Abstract
The tetrapod limb is a stunning example of evolutionary diversity, with dramatic variation not only among distantly related species, but also between the serially homologous forelimbs (FLs) and hindlimbs (HLs) within species. Despite this variation, highly conserved genetic and developmental programs underlie limb development and identity in all tetrapods, raising the question of how limb diversification is generated from a conserved toolkit. In some breeds of domestic pigeon, shifts in the expression of two conserved limb identity transcription factors, PITX1 and TBX5, are associated with the formation of feathered HLs with partial FL identity. To determine how modulation of PITX1 and TBX5 expression affects downstream gene expression, we compared the transcriptomes of embryonic limb buds from pigeons with scaled and feathered HLs. We identified a set of differentially expressed genes enriched for genes encoding transcription factors, extracellular matrix proteins, and components of developmental signaling pathways with important roles in limb development. A subset of the genes that distinguish scaled and feathered HLs are also differentially expressed between FL and scaled HL buds in pigeons, pinpointing a set of gene expression changes downstream of PITX1 and TBX5 in the partial transformation from HL to FL identity. We extended our analyses by comparing pigeon limb bud transcriptomes to chicken, anole lizard, and mammalian datasets to identify deeply conserved PITX1- and TBX5-responsive components of the limb identity program. Our analyses reveal a suite of predominantly low-level gene expression changes that are conserved across amniotes to regulate the identity of morphologically distinct limbs.
Introduction
The tetrapod limb is an extraordinary example of morphological variation. Dramatic evolutionary diversification of limb form and function has enabled animals to thrive in diverse terrestrial, fossorial, aquatic, and aerial environments. The limb is unusual in that phenotypic variation characterizes not only limbs of different species, but also the serially homologous forelimbs (FLs) and hindlimbs (HLs) within a species. Limb variation between FLs and HLs is arguably most striking in birds, in which divergent patterning of the muscles, tendons, and bones that form the limb is coupled with variation in the epidermal appendages that cover each limb type. For example, the elaborately feathered epidermis of the avian wing is dramatically different from the scaled epidermis that typically covers the feet.
Despite highly divergent adult limb morphologies among and within species, limb patterning and morphogenesis is orchestrated by deeply conserved genetic and developmental programs. As a result of decades of elegant descriptive and experimental studies in canonical model organisms (especially mouse and chicken), the cellular, genetic, and molecular regulators of limb development are relatively well defined (reviewed in Petit et al., 2017, Tickle, 2015, Rabinowitz and Vokes, 2012, Duboc and Logan, 2011b, Towers and Tickle, 2009, Tickle, 2004). These studies reveal that the integration of several key developmental signaling pathways – including Fibroblast Growth Factor (FGF), Bone Morphogenetic Protein (BMP), Wnt, Sonic Hedgehog (SHH), and Retinoic Acid (RA) pathways – is required for proper limb bud position, initiation, outgrowth, and patterning.
In contrast, FL and HL identity is associated with the reciprocal expression patterns of a smaller set of genes, including the transcription factors (TFs) TBX5 (FL-specific expression), TBX4 (HL-specific), and PITX1 (HL-specific) (Simon et al., 1997, Logan et al., 1998, Szeto et al., 1996, Gibson-Brown et al., 1996, Shang et al., 1997). Over the past 20 years, a combination of misexpression and genetic studies have illuminated essential roles for each of these TFs in limb development. The paralogs TBX5 and TBX4 direct the respective initiation of FL and HL bud development through activation of the FGF and Wnt signaling pathways (Takeuchi et al., 2003). In mice, Tbx5−/− mutants fail to form a FL bud, while Tbx4−/− mutants form a HL bud that arrests early in development due to a loss of FGF10 expression maintenance (Agarwal et al., 2003, Takeuchi et al., 1999, Rallis et al., 2003, Naiche and Papaioannou, 2003, Naiche and Papaioannou, 2007). Similarly, loss of tbx5 function in zebrafish results in a failure to form the pectoral fin (FL equivalent) (Ahn et al., 2002, Garrity et al., 2002), and disruption of tbx4 nuclear localization in the naturally occurring pelvic finless zebrafish strain is associated with arrested pelvic fin (HL equivalent) development (Don et al., 2016). Tbx5 and Tbx4 are not essential for development of limb-specific identities in mice (Hasson et al., 2007, Naiche and Papaioannou, 2007, Minguillon et al., 2005); however, misexpression of TBX4 in the FL or TBX5 in the HL of chick embryos causes a partial transformation in limb identity in a subset of embryos, including changes in digit number, limb flexure, and epidermal appendage type (Rodriguez-Esteban et al., 1999, Takeuchi et al., 1999). Although the basis for the discrepancies between mouse and chick experiments remains unclear, it is possible that TBX5 and TBX4 have subtly different functions in mammals and birds, which diverged more than 300 million years ago (Horton et al., 2008).
Unlike TBX4 and TBX5, PITX1 is not required for limb bud initiation or outgrowth, but instead regulates these processes indirectly via activation of TBX4 (Minguillon et al., 2005, Szeto et al., 1999, Logan and Tabin, 1999, Duboc and Logan, 2011a). In addition to TBX4-dependent regulation of HL outgrowth, PITX1 serves a second essential role in limb development via TBX4-independent regulation of HL identity (Duboc and Logan, 2011a). In Pitx1−/− mutant mice, HLs develop, but lack HL-specific morphological characteristics (Szeto et al., 1999). In both chick and mouse embryos, ectopic expression of PITX1 in the developing FL bud causes a partial transformation to a more HL-like identity (DeLaurier et al., 2006, Logan and Tabin, 1999). Similarly, mutations at the human PITX1 locus are associated with HL-like characteristics in the FL (Al-Qattan et al., 2013) and congenital HL defects (Gurnett et al., 2008). PITX1 regulatory mutations have also been discovered in natural populations of three-spine stickleback fish, in which loss of PITX1 gene expression contributes to the adaptive loss of pelvic (HL) structures (Shapiro et al., 2004, Chan et al., 2010).
Substantial research efforts over the past two decades have helped decipher the genetic networks regulated by PITX1, TBX4, and TBX5 during limb development (reviewed in Duboc and Logan, 2011b, Rabinowitz and Vokes, 2012, Tickle, 2015, Petit et al., 2017). More recently, contemporary genomic approaches have opened new avenues for defining downstream targets in an unbiased manner. Particular progress has been made in defining the network downstream of PITX1 by comparing limb buds from wild-type and PitxT−/− mutant mice using massively parallel sequencing of both RNA transcripts to identify differentially-expressed genes and DNA regions captured by chromatin immunoprecipitation to locate putative regulatory regions (RNA-seq and ChIP-seq, respectively) (Nemec et al., 2017, Infante et al., 2013, Wang et al., 2018). These approaches have resulted in a more comprehensive view of PITX1-dependent gene networks (and could be used to similarly reveal TBX5- and TBX4-dependent networks). Despite these important advances, we still have a limited understanding of the transcriptomic changes that result from less dramatic (non-binary) shifts in gene expression that are associated with evolutionary diversification of limb morphology.
The domestic pigeon (Columba livia) is an outstanding model to study the evolution of genetic and developmental programs that underlie limb diversification. Pigeons display striking variation in HL morphology within a single species and are amenable to genetic crosses, genomic analyses, and embryonic studies (Shapiro et al., 2013, Shapiro and Domyan, 2013, Domyan and Shapiro, 2017). While most pigeons have scaled HLs, in some breeds scaled epidermis is replaced by skin with a range of feather morphologies (Figure 1A-C). Classical genetic experiments show that large feather “muffs” are caused by the synergistic effects of mutant alleles at two loci, grouse (gr) and Slipper (Sl), which independently produce smaller foot feathers (Doncaster, 1912, Wexelsen, 1934, Hollander, 1937, Levi, 1986). We recently showed that gr and Sl are cis-regulatory alleles of the limb identity genes PITX1 and TBX5, respectively (Domyan et al., 2016), and proposed a model in which feathered feet are the result of a partial transformation from HL to FL identity. This genetically-encoded, partial transformation in identity provides a unique opportunity to determine how altered expression of conserved limb identity genes modifies the downstream HL identity transcriptional program.
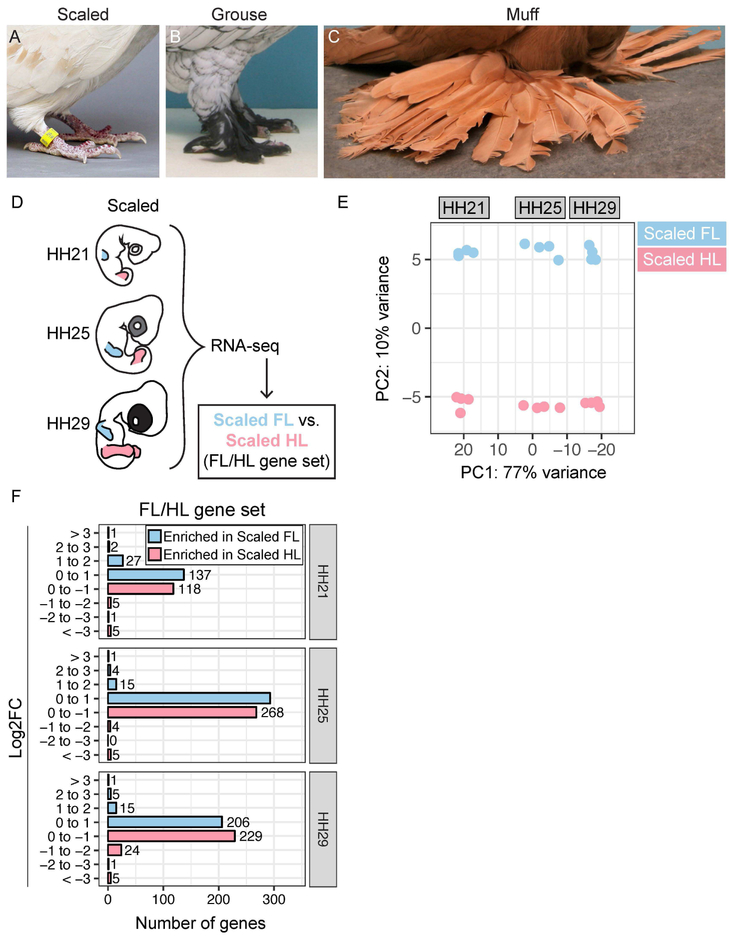
Figure 1. Comparative transcriptomics to identify gene expression changes associated with pigeon limb identity.
(A-C) Hindlimb phenotypes in domestic pigeon breeds, including scaled (A), grouse (B), and muff (C). (D) Experimental approach to identify differentially expressed genes at three stages (HH21, HH25, and HH29) of pigeon FL vs. scaled HL development. (E) PCA based on top 500 most variable genes in FL and scaled HLs. PC1 (77% of variance) separates samples by stage, PC2 (10% of variance) separates samples by limb type. (F) Number of genes that show FL-enriched or scaled HL-enriched expression pattern at HH21, HH25, or HH29. Genes are grouped based on Log2FC. FL = forelimb; HL = hindlimb; HH = Hamburger-Hamilton stage. Photo credits: A, Sydney Stringham; B and C, Thomas Hellmann.
The goal of this study is to identify genetic network changes that accompany shifts in limb identity. First, we identify FL- and HL-specific transcriptomic differences in pigeons. We then test for transcriptomic changes in pigeon HLs with partially altered limb identity attributable to changes in PITX1 and TBX5 regulation. Next, we compare our pigeon results with transcriptomic datasets from other amniotes to identify a set of genes whose expression typify FL and HL differences across broad phylogenetic distances. Finally, we find that a subset of the highly-conserved FL genetic program is also differentially expressed in mutant pigeon HLs with partial identity transformations.
Results
Gene expression changes associated with normal FL and HL development in pigeons
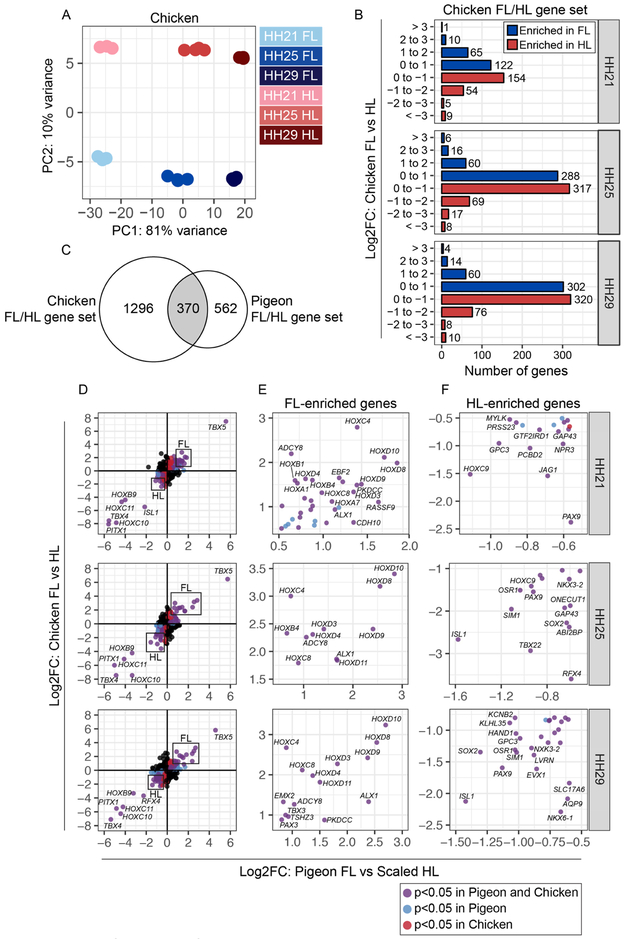
To identify differentially expressed transcripts associated with normal FL and HL development in pigeons, we performed RNA-seq on FL and HL buds from scale-footed pigeon embryos at three early stages of embryonic development (approximate pigeon equivalents of Hamburger-Hamilton (HH) stage 21, HH25, and HH29; Hamburger and Hamilton, 1951) (Figure 1D). Principal component analysis (PCA) based on the 500 most differentially expressed genes revealed that PC1 accounts for 77% of sample variation and corresponds to developmental stage, while PC2 accounts for 10% of variation and separates samples by limb type (FL vs. HL) (Figure 1E). Together, PC1 and PC2 group samples into six discrete clusters based on developmental stage and limb type.
