Abstract
Background:
There is substantial variation in breast cancer survival rates, even among patients with similar clinical and genomic profiles. New biomarkers are needed to improve risk stratification and inform treatment options. Our aim was to identify novel miRNAs associated with breast cancer survival, and quantify their prognostic value after adjusting for established clinical factors and genomic markers.
Methods:
Using the Women’s Healthy Eating and Living (WHEL) breast cancer cohort with > 15-years of follow-up and archived tumor specimens, we assayed PAM50 mRNAs and 25 miRNAs using the Nanostring nCounter platform.
Results:
We obtained high quality reads on 1253 samples (75% of available specimens), and used an existing research-use algorithm to ascertain PAM50 subtypes and risk scores (ROR-PT). We identified miRNAs significantly associated with breast cancer outcomes, and then tested these in independent TCGA samples. MiRNAs that were also prognostic in TCGA samples were further evaluated in multiple regression Cox models. We also used penalized regression for unbiased discovery.
Conclusions:
Two miRNAs, 210 and 29c, were associated with breast cancer outcomes in the WHEL and TCGA studies, and further improved risk stratification within PAM50 risk groups: 10-year survival was 62% in the node-negative high miRNA210-high ROR-PT group versus 75% in the low miRNA 210 -high ROR-PT group. Similar results were obtained for miRNA 29c. We identified three additional miRNAs, 187–3p, 143–3p, and 205–5p via penalized regression.
Impact:
Our findings suggest that miRNAs might be prognostic for long-term breast cancer survival, and might improve risk stratification. Further research to incorporate miRNAs into existing clinico-genomic signatures is needed.
Keywords: breast cancer survival, PAM50, miRNAs, prognostic models
INTRODUCTION
MicroRNAs (miRNA) are small non-coding RNAs that post-transcriptionally regulate the expression of their target genes (1). MiRNAs are involved in cell differentiation, regulation, and apoptosis, and miRNA dysregulation impacts development and progression of many cancers, including breast cancer (2–9). MiRNAs can have tumor suppressing or tumor promoting roles, and these functions may differ depending on tumor histopathology (8, 9).
In recent years, the potential of dozens of miRNAs for diagnosis, treatment response prediction, and prognosis of breast cancer have been studied (5, 8, 10–14); however, few have been examined in cohorts with long term follow-up. Currently, there are several well-established clinical prognostic factors for breast cancer survival, such as tumor size, nodal status, grade and histopathology (15), and a few genomic signatures e.g., Oncotype DX, Mammaprint (16–19), ProSigna (20, 21), and the research-based PAM50 signature (22–24). However, even with the existing clinico-genomic tools, an estimated 25–35% of breast cancer patients are classified as intermediate risk (20, 21, 25–27), and for these patients, the long-term risk of relapse remains uncertain. Thus, identifying novel biomarkers such as miRNAs that independently predict breast cancer outcomes could lead to better risk stratification and possibly improve treatment management strategies, potentially sparing aggressive chemotherapy for some, while offering targeted therapies for others.
In this study, we used >1200 archived tumor samples from the Women’s Healthy Eating and Living (WHEL) breast cancer cohort with 15+ years of follow-up (28), to investigate 25 miRNA predictors of long-term disease-free survival and breast cancer mortality. We developed models which included standard clinical factors and a research-use published PAM50 signature (29), as well as new miRNA features. We adopted two statistically rigorous complementary paradigms. First, we used an external validation approach in which we tested the prognostic value of the 25 miRNAs using the publicly available TCGA database (30); miRNAs that were prognostic in the WHEL samples and independently prognostic in TCGA database were considered hits, and evaluated for prognostic value over and above clinical factors and PAM50 subtypes. Second, we implemented an unbiased approach via penalized regression to select the most prognostic features from among clinical factors, PAM50 subtypes, and 25 individual mRNAs. In summary, the first approach is similar to the usual model building methods to evaluate the incremental effects of new candidate predictors, after adjusting for known prognosticators. However, we imposed stringent criteria for inclusion in multivariate models, namely requiring independent validation via TCGA. The second penalized regression method, by including all variables simultaneously rather than incrementally, has the potential to discover new features but avoids overfitting via a penalty term.
MATERIALS and METHODS
Study Sample
The Women’s Healthy Eating and Living (WHEL) Study was a randomized-controlled dietary secondary prevention trial of 3088 early stage breast cancer survivors (28, 31). Women were recruited between 1995 and 2000, and were eligible provided they were within four years of diagnosis of their primary operable invasive Stage I (≥ 1 cm), Stage II or Stage III breast carcinoma (28), were aged 18 to 70 years at diagnosis, and had completed primary treatment for breast cancer. Details on inclusion and exclusion criteria are provided in Pierce et al. (2002) (28).
An archival, formalin-fixed, paraffin-embedded (FFPE) block of breast tumor tissue was solicited from local hospitals for each WHEL participant. Blocks were sent to the WHEL Study Coordinating Center, where the Histology Core Lab at the UC San Diego Cancer Center cut ten unstained slides (5μm each) and one hematoxylin and eosin (H and E)-stained slide. Each unstained slide was dipped in paraffin for preservation before storage. The study pathologist reviewed each pathology report with the accompanying H and E slide to confirm that invasive tumor was present on the slide that corresponded to the tumor described in the participant’s pathology report. On occasion, the block sent to the coordinating center did not contain invasive tumor. In such cases, the unsuitable block was returned, the pathology report was reviewed and another block was requested. FFPE tissue from the primary tumor were available for 56% (n = 1723) of the WHEL cohort. The final analysis for this investigation was based on 1253 participants. As the dietary intervention produced no treatment effect (31), we treated the study population as a single cohort. We obtained IRB approval from participating institutions, and written informed consent from all participants, that included consent for genomic analysis.
Study Endpoints
In the current study, we evaluated two primary outcomes (i) a breast cancer event (locoregional recurrence, metastasis, or contralateral), and (ii) death from breast cancer. We also considered overall survival, i.e., death from any cause, as a secondary endpoint. Events were independently adjudicated by two breast oncologists. Carcinoma in situ was not counted as a breast cancer event. The WHEL study ceased active surveillance for cancer events in 2010, since then, deaths were ascertained via annual searches of the National Death Index. Time from diagnosis to a second breast cancer event defined the disease-free survival outcome; time from diagnosis to breast cancer death defined the breast-cancer-survival outcome. Time-to-event was censored at death (from non-breast-cancer causes), last contact, or end-of-follow up (2010 for breast cancer events, 2015 for death).
