Abstract
We report the emergence of the novel MEK1 V211D gatekeeper mutation in a patient with BRAF K601E colon cancer treated with the allosteric MEK inhibitor binimetinib and the anti-epidermal growth factor receptor (EGFR) antibody panitumumab. The MEK1 V211D mutation concurrently occurs in the same cell with BRAF K601E and leads to RAF-independent activity but remains regulated by RAF. The V211D mutation causes resistance to binimetinib by both increasing the catalytic activity of MEK1 and reducing its affinity for the drug. Moreover, the mutant exhibits reduced sensitivity to all the allosteric MEK inhibitors tested. Thus this mutation serves as a general resistance mutation for current MEK inhibitors; however, it is sensitive to a newly reported ATP-competitive MEK inhibitor, which therefore could be used to overcome drug resistance.
Introduction
The RAS/RAF/MEK/ERK pathway is a key driver of tumor growth in human cancers. Recurrent genomic alterations in this pathway occur most commonly in the KRAS, NRAS, and BRAF genes and activate the MEK (mitogen-activated protein kinase kinase) kinases to constitutively activate downstream signaling. Thus MEK represents a promising target for therapies directed against this pathway. Highly potent, allosteric MEK inhibitors that bind to MEK and keep it in a closed, inactive conformation are now clinically available. The MEK inhibitors trametinib, cobimetinib, and binimetinib, are all FDA approved together with RAF inhibitors to treat BRAF V600 mutant melanoma. Additionally, MEK inhibitors as single agents have been shown to enhance radioiodine uptake in advanced thyroid cancer (1) and to cause regression of neurofibromas in patients with neurofibromatosis type 1 (2) and of BRAF-mutant pediatric low-grade gliomas (3). Dramatic clinical responses have been observed with MEK inhibitors in a small number of patients with MEK1 mutations suggesting that MEK inhibitors may be an effective treatment in at least a subset of MEK1 mutant patients (4,5). While mechanisms of acquired resistance to RAF/MEK combinations have been extensively studied, mechanisms that limit the activity of MEK inhibitors in patients have yet to be defined.
Results
A MEK1 V211D mutation was detected in a colon cancer from a patient treated with binimetinib plus panitumumab
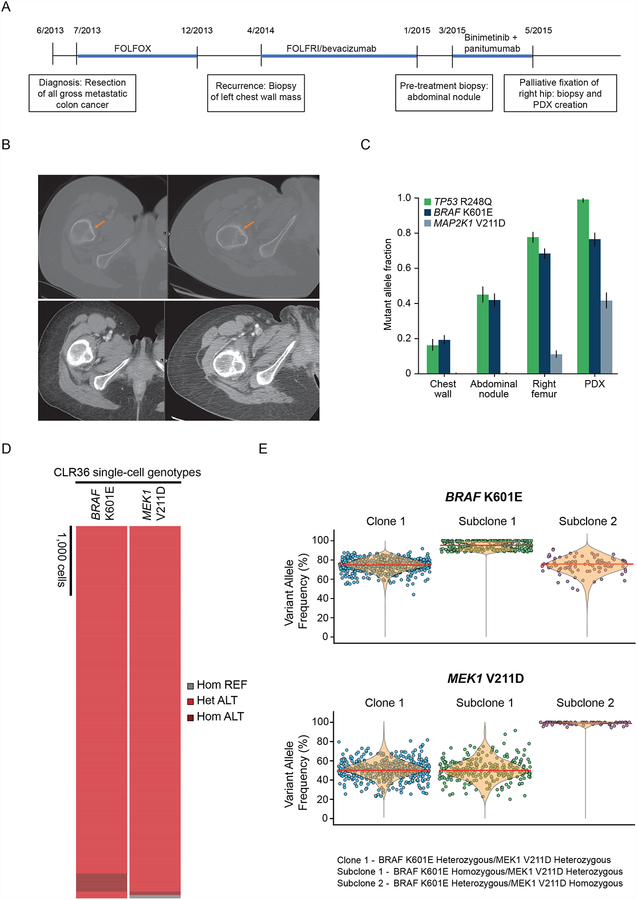
A 39-year old woman with a BRAF K601E-mutant metastatic colon cancer that involved the chest, abdominal wall, distant lymph nodes, and bones was treated with combined binimetinib and panitumumab for 6 weeks in a phase Ib/II trial sponsored by Novartis Pharmaceuticals and then Array BioPharma () (Fig. 1A). BRAF K601E is an activating, non-V600 BRAF mutation that is unresponsive to RAF inhibitors (6), unlike BRAF V600 alterations. Patients with colorectal cancers harboring activating non-V600 BRAF mutants do not clinically respond to anti-EGFR antibodies (manuscript under review). Reactivation of EGFR signaling has been shown to limit the clinical activity of ERK pathway inhibitors in colorectal cancers (7,8). In this patient, the clinical trial provided the opportunity to treat with the MEK inhibitor binimetinib to target ERK activation with the addition of the anti-EGFR antibody panitumumab to overcome reactivation of EGFR signaling after ERK inhibition. At 6 weeks, imaging showed a stable chest wall mass and an increase in the periosteal reaction and extraosseus soft tissue component anterior to the right femur, and she underwent palliative fixation of the right hip for persistent pain (Fig. 1B). Next-generation sequencing with MSK-IMPACT (9) of the right femur bone tissue, obtained while on treatment, revealed a new, subclonal MEK1 V211D mutation (Fig. 1C). The MEK1 V211D mutation was not identified in biopsy specimens collected either soon after diagnosis from the chest wall metastasis (0/824 reads) or immediately before starting this treatment from an abdominal wall nodule (0/870 reads). A section of the right femur tumor was implanted in a mouse to generate a patient-derived xenograft (PDX) model and sequencing suggested enrichment of the MEK1 V211D variant allelic fraction in the growing PDX (Fig. 1C).
