Abstract
Epigenetic gene regulation and metabolism are highly intertwined, yet little is known about whether altered epigenetics influence cellular metabolism during cancer progression. Here we show that EZH2 and NRasG12D mutations cooperatively induce progression of myeloproliferative neoplasms to highly penetrant, transplantable and lethal myeloid leukemias in mice. EZH1, an EZH2 homolog, is indispensable for EZH2-deficient leukemia-initiating cells and constitutes an epigenetic vulnerability. BCAT1, which catalyzes the reversible transamination of branched-chain amino acids (BCAAs), is repressed by EZH2 in normal hematopoiesis and aberrantly activated in EZH2-deficient myeloid neoplasms in mice and humans. BCAT1 reactivation cooperates with NRasG12D to sustain intracellular BCAA pools, resulting in enhanced mTOR signaling in EZH2-deficient leukemia cells. Genetic and pharmacological inhibition of BCAT1 selectively impairs EZH2-deficient leukemia-initiating cells and constitutes a metabolic vulnerability. Hence, epigenetic alterations rewire intracellular metabolism during leukemic transformation, causing epigenetic and metabolic vulnerabilities in cancer-initiating cells.
Keywords: Myeloproliferative Neoplasms, Leukemia, Epigenetics, Metabolism, Branched-Chain Amino Acids
INTRODUCTION
Epigenetic and metabolic alterations are highly intertwined in cancer cells. Many epigenetic enzymes catalyzing DNA or histone modifications are susceptible to changes in co-substrates of metabolism, thus providing a potential link between intracellular metabolism and epigenetic regulation (1–3). It has been postulated that epigenetic alterations may also impact metabolism to control disease progression, largely based on the assumption that altered epigenetics may lead to deregulated metabolic genes. However, few examples exist to demonstrate whether and how epigenetic alterations influence metabolism during cancer progression.
Myeloproliferative neoplasms (MPNs) are progressive blood cancers consisting of polycythemia vera (PV), essential thrombocythemia (ET), primary myelofibrosis (PMF), and prefibrotic PMF (4). The identification of driver mutations in JAK2, CALR or MPL gene has transformed our knowledge of MPN pathogenesis (5,6); however, patients with non-mutated JAK2, CALR and MPL (so-called ‘triple-negative’) have the highest incidence of leukemic transformation (7), indicating that other factors may also contribute to MPN progression. Mutations in NRAS, a member of the RAS GTPases, are found in 7~19% of post-MPN leukemias (5,8,9). Interestingly, NRAS mutations occurred exclusively in triple-negative MPNs (10), illustrating a unique role of oncogenic RAS in myeloid transformation. The molecular processes controlling MPN progression to leukemic transformation remain unknown. This poses a major barrier for developing target-based therapeutics to selectively eliminate mutant stem cells to prevent disease progression and/or relapse.
EZH2, the enzymatic subunit of the Polycomb Repressive Complex 2 (PRC2) that catalyzes H3-Lys27 methylation, is one of the most frequently mutated epigenetic regulators in hematologic malignancies. Loss-of-function EZH2 mutations are found in 12~25% of MPNs, 10~15% of myelodysplastic syndrome (MDS), and 20~33% of juvenile myelomonocytic leukemia (JMML) (11–14). Other common mutations in myeloid neoplasms including ASXL1 and SRSF2 also affect EZH2 function through impaired chromatin recruitment or aberrant mRNA splicing, suggesting that the frequency of EZH2 dysregulation may be under-estimated (15,16). Inactivating EZH2 mutations are associated with worse clinical outcomes in MPNs (17,18). Paradoxically, overexpression or gain-of-function mutations of EZH2 are also common in cancers (19,20), indicating that both hyper- and hypoactive EZH2 can be tumorigenic. PRC2 consists of EED, SUZ12, and the homologous methyltransferases EZH1 and EZH2. While loss of Ezh1 or Ezh2 has minimal effect on hematopoiesis in mice, complete loss of PRC2 by combined knockout (KO) of Ezh1 and Ezh2, or Eed KO, leads to loss of hematopoietic stem cells (HSCs) (21–23), suggesting that PRC2 regulates normal HSCs in a dose-dependent manner. Although studies have shown that Ezh2 loss in combination with other lesions such as Jak2V617F, Runx1 or Tet2 mutations promote myeloid or lymphoid malignancies (24–28), it remains unclear how different PRC2 dosages contribute to the development of hematopoietic malignancies under physiological conditions.
BCAAs (Valine, Isoleucine, and Leucine) are essential amino acids (29). BCAA levels are controlled at the first two steps in the BCAA metabolic pathway, catalyzed by the branched-chain aminotransferase isozymes (cytosolic BCAT1 and mitochondrial BCAT2) and branched-chain α-keto acid dehydrogenase (BCKDH) complex. BCAT1/2 catalyzes the reversible transamination that transfers an amino group from BCAA to α-ketoglutarate (α-KG), generating glutamate and the corresponding branched-chain α-keto acids (BCKAs). While BCAT2 is expressed in most cells, BCAT1 expression is confined to a few tissues. Increased BCAT1 expression was noted in various cancers, but distinct roles were proposed in each disease (30–34). Moreover, it remains unknown how BCAT1 is regulated in normal development and aberrantly activated in cancer cells.
Here we show that PRC2 mutations and NRasG12D cooperatively promote MPN progression to myelofibrosis and leukemic transformation in a dose-dependent manner. EZH1 is indispensable for EZH2-deficient LICs and constitutes an epigenetic vulnerability. We uncover a new molecular link between EZH2, BCAT1 and BCAA metabolism required for leukemogenesis. Distinct oncogenic drivers converge on the same metabolic pathway by modulating the enzyme and substrates for BCAA metabolism, thus providing a rationale for targeting the epigenetic and metabolic liabilities of leukemia-initiating cells.
RESULTS
PRC2 Loss Cooperates with NRasG12D to Promote Myeloid Neoplasms in a Dose-Dependent Manner
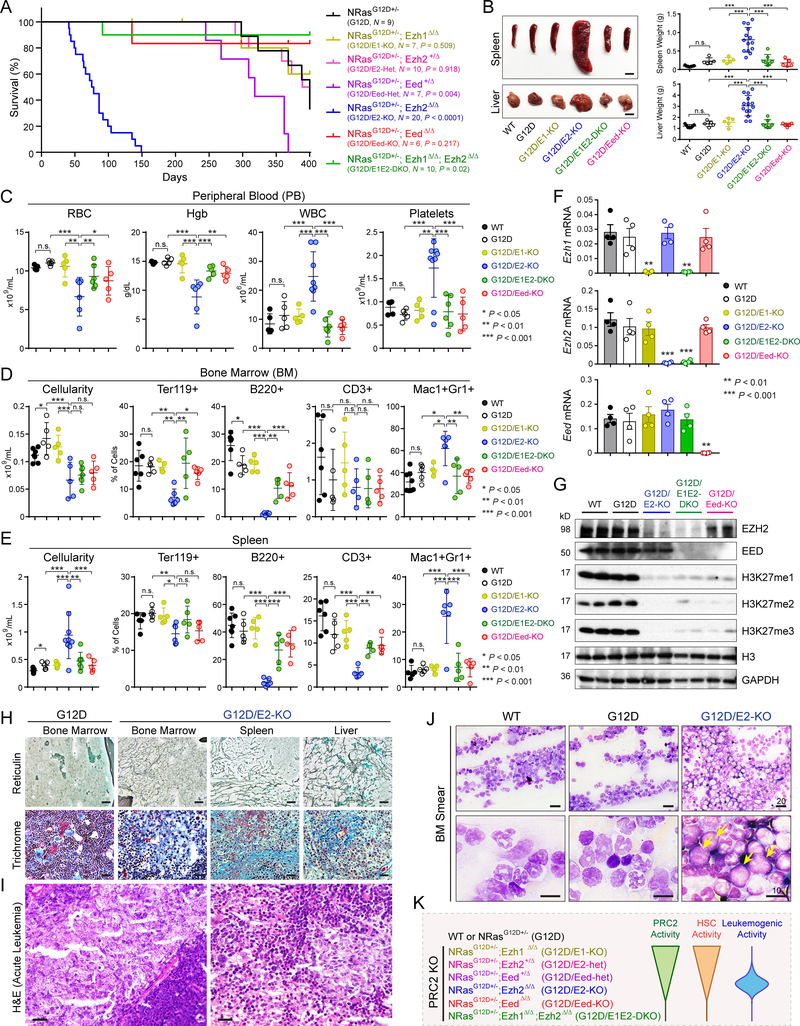
Since NRAS is a common target of oncogenic mutations in hematopoietic neoplasms and often co-occurs with mutations in epigenetic regulators (13,14,35), we sought to determine the cooperating alterations in EZH2 and NRAS in myeloid neoplasms. We used Mx1-Cre to activate heterozygous oncogenic RAS (NRasG12D+/−) and delete EZH2 (Ezh2f/f) alleles in hematopoietic cells. Activation of NRasG12D+/− alone (Mx1-Cre+;NRasG12D+/−, hereafter called G12D) led to chronic myeloproliferation with long latency (median survival >350 days), consistent with an MPN-like phenotype (4,36,37). By contrast, EZH2 KO together with G12D (Mx1-Cre+;NRasG12D+/−;Ezh2f/f, hereafter called G12D/E2-KO) markedly accentuated disease progression from indolent to highly penetrant lethal MPNs with a significantly shortened median survival (73 days, P < 0.001 vs G12D) (Fig. 1A).
Figure 1. PRC2 Loss Promotes NRasG12D-Induced Myeloid Neoplasms in a Dose-Dependent Manner.
A. Kaplan-Meier survival curves of control NRasG12D+/− mice (N = 9) and mice with combined NRasG12D+/− and various PRC2 KO mimicking different PRC2 dosages (N = 6 to 20). P values were calculated using log-rank (Mantel-Cox) test.
B. Representative pictures and quantification of spleen and liver of mice with indicated genotypes. Scale bars, 0.5 cm (top) and 1 cm (bottom).
C. Complete blood count (CBC) of peripheral blood (PB) red blood cells (RBC), hemoglobin (Hgb), white blood cells (WBC) and platelets in mice 10 weeks post-pIpC or moribund.
D. BM cellularity and frequencies of erythroid (Ter119+), B-lymphoid (B220+), T-lymphoid (CD3+) and myeloid (Mac1+Gr1+) cells in mice 10 weeks post-pIpC or moribund.
E. Spleen cellularity and frequencies of erythroid, B-lymphoid, T-lymphoid and myeloid cells in mice 10 weeks post-pIpC or moribund.
F. Expression of Ezh1, Ezh2 or Eed mRNA in BM cells 2 weeks post-pIpC.
G. Western blot of EZH2, EED, and H3K27 methylation (H3K27me1/2/3) in BM cells 2 weeks post-pIpC. Histone H3 and Gapdh were analyzed as loading controls.
H. EZH2 loss with NRasG12D led to development of myelofibrosis. Representative images are shown for reticulin and trichrome staining of BM, spleen or liver sections from G12D and G12D/E2-KO mice 8 to 12 weeks post-pIpC. Scale bars, 150μm.
I. Representative H&E staining of G12D/E2-KO BM sections shows sheets of blastic infiltrates (>20%) consistent with leukemic transformation. Scale bars, 25μm.
J. Histopathological analysis of Ezh2−/−NRasG12D-induced MPNs. Representative BM smears are shown in mice 10 weeks post-pIpC or moribund. The arrowheads indicate leukemic blasts with increased nucleus/cytoplasm ratio, fine chromatin, prominent nucleoli, and scant cytoplasm with fine eosinophilic granules. Scale bars, 20μm and 10μm.
K. Schematic of dose-dependent regulation of HSC or leukemogenic activity by PRC2. By KO of various PRC2 subunits (EZH1, EZH2 or EED), different levels of PRC2 activity were obtained. While partial loss of PRC2 by deletion of EZH1, EZH2 or EED heterozygous alleles had minimal impact on HSCs, complete loss of PRC2 by EZH1/2 DKO or EED KO led to HSC exhaustion. By contrast, while partial loss of PRC2 by EZH2 KO or EED heterozygous KO accentuated NRasG12D-induced MPN to leukemic transformation, complete loss of PRC2 was incompatible with leukemogenesis.
Results are mean ± SEM and analyzed by a repeated-measures one-way ANOVA with multiple comparisons, unless stated otherwise. *P < 0.05, **P < 0.01, ***P < 0.001, n.s. not significant.
To assess the role of PRC2 dosage in hematopoietic neoplasms, we established mouse models containing NRasG12D+/− and inactivation of various PRC2 subunits, including Ezh1 KO (Mx1-Cre+;NRasG12D+/−;Ezh1Δ/Δ or G12D/E1-KO), EZH2 heterozygous KO (Mx1-Cre+;NRasG12D+/−;Ezh2f/+ or G12D/E2-Het), EED heterozygous KO (Mx1-Cre+;NRasG12D+/−;Eedf/+ or G12D/Eed-Het), EED KO (Mx1-Cre+;NRasG12D+/−;Eedf/f or G12D/Eed-KO) and EZH1/2 double KO (Mx1-Cre+;NRasG12D+/−;Ezh1Δ/Δ;Ezh2f/f or G12D/E1E2-DKO) (Fig. 1A). Strikingly, while loss of EZH1 had no effect on hematopoiesis or MPNs, concurrent inactivation of EZH1 and EZH2 abolished MPN progression. G12D/Eed-KO phenocopied G12D/E1E2-DKO, consistent with the requirement for EED in both EZH1 and EZH2-containing PRC2 (23,38).
G12D/E2-KO (Mx1-Cre+;NRasG12D+/−;Ezh2f/f) mice developed splenomegaly and hepatomegaly with extramedullary hematopoiesis (EMH) and destructive myelodysplasia not seen in wild-type (WT, Mx1-Cre−) or either mutation alone within 300 days of recombination (Figs. 1B and S1A). Analysis of bone marrow (BM) and spleen revealed severe anemia, leukocytosis, thrombocytosis, increased Mac1+Gr1+ myeloid cells, and decreased Ter119+ erythroid cells, B220+ B-lymphoid and CD3+ T-lymphoid cells (Fig. 1C–E). Pathological examination of moribund G12D/E2-KO mice revealed severe leucoerythroblastic anemia and left shifted neutrophilia in the peripheral blood (PB), myelodysplasia, increased myeloid:erythroid (M:E) ratio, the presence of hypolobulated dysplastic megakaryocytes and osteosclerosis in BM, increased white:red pulp ratio in spleen, and the presence of infiltrative myeloid sarcoma in spleen and liver (Fig. S1A). None of these aberrations were observed in age-matched WT, G12D or G12D/E1-KO mice. Furthermore, the hematopoietic defects observed in G12D/E2-KO mice were largely normalized in G12D/E1E2-DKO or G12D/Eed-KO mice (Figs. 1C–E and S1A), suggesting that EZH2 loss promotes NRasG12D-induced MPNs in an EZH1-dependent manner.