We identified a set of 978 genes that are differentially expressed between FL and HL at HH21, HH25, and/or HH29 (“FL/HL” gene set; Table 1, Supplemental Figure 1, and Supplemental Table 1). Within the FL/HL gene set, only 12 genes are strongly enriched (defined here as an absolute value Log2FC > 2) in a FL- or HL-specific manner (Figure 1F), all of which are TFs that have been implicated in vertebrate limb development to varying degrees (Takeuchi et al., 1999, Rodriguez-Esteban et al., 1999, Szeto et al., 1999, Logan and Tabin, 1999, Zakany and Duboule, 2007, Tarchini et al., 2006, Capellini et al., 2006, Boulet and Capecchi, 2004, Wellik, 2003, Davis and Capecchi, 1994, Capellini et al., 2010, Narkis et al., 2012, Feenstra et al., 2012). In contrast, the majority (86-95%) of differentially expressed genes changed less than 2-fold (Log2FC < 1) between FL and HL (Figure 1F). These results suggest that, during early pigeon development, a small set of large gene expression changes is coupled with a large set of subtle gene expression changes to differentiate FL and HL identities. Notably, the number and level of gene expression changes that distinguish pigeon FL and HL is remarkably similar to what was recently reported for mouse FL and HL buds (Nemec et al., 2017), despite the radical differences between FL and HL morphology in pigeons and the more modest differences between limbs in mice.
Table 1.
Summary of pigeon differential expression gene sets.
| Differential expression gene sets | ||||
|---|---|---|---|---|
| FL/HL | grouse | muff | ||
| Comparison | FL vs. Scaled HL | Scaled HL vs. Grouse HL | Scaled HL vs. Muff HL | |
| Total | DE genes | 978 | 1425 | 3202 |
| Shared between grouse and muff | - | 42% (601/1425) | 19% (601/3202) | |
| Shared with FL/HL | - | 8% (108/1425) | 10% (334/3202) | |
| TFs | DE genes | 122 | 50 | 105 |
| Shared between grouse and muff | - | 42% (21/50) | 20% (21/105) | |
| Shared with FL/HL | - | 18% (9/50) | 38% (40/105) | |
| ECM | DE genes | 81 | 35 | 92 |
| Shared between grouse and muff | - | 43% (15/35) | 16% (15/92) | |
| Shared with FL/HL | - | 26% (9/35) | 29% (27/92) | |
Total = total number of significantly differentially expressed genes in each gene set; TFs = transcription factors; ECM = extracellular matrix genes; DE = differentially expressed. Percentage values indicate overlap of two gene sets (e.g., of the 1425 genes DE in scaled HL vs. grouse HL comparison, 601/1425 (42%) are also DE in scaled HL vs. muff HL comparison; 108/1425 (8%) are also DE in FL vs. scaled HL comparison).
Dynamic TBX5 and PITX1 expression differences characterize feathered HL development
We previously demonstrated that in pigeons, the grouse phenotype is associated with a cis-regulatory mutation that reduces PITX1 expression in the HH25 HL, while the muff phenotype is associated with cis-regulatory mutations that cause a combination of PITX1 reduction and ectopic TBX5 expression in the HH25 HL (Domyan et al., 2016, Boer et al., 2017). To determine if PITX1 and TBX5 expression differ in scaled, grouse, and muff HLs at other stages of development, we measured PITX1 and TBX5 expression in HH21, HH25, and HH29 HL buds by quantitative reverse-transcriptase PCR (qRT-PCR) (Supplemental Figure 2). We found that TBX5 is ectopically expressed in muff HLs relative to scaled and grouse HLs at all stages analyzed. PITX1 is significantly reduced in muff HLs relative to scaled HLs at all stages analyzed (Supplemental Figure 2). In contrast, PITX1 is significantly reduced in grouse HLs relative to scaled HLs only at HH25 and HH29 (Supplemental Figure 2), suggesting that the temporal dynamics of PITX1 HL expression is different between grouse and muff pigeon breeds.
PITX1- and TBX5-dependent gene expression during feathered HL development
We predicted that the divergent PITX1 and TBX5 expression profiles during development of scaled, grouse, and muff HLs could reveal gene expression changes downstream of TBX5 and/or PITX1 that are associated with limb diversification. To test this hypothesis, we performed RNA-seq on limb buds from grouse and muff embryos isolated at HH21, HH25, and HH29 (Figure 2A). We reasoned that comparison of differentially expressed gene sets from scaled vs. grouse HLs (“grouse” gene set) and scaled vs. muff HLs (“muff” gene set) would enable us to distinguish PITX1-responsive gene expression changes (misexpressed in both grouse and muff HLs) from the effects of TBX5 and/or PITX1 + TBX5 co-regulation (misexpressed only in muff HLs).
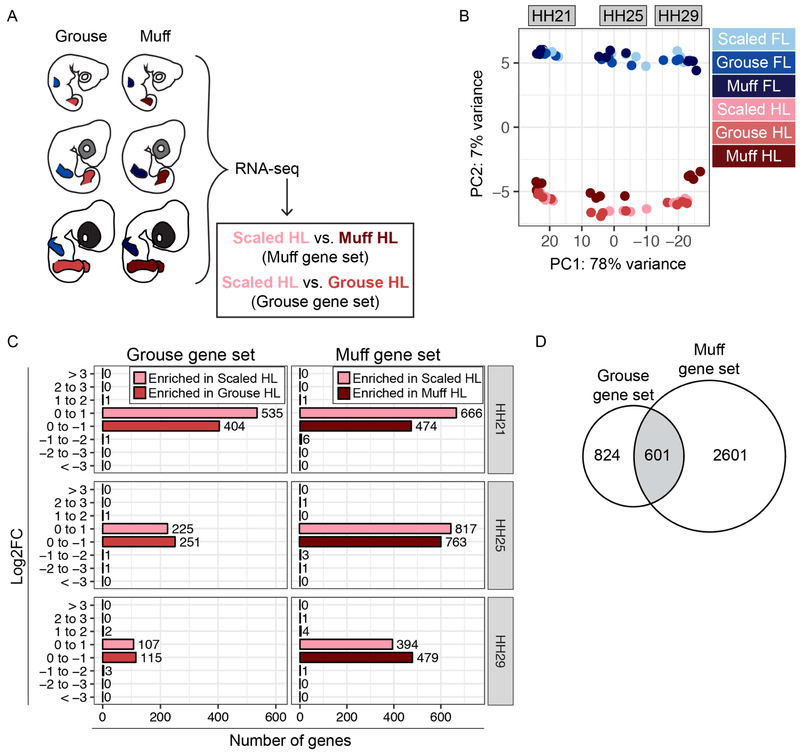
Figure 2. Comparison of hindlimb transcriptomes from scaled, grouse, and muff pigeon embryos.
(A) Experimental approach to identify differentially expressed genes from grouse and muff limb buds at HH21, HH25, and HH29. (B) PCA based on top 500 most variable genes from all FL and HL limb bud samples. PC1 (78% of variance) separates samples by developmental stage, PC2 (7% of variance) separates samples by FL vs. HL identity. (C) Number of genes enriched in scaled HL or feathered HL specific manner at HH21, HH25, or HH29. Genes are grouped based on Log2FC from scaled HL vs. grouse HL or scaled HL vs. muff HL comparisons. (D) Venn diagram comparing differentially expressed gene sets from scaled HL vs. grouse HL and scaled HL vs. muff HL comparisons (intersection p-value = 4.3e-83).
To test for consistency with our qRT-PCR results, we first examined the expression profiles of PITX1 and TBX5 in the RNA-seq samples from scaled, grouse, and muff limb buds. As expected, TBX5 is highly expressed in all FL samples at all developmental stages with no significant difference in expression among FLs from scaled, grouse, or muff embryos (Supplemental Figure 3). Within HL samples, TBX5 is significantly enriched in muff HLs relative to scaled and grouse HLs at all stages (Supplemental Figure 3). PITX1, which is expressed only in HLs, is significantly reduced in muff HLs relative to scaled HLs at all stages (Supplemental Figure 4). In contrast, PITX1 expression is reduced in grouse HLs relative to scaled HLs only at HH25 and HH29 (Supplemental Figure 4). Thus, our analyses of PITX1 and TBX5 expression by qRT-PCR and RNA-seq yielded qualitatively identical results.
To determine the relationship between scaled, grouse, and muff limb bud transcriptomes, we performed PCA as described above for limb buds from scale-footed pigeons. Addition of the grouse and muff datasets had a minimal effect on the overall structure of the results; FL and HL samples remain clustered by limb type and developmental stage (Figure 2B and Supplemental Figure 5). Within each HL cluster, muff HLs separated slightly from scaled and grouse HLs and clustered closer to their respective stage-matched FL clusters on PC2.
Next, we performed differential expression analyses to identify transcriptional changes associated with altered PITX1 and/or TBX5 expression in feathered HLs. Relative to scaled HLs, a total of 1425 genes are misexpressed in grouse HLs (grouse gene set) and 3202 genes are misexpressed in muff HLs (muff gene set) at HH21, HH25, and/or HH29 (Table 1, Supplemental Figure 6, and Supplemental Tables 2-3). Similar to the FL vs. scaled HL differential expression results, feathered HL development is characterized by extensive low-level gene expression changes, with nearly all differentially expressed genes (>99%) altered by less than 2-fold (Log2FC < 1, Figure 2C).
By comparing the grouse and muff gene sets, we identified 601 genes that are putatively PITX1-responsive, as they are misexpressed in both grouse and muff HLs (intersection p-value = 4.3e-83; Figure 2D, Table 1, and Supplemental Table 4). A subset (125/601) of the PITX1-responsive genes were also previously identified in transcriptomic analyses of mouse Pitx1−/− HLs (Wang et al., 2018, Nemec et al., 2017), including the TFs EMX2, HOXD11, RFX4, TBX15, and ZEB2. Other PITX1-responsive genes encode components of several important developmental signaling pathways, as well as a variety of extracellular matrix (ECM) proteins (see below). These independently identified pigeon and mouse PITX1-responsive genes show a significant degree of overlap (intersection p-value = 3.7e-05; Supplemental Figure 7 and Supplemental Table 4). The PITX1-responsive gene set also includes genes with known roles in limb development such as short stature homeobox (SHOX) (Decker et al., 2011, Tiecke et al., 2006) that, to our knowledge, were not known to be regulated by PITX1. Notably, Shox is not present in the mouse genome (but is present in other mammalian genomes, including human), precluding the identification of Shox as a Pitx1 target in previous genomic analyses of mouse Pitx1−/− HLs (Wang et al., 2018, Nemec et al., 2017).
In addition to the PITX1-responsive genes that are misexpressed in both grouse and muff HLs, we identified 824 genes that are misexpressed only in grouse HLs (Figure 2D). This finding raises the possibility that either 1) distinct genetic programs are activated depending on PITX1 expression level (PITX1 is differentially expressed between grouse and muff HLs; Supplemental Figure 4), 2) additional genetic changes modify the grouse phenotype but were not detected in our whole-genome scans for allele frequency differentiation (Domyan et al., 2016), and/or 3) there are breed-specific gene expression changes that distinguish Oriental Frills (grouse) from Racing Homers (scaled) and English Trumpeters (muff) and are unrelated to HL phenotypes. We also identified 2601 genes that are misexpressed only in muff HLs (Figure 2D). These genes are likely regulated by TBX5, although we cannot rule out co-regulation by TBX5 and PITX1. In summary, by comparing the transcriptional profiles associated with scaled, grouse, and muff HL development, we discovered sets of genes that are putative targets of PITX1 and/or TBX5 during pigeon limb development. While some of the genes we identified are known targets of PITX1 or TBX5, many were not previously linked to PITX1, TBX5, or limb development.
Partial co-option of pigeon FL genetic program during feathered HL development
We next wanted to know if feathered HL development is accompanied by FL-like transcriptional changes in genes other than PITX1 and TBX5. Therefore, we probed the grouse and muff gene sets for genes that are also differentially expressed between FL and scaled HL (i.e., in the FL/HL gene set).