Figure 1. MEK1 V211D mutation emerges in a patient with colon cancer treated with binimetinib plus panitumumab.
A, Timeline of the patient’s treatment showing when she was treated with binimetinib and panitumuab, the duration of each treatment regimen, and when biopsy specimens were obtained for sequencing. B, Representative computerized tomography (CT) images showing periosteal changes (top) and marrow involvement (bottom) in the right femur lesion immediately before and after 6 weeks of binimetinib plus panitumumab treatment. C, Mutant allele fraction detected by MSK-IMPACT sequencing for the truncal TP53 mutation and for BRAF K601E and MEK1 V211D in the indicated tissues. Error bars indicate 95% binomial confidence intervals on the variant allele frequencies. D, Heatmap depicting single-cell genotypes for the CLR36 sample. The presence of a heterozygous alternate (ALT) allele is shown in red. Homozygous alternate alleles are shown in dark red, and reference alleles are depicted in gray. E, Variant allele frequency (VAF) distribution of BRAF K601E (top) and MEK1 V211D (bottom) in the three clonal/subclonal populations. The median of each VAF is represented as a red line. For data representation simplicity, each dot in clone 1 represents 10% of the total number of cells.
To determine if the BRAF K601E and MEK1 V211D mutations arose in the same population of tumor cells, we performed single-cell DNA sequencing without whole-genome amplification of the cell line generated from the PDX (CLR36). A total of 5,895 cells were sequenced (Supplementary Table 1). The BRAF K601E and MEK1 V211D alterations were found to co-occur in 92% of all cells (n=5,423) with a median variant allelic frequency (VAF) of 75% and 50%, respectively, very similar to the VAFs identified from the bulk sequencing of the PDX (Fig. 1D and E; Supplementary Fig. 1A–E; Supplementary Table 2). Single-cell sequencing identified the concurrent mutations in three populations: a major clone (n=5,095 cells) heterozygous for both BRAF K601E and MEK1 V211D variants, a subclone (n=267 cells) homozygous for BRAF K601E and heterozygous for MEK1 V211D, and a subclone (n=61 cells) heterozygous for BRAF K601E and homozygous for MEK1 V211D.
Review of over 30,000 advanced tumors analyzed with MSK-IMPACT (http://cbioportal.mskcc.org) and over 250 colorectal cancers in The Cancer Genome Atlas (10) identified no cases with the MEK1 V211D mutation. Thus, this mutant rarely occurs in nature and emphasizes its emergence as the result of treatment exposure in this patient.
MEK1 V211D has elevated RAF-independent catalytic activity that is further stimulated by RAF
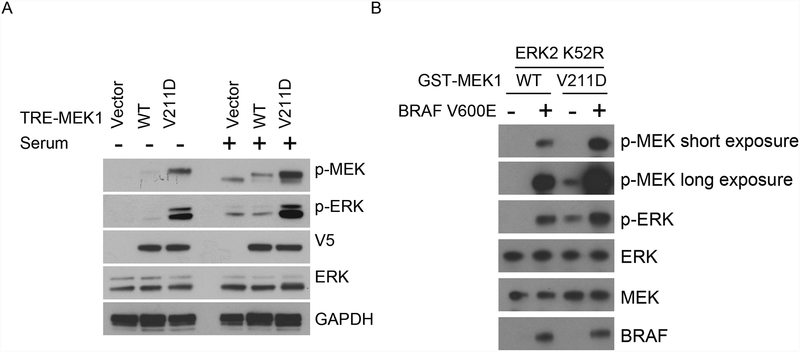
To characterize MEK1 V211D functionally, we first examined whether this mutant could activate ERK signaling as compared to wild-type (WT) MEK. In NIH-3T3 cells, expression of MEK1 V211D induced higher levels of p-MEK and p-ERK than WT MEK1 does in both serum-containing and serum-starved conditions (Fig. 2A). However, serum starvation reduced ERK activation in both WT and MEK1 V211D-mutant expressing cells. The decrease of MEK/ERK phosphorylation in the MEK1 V211D expressing cells could be due to the inhibition of endogenous MEK proteins or the mutant MEK1. We tested whether the phosphorylation and kinase activity of MEK1 V211D is still regulated by upstream RAF kinase. We purified GST-tagged WT MEK1 and MEK1 V211D-mutant proteins and performed an in vitro kinase assay in the absence or presence of active BRAF kinase. Purified WT MEK1 was not phosphorylated in the absence of RAF kinase, nor could it phosphorylate ERK. Addition of activated BRAF kinase induced both the phosphorylation and kinase activity of WT MEK1. In contrast, we found MEK1 V211D was phosphorylated and could phosphorylate ERK in the absence of activated BRAF, suggesting this mutant has acquired RAF-independent phosphorylation and basal activity. The phosphorylation and kinase activity of MEK1 V211D could be further enhanced with the addition of activated BRAF kinase (Fig. 2B). In addition, under the same reaction condition, MEK1 V211D was more effectively phosphorylated by RAF kinase than was the WT MEK protein. This could be responsible for its increased sensitivity to RAF-mediated kinase activation. Therefore, MEK1 V211D is among the class of RAF-regulated MEK1 mutants (11) that we recently defined as having autonomous kinase activity that can be further activated by RAF and more effectively transduces RAF activity downstream to ERK. In the patient, the MEK1 V211D mutant developed in a tumor with an activating BRAF K601E mutation that signals independently of RAS (Supplementary Fig. 2A). The MEK1 V211D mutant would be expected to further activate signaling in the setting of activated BRAF, and thus amplify ERK signaling in this tumor.
