Abstract
Pancreatic ductal adenocarcinoma (PDAC) is a highly aggressive disease with limited and very often, ineffective medical and surgical therapeutic options. The treatment of patients with advanced unresectable PDAC is restricted to systemic chemotherapy, a therapeutic intervention to which most eventually develop resistance. Recently, nab-paclitaxel has been added to the arsenal of first line therapies, and the combination of gemcitabine and nab-paclitaxel has modestly prolonged median overall survival. However, patients almost invariably succumb to the disease, and little is known about the mechanisms underlying nab-paclitaxel (n-PTX) resistance. Using the conditionally reprogrammed (CR) cell approach, we established and verified continuously growing cell cultures from treatment-naive PDAC patients. To study the mechanisms of primary drug resistance, nab-paclitaxel-resistant (n-PTX-R) cells were generated from primary cultures and drug resistance was verified in vivo, both in zebrafish and in athymic nude mouse xenograft models. Molecular analyses identified the sustained induction of c-MYC in the nab-paclitaxel-resistant cells. Depletion of c-Myc restored nab-paclitaxel sensitivity, as did treatment with either the MEK inhibitor, trametinib, or a small molecule activator of protein phosphatase 2a (SMAP).
Implications:
The strategies we have devised, including the patient-derived primary cells and the unique, drug resistant isogenic cells, are rapid and easily applied in vitro and in vivo platforms to better understand the mechanisms of drug resistance and for defining effective therapeutic options on a patient by patient basis.
Keywords: primary pancreatic cancer cells, conditional reprogramming, nab-paclitaxel, c-Myc
Summary.
Continuous cultures of genetically stable primary pancreatic cancer cells were used to define key mechanisms of drug resistance in vitro and in vivo
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is a highly aggressive disease, and patients with this malignancy have less than a 10% overall five-year survival rate. In addition, PDAC is the third leading cause of cancer-related deaths and is expected to become the second leading cause of cancer-related deaths in the next 15 years (1). The addition of nab-paclitaxel to gemcitabine, the standard first-line treatment for unresectable locally advanced and metastatic PDAC, has improved the median overall survival to 8.5–9.4 months vs. 6.7 months with gemcitabine alone (2,3). Unfortunately, nearly all patients succumb to the disease, and little is currently known about the mechanisms of resistance. This deficiency of mechanistic knowledge is partly due to a lack of clinically accurate resistance markers and a shortage of biologically relevant patient-derived cells.
Significant gains have been made in the generation of patient-derived primary cells for cancer research and drug development. In addition to the three-dimensional organoid models (e.g. (4-6)), we have reported extensively on a two-dimensional cell culture approach, the conditionally reprogrammed (CR) cell culture method, that induces a reversible yet indefinite proliferative state in primary mammalian epithelial cells, including those from the pancreas (7-12). This approach has enhanced the ability to rapidly screen for drug sensitivity, as highlighted in both clinical and preclinical studies (8,10,13-15), while preserving fundamentally important morphologic and genetic signatures of the tissues from which the cells were derived (10-12,15,16), including the genetic heterogeneity of the primary tumor (17).
In this report, we describe the generation and validation of continuous two-dimensional cultures of genetically stable PDAC CR cultures from treatment-naïve pancreatic cancer patients. These cells, currently in culture for over 60 passages, retain the tumor-defining KRAS mutations and are maintained and manipulated in a manner that is analogous to the current commercially available transformed cell lines. The responses of the PDAC CR cells to nab-paclitaxel were defined, and drug-selection strategies were used to generate nab-paclitaxel-resistant (n-PTX-R) cells. Importantly, when the CR cells were implanted as subcutaneous xenografts in athymic nude mice, the PDAC tumors self-assembled into histologically well-defined pancreatic adenocarcinomas, exhibiting glandular/ductal structures surrounded by intensely desmoplastic stroma, consistent with the human disease. The drug sensitivity profiles of the CR cells observed in vitro were retained in both mouse and zebrafish-based in vivo model systems.
Herein, we identified increased levels of c-Myc in the n-PTX-R cells that persisted for over 30 passages in the absence of nab-paclitaxel, and modulation of c-Myc levels in the CR cells impacted sensitivity to nab-paclitaxel. Strong links exist between the complex interactions of oncogenic KRAS and deregulated c-Myc in regulating PDAC tumor progression and aggressiveness (see (18)). Mutant KRAS induces phosphorylation of c-Myc on serine 62, leading to increased c-Myc stability and enhanced transactivation of c-Myc target genes (19). Additionally, c-Myc plays a major role in the metabolic plasticity of pancreatic cancer stem cells (20). Finally, while the MEK inhibitor Trametinib had only modest effects on n-PTX-sensitivity, treatment with SMAP2 (small molecule activator of protein phosphatase 2a-2 (SMAP2-DT061)) (21) resulted in a robust increase in n-PTX-sensitivity in the n-PTX-R cells, concomitant with decreases in the levels of ERK, total and phosphorylated c-Myc, and nuclear c-Myc immunopositivity.
Materials and Methods
Cell lines and cell culture.
The human cell line MiaPaCa was obtained from the ATCC and maintained in DMEM containing 10% FBS, L-glutamine, and 100 U/ml Penicillin-Streptomycin. Human pancreatic cancer samples were collected under the approval of the Thomas Jefferson University and Georgetown University Institutional Review Boards. Detailed pathology ensured that the tissue sections contained tumor cells.
Primary PDAC cultures were established at Georgetown using the conditional reprogramming (CR) of cells method as previously described (7).
Cell line authentication was performed via STR analysis by Genetica DNA Laboratories (Cincinnati, OH). Mycoplasma detection assay was performed by Lombardi Tissue Culture & Bio-banking Shared Resource (TCBSR) using MycoAlert detection kit (cat #LT-07118, Lonza Nottingham, LTD).
The PDAC CR cells were carried in culture for over 60 passages. All comparative studies were performed using the earliest and most comparable passages available.
Drug sensitivity studies were carried in conditioned media (CM) as described (9,10). All media were supplemented with 5 μM Y-27632.
Immunoblotting.
Protein extracts were separated on 4–12% Tris-glycine gels and electro-blotted onto PVDF membranes as previously described (10). Protein levels were assessed using antibodies against c-Myc (cat #9405, Cell Signaling, Danvers, MA 01923), p-Myc (cat #13748, Cell Signaling, Danvers, MA 01923), p-ERK½ (cat #4370, Cell Signaling, Danvers, MA 01923), total ERK½ (cat #9102, Cell Signaling, Danvers, MA 01923), GAPDH (cat #5174, Cell Signaling, Danvers, MA 01923), and β-actin (cat #3700, Cell Signaling, Danvers, MA 01923). Densitometry was performed using ImageJ (NIH, Bethesda, MD) as previously described (10).
c-Myc knockdown and overexpression.
For c-MYC knockdown, 5 × 105 n-PTX-R cells were seeded in 6-well plates and transfected with c-MYC siRNA (cat #4609, Dharmacon, Lafayette, CO) or scramble control (cat sc-37007, Santa Cruz Tech, Inc.) using Pepmute siRNA transfection reagents (cat #SL100566, SignaGen Lab, Rockville, MD). For c-Myc overexpression, 5 × 105 Parental cells per well were seeded in a 6-well plate and transfected with a pCMV-Myc-GFP (a gift from Wesley Sundquist (Addgene plasmid # 83375)) (22) or CMV-GFP control (a gift from Bo Huang (Addgene plasmid #70219)) (23) using Xtreme9 (cat # XTG9-RO, Sigma, St Louis, MO).
Cell viability and growth.
Cell viability and total cell counting were determined using trypan blue dye exclusion and a hemocytometer as previously described (10). For population doubling, the PDAC CRs were passed 1:10 in conditioned media (CM) once a week. Population doubling was calculated as: PD = 3.32 (log10 {number of cells harvested/number of cells seeded}).
Drug sensitivity assay experiments were performed seeding 3000 CR cells in CM per well of 96-well plates. Each PDAC CR line was seeded in triplicate. 24 hours after seeding, CM containing nab-paclitaxel, (Celgene, Summit, NJ), trametinib (GSK-1120212, LC Laboratory, Woburn, MA), SMAP-DT061 (cat # PC-70158, ProbeChem Biochemicals Co., Ltd., Shangai, P.R. China), gemcitabine (cat # G6423, Sigma Aldrich, St. Louis, MO), and 5FU (cat #F6627, Sigma Aldrich, St. Louis, MO) alone or in the combinations noted, was added. Cell survival was assessed after 72 hrs using the WST-1 reagent (Sigma #11644807001). Data were analyzed using Prism5 (GraphPad Software). Each experiment was performed three times.
