Abstract
The function of PTEN in the cytoplasm largely depends on its lipid-phosphatase activity by antagonizing the PI3K-Akt oncogenic pathway. However, molecular mechanism(s) underlying the role of PTEN in the nucleus remain largely elusive. Here, we report that DNA double-strand breaks (DSBs) promote PTEN interaction with MDC1, upon ATM-dependent phosphorylation of T/S398-PTEN. Importantly, DNA DSBs enhance NSD2 (MMSET/WHSC1)-mediated di-methylation of PTEN at K349, which is recognized by the tudor domain of 53BP1 to recruit PTEN into DNA damage sites, governing efficient repair of DSBs partly through dephosphorylation of γH2AX. Of note, inhibiting NSD2-mediated methylation of PTEN, either through expressing methylation-deficient PTEN mutants, or through inhibiting NSD2, sensitizes cancer cells to combinatorial treatment with PI3K inhibitor and DNA-damaging agents in both cell culture and in vivo xenograft models. Therefore, our study provides a novel molecular mechanism for PTEN regulation of DSBs repair in a methylation- and protein-phosphatase-dependent manner.
Keywords: PTEN, NSD2, 53BP1, methylation, DNA damage, DNA double strand breaks
INTRODUCTION
The lipid and protein dual-phosphatase, phosphatase and tensin homolog deleted on chromosome ten (PTEN), is a tumor suppressor frequently mutated, deleted, or epigenetically silenced in various types of human cancers (1,2). In the cytoplasm and membrane, PTEN primarily governs key cellular processes including cell survival, proliferation, aging, angiogenesis and metabolism through its lipid-phosphatase activity to antagonize the PI3K-Akt oncogenic pathway (3–5). However, compared to its well-studied lipid-phosphatase activity, the protein-phosphatase function of PTEN remains largely undefined. There is increasing evidence demonstrating that PTEN also possesses multiple important functions in the nucleus independent of its lipid-phosphatase activity, such as controlling chromosomal integrity (6), chromatin structure (7,8), DNA replication (9) and DNA damage repair (10,11). In support of a critical role for PTEN in DNA double-strand breaks (DSBs) repair, both large-scale proteomic analyses and in vitro specific biochemical kinase assays identified PTEN to be phosphorylated at T398 (S398 in mouse Pten, thereafter referred as T/S398) by the ataxia telangiectasia mutated (ATM) kinase, in response to DNA damage (11,12). However, the precise molecular mechanism by which ATM-dependent phosphorylation of PTEN T/S398 regulates its role in DNA damage repair remains largely elusive.
Protein methylation, especially histone methylation, is one of the key post-translational modifications in regulating chromatin properties and gene expression profiles, which governs various cellular processes including DNA replication and cell cycle progression (13,14). Beyond histone methylation, recent studies have revealed that methylation of non-histone proteins also play critical roles in controlling cellular signaling and functions (15,16). For example, methylation of the tumor suppressor p53 at K372 by the methyltransferase Set9 activates its transcriptional function, while the Smyd2-mediated methylation of p53 at K372 has an opposite role (17,18). It also has been reported that the tumor suppressor Rb is methylated by Smyd2 at K860, which is then recognized by the MBT domain of the transcriptional repressor L3MBTL1 (19). Moreover, the Set7/9 methyltransferase methylates Rb at K810, which is subsequently recognized by the tudor domain of 53BP1 to maintain Rb in its hypophosphorylated state and response to the DNA-damage signaling (20). However, whether and how the tumor suppressor PTEN can be regulated by methylation remains largely unexplored.
Here, we report that DNA DSBs promote NSD2 (also named MMSET/WHSC1)-mediated di-methylation of PTEN at K349, which is read by the tudor domain of 53BP1 to recruit PTEN into DNA damage sites to govern the timely repair of DSBs in part through dephosphorylating γH2AX. More importantly, inhibiting NSD2-mediated methylation of PTEN sensitizes cancer cells to combinatorial treatment with PI3K inhibitor and DNA-damaging agents in both cell culture and in vivo xenograft models.
RESULTS
ATM-mediated phosphorylation of PTEN is required for binding the BRCT domain of MDC1 upon DNA damage signaling
To further explore the role of PTEN in the nucleus to govern DSBs repair, we found that phosphorylation of PTEN could be readily detected using the phospho-(Ser/Thr) ATM/ATR substrate antibody upon various DNA damaging agents including etoposide, irradiation (IR) or N-methyl-N’-nitro-N-nitrosoguanidine (MNNG) treatment in different cell lines (Fig. 1A and Supplementary Fig. S1A–1D). Moreover, cellular fractionation assays showed that etoposide-induced phosphorylation of PTEN existed in both the cytoplasm and nucleus (Supplementary Fig. S1E). Furthermore, we found that inhibiting the DNA damage upstream kinase ATM, but not ATR using the pharmacological inhibitors or shRNAs, could dramatically decrease etoposide-induced phosphorylation of PTEN in cells (Supplementary Fig. S1F and S1G), suggesting that the ATM kinase is the major physiological kinase that phosphorylates PTEN at S398 in response to DNA damage (Supplementary Fig. S1H). However, the precise molecular mechanism governing ATM-dependent phosphorylation of PTEN at S398 to regulate its role in DNA damage repair remains largely elusive.
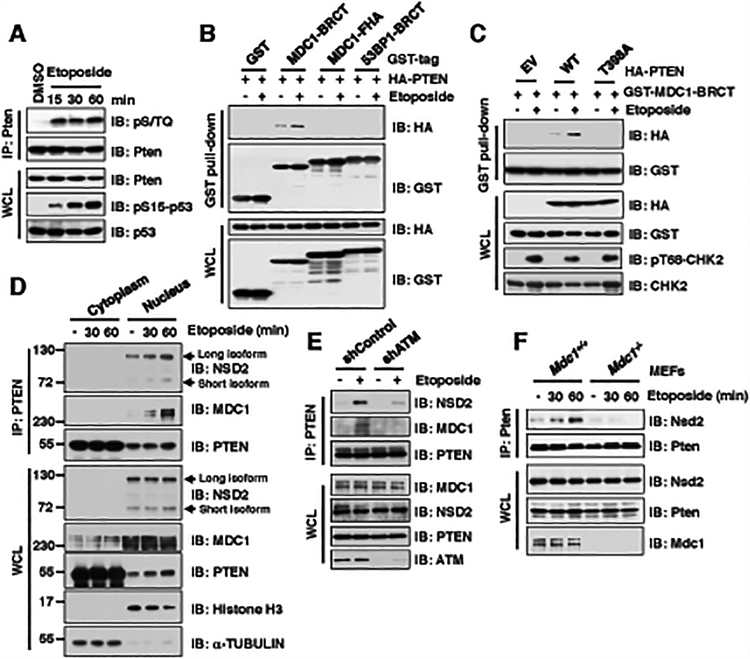
Figure 1. ATM-mediated phosphorylation of PTEN is required for its binding with the BRCT domain of MDC1 upon DNA damage signaling.
(A) Phosphorylation of Pten was detected using the phospho-(Ser/Thr) ATM/ATR substrate antibody (pS/TQ) upon etoposide treatment. Immunoblot (IB) analysis of anti-Pten immunoprecipitations (IPs) and whole cell lysates (WCL) derived from NIH3T3 cells treated with 30 μM etoposide as indicated time points before harvesting.
(B) Etoposide treatment promoted PTEN interaction with MDC1 BRCT domain, but neither MDC1 FHA nor 53BP1 BRCT domain. IB analysis of GST pull-down and WCL derived from U2OS cells transfected with indicated constructs and treatment with/without 30 μM etoposide for 30 min before harvesting.
(C) Etoposide treatment promoted wild type (WT), but not T398A mutant PTEN, interaction with MDC1 BRCT domain. IB analysis of GST pull-down and WCL derived from U2OS cells co-transfected with indicated constructs. 36 h after transfection, cells were treated with/without 30 μM etoposide for 30 min and harvested for IP assays.
(D) PTEN, MDC1, and NSD2 formed a tertiary complex in the nucleus upon etoposide treatment. IB analysis of anti-PTEN IPs from cytoplasm or nucleus as well as WCL derived from U2OS cells treated with 30 μM etoposide as indicated time points before harvesting.
(E) Depletion of ATM disrupted PTEN interaction with MDC1 and NSD2 upon etoposide treatment. IB analysis of anti-PTEN IPs and WCL derived from U2OS cell lines stably expressing shControl or shATM transfected with indicated constructs. 36 h after transfection, cells were treated with/without 30 μM etoposide for 30 min before harvesting.
(F) Depletion of Mdc1 impaired the interaction between Pten and Nsd2. IB analysis of anti-Pten IPs and WCL derived from Mdc1+/+ and Mdc1−/− MEFs treated with 30 μM etoposide as indicated time points before harvesting.
Previous reports have revealed that pS/pTQ events can be recognized by the BRCA1 carboxy-terminal (BRCT) or forkhead-associated (FHA) domains in DNA damage repair (DDR) proteins in order to facilitate the DNA damage repair process (Supplementary Fig. S1I) (21,22). Despite the subtle difference in the 398TQ399 phospho-motif in human PTEN versus the 398SQ399 motif in mouse Pten, they are both phospho acceptor and likely function in a similar manner (Supplementary Fig. S1J) (11,12). In keeping with this notion, we found that etoposide treatment enhanced both human PTEN and mouse Pten binding with the BRCT domain of MDC1, but not its FHA domain, nor the BRCT domain-derived from 53BP1 or BRCA1 (Fig. 1B; Supplementary Fig. S1K and S1L).
As further evidence in support of the notion that the 398pTQ399 motif is the primary residues in PTEN that interacts with MDC1, we observed that MDC1 strongly interacted with the C-terminal domain of PTEN containing the ATM-meditated pT398 site, but not the N-terminus of PTEN (Supplementary Fig. S1M and S1N). Moreover, the ATM-phosphorylation-deficient PTEN-T398A mutant lost its interaction with the MDC1-BRCT domain after etoposide treatment (Fig. 1C). Previous large-scale screening and biochemical studies identified that the ATM kinase also can phosphorylate the methyltransferase NSD2 at the S102 after DNA damage, leading to the formation of a complex with 53BP1 and MDC1 to involve in DNA DSBs repair process (12,23,24). In keeping with these findings, we found that etoposide treatment enhanced the interaction between MDC1 and NSD2 in cells (Supplementary Fig. S1O). Hence, we hypothesized that upon DNA damage, MDC1, NSD2, and PTEN might form a tertiary complex in cells. In support of this notion, we found that MDC1 and NSD2 existed in anti-PTEN immunoprecipitates after etoposide treatment (Supplementary Fig. S1P). Furthermore, we showed that the tertiary complex of MDC1, NSD2 and PTEN largely existed in the nucleus, but not in the cytoplasm (Fig. 1D and Supplementary Fig. S1Q), indicating that this complex might be involved in the DNA damage repair process in the nucleus.
Moreover, although there are several NSD2 isoforms reported (25), our results showed that only the long isoform of NSD2 has a high affinity binding with PTEN in the nucleus (Fig. 1D). Hence, in the remainder of this manuscript, we mainly focused on the long isoform of NSD2 in our study. Notably, the ATM deficiency largely disrupted etoposide-induced formation of tertiary complex among PTEN, MDC1 and NSD2 (Fig. 1E), which might be due to loss of ATM-mediated phosphorylation of PTEN and its interaction with MDC1 (Supplementary Fig. S1G and Fig. 1C). Moreover, the etoposide-induced interaction between PTEN and NSD2 was largely disrupted in Mdc1−/− MEFs (Fig. 1F), which suggests that MDC1 likely plays a crucial role in mediating the interaction of PTEN and NSD2 upon DNA damage signaling. Although ATM also phosphorylates MDC1 at the T4 sites to promote the dimer formation of MDC1 (26), the MDC1-Δ1–143 mutant with deleting the N-terminal region containing T4 and FHA domain still interacted with PTEN after etoposide treatment (Supplementary Fig. S1R), indicating that ATM-mediated phosphorylation of MDC1 likely does not play an important role in mediating PTEN/MDC1 interaction upon DNA damage. Altogether, these results support a model that in response to DSBs, PTEN, MDC1, and NSD2 could form a tertiary complex in an ATM-mediated PTEN-phosphorylation-dependent manner.