Figure 2. MEK1 V211D has increased basal activity that is further activated by RAF.
A, NIH-3T3 cells stably expressing doxycycline-inducible WT or indicated mutant MEK1 were plated for 12 hours to adhere and then serum was removed as indicated. Twelve hours later, cells were treated with doxycycline (300 ng/ml) for 24 hours. Cells were then collected, expression and phosphorylation of the indicated proteins were assayed by western blot. B, Purified GST fusion WT or V211D mutant MEK1 protein were incubated with recombinant inactive ERK2 K52R in the absence or presence of recombinant BRAF V600E at 30°C for 15 minutes. Western blot analysis was performed using antibodies against p-ERK, ERK, p-MEK, MEK, and BRAF.
We evaluated the effects of the V211D mutation on the interactions between MEK1 and its kinase and substrate (Supplementary Fig. 2B). We expressed either WT MEK1 or V211D MEK1 together with WT or K601E BRAF or with WT ERK1 or ERK2 in 293H cells and performed immunoprecipitation. The MEK1 V211D mutation did not affect MEK1 binding to WT or K601E BRAF. However, the binding of MEK1 to ERK was reduced by the MEK1 V211D mutation. This is likely due to the elevated kinase activity of the MEK1 V211D mutant versus WT MEK1.
MEK1 V211D causes resistance to allosteric MEK inhibitors
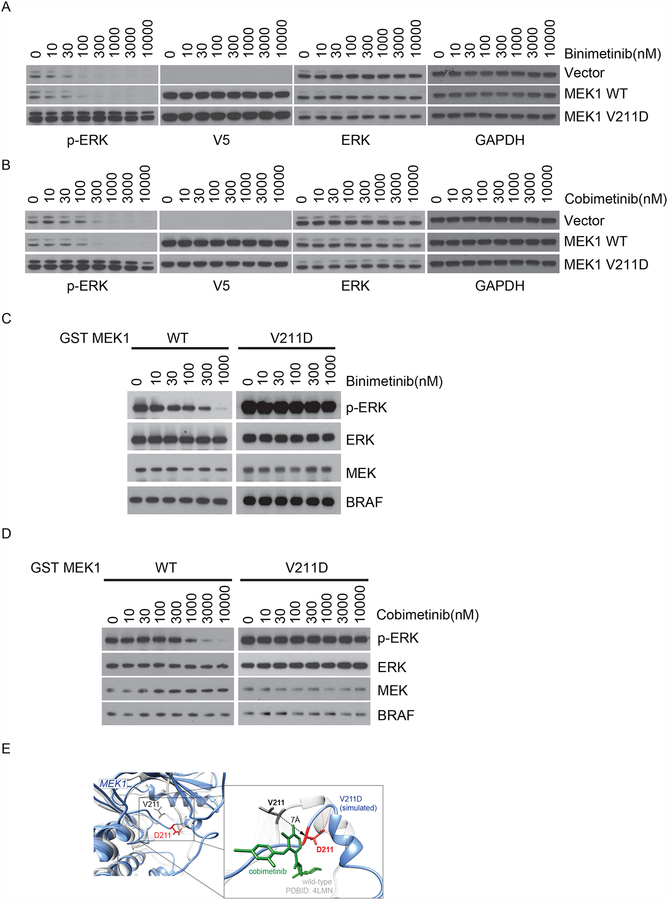
To determine whether the MEK1 V211D allele affects sensitivity to allosteric MEK inhibitors, we tested the effects of binimetinib in NIH-3T3 cells expressing WT or MEK1 V211D mutant. Binimetinib potently inhibited ERK activation at a dose of 0.1uM in vector expressing parental cells and cells with ectopic expression of WT MEK1, whereas p-ERK remained unaffected by 3uM binimetinib in cells expressing MEK1 V211D (Fig. 3A). These data suggest that, in cells, MEK1 V211D-driven ERK activation is insensitive to binimetinib treatment. Silencing the expression of MEK1 V211D in CLR36 cells sensitized the cells to treatment with binimetinib (Supplementary Fig. 3A and B). Similarly, this MEK1 mutation also decreased the sensitivity of ERK signaling to another allosteric MEK inhibitor cobimetinib (Fig. 3B). To understand the mechanism underlying this insensitivity to MEK inhibitors, we tested whether the V211D mutation causes resistance of MEK1 to these drugs in vitro. Purified GST-tagged WT MEK1 and MEK1 V211D mutant proteins were incubated with increasing doses of MEK inhibitors and their kinase activity was assessed by a in vitro kinase assay using inactive ERK2 as substrate. Consistent with what we observed in cells, the activity of MEK1 V211D, reflected in p-ERK levels, remained unchanged following increasing doses of either binimetinib or cobimetinib treatment, compared to potent inhibition of WT MEK1 activity by the above two inhibitors (Fig. 3C and D). These data suggest that MEK1 V211D is sufficient to cause resistance to multiple allosteric MEK inhibitors both in vitro and in cells.
