected, and IL10 was immediately measured using the IL10 Human ELISA kit following the manufacturer-recommended protocol (Thermo Fisher).
Flow cytometry
The effect of aptamers on IL2 signaling and the biological activity of IL2-CD25 complexes were determined by flow cytometry (14). To measure the effect of aptamers on IL2-induced phospho-STAT5 signaling, Tregs were incubated with 250 nM IL2 in the presence or absence of 100 nM aptamers at 37°C for 30 minutes. After washing the cells with 1 mL of PBS containing BSA (5 mg/mL), cells were fixed with 1X Foxp3 Fixation/Permeabilization buffer (eBiosciences), permeabilized with ice-cold methanol, and then stained for phosho-STAT5 (BD Biosciences). To determine the biological activity of IL2-CD25 complexes, primary human CD4+ T cells were cultured with IL2-CD25 complex-coated Dynabeads created at different fractional occupancies (89% to 0%) using His-CD25-coated Dynabeads, as above) and 0 nM to 160 nM of IL2. Cells were incubated for 30 minutes (for phospho-STAT5) or 3 days (for Treg induction and immunosuppression) at 37°C. Phospho-STAT5 was measured as described above. Treg induction was measured by staining the cells for CD4 (Clone SK3; BD Biosciences) and Foxp3 (Cline 236A/E7; eBiosciences) using the Foxp3/Transcription factor Staining Buffer set (eBiosciences). Immunosuppressive ability was determined by coculturing complex-treated CD4+ T cells with CFSE-labeled autologous CD8+ T cells (1:1 ratio) for 3 days. All samples were read in an LSR II flow cytometer (BD Biosciences) and analyzed using FlowJo® v10.2 (FlowJo®, LLC).
LIgand-REceptor Complex-binding APtamer (LIRECAP) assay to measure IL2 occupancy on CD25
A TaqMan RT-qPCR-based binding assay was developed using paired aptamers (one with enhanced binding to unoccupied CD25 and one with enhanced binding to the complex) to reflect the fraction of CD25 occupied by IL2. The primer and probe TaqMan RT-qPCR sequences are listed in Supplementary Table S5. Briefly, One μL (per reaction) of His-tag-binding Dynabeads coated with 25 nM human His-tagged CD25 were blocked with binding buffer containing BSA and tRNA (0.1 mg/mL each) and incubated with two-fold serially diluted IL2 concentrations (160 nM to 5 nM) to create various fractions of CD25 occupied by IL2. Fifty μL of supernatant containing unbound IL2 was removed and quantified using an IL2 ELISA kit (Thermo Fisher). Data was read with the 96-well plate ELISA reader (Molecular Devices) and the receptor occupancy was estimated, as described elsewhere (27). Dynabeads containing IL2-CD25 concentration at various receptor occupancy, as prepared above, was mixed with aptamers showing variable binding preferences for the unoccupied receptor or the ligand-receptor complex were mixed (100 nM of Tr-8 with Tr-1 or Tr-7). After washing, bound aptamers were extracted using Trizol, reverse transcribed with Superscript™ III RT and quantified with then TaqMan™ Universal qPCR master mix. Primers and probes used for the quantification are listed in Supplementary Table S5. The ratio of bound aptamers at each fractional occupancy of IL2 on CD25 was calculated by dividing the absolute quantity of bound Tr-1 or Tr-7 aptamer by the absolute quantity of bound Tr-8 aptamers. Standard curves were generated by plotting the binding ratio against the fraction of IL2-occupied CD25 (27).
LIRECAP assay for quantifying fractional occupancy of CD25 in serum from FL and DLBCL patients
Serum samples were immunoprecipitated with anti-CD25 (Clone M-A251; BD Biosciences)-coated Dynabeads. Protein-bound beads were washed and resuspended in binding buffer and incubated with equimolar concentrations (100 nM) of Tr-7 and Tr-8 at 40C for 45 minutes. Aptamer-bound beads were washed in binding buffer twice for 5 minutes at 40C, and bound aptamers were extracted and quantified, as mentioned under LIRECAP assay. Standard curves for quantifying fractional occupancy of CD25 in serum samples were prepared as outlined above. Bound aptamer ratios were determined, as described under LIRECAP assay and the fractional occupancy of CD25 by IL2 in each test specimen were derived by comparing the bound aptamer ratio from each sample to the standard curve.
DNA melt assay
The change in the complexity of aptamer pools was monitored after select rounds of enrichment using a DNA melt assay20. 20 pmol of PCR DNA generated from the Treg-bound RNA pool after SELEX (rounds 2, 4, 6, and 8) were individually mixed with equal amounts of 2X SYBR green PCR mix (Promega, USA). Samples were then subjected to a standard DNA melt curve analysis (95°C to 25°C, over 20 min ramp time). SYBR green fluorescence was plotted against the temperature, and the melt curve was generated. The complexity of nucleotide sequences in the aptamer library progressively decreased after each round of SELEX due to enrichment and increase in the copy number of Treg-specific sequences that results in a shift in the overall melting curve towards higher Tm (shift to right).
Statistical analysis
All experiments, unless specifically mentioned, were repeated at least twice to ensure reproducibility. Data points from multiple assays, done with similar conditions, were pooled and mean and standard error of mean (SEM) were calculated. Significance of the mean (p value) were analyzed using student t-test using the GraphPad Prism v8.0.1 and a p value of less than 0.05 was considered significant.
Results
Cell-based SELEX to select human T regulatory cell (Treg)–binding RNA aptamers
A Treg-based SELEX approach was designed to select RNA aptamers that bind to human Tregs (Figure 1A). The selection conditions used for each round of SELEX are described in the in Supplementary Table S2. The enrichment of the aptamer pool during SELEX was monitored using a DNA melt assay that inversely relates the melting temperature to the sequence complexity of the aptamer pool (24). As shown in Supplementary Figure S1A, sequential rounds of SELEX resulted in a rightward shift in the DNA melt curve, indicating a decrease in library sequence complexity and increased aptamer enrichment with advancing SELEX rounds.
The specificity of aptamer pools for Treg and Teff cells was assessed using a T cell–based binding assay. Aptamer pools from SELEX rounds 1, 4, and 7 were incubated with CD4+CD25high Tregs and CD4+CD25− Teff cells from the same donor, and aptamer binding was measured using SYBR green-based RT-qPCR. As illustrated in Figure 1B, a significantly increased binding of aptamer pools to Tregs was seen in Rd 7 compared to Rds 1 and 4, indicating enrichment of Treg-binding aptamers [p=0.002 (Rd 7 vs. Rd 1)]. These data indicated that the SELEX process enriched for RNA aptamers that bound preferentially to antigens expressed by Tregs relative to Teff cells.
