Abstract
Enzymes like uracil DNA glycosylase (UDG) can achieve ground state destabilization, by polarizing substrates to mimic rare tautomers. On the basis of computed nucleus independent chemical shifts, NICS(1)zz, and harmonic oscillator model of electron delocalization (HOMED) analyses, of quantum mechanics (QM) and quantum mechanics/molecular mechanics (QM/MM) models of the UDG active site, uracil is strongly polarized when bound to UDG and resembles a tautomer >12 kcal/mol higher in energy. Natural resonance theory (NRT) analyses identified a dominant O2 imidate resonance form for residue bound 1-methyluracil. This “tautomeric strain” raises the energy of uracil, making uracilate a better than expected leaving group. Computed gas-phase SN2 reactions of free and hydrogen bonded 1-methyl-uracil demonstrate the relationship between the degree of polarization in uracil and the leaving group ability of uracilate.
In this Communication, we present computational evidence showing that enzyme environments can polarize substrates to mimic rare tautomers and, in this way, achieve ground state destabilization1,2 through substrate strain. In the active site of uracil DNA glycosylase (UDG), uracil is polarized, and the resulting structure bears resemblance to a tautomeric form that is much higher in energy.
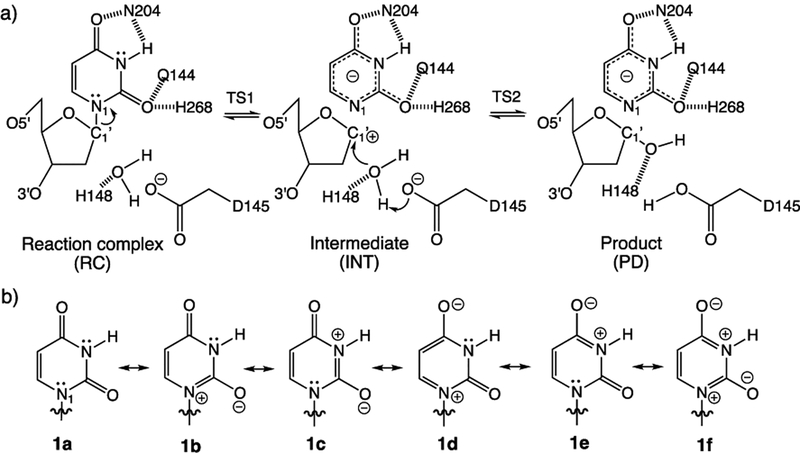
UDG is a base excision repair enzyme that removes mutagenic uracil from DNA by cleaving the glycosidic N1−C1′ bond of uridine. Among the family of base excision repair enzymes, UDG is the most efficient and well-characterized, showing an enormous catalytic rate acceleration of 1012-fold. Studies based on NMR, Raman, kinetic isotope effect measurements, and quantum mechanics/molecular mechanics (QM/MM) computations established a stepwise dissociative mechanism, involving uracilate as the leaving group3–17 (Figure 1a).
Figure 1.
(a) Proposed stepwise dissociative pathway for UDG. (b) Diketo (1a) and imidate (1b−1f) resonance forms of uracil.