Figure 3. MEK1 V211D causes resistance to allosteric MEK inhibitors.
A-B, Wild-type (WT) or mutant MEK1 tagged with V5 were expressed in NIH-3T3 cells upon culturing in medium containing doxycycline (300 ng/ml) for 24 hours. Cells were then treated for 1 hour with increasing concentrations of two different allosteric MEK1 inhibitors binimetinib (A) or cobimetinib (B). C-D, In the in vitro kinase assay, WT or mutant MEK1 protein were treated with either binimetinib (C) or cobimetinib (D) at increasing concentrations before incubation with recombinant inactive ERK2 K52R in the presence of recombinant BRAF V600E at 30°C for 15 minutes. Western blot analysis was performed using antibodies against p-ERK, ERK, p-MEK, MEK, and BRAF. E, Structural model with an overlay of WT MEK1 in grey and V211D MEK1 in blue. The zoomed in figure shows the interface of D211 with the allosteric MEK inhibitor cobimetinib.
Indeed, MEK1 V211D was previously implicated as a resistance allele to diarylamine MEK inhibitor (AZD6244 or CI-1040) in a random mutagenesis screen. Based on mapping the mutant allele within the three-dimensional structure of the full-length MEK1 kinase domain, Emery et al suggested that the V211D mutation, situated directly within the arylamine binding pocket, may cause resistance by direct interference with drug binding (12). To evaluate the structural effects of this mutant, structural models of MEK1 WT and V211D were generated using template-based modeling and molecular dynamics simulations (Fig. 3E). MEK1 D211 residue forms a hydrogen bond to nearby MEK1 residues, which does not occur in WT MEK1 and results in displacement of D211 from the WT position by 7 angstroms. A zoomed in image with cobimetinib shows that D211 MEK1 is pulled away from its WT position and faces away form the drug’s binding site. The hydrophobic carbon atoms of V211 which interact with cobimetinib are lost as D211 is not a hydrophobic residue. Our data indicate that V211D is a gatekeeper mutation for allosteric MEK inhibitors. Furthermore, our findings also suggest that MEK1 V211D displays enhanced kinase activity in addition to its effect in reducing drug binding, which promotes resistance to allosteric MEK inhibitors.
MEK1 V211D is sensitive to an ATP-competitive MEK inhibitor
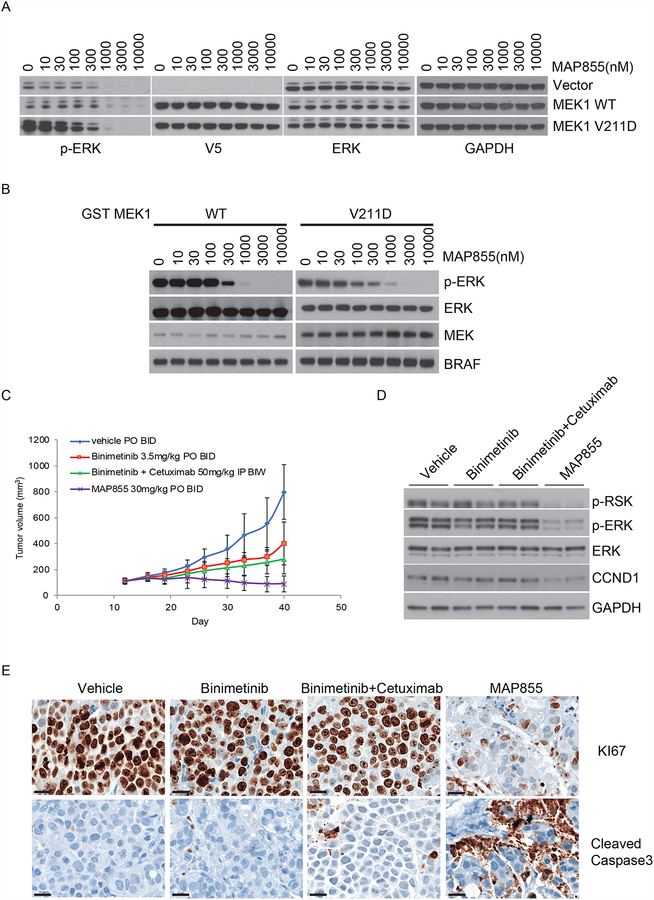
We previously reported that allosteric MEK inhibitor-insensitive MEK1 mutants which exhibits RAF-independent activity could be effectively treated by a selective ATP competitive MEK inhibitor, MAP855, through direct interference with ATP binding (11). We thus hypothesized that MAP855 could also inhibit MEK1 V211D-driven ERK sigaling by targeting its ATP site. We tested the activity of MAP855 in MEK1 V211D expressing NIH-3T3 cells and found that this drug inhibited ERK activation driven by either WT MEK or MEK1 V211D at similar doses although MEK1 V211D expressing cells had higher initial phospho-ERK (Fig. 4A). Using the additional ATP-competitive MEK inhibitor BI-847325 (13), we confirmed that these results were not compound specific and that WT and V211D MEK1 exhibited similar sensitivity to same type ATP-competitive MEK inhibitors (Supplementary Fig. 3C). Consistent with these findings, the kinase activtiy of MEK1 V211D was inhibited by MAP855 at equal potency compared to WT MEK1 in vitro (Fig. 4B). We tested whether the patient’s tumor might be sensitive to MAP855 in the PDX model derived from the progressing right femur lesion, which produces tumors that continued to grow with either binimetinib treatment alone or in combination with the EGFR antibody cetuximab. In contrast, MAP855 treatment at a non-toxic dose led to around 30% tumor regression (Fig. 4C; Supplementary Fig. 4). Consistently, MAP855 potently inhibited ERK signaling and tumor proliferation (Ki-67) and induced the apoptosis marker cleaved capsase-3 in the PDX tumors, which were resistant to either binimetinib alone or combined binimetinib/cetuximab treatment (Fig. 4D and E). Taken together, our data suggest that ATP competitive MEK inhibition represents a novel therapeutic strategy for tumors with acquired resistance to current allosteric MEK inhibitors.