Nucleic Acid Extraction
The H and E stained slide was used for histopathological review and to guide tumor macrodissection of sections from four unstained slides. The unstained slides from samples with ≥ 40% tumor cellularity were incubated at 65 °C for 30 minutes and deparaffinized using Citrisolv (Fisher Scientific, Pittsburgh, PA) followed by ethanol wash. Tumor tissues were macrodissected from the slides into RNAse-free microfuge tubes, and nucleic acids isolated using the Qiagen AllPrep FFPE kit (#80234). Manufacturer’s instructions were followed with the exception that the proteinase K digestion step was extended to an overnight incubation for the DNA isolation. Total RNA and DNA were quantified using the Invitrogen Qubit and corresponding quantification kits. Total RNA was used for the miRGE assay (see below). DNA pellets were stored at −80° C for future use.
mRNA and miRNA quantification
Transcript expression was quantified with 250 ng of total RNA using the NanoString nCounter analysis system with a custom miRGE CodeSet which included probes for 55 PAM50 genes (including 5 housekeepers) and 25 miRNA targets (Table S1). The choice of miRNA targets was based on a review of the literature for promising targets at the time of initiation of the WHEL-genomic substudy. In particular, 25 miRNAs (Table S1) were identified as prognostic for breast cancer in studies with moderate-to-large sample-sizes from published reports (1, 2, 12, 32–34). Probe selection (miRNA) for the custom miRGE Codeset was determined through screening a set of well-characterized breast cancer tumor samples using the Nanostring Human miRNA assay V2.1 panel that included 800 targets. Positive and negative controls were also included, and assay reactions were assembled per manufacturer’s specifications (NanoString Technologies, INC Seattle, WA).
Statistical approach
Normalization and data processing
MiRNA expression values were normalized using geometric means of the two miRNA positive controls to eliminate assay technical variation, and then subtracting the maximum value of eight mRNA negative controls to eliminate background effect. Expression values were log2 transformed to reduce skewness and quantile normalized to reduce batch effects. As sensitivity analysis, normalization was also conducted using three different sets of housekeepers: the five mRNA housekeepers used to normalize the mRNA values, five most highly expressed miRNAs, and also three putative housekeeper miRNAs with very high expression values and low standard deviations; results were consistent across normalization methods (35, 36).
Expression of the 50 PAM50 mRNAs were normalized to negative and positive controls, and standardized to five housekeepers, per standard practice (29). The published PAM50 algorithm (29) was used to classify each subject into an intrinsic subtype: Luminal A, Luminal B, basal-like, Her2-enriched, normal-like. Prior to implementing this algorithm, mRNA values were platform-adjusted (37). Risk-of-recurrence (ROR-PT) scores were calculated (29) and categorized into low, medium and high risk strata.
Individual miRNA and breast cancer outcomes
Associations between individual miRNAs and breast cancer outcomes were investigated in Cox models with and without adjustment for tumor characteristics (i.e., tumor stage, tumor grade, and age at diagnosis). Cox models with delayed entry were used to account for varying times from cancer diagnosis to study entry. Hazard ratios and 95% confidence interval were computed, and correspond to the increase in (log)-hazard per 1-unit change in log2 of the miRNA value or equivalently the change in (log)-hazard for doubling of the miRNA level. Proportional hazards assumptions were assessed using scaled Schoenfeld residuals. We also used Bonferroni correction for multiple comparisons when evaluating 25 miRNAs, and present the corrected and uncorrected results.
External validation of prognostic miRNA
To assess reproducibility of WHEL miRNAs findings, we downloaded TCGA miRNA data for patients with primary breast cancer through bioconductor TCGA Biolinks (30). Similar to WHEL samples, TCGA miRNAs were log2 transformed and normalized; delayed entry Cox models were used to assess the association between miRNAs and disease- free survival, time to breast cancer related death or time to overall survival within the TCGA samples.
Additional prognostic value of TCGA-validated miRNAs in clinico-genomic models
After adjusting for standard clinical features (i.e., tumor stage, tumor grade, and age at diagnosis), we identified miRNAs that were prognostic in both the WHEL and TCGA analysis. These miRNAs were then added to models that included PAM50 subtypes and standard clinical features in the WHEL cohort. To assess potential clinical utility, Kaplan-Meier curves and score tests were used to examine if these miRNAs further delineated PAM50 ROR-PT risk categories, using the median miRNA values to further divide the ROR-PT risk strata. Thus, rather than adjustment for multiple comparisons and the attendant loss in power, we implemented a rigorous external validation framework for testing predictive value of the miRNAs over and above PAM50 subtypes.
Unbiased selection and internal validation
To examine all the prognostic factors on an equal footing and implement unbiased variable selection, we used penalized regression. We included all variables: clinical factors, PAM50 subtypes, and 25 individual miRNAs in the initial model and used a lasso method for variable selection within the Cox regression (38). The tuning parameter λ was chosen by 10-fold cross-validation to minimize model deviance.
In summary, given the moderately large set of miRNAs, we used two complementary methodological approaches for selecting miRNAs for further multivariate analysis. The first method used external validation, and only those miRNAs that were also prognostic in TCGA samples, were evaluated for their incremental prognostic value, after adjusting for clinical features and PAM50 subtypes. The second approach used penalized regression (lasso) with all variables included in the model and used cross-validation for feature selection, thus implementing stringent variable selection within the WHEL cohort. Furthermore, the penalized approach examines all predictors simultaneously and could potentially discover new features.
RESULTS
FFPE Samples
Of the 1723 FFPE samples, 25% had low tumor cellularity or low RNA content prohibiting further analysis. Gene expression was obtained on 1291 samples; of these, 38 were eliminated due to outliers or poor quality reads, resulting in a final sample of N = 1253 for statistical analysis.
Clinical and Demographic characteristics
The distribution of demographic and clinical characteristics in the WHEL genomic substudy were similar to those in the parent study (N = 3088) (31). Women in the substudy were median 50 years at cancer diagnosis, 85% were White, 36% had Stage I and 46% Stage II tumors, three-quarters had ER+ histopathology, and 16% had triple negative histopathology (Table 1). There were 303 breast cancer events (locoregional recurrence, metastasis, or contralateral breast cancer), 219 deaths due to breast cancer, and 316 total deaths
Table 1.