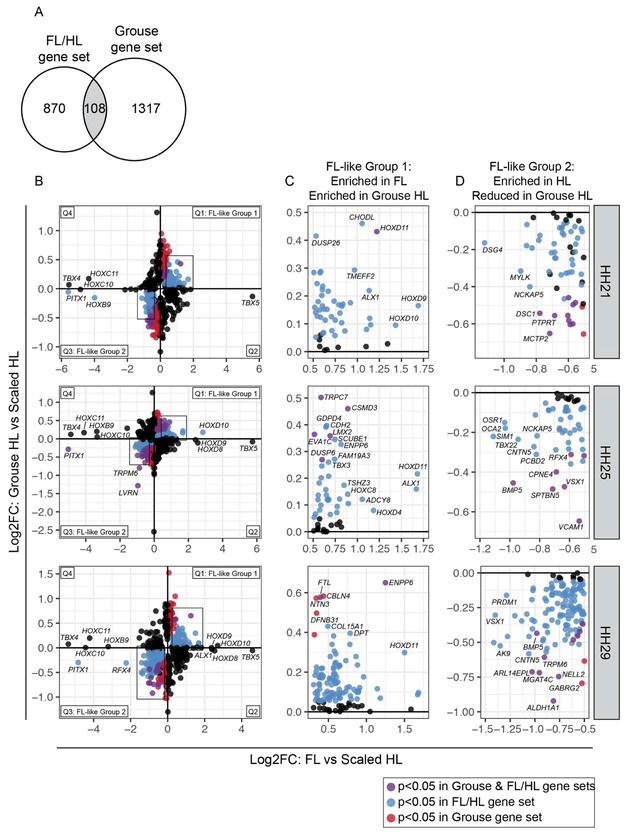
We first identified signatures of FL identity associated with reduced PITX1 expression by comparing the grouse and FL/HL gene sets. Of the genes that are misexpressed in grouse HLs, ~8% (108/1425) are also differentially expressed between FL and scaled HL (intersection p-value = 0.029; Figure 3A, Table 1, and Supplemental Table 2). Within this set of 108 genes, we identified two categories of genes with FL-like gene expression patterns: FL-like Group 1 genes are enriched in FLs and grouse HLs relative to scaled HLs, while FL-like Group 2 genes have reduced expression in FLs and grouse HLs relative to scaled HLs (Figure 3B-D). FL-like Group 1 includes the TFs HOXD11 and LMX2, as well as DUSP6, a negative regulator of FGF signaling (Figure 3B-C). FL-like Group 2 includes PITX1, other TFs such as VSX1 and RFX4, BMP5, and several genes that encode cell adhesion and cytoskeletal proteins (including LVRN, DSC1, VCAM1, and SPTBN5) (Figure 3B,D).
Figure 3. A subset of genes in the grouse HL transcriptome display FL-like expression patterns.
(A) Venn diagram comparing differentially expressed gene sets from FL vs. scaled HL and scaled HL vs. grouse HL comparisons (intersection p-value = 0.029). (B) Scatterplots of Log2FC values for all genes that are differentially expressed in FL vs. scaled HL or scaled HL vs. grouse HL at HH21, HH25, or HH29. Scatterplots are divided into four quadrants (Q); Q1 and Q3 highlight genes that show FL-like expression pattern in grouse HLs. Genes in Q1 (FL-like Group 1) and Q3 (FL-like Group 2) are color coded: blue dots denote genes that are significantly differentially expressed between FL and scaled HL; red dots denote genes that are significantly differentially expressed between scaled HL and grouse HL; purple dots represent genes that are significantly differentially expressed in both comparisons. Boxes indicate regions of scatterplot displayed in (C) or (D). (C) Detail of FL-like Group 1 genes. (D) Detail of FL-like Group 2 genes. Statistical significance = adjusted p-value < 0.05.
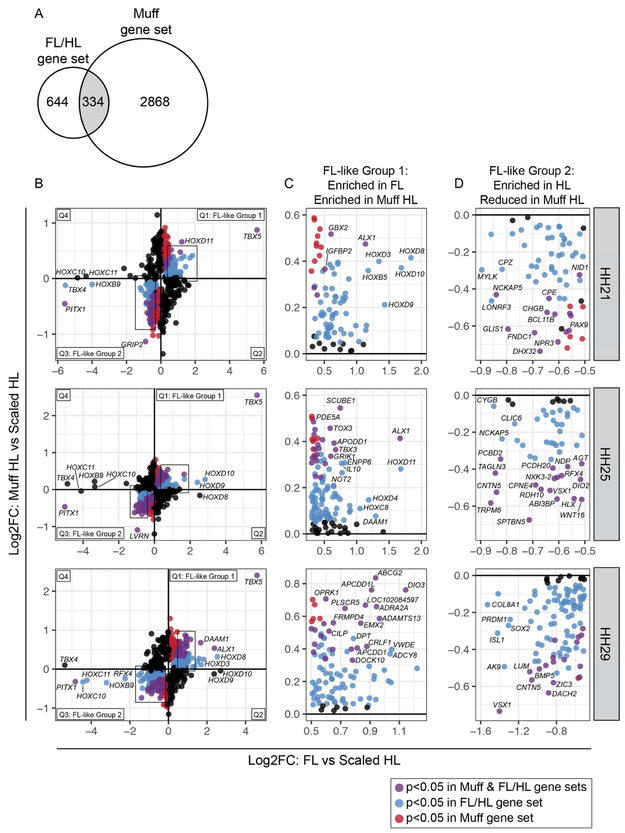
We next identified signatures of FL identity that are putatively regulated by TBX5 (or coregulated by TBX5 and PITX1) by comparing the muff and FL/HL gene sets. Of the genes that are misexpressed in muff HLs, ~10% (334/3202) are also differentially expressed between FL and scaled HL (intersection p-value = 7.5e-24; Figure 4A, Table 1, and Supplemental Table 3). Similar to the grouse gene set, we identified two categories of genes with FL-like gene expression patterns (Figure 4B-D). FL-like Group 1 (enriched in FL and muff HL relative to scaled HL) includes the TFs TBX5, HOXD11, GBX2, ALX1, and EMX2, as well as the Wnt-associated scaffolding gene DAAM1 (Figure 4B-C). FL-like Group 2 (reduced in FL and muff HL relative to scaled HL) includes the TFs PITX1, PAX9, VSX1, RFX4, NKX3-2, DACH2 and ZIC3; components of developmental signaling pathways with known roles in limb development such as WNT16, RDH10 and BMP5; and several genes encoding ECM proteins (Figure 4B,D). In addition, many other genes display non-significant FL-like expression patterns in muff HLs, including the TFs ISL1, SOX2, HOXB5, HOXD3, HOXD4, HOXC8, HOXD8, HOXD9, HOXD10, and HOXD11 (Figure 4B,D).
Figure 4. A subset of genes in the muff HL transcriptome display FL-like expression patterns.
(A) Venn diagram comparing differentially expressed gene sets from FL vs. scaled HL and scaled HL vs. muff HL comparisons (intersection p-value = 7.5e-24). (B) Scatterplots of Log2FC values for all genes that are differentially expressed in FL vs. scaled HL or scaled HL vs. muff HL at HH21, HH25, or HH29. Scatterplots are divided into four quadrants (Q); Q1 and Q3 highlight genes that show FL-like expression pattern in muff HLs. Genes in Q1 (FL-like Group 1) and Q3 (FL-like Group 2) are color coded: blue dots denote genes that are significantly differentially expressed between FL and scaled HL; red dots denote genes that are significantly differentially expressed between scaled HL and muff HL; purple dots represent genes that are significantly differentially expressed in both comparisons. Boxes indicate regions of scatterplot displayed in (C) or (D). (C) Detail of FL-like Group 1 genes. (D) Detail of FL-like Group 2 genes. Statistical significance = adjusted p-value < 0.05.
TFs are differentially expressed in feathered HLs
In our analyses of the grouse and muff gene sets, we noted an enrichment of FL-like expression of genes encoding TFs, ECM proteins, and components of developmental signaling pathways. Hence, these gene classes could be key parts of the co-opted FL limb identity program in feathered HLs. To test this prediction, we next focused in on differentially expressed genes encoding TFs, ECM proteins, and developmental signaling components.
To identify TFs that are normally differentially expressed between FL and HL, we compared the FL/HL gene set to a genome-wide list of 1473 TFs (AnimalTFDB 2.0 database; Zhang et al., 2015). From this list, 122 TFs are differentially expressed between FLs and scaled HLs (Figure 5A and Table 1). As expected, we detected strong FL enrichment of many known FL-specific TFs, including TBX5, HOXD10, HOXD8, HOXD9, and ALX1 (Figure 5A). Likewise, we detected HL enrichment of known HL-specific TFs, including TBX4, PITX1, HOXC11, HOXC10, HOXB9, and ISL1 (Figure 5A). Many other TFs showed FL or HL expression bias to a lesser degree, and some TFs displayed temporally dynamic FL/HL expression patterns (Figure 5A).
Figure 5. Differential expression of transcription factors associated with pigeon limb identity.
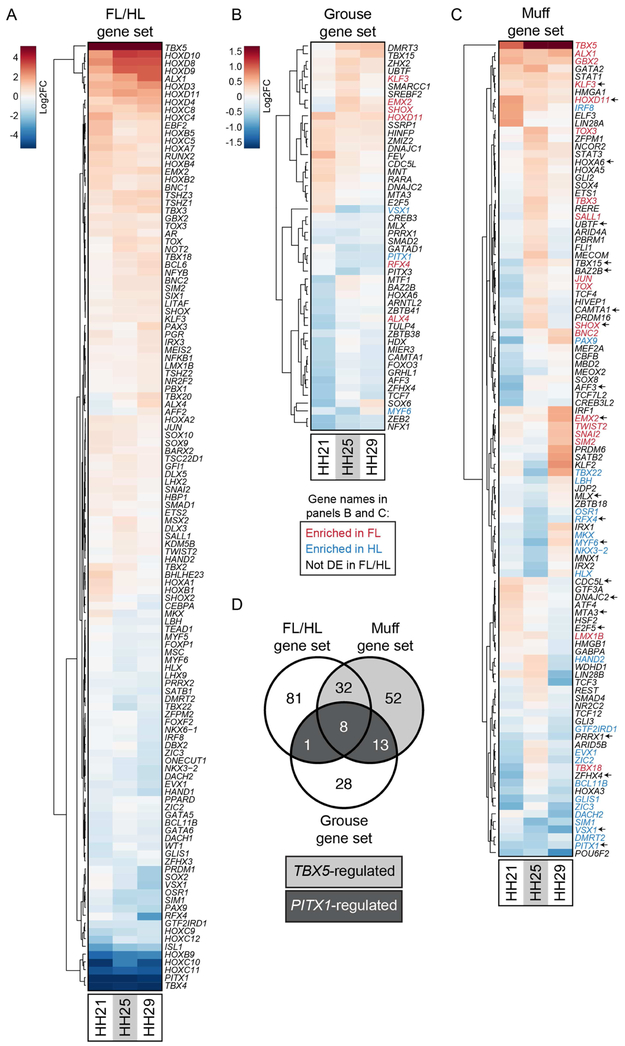
(A-C) Gene-wise hierarchical clustering heat maps of transcription factors (TFs) that are significantly differentially expressed between FL vs. scaled HL (A), scaled HL vs. grouse HL (B), or scaled HL vs. muff HL (C). For (A-C), all TFs that are significantly differentially expressed at one or more stages are included in the heat map. The scale for (A) is Log2FC −5 (HL-enriched) to 5 (FL-enriched); the scale for (B) and (C) is Log2FC −1.5 (scaled HL-enriched) to 1.5 (grouse HL-enriched). In (B) and (C), gene names highlighted in red or blue denote genes from FL vs. scaled HL comparison that are significantly FL-enriched or HL-enriched, respectively. In (C), arrows next to gene names highlight genes that are differentially expressed in both grouse and muff HLs relative to scaled HLs. (D) Venn diagram showing overlap of differentially expressed TFs from FL vs. scaled HL, scaled HL vs. grouse HL, and scaled HL vs. muff HL comparisons. Dark gray shading highlights TFs that are differentially expressed in all three comparisons and are putatively regulated by PITX1. Light gray shading denotes TFs that are differentially expressed only in FL vs. scaled HL and scaled HL vs. muff HL comparisons, suggesting regulation by TBX5.
We next identified TFs that are misexpressed during feathered HL development by comparing the grouse and muff gene sets to the genome-wide TF list (Figure 5B-C). We found 50 TFs that are misexpressed in grouse HLs and 105 TFs that are misexpressed in muff HLs (Figure 5B-C and Table 1). Comparison of the grouse and muff results revealed 21 TFs that are likely regulated by PITX1, as they are misexpressed in both grouse and muff HLs (Figure 5B-D and Table 1). A subset of the TFs in the grouse and muff gene sets were also identified in the FL/HL gene set, suggesting that these TFs are co-opted from the normal FL identity program (Figure 5B-D and Table 1). In addition, we found a set of TFs, including several HOX and T-box genes (HOXA3, HOXA5, HOXA6, TBX15, IRX2), that are misexpressed in feathered HLs but are not differentially expressed between FL and scaled HLs (Figure 5B-D). These results suggest that, in addition to a partial redeployment of the FL identity program, feathered HL development involves the activation of TFs not normally required for differentiation of FLs and scaled HLs.