To determine the molecular basis, we examined the expression of EZH2 and EED. At two weeks post-pIpC, we observed 98.1±0.4% or 94.5±1.9% excision efficiency of Ezh2 or Eed alleles, respectively (Fig. S1B), resulting in the near absence of EZH2 or EED mRNA and protein in BM (Fig. 1F,G). Highly efficient Ezh2 or Eed KO led to significantly decreased histone H3K27 mono-, di- and tri-methylation (H3K27me1/2/3) (Figs. 1G and S1C). The levels of EZH2, EED and H3K27me1/2/3 remained low or absent in G12D/E1E2-DKO or G12D/Eed-KO BM at 1 month post-pIpC, but were rapidly restored after 3 months (Fig. S1D). These data demonstrate that complete loss of PRC2 by EZH1/2 DKO or EED KO is incompatible with hematopoiesis, resulting in the elimination of G12D/E1E2-DKO or G12D/Eed-KO HSCs and MPN-initiating cells, and repopulation of cells that escaped recombination of Ezh2 or Eed alleles.
EZH2 Loss Cooperates with NRasG12D to Drive Leukemic Transformation
PMF is the most virulent form of MPNs characterized by abnormal proliferation of megakaryocytes, deposition of fibrous tissues in BM, osteosclerosis, and EMH (4). While no fibrosis was observed in G12D or E2-KO mice, EZH2 KO with G12D (G12D/E2-KO) accentuated megakaryocyte hyperproliferation, resulting in myelofibrosis and myelodysplasia in BM and spleen (Fig. 1H). G12D/E2-KO mice showed histopathological features of dense reticulin fibers with extensive interactions, coarse trichrome-positive collagen fibrosis and osteosclerosis in BM, spleen and liver (Fig. 1H), consistent with advanced PMF (4). More importantly, while none of G12D mice showed acute leukemias, 6 of 13 (or 46.2%) G12D/E2-KO mice displayed leukemic transformation consistent with post-MPN leukemias or blast-phase MPNs (MPN-BP) (4), including myelodysplasia, 20% or more c-Kit-positive leukemic blasts and evolving acute leukemic infiltration in BM and spleen (Figs. 1I,J and S1E,F). G12D/Eed-Het had intermediate phenotypes, whereas G12D/E2-Het had no impact on NRasG12D-induced MPNs (Fig. S2A,B). Additionally, E2-KO alone had no effect on MPNs or RAS signaling in hematopoietic stem/progenitor cells (HSPCs) (Fig. S2C,D), consistent with the oncogenic cooperation between EZH2 loss and NRasG12D.
Together, these results establish new genetic models that recapitulate progressive myelofibrosis and leukemic transformation in human MPNs. PRC2 inactivation cooperates with NRasG12D to promote MPN progression in a dose-dependent manner (Fig. 1K). EZH2 plays a tumor suppressive role in NRasG12D-induced MPNs and EZH2 insufficiency accelerates disease progression. Moreover, these results establish an essential role for EZH1 in EZH2-deficient MPNs and identify a selective epigenetic vulnerability for EZH2-deficiency-induced neoplasms.
Ezh2−/−NRasG12D Cells Propagate MPNs and Leukemogenesis in Transplant Recipients
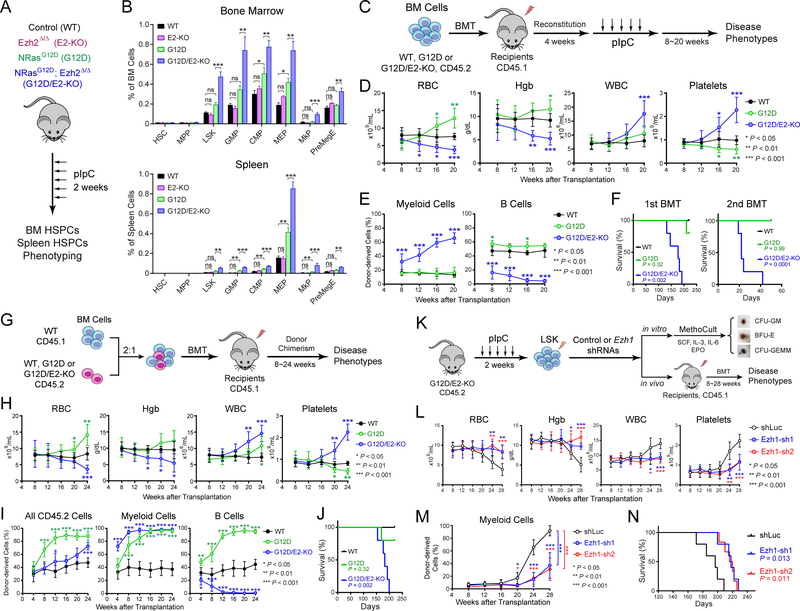
NRasG12D impairs HSC self-renewal and differentiation (36,37,39), whereas EZH2 loss minimally affects hematopoiesis (23). To determine the effect of combined EZH2 loss and NRasG12D on HSPCs, we analyzed WT, E2-KO, G12D, and G12D/E2-KO mice 2 weeks post-pIpC (Figs. 2A and S1F). Compared to WT, E2-KO alone had no significant impact on HSPCs in BM and spleen. G12D led to a trend toward increases in LSK (Lin−Sca1+c-Kit+), CMP (Lin−c-Kit+CD34+CD16/32−), GMP (Lin−c-Kit+CD34+CD16/32+), MEP (Lin−c-Kit+CD34−CD16/32−) populations in BM and/or spleen (Fig. 2B). By contrast, G12D/E2-KO displayed a marked expansion of HSPCs, including LSK, CMP, GMP, MEP, and megakaryocyte progenitors MkP (Lin−c-Kit+CD150+CD41+) in BM and spleen (Fig. 2B). Moreover, G12D/E2-KO had significantly decreased erythroid and lymphoid cells, and markedly increased granulocytes (Fig. 1D,E), indicating that EZH2 loss leads to a skewed lineage differentiation towards megakaryopoiesis and granulopoiesis at the expense of erythropoiesis and lymphopoiesis.
Figure 2. Ezh2−/−NRasG12D Cells Propagate MPN Progression and Leukemogenesis in an Intrinsic and EZH1-Dependent Manner.
A. Experimental design to access the effects of EZH2 KO and/or NRasG12D on HSPCs.
B. Frequencies of HSPCs in BM or spleen 2 weeks post-pIpC. N = 4 to 7 mice per group.
C. Schematic of bone marrow transplantation (BMT) assays.
D. CBC analysis of RBC, Hgb, WBC and platelets 8 to 20 weeks post-BMT.
E. Frequencies of donor-derived CD45.2+ myeloid and B-lymphoid cells 8 to 20 weeks post-BMT.
F. Survival curves of recipient mice transplanted with WT, G12D or G12D/E2-KO donor cells in first or secondary BMT (N = 5 per genotype). P values were calculated between G12D (or G12D/E2-KO) and WT using log-rank (Mantel-Cox) test.
G. Schematic of competitive BM repopulation assays.
H. CBC of RBC, Hgb, WBC and platelets 4 to 24 weeks post-BMT.
I. Frequencies of donor-derived CD45.2+ cells, myeloid and B-lymphoid cells in PB of recipients 4 to 24 weeks post-BMT.
J. Survival curves of recipients transplanted with WT, G12D or G12D/E2-KO donor cells (N = 5 per genotype).
K. Experimental design to determine the requirement of EZH1 in EZH2-deficient MPNs.
L. CBC in recipients of G12D/E2-KO HSPCs transduced with control (shLuc) or EZH1 shRNAs 8 to 28 weeks post-BMT.
M. Frequencies of donor-derived myeloid cells in PB 8 to 28 weeks post-BMT.
N. Survival curves of recipients transplanted with G12D/E2-KO HSPCs transduced with shLuc or EZH1 shRNAs (N = 5 per genotype). P values were calculated between Ezh1-sh1 (or Ezh1-sh2) and shLuc using log-rank (Mantel-Cox) test.
Results are mean ± SEM and analyzed by a one-way ANOVA with multiple comparisons, unless stated otherwise. *P < 0.05, **P < 0.01, ***P < 0.001, n.s. not significant.
To determine whether EZH2 loss and NRasG12D cooperate to promote leukemogenesis through cell-intrinsic mechanisms, we performed bone marrow transplantation (BMT) assays (Fig. 2C). CD45.2+ BM cells from WT, G12D or G12D/E2-KO mice were transplanted into CD45.1+ lethally irradiated recipient mice. After confirming reconstitution, the recipients were injected with pIpC to induce EZH2 loss and NRasG12D expression. All G12D/E2-KO recipients developed lethal hematopoietic diseases similar to primary mice (Fig. 2D). PB of G12D/E2-KO recipients had significantly increased donor-derived myeloid cells and decreased B cells (Fig. 2E), consistent with increased myeloid expansion with EZH2 loss and NRasG12D combination. G12D/E2-KO recipients had significantly impaired survival compared with recipients of WT or G12D cells (Fig. 2F). Pathological analyses revealed leucoerythroblastic anemia and neutrophilia in PB, splenomegaly, EMH, myelodysplasia and myelofibrosis in BM and spleen of G12D/E2-KO recipients at 20 weeks post-BMT (Fig. S3A,B). In secondary transplants, all G12D/E2-KO recipients developed lethal hematopoietic diseases and died within 6 weeks, whereas no WT or G12D recipients developed diseases (Fig. 2F).
To examine the role of EZH2 loss and NRasG12D on HSCs, we performed competitive repopulation assays (Fig. 2G). WT competitor (CD45.1+) BM cells were mixed with CD45.2+ WT, G12D or G12D/E2-KO BM cells at 2:1 ratio, and injected into lethally irradiated recipients (CD45.1+). Similar to primary mice, G12D/E2-KO recipients developed lethal hematopoietic diseases with impaired survival (Fig. 2H–J). G12D/E2-KO recipients displayed the highest chimerism in myeloid cells but not B-lymphoid cells (Fig. 2I). At 24 weeks post-BMT, G12D/E2-KO cells contributed competitively to HSCs and progenitors, but not B or T cells (Fig. S3C). Pathological examination revealed splenomegaly, EMH, myelodysplasia and myelofibrosis in G12D/E2-KO recipients (Fig. S3D,E). Together, these results demonstrate that EZH2 loss and NRasG12D enhanced the repopulation capability of MPN-initiating cells, resulting in efficient propagation of NRasG12D-induced MPNs and leukemogenesis in a cell-intrinsic manner.
EZH1 is Required for EZH2-Deficiency-Induced Myeloid Neoplasms
Since concurrent EZH1 and EZH2 KO abrogated MPNs, we explored whether EZH1 is an epigenetic liability of EZH2-deficient leukemia-initiating cells (LICs) (Fig. 2K). Compared to cells transduced with control shRNAs (shLuc), EZH1-depleted G12D/E2-KO HSPCs were significantly impaired in colony-forming activities, resulting in markedly decreased size and number of CFU-GM (colony-forming unit-granulocyte, macrophage) and the more immature CFU-GEMM (colony-forming unit-granulocyte, erythrocyte, monocyte, megakaryocyte) colonies (Fig. S4A,B). We next determined the effects of EZH1 depletion on G12D/E2-KO LICs by transplanting the control or EZH1-depleted G12D/E2-KO HSPCs into lethally irradiated recipients. While recipients of shLuc-transduced cells developed lethal hematopoietic diseases and early lethality, EZH1 depletion significantly ameliorated leukemic phenotypes and delayed disease onset (Figs. 2L–N and S4C,D). It is important to note that EZH1 KO alone or with NRasG12D had no effect on hematopoiesis or MPNs (Figs. 1A–E and S1A–C). These results demonstrate that EZH1 is dispensable for normal HSCs but required for EZH2-deficient LICs, thus highlighting a selective epigenetic vulnerability induced by EZH2 deficiency.
EZH2 Loss Reactivates BCAT1 and BCAA Metabolism in Leukemia-Initiating Cells
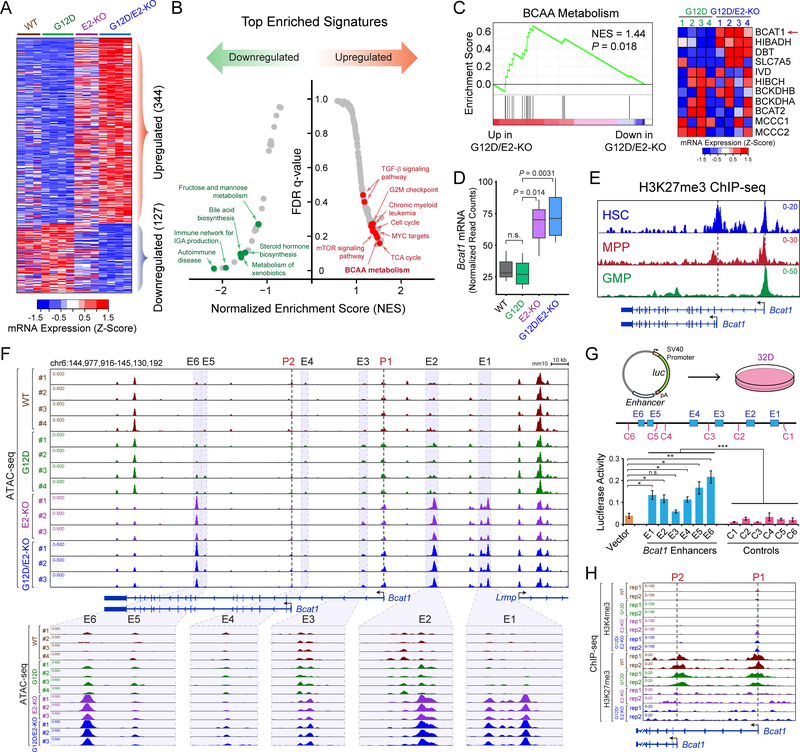
To determine the gene programs for aberrant HSC activities, we performed RNA-seq in LSK cells from WT, G12D, E2-KO and G12D/E2-KO mice 2 weeks post-pIpC. We chose the early time point to ensure that the observed changes were not due to differences in disease progression and/or leukemic transformation. We identified 344 and 127 significantly up- or downregulated genes in G12D/E2-KO relative to G12D cells (≥ 1.5-fold, adjusted P value ≤ 0.05; Fig. 3A, Tables S1 and S2). In addition to several pathways including cell cycle known to be regulated by PRC2, we discovered that BCAA metabolism was one of the top upregulated pathways in G12D/E2-KO relative to G12D cells (Fig. 3B). Importantly, BCAT1, the first enzyme catalyzing BCAA transamination, was specifically upregulated in EZH2-deficient LSK cells (Fig. 3C,D). BCAT2, a BCAT1 paralogue, did not show differential expression. Moreover, the Bcat1 promoters were highly enriched with H3K27me3, a histone mark catalyzed by EZH2-PRC2, in BM HSC, MPP and GMP cells (Fig. 3E), suggesting that Bcat1 is epigenetically silenced by EZH2-PRC2 in hematopoiesis but aberrantly activated upon EZH2 loss.