Demographic and clinical characteristics at study entry of the WHEL-genomics breast cancer substudy (N = 1253)
| Age at breast cancer diagnosis | |
| Median (range) | 50 (27–70) |
| Race/Ethnicity N (%) | |
| White | 1060 (84.6%) |
| Black | 45 (3.6%) |
| Hispanic | 85 (6.8%) |
| Asian | 31 (2.5%) |
| Other | 32 (2.6%) |
| Stage N (%) | |
| I | 453 (36.2%) |
| IIA | 432 (34.5%) |
| IIB | 144 (11.5%) |
| IIIA | 166 (13.2%) |
| IIIC | 58 (4.6%) |
| Nodal status N (%) | |
| Negative | 702 (56%) |
| Positive | 551 (44%) |
| Tumor size (cm) | |
| Mean (SD) | 2.3 (1.44) |
| Grade N (%) | |
| Poorly differentiated | 497 (39.7%) |
| Moderately differentiated | 496 (39.6%) |
| Well differentiated | 159 (12.7%) |
| Unspecified | 101 (8.1%) |
| Histopathology N (%) | |
| ER+ | 909 (73.7%) |
| PR+ | 809 (66.4%) |
| Her2+ | 217 (17.3%) |
| Triple negative | 199 (15.9%) |
| Years diagnosis to study entry | |
| Median (25th, 75th%-iles) | 1.8 (1.03, 2.8) |
| Chemotherapy and Anti-estrogen therapy N (%) | |
| Yes, Yes | 590 (47.1%) |
| Yes, No | 314 (25.1%) |
| No, Yes | 258 (20.6%) |
| No, No | 76 (6.1%) |
| Yes, Unknown | 5 (0.4%) |
| No, Unknown | 9 (0.7%) |
| Outcomes | |
| Breast cancer events (N) | 303 |
|
Disease-free survival (yrs) Median (25th, 75th)%-iles |
9.5 (6.7, 11.3) |
| Breast cancer deaths (N) | 219 |
|
Breast-cancer survival (yrs) Median (25th, 75th)%-iles |
16.8 (15.3, 18.2) |
Prognostic value of individual miRNAs for disease-free survival, breast cancer survival and overall survival
We examined associations between each of the 25 miRNAs and outcomes, after adjusting for stage, grade, and age at diagnosis (Table 2). The statistically significant results with HR (95% CI) were as follows: three miRNAs were significantly associated with disease-free survival, namely, miRNA 27b-3p: 1.13 (1.01, 1.27), miRNA 210–3p: 1.1 (1.01, 1.2), and miRNA 143–3p: 1.23 (1.08, 1.41); six miRNAs were associated with breast cancer free survival, namely, miRNA 150–5p: 0.91 (0.84, 0.99), miRNA 16–5p: 1.18 (1.02, 1.37), miRNA 205–5p: 0.93 (0.88, 0.99), miRNA 29c-3p: 1.2 (1.05, 1.37), miRNA 27b-3p: 1.2 (1.04, 1.38), miRNA 143–3p: 1.28 (1.1, 1.51); and four miRNAs were associated with overall survival, namely, miRNA 150–5p: 0.93 (0.87, 0.99), miRNA 29c-3p: 1.19 (1.07, 1.33), miRNA 187–3p: 1.09 (1.01, 1.17), miRNA 143–3p: 1.14 (1, 1.3)). Of these, the associations of miRNA 143–3p with disease-free and breast cancer survival, and miRNA 29c-3p with overall survival were still significant after Bonferroni correction for 25 miRNA tests. Unadjusted associations between each of the 25 miRNAs and each breast cancer outcome were similar to the adjusted analysis and are displayed in Figure S1.
Table 2.
Associations between individual miRNA and breast cancer outcomes+ in the WHEL-genomics breast cancer study (N = 1253)
| Disease-free survival (# of events 303) | Breast cancer survival (# of events 219) | Overall survival (# of events 316) | ||||
|---|---|---|---|---|---|---|
| miRNA | HR (95% CI) | P Valuea | HR (95% CI) | P Valuea | HR (95% CI) | P Valuea |
| 93.5p | 1.07 (0.96, 1.19) | 0.202 | 1.1 (0.96, 1.25) | 0.169 | 1.04 (0.95, 1.15) | 0.381 |
| 150.5p | 0.94 (0.87, 1.01) | 0.072 | 0.91 (0.84, 0.99) | 0.025 | 0.93 (0.87, 0.99) | 0.033 |
| 128.3p | 1 (0.94, 1.08) | 0.903 | 1.03 (0.94, 1.13) | 0.531 | 1.01 (0.94, 1.08) | 0.850 |
| 141.3p | 1.07 (0.97, 1.19) | 0.160 | 1.11 (0.99, 1.26) | 0.079 | 1.08 (0.98, 1.19) | 0.127 |
| 21.5p | 1.04 (0.93, 1.17) | 0.486 | 1.14 (0.98, 1.31) | 0.083 | 1.06 (0.94, 1.18) | 0.336 |
| 1246 | 1.12 (0.98, 1.28) | 0.101 | 1.03 (0.88, 1.2) | 0.722 | 1.06 (0.93, 1.22) | 0.353 |
| 16.5p | 1.11 (0.99, 1.25) | 0.082 | 1.18 (1.02, 1.37) | 0.028 | 1.12 (1, 1.26) | 0.057 |
| 205.5p | 0.96 (0.91, 1.02) | 0.173 | 0.93 (0.88, 0.99) | 0.021 | 0.95 (0.9, 1) | 0.056 |
| 342.3p | 0.98 (0.89, 1.07) | 0.649 | 1 (0.89, 1.12) | 0.949 | 1.01 (0.92, 1.1) | 0.862 |
| 29c.3p | 1.09 (0.97, 1.21) | 0.139 | 1.2 (1.05, 1.37) | 0.009 | 1.19 (1.07, 1.33) | 0.002b |
| 27b.3p | 1.13 (1.01, 1.27) | 0.037 | 1.2 (1.04, 1.38) | 0.012 | 1.05 (0.95, 1.17) | 0.326 |