Differentially expressed ECM components in feathered HLs
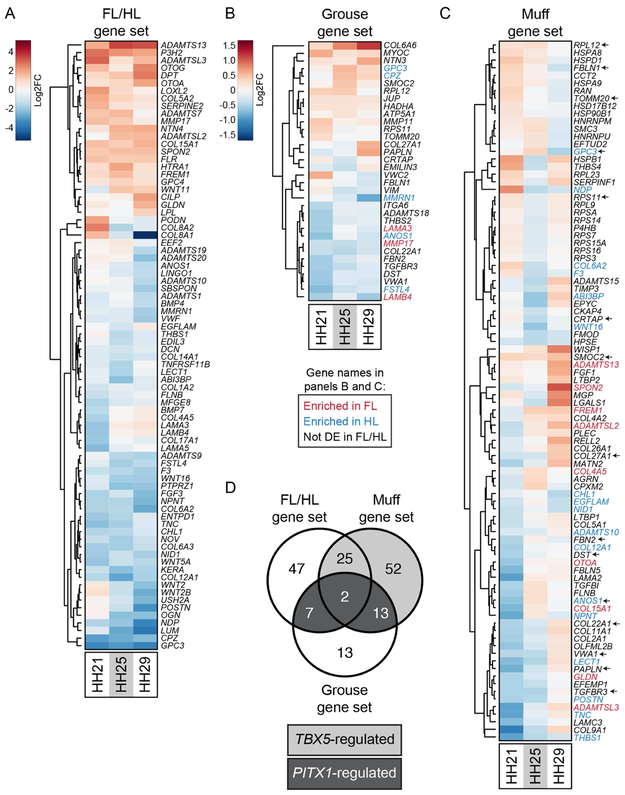
In birds, ECM composition differs between scaled and feathered epidermis (Sengel, 1990). Consistent with this observation, recent transcriptomic analyses have identified specific ECM genes that are differentially expressed between these epidermal types in developing archosaur embryos (Wu et al., 2018, Musser et al., 2018, Lai et al., 2018). With the importance of the ECM in mind, we identified ECM genes putatively regulated by PITX1 and/or TBX5 during pigeon limb development by cross-referencing our FL/HL, grouse, and muff gene sets to a list of 491 genes annotated as ECM components in the Gene Ontology database (GO:0031012) (Ashburner et al., 2000, The Gene Ontology, 2017, Carbon et al., 2009). From this list, we identified 81, 35, and 92 ECM genes in the FL/HL, grouse, and muff gene sets, respectively (Figure 6A-D and Table 1). By comparing the ECM genes in the grouse and muff gene sets, we pinpointed ECM genes that are downstream of PITX1 and/or TBX5 during early pigeon limb development, including ADAMTS proteases, collagens, and Wnt pathway ligands (Figure 6D). Notably, almost half (12/27) of the ECM genes shared between the FL/HL, grouse, and muff gene sets were also found to be differentially expressed between feather- and scale-forming epidermis in chick embryos (Wu et al., 2018), supporting the model that distinct ECM signatures accompany the development of divergent limb identities in birds.
Figure 6. Differential expression of genes encoding extracellular matrix components is associated with pigeon limb identity.
(A-C) Gene-wise hierarchical clustering heat maps of genes encoding extracellular matrix (ECM) components that are significantly differentially expressed between FL vs. scaled HL (A), scaled HL vs. grouse HL (B), or scaled HL vs. muff HL (C). For (A-C), all ECM-encoding genes that are significantly differentially expressed at one or more stages are included in the heat map. The scale for (A) is Log2FC −5 (HL-enriched) to 5 (FL-enriched); the scale for (B) and (C) is Log2FC −1.5 (scaled HL-enriched) to 1.5 (muff HL-enriched). In (B) and (C), gene names highlighted in red or blue denote genes from FL vs. scaled HL comparison that are significantly FL-enriched or HL-enriched, respectively. In (C), arrows next to gene names highlight genes that are differentially expressed in both grouse and muff HLs relative to scaled HLs. (D) Venn diagram showing overlap of differentially expressed ECM-encoding genes from FL vs. scaled HL, scaled HL vs. grouse HL, and scaled HL vs. muff HL comparisons. Dark gray shading highlights ECM-encoding genes that are differentially expressed in all three comparisons and are putatively regulated by PITX1. Light gray shading denotes ECM-encoding genes that are differentially expressed only in FL vs. scaled HL and scaled HL vs. muff HL comparisons, suggesting regulation by TBX5.
Modulation of developmental signaling pathways in feathered HLs
Normal limb development requires the integration of several key developmental signaling pathways including the SHH, FGF, BMP, RA, and Wnt pathways (reviewed in Duboc and Logan, 2011b, Tickle, 2015). Within the FL/HL gene set, we identified suites of genes involved in each of these pathways using Ingenuity Pathway Analysis (IPA) (Supplemental Figures 8-11), suggesting that fine-tuning of developmental pathways is a normal part of FL vs. HL development. To determine if misexpression of PITX1 and/or TBX5 in feathered HLs is associated with altered expression of genes within the same pathways, we also probed the grouse and muff gene sets using IPA. In both the grouse and muff gene sets, we detected differential expression of genes associated with FGF, RA, and Wnt signaling (Supplemental Figures 8-10), raising the possibility that TBX5 and/or PITX1 interacts with each of these pathways. We also identified a small number of genes within the muff gene set that are associated with BMP and SHH signaling, suggesting that TBX5 may also interact with these pathways during early stages of limb development (Supplemental Figure 11). Together, these results suggest that fine-tuning of developmental signaling pathways by PITX1 and TBX5 may be associated with the development of distinct limb morphologies and provide a foundation for further investigation of interactions between PITX1, TBX5, and signaling pathways involved in limb development.
Evidence for conservation of limb identity programs among birds
Despite highly variable adult limb morphologies, the genetic toolkit that regulates limb development among vertebrates is evolutionarily conserved. However, the degree to which regulation of these genes is conserved across distantly related species remains unclear. To determine if limb-specific transcriptional profiles are conserved across large phylogenetic distances, we first compared gene expression in limb buds from pigeons and chickens, two distantly related avian species. We generated an RNA-seq dataset from FL and HL buds from scale-footed chicken embryos at developmental stages comparable to our pigeon dataset (HH21, HH25, and HH29). Consistent with the pigeon dataset, PCA based on the 500 most differentially expressed genes separated samples by stage (PC1, 81% of variance explained) and limb type (PC2, 10%) (Figure 7A and Supplemental Figure 5). We identified a set of 2160 genes that are differentially expressed between chicken FL and HL buds at HH21, HH25, and/or HH29 (chicken FL/HL gene set, Supplemental Figure 12 and Supplemental Table 5). As in our pigeon analyses, we found that most of the gene expression changes between chicken FL and HL are relatively modest (Log2FC < 1) (Figure 7B). Similar to our analysis of pigeon FL and HL buds, we identified a suite of TFs, genes encoding ECM proteins, and components of developmental signaling pathways that are differentially expressed between chicken FL and HL buds (Supplemental Figure 13).
Figure 7. Identification of conserved gene expression patterns in embryonic limb buds from chicken and pigeon.
(A) PCA based on top 500 most variable genes in chicken FL and scaled HL buds at HH21, HH25, and HH29. PC1 (82% of variance) separates samples by stage; PC2 (11% of variance) separates samples by limb type. (B) Number of genes that show FL-enriched or scaled HL-enriched expression pattern in chicken embryos at HH21, HH25, or HH29. Genes are grouped based on Log2FC. (C) Venn diagram showing overlap of chicken FL vs. HL and pigeon FL vs. scaled HL gene sets (intersection p-value = 3.1e-61). (D) Scatterplots of Log2FC values for all genes that are differentially expressed in chicken FL vs. HL or pigeon FL vs. scaled HL at HH21, HH25, or HH29. Scatterplots are divided into four quadrants (Q); Q1 and Q3 highlight genes that show FL-enriched or HL-enriched expression in both avian species. Genes in Q1 (FL-enriched) and Q3 (HL-enriched) are color coded: blue dots denote genes that are significantly differentially expressed in pigeon FL vs. scaled HL; red dots denote genes that are significantly differentially expressed in chicken FL vs. HL; purple dots represent genes that are significantly differentially expressed in both comparisons. Boxes indicate regions of scatterplot displayed in (C) or (D). (E) Detail of FL-enriched genes. (F) Detail of HL-enriched genes. Statistical significance = adjusted p-value < 0.05.
To determine the degree to which the limb identity network is conserved between pigeons and chickens, we compared the FL/HL gene sets from each species. For this analysis, we included only the genes that are annotated in both species (based on Cliv_2.1; Holt et al., 2018 and Gallus_gallus-5.0; Warren et al., 2017), resulting in comparison of 933/978 pigeon and 1667/2160 chicken genes. Despite this limitation, we found 370 genes that are differentially expressed between FL and HL in both chicken and pigeon (intersection p-value = 3.1e-61; Figure 7C and Supplemental Table 6). In both species, TBX5 stood out as the most FL-enriched gene, while TFs HOXD8, HOXD9, HOXD10, and ALX1 were also highly FL-enriched (Log2FC > 2) in both species (Figure 7D-E). A small set of TFs (TBX4, PITX1, HOXB9, HOXC10, HOXC11, ISL1) was highly HL-enriched (Log2FC > 2) in both species (Figure 7D,F). In both chickens and pigeons, many other TFs were expressed in a FL- or HL-specific manner to a lesser degree (Figure 7D-F and Supplemental Table 6). Notably, 20% (76/370) of the genes identified in both the pigeon and chicken FL vs. HL datasets were TFs (Supplemental Table 6), further suggesting that TFs are highly conserved components of limb identity programs.
Evidence for conservation of limb identity programs among amniotes
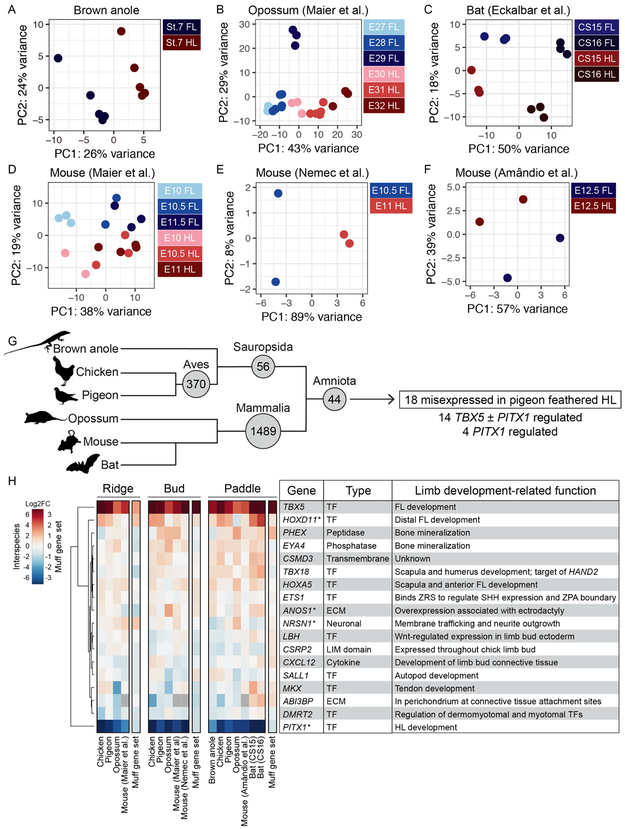
Recent studies have characterized embryonic FL and HL bud transcriptomes in several mammalian species, including mouse, opossum, pig, and bat (Taher et al., 2011, Maier et al., 2017, Sears et al., 2018, Wang et al., 2018, Gyurjan et al., 2011, Eckalbar et al., 2016, Shou et al., 2005, Nemec et al., 2017). However, similar analyses in other vertebrate classes are sparse, precluding broader comparative analyses of limb identity networks. We therefore extended our analyses by identifying differentially expressed genes in embryonic FLs and HLs of brown anole lizards (Anolis sagrei, Supplemental Figure 14). In addition, we re-analyzed several publicly available mammalian (mouse, opossum, and bat) limb bud datasets (Nemec et al., 2017, Maier et al., 2017, Amandio et al., 2016, Eckalbar et al., 2016). Together with our pigeon and chicken datasets, these additional data enabled us to identify conserved components of the limb identity network in Aves (pigeon and chicken), Sauropsida (pigeon, chicken, and anole), Mammalia (mouse, opossum, and bat), and Amniota (pigeon, chicken, anole, mouse, opossum, and bat) (Figure 8A). To facilitate comparisons among species, we categorized samples from each dataset into ridge, bud, or paddle developmental stages, which approximately correspond to chicken stages HH21, HH25, and HH29, respectively.
Figure 8. Identification of deeply conserved elements of limb identity program in amniotes.