High-throughput sequencing (HTS) to identify candidate Treg aptamers
An Illumina-based HTS platform was used to sequence aptamers from all rounds, including the starting aptamer library (Rd 0). HTS across the eight rounds of aptamer selection yielded 157.8 million reads representing 77.1 million different aptamer sequences, with an average of 3.9 million unique aptamers identified in each SELEX round. A steady decline in the number of unique aptamer sequences, indicative of enrichment of Treg-binding sequences (Figure 1C), was seen with early rounds of selection. Enrichment of 60% was seen by round four, with the degree of enrichment slowing at rounds five and six. A plateau was reached between rounds seven and eight (Figure 1D). This HTS data on library diversity was consistent with the data on Treg specificity observed in the T-cell RT-PCR-based binding, suggesting more than eight rounds of enrichment were unlikely to have additional benefit.
Aptamers with the potential to be specific for Tregs were identified as those sequences that increased in abundance (50 reads) and exhibited persistence (found within at least 4 rounds) above the unselected library (Supplementary Figures S1B and S1C). 4,457 unique aptamer sequences represented by 3.5 million reads met these criteria. To identify the most promising candidate aptamers, aptamer sequences were ranked by Log2 fold-enrichment and were assessed for a steady increase in abundance across sequential rounds of selection. Next, the aptamer sequences were clustered based on sequence similarity (edit distance) and structural similarity (tree distance) (24). Aptamers selected for the final panel had high copy numbers, demonstrated a stepwise increase in fold-enrichment over rounds of selection, and were from different sequence and structure families. The final panel consisting of eleven candidates were synthesized for further evaluation (Supplementary Table S6). An aptamer (C-248) that was only observed in the unselected starting aptamer library (Rd 0) and not in any other selection round was identified to serve as a non-selected control aptamer.
Aptamer specificity for Tregs and Teff cells
Synthesized aptamers were tested for their ability to bind to Tregs and Teff cells. Aptamers were incubated with enriched primary human Tregs and Teff cells, and bound aptamers were quantified using RT-qPCR. All selected aptamers bound in higher quantities to Tregs than to Teff cells (Figure 2A). The negative control aptamer (C-248) did not bind to either T-cell subset. These data confirmed all candidate aptamers bound more extensively to Tregs than to Teff cells.
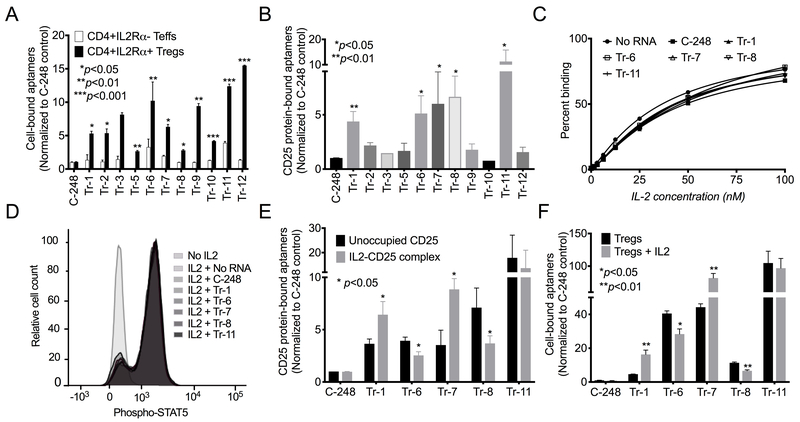
Figure 2: Select Treg-specific aptamers recognize human CD25 and demonstrate differential binding to unoccupied CD25 vs. IL2-CD25 ligand-receptor complexes.
(A) The top enriched synthesized aptamers were evaluated for their ability to bind to CD4+CD25high Tregs ((IL2Rα+) and CD4+CD25− Teff cells (IL2Rα−). Shown are representative data of two independent sets of experiments (Mean±SEM). (B) Binding of the twelve aptamers and C-248 control to recombinant human CD25. ((Mean±SEM); N=2). (C) An ELISA-based assay demonstrating the EC50 of the IL2-CD25 interaction (N=3). A representative plot from three independent sets of experiments is shown. (D) Flow cytometry of the phosphorylation of STAT5 (median fluorescent intensity) in human Tregs induced by IL2 and the influence of CD25 aptamers. A representative plot of three independent sets is shown. (E) Binding of aptamers to unoccupied CD25 and IL2-CD25 ligand-receptor complexes using CD25-coated beads incubated with and without IL2 (Mean±SEM; N=3). (F) Binding of aptamers to Tregs incubated with and without the addition of IL2. (Mean±SEM; N=2). All P values shown were calculated using the unpaired Student’s t-test.
Identification of aptamers that recognize CD25
Tregs used in the selection process were defined as CD4+CD25high cells (28), and thus, the lead aptamers were assessed for binding to CD25 protein. Five of the top eleven aptamers (Tr-1, Tr-6, Tr-7, Tr-8, and Tr-11) bound to recombinant human CD25-coated Dynabeads, demonstrating their target antigen was CD25 (Figure 2B, Supplementary Table S3).
Aptamers do not interfere with binding of IL2 to CD25 or IL2-induced signaling
Because IL2 is the natural ligand for CD25, CD25-binding aptamers were assessed for their ability to interfere with the binding of IL2 to CD25. Using an ELISA-based assay, the EC50 of IL2 binding to recombinant CD25 was assessed in the presence or absence of aptamers. As shown in Figure 2C, none of the CD25-binding aptamers significantly altered the ability of IL2 to bind CD25, irrespective of the order of the addition of aptamers (prior to or after addition of IL2 to CD25). Similar results were found in a cell-based system exploring the impact of aptamer on Treg signaling mediated by IL2, measured by tyrosine phosphorylation of STAT5 (14). Up to a 10-fold increase in phospho-STAT5 was observed 30 minutes after addition of IL2, as measured by flow cytometry. CD25-binding aptamers did not significantly alter phospho-STAT5 induced by IL2 in Tregs (Figure 2D). Thus, the CD25-binding aptamers did not alter IL2 binding or signaling through CD25.