Figure 3. Loss of EZH2 Reactivates BCAT1 in Myeloid Neoplasms.
A. Heatmap showed Z-scores of DEseq2 normalized read counts for differentially expressed genes identified by RNA-seq in BM LSK cells 2 weeks post-pIpC. N = 3, 4, 3 and 4 mice for WT, G12D, E2-KO and G12D/E2-KO, respectively.
B. Top enriched signatures in upregulated or downregulated genes in G12D/E2-KO LSK cells.
C. GSEA showed the upregulation of BCAA metabolism related genes in G12D/E2-KO LSK cells. Heatmap showed Z-scores of DEseq2 normalized read counts for genes in BCAA metabolism pathway.
D. Bcat1 mRNA was upregulated in E2-KO or G12D/E2-KO relative to WT or G12D LSK cells. Y axis showed normalized read counts of Bcat1 by DEseq2. P value was calculated by a two-sided t-test. Boxes showed median of the data and quartiles, and whiskers extend to 1.5x of the interquartile range.
E. Density maps for ChIP-seq of H3K27me3 at Bcat1 gene (chr6:144,975,000–145,095,000; mm10) in BM HSC, MPP and GMP cells. Dashed lines and arrowheads indicate Bcat1 transcriptional start sites (TSS).
F. EZH2 KO increased chromatin accessibility at Bcat1 promoters and distal elements. Results from ATAC-seq (chr6:144,977,916–145,130,192; mm10) in WT, G12D, E2-KO and G12D/E2-KO LSK cells are shown. N = 4, 4, 3, and 3 for WT, G12D, E2-KO and G12D/E2-KO, respectively. Bcat1 promoters (P1 and P2) and putative enhancers (E1 to E6) are indicated by dashed and shaded lines, respectively. The zoom-in view of each enhancer is shown on the bottom.
G. Bcat1 distal elements with increased ATAC-seq enhanced reporter expression relative to vector or neighboring control DNA sequences (C1 to C6). Schematic of enhancer reporter assays in 32D cells is shown. Results are mean ± SEM (N = 3 experiments) and analyzed by a two-sided t-test. *P < 0.05, **P < 0.01, ***P < 0.001, n.s. not significant.
H. EZH2 KO decreased H3K27me3 and increased H3K4me3 at Bcat1 promoters. Results from ChIP-seq (chr6:145,087,000–145,037,000; mm10) in WT, G12D, E2-KO and G12D/E2-KO LSK cells (N = 2 mice per genotype) are shown.
To elucidate the mechanisms for Bcat1 activation, we measured chromatin accessibility by ATAC-seq and epigenetic landscapes by ChIP-seq in LSK cells 4 weeks post-pIpC. ATAC-seq revealed significant increases in accessibility of multiple distal regulatory elements at Bcat1 in E2-KO or G12D/E2-KO relative to WT or G12D cells (E1 to E6; Fig. 3F), suggesting that they may act as transcriptional enhancers. Consistently, DNA sequences containing the ATAC-seq peaks displayed strong enhancer activities in reporter assays (Fig. 3G). Increased ATAC-seq signals at Bcat1 enhancers were accompanied by decreased H3K27me3 and increased H3K4me3 at Bcat1 promoters in E2-KO or G12D/E2-KO cells (Figs. 3H and S5A). No change in chromatin accessibility or histone marks was detected at Bcat2 (Fig. S5B). At the genome scale, EZH2 loss in E2-KO and G12D/E2-KO LSK led to profound changes in epigenetic landscapes including significantly increased ATAC-seq signals in gene-distal elements, and markedly decreased H3K27me3 in both promoters and gene-distal elements (Figs. S6A–E and S7A–F). The epigenetic profiles were similar in E2-KO and G12D/E2-KO but distinct from WT and G12D cells at both genomic and single gene levels, suggesting that EZH2 loss leads to altered epigenetic and transcriptomic profiles including BCAT1 activation independent of NRasG12D.
In addition, we identified MYC and CEBPA as the top enriched TF motifs at Bcat1 enhancers (Fig. S8A). MYC and CEBPA bind strongly to Bcat1 enhancers, whereas MYC or CEBPA depletion impaired Bcat1 expression in G12D/E2-KO HSPCs (Fig. S8B–D). We also measured Bcat1 expression in LSK cells of various PRC2 KO. Bcat1 is only slightly upregulated in G12D/E2-Het (1.8-fold) and modestly upregulated in G12D/Eed-Het (2.4-fold), but significantly upregulated in Ezh2 KO, Eed KO or Ezh1/2 DKO with or without NRasG12D (4.2 to 4.9-fold; Fig. S8E). Moreover, Ezh1 KO or Ezh2-Het KO did not significantly decrease H3K27me3 at Bcat1 promoters, in contrast to the near absence of H3K27me3 in Ezh2 or Eed KO cells (Fig. S8F). Bcat1 is also significantly upregulated upon EZH2 KO in JAK2V617F-expressing HSC and MEP cells (Fig. S8G) (24), suggesting that BCAT1 is directly regulated by EZH2 independent of oncogenic drivers (NRasG12D or JAK2V617F). Together, these results demonstrate that EZH2-PRC2 represses Bcat1 in normal hematopoiesis, and loss of PRC2 reactivates Bcat1 through alleviated promoter repression and activated enhancers.
EZH2 Regulates BCAT1 in Human MPNs and Myeloid Leukemia
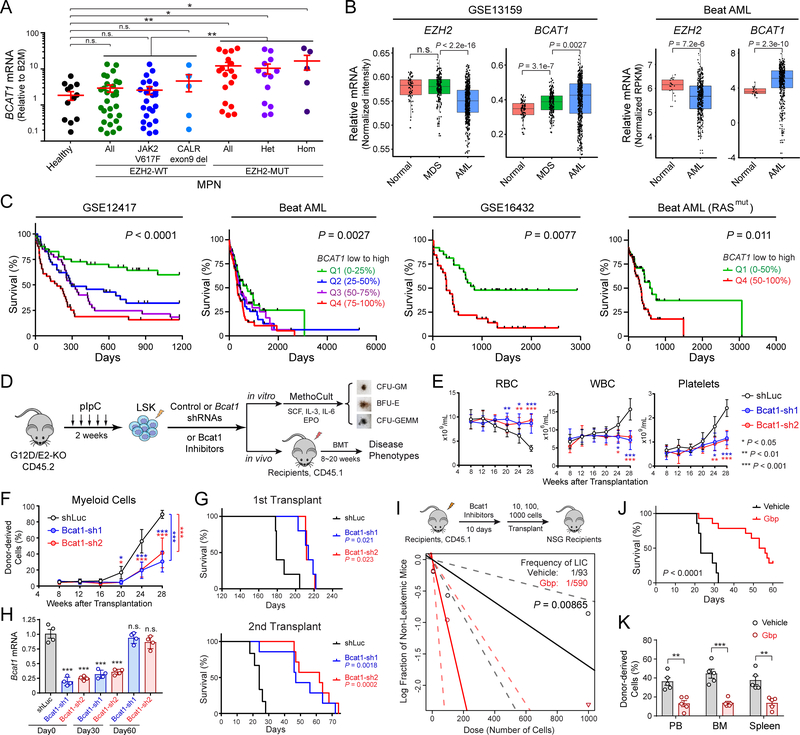
BCAT1 transfers BCAA nitrogen to α-KG to generate glutamate (Glu) and BCKAs (Fig. S9A). BCKAs can be reaminated by BCAT1/2 or oxidized to TCA cycle for ATP generation and/or macromolecule synthesis (29). Increased BCAT1 expression was noted in cancers yet distinct roles were proposed (Fig. S9B). To investigate the EZH2-BCAT1 axis in human myeloid malignancies, we surveyed the expression of BCAT1 mRNA in granulocytes from MPN patients (Fig. 4A). Compared to healthy donors, there was a trend toward increased BCAT1 in MPN samples with wild-type EZH2 (2.1-fold of controls, P = 0.29). The presence of the JAK2V617F or CALR deletion had no impact on BCAT1. Instead BCAT1 was markedly upregulated in MPNs harboring EZH2 mutations (8.0-fold of controls, P = 0.0068). By further classifying MPNs into EZH2 heterozygous and homozygous mutations, we observed significant upregulation of BCAT1 in both groups (6.9-fold and 10.9-fold, P = 0.013 and P = 0.0032, respectively), with a trend toward higher BCAT1 in patients with EZH2 homozygous mutations (1.6-fold, P = 0.38). BCAT2 expression was modestly downregulated in MPNs and EZH2 mutations had no impact on BCAT2 (Fig. S9C). It is important to note that EZH2 and RAS mutations are often seen in MPNs with leukemic transformation (9,10) (Fig. S9D), consistent with our mouse genetics that EZH2 loss and NRASG12D cooperate to drive leukemic transformation.
Figure 4. BCAT1 Is Required for EZH2-Deficient LICs and Human Myeloid Neoplasms.
A. EZH2 mutations in MPN patients increased BCAT1 expression. Each dot indicates an independent sample. All, all patients; Het, EZH2 heterozygous mutation; Hom, EZH2 homozygous mutation.
B. Expression of EZH2 and BCAT1 in CD34+ HSPCs or BM cells from healthy donors (normal), MDS and AML patients in GSE13159 (40–42) and Beat AML (43) cohorts. Each dot indicates an independent sample. Boxes show median of the data and quartiles, and whiskers extend to 1.5x of the interquartile range. P values were calculated by a two-sided t-test.
C. Increased BCAT1 negatively associates with overall survival in AML cohorts including GSE12417 (42), Beat AML (all or RAS mutant patients) (43), and GSE16432 (64). Patients were ranked by BCAT1 mRNA levels from low to high and divided to 4 or 2 quantiles. P value was calculated for all quantiles using log-rank (Mantel-Cox) test.
D. Experimental design to determine the role of BCAT1 in EZH2-deficient MPNs.
E. BCAT1 depletion impaired EZH2-deficient MPNs. CBC results are shown in recipients of G12D/E2-KO HSPCs with control (shLuc) or BCAT1 shRNAs 8 to 28 weeks post-BMT.
F. Frequencies of donor-derived myeloid (Mac1+Gr1+) cells in PB 8 to 28 weeks post-BMT.
G. Survival curves of mice transplanted with G12D/E2-KO HSPCs with shLuc or BCAT1 shRNAs in first or secondary transplantation (N = 5 per genotype). P values were calculated between Bcat1-sh1 (or Bcat1-sh2) and shLuc using log-rank (Mantel-Cox) test.
H. Validation of BCAT1 knockdown by qRT-PCR in donor-derived BM cells before (Day0) and after (Day30 and Day60) secondary transplantation.
I. LIC frequencies in vehicle or BCAT1 inhibitor (Gbp) treated mice were determined by limiting dilution at three doses by transplantation to NSG mice (N = 5 per group). LIC frequency fitted to each dilution series is shown by a solid line relating the log10 fraction of non-leukemic mice to the numbers of transplanted cells at 95% confidence intervals (dashed lines). P value was calculated by chi-squared test to evaluate the difference between vehicle and Gbp groups.
J. Survival curves of mice transplanted with G12D/E2-KO HSPCs treated with vehicle or BCAT1 inhibitor (Gbp) (N = 7 and 14). P values were calculated using log-rank (Mantel-Cox) test.
K. Frequencies of donor-derived (CD45.2+) cells in PB, BM and spleen in mice transplanted with G12D/E2-KO HSPCs with vehicle or Gbp treatment 30 days post-transplantation.
Results are mean ± SEM and analyzed by a repeated-measures one-way or two-way ANOVA with multiple comparisons, unless stated otherwise. *P < 0.05, **P < 0.01, ***P < 0.001, n.s. not significant.
We next compared EZH2 and BCAT1 expression in healthy donors (normal), MDS or acute myeloid leukemia (AML) patients (40–43) (Fig. 4B). EZH2 expression is comparable in normal and MDS but downregulated in AML HSPCs. By contrast, BCAT1 is progressively upregulated in MDS and AML samples. The reciprocal expression of EZH2 and BCAT1 is consistent with the role of EZH2 in silencing BCAT1. Importantly, increased BCAT1 is associated with significantly poorer overall survival in AML cohorts including patients with RAS alterations (Fig. 4C), suggesting that overexpression of BCAT1 plays an oncogenic role in AML. Moreover, the human BCAT1 promoter was enriched with H3K27me3 in normal CD34+ HSPCs, erythroblasts and monocytes, whereas the H3K27me3 level was decreased or absent in CD34+ HSPCs from AML patients (Fig. S10), suggesting that BCAT1 is also an EZH2-PRC2 target in human hematopoiesis. Together these results demonstrate that EZH2 mutations or decreased expression are associated with increased BCAT1 in human MPN and AML, illustrating an evolutionarily conserved role for EZH2-BCAT1 axis in myeloid malignancies.
