Abstract
Dietary proteins are crucial for oogenesis. The Target of Rapamycin (TOR) is a major nutrient sensor controlling organismal growth and fertility, but the downstream effectors of TOR signaling remain largely uncharacterized. We previously identified Drosophila Spargel/dPGC-1 as a terminal effector of the TOR-TSC pathway, and now report that Spargel connects nutrition to oogenesis. We found that Spargel is expressed predominantly in the ovaries of adult flies, and germline spargel knockdown inhibits cyst growth, ultimately leading to egg chamber degeneration and female sterility. In situ staining demonstrated nuclear localization of Spargel in the nurse cells and follicle cells of the ovariole. Furthermore, Spargel/dPGC-1 expression is influenced by dietary yeast concentration and TOR signaling, suggesting Spargel/dPGC-1 might transmit nutrient-mediated signals into ovarian growth. We propose that potentiating Spargel/dPGC-1 expression in the ovary is instrumental in nutrient-mediated regulation of oogenesis.
Keywords: Spargel, PGC-1, Drosophila, Nutrition, Insulin, TOR
Introduction
Nutrients from our diet are channeled proportionately to meet the needs of organismal growth, tissue maintenance, and reproduction (Goberdhan and Wilson, 2003; Hafen, 2004). Under limiting nutrient conditions, organismal growth and fertility are both compromised. The relationship between diet and reproduction appears to be ancient (Laws and Drummond-Barbosa, 2017) and to extend to various species including human, rodents, and invertebrates (Dupont and Scaramuzzi, 2016). Carbohydrates, essential amino acids, sterols, and vitamins are particularly important for reproduction (Mirth et al 2019). Specific nutrient sensors, like the InR/IGF-1/TOR signaling pathway, sense amino acids in the diet, are highly conserved from yeast and Drosophila to higher vertebrates, and essential for reproduction (Efeyan et al., 2015; Oldham and Hafen, 2003; Das & Arur 2017). The molecular mechanisms by which cells interpret nutritionally-derived signals to promote cell growth supporting reproduction, however, remain largely unclear.
As in other insects, egg production in Drosophila relies heavily on maternal diet and nutritionally-derived endocrine signals. The ability to manipulate adult diet composition and wealth of genetic tools make Drosophila an excellent model in which to study how diet promotes organismal fertility (Laws & Drummond-Barbosa 2017; Mirth et al 2019; Ables et al 2012). Adult ovaries consist of 16-20 strings of progressively developing egg chambers, called ovarioles (McLaughlin and Bratu, 2015). Oocyte production is supported by the activity of germline stem cells, which reside in the anterior tip of each ovariole. Self-renewing divisions of the stem cell give rise to a daughter committed to differentiation, which divides exactly four times with incomplete cytokinesis to form a 16-celled cyst. Among those 16 cells, one differentiates into an oocyte while the other 15 cells act as nurse cells to support the development of the oocyte. Concurrently, cysts are covered with a single layer of somatic follicle cells, which support egg chamber growth and eggshell formation. Egg chambers progress through 14 distinct developmental stages, growing 20-fold in length and 7-fold in width within 48 hours post-eclosion (Frydman and Spradling, 2001).
To meet such enormous growth rates, Drosophila oogenesis requires a continuous supply of nutrients, provided predominately by yeast in the diet (Ashburner, 1989; Bownes and Blair, 1986; King, 1970). Under limiting nutrients, stem cell division is slowed, egg chamber growth is slowed, and egg chambers are induced to enter programmed cell death. Oogenesis resumes within two days of yeast re-introduction to starved flies, albeit at slightly reduced rate (Drummond-Barbosa and Spradling, 2001). Egg chamber growth in response to nutrients is regulated, at least in part, via InR/TOR signaling (LaFever and Drummond-Barbosa 2005, Pritchett & McCall 2012, Burn et al 2015). Indeed, Insulin Receptor (InR) (Chen et al., 1996), Insulin receptor substrate (IRS) (Böhni et al., 1999; Drummond-Barbosa and Spradling, 2001), and Target of Rapamycin (Tor) (Zhang et al., 2006, LaFever et al 2010) mutants each have ovarian growth defects culminating in loss of fertility. It remains unclear, however, how InR/TOR signaling promotes egg chamber growth in response to nutrients.
Peroxisome gamma proliferator coactivator (PGC-1) is a family of transcriptional coactivators in mammals, consisting of PGC-1α, PGC-1β and PRC, that modulate the activity of many diet-related transcription factors (Puigserver and Spiegelman, 2003; Lin et al., 2005; Ventura-Clapier et al., 2008; Villena, 2015). Under extreme physiological conditions like cold exposure, fasting, and exercise, PGC-1 proteins activate these transcription factors to boost ATP production through mitochondrial biogenesis in energy-demanding tissues (Lin et al., 2005). More recently, knockout animal studies have revealed that PGC-1 proteins also regulate lipogenesis, lipoprotein secretion, muscle fiber type specification, angiogenesis, adipocyte differentiation, hematopoiesis, and the immune response (Villena, 2015). The PGC-1 family is not known to directly regulate fertility; however, a critical interactor of PGC-1α, PPARγ, is expressed in rodent and ruminant ovaries. PPARγ deletion affects follicular development, ovulation and oocyte maturation (Froment et al., 2006), leading to infertility.
The single Drosophila homolog of the PGC-1 family, called spargel/dPGC-1 (srl) (Tiefenböck et al., 2010; Mukherjee et al., 2014), is an attractive way to explore the biology of this transcriptional coactivator family, as it overcomes the challenges of redundancy of PGC1 genes in mammals. Like PGC-1 proteins, Spargel regulates the function of mitochondrial OXPHOS genes and a Spargel gain-of-function boosts mitochondrial O2 consumption (Tiefenbock et al., 2010). Spargel and PGC-1 proteins are nuclear, forming distinct punctate structures by associating with the splicing complex (Monsalve et al., 2000; Mukherjee and Duttaroy, 2013). Drosophila Spargel/dPGC-1 is involved in Insulin-Tor mediated cellular growth (Mukherjee and Duttaroy, 2013), gut tissue homeostasis (Rera et al., 2011) and cardiomyopathy (Diop et al., 2015). We and others have also demonstrated that females carrying a hypomorphic mutation in srl produce small ovaries and are infertile, suggesting a role for srl in oogenesis (Mukherjee et al 2014; Tiefenbock et al 2010). Here, we reveal that Spargel/dPGC-1 is required for female fertility and oogenesis.
Materials and Methods:
Drosophila strain:
Drosophila melanogaster stocks used: W1118 (w[1118]), UAS-srlRNAi_1 (y[1] sc[*] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.HMS00857}attP2; 33914), Maternal tubulin Gal4 (w[*]; P{w[+mC]=matalpha4-GAL-VP16}V37; 7063), Maternal Triple gal 4 (P{w[+mC]=otu-GAL4::VP16.R}1, w[*]; P{w[+mC]=GAL4-nos.NGT}40; P{w[+mC]=GAL4::VP16-nos.UTR}CG6325[MVD1]; 31777), NRE-EGFP (w[1118]; P{w[+m*]=NRE-EGFP.S}5A; 30727), UAS TOR RNAi (y[1] sc[*] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.GL00156}attP2l; 35578), y[1] w[*]; P{w[+mC]=Ubi-GFP.D}33 P{w[+mC]=Ubi-GFP.D}38P{ry[+t7.2]=neoFRT}40A, y[1] w[*]; Tor[DeltaP] P{ry[+t7.2]= neoFRT}40A/CyO, and w[1118]; MKRS, P{ry[+t7.2]=hsFLP}86E/TM6B, Tb[1] (Bloomington Drosophila Stock Center, Indiana University, IN, USA), UASp-srl GFP (w[1118]; P{w[+mC]=UASp-EGFP.Spargel}/CyO) and UAS-srlRNAi_2 (y1 w67c23; P{UAS-srlRNAi-2/ CyO}attP40) (Duttaroy lab). All experimental and control parents were fed the same diet and grown at constant temperature.
Clonal analysis:
TorΔP homozygous germline clones were generated by following the protocol described previously (Wei et al., 2014). Briefly, HS-FLP; Ubi-GFP FRT40A/TorΔP FRT40A females were heat shocked twice a day for 1 h in a 37°C water bath for 2 days. After heat shock, flies were fed yeast paste along with regular food for 6-7 days and dissected for ovaries. TorΔP homozygous clones were marked by the absence of GFP.