(A-F) PCAs based on top 500 most variable genes from brown anole (A), opossum (B), bat (C), and three different mouse (D-F) limb bud datasets. (B) and (D) are analyses of datasets from (Maier et al., 2017), (C) is analysis of dataset from (Eckalbar et al., 2016), (E) is analysis of dataset from (Nemec et al., 2017), and (F) is analysis of dataset from (Amandio et al., 2016). For (A-F), specific stages of samples are listed and color coded based on stage group. (G) Tree summarizing evolutionary relationships between species included in comparative transcriptomics analyses. Number of genes differentially expressed in FL vs. HL of Aves (chicken and pigeon), Mammalia (opossum, bat, and/or mouse), Sauropsida (brown anole and chicken or pigeon), and Amniota (brown anole, chicken or pigeon, and opossum, bat, or mouse) are denoted in gray circles on the tree. Of the 44 genes differentially expressed in FL vs. HL across amniotes, 18 are also misexpressed in pigeon feathered HLs. Of these, 4 genes are putative PITX1 targets (misexpressed in grouse and muff HLs relative to scaled HLs) and 14 are likely regulated by TBX5 or co-regulated by TBX5 and PITX1 (misexpressed in muff HLs, but not grouse HLs). (H) Gene-wise hierarchical clustering heat map of 18 genes that are differentially expressed in FL vs. HL across amniotes and scaled vs. feathered HL in pigeons. The scale for between-species comparisons is Log2FC −6 (HL-enriched) to 6 (FL-enriched); the scale for pigeon HL comparisons is Log2FC −3 (scaled HL-enriched) to 3 (muff HL-enriched). Gray boxes within the heat map denote no data (ABI3BP is not annotated in mouse genome). Table summarizes 18 deeply conserved genes, including type of gene and limb development-related function (if known). Asterisks (*) denote 4 putative PITX1 targets. Credits for animal silhouettes in G: Sarah Werning (anole and opossum), Steven Traver (chicken), Dori and Nevit Dilment (pigeon), David Liao (mouse), and Yan Wong (bat).
Similar to the avian PCA results, limb buds from anole, mouse, opossum, and bat datasets generally clustered based on limb type (FL vs. HL) and stage (for datasets that included multiple stages) (Figure 8B-F). For each dataset, we identified genes that are differentially expressed between stage-matched FL and HL buds (Supplemental Table 7). Consistent with our results from pigeon and chicken, in most data sets, almost all gene expression changes between FL and HL are low-level (Log2FC < 1), with only a small set of highly differentially expressed genes (Log2FC > 2) (Supplemental Figure 15).
To identify deeply conserved limb identity genes, we compared the differentially expressed gene sets from all six amniote species. Despite caveats associated with this type of meta-analysis that undoubtedly reduce our power to detect conserved genes (addressed in Discussion), we identified differentially expressed genes that are associated with FL/HL limb identity in Mammalia (1489 in mouse, opossum, and/or bat), Sauropsidia (56 in anole and pigeon or chicken), and Amniota (44 in anole, pigeon or chicken, and mouse, opossum, or bat) (Figure 8G and Supplemental Table 7). Within both the Sauropsidia and Amniota gene sets, ~50% (25/56 in Sauropsidia, 21/44 in Amniota) of the conserved differentially expressed genes are TFs (Supplemental Table 7), again highlighting that TFs are a highly conserved component of limb identity networks.
Finally, we asked if the genetic program associated with feathered HL development in pigeons represents a co-option of deeply conserved components of the FL identity program. We compared the grouse and muff gene sets to the 44 limb identity genes conserved in Amniota and found that 41% (18/44) of deeply conserved limb identity genes are also differentially expressed in grouse and/or muff pigeon HLs (Figure 8G-H). In addition to PITX1, we identified 3 deeply conserved limb identity genes (HOXD11, ANOS1, NRSN1) that are misexpressed in both grouse and muff HLs, suggesting that these genes are conserved downstream targets of PITX1 (Figure 8H). In addition to TBX5, 13 deeply conserved limb identity genes (PHEX, EYA4, CSMD3, TBX18, HOXA5, ETS1, LBH, CSRP2, CXCL12, SALL1, MKX, ABI3BP, DMRT2) are misexpressed only in muff HLs (Figure 8H). This set of 13 genes, which includes 7 TFs, is putatively regulated by TBX5 or co-regulated by TBX5 and PITX1. In particular, CXCL12, MKX, and DMRT2 may be co-regulated by TBX5 and PITX1, as these genes were recently identified as conserved regulatory targets of PITX1 in mouse and anole HLs (Wang et al., 2018). Of the 18 deeply conserved limb identity genes that are deployed during pigeon feathered HL development, most have been implicated in specific aspects of limb development with varying degrees of supporting evidence (TBX5, HOXD11, TBX18, HOXA5, ETS1, LBH, CSRP2, SALL1, CXCL12, MKX, PITX1) (Boulet and Capecchi, 2004, Zakany and Duboule, 2007, Sheeba and Logan, 2017, Haraguchi et al., 2015, Xu et al., 2013, Lettice et al., 2012, Kvon et al., 2016, Briegel and Joyner, 2001, Bonnin et al., 2002, Kawakami et al., 2009, Farrell and Munsterberg, 2000, Nassari et al., 2017, Odemis et al., 2005, Liu et al., 2010, Ito et al., 2010, Duboc and Logan, 2011b) (Figure 8H). For others (EYA4, ANOS1, PHEX, NRSN1, DMRT2, ABI3BP, CSMD3), a link to limb development has not been established. Furthermore, for most of the 18 deeply conserved limb identity genes, the precise regulatory connections to TBX5 and/or PITX1 remain open for discovery.
Discussion
We identified transcriptional changes associated with differences in FL, HL, and limbs with mixed identities. We took advantage of regulatory variants associated with pigeon foot feathering to specifically determine how shifts in the expression of two regulators of limb identity, PITX1 and TBX5, affect transcription of downstream genes during embryonic limb development. We found that most gene expression differences between FLs and HLs are subtle, and that only a small set of genes is highly differentially expressed. We identified suites of genes encoding TFs, ECM proteins, and components of developmental signaling pathways with known roles in limb development that are regulated downstream of TBX5 and/or PITX1. Finally, by comparing limb identity networks across amniotes, we found a small group of deeply conserved gene expression changes associated with limb identity. Some of the genes in this core group are also misexpressed in feathered pigeon HLs, consistent with a partial HL to FL transformation at both the phenotypic and transcriptomic level.
Embryonic limb identity networks
In birds, we found that limb type-specific expression of a small set of TFs is coupled with an accumulation of widespread low-level gene expression changes to form radically divergent FL and HL morphologies. Notably, transcriptomic studies of mouse embryonic limb development identified a similar number and level (i.e. fold-change) of gene expression changes between FL and HL (Nemec et al., 2017). These parallel findings are particularly interesting considering that variation in adult limb morphologies of mice (FL and HL are covered with hair and have similar skeletal proportions) is less dramatic than in birds (FLs are feather covered wings, HLs are scaled covered legs, and limb skeletons are highly divergent). Thus, the identity of the differentially expressed genes, and not the overall number or level of gene expression changes, ultimately determines adult limb phenotype. In our most phylogenetically inclusive analyses, we identified a core set of gene expression changes associated with amniote FL/HL limb identity, but we also found many genes that are differentially expressed between FLs and HLs in a species-specific manner. We predict that these unique sets of genes are associated with the development of species-specific limb phenotypes. In support of this idea, specific sets of genes initiate distinct developmental programs to form feathers or different scale types in archosaur embryos (Musser et al., 2018, Wu et al., 2018, Lai et al., 2018).
Parsing TBX5 vs. PITX1 -dependent regulation of the limb identity network
Pigeon foot feathering offers a unique opportunity to understand the independent and combinatorial effects of PITX and TBX5 expression changes on limb patterning. Both grouse and muff HLs are feathered, but the combination of reduced PITX1 and ectopic TBX5 expression (muff) has a more pronounced effect on the limb regulatory network than reduced PITX1 expression alone (grouse). Because our comparative analyses were performed using samples from one scaled breed (Racing Homer), one grouse breed (Oriental Frill), and one muff breed (English Trumpeter), it is possible that some of the differentially expressed genes we detected between scaled and feathered HLs represent breed-specific shifts in gene expression that are independent of the HL appendage phenotype. However, by comparing grouse and muff gene sets, we were able to infer a set of 601 putative PITX1-specific transcriptional targets that are differentially expressed in both grouse and muff HLs relative to scaled HLs. Validating specific genes of interest as direct downstream targets of PITX1 remains an important avenue of future research. In the present study, we were unable to separate TBX5-specific from TBX5/PITX1-synergistic effects in pigeon limb development. This shortcoming could be resolved by analyzing HL bud transcriptomes from feather-footed pigeons that carry the Slipper (TBX5) allele but not the grouse (PITX1) allele, although such breeds are rare (English and Pigmy Pouters only).
Partial co-option of pigeon FL identity program in feathered HLs
By comparing the differentially expressed genes from scaled and feather-footed pigeons, we found that the gene expression programs controlling feathered HL development are partially co-opted from the normal FL program. Thus, in feather-footed pigeons, a portion of the FL program is redeployed in the HL to form feathered HLs with partial anatomical and molecular FL identity. Spatiotemporal modulation of gene expression, particularly of genes like PITX1 and TBX5 that function at the top of gene regulatory hierarchies, is an important driver of evolutionary diversification (Davidson, 2006, Erwin and Davidson, 2009, Rebeiz et al., 2015, Infante et al., 2018). Generation of phenotypic diversity and novelty via co-option of genetic circuits or top-level network regulators is a repeated theme in evolution; notable examples include co-option of the adult skeletogenic network to form the larval skeleton in echinoderms, redeployment of the highly conserved arthropod appendage-patterning network to form beetle horns, and overlap of the genetic networks that control embryonic limb and external genital development in tetrapods (Rebeiz et al., 2015, Davidson, 2006, Erwin and Davidson, 2009, Infante et al., 2015, Infante et al., 2018, Gao and Davidson, 2008, Tschopp et al., 2014, Herrera and Cohn, 2014, Leal and Cohn, 2016, Leal and Cohn, 2018). Our comparative analyses in pigeons indicate that TFs are a major component of the FL identity program redeployed in feathered HLs; similarly, others have noted that genes at the top of regulatory networks are more evolutionarily stable than differentiation genes at the network periphery (Davidson, 2006, Erwin and Davidson, 2009, Rebeiz et al., 2015). Within the co-opted FL program in feathered HLs, we also identified genes encoding developmental signaling pathway components and ECM proteins, which link subcircuits of the regulatory network and function as differentiation genes at the network periphery, respectively (Erwin and Davidson, 2009, Davidson, 2006, Rebeiz et al., 2015). These findings suggest that changes in the expression of top-level regulators (PITX1 and/or TBX5) have widespread effects on all levels of the limb development network.
Despite a partial co-option of the FL identity program, it is important to note that pigeon feathered HLs are not FLs. They are not the consequence of a complete redeployment of the FL identity network onto a blank canvas. Instead, the ectopic FL program is superimposed onto the resident HL regulatory network, resulting in a HL with mixed identity. During normal limb bud development, ectodermal identity is determined in response to signals from the underlying mesoderm (Cairns and Saunders, 1954, Saunders et al., 1959, Sengel, 1990, Hughes et al., 2011). In muff HLs, ectopic and spatially-restricted mesodermal expression of TBX5 signals to the ectoderm to form feathered epidermis. The largest flight-like feathers of muffed pigeon feet develop superficial to the ectopic TBX5 expression domain in the posterior-dorsal limb bud. This spatially-restricted FL-like genetic program occurs within the context of HL identity program with reduced PITX1 expression. Together, changes in PITX1 and TBX5 expression cause a reorganization of the downstream limb identity network that results in a limb that is neither completely HL nor completely FL, as evident from the changes in both epidermal appendage type and musculoskeletal patterning (Domyan et al., 2016). This layering of multiple genetic networks represents an intriguing potential mechanism of phenotypic diversification.
Deeply conserved components of limb identity networks
By comparing FL and HL bud transcriptomes from several amniote species, we identified a core set of deeply conserved limb identity genes that is enriched for genes encoding TFs, ECM proteins, and signaling pathway components. We suspect that only a subset of the deeply conserved genes was identified in our analyses, due primarily to technical caveats commonly associated with genomic analyses (e.g., differences in sample preparation, sequencing method, quality of reference genome assembly and annotation). For example, in our analyses of brown anole (Anolis sagrei) limb buds, RNA-seq reads had to be aligned to the genome of the closely related green anole (Anolis carolinensis) because a brown anole genome assembly is not currently available. Analysis modifications like this could reduce our ability to detect differentially expressed genes. Nevertheless, our analyses suggest that the number of genes differentially expressed between FL and HL in a species-specific manner is considerably greater than the number of deeply conserved limb identity genes. This result suggests that relatively few genes are absolutely required for amniote FL vs. HL identity and raises the possibility that there is considerable evolutionary variation in the genes that were recruited to the limb development and identity programs. Another classic example of this paradigm is animal sex determination, in which a small set of master sex determination genes are highly conserved across species (e.g. DMRT1, SOX3), but the genetic players vary dramatically at the network periphery (Herpin and Schartl, 2015).