However, a puzzle regarding UDG is how it activates the removal of N1 deprotonated uracilate. In water (pH = 7), uracil N1–H has a pKa of 9.8 (i.e., only as acidic as ammonium), and N1 deprotonated uracilate is not considered to be a very good leaving group at all. Parikh and Tainer et al. suggested that, in UDG, uridine adopts a strained conformation that weakens the glycosidic N1–C1′ bond through anomeric effects.10 Drohat and Stivers showed, on the basis of NMR experiments, that uracil displayed increased acidity (pKa = 6.4) in the active site of UDG, indicating the appearance of an O2 imidate form.11,12 Wetmore et al. showed that uracil exhibited increased acidity when hydrogen bonded at the C=O and N–H sites.18,19 Nevertheless, Lee et al. pointed out that increased (thermodynamic) acidity in uracil cannot be used to reason the (kinetic) leaving group ability of uracilate in UDG.20–22 In the gas phase, uracil N1−H is as acidic as HCl, but the leaving group ability of N1 deprotonated uracilate is much poorer compared to that of Cl−.21
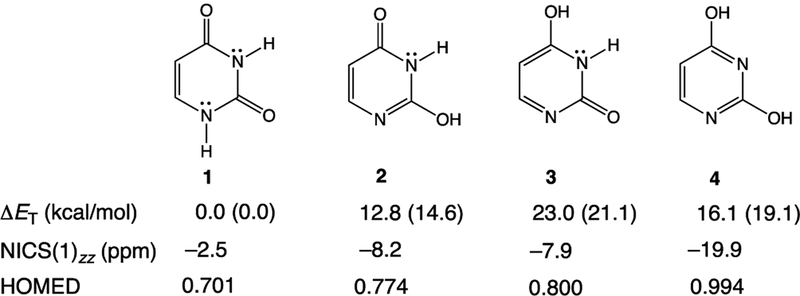
Here, we show that when uracil is bound to UDG, it does not exist in the canonical diketo form (1) but instead transforms to a structure akin to a rare tautomer of uracil.23–28 In UDG, uracil forms short hydrogen bonds to several nearby residues (His268, Asn204, Gln144, and the backbone of Gln144). These hydrogen bonding interactions polarize the π-electrons of uracil, increase cyclic [4n + 2] π-electron delocalization in the ring (see resonance forms 1b−1f, in Figure 1b), and the resulting polarized uracil mimics a higher energy keto−enol tautomer (2 and 3, see also the dienol form, 4, Figure 2). Notably, all of these tautomers are >12 kcal/mol higher in energy than the canonical diketo form (1) (see ΔET computed in the gas phase and at ε = 4, Figure 2). A dielectric constant (ε) of 4.0 was employed to simulate the enzyme active site environment.29–31 Computed relative energies for residue−tautomer complexes are provided in the Supporting Information, SI, for comparison (see Table S1). We now show that this “tautomeric strain” raises the energy of uracil in UDG, bringing it closer to the transition state structure for N1−C1′ bond cleavage. In this way, ground state destabilization helps reduce the activation barrier to expel uracilate.
Figure 2.
Computed tautomerization energies (ΔET) in the gas phase and at ε = 4 (values in parentheses), NICS(1)zz, and HOMED for the canonical diketo form of uracil (1), and its keto−enol (2, 3) and dienol (4) tautomers.
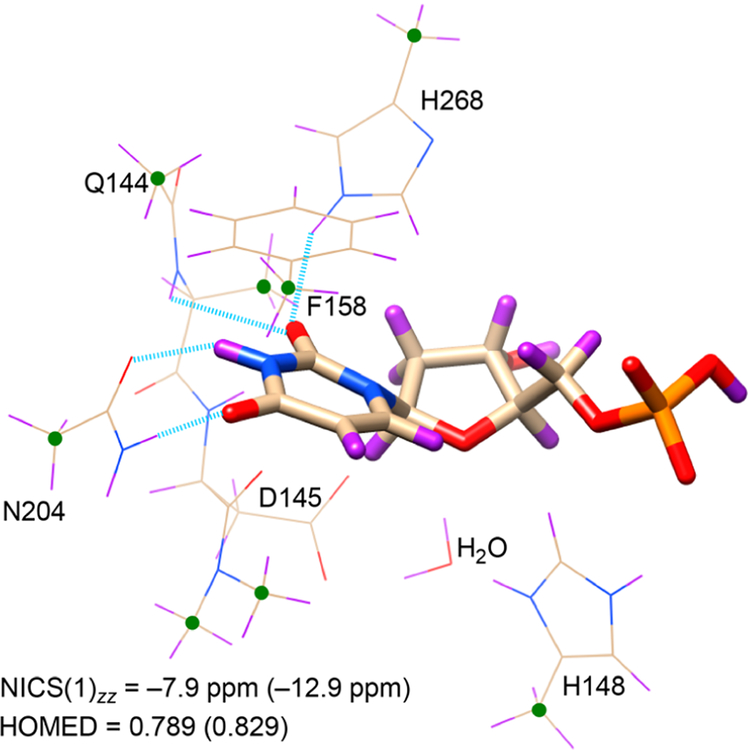
