SUMMARY.
Purpose:
Approximately 10% of mismatch repair proficient (MMRp) colorectal cancer (CRC) patients showed clinical benefit to anti-PD-1 monotherapy (). We sought to identify biomarkers that delineate patients with immunoreactive CRC and to explore new combinatorial immunotherapy strategies that can impact MMRp CRC.
Experimental Design:
We compared the expression of 44 selected immune-related gene expression in the primary colon tumor of 19 metastatic CRC patients who responded (n=13) versus those who did not (n=6) to anti PD-1 therapy (). We define a 10 gene-based immune signature that could distinguish responder from non-responder. Resected colon specimens (n=14) were used to validate the association of the predicted status (responder and non-responder) with the immune related gene expression, the phenotype and the function of tumor-infiltrating lymphocytes (TILs) freshly isolated from the same tumors.
Results:
Although both IL-17Low and IL17High immunoreactive MMRp CRC are associated with intratumor correlates of adaptive immunosuppression (CD8/IFNγ and PD-L1/IDO1 colocalization), only IL-17Low MMRp tumors (3 out of 14) have a tumor immune microenvironment (TiME) that resembles the TiME in primary colon tumors of metastatic CRC patients responsive to anti-PD1 treatment.
Conclusions:
The detection of a preexisting anti-tumor immune response in MMRp CRC (immunoreactive MMRp CRC) is not sufficient to predict a clinical benefit to T cell checkpoint inhibitors. Intra-tumoral IL-17-mediated signaling may preclude responses to immunotherapy. Drugs targeting the IL-17 signaling pathway are available in clinic and their combination with T cell checkpoint inhibitors could improve CRC immunotherapy.
Keywords: IL-17, IDO, MSS, PD-L1, immunotherapy, checkpoint blockade
INTRODUCTION
Therapeutic antibodies inhibiting immune checkpoints, cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) and programmed cell death protein 1 (PD-1), have provided remarkable durable clinical benefit in treating cancer across histologies(1). However, the impact of these targeted therapies on colorectal cancer (CRC) remains limited(2). In our clinical trial using the anti-PD1 drug pembrolizumab to treat patients with mismatch repair (MMR)-deficient (MMRd) versus MMR-proficient (MMRp) metastatic colorectal cancer (mCRC), the overall response rate was 52% for MMRd mCRC with a 2 year progression free survival (PFS) of 53%(3). However, advanced CRC with DNA MMR deficiency represents less than 5% of the total number of mCRC patients. Although MMRp mCRC did not exhibit either high mutational density or objective responses to anti-PD-1 therapy, the disease control rate (DCR) was 12% with 3 out of 25 patients stabilizing their disease (4). Additionally, although the phase III clinical trial IMblaze370 testing the efficacy of Atezolizumab (anti-PD-L1) / cobimetinib (MEK inhibitor) combination to treat MMRp mCRC did not meet its primary endpoints(5), Bendell et al. initially reported partial responses for 7 out of 84 patients in the phase 1b clinical trial(6). Overall, these clinical results together with the findings by our group and others that a subset of MMRp colorectal tumors is characterized by a MMRd-like tumor immune environment (TiME)(7, 8), support the concept that MMRp CRC with low tumor mutational burden can trigger anti-tumor immune responses and can benefit from immune checkpoint blockade-based immunotherapy. Extensive efforts are needed to identify biomarkers that delineate these patients with immunoreactive CRC and to explore new combinatorial immunotherapy strategies that can impact MMRp CRC(2).
Herein, we performed a comparative analysis of immune-related gene expression in the TiME of primary colon tumor specimens obtained from mCRC patients who responded versus those who did not respond to anti PD-1 therapy (; clinicaltrial.gov) in an attempt to define an immune signature associated with clinical response to immunotherapy and that could ultimately help delineate immunoreactive MMRp CRC from non-immunoreactive conventional CRC. We found that a subset of MMRp CRC patients characterized by immune correlates of intratumor immunosuppression (IFNγ+PD-1hiCD8+ T cell infiltration, programmed death-ligand 1 (PD-L1)/ Indoleamine-pyrrole 2, 3-dioxygenase (IDO)-1 counter-expression on tumor cells) may be limited in their capacity to derive clinical benefit from immune checkpoint blockade because of the concomitant presence of intratumoral Th17 immunity. Our results brought evidence that a high mutational burden and/or detection of preexisting anti-tumor cytotoxic T lymphocytes (CTL) in mCRC may not be sufficient to predict clinical response to immune checkpoint blockade and other features of the TiME, including IL-17 production, may negatively alter the clinical outcome of CRC patients.
METHODS
Clinical trial, Patient selection, tumor samples.
Patients with treatment refractory progressive mCRC were recruited from three centers for this phase 2 study using pembrolizumab as previously published (4). Patients are described in Table S1. 14 microsatellite stable (MSS, also called MMRp) and 6 microsatellite instable (MSI, also called MMRd) CRC specimen were randomly selected for in situ gene expression analysis. Fresh primary sporadic CRC tissue specimens along with patient-matched normal distal colon tissue were collected at Johns Hopkins Hospital (7). Specimen are described in Table S2. The MSI status was determined as previously described and according to the revised Bethesda guidelines(7). This study was approved by the Institutional Review Board of Johns Hopkins University and was conducted in accordance with the Declaration of Helsinki and the International Conference on Harmonization Good Clinical Practice guidelines. The patients described in this study provided written informed consent and all samples were obtained in accordance with the Health Insurance and Accountability Act.
Histopathology, immunohistochemistry and image analysis.
Formalin-fixed paraformaldehyde embedded (FFPE) specimens were stained with Hematoxylin and Eosin combination, CD3 (clone PS1, Leica Biosystems, Buffalo Grove, IL), CD8 (clone C8144B, Cell Marque, Rocklin, CA) and IDO1 (clone SP260, abcam/Spring Bio) according to standard protocols. Immunohistochemistry (IHC) for PD-L1 (clone 5H1) stain and scoring technic were previously described(7). For RORγT staining, slides were baked and dewaxed followed by high pH EDTA buffer antigen retrieval for 40 min at 100°C. Endogenous peroxidase was blocked, subsequently anti- Retinoic-acid-receptor-related orphan nuclear receptor gamma T (RORγT; clone 6F3.1, Biocare Medical, LLC, Pacheco, CA) was applied for 60 min at a concentration of 0.0835 µg/mL at room temperature. Detection was performed using the Bond Polymer Refine Kit (Leica Biosystems, Buffalo Grove, IL). Slides were counterstained, dehydrated through a graded-alcohol and coverslipped using Ecomount (Biocare Medical, Walnut Creek, CA). 20X whole slides scanning used Scanscope XT and were annotated by the pathologist (RA Anders) for invasive (IF) and tumor infiltrating (TIL) areas. Digital whole image analysis was performed via the Indica Labs HALO platform and immune cells densities are quantified accordingly.