Generation of srlRNAi transgenic fly:
The UAS srlRNAi_2 line was generated using the WALIUM 22 vector (Harvard Medical School) at the attp 40 landing site following the protocol for Transgenic RNAi Project (TRiP) (Ni et al., 2009). Briefly, the 21 nucleotide sense 5’CTGCGTATGCTAGAGATGAAA3’ and antisense 5’TTTCATCTCTAGCATACGCAG3’ siRNA sequences targeting the spargel transcript were designed using the Designer of Small Interfering RNA (DSIR) web tool. To construct a proper miR1 scaffold, additional nucleotides were added at the 5’ and 3’ ends as well as within the sense and antisense strands, which resulted in the following strand sequences:
Top:
ctagcagtCTGCGTATGCTAGAGATGAAAtagttatattcaagcataTTTCATCTCTAGCATACGCAGgcg,
Bottom:
aattcgcCTGCGTATGCTAGAGATGAAAtagttatattcaagcataTTTCATCTCTAGCATACGCAGactg.
These two strands were synthesized by Invitrogen and annealed by mixing 20 μM of each strand in annealing buffer (10 mM Tris-HCl, pH 7.5, 0.1 M NaCl, 1 mM EDTA). This mix was incubated at 95°C for 5 min and slowly cooled down to room temperature, creating a DNA fragment with Nhel and EcoR1 overhangs. The WALIUM 22 vector was also digested with Nhel and EcoR1, and linearized vector was purified from an agarose gel. The DNA fragment was cloned into the linearized WALIUM 22 vector (Reaction: 6 μl DNA fragment, 2 μl 10X ligation reaction mix, 1 μl T4 DNA ligase, 40 ng WALIUM 22 vector, 10 μl H2O). Then, 20μl DNA reaction mix was used to transform a 50μl aliquot of TOP10 competent cells. After following a standard transformation protocol, cells were grown on ampicillin nutrient agar plates. The PCR primers F: 5‘-GGTGATAGAGCCTGAACCAG-3‘ R: 5‘-TAATCGTGTGTGATGCCTACC-3‘ (TRIP) were used to select a positive clone. The positive clone was then sequenced to confirm the absence of any aberrant nucleotide substitutions. The TRIP primer (5’-GGTGATAGAGCCTGAACCAG-3’) was used for sequencing. Plasmid DNA from the sequenced clone was used to generate transgenic flies y1, w67c23; P{UAS-srlRNAi-2/ CyO}attP40 (Best Gene). The transgene was targeted to the second chromosome at polytene interval 25C6. To test the transgene’s ability to knockdown spargel transcripts, transgenic srlRNAi_2 flies were crossed with Maternal Tubulin Gal4 (MAT Gal4) to evaluate the ovarian phenotype. The TRiP project used the following sense 5’TACGACAAAGAAGATATTAAA3’ and antisense 5’TTTAATATCTTCTTTGTCGTA3’ strands against the spargel transcript to make the srlRNAi_1 (TRiP) line.
Yeast feeding.
Standard yeast/cornmeal agar was used to nurture flies at 23°C. Two-day-old flies were fed yeast paste for 2 days to allow the ovaries to grow properly. After yeast feeding, ovaries were dissected in Phosphate Buffered Saline (1XPBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4; PH 7.4) and processed for further experimentation. In yeast feeding experiments, No Yeast (NY) and Starvation indicate a completely yeast-free diet, while Regular Yeast (RY) signifies ~2% yeast and Yeast-Enriched (YE) means that 25% yeast paste was added to the vial containing regular food. For Rapamycin feeding, three-day-old flies were fed with either yeast paste, or yeast paste with 4 mM Rapamycin for six days.
Anti-Spargel antibody preparation.
The Spargel peptide (1-250 amino acids) was used as an immunogen to raise both polyclonal and monoclonal antibodies against Spargel. For production of the polyclonal antibody, rabbits were immunized with the Spargel antigen by Genescript, USA. Affinity-purified Spargel antibody was tested by Western blotting to confirm Spargel specificity. For the preparation of the monoclonal antibody, five mice were immunized with Spargel antigen. Serum from immunized mice was screened for positive results, while serum from nonimmunized mice was used as a negative control. Following the identification of a mouse carrying Spargel antibody, splenocytes were isolated and fused with myeloma cells to form hybridoma cells. Next, 20 hybridoma clones were screened for positive results. Clone 7A10 was identified as recognizing the Spargel protein very strongly. Therefore, clone 7A10 was selected and subcloned to generate an established cell line capable of producing anti-Spargel antibody.
Western Blot.
For Western blotting, thirty ovaries were dissected in 1X PBS and transferred to RIPA buffer (10 mM Tris/Cl pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.1% SDS, 1% Triton X-100; 1% Deoxycholate, Protease Inhibitor and Phosphatase Inhibitor) for protein extraction. Drosophila carcasses (without the ovaries) were also collected for protein extraction. Protein samples were mixed with 2 x SDS-sample buffer (120 mM Tris/Cl pH 6.8; 20% glycerol; 4% SDS, 0.04% bromophenol blue; 10% β-mercaptoethanol) and boiled for 10 minutes at 95°C. Proteins were then separated by electrophoresis on 4 to 20% SDS-PAGE gels (Bio-Rad). Samples were run alongside a protein standard (Bio-Rad Precision Plus 250, 10 kDa). Following electrophoresis, proteins were transferred to PVDF membranes (Millipore Immobilion-PSQ, 0.2 m pore size) using 1x Tris/Glycine/SDS buffer (25 mM Tris, 192 mM glycine, 0.1% SDS; pH 8.3) with 20% ethanol for 2 hours at 4°C and 100 V. The PVDF membrane was then blocked with 5% BSA in 1X TBST (50 mM Tris-Cl, 150 mM NaCl, 0.1% Tween 20; pH 7.5) for 1 hour at room temperature. The membrane was washed briefly with 1× TBST and probed with a primary antibody mixed in 1X TBST containing 3% BSA overnight at 4°C. The following day, the primary antibody solution was washed off by rinsing three times (5 min each) with 1× TBST. The membrane was then incubated with a secondary antibody mixed with TBST containing 3% BSA for 1 hour and rinsed three times with 1× TBST. To generate a chemiluminescent signal, ~4 ml of ECL (Amersham Biosciences) was added to each blot for 2 minutes. The signal was detected and the band intensity was calculated using a Li-cor machine.
RNA purification and quantitative real-time PCR.
Total RNA was extracted from ovaries (previtellogenic stages) using a RNeasy mini kit. Stage controlling was performed via hand dissection of ovaries under a microscope. Ovaries were stored in RNA later solution (Invitrogen) to prevent RNA breakdown. We used the iScript cDNA kit to synthesize cDNA from 1 μg of each RNA sample. The cDNA was diluted 10X and 1 μl of the diluted cDNA was mixed with 1 μl each of the forward and reverse primers to obtain a final primer concentration of 250 nM for each target gene. Quantitative PCR primers were designed using NCBI primer blast. Then, 10 μl of SYBR Green was added to each reaction mix. Quantitative PCR data analysis was performed using the Bio-Rad CFX Manager software. Each run was performed in triplicates representing three different biological replicates. Target gene expression was normalized to tubulin RNA levels. Primers used in this study are as follows: Tubulin (F 5’-TTCGTCCACTGGTACGTTGG-3’, R 5’-GCGTGACGCTTAGTACTCCT-3’).
Immunostaining.
Eight to ten pairs of ovaries were dissected in 1XPBS buffer and fixed for 20 minutes at room temperature in 4% formaldehyde in 1xPBS on a rotator. Following fixation, ovaries were washed with 1X PBST (1X PBST + 0.3% Triton X-100) for 15 minutes on a rotator. After washing two more times, ovaries were blocked in 1 ml 1X PBSTA (1X PBST + 3% BSA) for 1 hour at 4°C. Next, the ovaries were transferred into 500-800 μl primary antibody solution (Primary antibody + 1x PBSTA) and incubated overnight at 4 °C on a mild shaker. Most primary antibodies used were between 0.5 – 1 μg/ml concentration. The following day, ovaries were washed three times with 1 ml 1X PBSTA for 15 minutes and then incubated in 500-800 μl of 1X PBSTA with secondary antibody for 3-4 hours at room temperature. Then, 1 μl of phalloidin was added to each 100 μl secondary 1X PBSTA solution when actin staining was necessary. Finally, the ovaries were washed three times with 1 ml of 1X PBSTA for 15 minutes each and mounted in VECTASHIELD with DAPI. Mounted ovaries were stored at 4 °C prior to imaging. The primary and secondary antibodies used in this study are as follows: mouse anti-Spargel (7A10), mouse anti-tubulin, mouse anti-Egfr, anti-rabbit and anti-mouse HRP (Abcam), mouse anti-Gurken (DSHB), rabbit anti-GFP (TorreyPines Biolabs), Alexa 488-conjugated goat anti-mouse, Alexa 488-conjugated goat anti-rabbit secondary antibodies Rhodamine Red™-X goat anti-mouse (Molecular Probes, Life Technologies).