According to computed nucleus independent chemical shifts,32,33 NICS(1)zz, uracil, 1, is essentially nonaromatic (−2.5 ppm), and solvation in water, based on either implicit (−3.9 ppm, employing the IEF-PCM approach) or explicit (−3.3 ppm, including three water molecules) models, increases its aromaticity negligibly. This implies that solvating naked uracil in aqueous solution has a minor effect on its tautomeric form. However, when uracil is bound to the active site of UDG, it becomes markedly aromatic. NICS(1)zz values computed for uracil in truncated QM (−7.9 ppm) and QM/MM (−12.9 ppm) models of the UDG active site document significant aromaticity gain, suggesting a close resemblance of the electronic structure of the substrate to the rare tautomers of uracil. Despite being higher in energy, 2 (−8.2 ppm), 3 (−7.9 ppm), and 4 (−19.9 ppm) are all more aromatic than 1 (−2.5 ppm) (Figure 2). Differences in the estimated aromaticity gain of uracil based on QM and QM/MM models suggest that uracil is “aromatized” not only by nearby hydrogen bonding residues but also by other long-range electrostatic interactions in the active site of UDG. See the description of truncated QM and QM/MM models in the Computational Methods section.
Harmonic oscillator model of electron delocalization (HOMED) values,34 a geometric index for aromaticity, indicate significant aromaticity gain for uracil 1 (0.701) when its geometry is considered in truncated QM (0.789) and QM/MM (0.829) models of the UDG active site (cf. HOMED values for 2, 0.774; 3, 0.800; 4, 0.994). HOMED values close to 1 indicate fully aromatic rings. This “aromatizing” effect brings the uracil substrate closer in geometry to the N1 deprotonated uracilate (HOMED: 0.811). Natural resonance theory (NRT) analyses35–37 for 1-methyl-uracil reveal a dominant diketo resonance form 1a (31.3%), followed by smaller weights for the O2 imidate (16.5%) and O4 imidate (7.9%) forms. However, when 1-methyl-uracil is hydrogen bonded to His, Asn, a fragment of protein backbone, and freely optimized, the resonance weight of 1a decreases (15.0%), while those of O2 imidate (24.1%) and O4 imidate (19.5%) increase. Changes in these resonance weights suggest that, in UDG, the electronic structure of polarized uracil resembles its keto−enol tautomers (2 and 3), which are 12−23 kcal/mol higher in energy. We note that the dominant O2 imidate form of hydrogen bonded 1-methyl-uracil agrees with Drohat and Stivers’ finding of increased acidity of uracil due to the appearance of an O2 imidate.
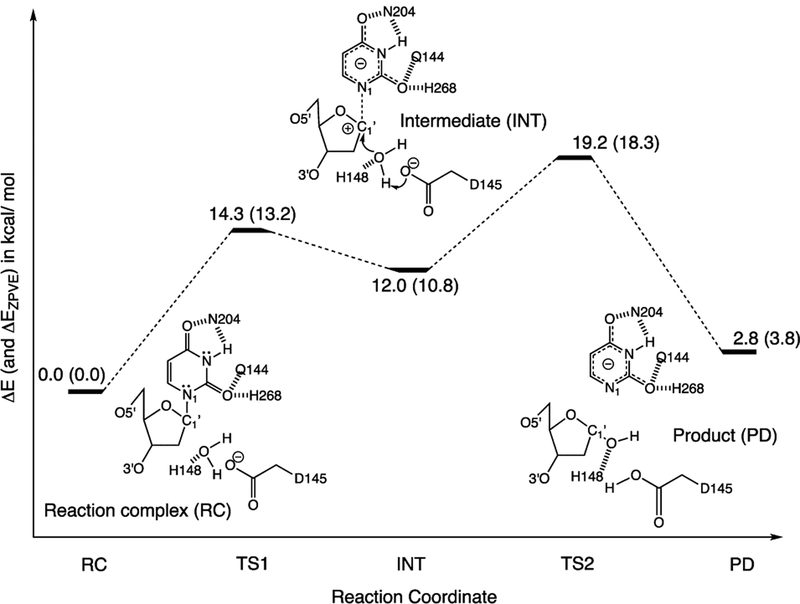
Remarkably, the majority of aromaticity gain in uracil happens as soon as it is transferred from a nonenzymic environment (i.e., in water) into the UDG active site. From there, the geometry and electronic distribution of uracil already resemble a higher energy keto−enol tautomer, and the incipient uracilate is set up to be a better than expected leaving group. We computed NICS(1)zz for uracil and uracilate rings at stationary points along the computed reaction potential energy surface and found that uracil continues to gain aromaticity throughout the stepwise dissociative reaction: RC (−7.9 ppm), TS1 (−9.4 ppm), INT (−10.8 ppm), TS2 (−11.3 ppm), PD (−11.2 ppm) (see Figure 4). All stationary points were computed on the basis of a constrained and truncated QM model of the UDG active site, and the relative energies closely follow prior QM/MM studies.15
Figure 4.