DNA damage promotes NSD2-mediated di-methylation of PTEN at K349
In support of a specific interaction between NSD2 and PTEN, we found that only NSD2, but none of the other methyltransferases we examined, including SET8, EZH2 or NSD3, interacted with PTEN in cells, under etoposide treatment condition (Supplementary Fig. S2A and S2B). We further identified that NSD2 interacted with PTEN largely through its C-terminal domain (784–1365) in cells (Supplementary Fig. S2C). Moreover, compared with the methyltransferase SET8 or EZH2, NSD2 could dramatically promote the methylation of PTEN in cells, as revealed by methyl-lysine motif antibodies (Supplementary Fig. S2D). Notably, we observed significant di-methylation of PTEN at endogenous levels after etoposide treatment, which largely correlated with an elevated interaction between NSD2 and PTEN (Fig. 2A). Furthermore, compared to Nsd2+/+ MEFs, the di-methylation of PTEN was dramatically reduced in Nsd2−/− MEFs upon etopside treatment (Fig. 2B). Hence, these results support a possible role of NSD2 in promoting PTEN methylation upon DNA damage.
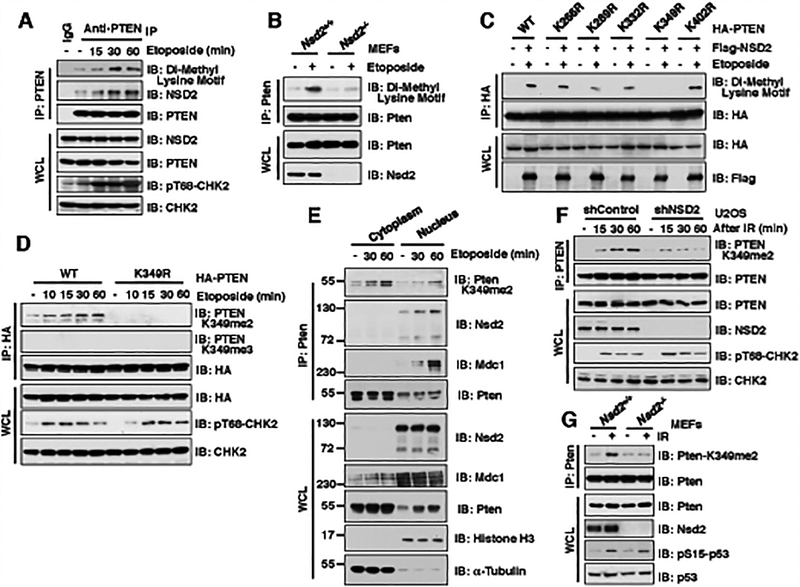
Figure 2. DNA damage promotes NSD2-mediated di-methylation of PTEN at K349.
(A) Methylation of PTEN was detected using the Di-Methyl Lysine motif antibody. IB analysis of anti-PTEN IPs and WCL derived from U2OS cells treated with 30 μM etoposide as indicated time points before harvesting.
(B) Depletion of Nsd2 impaired the di-methylation of Pten. IB analysis of anti-Pten IPs and WCL derived from Nsd2+/+ and Nsd2−/− MEFs treated with 30 μM etoposide for 30 min before harvesting.
(C) K349 was identified as the major di-methylation site on PTEN. IB analysis of anti-HA IPs and WCL derived from 293T cells transfected with the indicated constructs and treated with/without 30 μM etoposide for 30 min before harvesting.
(D) PTEN was detected by the specific K349 di-methylation (K349me2) antibody. IB analysis of anti-HA IPs and WCL derived from U2OS cells transfected with HA-PTEN WT or K349R mutant and treated with/without 30 μM etoposide at indicated time points.
(E) The K349 di-methylation of Pten existed in both cytoplasm and nucleus. IB analysis of anti-Pten IPs from cytoplasm or nucleus as well as WCL derived from NIH3T3 cells treatment with 30 μM etoposide as indicated time points before harvesting.
(F and G) NSD2 deficiency decreased the di-methylation of PTEN at K349. IB analysis of anti-PTEN IPs and WCL derived from U2OS cells stably expressing shNSD2 (F) or Nsd2−/− MEFs (G) that were treated with IR (5 Gy) at indicated time points before harvesting.
Consistent with the finding that MDC1 plays a crucial role in mediating the interaction of PTEN and NSD2 (Fig. 1F), etoposide-induced di-methylation of PTEN was also markedly decreased in Mdc1−/− MEFs compared to Mdc1+/+ MEFs (Supplementary Fig. S2E). Moreover, compared to WT-PTEN, the ATM phosphorylation-deficient T398A-PTEN mutant was impaired in associating with NSD2 in response to etoposide treatment (Supplementary Fig. S2F), thereby exhibiting a marked reduction in NSD2-mediated di-methylation of PTEN in cells (Supplementary Fig. S2G). Taken together, these results demonstrate that MDC1 plays a pivotal role in connecting the NSD2 methyltransferase to ATM-mediated phosphorylation of PTEN and subsequently promoting NSD2-mediated methylation of PTEN.
Next, we set out to identify NSD2-mediated methylation site(s) on PTEN. Consistent with our observation that PTEN interacted with NSD2 through its C-terminal domain (Supplementary Fig. S2H and S2I), major di-methylation sites were identified in the C-terminus of PTEN under etoposide treatment condition (Supplementary Fig. S2J). Using a mutagenesis approach to remove major lysine residues exposed at the PTEN protein surface (27), we found that the K349R mutant was largely deficient in undergoing NSD2-mediated methylation in cells, but not other lysine mutants we examined including K266R, K289R, K332R or K402R (Fig. 2C). Moreover, the K349 residue on PTEN was conserved among different species and located on the surface of crystal structure of PTEN (Supplementary Fig. S2K and S2L), further indicating K349 as a potential NSD2-mediated di-methylation site.
To further investigate the physiological role of di-methylation of PTEN-K349 during DSBs repair, we generated antibodies that could specifically recognize di-methylated PTEN on K349 (PTEN-K349me2) or tri-methylated on K349 (PTEN-K349me3). Using dot blot assays with synthetic PTEN peptides with K349 non-, mono-, di- and tri-methylation (Supplementary Fig. S2M), we validated that the PTEN-K349me2 antibody specifically recognized the di-methylated-K349-PTEN synthetic peptides, while the PTEN-K349me3 antibody specifically recognized the tri-methylated-K349-PTEN synthetic peptides (Supplementary Fig. S2N). However, upon etoposide treatment, we only detected di-methylation, but not tri-methylation, of PTEN at K349 in cells (Fig. 2D). More importantly, di-methylation of K349-PTEN was observed at endogenous levels upon etoposide or γ-irradiation treatment, a process that could be largely abolished following depletion of endogenous NSD2 (28) (Fig. 2E–2G and Supplementary Fig. S2O). Consistent with the results that ATM-mediated phosphorylation of PTEN is critical for PTEN methylation (Supplementary Fig. S2F and S2G), the ATM inhibitor also suppressed the di-methylation of PTEN at K349 after etoposide treatment (Supplementary Fig. S2P). Moreover, the 3-deazaneplanocin A (DZNep), a pan-inhibitor of S-adenosylmethionine (SAM)-dependent methyltransferases that includes NSD2 (29,30), could also dramatically suppress K349 di-methylation of PTEN in cells in a dose-dependent manner (Supplementary Fig. S2Q). Taken together, these results reveal NSD2 as a major physiological upstream methyltransferase that predominantly promotes K349 di-methylation of PTEN in response to DNA damaging signals.
NSD2-mediated di-methylation of PTEN is recognized by the tudor domain of 53BP1
Given that methylation signals at DNA damage sites can be recognized by tudor domain-containing proteins, such as 53BP1 or histone demethylases, to facilitate the recruitment of DDR effector proteins including p53 and Rb (Supplementary Fig. S3A) (20,31,32), we explored the physiological reader that can recognize NSD2-mediated K349 di-methylation of PTEN. To this end, we found that etoposide treatment could dramatically enhance PTEN interaction with 53BP1 largely through its tudor domain, but not other tudor domains we examined, including SETDB1 or KDM4B (Fig. 3A and 3B; Supplementary Fig. S3B and S3C), indicating a possible role for 53BP1 in mediating the recruitment of PTEN to DNA damage sites. We also observed that DNA damaging agent can lead to reduced protein abundance of 53BP1, a process that can be partially blocked by treatment with the 26S proteasome inhibitor, MG132 (Supplementary Fig. S3D). This indicates that the ubiquitin proteasome pathway might be involved in DNA damage-induced 53BP1 destruction but the underlying mechanism remains to be determined. Notably, the methyltransferase NSD2 promoted PTEN-WT, but not the PTEN-K349R, interaction with the tudor domain of 53BP1, indicating the involvement of methyl-K349-PTEN and its subsequent recognition by the 53BP1-tudor domain (Fig. 3C and Supplementary Fig. S3E). However, although the PTEN-K349R mutant failed to interact with 53BP1-tudor domain, PTEN-K349R still bound to NSD2 (Supplementary Fig. S3F). Moreover, the ATM-mediated phosphorylation deficient 53BP1-S/T28A also interacted with PTEN (Supplementary Fig. S3G), suggesting that 53BP1 interacts with PTEN largely through its tudor domain (Fig. 3B).
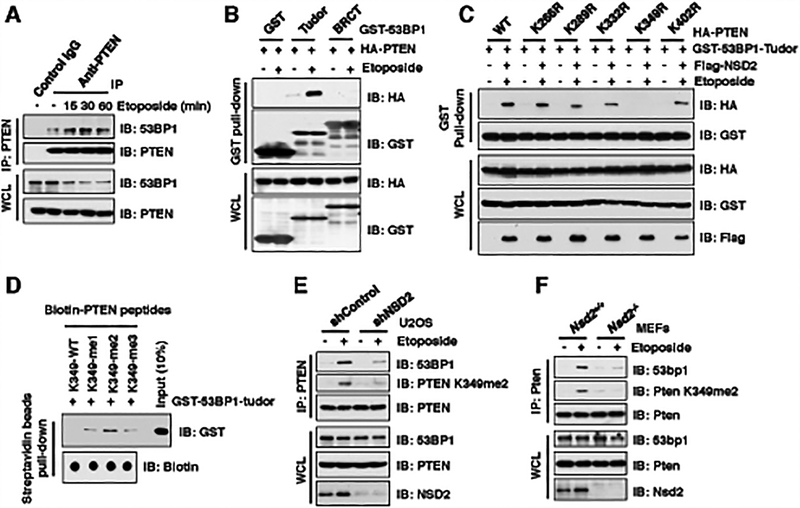
Figure 3. NSD2-mediated di-methylation of PTEN is recognized by the tudor domain of 53BP1.
(A) Etoposide treatment enhanced PTEN interaction with 53BP1. IB analysis of anti-PTEN IPs and WCL derived from U2OS cells treated with 30 μM etoposide as indicated time points before harvesting.
(B and C) PTEN-K349R mutant disrupted its binding with 53BP1 upon etoposide treatment. IB analysis of GST pull-down and WCL derived from U2OS cells transfected with the indicated constructs. 36 h after transfection, cells were treated with/without 30 μM etoposide for 30 min and harvested for GST pull-down assays.