Generation of isogenic nab-paclitaxel-resistant cells.
3000 cells/well were seeded in 96-well plates in CM. 24 hours after seeding, regular CM was replaced with CM containing the IC10 concentration of the drug. CM containing media was replaced every two days. The drug concentration was increased once a week from IC10, to IC20, IC50, and IC80. At IC80, CM without drug was added to allow the resistant cells to proliferate. Once the resistant cells reached confluence, they were passed, banked, and further expanded for analysis. Drug sensitivity assessment was performed as above to confirm the acquired resistance. Cell survival data were analyzed using Prism5 (GraphPad software).
Exome sequencing:
Exome sequence libraries were created using the Illumina TruSeq Exome Library and an Illumina NextSeq 500. The samples were aligned to the human genome version GRCh38 using the Burrows-Wheeler Alignment (BWA) tool, BWA-MEM (24,25). Single nucleotide variants (SNVs) and insertion-deletions (indels) were called using the Genome Analyses Tool Kit v3.5 (26). Variants were called individually for each sample in the GVCF mode of HaplotypeCaller. The identified SNVs were filtered with minimum read depth of 20 reads across all samples, a minimum mapping quality (MAQ) of 90, and a minor allele frequency of greater than 5%. The final sets of ‘high-confidence’ variants were functionally annotated using ANNOVAR (27) for allele frequencies of the 1000 genomes and the gnomAD (28) and for functional prediction with the dbNSFP3.3 database (29). All exome sequencing data are available under SRA accession number PRJNA544733.
High-confidence variants to determine the Jaccard index for each pair of samples measures the overlap of variants between any two samples versus the union of the two samples. The profiles identified missense, nonsense, frame shift, or putative splice-site variants in the drug-resistant line but absent from the ‘parental’ line and not annotated within the 1000 genomes or gnomAD databases with a MAF of greater than 0.01%.
Karyotype and STR analyses:
For metaphase spreads, flasks with CM containing parental or the drug-resistant cells were sent to KaryoLogic, Inc (Durham, NC) for karyotyping analysis. For STR analyses of human PDAC tissue, the parental PDAC and the drug-resistant CRs, DNA was extracted using a Qiagen DNA extraction kit (cat # 69506, Qiagen, Germantown, MD). STR analyses were provided by Genetica DNA Laboratories (Cincinnati, OH). Fifteen independent genetic sites and amelogenin were analyzed and used to authenticate and verify the CR cells. Final match profiles were evaluated using the Percentage Match Algorithm (ANSI/ATCC ASN-0002–2011).
RNA microarray:
RNA was extracted from parental and drug resistant cells using RNeasy-plus mini kit (cat #74134 Qiagen, Germantown, MD). Agilent Human Gene expression v2 arrays were performed and analyzed in the Georgetown-Lombardi Genomics and Epigenomics Shared Resource. All microarray data are available under GEO accession number GSE131762.
Pathway analysis:
Gene interaction networks, biofunctions, and pathway analysis were generated by Ingenuity Pathway Analysis (IPA) (Ingenuity Systems; Mountain View, CA, USA). Differentially expressed genes were mapped to molecular functions and genetic networks available from the Ingenuity database. The significance was set at a p-value and adjusted p-value of 0.05 and a fold change of 1.5. The significance of the association between the input data set and the functions or pathways was determined based on three parameters: (i) a ratio of the number of genes from the data set that map to the function/pathway divided by the total number of genes that map to the function/pathway (overlap percentage) and (ii) a p-value calculated using Fischer’s exact test, and (iii) a z-score as a statistical measure of the match between expected relationship direction and observed gene expression. The calculated z-score predicts activation or inhibition of a pathway based on a positive or negative z-score, respectively.
Reverse Phase Protein Microarray (RPPA):
RPPA was performed as previously described (30,31). Total protein was quantified on selected arrays using Sypro Ruby staining solution (Molecular Probes, Eugene, OR). Immunostaining was performed using an automated system (Dako Cytomation, Carpinteria, CA). Scanned slides were analyzed using MicroVigene 5.1.0.0 (Vigenetech, Carlislie, MA).
Zebrafish:
The zebrafish embryos were anesthetized with 0.0003016% tricaine (Pentair Aquatic Eco-Systems, Sigma-Aldrich, St. Louis, MO, USA) in the Georgetown-Lombardi Animal Shared Resource and positioned within our zebrafish stereotax on a proprietary microinjection plate. Approximately 150 cells per CR culture were labeled with the red fluorescent membrane stain, Dil, and subsequently injected into the yolk sac using an air driven Picospritzer IIa microinjector (General Valve/Parker Hannifin) under a stereoscope. After transplantation, embryos were allowed to recover for 1 h at 33°C. Confirmation of the presence of cells in the yolk sac, and not in the circulation, was followed by incubation at 33°C for up to 5 days. Treatment experiments were carried out at a constant temperature (33°C). Baseline images of the PDAC CR implants were acquired within 1 day of implantation, and embryos that were not successfully implanted were removed from the study. The embryos are imaged again 2–7 days post-implantation. Imaging was performed on an Olympus IX-71 inverted microscope with a color CCD camera in the Georgetown-Lombardi Microscopy Shared Resource. The ellipsoid formula (V = 4/3 × π × a × b × c) was used to estimate temporal volume changes. Student’s t-tests were used to establish statistical significance.
Xenografting.
All animal experiments conducted were fully approved by the Georgetown University Animal Use Committee. For flank tumor induction, 5×105 CR cells in sterile Matrigel and F media (1:1) were injected subcutaneously in 7–9 week old male and female NCI/nude mice using a 27-gauge needle. For Patient 1-Parental and P1-n-PTX-R7, two independent studies were conducted for a total of sixteen mice (32 tumors) for each cell line. For Patient 2-Parental, a total of nine mice (18 tumors) were evaluated. Four mice were used for P2-n-PTX-R6. The xenografts were grown to a maximum tumor volume of 1.0 cm3. Tumor measurements were taken twice weekly, and volumes calculated using the ellipsoid formula as previously described (32). Student’s t-tests were used to establish statistical significance.
Drug sensitivity test in vivo:
The anti-tumor activity of nab-PTX was investigated in the subcutaneous CR-PDX model. nab-paclitaxel was administrated by intraperitoneal injection at 10 mg/kg twice a week for 2.5 weeks. Saline was administrated to the control group. Tumor volumes were evaluated by caliper measurement using the ellipsoid formula as previously described (32). Percentage differences were evaluated between control and treated mice over time. Student’s t-tests were used to establish statistical significance.
Magnetic Resonance Imaging:
In vivo magnetic resonance imaging was performed in the Preclinical Imaging Research Laboratory of the Georgetown-Lombardi Animal Shared Resource and the Center for Translational Imaging in a Bruker Biospec 7T magnet running Paravision 5.1 as previously described (33). Anesthetized (1.5% isoflurane in a gas mixture of 30% oxygen and 70% nitrous oxide) mice were placed in a custom-manufactured (ASI Instruments, Warren, MI) stereotaxic device with built-in temperature and cardio-respiratory monitoring engineered to fit a 40 mm Bruker mouse volume coil. The sequence used to non-invasively identify metastatic tumors was a T2-weighted rapid acquisition with rapid enhancement (RARE) sequences with the following parameters: TR: 3000 ms, TE: 24 ms, RARE factor: 4, Matrix: 256 × 256.
Histology:
Formalin-fixed paraffin embedded samples were sectioned and stained with hematoxylin and eosin, Masson’s trichrome blue, and c-MYC (cat # ab32072, abcam, Cambridge, MA) in the Georgetown-Lombardi Histology and Tissue Shared Resources. All tumor samples were analyzed by a board-certified pathologist.
Results
Generation and validation of the patient-derived primary pancreatic cultures
Resected treatment-naive pancreatic cancer samples were collected under the approval of the Institutional Review Boards of Georgetown and Thomas Jefferson Universities. The PDAC cells were initiated and maintained as continuous cultures using the conditional reprogramming (CR) method essentially as described (7,10,16).