BCAT1 Inhibition Specifically Impairs EZH2-Deficient Leukemia-Initiating Cells
Given that BCAT1 is aberrantly upregulated in EZH2-deficient MPNs and associates with more aggressive phenotypes, BCAT1 may contribute to the malignant properties of EZH2-deficient LICs. To test this, we examined the effects of BCAT1 knockdown on G12D/E2-KO HSPCs (Fig. 4D). Compared to shLuc, BCAT1 depletion significantly impaired CFU-GM and CFU-GEMM colony-forming activities (Fig. S11A). Upon transplantation, recipients of shLuc cells developed progressive anemia, leukocytosis, thrombocytosis, myeloid expansion, EMH, myelofibrosis and early lethality. By contrast, BCAT1 depletion significantly ameliorated EZH2-deficiency-induced phenotypes and delayed disease onset in primary and secondary transplants (Figs. 4E–G and S11B,C). Moreover, BCAT1 expression was significantly depleted in donor cells before (day0) and 30 days after transplantation but restored at day60 post-transplantation (Fig. 4H). These results suggest that escape from shRNA-mediated BCAT1 suppression may underlie disease progression in BCAT1 shRNA groups, and that BCAT1 is functionally required for the propagation of LICs.
To determine whether BCAT1 enzymatic function is required, we inhibited BCAT1 activity using two inhibitors, BCAT1i (CAS No. 406191–34-2) (44) and gabapentin (Gbp) (45). BCAT1 inhibition significantly impaired G12D/E2-KO LICs, resulting in smaller colonies and marked reduction in colony-forming ability relative to DMSO-treated cells (Fig. S11D). G12D/E2-KO myeloid progenitors (CFU-GM) were nearly absent upon BCAT1 inhibition, whereas WT or G12D cells were minimally affected. More importantly, BCAT1 inhibition by Gbp significantly impaired EZH2-deficient LIC activity in limiting dilution assays, resulting in a 6.3-fold decrease in LIC frequency relative to vehicle controls (Fig. 4I). BCAT1 inhibition also significantly ameliorated leukemic burden and prolonged survival of recipient mice (Fig. 4J,K). Together, these results suggest that BCAT1 inhibition preferentially impairs the propagation of EZH2-deficient LICs without affecting normal HSPCs, indicating that EZH2 insufficiency creates a selective dependency on BCAT1 in myeloid malignancies.
BCAT1 Is Required to Maintain Intracellular BCAA Pools in EZH2-Deficient LICs
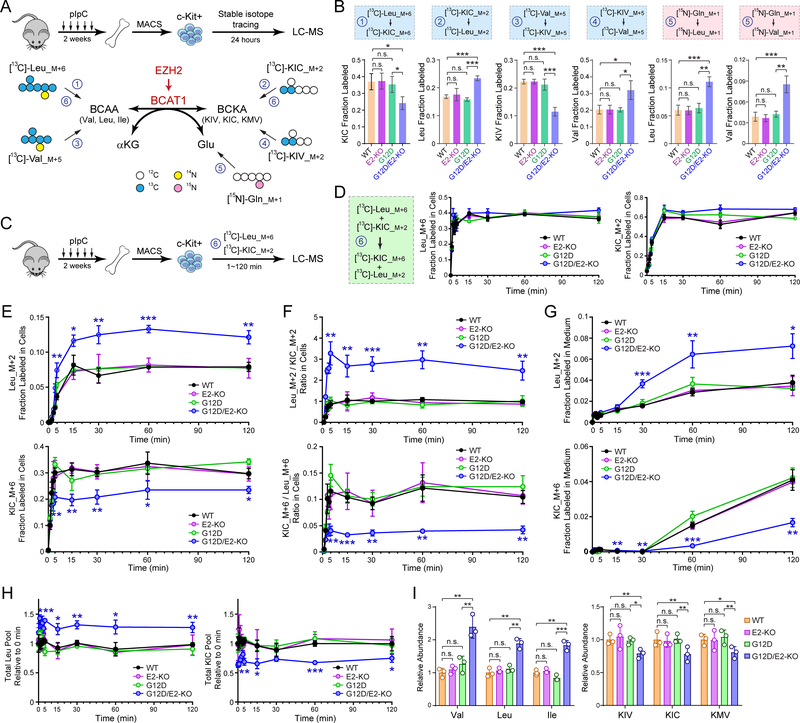
Since BCAT1 catalyzes reversible BCAA transamination (30–34,46), we sought to elucidate the metabolic role of BCAT1 in EZH2-deficient LICs. We first determined the concentrations of BCAAs (Val, 175.2 ± 12.5μM; Leu, 128.2 ± 14.1μM) and BCKAs (KIV, 34.9 ± 6.4μM; KIC, 29.0 ± 7.8μM) in the plasma of WT mice (Fig. S12A,B). We then used isotope tracing to assess the interconversion of BCAAs and BCKAs. First, c-Kit+ HSPCs from WT, G12D, E2-KO or G12D/E2-KO mice were incubated with [13C]-Leu or [13C]-KIC for 24 hours, followed by LC-MS analysis of 13C labeled metabolites (Fig. 5A). To determine whether Leu can be deaminated to KIC by BCAT1, cells were cultured with uniformly labeled [13C]-Leu_M+6 and non-labeled KIC at physiological concentrations (170 and 30 μM). We detected deaminated [13C]-KIC_M+6 in all four genotypes (Fig. 5B). Second, to determine whether KIC can be reaminated to Leu by BCAT1, cells were cultured with [13C]-KIC_M+2 and non-labeled Leu at physiological concentrations. We also detected [13C]-Leu_M+2 in all four genotypes (Fig. 5B). Third, we observed similar results using [13C]-Val and [13C]-KIV (Fig. 5A,B). Finally, since BCKA reamination requires Glu as the amino donor, we traced HSPCs with 2mM [α−15N]-glutamine (Gln), which is converted to [α−15N]-Glu by glutaminase (GLS). We also detected [15N]-labeled Val and Leu, indicating transfer of 15N from Glu to BCAAs. These results demonstrate that HSPCs interconvert BCAAs and BCKAs, consistent with the known function of BCAT1.
Figure 5. BCAT1 Promotes Intracellular BCAA Production in EZH2-Deficient LICs.
A. Schematic of isotope tracing in EZH2-deficient LICs. BM c-Kit+ HSPCs from 8~12 weeks WT, G12D, E2-KO or G12D/E2-KO mice were traced 24 hours with [13C]-Leu_M+6, [13C]-KIC_M+2, [13C]-Val_M+5, [13C]-KIV_M+5, or [15N]-Gln_M+1, followed by LC-MS analysis of 13C or 15N-labeled metabolites.
B. Fractional labeling of [13C] or [15N]-labeled BCAAs or BCKAs in BM HSPCs.
C. Schematic of isotope tracing to determine the kinetics of BCKA and BCAA interconversion.
D. The uptake of [13C]-Leu_M+6 and [13C]-KIC_M+2 tracers was comparable in all genotypes. Fractional labeling of Leu_M+6 or KIC_M+2 was determined by LC-MS at the indicated time points (0 to 120 min).
E. G12D/E2-KO increased intracellular Leu in HSPCs. Fractional labeling of Leu_M+2 or KIC_M+6 was determined at the indicated time points (0 to 120 min).
F. G12D/E2-KO increased Leu_M+2/KIC_M+2 and decreased KIC_M+6/Leu_M+6 ratio in HSPCs.
G. G12D/E2-KO increased exported Leu_M+2 in medium.
H. Leu and KIC pools were determined in HSPCs at the indicated time points (0 to 120 min).
I. Relative ion counts by means of LC-MS of intracellular BCAAs and BCKAs in HSPCs.
Results are mean ± SD (N = 3 independent experiments) and analyzed by a repeated measures one-way ANOVA, unless stated otherwise. *P < 0.05, **P < 0.01, ***P < 0.001. n.s. not significant.
To determine the exchange kinetics between BCAAs and BCKAs, we performed time-course analysis (Fig. 5C). WT, G12D, E2-KO or G12D/E2-KO HSPCs were traced with [13C]-Leu_M+6 and [13C]-KIC_M+2 (170 and 30 μM), sampled at time intervals (0, 1, 2, 3, 4, 5, 15, 30, 60 and 120 min), and analyzed for the resulting [13C]-KIC_M+6 and [13C]-Leu_M+2. We first noted that the uptake of [13C]-Leu_M+6 was faster than [13C]-KIC_M+2, although labeling reached steading for both tracers by 15 min without difference between genotypes (Fig. 5D). Importantly, we observed persistent increases in fractional labeling and abundance of [13C]-Leu_M+2, the BCAT1-catalyzed reamination product, in G12D/E2-KO relative to other genotypes (Figs. 5E and S12C). In contrast, the fractional labeling and abundance of [13C]-KIC_M+6, the deamination product of [13C]-Leu_M+6, was significantly lower in G12D/E2-KO. By calculating the ratios of the abundance for labeled species, we noted that G12D/E2-KO markedly increased Leu_M+2/KIC_M+2 and decreased KIC_M+6/Leu_M+6 (Fig. 5F). The exported Leu_M+2 in medium was significantly increased, whereas KIC_M+6 was decreased in E2-KO after 30~120 min (Figs. 5G and S12D). Notably, the total intracellular Leu and KIC pools remained unchanged, although the Leu pool was larger in G12D/E2-KO relative to other genotypes (Fig. 5H). Intracellular α-KG levels remained unchanged in all genotypes (Fig. S12E). Minimal or no detectable labeling of TCA intermediates or other amino acids was observed (Fig. S13A,B), suggesting that BCAAs did not significantly contribute to catabolic pathways in HSPCs.
Finally, we measured endogenous BCAA levels in freshly isolated HSPCs. The levels of all three BCAAs were significantly higher in G12D/E2-KO compared to other genotypes (Fig. 5I), suggesting that increased BCAT1 in G12D/E2-KO HSPCs is associated with net BCAA increases. Taken together, our results demonstrate that EZH2 loss with NRasG12D reactivates BCAT1 to enhance BCKA to BCAA conversion, resulting in increased BCAA pools in HSPCs.
EZH2 Deficiency and NRasG12D Cooperate to Increase Intracellular BCAAs in Leukemia
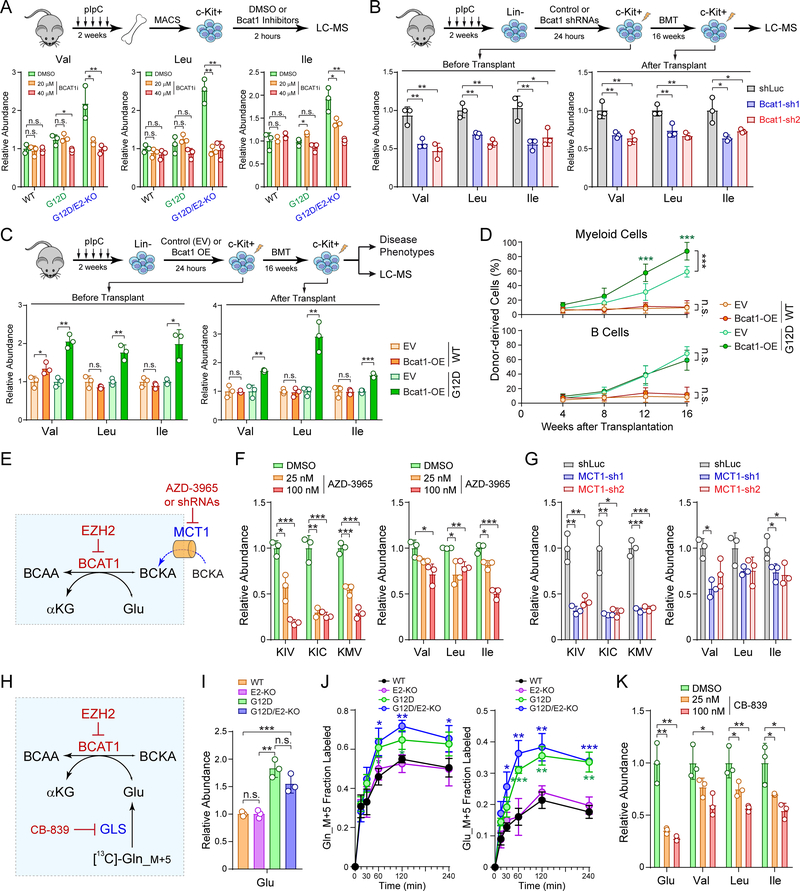
We next determined whether modulation of BCAT1 would impact BCAAs. We first noted that BCAT1 inhibition significantly decreased BCAAs in G12D/E2-KO HSPCs (Fig. 6A). Similarly, BCAT1 depletion by shRNAs significantly decreased BCAAs in EZH2-deficient HSPCs before and 16 weeks after transplantation in vivo (Fig. 6B). Conversely, BCAT1 overexpression (OE) increased BCAAs in G12D HSPCs before and after transplantation relative to empty vector (EV) control (Fig. 6C). Of note, BCAT1 OE enhanced myeloid expansion of G12D HSPCs (Fig. 6D), suggesting that increased BCAT1 in NRasG12D-expressing cells largely recapitulates the metabolic and functional changes as seen in G12D/E2-KO LICs. More importantly, the findings that blocking BCAT1 reduces BCAAs while overexpressing BCAT1 increases BCAAs in NRasG12D HSPCs provide strong evidence independent of tracing data that BCAT1 allows these cells to accumulate BCAAs.
Figure 6. Perturbations of BCAT1 Impairs BCAA Metabolism.
A. Inhibition of BCAT1 (BCAT1i) decreased BCAAs in EZH2-deficient LICs.
B. BCAT1 depletion by shRNAs decreased BCAAs in EZH2-deficient LICs before and post-BMT. BCAAs were determined by LC-MS in G12D/E2-KO HSPCs with shLuc or BCAT1 shRNAs.
C. BCAT1 overexpression (OE) increased BCAAs in G12D HSPCs before and post-BMT. BCAAs were determined in WT or G12D HSPCs with empty vector (EV) or BCAT1 OE.
D. BCAT1 OE enhanced myeloid expansion upon BMT. Frequencies of donor-derived myeloid and B-lymphoid cells in PB of recipients were analyzed at 4 to 16 weeks post-BMT. Results are mean ± SD (N = 5 per genotype) and analyzed by a two-way ANOVA.