(D) 53BP1 tudor domain had a high affinity with K349-me2-PTEN peptides. 1 μg of indicated biotin-labeled synthetic PTEN peptides were incubated with 250 ng purified recombinant GST-tagged 53BP1 tudor domain, respectively. Streptavidin beads were added to perform pull-down assays and precipitations were analyzed by IB. Dot blot assays were performed to show equal amount of biotinylated peptides was used for the pull-down assay.
(E and F) Depletion of NSD2 disrupted PTEN interaction with 53BP1. IB analysis of anti-PTEN IPs and WCL derived from U2OS cells stably expressing shNSD2 (E) or Nsd2−/− MEFs (F) that were treated with/without 30 μM etoposide for 30 min before harvesting.
In further support of a direct association between the 53BP1-tudor domain and methyl-K349-PTEN, using in vitro pull-down assay with synthetic PTEN peptides that are either unmodified K349 (K349-me0), mono-methylated (K349-me1), di-methylated (K349-me2) or tri-methylated (K349-me3) (Supplementary Fig. S2M), we found that the purified recombinant tudor domain of 53BP1 specifically interacted with methylated (K349-me1,2,3), but not with unmodified (K349-me0), synthetic PTEN peptides in vitro (Fig. 3D). Moreover, among all three methylated synthetic PTEN peptides, the di-methylated PTEN peptide (K349-me2) appeared to interact with the 53BP1-tudor domain with a relatively higher affinity (Fig. 3D). More importantly, depletion of NSD2 dramatically reduced endogenous PTEN binding with 53BP1 in cells, which was largely correlative to the reduced PTEN K349 di-methylation in NSD2-depleted cells (Fig. 3E and 3F). Taken together, these results demonstrate that NSD2-mediated K349 di-methylation of PTEN is required for efficient recognition by the methyl-lysine reading tudor domain of 53BP1 upon DNA damage signals (Supplementary Fig. S3H).
PTEN K349 di-methylation and protein phosphatase activity are required for efficient DSBs repair
To explore the physiological function of PTEN in DNA damage repair process, we investigated the DNA damage response in PTEN+/+ and PTEN−/− HCT116 cells (33) after exposure to γ-irradiation or etoposide. Notably, we found that the magnitude and duration of γH2AX were dramatically elevated, especially during late stages post γ-irradiation or etoposide treatment (4, 8 and 24-hour time points) in PTEN−/− cells, as compared with PTEN+/+ cells (Fig. 4A, 4B and Supplementary Fig. S4A). Although the PTEN K349R mutant still displayed lipid-phosphatase activity at levels comparable to WT-PTEN (Supplementary Fig. S4B), it was largely deficient in rescuing the prolonged γH2AX status in PTEN−/− HCT116 cells post γ-irradiation or etoposide treatment (Fig. 4C, 4D and Supplementary Fig. S4C), suggesting that the lipid-phosphatase activity of PTEN might not be involved in this process, at least in this experimental setting. Since PTEN also possesses protein phosphatase activity (34) to impact the biological functions of its protein substrates (35,36), we went on to explore whether the duration of γH2AX after irradiation in PTEN−/− cells is relevant to PTEN protein phosphatase activity. To this end, we re-introduced the PTEN-WT, C124S (lipid- and protein-phosphatase dead mutant), G129E (lipid-phosphatase dead mutant) or Y138L (protein-phosphatase dead mutant) into PTEN−/− HCT116 cells at comparable levels (Supplementary Fig. S4D and S4E). Consistent with previous reports (34,35), ectopic expression of PTEN WT and Y138L mutant, but not the lipid-phosphatase deficient C124S nor G129E mutants, could reduce the levels of pS473-AKT in PTEN−/− HCT116 (Supplementary Fig. S4D and S4E). Notably, we found that PTEN−/− HCT116 cells re-introduced WT or G129E, but not Y138L or C124S, were capable of rescuing the elevated and prolonged γH2AX status upon γ-irradiation (Fig. 4E and 4F). Although the protein phosphatase-deficient PTEN-Y138L mutant failed to rescue the γH2AX levels compared to the PTEN-WT, the PTEN-Y138L mutant could be still modified with etoposide-induced phosphorylation, methylation as well as interacted with MDC1 and NSD2 upon etoposide treatment (Supplementary Fig. S4F–S4I), which suggests that the PTEN protein phosphatase activity towards regulating γH2AX levels might be the late stage event in the process of DNA damage repair.
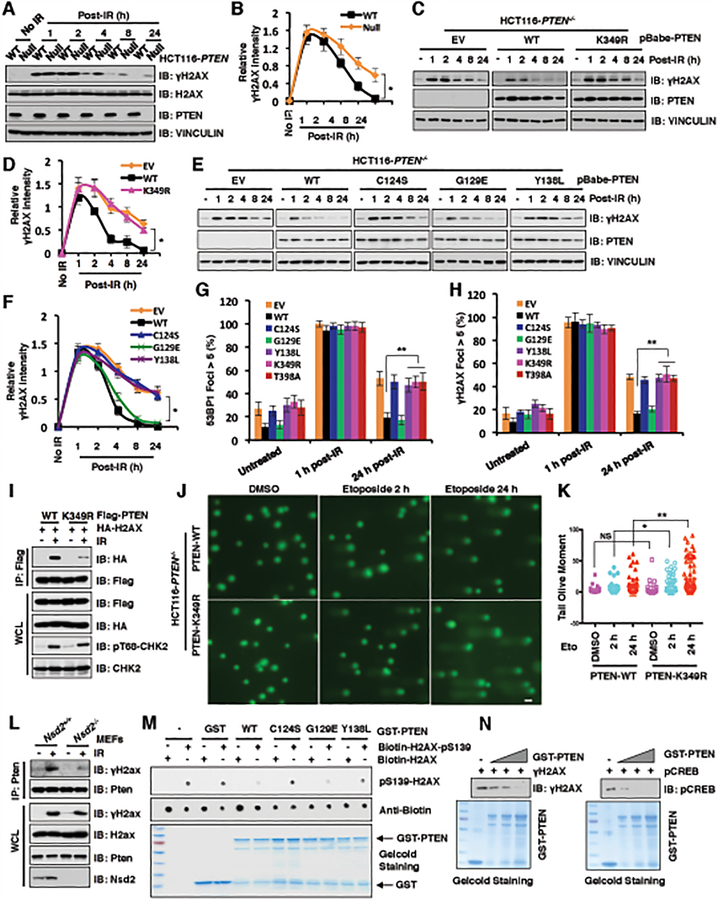
Figure 4. PTEN K349 di-methylation and protein phosphatase activity are required for efficient DSBs repair.
(A and B) PTEN deficiency elevated γH2AX after 4 h post-irradiation (IR) treatment. IB analysis of WCL derived from PTEN+/+ and PTEN−/− HCT116 cells after treatment with IR (5 Gy) as indicated time points (A). Quantification of protein intensity was performed using the ImageJ software (B). γH2AX immunoblot bands were normalized to VINCULIN, and then normalized to the control (no IR treatment). Data are represented as mean ± s.d., n = 3. *p < 0.05 (Student’s t-test).
(C and D) PTEN wild type (WT), but not K349R mutant, rescued PTEN deficiency mediated high levels of γH2AX after 4 h post-IR treatment. IB analysis of WCL derived from PTEN−/− HCT116 cells introducing PTEN WT, K349R as well as EV, which were treated with IR (5 Gy) at indicated time points before harvesting (C). Quantification of protein intensity was performed using the ImageJ software (D). γH2AX immunoblot bands were normalized to VINCULIN, and then normalized to the control (no IR treatment). Data are represented as mean ± s.d., n = 3. *p < 0.05 (Student’s t-test).
(E and F) PTEN protein phosphatase activity, but not lipid phosphatase activity, was required for regulating γH2AX levels. IB analysis of WCL derived from PTEN−/− HCT116 cells re-introducing PTEN WT, C124S, G129E, Y138L as well as EV, which were treated with IR (5 Gy) at indicated time points before harvesting (E). Quantification of protein intensity was performed using the ImageJ software (F). γH2AX immunoblot bands were normalized to VINCULIN, then normalized to the control (no IR treatment). Data are represented as mean ± s.d., n = 3. *p < 0.05 (Student’s t-test).
(G and H) PTEN−/− HCT116 cells reconstituted with the indicated PTEN constructs were treated with IR (5 Gy) and immunostained at the indicated times with anti-53BP1 or anti-γH2AX. Quantification of 53BP1 (G) or γH2AX (H) foci positive cells (foci > 5 per cell) was performed by counting a total of 100 cells per sample, respectively. Data are represented as mean ± s.d., n = 3, and **p < 0.01 (Student’s t-test).
(I) PTEN-K349R mutant decreased its interaction with H2AX. IB analysis of anti-Flag IPs and WCL derived from U2OS cells transfected with the indicated constructs. 36 h after transfection, cells were harvested for IP assays after treatment with IR (5 Gy) for 60 min.
(J and K) PTEN-K349R mutation attenuated DNA damage repair. PTEN-WT or PTEN-K349R mutant expressed HCT116 cells were treated with DMSO or etoposide for 2h and 24h, respectively. (J) Images were captured following Comet assay; and (K) Tail olive moment values were calculated using Casplab software. Error bars represent standard error of the mean. **p<0.01, *p<0.05. n=80. Scale bar: 50 μM.
(L) Nsd2 deficiency disrupted Pten interaction with γH2ax. IB analysis of anti-Pten IPs and WCL derived from Nsd2+/+ and Nsd2−/− MEFs treatment with/without IR (5 Gy) for 60 min before harvesting.
(M and N) In vitro dephosphorylation assays were performed with bacterially purified recombinant GST-tagged PTEN WT and the indicated PTEN mutants including C124S, G129E, and Y138L incubating with indicated H2AX synthetic peptides (M) or purified γH2AX (N) from HCT116 cells after etoposide treatment, then analyzed by immunoblot analyses.
To further examine the PTEN protein phosphatase activity in regulating DSBs repair, we detected the DNA damage response profiles in PTEN−/− HCT116 cells following re-introducing WT and various mutants including C124S, G129E, Y138L, K349R and T398A. The foci formations of these cell lines were monitored by immuno-staining against DSBs markers 53BP1 and γH2AX at the indicated time points post γ-irradiation (37–39). Notably, 1 hour post γ-irradiation, the 53BP1 foci could be observed in all cell lines (Fig. 4G and Supplementary Fig. S4J), a faithful marker indicative of double strand breaks generated by IR (40,41). However, 24 hours after irradiation, 53BP1 foci were largely resolved in PTEN−/− HCT116 cells stably expressing PTEN WT and G129E, but not in cells expressing C124S and Y138L that are deficient in PTEN protein-phosphatase activity, or in the K349R or S398A mutant that are deficient in undergoing NSD2-mediated di-methylation (Fig. 4G and Supplementary Fig. S4J). Consistently, cells expressing C124S, Y138L, K349R and S398A, but not WT or G129E mutants of PTEN, also exhibited deficiencies in resolving γH2AX foci at 24 hours post γ-irradiation (Fig. 4H and Supplementary Fig. S4K). More importantly, we monitored the foci formation of DSBs markers 53BP1 and γH2AX in multiple cell lines including PTEN-deficient U87MG and PC3 reconstituted with WT-PTEN or K349R-PTEN to validate that cells deficient in undergoing K349-PTEN methylation are defective in resolving DSBs foci induced by IR (Supplementary Fig. S4L–O).