Relative energies (ΔE, in kcal/mol) of computed stationary points along the stepwise dissociative pathway of UDG, based on a constrained and truncated QM model (see the Computational Methods section for details) of the UDG active site. Values corrected for zero-point energy vibration (ZPVE) are shown in parentheses (ΔEZPVE, in kcal/mol). Geometries of all stationary points were optimized at ωB97X-D/6-31+G(d).
We note that “aromatization” of uracilate can also contribute to preferential enzyme−substrate binding in the transition state. We have shown that hydrogen bonding interactions that polarize π-electrons to increase the aromatic character of heterocycles become stronger than expected.38,39 At the transition state, hydrogen bonding interactions between the UDG active site residues and uracil polarize the π-electrons of uracil to increase aromaticity; in this way, the corresponding hydrogen bonds are strengthened, and preferential binding can happen.
As a model to probe the effect of hydrogen bonding on barrier to N−C bond cleavage, we computed gas-phase SN2 reaction barriers for free and hydrogen bonded 1-methyl-uracil (with formate as the nucleophile). In the absence of hydrogen bonding interactions, the activation free energy barrier for N−CH3 bond cleavage is ΔGa = 37.8 kcal/mol, but the barrier reduces when 1-methyl-uracil is hydrogen bonded to three waters (35.7 kcal/mol) and even more so when 1-methyl-uracil is hydrogen bonded to two zwitterionic glycines (33.3 kcal/mol) (see Figure S1 in the SI). Computed NICS(1)zz values show that these changes correlate to increased aromaticity in 1-methyl-uracil in the reaction complex (free 1-methyl-uracil, −4.0 ppm; bound to three waters, −5.0 ppm; bound to two glycines, −5.8 ppm)–increased aromaticity in 1-methyl-uracil enhances the leaving group ability of uracilate. As the uracil ring becomes structurally more like its rare keto−enol tautomers, uracilate becomes a better leaving group.
Tautomeric strain is one way to raise the energy of substrates in enzymes, and recognizing this mode of ground state destabilization has important interpretive merit for understanding how enzymes work. Although examples of electronic strain in substrates have been reported for various enzymes, for example, substrates with polarized C=O, C=C, and C–H bonds or red-shifted UV absorptions,40 the possibility and impact of substrate tautomerization have not been fully appreciated. Other enzymes that catalyze the glycosidic hydrolysis or biosynthesis of purine or pyrimidine substrates may take advantage of a similar mechanistic trick.
Computational Methods.
All quantum mechanical (QM) geometries were optimized at ωB97X-D/6-31+G(d) in the gas phase employing Gaussian09.41 Vibrational frequency analyses verified the nature of the stationary points. Computed nucleus independent chemical shifts (NICS),32,33 harmonic oscillator model of electron delocalization (HOMED),34 and natural resonance theory (NRT)35,36 data were performed at the same level. Computations in implicit solvation employed the IEFPCM model.