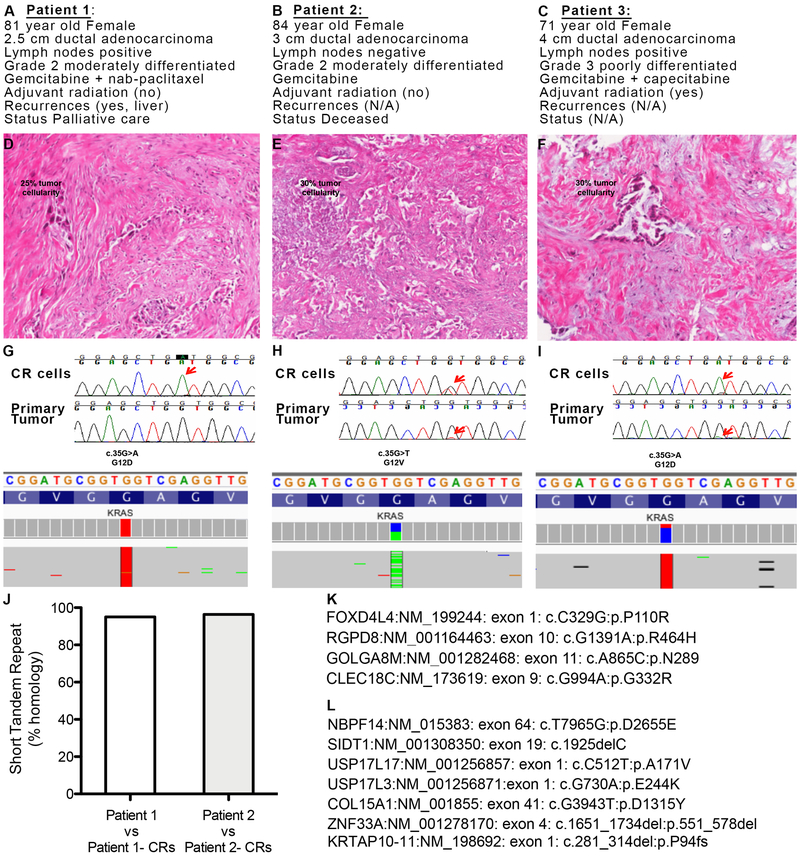
Four primary CR cultures (termed Patients 1–4) were generated, and KRAS mutations were confirmed (see Fig. 1 and data not shown). Formalin-fixed paraffin-embedded (FFPE) tumor samples for three of these patients were available (Patients 1–3), and their clinical characteristics are shown (Fig. 1A-C and Table S1). The FFPE samples were used to assess tumor positivity and cellularity, which were determined to be between 25 and 30% tumor as shown (Fig. 1D-F). Identical KRAS mutations were identified in the tissue samples and in the CR cells from Patients 2 and 3 by either Sanger sequencing or by whole exome sequencing (WES) (Fig. 1H,I); however due to the low cellularity, the KRAS mutation in Patient 1’s tumor sample is unclear (Fig. 1D). TP53 mutations were also detected: Patient 1 R273C in Exon 8; Patient 2 627_628delAA in Exon 6; Patient 3 R282W in Exon 8 (Fig. S1A).
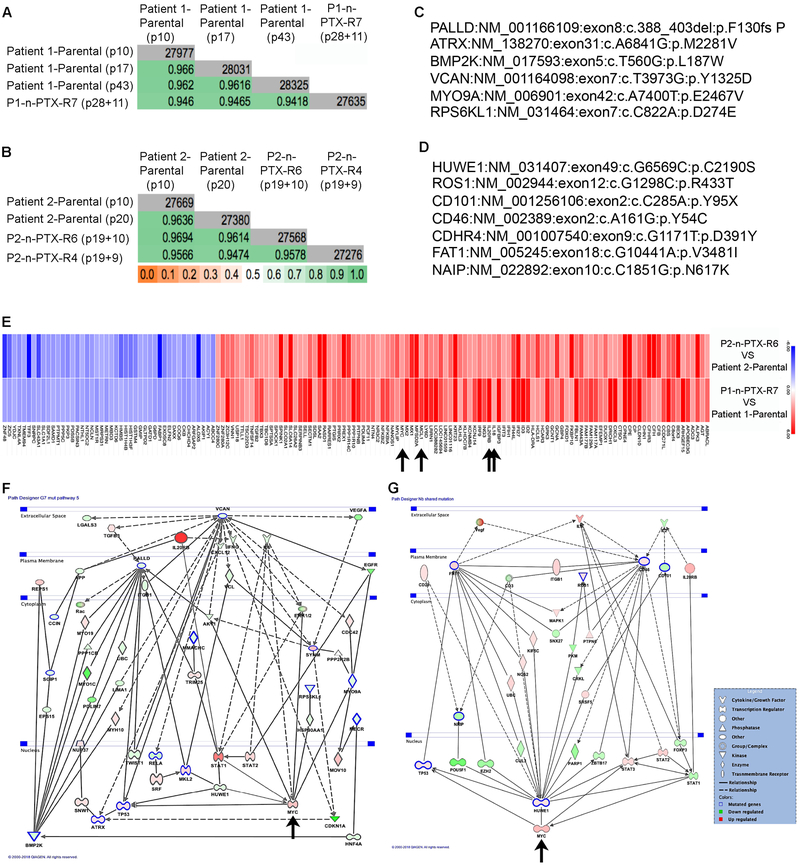
Fig. 1. Conditionally reprogrammed (CR) cells are genetically stable.
(A-C) Key clinical data from Patients 1, 2 and 3 respectively. N/A: not available. (D-F) Histopathological analysis of primary tumor samples from Patients 1, 2 and 3 respectively. The estimated tumor cellularity is shown. (G-I). Top panel: Sanger KRAS sequencing analysis of the PDAC CR cells and matched primary tumor sample from Patients 1-3, respectively. Red arrows indicate codon 12 sequencing reads. Bottom panel: Whole exome sequencing KRAS reads of the nucleotides and the predicted amino acids in the CR cells from Patients 1-3, respectively. (J) Short tandem repeat analysis of primary tumor samples from Patient 1 and Patient 2 vs. their respective CR cells. Non-synonymous somatic mutations observed in late vs. early passages in CR cells from (K) Patient 1 and (L) Patient 2.
In order to assess copy number variation (CNV) and loss of heterozygosity (B-allele frequency), the CR cells from Patients 2 and 3 were also interrogated using the OncoScan whole-genome copy number microarray (Affymetrix). Normal leukocytes, which exhibit a low degree of aneuploidy, and the MiaPaCa cell line, which exhibits a high degree of aneuploidy, served as negative and positive controls, respectively (Fig. S1B,C). Regions of loss of heterozygosity at 9p (p16/CDKN2A), 17p (TP53), and 18q (SMAD4), typically found in PDAC, were present in the CR cells (Fig. S1D,E). Short tandem repeat (STR) analysis performed on the primary tumor samples and corresponding CR cells established the fidelity of the CR cells to the tissue of origin (Fig. 1J). WES identified only four somatic mutations that differentiated late (p43) and early passage (p10) CR cells from Patient 1 (Fig. 1K) and only seven somatic mutations between late (p25) and early (p10) passage CR cells from Patient 2 (Fig. 1L). Karyotyping further demonstrated intra-patient heterogeneity and established that the intra-tumor heterogeneity was retained in culture (Fig. S2).
Collectively, the CR cultures exhibited long-term genetic stability while retaining key genetic features of the primary tissue, consistent with the previous studies using CR cells.
Generation of nab-paclitaxel resistant PDAC CR cultures
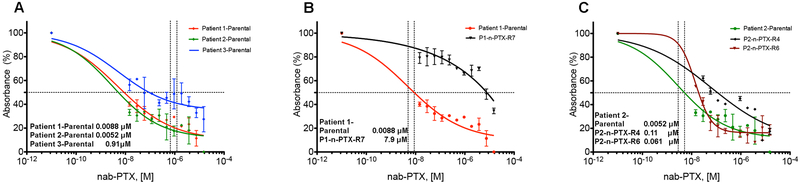
In order to begin to investigate the mechanisms associated with therapeutic failure, nab-paclitaxel (n-PTX) sensitivity profiles were performed in J-2 conditioned media (CM) as previously described (9), and the IC50 values ranged from approximately 5 nM to 900 nM (Fig. 2A and Fig. S4A). Cellular responses to gemcitabine and 5-FU were also determined, further demonstrating intra-patient heterogeneity in drug sensitivity (Fig. S3B,C).