| 187.3p | 1.03 (0.96, 1.11) | 0.348 | 1.08 (0.99, 1.17) | 0.085 | 1.09 (1.01, 1.17) | 0.021 |
| 26b.5p | 1.03 (0.89, 1.2) | 0.695 | 1.08 (0.9, 1.3) | 0.393 | 1.05 (0.91, 1.22) | 0.508 |
| 15a.5p | 1.06 (0.92, 1.22) | 0.449 | 1.08 (0.9, 1.29) | 0.423 | 1.08 (0.93, 1.24) | 0.301 |
| 221.3p | 1.09 (1, 1.19) | 0.062 | 1.1 (0.99, 1.23) | 0.071 | 1.01 (0.93, 1.09) | 0.809 |
| 494.3p | 0.95 (0.84, 1.09) | 0.497 | 0.93 (0.8, 1.09) | 0.390 | 0.99 (0.87, 1.12) | 0.845 |
| 30c.5p | 1.05 (0.95, 1.16) | 0.374 | 1.01 (0.9, 1.14) | 0.868 | 0.99 (0.9, 1.08) | 0.743 |
| 769.5p | 0.97 (0.88, 1.06) | 0.489 | 1.01 (0.9, 1.13) | 0.873 | 1.01 (0.92, 1.11) | 0.830 |
| 210.3p | 1.1 (1.01, 1.2) | 0.027 | 1.06 (0.96, 1.18) | 0.226 | 1.04 (0.96, 1.13) | 0.375 |
| 10b.5p | 1.07 (0.98, 1.16) | 0.138 | 1.1 (0.99, 1.23) | 0.065 | 1.05 (0.96, 1.14) | 0.286 |
| 143.3p | 1.23 (1.08, 1.41) | 0.002b | 1.28 (1.1, 1.51) | 0.002b | 1.14 (1, 1.3) | 0.046 |
| 7.5p | 1.03 (0.96, 1.11) | 0.392 | 0.99 (0.91, 1.08) | 0.772 | 1.02 (0.96, 1.1) | 0.490 |
| 200b.3p | 1.04 (0.94, 1.15) | 0.426 | 1.08 (0.95, 1.22) | 0.250 | 1.08 (0.97, 1.2) | 0.142 |
| 135a.5p | 0.98 (0.94, 1.02) | 0.315 | 0.99 (0.94, 1.04) | 0.595 | 0.98 (0.94, 1.03) | 0.480 |
| 126.3p | 1.06 (0.95, 1.2) | 0.307 | 1.11 (0.96, 1.3) | 0.166 | 1.01 (0.9, 1.12) | 0.922 |
adjusted for tumor stage, tumor grade, age at diagnosis
p-values are not adjusted for multiple comparisons
statistically significant after Bonferroni adjustment for 25 miRNAs
External validation with TCGA
We next evaluated the prognostic value of each of the 25 miRNAs for each breast cancer outcome using TCGA database (30). To ensure comparability with WHEL samples, we only included early stage (I, II, III) TCGA breast cancer samples, resulting in a total TCGA sample-size of 1034. The median age at diagnosis of TCGA participants was 58 (range: 26–90) years; 76.4% had stages I and II, 73.8% were ER+, 64% were PR+, 14.9% were Her2+ and 10.4% were triple negative. These characteristics were comparable to the WHEL genomics substudy (Table 1). In TCGA samples, there were 171 breast cancer recurrences, and 123 deaths with 63 known breast cancer specific deaths.
In TCGA analysis, after adjustment for age and stage, four miRNAs, namely 1246, 135a.2, 210, and 29c, were significantly associated with the three survival outcomes, and miRNA 342 was associated with breast-cancer- and overall-mortality at the 5% significance level (Table S2). Of these, miRNA 210 and miRNA 29c were also significantly prognostic in the WHEL cohort (Table 2). Complete results for all miRNAs are presented in Table S2 and Figure S1.
Added Prognostic value of TCGA-validated miRNAs in the WHEL Study
Using the WHEL cohort, we next examined TCGA-validated miRNAs, 210 and 29c, in multiple regression Cox models, after adjusting for clinical factors, and PAM50 subtypes; separate multiple regression models were fit for miRNA 210 and miRNA 29c (Table 3). As expected tumor stage was strongly associated with survival outcomes with hazard ratios increasing from 1.26 to 6.26 as stage increased. After adjusting for stage, grade, and age at diagnosis, the luminal B subtype remained a significant prognostic factor with hazard ratio > 1.5 in all models. The hazard ratio (95% CI) for miRNA 210 was 1.09 ((1, 1.2), p = 0.05) for disease-free survival, 1.07 ((0.96, 1.19), p=0.22) for breast cancer survival, and 1.04 ((0.95, 1.13), p=0.4) for overall survival after adjusting for clinical variables and PAM50 subtypes. The hazard ratio (95% CI) for miRNA 29c was 1.08 ((0.95, 1.22), p = 0.22) for disease-free survival, 1.17 ((1.01, 1.37), p=0.04) for breast cancer survival and 1.16 ((1.02, 1.31), p=0.02) for overall survival after adjusting for clinical variables and PAM50 subtypes. Thus even if not consistently significant at the 5% level, the hazard ratios for these miRNAs were similar to their hazard ratios in the models that did not include PAM50 subtypes (Table 2), suggesting that these miRNAs are likely independent predictors of breast cancer outcomes.