Microscopy.
Mounted ovaries were visualized and imaged using a Nikon Ti-E-PFS inverted microscope equipped with a Yokogawa CSU-X1 spinning disk confocal unit. The 40x and 20× 1.4 NA Plan Apo Lambda objective lenses were used to capture images. The system is also equipped with a self-contained 4-line laser module (excitation at 405, 488, 561, and 640 nm), and an Andor iXon 897 EMCCD camera. EGFP and the Alexa 488-conjugated secondary antibody were excited using the 488 nm laser and detected with the 525 nm emission filter. The Rhodamine Red™-X-conjugated secondary antibody and the FM® 4-64 Dye were excited at 540 nm and detected with the 570 nm filter. DAPI was excited at 358 nm and collected with the 461 nm filter. For each image, the Nikon Ti-E internal focus motor was used to take 12 z-series optical sections with a step size of 2 microns. Four to five images were taken above and below the mid-section of the ovary and were subsequently compiled together. The gamma, brightness, and contrast were adjusted (identically for compared image sets) using NIS Elements Ar imaging software. For whole-ovary fluorescent images, 64 images were taken with the 20x objective lens to cover the whole ovary. These pictures were automatically stitched together into a single image using the NIS Elements Ar imaging software. Multiple stage positions were collected using a Prior ProScan motorized stage.
Egg laying analysis and quantification.
For each genotype, three independent vials were set up with two days of conditioned yeast and five females and males for optimum egg laying. Eggs I were counted for five consecutive days. The average number of eggs per female was calculated and plotted on the y-axis while the x-axis was used to represent the genotypes.
Quantification and statistical analysis.
At least three independent samples were dissected, fixed, and stained in parallel under identical conditions, and the image acquisition settings were also exactly the same for all images used for the quantification. This ensured the consistency among samples for intensity measurements. All measurements were done using NIS Elements Ar imaging software for at least three independent experiments. Data were subjected to both the nonparametric Mann-Whitney U test and parametric student’s t test.
To measure Spargel expression between follicle and nurse cells, the mean fluorescence intensity of follicle and nurse cells nuclei from stage 7/8 egg chamber was measured. Thirty-five randomly selected fields of nuclei from three independent experiments were analyzed. To measure differential Spargel expression in response to yeast manipulation in the diet, the mean fluorescence intensity from nurse cells nuclei from stage 6/7 egg chamber was measured. Thirty-six randomly selected fields of nurse cell nuclei from three independent experiments were analyzed. To measure NRE:GFP expression, the mean fluorescence intensity from follicle cells nuclei from stage 6/7 egg chamber was measured. Forty-four randomly selected fields of nurse cell nuclei from three independent experiments were analyzed. To measure mitochondrial number, the mean intensity of ATP-5A was measured by drawing a region of interest (ROI) of 4.5 μm2 on Control and MAT Gal4>srlRNAi_1. Thirty-two ROI were drawn on Control and MAT Gal4>srlRNAi–1 germline where highest fluorescence intensity of ATP-5A was observed. Student’s t test was used to assess statistical significance between two groups of data. Statistically significant differences are as follows: *p < 0.05 and ***p < 0.001.
Software.
RNAi Designing tool: http://biodev.extra.cea.fr/DSIR/DSIR.html. Primer Blast Tool: https://www.ncbi.nlm.nih.gov/tools/primer-blast/
Results
Levels of the PGC1 ortholog Spargel (Srl) fluctuate with dietary yeast in the ovarian germline.
The Drosophila PGC-1 ortholog Spargel is a downstream effector of the InR/TOR signaling pathway (Mukherjee & Duttaroy 2013). Given the roles of InR/TOR in oogenesis, we sought to test whether Spargel might function to coordinate egg chamber growth in response to nutrients. We generated a Spargel polyclonal antibody that detected an endogenous protein of approximately 150 kD in size by Western blot analysis of Drosophila ovarian extracts (Fig. 1A). The Spargel antibody detected both the endogenous protein and a larger protein in ovarian extracts from MAT Gal4>UAS srl-GFP flies, which co-express a Spargel-GFP fusion protein (Fig. 1A). Although Spargel was primarily expressed in ovaries, some protein could be detected in the rest of the body (carcass) by loading double the amount of protein extract (Fig. 1B). Based on the enriched expression of Spargel in the ovaries, we hypothesized that Spargel is involved in female fertility. This is consistent with earlier observations that spargel hypomorphic females (srl1/srl1) are poorly fertile (Mukherjee et al., 2014; Tiefenbock et al., 2010).
Fig. 1. Spargel is highly expressed in the ovary localizing to nurse and follicle cell nuclei.
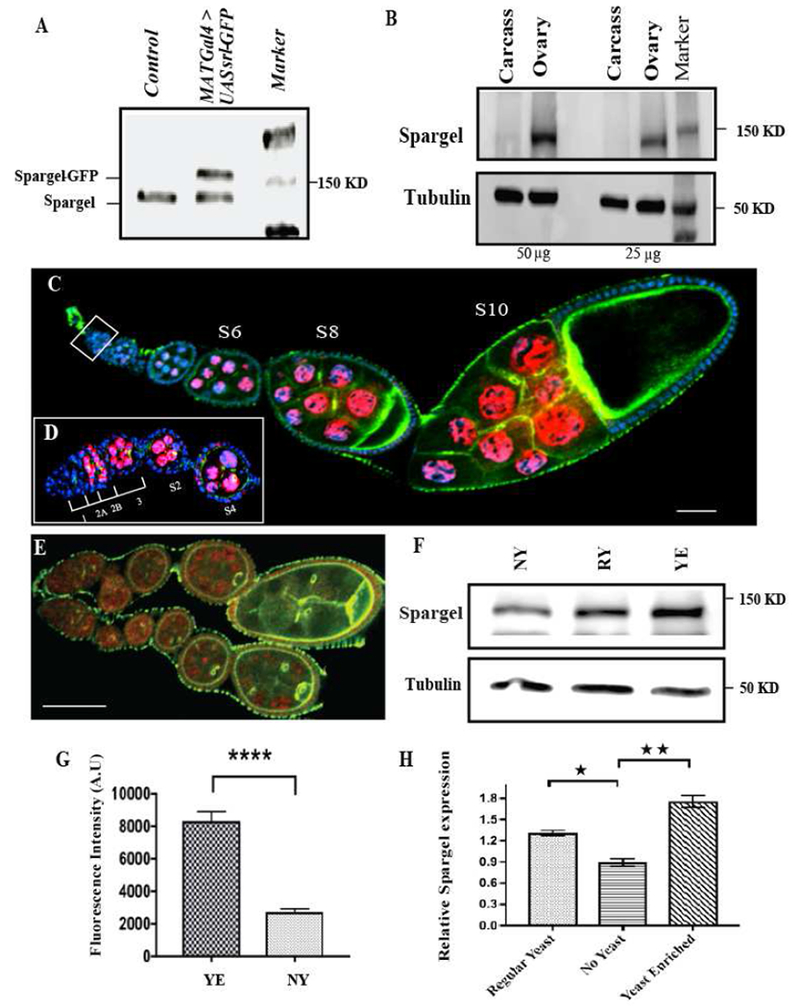
(A) Primary antibody against Spargel/dPGC-1 recognizes endogenous Spargel (<150 kD) by Western blot of the indicated genotypes (Control indicates W1118). (B) Western blot of protein extracts prepared separately from the ovaries and the carcass of same flies (C) Spargel monoclonal antibody (7A10) (Red anti-Spargel; blue DAPI) recognizes Spargel protein in germ cell and follicle cell nuclei. (D) Spargel expression begins in early stage egg chambers and persists until stage 10 egg chambers. A magnified view of the germarium indicates Spargel does not appear in germline stem cells but it is present from region 2b onwards, (E) In situ demonstration of reduced Spargel expression in ovarioles raised in no yeast (Starved) diet. (F) Flies fed a Regular Yeast diet (RY) express Spargel at a higher level than flies fed a No Yeast diet (NY). Spargel expression was highest in flies fed a Yeast Enriched (YE) diet. (G) Quantification of Spargel from the germ cell nuclei of YE vs. NY fed flies. Total fluorescence intensity calculated in arbitrary unit (H) Quantification of Western blots revealed relative Spargel expression in response to variation in yeast concentration. Spargel expression was normalized against tubulin. (*P ≤0.05; **P≤ 0.01). Statistical analysis was done with two tailed t test and error bar indicates s.e.m, (N=35, ****p≤ 0.0001). Scale bar, 50 μm.