Figure 4. MEK1 V211D is sensitive to an ATP-competitive MEK inhibitor.
A, WT or mutant MEK1 tagged with V5 were expressed in NIH-3T3 cells upon culturing in medium containing 300 ng/ml doxycycline for 24 hours. Cells were then treated for 1 hour with increasing concentrations of MAP855. Expression and phosphorylation of the indicated proteins were assayed by western blot. B, In the in vitro kinase assay, GST tagged WT or mutant MEK1 protein were treated with MAP855 at increasing concentrations before incubation with recombinant inactive ERK2 K52R in the presence of recombinant BRAF V600E at 30°C for 15 minutes. Western blot analysis was performed using antibodies against p-ERK, ERK, p-MEK, MEK, and BRAF. C, PDX made from the progression specimen of the patient was expanded into mice that were treated with vehicle, binimetinib (3.5 mg/kg orally twice daily), or binimetinib (3.5 mg/kg orally twice daily) plus cetuximab (50 mg/kg i.p. injection twice per week), or MAP855 (30 mg/kg orally twice daily). Tumor volumes (and standard deviation) are shown as a function of time on treatment. D, Tumors were collected at day 40, and two samples from each group were lysed for immunoblotting with the indicated antibodies. E, Representative imagines of immunohistochemistry of samples from each group for the proliferation marker Ki-67 and the apoptosis marker cleaved caspase-3.
Discussion
Previous studies in cell line models have identified multiple mechanisms for acquired resistance to allosteric MEK inhibitors, including amplification of upstream oncogenic drivers of the ERK pathway in BRAF or KRAS mutant colorectal cancer cells (14,15), or MEK1 mutations in both helix A and the allosteric binding pocket of MEK protein (12). In our recent work, we have shown that MEK1 mutations exhibit allelic-specific mechanisms of ERK activation (11). A subset of MEK1 mutants acquire RAF-independent kinase activity. The degree of autonomous ERK activation varies across mutants and can be further enhanced by RAF activation (RAF-regulated mutants) or totally independent of RAF. In addition, we showed that the RAF-independent activities of MEK1 mutants reduced their sensitivity to current MEK inhibitors. However, the clinical relevance of the proposed resistance mechanisms from cell line models needs validation in tumor samples from patients treated with MEK inhibitors. Our study reports the first case of a cancer patient who acquired a MEK1 V211D mutation in a progressing tumor. We further propose a strategy to overcome this resistance mechanism using a new class of ATP competitive MEK inhibitor and demonstrate its efficacy in the PDX made from this patient’s progressing tumor. Our results suggest that treatment with the ATP competitive MEK inhibitor is a rational therapeutic strategy for patients whose tumors exhibit acquired resistance to allosteric MEK inhibitors.
So far, we have not identified any cases of the MEK1 V211D mutation in a review of over 30,000 clinical specimens sequenced at Memorial Sloan Kettering and in TCGA, suggesting that this mutation does not arise in the absence of therapy. In this patient, MEK1 V211D occurred in the setting of a BRAF K601E activating mutation upon drug treatment and the two alterations are in the same cells, validating our finding that RAF-regulated MEK1 mutants can co-occur with upstream alterations to amplify BRAF signaling. Interestingly, the MEK1 V211D mutation was first discovered in a screen of resistance mechanisms to MEK inhibitors in the background of BRAF V600E melanoma cells (12). In the absence of drug, the resultant hyperactivation of ERK signaling may have led to a growth disadvantage in cells. This is also reflected by the low occurrence of the hyperactive RAF-independent MEK1 mutants (11). In clinical samples from this patient, the MEK1 V211D mutant was not detected, even with deep tumor sequencing, prior to targeted therapy treatment and emerged as a resistance alteration to treatment. These data suggest that in this patient, the MEK V211D was likely only present in a rare subclone that was then selected with drug exposure or acquired rapidly after treatment.
The genomic background of mutant BRAF K601E may have impacted the resistance alteration seen in this case. Our group has recently shown different functional properties of allele-specific BRAF alterations and have classified BRAF mutants into three groups (16,17). Class 1 BRAF mutants consist of BRAF V600 alterations, are highly activating, and can signal as monomers independent of RAS. Class 2 BRAF alterations, such as K601E, are activating, but often less so than V600E, and signal as RAS-independent dimers. This case suggests that in tumors with less activating alterations, such as non-V600 BRAF mutations, secondary mutations may develop to amplify ERK signaling and these alterations may attenuate the effect of ERK pathway inhibitors.