A QM model of the UDG active site was computed based on a modified, constrained, and geometry optimized crystal structure of a UDG-inhibitor complex (PDBID: 1EMH). In the pdb file, the positions of atoms C1 and N5 in the deoxypseudouridine substrate were exchanged to give a deoxyuridine substrate containing a N1−C1′ glycosidic bond. Residues and protein backbone fragments that formed hydrogen bonding (His268, Asn204, Gln144, and the backbone of Gln144) and π-stacking (Phe158) interactions to uracil, along with those relevant for activating (His148 and Asp145) the nucleophilic water molecule, were included to the model and truncated at selected carbon positions. Prior QM/MM studies document the important roles of these residues in UDG.15 The 5′ phosphate group was included because of its recognized importance for facilitating the dissociation of uracilate. The positions of selected atoms were frozen (see green dots in Figure 3) during geometry optimization of the RC, TS1, INT, TS2, and PD. Following prior QM/MM studies, bond distances for N1−C1′ were fixed to 2.04 Å (for TS1), 2.74 Å (for INT), and 2.95 Å (for TS2), and distances between C1′ and the O atom of water were fixed to 3.04 Å (for TS1), 2.85 Å (for INT), and 1.95 Å (for TS2).15
Figure 3.
Truncated QM model of the UDG active site: Green dots and blue dashed lines indicate fixed positions of atoms and bond distances during geometry optimization. Computed NICS(1)zz and HOMED values for uracil in the truncated QM model (values based on the QM/MM model are in parentheses).
All QM/MM calculations were performed using the LICHEM software package,42,43 which employs Gaussian09 for the quantum region (QM) and TINKER844 for the classical region (MM). The crystal structure of a UDG-inhibitor complex was not solvated for QM/MM calculations. The QM region (UDG: Gln144, Asp145, Pro146, Tyr147, His148, Phe158, Asn204, His268, deoxyuridine, and one water molecule) was calculated at ωB97X-D/6-31+G(d). The QM/MM system involved the cleavage of covalent bonds, and thus the pseudobond approach was employed.45 Two QM/MM simulation systems (see Figures S2 and S3 in the SI) were created on the basis of the crystal structure of a UDG-inhibitor complex: one to perform the QM/MM geometry optimization (126 QM atoms + 6 pseudobonds) and the other for the QM/MM single-point calculation (154 QM atoms + 8 pseudo-bonds). The MM region was modeled using the AMOEBABIO18 force field.46,47
Supplementary Material
ACKNOWLEDGMENTS
J.I.W. thanks the National Science Foundation (NSF) for support (CHE-1751370) and computational resources provided by the uHPC cluster managed by the University of Houston and acquired through support from the NSF (MRI-1531814). G.A.C. thanks the UNT Department of Chemistry for the use of the HPC Cluster CRUNTCh3, as well as support from the NSF (CHE-1531468) and NIH (R01GM108583). E.A.V.-M. wishes to acknowledge CONACyT for funding. We thank Dr. Chia-Hua Wu for helpful discussions.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.9b06447.