Aptamers display differential preference for unoccupied receptor and complexes
Although the aptamers did not interfere with IL2 binding to CD25, IL2 did have an impact on the ability of some CD25-binding aptamers to recognize CD25. Pre-incubation of recombinant CD25-coated Dynabeads with IL2 resulted in significantly enhanced binding of some aptamers to CD25 (Tr-1 and Tr-7). In contrast, addition of IL2 significantly reduced binding of other aptamers to CD25 (Tr-6 and Tr-8)(Figure 2E). Results obtained with a Treg-based system were similar. IL2 treatment of Tregs led to significantly higher binding of Tr-1 and Tr-7 and lower binding of Tr-6 and Tr-8 (Figure 2F). Tr-11 showed similar binding to both receptor and ligand-receptor complexes (Figures 2E–F). Thus, some aptamers bound preferentially to the IL2-CD25 ligand-receptor complex, while others bound preferentially to unoccupied CD25.
Binding kinetics of aptamers confirm differential binding to CD25 vs. IL2-CD25 complex
The binding kinetics of the aptamers towards CD25 and the IL2-CD25 complex, including association and dissociation kinetics, were determined using biolayer interferometry. Kd, Kon and Kdis rates for aptamer binding are summarized in Table 1 and the sensorgram in Supplementary Figure S2. Tr-1 and Tr-7 that bound preferentially to the complex displayed a stronger affinity (lower Kd) for the IL2-CD25 complex than for the unoccupied CD25, whereas Tr-6 and Tr-8 that bound preferentially to unoccupied CD25 showed weaker affinity (higher Kd) for the IL2-CD25 complex than for unoccupied CD25. The Tr-11 aptamer that bound to the receptor and the complex equally displayed similar affinities for both (Table 1). Thus, differential affinity of aptamers for IL2-CD25 complex versus unoccupied CD25 was consistent with their differential binding preferences.
Table 1:
Kinetics of aptamer binding to CD25 and IL2-CD25 complex, measured using biolayer interferometry
| CD25 binding | IL2-CD25 complex | |||||
|---|---|---|---|---|---|---|
| Aptamer | Kd (nM) | Kon (x103/Ms) |
Kdis (x10−3/s) |
Kd (nM) | Kon (x103/Ms) |
Kdis (x10−3/s) |
| Tr-1 | 122.0 +/− 5.23 | 42.7 +/− 1.4 | 5.23 +/−0.14 | 67.8 +/− 2.5 | 60.2 +/− 1.5 | 4.0 +/− 0.11 |
| Tr-6 | 96.9 +/− 5.77 | 40 +/− 1.7 | 3.9 +/−0.17 | 144.0 +/− 7.6 | 26 +/− 1.1 | 3.8 +/− 0.11 |
| Tr-7 | 68.2 +/− 2.73 | 150 +/− 5.1 | 10 +/−0.22 | 45.0 +/− 1.5 | 120 +/− 2.9 | 5.3 +/− 0.13 |
| Tr-8 | 35.7 +/− 1.56 | 144 +/− 4.6 | 5.2 +/−0.16 | 60.7 +/− 2.9 | 50 +/− 1.4 | 3.0 +/− 1.2 |
| Tr-11 | 46.1 +/− 1.76 | 172 +/− 5.3 | 7.99 +/−0.17 | 48.4+/−2.3 | 97.5 +/− 2.9 | 4.0 +/− 0.14 |
Quantification of fractional occupancy using paired aptamers with differential binding
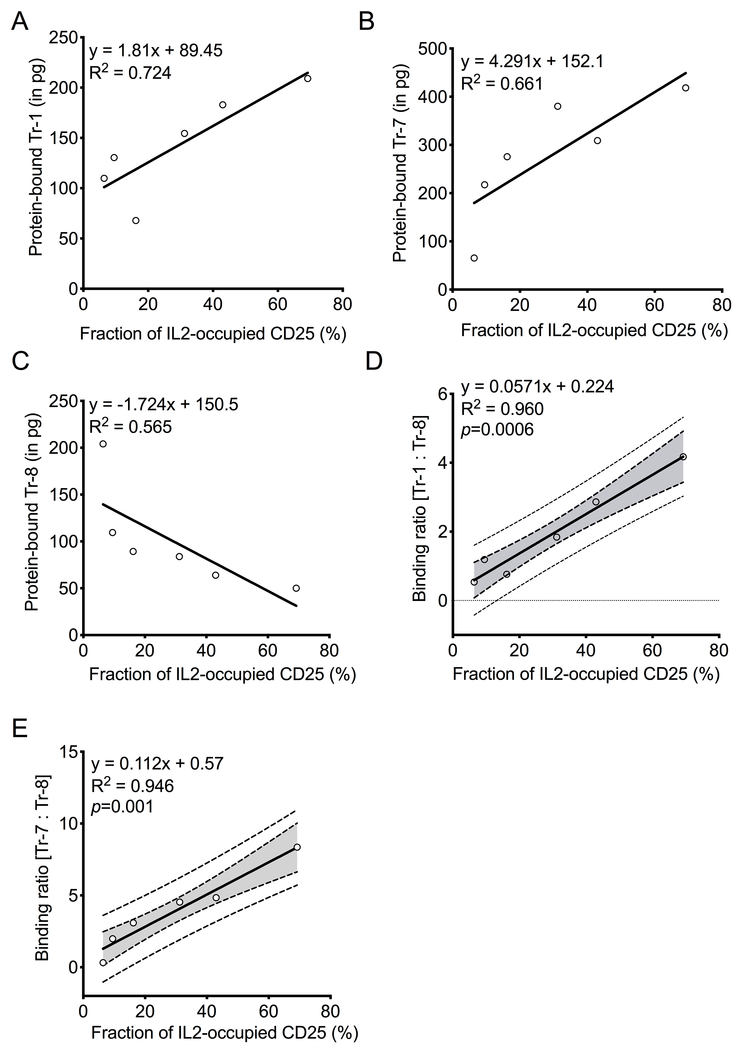
CD25-binding aptamers did not significantly alter IL2 binding to CD25, nor did they compete with each other for binding to CD25 (Supplementary Figure S3). Therefore, if an aptamer preferring IL2-CD25 complexes (e.g. Tr-1 or Tr-7) paired with an aptamer preferring unoccupied CD25 (e.g. Tr-8) are added together to a biospecimen predominantly containing the IL2-CD25 complex, more of the aptamer preferring the IL2-CD25 complex (Tr-1 or Tr-7) would be expected to bind. In contrast, if the biospecimen contained largely unoccupied CD25, more of the aptamer preferring the unoccupied receptor (e.g. Tr-8) should bind. Both aptamers can be amplified by the same primer set to determine their binding ratio. This provides a robust internal control. These characteristics form the basis for the LIRECAP assay concept that should allow for the measurement of the fraction of receptor (in this case CD25) occupied by ligand (in this case IL2) in a sample. To test this concept, samples with different fractional occupancies were created by incubating CD25-coated Dynabeads with various concentrations of IL2. Equimolar concentrations of a complex-preferring aptamer (e.g. Tr-1 or Tr-7) and an unoccupied receptor-preferring aptamer (e.g. Tr-8) were added to the samples, and bound aptamers were quantified using a TaqMan RT-qPCR (Supplementary Table S5). As shown in Figures 3A–B, binding levels of Tr-1 (R2=0.724) and Tr-7 (R2=0.661) correlated with increasing fractional occupancy of CD25. Conversely, Tr-8 displayed an inverse correlation with increasing fractional occupancy of CD25 (R2=0.565; Figure 3C). The ratio of bound Tr-1 or Tr-7 to bound Tr-8 was determined at each receptor occupancy level to create regression curves illustrating receptor occupancy. A significant linear relationship [R2=0.960; p=0.006 (Tr-1:Tr-8) and R2=0.946; p=0.001 (Tr-7:Tr-8)] was observed between aptamer binding ratios when plotted against the fraction of CD25 occupied by IL2 (Figures 3D–E).