RNA extraction from FFPE tissue and Taqman quantitative RT-PCR.
FFPE tissue was obtained by laser capture microdissection (LCM) using the Leica LMD 7000 system (Leica, Buffalo Grove, Il) or Pinpoint™ slide RNA isolation procedure (Zymo Research, Irvine, CA). RNA isolation, cDNA synthesis and preamplification for Taqman RT-PCR –based gene expression analysis was previously described (7). The selected genes were previously tested and validated to distinguish the TiME of MMRp from MMRd CRC(7). Data are expressed as 2−ΔCt where ΔCt=Ctgene-Ctctrl. For our calculation Ctctrl is the average of Ct for 2 ubiquitous genes (GAPDH, GUSB). When undetectable, a value of 40 was arbitrarily assigned as Ct (higher number of amplification cycles). Details of the taqman assays (Thermofisher, Waltham, MA) are shown in Tables S3A–B.
Tumor processing and flow cytometry.
TIL were isolated from freshly dissociated tissues using an enzymatic digestion cocktail (DNase I, 2500U/ml and Liberase 400u/ml, MilliporeSigma, St Louis, MO) and a Percoll density gradient (Thermofisher)(7). Multiparameter flow cytometry and cytokine intracellular staining for IFN-γ were performed following a 3-hour in vitro stimulation in the presence of stimulation cocktail (PMA and ionomycin; Ebioscience/Thermofisher) and GolgiStop (Monensin; BD Biosciences; San Jose, CA) according to the manufacturer instructions (7). Data were analyzed using DIVA 6.1 Software (BD Biosciences).
Statistical analysis.
The gene expressions were used to build a model to predict clinical responses to anti-PD-1 coded as R (complete response / partial response) (R) and NR (no response). Gene expression levels were on the log scale and were centered across patients for each gene. Summary statistics were calculated for all genes, and compared between response status using Wilcoxon-Mann-Whitney tests. Heat maps of gene expression levels were depicted for those genes with the 20 lowest p-values based on the Wilcoxon-Mann Whitney test comparing R vs. NR. An unsupervised Gaussian mixture model of response was performed with a limited top 10 smallest p value (RORC, IL23R, LAG3, IDO1, CD4, CD274, ICOSLG, VEGFA, CD8A, MMP9). The mixture model assumed a Multivariate Gaussian distribution for each of 2 groups and assumes equal volume and shape across the 2 groups as described in Scrucca et al. (38). Assessment of the model performance in predicting response includes the sensitivity/specificity. The mixture model was first developed from trial patients, and then was used to predict response status for those patients in the untreated primary CRC cohort. CRC patients’ gene expression levels were depicted in heat maps and separated by predicted response status (Fig. S1). Individual gene expressions were further compared between the predicted response status (R v NR) of the MMRp CRC cases using the Wilcoxon-Mann-Whitney test. Statistical analyses were performed using the R statistical package (version 3.4.0). P-values were not adjusted for multiplicity.
RESULTS
Identification of immunoreactive MMRp CRC.
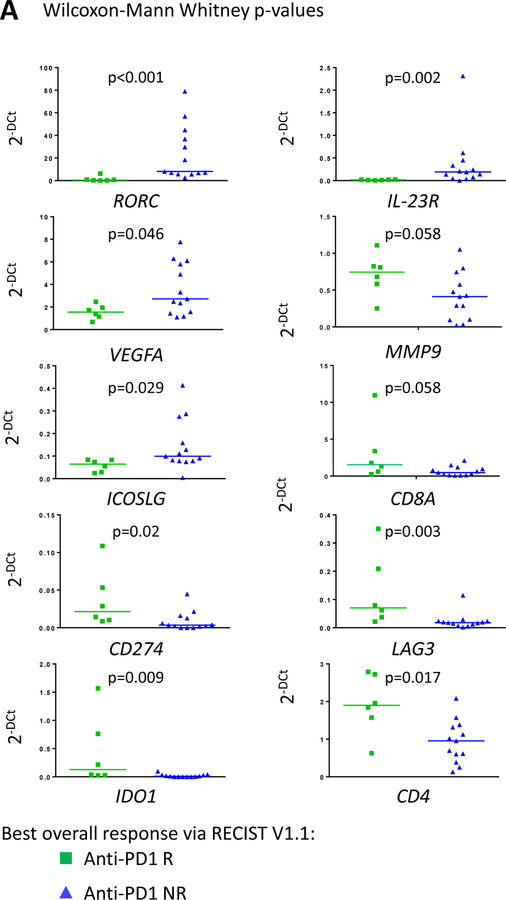
An immune related gene expression signature in the TiME of primary colon tumors of mCRC patients who responded to anti-PD-1 was used to define immunoreactive MMRp CRC patients who benefited from immune checkpoint blockade-based immunotherapy (7, 8). For this purpose, we tested the expression of 44 genes in the primary colon tumors (prior to standard of care therapy) of mCRC patients treated with pembrolizumab (Table S3A). Total RNA was extracted and analyzed in 19 primary colon tumor specimens obtained from mCRC patients treated with pembrolizumab under our study (; clinicaltrial.gov). Six patients have an objective response including complete response/partial response. Nine patients had progressive disease (PD) and 4 had stable disease (SD) (4). Samples were tested with this forty-four gene panel and then narrowed down to the top 10 differentially expressed genes (the lowest p values; median, Mann Whitney non parametric test) between responders (R) and non-responders (NR; PD and SD) mCRC patients. Significantly upregulated genes were CD4 and CD8A (p=0.017and 0.058, respectively), RORC and IL23R (p<0.001 and =0.002, respectively), CD274 (coding PD-L1), LAG3 and IDO1 (p=0.02, 0.003 and 0.009, respectively), VEGF and MMP9 (p=0.046 and 0.058, respectively), and ICOSLG (p=0.029) (Fig. 1A and Table S4 for detailed analysis). We observed that responder mCRC patients exhibited a stronger T cell infiltration (indicative of inflamed MMRp CRC) and checkpoint gene expression (indicative of adaptive immunosuppression) signatures as well as a lower Th17-associated gene expression compared to non-responder mCRC (Fig. 1A).