E. Schematic of MCT1-dependent transport of BCKAs in BCAA metabolism.
F. MCT1 inhibition by AZD-3965 (25nM and 100nM for 2 h) decreased BCKAs and BCAAs in G12D/E2-KO HSPCs.
G. MCT1 depletion by shRNAs decreased BCKAs and BCAAs relative to shLuc in G12D/E2-KO HSPCs.
H. Schematic of GLS-dependent Gln to Glu conversion in BCAA metabolism.
I. NRasG12D+/− activation increased intracellular Glu in G12D and G12D/E2-KO HSPCs.
J. [13C]-Gln_M+5 tracing showed increased Gln and Glu in G12D and G12D/E2-KO HSPCs. Fractional labeling of Gln_M+5 and Glu_M+5 was determined at the indicated time points.
K. GLS inhibition by CB-839 (25nM and 100nM for 2 h) decreased Glu and BCAA levels in G12D/E2-KO HSPCs.
Results are mean ± SD (N = 3 independent experiments) and analyzed by a two-sided t-test, unless sated otherwise. *P < 0.05, **P < 0.01, ***P < 0.001. n.s. not significant.
Another critical question relates to the sources of BCKAs and Glu, the two substrates for BCAT1 transamination. We hypothesized that BCKAs were imported from extra-cellular sources through monocarboxylate transporters (MCTs) (Fig. 6E), and noted by RNA-seq that MCT1 (or Slc16a1) was the predominant transporter expressed in HSPCs (Fig. S13C). Importantly, inhibition of MCT1 by AZD-3965 or shRNAs significantly decreased BCKAs and BCAAs in G12D/E2-KO HSPCs (Fig. 6F,G) and modestly decreased BCAAs in WT or G12D HSPCs (Fig. S13D,E). In addition, NRasG12D activation resulted in increased intracellular Glu in G12D and G12D/E2-KO HSPCs (Fig. 6H,I). We next determined whether NRasG12D increased Gln uptake and intracellular Glu pools through glutaminase (GLS)-mediated Gln to Glu conversion, which ‘fuels’ BCAT1-catalyzed BCKA reamination (Fig. 6H). Consistently, we found by [13C]-Gln_M+5 tracing that NRasG12D enhanced intracellular Gln to Glu conversion in G12D and G12D/E2-KO HSPCs (Fig. 6J), whereas GLS inhibition by CB-839 significantly decreased Glu and BCAAs in G12D/E2-KO HSPCs (Fig. 6K).
Together, these results demonstrate that MCT1-dependent BCKA transport and GLS-dependent Gln to Glu conversion are required for BCAT1 transamination in EZH2-deficient LICs. Our results demonstrate that distinct oncogenic drivers (EZH2 loss and RAS activation) converge on the same metabolic pathway to drive leukemic transformation by modulating the enzyme and metabolic substrates for BCAA metabolism.
BCAT1-Driven Leukemia Is Sensitive to mTOR Inhibition
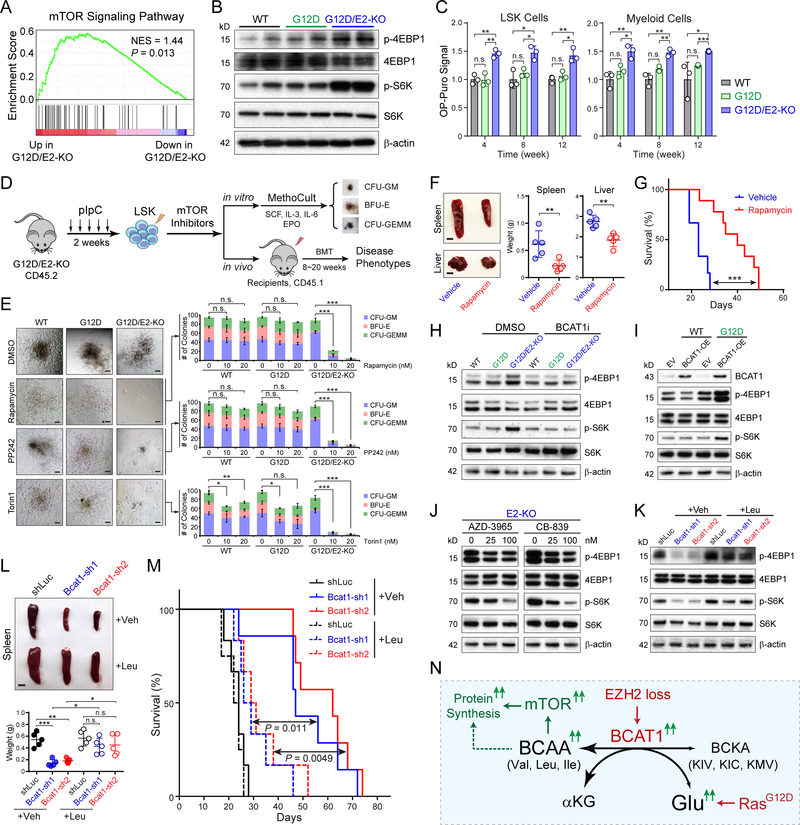
BCAAs, particularly Leu, activate mTORC1 to promote cell growth. Since activated BCAT1 increased BCAAs in EZH2-deficient LICs, we explored whether BCAT1 promotes leukemia through mTORC1 activation. We first noted that known mTOR targets were upregulated in G12D/E2-KO compared to WT or G12D LSK cells (Fig. 7A). Activated mTORC1 phosphorylates 4EBP1 and S6 kinase (S6K) to promote protein translation. Consistently, we observed increased phosphorylation of 4EBP1 (p-4EBP1) and S6K (p-S6K) in G12D/E2-KO LSK cells (Fig. 7B), and increased protein synthesis in G12D/E2-KO LSK and myeloid cells (Fig. 7C).
Figure 7. BCAT1-Driven Leukemia Are Sensitive to mTOR Inhibition.
A. Upregulation of mTOR signature genes in G12D/E2-KO relative to G12D LSK cells by GSEA.
B. Increased p-4EBP1 and p-S6K in G12D/E2-KO relative to WT or G12D LSK cells.
C. Protein synthesis rate was assessed by OP-puro incorporation in BM LSK and myeloid cells at 4, 8 and 12 weeks post-pIpC.
D. Schematic of experimental design to determine the effect of mTOR inhibition.
E. Inhibition of mTOR by rapamycin, PP242, or Torin1 abolished the colony-forming ability of G12D/E2-KO HSPCs. Representative photomicrographs for CFU-GM, BFU-E and CFU-GEMM colonies are shown. Scale bars, 100μm.
F. Representative pictures and quantification of spleen and liver of recipients transplanted with G12D/E2-KO HSPCs treated with vehicle or rapamycin. Scale bars, 0.5 cm (top) and 1 cm (bottom).
G. Survival curve of recipients transplanted with G12D/E2-KO HSPCs treated with vehicle or rapamycin (N = 5 and 8). P values were calculated using log-rank (Mantel-Cox) test.
H. BCAT1 inhibition by BCAT1i (40 μM) decreased mTOR activity (p-4EBP1 and p-S6K) in G12D/E2-KO HSPCs.
I. Overexpression of BCAT1 in WT or G12D HSPCs enhanced mTOR activity.
J. Inhibition of MCT1 by AZD-3965 or GLS by CB-839 impaired mTOR activity in G12D/E2-KO HSPCs.
K. Depletion of BCAT1 impaired mTOR activity in G12D/E2-KO HSPCs, which was restored by Leu supplementation (500 μM).
L. Leu supplementation promoted EZH2-deficient MPNs in vivo. Representative pictures are shown for spleens of recipients transplanted with BCAT1-depleted G12D/E2-KO HSPCs supplemented with vehicle or Leu. Scale bars, 0.5 cm.
M. Survival curve of recipients transplanted with BCAT1-depleted G12D/E2-KO HSPCs with vehicle or Leu. P values were calculated using log-rank (Mantel-Cox) test.
N. Model for the oncogenic cooperativity between EZH2 loss and RAS activation in myeloid neoplasms.
Results are mean ± SEM and analyzed by a repeated-measures one-way ANOVA with multiple comparisons, unless stated otherwise. *P < 0.05, **P < 0.01, ***P < 0.001, n.s. not significant.
To assess whether BCAT1-driven leukemia is sensitive to mTOR inhibition, we examined the clonogenic activity by treating HSPCs with rapamycin or active-site mTOR inhibitors PP242 and Torin1 (Fig. 7D). Strikingly, mTOR inhibition markedly impaired G12D/E2-KO HSPCs, resulting in a dose-dependent reduction in colony-forming ability (Fig. 7E). WT or G12D HSPCs were minimally affected, suggesting that BCAT1-driven LICs are more sensitive to mTOR inhibition. To assess the in vivo effects, we transplanted G12D/E2-KO splenic cells into lethally irradiated recipients followed by treatment with rapamycin or vehicle control (Fig. 7D). Vehicle-treated recipients developed lethal hematopoietic diseases similar to primary mice. By contrast, mTOR inhibition significantly ameliorated BCAT1-driven phenotypes and delayed disease onset (Fig. 7F,G). Since perturbations of BCAT1, MCT1 or GLS impaired BCAA metabolism, we examined whether mTOR activity was affected by changes in BCAT1 or BCAAs. BCAT1 inhibition decreased 4EBP1 and S6K phosphorylation in G12D/E2-KO HSPCs (Fig. 7H), whereas BCAT1 OE enhanced 4EBP1 and S6K phosphorylation in G12D HSPCs (Fig. 7I). Similarly, MCT1 or GLS inhibition impaired mTOR in G12D/E2-KO HSPCs (Fig. 7J). Leu supplementation enhanced mTOR and rescued the clonogenic activity of BCAT1-depleted G12D/E2-KO LICs (Figs. 7K and S13F). Leu supplementation in mice transplanted with BCAT1-depleted G12D/E2-KO LICs significantly promoted disease progression and shortened survival (Fig. 7L,M).
Taken together, these results demonstrate that loss of EZH2 reactivates BCAT1 to increase BCAAs in EZH2-deficient LICs (Figs. 7N and S14). Oncogenic RAS increases intracellular Glu, which cooperates with EZH2 loss to further enhance BCAT1 transamination. Increased BCAAs promote leukemia transformation through activated mTOR signaling. Moreover, EZH2-deficient LICs are sensitive to BCAT1 or mTOR inhibition, thus establishing new therapeutic strategies to selectively eradicate EZH2-deficient and/or BCAT1-driven hematopoietic malignancies.
DISCUSSION
PRC2 Regulates Hematopoiesis and Leukemia in a Dose-Dependent Manner
The presence of gain- and loss-of-function PRC2 mutations in cancers indicates that both hyper- and hypoactive PRC2 can be tumorigenic (11,19,20); however, how different PRC2 dosages impact cancer development remained unknown in vivo. Previous studies in mouse models demonstrated that PRC2 regulates HSCs in a dose-dependent manner. While partial loss of PRC2 has no or minimal effect on hematopoiesis, complete loss of PRC2 leads to HSC exhaustion (21–23). Here we show that partial loss of PRC2 by EZH2 KO or EED heterozygous KO cooperates with oncogenic NRAS to promote MPNs, whereas complete loss of PRC2 by EZH1/2 DKO or EED KO abolishes MPN progression. Compared with NRasG12D alone (36,37), combined EZH2 loss and NRasG12D led to more aggressive MPNs including myelofibrosis, myeloid expansion, leukemic transformation and reprogramming of BCAA metabolism, consistent with the oncogenic cooperation between PRC2 deficiency and RAS activation in leukemogenesis. Moreover, EZH1 inhibition selectively impairs the leukemogenic activity of EZH2-deficient LICs, highlighting a selective epigenetic vulnerability for EZH2-deficiency-induced neoplasms. Since EZH2 mutations are usually acquired somatically and EZH1 is dispensable for normal HSCs (21–23), our results raise the possibility of leveraging EZH1 as targeted therapies to specifically eradicate EZH2-mutant LICs. Thus, our findings provide a rationale for developing EZH1-targeting genetic and/or pharmacological tools to selectively eradicate EZH2-deficient hematopoietic neoplasms that may be generally applicable to other disorders caused by PRC2 dysregulation.
EZH2-BCAT1 Axis in Hematopoiesis and Leukemia
Here we describe a new mechanism connecting EZH2, BCAT1 and BCAA metabolism in hematopoiesis and leukemia. BCAT1 expression is restricted to a few tissues including brain and pancreas, while it is minimally or not expressed in most hematopoietic tissues (29,45,47). We show that BCAT1 is epigenetically silenced by EZH2 in mouse and human HSPCs, and EZH2 loss reactivates BCAT1 through alleviated promoter repression and activated enhancers. BCAT1 is markedly upregulated in MPN patients carrying EZH2 mutations, and increased BCAT1 associates with poorer survival in AML. It is important to note that, while heterozygous EZH2 mutations are more frequent in human MPNs, Ezh2-Het KO did not significantly promote NRasG12D-induced MPNs in mice in our experimental timeframe. Notably, BCAT1 was markedly upregulated in human MPNs with EZH2 heterozygous mutations (6.9-fold; Fig. 4A), but was only slightly upregulated in EZH2-Het KO (1.8-fold; Fig. S8E) mouse HSPCs. By contrast, BCAT1 was significantly upregulated in EZH2 homozygous KO HSPCs (4.2 to 4.9-fold). These results suggest that BCAT1 is repressed by EZH2-PRC2 in a dose-dependent manner, and EZH2-Het KO in mice is not sufficient to fully alleviate BCAT1 repression. Nonetheless, BCAT1 inhibition selectively impairs EZH2-deficient LICs without affecting normal HSPCs, establishing BCAT1 as a metabolic liability for EZH2-deficient LICs. Further, EZH2 loss-of-function mutations are frequently found in MPN and MDS patients but rarely in AML (11,12). However, the co-occurrence of BCAT1 activation and EZH2-PRC2 alterations in human AML may be under-estimated due to other mechanisms that do not involve EZH2 mutations. For instances, mutations of PRC2-associated proteins such as ASXL1 impair EZH2 chromatin recruitment (15). Mutations of SRSF2 cause aberrant EZH2 mRNA splicing in myeloid neoplasms (16). Other genomic abnormalities such as monosomy 7 or 7q deletion may also impair EZH2 function since EZH2 gene locates on chromosome 7q. Finally, EZH2 expression is significantly downregulated in AML (Fig. 4B), suggesting that additional mechanisms may underlie the deregulation of EZH2-PRC2 and activation of BCAT1 in AML. Although altered metabolism and metabolic substrates often affect epigenetics, little is known about how epigenetic alterations influence metabolism in cancer progression. Our results for the first time connect EZH2 dysregulation with altered metabolic pathways in cancer progression, establishing an example that epigenetic alterations rewire metabolic processes through reprogrammed expression of key metabolic genes in cancer-initiating cells.