Fig. 2. Nab-paclitaxel sensitivity in parental and resistant CR cells.
(A) Dose-response IC50 calculations performed on the CR cells from Patients 1-3 exposed to nab-paclitaxel (nab-PTX) for 72 hrs. [M]: Molar. (B,C) The IC50 calculations of nab-paclitaxel-resistant (n-PTX-R) cells from Patients 1 and 2, respectively, vs. their matched parental cells. All values are the average +/− SEM of three separate experiments performed in triplicate.
The CR cells from Patient 1 and Patient 2 were chosen to devise a longitudinal drug-dosing strategy for generating drug-resistant cells. The dose escalation methodology (see Methods) began at the empirically determined IC10 concentrations for the CR cells. Several resistant cultures were established, of which one nab-paclitaxel-resistant culture from Patient 1 (P1-n-PTX-R7) and two cultures from Patient 2 (P2-n-PTX-R4 and P2-n-PTX-R6) were selected for further analysis and were maintained in the absence of n-PTX throughout the study. The IC50 values of these n-PTX-R cells are shown (Fig. 2B,C). No significant differences in the growth kinetics of either the parental cultures (now termed Patient 1-Parental (P1-Parental) and Patient 2-Parental (P2-Parental)) or the n-PTX-R cells were found, suggesting that the differential drug sensitivities seen were not a function of cell proliferation (Fig. S4). The n-PTX sensitivity profiles for Patient 1-derived cells were also confirmed under three-dimensional spheroid conditions (Fig. S5), indicating that the inter-patient heterogeneity persisted with other drugs and under various culture conditions. Importantly, the CR cells remain reliant on KRAS for survival, with nearly an 80% reduction in cell viability within one week following KRAS knockdown (Fig. S6), as previously shown in KRAS mutant human PDAC cells (34).
In vivo validation of PDAC CR cells drug resistance in mouse and zebrafish PDX models
Murine and zebrafish xenograft models were used to determine the tumor-forming capacity of the CR cells and to assess whether the n-PTX resistance was retained in vivo.
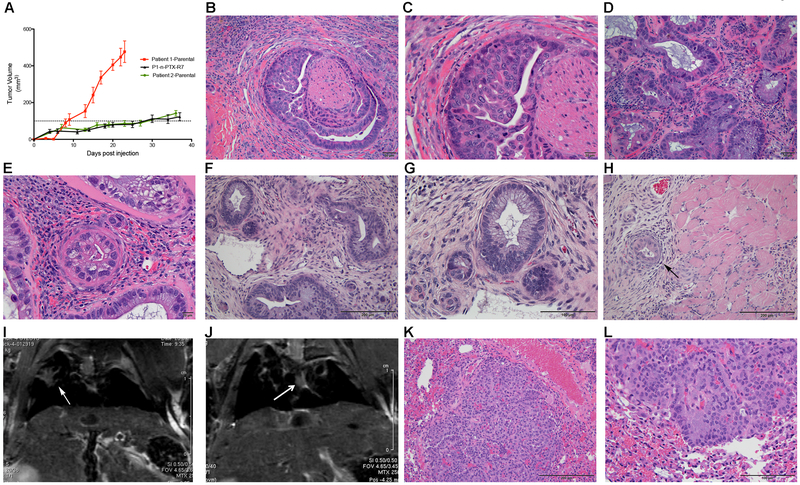
The parental and n-PTX-R cells were implanted subcutaneously into the flanks of female and male athymic nude mice. Ten males and six females were used in Parental 1 and P1-n-PTX-R7 experiments, and five females and six males in the Parental 2 experiments. Tumor growth was evaluated by palpation and caliper measurement. Tumors developed with >90% success rates from the P1-Parental and the P1-n-PTX-R7 cells. The P2-Parental cells generated tumors in over 50% of the implantations. The P1-Parental and corresponding P1-nab-PTX-R7 cells formed tumors within approximately 2–3 weeks, while the P2-Parental tumors took 3–4 weeks to develop. The growth rates of the P1-Parental, P1-nab-PTX-R7, and P2-Parental tumors are shown (Fig. 3A). No gender-related differences were seen. Histopathological evaluation confirmed that the CR cells self-assembled into tumors with the archetypical ductal acinar structures comprised of pleomorphic pancreatic epithelial cells and that the tumor microenvironment exhibited extensive areas of stromal desmoplasia, central histological features of human PDAC (Fig. 3B-G, Fig. S7A). The P1-n-PTX-R7 cells also progressed to a metastatic disease, invading the surrounding muscle by 6 weeks (Fig. 3H). Interestingly, while the P2-n-PTX-R6 cells did not form large flank tumors, Magnetic Resonance Imaging identified metastases in the lungs of the P1-n-PTX-R6 (Fig. 3I) and in P1-n-PTX-R7 mice (Fig. 3J) and was confirmed by histological evaluation of the P1-n-PTX-R7 lung tissue (Fig. 3K,L). The Parental 1- and Parental 2- based mice also presented with lung metastases (Fig. S7B). We term this model CR-PDX (Conditionally Reprogramed-Patient Derived Xenograft), as this is the first report on the use of long-term PDAC CR cells as well as the first description of subcutaneous xenograft tumors being derived from these cells. Additionally, the PDAC CR-based model has the unique characteristics of both retaining the ability to rapidly self-assemble into histologically accurate ductal structures and to induce the intense stromal desmoplasia seen in the human disease.
Fig. 3. CR cells form tumors in mice that exhibit histologically accurate features of PDAC.
(A) Tumor growth kinetics of Patient 1-Parental, Patient 2-Parental and P1-n-PTX-R7 (Patient 1-n-PTX-R7) CR-PDX tumors (N=3 separate experiments). The Patient 1-Parental tumors reached their endpoint at day 23. (B,C) Representative histological images of Patient 1-Parental tumors at 20x and 40x magnification, respectively. (D,E) Representative histological images of Patient 2-Parental tumors at 20x and 40x, respectively. (F,G) Representative histological images of P1-n-PTX-R7 tumors at 20x and 40x, respectively. (H) Local invasion of hind limb muscle by P1-n-PTX-R7 tumors (10x). (I,J) Representative T2-RARE (Rapid Acquisition with Refocused Echo) magnetic resonance images of lung tumors (arrows) in P2-nPTX-R6 and P1-n-PTX-R7 tumor-bearing mice. (K,L) Histological images (10x and 20x, respectively) of P1-n-PTX-R7 lung metastases.
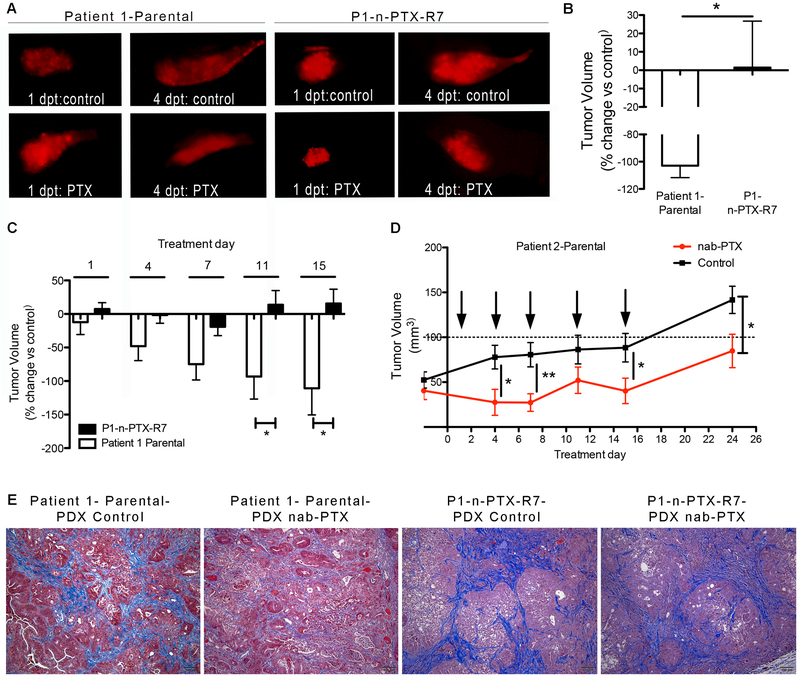
To assess whether the drug responses were maintained in vivo, diI-labeled Parental and n-PTX-R cells were implanted in two-day-old zebrafish embryos. Paclitaxel, used to avoid any complications associated with the PTX albumin coating, was added directly to the water 24 hrs after implantation at a final concentration of 15 uM (the highest concentration used in vitro). The tumors from Patient 1-based cells, evaluated by fluorescence microscopy at one and four days post-treatment (dpt), showed no significant differences in initial volumes between parental and resistant tumors (Fig. 4A). However, while the P1-Parental tumors showed marked reductions in volume following exposure to 15 uM paclitaxel vs. vehicle-treated embryos, the mean tumor size of the n-PTX-R7 tumors either increased or remained constant over the 4-day treatment period (Fig. 4B). Patient 2-based CR cells failed to form viable tumors.