Figure 3: Binding of aptamers correlates with IL2 occupancy of CD25.
Aptamer binding to His-tagged CD25-coated Dynabeads to determine (A) Tr-1, (B) Tr-7, and (C) Tr-8 binding to CD25 as its IL2 occupancy increases. This demonstrates binding to the IL2-CD25 complex (A-B) and to unoccupied CD25 (C). (D) The ration of Tr-1 to Tr-8 binding and (E) the ratio of Tr-7 to Tr-8 binding and their correlation with IL2 occupancy of CD25. A representative linear regression plot from four independent sets of experiment is shown. (A-C) Equation of the line, R2, and p values (where applicable) are shown. (D-E) Equation of the linear regression line (solid line), 95% confidence interval (dotted line with shaded area), and R2 are shown. P values for the slope of the regression line is shown.
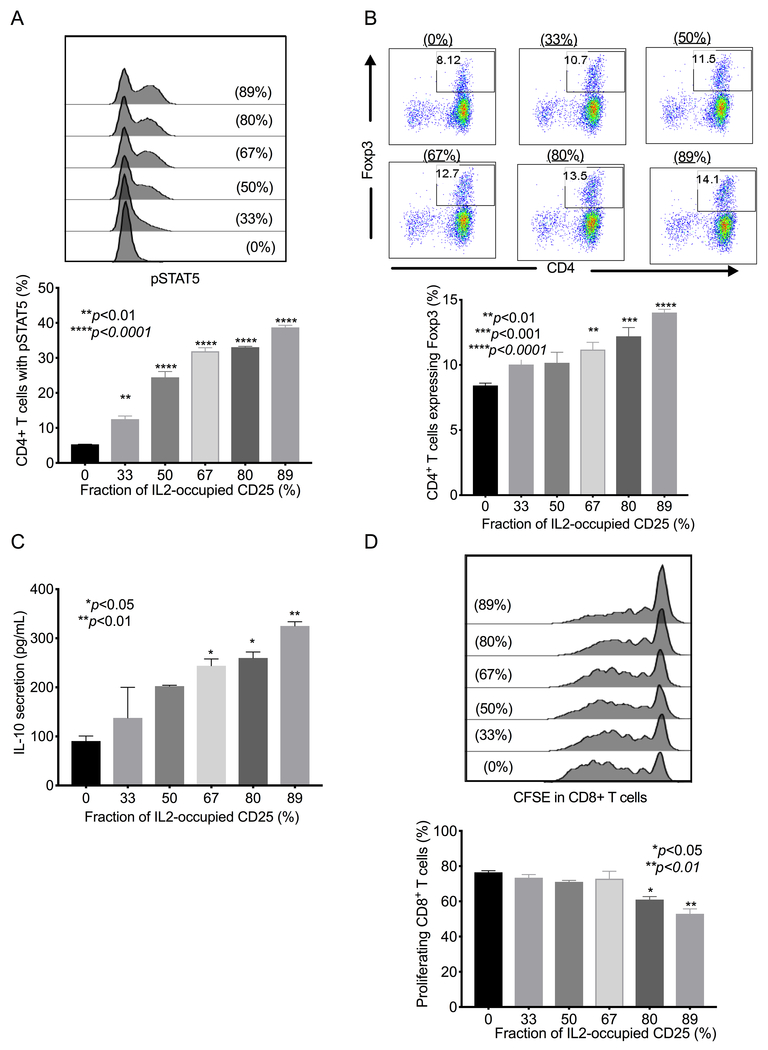
IL2 bound to CD25 is known to transactivate IL2Rβγ subunits expressed on CD4+ T cells, increase phospho-STAT5, and induce an immunosuppressive phenotype (14). To confirm that data obtained using the LIRECAP assay reflects the distinct biologic activity of the IL2-CD25 complex versus unoccupied soluble CD25, we measured the activity of various IL2-CD25 occupancies. Phospho-STAT5 signaling and Treg phenotype were determined in primary human CD4+ T cells treated with Dynabeads-coated with IL2-CD25 complexes, representing different fractional occupancies. As shown in Figure 4, increasing IL2 occupancy of CD25 resulted in a dose-dependent increase in the phospho-STAT5 (Figure 4A) and induction of the Foxp3+ Treg phenotype (Figure 4B). Complex-induced Tregs also showed a dose-dependent increase in their immunosuppressive activity, as evidenced by increased IL10 secretion (Figure 4C) and enhanced ability to inhibit the proliferation in cocultured autologous CD8+ T cells (Figure 4D).
Figure 4: Complex-induced immunosuppressive activity in CD4+ T cells correlates with IL2 occupancy of CD25.