Metabolic Roles of BCAT1 and BCAAs in Myeloid Leukemia
Although increased BCAT1 was noted in cancers and associated with more aggressive phenotypes, distinct roles for BCAT1 were proposed in different diseases (30–34). These mechanisms differ according to whether BCAT1 catalyzes BCAA deamination or BCKA reamination. In gliomas carrying wild-type IDH1, BCAT1 is activated to catalyze BCAA deamination and Glu production (30). IDH mutant gliomas produce 2-hydroxyglutarate to inhibit BCAT1/2 and impair Glu biosynthesis (46). NSCLC tumors increase BCAA uptake as a nitrogen source for proteins and nucleotides (34). In the context of leukemia, increased BCAT1 was shown to promote BCAA production in K562 CML cells by reaminating BCKAs (32), whereas elevated BCAT1 in human AML promoted cell growth through BCAA deamination and depleting α-KG (33). Our results suggest that activated BCAT1 cooperates with increased Glu in EZH2-deficient and NRasG12D-expressing LICs to enhance BCKA reamination, resulting in increased BCAAs and mTOR (Fig. S14). These findings highlight the oncogenic cooperativity between EZH2 loss and RAS activation by converging on the same metabolic pathway, and provide the mechanistic explanation that individual RAS or EZH2 mutations are not sufficient to promote malignant phenotypes. Rather, these mutations cooperate to cause maladaptive metabolic and signaling disturbances in cancer progression.
Therapeutic Implications of Targeting BCAA Metabolism
Approximately 5~10% of MPNs progress to acute leukemias within 10 years of diagnosis. Treatment options for post-MPN leukemia patients are limited and conventional approaches fail to offer long-term cure. BCAAs are essential amino acids that must be acquired from external sources. BCAA supplementation or BCAA-rich diets are associated with increased body weight, muscle protein synthesis and glucose homeostasis (48). Paradoxically, elevated circulating BCAAs correlate with obesity, insulin resistance, type 2 diabetes mellitus (T2DM) (48), and increased risk of pancreatic cancer (49). By contrast, decreased BCAA consumption promotes metabolic health and longevity in mice and humans by improving glucose tolerance and reducing fat accumulation (50). Here we show that BCAT1 is aberrantly activated to sustain BCAA pools in EZH2-deficient leukemia. Genetic and pharmacological inhibition of BCAT1 impairs EZH2-deficient cancer-initiating cells. Because normal HSPCs are unaffected by BCAT1 loss, the selective vulnerability raises the prospect of leveraging BCAT1 or BCAAs as targeted therapies to specifically eradicate EZH2-mutant cells. If validated, dietary BCAA restriction may be applicable for the treatment of hematologic malignancies, particularly for patients with EZH2-deficient myeloid neoplasms, through the use of diet plans or the prescription of medical foods lacking specific BCAAs. Our studies provide a strong rationale for the development of more specific and bioavailable BCAT1 inhibitors targeting metabolic liabilities of cancer-initiating cells that may be generally applicable to other neoplasms caused by EZH2 dysregulation.
MATERIALS AND METHODS
Cells and Cell Culture
Human AML cell lines NB4 and Kasumi1 were cultured in RPMI1640 medium containing 10% fetal bovine serum (FBS), 1% penicillin/streptomycin. Human AML cell line MV4–11 was cultured in IMDM medium containing 20% FBS and 1% penicillin/streptomycin. Mouse myeloblast cell line 32D was cultured in RPMI1640 containing 10% WEHI conditioned medium, 10% FBS and 1% penicillin/streptomycin. All cultures were incubated at 37°C in 5% CO2. No cell line used in this study was found in the database of commonly misidentified cell lines that is maintained by ICLAC and NCBI BioSample. All cell lines were tested for mycoplasma contamination.
Mice
Ezh2 and Eed floxed mice were generated by gene targeting as previously described (23,51). Ezh1 constitutive knockout mice were obtained from Thomas Jenuwein’s laboratory (Max Planck Institute of Immunology and Epigenetics, Freiburg, Germany) and will be described elsewhere. NRasG12D mice containing the LSL-NRasG12D+/− knockin allele (52) and Mx1-Cre mice (53) were obtained from the Jackson Laboratory. All mouse lines were maintained on a C57BL/6 background. All mouse experiments were performed under protocols approved by the Institutional Animal Use and Care Committee of UT Southwestern Medical Center (UTSW).
Primary Human MPN and AML Samples
The collection of blood samples from MPN patients was performed at the study center in Basel, Switzerland and approved by the local Ethics Committee (Ethik Kommission Beider Basel). Written informed consent was obtained from all patients in accordance with the Declaration of Helsinki. Granulocyte fraction for RNA isolation was prepared from the peripheral blood using Ficoll gradient followed by the erythrocyte lysis. Primary human AML samples were collected for diagnosis and de-identified for our study. This study was approved by the Institutional Review Board at UT Southwestern Medical Center (IRB STU 122013–023). Samples were frozen in FBS with 10% DMSO and stored in liquid nitrogen.
BCAT1 and BCAT2 Transcript Quantitation in Human MPN Samples
Total RNA from granulocytes was isolated using TriFast (VWR International) and 500ng was reverse-transcribed with High Capacity cDNA Reverse Transcription Kits (Applied Biosystems). cDNA was amplified with TaqMan Universal Master Mix II (Applied Biosystems) and TaqMan probes for human BCAT1 (Hs00398962_m1) and BCAT2 (Hs01553550_m1) on ViiA 7 Real-Time PCR System (Applied Biosystems). Relative gene expression change was calculated according to ΔΔCt method using human B2M probe (Hs00984230_m1) for normalization.
RNA Isolation and qRT-PCR Analysis
Total RNA was isolated using RNeasy Plus Mini Kit (Qiagen) and reverse-transcribed using iScript cDNA Synthesis Kit (Bio-Rad) following manufacturer’s protocols. Quantitative RT-PCR (qRT-PCR) was performed in triplicate with the iQ SYBR Green Supermix (Bio-Rad) using CFX384 Touch Real-Time PCR Detection System (Bio-Rad). PCR amplification parameters were 95°C (3 min) and 45 cycles of 95°C (15 sec), 60°C (30 sec), and 72°C (30 sec). Primer sequences are listed in Table S3.
RNA-seq Analysis
RNA-seq library was prepared using the Ovation RNA-seq system (NuGEN) or SMARTer stranded pico input total RNA-seq kit (Takara). Sequencing reads from all RNA-seq experiments were aligned to mouse reference genome (GENCODE Version M9) by STAR 2.5.2b (54) with the parameters: --outFilterMultimapNmax 1. GSEA was performed as previously described (55) using KEGG and hallmark gene sets in GSEA databases and signature genes of various metabolite pathways (total 313 gene sets) (56). Differentially expressed genes were identified by DESeq2 (57).
ChIP and ChIP-seq Analysis
ChIPmentation was performed as described (58) using antibodies for H3K4me3 (Millipore, Cat# 04–745), H3K27me3 (Millipore, Cat# 07–449), or H3K27ac (Abcam, Cat# ab4729) in FACS-sorted BM LSK cells from WT, G12D, E2-KO or G12D/E2-KO mice, respectively. ChIP-qPCR was performed as described (59) using primers listed in Table S3. ChIP-seq libraries were generated using NEBNext Ultra II DNA library prep kit following the manufacturer’s protocol (NEB), and sequenced on an Illumina NextSeq500 system using the 75bp high output sequencing kit. ChIP-seq raw reads were aligned to the mouse genome assembly (GENCODE Version M9) using Bowtie2 (60) with the default parameters. Only tags that uniquely mapped to the genome were used for further analysis. ChIP-seq peaks were identified using MACS2 (61). MAnorm (62) was used to compare ChIP-seq signal intensities between samples, and identify enriched or depleted peaks (|log2 fold change| ≥ 1) between G12D/E2-KO and G12D LSK cells.
ATAC-seq Analysis
ATAC-seq was performed as previously described with modifications (63). Briefly, 2 × 104 BM LSK cells were washed twice in PBS and resuspended in 500 μl lysis buffer (10 mM Tris-HCl, 10 mM NaCl, 3 mM MgCl2, 0.1% NP-40, pH 7.4). Nuclei were harvested by centrifuge at 500 x g for 10 min at 4°C. Nuclei were suspended in 50 μl of tagmentation mix (10 mM TAPS (Sigma), 5 mM MgCl2, pH 8.0 and 2.5 μl Τn5) and incubated at 37°C for 30 min. Tagmentation reaction was terminated by incubating nuclei at room temperature for 2 min followed by incubation at 55°C for 7 min after adding 10 μl of 0.2% SDS. Tn5 transposase-tagged DNA was purified using QIAquick MinElute PCR Purification kit (Qiagen), amplified using KAPA HiFi Hotstart PCR Kit (KAPA), and sequenced on an Illumina Nextseq500 system using the 75 bp high output sequencing kit. ATAC-seq raw reads were trimmed to remove adaptor sequence and aligned to mouse genome assembly (GENCODE Version M9) using Bowtie2 (60) with default parameters. Only tags that uniquely mapped to the genome were used for further analysis. ATAC-seq peaks were identified using MACS2 (61). MAnorm (62) was used to compare ATAC-seq signal intensities between samples, and identify enriched or depleted peaks (|log2 fold change| ≥ 2) between G12D/E2-KO and G12D LSK cells.
Gene Expression and Survival Analysis in Patient Cohorts
Expression correlation analysis of EZH2 and BCAT1 was performed using gene expression microarray or RNA-seq in public datasets including GSE12417 (42), GSE13159 (40,41), and the Beat AML cohort (http://www.vizome.org) (43). Survival analysis was performed using public datasets in GSE12417, Beat AML and the German AML (GSE16432) patient cohorts (64). In the Beat AML cohort, patients containing at least one of the following alterations in RAS signaling pathways were denoted as RAS mutant (RASmut), including NRAS and KRAS hotspot mutations, NF1 and PTPN11 mutations. In the GSE16432 AML cohort, survival analysis was performed on patients without IDH1/2, TET2 or NPM mutations.
Complete Blood Counts
Blood was collected via the retro-orbital plexus and complete blood counts (CBC) were performed on a HEMAVET HV950 (Drew Scientific) according to the manufacturer’s protocol.
Flow Cytometry and MACS Cell Separation
Bone marrow cells were obtained by flushing femurs and tibias with a 25G needle or by crushing femurs, tibias, vertebrae and pelvic bones with a mortar in Ca2+ and Mg2+-free Hank’s buffered salt solution (HBSS; Gibco) supplemented with 2% heat-inactivated bovine serum (HIBS, Gibco). Spleens and were dissociated by crushing followed by trituration. All cell suspensions were filtered through a 70 μm cell strainer. Cell numbers were determined with a Vi-CELL cell viability analyser (Beckman Coulter). For flow cytometric analysis and isolation, cells were incubated with combinations of antibodies listed in Table S4. Lineage markers for HSCs and progenitors were CD2, CD3, CD5, CD8, B220, Gr1 and Ter119. Antibody staining was performed at 4 °C for 30 min, or, when CD34 was included in the cocktail, on ice for 90 min. Biotinylated antibodies were visualized by incubation with PE/Cy7-conjugated streptavidin at 4°C for 30 min. DAPI (4,6-diamidino-2-phenylindole; 2 μg/ml in PBS) or PI (propidium iodide; 1 μg/ml) were used to exclude dead cells during flow cytometry. For isolation of c-Kit+ or lineage negative cell populations, cells were stained with c-Kit-APC780 or lineage-Biotin antibodies followed by microbeads conjugated anti-APC or anti-Biotin secondary antibodies. Cells then were enriched or depleted by autoMACS magnetic separator (Miltenyi Biotec) or manual separation using LS Columns (Miltenyi Biotec). Analysis and cell sorting were performed using a FACSAria or FACSCanto flow cytometer (BD Biosciences). Data were analyzed using FACSDiva (BD Biosciences).