Fig. 4. Nab-paclitaxel resistance profiles are retained in vivo.
(A) Representative images of zebrafish xenografts of diI-labeled P1-Parental (Patient 1-Parental) and P1-n-PTX-R7 (Patient 1-n-PTX-R7) CR cells treated with either 15 uM paclitaxel (PTX) or DMSO (control). (B) The average percent change in tumor volume at four days post treatment (dpt) in PTX-treated vs. DMSO control embryos. N>10. *, p<0.05 vs. Parental cells. (C) Responses of mouse CR-PDX tumors to nab-PTX. Volume changes in Patient 1-Parental and P1-n-PTX-R7 CR-PDX tumors treated with 10 mg/kg in n-PTX twice per week for 2.5 weeks. Differences in volumes are shown as average +/− SEM in nab-PTX-treated vs. control-treated mice. N≥6 mice in each group. *, p<0.05 n-PTX-R7 vs. Parental-1 tumors. (D) Tumor growth rates of Patient 2-Parental tumors treated with either 10 mg/kg nab-paclitaxel or vehicle twice per week for 2.5 weeks. Difference in tumor volumes are shown as average +/− SEM versus control treated mice with N≥6 mice in each group. *, p<0.05 vs. control treated tumors, **, p<0.01. (E) Masson's trichrome staining (collagen/fibrosis in blue) of representative histological images from P1-Parental and P1-n-PTX-R7 CR-PDX mouse tumors.
The responses of the P1-Parental, P1-n-PTX-R7, and P2-Parental CR cells to nab-paclitaxel were next assessed using the CR-PDX mouse models described above. Once the tumors reached approximately 0.5 cm3, nab-PTX was administered two times per week at 10 mg/kg for two-and-a-half weeks by intraperitoneal injection. The size of the P1-Parental (Fig. 4C) and P2-Parental (Fig. 4D, Fig. S7C) tumors decreased versus both their respective pretreatment volumes and versus vehicle-treated mice. Interestingly, while the nab-PTX effects on P2-Parental tumor volumes remained statistical significant throughout the majority of the study, the treated tumors appeared to begin to grow after their last treatment (Fig. 4D), suggesting the possibility that nab-PTX resistance may have begun to occur in vivo. Initial tumor sizes were not significantly different at the time of the first treatment. The P1-n-PTX-R7 tumors showed only modest responses to nab-PTX, and the tumors resumed growth by the fourth dose (Fig. 4C). Histologically, nab-PTX treatment of P1-Parental CR-PDX mice resulted in changes in both the epithelial and the stromal compartments. Masson’s trichrome stain, used to visualize the connective tissue/fibrous component (blue staining) and quantified by ImageJ, revealed that nab-PTX induced a significant disruption of the pancreatic ductal architecture, as well as an significant decrease in the density and amount of the fibrotic, desmoplastic stroma (Fig. S7D). Interestingly, while the P1-n-PTX-R7 tumors were no less stromal rich than the parental tumors in the untreated state (Fig. S7A), only modest alterations in the ductal epithelial compartment were observed in the PTX treated P1-n-PTX-R7 CR-PDX mice (Fig. 4E), and no significant changes were found in the density or amount of the stroma (Fig. S7D).
Genetic and molecular analyses of PDAC pathways associated with resistance
The genetic fidelity of the n-PTX-R cells with respect to their parental cells was determined by evaluating the WES data using the Jaccard Index (JI), with a JI value of 1.0 representing complete correlation (35). The JI values (green boxes), along with the number of genes (gray boxes) and the cell passage number (p) of the two parental cultures vs. one nab-PTX-R culture per parent culture, are shown (Fig. 5A,B). The JI values of the CR cells remained above 0.94, indicating sustained genetic stability despite extended time in culture and selection in nab-paclitaxel. We next used the WES data to identify mutations present unique to the n-PTX-R cells. Twenty-one non-synonymous single nucleotide polymorphisms (i.e. missense, nonsense, or frame-shift mutations) were identified in P1-n-PTX-R7 cells vs. the P1-Parental cells (Fig. 5C and Table S2), and twelve common mutations were found in the P2-n-PTX-R4 and P2-n-PTX-R6 cells vs. P2-Parental cells (Fig. 5D and Table S2). In addition, ten unique mutations were found in P2-n-PTX-R4 cells (Table S2). No mutations unique to P2-n-PTX-R6 were identified.
Fig. 5. n-PTX-R cells are genetically stable and have shared mechanisms of nab-paclitaxel resistance.
Jaccard indices (JI) of whole exome sequencing (WES) data of the CR cells from (A) Patient 1 and (B) Patient 2. Gray boxes: total number of genes, green boxes: JI value. The passage numbers at the time of analysis are shown in parentheses. For the n-PTX-R cells, the numbers represent the passage when cell selection began plus the post-selection passage when exome sequencing was performed. (C) Representative non-synonymous mutations in P1-n-PTX-R7 cells vs. Patient 1-parental cells by WES. (D) Shared non-synonymous mutations in P2-n-PTX-R4 (Patient 2-n-PTX-R4) and P2-n-PTX-R6 (Patient 2-n-PTX-R6) cells vs. P2-parental cells by WES. (E). The top 50% shared regulated genes in n-PTX-R cells identified by microarray are shown. Blue: repressed genes, red: induced genes. Arrows: IL20RB, IL18, MCL1 and Myc. Combined mutational and gene expression interactomes for (F) P1-n-PTX-R7 and (G) P2-PTX-R4/6 CR cells. Green shapes: repressed genes in resistant cells, red shapes: induced genes in resistant cells. Blue outlined shapes: mutated genes in the resistant cells, Arrows: Myc.
In the P1-n-PTX-R7 cells, changes were found in genes associated with the cytoskeleton (Palladin, CCIN, MLK2, and MYO9A), with differentiation and morphogenesis (BMP2K/BIKE and the Androgen Receptor, noteworthy as antiandrogenic therapy may provide therapeutic benefits in PDAC), and in telomere maintenance (ATRX) (Fig. 5C). In the P2-n-PTX-R4 and P2-n-PTX-R6 cells, HUWE1, an X-linked E3 ubiquitin ligase that plays a wide role in cellular and developmental processes (36), was mutated. HUWE1 mutations are linked to a number of cancers, and the genetic deletion of HUWE1 in Apc mutant mice lead to accelerated colorectal cancer initiation, in part through an induction in MYC expression (37).
Identification of c-MYC as an important driver of nab-paclitaxel resistance
Gene transcription microarrays were performed on the parental and their corresponding n-PTX-R cells at a similar passage number. The top 50% of the differentially regulated genes shared between the n-PTX-R cells are shown (Fig. 5E), establishing that the transcriptomic changes were highly durable and persisted in the absence of n-PTX for well over 25 passages. Interestingly, an induction of c-MYC expression was observed, as were increases in pro-inflammatory pathway(s) typified by expression of IL20RB and IL18, possibly contributing to the c-MYC elevation (Fig. 5E, arrows). The shared induction of MCL1 is also highlighted (Fig. 5E, arrow).