The biological effect of IL2-CD25 complex with varying occupancies was determined. Variable fractional occupancy of CD25 by IL2 (0% to 89% was created by mixing Dynabeads coated with recombinant His-tagged CD25 and various concentrations of IL2 (160 nM to 0 nM). Beads containing various concentrations of unoccupied and IL2-occupied CD25 were added to freshly isolated primary human CD4+ T cells. (A) Flow cytometry showing the percentage of CD4+ T cells’ STAT5 phosphorylation in response to IL2-CD25 complexes compared to un-complexed CD25. A representative histogram of two independent experiments is shown. (B) Flow cytometry plots of Foxp3+ cells (Tregs) induced with increasing IL2 occupancy of CD25. A representative dot blot from two independent experiments is shown. Percentage of CD4+ T cells expressing FoxP3 is indicated. (C) ELISA of IL10 production in conditioned media with increasing IL2 occupancy of CD25. Mean±SEM of two independent experiments is shown. (D) Flow cytometry plot of CD8+ T-cell proliferation, as determined by CFSE, with increasing IL2 occupancy of CD25, measured by analyzing the ability of induced Tregs to inhibit the proliferation of cocultured CD8+ T cells. A representative histogram of two independent experiments is shown. All P values shown were calculated using the unpaired Student’s t-test.
Circulating IL2-CD25 ligand-receptor complexes in serum of subjects with lymphoma
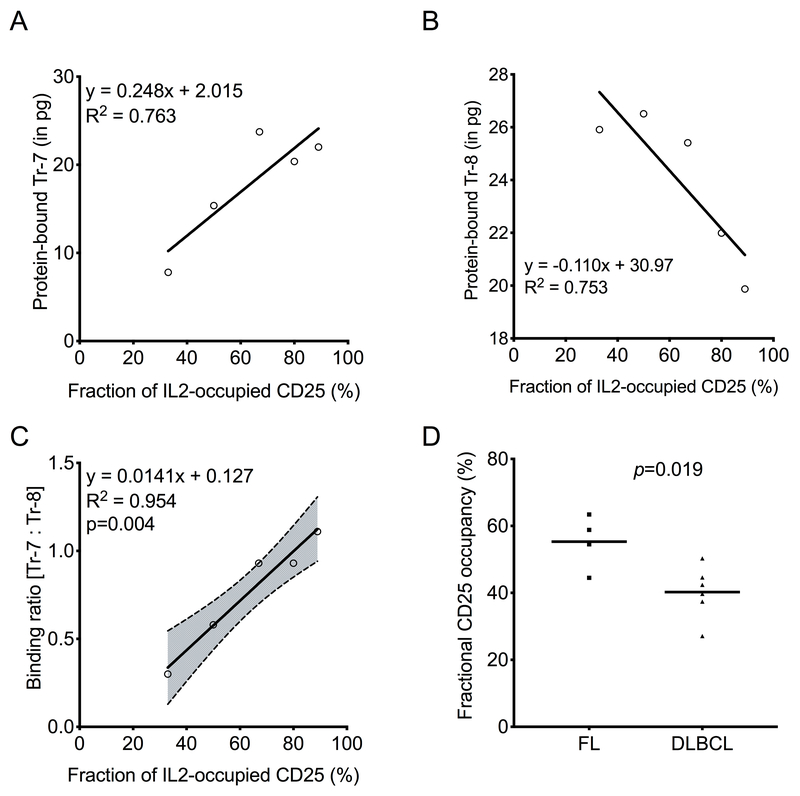
Serum from lymphoma patients contains significantly higher concentrations of soluble CD25 compared to normal individuals (14). It is not known to what extent the soluble CD25 in serum of lymphoma patients is complexed with IL2. The IL2-CD25 LIRECAP assay was used to assess the fraction of soluble CD25 occupied by IL2 in frozen serum samples from lymphoma subjects with known levels of soluble CD25 and IL2 (14). Soluble CD25 was immunoprecipitated from serum using Dynabeads coated with anti-human CD25, which neither interfered with the IL2-CD25 complex (29,30) nor competed with the aptamer binding to CD25 (Supplementary Figure S4). Dynabeads containing CD25 immunoprecipitated from the biospecimens, and the formed standards containing known fractional occupancies of IL2-occupied CD25 were evaluated using the LIRECAP assay. Results of standard curves in serum were similar to those seen in buffer, demonstrating that the LIRECAP assay can be conducted in serum effectively. As shown in Figure 5, binding of the Tr-7 aptamer showed a positive correlation with increasing IL2-CD25 complexes in standard samples (R2=0.763; Figure 5A), whereas binding of Tr-8 showed an inverse correlation (R2=0.753; Figure 5B) with increasing IL2-CD25 complexes. The ratio of Tr-7 to Tr-8 showed a linear relationship with the calculated fraction of IL2-occupied CD25 (R2=0.954, p=0.004; Figure 5C). Using this standard curve, the fraction of soluble CD25 occupied by IL2 in the clinical test samples was determined. Variability in the fractional occupancy from sample to sample was observed (Supplementary Table S1). On average, approximately half of the soluble CD25 was occupied by IL2 in lymphoma patient serum (Mean +/−SD = 46.3% +/− 10.8%; n=10). The fraction of CD25 occupied by IL2 did not correlate with either the total soluble CD25 (R2=0.127, p=0.311) or total IL2 (R2=0.111; p=0.347) (Supplementary Figure S5A–B). However, a significantly higher fraction of soluble CD25 was occupied by IL2 in serum from FL patients compared to serum from DLBCL patients (p=0.019; Figure 5D). Total CD25 and IL2 levels, measured using Luminex assays as described previously (14), did not correlate with lymphoma type (p=0.256 and 0.294, respectively) (Supplementary Figure S5C–D).
Figure 5: Soluble CD25 occupancy by IL2 is greater in serum from subjects with FL than with DLBCL.
LIRECAP assay was performed on serum samples from lymphoma subjects (FL n=4; DLBCL n=6). A standard curve was produced by adding soluble CD25 and various concentrations of IL2 to normal human serum. Fractional occupancy of CD25 waswas analyzed using LIRECAP assay, as described in the Methods section. Correlation between the fractional occupancy of standards and aptamer binding was determined by linear regression. (A) Correlation of Tr-7, and (B) Tr-8 binding as IL2 occupancy of soluble CD25 increased. (C) The ratio of Tr-7 to Tr-8 binding and correlation with IL2 occupancy of soluble CD25. Equation of the linear regression line (solid line), 95% confidence interval (dotted line with shaded area) and R2 are shown. P values for the slope of the regression line is shown. (D) Comparison of IL2 occupancy of soluble CD25 in FL and DLBCL patient serum. P values shown were calculated using the unpaired Student’s t-test.