Bone Marrow Transplantation
Recipient mice (CD45.1) were irradiated using an XRAD 320 X-ray irradiator (Precision X-Ray Inc.) with two doses of 540 rad (total 1,080 rad) delivered at least 3 hours apart. Cells were injected into the tail vein of anaesthetized recipients. After transplantation, mice were maintained on antibiotic water for 4 weeks. Blood was obtained from recipient mice every four weeks. Red blood cells were lysed with ammonium chloride potassium buffer. The remaining cells were stained with antibodies against CD45.2, CD45.1, B220, Mac1, CD3 and Gr1 and analyzed by flow cytometry. For non-competitive direct bone marrow transplantation, 5 × 105 CD45.2 whole bone marrow cells were transplanted into irradiated CD45.1 recipients and pIpC (20 mg/kg) was administrated 4 weeks after transplant. For competitive transplantation, mixed donor (CD45.2, 2 weeks post-pIpC) and competitor (CD45.1) whole bone marrow cells were transplanted into irradiated CD45.1 recipients. For secondary transplantation, 2 × 106 donor cells (CD45.2, lineage negative cells in bone marrow and spleen from moribund recipient of primary transplants) mixed with 2 ×105 competitor BM cells (CD45.1) were transplanted via tail vein injection into lethally irradiated CD45.1 recipient mice. For gabapentin and rapamycin drug trials, 2 × 106 whole donor cells (CD45.2, lineage negative cells in bone marrow and spleen from moribund recipient of primary transplants) mixed with 2 × 105 competitor BM cells (CD45.1) were transplanted via tail vein injection into lethally irradiated CD45.1 recipient. Gapapentin was reconstituted in PBS and administrated at 200 mg/kg by intraperitoneal injection. Rapamycin was reconstituted in absolute ethanol at 10 mg/ml and diluted in 5% Tween-80 (Sigma) and 5% PEG-400 (Sigma) before administration by intraperitoneal injection at 4 mg/kg per day. For transplantation with EZH1 or BCAT1 shRNAs, lineage negative cells (CD45.2, 2 weeks post-pIpC) were magnetically isolated and maintained in Prime-XV Mouse Hematopoietic Cell Medium (Irvine Scientific) supplemented with 50 μM β-Mercaptoethanol, 1% FBS, 50 ng/ml SCF and 50 ng/ml TPO. After two rounds of spin infection with shRNA viruses within 48 hours, 1,500 GFP+ LSK cells were sorted, mixed with 5 ×105 supporting BM cells (CD45.1), and transplanted into lethally irradiated CD45.1 recipient mice. For secondary transplant with Bcat1 shRNAs, lineage negative cells from donor bone marrow and splenic cells (CD45.2, from moribund recipients of primary transplants) were magnetically separated, transduced with two rounds of spin infection. 5×105 c-Kit+ and GFP+ double positive cells were sorted and mixed with 5 ×105 supporting BM cells (CD45.1) were transplanted via tail vein injection into lethally irradiated CD45.1 recipients. The recipient mice were maintained in regular drinking water or supplemented with 1.5 g/L Leucine. For metabolic analysis of cells with BCAT1 overexpression or knockdown experiments, lineage negative cells (CD45.2) from WT, G12D or G12D/E2-KO mice 2 weeks post-pIpC were isolated and transduced two rounds with BCAT1 overexpression or knockdown viruses. Sorted c-Kit+GFP+ (5 × 105) cells were mixed with 5 × 105 supporting BM cells (CD45.1) and transplanted via tail vein injection into lethally irradiated CD45.1 recipients. CD45.2+c-Kit+ cells were sorted from bone marrow of recipients 16 weeks after transplantation and subjected to metabolic analysis. To measure LIC activity by limiting dilution assays, recipient mice transplanted with G12D/E2-KO lineage negative cells in bone marrow and spleen (CD45.2, from moribund recipients of primary transplants) were treated with BCAT1 inhibitor (Gbp, 200 mg/kg) or vehicle control for 10 days. Then splenic CD45.2+ cells from recipient mice were isolated and transplanted into NSG (NOD-scid L2Rgnull) recipient mice at 10, 100 or 1000 cells per mouse via tail vein injection. A log-log plot and LIC frequencies were calculated using the ELDA tool.
Colony Formation Assays
A modified methylcellulose containing BCAAs (170 μM each) and BCKAs (30 μM each) were made based on amino acids deficient DMEM powder (D9800–27, USBiological Life Sciences) and StemXVivo Methylcellulose Concentrate (HSC011, R&D system). The final components are 1.4% methylcellulose (1500 cps), 25% fetal bovine serum, 2% bovine serum albumin, 2 mM L-Glutamine, 50 μM β-Mercaptoethanol, 16 μg/mL recombinant human insulin, 300 μg/mL human transferrin, 50 ng/ml recombinant mouse stem cell factor (SCF, human or mouse), 20 ng/ml recombinant mouse interleukin 3 (IL-3, human or mouse), recombinant granulocyte-macrophage colony-stimulating factor (GM-CSF, human or mouse), recombinant human interleukin 6 (IL-6), and recombinant human erythropoietin (EPO). LSK cells were sorted from mice 2 weeks post-pIpC. Five hundred LSK cells were seeded in the above modified methylcellulose medium in the presence of vehicle (DMSO), BCAT1i (20 and 40 μM), gabapentin (10 and 20 mM), rapamycin (10 and 20 nM), PP242 (10 and 20 nM) or Torin 1(10 and 20 nM). Colonies propagated in culture were counted at day 7. For colony formation with shRNAs, lineage negative cells (CD45.2, 2 weeks post-pIpC) were magnetically isolated and maintained in Prime-XV Mouse Hematopoietic Cell Medium (Irvine Scientific) supplemented with 50 μM β-Mercaptoethanol, 1% FBS, 50 ng/ml SCF and 50 ng/ml TPO. After two rounds of spin infection with shRNA viruses within 48 hours, 500 GFP+ LSK cells were sorted by flow cytometry, seeded in the modified methylcellulose medium, and counted at day 7. For Leu rescue experiments, 500 μM Leu was supplemented in the modified methylcellulose medium. For human AML cells, human mononuclear cells were seeded in the modified methylcellulose medium treated with DMSO or BCAT1 inhibitors as indicated.
Cytospin, Histology and Immunohistochemistry (IHC)
Cytospin preparations from peripheral blood or bone marrow cells stained with May-Grunwald-Giemsa as described previously (65). Tissue samples were fixed formalin, dehydrated and embedded in paraffin. Sectioned slides were rehydrated and followed by standard H&E staining protocol. For IHC, antigen was retrieved by boiling slides in 10mM sodium citrate buffer at 90–100°C for 20 minutes and then Cooled down to room temperature. After washing twice with PBST, slides were incubated in methanol with 3% H2O2 for 20 minutes followed by blocking with 5% goat serum. Primary antibody used was c-Kit (CD117) (Biolegend, Cat# 105802). Detection was performed with the Elite ABC Kit and DAB Substrate (Vector Laboratories), followed by hematoxylin counterstaining (Sigma). Reticulin and trichrome staining of BM, spleen or liver sections were performed by the Molecular Pathology Core facility at UTSW.
Measurement of Protein Synthesis
The rate of protein synthesis was determined as described previously (66) with modifications. Briefly, mice transplanted with WT, G12D or G12D/E2-KO donor cells (CD45.2, 2 weeks post-pIpC) were injected intraperitoneally with OP-Puro (O-propargyl-puromycin, pH 6.4–6.6 in PBS) at 50 mg/kg body mass one hour before euthanizing at indicated time points post-transplant. Bone marrow cells were harvested as above and 3 × 106 cells were stained with antibodies against cell surface markers (LSK, CD45.2+Lin−Sca1+c-Kit+; Myeloid cells, CD45.2+Mac1+Gr1+). Cells were then fixed in 0.5 ml of 1% paraformaldehyde (Affymetrix) in PBS for 15 minutes and washed with PBS, permeabilized in 200 μl permeabilization buffer (PBS supplemented with 3% fetal bovine serum and 0.1% saponin) for 5 min at room temperature. The azide-alkyne cycloaddition was performed using the Click-iT Cell Reaction Buffer Kit (Thermo-Fisher) and azide conjugated to Alexa Fluor 555 (Thermo-Fisher) at 5 μM final concentration for 30 min. Cells were then washed twice with permeabilization buffer and analyzed by flow cytometry.
Metabolic Analysis of BCAAs and BCKAs
Freshly isolated bone marrow c-Kit+ HSPCs, plasma or peripheral blood samples from mice were resuspended in 1.0 ml cold 80% methanol pre-chilled on dry ice. After overnight incubation at −80°C, lysates were harvested and centrifuged at 21,000 g for 20 min and supernatant was transferred to a new tube and lyophilized using a Speedvac (Thermo Scientific). Dried metabolites were reconstituted in 100 μl MPA and analyzed using liquid chromatography-tandem mass spectrometry (LC-MS/MS) as previously described (67). Analysis was performed on an AB Sciex 5500 QTRAP liquid chromatography/mass spectrometer (Applied Biosystems SCIEX) equipped with a vacuum degasser, a quaternary pump, an autosampler, a thermostatted column compartment, and a triple quadrupole/ion-trap mass spectrometer with electrospray ionization interface, and controlled by AB Sciex Analyst 1.6.1 Software. Phenomenex Luna C8 (150mm×2.0mm, 5μm) column was used for BCAA separation and Biodent C18 column was used for BCKA separation as described previously (68). Solvents for the mobile phase were 1mM aqueous ammonium acetate (A) and 5mM ammonium acetate in 90% acetonitrile (v/v) aqueous (B). The gradient elution was: 0–2.5 min, linear gradient 0–5% B; 2.5–6 min, linear gradient 5–40% B; and finally, washing the column with 100% B for 3 min before reconditioning it for 5 min using 0% B. The flow rate was 0.5 ml/min and the column was operated at 35°C. Multiple reaction monitoring (MRM) data were acquired with the transitions listed in Table S5. Chromatogram review and peak area integration were performed using MultiQuant software version 2.1 (AB SCIEX). For absolute quantification, the peak area for each metabolite was normalized against total cell number. The normalized area values were for statistical analysis. To determine physiological concentration of valine, leucine, α-ketoisovalerate and α-ketoisocaproate in mouse, plasma samples were spiked-in with 25μM 13C labeled valine, leucine, α-ketoisovalerate and α-ketoisocaproate at 1:4 ratio in 80% methanol. The final concentrations were calculated by normalizing to spiked-in isotopes. For ex vivo tracing experiment, c-Kit+ cells were isolated and cultured in Prime-XV Mouse Hematopoietic Cell Medium (Irvine Scientific) supplemented with 50 μM β- Mercaptoethanol, 1% FBS, 50 ng/ml SCF and 50 ng/ml TPO overnight. The next day, medium was replaced with a modified Dulbecco’s MEM (DMEM) Medium based on amino acids deficient DMEM (D9800–27, USBiological Life Sciences) supplemented with 50 μM β- Mercaptoethanol, 10% FBS, 50 ng/ml SCF and 50 ng/ml TPO, 2.2 g/L Bicarbonate, and 4.5 g /L D-Glucose. Cells were then treated with 13C-Valine, 13C-α-ketoisovalerate, 13C-Leucine, 13C-α-ketoisovalerate, or 15N-Glutamine for 24 hours in the presence of unlabeled substrate at indicated concentrations. 1× 106 cells were washed with cold saline twice and resuspended in 1.0 ml cold 80% methanol. Samples were then subjected to metabolomics analysis as described above. For time course tracing experiments, c-Kit+ cells were isolated and cultured in Prime-XV full Mouse Hematopoietic Cell Medium as described above overnight. The next day, medium was replaced with the modified DMEM medium supplemented with 2 mM Glutamine, 170 μM Leucine and 30 μM KIC for 2 hours. The culture medium was then replaced with the modified DMEM medium supplemented with 170 μM 13C6-Leucine and 30 μM 13C6-KIC or 2 mM 13C5-Glutamine accordingly. Samples were collected at indicated time and extracted in 1.0 ml cold 80% methanol spiked-in with 1 μM D3-Leucine and 1 μM D7-KIC. The abundance of natural 13C was corrected based on the composition in unlabeled standard samples. For metabolic analysis of cells with BCAT1 overexpression or knockdown experiments, the sorted cells were recovered in the modified DMEM medium supplemented with 50 μM β-Mercaptoethanol, 10% FBS, 50 ng/ml SCF, 50 ng/ml TPO, 2.2 g/L Bicarbonate, 4.5 g /L D-Glucose, 2 mM Glutamine, BCAAs (170 μM each) and BCKAs (30 μM each) or 2 mM Glutamine for 2 hours, and subjected to metabolic analysis. The same cell culture medium was used in the Western blot analysis of mTOR signaling ex vivo.
Quantification and Statistical Analysis
Statistical details including N, mean and statistical significance values are indicated in the text, figure legends, or Materials and Methods. Error bars in the experiments represent standard error of the mean (SEM) or standard deviation (SD) from either independent experiments or independent samples. All statistical analyses were performed using GraphPad Prism, and the detailed information about statistical methods is specified in figure legends or methods.
Data Availability
All raw and processed RNA-seq, ChIP-seq and ATAC-seq are available in the Gene Expression Omnibus (GEO): GSE112995.
Supplementary Material
SIGNIFIANCE.
EZH2 inactivation and oncogenic NRAS cooperate to induce leukemic transformation of myeloproliferative neoplasms by activating BCAT1 to enhance BCAA metabolism and mTOR signaling. We uncover a mechanism that epigenetic alterations rewire metabolism during cancer progression, causing epigenetic and metabolic liabilities in cancer-initiating cells that may be exploited as potential therapeutics.
ACKNOWLEDGMENTS
We thank Stuart Orkin for sharing of Ezh2 and Eed knockout mice, Mi Deng and Chengcheng Zhang for reagents and protocols, and Michalis Agathokleous for discussions. X. Liu was supported by the American Heart Association postdoctoral fellowship (18POST34060219). Y. Liu and K. Li were supported by the Cancer Prevention and Research Institute of Texas (CPRIT) training grant (RP160157). A. Tasdogan was supported by the Leopoldina Fellowship Program (LPDS 2016–16) from the German National Academy of Sciences Leopoldina. S.J. Morrison and R.J. DeBerardinis are Howard Hughes Medical Institute (HHMI) Investigators. This work was supported by the NIH grants R01DK111430 and R01CA230631, the CPRIT grants (RR140025, RP180504, RP180826 and RP190417), the Leukemia Texas Foundation research award, the Welch Foundation grant I-1942, the American Society of Hematology Scholar award, and the Leukemia and Lymphoma Society Scholar award (to J. Xu).
Footnotes
Disclosure of Potential Conflicts of Interest:
No potential conflicts of interest were disclosed.