Interactome maps that integrated the gene mutations (bold outline) with the mRNA expression profiles were generated (Fig. 5F,G), highlighting the notable differences and distinct similarities between the various independent n-PTX-R CR cells. Among the most prominent features of the drug-resistant phenotype were the induction of the MAPK/ERK and Janus kinase-signal transducer and activator of transcription (JAK/STAT) pathways and induction of c-MYC expression.
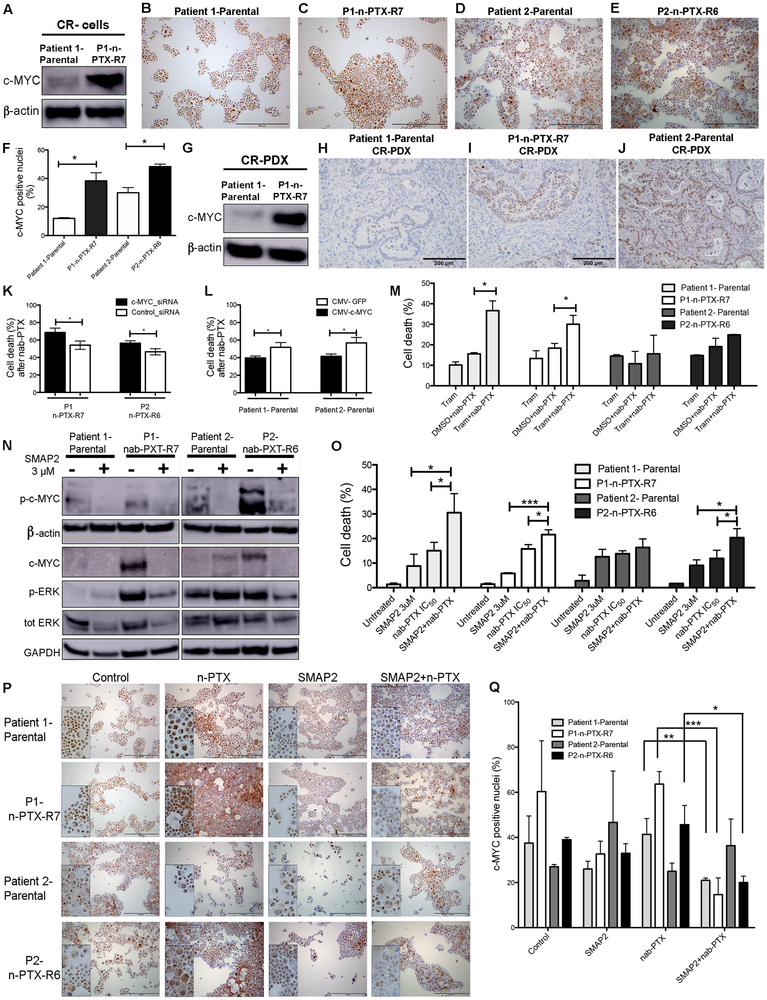
Reverse phase proteomic microarrays confirmed the persistent induction of components of the MAP kinase, JAK/STAT and NF-κB pathways in the n-PTX-R cells vs. either of the parental CR cells (Fig. S8). The increase in c-Myc was confirmed by western blotting (Fig. 6A). In addition, cytospin immunocytochemistry of c-Myc revealed an increase in the number of nuclear c-Myc-positive n-PTX-R CR cells (Fig. 6B-G) as quantified using ImageJ (Fig. 6F). Increased expression of c-Myc was also observed in the P1-n-PTX-R7 CR-PDX tumors (Fig. 6G-I), and the increased c-Myc positivity seen in Patient 2-Parental CR-PDX tumors (Fig. 6J) as compared to Patient 1-Parental tumors (Fig. 6H) is consistent with the cytospin data (Fig. 6F).
Fig. 6. Nab-paclitaxel resistance is regulated by c-MYC and ERK.
(A) Western blot of c-Myc expression in CR (conditionally reprogrammed) cells. (B-E) Immunocytochemical staining of c-Myc in the parental and nab-Paclitaxel resistant (n-PTX-R) cells. (F) Quantification of c-Myc nuclear-positivity in the cells in panels B-E. (G) Western blot of c-Myc expression in mouse CR-PDX (conditionally reprogrammed patient derived xenograft) tumors. (H-J) Immunohistochemical staining of c-Myc in mouse CR-PDX tumor samples. (K) Effect of c-Myc siRNA knockdown on cell survival in n-PTX-R cells treated with n-PTX at IC50 concentrations for 48 hrs, N=8 *, p<0.05. control-siRNA: scrambled-siRNA control. (L) Effects of c-Myc cDNA overexpression on survival in parental CR cells treated with n-PTX at IC50 concentrations for 48 hrs, N>8. *, p<0.05. (M) Effects of trametinib (Tram, 1 uM) alone or in combination with n-PTX (IC50) on cell survival in CR cells treated with n-PTX at IC50 concentrations for 48 hrs, N=3 *, p<0.05. Effects of SMAP2 on (N) c-MYC and ERK in parental and n-PTX-R cells. (O) Survival of CR cells exposed to SMAP2 with or without n-PTX (IC50); treated with n-PTX at IC50 concentrations for 48 hrs, N=3. *, p<0.05. ***, p<0.001. Effects of SMAP2 and n-PTX treatment for 48 hrs on c-Myc nuclear positivity by (P) Immunocytochemical staining for c-Myc. (Q). Quantification of c-Myc nuclear positivity in cells in panel O. *, p<0.05, **, p<0.01, ***, p<0.001.
The genetic suppression of c-MYC by siRNA in the n-PTX-R cells resulted in statistically significant (p<0.05) increases in n-PTX-induced cell death following treatment (Fig. 6K, Fig. S9A,B). Conversely, the transient overexpression of c-Myc in the parental CR cells significantly decreased n-PTX sensitivity (Fig. 6L, Fig. S9C,D).
Small molecule inducers of Protein Phosphatase 2a sensitize PDAC CR cells to nab-paclitaxel
Mutant KRAS remains an undruggable target and alternative methods of inhibiting its downstream targets (e.g. components of the MEK/ERK signaling pathway) are being tested in Ras-driven tumor models. Preclinical studies have shown that the MEK inhibitor, trametinib (Tram), can re-sensitize PDAC cell lines to nab-paclitaxel via inhibition of the RAS/MAPK pathway (38), unfortunately, the addition of Tram with gemcitabine did not improve overall outcomes in patients with previously untreated metastatic pancreatic cancer (39). Co-treatment with Tram and nab-paclitaxel increased cell death in the P1-Parental and the P1-n-PTX-R7 cells vs. nab-paclitaxel treatment alone (Fig. 6M) but not in the P2-Parental or the P2-n-PTX-R6 cells (Fig. 6M).
Deficiencies in Protein Phosphatase 2a (PP2a), a heterotrimeric enzyme complex with key roles in many cellular processes, are linked to the induction of ERK and Myc (reviewed in (40)). The orally bioavailable small molecule activators of PP2a (SMAPs) have been shown to augment ERK and c-Myc inhibition in k-Ras-mutant lung cancers (21,41). Dose-response curves were performed using SMAP2/DT1154 (Fig. S10A). SMAP2 at 3 uM was used for all additional studies. In the P1-Parental CR cells and in P1-n-PTX-R7 and P2-n-PTX-R6 CR cells, the levels of phospho-c-Myc were reduced in response to SMAP2 exposure, and the levels of p-ERK were reduced in P1-n-PTX-R7 and P2-n-PTX-R6 CR cells (Fig. 6N), consistent with our previous studies (41). In the P2-Parental CR cells, phospho c-Myc levels were reduced by SMAP2, while the levels of total c-Myc and phospho-Erk increased (Fig. 6N). SMAP2 addition to the IC50 concentrations of nab-paclitaxel significantly increased cell death in the P1-Parental, P1-n-PTX-R7, and, unlike Tram, in the P2-n-PTX-R6 cells versus either compound alone (Fig. 6O). Interestingly, while P2-Parental cells were sensitive to each drug individually, combination treatments did not significantly increase sensitivity (Fig. 6O). In addition, c-Myc knockdown abrogated the ability of SMAP2 to enhance n-PTX-induced cell death in the resistant cells (Fig S10B). Immunocytochemical staining for c-Myc established that the combined exposure of the CR cells to SMAP2 and nab-PTX significantly reduced c-MYC nuclear positivity in the P1-Parental cells, P1-n-PTX-R7, and P2-n-PTX-R6 cells but not in P2-Parental cells (Fig. 6P,Q), suggesting that the sustained c-Myc induction in P2-Parental cells may contribute to the sustained resistance. Of note, differences in c-Myc nuclear positivity in SMAP2 treated vs. co-treated CRs were only significant in P1-n-PTX-R7 cells (Fig. 6Q).