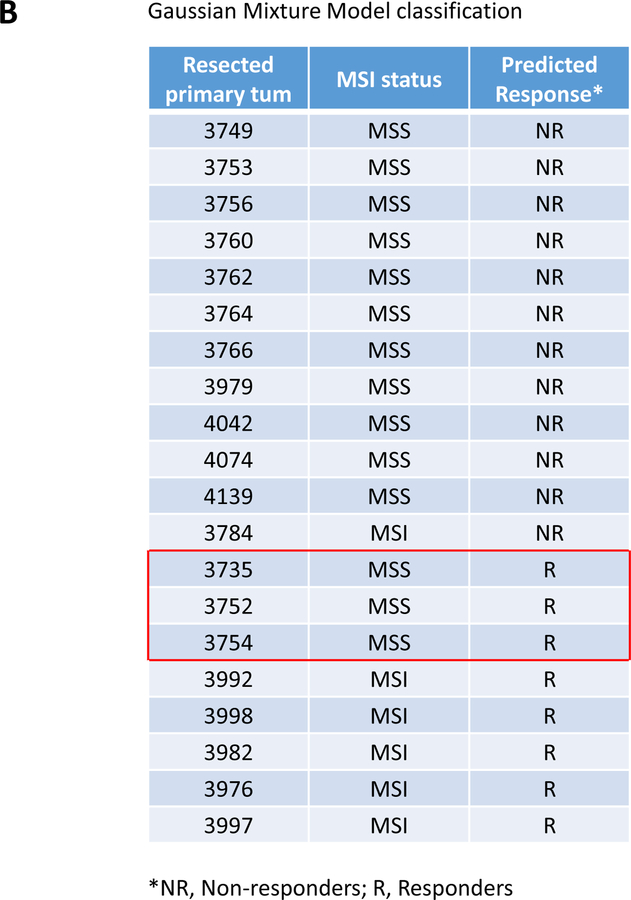
Figure 1. A 10 immune-related gene expression signature associated with clinical response to pembrolizumab treatment delineates a group of immunoreactive MMRP CRC among resected primary CRC patients.
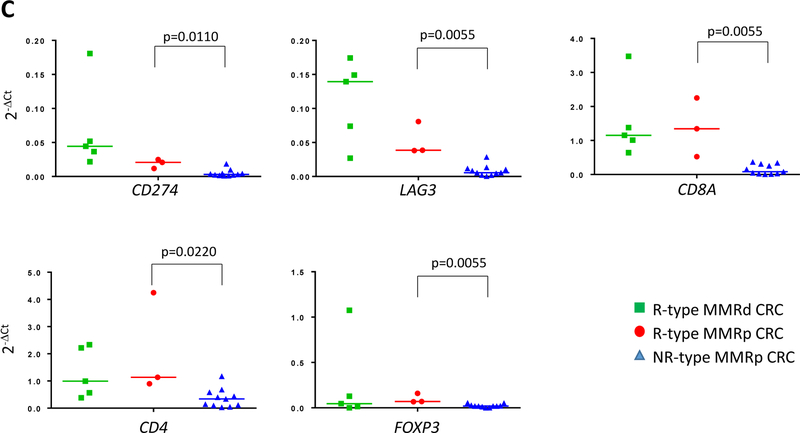
A, immune related gene expression comparison between responders (R; n=6, green squares) and non-responder (NR; n=13, blue triangles) mCRC to pembrolizumab treatment. 10/44 top genes with the lowest p values when comparing R and NR-derived primary colon tumor tissues of mCRC are shown. The full analysis including the 44 genes is shown in table S4. A Gaussian mixture model was used to model the relationship between the 10 genes and the R/NR status. The model achieved a sensitivity and specificity of 0.83 (0.36, 1.00) and 0.54 (0.25, 0.81), respectively (Fig. S1). B, the Gaussian mixture model using the 10 genes based on p-values was used to predict response status (R versus NR) in resected primary CRC specimens (untreated patients). The red open box indicates the MSS CRC (i.e MMRp) with an R-type immune signature. C, Differential gene expression between R-type MMRp (red circles) and NR-type MMRp (blue triangles) primary CRC. Gene expression analysis performed on FFPE tumor tissue showed a higher expression of T cell checkpoints (CD274, LAG3, and CTLA4), CTL (CD8A), and Treg (CD4 and FOXP3) associated genes in R-type MMRp (red circles) versus NR-type MMRp (blue triangles). MMRd CRC (green squares) are shown for representing highly immunogenic specimens. Graphs show 2−ΔCt where ΔCt are the gene of interest Ct normalized by GAPDH and GUSB Ct.
A Gaussian mixture model was built to define how this 10 gene-signature would separate responding from non-responding patients. The model achieved a sensitivity of 0.67 (95% confidence interval [CI]: 0.22–0.96) and and a specificity of 0.92 (95% CI: 0.64–1.00), respectively (Fig. S1). We further validated this model in a separate cohort of untreated patients primary colon tumors (n=20). We wanted to test whether we would be able to identify inflammed CRC including MMRd and immunoreactive MMRp colon tumors (4). Therefore, based on our immune gene signature untreated primary colon tumors were predicted to exhibit a R or NR phenotype based on the resemblance of their TiME with that of primary colon tumor in mCRC patients responsive (R-type TiME) or not responsive to anti-PD1(NR-type TiME) therapy. The resected colon specimens served as a validation set to test the association of the predicted status (R and NR) with the immune related gene expression, the phenotype and the function of tumor-infiltrating lymphocytes (TILs) freshly isolated from the same tumors. Three out of fourteen resected MMRp tumor specimens (~21%; #3735, #3752, #3754) exhibited an R-type TiME (Fig. 1B). Five out of the six MMRd resected tumor specimens (~83%) were associated with R-type TiME and therefore suggested the accuracy of the model used since it would be predicted that ~80% of MMRd CRC would show clinical response to anti-PD-1 therapy (4, 9).
R-type MMRp colon tumors from the validation set are MMRp tumors infiltrated with exhausted IFNγ-producing PD1high CD8+ T cells.