Discussion
Immunologic, diagnostic, and pharmacologic agents, including radiolabeled ligands, monoclonal antibodies, small molecules, and RNA aptamers, are used to study receptors and ligands in a broad range of scientific, diagnostic, and therapeutic applications in cancer and beyond. Technologies, such as mass spectrometry, FRET, BIAcore, etc., are used to assess ligand-receptor interactions, kinetics, and biology (31,32). In general, such technologies rely on the recognition of either the receptor or the ligand. Each of these technologies has an important role in cancer research, yet each has its limitations, such as lack of specificity, complexity, and cost. This limits their value when it comes to their high-throughput use as biomarkers. Here, we described a technology platform, designated as LIRECAP, for determining the fraction of receptors occupied by a given ligand. This assay is straight-forward, requires no specialized equipment, and has the potential to be applied in a high-throughput manner, thereby, leading to evaluation of novel biomarkers for cancer research and eventually translated to standard of care.
The reagents at the core of this platform were RNA aptamers with differential affinity to ligand-occupied vs. unoccupied receptors identified using a whole cell SELEX approach. It was not the initial intent of these studies to enrich for aptamers displaying such differential affinity. However, given the use of normal donor PBMCs as targets, it was not surprising in retrospect that some of the CD25 molecules expressed by the Tregs used in the selection process were occupied by IL2, whereas other CD25 molecules were unoccupied. This led to selection of some RNA aptamers that displayed stronger affinity and binding to complexes and some to unoccupied receptor. Once it was determined that RNA aptamers had been identified that bind to the same target, but have binding that is differentially influenced by the presence of ligand, it occurred to us that such ligand-receptor complex aptamers could be used for precise quantification of the fraction of receptors occupied by ligand.
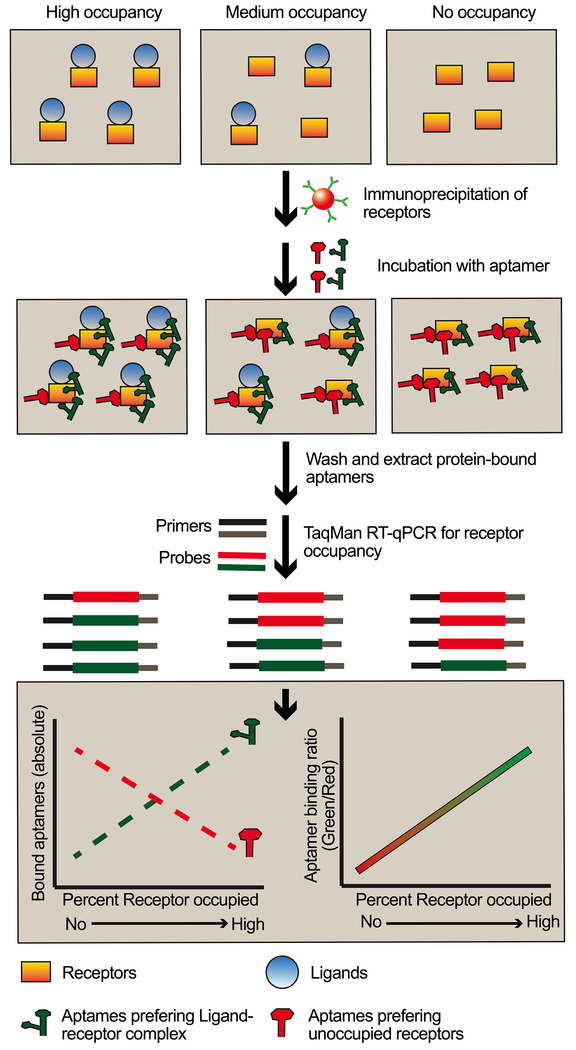
The LIRECAP assay involves an aptamer pair composed of one aptamer that binds preferentially to the complex (here, IL2-CD25) and the second that binds preferentially to unoccupied receptor (here, CD25). When they are added together to a biospecimen, the ratio of aptamer binding reflects the fraction of receptor occupied by ligand (Figure 6). In samples containing unoccupied receptors, the aptamer that preferentially binds to the unoccupied receptor should be predominant, whereas in samples with higher ligand-receptor complexes, aptamers that preferentially binds to the complex should be predominant. The aptamers that bind preferentially to unoccupied receptors and the aptamers that bind preferentially to complexes are of the same length and can be expanded using the same primers. Expansion should, therefore, be proportional. A standard curve linking the aptamer ratio to receptor occupancy can then be established based on values of the aptamer ratio obtained from samples with known receptor occupancy. The preference of aptamers for unoccupied receptor or complex does not need to be absolute, only relative, because adjustment for background binding results from using the ratio of two aptamers and a standard curve.
Figure 6: Schema demonstrating use of LIRECAP assay to estimate fraction of receptors occupied by ligand.
Differential binding of aptamers to unoccupied versus ligand-occupied receptor can be used to determine the fraction of receptor occupancy in biospecimens. After preparation of cells or immunoprecipitation of soluble receptors, aptamer pairs consisting of equimolar mix of aptamers preferring the complex and aptamers preferring the unoccupied receptors are incubated with samples with unknown levels of receptor occupancy. Aptamers preferring the complex bind to a greater degree in samples with higher receptor occupancy by ligand. Aptamers preferring the unoccupied receptors bind to a greater degree in samples with lower receptor occupancy by ligand. Aptamer levels are then quantified by probe-based RT-qPCR. The ratio of binding of aptamers to each sample is determined and compared to a standard curve to determine the percent of receptors occupied by ligand in the sample.