Table 3:
Multiple regression analysis of clinical factors, PAM50 subtype, each TCGA-validated miRNA and breast cancer outcomes in the WHEL-genomics breast cancer substudy
| Predictors | Disease free survival (N=295 relapse events)* |
Breast cancer survival (N=212 breast cancer deaths)* |
Overall survival (N=306 deaths)* |
|---|---|---|---|
| HR (95% CI) | HR (95% CI) | HR (95% CI) | |
| Model for miRNA-210 | |||
| I (Ref) IIA IIB IIIA IIIC |
1.0 1.72 (1.24, 2.38) 2.47 (1.67, 3.65) 3.52 (2.46, 5.02) 4.00 (2.52, 6.36) |
1.0 1.90 (1.25, 2.89) 3.09 (1.92, 4.97) 4.52 (2.92, 7) 6.26 (3.7, 10.6) |
1.0 1.26 (0.93, 1.71) 1.83 (1.26, 2.65) 2.36 (1.67, 3.33) 3.75 (2.44, 5.77) |
| Luminal A (Ref) Basal Her2 Luminal B |
1.0 1.14 (0.79, 1.65) 0.91 (0.60, 1.39) 1.55 (1.16, 2.08) |
1.0 0.94 (0.6, 1.47) 0.86 (0.52, 1.42) 1.64 (1.17, 2.31) |
1.0 0.9 (0.61, 1.32) 0.97 (0.64, 1.45) 1.58 (1.2, 2.09) |
| MiRNA 210 | 1.09 (1, 1.2) | 1.07 (0.96, 1.19) | 1.04 (0.95, 1.13) |
| Model for miRNA-29c | |||
| I (Ref) IIA IIB IIIA IIIC |
1.0 1.73 (1.25, 2.4) 2.47 (1.67, 3.65) 3.37 (2.35, 4.82) 3.95 (2.48, 6.3) |
1.0 1.87 (1.23, 2.85) 3.05 (1.89, 4.92) 4.27 (2.75, 6.62) 6.05 (3.57, 10.26) |
1.0 1.24 (0.92, 1.68) 1.8 (1.24, 2.61) 2.25 (1.59, 3.17) 3.61 (2.35, 5.56) |
| Luminal A (Ref) Basal Her2 Luminal B |
1.0 1.36 (0.92, 2.0) 1.01 (0.66, 1.54) 1.58 (1.18, 2.12) |
1.0 1.23 (0.77, 1.97) 0.99 (0.6, 1.62) 1.66 (1.18, 2.32) |
1.0 1.13 (0.75, 1.7) 1.08 (0.72, 1.63) 1.58 (1.2, 2.09) |
| MiRNA 29c | 1.08 (0.95, 1.22) | 1.17 (1.01, 1.37) | 1.16 (1.02, 1.31) |
Models also adjusted for age at diagnosis, grade; Patients with a Normal PAM50 subtype were excluded.
ROR-PT categories, externally validated miRNA, and survival
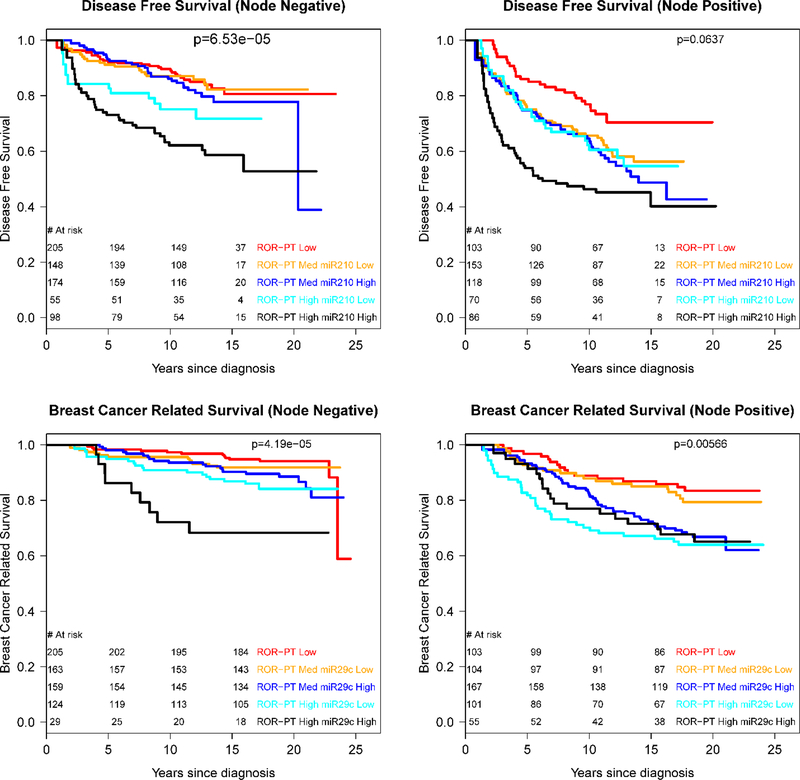
To assess potential clinical utility, we tested if TCGA-validated miRNAs could further discriminate PAM50 ROR-PT risk categories. For this analysis, we dichotomized each miRNA at the median value, and used Kaplan Meier curves and score statistics to compare survival rates in the ROR-PT by miRNA categories. Based on the model results (Table 3), we evaluated miRNA 210 for disease-free survival, and miRNA 29c for breast cancer survival. Figure 1 gives the Kaplan-Meier curves for ROR-PT*miRNA categories stratified by nodal status. In the node-negative stratum (Figure 1 top), miRNA 210*ROR-PT groups had significantly different survival rates (p < 0.001): women with high miRNA 210 levels and high ROR-PT scores had notably worse disease-free survival (10-year rate 62%) compared to those with low miRNA 210 levels and high ROR-PT scores (10-year survival 75%). Similar results were observed for the node-positive group (Fig 1b), although survival differences across ROR-PT*miRNA 210 strata were marginally significant (p = 0.06). Interestingly, in the node-positive group, the ROR-PT-high + miRNA 210-low subgroup had similar survival to the ROR-PT medium risk group. Thus, miRNA 210 expression delineated risk for the ROR-PT high risk category, identifying a subgroup with very poor prognosis.
Figure 1:
Kaplan-Meier survival plots of miRNA*ROR-PT categories by nodal status: miRNA 210 (top), miRNA 29c (bottom)
ROR-PT*miRNA 29c categories were significantly associated with breast cancer survival for node-negative (p < 0.001) and node-positive strata (p = 0.006). For the node-negative high ROR-PT group (Figure 1 bottom), low levels of miRNA 29c were associated with higher survival rates compared to those with high miRNA 29c levels (10-year survival rate 91% versus 72%). Interestingly, for the node-positive ROR-PT medium risk group, those with low miRNA29c levels had similar survival rates to the node-positive low-ROR-PT risk group, suggesting that miRNA 29c was able to identify a low-risk phenotype within the ROR-PT medium risk category.
Unbiased selection with internal validation Results
Variable selection models were built via lasso penalized regression to predict disease-free survival, breast cancer related survival and overall survival. Candidate predictors included age at diagnosis, tumor stage, tumor grade, PAM50 subtypes and 25 miRNAs. Tumor stage and PAM50 subtypes were consistently selected across the outcomes. No miRNAs were selected for disease free survival. For breast cancer survival the selected miRNAs with HR (95% CI) after adjusting for stage, grade, and PAM50 subtype were: miRNA 143 1.31 (1.12, 1.54), p < 0.001, and miRNA 205 0.95 (0.89, 1.01), p=0.09. For overall survival, miRNA 29c 1.16 (1.02, 1.32) p=0.02, and miRNA 187 1.09 (1.01, 1.17) p=0.02 were selected. We note that the 95% CIs do not account for the variable selection and should be interpreted with caution. The estimated HRs, however, do accurately reflect the predictive ability of the miRNA for breast cancer outcomes.