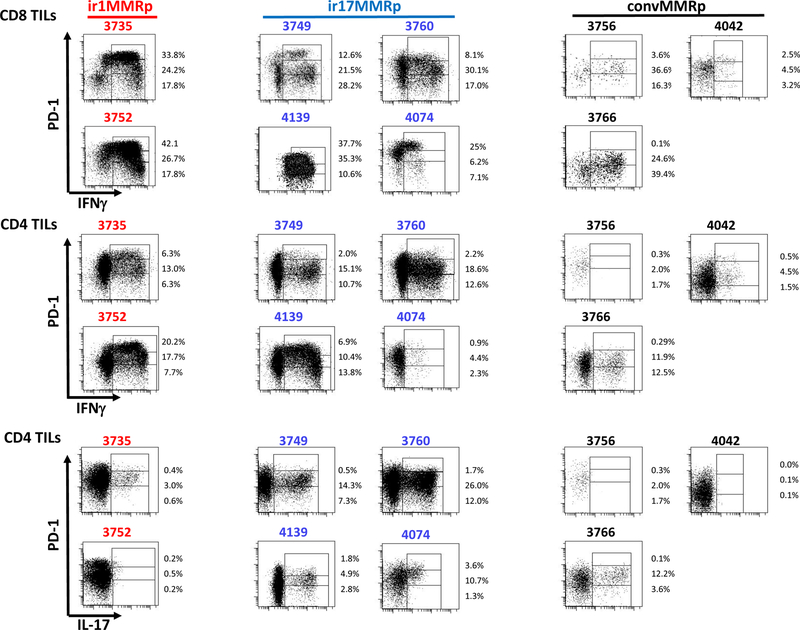
The R-type MMRp CRC from the validation cohort expressed a higher level of genes associated with immune infiltration (CD4, p=0.0220 and CD8A, p=0.0055) T cell checkpoints (CD274, p=0.0110 and LAG3, p=0.0055), Treg (FOXP3, p=0.0055), compared to other MMRp CRC cases (Fig. 1C and detailed in Table S5). TIL freshly isolated from resected specimens were evaluated by intracellular cytokine staining, multiparameter flow cytometry and cell-sorting of the populations of interest. Two out of the three R-type MMRp colon tumors were highly infiltrated with IFNγ+PD-1hi activated CD8+ T cells and considered to be immunoreactive (unavailable fresh TILs for 3754; Fig. 2). Notably, flow cytometry analysis of these immunoreactive MMRp CRC specimens showed a dramatic difference in IFNγ production by PD1hi versus PD1low CD8+ cells, with PD1low CD8+ T cells having systematically a higher IFNγ mean of fluorescence compared to PD1hi CD8+ cells (Fig. S2A). This suggests an exhausted function of PD1hiCD8+ T cells compared to PD1loCD8+ T cells (10). IFNγ+PD-1hi CD8+ T cells are, at the most, sparse in the corresponding patient-matched distal normal tissue (Fig. S2B). We then sorted the PD1hi, PD1lo and PD1neg CD8+ TIL (n=6) and compared gene expression between PD1hi and PD1lo to PD1neg CD8+ TIL using Taqman qRT-PCR on a defined immune related gene array (Tables S3B&S6). PD1+ TILs (PD1hi and Lo) exhibited an exhausted / effector memory gene expression profile since these cells lacked of effector cytokines (IL2, IL15 and TNFA) and had a lower CD28, CCR7, IL-7R and CD62L gene expressions than PD1neg CD8 cells (Fig. S3). We observed that CXCL13 expression was increased in PD1+CD8+ TILs versus PD1negCD8+ TILs. CXCL13 is a chemoattractant protein involved in the recruitment of immune cells and the formation of tertiary lymphoid structures(11, 12). PD1+CD8+ TILs also demonstrated higher expression of the T cell checkpoints CTLA4, LAG3, and HAVR2 (coding Tim3), costimulatory molecules, TNFRSF4 (coding OX40), TNFRSF9 (coding 4–1BB) and TNFRSF18 (coding GITR), as well as higher expression of EOMES and GZMB mRNA (Fig. S3). Importantly, genes involved in senescence (KLRG1) or anergy (EGR3) were not upregulated in the PD1hi fraction (Fig. S3). Our analysis therefore suggested that, higher PD-1 expression (negative < low < high PD-1 expression) is associated with a deeper functional exhaustion of CD8 TILs. MMRd CRC #3784, which was associated with a NR-type TiME (Fig. 1B), had no detectable IFNγ+PD1hi CD8+TILs by flow cytometry (Fig. S4). Therefore, TILs signature in immunoreactive MMRp colon tumors resembles the one recently described in MMRd tumors and characterized by a vigorous CD8+ T cell infiltration with a Th1/Tc1 polarization as well as T cell checkpoints expression(7). However, Fig. 2 showed that despite being associated with a NR-type TiME, four MMRp CRC cases (#3749, #3760, #4074, and #4139) were unexpectedly characterized by PD-1hiIFNγ+ CD4+ and/or CD8+ TILs (Fig. 2) suggesting that a subset of NR-type MMRp CRC are nevertheless immunoreactive. In conclusion, the sole detection of activated CD8+ T cells in MMRp colon tumors was not sufficient to define R-type TiME guaranteeing the efficacy of immune checkpoint blockade treatment.
Figure 2. Flow cytometry analysis of IFNγ and IL-17 expression by freshly isolated immunoreactive and conventional MMRp colon tumor-infiltrating lymphocytes.
The figure shows PD-1 expression versus IFNγ production by CD8 (top 2 rows) and CD4 (2 middle rows), and PD-1 expression versus IL-17A by CD4 cells (bottom 2 rows) in type 1 (#3735 and 3752), type 17 (#3749, 3760, 4139 and 4074) immunoreactive (ir1MMRp and ir17MMRp, respectively) and conventional (convMMRp; # 3756, 3766, 4042) MMRp colon tumors. CD4 and CD8 cells are distinguished by their level of PD-1 expression, high, low and negative (Hi, Lo and Neg gates in each plot) and % in each gate are indicated. The PD-1 gates are defined relatively to the level of PD-1 in the patient-matched normal colon.
NR-type immunoreactive MMRp colon tumors are infiltrated with IL-17+ T cells.