To determine the in situ profile of Spargel expression in the ovariole, we raised a monoclonal antibody that, unlike the polyclonal antibody, detects Spargel in tissue immunofluorescence. The monoclonal antibody detected Spargel expression in nurse and follicle cell nuclei (Fig. 1C–D), consistent with the known role of Spargel/PGC-1 as a transcriptional co-activator (Mukherjee et al., 2013). Closer examination of Spargel in the ovariole revealed that Spargel expression in germ cells initiated in the germarium in 16-cell cysts, with continued expression outside of the germarium through Stage 10 (Fig. 1D). Oocyte nuclei did not show Spargel expression (Fig. 1C). In addition to the nurse cell nuclei, follicle cells also appear to express Spargel (Fig. 1C), but at a much lower level. The spatial distribution suggests that Spargel may control egg chamber growth and development.
Prior studies indicated that spargel mRNA expression is upregulated immediately when starved flies are fed a yeast-enriched diet (Gershman, 2007). We postulated that dietary yeast concentration would influence Spargel protein levels in the ovary. Using immunoblots and immunofluorescence, we observed substantially reduced levels of Spargel in the ovaries of adult flies fed a yeast-free diet (No Yeast=NY) for 2 days compared to flies fed a regular yeast diet (Regular Yeast=RY) for 2 days (Fig. 1E–G). Flies transferred from a NY diet to a Yeast Enriched diet (Yeast Enriched=YE) for the same length of time also showed significantly elevated Spargel levels in ovaries. Levels of srl transcript were similarly modulated in response to dietary yeast (Fig. 1H). Together, these data confirm that dietary yeast elevates Spargel levels in germ cell nuclei.
Spargel is essential in germ cells to sustain oocyte development.
To investigate the role of Spargel in nurse cells, we used short hairpin interfering RNA (RNAi) to specifically reduce srl function via the UAS/Gal4 system. We generated two germline-enhanced RNAi lines (srlRNAi-1 and srlRNAi-2) and selected Maternal Tubulin Gal4 (MAT Gal4) and Maternal Triple Gal4 (MTD Gal4) drivers (Staller and Perrimon, 2013) to specifically knockdown spargel in the germline. Germline-specific depletion of srl was sufficient to decrease, but not eliminate, the total amount of Srl protein in ovarian extracts, suggesting that somatic Srl expression was unaffected (Fig. 2A). Females in which MAT Gal4 drove expression of srlRNAi-1 or srlRNAi-2 were completely sterile (Fig. 2B). MTD Gal4>srlRNAi-1 females laid a few eggs (Fig. 2B); however, none hatched, suggesting these eggs were not able to sustain embryonic development.
Fig. 2. Spargel is essential for ovarian growth and oogenesis.
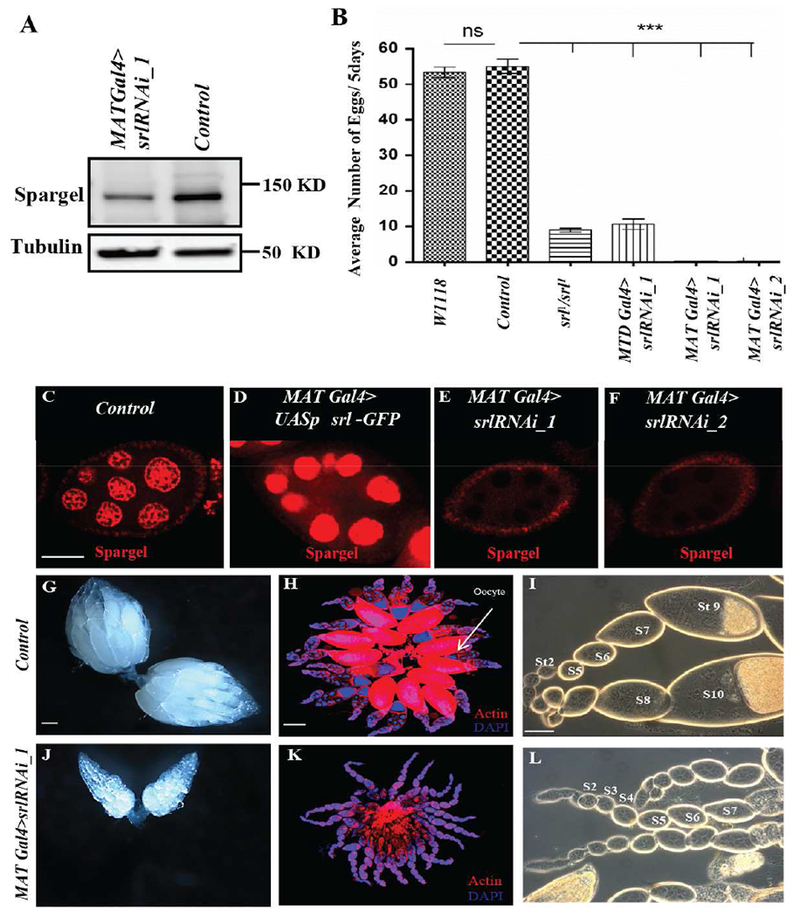
(A) Reduction in Spargel expression in spargel knockdown ovaries compared to the control (Parent srlRNAi_1 line without Gal4 drivers). (B) Spargel is essential for female fertility. Number of eggs laid is plotted for each genotype. Compared to w1118 and control, significantly fewer eggs were laid by srl1/srl1 (Spargel hypomorph), MTD Gal4>srlRNAi_1, MATGal4>srlRNAi_1, MATGal4>srlRNAi_2 females. (C) Spargel expression in MAT-GAL4 control ovaries. (D) Anti-Spargel antibody recognized the overexpressed Spargel protein (red) with greater intensity. (E-F) Spargel expression is virtually eliminated from the germ cell nuclei of MAT GAL4> srlRNAi_1 and MAT GAL4> srlRNAi_2 females. (G-I) Control ovaries. Control genotype is srlRNAi_1 line without Gal4 drivers. (J-L) spargel-depleted ovarioles in MATGal4>srlRNAi_1 failed to form mature oocytes. MATGal4>srlRNAi_1 egg chambers degenerate before exiting the previtellogenic stages (stage 1-7). Phase contrast images indicate the absence of vitellogenic egg chambers ofMATGal4>srlRNAi_1. Scale bar, 50 μm.
To confirm germline specificity in our RNAi depletion models, we performed anti-Srl immunofluorescence in control and srl RNAi mutant ovaries (Fig. 2C–F). As expected, Srl protein was expressed at higher levels in nurse cells than in surrounding follicle cells in control egg chambers (Fig. 2C). A stronger signal was observed in ovaries that overexpress srl (MAT Gal4>UASp srl-GFP) (Fig. 2D), but was absent in nurse cells in the presence of the germ cell-specific srl RNAi (MAT Gal4>srlRNAi-1 and MAT Gal4>srlRNAi-2) (Fig. 2E–F). Because both srlRNAi-1 and srlRNAli-2 lines showed virtually identical effects on female fertility upon activation with either MAT Gal4 or MTD Gal4 drivers, most of the subsequent experiments were performed in MAT Gal4>srlRNAi-1 females.
Germline-specific depletion of srl resulted in small ovaries with a complete loss of vitellogenic and mature eggs (Fig. 2, compare G-H with J-K). Phase contrast images of MAT Gal4>srlRNAi-1 ovarioles indicated an increased number of previtellogenic egg chambers (Fig. 2L), as compared to controls (Fig. 2I). While most well-fed wild-type ovarioles contain a mixed representation of stages of oogenesis (Spradling 1993), most MAT Gal4>srlRNAi-1 ovarioles contained a single egg chamber at every stage of development through Stage 8, suggesting that depletion of srl in germ cells delays egg chamber growth. Females with global reductions in srl (srl1 /srl1) display a similar phenotype, with elevated previtellogenic egg chambers due to retarded growth rates (Mukherjee et al., 2014). Interestingly, although reintroduction of yeast-enriched diet can restore oogenesis in yeast-starved females (Bownes and Blair 1986), a YE diet did not restore ovarian growth in MAT Gal4>srlRNAi-1 ovarioles (not shown). Taken together, these data indicate that srl is essential in germ cells for oogenesis.