Limitations to our study include that only one patient with resistance to MEK inhibitor treatment was studied and biopsy specimens were analyzed so multi-regional samples for each metastatic site were not available. However, consistent with our finding that alterations that amplify BRAF signaling can confer resistance to MEK inhibitors in the clinic, MEK mutations were identified at resistance to MEK inhibitors in two patients with BRAF V600E melanoma (12,18). In the first patient treated with selumetinib, post-progression tissue harbored MEK1 P124L, a RAF-dependent MEK mutant that amplifies ERK signaling from activated BRAF (11). The other patient was treated with trametinib and developed concurrent BRAF amplification and MEK2 Q60P, an alteration analogous to the RAF-regulated MEK1 Q56P mutation (11), at progression. Together these data suggest that MEK alterations that increase ERK pathway activation represent a clinically relevant, recurrent mechanism of resistance to allosteric MEK inhibitors, and these alterations would still be sensitive to the ATP competitive MEK inhibitor (11).
In summary, we report and functionally characterize a mechanism of acquired resistance to MEK inhibitors in the clinic. We find in a colon cancer patient that MEK1 V211D emerged with treatment and caused resistance by amplifying ERK activation and interfering with allosteric inhibitor drug binding. Our data suggest that this resistance to current MEK inhibitors could be overcome by a selective ATP competitive inhibitor by its binding to a different site on MEK protein.
Methods
Clinical specimens
The patient provided written informed consent to treatment in the clinical trial. Progression biopsies and collection of patient samples were conducted under appropriate Institutional Review Board protocols (#06–107, 14–019). DNA from pre-treatment and disease progression specimens were analyzed using MSK-IMPACT (Integrated Mutation Profiling of Actionable Cancer Targets), a targeted exome capture assay with deep sequencing coverage. Target specific-probes for hybrid selection were designed as previously described to capture all protein-coding exons of greater than 300 oncogenes, tumor suppressor genes, and components of pathways deemed actionable by targeted therapies (9). All studies were conducted in accordance with the Declaration of Helsinki.
Patient-derived xenograft models
Patient derived tumor models were generated by mincing about 1 g of tumor tissue, mixing it with matrigel (50%), and implanting subcutaneously into NSG (NOD scid gamma) mice (Institutional Review Board protocols 06–107, 14–091). The PDX generated was sequenced to confirm the genomic alterations present. A cell line was generated from the PDX by growing about 1 g of tumor tissue from the PDX in McCoy’s media.
Single cell sequencing
The cell line generated from the PDX was subjected to single cell sequencing (please see Supplementary methods for full details). A total of 250,000 cells were used for the barcoding run. The droplet workflow for genomic DNA amplification and barcoding was done as previously described (19). Libraries were analyzed on a DNA 1000 assay chip with a Bioanalyzer (Agilent Technologies) and sequenced on a Illumina HiSeq 2500 instrument (Illumina, Inc., San Diego, CA, USA). Sequence data were analyzed using the proprietary software provided by Mission Bio (19).
Cell culture
NIH-3T3 and Phoenix AMPHO cells were purchased from ATCC (American Type Culture Collection) between 2013 and 2015. Cells were grown in Dulbecco’s modified Eagle’s medium with glutamine, antibiotics, and 10% FBS. Cell lines were validated by STR profiling at the Integrated Genomics Operation of MSKCC and screened for Mycoplasma using MycoAlert™ Plus Mycoplasma Detection Kit from Lonza.
NIH-3T3 cells were used to construct stable lines with inducible expression of mutant MEK1s to study MEK1 mutant-driven ERK signaling and their response to different types of MEK inhibitors. Cell lines were used within 3 months of passages post receipt for the above experiments.
Antibodies
Western blot, immunoprecipitation, and in vitro kinase assays were performed as previously described (11). The following antibodies were used: anti-p217/p221-MEK1/2 (p-MEK1/2) (#9154), anti-p202/p204-ERK1/2 (p-ERK1/2) (#4370), anti-MEK1/2 (#4694), anti-ERK1/2 (#4696), anti-p380-p90RSK(p-RSK) (#9341), GAPDH (#2118) from Cell Signaling, anti-V5 (R960–25) from Thermo Fisher Scientific and anti-BRAF (sc-5284) and anti-cyclin D1 (M-20) from Santa Cruz Biotechnology.
Plasmids
The MEK1 gene was sub-cloned into pGEX6P1 (Addgene) for in vitro protein purification. Plasmids TTIGFP-MLUEX and pMSCV-rtTA3-PGK-Hygro for inducible gene expression were provided by Scott Lowe’s laboratory at MSKCC. The MEK1 gene was sub-cloned into TTIGFP-MLUEX vector harboring the Tet-responsive promoter. Mutations were introduced by using the site-directed Mutagenesis Kit (Stratagene).
Compounds
Binimetinib and cobimetinib were obtained from Selleckchem. MAP855((1-((3S,4S)-4-(8-(2-chloro-4-(pyrimidin-2-yloxy)phenyl)-7-fluoro-2-methyl-1H-imidazo[4,5-c]quinolin-1-yl)-3-fluoropiperidin-1-yl)-2-hydroxyethanone)) (11) was obtained from Novartis (compound No. 1, WO2015022662). These drugs were dissolved in DMSO to yield 10 mM stock and stored at −20°C. Cetuximab for in vivo experiments was purchased from the hospital pharmacy.