Details of the HOMED method, optimized Cartesian coordinates for all QM and QM/MM structures, and computed gas-phase N−CH3 cleavage barriers for model SN2 reactions (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Jencks WP Catalysis in Chemistry and Enzymology; McGraw-Hill: New York, 1969; p 282. [Google Scholar]
- (2).Menger FM Analysis of ground-state and transition-state effects in enzyme catalysis. Biochemistry 1992, 31, 5368–5373. [DOI] [PubMed] [Google Scholar]
- (3).Savva R; McAuley-Hecht K; Brown T; Pearl L The structural basis of specific base-excision repair by uracil–DNA glycosylase. Nature 1995, 373, 487–493. [DOI] [PubMed] [Google Scholar]
- (4).Mol CD; Arvai AS; Slupphaug G; Kavli B; Alseth I; Krokan HE; Tainer JA Crystal structure of human uracil−DNA glycosylase in complex with a protein inhibitor: Protein mimicry of DNA. Cell 1995, 80, 869–878. [DOI] [PubMed] [Google Scholar]
- (5).Slupphaug G; Mol CD; Kavli B; Arvai AS; Krokan HE; Tainer JA A nucleotide-flipping mechanism from the structure of human uracil–DNA glycosylase bound to DNA. Nature 1996, 384, 87–92. [DOI] [PubMed] [Google Scholar]
- (6).Kimura E; Kitamura H; Koike T; Shiro M Facile and selective electrostatic stabilization of uracil N(1)− anion by a proximate protonated amine: A chemical implication for why uracil N(1) is chosen for glycosylation site. J. Am. Chem. Soc 1997, 119, 10909–10919. [Google Scholar]
- (7).Drohat AC; Jagadeesh J; Ferguson E; Stivers JT Role of electrophilic and general base catalysis in the mechanism of Escherichia coli uracil DNA glycosylase. Biochemistry 1999, 38, 11866–11875. [DOI] [PubMed] [Google Scholar]
- (8).Shroyer MJN; Bennett SE; Putnam CD; Tainer JA; Mosbaugh DW Mutation of an active site residue in Escherichia coli uracil DNA glycosylase: effect on DNA binding, uracil inhibition and catalysis. Biochemistry 1999, 38, 4834–4845. [DOI] [PubMed] [Google Scholar]
- (9).Drohat AC; Xiao G; Tordova M; Jagadeesh J; Pankiewicz KW; Watanabe KA; Gilliland GL; Stivers JT Heteronuclear NMR and crystallographic studies of wild-type and H187Q Escherichia coli uracil DNA glycosylase: electrophilic catalysis of uracil expulsion by a neutral histidine 187. Biochemistry 1999, 38, 11876–11886. [DOI] [PubMed] [Google Scholar]
- (10).Parikh SS; Walcher G; Jones GD; Slupphaug G; Krokan HE; Blackburn GM; Tainer JA Uracil-DNA glycosylase-DNA substrate and product structures: conformational strain promotes catalytic efficiency by coupled stereoelectronic effects. Proc. Natl. Acad. Sci. U. S. A 2000, 97, 5083–5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Drohat AC; Stivers JT Escherichia coli uracil DNA glycosylase: NMR characterization of the short hydrogen bond from His187 to uracil O2. Biochemistry 2000, 39, 11865–11875. [DOI] [PubMed] [Google Scholar]
- (12).Drohat AC; Stivers JT NMR evidence for an unusually low N1 pKa for uracil bound to uracil DNA glycosylase: implications for catalysis. J. Am. Chem. Soc 2000, 122, 1840–1841. [Google Scholar]
- (13).Werner RM; Stivers JT Kinetic isotope effect studies of the reaction catalyzed by uracil DNA glycosylase: evidence for an oxocarbenium ion−uracil anion intermediate. Biochemistry 2000, 39, 14054–14064. [DOI] [PubMed] [Google Scholar]
- (14).Dong J; Drohat AC; Stivers JT; Pankiewicz KW; Carey PR Raman spectroscopy of uracil DNA glycosylase-DNA complexes: insights into DNA damage recognition and catalysis. Biochemistry 2000, 39, 13241–13250. [DOI] [PubMed] [Google Scholar]
- (15).Dinner AR; Blackburn GM; Karplus M Uracil-DNA glycosylase acts by substrate autocatalysis. Nature 2001, 413, 752–755. [DOI] [PubMed] [Google Scholar]