Spargel depletion does not trigger cell death during previtellogenic stages
Starvation is known to induce cell death in Stage 8 egg chambers via Caspase 3 activation (Pritchett and McCall, 2012). We reasoned that loss of vitellogenic egg chambers in srl mutants could be caused by premature induction of cell death. As expected, starved egg chambers (stage 8) showed Cleaved caspase 3 activation and punctate nurse cell nuclei (Fig. 3A–B). In contrast, loss of srl from germ cells did not induce Cleaved caspase 3 expression (Fig. 3C–D). Similarly, Lamin staining revealed that the germ cell nuclear boundary remains intact upon depletion of Spargel, arguing against nuclear membrane degradation, a hallmark of yeast deprivation in germ cells (Fig. 3E–H). These results suggest that in the absence of srl, egg chamber development is slowed or halted, rather than prematurely terminated.
Fig. 3. Spargel depletion does not induce cell death.
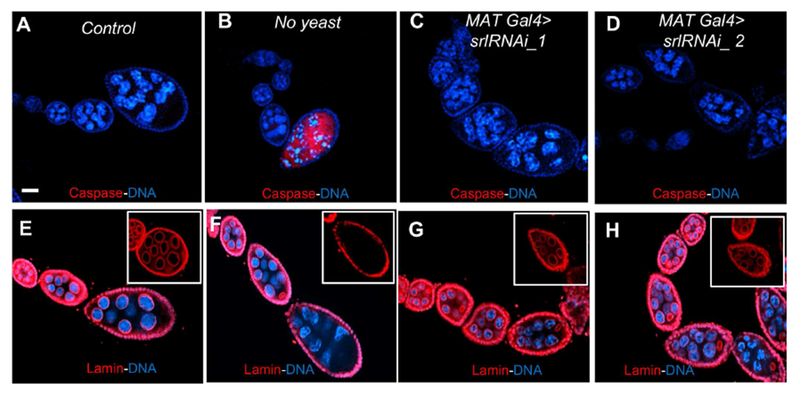
(A) Cleaved caspase 3 is not activated in control egg chambers of srlRNAi_1 line without Gal4 drivers. (B) Dying egg chambers in the starved ovary activate cleaved caspase 3 (Red). (C, D) MATGal4>srlRNAi_1 and MATGal4>srlRNAi_2 ovariole without the activation of Cleaved caspase 3. (E, F) Nuclear Lamin (Red) staining show Lamin remains intact in control nurse cell nuclei, but Lamin disappears in starved egg chambers due to starvation induced cell death. Insets represent stage 8 egg chamber in 40x magnification. (G, H) Lamin stays intact in the nurse and follicle cell nuclear membranes of MATGal4>srlRNAi_1 and MATGal4>srlRNAi_2 ovarioles. Insets represent stage 7/8 egg chamber in 40x magnification. Control genotype is srlRNAi_1 line without Gal4 drivers. Scale bar, 50 μm.
Spargel is essential in germ cells for pre-vitellogenic egg chamber growth.
To better understand why pre-vitellogenic egg chambers are over-represented in germline-specific srl mutant ovarioles, we looked for specific molecular hallmarks of egg chamber development. We first asked whether loss of srl affected localization of the EGFR ligand Gurken. In Drosophila, Gurken controls the dorsal-ventral axis of the oocyte (Stein and Stevens, 2014). During mid-late stages of oogenesis, grk mRNA and protein are localized to the dorsal-anterior of the oocyte in a crescent shape (Fig. 4A). This localization activates EGFR signaling in follicle cells overlying the oocyte. Follicle cells then, in turn, trigger movement of the oocyte nucleus dorsally, shifting Gurken protein localization and specifying the dorsal side of the egg chamber (Fig. 4B–C). In contrast, we observed unusual Gurken localization in MAT Gal4>srlRNAi-1 mutant egg chambers (Fig. 4D–F). Gurken protein was distributed throughout the apical margin of srl mutant oocytes, and occasionally detected in neighboring nurse cells (Fig. 4D–F). Mutant oocytes failed to migrate dorsally and failed to take on yolk (Fig. 4F). Moreover, we observed a double layer of somatic follicle cells overlying srl mutant oocytes (Fig. 4F). We speculate that the rates of follicle cell proliferation outpace srl mutant cyst growth, resulting in extra follicle cells.
Fig. 4. Oocyte development is perturbed in Spargel-depleted germline.
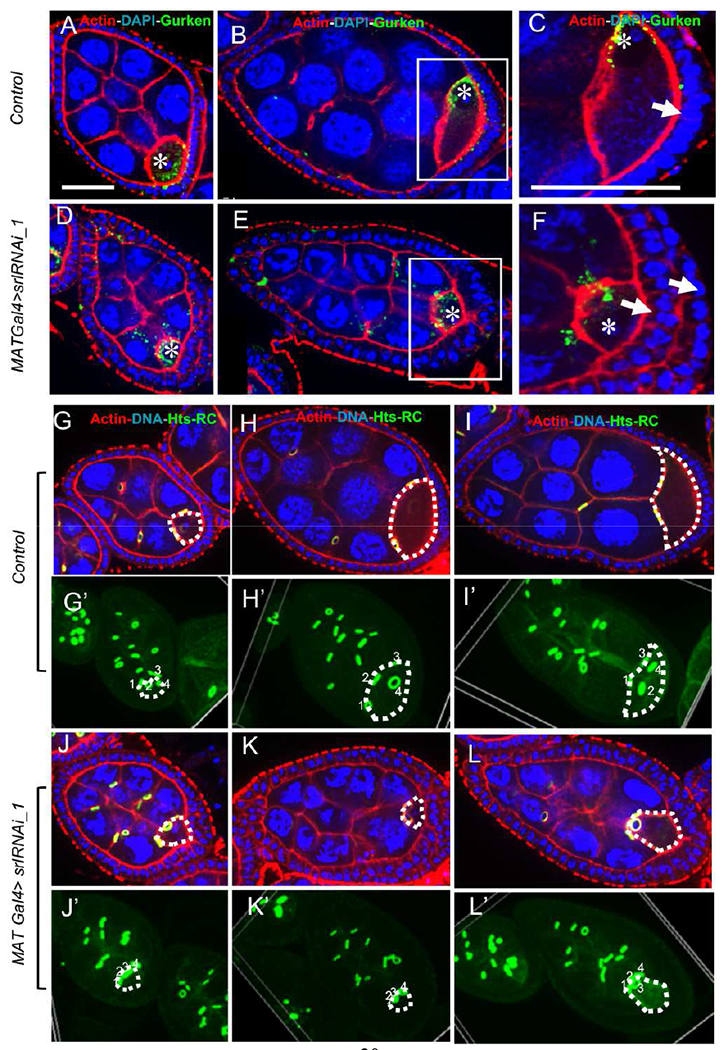
(A-B). Control oocyte in stage 6 and 7. Oocyte nuclei is marked as asterisk (*). Gurken (Green) initially remains at the posterior side of the oocyte nuclei but changes its location to the anterior site of the oocyte as a crescent shape in wild-type flies. (C) Magnified view of oocyte where a single layer of follicular epithelium is observed at its posterior site. Single layer of follicle is indicated with arrow (D-E) MATGal4>srlRNAi_1 oocytes show aberrant localization of Gurken. (F) Magnified view of spargel depleted egg chamber’s oocyte where a two layer of follicular epithelium is observed at its posterior site. Two layers of follicle are indicated by two arrows. (G-I) Egg chamber’s and Oocyte’s (oocyte is marked by white dashed line) growth from stage 6 to 8. Nucleus, actin and ring canal were stained with DAPI (Blue), Phalloidin (Red) and anti- hu-li tai shao-ring canal (hts-RC, green) respectively. (G’-I) Volume view shows that the oocyte grows from stage 6 to 8, the four ring canals that connect the nurse cells to the oocyte spread apart. Top to bottom of the egg chamber were imaged at 2 μm section, which were used to create a volume view of the egg chamber where all the ring canal is visible as green. (J-L’) MAT Gal4> srlRNAi_1 oocytes do not substantially increase in size and the four ring canals do not appear to spread apart, but the size of the egg chamber is increased. J’ to L’ is volume view. Control genotype is srlRNAi_1 line without Gal4 drivers. Scale bar, 50 μm
Lastly, we compared the structure and spacing of the germline cyst ring canals as an indicator of cyst growth. During stage 6, the four ring canals that connect nurse cells to the oocyte remain close to each other (Fig. 4G,G’), but as the egg chamber grows, they spread apart in wild-type flies (Fig. 4H–I’). In contrast, MAT Gal4>srlRNAi-1 oocytes did not increase in size and the four ring canals did not appear to spread out from each other (Fig. 4J–L’), likely due to reduced growth. Taken together, our results support the conclusion that Spargel is necessary in germ cells for cyst growth.