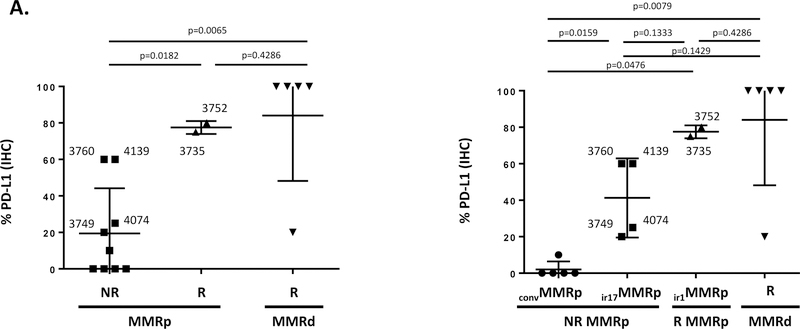
Since MMRp CRC cases #3735, #3752 and #3754 were characterized by a significantly higher expression of the CD274 gene (coding PD-L1) compared to other MMRp CRC specimens (Fig. 1C) and by the presence of PD-1hiIFNγ+ TILs (Fig. 2), we sought first to confirm the expression of PD-L1 protein by IHC on FFPE tumor tissue sections. Figure 3A shows that two of the R-type MMRp CRC (#3735 and #3752) exhibited a remarkable high expression of PD-L1 (80 and 100% respectively; p=0.0182 compared with NR-type MMRp tumor). The third specimen (#3754) did not have an identifiable invasive front region in the examined specimen and therefore could not be scored for PD-L1 expression. However, four MMRp CRC cases (#3749, #3760, #4074, and #4139) with a NR-type TiME (Fig. 1), and reported earlier to be infiltrated with IFNγ+ PD-1hiCD8+ T cells (Fig. 2) also exhibited a strong PD-L1 staining (>20%; Fig. 3A). This finding reinforced the concept that CTL infiltration and activation of the PD-1 / PD-L1 axis in CRC may not be sufficient to grant efficacy to immune checkpoint blockade (or R-type immune signature in primary colon tumor) and that other components of the TiME may interfere with the clinical response. When testing the TILs freshly isolated from the CRC specimens, we found that the four PD-L1hi MMRp specimens (3760, 4139, 4074 and 3749) which were modeled as non-responder in Fig. 1, produced IFNγ (55%, 84%, 38% and 63% of CD8+ T cells; 33%, 31%, 8% and 28% of CD4+ T cells, respectively) and IL-17 (40%, 10%, 16% and 22% of CD4+ T cells, respectively) upon PMA/Ionomycin stimulation (Fig. 2). With the exception of patient #3749, detection of IL-17-producing TILs was associated with the in situ expression of IL-17-associated genes (IL17A, RORC and/or IL23R) for three of these patients (Table 1). Therefore, although these MMRp specimens were immunoreactive and characterized by IFNγ+PD1hi CD8+ TIL (Fig. 2) along with a high level of PD-L1 expression (Fig. 3A), the detection of IL-17-producing cells (4/4) and/or an IL-17 associated transcriptomes (3/4, Table 1) prevailed over their classification as possible R-type immunoreactive MMRp cases (Fig. 1B). None or low IL-17 protein expression was found for the MMRp CRC cases 3752 and 3735 (1% and 4% of CD4+ T cells, respectively; Fig. 2). In sum, our findings predict a deleterious impact of IL-17 in immunoreactive MMRp colon tumors interfering with their clinical response to immune checkpoint blockade. Altogether, we propose to distinguish conventional MMRp CRC (PD-L1−, IFNγ−IL17−) and type 17 (PD-L1+, IFNγ+IL17hi) from the type 1 (PD-L1+, IFNγ+IL17lo) immunoreactive MMRp CRC group, the latter being expected to benefit from immune checkpoint blockade. Conventional MMRp and type 17 immunoreactive MMRp CRC would be expected to progress upon checkpoint blockade monotherapy. Both type 1 and 17 immunoreactive MMRp tumor specimens had significantly higher expression of PD-L1 at the IF compared to conventional MMRp CRC (p=0.0476 and 0.0159; Fig. 3A). Both also expressed higher levels of genes associated with a type 1 immune signature, including IFNG (p=0.0354 and 0.0079, respectively), CD8A (p=0.0357 and 0.0159, respectively) and genes coding T cell checkpoints, including CD274 (p=0.035 for ir1MMRp but not significant for ir17MMRp) and LAG3 (p=0.0357 and 0.0159, respectively) (Fig. S5).
Figure 3. PD-L1, IDO-1 and RORγt protein expressions in immunoreactive MMRp and conventional MMRp CRC.
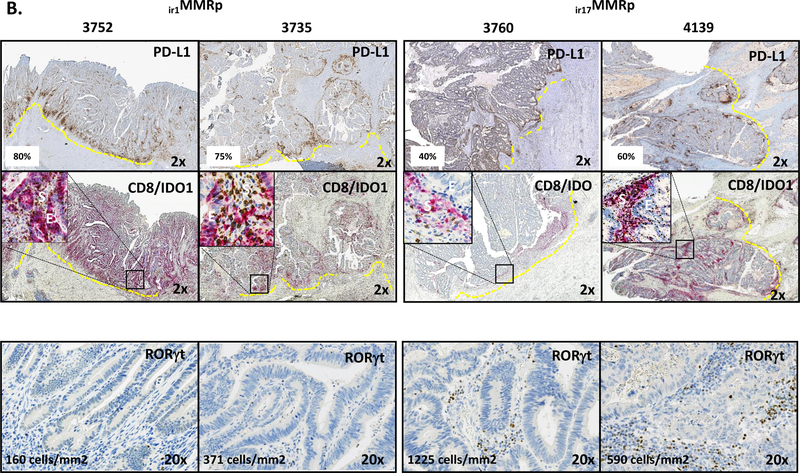
A, the left graph shows PD-L1 IHC performed on FFPE tumor tissue sections of R-type MMRd (R MMRd; inversed triangles), R-type MMRp (R MMRp; triangles), and NR-type MMRp (NR MMRp; squares) primary CRC specimens. The graph in the right shows the PD-L1 expression when distinguishing NR MMRp as conventional MMRp (convMMRp; circles) and type 17 immunoreactive MMRp (ir17MMRp: squares) colon tumors. R-type MMRp are type 1 immunoreactive MMRp (ir1MMRp; triangles) colon tumors. R MMRd are represented as inversed triangles. PD-L1 expressing ir17 MMRp specimens (#3760, #3749, #4139 and #4074) were found to be infiltrated with CD8+PD-1hiIFNγ+ TIL in Fig. 2. B, IHC staining for PD-L1, CD8/IDO1 (dual staining) and RORγT in representative R-type ir1MMRp CRC (#3735, #3752) and NR-type ir17MMRp (#3760 and 4139) colon tumors. RORγT cells densities expressed as number of cells/mm2 in annotated tumor invasive front. Insets in CD8/IDO-1 pictures represent 20X magnification of epithelial (E) versus stromal (S) patterns of expression of IDO1.
Table 1.
Characteristics of the untreated primary MMRp CRC resections
| id | TiME signature | MMRp groupsa | Genomics | IHC | MFC | gene expression signatured | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MSI | # mutations | CD8 | IDO1b | PD-L1c | Th1 | Th17 | Th1 | Th17 | |||||
| 3735 | R | ir1MMRp | Neg | 72 | 290 | + | 75% | 26% | 4% | +++ | IFNG/TBX21/IDO1 | +++ | IL17A/RORC/IL23R |
| 3752 | R | ir1MMRp | Neg | 76 | 1563 | + | 80% | 46% | <1% | +++ | IFNG/TBX21/IDO1 | - | NA |
| 3754 | R | ir1MMRp | Neg | nd | 25 | nd | 10% | nd | nd | + | IDO1 | ++ | RORC/IL23R |
| 3749 | NR | ir17MMRp | Neg | 120 | 325 | - | 20% | 26% | 22% | - | NA | - | NA |
| 3760 | NR | ir17MMRp | Neg | 95 | 170 | - | 50% | 33% | 40% | ++ | IFNG/TBX21 | +++ | IL17A/RORC/IL23R |
| 4074 | NR | ir17MMRp | Neg | 122 | 336 | - | 10% | 8% | 15% | +++ | IFN γ/TBX21/IDO1 | ++ | RORC/IL23R |
| 4139 | NR | ir17MMRp | Neg | 188 | 159 | - | 50% | 31% | 9% | ++ | IFNG/TBX21 | +++ | IL174/RORC/IL23R |
| 3756 | NR | convMMRp | Neg | nd | 18 | - | 1% | 4% | 4% | + | TBX21 | + | IL23R |
| 3762 | NR | convMMRp | Neg | nd | 138 | nd | 5% | nd | nd | - | NA | ++ | IL17A/IL23R |
| 3764 | NR | convMMRp | Neg | nd | 182 | nd | 0% | nd | nd | ++ | TBX21/IDO1 | ++ | IL17A/IL23R |
| 3766 | NR | convMMRp | Neg | nd | 130 | - | 1% | 25% | 16% | - | NA | ++ | IL17A/IL23R |
| 4042 | NR | convMMRp | Neg | 127 | 48 | nd | 10% | 6% | 0% | +++ | IFN γ/TBX21/IDO1 | ++ | RORC/IL23R |
ir1MMRp, type 1 immunoreactive MMRp; ir17MMRp, type 17 immunoreactive MMRp; convMMRp, conventional MMRp
detection of IDO1 on epithelial cells
scoring of PD-L1 at the invasive front
+, one gene expressed; ++, two genes expressed; +++, three genes expressed; -, no genes expressed
Immunopathological definition of type 1 and 17 immunoreactive MMRp CRC.