Of the deeply conserved FL/HL identity genes we identified, a subset was also differentially expressed between scaled and feathered pigeon HLs, suggesting that the genetic program underlying pigeon foot feathering is not completely novel, but instead incorporates elements of the deeply conserved FL/HL limb identity network. In the future, it will be exciting to determine if similar mechanisms of network co-option and/or overlaying of multiple regulatory networks are broadly associated with vertebrate limb diversification.
Materials and methods
Animal husbandry, isolation of embryonic tissue and sample sex determination
Columba livia were housed in accordance with the University of Utah Institutional Animal Care and Use Committees of University of Utah (16-03010). Pigeon eggs were collected from Racing Homer (scaled), Oriental Frill (grouse) and English Trumpeter (muff) breeding pairs and incubated to the desired embryonic stage. White Leghorn chicken eggs were obtained from AA Lab Eggs (Westminster, California) and incubated to the desired stage. For avian samples, pairs of forelimb (FL) and hindlimb (HL) buds were dissected from scaled, grouse and muff pigeon embryos or chicken embryos at HH21 (embryonic day E3.5), HH25 (E4.5), and HH29 (E6) and stored in RNAlater (ThermoFisher Scientific) at −80°C. Additional tissue was harvested from each embryo and stored at −80°C for DNA extraction and genotyping. Genomic DNA was isolated from each embryonic tissue sample using a DNeasy Blood and Tissue Kit (Qiagen). For each pigeon sample, sex was determined using a previously published PCR-based assay (Fridolfsson and Ellegren 1999).
Anolis sagrei were maintained at the University of Georgia following published guidelines (Sanger et al., 2008a). Animals were wild individuals captured at the Fairchild Tropical Botanic Gardens in Miami, FL. Breeding cages housed up to 4 adult females and 1 adult male together. Eggs were collected from nest boxes weekly and incubated at 26°C. Pairs of FL and HL buds were dissected from Stage 7 A. sagrei embryos and stored in RNAlater at −80°C. Embryos were staged according to (Sanger et al., 2008b). All experiments followed the National Research Council’s Guide for the Care and Use of Laboratory Animals and were performed with the approval and oversight of the University of Georgia Institutional Animal Care and Use Committee (A2015 02-020-Y3-A13).
RNA isolation, cDNA synthesis, and qRT-PCR analysis
Total RNA was extracted from FL and HL bud samples using the RNeasy Mini Kit with RNase-Free DNAse Set and a TissueLyser LT (Qiagen). For each sample, a cDNA library was prepared using M-MLV Reverse Transcriptase with oligo(dT) primer (ThermoFisher Scientific). Intron-spanning amplicons from pigeon PITX1, TBX5, and ACTB were amplified from cDNA libraries using a CFX96 instrument and iTaq Universal Sybr Green Supermix (BioRad). Primer sequences were published previously (Domyan et al., 2016). For a single qRT-PCR experiment, three technical replicates were performed for each sample and the mean value was determined. Each experiment was repeated three times. Statistical significance was determined using a pairwise Wilcoxon rank sum test in R version 3.3.3 (R Core Team, 2018).
Library preparation and RNA sequencing of avian samples
RNA-sequencing libraries were prepared and sequenced by the High-Throughput Genomics and Bioinformatic Analysis Shared Resource at the University of Utah. RNA sample quality was assessed using the RNA ScreenTape Assay (Agilent); RNA integrity (RIN) scores are presented in Supplemental Table 8. For each sample, a sequencing library was prepared using the TruSeq Stranded mRNA Sample Prep Kit with oligo(dT) selection (Illumina). 125-cycle paired-end sequencing was performed on an Illumina HiSeq 2500 instrument (12 libraries/lane). An average of 21 million reads were generated for each pigeon sample and 25 million reads for each chicken sample.
Analysis of avian RNA-seq data
Sequencing read quality was assessed with FastQC (Babraham Bioinformatics). Read alignment was performed using STAR version 2.5.0a (Dobin et al., 2013). Illumina adapters were trimmed and reads were aligned to the pigeon Cliv_2.1 reference assembly (Holt et al., 2018) or chicken Gallus_gallus-5.0 genome assembly (Warren et al., 2017) with STAR using the 2-pass mode. GTF annotation files were used to guide spliced read alignments. The average percentage of uniquely mapped reads was 84.07% for pigeon samples and 91.45% for chicken samples. Mapped reads were assigned to genes using featureCounts from the Subread package version 1.5.1 (Liao et al., 2014), with an average of 72.36% of reads assigned in pigeon samples and 74.48% in chicken samples. Principal component analyses (PCA) and differential expression analyses were performed with the R package DESeq2 version 1.12.4 (Love et al., 2014). Read count files were imported into DESeq2 and a DESeqDataSet object was generated that included sample sex and sequencing lane as covariates. Prior to running any DESeq2 functions, datasets were pre-filtered to remove genes with a total of 0 reads across all samples. For PCA, regularized log (rlog) transformation (RLT) or variance stabilized transformation (VST) was applied to the entire pigeon or chicken dataset using built-in DESeq2 commands. To generate differentially expressed gene lists, we employed a Benjamini & Hochberg adjusted p-value cutoff of p<0.05 because in some cases, more conservative cutoffs resulted in gene sets that did not include PITX1 or other known limb identity factors in the FL vs. scaled HL comparisons. The R package GeneOverlap version 1.18.0 (Shen and Sinai, 2018) was used with default settings to test for statistically significant intersections between sets of differentially expressed genes. Background genome size was set to 15392 (pigeon vs. pigeon comparisons), 22604 (mouse vs. mouse comparison), or 11202 (pigeon vs. mouse comparisons). In order to perform downstream comparative analyses, differentially expressed gene lists were annotated with human gene names using a custom R script (available as a Supplemental File).
Analysis of transcription factors, extracellular matrix components, and signaling pathways
To identify transcription factors (TFs) within our differentially expressed gene lists, we cross-referenced gene lists to a list of ~1400 genome-wide TFs compiled from AnimalTFDB 2.0 (Zhang et al., 2015). Extracellular matrix genes were identified by comparing differential expression results to a list of genes associated with the GO term “extracellular matrix” (GO:0031012). Ingenuity Pathway Analysis 2.2 (IPA, Qiagen) was used to identify differentially expressed components of specific developmental signaling pathways with roles in limb development. We specifically searched for significant enrichment of the following canonical pathways in IPA: Wnt signaling (Wntβ-catenin signaling, PCP pathway, Wnt/Ca+ signaling), BMP signaling, TGF-β signaling, Sonic Hedgehog signaling, and RAR activation.
RNA-seq and analysis of brown anole (Anolis sagrei) limb buds
A. sagrei FL and HL RNA-seq libraries were prepared using the TruSeq Stranded mRNA Sample Prep Kit with oligo(dT) selection (Illumina). Five separate FL and HL library sets were prepared from limb RNA collected from five different embryos. 75-cycle paired-end sequencing was performed on an Illumina NextSeq 500 instrument (10 libraries/lane). An average of 43 million reads were generated for each A. sagrei library. Read quality assessment, adapter trimming, and mapping to the Green anole (Anolis carolinensis) AnoCar2.0 genome assembly were performed as described above with modified alignment parameters (-- outFilterScoreMinOverLread 0.3 --outFilterMatchNminOverLread 0.3 --outFilterMultimapNmax 15) to allow for mismatches due to species-specific sequence differences. The average percentage of uniquely mapped reads was 79.19% for brown anole samples. Mapped reads were assigned as described above, with an average of 61.23% of reads assigned. PCA was performed using the top 100 most differentially expressed genes and FL vs. HL differential expression analyses were performed using DESeq2 as described above.
Analysis of publicly available mammalian RNA-seq datasets and comparative transcriptomics between species
Raw RNA-seq reads from three limb bud datasets from mouse (Mus musculus; Maier et al., 2017, Nemec et al., 2017, Amandio et al., 2016), one limb bud dataset from opossum (Monodelphis domestica; Maier et al., 2017), and one limb bud dataset from bat (Miniopterus natalensis; Eckalbar et al., 2016) were downloaded from the GEO (GSE71390, GSE79028, GSE100734) and BioProject (PRJNA270639) databases. Read quality assessment, adapter trimming, and read mapping to mouse GRCm38.p6, opossum monDom5, or bat Mnat.v1 genome assemblies were performed as described above, with an average alignment percentage of 86.90% (mouse; Maier et al., 2017), 76.46% (mouse; Nemec et al., 2017), 88.43% (mouse; Amandio et al., 2016), 84.76% (opossum; Maier et al., 2017), and 92.53% (bat; Eckalbar et al., 2016). Aligned reads were assigned to genes as described above, with an average of 66.47% (mouse; Maier et al., 2017), 48.80% (mouse; Nemec et al., 2017), 70.90% (mouse; Amandio et al., 2016), 58.93% (opossum; Maier et al., 2017), and 50.40% (bat; Eckalbar et al., 2016) successfully assigned reads. For each dataset, PCA and FL vs. HL differential expression analyses were performed using DESeq2 as described above. A master list of differentially expressed genes in mouse limb buds was generated by combining the gene sets obtained from the mouse datasets.
To facilitate comparative transcriptomic analyses, gene lists from all species were annotated with human gene names using a custom R script. To identify differentially expressed genes conserved in Aves, we intersected FL vs. HL gene lists from pigeon and chicken. To find genes conserved in Mammalia, we intersected the mouse, opossum, and bat gene sets and identified genes differentially expressed in at least two species. FL vs. HL genes conserved in Sauropsida were determined by intersecting gene lists from pigeon, chicken, and brown anole and identifying genes that are differentially expressed in brown anole and at least one member of Aves (pigeon and/or chicken). Similarly, FL vs. HL genes conserved in Amniota were determined by identifying genes that are differentially expressed in brown anole, at least one member of Aves (pigeon and/or chicken) and at least one member of Mammalia (mouse, opossum, and/or bat). Finally, we cross-referenced the Amniota gene list to the pigeon scaled HL vs. grouse HL and scaled HL vs. muff HL gene lists to identify deeply conserved FL vs. HL genes that are also misexpressed during grouse and/or muff HL development.
Photo and image credits
Photos used in Figure 1 were taken by Sydney Stringham (A, Racing Homer) and Thomas Hellmann (B, Oriental Frill and C, English Trumpeter). Animal silhouettes used in Figure 8G were downloaded as vector images from PhyloPic (phylopic.org). The silhouettes were created by Sarah Werning (anolis and opossum), Steven Traver (chicken), Dori and Nevit Dilmen (pigeon), David Liao (mouse), and Yan Wong (bat) and reused under the Creative Commons Attribution-ShareAlike 3.0 Unported license (mouse and pigeon; https://creativecommons.org/licenses/by-sa/3.0/), the Creative Commons Attribution 3.0 Unported license (anolis and opossum; https://creativecommons.org/licenses/by/3.0/), or the Public Domain Dedication 1.0 license (chicken and bat).
Supplementary Material
Highlights.
Feathered feet of pigeons result from a partial change in hindlimb identity.
PITX1- and TBX5-responsive genes are misexpressed in feathered feet of pigeons.
Some misexpressed “feathered feet” genes also distinguish forelimbs and hindlimbs.
Differentially expressed genes include TFs and other gene classes.
Comparisons among amniotes reveals conserved PITX1 and TBX5-responsive genes.
Acknowledgements
We thank Robert Greenhalgh and Timothy Mosbruger for their help with genomic analyses; Timothy Parnell and the Huntsman Cancer Institute Bioinformatic Analysis Shared Resource for assistance with and access to Ingenuity Pathway Analysis; and Anna Vickrey and Robert Greenhalgh for comments on the manuscript. We acknowledge the laboratory of Mark Yandell and the Center for High Performance Computing at the University of Utah for support and computing resources.
Funding
This work was supported by National Institutes of Health (NIH) grant F32DE028179 to EFB; National Science Foundation (NSF) grant IOS1149453 and NIH grant R01HD081034 to DBM; and NSF grant DEB1149160 and NIH grants R01GM115996 and R35GM131787 to MDS.
Footnotes
Competing interests
No competing interests declared.