DISCUSSION
Despite major advances in clinico-genomic risk classification tools, there is still substantial variation in breast cancer relapse and survival rates in subgroups with similar risk profiles. New biomarkers are needed to improve risk stratification and inform treatment options, especially for women who are currently classified as having an intermediate-to-high risk of relapse. In this work, we identified two miRNAs, miRNA 210 and miRNA 29c, that were associated with breast cancer outcomes, after adjusting for tumor characteristics and the PAM50 molecular subtypes, in a large breast cancer cohort with > 15-years of follow-up.
MiRNA 210–3p has been extensively studied and is involved in breast cancer cell migration, proliferation and invasion (39). Concordant with our findings, miRNA 210 over-expression was associated with worse prognosis in multiple studies and systematic reviews (4, 5, 40–42) and likely characterizes an aggressive phenotype. The miRNA 29 family is reported to exhibit both tumor suppressing and promoting roles in breast cancer (43–46). In our study, higher levels of miRNA 29c were associated with worse prognosis, in contrast to published findings in triple negative breast cancer, where higher miRNA 29c levels were associated with better prognosis (47–50). However, it is notable that < 16% of our sample had triple negative tumors which could explain discrepancies. Importantly, we found that both miRNA 210 and miRNA 29c were able to improve risk stratification within ROR-PT categories, with miRNA 210 identifying a very high risk subgroup within the ROR-PT high risk stratum, and miRNA 29c delineating a low risk group in the ROR-PT medium risk category. These findings could inform treatment guidelines and research, e.g., developing new and targeted therapies for women with high miRNA 210-high ROR-PT disease, while possibly sparing aggressive therapies for women with low miRNA 29c-medium ROR-PT disease.
We also implemented an unbiased approach to discover novel markers. To maintain rigor and reduce overfitting, we used internally cross-validated, penalized regression in which all clinical variables, miRNAs, and PAM50 subtypes were included as predictors. This analysis identified a robust, parsimonious prognostic set that included three additional miRNAs: miRNA 187–3p, miRNA 143–3p, and miRNA 205–5p. Higher levels of miRNAs 187–3p and 143–3p, and lower levels of 205–5p were associated with shorter survival times. MiRNA 205, an oncosuppressor, was previously reported to reduce invasion and was associated with better prognosis (5, 10). This is consistent with the marginally significant protective effect (HR=0.94) for breast cancer survival observed in our study. Also, similar to our results, miRNA 187 has been associated with breast cancer progression and worse survival (5, 51). Reports on the role of miRNA 143 in breast cancer are mixed (52), with several laboratory studies indicating a tumor suppressive effect (53–57), while others suggest tumorigenic effects (58). To our knowledge, our finding that miRNA 143 is associated with worse prognosis in long-term breast cancer survival after adjusting for clinical factors and PAM50 subtypes is novel and has not been reported previously.
We note that miRNA-outcome associations were not consistently statistically significant across the three outcomes in our WHEL analysis: e.g., miRNA 150 was significantly associated with breast-cancer and overall mortality, but not disease-free survival, whereas miRNA 27b was significantly associated with disease-free and breast-cancer survival but not overall survival. While determining reasons for these discrepancies is beyond our scope, we note that the estimated hazard ratios for all three outcomes were in the same direction, with similar effect-sizes in most cases. Also, there were discrepancies between the lists of prognostic miRNAs in the WHEL and TCGA samples. We conjecture that these could be due to different cohort characteristics (although we tried to match on key variables), and assay methodologies. More importantly, by focusing only on features that were prognostic in both cohorts, we required a higher level of replication, which should reduce spurious cohort-specific findings. On the other hand, the penalized regression approach allowed us to discover new features in the WHEL cohort, which would need to be validated in the future. Given that there are not many studies with long-term follow-up that have evaluated miRNAs, we believe that both approaches provide useful and important information and add to the body of literature in this emerging field.
Our study has many strengths. The study sample comprised a large well-characterized clinical cohort with over 15 years follow-up. We obtained high-quality assays using the validated Nanostring platform. In addition, we used rigorous statistical approaches for identifying miRNA hits, model development and comparison. First, rather than using multiple testing correction, we used stringent external validation, and only considered miRNAs significant if they were also prognostic in the TCGA dataset. This should enhance reproducibility of our results, and reduce the chance of spurious findings. Second, we used cross-validated penalized regression methods for unbiased variable selection in our statistical models, which allows discovery of new features while at the same time reducing overfitting.
There are also limitations in our study. Our study cohort was diagnosed with breast cancer between 1991 and 2000, and did not receive current standard of care: women with Her2+ tumors did not receive adjuvant trastuzumab, and few postmenopausal women received adjuvant aromatase inhibitors. Additionally, the average interval from diagnosis to entry into the WHEL Study was two years suggesting that women susceptible to early relapse, e.g., those with basal tumors, may have been under-represented in the WHEL sample. Selection against the basal subtype, which comprise primarily triple negative breast cancer, could partially explain why we did not observe a protective effect for miRNA 29c, a finding reported primarily for triple negative breast cancer (47).
In summary, using a large breast cancer cohort with > 15 years of follow-up, we identified five miRNAs that might be prognostic for breast cancer survival. In addition, our results suggest that miRNAs might identify high (or low) risk groups within PAM50-clinical risk score categories. If replicated in future studies, adding these miRNA targets to existing prognostic tools could lead to improved risk stratification, and ultimately, to better informed treatment decisions and clinical management for breast cancer patients.
Supplementary Material
Acknowledgments
Supported by National Cancer Institute, National Institutes of Health, Awards No. R01CA166293 (KM, LN, MP, JP, BP, SF, ERM, ME, SD, TV), P30CA023100 (KM, LN, MP, EP) and F32CA220859 (CRM).
Footnotes
Publisher's Disclaimer: Disclaimers: SD has stock or other ownership in NanoString Techologies. BAP has a consulting or advisory Role with Bioalta; Research Funding from Genetech, Glaxo Smith Kline, and Novartis; and Patents, Royalties, Other Intellectual Property with Salk Institute Licensed Technology. MJE has a leadership role, Employment of Immediate Family Member, and stock or other ownership, consulting or advisory role, and Honoraria with Bioclassifier, Prosigna, and NanoString Technologies. ERM has a leadership role and stock with Qiagen N.V., and a consulting or advisory role with Regeneron. The remaining authors declare no conflict of interest.