Type 1 and 17 MMRp colon tumors were further distinguished by CD8/ IDO1 /PD-L1 and RORγt IHC studies. Representative cases of both groups of immunoreactive and conventional MMRp colon tumors are shown in Fig. 3B and S6–7. IDO1 and CD274 (coding PD-L1) are both IFNγ-target genes, and the co-localization of encoded proteins IDO1 and PD-L1 with CD8 cells (in 3752, 3735, 3760, 3749) is thought to reflect the intratumor adaptive immunosuppression phenomenon(13). Importantly, we found a remarkable correlation between the proportions of Th1 but not of Th17 cells in freshly isolated TIL and the PD-L1 scoring obtained via IHC (Fig. S8) validating the use of PD-L1 IHC as a correlate of intratumoral type-1 immunity. RORγt is a transcription factor critical for IL-17 expression and we used its detection in tumor tissue as a correlate of intratumoral type 17-immunity. Fig. 3B and S6–7 show that the detection of PD-L1 and RORγt allows the distinction between type 1 (PD-L1 high, RORγT low) and type 17 (PD-L1 high, RORγT high) MMRp CRC. Conventional MMRp CRC exhibited no or low PD-L1 and RORγT staining (Fig. S7). Although the increased gene expression of IDO1 in types 1 and 17 immunoreactive MMRp tumors compared with conventional MMRp tumors was not significant, we found that the IDO1 protein IHC staining was not detected in conventional MMRp colon tumors pointing out a discrepancy between mRNA and protein expression (Fig. S7). Furthermore, the pattern of expression of IDO1 in tumor tissue distinguished types 1 and 17 immunoreactive MMRp colon tumors, IDO1 was strongly expressed in epithelial cells of type 1 immunoreactive MMRp tumors and conversely, mainly expressed in stromal cells of type 17 immunoreactive MMRp tumors (Fig. 3B). Whereas number of mutations and CD8 density in the invasive front and tumor area are significantly higher in MMRd colon tumors, in our case, these biomarkers did not clearly distinguish conventional MMRp from type 1 and type 17 immunoreactive MMRp tumors (Fig. S9). Neither immunoreactive MMRp (resected primary tumors from untreated patients)nor MMRp mCRC patients with durable SD upon anti-PD-1 therapy, showed POLD or POLE mutations (not shown) or displayed high numbers of mutations (table S1 & S2).
DISCUSSION
It is predicted that about 101,420 new cases of CRC will be diagnosed in 2019. Approximately 20% will be metastatic and 51,020 deaths will occur due to metastatic CRC (American Cancer Society. “Key statistic for colorectal cancer”, www.Cancer.org). Only 2–3% of mCRC are MMRd and currently eligible for pembrolizumab treatment as the standard of care (3). We established that MMRp CRC which express high levels of PD-L1, infiltrated by CD8+PD-1hiIFNγ+ and no IL17 producing TIL have a TiME which resembles that of advanced MMRd mCRC responsive to anti-PD-1 therapy in the setting of our pembrolizumab trial (4). PD1hiCD8+ TILs detected in MMRp CRC patients were characterized by an exhausted/memory transcriptome suggesting the presence of an anti-tumor T cell repertoire, as previously reported in melanoma and digestive system cancers(10). Endogenous anti-tumor T cell immunity is largely restricted to PD1hi TILs and infiltration of non-small cell lung carcinoma with such PD1hiCD8+ T cells has recently been associated with clinical response to anti-PD-1(10, 11). Our findings, therefore, point out that low mutational density cancers may still exhibit one or several immunogenic mutations that can be recognized by the patient’s immune system(14). On that note, we recently reported that one of the MMRp mCRC patients who developed a durable SD upon anti-PD-1 treatment in the clinical trial was indeed characterized by an immune response targeting the hotspot mutation AKT1 E17K(15). Unfortunately, immunoreactive MMRp mCRC patients are not captured by the currently FDA approved microsatellite-high/DNA mismatch repair-deficient biomarker and are therefore unable to receive immune checkpoint blockade as part of their care despite being highly immunogenic and having benefited from pembrolizumab treatment in our phase 2 clinical trial(4).