With respect to potential clinical applicability, soluble CD25 has been shown to contribute to the pathophysiology of several tumors, including B-cell lymphomas, and can predict the survival in lymphoma patients (14,33–35). The source of elevated soluble CD25 in the circulation of cancer patients is not fully understood, although it could arise from tumor-infiltrating T cells (including Tregs and activated Teff cells) through cleavage by macrophage-associated metalloproteinases, such as MMP9 (35). The tumor microenvironment in FL displays increased Treg infiltration compared to DLBCL tumors (36). Our observation that the IL2-CD25 complexes were higher in the sera of follicular lymphoma patients compared to the sera of DLBCL patients aligned with the hypothesis that much of the IL2-CD25 complexes in the sera of lymphoma patients was shed by Tregs. However, evaluation of this hypothesis will require further investigation. Irrespective of its source, soluble CD25 can suppress antitumor immune responses by promoting the induction of Foxp3+ Tregs in the tumor microenvironment (14,33,34), particularly if it is complexed with IL2. If soluble CD25 exists as IL2-bound complexes in the circulation, it could provide strong transactivation signals through the intermediate affinity IL2Rs (IL2Rβγ heterodimers) expressed on tumor-infiltrating Teff cells and promote Treg conversion in the lymphoma microenvironment (26). Thus, the fraction of CD25 occupied by IL2 could serve as a powerful biomarker that reflects on suppression of the antitumor response. Further studies are ongoing to assess whether the fraction of soluble CD25 occupied by IL2 is a predictor of outcome in FL lymphoma patients.
The studies reported here were conducted using recombinant CD25 and IL2 as a model, and lymphoma as a tumor type. However a similar approach could be applied to other ligand-receptor systems that may be relevant in cancer biology and cancer therapy. Although our current efforts are focused on evaluating the LIRECAP assay to measure occupancy of soluble CD25 in serum, a similar assay could be tailored to quantify the percent of membrane CD25 occupied by IL2 on circulating T cells and those infiltrating solid tissues. With further refinement, this approach might be used to visualize receptor occupancy using in situ PCR. It also could be used to assess molecular complexes beyond cytokine-receptor interactions. For example, this technology could be applied to determine the percent of PD-1 molecules occupied by PD-L1 or PD-L2, or to differentiate receptor homodimers from heterodimers, such as the EGFR family of receptors. Using a modification of the SELEX system reported here and with purified receptors and ligands, we are currently working to identify such pairs of aptamers for a variety of different multimolecular complexes.
There are potential technical challenges that need to be considered when applying the LIRECAP technology more broadly. It may be difficult to create aptamers against some target molecules or molecular complexes. In the studies reported here, the aptamers did not interfere with ligand receptor binding. This might not be the case with other systems, particularly where ligand-receptor affinity is relatively low. The standard curves based on ligand concentrations for aptamers specific for unoccupied receptor or complexes may vary and need to be considered individually for each system. Despite these potential limitations, this technology could serve to significantly expand our use of molecular complexes as biomarkers.
In summary, this report described an assay platform, designated the LIRECAP assay, that is based on pairs of RNA aptamers that bind differentially based on the presence or absence of ligand. Such assays can be used to quantify the fraction of receptors occupied by ligand in a variety of biospecimens. Here, the IL2-CD25 complex as a biomarker for lymphoma was used in these proof-of-principle studies. However, a similar approach could be applied to a broad variety of molecular complexes and disease conditions.
Supplementary Material
Acknowledgements
The authors would like to thank the University of Iowa Flow Cytometry Core Facility, Carver College of Medicine’s Protein and Crystallography Facility, Genomics Division of the Iowa Institute for Human Genetics and Sarah Bell, HCCC Biostatistics core for invaluable assistance.
Financial Support: These studies were supported in part by NIH grants P30 CA86862 and P50 CA97274.
Footnotes
Conflict of interests: The authors declare no potential conflict of interests.
Competing financial interests
The authors declare no competing financial interests.
References:
- 1.Waldmann TA. The biology of interleukin-2 and interleukin-15: implications for cancer therapy and vaccine design. Nat Rev Immunol 2006;6(8):595–601 doi 10.1038/nri1901. [DOI] [PubMed] [Google Scholar]
- 2.Ito M, Zhao N, Zeng Z, Zhou X, Chang CC, Zu Y. Interleukin-2 Functions in Anaplastic Large Cell Lymphoma Cells through Augmentation of Extracellular Signal-Regulated Kinases ½ Activation. Int J Biomed Sci 2011;7(3):181–90. [PMC free article] [PubMed] [Google Scholar]
- 3.Malek TR, Castro I. Interleukin-2 receptor signaling: at the interface between tolerance and immunity. Immunity 2010;33(2):153–65 doi 10.1016/j.immuni.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peggs KS, Quezada SA, Chambers CA, Korman AJ, Allison JP. Blockade of CTLA-4 on both effector and regulatory T cell compartments contributes to the antitumor activity of anti-CTLA-4 antibodies. J Exp Med 2009;206(8):1717–25 doi 10.1084/jem.20082492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whiteside TL, Schuler P, Schilling B. Induced and natural regulatory T cells in human cancer. Expert Opin Biol Ther 2012;12(10):1383–97 doi 10.1517/14712598.2012.707184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Binder M, O’Byrne MM, Maurer MJ, Ansell S, Feldman AL, Cerhan J, et al. Associations between elevated pre-treatment serum cytokines and peripheral blood cellular markers of immunosuppression in patients with lymphoma. Am J Hematol 2017;92(8):752–8 doi 10.1002/ajh.24758. [DOI] [PubMed] [Google Scholar]
- 7.Mir MA, Maurer MJ, Ziesmer SC, Slager SL, Habermann T, Macon WR, et al. Elevated serum levels of IL-2R, IL-1RA, and CXCL9 are associated with a poor prognosis in follicular lymphoma. Blood 2015;125(6):992–8 doi 10.1182/blood-2014-06-583369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Melero I, Berman DM, Aznar MA, Korman AJ, Perez Gracia JL, Haanen J. Evolving synergistic combinations of targeted immunotherapies to combat cancer. Nat Rev Cancer 2015;15(8):457–72 doi 10.1038/nrc3973. [DOI] [PubMed] [Google Scholar]