Additionally, we performed the immuno-staining for the homologous recombination (HR) markers of RAD51 and BRCA1 after γ-irradiation treatment in the PTEN−/− HCT116 cells with reconstituting PTEN-WT, PTEN-K349R as well as empty vector (EV) as a negative control. Our results showed that compared to putting back both of PTEN-WT and PTEN-K349R, PTEN−/− HCT116 cells stably expressing EV exhibited the decreased number of RAD51 foci 4 hours after IR treatment (Supplementary Fig. S4P), which is consistent with previous reports that PTEN could recruit the RAD51 into the DNA damage sites (11,42,43). However, 24 hours post-IR treatment, foci of RAD51 and BRCA1 were largely resolved in PTEN−/− HCT116 cells with re-introducing both of PTEN-WT and K349R (Supplementary Fig. S4P–Q). Moreover, we sought to directly measure HR efficiency using a reporter assay in PTEN-deficient DR-GFP U87MG cells with reconstituting PTEN-WT, PTEN-K349R as well as EV as a negative control. Compared with the EV, both PTEN-WT and K349R mutant could enhance the efficiency of HR-mediated DSBs repair (Supplementary Fig. S4R). Together, these results demonstrated that NSD2-mediated methylation of PTEN might rely on 53BP1 to preferentially use NHEJ over the HR for DSBs repair.
Consistent with a previous report (44), we found that PTEN co-localized with γH2AX in DNA damage sites generated by laser micro-irradiation (Supplementary Fig. S4S). Moreover, the interaction between PTEN and H2AX appears to be dependent on NSD2-mediated methylation of PTEN at K349 as the interaction between H2AX and K349R-PTEN was dramatically decreased in comparison to WT-PTEN, after γ-irradiation treatment (Fig. 4I). Moreover, COMET assays showed that compared to PTEN-WT cells, cells expressing PTEN-K349R exhibited significant longer tail moment after etoposide treatment, which suggests the DNA damage repair process was dramatically suppressed in the context of K349R mutation (Fig. 4J and 4K). Furthermore, depletion of the Nsd2 methyltransferase impaired irradiation-induced interaction of γH2AX with PTEN (Fig. 4L). Moreover, we found that the purified recombinant GST-PTEN protein could pull-down H2AX, but not H2A containing nucleosome core particles in vitro (Supplementary Fig. S4T). These results together indicate that upon DNA damaging signals, a portion of PTEN could be recruited into nucleus, where interacts with H2AX and involves in DNA damage repair.
As our results showed that the PTEN protein phosphatase activity plays a critical role in resolving the foci of γH2AX, we want to explore whether PTEN could directly dephosphorylate γH2AX. To this end, we performed in vitro dephosphorylation assays using bacterially purified recombinant PTEN-WT, C124S, G129E and Y138L proteins incubating with biotin-labeled H2AX peptides with/without phosphorylation of Ser139. Notably, PTEN WT and G129E, but not the protein-phosphatase dead mutants C124S and Y138L, could dramatically dephosphorylate biotin-labeled γH2AX in vitro (Fig. 4M). To further mimic the reaction condition in cells, we purified the γH2AX from HCT116 cells after etoposide treatment and the positive control pS133-CREB (43) through using the anti-γH2AX or anti-pS133-CREB antibody, respectively. The in vitro dephosphorylation assays showed that PTEN could dephosphorylate the γH2AX or pS133-CREB in a dose-dependent manner (Fig. 4N). Although both our results and previous study (12) showed that PTEN could be recruited into the DNA damage sites, which factor(s) playing a critical role in recruiting PTEN into DNA damage sites remains elusive. Based on our results, we hypothesized that the DDR proteins MDC1, 53BP1 or γH2AX might be involved in this recruiting process upon DNA damage signaling. To address this question, we used the Mdc1−/−, 53BP1−/− or H2ax−/− as well as their counterpart WT cells to perform the cellular fractionation assays with/without etoposide treatment. Our results demonstrated that PTEN recruitment into the DNA damage sites might be largely dependent on MDC1 and 53BP1, but not H2AX because upon etoposide treatment the accumulation of PTEN on chromatin was reduced in the Mdc1−/− or 53BP1−/−, but not in H2ax−/− cells, compared to their counterpart WT cells (Supplementary Fig. S4U–S4W). Moreover, the di-methylation of PTEN at K349 was also critical for the localization of PTEN on chromatin upon etoposide treatment because compared to the PTEN-WT, the PTEN-K349R mutant failed to localize on the chromatin (Supplementary Fig. S4X). Notably, we also found that the ATM-mediated phosphorylation of 53BP1 at S25/29, two of 28 amino-terminal Ser/Thr-Gln (S/T-Q) sites that are required for recruiting DSB-responsive factors including PTIP and RIF1 (45), were dramatically reduced in the nucleus of PTEN KO cells compared to PTEN WT cells (Supplementary Fig. S4Y and S4Z). This result suggested that PTEN might also function as a scaffold protein for amplifying the ATM-mediated phosphorylation signaling on 53BP1 to help recruit other DDR response factors participating in the DNA damage repair process. Taken together, these results support our model that upon DNA damage, a portion of methylated PTEN species were recruited on DNA damage sites along with other factors to help DSBs repair largely through its protein-phosphatase activity in the nucleus.
The protein phosphatase activity of PTEN is required for DSBs repair in vivo
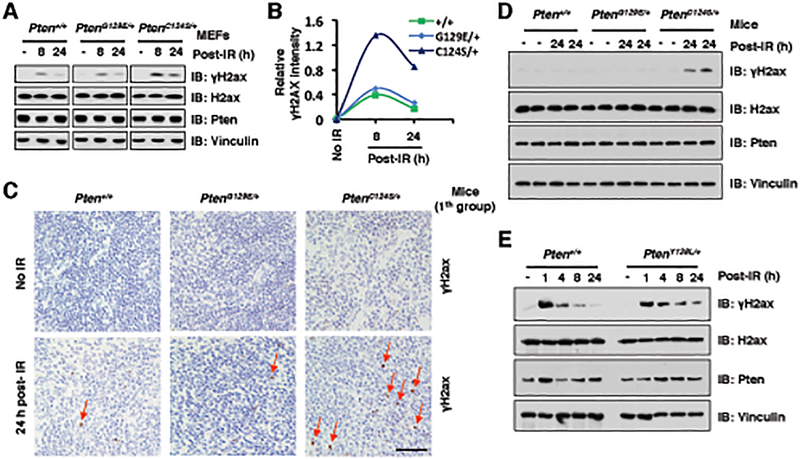
To study the physiological role of PTEN protein phosphatase activity in regulating γH2AX levels in vivo, we used Pten knock-in mice harboring PtenC124S or PtenG129E mutants (46). Compared with Pten+/+ and PtenG129E/+ MEFs, the levels of γH2ax were elevated in PtenC124S/+ MEFs at late stages of DSBs repair post γ-irradiation (8 and 24-hour time points) (Fig. 5A and 5B). Furthermore, PtenC124S/+ mice also displayed high levels of γH2ax at 24 h post γ-irradiation in relative to Pten+/+ and PtenG129E/+ mice (Fig. 5C–5D and Supplementary Fig. S5A). More importantly, to dissect the critical physiological role of the protein phosphatase activity of PTEN in DSBs repair, we isolated PtenY138L/+ MEFs and found that in comparison to WT MEFs, heterozygous PtenY138L/+ MEFs also displayed relatively higher levels of γH2AX at 8 and 24-hour time points post γ-irradiation (Fig. 5E and Supplementary Fig. S5B). Although our previous study showed that PTEN-C124S or G129E mutant affects their dimerization to regulate PTEN lipid phosphatase activity (46), the protein phosphatase-deficient Y138L or methylation-deficient K349R mutant did not exert significant effects on PTEN dimerization in cells (Supplementary Fig. S5C and S5D). Taken together, these results suggest that the protein-phosphatase activity, but not lipid-phosphatase activity, of PTEN appears to play a crucial role in regulation of γH2AX status in response to DNA damage in vivo.
Figure 5. The protein phosphatase activity of PTEN is required for DSBs repair in vivo.
(A and B) PtenC124S/+ MEFs displayed higher levels of γH2ax at 24 h post γ-irradiation (IR) treatment compared with Pten+/+ and PtenG129E/+ MEFs. IB analysis of WCL derived from Pten+/+, PtenG129E/+ and PtenC124S/+ MEFs, which were treated with IR (5 Gy) at indicated time points before harvesting (A). Quantification of protein intensity was performed using the ImageJ software (B). γH2ax immunoblot bands were normalized to Vinculin, and then normalized to the control (no IR treatment).
(C and D) Representative immunohistochemistry (IHC) analysis of spleen tissues derived from Pten+/+, PtenG129E/+ and PtenC124S/+ mice (1th group), which were treated with IR (3 Gy) and sacrificed at 24 h after irradiation (C). Scale bar, 50 μm. IB analysis of the sample was performed using indicated antibodies (D). Four mice each group.
(E) PtenY138L/+ MEFs displayed higher levels of γH2ax at 8 or 24 h post IR treatment compared with Pten+/+ MEFs. IB analysis of WCL derived from Pten+/+ and PtenY138L/+ MEFs, which were treated with IR (5 Gy) at indicated time points before harvesting.
NSD2-mediated methylation of PTEN at K349 dictates cellular sensitivity to DNA-damaging agents
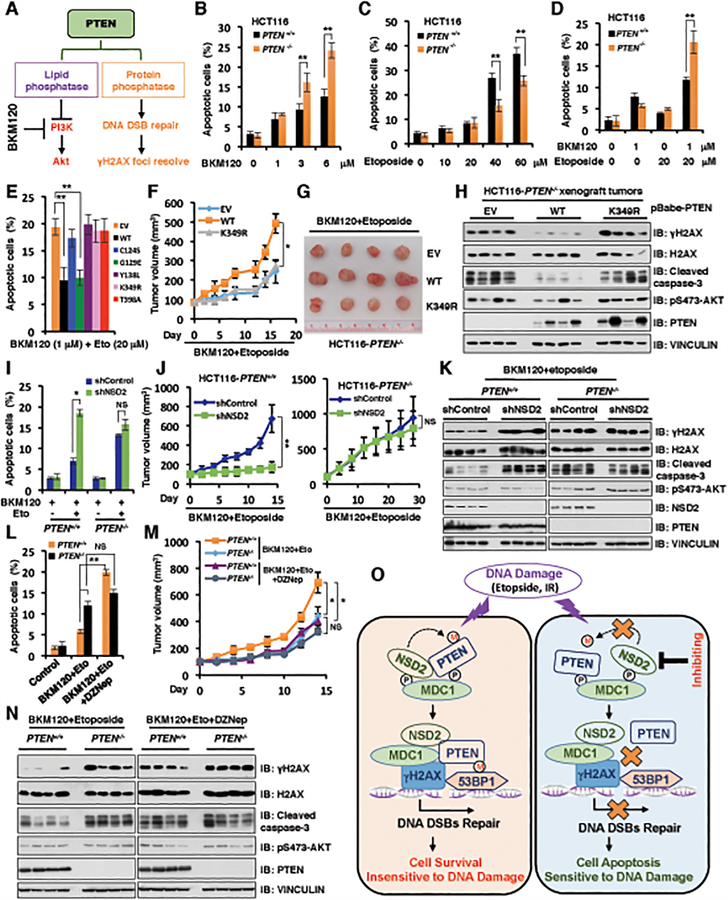
Given the critical role of DSBs repair in conferring cell survival and apoptosis signaling, we further explored if the di-methylation status or protein phosphatase activity of PTEN plays important roles in regulating cell survival upon DNA-damaging agents. As PTEN has both lipid and protein phosphatase activities, to study PTEN protein phosphatase activity on DNA damage-induced cell survival and apoptosis, we utilized BKM120 (47), a pan-PI3K inhibitor to block the PTEN lipid phosphatase activity pathway (Fig. 6A). To identify the lowest dose for BKM120 combination with etoposide or IR treatment, we used a gradient dose for BKM120, etoposide or IR treatment for HCT116 with PTEN+/+ and PTEN−/− cells. Compared to PTEN+/+ cells, although PTEN−/− cells were relatively more sensitive to the PI3K inhibitor, BKM120 at high doses (3 and 6 μM) for 72 hours treatment, we did not observe significant apoptotic differences between PTEN+/+ and PTEN−/− HCT116 cells upon low dose BKM120 (1μM) single reagent treatment (Fig. 6B). Moreover, there was no significant difference in the sensitivity to the low dose of etoposide (10 and 20 μM) or IR (2 Gy) treatment between PTEN+/+ and PTEN−/− HCT116 cells (Fig. 6C and Supplementary Fig. S6A). However, compared to PTEN+/+ HCT116 cells, PTEN−/− HCT116 cells became more resistant to the high dose of etoposide (40 or 60 μM) or IR treatment (5 or 10 Gy) (Fig. 6C and Supplementary Fig. S6A), which might be due to the elevated Akt oncogenic activity in PTEN deficient cells.