Inducible gene expression in cells
Retroviruses encoding rtTA or MEK1 genes were packaged in Phoenix-AMPHO cells. The supernatant-containing virus was filtered with 0.45 μM PVDF membrane. The target cells were infected with virus for 8 hours. Forty-eight hours later, cells were selected in medium containing Puromycin (2 μg/ml) or Hygromycin (100 μg/ml) for 3 days. The positive infected cell populations were further sorted using GFP as a marker after overnight exposure to 1μg/ml doxycycline. GFP positive cells were then cultured and expanded in medium with doxycycline along with antibiotics.
Expression and purification of recombinant MEK1
Human wild type MEK1, as well as V211D mutant used in this study, were subcloned into pGEX6P1, expressed as glutathione-S-transferase fusions and purified by Pierce™ Glutathione Agarose (Thermo Fisher Scientific).
In vitro kinase assay
In vitro kinase assays were conducted in the presence of 200 μM ATP, at 30°C for 15 minutes. Briefly, GST-MEK1 or mutants were incubated in the absence or presence of active BRAF (V600E) Protein (Upstate). Changes in MEK1 phosphorylation were estimated by immunoblotting for p-MEK. To test the kinase activity of WT or mutant MEK1 protein, recombinant inactive ERK2 protein (GenWay Biotech) was used as a substrate and the reaction was terminated with the addition of 1X SDS loading buffer and boiling. Kinase activity was estimated by immunoblotting for p-ERK.
In vivo studies
The patient-derived tumor was implanted as subcutaneous xenografts into 6 weeks old NSG mice (Jackson Laboratories), and treatments started when tumors reached 100 mm3 volumes. Mice (5/group) were randomized to each treatment arm and observed daily throughout the treatment period for signs of morbidity/mortality. Body weights were recorded twice weekly. Tumors were measured twice weekly using calipers, and volume was calculated using the formula length × width2 × 0.52. All studies were performed in compliance with institutional guidelines under an Institutional Animal Care and Use Committee (IACUC) approved protocol. Investigators were not blinded when assessing the outcome of in vivo experiments.
Protein structures
The structure of MEK1 WT was generated using I-Tasser (20) (v5.1) with published MEK1 structure PDB:5KKR (21) as a template and stabilized by 10 ns of molecular dynamics simulation using GROMACS (22) (v5.1.4). To generate the mutant structure, UCSF Chimera (v1.12) was used to mutate V211D, before simulating for additional 10 ns to re-stabilize. See Supplementary methods for complete details.
Statistical analysis
Results are mean values ± standard deviations. All cellular experiments were repeated at least three times.
Supplementary Material
Statement of significance.
We report a resistance mechanism to allosteric MEK inhibitors in the clinic. A MEK1 V211D mutation developed in a BRAF K601E colon cancer patient on MEK and EGFR inhibitors. This mutant increases the catalytic activity of MEK1 and reduces its affinity for binimetinib, but remains sensitive to ATP-competitive MEK inhibitors.
Acknowledgments
Supported by the Byrne Fund (R.Y.), the Sarah Jenkins Fund and Breast Cancer Research Foundation (J.S.R-F.), and the National Institutes of Health R01 CA233736 (N.R., R.Y.), U54 OD020355 (E. dS.), R01 CA204749 (B.S.T.), and Cancer Center Core Grant P30 CA 008748. This research is the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Also supported by a Stand Up To Cancer Colorectal Dream Team Translational Research Grant (SU2C-AACR-DT22–17). Stand Up To Cancer is a division of the Entertainment Industry Foundation. Research grants are administered by the American Association for Cancer Research, the Scientific Partner of SU2C.
Footnotes
Disclosure of Potential Conflicts of Interest:
J.S.R-F. is a member of the scientific advisory board (with paid honoraria) of Volition Rx, Paige.AI, Invicro, Roche, Genentech, and Ventana, and a consultant with paid fees of Goldman Sachs Merchant Banking. N.R. is on the scientific advisory board (SAB) of Chugai, BeiGene, Fortress Biotech, Daiichi-Sankyo, AstraZeneca, F-Prime, Zai Lab, Arvinas, and Array BioPharma; and he is a past SAB member of Millennium-Takeda, Kadmon, Kura Oncology, and Araxes. N.R. is also a consultant to Novartis Biomed, Boehringer Ingelheim, Tarveda, Foresite Capital, Array BioPharma, and Revolution Medicines; and in recent years has also consulted with Eli Lilly, Merrimack, Kura Oncology, Araxes, and Kadmon. N.R. owns equity in BeiGene, Zai Lab, Fortress Biotech, Kura Oncology, Araxes, Kadmon, and Effector. N.R. collaborates with Plexxikon; he receives research support from Chugai. R.Y. has received research funding from Array BioPharma, Genentech, GlaxoSmithKline, and Novartis and has served as an advisory board member for GlaxoSmithKline.