- (16).Stivers JT; Jiang YL A mechanistic perspective on the chemistry of DNA repair glycosylases. Chem. Rev 2003, 103, 2729–2759. [DOI] [PubMed] [Google Scholar]
- (17).Parker JB; Stivers JT Uracil DNA glycosylase: revisiting substrate-assisted catalysis by DNA phosphate anions. Biochemistry 2008, 47, 8614–8622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Whittleton SR; Hunter KC; Wetmore SD Effects of hydrogen bonding on the acidity of uracil derivatives. J. Phys. Chem. A 2004, 108, 7709–7718. [Google Scholar]
- (19).Di Laudo M; Whittleton SR; Wetmore SD Effects of hydrogen bonding on the acidity of uracil. J. Phys. Chem. A 2003, 107, 10406–10413. [Google Scholar]
- (20).Kurinovich MA; Lee JK The acidity of uracil from the gas phase to solution: The coalescence of the N1 and N3 sites and implications for biological glycosylation. J. Am. Chem. Soc 2000, 122, 6258–6262. [Google Scholar]
- (21).Zhachkina A; Lee JK Uracil and thymine reactivity in the gas phase: The SN2 reaction and implications for electron delocalization in leaving groups. J. Am. Chem. Soc 2009, 131, 18376–18385. [DOI] [PubMed] [Google Scholar]
- (22).Chen M; Lee JK Computational studies of the gas-phase thermochemical properties of modified nucleobases. J. Org. Chem 2014, 79, 11295–11300. [DOI] [PubMed] [Google Scholar]
- (23).Daniels M Tautomerism of uracil and thymine in aqueous solution: spectroscopic evidence. Proc. Natl. Acad. Sci. U. S. A 1972, 69, 2488–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Tsuchiya Y; Tamura T; Fujii M; Ito M Keto-enol tautomer of uracil and thymine. J. Phys. Chem 1988, 92, 1760–1765. [Google Scholar]
- (25).Rejnek J; Hanus M; Kabelac M; Ryjacek F; Hobza P Correlated ab initio study of nucleic acid bases and their tautomers in the gas phase, in a microhydrated environment and in aqueous solution. Part 4. Uracil and thymine. Phys. Chem. Chem. Phys 2005, 7, 2006–2017. [DOI] [PubMed] [Google Scholar]
- (26).Kryachko ES; Nguyen MT; Zeegers-Huyskens T Theoretical study of tautomeric forms of uracil. 1. Relative order of stabilities and their relation to proton affinities and deprotonation enthalpies. J. Phys. Chem. A 2001, 105, 1288–1295. [Google Scholar]
- (27).Kurinovich MA; Lee JK The acidity of uracil and uracil analogs in the gas phase: four surprisingly acidic sites and biological implications. J. Am. Soc. Mass Spectrom 2002, 13, 985–995. [DOI] [PubMed] [Google Scholar]
- (28).Colasurdo DD; Pila MN; Iglesias DA; Laurella SL; Ruiz DL Tautomerism of urail and related compounds: A mass spectrum study. Eur. J. Mass Spectrom 2018, 24, 214–224. [DOI] [PubMed] [Google Scholar]
- (29).DeYonker NJ; Webster CE A theoretical study of phosphoryl transfers of Tyrosyl-DNA Phosphodiesterase I (Tdp1) and the possibility of a “Dead-End” phosphohistidine intermediate. Biochemistry 2015, 54, 4236–4247. [DOI] [PubMed] [Google Scholar]
- (30).Blomberg MRA; Borowski T; Himo F; Liao R-Z; Siegbahn PEM Quantum chemical studies of mechanisms for metalloenzymes. Chem. Rev 2014, 114 (7), 3601–3658. [DOI] [PubMed] [Google Scholar]
- (31).Himo F Recent trends in quantum chemical modeling of enzymatic reactions. J. Am. Chem. Soc 2017, 139, 6780–6786. [DOI] [PubMed] [Google Scholar]
- (32).Schleyer P. v. R.; Maerker C; Dransfeld A; Jiao H; Hommes N. J. R. v. E. Nucleus-independent chemical shifts: A simple and efficient aromaticity probe. J. Am. Chem. Soc 1996, 118, 6317–6318. [DOI] [PubMed] [Google Scholar]
- (33).Chen Z; Wannere CS; Corminboeuf C; Puchta R; Schleyer P. v. R. Nucleus-independent chemical shifts (NICS) as an aromaticity criterion. Chem. Rev 2005, 105, 3842–3888. [DOI] [PubMed] [Google Scholar]