Figure 6. NSD2-mediated methylation of PTEN at K349 dictates cellular sensitivity to DNA-damaging agents.
(A) A schematic model to illustrate that PTEN has lipid phosphatase and protein phosphatase activity, which involves in PI3K/Akt signaling pathway in cytoplasm and DNA DSB repair/γH2AX pathway in nucleus, respectively.
(B and C) PTEN+/+ and PTEN−/− HCT116 cells were treated with increased concentrations of BKM120 (B) for 72 h or etoposide (C) for 48 h, cells were harvested for cell apoptosis assays. Data are represented as mean ± s.d., n = 3, and **p < 0.01 (Student’s t-test).
(D) PTEN+/+ and PTEN−/− HCT116 cells were pre-treated with 1 μM BKM120 for 24 h followed by additional etoposide (20 μM) treatment for 48 h, cells were harvested for cell apoptosis assays. Data are represented as mean ± s.d., n = 3, and **p < 0.01 (Student’s t-test).
(E) PTEN−/− HCT116 cells reconstituted with the indicated PTEN constructs were pre-treated with 1 μM BKM120 for 24 h followed by additional etoposide (20 μM) treatment for 48 h. Cells were harvested for cell apoptosis assays. Data are represented as mean ± s.d., n = 3, and **p < 0.01 (Student’s t-test).
(F-H) Tumor xenograft assays were performed by subcutaneous injection of PTEN−/− HCT116 cells stably expressing PTEN WT, K349R and empty vector (EV). Tumor growth rate in nude mice treated every other day with combination of etoposide (20 mg/kg) and BKM120 (25 mg/kg) was shown (F). Tumors were dissected and recorded after euthanizing the mice (G). IB analysis of the samples was performed using indicated antibodies (H). Four mice each group. *p < 0.05 (Student’s t-test).
(I) PTEN+/+ and PTEN−/− HCT116 cells stably depleting NSD2 by shRNA (with shControl as a negative control) were pre-treated with 1 μM BKM120 for 24 h followed by additional etoposide (20 μM) treatment. 48 h post-treatment, cells were harvested for cell apoptosis assays. Data are represented as mean ± s.d., n = 3, and *p < 0.05 (Student’s t-test).
(J and K) Tumor xenograft assays were performed by subcutaneous injection of PTEN+/+ and PTEN−/− HCT116 cells stably expressing shRNA against NSD2 or shControl as a negative control. Tumor growth rate in nude mice treated every other day with a combination of etoposide (20 mg/kg) and BKM120 (25 mg/kg) was shown in (J). Tumors were dissected after euthanizing the mice and were analyzed by IB with indicated antibodies (K). Statistical analysis of tumor volumes showed significant differences in mean tumor volumes between the shNSD2 and the shcontrol groups. Four mice each group. *p < 0.05, **p < 0.01, NS indicates no significant difference (Student’s t-test).
(L) PTEN+/+ and PTEN−/− HCT116 cells were pre-treated with 1 μM BKM120 for 24 h followed by additional etoposide (20 μM) treatment. 48 h post-treatment, cells were harvested for cell apoptosis assays. Data are represented as mean ± s.d., n = 3. *p < 0.05, **p < 0.01, NS indicates no significant difference (Student’s t-test).
(M and N) Tumor xenograft assays were performed by subcutaneously implanting PTEN+/+ and PTEN−/− HCT116 cells. Tumor growth rate in nude mice treated every other day with a combination of etoposide (20 mg/kg) and BKM120 (25 mg/kg) with DZNep (1 mg/kg) (or with vehicle as a negative control) was shown (M). Tumors were dissected after euthanizing the mice and were analyzed by IB using indicated antibodies (N). Four mice each group. *p < 0.05, NS indicates no significant difference (Student’s t-test).
(O) A schematic model to show the molecular mechanism of NSD2-mediated methylation of PTEN involving in DNA damage repair. DNA damaging agents promote NSD2-mediated di-methylation of PTEN, which is recognized by the 53BP1 tudor domain to recruit PTEN on DNA damage sites and help DNA DSBs repair largely through dephosphorylating γH2AX (left panel). However, inhibiting NSD2 could suppress PTEN di-methylation at K349 and its recruitment on DNA damage sites, which subsequently leads to DSBs repair deficiency and sensitizes cancer cells to DNA damaging agents (right panel). P: phosphorylation; M: methylation.
In keeping with this notion, we found that although low concentration of BKM120 (1 μM), etoposide (20 μM) or IR (2 Gy) single reagent treatment for 72 hours did not cause significant apoptotic difference between PTEN+/+ and PTEN−/− HCT116 cells, PTEN−/− HCT116 cells displayed an elevated sensitivity to low dose treatment of BKM120 (1 μM) in combination with etoposide (20 μM) or IR (2 Gy) as shown in both apoptosis and cell viability assays (Fig. 6D and Supplementary Fig. S6B–S6C). However, the low concentration of BKM120 (1 μM) did not affect the etoposide-induced phosphorylation and methylation of PTEN as well as PTEN interaction with MDC1 or NSD2 in cells (Supplementary Fig. S6D–S6G). Moreover, BKM120 treatment could not affect the PTEN protein phosphatase activity towards γH2AX in vitro (Supplementary Fig. S6H). However, BKM120 (1 μM) could slightly induce the nuclear localization of PTEN (Supplementary Fig. S6I and S6J). Hence, in the remainder of experiments we choose the low concentration of BKM120 (1μM) in combination with etoposide (20 μM) or IR (2 Gy) treatment.
To further pinpoint the methylation status of PTEN as well as the PTEN protein-phosphatase activity involvement in this process, we monitored the chemo- or radio-sensitivity of PTEN−/− HCT116 cells stably expressing WT, C124S, G129E, Y138L, K349R, T398A, as well as the empty vector as a negative control. Notably, consistent with their deficiency in resolving DSBs foci (Fig. 4G and 4H), compared to cells with expression of WT-PTEN or a lipid-phosphatase dead G129E mutant, cells expressing the protein-phosphatase dead mutant C124S or Y138L, or methylation-deficient mutant K349R or T398A, were more sensitive to combined treatment of BKM120 with DNA damaging agents including etoposide or IR in different cancer cell lines including HCT116, U87MG and PC3 (Fig. 6E; Supplementary Fig. S6K–P).
More importantly, compared with PTEN-WT expressing cells, combined treatment with BKM120 and etoposide displayed a greater inhibition of xenograft tumor growth in tumors bearing PTEN−/− HCT116 cells stably expressing the methylation-deficient mutant K349R, accompanied with elevated levels of γH2AX and cleaved caspase 3 in tumors with the PTEN-K349R (Fig. 6F–6H). In further support of a critical role of NSD2-mediated PTEN di-methylation in dictating cellular sensitivity to DSB agents in this experimental setting, we observed that in PTEN+/+, but not PTEN−/− HCT116 cells, additional depletion of NSD2 could greatly retard cancer cell growth in in vitro cell culture system as well as in vivo xenograft tumor growth following combined treatment of BKM120 with etoposide, coupled with increased γH2AX and caspase-3 cleavage (Fig. 6I–6K; Supplementary Fig. S6Q–S6S). We further observed that inhibition of NSD2 using the pan-methyltransferase inhibitor, DZNep (29,30), also led to a marked elevation in cellular sensitivity to treatment with BKM120 together with etoposide or IR in PTEN+/+, but not in PTEN−/− HCT116 cells (Fig. 6L–6N; Supplementary Fig. S6T–S6X). Taken together, these results coherently demonstrate that NSD2-mediated PTEN methylation plays a critical role in dictating cellular sensitivity to combination treatment with DSB inducing agents and PI3K inhibitors (Fig. 6O and Supplementary Fig. S6Y).
DISCUSSION
Beyond the well-studied function of PTEN in the cytoplasm, increasing evidence demonstrated that PTEN also plays an important role in the extracellular environment through secretion (48,49) and regulating DNA damage repair in the nucleus (10,11). Specifically, DNA DSBs induced the phosphorylation of PTEN at T/S398 by the ATM kinase (11). However, the physiologic consequence of this event remains largely elusive. In this study, we identified that phosphorylation of PTEN at T/S398 is required for binding with MDC1 BRCT domain, which also recognized γH2AX upon DNA damage signaling (50). Furthermore, we found that DNA damaging agents could induce a complex formation between PTEN and the NSD2 methyltransferase, which is largely dependent on MDC1. Subsequently, our results showed that NSD2 promotes PTEN di-methylation at K349, which is read by the tudor domain of 53BP1 to recruit PTEN into DNA damage sites.
Although it has been reported that PTEN has lipid and protein dual phosphatase activity (34,51), previous studies have been predominantly focused on its lipid phosphatase activity toward phosphatidylinositol-3,4,5-trisphosphate (PIP3) to antagonize the PI3K-AKT oncogenic signaling pathway. The protein phosphatase activity of PTEN has not been well recognized. However, genetic evidence revealed that mice with Pten deficiency and transgenic expression of activated AKT displayed development of different tumor types (52,53). These reports indicate that in addition to the lipid phosphatase activity, PTEN should have other critical biological functions, such as protein phosphatase activity. In support of this notion, we found that the protein phosphatase activity of PTEN is critical for DNA DSBs repair. Compared to PTEN+/+ cells, the magnitude and duration of γH2AX in PTEN−/− cells are dramatically elevated, especially at the late stage post γ-irradiation, which can be largely rescued by re-introducing WT or lipid-phosphatase deficient mutant (G129E), but not protein-phosphatase deficient mutants (C124S and Y138L).
Consistent with these findings, compared with Pten+/+ and PtenG129E/+ mice, the levels of γH2ax were upregulated in PtenC124S/+ mice at 24 hours post γ-irradiation. More importantly, we also showed that PTEN could interact with and dephosphorylate γH2AX after treatment with DNA damaging agents, which is required for NSD2-mediated di-methylation of PTEN at K349. As previous reports demonstrated that the phosphatases PP2A and PP4 directly dephosphorylate γH2AX to facilitate DNA damage repair (54,55), it warrants further in-depth study to explore how these different protein phosphatases are involved in dissolving γH2AX foci to control the process of DSBs repair. Previous studies showed that PTEN could be modified by other post-translational modifications including the mono-ubiquitination at K289 and SUMOylation at the K254 and K266 to control PTEN nuclear import or retention (11,46,56). Moreover, Bassi C. et al found that SUMOylation of PTEN at K254 is critical for its nuclear retention and the SUMOylated PTEN was rapidly excluded from the nucleus upon activation of DNA damage signaling (11). On the other hand, the studies from other two groups showed that the SUMOylation of PTEN at K254 and K266 sites are required for PTEN’s plasma membrane localization to suppress the PI3K-Akt signaling pathway (56,57). Our results showed that the mono-ubiquitination deficient PTEN-K289R mutant, but not the SUMOylation deficient PTEN-K266R could slightly decrease the NSD2-mediated di-methylation of PTEN and subsequently reduce PTEN interaction with the 53BP-tudor domain (Fig. 2C and Fig. 3C). Additionally, recent reports demonstrated that WWP1-mediated K27-linked ubiquitination and PRMT6-mediated arginine methylation of PTEN regulate PTEN lipid phosphatase activity towards the PI3K-Akt signaling oncogenic pathway and tumorigenesis (5,58), whereas FGFR2-mediated tyrosine phosphorylation of PTEN controls DNA damage repair and radiation sensitivity in glioblastoma in part via governing the chromatin localization of PTEN (59). However, the cross-talk between our finding that PTEN lysine methylation and other post-translational modifications including SUMOylation and tyrosine phosphorylation under the DNA damaging conditions remains elusive.