References
- 1.Ho AL, Grewal RK, Leboeuf R, Sherman EJ, Pfister DG, Deandreis D, et al. Selumetinib-enhanced radioiodine uptake in advanced thyroid cancer. N Engl J Med 2013;368(7):623–32 doi 10.1056/NEJMoa1209288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dombi E, Baldwin A, Marcus LJ, Fisher MJ, Weiss B, Kim A, et al. Activity of Selumetinib in Neurofibromatosis Type 1-Related Plexiform Neurofibromas. N Engl J Med 2016;375(26):2550–60 doi 10.1056/NEJMoa1605943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerjee A, Jakacki RI, Onar-Thomas A, Wu S, Nicolaides T, Young Poussaint T, et al. A phase I trial of the MEK inhibitor selumetinib (AZD6244) in pediatric patients with recurrent or refractory low-grade glioma: a Pediatric Brain Tumor Consortium (PBTC) study. Neuro Oncol 2017;19(8):1135–44 doi 10.1093/neuonc/now282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diamond EL, Durham BH, Haroche J, Yao Z, Ma J, Parikh SA, et al. Diverse and Targetable Kinase Alterations Drive Histiocytic Neoplasms. Cancer discovery 2016;6(2):154–65 doi 10.1158/2159-8290.CD-15-0913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grisham RN, Sylvester BE, Won H, McDermott G, DeLair D, Ramirez R, et al. Extreme Outlier Analysis Identifies Occult Mitogen-Activated Protein Kinase Pathway Mutations in Patients With Low-Grade Serous Ovarian Cancer. J Clin Oncol 2015;33(34):4099–105 doi 10.1200/JCO.2015.62.4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yao Z, Torres NM, Tao A, Gao Y, Luo L, Li Q, et al. BRAF Mutants Evade ERK-Dependent Feedback by Different Mechanisms that Determine Their Sensitivity to Pharmacologic Inhibition. Cancer Cell 2015;28(3):370–83 doi 10.1016/j.ccell.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corcoran RB, Ebi H, Turke AB, Coffee EM, Nishino M, Cogdill AP, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer discovery 2012;2(3):227–35 doi 10.1158/2159-8290.CD-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Misale S, Arena S, Lamba S, Siravegna G, Lallo A, Hobor S, et al. Blockade of EGFR and MEK intercepts heterogeneous mechanisms of acquired resistance to anti-EGFR therapies in colorectal cancer. Science translational medicine 2014;6(224):224ra26 doi 10.1126/scitranslmed.3007947. [DOI] [PubMed] [Google Scholar]
- 9.Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn 2015;17(3):251–64 doi 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487(7407):330–7 doi 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao Y, Chang MT, McKay D, Na N, Zhou B, Yaeger R, et al. Allele-Specific Mechanisms of Activation of MEK1 Mutants Determine Their Properties. Cancer discovery 2018;8(5):648–61 doi 10.1158/2159-8290.CD-17-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emery CM, Vijayendran KG, Zipser MC, Sawyer AM, Niu L, Kim JJ, et al. MEK1 mutations confer resistance to MEK and B-RAF inhibition. Proc Natl Acad Sci U S A 2009;106(48):20411–6 doi 10.1073/pnas.0905833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phadke MS, Sini P, Smalley KS. The Novel ATP-Competitive MEK/Aurora Kinase Inhibitor BI-847325 Overcomes Acquired BRAF Inhibitor Resistance through Suppression of Mcl-1 and MEK Expression. Mol Cancer Ther 2015;14(6):1354–64 doi 10.1158/1535-7163.MCT-14-0832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corcoran RB, Dias-Santagata D, Bergethon K, Iafrate AJ, Settleman J, Engelman JA. BRAF gene amplification can promote acquired resistance to MEK inhibitors in cancer cells harboring the BRAF V600E mutation. Sci Signal 2010;3(149):ra84 doi 10.1126/scisignal.2001148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Little AS, Balmanno K, Sale MJ, Newman S, Dry JR, Hampson M, et al. Amplification of the driving oncogene, KRAS or BRAF, underpins acquired resistance to MEK1/2 inhibitors in colorectal cancer cells. Sci Signal 2011;4(166):ra17 doi 10.1126/scisignal.2001752. [DOI] [PubMed] [Google Scholar]
- 16.Yaeger R, Corcoran RB. Targeting Alterations in the RAF-MEK Pathway. Cancer discovery 2019;9(3):329–41 doi 10.1158/2159-8290.CD-18-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yao Z, Yaeger R, Rodrik-Outmezguine VS, Tao A, Torres NM, Chang MT, et al. Tumours with class 3 BRAF mutants are sensitive to the inhibition of activated RAS. Nature 2017;548(7666):234–8 doi 10.1038/nature23291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Villanueva J, Infante JR, Krepler C, Reyes-Uribe P, Samanta M, Chen HY, et al. Concurrent MEK2 mutation and BRAF amplification confer resistance to BRAF and MEK inhibitors in melanoma. Cell reports 2013;4(6):1090–9 doi 10.1016/j.celrep.2013.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pellegrino M, Sciambi A, Treusch S, Durruthy-Durruthy R, Gokhale K, Jacob J, et al. High-throughput single-cell DNA sequencing of acute myeloid leukemia tumors with droplet microfluidics. Genome Res 2018;28(9):1345–52 doi 10.1101/gr.232272.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 2008;9:40 doi 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dhawan NS, Scopton AP, Dar AC. Small molecule stabilization of the KSR inactive state antagonizes oncogenic Ras signalling. Nature 2016;537(7618):112–6 doi 10.1038/nature19327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pronk S, Pall S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013;29(7):845–54 doi 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.