Recent advances in the understanding of the role of the cancer associated immune contexture into clinical prognosis have fueled efforts at stratifying primary and metastatic CRC patients into different subsets with the final aim of defining eligibility of CRC patients for immunotherapeutic interventions(16). Extensive and sophisticated gene expression profiling and Immunoscore® analysis of CRC recently helped to predict clinical outcomes of CRC patients (8, 17). However, none of these complex characterization approaches of CRC have thus far been tested as predictive biomarkers of response to immune checkpoint blockade. Herein, taking advantage of a landmark clinical trial comparing the clinical response of MMRp and MMRd mCRC patients to pembrolizumab (4), we found ourselves in a unique position to interrogate the TiME of the primary colon tumors of CRC patients who responded versus those who did not. By associating the clinical response to anti-PD1 with the nature of the TiME, we further identified a significant proportion (3/14 or ~21%) of MMRp CRC patients (type 1 immunoreactive MMRp CRC) who may be targeted via immune checkpoint blockade-based immunotherapy. Most importantly, our findings also highlight that the detection of IL-17 and/or an IL-17 gene expression signature should be investigated in cohorts of CRC patients treated with PD-1 blockade as a critical component of the colon TiME which might be able to distinguish R from NR patients. Th17 cells were recently found to be enriched in MMRp compared to MMRd colon tumors and their detection was associated with a worse clinical outcome, but they have not yet been evaluated for their impact on the clinical response to checkpoint inhibitors(18, 19). Our findings in untreated primary MMRp CRC confirmed that despite the fact that several MMRp cases were characterized by a strong PD-L1 expression and an IFNγ signature, their corresponding IL-17 signature distinguished them from MMRp predicted to respond to immune checkpoint blockade treatment. In sum, we have defined on resected CRC specimens the analytical tools that, when validated on larger cohort of treatment refractory mCRC receiving immune checkpoint inhibitors, could help identify a broader population of mCRC more likely to benefit from T cell checkpoint blockade. Mining MMRp CRC RNA sequencing in The Cancer Genome Atlas (TCGA) datasets, we found that about 10% of primary MMRp CRC patients are characterized by a type-1 immune signature without IL-17-related gene expression (Fig.S10). Furthermore, Tosolini et al. found that 4% of CRC patients had a high Th1/cytotoxic gene signature with low expression of gene from the Th17 cluster (IL17A/RORC) with the best disease free survival at 5years (19). The perspective that up to 10% of immunoreactive MMRp mCRC could also benefit from anti-PD1 as monotherapy or as combinations could more than double the impact of immunotherapy on CRC treatment. About 40% of MMRp CRC have a mixed Th1/Th17 signature (Supplementary Fig.S10). The mechanisms associated with the generation of an intratumoral IL-17 response in certain subsets of colon tumors remain unclear. The microbiota (including biofilm formation) and the tumor metabolism (including IDO/Kynurenine/AHR pathway), two critical components of the TME are thought to be key in shaping the intratumoral immunity and responses to immunotherapy in CRC by switching on/off intratumoral IL-17 production(20, 21). Although the contribution of IL-17 into the mechanisms of resistance to immunotherapy is largely unknown, IL-17 and Th17 cells are critical inflammatory components for the defense against invading pathogens (recruitment of phagocytic and granulocytic immune cells or induction of IL-22-dependent bactericidal peptides) as well as for maintaining the integrity of the intestinal barrier by promoting epithelial regeneration upon tissue damage (IL-22 and Stat3-dependent epithelial cells proliferation and survival), features that all could contribute to tumor growth when unchecked (22).
In conclusion, our analysis of the TiME of MMRp CRC patients highlights two critical findings for the selection of cancer patients for immune checkpoint blockade. First, a substantial number of MMRp CRC exhibit a TiME similar to MMRd CRC despite their low tumor mutation number. Second, the detection of an IL-17 immune signature is a predominant element in the TiME of CRC patients that may impact their response to immune checkpoint blockade and which should be taken into account for guiding immunotherapy decisions as previously suggested (23, 24). If validated in a larger cohort of patients as part of a biomarker-driven clinical trial, the association of IL-17 detection with resistance to immune checkpoint blockade could provide one mechanistic explanation for the large proportion of patients who still remain resistant to anti-PD-1 therapy despite exhibiting a high level of PD-L1 and a strong CD8 infiltration in their tumors, both canonical hallmarks of a preexisting anti-tumor immune response(13). This could also be the foundation for a new avenue of immunotherapy in CRC aiming at combining T cell checkpoint blockade with inhibitors of the IL23/Stat3/IL17 signaling axis known to be detrimental in CRC clinical outcomes(19). Previous experimental data showed that tumor bearing mice treated with 5-fluouracil had increased intratumoral IL17 and IL-17A-neutralizing antibodies improved tumor reduction(25). Drugs targeting the IL-23 / IL-17 have already proven clinical efficacy or are currently tested in clinical trials in the treatment of autoimmune disorders, such as psoriasis, rheumatoid arthritis or spondyloarthritis(26, 27) and could therefore be tested in combination with immune checkpoint blockade to improve the clinical responses of CRC to immunotherapy.
Supplementary Material
Statement of translational relevance.
Less than 5% of metastatic CRC patients are MMRd and thus candidates for anti-PD1 as standard of care. The finding that ~10% of immunoreactive MMRp mCRC could also benefit from anti-PD1 as monotherapy or as combinations could more than double the impact of immunotherapy on CRC treatment. Extensive efforts are underway to identify biomarkers that may define tumor immune microenvironments in CRC patients developing clinical benefit upon anti-PD1 treatment and therefore may delineate CRC patients eligible for immunotherapy. The association of IL-17 detection with resistance to immune checkpoint blockade could provide one mechanistic explanation for the large proportion of patients who still remain resistant to anti-PD-1 therapy despite exhibiting preexisting anti-tumor immune response. This could also be the foundation for a new avenue of immunotherapy in CRC aiming at combining T cell checkpoint blockade with inhibitors of the IL23/Stat3/IL17 signaling axis.
Acknowledgments
All the authors are supported by the Bloomberg~Kimmel Institute for Cancer Immunotherapy, Bloomberg Philanthropies and NIH grants P30 DK089502, P30 CA006973. This work is also supported by The Swim Across America foundation (FH and LAD), The Mark foundation for Cancer Research (KNS, JZ and DMP), the Commonwealth foundation (FH and CLS), a Cancer Research Institute/FightColorectalCancer I/O grant (FH and CLS), NIH grants R01 CA203891 (FH) and R01 CA142779 (JMT and DMP). This work is also supported by NIH Gastrointestinal Specialized Programs of Research Excellence (SPORE) grant (P50 CA062924, FH), T32 CA193145, Merck, Bristol Myers Squibb, the Stand Up to Cancer Immunology Dream Team Translational Research Grant (SU2C-AACR-DT1012), and the Colorectal Cancer Dream Team Translational Research Grant (SU2C-AACR-DT22-17). Stand Up to Cancer is a division of the Entertainment Industry Foundation. Research grants are administered by the American Association for Cancer Research, the scientific partner of SU2C.
Conflicts of interest
NJL: Master Nonclinical Research Agreement between Johns Hopkins University and Bristol-Myers Squibb Company; Pediatric Advisory Council BMS; FH and CLS : research grant provided by Bristol Myer Squibb; DMP: Research funding: Bristol- Myers Squibb, Merck, Medimmune/Astra Zeneca. JMT, research grant through BMS; advisory board/consultant for Merck, BMS, Astra Zeneca and Amgen. JMT received equipment and reagents from Akoya Biosciences. LAD is a member of the board of directors of Personal Genome Diagnostics (PGDx) and Jounce Therapeutics. LAD holds equity in PapGene, Personal Genome Diagnostics (PGDx) and Phoremost. LAD is a paid consultant for PGDx and Neophore and is an unpaid consultant for Merck. LAD is an inventor of licensed intellectual property related to technology for circulating tumor DNA analyses and mismatch repair deficiency for diagnosis and therapy (WO2016077553A1) from Johns Hopkins University. These licenses and relationships are associated with equity or royalty payments to LAD. The terms of all these arrangements are being managed by Johns Hopkins and Memorial Sloan Kettering in accordance with their conflict of interest policies. In addition, in the past 5 years, LAD has participated as a paid consultant for one-time engagements with Caris, Lyndra, Genocea Biosciences, Illumina and Cell Design Labs. DTL serves on advisory boards for Merck and Bristol Myers Squibb and has received research funding from Merck, Bristol Myers Squibb, Aduro Biotech, Curegenix, and Medivir. She has received speaking honoraria from Merck and is an inventor of licensed intellectual property related to technology for mismatch repair deficiency for diagnosis and therapy (WO2016077553A1) from Johns Hopkins University. The terms of these arrangements are being managed by Johns Hopkins. RAA is a consultant/advisory board member for Bristol Myers Squibb, Merck, AstraZeneca, Adaptive Biotechnologies and has received research funding from Bristol Myers Squibb, Stand-up to Cancer and contractual work from Five Prime Therapeutics and FLX Bio.