Given that NSD2 is frequently overexpressed or mutated to enhance its catalytic activity in many types of human cancers (25,60), and overexpression of NSD2 facilitates tumorigenesis in mouse models, emerging evidence suggests NSD2 as a potential therapeutic target in cancer (61,62). Moreover, previous studies demonstrated that the NSD2 enhances the DNA DSBs repair, Ig class switch recombination (CSR) and resistance to DNA damaging agents largely through the NSD2 methyltransferase activity and its ability of recruiting DNA damaging factors into DNA damage sites (23,24,32,63), which indicates that pharmacological suppression of NSD2 might synergize with DNA damaging agents for cancer therapy. Our results also suggest that the development of NSD2 specific inhibitor(s), which if paired in combination with PI3K inhibitors, could efficiently sensitize cancer cells with wild-type PTEN genetic status to chemo- or radio-therapeutics (Supplementary Fig. S6Y). However, although two NSD2 specific inhibitors LEM-06 and LEM-14 were developed for suppressing the NSD2 activity in vitro (64,65), we did not confirm the biological effects of both inhibitors in cell culture system as evidenced by no obvious changes in H3K36me2 upon treatment with either LEM-06 or LEM-14 in U2OS or HCT116 cells, which warrants further study to develop and optimize NSD2 selective inhibitors for cancer therapy.
Taken together, our study uncovers a critical role of NSD2-mediated PTEN methylation in regulating DNA DSBs repair, which dictates cellular sensitivity to DNA damaging-agents including both chemo- and radio-therapeutics, in part by governing the DSB repair process via dephosphorylating γH2AX. DNA damaging agents induce MDC1, NSD2 and PTEN forming a tertiary complex and promote NSD2-mediated di-methylation of PTEN, which can be recruited to DNA damage sites by the 53BP1 tudor domain and help DNA DSB repair largely through dephosphorylating gH2AX (Fig. 6O, left panel). However, inhibiting NSD2 methyltransferase activity using shRNA or inhibitor can decrease PTEN di-methylation at K349 and subsequently retard 53BP1-mediated recruitment to DNA damage sites, which causes DNA DSBs repair deficiency and sensitizes cancer cells to DNA damaging agents (Fig. 6O, right panel). This work therefore sheds new light into the precise molecular mechanism by which nuclear PTEN controls DNA damage pathway in a methylation- and protein phosphatase-dependent manner.
METHODS
Cell culture, transfections and viral infections
HEK293T, HEK293, U2OS, U87MG, Mouse embryonic fibroblasts (MEFs) and NIH3T3 were cultured in DMEM medium supplemented with 10% FBS (Gibco), 100 units of penicillin and 100 μg/ml streptomycin (Gibco), which were obtained from Dr. William G. Kaelin, Jr. (Dana-Farber Cancer Institute), in June 2006. HCT116, HCT116 PTEN+/+ and PTEN−/− cells were obtained from Dr. Todd Waldman (School of Medicine, Georgetown University) in June 2014. MEFs Mdc1+/+ and Mdc1−/− cells were obtained from Dr. Anyong Xie (The Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, China) in July 2015. MEFs Nsd2+/+ and Nsd2−/− (also named Whsc1+/+ and Whsc1−/− cells) were obtained from Dr. Keiko Ozato (NICHD, National Institutes of Health) in July 2016. DU145, PC3, Pten+/+, PtenG129E/+ and PtenC124S/+ MEFs were from the lab of Dr. Pier Paolo Pandolfi in 2018. Pten+/+ and PtenY138L/+ MEFs were obtained from the lab of Dr. Nicholas R. Leslie in October 2017. H2ax−/− cells were obtained from the lab of Dr. Yang Shi in November 2018. DR-GFP-U87MG cells were gifts from the lab of Dr. Vuk Stambolic at University of Toronto in January 2015. Cell lines were obtained between 2006 and 2018, which were routinely tested negative for Mycoplasma. However, the cell line authentication was not routinely performed. Cell transfection was performed using lipofectamine and plus reagents. Packaging of lentiviral, retroviral cDNA expressing viruses, as well as subsequent infection of various cell lines were performed according to the routine protocols in our lab. Following viral infection, cells were selected in the presence of hygromycin B (200 μg/ml) or puromycin (1 μg/ml) for 3 days.
Plasmids and NSD2 shRNAs
HA-PTEN and HA-Pten were generated by inserting the corresponding cDNAs into pcDNA3-HA vector. Flag-NSD2, Flag-EZH2 and Flag-Set8 were constructed by cloning corresponding cDNAs into pFlag-CMV vector. pCMV-GST-MDC1-BRCT, pCMV-GST-53BP1-BRCT, pCMV-GST-BRCA1-BRCT and pCMV-GST-53BP1 tudor domain were cloned into mammalian expression GST-fusion vectors. GST-PTEN was constructed by inserting the cDNA into pGEX-4T-1 vector. pBabe-Super-HA-PTEN was constructed by subcloning the PTEN cDNA into pLenti-HA-puro and pBabe-Super-HA-hygro vector. Various PTEN mutants were generated using the QuikChange XL Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer’s instructions. All mutants were generated using mutagenesis PCR and the sequences were verified by DNA sequencing. pcDNA5-FRT/T0-Flag-53BP1 (#52507), 3xFlag-pcDNA5-FRT/T0–53BP1 28A (#52505) and pSpCas9(BB)-T2A-Puro_h53BP1_gRNA_D were purchased from Addgene. Flag-MDC1 WT and Δ1–143 mutant were gifts from the lab of Dr. Xingzhi Xu at the Shenzhen University. pLentilox3.7-shRNA vectors to deplete endogenous NSD2 were kind gifts from Dr. Zhenkun Luo in the Department of Oncology at Mayo Clinic. The sequences for NSD2 shRNA were listed below: NSD2 shRNA 1: 5’-GCACGCTACAACACCAAGTTT; NSD2 shRNA 2: 5’-GCACAGTCTTCGGAAGAGAGACACAATCA. SgPTEN-F: CACCGGCATATTTATTACATCG; sgPTEN-R: AAACCGATGTAATAAATATGCC.
Antibodies and Reagents
All antibodies were used at a 1:1000 dilution in TBST buffer with 5% non-fat milk for western blotting. Anti-phospho-ATM/ATR Substrate (S*Q) (9607), anti-phospho-ATM/ATR Substrate (6966), anti-pS15-p53 (9284), anti-pT68-Chk2 (2661), anti-Chk2 antibody (3440), anti-Di-Methyl Lysine Motif (14117), anti-Tri-Methyl Lysine Motif (14680), anti-His tag (2366), anti-γH2AX (9718), anti-pS473-Akt (4060), anti-Akt1 antibody (2938), anti-PTEN (9188), anti-Cleaved caspase3 (9661), anti-Mek1/2 (8727), anti-pS25/29–53BP1 (2674) and anti-H2AX (7631) antibodies were purchased from Cell Signaling Technology. Anti-PTEN (sc-7974), anti-MDC1 (sc-27737), anti-p53 (sc-6243/126), anti-ATM (sc-23921), anti-ATR (sc-1887), anti-pCREB (sc-81486), anti-Lamin A/C (sc-7293), anti-GAPDH (sc-47724), anti-BRCA1 (sc-6954), anti-53BP1 (sc-22760), anti-Rad51 (sc-398587), anti-HA (sc-805) and anti-GST (sc-459) antibodies were obtained from Santa Cruz. Anti-Flag (F-2425), anti-Flag (F-3165, clone M2), anti-Tubulin antibody (T-5168), anti-Vinculin (V9131), anti-BRCA1 (07434), anti-Flag agarose beads (A-2220), anti-HA agarose beads (A-2095), peroxidase-conjugated anti-mouse secondary antibody (A-4416) and peroxidase-conjugated anti-rabbit secondary antibody (A-4914) were purchased from Sigma. Anti-HA (MMS-101P) was obtained from Covance. Anti-MMSET/NSD2 (ab75359) and anti-MDC1 (ab41951 and ab11171) were purchased form the Abcam. Anti-53BP1 (612523) was obtained from the BD Biosciences.
The polyclonal anti-PTEN-K349-me2 and anti-PTEN-K349-me3 antibodies generated by Cell Signaling Technology were derived from rabbit. The antigen peptide sequence comes from 10 amino acids surrounding the modification site (K349) of human PTEN. The antibodies were affinity purified using the antigen peptide column, but they were not counter selected on unmodified antigen. Therefore, we validated these antibodies by different methods, including dot blotting assays with synthetic PTEN peptides with mono-methylation, di-methylation and tri-methylation modification at K349, PTEN K349R mutant, as well as reaction products derived from methylation assays in cells.
Etoposide (E1383) was purchased from Sigma. KU-55933 (ATM Kinase Inhibitor, S1092), VE-821 (S8007), BKM120 (S2247) and 3-deazaneplanocin A (DZNeP) HCl (S7120) were purchased from Selleckchem.
Immunoblot (IB) and immunoprecipitations (IP) analyses
Cells with indicated treatments were lysed in EBC buffer (50 mM Tris pH 7.5, 120 mM NaCl, 0.5% NP-40) supplemented with protease inhibitors (Complete Mini, Roche) and phosphatase inhibitors (phosphatase inhibitor cocktail set I and II, Calbiochem). The protein concentrations of whole cell lysates were measured by the Beckman Coulter DU-800 spectrophotometer using the Bio-Rad protein assay reagent. Equal amounts of whole cell lysates were resolved by SDS-PAGE and immunoblotted with indicated antibodies. For immunoprecipitations analysis, 1000 μg lysates were incubated with the indicated antibody-conjugated beads for 4 h at 4 °C. The recovered immuno-complexes were washed four times with NETN buffer (20 mM Tris, pH 8.0, 150 mM NaCl, 1 mM EDTA and 0.5% NP-40) before being resolved by SDS-PAGE and immunoblotted with indicated antibodies. For the endogenous PTEN methylation assays, EBC and NETN buffer containing 300 mM NaCl were used to disrupt the non-specific interacting proteins.
In vitro dephosphorylation assays
For measuring the dephosphorylation of γH2AX by PTEN, the GST-fusion PTEN WT, C124S, G129E and Y138L, as well as GST protein were purified from E. coli BL21 as described previously (34). γH2AX (Cat # 12–0040), H2AX (Cat # 12–0039) peptides from EpiCypher or full length γH2AX purified from the HCT116 cells after etoposide treatment as well as the positive control pS133-CREB from HCT116 cells through using the anti-γH2AX or anti-pS133-CREB antibody served as substrates. The dephosphorylation assays were performed in phosphatase assay buffer (20 mM HEPES, pH 7.2, 100 mM NaCl and 3 mM DTT). The reactions were incubated at 37 °C for 30 min with or without the addition of recombinant GST-fusion PTEN WT, C124S, G129E and Y138L, as well as GST protein as negative control, and were stopped by adding 3 × SDS loading buffer for western blot.
Peptide-binding assays
PTEN peptides with/without methylation modifications were synthesized at Tufts Medical School. Each contained an N-terminal biotin and free C-terminus and was synthesized in 0.1 mM scale. Peptides were diluted into 1 mg/ml for further biochemical assays. The sequences were listed below:
PTEN WT: Biotin-NFKVKLYFTKTVEEPSNPE; PTEN K349_mono-methylation (K-me1): Biotin-NFKVKLYFTK(me1)TVEEPSNPE; PTEN K349_Di-methylation (K-me2): Biotin-NFKVKLYFTK(me2)TVEEPSNPE; PTEN K349_Tri-methylation (K-me3): Biotin-NFKVKLYFTK(me3)TVEEPSNPE.