REFERENCES
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116(2):281–97. [DOI] [PubMed] [Google Scholar]
- 2.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res 2005;65(16):7065–70. [DOI] [PubMed] [Google Scholar]
- 3.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med 2017;9(6):852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Volinia S, Croce CM. Prognostic microRNA/mRNA signature from the integrated analysis of patients with invasive breast cancer. Proc Natl Acad Sci U S A 2013;110(18):7413–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Schooneveld E, Wildiers H, Vergote I, Vermeulen PB, Dirix LY, Van Laere SJ. Dysregulation of microRNAs in breast cancer and their potential role as prognostic and predictive biomarkers in patient management. Breast Cancer Res 2015;17:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertoli G, Cava C, Castiglioni I. MicroRNAs: New Biomarkers for Diagnosis, Prognosis, Therapy Prediction and Therapeutic Tools for Breast Cancer. Theranostics 2015; 5(10):1122–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang JT, Wang F, Chapin W, Huang RS. Identification of MicroRNAs as Breast Cancer Prognosis Markers through the Cancer Genome Atlas. Plos One 2016;11(12):e0168284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Bryan S, Dong S, Mathis JM, Alahari SK. The roles of oncogenic miRNAs and their therapeutic importance in breast cancer. Eur J Cancer 2017;72:1–11. [DOI] [PubMed] [Google Scholar]
- 9.Dvinge H, Git A, Graf S, Salmon-Divon M, Curtis C, Sottoriva A, et al. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature 2013; 497(7449):378–82. [DOI] [PubMed] [Google Scholar]
- 10.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med 2012;4(3):143–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buffa FM, Camps C, Winchester L, Snell CE, Gee HE, Sheldon H, et al. microRNA-Associated Progression Pathways and Potential Therapeutic Targets Identified by Integrated mRNA and microRNA Expression Profiling in Breast Cancer. Cancer Research 2011; 71(17):5635–45. [DOI] [PubMed] [Google Scholar]
- 12.Foekens JA, Sieuwerts AM, Smid M, Look MP, de Weerd V, Boersma AW, et al. Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proc Natl Acad Sci U S A 2008;105(35):13021–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang G-L, Zhang X-H, Guo G-L, Huang K-T, Yang K-Y, Shen X, et al. Clinical significance of miR-21 expression in breast cancer: SYBR-Green I-based real-time RT-PCR study of invasive ductal carcinoma. Oncology Reports 2009;21(3):673–9. [PubMed] [Google Scholar]
- 14.Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, et al. The micrornas mir-373 and mir-520c promote tumor migration, invasion and metastasis. Proc American Assoc Cancer Research Annual Meeting 2008;49:1196–7. [Google Scholar]
- 15.Ravdin PM, Siminoff LA, Davis GJ, Mercer MB, Hewlett J, Gerson N, et al. Computer program to assist in making decisions about adjuvant therapy for women with early breast cancer. J Clin Oncol 2001;19(4):980–91. [DOI] [PubMed] [Google Scholar]
- 16.Buyse M, Loi S, van’t Veer L, Viale G, Delorenzi M, Glas AM, et al. Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J Natl Cancer Inst 2006;98(17):1183–92. [DOI] [PubMed] [Google Scholar]
- 17.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 2004;351(27):2817–26. [DOI] [PubMed] [Google Scholar]
- 18.Paik S, Tang G, Shak S, Kim C, Baker J, Kim W, et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol 2006;24(23):3726–34. [DOI] [PubMed] [Google Scholar]
- 19.van’t Veer LJ, Dai HY, van de Vijver MJ, He YDD, Hart AAM, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002; 415(6871):530–6. [DOI] [PubMed] [Google Scholar]
- 20.Nielsen T, Wallden B, Schaper C, Ferree S, Liu SZ, Gao DX, et al. Analytical validation of the PAM50-based Prosigna Breast Cancer Prognostic Gene Signature Assay and nCounter Analysis System using formalin-fixed paraffin-embedded breast tumor specimens. BMC Cancer 2014;14:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wallden B, Storhoff J, Nielsen T, Dowidar N, Schaper C, Ferree S, et al. Development and verification of the PAM50-based Prosigna breast cancer gene signature assay. BMC Medical Genomics 2015;8:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glas AM, Floore A, Delahaye L, Witteveen AT, Pover RCF, Bakx N, et al. Converting a breast cancer microarray signature into a high-throughput diagnostic test. BMC Genomics. 2006;7:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nielsen TO, Parker JS, Leung S, Voduc D, Ebbert M, Vickery T, et al. A Comparison of PAM50 Intrinsic Subtyping with Immunohistochemistry and Clinical Prognostic Factors in Tamoxifen-Treated Estrogen Receptor-Positive Breast Cancer. Clinical Cancer Research 2010;16(21):5222–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prat A, Ellis MJ, Perou CM. Practical implications of gene-expression-based assays for breast oncologists. Nat Rev Clin Oncol 2012;9(1):48–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sestak I, Buus R, Cuzick J, Dudsky P, Kronenwett R, Ferree S, et al. Comprehensive Comparison of Prognostic Signatures for Breast Cancer Recurrence San Antonio Breast Cancer Symposium 2016; Abstract S6–05; San Antonio, TX [Google Scholar]
- 26.Sestak I, Buus R, Cuzick J, Dubsky P, Kronenwett R, Denkert C, et al. Comparison of the Performance of 6 Prognostic Signatures for Estrogen Receptor-Positive Breast Cancer: A Secondary Analysis of a Randomized Clinical Trial. JAMA Oncol 2018;4(4):545–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buus R, Sestak I, Kronenwett R, Denkert C, Dubsky P, Krappmann K, et al. Comparison of EndoPredict and EPclin With Oncotype DX Recurrence Score for Prediction of Risk of Distant Recurrence After Endocrine Therapy. J Natl Cancer Inst 2016;108(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pierce JP, Faerber S, Wright FA, Rock CL, Newman V, Flatt SW, et al. A randomized trial of the effect of a plant-based dietary pattern on additional breast cancer events and survival: the Women’s Healthy Eating and Living (WHEL) Study. Control Clin Trials 2002;23(6):728–56. [DOI] [PubMed] [Google Scholar]