REFERENCES
- 1.Kaelin WG Jr., McKnight SL. Influence of metabolism on epigenetics and disease. Cell 2013;153(1):56–69 doi 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cantor JR, Sabatini DM. Cancer cell metabolism: one hallmark, many faces. Cancer discovery 2012;2(10):881–98 doi 10.1158/2159-8290.cd-12-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kinnaird A, Zhao S, Wellen KE, Michelakis ED. Metabolic control of epigenetics in cancer. Nature reviews Cancer 2016;16(11):694–707 doi 10.1038/nrc.2016.82. [DOI] [PubMed] [Google Scholar]
- 4.Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016;127(20):2391–405 doi 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 5.Vainchenker W, Delhommeau F, Constantinescu SN, Bernard OA. New mutations and pathogenesis of myeloproliferative neoplasms. Blood 2011;118(7):1723–35 doi 10.1182/blood-2011-02-292102. [DOI] [PubMed] [Google Scholar]
- 6.Klampfl T, Gisslinger H, Harutyunyan AS, Nivarthi H, Rumi E, Milosevic JD, et al. Somatic mutations of calreticulin in myeloproliferative neoplasms. The New England journal of medicine 2013;369(25):2379–90 doi 10.1056/NEJMoa1311347. [DOI] [PubMed] [Google Scholar]
- 7.Rumi E, Pietra D, Pascutto C, Guglielmelli P, Martinez-Trillos A, Casetti I, et al. Clinical effect of driver mutations of JAK2, CALR, or MPL in primary myelofibrosis. Blood 2014;124(7):1062–9 doi 10.1182/blood-2014-05-578435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beer PA, Delhommeau F, LeCouedic JP, Dawson MA, Chen E, Bareford D, et al. Two routes to leukemic transformation after a JAK2 mutation-positive myeloproliferative neoplasm. Blood 2010;115(14):2891–900 doi 10.1182/blood-2009-08-236596. [DOI] [PubMed] [Google Scholar]
- 9.Lasho TL, Mudireddy M, Finke CM, Hanson CA, Ketterling RP, Szuber N, et al. Targeted next-generation sequencing in blast phase myeloproliferative neoplasms. Blood advances 2018;2(4):370–80 doi 10.1182/bloodadvances.2018015875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Menghrajani K, Rapaport F, McNamara CJ, Devlin SM, Famulare C, Patel M, et al. SETBP1 and NRAS mutations are frequent events in post-myeloproliferative neoplasm acute myeloid leukemia (post-MPN AML) lacking JAK-STAT activating mutations. Blood 2017;130:203. [Google Scholar]
- 11.Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nature genetics 2010;42(8):722–6 doi 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 12.Nikoloski G, Langemeijer SM, Kuiper RP, Knops R, Massop M, Tonnissen ER, et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nature genetics 2010;42(8):665–7 doi 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- 13.Stieglitz E, Taylor-Weiner AN, Chang TY, Gelston LC, Wang YD, Mazor T, et al. The genomic landscape of juvenile myelomonocytic leukemia. Nature genetics 2015;47(11):1326–33 doi 10.1038/ng.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caye A, Strullu M, Guidez F, Cassinat B, Gazal S, Fenneteau O, et al. Juvenile myelomonocytic leukemia displays mutations in components of the RAS pathway and the PRC2 network. 2015;47(11):1334–40 doi 10.1038/ng.3420. [DOI] [PubMed] [Google Scholar]
- 15.Abdel-Wahab O, Adli M, LaFave LM, Gao J, Hricik T, Shih AH, et al. ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer cell 2012;22(2):180–93 doi 10.1016/j.ccr.2012.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim E, Ilagan JO, Liang Y, Daubner GM, Lee SC, Ramakrishnan A, et al. SRSF2 Mutations Contribute to Myelodysplasia by Mutant-Specific Effects on Exon Recognition. Cancer cell 2015;27(5):617–30 doi 10.1016/j.ccell.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guglielmelli P, Biamonte F, Score J, Hidalgo-Curtis C, Cervantes F, Maffioli M, et al. EZH2 mutational status predicts poor survival in myelofibrosis. Blood 2011;118(19):5227–34 doi 10.1182/blood-2011-06-363424. [DOI] [PubMed] [Google Scholar]
- 18.Tefferi A, Lasho TL, Guglielmelli P, Finke CM, Rotunno G, Elala Y, et al. Targeted deep sequencing in polycythemia vera and essential thrombocythemia. Blood advances 2016;1(1):21–30 doi 10.1182/bloodadvances.2016000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nature genetics 2010;42(2):181–5 doi 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 2002;419(6907):624–9 doi 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 21.Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Molecular cell 2008;32(4):491–502 doi 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mochizuki-Kashio M, Mishima Y, Miyagi S, Negishi M, Saraya A, Konuma T, et al. Dependency on the polycomb gene Ezh2 distinguishes fetal from adult hematopoietic stem cells. Blood 2011;118(25):6553–61 doi 10.1182/blood-2011-03-340554. [DOI] [PubMed] [Google Scholar]
- 23.Xie H, Xu J, Hsu JH, Nguyen M, Fujiwara Y, Peng C, et al. Polycomb repressive complex 2 regulates normal hematopoietic stem cell function in a developmental-stage-specific manner. Cell stem cell 2014;14(1):68–80 doi 10.1016/j.stem.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shimizu T, Kubovcakova L, Nienhold R, Zmajkovic J, Meyer RJ, Hao-Shen H, et al. Loss of Ezh2 synergizes with JAK2-V617F in initiating myeloproliferative neoplasms and promoting myelofibrosis. The Journal of experimental medicine 2016;213(8):1479–96 doi 10.1084/jem.20151136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sashida G, Wang C, Tomioka T, Oshima M, Aoyama K, Kanai A, et al. The loss of Ezh2 drives the pathogenesis of myelofibrosis and sensitizes tumor-initiating cells to bromodomain inhibition. The Journal of experimental medicine 2016;213(8):1459–77 doi 10.1084/jem.20151121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Y, Akada H, Nath D, Hutchison RE, Mohi G. Loss of Ezh2 cooperates with Jak2V617F in the development of myelofibrosis in a mouse model of myeloproliferative neoplasm. Blood 2016;127(26):3410–23 doi 10.1182/blood-2015-11-679431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muto T, Sashida G, Oshima M, Wendt GR, Mochizuki-Kashio M, Nagata Y, et al. Concurrent loss of Ezh2 and Tet2 cooperates in the pathogenesis of myelodysplastic disorders. The Journal of experimental medicine 2013;210(12):2627–39 doi 10.1084/jem.20131144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Booth CAG, Barkas N, Neo WH, Boukarabila H, Soilleux EJ, Giotopoulos G, et al. Ezh2 and Runx1 Mutations Collaborate to Initiate Lympho-Myeloid Leukemia in Early Thymic Progenitors. Cancer cell 2018;33(2):274–91.e8 doi 10.1016/j.ccell.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 29.Hutson SM, Sweatt AJ, Lanoue KF. Branched-chain [corrected] amino acid metabolism: implications for establishing safe intakes. The Journal of nutrition 2005;135(6 Suppl):1557s–64s. [DOI] [PubMed] [Google Scholar]
- 30.Tonjes M, Barbus S, Park YJ, Wang W, Schlotter M, Lindroth AM, et al. BCAT1 promotes cell proliferation through amino acid catabolism in gliomas carrying wild-type IDH1. Nature medicine 2013;19(7):901–8 doi 10.1038/nm.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshikawa R, Yanagi H, Shen CS, Fujiwara Y, Noda M, Yagyu T, et al. ECA39 is a novel distant metastasis-related biomarker in colorectal cancer. World journal of gastroenterology 2006;12(36):5884–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hattori A, Tsunoda M, Konuma T, Kobayashi M, Nagy T, Glushka J, et al. Cancer progression by reprogrammed BCAA metabolism in myeloid leukaemia. Nature 2017;545:500–4 doi 10.1038/nature22314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raffel S, Falcone M, Kneisel N, Hansson J, Wang W, Lutz C, et al. BCAT1 restricts alphaKG levels in AML stem cells leading to IDHmut-like DNA hypermethylation. Nature 2017;551(7680):384–8 doi 10.1038/nature24294. [DOI] [PubMed] [Google Scholar]
- 34.Mayers JR, Torrence ME, Danai LV, Papagiannakopoulos T, Davidson SM, Bauer MR, et al. Tissue of origin dictates branched-chain amino acid metabolism in mutant Kras-driven cancers. Science (New York, NY) 2016;353(6304):1161–5 doi 10.1126/science.aaf5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bacher U, Haferlach T, Schoch C, Kern W, Schnittger S. Implications of NRAS mutations in AML: a study of 2502 patients. Blood 2006;107(10):3847–53 doi 10.1182/blood-2005-08-3522. [DOI] [PubMed] [Google Scholar]
- 36.Li Q, Haigis KM, McDaniel A, Harding-Theobald E, Kogan SC, Akagi K, et al. Hematopoiesis and leukemogenesis in mice expressing oncogenic NrasG12D from the endogenous locus. Blood 2011;117(6):2022–32 doi 10.1182/blood-2010-04-280750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J, Kong G, Liu Y, Du J, Chang YI, Tey SR, et al. Nras(G12D/+) promotes leukemogenesis by aberrantly regulating hematopoietic stem cell functions. Blood 2013;121(26):5203–7 doi 10.1182/blood-2012-12-475863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu J, Shao Z, Li D, Xie H, Kim W, Huang J, et al. Developmental control of polycomb subunit composition by GATA factors mediates a switch to non-canonical functions. Molecular cell 2015;57(2):304–16 doi 10.1016/j.molcel.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Q, Bohin N, Wen T, Ng V, Magee J, Chen SC, et al. Oncogenic Nras has bimodal effects on stem cells that sustainably increase competitiveness. Nature 2013;504(7478):143–7 doi 10.1038/nature12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mills KI, Kohlmann A, Williams PM, Wieczorek L, Liu WM, Li R, et al. Microarray-based classifiers and prognosis models identify subgroups with distinct clinical outcomes and high risk of AML transformation of myelodysplastic syndrome. Blood 2009;114(5):1063–72 doi 10.1182/blood-2008-10-187203. [DOI] [PubMed] [Google Scholar]
- 41.Haferlach T, Kohlmann A, Wieczorek L, Basso G, Kronnie GT, Bene MC, et al. Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: report from the International Microarray Innovations in Leukemia Study Group. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2010;28(15):2529–37 doi 10.1200/jco.2009.23.4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Metzeler KH, Hummel M, Bloomfield CD, Spiekermann K, Braess J, Sauerland MC, et al. An 86-probe-set gene-expression signature predicts survival in cytogenetically normal acute myeloid leukemia. Blood 2008;112(10):4193–201 doi 10.1182/blood-2008-02-134411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tyner JW, Tognon CE, Bottomly D, Wilmot B, Kurtz SE, Savage SL, et al. Functional genomic landscape of acute myeloid leukaemia. Nature 2018;562(7728):526–31 doi 10.1038/s41586-018-0623-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu LY, Boxer PA, Kesten SR, Lei HJ, Wustrow DJ, Moreland DW, et al. The design and synthesis of human branched-chain amino acid aminotransferase inhibitors for treatment of neurodegenerative diseases. Bioorganic & medicinal chemistry letters 2006;16(9):2337–40 doi 10.1016/j.bmcl.2005.07.058. [DOI] [PubMed] [Google Scholar]
- 45.Hutson SM, Berkich D, Drown P, Xu B, Aschner M, LaNoue KF. Role of branched-chain aminotransferase isoenzymes and gabapentin in neurotransmitter metabolism. Journal of neurochemistry 1998;71(2):863–74. [DOI] [PubMed] [Google Scholar]
- 46.McBrayer SK, Mayers JR, DiNatale GJ, Shi DD, Khanal J, Chakraborty AA, et al. Transaminase Inhibition by 2-Hydroxyglutarate Impairs Glutamate Biosynthesis and Redox Homeostasis in Glioma. Cell 2018;175(1):101–16.e25 doi 10.1016/j.cell.2018.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ananieva EA, Patel CH, Drake CH, Powell JD, Hutson SM. Cytosolic branched chain aminotransferase (BCATc) regulates mTORC1 signaling and glycolytic metabolism in CD4+ T cells. The Journal of biological chemistry 2014;289(27):18793–804 doi 10.1074/jbc.M114.554113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lynch CJ, Adams SH. Branched-chain amino acids in metabolic signalling and insulin resistance. Nature reviews Endocrinology 2014;10(12):723–36 doi 10.1038/nrendo.2014.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mayers JR, Wu C, Clish CB, Kraft P, Torrence ME, Fiske BP, et al. Elevation of circulating branched-chain amino acids is an early event in human pancreatic adenocarcinoma development. Nature medicine 2014;20(10):1193–8 doi 10.1038/nm.3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fontana L, Partridge L. Promoting health and longevity through diet: from model organisms to humans. Cell 2015;161(1):106–18 doi 10.1016/j.cell.2015.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, et al. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell 2009;139(7):1303–14 doi 10.1016/j.cell.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haigis KM, Kendall KR, Wang Y, Cheung A, Haigis MC, Glickman JN, et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nature genetics 2008;40(5):600–8 doi 10.1038/ng.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science (New York, NY) 1995;269(5229):1427–9. [DOI] [PubMed] [Google Scholar]
- 54.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics (Oxford, England) 2009;25(9):1105–11 doi 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 2005;102(43):15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature 2011;476(7360):346–50 doi 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology 2014;15(12):550 doi 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmidl C, Rendeiro AF, Sheffield NC, Bock C. ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors. Nature methods 2015;12(10):963–5 doi 10.1038/nmeth.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang J, Liu X, Li D, Shao Z, Cao H, Zhang Y, et al. Dynamic Control of Enhancer Repertoires Drives Lineage and Stage-Specific Transcription during Hematopoiesis. Developmental Cell 2016;36(1):9–23 doi 10.1016/j.devcel.2015.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology 2009;10(3):R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS). Genome biology 2008;9(9):R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shao Z, Zhang Y, Yuan GC, Orkin SH, Waxman DJ. MAnorm: a robust model for quantitative comparison of ChIP-Seq data sets. Genome biology 2012;13(3):R16 doi 10.1186/gb-2012-13-3-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu X, Zhang Y, Chen Y, Li M, Zhou F, Li K, et al. In Situ Capture of Chromatin Interactions by Biotinylated dCas9. Cell 2017;170(5):1028–43.e19 doi 10.1016/j.cell.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kharas MG, Lengner CJ, Al-Shahrour F, Bullinger L, Ball B, Zaidi S, et al. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nature medicine 2010;16(8):903–8 doi 10.1038/nm.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu J, Peng C, Sankaran VG, Shao Z, Esrick EB, Chong BG, et al. Correction of sickle cell disease in adult mice by interference with fetal hemoglobin silencing. Science (New York, NY) 2011;334(6058):993–6 doi 10.1126/science.1211053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Signer RA, Magee JA, Salic A, Morrison SJ. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature 2014;509(7498):49–54 doi 10.1038/nature13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu X, Zhang Y, Ni M, Cao H, Signer RAJ, Li D, et al. Regulation of mitochondrial biogenesis in erythropoiesis by mTORC1-mediated protein translation. Nature cell biology 2017;19(6):626–38 doi 10.1038/ncb3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang Y, Yin B, Li R, He P. Determination of Branched-Chain Keto Acids in Serum and Muscles Using High Performance Liquid Chromatography-Quadrupole Time-of-Flight Mass Spectrometry. Molecules (Basel, Switzerland) 2018;23(1) doi 10.3390/molecules23010147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw and processed RNA-seq, ChIP-seq and ATAC-seq are available in the Gene Expression Omnibus (GEO): GSE112995.