Peptides (2 μg) were incubated with 1 mg of whole cell lysates in a total volume of 500 μl EBC buffer. After incubation for 4 h at 4°C, 10 μl Streptavidin agarose (Thermo Scientific 20353) was added in the sample for another 1 h. The agarose was washed four times with NETN buffer. Bound proteins were added in 2 × SDS loading buffer and resolved by SDS-PAGE for immunoblot analysis.
Immunohistochemistry (IHC) assays
Spleen tissues from 4 month-old mice (Pten+/+, PtenG129E/+ and PtenC124S/+ mice) were dissected and fixed in 4% paraformaldehyde for IHC analysis. For staining, tissues were fixed in 4% paraformaldehyde overnight, paraffin embedded, and then sectioned at 5 μm. After deparaffinization and rehydration, antigen retrieval was performed in a pressure cooker with sodium citrate buffer at 95°C for 25 minutes. Sections were incubated in a 0.3% H2O2 solution in 1× PBS, and then a 10% serum solution in 1 × PBS for 30 minutes each solution was used to block endogenous peroxidase and background from the secondary antibody, respectively. The sections were stained with the γH2ax (Cell Signaling #9718, 1:500) in 1 × PBS at 4°C overnight, and incubated in a biotinylated anti-rabbit secondary antibody in 1 × PBS (1:1000) at room temperature for 30 minutes. The Vectastain ABC Elite kit was used to enhance specific staining, and the staining was visualized using a 3’-diaminobenzidine (DAB) substrate. Stained sections were counterstained using hematoxylin and dehydrated before they were sealed with a coverslip with Richard-Allan Scientific® Cytoseal™ XYL Mounting Medium. Stained slides were visualized by a bright-field microscope.
γ-irradiation (IR) treatments
Cells with 60% confluence were treated with irradiation (5 Gy). After IR treatment, cells were left to recover at the incubator for the indicated times. Total-body irradiation (3 Gy) of 4 month-old mice (Pten+/+, PtenG129E/+ and PtenC124S/+) was performed, after which the mice were caged separately before being sacrificed at 24 h after treatment. Moreover, these mice were carefully monitored during the recovery period.
COMET assays
The COMET assay was performed by following the manufacture’s instruction (Comet SCGE Assay Kit ADI-900–166). Briefly, HCT116-PTEN-WT and K349R cells were treated with DMSO or Etoposide for the indicated time points, respectively. Cells were then gently trypsinized and pelleted by centrifuge. Cells were resuspended at 1 × 105 cells/ml in cold PBS, followed by combining cells with molten LMAgarose at a ratio of 1:10 (V/V) and immediately pipetting 75 μl onto Comet Slide. Slide was placed flat at 4°C in the dark for 10 min, and immersed in prechilled lysis solution, and left on ice for 30 min. Excess buffer was tapped off from slide. The slide was immersed in freshly prepared Alkaline solution. Comet Slide was left in Alkaline solution for 30 min at room temperature, then transferred to a horizontal electrophoresis apparatus to perform TBE electrophoresis. Samples were dried and placed in 100 μl of diluted stain onto each circle of dried agarose and stained for 30 min in the dark. Images were recorded by epifluorescence microscopy and analyze the data by CometScore.
Cellular fractionation
U2OS, NIH3T3 or MEFs cultured in the 6 cm dishes with 70%−80% confluence were harvested. Cell pellets were resuspended in 8 volumes of the Buffer A (hypotonic: 10 mM Hepes pH7.9, 0.2 mM EDTA, 0.2 mM EGTA, 1 mM DTT, 1 × protease inhibitors and 1 × protein phosphatase inhibitors). The cells in Buffer A were incubated on ice for 10 min and added 10% Triton X-100 to the final concentration of 0.1%, followed by vortex 15 seconds. After centrifuge at 7500 rpm for 5 min, the supernatant was transferred into new tube as the cytoplasmic part. The remaining pellets were washed twice with the Buffer A and resuspended in 2–5 volumes of the Buffer B (40 mM Hepes pH7.9, 0.7 M KCl, 10% glycerol, 2 mM EDTA, 2 EGTA, 1 mM DTT, 1xprotease inhibitors and 1xprotein phosphatase inhibitors). The pellets in Buffer B were incubated at 4°C onto shaking platform for 15 min. After centrifuge at 14000 rpm for 10 min, the supernatant was transferred into new tube as the nucleoplasmic part. The remaining pellets were sonicated as the chromatin-associated part.
Cell viability and apoptosis assays
For cell viability assays, 2000 cells per well were plated in 96-well plates, and incubated with complete DMEM medium containing different treatments as indicated. Assays were performed with the Cell Titer-Glo Luminescent Cell Viability Assay Kit according to the manufacturer’s instructions (Promega). For detection of apoptosis, cells treated with indicated treatment were co-stained with Annexin-V-PE and 7-AAD (Annexin V-PE Apoptosis Detection Kit I, BD Bioscience) and analyzed using BD FACSCanto flow cytometer according to the manufacturer’s instructions.
Murine models
The Beth Israel Deaconess Medical Center IACUC Committee on Animal Research approved all animal experiments. The transgenic mice used in this study (Pten+/+, PtenC124S/+, and PtenG129E/+) were reported previously (46). Four mice per genotype were randomly chosen and analyzed at the indicated age.
Xenograft tumor growth
For the xenograft model, 6-week-old female nude mice housed in specific pathogen-free environments were injected s.c. with 1.0 × 106 HCT116 derivatives mixed with DMEM medium and Matrigel (vol/vol, 1:1). Treatments were initiated when the mean tumor volume in each randomized group reached around 100 mm3. NVP-BKM120 (MedChem Express) was formulated in 0.5% methylcellulose/0.5% Tween 80) at 6 mg/ml. Etoposide (Selleckchem) was formulated in Saline at 10 mg/ml. For BKM120, treatments were carried out orally, every other day, using an application volume 25 mg/kg. For etoposide, treatments were carried out by I.P. injection with 20 mg/kg every other day. DZNep treatments were carried out by I.P. injection with 1 mg/kg body weight twice a week for 2 weeks. Animals were sacrificed when the tumor size in the vehicle control group reached 500 to 600 mm3. Tumor volumes were determined using calipers. All experimental procedures strictly complied with the IACUC guidelines.
Statistical Analysis
Group data are presented as mean ± s.d. The statistical significance between experimental groups was determined by Student’s t-test. p<0.05 was considered to be significant. Analyses were performed using the Microsoft Excel 2011 and GraphPad Prism V5.
Supplementary Material
STATEMENT OF SIGNIFICANCE.
NSD2-mediated di-methylation of PTEN is recognized by the 53BP1 tudor domain to facilitate PTEN recruitment into DNA damage sites, governing efficient repair of DNA DSBs. Importantly, inhibiting PTEN methylation sensitizes cancer cells to combinatorial treatment of PI3K inhibitor with DNA-damaging agents in both cell culture and in vivo xenograft models.
ACKNOWLEDGEMENTS
We thank Drs. Brian North, Jianping Guo, Xiangpeng Dai, Tao Zhang and other Wei lab members for critical reading of the manuscript, and members of the Wei and Pandolfi laboratories for discussions. We thank the Drs. Yasheng Zhu and Zhiqiang Ma for helping to prepare the buffers and gels. We also sincerely thank Dr. Nicholas Willis in Dr. Ralph Scully lab for technical support. W.W. is a Leukemia & Lymphoma Society (LLS) research scholar.
This work was supported in part by the NIH grants (W.W., GM094777 and CA177910; J.M.A., 5P01CA120964 and 1S10OD010612; J.Z., 1K99CA212292–01; W.G., R00CA207867).
Financial support: J.Z. is supported by 1K99CA212292–01. W.G. is supported by R00CA207867. W.W. is supported by R01CA177910 and R01GM094777. J.M.A. is supported by 1S10OD010612 and 5P01CA120964.
Abbreviations:
- PTEN
phosphatase and tensin homolog deleted on chromosome ten
- DSBs
DNA double-strand breaks
- PTEN-K349me2
di-methylation of K349 on PTEN
Footnotes
Conflict of interest: W.W. and P.P.P. are co-founder and stock holder of the Rekindle Therapeutics.