Data availability
All RNA-seq data generated for this study have been deposited at Gene Expression Omnibus under accession numbers GSE127775 (C. livia and G. gallus) and GSE128151 (A. sagrei).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- AGARWAL P, WYLIE JN, GALCERAN J, ARKHITKO O, LI C, DENG C, GROSSCHEDL R & BRUNEAU BG 2003. Tbx5 is essential for forelimb bud initiation following patterning of the limb field in the mouse embryo. Development, 130, 623–33. [DOI] [PubMed] [Google Scholar]
- AHN DG, KOURAKIS MJ, ROHDE LA, SILVER LM & HO RK 2002. T-box gene tbx5 is essential for formation of the pectoral limb bud. Nature, 417, 754–8. [DOI] [PubMed] [Google Scholar]
- AL-QATTAN MM, AL-THUNAYAN A, ALABDULKAREEM I & AL BALWI M 2013. Liebenberg syndrome is caused by a deletion upstream to the PITX1 gene resulting in transformation of the upper limbs to reflect lower limb characteristics. Gene, 524, 65–71. [DOI] [PubMed] [Google Scholar]
- AMANDIO AR, NECSULEA A, JOYE E, MASCREZ B & DUBOULE D 2016. Hotair Is Dispensible for Mouse Development. PLoS Genet, 12, e1006232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ASHBURNER M, BALL CA, BLAKE JA, BOTSTEIN D, BUTLER H, CHERRY JM, DAVIS AP, DOLINSKI K, DWIGHT SS, EPPIG JT, HARRIS MA, HILL DP, ISSEL-TARVER L, KASARSKIS A, LEWIS S, MATESE JC, RICHARDSON JE, RINGWALD M, RUBIN GM & SHERLOCK G 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet, 25, 25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOER EF, VAN HOLLEBEKE HF & SHAPIRO MD 2017. Genomic determinants of epidermal appendage patterning and structure in domestic birds. Dev Biol, 429, 409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BONNIN MA, EDOM-VOVARD F, KEFALAS P & DUPREZ D 2002. CRP2 transcript expression pattern in embryonic chick limb. Mech Dev, 116, 151–5. [DOI] [PubMed] [Google Scholar]
- BOULET AM & CAPECCHI MR 2004. Multiple roles of Hoxa11 and Hoxd11 in the formation of the mammalian forelimb zeugopod. Development, 131, 299–309. [DOI] [PubMed] [Google Scholar]
- BRIEGEL KJ & JOYNER AL 2001. Identification and characterization of Lbh, a novel conserved nuclear protein expressed during early limb and heart development. Dev Biol, 233, 291–304. [DOI] [PubMed] [Google Scholar]
- CAIRNS JM & SAUNDERS JW 1954. The influence of embryonic mesoderm on the regional specification of epidermal derivatives in the chick. Journal of Experimental Zoology, 127, 221–248. [Google Scholar]
- CAPELLINI TD, DI GIACOMO G, SALSI V, BRENDOLAN A, FERRETTI E, SRIVASTAVA D, ZAPPAVIGNA V & SELLERI L 2006. Pbx1/Pbx2 requirement for distal limb patterning is mediated by the hierarchical control of Hox gene spatial distribution and Shh expression. Development, 133, 2263–73. [DOI] [PubMed] [Google Scholar]
- CAPELLINI TD, VACCARI G, FERRETTI E, FANTINI S, HE M, PELLEGRINI M, QUINTANA L, DI GIACOMO G, SHARPE J, SELLERI L & ZAPPAVIGNA V 2010. Scapula development is governed by genetic interactions of Pbx1 with its family members and with Emx2 via their cooperative control of Alx1. Development, 137, 2559–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CARBON S, IRELAND A, MUNGALL CJ, SHU S, MARSHALL B, LEWIS S, AMI GOH & WEB PRESENCE WORKING, G. 2009. AmiGO: online access to ontology and annotation data. Bioinformatics, 25, 288–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHAN YF, MARKS ME, JONES FC, VILLARREAL G JR., SHAPIRO MD, BRADY SD, SOUTHWICK AM, ABSHER DM, GRIMWOOD J, SCHMUTZ J, MYERS RM, PETROV D, JONSSON B, SCHLUTER D, BELL MA & KINGSLEY DM 2010. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science, 327, 302–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAVIDSON EH 2006. The regulatory genome : gene regulatory networks in development and evolution, Burlington, MA ; San Diego, Academic. [Google Scholar]
- DAVIS AP & CAPECCHI MR 1994. Axial homeosis and appendicular skeleton defects in mice with a targeted disruption of hoxd-11. Development, 120, 2187–98. [DOI] [PubMed] [Google Scholar]
- DECKER E, DURAND C, BENDER S, RODELSPERGER C, GLASER A, HECHT J, SCHNEIDER KU & RAPPOLD G 2011. FGFR3 is a target of the homeobox transcription factor SHOX in limb development. Hum Mol Genet, 20, 1524–35. [DOI] [PubMed] [Google Scholar]
- DELAURIER A, SCHWEITZER R & LOGAN M 2006. Pitx1 determines the morphology of muscle, tendon, and bones of the hindlimb. Dev Biol, 299, 22–34. [DOI] [PubMed] [Google Scholar]
- DOBIN A, DAVIS CA, SCHLESINGER F, DRENKOW J, ZALESKI C, JHA S, BATUT P, CHAISSON M & GINGERAS TR 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics, 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOMYAN ET, KRONENBERG Z, INFANTE CR, VICKREY AI, STRINGHAM SA, BRUDERS R, GUERNSEY MW, PARK S, PAYNE J, BECKSTEAD RB, KARDON G, MENKE DB, YANDELL M & SHAPIRO MD 2016. Molecular shifts in limb identity underlie development of feathered feet in two domestic avian species. Elife, 5, e12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOMYAN ET & SHAPIRO MD 2017. Pigeonetics takes flight: Evolution, development, and genetics of intraspecific variation. Dev Biol, 427, 241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DON EK, DE JONG-CURTAIN TA, DOGGETT K, HALL TE, HENG B, BADROCK AP, WINNICK C, NICHOLSON GA, GUILLEMIN GJ, CURRIE PD, HESSELSON D, HEATH JK & COLE NJ 2016. Genetic basis of hindlimb loss in a naturally occurring vertebrate model. Biol Open, 5, 359–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DONCASTER L 1912. Notes on inheritance of colour and other characters in pigeons. Journal of Genetics, 2, 89–98. [Google Scholar]
- DUBOC V & LOGAN MP 2011a. Pitx1 is necessary for normal initiation of hindlimb outgrowth through regulation of Tbx4 expression and shapes hindlimb morphologies via targeted growth control. Development, 138, 5301–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DUBOC V & LOGAN MP 2011b. Regulation of limb bud initiation and limb-type morphology. Dev Dyn, 240, 1017–27. [DOI] [PubMed] [Google Scholar]
- ECKALBAR WL, SCHLEBUSCH SA, MASON MK, GILL Z, PARKER AV, BOOKER BM, NISHIZAKI S, MUSWAMBA-NDAY C, TERHUNE E, NEVONEN KA, MAKKI N, FRIEDRICH T, VANDERMEER JE, POLLARD KS, CARBONE L, WALL JD, ILLING N & AHITUV N 2016. Transcriptomic and epigenomic characterization of the developing bat wing. Nat Genet, 48, 528–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ERWIN DH & DAVIDSON EH 2009. The evolution of hierarchical gene regulatory networks. Nat Rev Genet, 10, 141–8. [DOI] [PubMed] [Google Scholar]
- FARRELL ER & MUNSTERBERG AE 2000. csal1 is controlled by a combination of FGF and Wnt signals in developing limb buds. Dev Biol, 225, 447–58. [DOI] [PubMed] [Google Scholar]
- FEENSTRA JM, KANAYA K, PIRA CU, HOFFMAN SE, EPPEY RJ & OBERG KC 2012. Detection of genes regulated by Lmx1b during limb dorsalization. Dev Growth Differ, 54, 451–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GAO F & DAVIDSON EH 2008. Transfer of a large gene regulatory apparatus to a new developmental address in echinoid evolution. Proc Natl Acad Sci U S A, 105, 6091–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GARRITY DM, CHILDS S & FISHMAN MC 2002. The heartstrings mutation in zebrafish causes heart/fin Tbx5 deficiency syndrome. Development, 129, 4635–45. [DOI] [PubMed] [Google Scholar]
- GIBSON-BROWN JJ, AGULNIK SI, CHAPMAN DL, ALEXIOU M, GARVEY N, SILVER LM & PAPAIOANNOU VE 1996. Evidence of a role for T-box genes in the evolution of limb morphogenesis and the specification of forelimb/hindlimb identity. Mech Dev, 56, 93–101. [DOI] [PubMed] [Google Scholar]
- GURNETT CA, ALAEE F, KRUSE LM, DESRUISSEAU DM, HECHT JT, WISE CA, BOWCOCK AM & DOBBS MB 2008. Asymmetric lower-limb malformations in individuals with homeobox PITX1 gene mutation. Am J Hum Genet, 83, 616–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GYURJAN I, SONDEREGGER B, NAEF F & DUBOULE D 2011. Analysis of the dynamics of limb transcriptomes during mouse development. BMC Dev Biol, 11, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAMBURGER V & HAMILTON HL 1951. A series of normal stages in the development of the chick embryo. J Morphol, 88, 49–92. [PubMed] [Google Scholar]
- HARAGUCHI R, KITAZAWA R & KITAZAWA S 2015. Epigenetic regulation of Tbx18 gene expression during endochondral bone formation. Cell Tissue Res, 359, 503–512. [DOI] [PubMed] [Google Scholar]
- HASSON P, DEL BUONO J & LOGAN MP 2007. Tbx5 is dispensable for forelimb outgrowth. Development, 134, 85–92. [DOI] [PubMed] [Google Scholar]
- HERPIN A & SCHARTL M 2015. Plasticity of gene-regulatory networks controlling sex determination: of masters, slaves, usual suspects, newcomers, and usurpators. EMBO Rep, 16, 1260–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HERRERA AM & COHN MJ 2014. Embryonic origin and compartmental organization of the external genitalia. Sci Rep, 4, 6896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOLLANDER WF 1937. Hereditary Interrelationships of Certain Factors in Pigeons. University of Wisconsin-Madison. [Google Scholar]
- HOLT C, CAMPBELL M, KEAYS DA, EDELMAN N, KAPUSTA A, MACLARY E, E TD, SUH A, WARREN WC, YANDELL M, GILBERT MTP & SHAPIRO MD 2018. Improved Genome Assembly and Annotation for the Rock Pigeon (Columba livia). G3 (Bethesda), 8, 1391–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HORTON AC, MAHADEVAN NR, MINGUILLON C, OSOEGAWA K, ROKHSAR DS, RUVINSKY I, DE JONG PJ, LOGAN MP & GIBSON-BROWN JJ 2008. Conservation of linkage and evolution of developmental function within the Tbx2/¾/5 subfamily of T-box genes: implications for the origin of vertebrate limbs. Dev Genes Evol, 218, 613–28. [DOI] [PubMed] [Google Scholar]
- HUGHES MW, WU P, JIANG TX, LIN SJ, DONG CY, LI A, HSIEH FJ, WIDELITZ RB & CHUONG CM 2011. In search of the Golden Fleece: unraveling principles of morphogenesis by studying the integrative biology of skin appendages. Integr Biol (Camb), 3, 388–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- INFANTE CR, MIHALA AG, PARK S, WANG JS, JOHNSON KK, LAUDERDALE JD & MENKE DB 2015. Shared Enhancer Activity in the Limbs and Phallus and Functional Divergence of a Limb-Genital cis-Regulatory Element in Snakes. Dev Cell, 35, 107–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- INFANTE CR, PARK S, MIHALA AG, KINGSLEY DM & MENKE DB 2013. Pitx1 broadly associates with limb enhancers and is enriched on hindlimb cis-regulatory elements. Dev Biol, 374, 234–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- INFANTE CR, RASYS AM & MENKE DB 2018. Appendages and gene regulatory networks: Lessons from the limbless. Genesis, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ITO Y, TORIUCHI N, YOSHITAKA T, UENO-KUDOH H, SATO T, YOKOYAMA S, NISHIDA K, AKIMOTO T, TAKAHASHI M, MIYAKI S & ASAHARA H 2010. The Mohawk homeobox gene is a critical regulator of tendon differentiation. Proceedings of the National Academy of Sciences, 107, 10538–10542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAWAKAMI Y, UCHIYAMA Y, RODRIGUEZ ESTEBAN C, INENAGA T, KOYANO-NAKAGAWA N, KAWAKAMI H, MARTI M, KMITA M, MONAGHAN-NICHOLS P, NISHINAKAMURA R & IZPISUA BELMONTE JC 2009. Sall genes regulate region-specific morphogenesis in the mouse limb by modulating Hox activities. Development, 136, 585–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KVON EZ, KAMNEVA OK, MELO US, BAROZZI I, OSTERWALDER M, MANNION BJ, TISSIERES V, PICKLE CS, PLAJZER-FRICK I, LEE EA, KATO M, GARVIN TH, AKIYAMA JA, AFZAL V, LOPEZ-RIOS J, RUBIN EM, DICKEL DE, PENNACCHIO LA & VISEL A 2016. Progressive Loss of Function in a Limb Enhancer during Snake Evolution. Cell, 167, 633–642 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAI YC, LIANG YC, JIANG TX, WIDELITZ RB, WU P & CHUONG CM 2018. Transcriptome analyses of reprogrammed feather / scale chimeric explants revealed co-expressed epithelial gene networks during organ specification. BMC Genomics, 19, 780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEAL F & COHN MJ 2016. Loss and Re-emergence of Legs in Snakes by Modular Evolution of Sonic hedgehog and HOXD Enhancers. Curr Biol, 26, 2966–2973. [DOI] [PubMed] [Google Scholar]