Spargel regulates mitochondrial density in egg chambers
The regulation of mitochondrial biogenesis, mitochondrial oxidative phosphorylation (OXPHOS) and ATP production are ancestral functions of the PGC-1 group of proteins, including Spargel (Lin et al., 2005; Tiefenböck et al., 2010, Rera et al., 2011). Mitochondrial activity is a major energy source for cellular active transport, and mitochondria likely regulate egg chamber growth (Tourmente, et al., 1990). Mitochondria significantly increase in number in follicle and nurse cells, up to stage 7. To monitor the status of mitochondria, we stained for ATP5A synthase (Cox and Spradling, 2003). Relative to control egg chambers, ATP5A synthase staining almost disappears in stage 6/7 MAT Gal4> srlRNAi-1 egg chambers (Fig. 5). These data suggest that mitochondrial density is reduced in egg chambers upon depletion of Spargel.
Fig. 5. Loss of Spargel affects mitochondrial number.
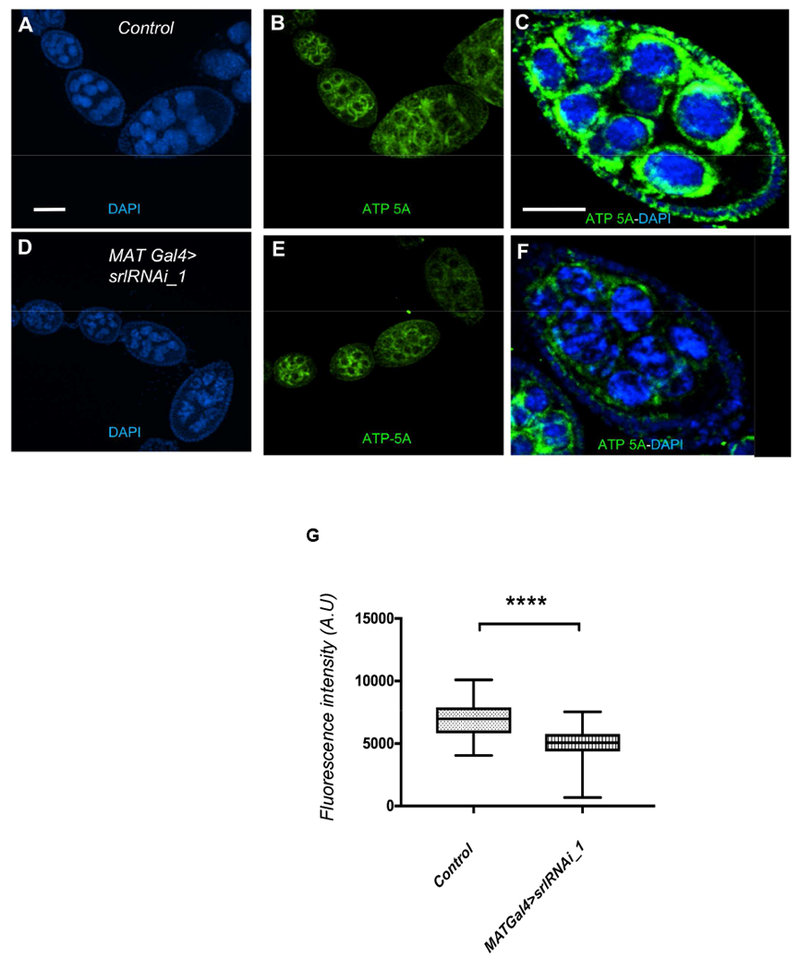
(A, B) From the very early stages of oogenesis, mitochondria accumulate in large numbers (green, ATP5A synthase), surrounding the nurse cell nuclei (Blue). (C) A stage 7 egg chamber is packed with mitochondria surrounding each nurse cell nuclei. (D, E) In MAT Gal4>srlRNAi_1 ovariole, mitochondria appeared in early stage egg chambers but their intensity decrease from stage 6 onward. (F) A stage 7 egg chamber of MATGal4>srlRNAi_1 showed less mitochondrial signals. Control genotype is srlRNAi_1 line without Gal4 drivers. Scale bar, 50 μm. (G) Quantification of ATP-5A fluorescence intensity revealed that the control germlines have significantly a greater number of mitochondria than the MATGal4>srlRNAi_1 germline. Statistical analysis was done with Mann Whitney U test. N=32, ****P≤ 0.0001).
Depletion of the nutrient sensor TOR triggers loss of Spargel expression
The Target of Rapamycin (TOR) is a well-known nutrient sensor. Genetic epistasis analysis suggests that TOR acts upstream of Spargel in the insulin signaling pathway (Mukherjee et al., 2013). Inactivation of Tor causes a dramatic delay in cyst growth, leading to infertility (Zhang et al., 2006; LaFever et al., 2010). TOR signaling also mediates the effects of Insulin Receptor signaling on cyst growth (LaFever et al., 2010). Since srl mutants phenocopy Tor mutant cysts, we hypothesized that TOR signaling might regulate Spargel expression in cysts to control cyst growth. To test this hypothesis, we used the TOR inhibitor, Rapamycin, to block TOR signaling in wild-type flies. As expected, female flies fed Rapamycin had small, disorganized ovaries (Fig. 6A). Interestingly, blocking TOR signaling via Rapamycin also resulted in significantly decreased levels of Spargel protein in nurse cells (Fig. 6B–D), reminiscent of nutrient-deprived flies (Fig. 1E–H). Moreover, loss of Tor in germline clones (TorΔP mutants) reduced Spargel protein levels compared to neighboring wild-type cysts (Fig. 6E–G). Importantly, Spargel protein levels were decreased in TorΔP mutant germ cells prior to hallmarks of cell death, suggesting that Spargel and TOR signaling promote cyst growth independently of cell survival. We cannot rule out the possibility, however, that TOR also promotes cell survival via Spargel-independent mechanisms. Indeed, additional nutrient-dependent pathways likely influence egg chamber degeneration in response to starvation at midoogenesis (Pritchett & McCall 2012). Together, these data support the model that TOR signaling promotes Spargel expression in ovarian germ cells to promote cyst growth.
Figure 6: Downregulation of Tor reduces Spargel expression in the ovary.
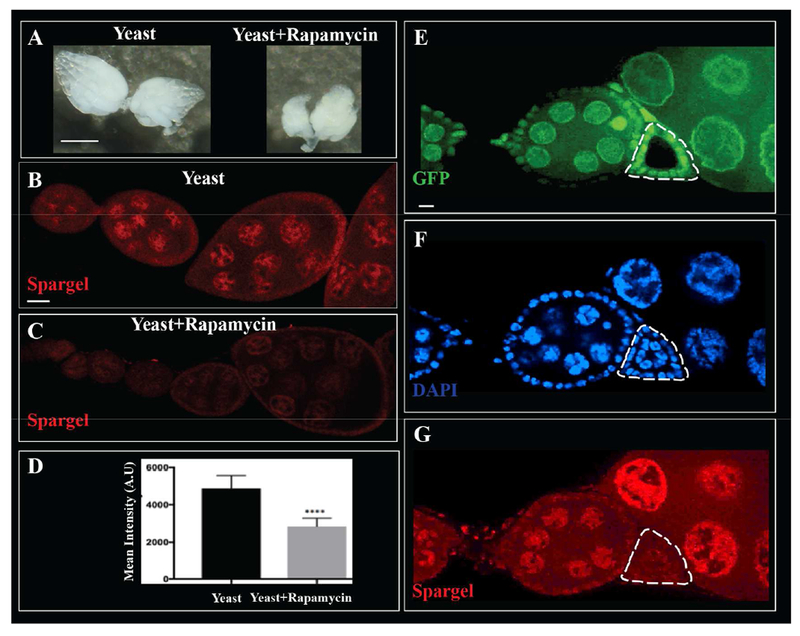
(A) In comparison to yeast fed flies (Left) ovaries in yeast + Rapamycin (an inhibitor of Tor) fed flies (Right) resulted in smaller ovaries. (B-C) Rapamycin feeding reduced Spargel expression in the nuclei of follicles. (D) Quantification of Spargel expression in the germ cell nuclei of control vs rapamycin fed ovaries. Total fluorescence intensity is calculated in arbitrary unit. (E) A germline clone of TorΔP (Delta-P) is GFP negative, (F) The germ cell nuclei (DAPI) of the TorΔP/ TorΔP clone appears healthy yet (G) TorΔP homozygous clone in the germline expresses less Spargel than wild type egg chambers.