Abbreviation list
- CRC
colorectal cancer
- CTL
cytotoxic T cells
- CTLA-4
cytotoxic T-lymphocyte-associated protein 4
- DCR
disease control rate
- FFPE
formaldehyde-fixed paraffin embedded
- IDO
indoleamine-pyrrole 2,3-dioxygenase
- IF
invasive front
- IHC
immunohistochemistry
- LCM
laser capture microdissection
- mCRC
metastatic CRC
- MMR
mismatch repair
- MMRd
MMR deficient
- MMRp
MMR proficient
- MSI
microsatellite instable
- MSS
microsatellite stable
- NR
non-responder
- PD
progressive disease
- PD-1
programmed cell death protein 1
- PD-L1
programmed death-ligand 1
- PFS
progression-free survival
- R
responder
- RORγT
retinoic-acid-receptor-related orphan nuclear receptor gamma T
- SD
stable disease
- TCGA
the cancer genome atlas
- TIL
tumor-infiltrating lymphocytes
- TiME
tumor immune microenvironment
REFERENCES
- 1.Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell 2015;27:450–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Overman MJ, Ernstoff MS, Morse MA. Where We Stand With Immunotherapy in Colorectal Cancer: Deficient Mismatch Repair, Proficient Mismatch Repair, and Toxicity Management. Am Soc Clin Oncol Educ Book 2018:239–47. [DOI] [PubMed]
- 3.Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357:409–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372:2509–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bendell J, Segal N, Sobrero A, Uyei A, Chang I, Roberts L, et al. LBA-004 Efficacy and safety results from IMblaze370, a randomised Phase III study comparing atezolizumab+cobimetinib and atezolizumab monotherapy vs regorafenib in chemotherapy-refractory metastatic colorectal cancer. Ann Oncol 2018;29:suppl_5. [Google Scholar]
- 6.Bendell JC, Bang Y-J, Chee CE, Ryan DP, McRee AJ, Chow LQ, et al. A phase Ib study of safety and clinical activity of atezolizumab (A) and cobimetinib (C) in patients (pts) with metastatic colorectal cancer (mCRC). J Clin Oncol 2018;36:560. [Google Scholar]
- 7.Llosa NJ, Cruise M, Tam A, Wicks EC, Hechenbleikner EM, Taube JM, et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov 2015;5:43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mlecnik B, Bindea G, Angell HK, Maby P, Angelova M, Tougeron D, et al. Integrative Analyses of Colorectal Cancer Show Immunoscore Is a Stronger Predictor of Patient Survival Than Microsatellite Instability. Immunity 2016;44:698–711. [DOI] [PubMed] [Google Scholar]
- 9.Overman MJ, Lonardi S, Wong KYM, Lenz HJ, Gelsomino F, Aglietta M, et al. Durable Clinical Benefit With Nivolumab Plus Ipilimumab in DNA Mismatch Repair-Deficient/Microsatellite Instability-High Metastatic Colorectal Cancer. J Clin Oncol 2018;36:773–9. [DOI] [PubMed] [Google Scholar]
- 10.Gros A, Robbins PF, Yao X, Li YF, Turcotte S, Tran E, et al. PD-1 identifies the patient-specific CD8(+) tumor-reactive repertoire infiltrating human tumors. J Clin Invest 2014;124:2246–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thommen DS, Koelzer VH, Herzig P, Roller A, Trefny M, Dimeloe S, et al. A transcriptionally and functionally distinct PD-1(+) CD8(+) T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat Med 2018;24:994–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013;39:782–95. [DOI] [PubMed] [Google Scholar]
- 13.Taube JM, Anders RA, Young GD, Xu H, Sharma R, McMiller TL, et al. Colocalization of inflammatory response with B7-h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci Transl Med 2012;4:127ra37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tran E, Ahmadzadeh M, Lu YC, Gros A, Turcotte S, Robbins PF, et al. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science 2015;350:1387–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith KN, Llosa NJ, Cottrell TR, Siegel N, Fan H, Suri P, et al. Persistent mutant oncogene specific T cells in two patients benefitting from anti-PD-1. J Immunother Cancer 2019;7:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fridman WH, Pages F, Sautes-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer 2012;12:298–306. [DOI] [PubMed] [Google Scholar]
- 17.Guinney J, Dienstmann R, Wang X, de Reynies A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med 2015;21:1350–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L, Yu X, Zheng L, Zhang Y, Li Y, Fang Q, et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature 2018;564:268–72. [DOI] [PubMed] [Google Scholar]
- 19.Tosolini M, Kirilovsky A, Mlecnik B, Fredriksen T, Mauger S, Bindea G, et al. Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer Res 2011;71:1263–71. [DOI] [PubMed] [Google Scholar]
- 20.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009;139:485–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prendergast GC, Mondal A, Dey S, Laury-Kleintop LD, Muller AJ. Inflammatory Reprogramming with IDO1 Inhibitors: Turning Immunologically Unresponsive ‘Cold’ Tumors ‘Hot’. Trends Cancer 2018;4:38–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hurtado CG, Wan F, Housseau F, Sears CL. Roles for Interleukin 17 and Adaptive Immunity in Pathogenesis of Colorectal Cancer. Gastroenterology 2018;155:1706–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018;359:97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duffield AS, Ascierto ML, Anders RA, Taube JM, Meeker AK, Chen S, et al. Th17 immune microenvironment in Epstein-Barr virus-negative Hodgkin lymphoma: implications for immunotherapy. Blood Adv 2017;1:1324–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang K, Kim MK, Di Caro G, Wong J, Shalapour S, Wan J, et al. Interleukin-17 receptor a signaling in transformed enterocytes promotes early colorectal tumorigenesis. Immunity 2014;41:1052–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frieder J, Kivelevitch D, Menter A. Secukinumab: a review of the anti-IL-17A biologic for the treatment of psoriasis. Ther Adv Chronic Dis 2018;9:5–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paine A, Ritchlin CT. Targeting the interleukin-23/17 axis in axial spondyloarthritis. Curr Opin Rheumatol 2016;28:359–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.