- (34).Raczyñska ED; Hallman M; Kolczyñska K; Stêpniewski TM On the harmonic oscillator model of electron delocalization (HOMED) index and its application to heteroatomic π-electron systems. Symmetry 2010, 2, 1485–1509. [Google Scholar]
- (35).Glendening ED; Badenhoop JK; Reed AE; Carpenter JE; Bohmann JA; Morales CM; Karafiloglou P; Landis CR; Weinhold F NBO, 7.0 Theoretical Chemistry Institute, University of Wisconsin: Madison, 2018. [Google Scholar]
- (36).Glendening ED; Badenhoop JK; Weinhold F Natural resonance theory: III. chemical applications. J. Comput. Chem 1998, 19, 628–646. [Google Scholar]
- (37).Glendening ED; Landis CR; Weinhold F Resonance theory reboot. J. Am. Chem. Soc 2019, 141, 4156–4166. [DOI] [PubMed] [Google Scholar]
- (38).Wu JI; Jackson JE; Schleyer P. v. R. Reciprocal hydrogen bonding-aromaticity relationships. J. Am. Chem. Soc 2014, 136, 13526–13529. [DOI] [PubMed] [Google Scholar]
- (39).Kakeshpour T; Wu JI; Jackson JE AMHB: (anti-)aromaticity-modulated hydrogen bonding. J. Am. Chem. Soc 2016, 138, 3427–3432. [DOI] [PubMed] [Google Scholar]
- (40).Anderson VE Ground state destabilization In Encyclopedia of Life Sciences; John Wiley & Sons, Ltd., 2001; www.els.net, and references therein. [Google Scholar]
- (41).Frisch MJ; Trucks GW; Schlegel HB; Scuseria GE; Robb MA; Cheeseman JR; Scalmani G; Barone V; Mennucci B; Petersson GA; Nakatsuji H; Caricato M; Li X; Hratchian HP; Izmaylov AF; Bloino J; Zheng G; Sonnenberg JL; Hada M; Ehara M; Toyota K; Fukuda R; Hasegawa J; Ishida M; Nakajima T; Honda Y; Kitao O; Nakai H; Vreven T; Montgomery JA Jr.; Peralta JE; Ogliaro F; Bearpark M; Heyd JJ; Brothers E; Kudin KN; Staroverov VN; Kobayashi R; Normand J; Raghavachari K; Rendell A; Burant JC; Iyengar SS; Tomasi J; Cossi M; Rega N; Millam JM; Klene M; Knox JE; Cross JB; Bakken V; Adamo C; Jaramillo J; Gomperts R; Stratmann RE; Yazyev O; Austin AJ; Cammi R; Pomelli C; Ochterski JW; Martin RL; Morokuma K; Zakrzewski VG; Voth GA; Salvador P; Dannenberg JJ; Dapprich S; Daniels AD; Farkas O; Foresman JB; Ortiz JV; Cioslowski J; Fox DJ Gaussian 09, revision D.01; Gaussian, Inc.: Wallingford, CT, 2009. [Google Scholar]
- (42).Kratz EG; Walker AR; Lagardère L; Lipparini F; Piquemal J-P; Andrés Cisneros G LICHEM: A QM/MM program for simulations with multipolar and polarizable force fields. J. Comput. Chem 2016, 37, 1019–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Gökcan H; Vázquez-Montelongo EA; Cisneros GA LICHEM 1.1: recent improvements and new capabilities. J. Chem. Theory Comput 2019, 15, 3056–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Rackers JA; Wang Z; Lu C; Laury ML; Lagardère L; Schnieders MJ; Piquemal J-P; Ren P; Ponder JW Tinker 8: Software Tools for Molecular Design. J. Chem. Theory Comput 2018, 14, 5273–5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Parks JM; Hu H; Cohen AJ; Yang W A pseudobond parametrization for improved electrostatics in quantum mechanical/molecular mechanical simulations of enzymes. J. Chem. Phys 2008, 129, 154106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Shi Y; Xia Z; Zhang J; Best R; Ponder JW; Ren P Polarizable Atomic Multipole-Based AMOEBA Force Field for Proteins. J. Chem. Theory Comput 2013, 9, 4046–4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Zhang C; Lu C; Jing Z; Wu C; Piquemal J-P; Ponder JW; Ren P AMOEBA Polarizable Atomic Multipole Force Field for Nucleic Acids. J. Chem. Theory Comput 2018, 14, 2084–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.