Discussion
We have discovered a previously unappreciated, essential role for Spargel/dPGC-1 in oogenesis that was not uncovered by various forward genetic screens (Robinson and Cooley, 1997; Jagut et al., 2103). In mature adults, Spargel/dPGC-1 is expressed almost exclusively in the ovary. Germline specific knockdown of Spargel stops the growth of egg chambers before vitellogenic stages, causing complete sterility. This is consistent with previous observations that a spargel hypomorphic mutant appeared almost sterile (Mukherjee et al., 2014). With regard to Spargel expression in the ovary, a Spargel monoclonal antibody confirmed that Spargel is not expressed in germline stem cells. In fact, Spargel expression in the germarium appeared after the 1a region, indicating that Spargel is not required for 16 cell cyst formation. However, Spargel expression in the early stage egg chambers persists until stage 10, and loss of Spargel in germ cells delays egg chamber growth, leading to an accumulation of previtellogenic egg chambers in the ovariole. Thus, Spargel function is essential in egg chambers, especially in previtellogenic stages.
The slower growth rate of cells lacking Spargel was reported earlier in isolated somatic cell clones of fat body cells (Mukherjee et al., 2013). Here we discovered that Spargel is expressed mostly in the ovaries of mature adults, though a very low level of Spargel expression persists in somatic cells. The developmental profile of Spargel expression is poorly understood. Gene array data (Fly Base) shows that spargel mRNA is expressed in many tissues during development. It is possible that somatic cell clones without Spargel suffers a slower growth rate because these clones were made during development (Mukherjee et al., 2013). Developmental requirement of Spargel might also explain the reduced body size and slow growth rate of Spargel hypomorphs (Tiefenbock et al., 2010; Mukherjee et al., 2014). Alternatively, low levels of Spargel in somatic cells might play a significant role in cell growth. Efforts are underway to tease apart the developmental requirement(s) of Spargel.
The intricate crosstalk between nutrition, nutrient signaling, and oogenesis was established first in Drosophila mutants of insulin receptor (InR), Insulin receptor substrate Chico, dTOR, and recently in the same set of genetic mutants identified in mosquitoes (Hansen et al., 2005) and Red Flour beetles (Sheng et al., 2011). In each case, improper nutrient signaling halted oocytes from entering vitellogenesis (Chen et al., 1996; Drummond-Barbosa and Spradling, 2001; Bohni et al., 1999; Montagne et al., 1999; Zhang et al., 2006). The cellular overgrowth resulting from overexpression of InR is suppressed in spargel hypomorphs (Tefenbock et al., 2010), and Spargel overexpression can epistatically suppress the reduced cell size phenotypes of InR, dTOR and S6K mutant cell clones (Mukherjee et al., 2013). Based on these observations, we proposed that Spargel could be a new terminal effector in the Insulin/Tsc/TOR nutrient signaling pathway (Mukherjee et al., 2013). Our current findings reinforce this conclusion, because Spargel’s action on oogenesis recapitulates the effects of other nutrient-sensing genes belonging to the TOR pathway. Loss of Spargel phenocopies the slowed growth of TOR and InR mutants, and loss of TOR signaling reduces Spargel protein levels. Since rapamycin specifically effects the TORC1 complex, which is involved in growth regulation via nutrient uptake, ribosome biogenesis, and lipid synthesis, we propose that Spargel promotes cyst growth downstream of TOR. Our data do not, however, support a role for Spargel in germ cell survival. Thus, although our genetic experiments place Spargel downstream of TOR in the regulation of cyst growth, we cannot exclude the possibility that TOR promotes cell survival independently of Spargel. Future experiments should test the relationship between Spargel, InR, TOR, and nutrients at specific stages of oogenesis, as these pathways may control discrete cell processes as oocyte development proceeds.
Various environmental stimuli including cold, exercise and fasting are known to regulate the PGC1 group of genes (Lin et al., 2005; Villena, 2015). However, the regulation of Spargel/dPGC-1 by dietary yeast is quite novel. The TOR-Tsc complex is primarily engaged in sensing the available amino acids in the cell (Sancak et al., 2008). TOR might activate Spargel expression after sensing the dietary protein content, to regulate the metabolic needs of dynamic oocyte growth. Amino acids are important metabolic substrates for a variety of anabolic and catabolic needs during oocyte growth, serving as substrates for the synthesis of proteins, nucleotides, Glutathione (GSH), glycoproteins, hyaluronic acid and signaling molecules such as nitric oxide (Dumollard et al., 2007; Sturmey et al., 2008). Hence, the withdrawal of proteins from the fly diet causes degeneration of egg chambers at the onset of vitellogenesis, whereas dietary protein supplementation essentially enhances the rate of egg production almost 60-fold (Drummond-Barbosa and Spradling, 2001). Amino acid supplementation with In Vitro Maturation (IVM) medium also appears to have beneficial effects on oocyte maturation and development in bovine (Watson et al., 2000; Bilodeau-Goeseels, 2006), suggesting a potential parallel between vertebrate and invertebrate systems. Amino acids are the most abundant nutrient in dietary yeast (Schultze, 1995). The near exclusive expression of Spargel in the Drosophila ovary, accompanied by its regulation through yeast supplementation, suggest that this system can be utilized to further our understanding of nutrient-mediated regulation of oogenesis at the molecular level.
Highlights.
-
(1)
PGC-1 ortholog, Spargel has been identified as a potential mediator of Drosophila oogenesis for the first time,
-
(2)
Spargel expression is extremely high in the ovary that correlates with the concentration of yeast in the female fly’s diet.
-
(3)
Spargel acts downstream of Tor to mediate nutritional response to support ovarian growth.
-
(4)
Using germline-specific RNAi, we confirm that Spargel is necessary in the germline for egg chamber development and female fertility.
Acknowledgements
We thankfully acknowledge BDSC for all the fly stocks. Drosophila Genomics Resource Center supported by NIH grant 2P40OD010949. Thanks are due to the spinning disc confocal facility funded by the DOD to Howard University (P.I: Anna Allen). We would like to thank Life Science Editors for editorial assistance.