Discussion
The CR approach enabled the rapid generation of primary cultures of PDAC cells that were manipulated in a manner similar to conventional transformed cell lines. Importantly, the CR cells preserved the features of the primary tumor of origin as shown by the maintenance of genetic stability, retention of tumor heterogeneity, and the recapitulation of key genetic and molecular hallmarks linked with this cancer. In addition, we found that the CR conditions supported long-term genomic stability even after the establishment of drug-resistant cells. This is a particularly important finding given the extensive genetic and morphologic instability and heterogeneity documented in transformed cell lines; a clear example of this are MCF-7 breast cancer cell lines, in which cell culture conditions induced artificial heterogeneity, culminating in large discrepancies in gene expression patterns and widely differing drug responses (42).
Our observations that CR cells exhibited the capacity for retaining pathognomonic histologic and molecular hallmarks of PDAC, including the ability to metastasize, are of particular importance in light of serious issues encountered with traditional PDX models, where up to 12% of the genome is differentially altered by the fourth passage of the tumors through mice (reviewed in (43)).
The use of treatment-naive CR cells in our study of the underlying mechanisms of drug resistance identified an induction of pro-inflammatory pathways, changes in components of the JAK-STAT signaling pathway, and induction of c-Myc that persisted long after the cells were removed from selective pressure. However, common genetic mutations were not found, further underscoring the phenotypic complexities of both PDAC and deregulated c-MYC, and further highlight the potential of using agents that impact MYC for the treatment of chemoresistant PDAC (44). The combination of SMAP2 and nab-paclitaxel reduced total- and phospho-c-MYC levels in the CR cells and resulted in the redistribution of c-MYC from the nucleus to the cytoplasm, and are fully consistent with our previous SMAP studies (41,45) and with other studies in tumors with a “MYC-high” signature (46,47). While some differing studies exist (48,49), these data strongly imply that potential benefits exist by including SMAPS in the chemotherapeutic regimens for PDAC. Future studies in the PDAC CR cells further define the molecular signatures of responses to SMAP treatment.
Interestingly, the CR cells exhibited different behaviors in vivo that appeared unrelated to c-Myc expression levels. For example, the P1-n-PTX-R7 and Patient 2-Parental tumors grew more slowly vs. Patient 1-Parental tumors, while the P2-n-PTX-R6 cells apparently disseminated from the site of implantation, exhibiting no growing tumor mass during the coarse of the experiment. Interestingly, all CR-PDX models including, P2-n-PTX-R6, progressed to a metastatic disease. Takahasi et al (50) reported that only orthotopic implantation of SUIT-2 or Panc-1 cells progressed to metastatic tumors, suggesting an “education” occurred within the pancreas; however, this was clearly not the case in the CR-PDX models. Future studies will investigate the potential genetic and molecular mechanisms underlying the differences in cell/tumor behavior. Unlike traditional PDX models, the CR cells are highly manipulable via transfection, infection or gene editing by CRISP-R (EP, CA, not shown), making modification of key genes highly feasible, further highlighting the utility of this model system.
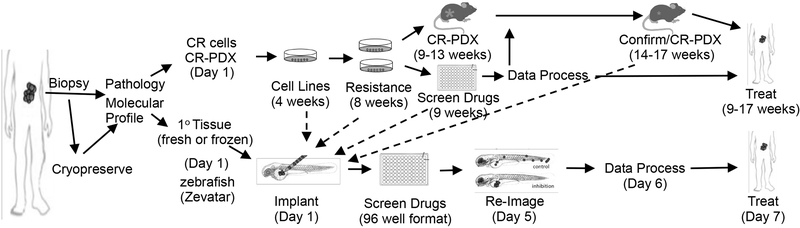
While our CR-PDX models are important new resources, they still require a significant investment of time and resources that, like conventional PDX models, may preclude them from routine adoption in drug sensitivity screens of individual patient cancer cells. In contrast, our zebrafish embryo model is rapid (5 days) and can provide a faithful readout of drug sensitivity. The multi-dimensional CR-based approaches described, in combination with direct implantation of patient tissue samples in zebrafish (MP, SB, CA unpublished), will significantly advance the use of patient-derived tissue for research with the potential for improving precision medicine-driven approaches for the treatment of PDAC (Fig. 7).
Fig. 7. Applications of fresh, frozen and CR-based PDAC samples.
Schematic and timelines of development of CR cells and of zebrafish and mouse CR-PDX platforms for translation of experimental data into the clinic. The times stated in the parentheses represent the estimated total time from data initiation to data analysis for each model system. Note that all drug outcomes can be rapidly confirmed in zebrafish.
Supplementary Material
Acknowledgments:
We thank R. Schlegel, X. Liu, S. Agarwal and P. Sripadhan for advice and technical support and C. Der and A. Cox for in depth KRAS advice.
Funding: C.A., J.R.B., J.M.W., and M.J.P. were supported by the 2015 Pancreatic Cancer Action Network-AACR Research Acceleration Network Grant, Grant Number 15–90-25-BROD. C.A., O.R., E.P., S.B. E.G and G.R. were funded by P30 CA051008. CA, SB and E.P were funded by the Department of Energy/Lawrence Livermore National Laboratory - DE-AC52–07NA27344. C.A. and D.K. were funded by U01 CA194730. C.I. was funded by NIH KL2TR001432. J.B. and E.L. were funded by P30 CA056036. S.M was funded by T32 CA009686. AS and GGL performed work under the auspices of the U.S. Department of Energy by Lawrence Livermore National Laboratory under Contract DE-AC52–07NA27344, and were supported by LLNL-LDRD SI-17–002.
Footnotes
Conflicts of interests: None. The CR cells can be obtained from C.A. under a collaborative material transfer agreement (MTA) with Georgetown University.