- LEAL F & COHN MJ 2018. Developmental, genetic, and genomic insights into the evolutionary loss of limbs in snakes. Genesis, 56. [DOI] [PubMed] [Google Scholar]
- LETTICE LA, WILLIAMSON I, WILTSHIRE JH, PELUSO S, DEVENNEY PS, HILL AE, ESSAFI A, HAGMAN J, MORT R, GRIMES G, DEANGELIS CL & HILL RE 2012. Opposing functions of the ETS factor family define Shh spatial expression in limb buds and underlie polydactyly. Dev Cell, 22, 459–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEVI WM 1986. The Pigeon, Sumter, S.C, Levi Publishing Co., Inc. [Google Scholar]
- LIAO Y, SMYTH GK & SHI W 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics, 30, 923–30. [DOI] [PubMed] [Google Scholar]
- LIU W, WATSON SS, LAN Y, KEENE DR, OVITT CE, LIU H, SCHWEITZER R & JIANG R 2010. The atypical homeodomain transcription factor Mohawk controls tendon morphogenesis. Mol Cell Biol, 30, 4797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOGAN M, SIMON HG & TABIN C 1998. Differential regulation of T-box and homeobox transcription factors suggests roles in controlling chick limb-type identity. Development, 125, 2825–35. [DOI] [PubMed] [Google Scholar]
- LOGAN M & TABIN CJ 1999. Role of Pitx1 upstream of Tbx4 in specification of hindlimb identity. Science, 283, 1736–9. [DOI] [PubMed] [Google Scholar]
- LOVE MI, HuBER W & ANDERS S 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol, 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAIER JA, RIVAS-ASTROZA M, DENG J, DOWLING A, OBOIKOVITZ P, CAO X, BEHRINGER RR, CRETEKOS CJ, RASWEILER JJT, ZHONG S & SEARS KE 2017. Transcriptomic insights into the genetic basis of mammalian limb diversity. BMC Evol Biol, 17, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MINGUILLON C, DEL BUONO J & LOGAN MP 2005. Tbx5 and Tbx4 are not sufficient to determine limb-specific morphologies but have common roles in initiating limb outgrowth. Dev Cell, 8, 75–84. [DOI] [PubMed] [Google Scholar]
- MUSSER JM, WAGNER GP, LIANG C, STABILE FA, CLOUTIER A, BAKER AJ & PRUM RO 2018. Subdivision of ancestral scale genetic program underlies origin of feathers and avian scutate scales. bioRxiv, 377358. [Google Scholar]
- NAICHE LA & PAPAIOANNOU VE 2003. Loss of Tbx4 blocks hindlimb development and affects vascularization and fusion of the allantois. Development, 130, 2681–93. [DOI] [PubMed] [Google Scholar]
- NAICHE LA & PAPAIOANNOU VE 2007. Tbx4 is not required for hindlimb identity or post-bud hindlimb outgrowth. Development, 134, 93–103. [DOI] [PubMed] [Google Scholar]
- NARKIS G, TZCHORI I, COHEN T, HOLTZ A, WIER E & WESTPHAL H 2012. Isl1 and Ldb co-regulators of transcription are essential early determinants of mouse limb development. Dev Dyn, 241, 787–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NASSARI S, BLAVET C, BONNIN M-A, STRICKER S, DUPREZ D & FOURNIER-THIBAULT C 2017. The chemokines CXCL12 and CXCL14 differentially regulate connective tissue markers during limb development. Scientific Reports, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NEMEC S, LUXEY M, JAIN D, HUANG SUNG A, PASTINEN T & DROUIN J 2017. Pitx1 directly modulates the core limb development program to implement hindlimb identity. Development, 144, 3325–3335. [DOI] [PubMed] [Google Scholar]
- ODEMIS V, LAMP E, PEZESHKI G, MOEPPS B, SCHILLING K, GIERSCHIK P, LITTMAN DR & ENGELE J 2005. Mice deficient in the chemokine receptor CXCR4 exhibit impaired limb innervation and myogenesis. Mol Cell Neurosci, 30, 494–505. [DOI] [PubMed] [Google Scholar]
- PETIT F, SEARS KE & AHITUV N 2017. Limb development: a paradigm of gene regulation. Nat Rev Genet, 18, 245–258. [DOI] [PubMed] [Google Scholar]
- R CORE TEAM 2018. R: A Language and Environment for Statistical Computing. 3.5.1 ed. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- RABINOWITZ AH & VOKES SA 2012. Integration of the transcriptional networks regulating limb morphogenesis. Dev Biol, 368, 165–80. [DOI] [PubMed] [Google Scholar]
- RALLIS C, BRUNEAU BG, DEL BUONO J, SEIDMAN CE, SEIDMAN JG, NISSIM S, TABIN CJ & LOGAN MP 2003. Tbx5 is required for forelimb bud formation and continued outgrowth. Development, 130, 2741–51. [DOI] [PubMed] [Google Scholar]
- REBEIZ M, PATEL NH & HINMAN VF 2015. Unraveling the Tangled Skein: The Evolution of Transcriptional Regulatory Networks in Development. Annu Rev Genomics Hum Genet, 16, 103–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RODRIGUEZ-ESTEBAN C, TSUKUI T, YONEI S, MAGALLON J, TAMURA K & IZPISUA BELMONTE JC 1999. The T-box genes Tbx4 and Tbx5 regulate limb outgrowth and identity. Nature, 398, 814–8. [DOI] [PubMed] [Google Scholar]
- SANGER TJ, HIME PM, JOHNSON MA, DIANI J & LOSOS JB 2008a. Laboratory protocols for husbandry and embryo collection of Anolis lizards. Herpetol Rev, 39, 58–63. [Google Scholar]
- SANGER TJ, LOSOS JB & GIBSON-BROWN JJ 2008b. A developmental staging series for the lizard genus Anolis: a new system for the integration of evolution, development, and ecology. J Morphol, 269, 129–37. [DOI] [PubMed] [Google Scholar]
- SAUNDERS JW, GASSELING MT & CAIRNS JM 1959. The differentiation of prospective thigh mesoderm grafted beneath the apical ectodermal ridge of the wing bud in the chick embryo. Developmental Biology, 1, 281–301. [Google Scholar]
- SEARS K, MAIER JA, SADIER A, SORENSEN D & URBAN DJ 2018. Timing the developmental origins of mammalian limb diversity. Genesis, 56. [DOI] [PubMed] [Google Scholar]
- SENGEL P 1990. Pattern formation in skin development. Int J Dev Biol, 34, 33–50. [PubMed] [Google Scholar]
- SHANG J, LUO Y & CLAYTON DA 1997. Backfoot is a novel homeobox gene expressed in the mesenchyme of developing hind limb. Dev Dyn, 209, 242–53. [DOI] [PubMed] [Google Scholar]
- SHAPIRO MD & DOMYAN ET 2013. Domestic pigeons. Curr Biol, 23, R302–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHAPIRO MD, KRONENBERG Z, LI C, DOMYAN ET, PAN H, CAMPBELL M, TAN H, HUFF CD, HU H, VICKREY AI, NIELSEN SC, STRINGHAM SA, HU H, WILLERSLEV E, GILBERT MT, YANDELL M, ZHANG G & WANG J 2013. Genomic diversity and evolution of the head crest in the rock pigeon. Science, 339, 1063–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHAPIRO MD, MARKS ME, PEICHEL CL, BLACKMAN BK, NERENG KS, JONSSON B, SCHLUTER D & KINGSLEY DM 2004. Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature, 428, 717–23. [DOI] [PubMed] [Google Scholar]
- SHEEBA CJ & LOGAN MP 2017. The Roles of T-Box Genes in Vertebrate Limb Development. Curr Top Dev Biol, 122, 355–381. [DOI] [PubMed] [Google Scholar]
- SHEN L & SINAI M 2018. GeneOverlap: Test and visualize gene overlaps. [Google Scholar]
- SHOU S, SCOTT V, REED C, HITZEMANN R & STADLER HS 2005. Transcriptome analysis of the murine forelimb and hindlimb autopod. Dev Dyn, 234, 74–89. [DOI] [PubMed] [Google Scholar]
- SIMON HG, KITTAPPA R, KHAN PA, TSILFIDIS C, LIVERSAGE RA & OPPENHEIMER S 1997. A novel family of T-box genes in urodele amphibian limb development and regeneration: candidate genes involved in vertebrate forelimb/hindlimb patterning. Development, 124, 1355–66. [DOI] [PubMed] [Google Scholar]
- SZETO DP, RODRIGUEZ-ESTEBAN C, RYAN AK, O’CONNELL SM, LIU F, KIOUSSI C, GLEIBERMAN AS, IZPISUA-BELMONTE JC & ROSENFELD MG 1999. Role of the Bicoid-related homeodomain factor Pitx1 in specifying hindlimb morphogenesis and pituitary development. Genes Dev, 13, 484–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SZETO DP, RYAN AK, O’CONNELL SM & ROSENFELD MG 1996. P-OTX: a PIT-1- interacting homeodomain factor expressed during anterior pituitary gland development. Proc Natl Acad Sci U S A, 93, 7706–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAHER L, COLLETTE NM, MURUGESH D, MAXWELL E, OVCHARENKO I & LOOTS GG 2011. Global gene expression analysis of murine limb development. PLoS One, 6, e28358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAKEUCHI JK, KOSHIBA-TAKEUCHI K, MATSUMOTO K, VOGEL-HOPKER A, NAITOH-MATSUO M, OGURA K, TAKAHASHI N, YASUDA K & OGURA T 1999. Tbx5 and Tbx4 genes determine the wing/leg identity of limb buds. Nature, 398, 810–4. [DOI] [PubMed] [Google Scholar]
- TAKEUCHI JK, KOSHIBA-TAKEUCHI K, SUZUKI T, KAMIMURA M, OGURA K & OGURA T 2003. Tbx5 and Tbx4 trigger limb initiation through activation of the Wnt/Fgf signaling cascade. Development, 130, 2729–39. [DOI] [PubMed] [Google Scholar]
- TARCHINI B, DUBOULE D & KMITA M 2006. Regulatory constraints in the evolution of the tetrapod limb anterior-posterior polarity. Nature, 443, 985–8. [DOI] [PubMed] [Google Scholar]
- THE GENE ONTOLOGY, C. 2017. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res, 45, D331–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TICKLE C 2004. The contribution of chicken embryology to the understanding of vertebrate limb development. Mech Dev, 121, 1019–29. [DOI] [PubMed] [Google Scholar]
- TICKLE C 2015. How the embryo makes a limb: determination, polarity and identity. J Anat, 227, 418–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TIECKE E, BANGS F, BLASCHKE R, FARRELL ER, RAPPOLD G & TICKLE C 2006. Expression of the short stature homeobox gene Shox is restricted by proximal and distal signals in chick limb buds and affects the length of skeletal elements. Dev Biol, 298, 585–96. [DOI] [PubMed] [Google Scholar]
- TOWERS M & TICKLE C 2009. Growing models of vertebrate limb development. Development, 136, 179–90. [DOI] [PubMed] [Google Scholar]
- TSCHOPP P, SHERRATT E, SANGER TJ, GRONER AC, ASPIRAS AC, HU JK, POURQUIE O, GROS J & TABIN CJ 2014. A relative shift in cloacal location repositions external genitalia in amniote evolution. Nature, 516, 391–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG JS, INFANTE CR, PARK S & MENKE DB 2018. PITX1 promotes chondrogenesis and myogenesis in mouse hindlimbs through conserved regulatory targets. Dev Biol, 434, 186–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WARREN WC, HILLIER LW, TOMLINSON C, MINX P, KREMITZKI M, GRAVES T, MARKOVIC C, BOUK N, PRUITT KD, THIBAUD-NISSEN F, SCHNEIDER V, MANSOUR TA, BROWN CT, ZIMIN A, HAWKEN R, ABRAHAMSEN M, PYRKOSZ AB, MORISSON M, FILLON V, VIGNAL A, CHOW W, HOWE K, FULTON JE, MILLER MM, LOVELL P, MELLO CV, WIRTHLIN M, MASON AS, KUO R, BURT DW, DODGSON JB & CHENG HH 2017. A New Chicken Genome Assembly Provides Insight into Avian Genome Structure. G3 (Bethesda), 7, 109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WELLIK DM 2003. Hox10 and Hox11 Genes Are Required to Globally Pattern the Mammalian Skeleton. Science, 301, 363–367. [DOI] [PubMed] [Google Scholar]
- WEXELSEN H 1934. Types of leg feathering in pigeons. Hereditas, 18, 192–198. [Google Scholar]
- WU P, YAN J, LAI YC, NG CS, LI A, JIANG X, ELSEY RM, WIDELITZ R, BAJPAI R, LI WH & CHUONG CM 2018. Multiple Regulatory Modules Are Required for Scale-to-Feather Conversion. Mol Biol Evol, 35, 417–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XU B, HRYCAJ SM, MCINTYRE DC, BAKER NC, TAKEUCHI JK, JEANNOTTE L, GABER ZB, NOVITCH BG & WELLIK DM 2013. Hox5 interacts with Plzf to restrict Shh expression in the developing forelimb. Proc Natl Acad Sci U S A, 110, 19438–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZAKANY J & DUBOULE D 2007. The role of Hox genes during vertebrate limb development. Curr Opin Genet Dev, 17, 359–66. [DOI] [PubMed] [Google Scholar]
- ZHANG HM, LIU T, LIU CJ, SONG S, ZHANG X, LIU W, JIA H, XUE Y & GUO AY 2015. AnimalTFDB 2.0: a resource for expression, prediction and functional study of animal transcription factors. Nucleic Acids Res, 43, D76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.