- 29.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 2009;27(8):1160–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res 2016;44(8):e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pierce JP, Natarajan L, Caan BJ, Parker BA, Greenberg ER, Flatt SW, et al. Influence of a diet very high in vegetables, fruit, and fiber and low in fat on prognosis following treatment for breast cancer: the Women’s Healthy Eating and Living (WHEL) randomized trial. JAMA 2007;298(3):289–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buffa FM, Camps C, Winchester L, Snell CE, Gee HE, Sheldon H, et al. microRNA-associated progression pathways and potential therapeutic targets identified by integrated mRNA and microRNA expression profiling in breast cancer. Cancer Res 2011;71(17):5635–45. [DOI] [PubMed] [Google Scholar]
- 33.Huang GL, Zhang XH, Guo GL, Huang KT, Yang KY, Shen X, et al. Clinical significance of miR-21 expression in breast cancer: SYBR-Green I-based real-time RT-PCR study of invasive ductal carcinoma. Oncol Rep 2009;21(3):673–9. [PubMed] [Google Scholar]
- 34.Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol 2008;10(2):202–10. [DOI] [PubMed] [Google Scholar]
- 35.Fortina P, Surrey S. Digital mRNA profiling. Nat Biotechnol 2008;26(3):293–4. [DOI] [PubMed] [Google Scholar]
- 36.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol 2008;26(3):317–25. [DOI] [PubMed] [Google Scholar]
- 37.Zhao X, Rodland EA, Tibshirani R, Plevritis S. Molecular subtyping for clinically defined breast cancer subgroups. Breast Cancer Research 2015;17:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tibshirani R The lasso method for variable selection in the Cox model. Stat Med 1997;16(4):385–95. [DOI] [PubMed] [Google Scholar]
- 39.Rothe F, Ignatiadis M, Chaboteaux C, Haibe-Kains B, Kheddoumi N, Majjaj S, et al. Global microRNA expression profiling identifies MiR-210 associated with tumor proliferation, invasion and poor clinical outcome in breast cancer. Plos One 2011;6(6):e20980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y, Wang Y, Xu Q, Zhou X, Qin Z, Chen C, et al. Prognostic evaluation of microRNA-210 in various carcinomas: Evidence from 19 studies. Medicine (Baltimore) 2017;96(43):e8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Block I, Burton M, Sorensen KP, Andersen L, Larsen MJ, Bak M, et al. Association of miR-548c-5p, miR-7–5p, miR-210–3p, miR-128–3p with recurrence in systemically untreated breast cancer. Oncotarget 2018;9(10):9030–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Volinia S, Galasso M, Sana ME, Wise TF, Palatini J, Huebner K, et al. Breast cancer signatures for invasiveness and prognosis defined by deep sequencing of microRNA. Proc Natl Acad Sci U S A 2012;109(8):3024–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang H, Zhang G, Wu JH, Jiang CP. Diverse roles of miR-29 in cancer (review). Oncol Rep 2014;31(4):1509–16. [DOI] [PubMed] [Google Scholar]
- 44.Muluhngwi P, Alizadeh-Rad N, Vittitow SL, Kalbfleisch TS, Klinge CM. The miR-29 transcriptome in endocrine-sensitive and resistant breast cancer cells. Sci Rep 2017;7(1):5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmitt MJ, Margue C, Behrmann I, Kreis S. MiRNA-29: a microRNA family with tumor-suppressing and immune-modulating properties. Curr Mol Med 2013;13(4):572–85. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y, Zhang X, Li H, Yu J, Ren X. The role of miRNA-29 family in cancer. Eur J Cell Biol 2013;92(3):123–8. [DOI] [PubMed] [Google Scholar]
- 47.Bhardwaj A, Singh H, Rajapakshe K, Tachibana K, Ganesan N, Pan Y, et al. Regulation of miRNA-29c and its downstream pathways in preneoplastic progression of triple-negative breast cancer. Oncotarget 2017;8(12):19645–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lanczky A, Nagy A, Bottai G, Munkacsy G, Szabo A, Santarpia L, et al. miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat 2016;160(3):439–46. [DOI] [PubMed] [Google Scholar]
- 49.Lu L, Mao X, Shi P, He B, Xu K, Zhang S, et al. MicroRNAs in the prognosis of triple-negative breast cancer: A systematic review and meta-analysis. Medicine (Baltimore) 2017;96(22):e7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nygren MK, Tekle C, Ingebrigtsen VA, Makela R, Krohn M, Aure MR, et al. Identifying microRNAs regulating B7-H3 in breast cancer: the clinical impact of microRNA-29c. Br J Cancer 2014;110(8):2072–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mulrane L, Madden SF, Brennan DJ, Gremel G, McGee SF, McNally S, et al. miR-187 is an independent prognostic factor in breast cancer and confers increased invasive potential in vitro. Clin Cancer Res 2012;18(24):6702–13. [DOI] [PubMed] [Google Scholar]
- 52.Almeida MI, Calin GA. The miR-143/miR-145 cluster and the tumor microenvironment: unexpected roles. Genome Med 2016;8(1):29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johannessen C, Moi L, Kiselev Y, Pedersen MI, Dalen SM, Braaten T, et al. Expression and function of the miR-143/145 cluster in vitro and in vivo in human breast cancer. Plos One 2017;12(10):e0186658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li D, Hu J, Song H, Xu H, Wu C, Zhao B, et al. miR-143–3p targeting LIM domain kinase 1 suppresses the progression of triple-negative breast cancer cells. Am J Transl Res 2017;9(5):2276–85. [PMC free article] [PubMed] [Google Scholar]
- 55.Wang S, Liu JC, Ju Y, Pellecchia G, Voisin V, Wang DY, et al. microRNA-143/145 loss induces Ras signaling to promote aggressive Pten-deficient basal-like breast cancer. JCI Insight 2017. August 3;2(15). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu X, Zhang X, Dhakal IB, Beggs M, Kadlubar S, Luo D. Induction of cell proliferation and survival genes by estradiol-repressed microRNAs in breast cancer cells. BMC Cancer 2012;12:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou LL, Dong JL, Huang G, Sun ZL, Wu J. MicroRNA-143 inhibits cell growth by targeting ERK5 and MAP3K7 in breast cancer. Braz J Med Biol Res 2017;50(8):e5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Donnarumma E, Fiore D, Nappa M, Roscigno G, Adamo A, Iaboni M, et al. Cancer-associated fibroblasts release exosomal microRNAs that dictate an aggressive phenotype in breast cancer. Oncotarget 2017;8(12):19592–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.