References:
- 1.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 2014;74:2913–21 [DOI] [PubMed] [Google Scholar]
- 2.Von Hoff DD, Ervin T, Arena FP, Chiorean EG, Infante J, Moore M, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med 2013;369:1691–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blomstrand H, Scheibling U, Bratthall C, Green H, Elander NO. Real world evidence on gemcitabine and nab-paclitaxel combination chemotherapy in advanced pancreatic cancer. BMC Cancer 2019;19:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boj SF, Hwang CI, Baker LA, Chio II, Engle DD, Corbo V, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell 2014;160:324–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang L, Holtzinger A, Jagan I, BeGora M, Lohse I, Ngai N, et al. Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell- and patient-derived tumor organoids. Nat Med 2015;21:1364–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolff RA, Wang-Gillam A, Alvarez H, Tiriac H, Engle D, Hou S, et al. Dynamic changes during the treatment of pancreatic cancer. Oncotarget 2018;9:14764–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu X, Ory V, Chapman S, Yuan H, Albanese C, Kallakury B, et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. American Journal of Pathology 2012;180:590–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan H, Myers S, Wang J, Zhou D, Woo JA, Kallakury B, et al. Use of reprogrammed cells to identify therapy for respiratory papillomatosis. N Engl J Med 2012;367:1220–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palechor-Ceron N, Suprynowicz FA, Upadhyay G, Dakic A, Minas T, Simic V, et al. Radiation Induces Diffusible Feeder Cell Factor(s) That Cooperate with ROCK Inhibitor to Conditionally Reprogram and Immortalize Epithelial Cells. Am J Pathol 2013;183:1862–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ringer L, Sirajuddin P, Tricoli L, Waye S, Parasido E, Lee RJ, et al. The Induction of the p53 Tumor Suppressor Protein Bridges the Apoptotic and Autophagic Signaling Pathways to Regulate Cell Death in Prostate Cancer Cells. Oncotarget 2014;5:10678–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Timofeeva OA, Palechor-Ceron N, Li G, Yuan H, Krawczyk E, Zhong X, et al. Conditionally reprogrammed normal and primary tumor prostate epithelial cells: a novel patient-derived cell model for studies of human prostate cancer. Oncotarget 2017;8:22741–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan H, Krawczyk E, Blancato J, Albanese C, Zhou D, Wang N, et al. HPV positive neuroendocrine cervical cancer cells are dependent on Myc but not E6/E7 viral oncogenes. Sci Rep 2017;7:45617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crystal AS, Shaw AT, Sequist LV, Friboulet L, Niederst MJ, Lockerman EL, et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science 2014;346:1480–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pollock CB, McDonough S, Wang VS, Lee H, Ringer L, Li X, et al. Strigolactone analogues induce apoptosis through activation of p38 and the stress response pathway in cancer cell lines and in conditionally reprogramed primary prostate cancer cells. Oncotarget 2014;5:1683–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tricoli L, Naeem A, Parasido E, Mikhaiel JP, Choudhry MU, Berry DL, et al. Characterization of the effects of defined, multidimensional culture conditions on conditionally reprogrammed primary human prostate cells. Oncotarget 2018;9:2193–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu X, Krawczyk E, Suprynowicz FA, Palechor-Ceron N, Yuan H, Dakic A, et al. Conditional reprogramming and long-term expansion of normal and tumor cells from human biospecimens. Nat Protoc 2017;12:439–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Correa BRS, Hu J, Penalva LOF, Schlegel R, Rimm DL, Galante PAF, et al. Patient-derived conditionally reprogrammed cells maintain intra-tumor genetic heterogeneity. Sci Rep 2018;8:4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evan GI, Hah N, Littlewood TD, Sodir NM, Campos T, Downes M, et al. Re-engineering the Pancreas Tumor Microenvironment: A “Regenerative Program” Hacked. Clin Cancer Res 2017;23:1647–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev 2000;14:2501–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sancho P, Burgos-Ramos E, Tavera A, Bou Kheir T, Jagust P, Schoenhals M, et al. MYC/PGC-1alpha Balance Determines the Metabolic Phenotype and Plasticity of Pancreatic Cancer Stem Cells. Cell Metab 2015;22:590–605 [DOI] [PubMed] [Google Scholar]
- 21.Sangodkar J, Perl A, Tohme R, Kiselar J, Kastrinsky DB, Zaware N, et al. Activation of tumor suppressor protein PP2A inhibits KRAS-driven tumor growth. J Clin Invest 2017;127:2081–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Votteler J, Ogohara C, Yi S, Hsia Y, Nattermann U, Belnap DM, et al. Designed proteins induce the formation of nanocage-containing extracellular vesicles. Nature 2016;540:292–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamiyama D, Sekine S, Barsi-Rhyne B, Hu J, Chen B, Gilbert LA, et al. Versatile protein tagging in cells with split fluorescent protein. Nat Commun 2016;7:11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010;26:589–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 2013;43:11 0 1–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu X, Wu C, Li C, Boerwinkle E. dbNSFP v3.0: A One-Stop Database of Functional Predictions and Annotations for Human Nonsynonymous and Splice-Site SNVs. Hum Mutat 2016;37:235–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parasido EM, Silvestri A, Canzonieri V, Belluco C, Diodoro MG, Milione M, et al. Protein drug target activation homogeneity in the face of intra-tumor heterogeneity: implications for precision medicine. Oncotarget 2017;8:48534–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baldelli E, Calvert V, Hodge A, VanMeter A, Petricoin EF 3rd, Pierobon M Reverse Phase Protein Microarrays. Methods Mol Biol 2017;1606:149–69 [DOI] [PubMed] [Google Scholar]
- 32.Fricke S, Rodriguez O, Vanmeter J, Dettin L, Casimiro M, Chien C, et al. In Vivo Magnetic Resonance Volumetric and Spectroscopic Analysis of Mouse Prostate Cancer Models. Prostate 2006;66:708–17 [DOI] [PubMed] [Google Scholar]
- 33.Tian Y, Wang S, Zhang Z, Rodriguez O, Petricoin EF 3rd, Avantaggiati ML, et al. Intergration of network biology and imaging to study cancer phenotypes and responses. IEEE Trans on Comp Biol and Bioinformatics 2014;11:1009–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vaseva AV, Blake DR, Gilbert TSK, Ng S, Hostetter G, Azam SH, et al. KRAS Suppression-Induced Degradation of MYC Is Antagonized by a MEK5-ERK5 Compensatory Mechanism. Cancer Cell 2018;34:807–22 e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fuxman Bass JI, Diallo A, Nelson J, Soto JM, Myers CL, Walhout AJ. Using networks to measure similarity between genes: association index selection. Nat Methods 2013;10:1169–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kao SH, Wu HT, Wu KJ. Ubiquitination by HUWE1 in tumorigenesis and beyond. J Biomed Sci 2018;25:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Myant KB, Cammareri P, Hodder MC, Wills J, Von Kriegsheim A, Gyorffy B, et al. HUWE1 is a critical colonic tumour suppressor gene that prevents MYC signalling, DNA damage accumulation and tumour initiation. EMBO Mol Med 2017;9:181–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Awasthi N, Monahan S, Stefaniak A, Schwarz MA, Schwarz RE. Inhibition of the MEK/ERK pathway augments nab-paclitaxel-based chemotherapy effects in preclinical models of pancreatic cancer. Oncotarget 2018;9:5274–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Infante JR, Somer BG, Park JO, Li CP, Scheulen ME, Kasubhai SM, et al. A randomised, double-blind, placebo-controlled trial of trametinib, an oral MEK inhibitor, in combination with gemcitabine for patients with untreated metastatic adenocarcinoma of the pancreas. Eur J Cancer 2014;50:2072–81 [DOI] [PubMed] [Google Scholar]
- 40.Narla G, Sangodkar J, Ryder CB. The impact of phosphatases on proliferative and survival signaling in cancer. Cell Mol Life Sci 2018;75:2695–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kauko O, O’Connor CM, Kulesskiy E, Sangodkar J, Aakula A, Izadmehr S, et al. PP2A inhibition is a druggable MEK inhibitor resistance mechanism in KRAS-mutant lung cancer cells. Sci Transl Med 2018;10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ben-David U, Siranosian B, Ha G, Tang H, Oren Y, Hinohara K, et al. Genetic and transcriptional evolution alters cancer cell line drug response. Nature 2018;560:325–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Willyard C The mice with human tumours: Growing pains for a popular cancer model. Nature 2018;560:156–7 [DOI] [PubMed] [Google Scholar]
- 44.Wolf E, Lin CY, Eilers M, Levens DL. Taming of the beast: shaping Myc-dependent amplification. Trends Cell Biol 2015;25:241–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Allen-Petersen BL, Risom T, Feng Z, Wang Z, Jenny ZP, Thoma MC, et al. Activation of PP2A and Inhibition of mTOR Synergistically Reduce MYC Signaling and Decrease Tumor Growth in Pancreatic Ductal Adenocarcinoma. Cancer Res 2019;79:209–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Farrell AS, Allen-Petersen B, Daniel CJ, Wang X, Wang Z, Rodriguez S, et al. Targeting inhibitors of the tumor suppressor PP2A for the treatment of pancreatic cancer. Mol Cancer Res 2014;12:924–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chien W, Sun QY, Lee KL, Ding LW, Wuensche P, Torres-Fernandez LA, et al. Activation of protein phosphatase 2A tumor suppressor as potential treatment of pancreatic cancer. Mol Oncol 2015;9:889–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li W, Chen Z, Gong FR, Zong Y, Chen K, Li DM, et al. Growth of the pancreatic cancer cell line PANC-1 is inhibited by protein phosphatase 2A inhibitors through overactivation of the c-Jun N-terminal kinase pathway. Eur J Cancer 2011;47:2654–64 [DOI] [PubMed] [Google Scholar]
- 49.Hein AL, Seshacharyulu P, Rachagani S, Sheinin YM, Ouellette MM, Ponnusamy MP, et al. PR55alpha Subunit of Protein Phosphatase 2A Supports the Tumorigenic and Metastatic Potential of Pancreatic Cancer Cells by Sustaining Hyperactive Oncogenic Signaling. Cancer Res 2016;76:2243–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takahashi K, Ehata S, Koinuma D, Morishita Y, Soda M, Mano H, et al. Pancreatic tumor microenvironment confers highly malignant properties on pancreatic cancer cells. Oncogene 2018;37:2757–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.