REFERENCES
- 1.Worby CA, Dixon JE. Pten. Annual review of biochemistry 2014;83:641–69 doi 10.1146/annurev-biochem-082411-113907. [DOI] [PubMed] [Google Scholar]
- 2.Lee YR, Chen M, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor: new modes and prospects. Nat Rev Mol Cell Biol 2018;19(9):547–62 doi 10.1038/s41580-018-0015-0. [DOI] [PubMed] [Google Scholar]
- 3.Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol 2012;13(5):283–96 doi 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- 4.Ortega-Molina A, Serrano M. PTEN in cancer, metabolism, and aging. Trends in endocrinology and metabolism: TEM 2013;24(4):184–9 doi 10.1016/j.tem.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee YR, Chen M, Lee JD, Zhang J, Lin SY, Fu TM, et al. Reactivation of PTEN tumor suppressor for cancer treatment through inhibition of a MYC-WWP1 inhibitory pathway. Science 2019;364(6441) doi 10.1126/science.aau0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen WH, Balajee AS, Wang J, Wu H, Eng C, Pandolfi PP, et al. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell 2007;128(1):157–70 doi 10.1016/j.cell.2006.11.042. [DOI] [PubMed] [Google Scholar]
- 7.Chen ZH, Zhu M, Yang J, Liang H, He J, He S, et al. PTEN interacts with histone H1 and controls chromatin condensation. Cell reports 2014;8(6):2003–14 doi 10.1016/j.celrep.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gong L, Govan JM, Evans EB, Dai H, Wang E, Lee SW, et al. Nuclear PTEN tumor-suppressor functions through maintaining heterochromatin structure. Cell cycle 2015;14(14):2323–32 doi 10.1080/15384101.2015.1044174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He J, Kang X, Yin Y, Chao KS, Shen WH. PTEN regulates DNA replication progression and stalled fork recovery. Nature communications 2015;6:7620 doi 10.1038/ncomms8620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ming M, Feng L, Shea CR, Soltani K, Zhao B, Han W, et al. PTEN positively regulates UVB-induced DNA damage repair. Cancer research 2011;71(15):5287–95 doi 10.1158/0008-5472.CAN-10-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bassi C, Ho J, Srikumar T, Dowling RJ, Gorrini C, Miller SJ, et al. Nuclear PTEN controls DNA repair and sensitivity to genotoxic stress. Science 2013;341(6144):395–9 doi 10.1126/science.1236188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER 3rd, Hurov KE, Luo J, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 2007;316(5828):1160–6 doi 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 13.Audia JE, Campbell RM. Histone Modifications and Cancer. Cold Spring Harb Perspect Biol 2016;8(4):a019521 doi 10.1101/cshperspect.a019521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones PA, Issa JP, Baylin S. Targeting the cancer epigenome for therapy. Nat Rev Genet 2016;17(10):630–41 doi 10.1038/nrg.2016.93. [DOI] [PubMed] [Google Scholar]
- 15.Carlson SM, Gozani O. Nonhistone Lysine Methylation in the Regulation of Cancer Pathways. Cold Spring Harb Perspect Med 2016;6(11) doi 10.1101/cshperspect.a026435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biggar KK, Li SS. Non-histone protein methylation as a regulator of cellular signalling and function. Nat Rev Mol Cell Biol 2015;16(1):5–17 doi 10.1038/nrm3915. [DOI] [PubMed] [Google Scholar]
- 17.Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS, et al. Regulation of p53 activity through lysine methylation. Nature 2004;432(7015):353–60 doi 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- 18.Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA, et al. Repression of p53 activity by Smyd2-mediated methylation. Nature 2006;444(7119):629–32 doi 10.1038/nature05287. [DOI] [PubMed] [Google Scholar]
- 19.Saddic LA, West LE, Aslanian A, Yates JR 3rd, Rubin SM, Gozani O, et al. Methylation of the retinoblastoma tumor suppressor by SMYD2. J Biol Chem 2010;285(48):37733–40 doi 10.1074/jbc.M110.137612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carr SM, Munro S, Zalmas LP, Fedorov O, Johansson C, Krojer T, et al. Lysine methylation-dependent binding of 53BP1 to the pRb tumor suppressor. Proc Natl Acad Sci U S A 2014;111(31):11341–6 doi 10.1073/pnas.1403737111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reinhardt HC, Yaffe MB. Phospho-Ser/Thr-binding domains: navigating the cell cycle and DNA damage response. Nat Rev Mol Cell Biol 2013;14(9):563–80 doi 10.1038/nrm3640. [DOI] [PubMed] [Google Scholar]
- 22.Sirbu BM, Cortez D. DNA damage response: three levels of DNA repair regulation. Cold Spring Harb Perspect Biol 2013;5(8):a012724 doi 10.1101/cshperspect.a012724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shah MY, Martinez-Garcia E, Phillip JM, Chambliss AB, Popovic R, Ezponda T, et al. MMSET/WHSC1 enhances DNA damage repair leading to an increase in resistance to chemotherapeutic agents. Oncogene 2016;35(45):5905–15 doi 10.1038/onc.2016.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hajdu I, Ciccia A, Lewis SM, Elledge SJ. Wolf-Hirschhorn syndrome candidate 1 is involved in the cellular response to DNA damage. Proc Natl Acad Sci U S A 2011;108(32):13130–4 doi 10.1073/pnas.1110081108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bennett RL, Swaroop A, Troche C, Licht JD. The Role of Nuclear Receptor-Binding SET Domain Family Histone Lysine Methyltransferases in Cancer. Cold Spring Harb Perspect Med 2017;7(6) doi 10.1101/cshperspect.a026708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J, Luo S, Zhao H, Liao J, Li J, Yang C, et al. Structural mechanism of the phosphorylation-dependent dimerization of the MDC1 forkhead-associated domain. Nucleic Acids Res 2012;40(9):3898–912 doi 10.1093/nar/gkr1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee JO, Yang H, Georgescu MM, Di Cristofano A, Maehama T, Shi Y, et al. Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell 1999;99(3):323–34. [DOI] [PubMed] [Google Scholar]
- 28.Sarai N, Nimura K, Tamura T, Kanno T, Patel MC, Heightman TD, et al. WHSC1 links transcription elongation to HIRA-mediated histone H3.3 deposition. EMBO J 2013;32(17):2392–406 doi 10.1038/emboj.2013.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miranda TB, Cortez CC, Yoo CB, Liang G, Abe M, Kelly TK, et al. DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Mol Cancer Ther 2009;8(6):1579–88 doi 10.1158/1535-7163.MCT-09-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fujiwara T, Saitoh H, Inoue A, Kobayashi M, Okitsu Y, Katsuoka Y, et al. 3-Deazaneplanocin A (DZNep), an inhibitor of S-adenosylmethionine-dependent methyltransferase, promotes erythroid differentiation. J Biol Chem 2014;289(12):8121–34 doi 10.1074/jbc.M114.548651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 2006;127(7):1361–73 doi 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pei H, Zhang L, Luo K, Qin Y, Chesi M, Fei F, et al. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature 2011;470(7332):124–8 doi 10.1038/nature09658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim JS, Lee C, Bonifant CL, Ressom H, Waldman T. Activation of p53-dependent growth suppression in human cells by mutations in PTEN or PIK3CA. Molecular and cellular biology 2007;27(2):662–77 doi 10.1128/MCB.00537-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Myers MP, Stolarov JP, Eng C, Li J, Wang SI, Wigler MH, et al. P-TEN, the tumor suppressor from human chromosome 10q23, is a dual-specificity phosphatase. Proc Natl Acad Sci U S A 1997;94(17):9052–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tibarewal P, Zilidis G, Spinelli L, Schurch N, Maccario H, Gray A, et al. PTEN protein phosphatase activity correlates with control of gene expression and invasion, a tumor-suppressing phenotype, but not with AKT activity. Science signaling 2012;5(213):ra18 doi 10.1126/scisignal.2002138. [DOI] [PubMed] [Google Scholar]
- 36.Shi Y, Wang J, Chandarlapaty S, Cross J, Thompson C, Rosen N, et al. PTEN is a protein tyrosine phosphatase for IRS1. Nature structural & molecular biology 2014;21(6):522–7 doi 10.1038/nsmb.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zimmermann M, Lottersberger F, Buonomo SB, Sfeir A, de Lange T. 53BP1 regulates DSB repair using Rif1 to control 5’ end resection. Science 2013;339(6120):700–4 doi 10.1126/science.1231573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Escribano-Diaz C, Orthwein A, Fradet-Turcotte A, Xing M, Young JT, Tkac J, et al. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Molecular cell 2013;49(5):872–83 doi 10.1016/j.molcel.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 39.Chapman JR, Barral P, Vannier JB, Borel V, Steger M, Tomas-Loba A, et al. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Molecular cell 2013;49(5):858–71 doi 10.1016/j.molcel.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J Cell Biol 2000;151(7):1381–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ward IM, Minn K, van Deursen J, Chen J. p53 Binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Molecular and cellular biology 2003;23(7):2556–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dedes KJ, Wetterskog D, Mendes-Pereira AM, Natrajan R, Lambros MB, Geyer FC, et al. PTEN deficiency in endometrioid endometrial adenocarcinomas predicts sensitivity to PARP inhibitors. Sci Transl Med 2010;2(53):53ra75 doi 10.1126/scitranslmed.3001538. [DOI] [PubMed] [Google Scholar]
- 43.Mendes-Pereira AM, Martin SA, Brough R, McCarthy A, Taylor JR, Kim JS, et al. Synthetic lethal targeting of PTEN mutant cells with PARP inhibitors. EMBO Mol Med 2009;1(6–7):315–22 doi 10.1002/emmm.200900041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Izhar L, Adamson B, Ciccia A, Lewis J, Pontano-Vaites L, Leng Y, et al. A Systematic Analysis of Factors Localized to Damaged Chromatin Reveals PARP-Dependent Recruitment of Transcription Factors. Cell reports 2015;11(9):1486–500 doi 10.1016/j.celrep.2015.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Panier S, Boulton SJ. Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol 2014;15(1):7–18 doi 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- 46.Papa A, Wan L, Bonora M, Salmena L, Song MS, Hobbs RM, et al. Cancer-associated PTEN mutants act in a dominant-negative manner to suppress PTEN protein function. Cell 2014;157(3):595–610 doi 10.1016/j.cell.2014.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maira SM, Pecchi S, Huang A, Burger M, Knapp M, Sterker D, et al. Identification and characterization of NVP-BKM120, an orally available pan-class I PI3-kinase inhibitor. Mol Cancer Ther 2012;11(2):317–28 doi 10.1158/1535-7163.MCT-11-0474. [DOI] [PubMed] [Google Scholar]
- 48.Hopkins BD, Fine B, Steinbach N, Dendy M, Rapp Z, Shaw J, et al. A secreted PTEN phosphatase that enters cells to alter signaling and survival. Science 2013;341(6144):399–402 doi 10.1126/science.1234907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Putz U, Howitt J, Doan A, Goh CP, Low LH, Silke J, et al. The tumor suppressor PTEN is exported in exosomes and has phosphatase activity in recipient cells. Science signaling 2012;5(243):ra70 doi 10.1126/scisignal.2003084. [DOI] [PubMed] [Google Scholar]
- 50.Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 2005;123(7):1213–26 doi 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 51.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 1997;275(5308):1943–7. [DOI] [PubMed] [Google Scholar]
- 52.Suzuki A, de la Pompa JL, Stambolic V, Elia AJ, Sasaki T, del Barco Barrantes I, et al. High cancer susceptibility and embryonic lethality associated with mutation of the PTEN tumor suppressor gene in mice. Curr Biol 1998;8(21):1169–78. [DOI] [PubMed] [Google Scholar]
- 53.Blanco-Aparicio C, Renner O, Leal JF, Carnero A. PTEN, more than the AKT pathway. Carcinogenesis 2007;28(7):1379–86 doi 10.1093/carcin/bgm052. [DOI] [PubMed] [Google Scholar]
- 54.Lee DH, Pan Y, Kanner S, Sung P, Borowiec JA, Chowdhury D. A PP4 phosphatase complex dephosphorylates RPA2 to facilitate DNA repair via homologous recombination. Nature structural & molecular biology 2010;17(3):365–72 doi 10.1038/nsmb.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chowdhury D, Xu X, Zhong X, Ahmed F, Zhong J, Liao J, et al. A PP4-phosphatase complex dephosphorylates gamma-H2AX generated during DNA replication. Molecular cell 2008;31(1):33–46 doi 10.1016/j.molcel.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang J, Yan J, Zhang J, Zhu S, Wang Y, Shi T, et al. SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane. Nature communications 2012;3:911 doi 10.1038/ncomms1919. [DOI] [PubMed] [Google Scholar]
- 57.Gonzalez-Santamaria J, Campagna M, Ortega-Molina A, Marcos-Villar L, de la Cruz-Herrera CF, Gonzalez D, et al. Regulation of the tumor suppressor PTEN by SUMO. Cell Death Dis 2012;3:e393 doi 10.1038/cddis.2012.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Feng J, Dang Y, Zhang W, Zhao X, Zhang C, Hou Z, et al. PTEN arginine methylation by PRMT6 suppresses PI3K-AKT signaling and modulates pre-mRNA splicing. Proc Natl Acad Sci U S A 2019;116(14):6868–77 doi 10.1073/pnas.1811028116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ma J, Benitez JA, Li J, Miki S, Ponte de Albuquerque C, Galatro T, et al. Inhibition of Nuclear PTEN Tyrosine Phosphorylation Enhances Glioma Radiation Sensitivity through Attenuated DNA Repair. Cancer Cell 2019;35(3):504–18 e7 doi 10.1016/j.ccell.2019.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vougiouklakis T, Hamamoto R, Nakamura Y, Saloura V. The NSD family of protein methyltransferases in human cancer. Epigenomics 2015;7(5):863–74 doi 10.2217/epi.15.32. [DOI] [PubMed] [Google Scholar]
- 61.Kuo AJ, Cheung P, Chen K, Zee BM, Kioi M, Lauring J, et al. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Molecular cell 2011;44(4):609–20 doi 10.1016/j.molcel.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mirabella F, Wu P, Wardell CP, Kaiser MF, Walker BA, Johnson DC, et al. MMSET is the key molecular target in t(4;14) myeloma. Blood Cancer J 2013;3:e114 doi 10.1038/bcj.2013.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nguyen HV, Dong J, Panchakshari RA, Kumar V, Alt FW, Bories JC. Histone methyltransferase MMSET promotes AID-mediated DNA breaks at the donor switch region during class switch recombination. Proc Natl Acad Sci U S A 2017;114(49):E10560–E7 doi 10.1073/pnas.1701366114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.di Luccio E Inhibition of Nuclear Receptor Binding SET Domain 2/Multiple Myeloma SET Domain by LEM-06 Implication for Epigenetic Cancer Therapies. J Cancer Prev 2015;20(2):113–20 doi 10.15430/JCP.2015.20.2.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shen Y, Morishita M, Lee D, Kim S, Lee T, Mevius D, et al. Identification of LEM-14 inhibitor of the oncoprotein NSD2. Biochem Biophys Res Commun 2019;508(1):102–8 doi 10.1016/j.bbrc.2018.11.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.