- 9.Goyonlo VMEH, Nordlind K. Interleukin-2 expression in lupoid and usual types of old world cutaneous leishmaniasis. Iran Red Crescent Med J 2014;16(11):5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones D, Ibrahim S, Patel K, Luthra R, Duvic M, Medeiros LJ. Degree of CD25 expression in T-cell lymphoma is dependent on tissue site: implications for targeted therapy. Clin Cancer Res 2004;10(16):5587–94 doi 10.1158/1078-0432.CCR-0721-03. [DOI] [PubMed] [Google Scholar]
- 11.Kasprzak A, Spachacz R, Wachowiak J, Stefanska K, Kaczmarek E, Zabel M. Tissue expression of interleukin 2 (IL-2) and IL-2 receptor (IL-2R(alpha)/CD25) in non-Hodgkin B-cell lymphomas in children: correlations with clinical data. J Pediatr Hematol Oncol 2010;32(6):462–71 doi 10.1097/MPH.0b013e3181e33f9c. [DOI] [PubMed] [Google Scholar]
- 12.Miyamoto C, Mattos Neto RB, Cesare SD, Belfort Junior R, Burnier MN Jr. Use of CD25 as an immunohistochemical marker for acquired ocular toxoplasmosis. Arq Bras Oftalmol 2010;73(5):443–6. [DOI] [PubMed] [Google Scholar]
- 13.Wargo JARS, Reuben A, Sharma P. Monitoring immune responses in the tumor microenvironment. Curr Opin Immunol 2016;41:23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang ZZ, Grote DM, Ziesmer SC, Manske MK, Witzig TE, Novak AJ, et al. Soluble IL-2Ralpha facilitates IL-2-mediated immune responses and predicts reduced survival in follicular B-cell non-Hodgkin lymphoma. Blood 2011;118(10):2809–20 doi 10.1182/blood-2011-03-340885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma H, Liu J, Ali MM, Mahmood MA, Labanieh L, Lu M, et al. Nucleic acid aptamers in cancer research, diagnosis and therapy. Chem Soc Rev 2015;44(5):1240–56 doi 10.1039/c4cs00357h. [DOI] [PubMed] [Google Scholar]
- 16.Zhou J, Rossi J. Aptamers as targeted therapeutics: current potential and challenges. Nat Rev Drug Discov 2017;16(6):440 doi 10.1038/nrd.2017.86. [DOI] [PubMed] [Google Scholar]
- 17.Sun H Z Y A Highlight of Recent Advances in Aptamer Technology and Its Application. Molecules 2015;20(7):11959–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature 1990;346(6287):818–22 doi 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 19.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990;249(4968):505–10. [DOI] [PubMed] [Google Scholar]
- 20.Stoltenburg R, Reinemann C, Strehlitz B. SELEX--a (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol Eng 2007;24(4):381–403 doi 10.1016/j.bioeng.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 21.Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nat Rev Drug Discov 2010;9(7):537–50 doi 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dassie JP, Hernandez LI, Thomas GS, Long ME, Rockey WM, Howell CA, et al. Targeted inhibition of prostate cancer metastases with an RNA aptamer to prostate-specific membrane antigen. Mol Ther 2014;22(11):1910–22 doi 10.1038/mt.2014.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thiel WH, Esposito CL, Dickey DD, Dassie JP, Long ME, Adam J, et al. Smooth Muscle Cell-targeted RNA Aptamer Inhibits Neointimal Formation. Mol Ther 2016;24(4):779–87 doi 10.1038/mt.2015.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thiel WH, Bair T, Peek AS, Liu X, Dassie J, Stockdale KR, et al. Rapid identification of cell-specific, internalizing RNA aptamers with bioinformatics analyses of a cell-based aptamer selection. PLoS One 2012;7(9):e43836 doi 10.1371/journal.pone.0043836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McNamara JO 2nd, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E, et al. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol 2006;24(8):1005–15 doi 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- 26.Tobias RaK S Biomolecular Binding Kinetics Assays on the Octet Platform. ForteBio, Inc; Volume Application Note 142013. p 1–22. [Google Scholar]
- 27.Friguet B, Chaffotte AF, Djavadi-Ohaniance L, Goldberg ME. Measurements of the true affinity constant in solution of antigen-antibody complexes by enzyme-linked immunosorbent assay. J Immunol Methods 1985;77(2):305–19. [DOI] [PubMed] [Google Scholar]
- 28.Yu N, Li X, Song W, Li D, Yu D, Zeng X, et al. CD4(+)CD25 (+)CD127 (low/-) T cells: a more specific Treg population in human peripheral blood. Inflammation 2012;35(6):1773–80 doi 10.1007/s10753-012-9496-8. [DOI] [PubMed] [Google Scholar]
- 29.Bielekova B, Catalfamo M, Reichert-Scrivner S, Packer A, Cerna M, Waldmann TA, et al. Regulatory CD56(bright) natural killer cells mediate immunomodulatory effects of IL-2Ralpha-targeted therapy (daclizumab) in multiple sclerosis. Proc Natl Acad Sci U S A 2006;103(15):5941–6 doi 10.1073/pnas.0601335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin JF, Perry JS, Jakhete NR, Wang X, Bielekova B. An IL-2 paradox: blocking CD25 on T cells induces IL-2-driven activation of CD56(bright) NK cells. J Immunol 2010;185(2):1311–20 doi 10.4049/jimmunol.0902238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abu-Farha M, Elisma F, Figeys D. Identification of protein-protein interactions by mass spectrometry coupled techniques. Adv Biochem Eng Biotechnol 2008;110:67–80 doi 10.1007/10_2007_091. [DOI] [PubMed] [Google Scholar]
- 32.Stoddart LA, White CW, Nguyen K, Hill SJ, Pfleger KD. Fluorescence- and bioluminescence-based approaches to study GPCR ligand binding. Br J Pharmacol 2016;173(20):3028–37 doi 10.1111/bph.13316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cabrera R, Ararat M, Eksioglu EA, Cao M, Xu Y, Wasserfall C, et al. Influence of serum and soluble CD25 (sCD25) on regulatory and effector T-cell function in hepatocellular carcinoma. Scand J Immunol 2010;72(4):293–301 doi 10.1111/j.1365-3083.2010.02427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang ZZ, Ansell SM. The tumor microenvironment in follicular lymphoma. Clin Adv Hematol Oncol 2012;10(12):810–8. [PubMed] [Google Scholar]
- 35.Yoshida N, Oda M, Kuroda Y, Katayama Y, Okikawa Y, Masunari T, et al. Clinical significance of sIL-2R levels in B-cell lymphomas. PLoS One 2013;8(11):e78730 doi 10.1371/journal.pone.0078730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carreras J, Lopez-Guillermo A, Fox BC, Colomo L, Martinez A, Roncador G, et al. High numbers of tumor-infiltrating FOXP3-positive regulatory T cells are associated with improved overall survival in follicular lymphoma. Blood 2006;108(9):2957–64 doi 10.1182/blood-2006-04-018218. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.