Funding
This work is supported by NIH grant R25AG047843 awarded to AD. Work in the Ables Laboratory is supported by NIH grant R15-GM117502 (ETA).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interest
No competing interest
References
- Ables ET, Laws KM, and Drummond-Barbosa D (2012) Control of adult stem cells in vivo by a dynamic physiological environment: diet dependent systemic factors in Drosophila and beyond. Wiley Interdiscip Rev Dev Biol. 1(5) 657–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M (1989). Drosophila. A laboratory handbook. Cold Spring Harbor Laboratory Press. [Google Scholar]
- Bilodeau-Goeseels S (2006). Effects of culture media and energy sources on the inhibition of nuclear maturation in bovine oocytes. Theriogenology 66, 297–306. [DOI] [PubMed] [Google Scholar]
- Böhni R, Riesgo-Escovar J, Oldham S, Brogiolo W, Stocker H, Andruss BF, Beckingham K and Hafen E (1999). Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1–4. Cell 97, 865–875. [DOI] [PubMed] [Google Scholar]
- Bownes M and Blair M (1986). The effects of a sugar diet and hormones on the expression of the Drosophila yolk-protein genes. Journal of Insect Physiology 32, 493–501. [Google Scholar]
- Burn KM, Shimada Y, Ayers K, Lu Feiyue., Hudson AM, and Cooley L (2015) Somatic insulin signaling regualtes a germline starvation response in Drosophila egg chambers. Developmental Biology 398(2): 206–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Jack J and Garofalo RS (1996). The Drosophila insulin receptor is required for normal growth. Endocrinology 137, 846–856. [DOI] [PubMed] [Google Scholar]
- Cox RT and Spradling AC (2003). A Balbiani body and the fusome mediate mitochondrial inheritance during Drosophila oogenesis. Development 130, 1579–1590. [DOI] [PubMed] [Google Scholar]
- Das D and Arur S (2017) Conserved insulin signaling in the regulation of oocyte growth, Development and Maturation. Mol Reprod Dev 84: 444–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diop SB, Bisharat-Kernizan J, Birse RT, Oldham S, Ocorr K and Bodmer R (2015). PGC-1/spargel counteracts high-fat-diet-induced obesity and cardiac lipotoxicity downstream of TOR and brummer ATGL lipase. Cell Reports 10, 1572–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond-Barbosa D and Spradling AC (2001). Stem cells and their progeny respond to nutritional changes during Drosophila oogenesis. Developmental Biology 231, 265–278. [DOI] [PubMed] [Google Scholar]
- Dumollard R, Duchen M and Carroll J (2007). The role of mitochondrial function in the oocyte and embryo. Current Topics in Developmental Biology 77, 21–49. [DOI] [PubMed] [Google Scholar]
- Dupont J and Scaramuzzi RJ (2016). Insulin signalling and glucose transport in the ovary and ovarian function during the ovarian cycle. Biochemical Journal 473, 1483–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman HM, & Spradling AC (2001). The receptor-like tyrosine phosphatase Lar is required for epithelial planar polarity and for axis determination with Drosophila ovarian follicles. Development 128, 3209–3220. [DOI] [PubMed] [Google Scholar]
- Froment P, Gizard F, Defever D, Staels B, Dupont J and Monget P (2006). Peroxisome proliferator-activated receptors in reproductive tissues: from gametogenesis to parturition. Journal of Endocrinology 189, 199–209. [DOI] [PubMed] [Google Scholar]
- Gershman B, Puig O, Hang L, Peitzsch RM, Tatar M and Garofalo RS (2007). High-resolution dynamics of the transcriptional response to nutrition in Drosophila: a key role for dFOXO. Physiological Genomics 29, 24–34. [DOI] [PubMed] [Google Scholar]
- Goberdhan DC and Wilson C (2003). The functions of insulin signaling: size isn’t everything, even in Drosophila. Differentiation 71, 375–397. [DOI] [PubMed] [Google Scholar]
- Hafen E (2004). Cancer, type 2 diabetes, and ageing: news from flies and worms. Swiss Med Wkly 134, 711–719. [DOI] [PubMed] [Google Scholar]
- Hansen IA, Attardo GM, Roy SG and Raikhel AS (2005). Target of rapamycin-dependent activation of S6 kinase is a central step in the transduction of nutritional signals during egg development in a mosquito. Journal of Biological Chemistry 280, 20565–20572. [DOI] [PubMed] [Google Scholar]
- Jagut M, Mihaila-Bodart L, Molla-Herman A, Alin MF, Lepesant JA and Huynh JR (2013) A mosaic genetic screen for genes involved inearly stages of oogenesis. G3 3: 409–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RC (1970). Ovarian development in Drosophila melanogaster (No. 595.774 K5). [Google Scholar]
- LaFever L and Drummond-Barbosa D (2005). Direct control of germline stem cell division and cyst growth by neural insulin in Drosophila. Science 309, 1071–1073. [DOI] [PubMed] [Google Scholar]
- LaFever L, Feoktistov A, Hsu H-Jan., Drummond-Barbosa D (2010) Specific roles of targte of rapamycin in the control of stem cells and their progeny in the Drosophila ovary. Development 137, 2117–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laws KM and Drumond-Barbosa D (2017) Control of germline stem cell lineages by diet physiology. Results Probl Cell Differentiation 59, 67–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Handschin C and Spiegelman BM (2005). Metabolic control through the PGC-1 family of transcription coactivators. Cell Metabolism 1, 361–370. [DOI] [PubMed] [Google Scholar]
- McLaughlin JM, and Bratu DP (2015). Drosophila melanogaster oogenesis: an overview In Drosophila Oogenesis (pp. 1–20). Humana Press, New York, NY. [DOI] [PubMed] [Google Scholar]
- Mirth CK, Alves AN, and Piper MW (2019) Insights from nutritional biology and developmental biology of Drosophila. Current Opinions in Insect Science 31, 49–57. [DOI] [PubMed] [Google Scholar]
- Monsalve M, Wu Z, Adelmant G, Puigserver P, Fan M and Spiegelman BM (2000). Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Molecular Cell 6, 307–316. [DOI] [PubMed] [Google Scholar]
- Montagne J, Stewart MJ, Stocker H, Hafen E, Kozma SC and Thomas G (1999). Drosophila S6 kinase: a regulator of cell size. Science 285, 2126–2129. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Basar MA, Davis C and Duttaroy A (2014). Emerging functional similarities and divergences between Drosophila Spargel/dPGC-1 and mammalian PGC-1 protein. Frontiers in Genetics 5, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S and Duttaroy A (2013). Spargel/dPGC-1 Is a New Downstream Effector in the Insulin–TOR Signaling Pathway in Drosophila. Genetics 195, 433–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JQ, Liu LP, Binari R, Hardy R, Shim HS, Cavallaro A, Booker M, Pfeiffer BD, Markstein M, Wang H, Villalta C, Laverty TR, Perkins LA, and Perrimon N (2009) A Drosophila resource of transgenic RNAi lines for neurogenetics. Genetics 182: 1089–100.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham S and Hafen E (2003). Insulin/IGF and target of rapamycin signaling: a TOR de force in growth control. Trends in Cell Biology 13, 79–85. [DOI] [PubMed] [Google Scholar]
- Pritchett TL & McCall K (2012). Role of the insulin/Tor signaling network in starvation-induced programmed cell death in Drosophila oogenesis. Cell Death & Differentiation 19, 1069–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P, and Spiegelman BM (2003). Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1 alpha): transcriptional coactivator and metabolic regulator. Endocrine Review 24, 78–90. [DOI] [PubMed] [Google Scholar]
- Rera M, Bahadorani S, Cho J, Koehler CL, Ulgherait M, Hur JH, Ansari WS, Lo T, Jones DL and Walker DW (2011). Modulation of longevity and tissue homeostasis by the Drosophila PGC-1 homolog. Cell Metabolism 14, 623–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DN and Cooley L (1997) Genetic analysis of actin cytoskeleton in the Drosophila ovary. Annual Review Cell Developmental Biology 13:147–70. [DOI] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L and Sabatini DM (2008). The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze U (1995). Anaerobic physiology of Saccharomyces cerevisiae. Ph.D. Thesis. Technical University of Denmark. [Google Scholar]
- Sheng Z, Xu J, Bai H, Zhu F and Palli SR (2011). Juvenile hormone regulates vitellogenin gene expression through insulin-like peptide signaling pathway in the red flour beetle, Tribolium castaneum. Journal of Biological Chemistry 286, 41924–41936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling AC (1993). Developmental genetics of oogenesis In The Development of Drosophila melanogaster, Vol 1 (ed. Bate M and Martinez-Arias A), pp. 1–70. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Stein DS, and Stevens LM (2014) Maternal control of Drosophila Dorsal-Ventral body axis. Wiley Interdiscip Rev Dev Biol. 3:301–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturmey RG, Brison DR and Leese HJ (2008). Assessing embryo viability by measurement of amino acid turnover. Reproductive Biomedicine Online 17, 486–496. [DOI] [PubMed] [Google Scholar]
- Tiefenböck SK, Baltzer C, Egli NA and Frei C (2010). The Drosophila PGC-1 homologue Spargel coordinates mitochondrial activity to insulin signalling. The EMBO Journal 29, 171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourmente S, Savre-Train I, Berthier F, Renaud M (1990) Expression of six mitochondrial genes during Drosophila oogenesis: analysis by in situ hybridization. Cell Differ Dev 31: 137–49. [DOI] [PubMed] [Google Scholar]
- Villena JA (2015). New insights into PGC-1 coactivators: redefining their role in the regulation of mitochondrial function and beyond. FEBS 282, 647–672. [DOI] [PubMed] [Google Scholar]
- Watson AJ, De Sousa P, Caveney A, Barcroft LC, Natale D, Urquhart J and Westhusin ME (2000). Impact of Bovine Oocyte Maturation Media on Oocyte Transcript Levels, Blastocyst Development, Cell Number, and Apoptosis 1. Biology of Reproduction 62, 355–364. [DOI] [PubMed] [Google Scholar]
- Wei Y, Reveal B, Reich J, Laursen WJ, Senger S, Akbar T, Iida-Jones T, Cai W, Jarnik M and Lilly MA (2014). TORC1 regulators Iml1/GATOR1 and GATOR2 control meiotic entry and oocyte development in Drosophila. Proceedings of the National Academy of Sciences 111, pp.E5670–E5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Billington CJ, Pan D and Neufeld TP (2006). Drosophila target of rapamycin kinase functions as a multimer. Genetics 172, 355–362. [DOI] [PMC free article] [PubMed] [Google Scholar]


